Abstract
Many virulence factors are required for Salmonella enterica serovar Typhimurium to replicate intracellularly and proliferate systemically within mice. In this work, we have carried out genetic analyses in vivo to determine the functional relationship between two major virulence factors necessary for systemic infection by S. enterica serovar Typhimurium: the Salmonella pathogenicity island 2 (SPI-2) type III secretion system (TTSS) and the PhoP-PhoQ two-component regulatory system. Although previous work suggested that PhoP-PhoQ regulates SPI-2 TTSS gene expression in vitro, in vivo competitive analysis of mutant strains indicates that these systems contribute independently to S. typhimurium virulence. Our results also suggest that mutation of phoP may compensate partially for defects in the SPI-2 TTSS by deregulating SPI-1 TTSS expression. These results provide an explanation for previous reports showing an apparent functional overlap between these two systems in vitro.
Salmonella enterica serovar Typhimurium is an intracellular pathogen that causes gastroenteritis in humans and a typhoid-like infection in mice, which is frequently used as a model for human typhoid fever (21). Intramacrophage survival and replication have been shown to correlate with S. enterica serovar Typhimurium colonization of the mouse spleen and liver (20). In recent years, many factors involved at various stages of the infectious process have been identified (26). However, little is known about their regulation in vivo or how they interact during the infection.
Salmonella pathogenicity islands 1 (SPI-1) and 2 (SPI-2) encode structurally similar type III secretion systems (TTSSs) (47, 48, 54). TTSSs consist of a secreton, which exports proteins across the bacterial cell membranes; a translocon, which translocates them into the host cell; and transcriptional regulators, chaperones, and effector proteins, which can have a variety of effects on the host cell. Unlike structural components, the effectors generally share little or no similarity and confer functional specificity upon each system (34). The SPI-1 TTSS (also known as Inv/Spa) (47) is involved in invasion of epithelial cells (23, 24), whereas the SPI-2 TTSS (48, 54) is necessary for Salmonella proliferation within macrophages (11, 31, 48) and bacterial growth during systemic infection (29, 54). Some SPI-1 and SPI-2 effectors are not encoded within the islands, but elsewhere on the chromosome (8, 33, 43, 44, 58, 59).
Transcription of SPI-1 TTSS genes is regulated in vitro by a variety of factors, such as osmolarity, growth phase, and pH (12, 23). Specific regulation of SPI-1 genes is mediated by HilA, a transcriptional regulator of the OmpR-ToxR family encoded within SPI-1 (3, 4). HilA-regulated genes include invF, which encodes a transcriptional regulator of the AraC family and is located within SPI-1 (37), and the components of the secretion machinery (3, 4). HilA also participates with InvF in regulating the expression of the effectors encoded within SPI-1 (13, 17), but only InvF appears to regulate the expression of those effectors encoded outside SPI-1 (13, 17).
HilA expression is itself regulated by the PhoP-PhoQ two-component system (4, 46), which regulates the expression of at least 40 virulence genes (45, 46). Some of these are involved in intramacrophage survival (20) and resistance to host antimicrobial peptides (19, 27). When PhoP is phosphorylated by PhoQ, it becomes active, functioning as a transcriptional regulator of PhoP-activated genes (pags) (25, 45), and PhoP-repressed genes (prgs), including the Inv/Spa secreton components (6, 46). The PhoP-PhoQ system is induced when Salmonella reaches the phagosome (1, 9). PhoP seems to repress the expression of SPI-1 TTSS genes inside the macrophage phagosome (1, 4, 6, 46). Mutations in other genes have also been shown to affect HilA expression in vitro, suggesting that additional factors modulate the expression of SPI-1 TTSS genes (2, 4, 18, 35, 41, 50, 51).
The expression of SPI-2 TTSS genes is induced inside host cells and is completely dependent on SsrA-SsrB, a two-component regulatory system encoded within SPI-2 (11, 14, 57). ssrA transcription is in turn regulated by the OmpR-EnvZ two-component system (40), which is responsible for both activation and repression of gene expression in response to changes in osmolarity and pH (32, 56). The OmpR-EnvZ system is important for Salmonella replication and survival within macrophages (8, 40) and is necessary for full virulence in mice (16).
Several studies have analyzed the role of the PhoP-PhoQ regulatory system on SPI-2 TTSS gene expression, but the results are contradictory. Two studies have reported that mutations in phoP decrease the expression of SPI-2 TTSS genes in vitro (14, 60). Lee and coworkers (40) have reported similar results, albeit only under certain growth conditions. On the other hand, expression of SPI-2 TTSS genes within infected cultured macrophages appears to be largely independent of PhoP-PhoQ (57).
To determine if the PhoP-PhoQ system has a relevant role on the expression of SPI-2 TTSS genes in vivo, we have applied a method recently developed by our group that allows genetic analysis of functional relationships between different virulence genes (8, 53). The method uses the S. enterica serovar Typhimurium mouse model of systemic infection, which supports the simultaneous growth of two or more different strains within the same animal (42). The method is a modification of the competitive tests carried out by Baümler and coworkers (5) to study pathways of intestinal invasion by S. enterica serovar Typhimurium. In our method, combinations of single and double mutants are used to inoculate mice. In the ensuing mixed infection, the virulence attenuation of a strain carrying mutations affecting independent virulence functions would correspond to the sum of the attenuation caused by each individual mutation (5, 8, 53). In the case of two genes that contribute equally to the same specific function (e.g., essential components of a macromolecular structure), the double mutant strain would be no more attenuated than strains carrying single mutations. Using this method, we have previously demonstrated that the proteins encoded by the SPI-2 and spv loci (28) contribute independently to S. enterica serovar Typhimurium virulence (53). We have also shown that there is a functional dependence between the SPI-2 TTSS and OmpR-EnvZ, supporting an in vivo role for OmpR regulation of SsrA, as well as between SPI-2 and SifA. Further research into the latter relationship demonstrated that SifA is an SPI-2 effector protein responsible for the integrity of the Salmonella-containing vacuole (8).
In this study, we demonstrate that the SPI-2 TTSS and the PhoP-PhoQ regulatory system contribute to systemic infection of mice by S. enterica serovar Typhimurium through independent mechanisms. Furthermore, we show that although mutations in SPI-1 TTSS genes alone have no measurable effect on systemic growth, triple mutant strains lacking a functional SPI-1 TTSS, PhoP-PhoQ, and SPI-2 TTSS are significantly more attenuated than an isogenic strain lacking PhoP-PhoQ and the SPI-2 TTSS. These results suggest that in the absence of a functional PhoP-PhoQ system, deregulation of expression of the SPI-1 secretion and translocation machinery inside the cell partially trans-complements a defect in SPI-2-mediated secretion and translocation in vivo.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. enterica serovar Typhimurium strains used in this study are listed in Table 1. Bacteria were grown at 37°C with aeration in Luria-Bertani (LB) medium supplemented with ampicillin (50 μg/ml), kanamycin (50 μg/ml), tetracycline (25 μg/ml), or chloramphenicol (35 or 10 μg/ml for strains carrying the ssaV::cat mutation) as appropriate. Double and triple mutant strains were constructed by phage transduction, carried out with P22 HT 105/1 int201 (52). HH114 and HH124 were constructed by phage transduction from CS105 (45) and EE656 (4), respectively. Phage-free isolates were obtained by streaking the transductants onto green plates (10). All strains are derivatives of wild-type S. enterica serovar Typhimurium strain 12023, except for TA2367 (phoP-24) and HH196 (phoP-24 ssaV::aphT), which are derived from wild-type strain SL1344. pho-24 is a point mutation lacking any means of selection and therefore cannot be transferred by transduction. EE656 was kindly provided by C. Lee. CS105 and TA2367 were kindly provided by S. Miller.
TABLE 1.
Strains used in this study
| Name | Description | Source or reference |
|---|---|---|
| 12023s | Wild type | NCTCa |
| P3F4 | ssrA::mTn5 (Kmr) | 29 |
| HH114 | phoP102::Tn10dCm (Cmr) | This study |
| HH102 | ΔsseB::aphT (Kmr) | 31 |
| HH109 | ssaV::aphT (Kmr) | 15 |
| HH110 | ssaV::cat (Cmr) | 53 |
| P11D10 | ssaJ::mTn5 (Kmr) | 54 |
| HH124 | prgH020::Tn5lacZY (Tetr) | This study |
| TA2367 | pho-24 (PhoPc) | 38 |
| HH121 | phoP102::Tn10dCm ssaV::aphT (Cmr Kmr) | This study |
| HH127 | ssrA::mTn5 ssaV::cat (Kmr Cmr) | This study |
| HH129 | hilA::Tn10 (Tetr) | 7 |
| HH131 | phoP102::Tn10dCm ssrA::mTn5 (Cmr Kmr) | This study |
| HH175 | phoP102::Tn10dCm ssaJ::mTn5 (Cmr Kmr) | This study |
| HH176 | phoP102::Tn10dCm ΔsseB::aphT (Cmr Kmr) | This study |
| HH180 | ssaV::aphT hilA::Tn10 (Kmr Tetr) | This study |
| HH191 | phoP102::Tn10dCm ssaV::aphT hilA::Tn10 (Cmr Kmr Tetr) | This study |
| HH192 | phoP102::Tn10dCm hilA::Tn10 (Cmr Tetr) | This study |
| HH193 | ssaV::cat prgH020::Tn5lacZY (Cmr Tetr) | This study |
| HH194 | phoP102::Tn10dCm prgH020::Tn5lacZY (Cmr Tetr) | This study |
| HH195 | phoP102::Tn10dCm ssaV::aphT prgH020::Tn5lacZY (Cmr Kmr Tetr) | This study |
| HH196 | pho-24 ssaV::aphT (Kmr) | This study |
NCTC, National Collection of Type Cultures.
Mouse mixed infections.
Female BALB/c mice (20 to 25 g) were inoculated intraperitoneally (i.p.) with a 0.2-ml volume of physiological saline containing 105 bacteria. Bacteria were grown overnight at 37°C in LB medium with aeration, diluted into fresh medium (1:100), and grown until an optical density at 550 nm (OD550) of 0.35 to 0.6 was reached. The mixed inoculum was prepared in physiological saline at a concentration of 2.5 × 105 bacteria/strain/ml (input). The CFU of each strain in the input were enumerated by plating a dilution series of the inoculum and using the appropriate antibiotic to distinguish between the strains.
Mice were sacrificed 48 h after inoculation by carbon dioxide inhalation. The spleens were removed, placed in sterile water, and homogenized by mechanical disruption. After homogenization, the samples were allowed to settle on ice for 5 min before transfer of the supernatants into a fresh tube. Bacteria were then pelleted by centrifugation at 15,000 × g and resuspended in sterile water. Bacterial CFU were enumerated by plating a dilution series onto LB agar and LB agar with the corresponding antibiotic.
Determination of CI and COIs.
The competitive index (CI) is defined as the ratio between the mutant and wild-type strains within the output (bacteria recovered from the host after infection) divided by their ratio within the input (initial inoculum) (22, 55). For clarity, we have renamed the CIs corresponding to mixed infections of double or triple mutants with corresponding single mutant strains (8, 53) as the “cancelled-out index” (COI). We define COI as the ratio between a double or triple mutant strain and the corresponding single mutant in the output divided by their ratio in the input.
Expression of antibiotic resistance genes could potentially be affected by passage through the animal. To confirm that the antibiotic resistance of the strains used was not altered, control experiments were carried out. Representative strains carrying antibiotic resistance genes expressed from their own promoters were selected. In each case, the strain was injected i.p. in a mixture with the wild-type strain, and the spleens from the infected animals were processed 48 h after inoculation as described. The bacteria were grown on LB plates first (to allow expression of antibiotic resistance) and then patched out onto LB plates with and without the antibiotic selection. For each antibiotic resistance gene, the ratio between the two strains in the output, calculated from the patched-out colonies, was not significantly different from the results calculated from direct plating onto LB plates with and without antibiotic selection. The cat gene inserted in ssaV (ssaV::cat) is expressed from a SPI-2 promoter. Therefore, its expression could be altered by mutation of other genes affecting expression from this promoter. When strains carrying this marker were used, the controls described above were included in every experiment.
Statistical analysis.
Each CI or COI value is the mean of at least three independent infections ± the standard error. Student's t test was used to analyze every COI (e.g., COI corresponding to the mixed infection of strain a versus double mutant strain a b) with two null hypotheses: (i) mean COI is not significantly different from 1.0, and (ii) mean COI is not significantly different from the CI of the single mutant strain relevant in each case (i.e., CI of strain b for the case presented above). P values of 0.05 or less are considered significant.
RESULTS AND DISCUSSION
In vivo genetic analysis of the functional relationship between the SPI-2 TTSS and phoP.
Various studies of the regulation of SPI-2 TTSS gene expression by PhoP have produced somewhat confusing results (11, 14, 57, 60). Mutations in phoP or ompR can affect SseB expression depending on the formulation of the growth media (C. R. Beuzón and D. W. Holden, unpublished results). Consistent with these results, expression of ssaH, which encodes a putative component of the secreton (11), is affected by a phoP mutation only under certain growth conditions (40). On the other hand, in infected RAW macrophages, expression of SseB is largely independent of PhoP and only partially dependent on OmpR (Beuzón and Holden, unpublished), consistent with results obtained for other SPI-2 genes (11, 40). These conflicting results prompted us to study the role of the PhoP-PhoQ system in the regulation of SPI-2 gene expression in vivo.
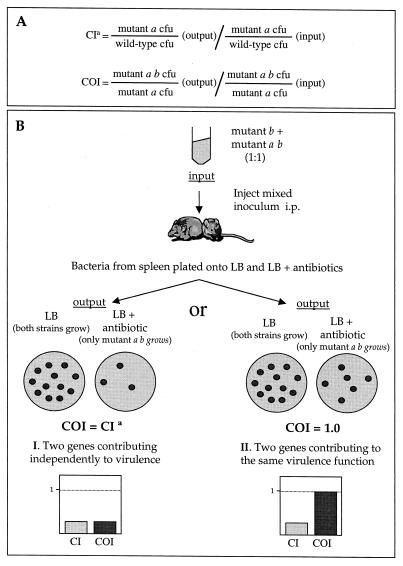
An outline of the method used and a simplified analysis of the possible outcomes is represented in Fig. 1. Briefly, mice are injected i.p. with a mixed inoculum containing equal quantities of single (mutation in gene a) and double (mutations in genes a and b) mutant strains. Bacteria are recovered from the spleen 48 h after inoculation and differentiated on the basis of antibiotic resistance. For clarity, we have termed the index obtained with the CFU from a mixed infection of single and double mutant strains, the “cancelled-out index” (COI), to differentiate it from the competitive index (CI), calculated from a mixed infection involving wild-type and single mutant strains. In a COI test, the attenuation caused by the mutation in gene a would affect both strains equally and would be “cancelled out.” In the case of two genes responsible for independent virulence functions, the COI would be similar to the CI of the strain carrying a mutation in gene b (Fig. 1B, I), whereas in the case of two genes involved in the same function (e.g., structural components of a secretion system), the double mutant would be no more attenuated than either of the single mutants and the resulting COI would be equal to 1.0 (Fig. 1B, II). There are clearly other scenarios, such as partial redundancy of function and a different degree of contribution of two proteins to the same function. In these cases, the COIs would be different from those shown in Fig. 1.
FIG. 1.
Theoretical representation of COI analysis of the interaction between two hypothetical genes, a and b. (A) CI is defined as the CFU ratio of mutant and wild-type strains recovered from the infected animal divided by their CFU ratio in the input. COI is defined as the CFU ratio of double and single mutant strains recovered from the infected animal divided by their CFU ratio in the input. (B) Determination and analysis of COI. A mixed inoculum containing equal CFU of single and double mutant strains is inoculated into mice. Bacteria are recovered from spleens 48 h after infection. Double mutant CFU are determined from colony counts of serial dilutions grown on selective media. Total CFU are determined from colony counts of serial dilutions grown on LB medium. Single mutant CFU are calculated by subtracting double mutant CFU from total CFU. I and II represent two possible outcomes of the analysis. aCI of a strain carrying a mutation in gene a. The COI corresponding to a mixed infection of the a single mutant and a b double mutant strains (not represented in the figure) would be equal to the CI of a strain carrying the mutation in gene b.
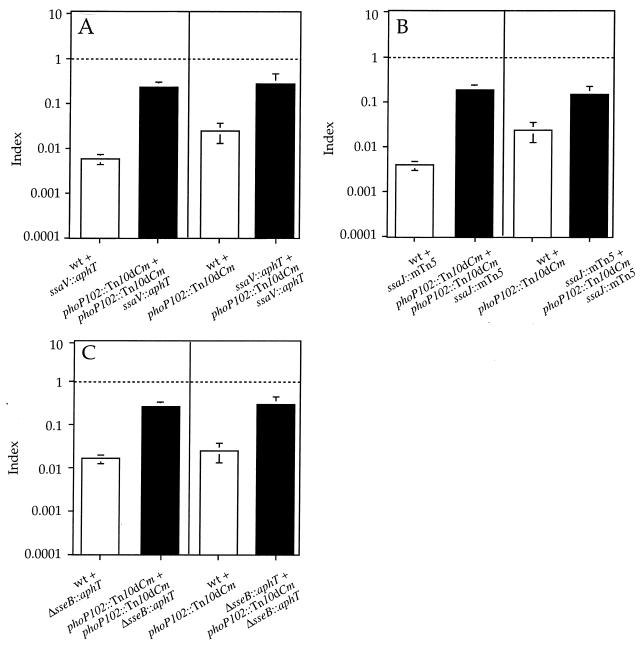
In order to analyze the relationship between the SPI-2 TTSS and the PhoP-PhoQ system in vivo, we constructed strains carrying both a null mutation in phoP (phoP102::Tn10dCm) (45) and one of several mutations in genes encoding the TTSS secretion and translocation machinery that prevent SPI-2 function. The first SPI-2-null mutant strain analyzed carries a nonpolar disruption of ssaV (ssaV::aphT) (15). SsaV is predicted to be a component of the secretion machinery (54), and its mutation has previously been shown to prevent secretion of SseB (7). A strain carrying mutations in phoP and ssaV was analyzed in mixed infections with either phoP or ssaV single mutant strains. The COIs obtained were significantly different from 1.0, indicating that the proteins encoded by these two genes have different functions. However, the COIs were also different from the CIs of the corresponding single mutant strains, suggesting that there may be some overlap in the in vivo functions of the proteins encoded by phoP and ssaV (Fig. 2A).
FIG. 2.
Graphical representation of COI analysis of strains carrying mutations in both phoP and genes encoding structural components of the SPI-2 TTSS. (A) Analysis of phoP ssaV double mutant strain (HH121) in mixed infections with either phoP or ssaV single mutant strains (HH114 and HH109, respectively). (B) Analysis of phoP ssaJ double mutant strain (HH175) in mixed infections with either phoP or ssaJ single mutant strains (HH114 and P11D10, respectively). (C) Analysis of phoP sseB double mutant strain (HH176) in mixed infections with either phoP or sseB single mutant strains (HH114 and HH102, respectively). The indices represented are CI (white bars) and COI (black bars). The strains used in each mixed infection are represented by the relevant mutations for simplicity and are indicated under the corresponding bar. COIs are compared to 1.0 (indicated by a horizontal dotted line) and to the value of the CI relevant in each case. COIs are significantly different from 1.0 and from the corresponding CIs (P ≤ 0.05). wt, wild type.
Mutation of ssaJ (ssaJ::mTn5), encoding a predicted component of the SPI-2 secreton (30), also prevents secretion of SseB (7). A strain carrying mutations in both phoP and ssaJ was analyzed in mixed infections with either single mutant strain, resulting in COIs that were intermediate between 1.0 and the corresponding CIs of the phoP and ssaJ mutant strains (Fig. 2B) and similar to the COIs of the phoP ssaV double mutant (Fig. 2A). Furthermore, the COIs of a strain carrying mutations in phoP and sseB (sseB::aphT) were also similar to the COIs corresponding to the phoP ssaV or phoP ssaJ mutant strains (Fig. 2C). A possible explanation of these results is that PhoP could modulate the expression of SPI-2 TTSS genes to some extent and regulate the expression of other functionally independent virulence factors.
PhoP and SsrA act independently.
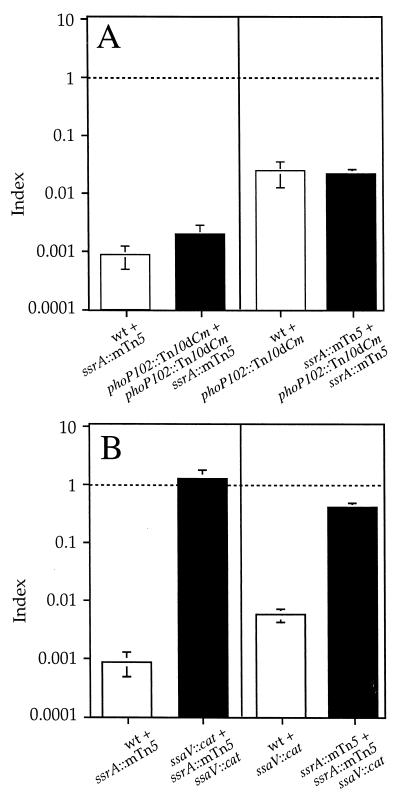
To further investigate the interaction between the two systems, we analyzed a strain carrying mutations in phoP and ssrA, which encodes the sensor component of the SPI-2 two-component regulatory system (ssrA::mTn5), in mixed infections with either phoP or ssrA single mutants. The mutation in ssrA prevents the expression of SPI-2 TTSS genes (7, 8, 11, 43) and causes strong defects in intracellular replication (11, 31) and virulence in mice (54). Surprisingly, in this case, the COIs from either mixed infection were not statistically different from the CI of the corresponding single mutant (Fig. 3A). These results can only be explained if the products of ssrA and phoP contribute independently to S. enterica serovar Typhimurium virulence. Therefore, it is unlikely that they coregulate the same virulence genes. This seems paradoxical in view of the finding that a strain carrying mutations in both ssrA and ssaV is not more attenuated than ssrA or ssaV single mutants (Fig. 3B) (indicating that SsrA does not appear to regulate virulence determinants that act independently of the SPI-2 TTSS), as well as the results of the COI analyses involving ssaV, ssaJ, and sseB with phoP. This apparent contradiction can be explained if the phoP mutation can partially rescue the virulence attenuation caused by the ssaV, ssaJ, and sseB mutations.
FIG. 3.
Graphical representation of COI analysis of strains carrying a mutation in ssrA. (A) Analysis of phoP ssrA double mutant strain (HH131) in mixed infections with either phoP or ssrA single mutant strains (HH114 and P3F4, respectively). (B) Analysis of ssrA ssaV double mutant strain (HH127) in mixed infections with either the ssrA or ssaV single mutant strain (P3F4 and HH110, respectively). The indices represented are CI (white bars) and COI (black bars). The strains used in each mixed infection are represented by the relevant mutation for simplicity and are indicated under the corresponding bar. COIs were compared to 1.0 (indicated by a horizontal dotted line) and to the value of the CI relevant in each case. (A) COIs are not significantly different from the corresponding CIs (P = 0.05). (B) COIs are not significantly different from 1.0 (P ≥ 0.05). wt, wild type.
SPI-2 defects can be partially compensated for by derepression of SPI-1 gene expression in the absence of PhoP.
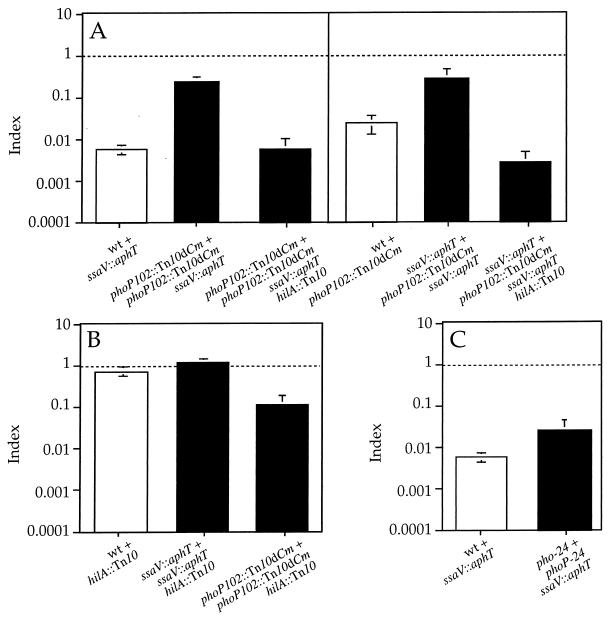
In the absence of a functional PhoP-PhoQ system repressing their expression, SPI-1 TTSS genes are likely to be expressed inside macrophages (4, 46). Since some SPI-1 and SPI-2 structural components are very similar (34), defects in one apparatus could potentially be trans-complemented by the other. If that was the case, it could explain the results obtained with strains carrying mutations in both phoP and genes encoding structural components of the SPI-2 TTSS. On the other hand, a phoP ssrA double mutant strain would not only lack SPI-2 secretion and translocation machinery, but also SPI-2 effectors, and could not be trans-complemented by the SPI-1 TTSS. If derepression of SPI-1 TTSS gene expression in a null phoP mutant strain is the basis for a partial rescue of the defects of the ssaV, ssaJ, or sseB mutant strains, introduction of a mutation preventing SPI-1 gene expression in any of the double mutant strains should prevent this rescue. In this case, the COIs of the triple mutant strain would be lower than that of the double mutant and similar to the corresponding CIs. Indeed, the COIs obtained with a strain carrying mutations in hilA (hilA::Tn10), ssaV, and phoP in mixed infections with either phoP or ssaV single mutants were lower than the COIs of the phoP ssaV double mutant and close to the CIs of ssaV or phoP single mutants (Fig. 4A). The introduction of the hilA mutation in strains carrying a single mutation in either ssaV or phoP did not cause a increase in attenuation that could account for the strong attenuation of the triple mutant (Fig. 4B). As expected, the hilA single mutant strain was as virulent as the wild-type strain when administered by the i.p. route, since SPI-1 TTSS function seems to be important for, but restricted to, bacterial translocation across the gut epithelium (24) (Fig. 4B). These results support previous work suggesting that SPI-1 TTSS gene expression is repressed by the PhoP-PhoQ system inside macrophages (1, 4, 6, 46).
FIG. 4.
Graphical representation of COI analysis of strains carrying a mutation in hilA. (A) Analysis of phoP ssaV hilA triple mutant strain (HH191) in mixed infections with either phoP or ssaV single mutant strains (HH114 and HH109, respectively). (B) CI corresponding to hilA single mutant strain (HH129) and COIs corresponding to ssaV hilA (HH180) and phoP hilA (HH192) double mutant strains, in mixed infections with either the ssaV or phoP single mutant strain (HH110 and HH114, respectively). (C) Analysis of the phoPc ssaV double mutant (HH196) in mixed infection with the phoPc single mutant strain (TA2367). The indices represented are CI (white bars) and COI (black bars). The strains used in each mixed infection are represented by the relevant mutation for simplicity and are indicated under the corresponding bar. COIs were compared to 1.0 (indicated by a horizontal dotted line) and to the value of the CI relevant in each case. (A and C) COIs are not significantly different from the corresponding CIs (P ≥ 0.05). (B) COIs are not significantly different from 1.0 (P ≥ 0.05). wt, wild type.
Analysis of a strain carrying both an ssaV mutation and a pho-24 mutation (phoPc) (which causes constitutive repression of SPI-1 gene expression) (4, 46, 49) supported the results obtained with the phoP ssaV hilA triple mutant. The COI of the phoPc ssaV double mutant, in a mixed infection with the phoPc single mutant, was similar to the CI of the ssaV mutant strain (Fig. 4C).
A functional SPI-1 TTSS can partially trans-complement SPI-2 secretion and translocation defects.
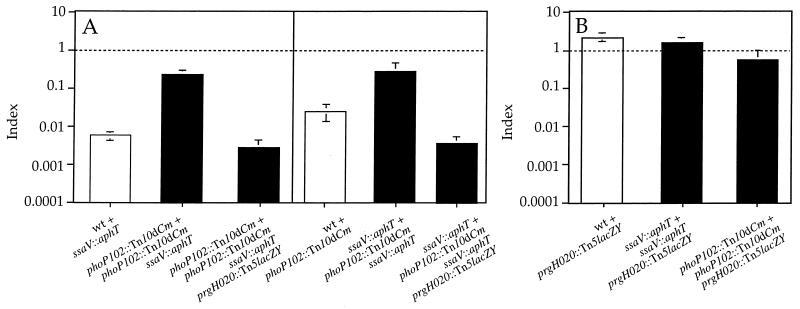
Derepression of SPI-1 TTSS could potentially rescue the SPI-2 defect by two mechanisms: direct secretion and translocation of SPI-2 effectors through the SPI-1 apparatus or substitution of the missing component of the SPI-2 TTSS with its homologue, forming a hybrid secretion system that could secrete and translocate SPI-2 effectors. Since a mutation in sseB, which has no homolog in SPI-1 (31), can be partially trans-complemented by a null phoP mutation, secretion and translocation of SPI-2 effectors through the SPI-1 secretion apparatus seem the more likely explanation. To test this hypothesis, we transduced a mutation in prgH (prgH020::Tn5lacZY) into the phoP ssaV double mutant strain. PrgH is a structural component of the SPI-1 TTSS (39) necessary for secretion (49), and it shares no homology with SsaV, SsaJ, or SseB. The COIs obtained with the phoP ssaV prgH triple mutant in mixed infections with either phoP or ssaV single mutants were significantly lower than the COIs of the phoP ssaV double mutant and were similar to the CIs of ssaV or phoP single mutants (Fig. 5A). The introduction of the prgH mutation into strains carrying a single mutation in either ssaV or phoP did not cause a decrease in attenuation that could account for the strong attenuation of the triple mutant (Fig. 5B). These results indicate that the absence of a structural component of the SPI-1 TTSS is sufficient to prevent the ssaV mutant from being trans-complemented by the SPI-1 TTSS. Therefore, the most likely explanation for the intermediate attenuation of virulence obtained with strains carrying mutations in both phoP and genes encoding structural components of the SPI-2 TTSS is that SPI-2 effectors can be translocated through a derepressed SPI-1 TTSS.
FIG. 5.
Graphical representation of COI analysis of strains carrying a mutation in prgH. (A) Analysis of phoP ssaV prgH triple mutant strain (HH195) in mixed infection with either phoP or ssaV single mutant strains (HH114 and HH109, respectively). (B) CI corresponding to prgH single mutant strain (HH124) and COIs corresponding to ssaV prgH (HH193) and phoP prgH (HH194) double mutant strains, in mixed infections with either the ssaV or phoP single mutant strain, respectively (HH110 and HH114). The indices represented are CI (white bars) and COI (black bars). The strains used in each mixed infection are represented by the relevant mutation for simplicity and are indicated under the corresponding bar. COIs were compared to 1.0 (indicated by a horizontal dotted line) and to the value of the CI relevant in each case. (A) COIs are not significantly different from the corresponding CIs (P ≤ 0.05). (B) COIs are not significantly different from 1.0 (P ≥ 0.05). wt, wild type.
These data provide an explanation for the contradictory results obtained in some analyses of SPI-2 regulation of expression and secretion in vitro, as discussed above. Certain combinations of growth conditions that are not encountered in vivo might induce simultaneous expression of both TTSSs and could generate artifactual results.
Our results also suggest that the SPI-1 secretion and translocation machinery is capable, at least to some extent, of secreting SPI-2 effectors across the vacuolar membrane. It is interesting that recent reports have shown that two proteins, SspH1 and SlrP, are likely to be translocated by both TTSSs (43, 44). These proteins contain a conserved N-terminal sequence that has been recently identified in several Salmonella effectors as necessary for translocation through the SPI-2 TTSS (43). In addition, some proteins secreted by the SPI-1 TTSS, such as SopD (36), have been found to contain this signal (8). It is possible that both systems can recognize similar secretion and translocation signals, and the specificity of translocation for each effector protein is determined by the profile of expression of each effector in relation to the expression of the SPI-1 and SPI-2 TTSSs. Thus, an effector under SsrAB regulation would only be translocated in vivo by the SPI-2 TTSS. Supporting this hypothesis, SspH1 and SlrP seem to be expressed constitutively (43) and could therefore be translocated by both TTSSs, unlike other effectors, the expression of which is coordinated with that of the TTSS that secretes them (8, 17, 43).
Finally, the results presented here emphasize the value of classical genetic analysis to advance the understanding of the regulation of virulence factors and their functional relationships in S. enterica serovar Typhimurium infection of mice.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council (United Kingdom) to David W. Holden.
We thank Herb Arst for valuable discussion. We also thank Javier Ruiz-Albert and Steve Garvis for critical review.
REFERENCES
- 1.Alpuche-Aranda C M, Swanson J A, Loomis W P, Miller S I. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altier C, Suyemoto M, Lawhon S D. Regulation of Salmonella enterica serovar Typhimurium invasion genes by csrA. Infect Immun. 2000;68:6790–6797. doi: 10.1128/iai.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 4.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 5.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beuzón C R, Banks G, Deiwick J, Hensel M, Holden D W. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 8.Beuzón C R, Meresse S, Unsworth K E, Ruiz-Albert J, Garvis S, Waterman S R, Ryder T A, Boucrot E, Holden D W. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. . (Erratum, 19:4191.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchmeier N A, Heffron F. Induction of Salmonella stress proteins upon infection of macrophages. Science. 1990;248:730–732. doi: 10.1126/science.1970672. [DOI] [PubMed] [Google Scholar]
- 10.Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972;50:883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- 11.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 12.Daefler S. Type III secretion by Salmonella typhimurium does not require contact with a eukaryotic host. Mol Microbiol. 1999;31:45–51. doi: 10.1046/j.1365-2958.1999.01141.x. [DOI] [PubMed] [Google Scholar]
- 13.Darwin K H, Miller V L. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol. 1999;181:4949–4954. doi: 10.1128/jb.181.16.4949-4954.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- 15.Deiwick J, Nikolaus T, Shea J E, Gleeson C, Holden D W, Hensel M. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol. 1998;180:4775–4780. doi: 10.1128/jb.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorman C J, Chatfield S, Higgins C F, Hayward C, Dougan G. Characterization of porin and ompR mutants of a virulent strain of Salmonella typhimurium: ompR mutants are attenuated in vivo. Infect Immun. 1989;57:2136–2140. doi: 10.1128/iai.57.7.2136-2140.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichelberg K, Galán J E. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun. 1999;67:4099–4105. doi: 10.1128/iai.67.8.4099-4105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichelberg K, Hardt W D, Galan J E. Characterization of SprA, an AraC-like transcriptional regulator encoded within the Salmonella typhimurium pathogenicity island 1. Mol Microbiol. 1999;33:139–152. doi: 10.1046/j.1365-2958.1999.01458.x. [DOI] [PubMed] [Google Scholar]
- 19.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 20.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay B B. Molecular and cellular mechanisms of Salmonella pathogenesis. Curr Topics Microbiol Immunol. 1994;192:163–185. doi: 10.1007/978-3-642-78624-2_8. [DOI] [PubMed] [Google Scholar]
- 22.Freter R, O'Brien P C M, Macsai M S. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect Immun. 1981;34:234–240. doi: 10.1128/iai.34.1.234-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galán J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 24.Galán J E, Curtiss R. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groisman E A, Chiao E, Lipps C J, Heffron F. Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc Natl Acad Sci USA. 1989;86:7077–7081. doi: 10.1073/pnas.86.18.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Groisman E A, Ochman H. How Salmonella became a pathogen. Trends Microbiol. 1997;5:343–349. doi: 10.1016/S0966-842X(97)01099-8. [DOI] [PubMed] [Google Scholar]
- 27.Groisman E A, Parra C A, Salcedo M, Lipps C J, Heffron F. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11939–11943. doi: 10.1073/pnas.89.24.11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gulig P A, Doyle T J. The Salmonella typhimurium virulence plasmid increases the growth rate of salmonellae in mice. Infect Immun. 1993;61:504–511. doi: 10.1128/iai.61.2.504-511.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 30.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaJ and the ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 31.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F C, Holden D W. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 32.Heyde M, Portalier R. Regulation of major outer membrane porin proteins of Escherichia coli K 12 by pH. Mol Gen Genet. 1987;208:511–517. doi: 10.1007/BF00328148. [DOI] [PubMed] [Google Scholar]
- 33.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 36.Jones M A, Wood M W, Mullan P B, Watson P R, Wallis T S, Galyov E E. Secreted effector proteins of Salmonella dublin act in concert to induce enteritis. Infect Immun. 1998;66:5799–5804. doi: 10.1128/iai.66.12.5799-5804.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaniga K, Bossio J C, Galan J E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 38.Kier L D, Weppelman R M, Ames B N. Regulation of nonspecific acid phosphatase in Salmonella: phoN and phoP genes. J Bacteriol. 1979;138:155–161. doi: 10.1128/jb.138.1.155-161.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kubori T, Sukhan A, Aizawa S I, Galan J E. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci USA. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee A K, Detweiler C S, Falkow S. OmpR regulates the two-component system SsrA-SsrB in Salmonella pathogenicity island 2. J Bacteriol. 2000;182:771–781. doi: 10.1128/jb.182.3.771-781.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas R L, Lostroh C P, DiRusso C C, Spector M P, Wanner B L, Lee C A. Multiple factors independently regulate hilA and invasion gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2000;182:1872–1882. doi: 10.1128/jb.182.7.1872-1882.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meynell G G, Stocker B A D. Some hypotheses on the aetiology of fatal infections in partially resistant hosts and their application to mice challenged with Salmonella paratyphi-B or Salmonella typhimurium by intraperitoneal injection. J Gen Microbiol. 1957;16:38–58. doi: 10.1099/00221287-16-1-38. [DOI] [PubMed] [Google Scholar]
- 43.Miao E A, Miller S I. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao E A, Scherer C A, Tsolis R M, Kingsley R A, Adams L G, Bäumler A J, Miller S I. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol. 1999;34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 45.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller S I, Mekalanos J J. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mills D M, Bajaj V, Lee C A. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 48.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 50.Rakeman J L, Bonifield H R, Miller S I. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol. 1999;181:3096–3104. doi: 10.1128/jb.181.10.3096-3104.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schechter L M, Damrauer S M, Lee C A. Two AraC/XylS family members can independently counteract the effect of repressing sequences upstream of the hilA promoter. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 52.Schmieger H, Galan J E, Curtiss R. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 53.Shea J E, Beuzón C R, Gleeson C, Mundy R, Holden D W. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thomas A D, Booth I R. The regulation of expression of the porin gene ompC by acid pH. J Gen Microbiol. 1992;138:1829–1835. doi: 10.1099/00221287-138-9-1829. [DOI] [PubMed] [Google Scholar]
- 57.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 58.Wood M W, Jones M A, Watson P R, Hedges S, Wallis T S, Galyov E E. Identification of a pathogenicity island required for Salmonella enteropathogenicity. Mol Microbiol. 1998;29:883–891. doi: 10.1046/j.1365-2958.1998.00984.x. [DOI] [PubMed] [Google Scholar]
- 59.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 60.Worley M J, Ching K H, Heffron F. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000;36:749–761. doi: 10.1046/j.1365-2958.2000.01902.x. [DOI] [PubMed] [Google Scholar]