Abstract
Mycobacterium avium and Mycobacterium intracellulare are closely related organisms and comprise the Mycobacterium avium complex. These organisms share many common characteristics, including the ability to cause life-threatening respiratory infections in people with underlying lung pathology or immunological defects and occasionally in those with no known predisposing conditions. However, the ability to invade the mucosa of the gastrointestinal tract and cause disseminated disease in AIDS patients has not been epidemiologically linked to M. intracellulare and appears to be unique to M. avium. We compared the abilities of M. avium and M. intracellulare to tolerate the acidic conditions of the stomach, to resist the membrane-disrupting activity of cationic peptides, and to invade intestinal epithelial cells in vitro and in vivo. We observed that M. avium and M. intracellulare were both tolerant to the acidic conditions encountered in the stomach and resistant to cationic peptides. However, when strains of M. avium and M. intracellulare were examined for their ability to enter cultured human intestinal cells or mouse intestinal mucosa, we observed that M. avium could invade more efficiently than M. intracellulare. To elucidate the basis of this pathogenic difference and identify genes involved in the invasion of the intestinal mucosa, we performed chromosomal DNA subtractive hybridization using M. avium and M. intracellulare chromosomal DNAs. In all, 21 genes that were present in M. avium but absent in M. intracellulare were identified, including some that may be associated with the ability of M. avium to invade the intestinal mucosa.
Mycobacterial diseases have been known for over 1,000 years and still pose serious health problems in both industrialized and underdeveloped parts of the world (34). Although Mycobacterium tuberculosis is the most aggressive species of this genus, infecting over one-third of the world's population and causing the death of more people than any other single infectious agent (8), other mycobacteria also cause serious disease. For example, the Mycobacterium avium complex (MAC) organisms are nontuberculosis mycobacteria associated with life-threatening infections in people with underlying lung pathology (e.g., chronic obstructive lung disease) or immune system defects (e.g., AIDS) and, on occasion, in those without apparent predisposing conditions. MAC consists of two closely related but genetically distinct species (M. avium and Mycobacterium intracellulare) that have been grouped into 21 serotypes, with serotypes 1 to 6, 8 to 11, and 21 designated M. avium and serotypes 7 and 12 to 20 designated M. intracellulare (2, 9, 33).
M. avium and M. intracellulare have both been isolated from AIDS patients but differ in the frequency of isolation, the route of infection, and the type of disease caused. Both organisms have been isolated from the sputa of AIDS patients with active pulmonary disease (24, 39); however, M. avium is by far the more common of the two organisms to infect this population (4, 39). In AIDS patients, M. avium is acquired predominantly via the gastrointestinal tract, where it is able to invade and translocate the intestinal mucosa, infect and multiply within submucosal macrophages, and cause bacteremia leading to the dissemination of the organism to the liver, spleen, and bone marrow (13). It is not clear if M. intracellulare can also infect the gastrointestinal tracts of AIDS patients, but to date no report of this route of infection has been documented. To elucidate the nature of this phenotypic difference, we examined the abilities of both species to survive the acidic conditions encountered in the stomach, to resist cationic peptides associated with the antimicrobial defense of the gastrointestinal tract, and to invade intestinal epithelial cells in vitro and in vivo. We also compared the genomes of these species via genomic DNA subtractive hybridization (1, 10, 15, 28) and identified several possible virulence factors unique to M. avium.
MATERIALS AND METHODS
Bacterial strains, epithelial cell lines, plasmids, and growth conditions.
M. avium strains 101, 104, and 109 were isolated from the blood of AIDS patients. M. intracellulare strains 83-8705, 84-8739, and 86-8953 were clinical isolates provided by Robert Good (Centers for Disease Control and Prevention, Atlanta, Ga.). M. intracellulare ATCC 13950 was acquired from the American Type Culture Collection (Manassas, Va.). Mycobacterium smegmatis mc2155 was the kind gift of W. Jacobs. HEp-2 human pharyngeal epithelial cells and HT-29 human intestinal epithelial cells were obtained from the American Type Culture Collection and grown as described previously (6). Mycobacteria were cultured on Middlebrook 7H11 agar supplemented with oleic acid, albumin, dextrose, and catalase (OADC) (Difco, Detroit, Mich.) at 37°C. For invasion assays, transparent colonies of M. avium or M. intracellular were transferred to Middlebook 7H9 broth supplemented with OADC, grown to log phase, washed in Hanks' buffered salt solution (HBSS), and vortexed and agitated for 2 min to disperse any clumps. The bacterial suspension was allowed to sit for 5 min, and the top 1 ml was removed and stained by the Ziehl-Neelsen method to confirm that the suspensions contained dispersed bacterial cells. Bacteria were plated in triplicate onto agar plates and enumerated for each experiment. Escherichia coli XL1Blue MRF′ (Stratagene, La Jolla, Calif.) was cultured onto Luria broth supplemented with kanamycin (50 μg/ml) or ampicillin (25 μg/ml) when appropriate. DNA fragments obtained by subtractive hybridization were cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.).
Acid tolerance assay.
Mycobacteria were grown to a cell density of approximately 105 bacteria/ml in 7H9 broth (pH 6.8) supplemented with OADC. One milliliter of this culture was centrifuged, and the pellet was suspended in 7H9 broth adjusted to either pH 2.2 or 6.8 with 1 N HCl. The suspensions at pH 2.2 were incubated at 37°C for 2 or 24 h and serially diluted in HBSS, and viable bacteria were quantified by heterotrophic plate counts. The suspensions at pH 6.8 were immediately quantified to obtain the number of viable bacteria at the beginning of the experiment. The percent survival was determined by dividing the number of bacteria present at the end of the experiment by the number of bacteria present at the beginning of the experiment and multiplying by 100.
Polymyxin B resistance assay.
The MICs of polymyxin B for M. avium, M. intracellulare, and M. smegmatis were identified by inoculating ca. 10,000 bacteria into 3 ml of 7H9 broth supplemented with OADC and polymyxin B, ranging in concentration from 0.5 to 500 μg/ml in twofold increments. The cultures were incubated at 37°C for 10 days and examined for growth by visual turbidity. The lowest concentration of polymyxin B that did not support growth was considered the MIC.
In vitro invasion assay.
To evaluate the ability of M. avium and M. intracellulare to invade epithelial cells in vitro, we carried out invasion assays using HEp-2 (pharyngeal) and HT-29 (intestinal) epithelial cell lines as described previously (6).
Intestinal invasion assay.
To determine the relative abilities of M. avium and M. intracellulare to invade the intestinal mucosa, we used a mouse intestinal loop model. C57BL/6 bg+/bg− black mice (female, 6 to 8 weeks old, weighing an average of 20 g) were obtained from Jackson Laboratories (Bar Harbor, Maine) and used after 1 to 2 weeks of quarantine. Mice were anesthetized using intraperitoneally administered phenobarbital and halothane by inhalation, which maintained them under anesthesia throughout the procedure. An incision in the abdominal cavity was made, and a segment of the small intestine ca. 3 cm in length above the ileo-cecal area was tied at both ends with a suture tight enough to close the intestinal lumen but not tight enough to restrict blood flow. Approximately 105 bacteria in HBSS were injected into the proximal portion of the segment, and the animals were maintained for 1, 1.5, and 2 h before the segment was removed, opened longitudinally, and washed extensively with HBSS to remove unbound bacteria. Washed intestines were placed in 5 ml of 7H9 broth, homogenized, serially diluted, plated onto 7H11 agar supplemented with antibiotics inhibitory to the intestinal biota but not the mycobacteria (polymyxin B, 5 μg/ml; amphotericin B, 4.5 μg/ml; carbenicillin, 20 μg/ml; and trimethoprim, 2.0 μg/ml), and quantified for mycobacteria via heterotrophic plate counts.
General molecular techniques.
Plasmid DNA was prepared using the Quigen Plasmid Mini Kit (Qiagen, Valencia, Calif.). Genomic DNA was prepared as described previously (31). Digoxigenin (DIG)-labeled probes were generated using the DIG Chem-Link System (Boehringer Mannheim, Indianapolis, Ind.) as per the manufacturer's instructions. Southern blot hybridization was performed as described previously (31). PCR was performed using the GC Rich Kit (Roche Diagnostics, Indianapolis, Ind.). Nucleotide sequence analysis was performed at the University of California at San Francisco Biomolecular Resource Center.
Genomic DNA subtractive hybridization.
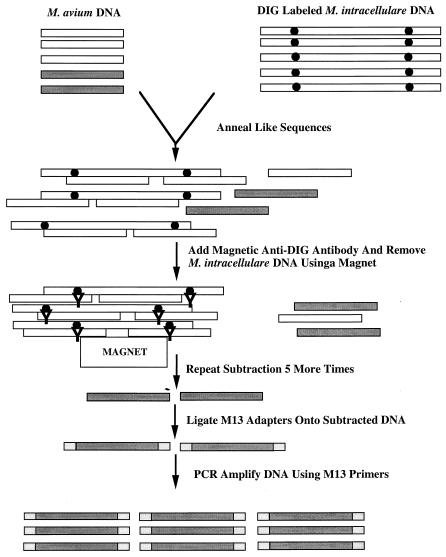
M. intracellulare genomic DNA was sheared to an average size of 2 to 5 kbp using a sonicator (Heat Systems-Ultrasonics Inc., Plainview, N.Y.) and DIG labeled as described above. M. avium strain 104 genomic DNA was digested with Sau3AI to produced fragments of ca. 200 to 500 bp. One microgram of M. avium DNA was mixed with 10 μg of M. intracellulare DNA in subtraction buffer (50 mM HEPES [pH 7.5], 0.5 M NaCl, 0.1% sodium dodecyl sulfate, 1 mM EDTA) and denatured in a thermal cycler (Hybaid, Middlesex, United Kingdom) at 95°C for 45 min. The temperature was lowered at a rate of 4°C/h until the mixture reached 40°C. The mixture was removed and allowed to cool to room temperature. Anti-DIG antibodies bound to magnetic particles (Boehringer Mannheim) were added, and the M. intracellulare DNA fragments, along with any M. avium fragments annealed to them, were removed using a magnet. The subtraction protocol was repeated five more times, after which any remaining M. avium sequences were ligated into M13 reverse Sau3AI adapters consisting of equimolar amounts of GATCCAGGAAACAGCTATGAC and GTCATAGCTGTTTCCTG. The ligation products were then ethanol precipitated to remove unligated adapters and PCR amplified using the M13R primer. PCR products were ligated into pCR2.1 and transformed into E. coli XL1Blue MRF′. Genomic DNA fragments generated by the subtraction protocol were used as probes in Southern hybridization experiments against M. avium and M. intracellulare genomic DNAs to ensure that they hybridized with M. avium but not M. intracellulare.
Electron microscopy.
Portions of the inoculated and control mouse ileal loops described above were cut into 1-mm pieces and fixed in ice-cold 1% glutaraldehyde in phosphate buffer for 1 h. The segments were immersed in 1% OsO4 for 1 h at room temperature, dehydrated through a graded ethanol series, embedded in L. R. White resin, and polymerized at 52°C. Thin sections were cut, stained with uranyl acetate and lead citrate, and examined with an electron microscope.
Nucleotide sequence accession numbers.
The nucleotide sequences of subtracted M. avium DNA products can be obtained from GenBank under accession numbers AF320114, AF320115, AF320116, AF320117, AF320118, AF320119, AF320120, AF320121, AF320122, AF320123, and AF321121.
RESULTS
Acid tolerance
To simulate the acidic conditions encountered in the stomach, we exposed cultures of M. avium, M. intracellulare, and M. smegmatis to pH 2.2 for 2 and 24 h (Table 1). All cultures exposed to pH 2.2 showed a decrease in viable cell numbers at both 2 and 24 h. M. avium was the most acid tolerant species, showing 91 and 34% survival after 2 and 24 h, respectively. M. intracellulare was intermediately acid tolerant, showing 48 and 17% survival, and M. smegmatis was the most acid sensitive, showing only 40 and <1% survival after 2 and 24 h, respectively.
TABLE 1.
Mycobacterial acid tolerance and polymyxin B resistance
| Species | Acid tolerancea (mean % survival ± SD)
|
Polymyxin B resistanceb (MIC, μg/ml) | |
|---|---|---|---|
| 2 h | 24 h | ||
| M. avium | 91 ± 0.7 | 34 ± 9.1 | >500 |
| M. intracellulare | 48 ± 2.8 | 17 ± 6.3 | >500 |
| M. smegmatis | 40 ± 2.9 | <1 ± 0.6 | 32 |
Bacteria were exposed to pH 2.2 for 2 and 24 h as described in Materials and Methods.
The MIC of polymyxin B was assayed as described in Materials and Methods.
Resistance to polymyxin B
Polymyxin B is a small cationic lipoprotein that has been used to model the membrane-disrupting activities of many cationic peptides produced by intestinal epithelial cells (5). The level of resistance to polymyxin B was determined for M. avium, M. intracellulare, and M. smegmatis at concentrations ranging from 0.5 to 500 μg/ml (Table 1). M. smegmatis was most susceptible to polymyxin B, which had an MIC of 32 μg/ml. M. avium and M. intracellulare were both resistant to polymyxin B and grew in medium containing >500 μg of polymyxin B per ml.
Invasion of cultured epithelial cells.
The ability of M. avium and M. intracellulare to invade cultured human epithelial cells was assayed using pharyngeal (HEp-2) and intestinal (HT-29) epithelial cells (Table 2). The percentages of M. avium and M. intracellulare that were able to invade the pharyngeal cells after 1 h were not significantly different (within twofold). However, when we compared the abilities of these strains to invade intestinal epithelial cells, we observed that M. avium was able to invade significantly more efficiently after 1 h (>5-fold).
TABLE 2.
In vitro invasion assays
| Species | Strain | Mean inoculum (106) ± SD | No. of repetitions | Invasion (mean % inoculum ± SD)a
|
|
|---|---|---|---|---|---|
| HEp-2 cells | HT-29 cellsb | ||||
| M. avium | 101 | 2.4 ± 0.3 | 10 | 3.6 ± 0.4 | 2.9 ± 0.2 |
| 104 | 4.1 ± 0.2 | 10 | 3.2 ± 0.3 | 3.3 ± 0.1 | |
| 109 | 3.4 ± 0.2 | 10 | 3.1 ± 0.4 | 2.8 ± 0.4 | |
| M. intracellulare | 83-8705 | 5.7 ± 0.3 | 3 | 2.5 ± 0.4 | 0.5 ± 0.07 |
| 84-8739 | 3.1 ± 0.3 | 5 | 1.6 ± 0.2 | 0.4 ± 0.08 | |
| 86-8953 | 4.1 ± 0.2 | 5 | 2.7 ± 0.4 | 0.3 ± 0.03 | |
The assay was performed as described in Materials and Methods. Invasion was allowed to occur for 1 h.
P < 0.05 for all comparisons between M. avium and M. intracellulare invasion of HT-29 intestinal cells.
Invasion of the intestinal mucosa
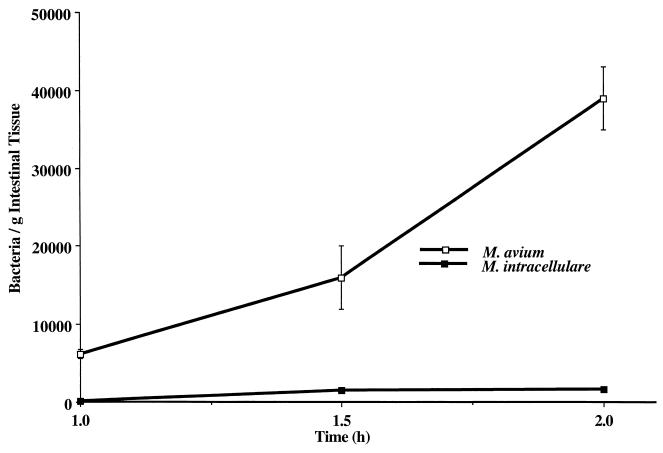
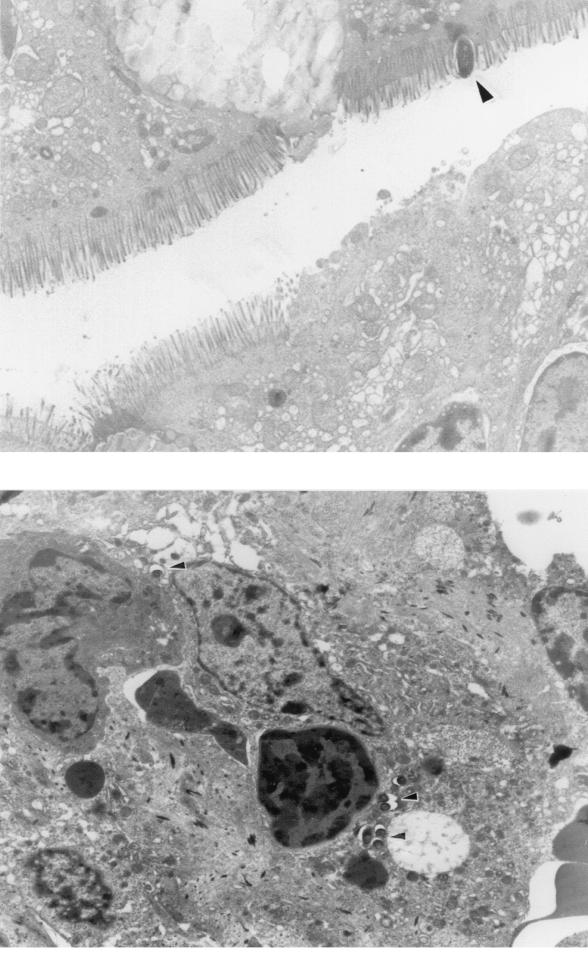
The ability of M. avium and M. intracellulare to interact with the intestinal mucosa was assayed in a mouse intestinal loop model. Approximately 105 bacteria were injected into a 3-cm segment of the intestine above the ileo-cecal area that was closed via two suture lines that blocked flow through the intestinal lumen but did not hinder blood circulation. M. avium was able to invade the intestinal mucosa and reached >6 × 103 bacteria/g of intestinal tissue after 1 h (Fig. 1). The number of bacteria in the intestinal tissues rapidly increased and reached >3.9 × 104/g of intestinal tissue after 2 h. M. intracellulare was substantially less efficient in invading the intestinal mucosa and reached only 1.4 × 102 bacteria/g of intestinal tissue after 1 h (>40-fold fewer than M. avium) and only 1.7 × 103 bacteria/g of intestinal tissue after 2 h (>22-fold fewer than M. avium). To ensure that M. avium was invading the mucosal cells and not just adhering to them, we performed transmission electron microscopy of representative samples after 1 h and observed that M. avium was either in the process of invading the cells or was already intracellular (Fig. 2). Although we believe that the vast majority of the bacteria counted in these assays were intracellular, we cannot exclude the possibility that some were extracellular.
FIG. 1.
Invasion of the mouse intestinal mucosa by M. avium or M. intracellulare. Ligated ileal loops were inoculated with ca. 105 CFU of M. avium or M. intracellulare and left for 1, 1.5, or 2 h, after which the loops were removed and quantified for mycobacteria as described in Materials and Methods. The experiment was repeated five times, and the data shown are the average numbers of bacteria per gram of intestine. For the comparison between M. avium and M. intracellulare, the P value was <0.001. Error bars represent standard deviations from five experiments; the error bars for M. intracellulare are too small to be visible.
FIG. 2.
Invasion of mouse intestinal tissues by M. avium. Mouse ligated ileal loops were inoculated with ca. 105 CFU of M. avium and left for 1 h, after which the ileal loops were removed, washed extensively, and prepared for electron microscopy as described in Materials and Methods. (Top) M. avium in the brush border of an enterocyte. (Bottom) M. avium inside an enterocyte. Arrowheads indicate bacterial cells.
Subtraction of M. avium genomic DNA with DNA from M. intracellulare
We hypothesized that M. avium contains genes, absent in M. intracellulare, that allow it to efficiently invade the intestinal mucosa. To identify these genes, we performed genomic DNA subtractive hybridization of M. avium strain 104 genomic DNA with M. intracellulare ATCC 13950 genomic DNA as described in Materials and Methods and diagrammed in Fig. 3. To ensure that DNA fragments obtained via this protocol were not present in M. intracellulare, we performed Southern analysis under conditions of low stringency with both M. avium and M. intracellulare genomic DNAs, which confirmed that the fragments were unique to M. avium (data not shown). M. avium genomic DNA was also subtracted against itself and PCR amplified, but no products were produced (data not shown).
FIG. 3.
Schematic diagram of the genomic DNA subtractive hybridization technique used to identify genes present in M. avium strain 104 but absent in M. intracellulare. Shaded rectangles, M. avium unique sequences; black circles, DIG label; Y, anti-DIG antibody; hatched rectangles, M13R-Sau3AI adapters.
Identification of M. avium-specific genes.
To identify the genes that the subtracted DNA fragments came from, we compared their sequences to the published M. avium genome sequence using the BLAST program (http://www.tigr.org/cgi-bin/BlastSearch/blast.cgi?organism=m_avium). In all, we identified 21 genes present in M. avium that were absent in M. intracellulare (Table 3). The JAM1 and JAM2 products are homologous to the products of the M. tuberculosis genes Rv0227 and Rv0226, respectively. These genes are predicted to be in an operon but have no known function. The JAM3, JAM6, and JAM14 products are homologous to M. tuberculosis PPE proteins that have no known function. The JAM4 product has homology with Zwf of M. tuberculosis, one of three glucose-6-phosphate dehydrogenase (G6PD) isoenzymes. The JAM5 product has homology with the M. tuberculosis protein encoded by Rv0106, which has no known function, and YciC, a membrane-bound protein that is involved in zinc uptake in Bacillus subtilis (18). The JAM7 product has homology with LipP from M. tuberculosis and EstA from Streptomyces chrysomallus. The JAM8 product has homology with the M. tuberculosis and Streptomyces coelicolor protein Tig, a chaperon/prolylisomerase. The JAM9 product is a homolog of an ABC-type transporter ATP-binding protein from M. tuberculosis (Rv1463) and Mycobacterium leprae (CAB16169). The JAM10 product is a NifS-like protein that is likely involved in the assembly of iron-sulfur clusters. The JAM11 product has homology to the M. tuberculosis Rv1871c gene product of unknown function. The JAM12 product has a high degree of homology with LonA, an ATP-dependent protease. The JAM13 product has homology to the membrane-bound lipoproteins LprL and LprK of M. tuberculosis. The JAM15 product has homology with the S. coelicolor regulatory protein CAB88970, a putative repressor of secondary metabolism and a member of the LuxR family of transcriptional regulators. The JAM16 product has homology to the M. tuberculosis protein encoded by Rv3254, which has no know function. The JAM17 product has homology with a peptidase of S. coelicolor and hydrolases from Pseudomonas aeruginosa and Campylobacter jejuni. The JAM18 product has homology to an S. coelicolor Na/H exchanger (CAB5180). The JAM19 product has homology to an M. leprae oxidoreductase. The JAM20 product has homology to an M. tuberculosis probable methyltransferase, and the JAM21 product has homology with an S. lividans protein of unknown function.
TABLE 3.
Gene products unique to M. avium identified via subtractive hybridization
| Gene product | Homolog | % Identity/% similarity/ no. of amino acids | Description/function |
|---|---|---|---|
| JAM1 | M. tuberculosis Rv0227c | 74/80/389 | Possible membrane protein/unknown |
| M. leprae CAA18554.1 | 67/76/446 | Putative membrane protein/unknown | |
| JAM2 | M. tuberculosis Rv0226c | 58/61/259 | Probable membrane protein/unknown |
| M. leprae CAA18553.1 | 50/55/276 | Putative membrane protein/unknown | |
| JAM3 | M. tuberculosis Rv1779c | 35/41/289 | PPE family of proteins/unknown |
| JAM4 | M. tuberculosis Zwf | 91/95/483 | GGPD/oxidative stress |
| S. coelicolor CAB50762.1 | 69/79/483 | G6PD/oxidative stress | |
| JAM5 | M. tuberculosis Rv0160 | 65/75/378 | Unknown/unknown |
| B. subtilis YciC | 25/41/359 | Zn Uptake/Zn homeostasis | |
| JAM6 | M. tuberculosis Rv0286 | 32/40/148 | PPE family of protens/unknown |
| JAM7 | M. tuberculosis LipP | 78/83/320 | Probable esterase/lipid biosynthesis |
| S. chrysomallus EstA | 45/55/317 | Esterase/unknown | |
| JAM8 | M. tuberculosis Tig | 96/97/59 | Chaperone-proylisomerase/protein folding and secretion |
| S. coelicolor Tig | 54/74/59 | Chaperone-proylisomerase/protein folding and secretion | |
| JAM9 | M. tuberculosis Rv1463 | 67/72/253 | ABC transporter/ATP-binding protein |
| M. leprae CAB16169.1 | 64/69/253 | ABC transporter/ATP-binding protein | |
| JAM10 | M. tuberculosis NifS | 88/91/100 | Protein modification/Fe-S cluster formation |
| M. leprae NifS | 82/94/100 | Protein modification/Fe-S cluster formation | |
| JAM11 | M. tuberculosis Rv1871c | 77/81/109 | Unknown/unknown |
| S. coelicolor CAB88434.1 | 56/66/112 | Unknown/unknown | |
| JAM12 | M. smegmatis LonA | 85/92/227 | ATP-dependent protease/protein degradation |
| M. xanthus LonA | 49/64/276 | ATP-dependent protease/protein degradation | |
| JAM13 | M. tuberculosis LprL | 28/47/374 | Membrane lipoprotein/unknown |
| M. tuberculosis LprK | 25/42/261 | Membrane lipoprotein/unknown | |
| JAM14 | M. tuberculosis Rv0453 | 30/37/147 | PPE family of proteins/unknown |
| JAM15 | S. coelicolor CAB88970 | 34/52/292 | Regulatory protein/regulation of secondary metabolites |
| M. tuberculosis Rv1204 | 25/36/526 | Possible secreted protein/unknown | |
| JAM16 | M. tuberculosis Rv3254 | 40/45/344 | Unknown/unknown |
| S. coelicolor CAA20504 | 36/50/357 | Possible secreted protein/unknown | |
| JAM17 | S. coelicolor CAB56384 | 57/67/403 | Probable peptidase/protein degradation |
| P. aeruginosa AAG06310 | 38/51/367 | Probable hydrolase/unknown | |
| JAM18 | S. coelicolor CAB51980 | 26/37/412 | Putative Na/H exchanger/unknown |
| M. tuberculosis YjcE | 24/37/538 | Putative Na/H exchanger/unknown | |
| JAM19 | M. leprae CAB09639 | 81/83/285 | Oxidoreductase/unknown |
| D. radiodurans AAF11444.1 | 57/72/280 | Oxidoreductase/unknown | |
| JAM20 | M. tuberculosis Rv0560c | 68/87/250 | Probable methyltransferase/unknown |
| M. tuberculosis Rv3699 | 52/64/108 | Probable methyltransferase/unknown | |
| JAM21 | S. lividans AAC25767 | 61/72/145 | Unknown/unknown |
| M. tuberculosis Rv1558 | 56/71/139 | Unknown/unknown |
DISCUSSION
MAC is composed of two closely related species, M. avium and M. intracellulare (2). However, there are significant differences in the pathologies caused by these organisms and the type of hosts they infect. M. avium infections are more common in AIDS patients, are acquired primarily via the gastrointestinal tract, and often result in disseminated disease (4, 39). M. intracellulare infections are more common in people with underlying lung pathology, are acquired via the respiratory tract, and usually remain limited to the pulmonary tissue (4, 39). Because of this information, we decided to compare the abilities of M. avium and M. intracellulare to evade the gastrointestinal defense barriers, such as the acidic conditions encountered in the stomach and the membrane-disrupting activities of cationic peptides in the intestine. We also examined the ability of the bacteria to invade intestinal epithelial cells in vitro and in vivo.
Enteropathogenic bacteria must be able to survive the acidic pH encountered in the stomach (pH of <3 for 2 h) (20, 21). We examined the abilities of M. avium, M. intracellulare, and M. smegmatis to survive pH 2.2 for 2 or 24 h and found that the order of acid tolerance was M. avium > M. intracellulare > M. smegmatis. Although there were differences in the level of acid tolerance among the mycobacteria, all three organisms were able to tolerate the acidic conditions encountered in the stomach better than many common enteropathogenic bacteria, including Salmonella enterica serovar Typhimurium (3), Listeria monocytogenes (26), and Vibrio cholerae (27). Thus, it does not appear that the level of acid tolerance of M. avium versus M. intracellulare is a limiting factor for gastrointestinal disease, although the difference observed may be clinically significant with low cell numbers.
The intestinal tracts of mammals are constantly exposed to potentially pathogenic microorganisms, but they remain disease free due in part to antimicrobial cationic peptides released from intestinal epithelial cells (7, 19, 30). The ability of the mycobacterial species to resist the membrane-disrupting activities of cationic peptides was modeled using polymyxin B (5). M. smegmatis was most susceptible to polymyxin B, which had a MIC of 32 μg/ml. Both M. avium and M. intracellulare were highly resistant and grew at concentrations of >500 μg of polymyxin B per ml. However, the polymyxin B MICs for many enteric bacteria, including E. coli (37), S. enterica serovar Typhimurium (37), and S. enterica (40), are less than 1 μg/ml. Thus, it does not seem likely that resistance to cationic peptides, as measured by resistance to polymyxin B, is a factor that limits the ability of M. avium or M. intracellulare to colonize the intestinal mucosa.
When assayed in a mouse intestinal loop model, M. avium invaded the intestinal mucosa >40-fold more efficiently than M. intracellulare after 1 h. To ensure that the bacteria were invading the mucosal cells, we performed transmission electron microscopy and observed that the bacteria either were in the process of invasion or were already internalized; however, we cannot exclude the possibility that some bacteria were only adherent. Because M. avium and M. intracellulare multiply so slowly (ca. one division per 20 h) (12), the percentage of bacteria that exit the lumen and invade the intestinal mucosa over short periods of time can be calculated by quantifying the number of bacteria in the intestinal tissues. In intestinal segments inoculated with M. avium, approximately 39% of the bacteria entered the intestinal mucosa after 2 h, compared with only 1.7% for M. intracellulare. These data are consistent with the in vitro data obtained using cultured intestinal epithelial cells and multiple strains of M. avium and M. intracellulare (Table 2) and confirm our hypothesis that only M. avium is able to efficiently invade the intestinal mucosa.
Genomic DNA subtractive hybridization of M. avium strain 104 genomic DNA with M. intracellulare chromosomal DNA (Fig. 3) revealed several genes present in M. avium that were absent in M. intracellulare. This list of genes should not be considered complete, as the subtractive hybridization protocol likely missed several genes which may play important roles in invasion; furthermore, we cannot conclude that all or any of the genes identified are absolutely necessary for invasion. Many of these genes that we identified encode proteins with no known or predicted function, including PPE proteins (JAM3, JAM6, and JAM14), membrane proteins of unknown function (JAM1 and JAM2), and other unknown proteins (JAM11, JAM16, and JAM21) (Table 3). However, some of the identified genes have the potential to participate in intestinal pathogenesis. One such gene encodes a protein with predicted G6PD activity. G6PD catalyzes the first step in the pentose phosphate pathway, which produces ribose for DNA synthesis and reducing equivalents in the form of NADPH that can be used for anabolic reactions and oxidative damage repair (16, 35). In M. avium there are at least three G6PD isoenzymes; one is homologous to the developmentally regulated DevB of Anabaena sp. (C. C. Bauer, unpublished data [GenBank accession no. U14553]), and the other two have various amounts of homology to Zwf of E. coli. In M. avium it is likely that each of the isoenzymes is regulated independently in response to factors such as growth rate, physiological state, and level of oxidative stress. Although nothing is known about the role that zwf plays in the pathogenicity of M. avium, zwf mutants of S. enterica serovar Typhimurium are avirulent (25). In P. aeruginosa the soxR regulon, including zwf, is required for the colonization of burn wounds, and soxR mutants which do not express multiple genes, including zwf, are unable to cause bacteremia and systemic disease in a burned mouse model (22).
Another gene that is possibly involved in intestinal epithelial cell invasion has homology with the chaperone/peptidyl-prolyl-cis-trans-isomerase (PPIase) Tig. This protein likely functions in the folding and secretion of proteins (11, 14). Enzymes with PPIase activity have been implicated as virulence factors in several pathogens, including Legionella pneumophila (17, 38), Mycoplasma pneumoniae (32), and S. enterica serovar Typhimurium (36). In S. enterica SurA functions as a PPIase and plays a role in the organism's ability to adhere to and invade HEp-2 cells (36). S. enterica surA mutants are attenuated for virulence in mice and can be used as an effective oral vaccine against virulent salmonellae (36).
An ABC-type transporter ATP-binding protein and a NifS homolog have also been identified and may play a role in intestinal invasion. ABC-type transporters are associated with a variety of processes that involve the translocation of small molecules across membranes (23), and NifS homologs are involved in the assembly of iron-sulfur clusters in proteins (41, 42). These genes have been implicated as M. smegmatis stress response genes and are upregulated during anaerobic stationary phase (29). Interestingly, these are the same conditions encountered in the human host after ingestion, and we have previously demonstrated that these conditions stimulate M. avium to invade intestinal epithelial cells with increased efficiency. It is plausible that these proteins are part of an anaerobic regulon required for gastrointestinal invasion. We are currently investigating the roles that these and other anaerobically induced proteins play in intestinal invasion.
Homologs of the M. smegmatis protease LonA and an S. coelicolor probable peptidase were also identified. Proteases mediate the degradation of damaged and short-lived proteins and provide amino acids for new protein synthesis. It is likely that the rapid elimination of key metabolic and regulatory proteins and the production of others are essential for M. avium to alternate from the saprophytic to the pathogenic lifestyle in the intestine. It is also likely that M. avium in the environment is in stationary phase and produces these and other stationary-phase-induced proteins, such as NifS and the ABC-type transporter ATP-binding protein mentioned above, and that this arsenal may be important for gastrointestinal invasion.
The ability to invade the gastrointestinal tract and cause disseminated disease is a complex trait and likely requires the coordinate production of multiple virulence factors in response to environmental cues. We have demonstrated that the invasion of the intestinal mucosa is an important phenotypic difference between virulent strains of M. avium and M. intracellulare. Although the subtractive hybridization protocol likely missed several genes present in M. avium but absent in M. intracellulare, it did identify several potential virulence factors that are unique to M. avium that may allow it to invade the intestinal mucosa more efficiently than M. intracellulare. Unfortunately to date there is no methodology to create site-specific mutations in M. avium, and thus we cannot examine the roles that individual genes play in intestinal pathogenesis. However, we are currently working towards developing such techniques in order to delineate the roles that these genes play in invasion and dissemination.
ACKNOWLEDGMENTS
This work was supported by grant AI-43199 from the National Institute of Allergy and Infectious Diseases.
We thank Jeff Cirillo, Merrill Hines, Dirk Wagner, and Lowell Young for critical reading of the manuscript.
REFERENCES
- 1.Akopyants N S, Fradkov A, Diatchenko L, Hill J E, Siebert P D, Lukyanov S A, Sverdlov E D, Berg D E. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95:13108–13113. doi: 10.1073/pnas.95.22.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baess I. Deoxyribonucleic acid relatedness among species of slowly-growing mycobacteria. Acta Pathol Microbiol Scand Sect B. 1979;87:221–226. doi: 10.1111/j.1699-0463.1979.tb02430.x. [DOI] [PubMed] [Google Scholar]
- 3.Bearson B L, Wilson L, Foster J W. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J Bacteriol. 1998;180:2409–2417. doi: 10.1128/jb.180.9.2409-2417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beggs M L, Stevanova R, Eisenach K D. Species identification of Mycobacterium avium complex isolates by a variety of molecular techniques. J Clin Microbiol. 2000;38:508–512. doi: 10.1128/jcm.38.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengoechea J A, Lindner B, Seydel U, Diaz R, Moriyon I. Yersinia pseudotuberculosis and Yersinia pestis are more resistant to bactericidal cationic peptides than Yersinia enterocolitica. Microbiology. 1998;144:1509–1515. doi: 10.1099/00221287-144-6-1509. [DOI] [PubMed] [Google Scholar]
- 6.Bermudez L E, Young L S. Factors affecting invasion of HT-29 and HEp-2 epithelial cells by organisms of the Mycobacterium avium complex. Infect Immun. 1994;62:2021–2026. doi: 10.1128/iai.62.5.2021-2026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bevins C L, Martin-Porter E, Ganz T. Defensins and innate host defence of the gastrointestinal tract. Gut. 1999;45:911–915. doi: 10.1136/gut.45.6.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bloom B R, Murray C J. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 9.Boddinghaus B, Wolters J, Heikens W, Bottger E C. Phylogenetic analysis and identification of different serovars of Mycobacterium intracellulare at the molecular level. FEMS Microbiol Lett. 1990;58:197–203. doi: 10.1111/j.1574-6968.1990.tb13978.x. [DOI] [PubMed] [Google Scholar]
- 10.Brown P K, Curtiss R., III Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc Natl Acad Sci USA. 1996;93:11149–11154. doi: 10.1073/pnas.93.20.11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston E J, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Crowle A J, Ross E R, Cohn D L, Gilden J, May M H. Comparison of the abilities of Mycobacterium avium and Mycobacterium intracellulare to infect and multiply in cultured human macrophages from normal and human immunodeficiency virus-infected subjects. Infect Immun. 1992;60:3697–3703. doi: 10.1128/iai.60.9.3697-3703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damsker B, Bottone E J. Mycobacterium avium-Mycobacterium intracellulare from the intestinal tracts of patients with the acquired immunodeficiency syndrome: concepts regarding acquisition and pathogenesis. J Infect Dis. 1985;151:179–181. doi: 10.1093/infdis/151.1.179. [DOI] [PubMed] [Google Scholar]
- 14.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 15.Emmerth M, Goebel W, Miller S I, Hueck C J. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J Bacteriol. 1999;181:5652–5661. doi: 10.1128/jb.181.18.5652-5661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer G, Bang H, Ludwig B, Mann K, Hacker J. Mip protein of Legionella pneumophila exhibits peptidyl-prolyl-cis/trans isomerase (PPlase) activity. Mol Microbiol. 1992;6:1375–1383. doi: 10.1111/j.1365-2958.1992.tb00858.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaballa A, Helmann J D. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol. 1998;180:5815–5821. doi: 10.1128/jb.180.22.5815-5821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganz T, Lehrer R I. Antimicrobial peptides of vertebrates. Curr Opin Immunol. 1998;10:41–44. doi: 10.1016/s0952-7915(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 20.Giannella R A, Broitman S A, Zamcheck N. Gastric acid barrier to ingested microorganisms in man: studies in vivo and in vitro. Gut. 1972;13:251–256. doi: 10.1136/gut.13.4.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannella R A, Broitman S A, Zamcheck N. Influence of gastric acidity on bacterial and parasitic enteric infections. A perspective. Ann Intern Med. 1973;78:271–276. doi: 10.7326/0003-4819-78-2-271. [DOI] [PubMed] [Google Scholar]
- 22.Ha U, Jin S. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect Immun. 1999;67:5324–5331. doi: 10.1128/iai.67.10.5324-5331.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins C F. ABC transporters: from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson M A, Hopewell P C, Yajko D M, Hadley W K, Lazarus E, Mohanty P K, Modin G W, Feigal D W, Cusick P S, Sande M A. Natural history of disseminated Mycobacterium avium complex infection in AIDS. J Infect Dis. 1991;164:994–998. doi: 10.1093/infdis/164.5.994. [DOI] [PubMed] [Google Scholar]
- 25.Lundberg B E, Wolf R E, Jr, Dinauer M C, Xu Y, Fang F C. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect Immun. 1999;67:436–438. doi: 10.1128/iai.67.1.436-438.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marron L, Emerson N, Gahan C G, Hill C. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl Environ Microbiol. 1997;63:4945–4947. doi: 10.1128/aem.63.12.4945-4947.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrell D S, Camilli A. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol. 2000;182:5342–5350. doi: 10.1128/jb.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrow B J, Graham J E, Curtiss R., III Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect Immun. 1999;67:5106–5116. doi: 10.1128/iai.67.10.5106-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murugasu-Oei B, Tay A, Dick T. Upregulation of stress response genes and ABC transporters in anaerobic stationary-phase Mycobacterium smegmatis. Mol Gen Genet. 1999;262:677–682. doi: 10.1007/s004380051130. [DOI] [PubMed] [Google Scholar]
- 30.Ouellette A J. IV. Paneth cell antimicrobial peptides and the biology of the mucosal barrier. Am J Physiol. 1999;277:G257–G261. doi: 10.1152/ajpgi.1999.277.2.G257. [DOI] [PubMed] [Google Scholar]
- 31.Parker A E, Bermudez L E. Sequence and characterization of the glyceraldehyde-3-phosphate dehydrogenase of Mycobacterium avium: correlation with an epidermal growth factor binding protein. Microb Pathog. 2000;28:135–144. doi: 10.1006/mpat.1999.0335. [DOI] [PubMed] [Google Scholar]
- 32.Reddy S P, Rasmussen W G, Baseman J B. Correlations between Mycoplasma pneumoniae sensitivity to cyclosporin A and cyclophilin-mediated regulation of mycoplasma cytadherence. Microb Pathog. 1996;20:155–169. doi: 10.1006/mpat.1996.0014. [DOI] [PubMed] [Google Scholar]
- 33.Saito H, Tomioka H, Sato K, Tasaka H, Tsukamura M, Kuze F, Asano K. Identification and partial characterization of Mycobacterium avium and Mycobacterium intracellulare by using DNA probes. J Clin Microbiol. 1989;27:994–997. doi: 10.1128/jcm.27.5.994-997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salo W L, Aufderheide A C, Buikstra J, Holcomb T A. Identification of Mycobacterium tuberculosis DNA in a pre-Columbian Peruvian mummy. Proc Natl Acad Sci USA. 1994;91:2091–2094. doi: 10.1073/pnas.91.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storz G, Tartaglia L A, Farr S B, Ames B N. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 36.Sydenham M, Douce G, Bowe F, Ahmed S, Chatfield S, Dougan G. Salmonella enterica serovar Typhimurium surA mutants are attenuated and effective live oral vaccines. Infect Immun. 2000;68:1109–1115. doi: 10.1128/iai.68.3.1109-1115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaara M, Vaara T. Ability of cecropin B to penetrate the enterobacterial outer membrane. Antimicrob Agents Chemother. 1994;38:2498–2501. doi: 10.1128/aac.38.10.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wintermeyer E, Ludwig B, Steinert M, Schmidt B, Fischer G, Hacker J. Influence of site specifically altered Mip proteins on intracellular survival of Legionella pneumophila in eukaryotic cells. Infect Immun. 1995;63:4576–4583. doi: 10.1128/iai.63.12.4576-4583.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakrus M A, Good R C. Geographic distribution, frequency, and specimen source of Mycobacterium avium complex serotypes isolated from patients with acquired immunodeficiency syndrome. J Clin Microbiol. 1990;28:926–929. doi: 10.1128/jcm.28.5.926-929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yethon J A, Gunn J S, Ernst R K, Miller S I, Laroche L, Malo D, Whitfield C. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect Immun. 2000;68:4485–4491. doi: 10.1128/iai.68.8.4485-4491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng L, Dean D R. Catalytic formation of a nitrogenase iron-sulfur cluster. J Biol Chem. 1994;269:18723–18726. [PubMed] [Google Scholar]
- 42.Zheng L, White R H, Cash V L, Dean D R. Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry. 1994;33:4714–4720. doi: 10.1021/bi00181a031. [DOI] [PubMed] [Google Scholar]