Abstract
Esophageal squamous cell carcinoma (ESCC) is a major histological subtype of esophageal cancer with a poor prognosis. Although several serum metabolomic investigations have been reported, ESCC tumor-associated metabolic alterations and predictive biomarkers in sera have not been defined. Here, we enrolled 34 treatment-naive patients with ESCC and collected their pre- and post-esophagectomy sera together with the sera from 34 healthy volunteers for a metabolomic survey. Our comprehensive analysis identified ESCC tumor-associated metabolic alterations as represented by a panel of 12 serum metabolites. Notably, postoperative abrosia and parenteral nutrition substantially perturbed the serum metabolome. Furthermore, we performed an examination using sera from carcinogen-induced mice at the dysplasia and ESCC stages and identified three ESCC tumor-associated metabolites conserved between mice and humans. Notably, among these metabolites, the level of pipecolic acid was observed to be progressively increased in mouse sera from dysplasia to cancerization, and it could be used to accurately discriminate between mice at the dysplasia stage and healthy control mice. Furthermore, this metabolite is essential for ESCC cells to restrain oxidative stress-induced DNA damage and cell proliferation arrest. Together, this study revealed a panel of 12 ESCC tumor-associated serum metabolites with potential for monitoring therapeutic efficacy and disease relapse, presented evidence for refining parenteral nutrition composition, and highlighted serum pipecolic acid as an attractive biomarker for predicting ESCC tumorigenesis.
Keywords: Esophageal squamous cell carcinoma, Serum metabolome, Esophagectomy, Predictive potential, Pipecolic acid
Introduction
Esophageal cancer (EC) is the ninth most common cancer and the sixth leading cause of cancer-related deaths globally, and its incidence continues to increase steadily [1], [2]. Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma are the two main histological types of EC. In the highest-risk region from northern Iran through central Asia to north-central China, ESCC is the most prevalent type accounting for 90% of EC, and patients with ESCC in China account for more than half of the global ESCC cases [2], [3], [4]. Esophagectomy is the primary treatment for initially diagnosed patients [5]. Chemotherapy or chemoradiotherapy is required for patients with locally advanced or clinical N1–N3 (N1, metastasis in one or two regional lymph nodes; N3, metastasis in seven or more regional lymph nodes) [2]. Due to the lack of specific signs or symptoms at an early stage, the majority of patients diagnosed with ESCC are at an advanced stage [6], [7]. Therefore, the prognosis and 5-year survival of these patients are unsatisfactory [8], [9], [10]. Early diagnosis is key to improve the therapeutic outcomes of patients with ESCC. Hence, it is urgently necessary to identify new biomarkers with predictive potential for ESCC tumorigenesis.
Metabolic reprogramming is a core hallmark of cancer [11], [12]. Neoplastic cells require a large amount of energy and raw materials in the process of cancer occurrence, development, and metastasis, thus, disturbing the global metabolism, including metabolic signatures of circulating blood in patients [11], [12], [13], [14], [15], [16]. Abnormal metabolites in the peripheral blood can be used for cancer diagnosis, treatment efficacy assessment, and patient prognosis prediction [14], [15], [17]. Hence, systemic measurement of metabolite composition in sera would deepen our understanding of ESCC molecular features and yield new diagnostic opportunities for patients with ESCC. Several metabolomic studies using the serum or plasma specimens of patients with ESCC have been conducted, and significant variations in lipid, glucose, and amino acid metabolic levels in patients with ESCC as compared to healthy controls (HCs) have been identified [7], [18], [19], [20], [21], [22]. However, due to metabolic modifications induced by ESCC indirectly as confounding factors in these data, ESCC tumor-associated metabolic alterations cannot be properly discerned. A recent study collected pre- and post-operative serum samples of patients with ESCC and performed metabolomic investigations to identify diagnostic biomarkers [23]. Nevertheless, these pre- and post-operative serum samples were derived from different ESCC patient cohorts, and the measurement platform using nuclear magnetic resonance spectroscopy had limited resolution [23]. Therefore, this study could not precisely and comprehensively capture ESCC tumor-associated metabolic alterations.
In this study, we enrolled 34 treatment-naive patients with ESCC and 34 healthy volunteers. Pre- and post-esophagectomy sera from patients with ESCC along with sera from healthy volunteers were collected for metabolomic investigation using gas chromatography-time-of-flight mass spectrometry (GC-TOFMS), with the aim of discovering ESCC tumor-associated metabolic alterations. Subsequently, we recruited control mice and carcinogen-induced mice at the dysplasia and ESCC stages. The sera of these mice were examined to validate ESCC tumor-associated metabolic alterations and to identify metabolite biomarkers with predictive potential. Finally, we conducted functional assays to explore the role of identified metabolite biomarkers in ESCC tumorigenesis.
Results
A distinct serum metabolic signature of preoperative patients with ESCC
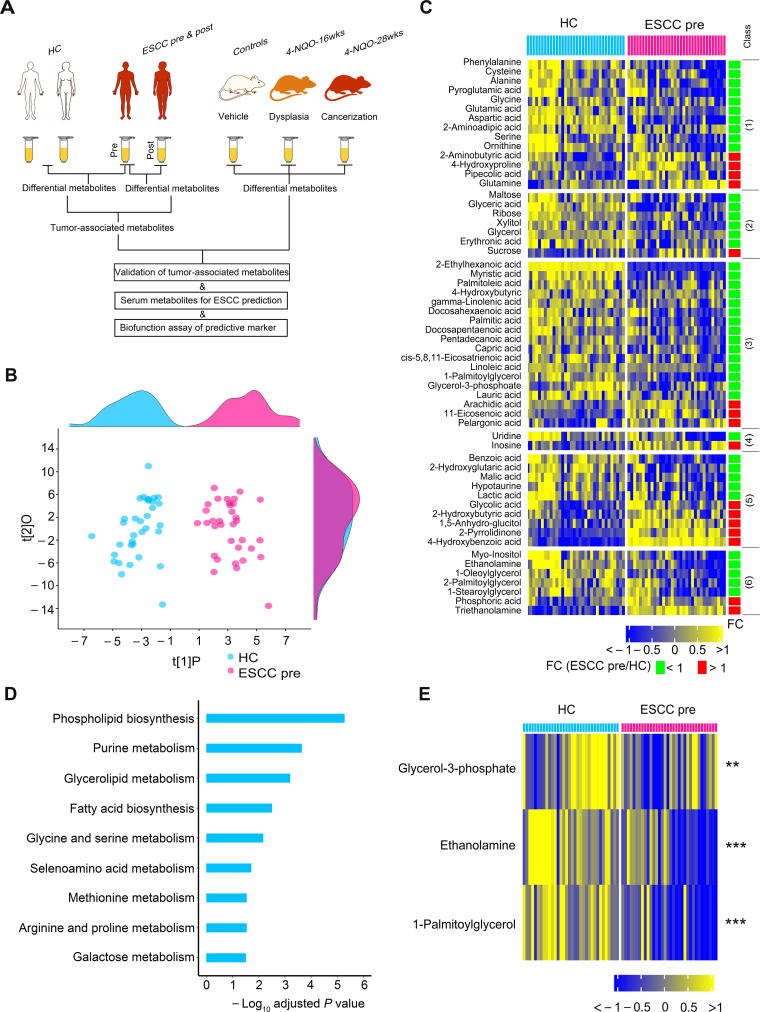
The design of this study is illustrated in Figure 1A. Serum samples were harvested from 34 patients with ESCC (pre-esophagectomy versus post-esophagectomy) and HCs to identify ESCC tumor-associated serum metabolites. Meanwhile, we collected serum samples from control mice and 4-nitroquinoline 1-oxide (4-NQO)-treated mice at the dysplasia and ESCC stages to identify ESCC tumor-associated metabolites conserved between mice and humans and to identify serum metabolite markers for ESCC prediction. Finally, we performed functional assays of the identified predictive metabolite markers. There were no significant differences in age, sex, serum alanine aminotransferase, serum aspartate aminotransferase, and serum creatinine between HCs and preoperative ESCC patients or between preoperative and postoperative ESCC patients (Table 1). Before metabolomic data analysis, we first assessed data quality and observed low relative standard deviation values of chemical standards in quality control samples, revealing the high stability of the measurement system (Table S1). A total of 161 metabolites were identified after excluding internal standards and unknown peaks. The serum metabolomic features of preoperative ESCC patients were remarkably altered relative to those of HCs, as demonstrated by the robust model of orthogonal partial least squares-discriminant analysis (OPLS-DA; R2Y = 0.88, Q2 = 0.81) (Figure 1B). Among the 161 metabolites, 58 were significantly changed in preoperative ESCC sera (Bonferroni-adjusted P < 0.05), including 14 amino acids, 7 carbohydrates, 18 lipids, 2 nucleosides, 10 organic acids, and 7 unclassified metabolites (Figure 1C). Furthermore, among these 58 changed metabolites, the levels of 16 of them were up-regulated, including those of 4 amino acids, 1 carbohydrate, 3 lipids, 1 nucleoside, 5 organic acids, and 2 unclassified metabolites; while the levels of the remaining metabolites were down-regulated, including those of 10 amino acids, 6 carbohydrates, 15 lipids, 1 nucleoside, 5 organic acids, and 5 unclassified metabolites. In agreement with the results of previous studies [19], [24], our results revealed that the abundance of glutamine was up-regulated in ESCC sera, while the concentrations of glutamic acid, alanine, glycine, serine, ornithine, ribose, glyceric acid, docosahexaenoic acid, and linoleic acid were restrained in ESCC sera.
Figure 1.
Serum metabolic signature was altered in preoperative patients withESCCas relative to HCs
A. Study design. Serum samples from patients with ESCC (pre-esophagectomy versus post-esophagectomy) and HCs were harvested for the identification of ESCC tumor-associated serum metabolites. Meanwhile, serum samples from control mice and 4-NQO-treated mice at the dysplasia and ESCC stages were collected for the discovery of ESCC tumor-associated metabolites conserved between mice and humans as well as serum metabolite markers for ESCC prediction. Finally, functional assays of the predictive metabolite marker were conducted. B. OPLS-DA score plot showing a global metabolic difference in sera between preoperative ESCC patients and HCs. The density plots of principal component 1 and principal component 2 are shown on the top and right-hand side of the OPLS-DA score plots, respectively. C. Heatmap showing 58 differentially expressed serum metabolites in preoperative ESCC patients compared to HCs. The metabolites were subclassified as follows: 1) amino acids, 2) carbohydrates, 3) lipids including fatty acids, 4) nucleotides, 5) organic acids, and 6) unclassified. D. Metabolic pathway enrichment analysis using differential metabolites between preoperative ESCC patients and HCs. E. Heatmap showing the differential serum metabolites involved in the most disturbed metabolic pathway phospholipid biosynthesis between preoperative patients with ESCC and HCs. ESCC, esophageal squamous cell carcinoma; HC, healthy control; 4-NQO, 4-nitroquinoline 1-oxide; OPLS-DA, orthogonal partial least squares-discriminant analysis; ESCC pre, ESCC pre-esophagectomy; ESCC post, ESCC post-esophagectomy; FC, fold change.
Table 1.
Basic characteristics of enrolled healthy volunteers and patients with ESCC
| Variable | HC (n = 34) | ESCC pre (n = 34) | ESCC post (n = 34) | P value (HC vs. ESCC pre) | P value (ESCC pre vs. ESCC post) |
|---|---|---|---|---|---|
| Age(year) | 0.745 | — | |||
| Median | 67 | 67 | 67 | ||
| Range | 56–78 | 54–77 | 54–77 | ||
| Gender[No. (%)] | 0.291 | — | |||
| Male | 21 (64.7) | 26 (76.5) | 26 (76.5) | ||
| Female | 13 (35.3) | 8 (23.5) | 8 (23.5) | ||
| ALT(U/l) | 0.367 | 0.104 | |||
| Medium | 19.5 | 18 | 24 | ||
| Range | 9–43 | 10–51 | 12–132 | ||
| AST(U/l) | 0.059 | 0.216 | |||
| Medium | 24 | 21 | 22.5 | ||
| Range | 15–36 | 12–43 | 9–62 | ||
| Creatinine (μmol/l) | 0.267 | 0.031 | |||
| Medium | 78.65 | 70 | 67.5 | ||
| Range | 50.9–108.3 | 48–129 | 39–102 | ||
| Hepatic or renal function(No.) | |||||
| Normal | 34 | 34 | 33 | — | 0.314 |
| Abnormal | 0 | 0 | 1 | ||
| TNM stage [No. (%)] | |||||
| Ⅰ | 9 (26.5) | ||||
| Ⅱ | 18 (52.9) | ||||
| III | 6 (17.6) | ||||
| VI | 1 (2.9) | ||||
| Tumor grade [No. (%)] | |||||
| 1 | 7 (18.4) | ||||
| 2 | 18 (47.4) | ||||
| 3 | 3 (7.9) | ||||
| Unclassified | 6 (15.8) | ||||
| Lymph node metastasis [No. (%)] | |||||
| Positive | 12 (35.3) | ||||
| Negative | 22 (64.7) |
Note: Hepatic abnormality as defined by ALT > 2.5× normal value or AST > 2.5× normal value, while renal abnormality as defined by creatinine > 2.5× normal value. HC, healthy control; ESCC, esophageal squamous cell carcinoma; ESCC pre, ESCC pre-esophagectomy; ESCC post, ESCC post-esophagectomy; ALT, alanine aminotransferase; AST, aspartate aminotransferase; —, not applicable.
Next, metabolite set enrichment analysis (MSEA) was performed using the 58 changed metabolites mentioned above to observe which metabolic pathways were perturbed in preoperative ESCC sera. The results showed that a total of nine metabolic pathways were significantly altered (Holm-adjusted P < 0.05) (Figure 1D). For the most disturbed metabolic pathway, phospholipid biosynthesis, the levels of all detected metabolites involved in this pathway, including glycerol-3-phosphate, ethanolamine, and 1-palmitoylglycerol, consistently declined in preoperative ESCC sera (Figure 1E).
A panel of ESCC tumor-associated metabolites were restored toward HCs by esophagectomy
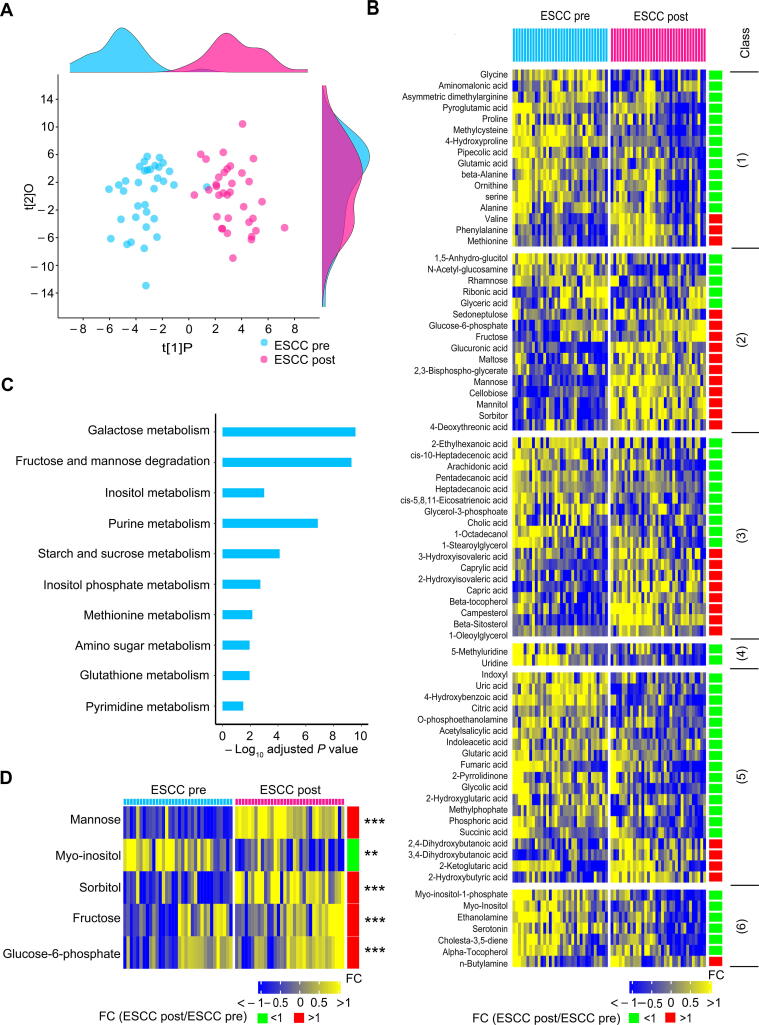
All enrolled patients with ESCC underwent esophagectomy to completely remove their tumors, and then were given abrosia and persistent parenteral nutrition from day 1 after the surgery. At day 3 after esophagectomy, serum samples from these patients were harvested for metabolomic measurements. First, we performed a comparative analysis between the preoperative and postoperative ESCC serum samples. Compared to the preoperative sera, postoperative sera exhibited a remarkably modified metabolic profile as illustrated by the robust OPLS-DA model (R2Y = 0.84, Q2 = 0.76) (Figure 2A), demonstrating that esophagectomy together with postoperative abrosia and parenteral nutrition replenishment could result in an overt impact on the global metabolism of patients with ESCC. The levels of 78 metabolites were found to be significantly changed in postoperative ESCC sera relative to preoperative specimens (Bonferroni-adjusted P < 0.05), including those of 16 amino acids, 16 carbohydrates, 18 lipids, 2 nucleosides, 19 organic acids, and 7 unclassified metabolites (Figure 2B). Subsequently, MSEA was conducted using these 78 metabolites, and the activities of 10 metabolic pathways were significantly fluctuated in postoperative ESCC sera when compared to those of preoperative ESCC sera (Holm-adjusted P < 0.05) (Figure 2C). Notably, galactose metabolism was the most disturbed pathway. At the metabolite level, the levels of four of five metabolites involved in this pathway were increased, while the abundance of the remaining metabolite was restrained in postoperative sera (Figure 2D).
Figure 2.
Serum metabolic feature was disturbed in patients with ESCC at the postoperative stage relative to the preoperative stage
A. OPLS-DA score plot showing modified global metabolism in postoperative ESCC patients compared to preoperative patients. The density plots of principal component 1 and principal component 2 are displayed on the top and right-hand side of the OPLS-DA score plots, respectively. B. Heatmap exhibiting 78 differential metabolites in postoperative ESCC patients relative to preoperative ESCC patients. The metabolites were subclassified as follows: 1) amino acids, 2) carbohydrates, 3) lipids including fatty acids, 4) nucleotides, 5) organic acids, and 6) unclassified. C. Metabolic pathway enrichment analysis using differential metabolites between preoperative and postoperative patients with ESCC. D. Heatmap showing the differential serum metabolites involved in the most disturbed metabolic pathway galactose metabolism between preoperative and postoperative patients with ESCC. **, P < 0.01; ***, P < 0.001.
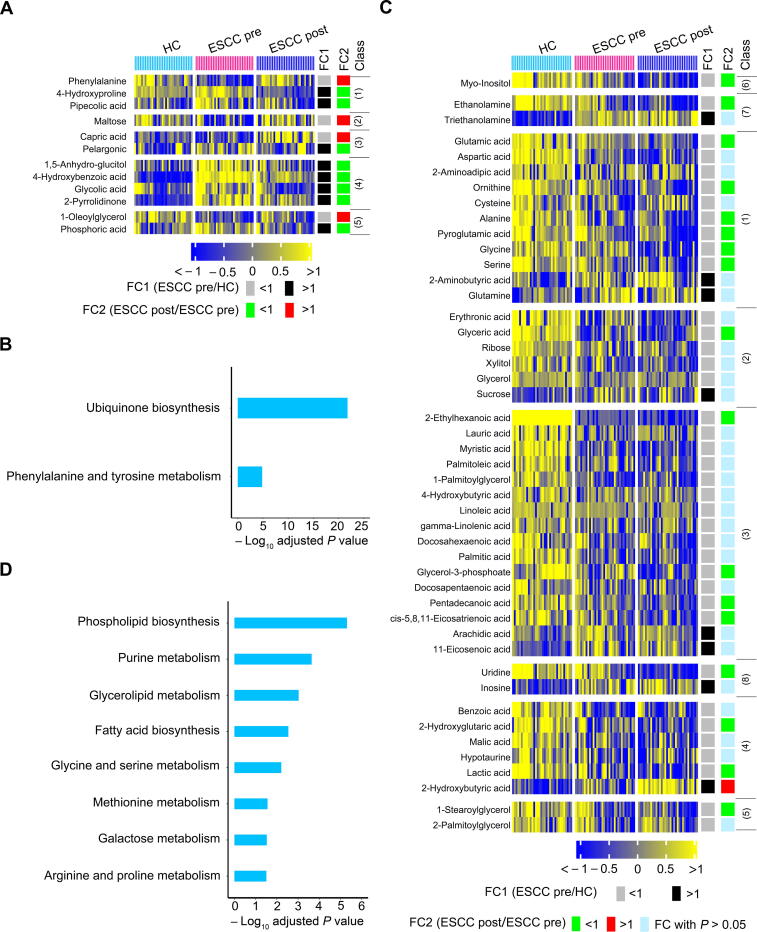
To determine the ESCC tumor-associated metabolic alterations, a comprehensive analysis was performed using HC, preoperative, and postoperative ESCC groups. Logically, 58 differential metabolites in preoperative ESCC sera as compared to HC sera (Figure 1C) contained metabolic signals altered by ESCC directly and indirectly. Therefore, we selected these metabolites to extract the ESCC tumor-associated metabolic alterations. As shown in Figure 3A, 12 metabolites in postoperative ESCC sera were significantly restored in the HC direction. This indicated that these serum metabolites were specifically modulated by ESCC tumors in patients. These metabolites were enriched in two pathways, including ubiquinone biosynthesis and phenylalanine and tyrosine metabolism (Holm-adjusted P < 0.05) (Figure 3B). In addition, the heatmap of Figure 3C showed that the levels of the remaining 46 metabolites in postoperative ESCC sera changed in the opposite HC direction or were not significantly altered relative to preoperative ESCC sera. We concluded that these 46 metabolites were not specifically modulated by ESCC tumors. Of note, these metabolites were enriched in eight pathways (Holm-adjusted P < 0.05) (Figure 3D).
Figure 3.
A few of 58 differential serum metabolites between preoperative patients and HCs were recovered in postoperative patientsinthe HC direction
Twelve metabolites were significantly restored in postoperative ESCC sera in the HC direction (A), and metabolic pathway enrichment analysis of these metabolites (B). The remaining 46 metabolites changed in postoperative ESCC sera in an opposite HC direction or not significantly altered in postoperative ESCC sera relative to preoperative ESCC sera (C), and metabolic pathway enrichment analysis of these metabolites (D). The metabolites in the heatmaps were subclassified as follows: 1) amino acids, 2) carbohydrates, 3) lipids including fatty acids, 4) organic acids, 5) unclassified, 6) alcohols, 7) amines, and 8) nucleotides.
Serum metabolism was perturbed by postoperative abrosia and parenteral nutrition
There were 58 differential metabolites in preoperative ESCC sera relative to HC sera, whereas there were 78 differential metabolites in postoperative ESCC sera relative to preoperative ESCC sera. This suggests that postoperative abrosia and parenteral nutrition might overtly disturb the serum metabolic profile of patients with ESCC. To verify this speculation, we analyzed 108 serum metabolites that were not altered in preoperative ESCC relative to HC. As shown in Figure 4A, the levels of 27 of these metabolites (25.00%) were significantly lower in postoperative ESCC than in preoperative ESCC or HCs. These metabolites included 4 amino acids, 3 carbohydrates, 5 lipids, 1 nucleotide, 10 organic acids, and 4 unclassified metabolites. Furthermore, the levels of 22 of 108 metabolites (20.37%) remarkably increased in postoperative ESCC relative to preoperative ESCC patients or HCs (Figure 4B). These up-regulated metabolites included 2 amino acids, 10 carbohydrates, 6 lipids, 3 organic acids, and 1 unclassified metabolite.
Figure 4.
Metabolic fluctuation induced by postoperative abrosia and parenteral nutrition
For the 108 serum metabolites not altered between preoperative patients with ESCC and HCs, the levels of 27 of them (25.00%) were significantly down-regulated in postoperative ESCC patients as compared to preoperative patients with ESCC or HCs (A), while the levels of 22 of them (20.37%) were remarkably increased in postoperative ESCC patients relative to preoperative ESCC patients or HCs (B). These 49 changed metabolites were subclassified as follows: 1) amino acids, 2) carbohydrates, 3) lipids including fatty acids, 4) nucleotides, 5) organic acids, and 6) unclassified. C. Metabolic pathway enrichment analysis of the above 49 altered metabolites.
MSEA using the above 49 altered metabolites in the sera of postoperative ESCC patients showed that these compounds were enriched in seven metabolic pathways (Figure 4C). Notably, carbohydrate anabolism was dramatically stimulated, as shown by the up-regulation of the abundance of 10 carbohydrates (Figure 4B and C). Collectively, among the 108 serum metabolites that were not modified between preoperative ESCC and HC, 45.37% were significantly perturbed in postoperative ESCC sera, indicating that postoperative abrosia and parenteral nutrition resulted in an overt fluctuation of serum metabolome.
Progressively modified serum metabolic signature of carcinogen-induced mice from dysplasia to ESCC
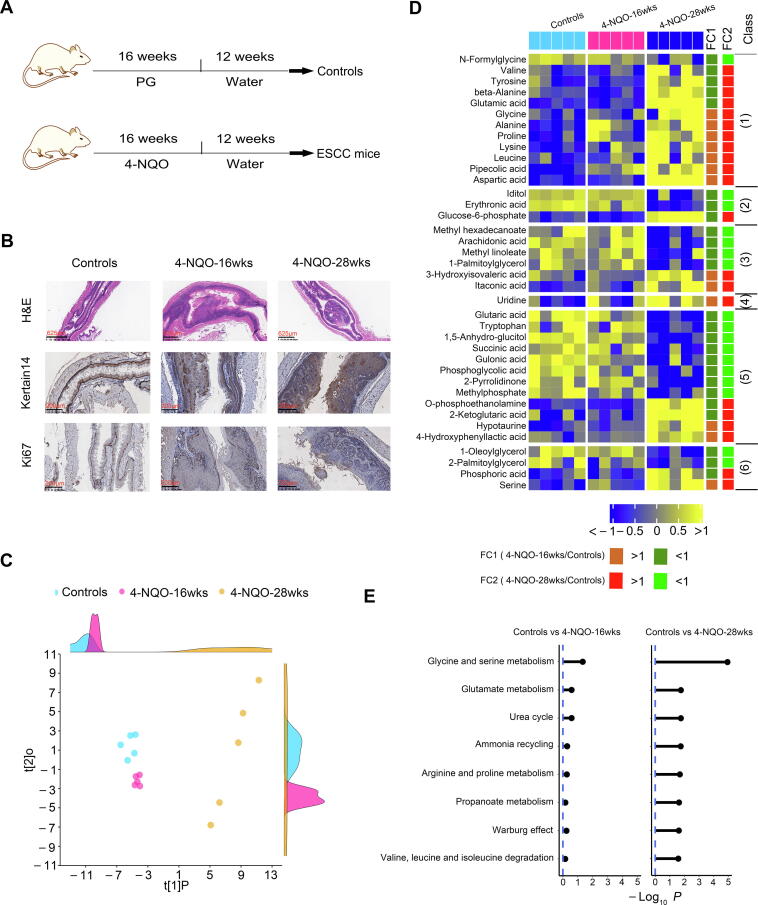
To validate the serum metabolic signature of preoperative ESCC patients and to discover metabolite biomarkers for the prediction of ESCC tumorigenesis, we employed a carcinogen-induced ESCC mouse model, as reported previously [24]. Mice were administered drinking water containing 100 μg/ml carcinogen 4-NQO or propylene glycol (PG) as a vehicle for 16 weeks and then were followed up for 12 weeks with pure drinking water. Thus, three groups of mice were formed, control, 4-NQO-16wks, and 4-NQO-28wks groups (Figure 5A). Disease progression was monitored by hematoxylin and eosin (H&E) staining of esophagi and by immunohistochemistry (IHC) assays for the ESCC molecular biomarker keratin 14 (K14) along with the cell proliferation marker Ki67 (Figure 5B). At week 16, esophageal squamous epithelia of the 4-NQO group showed increased squamous basal cell layers, thickened spinous cells, and disordered cell arrangement in each layer (Figure 5B, H&E image). Additionally, the esophagi of the 4-NQO-16wks group revealed cell atypia characterized by an increase in cell size and the ratio of nucleo-plasma, as well nuclear hyperchromatism and pathological nuclear fission (Figure 5B, H&E image). Furthermore, the expression of K14 and Ki67 was enhanced and extended to superficial cells as well as heteromorphic large cells (Figure 5B, IHC images). Together, these abnormal features demonstrated that mice in the 4-NQO-16wks group were at the dysplasia stage. At week 28, esophageal squamous epithelia of the 4-NQO group exhibited high-grade dysplasia, cancerization, and microstromal infiltration (Figure 5B, H&E image). Notably, the cells in the esophageal epithelial layer showed severe atypia. Additionally, heterotypical cells among these cells locally invaded the basement membranes (Figure 5B, H&E image). Moreover, the expression of K14 and Ki67 was diffuse and very strong (Figure 5B, IHC images). Collectively, these neoplastic characteristics indicated that mice in the 4-NQO-28wks group were at the cancerization stage.
Figure 5.
Progressively modified serum metabolome of carcinogen-induced mice from dysplasia to ESCC
A. A scheme depicting 4-NQO induced ESCC mouse model from dysplasia to ESCC. B. Representative images of H&E and IHC staining of esophageal tissues of control mice and 4-NQO-treated mice. C. PLS score plot of serum metabolic profiles of control mice, 4-NQO-induced mice at dysplasia stage (4NQO-16wks), and 4-NQO-induced mice at cancerization stage (4NQO-28wks). The density plots of principal component 1 and principal component 2 are displayed on the top and right-hand sides of the PLS score plots, respectively. Heatmap displaying progressive changes of serum metabolites of mice from normal control to cancerization stage (D) and metabolic pathway enrichment analysis of these metabolites (E). Progressively changed metabolites in the heatmap were subclassified as follows: 1) amino acids, 2) carbohydrates, 3) lipids including fatty acids, 4) nucleotides, 5) organic acids, and 6) unclassified. PG, propylene glycol; 4-NQO, 4-nitroquinoline 1-oxide; PLS, partial least-squares regression.
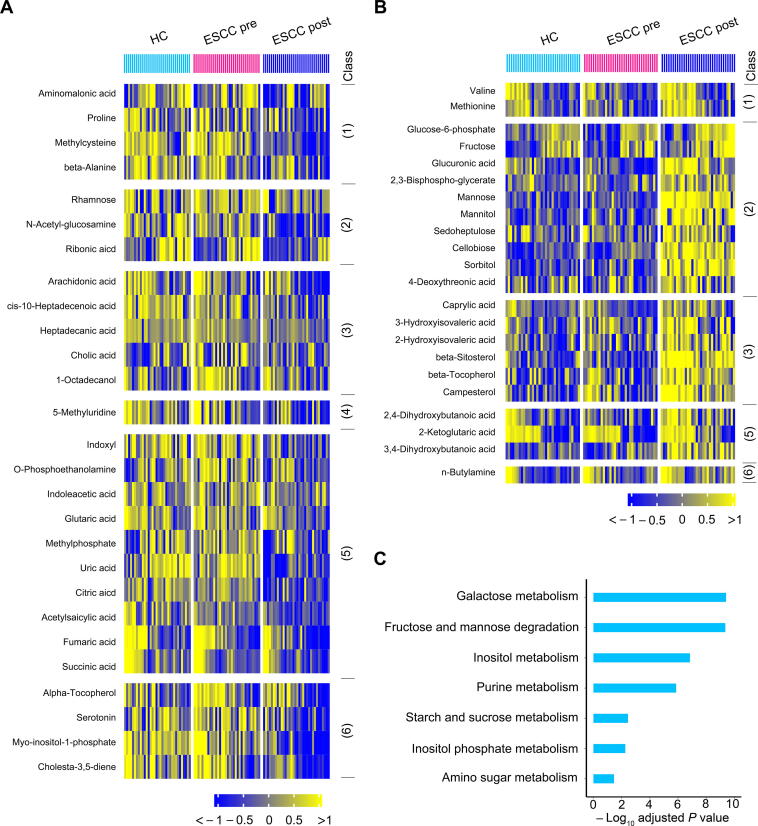
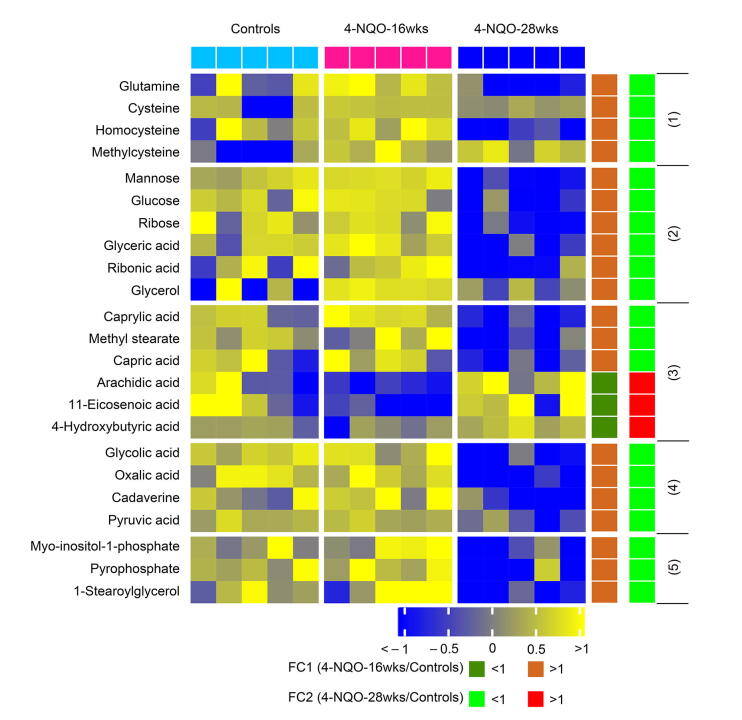
When compared to the control group, both 4-NQO-16wks and 4-NQO-28wks groups showed obvious alterations in their serum metabolic profiles, as illustrated by the robust model of partial least-squares (PLS) regression (R2Y = 0.98, Q2 = 0.80) (Figure 5C). A total of 61 serum metabolites were significantly altered among the three groups. Notably, 38 of these disturbed metabolites were progressively modified from the dysplasia stage to the cancerization stage as compared to the control group (Figure 5D), while the remaining 23 disturbed metabolites did not show this trend from normal control to cancerization (Figure S1).
Subsequently, we used the above 38 progressively modified metabolites in Figure 5D to conduct the MSEA. The results showed that eleven pathways were significantly disturbed in mouse sera at the cancerization stage as compared to controls (Holm-adjusted P < 0.05) (Figure 5E). Notably, these metabolic pathways were slightly altered in the mouse sera at the dysplasia stage (Figure 5E).
ESCC tumor-associated serum metabolites conserved between mice and humans with predictive potential for tumorigenesis
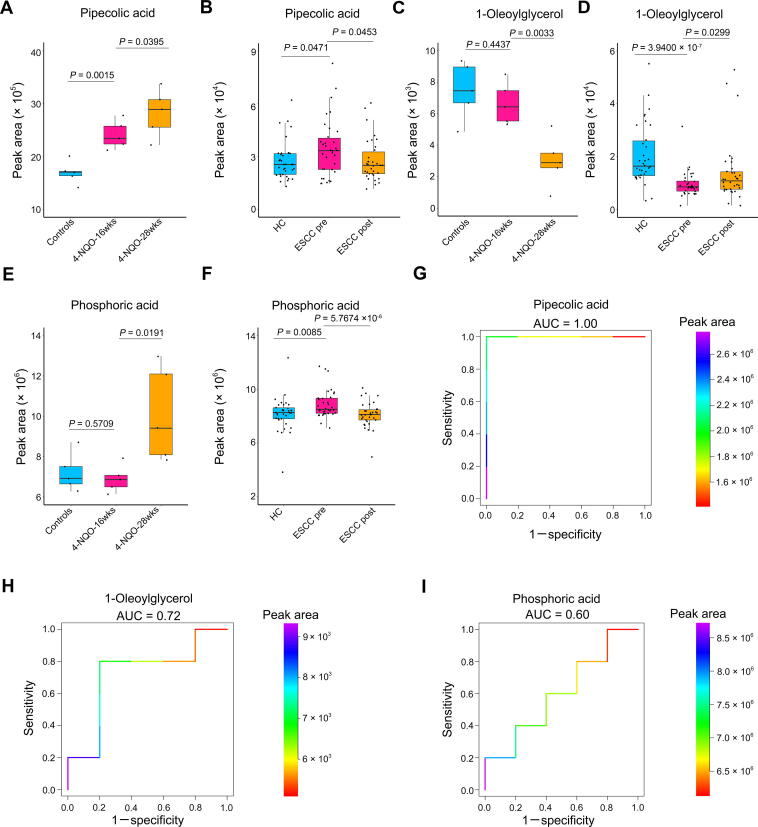
Subsequently, we wondered whether there were ESCC tumor-associated serum metabolites conserved between mice and humans with predictive potential for tumorigenesis. By comprehensively analyzing serum metabolomic data from ESCC mice and patients, we identified three metabolites conserved between mice and humans. First, the concentration of pipecolic acid was progressively increased in mouse sera from normal controls, dysplasia, and cancerization (Figure 6A). Consistently, the level of this serum metabolite was dramatically elevated in preoperative patients but down-regulated in postoperative cases (Figure 6B). Next, the abundance of 1-oleoylglycerol was progressively reduced in mouse sera from normal controls to cancerization (Figure 6C). In line with this finding, the level of this serum metabolite was overtly suppressed in preoperative patients but up-regulated in postoperative cases (Figure 6D). In addition, the abundance of phosphoric acid in mouse sera was not disturbed at the dysplasia stage as compared to the control, whereas it was remarkably elevated at the cancerization stage (Figure 6E). Correspondingly, the level of this metabolite was markedly increased in preoperative patients but declined in postoperative cases (Figure 6F).
Figure 6.
ESCC tumor-associated serum metabolites conserved between mice and humans with predictive potential for carcinogenesis
A. Abundance of serum pipecolic acid among control mice (n = 5), 4-NQO-induced mice at the dysplasia stage (4NQO-16wks) (n = 5), and 4-NQO-induced mice at the cancerization stage (4NQO-28wks) (n = 5). B. Abundance of serum pipecolic acid among HCs (n = 34), preoperative ESCC patients (n = 34), and postoperative ESCC patients (n = 34). C. Abundance of serum 1-oleoylglycerol among control mice (n = 5), 4-NQO-induced mice at the dysplasia stage (4-NQO-16wks) (n = 5) and 4-NQO-induced mice at the cancerization stage (4-NQO-28-wks) (n = 5). D. Abundance of serum 1-oleoylglycerol among HCs (n = 34), preoperative ESCC patients (n = 34), and postoperative ESCC patients (n = 34). E. Abundance of serum phosphoric acid among control mice (n = 5), 4-NQO-induced mice at the dysplasia stage (4-NQO-16-wks) (n = 5), and 4-NQO-induced mice at the cancerization stage (4-NQO-28wks) (n = 5). F. Abundance of serum phosphoric acid among HCs (n = 34), preoperative ESCC patients (n = 34) and postoperative ESCC patients (n = 34). ROC curves exhibiting the diagnostic capabilities of serum pipecolic acid (G), 1-oleoylglycerol (H), and phosphoric acid (I) to discriminate between healthy controls (n = 5) and precancerous mice (n = 5). Color key indicating metabolite abundance. AUC, area under the curve.
To ascertain whether the above serum metabolites possessed the predictive potential to accurately discriminate between healthy and precancerous cases, we performed receiver operating characteristic (ROC) curve analysis using control mice and 4-NQO-induced mice with epithelial dysplasia. The area under the curve values (AUC) were 1.00, 0.72, and 0.60 for pipecolic acid, 1-oleoylglycerol, and phosphoric acid, respectively, demonstrating that pipecolic acid possessed the excellent potential to be used as a biomarker for ESCC carcinogenesis (Figure 6G–I).
The protective effect of the predictive biomarker pipecolic acid on ESCC cells against oxidative stress
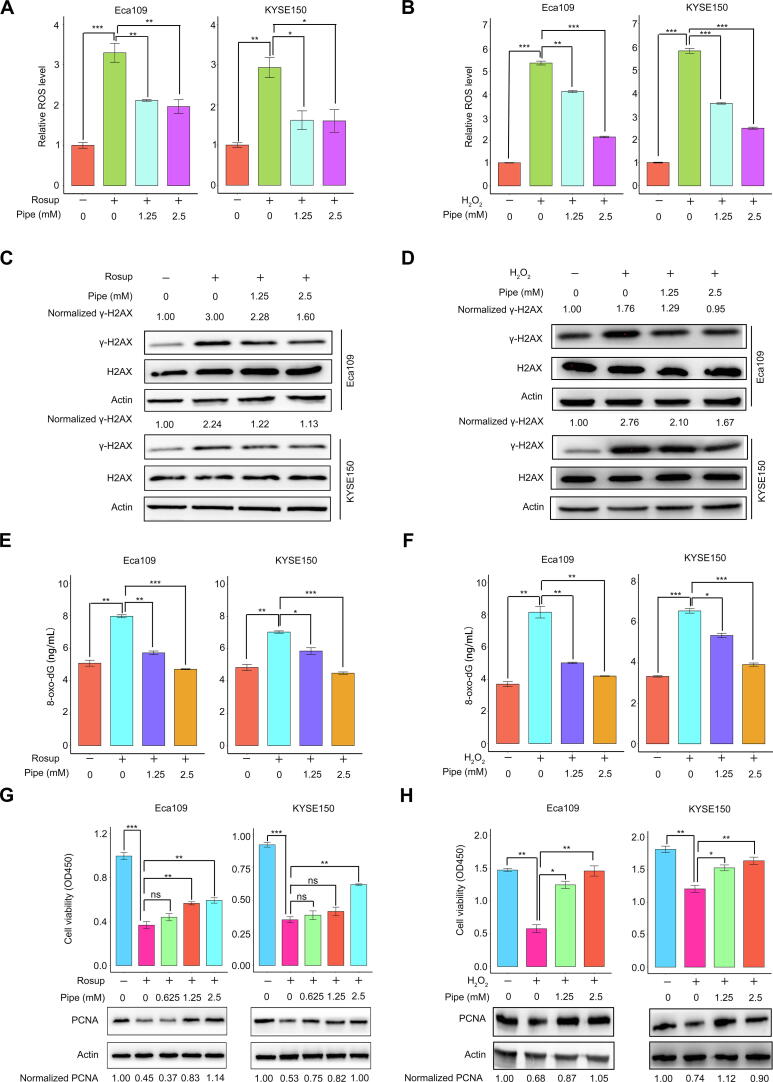
Finally, we explored whether the new biomarker identified in this study, pipecolic acid, possessed a tumor-promoting role. Elevated oxidative stress is extensively found in cancers, and the sustained increase in reactive oxygen species (ROS) promotes cell growth and facilitates carcinogenesis [25], [26]. However, overaccumulation of oxidative stress, once exceeding the toxic threshold, would cause damage to lipids, proteins, and DNA and induce cell death [26], [27]. Hence, cancer cells require efficient antioxidation systems to limit oxidative stress overaccumulation, thus, supporting cell survival and growth [26]. Pipecolic acid can enhance the antioxidant capability of mammalian cells [28]. Therefore, we assumed that increased levels of pipecolic acid in the sera of preoperative patients and 4-NQO-treated mice at the dysplasia and cancerization stages could enhance the ability of ESCC cells to oppose oxidative stress in the tumor microenvironment. Rosup is a compound mixture used for ROS generation [29]. Hence, Rosup was used to induce ROS in two human ESCC cell lines, Eca109 and KYSE150, in the current study. H2O2, one of the ROS, was used as a positive control.
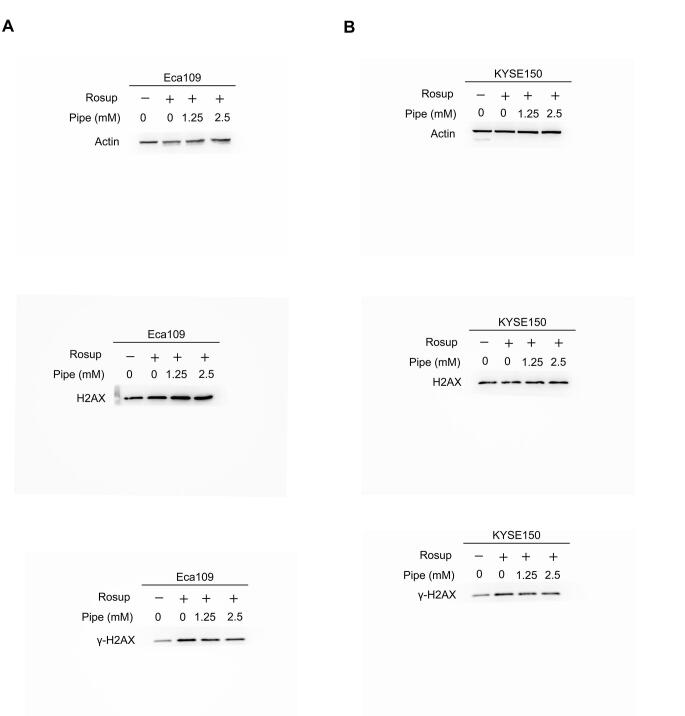
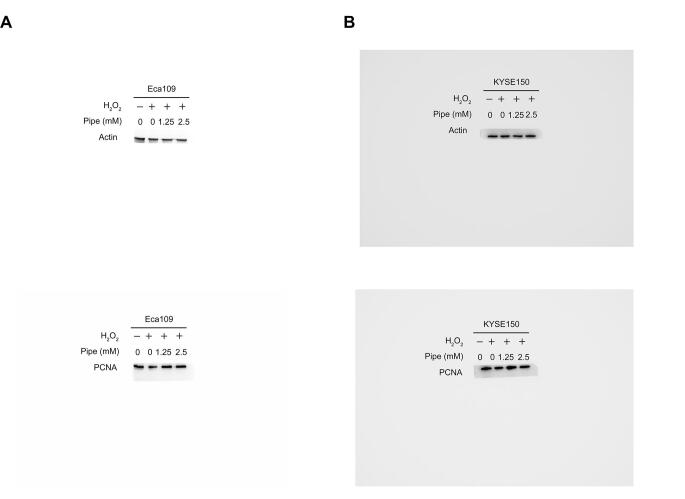
We observed that the addition of pipecolic acid dramatically reduced ROS production induced by Rosup and H2O2 in ESCC cells (Figure 7A and B). Moreover, the addition of pipecolic acid overtly reversed the DNA damage induced by Rosup and H2O2, as shown by the decreased expression of an established DNA damage marker phosphorylated H2AX (γ-H2AX) and by the down-regulation of the DNA oxidative damage marker 8-oxo-7,8-dihydro-20-deoxyguanosine (8-oxo-dG) (Figure 7C–F). Consequently, pipecolic acid administration restored ESCC cell growth impaired by Rosup and H2O2 treatment in a dose-dependent manner (Figure 7G and H). In conclusion, pipecolic acid is essential for ESCC cells to sustain redox homeostasis and to antagonize oxidative stress-induced DNA damage and cell proliferation arrest.
Figure 7.
The antioxidant activity of the predictive biomarker pipecolic acid
Impact of pipecolic acid on ROS production induced by Rosup (A) or H2O2 (B) in Eca109 and KYSE150 cells. The influence of pipecolic acid on the expression of the DNA damage marker γ-H2AX induced by Rosup (C) or H2O2 (D) in Eca109 and KYSE150 cells, respectively. The influence of pipecolic acid on the production of the DNA oxidative damage marker 8-oxo-dG induced by Rosup (E) or H2O2 (F) in Eca109 and KYSE150 cells, respectively. The impact of pipecolic acid on the proliferation and PCNA expression of Eca109 and KYSE150 cells treated with Rosup (G) or H2O2 (H). Error bars represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, no significance (two-tailed Student’s t-test). Pipe, pipecolic acid; ROS, reactive oxygen species; 8-oxo-dG, 8-oxo-7,8-dihydro-20-deoxyguanosine; PCNA, proliferating cell nuclear antigen.
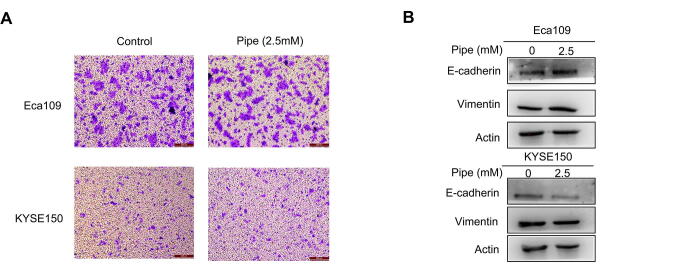
The circulating system is a crucial pathway for the distal metastasis of cancer cells [30]. Because pipecolic acid is a serum-containing metabolite in our study, we wondered whether it could affect the metastatic ability of ESCC cells. Treatment with pipecolic acid did not alter the migratory tendency or the expression of protein markers of epithelial-mesenchymal transition (an essential mechanism to drive cell migration) of ESCC cells (Figure S2), demonstrating that this metabolite did not perturb the metastatic ability of these malignant cells.
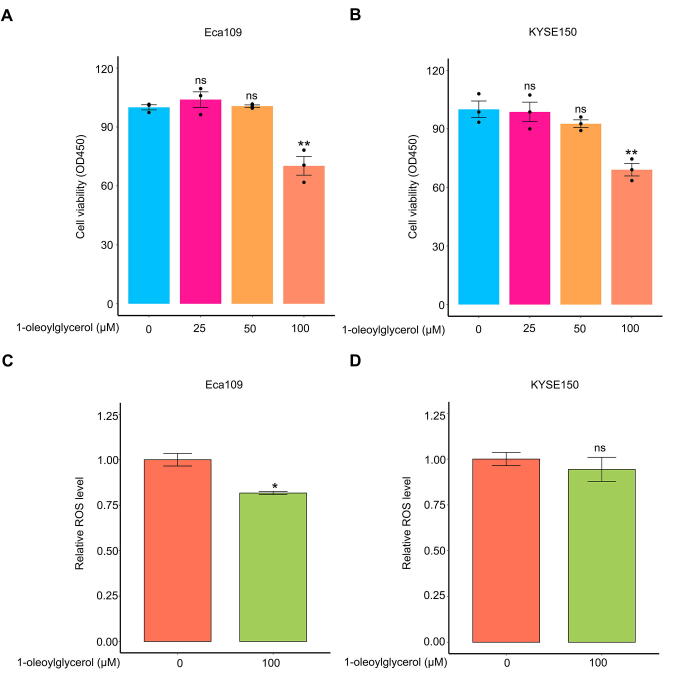
As another potential predictive marker 1-oleoylglycerol, shown in Figure 6, may have potential anticancer activity since this metabolite was negatively associated with the existence of ESCC tumors. Indeed, administration of 100 μM 1-oleoylglycerol significantly inhibited ESCC cell growth (Figure S3A and B). However, administration of 100 μM of this metabolite did not elevate intracellular ROS generation, indicating that it repressed ESCC cell growth, but not by increasing oxidative stress (Figure S3C and D).
Discussion
As stated in the introduction section, previous studies on ESCC serum metabolome are case-control studies [7], [18], [19], [20], [21], [22]. A recent serum metabolomic study used pre- and post-operative serum samples of ESCC patients, but the pre- and post-operative patients belonged to distinct cohorts [23]. Therefore, ESCC tumor-associated metabolic alterations in sera could not be exactly extracted from the published data. In the current study, we collected pre- and post-esophagectomy sera from the same ESCC patient cohort along with sera from healthy volunteers to facilitate the identification of ESCC tumor-associated metabolic changes. A total of 58 metabolites were found to be dramatically disturbed in the sera of preoperative ESCC patients compared to HCs. Importantly, our comprehensive analysis revealed that 12 of these serum metabolites were specifically linked to ESCC tumors, thus, providing new biomarkers with potential for monitoring therapeutic efficacy and disease relapse. Of note, these 12 metabolites were enriched in ubiquinone biosynthesis, phenylalanine, and tyrosine metabolism. A recent study found that ubiquinone is essential for the growth of colon cancer organoids with p53 deficiency [31]. In our study, the concentration of a biosynthetic precursor of ubiquinone biosynthesis, 4-hydroxybenzoic acid, was remarkably elevated in preoperative ESCC sera with a fold change of 4.30 (Figure 3A), indicating that this pathway was enhanced in patients with ESCC. Dietary phenylalanine and tyrosine are required for metastasis of melanoma, lung cancer, and hepatocarcinoma cells [32]. The substrate of phenylalanine and tyrosine metabolism, phenylalanine, was restrained in preoperative ESCC sera with a fold-change of −1.28 (Figure 3A), suggesting that this pathway was expedited in patients with ESCC. However, the roles of ubiquinone biosynthesis and phenylalanine and tyrosine metabolism in ESCC remain to be elucidated.
An intriguing finding of the current study is the metabolic perturbations in the sera of postoperative ESCC patients caused by the administration of abrosia and parenteral nutrition. The levels of a panel of carbohydrates were markedly up-regulated in postoperative sera, while there was only one known carbohydrate, glucose, in the parenteral nutrition solution. Of note, there were only two amino acids whose concentrations were increased in postoperative sera, while there were 20 amino acids in the parenteral nutrition solution (Table S2). Together, these results indicated that postoperative ESCC patients required substantial amounts of carbohydrates, and the components in parenteral nutrition solution were possibly used for conversion to these metabolites. Two essential amino acids, valine, and methionine, are crucial for pancreatic cancer growth and lung cancer cell stemness, respectively [33], [34]. These two amino acids were in parenteral nutrition solution, and their levels were remarkably elevated in postoperative ESCC sera. Thus, it would be reasonable to reduce the amount of these two amino acids in parenteral nutrition solutions to avoid activation of minimal residual disease. Moreover, it would be interesting to comprehensively ascertain the impact of metabolic perturbations induced by the administration of postoperative abrosia and parenteral nutrition on the therapeutic outcomes of patients with ESCC.
By enrolling a carcinogen-induced ESCC mouse model in the present study, we were able to delineate progressively modified serum metabolic signatures from the normal stage to the cancerization stage. Furthermore, ESCC tumor-associated serum metabolites conserved between mice and humans have been discovered, including pipecolic acid, 1-oleoylglycerol, and phosphoric acid. To the best of our knowledge, there are no predictive biomarkers for ESCC carcinogenesis. Our results demonstrated that serum pipecolic acid is a new biomarker with predictive potential for ESCC tumorigenesis, characterized by its high accuracy in discriminating between mice at the dysplasia stage and control mice. Pipecolic acid increases the antioxidant capability of mammalian cells [28]. In line with this, we found that this metabolite hindered oxidative stress-induced DNA damage and cell growth arrest, indicating that it was potentially involved in ESCC tumorigenesis. Another interesting metabolite is 1-oleoylglycerol, which is negatively linked to ESCC tumorigenesis. This metabolite impeded ESCC cell growth, but not by up-regulating intracellular ROS.
Admittedly, there are several limitations of the current GC-TOFMS-based study. First, the ESCC tumor-associated metabolic alterations in sera should be validated in independent patient cohorts from other cancer centers. Second, the predictive potential of serum pipecolic acid for ESCC tumorigenesis should be verified using samples from patients with squamous dysplasia of the esophageal epithelia. To better measure pipecolic acid in serum, the current protocol should be refined because of the low level of this metabolite in circulating blood, as previously reported [35]. The strategies include the use of a highly selective and sensitive triple quadruple instrument and pre-concentration of samples. Of note, when we apply serum pipecolic acid levels to predict ESCC tumorigenesis, several diseases such as Zellweger cerebro-hepato-renal syndrome and neonatal adrenoleukodystrophy, should be excluded because the level of this metabolite in the circulating blood of patients with these diseases is also increased [35]. Third, the separation and identification protocols for other differential metabolites with low abundance in serum, such as capric acid and 1-oleoylglycerol, should also be improved. These metabolites are more likely to interfere GC-TOFMS measurements, thus, resulting in low quality of mass spectra for these compounds. Finally, a metabolomic investigation of spent media of in vitro ESCC cell lines and ex vivo fresh ESCC tissues from patients should be performed to determine whether the abnormal serum metabolites discovered in this study are generated by ESCC cells.
Materials and methods
Enrollment of patients and healthy volunteers and collection of serum samples
A total of 34 patients with no prior treatment for their disease were enrolled in this study from the Affiliated Tumor Hospital of Nantong University (Nantong, China). All patients were diagnosed as ESCC by histopathologic examination and staged according to the American Joint Committee on Cancer 8th edition staging system [36]. From day 1 after esophagectomy, all cases were given abrosia and intravenous parenteral nutrition at a dosage of 3000 ml/day. One dosage of parenteral nutrition contained 1000 ml 5% glucose solution, 1000 ml physiological saline, 500 ml 10% fat emulsion, and 500 ml 10% amino acid solution (Table S2). In addition, age- and gender-matched healthy volunteers were enrolled from Longhua Hospital (Shanghai, China).
Overnight fasting peripheral blood samples were collected in the morning from healthy volunteers and ESCC patients before esophagectomy and after 3 days of surgery and then transferred into vacuum blood collection tubes without any anticoagulants. All blood samples were clotted at room temperature for < 2 h and centrifuged at 1500 g for 10 min. Serum samples were obtained and stored at −80 °C until analysis.
Carcinogen-induced ESCC mice and sample harvest
We purchased six-week-old C57BL/6 female mice and used a chemical carcinogen 4-NQO (Catalog No. N8141, Sigma-Aldrich, St. Louis, MO) to induce ESCC as previously reported [24]. Briefly, the 4-NQO stock solution was prepared in PG at a concentration of 6 mg/ml and then divided into aliquots for −20 °C storage. Two groups of mice were recruited. The experimental group was provided with the chemical carcinogen via drinking water containing 4-NQO with a final concentration of 100 μg/ml. While the control group was supplied with drinking water with no 4-NQO. Two groups received the same volume of PG in the drinking water, and drinking water was changed once a week. During the treatment, the mice were allowed to drink water whenever they wanted.
After a 16-week carcinogen or vehicle treatment, five control mice and five 4-NQO-induced mice were randomly selected and fasted overnight. The next morning, these mice were killed via cervical dislocation, and their serum specimens and esophageal tissues were collected. Serum samples were quickly moved into −80 °C refrigerator for storage, while esophageal tissues were either flash-frozen in liquid nitrogen or formalin-fixed paraffin-embedded and H&E stained.
Notably, the remaining five 4-NQO-induced mice were followed up for another 12 weeks. At the end of the experiment, these mice were fasted overnight and killed in the next morning. Serum specimens and esophageal tissues were harvested and treated using the same protocol for the previous batch of mice. Moreover, IHC staining assays were conducted for all paraffin-embedded tissues to measure the expression of ESCC biomarker K14 and cell proliferation marker Ki67.
Metabolomic profiling with GC-TOFMS
A total of 102 human sera together with 15 mouse sera were enrolled for metabolomic profiling using the GC-TOFMS platform. Metabolite extraction and measurement were executed using our previous protocol [14], [37], [38], [39]. Briefly, a 100 μl aliquot of each sample was used for metabolite extraction by adding 300 μl organic mixture (methanol:chloroform = 3:1, v/v). The mixture of organic solvent and serum was vortexed for 30 s, stored at −20 °C for 10 min, and centrifuged at 10,000 g for 10 min. Subsequently, a 300 μl supernatant was transferred to a gas chromatography (GC) vial, spiked with internal standards including 10 μl of heptadecanoic acid (1 mg/ml) and 4-chlorophenylalanine (0.3 mg/ml), and then dried at −20 °C under vacuum. Subsequently, 80 μl of methoxyamine (15 mg/ml in pyridine) was added to the residue to react at 30 °C for 90 min. Then the residue was derivatized by adding 80 μl N,O-bis-trimethylsilyl-trifluoroacetamide (with 1% trimethylchlorosilane) to react at 70 °C for 60 min.
Finally, derivatized samples were analyzed using the Pegasus HT system (Leco Corporation) coupled with an Agilent 7890B gas chromatography. For each sample, 1 μl volume was injected at a splitless mode with injector temperature at 270 °C. The flow rate for the carrier gas helium was set at 1.0 ml/min. The oven temperature program started at 70 °C for 2 min, and then increased to 180 °C with a rate of 10 °C/min, to 230 °C with a rate of 6 °C/min, finally to 295 °C with a rate of 40 °C/min. The temperature of 295 °C was held for 5 min. The temperatures of transferline interface and ion source were set at 270 °C and 220 °C, respectively. Of note, quality control samples were prepared with a mix of 17 standards and injected at a regular interval to monitor the data quality.
Data were obtained with an m/z range from 50 to 550 at an acquisition rate of 20 spectra per second. Raw data were undergone pre-treatment of baseline correction, de-noising, smoothing, alignment, time-window splitting, and multivariate curve resolution. Metabolites were identified by comparison with the internal library built with the standard reference chemicals. Particularly, a previously reported approach for mass spectral deconvolution [40] was used to determine the peak identities. To validate the identities of differential metabolites, raw chromatograms, and true mass spectrometry spectra of representative differential metabolites, as well as the mass spectrometry spectra in our reference library, were shown in File S1.
IHC staining and Western blot assays
IHC staining was conducted according to our previous description [39]. Tissue samples were stained with antibody against K14 (Catalog No. ab7800, Abcam, Cambridge, United Kingdom), Ki67 (Catalog No. ab245113, Abcam), or nonspecific IgG as a negative control.
Western blot was performed as previously described [38], [39], [41]. Cells cultured in vitro were digested with 0.25% trypsin and lysed using RIPA buffer (Catalog No. R0278, Sigma-Aldrich) containing 1% (v/v) protease inhibitor cocktail (Catalog No. P8340, Sigma-Aldrich) on ice. Supernatants of cell lysates containing total proteins were obtained by centrifugation at 12,000 g for 10 min, and their concentrations were determined by using a BCA assay kit (Catalog No. 23225, ThermoFisher Scientific, Waltham, MA). Protein extracts mixed with sample loading buffer (Catalog No. 1610747, Bio-rad, Hercules, CA) were boiled for 10 min, resolved by SDS-PAGE, and then transferred to PVDF membranes. After incubation with primary antibodies individually overnight at 4 °C, including proliferating cell nuclear antigen (PCNA) (Catalog No. 2586S, Cell Signaling Technology, Boston, MA), H2A histone family member X (H2AX)(Catalog No. 7631S, Cell Signaling Technology), phospho-histone H2AX (Catalog No. 9718S, Cell Signaling Technology), E-cadherin (Catalog No. 3195T, Cell Signaling Technology), N-cadherin (Catalog No. 13116T, Cell Signaling Technology), and Vimentin (Catalog No. 5741T, Cell Signaling Technology), the membranes were washed and then incubated with secondary antibody conjugated with IgG-HRP (Catalog No. 7074, Cell Signaling Technology). Images were captured by ChemiDoc Touch Imaging System (Bio-rad). The original gels for Western blot assays are provided in Figures S4–S8.
Biological function assays of pipecolic acid and 1-oleoylglycerol
Two human ESCC cell lines, KYSE150 (Stem Cell Bank, Chinese Academy of Sciences) and Eca109 (Stem Cell Bank, Chinese Academy of Sciences), were enrolled in this study. Firstly, the biological functions of pipecolic acid were investigated. Cells were seeded and grown to 80% confluence in 96-well plates for ROS measurement and cell viability analysis or in 6-well plates for analysis of H2AX and γ-H2AX and 8-oxo-dG production. Cells were then pretreated with or without pipecolic acid (Catalog No. HY-W012734, MedChemExpress, Princeton, NJ) in a complete medium for 12 h. Subsequently, Rosup (Catalog No. S0033S, Beyotime, Shanghai, China) was added into the medium at a concentration of 0.2 mg/ml for 6 h, while H2O2 (Catalog No. l1522097, Aladdin, Shanghai, China) was added into the medium at a concentration of 1 μM for 4 h. For analysis of ROS and cell viability, cells were treated in the plates using the corresponding kits. For analysis of H2AX and γ-H2AX and 8-oxo-dG production, cells were digested with trypsin and harvested for measurement.
In addition, for exploration of the impact of pipecolic acid on cell migration, a transwell assay was conducted, and pipecolic acid was added into the upper and lower chambers at a concentration of 2.5 mM. Furthermore, for investigation of the influence of pipecolic acid on epithelial-mesenchymal transition (EMT) protein marker expression, cells were seeded in a 6-well plate, treated with 2.5 mM pipecolic acid for 24 h, and harvested after trypsin digestion for Western blot assay.
Secondly, the biological functions of 1-oleoylglycerol (Catalog No. M7765, Sigma-Aldrich) were studied. Cells were seeded in 96-well plates. For cell viability assay, different concentrations of 1-oleoylglycerol, as indicated in the figure, were added into the wells. After 72 h, cell viability was measured. For the ROS generation assay, 100 μM 1-oleoylglycerol were added to the wells. After treatment for 4 h, intracellular ROS generation was assessed.
Measurement of ROS, 8-oxo-dG, cell viability, and cell migration
Production of ROS was measured by 2′,7′-dichlorofluorescein diacetate (DCFH-DA; Catalog No. S0033S-1, Beyotime). Briefly, ESCC cells (1 × 104/well) were incubated in a medium containing 10 μM DCFH-DA for 30 min at 37 °C, and then washed with serum-free medium three times. The fluorescence was analyzed using the fluorescence spectrophotometer (Excitation 488 nm, emission 525 nm).
Generation of 8-oxo-dG was assessed by using a modified ELISA kit (Catalog No. JL11850, Jianglaibio, Shanghai, China) according to the manufacturer’s instructions. Briefly, ESCC cells were crushed by ultrasound, centrifuged at 1200 g for 10 min, and the supernatant was collected for detection. For each sample, 50 μl supernatant was added to each well pre-coated with an 8-oxo-dG trapping antibody, and then 100 μl horseradish peroxidase-labeled antibody was added into each well. In the meantime, the calibration curve was executed by adding distinct concentrations of standard compound solution into wells (50 μl/well) with the same treatment protocol. Next, the 8-oxo-dG measurement plate was incubated at 37 °C for 1 h. After washing five times, 100 μl tetramethylbenzidine was added to each well, and the measurement plate was incubated at 37 °C for 15 min in the dark. Subsequently, 50 μl reaction termination solution was added to each well and mixed thoroughly. Finally, optical density values at 450 nm of wells of the measurement plate were recorded for quantifying 8-oxo-dG levels.
Cell viability was assessed by using Cell Counting Kit-8 (Catalog No. CK04, Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions.
Cell migration was analyzed in Corning Transwell Permeable Supports with polycarbonate membranes (24-well plate, 8 µm pore size) (Catalog No. 18320044, Corning, Corning, NY). ESCC cells were harvested after trypsin digestion, rinsed twice in phosphate buffered saline (PBS), resuspended in RPMI 1640 medium with 2.5 mM pipecolic acid or without pipecolic acid as indicated in the figure, and then seeded into the upper chambers of the transwell plates at a density of 1 × 105/well. The lower chamber of each well was filled with a complete medium with 20% fetal bovine serum containing 2.5 mM pipecolic acid or no pipecolic acid, as indicated in the figure. After incubation in the cell incubator for 24 h, the migrating ESCC cells were observed by crystal violet staining.
Statistical and bioinformatic analyses
To decipher the difference in serum metabolomic profiles between HCs and preoperative ESCC patients, or between preoperative and postoperative ESCC patients, a supervised multivariate regression model OPLS-DA was constructed with the software SIMCA-P+ (version 11.0, Umetric, Umea, Sweden). To identify the difference in global metabolism of sera among control mice, 4-NQO-induced mice for 16 weeks, and 4-NQO-induced mice for 28 weeks, supervised PLS regression was fitted using the software SIMCA-P+. The model parameters R2Y and Q2 were recorded. R2Y indicated the ‘goodness of fit’ in the data. Q2, calculated by a cross-validation procedure, indicated the predictability of the model.
Subsequently, univariate statistical analysis was performed to identify significantly altered metabolites. For the metabolomic data of human sera, a nonparametric Wilcoxon rank-sum test with Bonferroni adjustment was used to compare every two groups. For the metabolomic data of control mice, 4-NQO-induced mice for 16 weeks, and 4-NQO-induced mice for 28 weeks, one-way analysis of variance (ANOVA) was executed, and a post hoc test for multiple comparisons was implemented using least significant difference (LSD) method. For the data from cell experiments, Student’s t-test was used for statistical analysis. All univariate tests were two-tailed, and P values < 0.05 were considered statistically significant. Differential metabolites were displayed by heatmaps, which were generated by using an R package ComplexHeatmap [42].
MSEA for the differentially expressed metabolites was performed using a web-based tool MetaboAnalyst 4.0 [43], [44], and an algorithm of quantitative enrichment analysis was selected. Pathway-associated metabolite sets (SMPDB) were selected as the metabolite set library for MSEA analysis.
Ethical statement
All participants provided written informed consent in accordance with the regulation of the Institutional Review Board of the Affiliated Tumor Hospital of Nantong University (Approval No. 2019-022) in agreement with the Declaration of Helsinki. The mouse study was approved and conducted by Wuhan Servicebio Technology Co., Ltd (Approval No. 2018015).
Data availability
The mass spectrometry data of serum metabolome of healthy volunteers, ESCC patients, and mice have been deposited in The National Omics Data Encyclopedia (NODE) database at Bio-Med Big Data Center affiliated with Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences (Project ID: OEP001955), and are publicly accessible at https://www.biosino.org/node/project/detail/OEP001955.
CRediT author statement
Lei Liu: Resources, Investigation, Supervision. Jia Wu: Validation, Formal analysis, Data curation, Writing - original draft, Visualization. Minxin Shi: Resources. Fengying Wang: Resources. Haimin Lu: Resources. Jibing Liu: Resources. Weiqin Chen: Resources. Guanzhen Yu: Investigation, Formal analysis. Dan Liu: Investigation. Jing Yang: Investigation. Qin Luo: Investigation. Yan Ni: Investigation, Data curation. Xing Jin: Conceptualization, Supervision, Validation. Xiaoxia Jin: Conceptualization, Formal analysis. Wen-Lian Chen: Conceptualization, Supervision, Formal analysis, Data curation, Visualization, Writing - original draft, Writing - review & editing. All authors have read and approved the final manuscript.
Competing interests
The authors have stated that no competing interests exist.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31970708, 81770147, 81802891, and 82002953), the National Scientific and Technological Major Special Project of China (Grant No. 2019ZX09201004-002-013), the National Thirteenth Five-Year Science and Technology Major Special Project for New Drug Innovation and Development (Grant No. 2017ZX09304001), the Research fund of Shanghai Municipal Commission of Health (Grant No. 20174Y0090), the Shanghai Rising-Star Program (Grant No. 18QA1404100), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning, the Shanghai Youth Talent Program, the Shanghai Municipal Key Clinical Specialty (Grant No. shslczdzk03701), the Three-Year Plan of Shanghai Municipality for Further Accelerating The Development of Traditional Chinese Medicine [Grant No. ZY(2018-2020)-CCCX-1016], the Shanghai Chenguang Program (Grant No. 18CG47), the grant from Nantong Tumor Hospital (Grant No. BS201909), the Gaofeng Clinical Medicine Grant of Shanghai Municipal Education Commission, the Health Commission of Pudong New Area Health and Family Planning Scientific Research Project (Grant No. PW2019E-1), and the Xinling Scholar Program of Shanghai University of Traditional Chinese Medicine, China.
Handled by Zheng-Jiang Zhu
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2021.08.016.
Contributor Information
Xing Jin, Email: 16111230003@fudan.edu.cn.
Xiaoxia Jin, Email: 14111010068@fudan.edu.cn.
Wen-Lian Chen, Email: chenwl8412@shutcm.edu.cn.
Supplementary material
The following are the Supplementary data to this article:
Supplementary figure S1.
Heatmap displaying differential serum metabolites among control mice, carcinogen-induced mice at dysplasia stage (4-NQO-16wks), and carcinogen-induced mice at ESCC stage (4-NQO-28wks) with no progressive changes.
Supplementary figure S2.
The impact of pipecolic acid on ESCC cell migration and expression of EMT protein markers. A. Transwell assay revealing the influence of pipecolic acid on ESCC cell migration. B. Western blot showing the impact of pipecolic acid on the expression of EMT protein markers in ESCC cells. Pipe, pipecolic acid.
Supplementary figure S3.
The impact of 1-oleoylglycerol on ESCC cell growth and ROS generation. A. and B. The inhibitory effect of 1-oleoylglycerol on ESCC cell growth. C. and D. The influence of 1-oleoylglycerol on ROS production of ESCC cells. Error bars represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, by comparison with the group treated with 0 μM 1-oleoylglycerol, two-tailed Student’s t-test. Ns, no significance; ROS, reactive oxygen species.
Supplementary figure S4.
The original gels for western blot assays of Figure 7C.
Supplementary figure S5.
The original gels for western blot assays of Figure 7D.
Supplementary figure S6.
The original gels for western blot assays of Figure 7G.
Supplementary figure S7.
The original gels for western blot assays of Figure 7H.
Supplementary figure S8.
The original gels for western blot assays of Figure S2B.
Identity determination of differential metabolites.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Lagergren J., Smyth E., Cunningham D., Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 3.Arnold M., Soerjomataram I., Ferlay J., Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381–387. doi: 10.1136/gutjnl-2014-308124. [DOI] [PubMed] [Google Scholar]
- 4.Wang A.H., Liu Y., Wang B., He Y.X., Fang Y.X., Yan Y.P. Epidemiological studies of esophageal cancer in the era of genome-wide association studies. World J Gastrointest Pathophysiol. 2014;5:335–343. doi: 10.4291/wjgp.v5.i3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raghu Subramanian C., Triadafilopoulos G. Diagnosis and therapy of esophageal squamous cell dysplasia and early esophageal squamous cell cancer. Gastroenterol Rep. 2017;5:247–257. [Google Scholar]
- 6.Jin H., Qiao F., Chen L., Lu C., Xu L., Gao X. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J Proteome Res. 2014;13:4091–4103. doi: 10.1021/pr500483z. [DOI] [PubMed] [Google Scholar]
- 7.Ma H., Hasim A., Mamtimin B., Kong B., Zhang H.P., Sheyhidin I. Plasma free amino acid profiling of esophageal cancer using high-performance liquid chromatography spectroscopy. World J Gastroenterol. 2014;20:8653–8659. doi: 10.3748/wjg.v20.i26.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng H., Chen W., Zheng R., Zhang S., Ji J.S., Zou X., et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6:e555–e567. doi: 10.1016/S2214-109X(18)30127-X. [DOI] [PubMed] [Google Scholar]
- 9.Gavin A.T., Francisci S., Foschi R., Donnelly D.W., Lemmens V., Brenner H., et al. Oesophageal cancer survival in Europe: a EUROCARE-4 study. Cancer Epidemiol. 2012;36:505–512. doi: 10.1016/j.canep.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Njei B., McCarty T.R., Birk J.W. Trends in esophageal cancer survival in United States adults from 1973 to 2009: a SEER database analysis. J Gastroenterol Hepatol. 2016;31:1141–1146. doi: 10.1111/jgh.13289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Ward P.S., Thompson C.B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W.L., Wang J.H., Zhao A.H., Xu X., Wang Y.H., Chen T.L., et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood. 2014;124:1645–1654. doi: 10.1182/blood-2014-02-554204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics 2011;10:M110.004945. [DOI] [PMC free article] [PubMed]
- 16.Faubert B., Solmonson A., DeBerardinis R.J. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473. doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiNardo C.D., Propert K.J., Loren A.W., Paietta E., Sun Z., Levine R.L., et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood. 2013;121:4917–4924. doi: 10.1182/blood-2013-03-493197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mir S.A., Rajagopalan P., Jain A.P., Khan A.A., Datta K.K., Mohan S.V., et al. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J Proteomics. 2015;127:96–102. doi: 10.1016/j.jprot.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X., Wang K., Liu G., Wang Y., Xu J., Liu L., et al. Metabolic perturbation and potential markers in patients with esophageal cancer. Gastroenterol Res Pract. 2017;2017:5469597. doi: 10.1155/2017/5469597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J., Chen Y., Zhang R., Song Y., Cao J., Bi N., et al. Global and targeted metabolomics of esophageal squamous cell carcinoma discovers potential diagnostic and therapeutic biomarkers. Mol Cell Proteomics. 2013;12:1306–1318. doi: 10.1074/mcp.M112.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Z.J., Qi Z., Zhang J., Xue W.H., Li L.F., Shen Z.B., et al. Untargeted metabolomics analysis of esophageal squamous cell carcinoma discovers dysregulated metabolic pathways and potential diagnostic biomarkers. J Cancer. 2020;11:3944–3954. doi: 10.7150/jca.41733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Zhang T., Shen X., Liu J., Zhao D., Sun Y., et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics. 2016;12:116–125. [Google Scholar]
- 23.Yang Z., Liu Y., Ma L., Wen X., Ji H., Li K. Exploring potential biomarkers of early stage esophageal squamous cell carcinoma in pre- and post-operative serum metabolomic fingerprint spectrum using 1H-NMR method. Am J Transl Res. 2019;11:819–831. [PMC free article] [PubMed] [Google Scholar]
- 24.Tang X.H., Knudsen B., Bemis D., Tickoo S., Gudas L.J. Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice. Clin Cancer Res. 2004;10:301–313. doi: 10.1158/1078-0432.ccr-0999-3. [DOI] [PubMed] [Google Scholar]
- 25.Liou G.Y., Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge W., Zhao K., Wang X., Li H., Yu M., He M., et al. iASPP is an antioxidative factor and drives cancer growth and drug resistance by competing with Nrf2 for Keap1 binding. Cancer Cell. 2017;32 doi: 10.1016/j.ccell.2017.09.008. 561–73.e6. [DOI] [PubMed] [Google Scholar]
- 27.Schieber M., Chandel N.S. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–R462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natarajan S.K., Muthukrishnan E., Khalimonchuk O., Mott J.L., Becker D.F. Evidence for pipecolate oxidase in mediating protection against hydrogen peroxide stress. J Cell Biochem. 2017;118:1678–1688. doi: 10.1002/jcb.25825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang Y., Lai F., He G., Li Y., Yang L., Shen W., et al. Alleviation of rosup-induced oxidative stress in porcine granulosa cells by anthocyanins from red-fleshed apples. PLoS One. 2017;12:e0184033. doi: 10.1371/journal.pone.0184033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 31.Kaymak I., Maier C.R., Schmitz W., Campbell A.D., Dankworth B., Ade C.P., et al. Mevalonate pathway provides ubiquinone to maintain pyrimidine synthesis and survival in p53-deficient cancer cells exposed to metabolic stress. Cancer Res. 2020;80:189–203. doi: 10.1158/0008-5472.CAN-19-0650. [DOI] [PubMed] [Google Scholar]
- 32.Abdallah R.M., Starkey J.R., Meadows G.G. Dietary restriction of tyrosine and phenylalanine: inhibition of metastasis of three rodent tumors. J Natl Cancer Inst. 1987;78:759–769. [PubMed] [Google Scholar]
- 33.Li J.T., Yin M., Wang D., Wang J., Lei M.Z., Zhang Y., et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat Cell Biol. 2020;22:167–174. doi: 10.1038/s41556-019-0455-6. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z., Yip L.Y., Lee J.H.J., Wu Z., Chew H.Y., Chong P.K.W., et al. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med. 2019;25:825–837. doi: 10.1038/s41591-019-0423-5. [DOI] [PubMed] [Google Scholar]
- 35.Watkins P.A., Chen W.W., Harris C.J., Hoefler G., Hoefler S., Blake D.C., Jr, et al. Peroxisomal bifunctional enzyme deficiency. J Clin Invest. 1989;83:771–777. doi: 10.1172/JCI113956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin 2017;67:93−9. [DOI] [PubMed]
- 37.Wang J.H., Chen W.L., Li J.M., Wu S.F., Chen T.L., Zhu Y.M., et al. Prognostic significance of 2-hydroxyglutarate levels in acute myeloid leukemia in China. Proc Natl Acad Sci U S A. 2013;110:17017–17022. doi: 10.1073/pnas.1315558110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W.L., Wang Y.Y., Zhao A., Xia L., Xie G., Su M., et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30:779–791. doi: 10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen W.L., Jin X., Wang M., Liu D., Luo Q., Tian H., et al. GLUT5-mediated fructose utilization drives lung cancer growth by stimulating fatty acid synthesis and AMPK/mTORC1 signaling. JCI Insight. 2020;5 doi: 10.1172/jci.insight.131596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni Y., Su M., Qiu Y., Jia W., Du X. ADAP-GC 3.0: improved peak detection and deconvolution of co-eluting metabolites from GC/TOF-MS data for metabolomics studies. Anal Chem. 2016;88:8802–8811. doi: 10.1021/acs.analchem.6b02222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin X., Liang Y., Liu D., Luo Q., Cai L., Wu J., et al. An essential role for GLUT5-mediated fructose utilization in exacerbating the malignancy of clear cell renal cell carcinoma. Cell Biol Toxicol. 2019;35:471–483. doi: 10.1007/s10565-019-09478-4. [DOI] [PubMed] [Google Scholar]
- 42.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 43.Xia J., Wishart D.S. MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res. 2010;38:W71–W77. doi: 10.1093/nar/gkq329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486–W494. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identity determination of differential metabolites.
Data Availability Statement
The mass spectrometry data of serum metabolome of healthy volunteers, ESCC patients, and mice have been deposited in The National Omics Data Encyclopedia (NODE) database at Bio-Med Big Data Center affiliated with Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences (Project ID: OEP001955), and are publicly accessible at https://www.biosino.org/node/project/detail/OEP001955.