Abstract
Clear cell renal cell carcinoma (ccRCC) is a frequently occurring renal cancer. The Von Hippel-Lindau disease tumor suppressor VHL, a known tumor suppressor gene, is frequently mutated in about 50% of patients with ccRCC. However, it is unclear whether VHL influences the progression of ccRCC tumors expressing wild-type VHL. In the present study, we found that higher expression of VHL was correlated with the better disease-free survival (DFS) in ccRCC patients using The Cancer Genome Atlas (TCGA) datasets. We revealed that VHL overexpression in ccRCC cells inhibited epithelial-mesenchymal transition (EMT), sterol regulatory element-binding protein 1 (SREBP1)-regulated triglyceride synthesis, and cell proliferation. Proteomic analysis provided us a global view that VHL regulated four biological processes, including metabolism, immune regulation, apoptosis, and cell movement. Importantly, we found that VHL overexpression led to up-regulated expression of proteins associated with antigen processing and interferon-responsive proteins, thus rendering ccRCC cells more sensitive to interferon treatment. We defined an interferon-responsive signature (IRS) composed of ten interferon-responsive proteins, whose mRNA expression levels were positively correlated with DFS in ccRCC patients. Taken together, our results propose that the subset of ccRCC patients with high VHL expression benefit from immunotherapy.
Keywords: Renal cancer, VHL, Immunogenicity, Multi-omics, Interferon
Introduction
World Health Organization (WHO; http://gco.iarc.fr/today/online-analysis-table) has estimated that there are 400,000 new cases of renal cancer in 2018. Clear cell renal cell carcinoma (ccRCC) characterized by high glycogen and lipid contents accounts for 75% of all renal cell carcinoma (RCC) [1], [2]. The lack of ccRCC sensitivity to chemotherapy and radiation therapy has prompted the development of targeted therapies such as inhibitors for tyrosine kinases and mammalian target of rapamycin (mTOR) [3]. Immune checkpoint inhibitors have also emerged as effective treatment options against advanced ccRCC [4]. Nevertheless, 30% of ccRCC patients will develop recurrence or metastasis after surgical resection of their tumor [5]. It is critical to further investigate tumor-associated proteins and their functions over disease progression.
The Von Hippel-Lindau disease tumor suppressor (VHL), known as the most commonly mutated gene in ccRCC, exerts its canonical function via the oxygen-dependent ubiquitin-mediated degradation of hypoxia-inducible factor (HIF) [6], [7]. The stabilization and hyper activation of HIF1α and HIF2α play essential roles in tumorigenesis and progression of ccRCC. HIF-independent functions of VHL have also been intensively investigated, showing that VHL regulates Wnt signaling, NFκB signaling, Akt activity, microtubule stability, extracellular matrix, and maintenance of primary cilium [8]. Using isotope tracing, a recent study revealed that the glycolysis was increased and tricarboxylic acid cycle (TCA) was decreased in ccRCC patients, demonstrating that ccRCC patients display the classic Warburg phenotype which is specified as the transition from respiration to glycolysis [9]. As described earlier, the RCC clear-cell phenotype suggests dysregulated lipid and cholesterol metabolism and TRC8 has been reported as a hereditary RCC genes inhibiting cholesterol and lipid biosynthesis [10], [11].
Most VHL-related studies have focused on the effects of VHL mutations on ccRCC progression. However, there are a subset of ccRCC patients with wild-type VHL. It has been reported that ccRCC patients with wild-type VHL are more aggressive than those with inactive VHL, and PD-L1 expression is associated with wild-type VHL expression. Studies also proposed that tumors with wild-type VHL expression were distinctive from conventional ccRCC [12], [13], [14]. It is, therefore, important to characterize the effects of VHL expression on the pathological or prognostic significance in ccRCC patients carrying wild-type VHL.
In this study, we investigated the relationship between the VHL expression levels and disease-free survival (DFS) in ccRCC patients. A global proteomic analysis was conducted to determine the effects of VHL overexpression on the main biological processes in ccRCC cells. We revealed that VHL overexpression up-regulated interferon-responsive proteins via STAT1-mediated signaling. Collectively, our findings suggest that patients with high VHL expression will be more responsive to immunotherapy.
Results
Higher VHL expression is positively correlated with better DFS in ccRCC patients
Given the molecular heterogeneity of ccRCC, we analyzed VHL mutation frequencies in ccRCC from four different clinical datasets [6], [15], [16], [17], [18], [19]. VHL mutation frequencies were determined to be 25.5% (25 of 98), 40.6% (43 of 106), 51.2% (218 of 426), and 79.5% (62 of 78) in these four datasets (Figure 1A). The average frequency of VHL mutation in USA and Europe was 55.6% (280 of 504), which is in consistent with an earlier report [20]. These analyses also found that 44.4% of ccRCC patients carried wild-type VHL in USA and Europe, while 66.7% of ccRCC patients in China and Japan carried wild-type VHL, suggesting that VHL mutation frequencies were higher in Western ccRCC patients than those in Asia.
Figure 1.
High VHL expression was correlated with better DFS in ccRCC
A. Analysis of the mutation frequency of VHL from four different clinical datasets “China 2012” [15], “Japan 2013” [16], “USA 2013” [6], and “Europe 2014” [17]. B. Correlation analysis between the DFS and VHL expression levels of different cancer patients using datasets from TCGA with GEPIA (http://gepia2.cancer-pku.cn/#index). Blue-coded rectangle represents that high VHL expression is positively correlated with better DFS, whereas red-coded rectangle represents that high VHL expression is correlated with poor DFS. C. Higher VHL mRNA expression levels correlated with longer DFS of ccRCC patients using the KIRC datasets. n = 129, each in low (blue) or high (red) group. Log-rank test was used to determine the survival differences (P = 0.043). D. RT-PCR analysis of VHL expression in the control and VHL-OE cells. E. Western blotting analysis of HA-tagged VHL expression in the control and VHL-OE cells. F.VHL overexpression decreased cell proliferation in ccRCC cells. G.VHL overexpression decreased the colony formation in ccRCC cells. H. Representative images of the scratch assay of the control and VHL-OE cells. I. Quantitative result of the scratch assay shown in (H) by using ImageJ software. J.VHL overexpression decreased cellular ROS level in ccRCC cells. VHL, Von Hippel-Lindau disease tumor suppressor; DFS, disease-free survival; TCGA, The Cancer Genome Atlas; ccRCC, clear cell renal cell carcinoma; ROS, reactive oxygen species; OE, overexpression; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Next, we assessed the DFS of patients with different VHL expression levels using The Cancer Genome Atlas (TCGA) datasets. DFS analyses were based on the expression of VHL in all 32 TCGA datasets of different cancer types and revealed that VHL expression was correlated with patients’ survival in adrenocortical carcinoma (ACC), liver hepatocellular carcinoma (LIHC), and kidney renal clear cell carcinoma (KIRC) (Figure 1B). Higher VHL expression was correlated with poorer DFS in ACC and LIHC (Figure S1A and B), whereas it was positively correlated with better DFS in KIRC (Figure 1C). ccRCC patients with low VHL expression experienced significantly shorter survival, with a median DFS of 78 months compared to the subgroup with high VHL expression (Figure 1C). To determine if there was any covariant with cofounding effects, such as gender, age, and race, we conducted survival analysis by using the cox proportional hazards regression model. The results showed that factors including age and race had no effects on patients’ survival, and male patients tended to have worse survival as compared to female patients (Figure S1C). In consistent with the univariate analysis, we found that higher VHL expression was correlated with better DFS in ccRCC patients (Figure S1C). This suggests that VHL is a tumor suppressor in ccRCC, and patients with higher VHL expression tend to have a significantly longer DFS than those with lower VHL expression.
To understand the effects of VHL expression on ccRCC progression, we established VHL-overexpressing (VHL-OE) cell lines in 769-P and 786-O cells. 769-P and 786-O are VHL-deficient cell lines. VHL expression levels in 769-P cells were examined by real-time PCR (RT-PCR) and Western blotting, confirming that the mRNA expression level of VHL was 233.4-fold higher in VHL-OE cells compared with the control cells (P = 7.27E−4) (Figure 1D) and the HA-tagged VHL was only detected in VHL-OE cells (Figure 1E). The effects of VHL overexpression on cell proliferation and vitality were examined using cell counting kits and the colony formation assay. The VHL overexpression led to a decreased growth rate (P = 2E−5) (Figure 1F) and reduced viability in VHL-OE cells compared with the control cells (Figure 1G). The VHL overexpression also reduced cell motility (Figure 1H), and the migration rate of VHL-OE cells was 0.4-fold lower than that of control cells (P = 1.53E−7) (Figure 1I). The reactive oxygen species (ROS) level in VHL-OE cells was 0.7-fold lower than that in control cells (P = 3E−4) (Figure 1J). VHL overexpression caused similar phenotypic changes in 786-O cells (Figure S2A–E). These data indicate that VHL overexpression significantly inhibits cell proliferation and migration.
VHL overexpression reverses the Warburg phenotype and decreases cellular triglyceride by inhibiting SREBP1 maturation
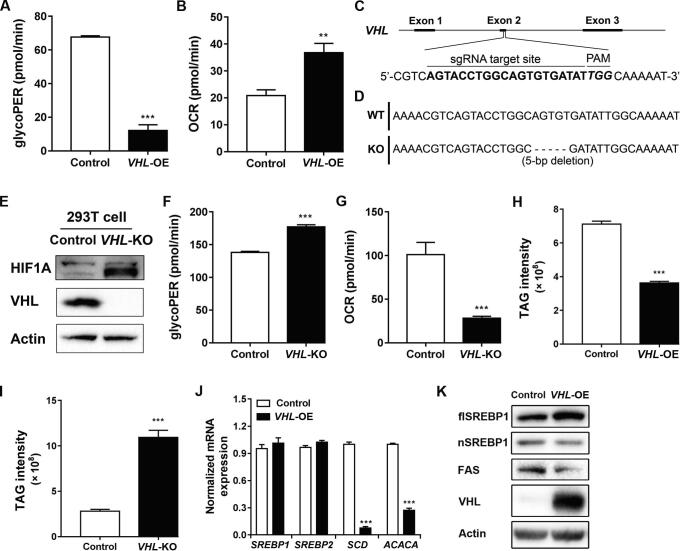
A recent study indicated that the glycolysis was increased and TCA cycle was decreased in ccRCC patients [9]. To explore the effects of VHL overexpression on cellular energy metabolism, the glycolytic and respiratory rates were measured using a Seahorse extracellular flux analyzer. An analysis of Seahorse data revealed that the basal glycolytic rate was 0.2-fold lower in VHL-OE cells than that in control cells (P = 9.69E−5) (Figure 2A), while the basal respiratory rate was 1.8-fold higher than that in control cells (P = 2.69E−3) (Figure 2B). Next, we established the stable VHL-knockout (VHL-KO) cell line using immortalized human embryonic kidney 293T cells by CRISPR/Cas9 method (Figure 2C). Genomic DNA sequencing showed that a 5-bp deletion was introduced in the exon 2 of VHL in VHL-KO cells leading to frameshift and a premature stop codon (Figure 2D). Western blotting showed that VHL was completely depleted in VHL-KO cells (Figure 2E). In consistent with the results described in the previous section, we found that VHL knockout increased cell proliferation (Figure S3A and B) and migration (Figure S3C and D). In contrast with VHL overexpression, the basal glycolytic rate in VHL-KO 293T cells was 1.3-fold higher than that in control cells (P = 3.74E−4) (Figure 2F), while the basal respiration rate was 0.3-fold lower than that in control cells (P = 6.12E−4) (Figure 2G). These findings demonstrate that VHL overexpression reverses the classic Warburg phenotype in ccRCC cells, while VHL knockout promotes a switch from oxidative phosphorylation to glycolysis.
Figure 2.
VHL overexpression reversed the Warburg phenotype and decreased cellular TAG by inhibiting SREBP1 maturation
A. and B.VHL overexpression decreased basal glycolysis (A) but increased basal respiration (B) in ccRCC cells. C. Diagram showed the sequence of sgRNA targeting VHL. D. Sequence alignment of the exon 2 in VHL in control and VHL-KO cells. E. Western blotting analysis of VHL and HIF1A expression in control and VHL-KO cells. F. and G.VHL knockout increased the basal glycolysis (F) but decreased the basal respiration (G) in 293T cells. H.VHL overexpression decreased the cellular TAG levels in ccRCC cells. I.VHL knockout increased the cellular TAG levels in 293T cells. J. RT-PCR analysis of SREBP1, SREBP2, SCD, and ACACA in VHL-OE and control cells. K. Western blotting analysis showing the expression of flSREBP1, nSREBP1, FAS, and VHL in VHL-OE and control cells. TAG, triglyceride; flSREBP1, full-length SREBP1; nSREBP1, nuclear SREBP1; KO, knockout.
Lipid accumulation is one of the remarkable histopathologic features in ccRCC, rendering the “clear” cytoplasm. Therefore, we measured the cellular triglyceride (TAG) level in ccRCC cells using an LC-MS/MS method. The TAG level was 0.5-fold lower in VHL-OE cells than that in control cells (P = 4E−4) (Figure 2H), whereas it was 3.9-fold higher in VHL-KO cells than that in control cells (P = 5E−4) (Figure 2I). Cellular lipid accumulation is controlled by extracellular lipid uptake, lipogenesis, and lipid oxidation. Cancer cells preferred de novo lipogenesis over extracellular lipid uptake [21]. Sterol regulatory element-binding protein 1 (SREBP1) is the master regulator in de novo lipogenesis. Its downstream targets including acetyl-CoA carboxylase (ACACA) and stearoyl-CoA desaturase (SCD) are both enzymes that catalyze the rate-limiting steps in fatty acid synthesis [22]. RT-PCR analysis showed that the mRNA levels of SREBP1 and SREBP2 were unchanged in VHL-OE and control cells, but the mRNA levels of SCD and ACACA were down-regulated in VHL-OE cells, being 0.1-fold (P = 5.59E−6) and 0.3-fold (P = 1.08E−5) lower than those in control cells, respectively (Figure 2J). Western blotting showed that the amounts of active nuclear-localized SREBP1 (nSREBP1) and fatty acid synthase (FAS) were decreased, being 0.7-fold and 0.3-fold lower in VHL-OE cells than those in control cells, respectively, and that the amount of full-length SREBP1 (flSREBP1) was increased, being 1.6-fold higher in VHL-OE cells than that in control cells (Figure 2K). We extracted the nuclear and cytoplasmic proteins separately from VHL-OE cells and control cells and detected the levels of flSREBP1 and nSREBP1. The results showed that the level of flSREBP1 was higher in cytosolic fraction in VHL-OE cells, whereas the level of nSREBP1 was lower in VHL-OE cells (Figure S4A). Furthermore, we conducted the rescue experiment in which either flSREBP1 or nSREBP1 was overexpressed in VHL-OE cells. The RT-PCR results showed that the expression levels of SREBP1 downstream genes SCD, ACACA, and FASN were up-regulated only after the reintroduction of nSREBP1 (Figure S4B). These results demonstrate that VHL overexpression decreases cellular lipid accumulation by inhibiting TAG synthesis via regulating SREBP1 maturation.
VHL overexpression alters protein expression profiling in ccRCC cells
Quantitative proteomics analysis was conducted on VHL-OE and control cells in biological triplicates for identifying differentially expressed proteins (DEPs). We identified 9459 proteins and 7037 proteins separately with two and more unique peptides quantified. The quantification ratios of biological replicates showed a high correlation, indicating that the results were reproducible (R2 = 0.9) (Figure 3A). The quantitative ratios were filtered by population statistics to calculate the threshold cut-off as 50% variation for DEPs (Figure S5A) [23]. Volcano diagram showed that 404 proteins were up-regulated [VHL-OE/Control > 1.5; false discovery rate (FDR) adjusted P value < 0.05] (Table S1), and 228 proteins were down-regulated (VHL-OE/Control < 0.67; FDR adjusted P value < 0.05) (Table S2; Figure 3B). VHL expression was significantly up-regulated, and its substrate HIF1α was significantly down-regulated (Figure 3B). The expression levels of proteins involved in glycolysis including Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Triosephosphate isomerase (TPI1), Enolase 2 (ENO2), Pyruvate kinase PKLR (PKLR), and Pyruvate kinase PKM (PKM) were down-regulated in VHL-OE cells compared with those in control cells (Figure 3C), whereas the expression levels of many subunits involved in the oxidative phosphorylation pathway, including subunits of Complex I, Complex II, Complex III, and Complex IV, were up-regulated in VHL-OE cells (Figure 3C). The changes of GLUT1, ENO2, and HK2 were validated by RT-PCR (Figure 3D), while the changes of GAPDH, NDUFV2, and NDUFA4 were further validated by Western blotting (Figure 3E). In consistent with the Seahorse measurement, most oxidative phosphorylation-related proteins were up-regulated, whereas glycolysis-related proteins were down-regulated, further confirming that VHL overexpression reverses the Warburg phenotype (Figure 2A and B).
Figure 3.
VHL overexpression altered protein expression profiling in ccRCC cells
A. The correlation of two biological replicates from proteomes exhibited a good repeatability. B. A volcano diagram of up-regulated (red) and down-regulated (blue) proteins between the VHL-OE and control cells based on fold change (x-axis) and adjusted P value (y-axis). Each point represents a single quantified protein. The significantly changed proteins were labeled with the gene symbols. C. A heatmap showing the changes of the proteins involved in glycolysis and oxidative phosphorylation. D. RT-PCR analysis of GLUT1, PGK, ENO2, and HK2 in VHL-OE and control cells. E. Western blotting analysis confirmed VHL overexpression-induced changes in GAPDH, NDUFV2, and NDUFA4 in VHL-OE cells. F. The top 20 canonical pathways can be grouped into four biological processes analyzed by IPA software. G. RT-PCR analysis revealed that VHL overexpression significantly up-regulated HLA-related genes but down-regulated PD-L1. H. Western blotting analysis confirmed VHL overexpression-induced changes in HLA-A, HLA-DR, and PD-L1 in VHL-OE cells. I. Antigen processing and presentation (red) and antigen processing and presentation via MHC class I (blue) were all highly up-regulated by GSEA (using “C5” signature from the MSigDB). The plots indicated a significant (FDR adjusted P value < 0.05) enrichment after VHL overexpression. IPA, Ingenuity Pathway Analysis; GSEA, gene set enrichment analysis; NES, normalized enrichment score.
To explore the biological relevance of the DEPs, functional enrichment analysis was conducted by using Ingenuity Pathway Analysis (IPA) software. The top 20 pathways enriched by IPA revealed that VHL regulated four major biological processes including metabolism, immune regulation, apoptosis, and cell movement (Figure 3F). The proteins involved in oxidative phosphorylation, fatty acid oxidation, mitochondrial L-carnitine shuttling, cholesterol biosynthesis, mevalonate pathway, and geranylgeranyl-diphosphate biosynthesis were mostly up-regulated. The number of up-regulated proteins involved in apoptosis and cell movement-related pathways was more than that of the down-regulated proteins (Figure 3F). Furthermore, we calculated the Z-scores of enriched biological pathways by using the core analysis method in IPA, which assesses the match of observed and predicted up- or down-regulation patterns and represents the non-randomness of directionality within a gene set. The results showed that the apoptotic pathways had Z-scores above 2, suggesting that apoptotic pathways were activated after VHL overexpression. On the other hand, the cell motility-related pathways had Z-scores below −2, suggesting that cell movement-related pathways were inhibited after VHL overexpression. These results indicated that VHL overexpression reduced cell viability and motility (Figure S5B).
Proteomics analysis also identified that major histocompatibility antigens HLA-A, HLA-B, HLA-C, HLA-E, and HLA-H were up-regulated in VHL-OE cells (Figure 3B). The changes were further validated by RT-PCR analysis, showing that the mRNA levels of HLA-A, HLA-B, and HLA-E were also up-regulated (Figure 3G). This was confirmed by Western blotting, in which both MHC class I molecule HLA-A and MHC class II molecule HLA-DR were up-regulated (3.2-fold and 4.5-fold higher in VHL-OE cells than in control cells, respectively) (Figure 3H). Intriguingly, PD-L1, an immune checkpoint protein, was down-regulated in VHL-OE cells at both mRNA and protein levels (Figure 3G and H). Consistently, the gene set enrichment analysis (GSEA) [24] revealed that the expression of antigen processing and presentation signature genes was increased in VHL-OE cells compared with that in control cells (Figure 3I). These results suggest that VHL overexpression up-regulates both MHC class I and class II molecules, thus enhancing antigen processing and presentation.
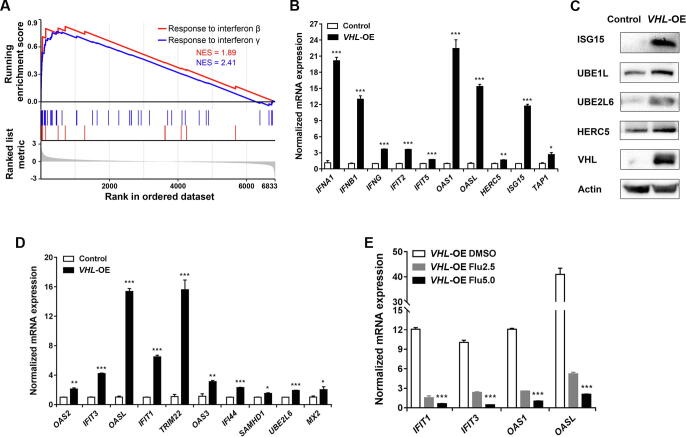
VHL overexpression up-regulates interferon-responsive proteins via STAT1-mediated signaling
Interestingly, the proteins involved in the interferon signaling pathway were significantly up-regulated (Figure 3F). Using GSEA, we confirmed that VHL overexpression enhanced the expression signatures for both interferon-γ-mediated signaling pathway and response to type I interferon (Figure 4A). RT-PCR analysis showed that VHL overexpression increased the mRNA levels of IFNA1, IFNB1, IFNG, IFIT2, IFIT5, OAS1, OASL, HERC5, ISG15, and TAP1 (Figure 4B). ISGylation was a representative event in responsive to interferon treatment [25], [26], and Western blotting showed that not only free ISG15 but also the three enzymes involved in ISGylation (UBE1L, UBE2L6, and HERC5) were all up-regulated in VHL-OE cells (Figure 4C). These results demonstrate that VHL overexpression activates the interferon signaling pathway in ccRCC cells.
Figure 4.
VHL overexpression up-regulated interferon-responsive proteins via STAT1-mediated signalling
A. Responses to interferon-β (red) and interferon-γ (blue) genes were highly up-regulated by GSEA. Plots indicated a significant (FDR adjusted P value < 0.05) enrichment after VHL overexpression. B. RT-PCR analysis showing the significantly up-regulated mRNA levels of interferon and interferon-responsive genes in VHL-OE cells. C. Western blotting analysis showing the up-regulated protein levels of ISG15, UBE1L, UBE2L6, HERC5, and VHL in VHL-OE cells. D. RT-PCR analysis showing the significantly up-regulated mRNA levels of the IRS genes in VHL-OE cells. E. RT-PCR analysis showing the down-regulated expression of interferon-responsive genes in VHL-OE cells treated with 2.5 µM or 5 µM fludarabine. IRS, interferon-responsive signature.
We designated an interferon-responsive signature (IRS) composed of the ten most up-regulated interferon-responsive proteins from the proteomic analysis, including OAS2, IFIT3, OASL, IFIT1, TRIM22, OAS3, IFI44, SAMHD1, UBE2L6, and MX2 (Figure 3B). Using RT-PCR analysis, we determined that the mRNA expression levels for the IRS genes were also markedly up-regulated (Figure 4D), which was consistent with the proteomic results (Figure 3B). This indicated that VHL overexpression transcriptionally up-regulated the IRS. A recent study showed that STAT1, STAT2, and IRF9 formed a complex termed interferon-stimulated gene factor 3 (ISGF3), which regulated the expression of interferon-stimulated genes [27]. All three proteins were up-regulated in our proteomic data (Figure 3B), and the level of STAT1-pSer727 which was the active form of STAT1 protein was higher in VHL-OE cells than that in control cells (Figure S6), suggesting that ISGF3 up-regulated interferon-stimulated genes. Fludarabine is a known cell-permeable p-STAT1 inhibitor [28]. We treated cells with 2.5 µM or 5 µM of fludarabine and found that the treatment significantly inhibited the transcription of the interferon-responsive genes in a dosage-dependent manner (Figure 4E), suggesting that VHL overexpression up-regulates the interferon-responsive gene expression via STAT1-mediated signaling.
VHL overexpression renders ccRCC cells sensitive to interferon treatment
The aforementioned results demonstrated that VHL overexpression up-regulated HLAs and interferon-responsive proteins, suggesting that cells with wild-type VHL expression are more sensitive to immunotherapy. Indeed, after treated with 4000 U/ml interferon-β for 24 h, the survival rate in VHL-OE cells was 50%, significantly lower than the 90% survival rate in control cells (P = 1.23E−5) (Figure 5A). After the treatment with 800 μg/ml interferon-γ for 24 h, the survival rate in VHL-OE cells was 45%, significantly lower than the 60% survival rate in control cells (P = 2.04E−5) (Figure 5B). These results demonstrated that VHL-OE cells were more sensitive to interferon-β and interferon-γ treatments compared to control cells.
Figure 5.
VHL overexpression rendered ccRCC cells sensitive to interferon treatment
A. and B.VHL overexpression sensitized ccRCC cells to interferon treatment. VHL-OE cells and control cells were treated with different concentrations of interferon-β (A) or interferon-γ (B) for 24 h. C.VHL expression was positively correlated with IRS expression (R = 0.78, P < 1E−5). Data were from the KIRC datasets from TCGA, and normalized by GAPDH. D. IRS expression was positively correlated with DFS in ccRCC patients using the KIRC datasets. n = 129, each in low (blue) or high (red) group. Log-rank test was used for comparing the survival curves (P = 0.0025).
Furthermore, a positive correlation of the VHL expression and the IRS expression was derived from the KIRC datasets in TCGA (Pearson correlation, R = 0.78) (Figure 5C). Survival analysis showed that ccRCC patients with higher IRS expression tended to have longer DFS than the subgroup with lower IRS expression (P = 0.0025) (Figure 5D).
Discussion
Energy metabolism reprogramming is a hallmark of cancer [29]. Metabolic reprogramming in ccRCC represents classic Warburg phenotype, specified as the transition from respiration to glycolysis [9], [30]. It has been known that Warburg phenotype in ccRCC is largely regulated by VHL-HIF1α pathway, in which VHL mutation activates HIF1α and its targeted signaling pathways. However, VHL mutation frequencies were about 50% in ccRCC patients, and even lower in Asian population. In the present study, we applied a multi-omics approach to determine the effects of VHL on ccRCC progression. Analysis of datasets in TCGA revealed that higher VHL expression was positively correlated with the better DFS in ccRCC patients. Using quantitative proteomics and functional studies, we found that VHL overexpression reversed Warburg phenotype in ccRCC cells by up-regulating most subunits of oxidative phosphorylation (Figure 3C). We also found that the expression level of VHL was negatively correlated with DFS in ACC and LIHC, suggesting that VHL may play different roles in different cell types and tissues. The two most frequently mutated genes in both ACC and LIHC are TP53 and CTNNB1 [31], [32], [33], whereas TP53 gene mutation is less common and CTNNB1 gene mutation is rare in KIRC [6]. The different genetic contexts in ACC and LIHC from KIRC may explain the different relationship between VHL expression level and DFS, whereas the underlying mechanisms remain to be further elucidated.
Cytoplasmic glycogen and lipid deposits in ccRCC defined the “clear cell” phenotype histologically, which emphasized the importance of dysregulated lipid metabolism in ccRCC. It remains unclear whether lipid storage promotes cancer development, or excessive lipid accumulation is caused by altered metabolism in ccRCC. The Harris group and Simon group individually reported that the lipid droplet structural protein Perilipin-2 regulated droplet formation, and proposed that lipid storage offers protection against oxidative stress and ER-stress [34], [35]. The Welford group identified a transcriptional repression target of HIF, carnitine palmitoyltransferase 1A (CPT1A) [36]. Consistent with these findings, we found that the level of CPT1A was also up-regulated in VHL-OE cells (Table S1). Unlike other studies, the current work demonstrates that VHL overexpression decreases TAG synthesis by inhibiting SREBP1 maturation. SREBP1 is the master regulator of lipid synthesis, and active SREBP1 form needs a maturation process including transportation to the Golgi, release by a two-step proteolytic cleavage, and shuttle into the nucleus to induce the expression of cholesterol and fatty acid synthesis-related genes [22]. Further studies are needed to examine the detailed mechanisms underlying VHL-regulated SREBP1 maturation.
More strikingly, we found that VHL overexpression markedly up-regulated the expression levels of HLAs and interferon-responsive proteins by using quantitative proteomics. RT-PCR analyses verified the proteomic results and suggested the regulation occurring at the transcriptional level. Three subunits of ISGF3 complex, which controls the expression of interferon-responsive genes, were all up-regulated. Fludarabine treatment significantly inhibited the transcription of the interferon-responsive genes. Up-regulation of proteins involved in antigen processing and interferon-responsive pathways rendered ccRCC cells sensitive to interferon treatment. Interferon-α and interferon-γ are currently used to treat ccRCC patients with metastatic tumors. The IRS designated in the present work is positively correlated with the VHL expression level as well as the DFS in ccRCC patients.
Conclusion
Taken together, this study demonstrates that high VHL expression is correlated with better prognosis in ccRCC patients. VHL overexpression reverses the Warburg phenotype in ccRCC cells and enhances STAT1-regulated immunogenicity. Our results suggest that patients with high VHL expression will benefit from immunotherapy.
Materials and methods
Lentivirus production and generation of the VHL-OE cell lines
The VHL-OE cell line was established as previously described [37]. Briefly, the cDNA of human VHL gene was cloned from HEK293T cells. N-terminal HA tag, a Xho I restriction site, and a BamH I restriction site were added to VHL cDNA by PCR. The obtained HA-VHL sequence was ligated into linearized lentivirus vector pLVX-IRES-ZsGreen1 by a ligation reaction. Vectors were transfected into HEK293T cells along with packing plasmids for lentivirus production. The supernatant containing viral particles was collected and then concentrated. The viral particles were added into the 769-P and 786-O cell cultures along with polybrene reagent. Cells expressing GFP were sorted by flow cytometer and seeded into 96-well plates for a monoclonal cell. The cells infected with viruses produced from the empty vector were used as a control.
Construction of the nSREBP1-OE plasmid
The plasmid for nSREBP1 overexpression was generated by cloning the SREBP1 fragment (encoding 2–490 aa) from the cDNA clone plasmid (Catalog No. HG17512-UT, Sino Biological, Beijing, China) into the pLVX-IRES-mCherry plasmid [38]. The nSREBP1-OE plasmid was generated from the LVX-IRES-ZsGreen1 plasmid by replacing the ZsGreen1 sequence with the mCherry sequence.
Generation of a VHL-KO cell line using CRISPR/Cas9
pX458 (Catalog No. 48138, Addgene) was a gift from Dr. Yongzhang Luo (Tsinghua University, China). pX458 was digested with Bbs I. sgRNA sequences for human VHL were selected from GECKO library [39] and oligonucleotides were ordered from Ruibiotech (Beijing, China). sgRNAs were cloned into pX458 as described in Zhang lab’s protocol [40]. sgRNA sequences are listed in Table S3.
Determination of cell proliferation rate
Cells were placed into 96-well plates. Cell counting kit-8 (Catalog No. CK04, Dojindo Molecular Technologies, Rockville, MD) was added into culture media every 24 h followed by 2-h incubation. Absorbance at 450 nm was measured.
Colony formation assay
Cells were seeded into 6-well tissue plates with 250 or 500 cells per well. Plates were incubated to allow colony growth for 7–14 days. Colonies were then fixed with ethanol and stained with crystal violet. The stained plates were pictured with Image Lab 4.0 (Bio-Rad Laboratories, Berkeley, CA).
Scratch assay
Seed cells into 6-well tissue culture plates containing RPMI1640 media supplemented with 10% FBS, at a density that it can reach 95% confluence as a monolayer after 24 h. Scratch a straight line in the cell monolayer. Replenish the well with medium without FBS. Grow cells for 24 h or 48 h. Take photos for the monolayer on a microscope at 12 h, 24 h, 48 h, and 96 h, respectively. The gap distance and area were quantitatively evaluated with ImageJ software.
Measurement of intracellular ROS
Cells were incubated with CellROX Deep Red Reagent (Catalog No. C10491, ThermoFisher Scientific, Waltham, MA) for 30 min. Then, the media were discarded and cells were detached. Flow cytometer was used to measure the fluorescence intensity.
Mitochondrial respiration analysis
Basal glycolysis and basal respiration of cells were measured with the Seahorse XFe96 Analyzer (Agilent TechnologiesSanta Clara, CA). Oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, and antimycin premixed with rotenone were sequentially added into the reaction wells. After the measurement, the cells were scratched and lysed for protein quantification by BCA kit. Results were normalized by protein amount and visualized using Wave 2.6 software.
Lipid analysis based on LC-MS/MS
According to Bligh and Dyer’s method, lipids were extracted from cells [41]. Briefly, 5 × 106 cells were suspended with PBS buffer, followed by addition of 3 ml of methanol and chloroform (V:V = 1:2). The sample was vortexed and centrifuged, and then the lower layer was collected and dried with nitrogen blow. Then, the sample was dissolved with methanol and chloroform solution (V:V = 1:2), separated with C18 column, and analyzed by using the Q Exactive HF-X mass spectrometer (ThermoFisher Scientific). Data were acquired in both negative and positive modes by using Xcalibur 3.0 software (ThermoFisher Scientific). Each scan was followed by the dissociation of the top 10 peaks with the highest intensity. 15%, 30%, and 45% were the stepped normalized collision energy used for dissociation. LipidSearch software (ThermoFisher Scientific) was used for database searching, with mass tolerance settings including 8 ppm for precursor ions and 15 ppm for the fragment ions. A mixture containing aliquots of all samples was used as quality control between runs.
Sample preparation and quantitative proteomics analysis
Samples for quantitative proteomics analysis were prepared as previously described [42]. Briefly, cells were lysed by sonication in RIPA (Catalog No. R0020, Solarbio, Beijing, China). The supernatant after centrifugation was collected and quantified with BCA kit. Equal amount of proteins was reduced with DTT reagent, alkylated with IAA reagent, and precipitated with precooled acetone in four volumes, and then the precipitation was resolved with 8 M urea. The protein concentration was measured with BCA kit. Then, equal amount of proteins was diluted with PBS buffer to 1.6 M urea and digested with trypsin (Catalog No. V5280, Promega, Madison, WI) for 16 h. The peptides were desalted with Sep-Pak C18 columns and labeled with TMT 6-plex reagents (Catalog No. 90061, ThermoFisher Scientific). TMT labeling reactions were quenched with 5% hydroxylamine. The TMT-labeled peptides were combined and desalted again. Desalted peptides were separated with basic RPLC. The mobile phase A for RPLC was 2% acetonitrile with ammonium hydroxide. The mobile phase B for RPLC was acetonitrile with ammonium hydroxide. The gradient of mobile phase B for RPLC was: 8% to 18%, 30 min; 18% to 32%, 22 min. The labeled peptides were firstly separated with a C18 column with an HPLC system. The HPLC system was connected to the Obitrap Fusion Lumos Tribrid mass spectrometer (ThermoFisher Scientific). Data acquisition was in data-dependent mode with Xcalibur 3.0 software. The m/z survey scan was set as 350 to 1550 (120,000 resolution), and each scan was followed by the dissociation of the top 30 peaks with the highest intensity and charge state two or above. The normalized collision energy for dissociation was 35%. The isolation window width was 0.7 Da, and the dynamic exclusion time was set to 8 s. Acquisition settings for MS2 spectra were 30,000 for resolution, 2 × 105 for AGC target, and 60 ms for maximum injection time.
The Human database (UniProt, released on March 17, 2017; containing 21,042 entries) was used by SequestHT in Proteome Discoverer 2.1 software (ThermoFisher Scientific) for searching the generated spectra. The searching process was based on the following criteria: one missed cleavage tolerance, full tryptic specificity required, and 10 ppm and 20 mmu for precursor and fragment ion mass tolerance, respectively. The fixed modifications were carbamidomethylation and TMT 6-plex, and the variable modification was oxidation. Peptide spectrum matches (PSMs) were filtered by using the Percolator calculator embedded in the Proteome Discoverer software based on 1% false discovery rate (FDR). Biological triplicates of the quantitative proteomics analysis were performed.
Western blotting
Lyse the cells with RIPA buffer, and then centrifuge and collect the supernatant. BCA method was used for protein concentration determination. Proteins were separated by electrophoresis, electro blotted to a membrane, blocked, incubated with a primary antibody, and then incubated with a secondary antibody. Antibodies including primary antibodies for VHL (Catalog No. CST2738), ISG15 (Catalog No. CST2758S), STAT1 (Catalog No. CST9172S), phospho-STAT1 (Ser727) (Catalog No. CST8826S), PD-L1 (Catalog No. CST13684T), and secondary antibodies for mouse or rabbit were from Cell Signaling Technology (Danvers, MA). Antibodies for HLA-A (Catalog No. Ab52922), HLA-DR (Catalog No. Ab213305), and UBE1L (Catalog No. ab133479) were from Abcam (Cambridge, MA). Antibodies for SREBP1 (Catalog No. SAB4502850), FAS (Catalog No. SAB4501532), NDUFV2 (Catalog No. SAB2107279), and NDUFA4 (Catalog No. SAB4501963) were from Sigma-Aldrich (St. Louis, MO). Antibodies for HIF1A (Catalog No. 20960-AP), UBE2L6 (Catalog No. 17278-I-AP), and GAPDH (Catalog No. 10494-1-AP) were from Proteintech (Rosemont, IL). HERC5 antibody (Catalog No. 41836) was from GeneTex (Irvine, CA). ACTIN antibody (Catalog No. CW0096M) was from CWBiotech (Beijing, China). HA antibody (Catalog No. BE2007-100) was from Easybio (Beijing, China).
Extraction of cytoplasmic and nuclear proteins
Nuclear and cytoplasmic proteins were prepared according to a previously reported method [43]. The control and VHL-OE culture cells (2 × 106 cells each) were collected and washed with cold PBS twice. The cell pellets were suspended in 5 volumes of hypotonic lysis buffer (1.5 mM MgCl2, 0.1 M DTT, 10 mM KCl, 10 mM HEPES pH 7.9, 1× protease inhibitor), and mixed gently to promote swelling of cells. The samples were centrifuged. Then, the pellets were collected, suspended in 2 volumes of hypotonic lysis buffer, and aspirated 5–10 times with a syringe and trypan blue staining to check the integrity of cell membrane. The samples were centrifuged, and the supernatants were then collected, which are the cytoplasmic extracts. The pellets were also collected, washed with hypotonic lysis buffer twice, suspended in RIPA lysis buffer, and centrifuged. Finally, the supernatants were collected as the nuclear extracts.
RT-PCR
RNA extraction kit (Catalog No. LS1040, Promega) was used for RNA extraction. Reverse transcription kit (Catalog No. A2800, Promega) was used for reverse transcription. RT-PCR was carried out by using the LightCycler 96 (Roche, Basel, Switzerland) with SYBR green reaction mixture (Catalog No. CW0957, CWBiotech, Beijing, China). 2−ΔΔCt method was used for quantitative analysis. The sequences of the RT-PCR primers are listed in Table S4.
Bioinformatics and statistical analyses
The mutation data and clinical information were obtained from TCGA datasets. All the quantified proteins from proteomes were used for canonical pathway analysis using IPA software. The survival analysis of TCGA data was performed by using R packages “survminer” and “survival”, or with GEPIA web tools [44]. The Pearson correlation coefficient (R) between tested genes was calculated in the correlation analysis. The gene set collections “c5” (GO gene sets) from MsigDB were used as the functional annotation tool for GSEA. The R packages “clusterProfiler” and “enrichplot” were used for GSEA [45]. RStudio (version 1.1, base R 3.5) and GraphPad Prism were used for plotting and statistical analysis. Student’s t-test was used to determine the differences between two independent groups. P < 0.05 was considered to be statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Data availability
The obtained proteomic data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (PRIDE: PXD012665), which are publicly accessible at http://proteomecentral.proteomexchange.org.
CRediT author statement
Songbiao Zhu: Conceptualization, Formal analysis, Investigation, Writing - original draft, Writing - review & editing, Visualization, Project administration. Wenxi Ding: Investigation. Yuling Chen: Formal analysis. Weixuan Wang: Resources. Renhua Xu: Resources. Chongdong Liu: Resources. Xiaohui Liu: Formal analysis, Investigation. Haiteng Deng: Conceptualization, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition. All authors have read and approved the final manuscript.
Competing interests
All authors declare no conflicts of interest.
Acknowledgments
This study was supported by the Chinese Ministry of Science and Technology (Grant Nos. 2017ZX10201101 and 2014CBA02005), the China Postdoctoral Science Foundation (Grant No. 2017M610080), the National Key R&D Program (Grant No. 2017YFA0505103), the National Nature Science Foundation of China (Grant No. 21877068), and the Shandong Science and Technology Committee (Grant No. 2018GSF118159). The Protein Chemistry and Proteomics Facility at Tsinghua University helped with MS analysis.
Handled by Zhenhai Zhang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2019.12.002.
Supplementary material
The following are the Supplementary data to this article:
Lower VHL mRNA expression levels correlated with longer DFS in ACC and LIHC A. Lower VHL mRNA expression levels correlated with longer DFS within ACC patients using the ACC datasets. There are 19 samples each in low (blue) and high (red) groups. Log-rank test was used for comparing the survival curves (P = 0.0013). B. Lower VHL mRNA expression levels correlated with longer DFS within LIHC patients using the LIHC datasets. There are 91 samples each in low (blue) and high (red) groups. Log-rank test was used for comparing the survival curves (P = 0.043). C. Cox proportional hazards regression analysis of gender, age, race and VHL expression level on ccRCC patients’ survival. ACC, adrenocortical carcinoma. LIHC, liver hepatocellular carcinoma
VHL overexpression decreased cell proliferation, vitality and cellular ROS level in 786-O cells A. RT-PCR analysis of VHL expression in the 786-O VHL-OE and control cells. B. Western blotting analysis of HA-tagged VHL expression in the 786-O VHL-OE and control cells. C. VHL-overexpression decreased cell proliferation rate in 786-O cells. D. VHL-overexpression decreased the colony formation in 786-O cells. E. VHL-overexpression decreased cellular ROS level in 786-O cells
VHL-knockout increased cell proliferation, vitality and migration A. VHL-knockout increased cell proliferation rate in 293T cells. B. VHL-knockout increased the colony formation in 293T cells. C. Representative images of the scratch assay of the 293T-control and 293T-VHL-KO cells. D. Quantitative analysis of the scratch assay of the 293T-control and 293T-VHL-KO cells by using ImageJ software
VHL overexpression decreased nuclear-localized SREBP1 level A. Western blotting analysis of subcellular localization of SREBP1 in VHL overexpressing cells and control cells. B. RT-PCR analysis of SREBP1 downstream genes after rescuing expression of full-length SREBP1 or nuclear SREBP1 in VHL overexpressing cells
Variation and pathway enrichment analysis of the proteomic result A. Determination of the experimental variation for the quantified ratios from proteomic result. B. The Z-score calculation results of proteomic data by using IPA software
Western blotting analysis of the level of STAT1-pSer727 Western blotting analysis of STAT1-pSer727 in VHL overexpressing cells and control cells
The list of 404 up-regulated proteins in 769-P VHL-OE cells compared to control cells
The list of 228 down-regulated proteins in 769-P VHL-OE cells compared to control cells
Sequences of oligoes used for CRISPR/Cas9 in this work
Primers used in this work
References
- 1.Ericsson J.L., Seljelid R., Orrenius S. Comparative light and electron microscopic observations of cytoplasmic matrix in renal carcinomas. Virchows Arch Pathol Anat Physiol Klin Med. 1966;341:204–223. doi: 10.1007/BF00961071. [DOI] [PubMed] [Google Scholar]
- 2.Jonasch E., Gao J., Rathmell W.K. Renal cell carcinoma. BMJ. 2014;349 doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zarrabi K., Fang C., Wu S. New treatment options for metastatic renal cell carcinoma with prior anti-angiogenesis therapy. J Hematol Oncol. 2017;10:1–38. doi: 10.1186/s13045-016-0374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barata P.C., Rini B.I. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67:507–524. doi: 10.3322/caac.21411. [DOI] [PubMed] [Google Scholar]
- 5.Motzer R.J., Bander N.H., Nanus D.M. Renal cell carcinoma. N Engl J Med. 1996;335:865–875. doi: 10.1056/NEJM199609193351207. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin W.G., Jr The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–873. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 8.Kaelin W.G., Jr The von Hippel-Lindau tumour suppressor protein. Annu Rev Cancer Biol. 2018;2:91–109. doi: 10.1038/nrc2502. [DOI] [PubMed] [Google Scholar]
- 9.Courtney K.D., Bezwada D., Mashimo K., Pichumani K., Vemireddy V., Funk A.M., et al. Isotope tracing of human clear cell renal cell carcinomas demonstrates suppressed glucose oxidation in vivo. Cell Metab. 2018;28:793–800. doi: 10.1016/j.cmet.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jo Y., Lee P.C., Sguigna P.V., DeBose Boyd R.A. Sterol-induced degradation of HMG CoA reductase depends on interplay of two Insigs and two ubiquitin ligases, gp78 and trc8. Proc Natl Acad Sci U S A. 2011;108:20503–20508. doi: 10.1073/pnas.1112831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drabkin H.A., Gemmill R.M. Cholesterol and the development of clear cell renal carcinoma. Curr Opin Pharmacol. 2012;12:742–750. doi: 10.1016/j.coph.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Dagher J., Kammerer Jacquet S.F., Brunot A., Pladys A., Patard J.J., Bensalah K., et al. Wild-type VHL clear cell renal cell carcinomas are a distinct clinical and histologic entity: a 10-year follow-up. Eur Urol Focus. 2016;3:284–290. doi: 10.1016/j.euf.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Kammerer Jacquet S.F., Crouzet L., Brunot A., Dagher J., Pladys A., Edeline J., et al. Independent association of PD-L1 expression with noninactivated VHL clear cell renal cell carcinoma-a finding with therapeutic potential. Int J Cancer. 2017;140:142–148. doi: 10.1002/ijc.30429. [DOI] [PubMed] [Google Scholar]
- 14.Batavia A.A., Schraml P., Moch H. Clear cell renal cell carcinoma with wild-type von Hippel-Lindau gene: a non-existent or new tumour entity? Histopathology. 2019;74:60–67. doi: 10.1111/his.13749. [DOI] [PubMed] [Google Scholar]
- 15.Guo G., Gui Y., Gao S., Tang A., Hu X., Huang Y., et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2011;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., et al. Integrated molecular analysis of clear cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 17.Gerlinger M., Horswell S., Larkin J., Rowan A.J., Salm M.P., Varela I., et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turajlic S., Larkin J., Swanton C. SnapShot: renal cell carcinoma. Cell. 2015;163:1556. doi: 10.1016/j.cell.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.H., Jeon Y.G., Lee K.H., Lee H.W., Park J., Jang H., et al. RNF20 suppresses tumorigenesis by inhibiting the SREBP1c-PTTG1 axis in kidney cancer. Mol Cell Biol. 2017;37:e00265–17. doi: 10.1128/MCB.00265-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown M.S., Goldstein J.L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 23.Gan C., Chong P.K., Pham T.K., Wright P.C. Technical, experimental and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2006;6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S.J., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang D., Zhang D.E. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res. 2011;31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogunovic D., Boisson Dupuis S., Casanova J.L. ISG15: leading a double life as a secreted molecule. Exp Mol Med. 2013;45:e18. doi: 10.1038/emm.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheon H., Holvey Bates E.G., Schoggins J.W., Forster S., Hertzog P., Imanaka N., et al. IFNβ-dependent increases in STAT1, STAT2 and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szelag M., Sikorski K., Czerwoniec A., Szatkowska K., Wesoly J., Bluyssen H.A. In silico simulations of STAT1 and STAT3 inhibitors predict SH2 domain cross-binding specificity. Eur J Pharmacol. 2013;720:38–48. doi: 10.1016/j.ejphar.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 29.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;5:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A., et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tissier F., Cavard C., Groussin L., Perlemoine K., Fumey G., Hagneré A.M., et al. Mutations of beta-catenin in adrenocortical tumours: activation of the Wnt signaling pathway is a frequent event in both benign and malignant adrenocortical tumors. Cancer Res. 2005;65:7622–7627. doi: 10.1158/0008-5472.CAN-05-0593. [DOI] [PubMed] [Google Scholar]
- 32.Zheng S., Cherniack A.D., Dewal N., Moffitt R.A., Danilova L., Murray B.A., et al. Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell. 2016;5:723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guichard C., Amaddeo G., Imbeaud S., Ladeiro Y., Pelletier L., Maad I.B., et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bensaad K., Favaro E., Lewis C.A., Peck B., Lord S., Collins J.M., et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 35.Qiu B., Ackerman D., Sanchez D.J., Li B., Ochocki J.D., Grazioli A., et al. HIF2alpha-dependent lipid storage promotes endoplasmic reticulum homeostasis in clear cell renal cell carcinoma. Cancer Discov. 2015;5:652–667. doi: 10.1158/2159-8290.CD-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W., Zhang L., Brett Morris A., Aguila B., Kerner J., Hoppel C.L., et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nat Commun. 2017;8:e1769. doi: 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Wang H., Wang Q., Deng H. Overexpression of CD38 decreases cellular NAD levels and alters the expression of proteins involved in energy metabolism and antioxidant defense. J Proteome Res. 2014;13:786–795. doi: 10.1021/pr4010597. [DOI] [PubMed] [Google Scholar]
- 38.Giandomenico V., Simonsson M., Grönroos E., Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP Family of transcription factors. Mol Cell Bio. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanjana N.E., Shalem Q., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 42.Tang H., Li J., Liu X., Wang G., Luo M., Deng H. Down-regulation of HSP60 suppresses the proliferation of glioblastoma cells via the ROS/AMPK/mTOR pathway. Sci Rep. 2016;6:e28388. doi: 10.1038/srep28388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shechter D., Dormann H.L., Allis C.D., Hake S.B. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 44.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu G., Wang L., Han Y., He Q. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lower VHL mRNA expression levels correlated with longer DFS in ACC and LIHC A. Lower VHL mRNA expression levels correlated with longer DFS within ACC patients using the ACC datasets. There are 19 samples each in low (blue) and high (red) groups. Log-rank test was used for comparing the survival curves (P = 0.0013). B. Lower VHL mRNA expression levels correlated with longer DFS within LIHC patients using the LIHC datasets. There are 91 samples each in low (blue) and high (red) groups. Log-rank test was used for comparing the survival curves (P = 0.043). C. Cox proportional hazards regression analysis of gender, age, race and VHL expression level on ccRCC patients’ survival. ACC, adrenocortical carcinoma. LIHC, liver hepatocellular carcinoma
VHL overexpression decreased cell proliferation, vitality and cellular ROS level in 786-O cells A. RT-PCR analysis of VHL expression in the 786-O VHL-OE and control cells. B. Western blotting analysis of HA-tagged VHL expression in the 786-O VHL-OE and control cells. C. VHL-overexpression decreased cell proliferation rate in 786-O cells. D. VHL-overexpression decreased the colony formation in 786-O cells. E. VHL-overexpression decreased cellular ROS level in 786-O cells
VHL-knockout increased cell proliferation, vitality and migration A. VHL-knockout increased cell proliferation rate in 293T cells. B. VHL-knockout increased the colony formation in 293T cells. C. Representative images of the scratch assay of the 293T-control and 293T-VHL-KO cells. D. Quantitative analysis of the scratch assay of the 293T-control and 293T-VHL-KO cells by using ImageJ software
VHL overexpression decreased nuclear-localized SREBP1 level A. Western blotting analysis of subcellular localization of SREBP1 in VHL overexpressing cells and control cells. B. RT-PCR analysis of SREBP1 downstream genes after rescuing expression of full-length SREBP1 or nuclear SREBP1 in VHL overexpressing cells
Variation and pathway enrichment analysis of the proteomic result A. Determination of the experimental variation for the quantified ratios from proteomic result. B. The Z-score calculation results of proteomic data by using IPA software
Western blotting analysis of the level of STAT1-pSer727 Western blotting analysis of STAT1-pSer727 in VHL overexpressing cells and control cells
The list of 404 up-regulated proteins in 769-P VHL-OE cells compared to control cells
The list of 228 down-regulated proteins in 769-P VHL-OE cells compared to control cells
Sequences of oligoes used for CRISPR/Cas9 in this work
Primers used in this work
Data Availability Statement
The obtained proteomic data have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database partner repository (PRIDE: PXD012665), which are publicly accessible at http://proteomecentral.proteomexchange.org.