Abstract
Neuroinflammation caused by COVID‐19 negatively impacts brain metabolism and function, while pre‐existing brain pathology may contribute to individuals' vulnerability to the adverse consequences of COVID‐19. We used summary statistics from genome‐wide association studies (GWAS) to perform Mendelian randomization (MR) analyses, thus assessing potential associations between multiple sclerosis (MS) and two COVID‐19 outcomes (severe acute respiratory syndrome coronavirus 2 [SARS‐CoV‐2] infection and COVID‐19 hospitalization). Genome‐wide risk genes were compared between the GWAS datasets on hospitalized COVID‐19 and MS. Literature‐based analysis was conducted to construct molecular pathways connecting MS and COVID‐19. We found that genetic liability to MS confers a causal effect on hospitalized COVID‐19 (odd ratio [OR]: 1.09, 95% confidence interval: 1.03−1.16) but not on SARS‐CoV‐2 infection (1.03, 1.00−1.05). Genetic liability to hospitalized COVID‐19 confers a causal effect on MS (1.15, 1.02−1.30). Hospitalized COVID‐19 and MS share five risk genes within two loci, including TNFAIP8, HSD17B4, CDC37, PDE4A, and KEAP1. Pathway analysis identified a panel of immunity‐related genes that may mediate the links between MS and COVID‐19. Our study suggests that MS was associated with a 9% increased risk for COVID‐19 hospitalization, while hospitalized COVID‐19 was associated with a 15% increased risk for MS. Immunity‐related pathways may underlie the link between MS on COVID‐19.
Keywords: COVID‐19, Mendelian randomization, multiple sclerosis
1. INTRODUCTION
Since the inception of the pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), a myriad of risk and protective factors have been reported for their association with the susceptibility or severity of COVID‐19. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 Meanwhile, a subpopulation of individuals with COVID‐19 may suffer from post‐COVID‐19 syndrome after they recover from acute illness, which is known as long COVID‐19. 9 , 10 , 11 , 12 , 13 Neurological manifestations are common among individuals with COVID‐19 infection. 14 , 15 It is well documented that the coronavirus SARS‐CoV‐2 invades the central nervous system, impacting the structure, metabolism, and function of the brain. 16 The neurotropic and neuroinvasive properties of COVID‐19 pose a remarkable threat to the brain health of affected individuals.
Many of the symptoms of “long COVID” are of distinct neurological origins. Pre‐existing CNS pathology, especially those affecting the metabolism and integrity of neurons and glia, may make individuals more vulnerable to the consequences of coronavirus invasion and, therefore, may adversely influence COVID‐19 outcomes. Multiple sclerosis (MS) is an acquired inflammatory demyelinating disease, possibly triggered by viral infections or reactivation episodes through stimulating immune responses. 17 , 18 Experimental studies show that coronaviruses may cause demyelination in animal models 19 and, at least in some cases, provoke a relapse of MS in humans. 20
The Mendelian randomization (MR) framework can be used to infer causative links between genetically determined exposure phenotypes and subsequent disease outcomes; this is done by utilizing genetic variants as instrumental variables. MR analysis has been widely employed in recent studies to explore relationships between related traits. 21 , 22 , 23 It is not known whether the brain pathological changes in MS may aggravate the severity of COVID‐19 or MS could be triggered by various COVID‐19 outcomes. We sought to explore shared genetic variation and the potential mutual causal associations between MS and two COVID‐19 outcomes (SARS‐CoV‐2 infection and COVID‐19 hospitalization). Exploring genetic links between MS and COVID‐19 may help to improve the management of MS in the context of coronavirus infection.
2. METHODS
2.1. GWAS summary datasets
The study utilized publicly available GWAS summary results. The summary statistics for the outcome of COVID‐19 were obtained from the COVID‐19 Host Genetics Initiative (HGI) GWAS meta‐analysis round 7 (release date: April 8, 2022, without the 23andMe cohort), including SARS‐CoV‐2 infection (122 616 cases and 2 475 240 controls) and hospitalized COVID‐19 (32 519 cases and 2 062 805 controls). 24 The MS GWAS data set included 47 429 cases and 68 374 controls. 25 All participants were of European origin. Ethical approval had been obtained in all the original studies. The SARS‐CoV‐2 infection data set mainly reflects the overall susceptibility to the virus, whereas hospitalized COVID‐19 represents the severity of the disease. We called hospitalized COVID‐19 “severe COVID‐19.”
2.2. Genetic correlation analysis
The genetic correlations between MS and the COVID‐19 outcomes were calculated using LD score regression. 26 , 27 The 1000 Genome Project phase 3 was used to estimate the LD structure for European populations. Single‐nucleotide polymorphisms (SNPs) were filtered by 1.1 million variants, a subset of 1000 Genomes and HapMap3, with MAF above 0.05.
2.3. MR analysis
MR demands three main assumptions on an instrumental variable (IV): (1) it is associated with the exposure; (2) it is not associated with confounding factors influencing the relationship between the exposure and the outcome; and (3) it cannot be associated with the outcome directly (but can only indirectly impact the outcome by its effect on the exposure). The analyses were performed using three complementary methods implemented in TwoSampleMR, 28 including inverse variance weighted (IVW), weighted median, and MR‐Egger. They have different assumptions about horizontal pleiotropy. 29 We used the IVW model as our primary MR method. The IVW model assumes an intercept of zero and provides a consistent estimate of the causality by a fixed‐effect meta‐analysis. The weighted median model places more weight on precise IVs; therefore, the estimate remains consistent even if up to 50% of the IVs are invalid or weak. The MR‐Egger model assumes that the pleiotropic effects are independent and applies a weighted linear regression of the outcome coefficient on the exposure coefficient. 29 The weighted median and MR‐Egger models are less statistically powerful than the IVW model but more robust to horizontal pleiotropy or invalid instruments. The intercept from the MR‐Egger regression was utilized to evaluate the average horizontal pleiotropy. 29 The IVs are not all valid when the MR Egger intercept significantly differs from zero. The significant associations between MS and the COVID‐19 outcomes were determined by IVW‐based FDR < 0.05.
For each exposure phenotype, SNPs with genome‐wide significance (p < 5 × 10–8) were selected as IVs and further pruned using a clumping r 2 cutoff of 0.01 within a 10 Mb window, using the 1000 Genomes Project Phase 3 (EUR) as the reference panel. For each MR analysis, we removed SNPs not present in the outcome data set and palindromic SNPs with intermediate allele frequencies. We harmonized each pair of the exposure and outcome datasets by aligning the effect allele for exposure and outcome and obtained variant effects and standard errors of each data set.
2.4. Shared genomic loci between COVID‐19 and MS
To identify overlapping risk genes between COVID‐19 and MS, we retrieved genome‐wide risk genes for the two traits from the two GWAS datasets. Functional mapping and annotation (FUMA) software was used to map SNPs to genes and identify LD‐independent genomic regions. 30 All genes located within the 10 kb vicinity of each variant were mapped. Independent significant SNPs (IndSigSNPs) were extracted when their p value was genome‐wide significant (p ≤ 5.0E‐08) and independent of each other (r 2 < 0.6). Lead SNPs were identified as a subset of the independent significant SNPs that were in LD with each other at r 2 < 0.1 within a 500 kb window. Genomic risk loci were identified by merging lead SNPs located at a distance of less than 500 kb from each other. Clumping procedures were carried out based on the European 1000 Genomes Project phase 3 reference panel. Due to extensive LD, the entire major histocompatibility complex locus was merged into one region (chr6: 25−35 Mb).
To identify the tissue specificity of a phenotype, SNP‐based tissue enrichment analysis was conducted by FUMA, 30 which utilizes gene‐property analyses to test associations between tissue‐specific gene expression profiles in general GTEx V8 tissues and GWAS hits.
2.5. Knowledge‐based analysis
To explore the potential connection between MS and COVID‐19 at the molecular level, large‐scale literature data mining was executed in the Pathway Studio (www.pathwaystudio.com) environment, 31 containing approximately 14 million unique associations from >40 million scientific references. Then, a set of molecular pathways connecting MS and COVID‐19 was constructed. First, the downstream targets and upstream regulators of MS and COVID‐19 were identified, followed by a manual review of the references and related sentences for quality control of each extracted relationship. The relationships with no polarity or indirectly related to COVID‐19 or MS were removed. The remaining relationships were employed to build a map of the molecular pathways connecting MS and COVID‐19.
Protein‐protein interactions (PPIs) among the genes mediating the effect of MS on COVID‐19 were derived using STRING v11. 32 KEGG‐based pathway enrichment analyses of the genes mediating the effect of MS on COVID‐19 were conducted using FUMA. 30
3. RESULTS
3.1. Genetic correlation analysis
Genetic correlation analyses indicated that MS had a nominal positive genetic correlation with hospitalized COVID‐19 (r g = 0.12 ± 0.06, p = 0.038, FDR = 0.076). However, MS did not have genetic correlations with SARS‐CoV‐2 infection (r g = 0.06 ± 0.06, p = 0.321).
3.2. MR analysis
Since each COVID‐19 data set had different sets of SNPs and the SNPs not present in the outcome data set were removed before the MR analysis, different numbers of IVs were yielded for each MR analysis. A total of 82 and 83 IVs were obtained from the MS data set for the MR analysis of MS on SARS‐CoV‐2 infection and hospitalized COVID‐19, respectively. We detected that genetic liability to MS confers a causal effect on hospitalized COVID‐19 (odd ratio: 1.09, 95% confidence interval: 1.03−1.16) but not on SARS‐CoV‐2 infection (1.03, 1.00−1.05, p = 0.038, FDR = 0.051) (Table 1 and Figure 1).
Table 1.
Causal associations between MS and COVID‐19
| Exposure | Outcome | b (se) | OR [95%CI] | N_IV | Egger_intercept | P_pleiotropy | p | FDR |
|---|---|---|---|---|---|---|---|---|
| MS | SARS‐CoV‐2 infection | 0.026 (0.012) | 1.03 [1.00−1.05] | 82 | 0.003 | 0.073 | 0.038 | 0.051 |
| MS | Hospitalized COVID‐19 | 0.090 (0.031) | 1.09 [1.03−1.16] | 83 | −0.000 | 0.922 | 3.45E‐03 | 0.014 |
| SARS‐CoV‐2 infection | MS | 0.165 (0.244) | 1.18 [0.73−1.90] | 21 | 0.023 | 0.387 | 0.500 | 0.500 |
| Hospitalized COVID‐19 | MS | 0.143 (0.062) | 1.15 [1.02−1.30] | 36 | 0.022 | 0.083 | 0.022 | 0.044 |
Abbreviations: b, effect size; CI, confidence interval; MS, multiple sclerosis; N_IV, number of instrumental variables; OR, odds ratio; se, standard error.
Figure 1.
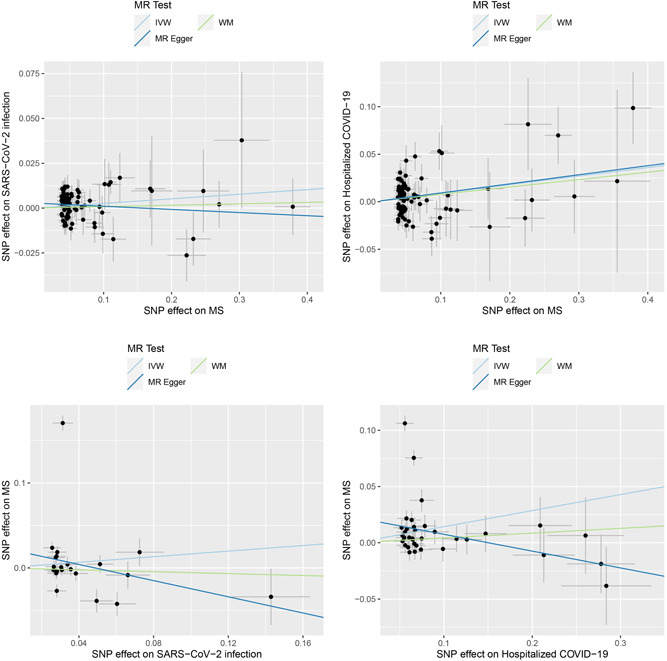
Causal associations between COVID‐19 outcomes and MS. The upper panel shows the causal effects of MS on COVID‐19 outcomes. The lower panel shows the causal effects of COVID‐19 outcomes on MS. IVW, inverse variance weighted; MS, multiple sclerosis; WM, weighted median.
In the MR analysis of the causal effects of the COVID‐19 outcomes on MS, the numbers of IVs were 21 for SARS‐CoV‐2 infection and 36 for hospitalized COVID‐19. We found that genetic liability to hospitalized COVID‐19 conferred a causal effect on MS (1.15, 1.02−1.30, p = 0.022, FDR = 0.044) (Table 1 and Figure 1).
The sensitivity analyses showed that the directions of causal effect estimates across the methods were largely the same (Supporting Information: Table 1). Notably, tests of MR‐Egger regression did not support directional pleiotropy in this MR analysis (MR‐Egger intercept < 0.03, p > 0.05).
3.3. Shared genomic loci between MS and hospitalized COVID‐19
In FUMA analysis, a total of 74 and 32 genomic loci were associated with MS and hospitalized COVID‐19, respectively (Supporting Information: Tables 2−3). MS and COVID‐19 had two overlapping loci, residing in 5q23.1 and 19p13.2 (Table 2 and Figure 2A). These two loci contained 5 protein‐coding genes shared between MS and COVID‐19, including TNFAIP8, HSD17B4, CDC37, PDE4A, and KEAP1.
Table 2.
Shared genomic loci between MS and hospitalized COVID‐19
| Trait | SNP | CHR | BP | Start:End | A1/A2 | p | Genes |
|---|---|---|---|---|---|---|---|
| Hospitalized COVID‐19 | rs564985937 | 5 | 118802956 | 118734448:118803517 | T/C | 2.40E‐08 | TNFAIP8; HSD17B4 |
| MS | rs28762138 | 5 | 118815815 | 118313421:119013643 | T/G | 1.46E‐08 | DTWD2; DMXL1; TNFAIP8; HSD17B4 |
| Hospitalized COVID‐19 | rs34536443 | 19 | 10463118 | 10392638:10662236 | C/G | 3.83E‐20 | RAVER1; ICAM1; ICAM5; ICAM4; FDX1L; ICAM3; TYK2; ZGLP1; CDC37; PDE4A; KEAP1;S1PR5; ATG4D |
| MS | rs28834106 | 19 | 10592144 | 10499002:10616303 | T/C | 3.79E‐11 | CDC37; PDE4A; KEAP1 |
Abbreviations: CHR, chromosome; BP, base pair.
Figure 2.
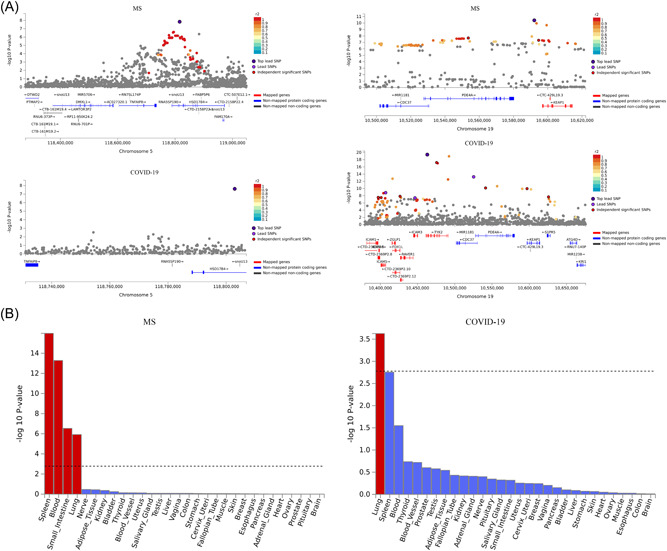
Genomic loci and tissue specificity of the two GWAS datasets. (A) Two overlapping loci between MS and hospitalized COVID‐19. (B) Tissue‐specific expression analysis for MS and hospitalized COVID‐19. Significantly enriched tissues are highlighted in red. MS, multiple sclerosis; WM, weighted median.
Tissue‐specific expression analysis showed that GWAS hits for hospitalized COVID‐19 were significantly enriched in the lung (Figure 2B), while GWAS hits for MS were significantly enriched in the spleen, blood, small intestine and lung (Figure 2B).
3.4. Knowledge‐based analysis
Mining of the molecular relationships and subsequent analysis of the reconstructed pathways revealed a total of 30 genes mediating the effect of MS on COVID‐19 (Figure 3A and Supplementary Table 4). A set of 24 MS‐driven genes that quantitatively change in MS and enhance COVID‐19 phenotypes included ADAMTS13, ALB, CCL19, CMA1, CRP, CSF2, CTLA4, CTSB, CXCL10, IL6, ACE, CXCL8, DPP4, F12, FN1, HRH1, IFNB1, IL2, KLF2, NCAM1, PADI2, SELE, TNF, and TNFSF12. On the other hand, 6 MS‐driven genetic changes suppress the phenotypes of COVID‐19, including CASP3, ERBB2, IFNA2, MAPK14, PLG, and TRPV4.
Figure 3.
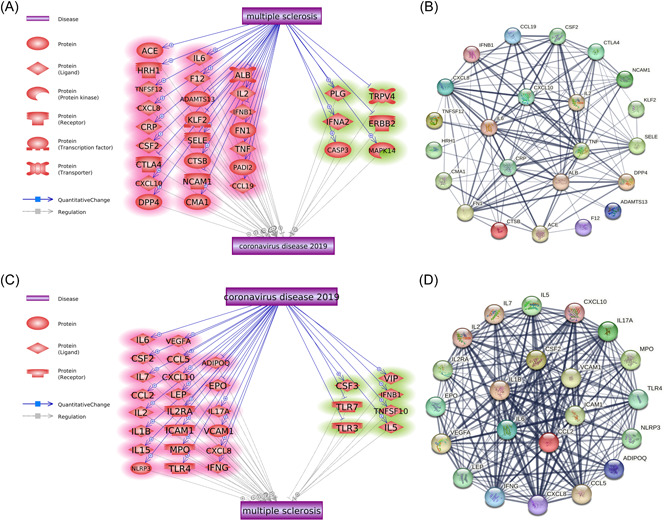
Molecular pathways connecting MS and COVID‐19. (A) Molecular pathways from MS to COVID‐19. Promoting effects are highlighted in red, and inhibitory effects are highlighted in green. Quantitative genetic changes driven by MS exert more promoting than inhibitory effects on COVID‐19. (B) Protein−protein interactions among the 24 COVID‐19‐promoting genes. Line sizes are proportional to the combined scores of the interactions. (C) Molecular pathways from COVID‐19 to MS. Quantitative genetic changes driven by COVID‐19 exert more promoting than inhibitory effects on MS. (D) Protein−protein interactions among the 22 MS‐promoting genes. Line sizes are proportional to the combined scores of the interactions. MS, multiple sclerosis.
A total of 29 genes were identified as mediating the effect of COVID‐19 on MS (Figure 3C and Supporting Information: Table 5). Among these genes, 22 COVID‐19‐driven genes can promote the development of MS, including ADIPOQ, CCL2, CCL5, CSF2, CXCL10, CXCL8, EPO, ICAM1, IFNG, IL17A, IL1B, IL2, IL2RA, IL5, IL6, IL7, LEP, MPO, NLRP3, TLR4, VCAM1, and VEGFA. Meanwhile, seven COVID‐19‐related genes can protect against MS, including CSF3, IFNB1, IL15, TLR3, TLR7, TNFSF10, and VIP.
Interestingly, five genes overlapped between the 24 MS‒COVID‐19‐promoting genes and 22 COVID‐19‒MS‐promoting genes, including CSF2, CXCL10, IL6, CXCL8, and IL2.
PPI analysis using the STRING database showed that both the 24 COVID‐19‐promoting proteins and the 22 MS‐promoting proteins formed a tightly interconnected network (Figure 3B,D). KEGG‐based pathway enrichment analysis in FUMA showed that both gene sets were enriched in immunity‐related molecular pathways, including cytokine receptor interaction, Toll‐like receptor signaling, NOD‐like receptor signaling, and JAK/STAT signaling pathways (Supporting Information: Figures 1−2).
4. DISCUSSION
Neurological diseases and COVID‐19 have been previously identified as risk factors for each other, with the mechanisms underlying their mutual influences being largely unknown. In this study, we conducted MR analysis to explore the connection between an important neurological condition of autoimmune nature, MS, and COVID‐19.
Here, we show the causal effect of MS on severe outcomes of COVID‐19. MS, especially in its progressive form, is recognized as a significant contributor to adverse COVID‐19 outcomes. 33 Garjani et al. found that patients with MS are more likely to experience a prolonged period of acute COVID‐19 symptoms and are vulnerable to the effects of the long COVID‐19. 34 These results agree with our finding of the positive causal effect of MS on COVID‐19 hospitalization. Our results suggest that neural lesions in MS patients contribute to the severity of COVID‐19.
On the other hand, genetic liability to MS may not increase the risk for SARS‐CoV‐2 infection as such. It seems that the differences in vulnerability to SARS‐CoV‐2 infection may be due to some prevention disparity between healthy individuals and those suffering from MS, possibly related to vaccine hesitancy in the MS cohort, 35 rather than to underlining genetics. On the other hand, the severity of the resultant disease may differ in these two groups. This observation is well aligned with the current understanding of the earliest stages of COVID‐19 and with commonly observed age‐ and disease‐dependent differential risks for the development of cytokine storms. 36
It is surprising to note that GWAS hits for MS were enriched in the spleen, blood, small intestine, and lung, instead of the brain, which supports the pivotal role of the inflammatory process in the development of MS. Lung, blood, and spleen are three tissues considered as most relevant to COVID‐19. The enriched tissues provide another layer of evidence for the MS‐COVID‐19 connection.
Comparison of GWAS hits of COVID‐19 and MS revealed two loci harboring five overlapping protein‐coding genes, which possibly contribute to the shared pathophysiology of these two diseases. The list of pleiotropic risk factors underpinning the association between MS and COVID‐19 severity includes CDC37, PDE4A and KEAP1 on chromosome 19p13.2, as well as TNFAIP8 and HSD17B4 on chromosome 5q23.1.
The most interesting candidate is KEAP1, which encodes Kelch‐like ECH‐associated protein 1, which, along with nuclear factor erythroid 2‐related factor 2 (NRF2), constitutes a central redox‐sensitive pathway that controls antioxidant, inflammatory, and immune system responses, facilitating GSH activity. 37 GSH depletion plays a prominent role in both MS 38 and COVID‐19. 39 Another interesting candidate in the KEAP1 neighborhood is PDE4A, a phosphodiesterase implicated in the degradation of cAMP in most, if not all, immune and inflammatory cells, including the cells in the lungs. 40 PDE4 inhibitors are approved for the treatment of chronic obstructive pulmonary disorder and atopic diseases while gaining attention for various neurological applications, including MS and COVID‐19. 41
On chromosome 5q23.1, HSD17B4 encodes a bifunctional enzyme that is involved in the peroxisomal beta‐oxidation pathway for both straight‐chain and 2‐methyl‐branched‐chain fatty acids. Mutations in this locus lead to a distinct neurological condition due to the inherent defect of beta‐oxidation. 42 While no specific studies of HSD17B4 have been reported in MS or COVID‐19 thus far, peroxisome pathways are disturbed in each of these two disorders. 43 , 44
A total of 24 protein‐coding genes were implicated by the analysis of molecular relationships within the reconstructed pathways mediating the synergistic link between MS and COVID‐19. A majority of these genes participate in various branches of cytokine signaling and danger sensing/pattern recognition, including Toll‐like receptors, T‐cell receptors, and the Jak/Stat machinery. Notably, many of the molecules highlighted by our analysis were widely discussed either as targets of COVID‐19 therapy (CCL19, CXCL10, IL6, CXCL8, DPP4, IL2, and TNF) 45 , 46 , 47 or as biomarkers of COVID‐19 severity (CRP, ALB, and F12). 48 , 49 Our results strengthen the proposed viewpoint that MS‐related processes may hasten COVID‐19 progression by overactivating innate immunity and the resultant “cytokine storm.”
The main strength of the study is that MR analysis is less affected by causality pitfalls, which are common in traditionally designed observational studies due to confounding factors and reverse causation. The largest available GWAS summary datasets were utilized for tracing the causative association between COVID‐19 and MS. All the participants in the GWAS datasets were of European ancestry, reducing the potential population heterogeneity. Our study has several limitations. In particular, we assessed only genetic liability for both diseases with no regard to the effects of the environment, which are critical for both MS and COVID‐19. We acknowledge that MR analyses may be biased due to pleiotropy, especially in nonhomogenous datasets. Therefore, we have tested the MR assumptions using various models.
5. CONCLUSIONS
In summary, our study supports that MS may augment the severity of COVID‐19, while hospitalized COVID‐19 may causally increase the risk for MS. Immunity‐related, inflammation‐driven pathways may underlie the link between MS and COVID‐19.
AUTHOR CONTRIBUTIONS
Fuquan Zhang conceived the project and supervised the study. Fuquan Zhang and Ancha Baranova analyzed the data. Ancha Baranova, Fuquan Zhang, and Hongbao Cao wrote the manuscript. Shaolei Teng and Kuan‐Pin Su revised the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Supplementary information.
Supplementary information.
ACKNOWLEDGMENTS
The authors thank all investigators and participants from the COVID‐19 Host Genetics Initiative and International Multiple Sclerosis Genetics Consortium for sharing these data.
Baranova A, Cao H, Teng S, Su K‐P, Zhang F. Shared genetics and causal associations between COVID‐19 and multiple sclerosis. J Med Virol. 2022;95:e28431. 10.1002/jmv.28431
Ancha Baranovaand Hongbao Cao are co‐first authors.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in The Covid19 Host Genetics Initiative (https://www.covid19hg.org/results/r7/) and The International Multiple Sclerosis Genetics Consortium (https://imsgc.net/).
REFERENCES
- 1. Cao H, Baranova A, Wei X, Wang C, Zhang F. Bidirectional causal associations between type 2 diabetes and COVID‐19. J Med Virol. 2022. 10.1002/jmv.28100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rao S, Baranova A, Cao H, Chen J, Zhang X, Zhang F. Genetic mechanisms of COVID‐19 and its association with smoking and alcohol consumption. Brief Bioinform. 2021;22(6):bbab284. [DOI] [PubMed] [Google Scholar]
- 3. Yang J, Tian C, Chen Y, Zhu C, Chi H, Li J. Obesity aggravates COVID‐19: an updated systematic review and meta‐analysis. J Med Virol. 2021;93(5):2662‐2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baranova A, Song Y, Cao H, Zhang F. Causal associations between basal metabolic rate and COVID‐19. Diabetes. 2023;72(1):149‐154. [DOI] [PubMed] [Google Scholar]
- 5. Baranova A, Xu Y, Cao H, Zhang F. Associations between pulse rate and COVID‐19. J Med Virol. 2022. 10.1002/jmv.28194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baranova A, Cao H, Chen J, Zhang F. Causal association and shared genetics between asthma and COVID‐19. Front Immunol. 2022;13:705379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang F, Baranova A. Smoking quantitatively increases risk for COVID‐19. Eur Respir J. 2022;60(6):2101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baranova A, Cao H, Teng S, Zhang F. A phenome‐wide investigation of risk factors for severe COVID‐19. J Med Virol. 2022:e28264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Islam MK, Molla MMA, Hasan P, et al. Persistence of sleep disturbance among post‐COVID patients: findings from a 2‐month follow‐up study in a Bangladeshi cohort. J Med Virol. 2022;94(3):971‐978. [DOI] [PubMed] [Google Scholar]
- 10. Asadi‐Pooya AA, Akbari A, Emami A, et al. Long COVID syndrome‐associated brain fog. J Med Virol. 2022;94(3):979‐984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. AM C, Singh AK, Roy P, et al. Long COVID following Omicron wave in Eastern India—a retrospective cohort study. J Med Virol. 2022. 10.1002/jmv.28214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paterson C, Davis D, Roche M, et al. What are the long‐term holistic health consequences of COVID‐19 among survivors? An umbrella systematic review. J Med Virol. 2022. 10.1002/jmv.28086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tao SS, Wang XY, Yang XK, et al. COVID‐19 and inflammatory bowel disease crosstalk: from emerging association to clinical proposal. J Med Virol. 2022;94(12):5640‐5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bodnar B, Patel K, Ho W, Luo JJ, Hu W. Cellular mechanisms underlying neurological/neuropsychiatric manifestations of COVID‐19. J Med Virol. 2021;93(4):1983‐1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Forero‐Peña DA, Hernandez MM, Mozo Herrera IP, et al. Remitting neuropsychiatric symptoms in COVID‐19 patients: viral cause or drug effect? J Med Virol. 2022;94(3):1154‐1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu JM, Tan BH, Wu S, Gui Y, Suo JL, Li YC. Evidence of central nervous system infection and neuroinvasive routes, as well as neurological involvement, in the lethality of SARS‐CoV‐2 infection. J Med Virol. 2021;93(3):1304‐1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lundström W, Gustafsson R. Human herpesvirus 6A is a risk factor for multiple sclerosis. Front Immunol. 2022;13:840753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sollid LM. Epstein‐Barr virus as a driver of multiple sclerosis. Sci Immunol. 2022;7(70):eabo7799. [DOI] [PubMed] [Google Scholar]
- 19. Brison E, Jacomy H, Desforges M, Talbot PJ. Glutamate excitotoxicity is involved in the induction of paralysis in mice after infection by a human coronavirus with a single point mutation in its spike protein. J Virol. 2011;85(23):12464‐12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florea A, Sirbu C, Ghinescu M, et al. SARS‐CoV‐2, multiple sclerosis, and focal deficit in a postpartum woman: a case report. Exp Ther Med. 2020;21(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang F, Rao S, Cao H, et al. Genetic evidence suggests posttraumatic stress disorder as a subtype of major depressive disorder. J Clin Invest. 2022;132(3):e145942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rao S, Baranova A, Yao Y, Wang J, Zhang F. Genetic relationships between attention‐deficit/hyperactivity disorder, autism spectrum disorder, and intelligence. Neuropsychobiology. 2022;81:484‐496. [DOI] [PubMed] [Google Scholar]
- 23. Baranova A, Wang J, Cao H, et al. Shared genetics between autism spectrum disorder and attention‐deficit/hyperactivity disorder and their association with extraversion. Psychiatry Res. 2022;314:114679. [DOI] [PubMed] [Google Scholar]
- 24. Initiative C‐HG . The COVID‐19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS‐CoV‐2 virus pandemic. Eur J Human Genet. 2020;28(6):715‐718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Multiple Sclerosis Genetics C . Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science. 2019;365(6460):eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bulik‐Sullivan BK, Loh PR, Finucane HK, et al. LD score regression distinguishes confounding from polygenicity in genome‐wide association studies. Nature Genet. 2015;47(3):291‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bulik‐Sullivan B, Finucane HK, Anttila V, et al. An Atlas of genetic correlations across human diseases and traits. Nature Genet. 2015;47(11):1236‐1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hemani G, Zheng J, Elsworth B, et al. The MR‐Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512‐525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio‐‐the analysis and navigation of molecular networks. Bioinformatics. 2003;19(16):2155‐2157. [DOI] [PubMed] [Google Scholar]
- 32. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein‐protein association networks with increased coverage, supporting functional discovery in genome‐wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607‐D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prosperini L, Tortorella C, Haggiag S, Ruggieri S, Galgani S, Gasperini C. Determinants of COVID‐19‐related lethality in multiple sclerosis: a meta‐regression of observational studies. J Neurol. 2022;269:2275‐2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garjani A, Middleton RM, Nicholas R, Evangelou N. Recovery from COVID‐19 in multiple sclerosis: a prospective and longitudinal cohort study of the United Kingdom multiple sclerosis register. Neurol Neuroimmunol Neuroinflamm. 2022;9(1):e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rakusa M, Ozturk S, Moro E, et al. COVID‐19 vaccination hesitancy among people with chronic neurological disorders: a position paper. Eur J Neurol. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gautret P, Million M, Jarrot PA, et al. Natural history of COVID‐19 and therapeutic options. Expert Rev Clin Immunol. 2020;16(12):1159‐1184. [DOI] [PubMed] [Google Scholar]
- 37. Abdalkader M, Lampinen R, Kanninen KM, Malm TM, Liddell JR. Targeting Nrf2 to suppress ferroptosis and mitochondrial dysfunction in neurodegeneration. Front Neurosci. 2018;12:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tavassolifar M, Vodjgani M, Salehi Z, Izad M. The influence of reactive oxygen species in the immune system and pathogenesis of multiple sclerosis. Autoimmune Dis. 2020;2020:1‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Labarrere CA, Kassab GS. Glutathione deficiency in the pathogenesis of SARS‐CoV‐2 infection and its effects upon the host immune response in severe COVID‐19 disease. Front Microbiol. 2022;13:979719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Contreras S, Milara J, Morcillo E, Cortijo J. Selective inhibition of phosphodiesterases 4A, B, C and D isoforms in chronic respiratory diseases: current and future evidences. Curr Pharm Des. 2017;23(14):2073‐2083. [DOI] [PubMed] [Google Scholar]
- 41. Crocetti L, Floresta G, Cilibrizzi A, Giovannoni MP. An overview of PDE4 inhibitors in clinical trials: 2010 to early 2022. Molecules. 2022;27(15):4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen S, Du L, Lei Y, Lin Y, Chen S, Liu Y. Two novel HSD17B4 heterozygous mutations in association with D‐Bifunctional protein deficiency: a case report and literature review. Front Pediatr. 2021;9:679597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gray E, Rice C, Hares K, et al. Reductions in neuronal peroxisomes in multiple sclerosis grey matter. Mult Scler J. 2014;20(6):651‐659. [DOI] [PubMed] [Google Scholar]
- 44. Yang J, Song H, Hao X. Whole‐transcriptome sequencing data reveals a disparate cognitive and immune signature in COVID‐19 patients with and without dementia. J Med Virol. 2022. 10.1002/jmv.28177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao Q, Zhang W, Li T, et al. Interrelationship between 2019‐nCov receptor DPP4 and diabetes mellitus targets based on protein interaction network. Sci Rep. 2022;12(1):188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kojima Y, Nakakubo S, Takei N, et al. Comparative efficacy of tocilizumab and baricitinib administration in COVID‐19 treatment: a retrospective cohort study. Medicina. 2022;58(4):513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levy G, Guglielmelli P, Langmuir P, Constantinescu SN. JAK inhibitors and COVID‐19. J Immunother Cancer. 2022;10(4):e002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y, Li H, Song C, et al. Early prediction of disease progression in patients with severe COVID‐19 using C‐reactive protein to albumin ratio. Dis Markers. 2021;2021:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Calderon‐Lopez MT, Garcia‐Leon N, Gomez‐Arevalillo S, Martin‐Serrano P, Matilla‐Garcia A. Coronavirus disease 2019 and coagulopathy: other prothrombotic coagulation factors. Blood Coagul Fibrinolysis. 2021;32(1):44‐49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information.
Supplementary information.
Data Availability Statement
The data that support the findings of this study are openly available in The Covid19 Host Genetics Initiative (https://www.covid19hg.org/results/r7/) and The International Multiple Sclerosis Genetics Consortium (https://imsgc.net/).


