Abstract
Background: A variety of emerging medical imaging technologies based on artificial intelligence have been widely applied in many diseases, but they are still limitedly used in the cerebrovascular field even though the diseases can lead to catastrophic consequences.
Objective: This work aims to discuss the current challenges and future directions of artificial intelligence technology in cerebrovascular diseases through reviewing the existing literature related to applications in terms of computer-aided detection, prediction and treatment of cerebrovascular diseases.
Methods: Based on artificial intelligence applications in four representative cerebrovascular diseases including intracranial aneurysm, arteriovenous malformation, arteriosclerosis and moyamoya disease, this paper systematically reviews studies published between 2006 and 2021 in five databases: National Center for Biotechnology Information, Elsevier Science Direct, IEEE Xplore Digital Library, Web of Science and Springer Link. And three refinement steps were further conducted after identifying relevant literature from these databases.
Results: For the popular research topic, most of the included publications involved computer-aided detection and prediction of aneurysms, while studies about arteriovenous malformation, arteriosclerosis and moyamoya disease showed an upward trend in recent years. Both conventional machine learning and deep learning algorithms were utilized in these publications, but machine learning techniques accounted for a larger proportion.
Conclusion: Algorithms related to artificial intelligence, especially deep learning, are promising tools for medical imaging analysis and will enhance the performance of computer-aided detection, prediction and treatment of cerebrovascular diseases.
Keywords: Cerebrovascular diseases, artificial intelligence, machine learning, computer-aided detection, computer-aided prediction, computer-aided treatment decision
1. INTRODUCTION
As one of the leading causes of death and disability worldwide, cerebrovascular diseases (CVDs) often result in catastrophic consequences for patients and their families and place a significant burden on the healthcare system and socioeconomic economy [1, 2]. According to the clinical outcomes, CVDs are normally divided into hemorrhagic and ischemic types. Broadly speaking, intracranial aneurysm (IA) and arteriovenous malformation (AVM) are two common hemorrhagic CVDs, while arteriosclerosis (AS) and moyamoya disease (MMD) are two representative ischemic CVDs. Although these CVDs have different etiologies, pathophysiologies, treatment strategies, and prognoses, they have several features in common. Primarily, the diagnosis can only be confirmed through angiography, such as computer tomography angiography (CTA), magnetic resonance angiography (MRA) and digital subtraction angiography (DSA), among which DSA is considered the gold standard. Second, accurate and rapid diagnosis is based on sharp images and clinical knowledge of vascular anatomy rather than postoperative pathological results. Next, CVD prognosis depends on accidental hemorrhagic or ischemic events rather than the malignancy grade. Finally, several therapeutic strategies are optional, including open surgery, endovascular treatment, and medical treatment.
Artificial intelligence (AI) is an umbrella term that encompasses many techniques of varying complexity that can be used to solve different problems. One notable subfield of AI that has recently gained significant interest is machine learning (ML), including advanced deep learning (DL) methods. In recent decades, AI has made great strides and has been widely used in medical imaging. Among these studies, the diagnosis of cancers based on a combination of imaging and pathology has achieved remarkable success in assisting clinicians due to its exceptionally high accuracy. For example, all of the accuracies in lung cancer detection [3-5] were over 90% using classifiers such as support vector machine (SVM) [3], artificial neural network (ANN) [4] and 3D convolutional neural network (CNN) [5]. In metastatic breast cancer detection, two works [6, 7] achieved prominent performance on whole slide image (WSI) classification with DL methods, which was close to or even higher than the pathologist’s diagnosis. In the field of CVDs, however, most advanced machine learning approaches have not been fully extended yet not only because of the complexity of the disease but also due to different demands for AI-based applications between cancer and CVDs. Primarily, accurate CVD diagnosis occurs through preoperative cerebral angiography, while the cancer diagnosis can only be confirmed by postoperative pathology. Therefore, AI models that combine medical imaging with pathology are not helpful for diagnosing CVDs. Second, most CVDs can be accurately identified from standard imaging by experienced clinicians. Thus, computer-aided detection models can help in rapid diagnosis, shorten the learning curve of inexperienced clinicians, and reduce the missed diagnosis rate. Third, most cancer progresses regardless of whether it is quick or slow, but CVDs may remain asymptomatic until the sudden onset of serious hemorrhage or infarction. Therefore, computer-aided prediction of stroke events is of clinical significance. Finally, the little knowledge of the stroke risk in CVDs makes it difficult to determine the time of treatment. Since guidelines and clinical experiences are highly dependent here, computer-aided models may be proposed to learn prior knowledge and help create treatment strategies.
In this review, we survey the existing literature on the application of AI-based techniques in four main CVDs: aneurysm, AVM, AS and MMD. The methods and procedures that were used in reviewing systematic literature on the topic are presented in Section 2. The AI approaches are depicted and evaluated in three fields including detection, prediction, and treatment assistance for CVDs according to the selected studies in Section 3. Limited by the late development and few publications of AI application in CVDs, several AI techniques in cardiovascular imaging are also discussed for reference since methods for the diagnosis or treatment of CVDs were mainly developed based on cardiovascular research. Then, the current challenges and future directions of improvement for these AI approaches are critiqued in Section 4 and 5, which can shed light on AI applications in CVD medical imaging, followed by a summary of the review in Section 6.
2. METHODS
This study was conducted based on a systematic review protocol that helped achieve a comprehensive understanding of the research interest while providing further information for future studies. To achieve a supplementary search, five databases were selected and searched: (1) Elsevier Science Direct (SD), which offers broad access to scientific papers across different academic disciplines; (2) IEEE Xplore Digital Library, which covers all the technical and scientific literature in the fields of computer science, electrical and electronics engineering; (3) Springer Link, which is the world's most comprehensive online collection of scientific, technological and medical journals, books and reference works; (4) Web of Science (WoS), which consists of several literature search databases designed to support scientific and scholarly research; and (5) National Center for Biotechnology Information (mainly using PubMed), which is one of the most popular databases in health and biomedical science. Generally, these databases provided rigorous insights for both researchers and scholars by covering all the research disciplines from scientific and technological perspectives.
2.1. Search Strategy
The literature search was comprehensively conducted on the above databases on a span of 15 years between January 2006 and May 2021, which was considered sufficient for coverage on artificial intelligence applications on CVD medical imaging. Boolean operators such as ‘OR’ and ‘AND’ were utilized during the process to gather as much relevant literature as possible. The first group of keywords was the four major CVDs, and the second group was related to applications such as detection and prediction. Since there were various modalities in CVD-related medical imaging, we did not take certain modalities as keywords in the first step of the search to obtain more relevant literature, and the identified studies were reviewed in the following step to select those using medical images.
2.2. Inclusion and Exclusion Criteria
The inclusion criteria were as follows: (1) the paper was written in the English language and was submitted to a journal or conference as a research article, (2) the paper addressed artificial intelligence application on CVD medical imaging including computer-aided detection, prediction, and treatment; and (3) the paper used some machine learning-related methods such as conventional image processing algorithms, traditional classifier and advanced deep learning models.
Regarding the exclusion criteria, apart from language, studies that did not present a clear scientific method for development were also excluded. For example, papers that mainly focused on clinical or statistical analysis without any AI techniques could be excluded. In addition, there was little difference among the four diseases when scanning. For intracranial aneurysms that had more relevant papers, studies that did not utilize enough medical images were excluded. However, for diseases with fewer papers, this criterion was relaxed slightly for further analysis.
2.3. Study Selection and Data Extraction
There were three steps for study refinement. The first step excluded duplicate papers published between January 2006 and May 2021 in English. Next, the titles and abstracts of the publications were scanned to exclude papers not covered by the study scope. In particular, careful effort was taken for AVM and AS because the location of their lesions is confusing. For example, many papers focused on pulmonary AVM or cardiac AS instead of the brain after the first refinement process, and they needed to be discarded in the second step. The remaining articles underwent the third screening step through full-text reading to investigate the relevance of the selected articles and exclude papers that did not meet the eligibility criteria and study domain. After these refinement steps, 70 papers were included for detailed analysis and extraction. Generally speaking, the final selected studies had advantages in either novel technical model or convincing performance based on huge dataset. The flowchart of this systematic review protocol is presented in Fig. (1).
Fig. (1).
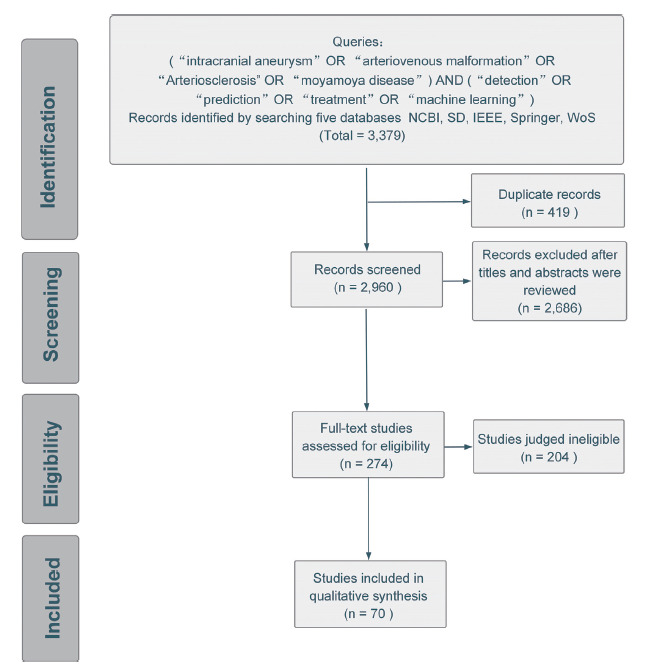
Flowchart of the systematic review to identify, screen and include relevant studies.
Since the aim of this review was to investigate the application of artificial intelligence on medical imaging of CVDs, some technical attributes were focused on and collected during the data extraction process. For instance, two medical data elements, modality and the number of subjects, were extracted to determine the distribution of different modalities and their corresponding volume, which may influence the performance. Other attributes that reflected algorithm information, such as feature type and decision criteria, were also considered, which could present the commonly used features (e.g., morphology, hemodynamics) and algorithms in this area. This procedure was followed by a summary and table with descriptions of the main findings in the next section.
3. RESULTS
This section presents our taxonomy, which summarizes the final results (i.e., 70 articles) of the search process. For the application categories, we first classified four CVDs into hemorrhagic and ischemic groups, and in each of them, great utility of AI was found in three main clinical applications on CVD medical imaging: computer-aided detection, prediction and treatment assistance as shown in Fig. (2). Detection refers to the localization of objects of interest in medical images, and computer-aided detection tools can be regarded as an initial screen against errors of omission by reducing observational oversights [8]. Since the shape of some vascular lesion-like aneurysms is relatively regular and similar to a sphere or ellipsoid, their detection may also realize segmentation to some extent. Once the suspicious region has been determined, it is imperative that some underlying risk be balanced before making decisions on intervention or treatment, especially for asymptomatic cases. Therefore, computer-aided prediction aims to provide robust descriptors to capture the risk of rupture, hemorrhage and infarction by integrating different kinds of features ranging from clinical and demographic information to morphological and hemodynamic characteristics. In addition to the aforementioned tasks, AI can also play a role in auxiliary treatment, such as real-time catheter segmentation and tracking, to provide a great convenience for the subsequent delivery of guidewires or image fusion to better guide bypass surgery. However, since this field requires high precision and multidisciplinary knowledge, it is still in its infancy, with few identified studies.
Fig. (2).
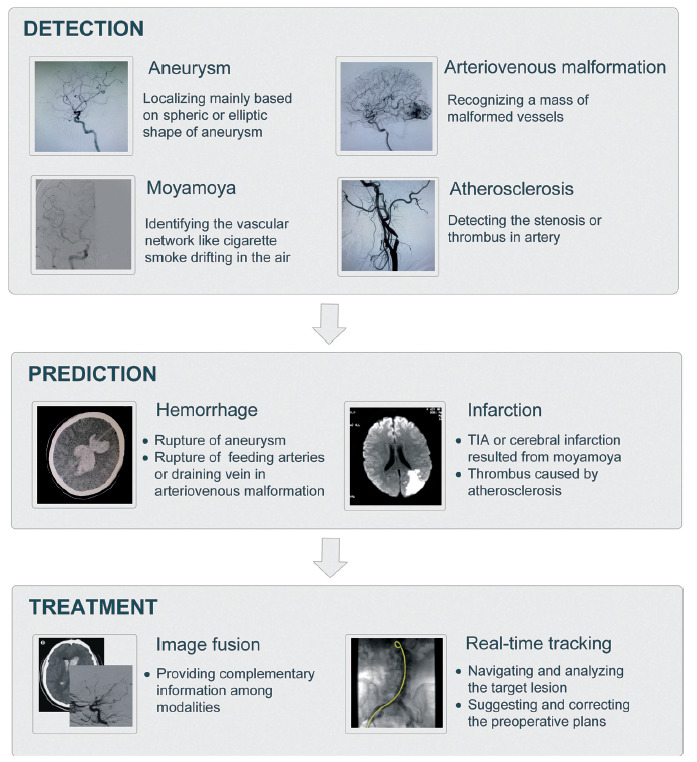
AI-based applications in CVDs.
Most medical imaging studies involved in the above three scenarios aim to extract some features from one or more imaging modalities to realize certain functions automatically or semiautomatically. Typically, after feature extraction, the realization of these functions can be implemented by either rule-based or classifier-based algorithms. The rule-based method usually refers to some threshold and specific conditions from professionals or empirical knowledge as the decision criteria, while the classifier-based method learns a rule automatically and separates the underlying classes optimally. Since the classifier-based scheme affords high objectivity through its ability to characterize complex patterns with less interobserver variability associated with assessment by human experts, the fundamental concept and corresponding characteristics of some common classification algorithms on medical imaging analysis are introduced first followed by the related literature explanation according to different categories.
3.1. Classification Algorithms
An overview of the common machine learning algorithms used in medical imaging analysis is given in Fig. (3). Even though only a few of them have been applied in CVDs, they still range from conventional machine learning algorithms to emerging deep learning. Generally, it seems that conventional algorithms are more popular in CVD applications, but DL has been noticed with its greater generalizability and robustness in other clinical scenarios, such as the diagnosis of lung cancer [9-11] or glioma [12-15], and this trend is gaining momentum in CVDs. The truly transformative aspect of DL methods is that they identify features implicitly instead of requiring a priori assumptions of what image features are important. Typically, DL works well with large quantities of data, while classical ML methods have advantages for smaller datasets [16]. Thus, it is important to use an appropriate classification algorithm contingent on the specific problem, and the respective characteristics of each algorithm are discussed in the following section.
Fig. (3).
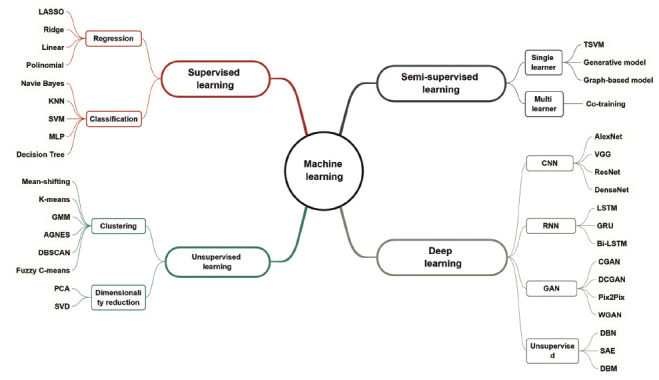
Framework of common machine learning algorithms.
3.1.1. Conventional Machine Learning Algorithms
3.1.1.1. K-Nearest Neighbor
As the simplest form of supervised classifier, K-nearest neighbor (KNN) is a typical representation of lazy learning since it does not require explicit classification function training. After storing some classified training data, classification of an independent test sample is performed by identifying the most similar measures, for example, the lowest Euclidean distance, between training and the test samples and then assigning the most frequent label of the K-nearest training neighbors to the test sample [17]. It is obvious that the selection of K is quite important and even accounts for significant differences among classification results with different K, as shown in Fig. (4). Even though KNN is a nonparametric estimation that is easy to implement and insensitive to outliers, it has large computational complexity and tends to be less effective when dealing with class-imbalanced problems.
Fig. (4).
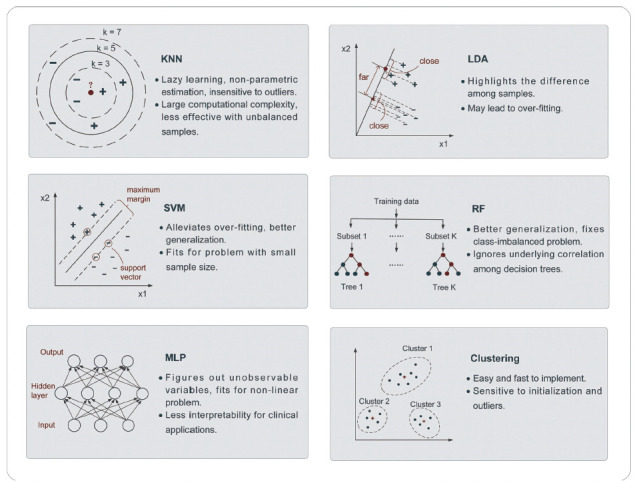
Conventional machine learning algorithms in CVD applications.
3.1.1.2. Linear Discriminant Analysis
A linear classifier can be viewed as learning a line or boundary (i.e., decision boundary) that separates points within the two classes and discriminates their labels. Linear discriminant analysis (LDA), namely, Fisher discriminant analysis [18], is one of the classical linear methods that aims to project high-dimensional samples to an optimal one-dimensional space by maximizing the variance in the between-class to within-class ratio. This principle highlights the difference among samples for powerful classification ability, and some nonlinear classifiers, such as quadratic discriminant analysis (QDA), have been developed. However, it may lead to overfitting because it has to consider all samples to determine the optimal boundary.
3.1.1.3. Support Vector Machine
Unlike LDA, which requires the whole dataset to build the optimal decision boundary, SVM only utilizes a set of training samples that lie closest to the boundary, known as support vectors [19], and then maximizes the margin between the boundary and support vectors from both sides. Despite its linear background, SVM algorithms have been extended into nonlinear problems with the help of a kernel function. In addition, the introduction of soft margins to alleviate overfitting also contributes to the stable performance of SVMs, making them one of the most popular methods. However, considering the complexity of solving quadratic programming, SVM is more applicable to problems with small sample sizes.
3.1.1.4. Random Forest
Another approach that can mitigate overfitting to some extent is random forest (RF), one of the ensemble learning algorithms based on decision trees, and multiple levels of randomization are the leading approaches [20], including randomness from training samples and features. Not only can the generalization accuracy be improved but also, the class-imbalance problem can be resolved properly with RF. It should be noted that even though independent and parallel relationships among decision trees contribute to higher generalization, they may ignore the influence of their underlying correlation.
3.1.1.5. Multilayer Perceptron
The multilayer perceptron (MLP), belonging to the ANN, is an extension of the linear perceptron classifier, which can also yield complex nonlinear decision boundaries. Typically, MLP consists of a few interconnected neurons to form a structure, including an input layer, hidden layer(s) and an output layer. A variety of nonlinear activation functions of the hidden layer neurons can be used to enhance the expressivity of a model (e.g., sigmoid function). During the training phase, the weights across a set of connected neurons are learned and updated according to the training data by using the back propagation technique [21] and then used for test sample classification. On the one hand, ANN or MLP considers unobservable variables and performs well on nonlinear datasets, especially datasets with large sizes. On the other hand, such a classifier lacks interpretability for most clinical applications.
3.1.1.6. Clustering
As a typical unsupervised learning branch, clustering is targeted at revealing the inherent relationship and similarity rule from unlabeled training data after a few iterations. There is a wide assortment of clustering algorithms, including partition clustering, density clustering and so on. For medical imaging of CVDs, K-means and fuzzy c-means (FCM), which belong to partition clustering, are generally used. K-means requires k initial clustering centers and assigns other samples to their nearest center according to certain distance metrics, such as Euclidean distance, followed by the update of new clustering centers [22]. The algorithm is easy and fast but sensitive to the number of initial clusters and some outliers. In contrast with the primitive K-means algorithm, each sample in FCM can have a membership degree to each cluster using a new distance metric, which is the product of the Euclidean distance and weight coefficient [23]. The introduction of fuzziness makes clustering a soft algorithm for boundary-ambiguous problems, such as the extraction of vessels.
3.1.2. Deep Learning Algorithms
3.1.2.1. Convolutional Neural Network
One of the most popular and typical networks in DL is CNN [24], which is designed to better utilize spatial and configuration information by taking 2D or 3D images as input rather than feature vectors. Structurally, CNNs have some convolutional layers for feature extraction, interspersed with several pooling layers for downsampling followed by a few fully connected layers. The first subgraph in Fig. (5) shows the basic CNN architecture, which automatically extracts image attributes hierarchically from local to global image representations through convolution kernels of different sizes. Recently, some CNN variants have proven to be especially useful in medical imaging analysis, such as ResNet-101 in brain tumor classification [25] and U-net in cell tracking [26], and similar applications in CVDs are also discussed in the following pertinent sections. However, as a data-driven method, not only for CNNs but also other DL algorithms, a large data size is the premise of reliable performance, which is challenging for medical images.
Fig. (5).
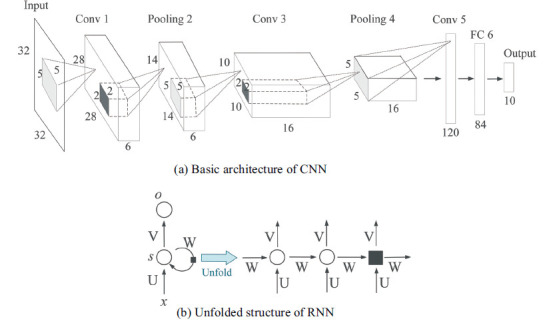
Architecture of common DL algorithms.
3.1.2.2. Recurrent Neural Network
Different from CNNs, which focus more on spatial information, recurrent neural networks (RNNs) deal with sequential data, namely, temporal information. As shown in Fig. (5b), hidden units in the same layer are no longer independent but connected, and the current output depends not only on the current input but also on the output from the previous moment (unit). RNNs have demonstrated great success in sequence labeling and prediction tasks such as speech recognition [27] and language modeling [28]. In regard to medical imaging analysis, it seems that RNNs have not been widely used, but their characteristic integration of time-varying information can play a significant role in CVD medical imaging, especially in 2D-DSA, which has dynamic flow changes among frames. To address gradient vanishing and gradient explosion problems in the training phase of some long sequences, long short-term memory (LSTM) [29] and its variants have been proposed with better performance than naïve RNNs.
3.2. Validation and Evaluation
Once the decision criteria are established in either rule-based or classifier-based ways, it is critical to verify the feasibility and effectiveness of the model with some validation strategies and evaluation metrics. To achieve better performance, a model should be trained with as much training data as possible, which is often a challenging issue in medical imaging studies. Therefore, for validation strategies, cross-validation (CV), which repeatedly evaluates model performance using multiple training and testing partitions, is commonly used. A popular CV approach is k-fold cross-validation [30], which splits the whole dataset into k mutually exclusive folds of approximately equal size. The model is trained on k-1 folds and tested on the remainder until all subsets have been traversed k times. The setting of the parameter k is empirical, depending on many factors such as the dataset distribution and model stability, and the common choices are ten or five at present. As a special case of k-fold CV, leave-one-out (LOOCV) has a similar procedure to standard k-fold CV except that the value of k in LOOCV equals the number of samples. Overall, CV approaches help ensure the generalization of the model, and compared with k-fold CV, LOOCV is typically used on smaller sample sizes, but leaving each sample out can be slightly computationally expensive.
For evaluation, metrics including accuracy, sensitivity, specificity, receiver operating characteristic (ROC) curve and free-response receiver operating characteristic (FROC) curve are commonly used in medical imaging analysis. Accuracy indicates how accurately the model classifies the positive and negative samples, sensitivity shows the proportion of true positives correctly identified and specificity demonstrates the proportion of true negatives correctly identified by the model. The overall performance of the model can be assessed by the ROC curve, which shows the balance between the true positive rate and the false positive rate across a range of decision thresholds within the model and provides a summary of the area under the curve (AUC). In addition, in some CVD-related tasks, such as lesion location and detection, FROC [31] is widely accepted for addressing the problem of evaluating one or more abnormalities in the same subject (e.g., detecting multiple aneurysms). Not only the accuracy of ratings but also the number of marks is considered one of the integral system characteristics, which is the distinctive feature of FROC.
3.3. Current Clinical Applications
With the aforementioned algorithms applied in medical imaging analysis, several AI-based progress in CVDs has been made even though it is still in the preliminary exploration stage compared with other diseases, such as cancer or psychiatric disorders. Here, recent studies related to computer-aided detection, prediction and treatment in four CVDs are reviewed and listed in Tables 1 and 2.
Table 1.
Applications on hemorrhagic CVDs.
| References | Application | Modality |
No. of
Subjects |
Feature Type |
Decision
Criteria |
Validation Type | Highest Results |
|---|---|---|---|---|---|---|---|
| Uchiyama, Y. et al. (2006) [33] | Aneurysm, detection of IA | 3D-MRA | 20 | - Shape (e.g., size, sphericity) - intensity |
QDA | / | SEN (100%), 1.85FPs |
| Arimura, H. et al. (2006) [34] | Aneurysm, detection of IA | 3D-MRA | 115 | - Gray-level - morphology - human-designed (e.g., SBD) |
LDA | LOOCV | SEN (97%), 3.8FPs |
| Lauric, A. et al. (2010) [35] | Aneurysm, detection of IA | - 3D-RA - CTA |
20 | Human-designed (e.g., writhe number) |
Rule-based | / | - SEN (100%), 0.66FPs and 5.36FPs for RA and CTA |
| Yang, X. et al. (2011) [36] | Aneurysm, detection of IA | 3D TOF MRA | 222 | Geometrics (e.g., position, radius) |
Rule-based | / | SEN (95%), 9 FPs |
| Macía, I. et al. (2011) [37] | Aneurysm, detection of endoleaks | CTA | 5 | - Geometrics - human-designed (e.g., NTD) |
MLP | 10-fold CV | ACC (93.65%) |
| Nakao, T. et al. (2018) [38] | Aneurysm, detection of IA | 3D TOF MRA | 450 | High-level features from DL | CNN (self-designed) | Independent validation | SEN (94.2%), 2.9 FPs |
| Malik, K.M. et al. (2018) [39] | Aneurysm, detection of IA, prediction of rupture | 2D-DSA | 59 | Texture (e.g., contrast, entropy) | - MLP - SVM - Naïve Bayes - LR |
10-fold CV | - SEN (98%) for detection - ACC (86%) for prediction |
| Rahmany, I. et al. (2018) [40] | Aneurysm, detection of IA | 2D-DSA | 30 | Robust feature (e.g., SURF, SIFT) | Rule-based | Independent validation | SEN (100%) |
| Chandra, A. et al. (2014) [41] | Aneurysm, detection of IA | 2D-DSA | 3 | / | Rule-based | / | ACC (99%) |
| Sulayman, N. et al. (2016) [42] | Aneurysm, detection of IA | 2D-DSA | 10 | Shape (e.g., circularity,solidity) |
Rule-based | / | SEN (89.47%) |
| Kohout, J. et al. (2013) [44] | Aneurysm, detection of IA | CTA | / | / | Rule-based | / | SEN (100%) |
| Suniaga, S. et al. (2012) [45] | Aneurysm, detection of IA | 3D-MRA | 20 | - Morphology (e.g., blobness, vesselness) - structure (e.g., bifurcation distance) |
SVM | LOOCV | SEN (100%), 3.86FPs |
| Jerman, T. et al. (2015) [46] | Aneurysm, detection of IA | 3D-DSA | 25 | Rotation invariant and scale normalized feature | RF | Independent validation | SEN (100%), 0.4FPs |
| Hentschke, C.M. et al. (2011) [47] | Aneurysm, detection of IA | - 3D-RA - CTA - MRA |
65 | - Morphology (e.g., blobness, vesselness) - intensity |
Rule-based | / | - SEN (96%), 2.6FPs for RA - SEN (94%), 8FPs for MRA - SEN (90%), 28FPs for CTA |
| Hentschke, C.M. et al. (2012) [48] | Aneurysm, detection of IA | - 3D-TOF MRA - CE MRA - CTA |
66 | - Gray-level - morphology - human-designed (e.g., aneurysmness) |
Rule-based | / | - SEN (100%), 8.75FPs for CE MRA - SEN (93%), 11.3FPs for TOF MRA - SEN (96%), 20.94FPs for CTA |
| Hentschke, C.M. et al. (2014) [49] | Aneurysm, detection of IA | - 3D-TOF MRA - CE MRA - CTA |
151 | - Gray-level - morphology (e.g., blobness, vesselness) |
- LDF - SVM - RF - ADtree - Logit-Boost |
4-fold CV | - SEN (95%), 8.2FPs for CE MRA - SEN (95%), 11.3FPs for TOF MRA - SEN (95%), 22.8FPs for CTA |
| Hanaoka, S. et al. (2015) [52] | Aneurysm, detection of IA | MRA | 300 | Human-designed (e.g., HoTPiG) |
SVM | 3-fold CV | SEN (81.8%), 3FPs |
| Rahmany, I. et al. (2014) [53] | Aneurysm, detection of IA | 2D-DSA | / | Fuzzy morphology | Rule-based | / | SEN (100%) |
| Geng, C. et al. (2020) [54] | Aneurysm, detection of IA | TOF MRA | 131 | High-level features from DL | CNN (3D-Unet) | Independent validation | SEN (82.9%), 0.86FPs |
| Park, A. et al. (2019) [55] | Aneurysm, detection of IA | CTA | 662 | High-level features from DL | CNN (3D-HeadXNet) | Independent validation | SEN (94.9%) |
| Sichtermann,T. et al. (2018) [56] | Aneurysm, detection of IA | TOF MRA | 85 | High-level features from DL | CNN (3D-Deep-Medic) | Independent validation | SEN (90%) |
| Shahzad, R. et al. (2020) [57] | Aneurysm, detection of IA | CTA | 253 | High-level features from DL | CNN (3D-Deep-Medic) | - 5-fold CV - Independent validation |
SEN (87%) |
| Shi, Z. et al. (2020) [58] | Aneurysm, detection of IA | CTA | 1177 | High-level features from DL | CNN (3D-Dual Attention ResUnet) | Independent validation | SEN (97.3%), 0.29FPs |
| Joo, B. et al. (2020) [59] | Aneurysm, detection of IA | TOF MRA | 744 | High-level features from DL | CNN (3D-ResNet) | Independent validation | SEN (87.1%) |
| Dai, X.L. et al. (2020) [60] | Aneurysm, detection of IA | CTA | 311 | High-level features from DL | CNN (faster-RCNN) | Independent validation | SEN (91.8%) |
| Jerman, T. et al. (2017) [61] | Aneurysm, detection of IA | 3D-DSA | 15 | High-level features from DL | CNN (self-designed) | 3-fold CV | SEN (100%), 2.4FPs |
| Podgorsak, A. et al. (2020) [62] | Aneurysm, detection of IA | DSA | 350 | High-level features from DL | CNN (U-shaped) | Independent validation | AUC (79.1%) |
| Rahmany, I. et al. (2018) [63] | Aneurysm, detection of IA | 2D-DSA | 30 | High-level features from DL | CNN (pre-trained Inception-V3) | Independent validation | SEN (100%) |
| Ueda, D. et al. (2019) [64] | Aneurysm, detection of IA | TOF MRA | 1271 | High-level features from DL | CNN (untrained ResNet-18) |
- 5-foldCV - Independent validation |
- SEN (91%) for internal data - SEN (93%) for external data |
| Jin, H. et al. (2020) [67] | Aneurysm, detection of IA | 2D-DSA | 493 | Spatial and temporal features from DL | - CNN (U-shaped) - Biconv-LSTM |
Independent validation | SEN (89.3%), 3.77FPs |
| Duan, H.H. et al. (2019) [68] | Aneurysm, detection of IA | 2D-DSA | 281 | High-level features from DL | CNN (self-designed) | Independent validation | AUC (94.2%) |
| Stember, J.N. et al. (2019) [69] | Aneurysm, detection of IA | MRA | 302 | High-level features from DL | CNN (U-shaped) | Independent validation | SEN (98.8%) |
| Zeng, Y. et al. (2020) [70] | Aneurysm, detection of IA | 3D-RA | 300 | High-level features from DL | CNN (modified VGG16) | Independent validation | ACC (98.9%) |
| Hahn, S. et al. (2019) [71] | Aneurysm, detection of endoleaks | CTA | 334 | High-level features from DL | CNN (RetinaNet, modified ResNet-50) | 5-fold CV | ACC (89%) |
| Yang, X. et al. (2020) [72] | Aneurysm, detection, segmentation of IA | TOF MRA | 103 | High-level features from DL | CNN (PointNet, DGCNN) | 5-fold CV | Dice (88.7%) |
| Ye, H. et al. (2019) [73] | Aneurysm, recognition of aSAH | CT | 2836 | High-level features from DL | CNN (3D-RCNN) | Independent validation | AUC (88.2%) |
| Danilov, G. et al. (2020) [74] | Aneurysm, recognition of aSAH | CT | 401 | High-level features from DL | CNN (modified ResNexT) | / | ACC (81%) |
| Cho, J. et al. (2019) [75] | Aneurysm, recognition of aSAH | CT | 5702 | High-level features from DL | - CNN (modified GoogleNet) - FCN |
5-fold CV | SEN (97.9%) |
| Lahmiri, S. et al. (2014) [76] | AVM, detection of AVM | MRI | 28 | Human-designed (e.g., fractal dimension) |
SVM | 10-fold CV | ACC (100%) |
| Lahmiri, S. et al. (2014) [77] | AVM, detection of AVM | MRI | 56 | - Hurst's exponent - human-designed (e.g., energy of flunctuation) |
SVM | 10-fold CV | ACC (98%) |
| Babin, D. et al. (2014) [78] | AVM, segmenta- tion of AVM |
3D-CTA | 3 | Human-designed (e.g., node degree) |
Rule-based | / | Dice (80.3%) |
| Babin, D. et al. (2014) [79] | AVM, segmenta- tion of AVM |
3D-CTA | 3 | Human-designed (e.g., density) | Rule-based | / | Dice (79%) |
| Lian, Y.X. et al. (2015) [80] | AVM, segmenta- tion of AVM |
DSA | / | / | Rule-based | / | / |
| Peng, S.J. et al. (2019) [81] | AVM, segmentation of AVM-related region |
T2W MRI | 25 | Voxel intensity | FCM | / | SEN (81.9%) |
| Lee, C.C. et al. (2019) [82] | AVM, segmentation of AVM-related region | T2W MRI | 39 | Voxel intensity | FCM | / | Dice (79.5%) |
| Wang, T.H. et al. (2018) [83] | AVM, segmenta- tion of AVM |
CT | 80 | High-level features from DL | CNN (3D-Vnet) | 4-fold CV | Dice (85%) |
| Chen, G. et al. (2020) [90] | Aneurysm, prediction of rupture risk | CTA | 1115 | - Morphology - clinical (e.g., hypertension, diabetes) - hemodynamics (e.g., flow complexity, stability) |
- LR - RF - MLP - SVM |
- 10-fold CV - Independent validation |
AUC (88.6%) |
| Liu, J. et al. (2018) [91] | Aneurysm, prediction of rupture risk | CTA | 594 | - Morphology - demographics (e.g., sex) - smoking and hypertension |
MLP | Independent validation | ACC (94.8%) |
| Ou, C.B. et al. (2020) [92] | Aneurysm, prediction of rupture risk | 3D-DSA | 374 | - Morphology - demographics - medical history - lifestyle |
- LR - SVM - ANN - XGBoost |
Independent validation | AUC (88.2%) |
| Shi, Z. et al. (2021) [96] | Aneurysm, prediction of rupture risk | CTA | 681 | - Morphology - hemodynamics - clinical data |
- LR - RF - SVM - MLP |
Independent validation | AUC (88%) |
| Lauric, A. et al. (2011) [97] | Aneurysm, prediction of rupture risk | 3D-RA | 154 | Human-designed (e.g., entropy of centroid-radii model) | LR | 10-fold CV | ACC (80.3%) |
| Niemann, U. et al. (2018) [98] | Aneurysm, prediction of rupture risk | 3D-RA | 74 | Morphology | - Naïve Bayes - SVM - KNN - GBT - RF |
10-fold stratified CV | ACC (69%) |
| Kim, H.C. et al. (2019) [99] | Aneurysm, prediction of rupture risk | 3D-DSA | 640 | High-level features from DL | CNN (AlexNet-V2) | Independent validation | ACC (76.84%) |
| Liu, Q. et al. (2019) [100] | Aneurysm, prediction of stability | DSA | 368 | - Morphology (e.g., compactness, surface volume ratio) - clinical (e.g., hyper-lipemia) |
- Lasso regression - Ridge regression |
/ | AUC (85.6%) |
| Paliwal, N. et al. (2018) [101] | Aneurysm, prediction of treatment outcome | 3D-DSA | 80 | - Morphology - hemodynamics - Flow diverter-based |
- LR - SVM - KNN |
- 4-fold CV - Independent validation |
AUC (96.7%) |
| de Toledo, P. et al. (2009) [102] | Aneurysm, prediction of SAH outcome | CT | 634 | - WFNS - Fisher’s scale - clinical (e.g., age) |
Decision Trees | Independent validation | AUC (84%) |
| Xia, N.Z. et al. (2020) [103] | Aneurysm, prediction of SAH outcome | CTA | 809 | Morphology(e.g., size, height, aspect ratio) | RF | Independent validation | AUC (90%) |
| Hong, J.S. et al. (2020) [104] | AVM, prediction of AVM nidus | DSA | 171 | Hemodynamics (e.g., peak density, time to peak) |
- Naïve Bayes - SVM - RF |
Independent validation | ACC (85.7%) |
| Asadi, H. et al. (2016) [105] | AVM, prediction of treatment outcome | / | 199 | / | - ANN - SVM |
- Nested CV - Independent validation |
ACC (97.5%) |
| Oermann, E.K. et al. (2016) [106] | AVM, prediction of treatment outcome | DSA | 1502 | - Clinical (e.g., sex, age) - treatment (e.g., isodose) - angioarchi-tectural (e.g., location) |
- LR - RF - SVM - Gradient Boosting - Extra trees |
Independent validation | ACC (74%) |
| Zhou, Y.J. et al. (2020) [107] | Aneurysm, Real-time tracking of catheter | 2D X-ray sequence | 30 | Spatial and temporal features from DL | - RCNN - CNN (pre-trained MobileNetV2) |
Independent validation | SEN (95.8%) |
Abbreviations: TOF, Time of flight; CE, Contrast enhancement; RA, Rotational angiography; LR, Logistic regression; ADtree, Alternating decision tree; GBT, Gradient boosted trees; aSAH, Aneurysmal subarachnoid hemorrhage; WFNS, World federation ofneurological surgeons; T2W, T2-Weighted.
Table 2.
Applications on ischemic CVDs.
| References | Application | Modality |
No. of
Subjects |
Feature Type |
Decision
Criteria |
Validation Type | Highest Results |
|---|---|---|---|---|---|---|---|
| Gao, W. et al. (2009) [108] | AS, estimation of AS degree | / | 50 | Clinical (e.g., hypertension, cholesterol, age) | FSVM | Independent validation | Average error (11.2%) |
| Terrada, O. et al. (2020) [109] | AS, diagnosis of AS | / | 636 | Clinical (e.g., age, sex, chest pain type) | - ANN - KNN |
Independent validation | ACC (96.4%) |
| Ding, L. et al. (2010) [110] | AS, classification of AS degree | Pulse wave | 50 | Wave features in time, frequency domain | Fuzzy pattern recognition | / | ACC (90.0%) |
| Zhang, Y. et al. (2018) [111] | AS, diagnosis of AS | PPG | 235 | Wave features in time, frequency domain | SVM | Independent validation | ACC (98.4%) |
| Kim, T. et al. (2019) [112] | MMD, detection of MMD | X-ray skull image | 753 | High-level features from DL | CNN (self-designed) | Independent validation | AUC (91.0%) |
| Akiyama, Y. et al. (2020) [113] | MMD, detection of MMD | T2W MRI | 210 | High-level features from DL | CNN (pre-trained VGG16) | Independent validation | ACC (92.8%) |
| Hu, T. et al. (2021) [114] | MMD, detection of MMD | DSA | 630 | High-level features from DL | CNN (3D-CNN, BiConv-GRU) | Independent validation | ACC (98.6%) |
| Lei, Y. et al. (2021) [115] | MMD, prediction of hemorrhagic risk | DSA | 878 | High-level features from DL | CNN (pre-trained ResNet-152, self-designed) | - 5-fold CV - Independent validation |
ACC (90.7%) |
| Lei, Y. et al. (2020) [116] | MMD, recognition of VCI | Resting- state fMRI |
105 | Local clustering coefficient of low-/high-order FC networks |
SRC | 10-fold CV | AUC (91.0%) |
Abbreviations: PPG, Photoplethysmogram; FSVM, Fuzzy support vector machine; T2W, T2-Weighted; VCI, Vascular cognitive impairment; FC, Functional connectivity; SRC, Sparse representation-based classification.
3.3.1. Hemorrhagic Cerebrovascular Diseases
3.3.1.1. Computer-aided Detection
As mentioned previously, hemorrhagic CVDs mainly refer to aneurysms and AVMs. As pathological bulges in weak arterial walls, the majority of aneurysms are saccular, and the detection of unruptured intracranial aneurysms (UIAs) is a key subject in aneurysm-related detection since most UIAs are asymptomatic, paving the way for risk prediction. The general detection workflow in most of the literature is shown in Fig. (6), including vessel segmentation, candidate detection, feature extraction and false positive (FP) reduction. In the segmentation step, the region-growing technique [32] is the most common tactic to segment vasculatures in several studies [33-38] and usually performs well without prior knowledge. Filter enhancement combined with the thresholding method also appears in some articles [39-41]. For decisions regarding the optimal threshold, Rahmany, et al. [40] and Sulayman, et al. [42] adopted Otsu’s method [43] for the research by minimizing the within-class variances and maximizing the between-class variance in the segmented image. Chandra, et al. [41] used a simpler method to directly obtain the optimal threshold from the histogram of grayscale images. It is believed that the quality of vessel segmentation impacts the accuracy of the detection method; thus, utilizing the proper segmentation approach according to the characteristics of the imaging modality and corresponding disease is crucial for further implementation. For example, coarse thresholding based on the voxel value of an image is easily implemented, but it may result in incorrect segmentation if the image has low resolution with high noise disturbance. In regard to detecting candidates, skeletonization (namely, the thinning algorithm) [34-36, 44, 45] and blob enhancement filters (namely, dot-enhancement filters) [46-49] are widely used based on different assumptions. The former presumes that a medial axis of the vessel can be computed after skeletonization, and aneurysms appear as small branches along the medial axis [50], while the latter assumes that most of the aneurysms are blob-like or spherical structures; thus, bright spherical objects can be selected on a dark background by analyzing Hessian eigen values [51]. However, the ‘short hair’ problem (i.e., a large number of false short branches where the arterial wall has small lumps) is common in skeletonization or centerline extraction, making a post-process to remove or classify short hairs indispensable. Therefore, a novel feature set named HoTPiG (histogram of triangular paths in graph) was proposed to address this problem graphically [52]. For feature extraction, morphological characteristics such as size, sphericity and effective diameter [33] and gray-level characteristics such as average and standard deviation of voxel value [49] account for most of the extracted aneurysm features. In addition, there are also some user-defined features, such as writhe numbers proposed by Lauric, et al. [35], to detect perturbations along the vasculature or shape-based difference (SBD) features suggested by Arimura, et al. [34] to describe the local changes in vessel thicknesses caused by aneurysms. Although these user-defined features achieved good performance in the corresponding study, their reliability and reproducibility remain verified. Conventional classifiers involved in the last step are mainly MLP [37, 39], SVM [45, 49], LDA (QDA) [33, 34] and fuzzy methods [53]. For example, both studies [37, 39] validated their corresponding model in the MLP classifier with tenfold CV and reached accuracies of 93.65% and 98%, respectively.
Fig. (6).
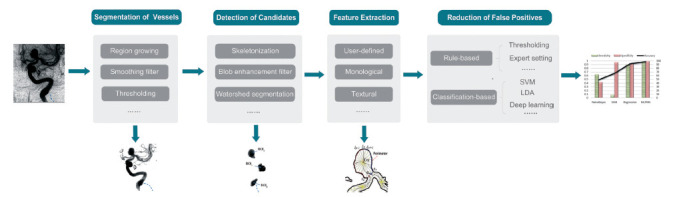
General aneurysm detection workflow.
Even though (Fig. 6) displays a general and complete aneurysm detection pipeline, it is not necessary to complete all of these procedures, especially for studies employing deep learning algorithms. In a DL model, the dimension of input images is worthy of consideration because 3D input can provide more useful information, but it requires more memory and training time than 2D input. For 3D raw data, Geng, et al. [54] and Park, et al. [55] developed a 3D model with an encoder and decoder similar to traditional U-net. Sichtermann, et al. [56] and Shahzad, et al. [57] established a 3D CNN architecture with two identical pathways that apply different image resolutions to capture contextual information. Different from utilizing the whole 3D raw data directly, some tactics were proposed to reduce the computational complexity, including splitting cube patches [58, 59] and transforming the 3D problem into the 2D domain in [38, 60, 61]. For example, Shi, et al. [58] developed a 3D CNN with an encoder-decoder architecture for 3D patch segmentation, and a dual attention block was embedded in their model to learn long-range contextual information and obtain more reliable feature representations. In addition to the introduction of attention blocks, another strength of this article is that the prediction result of those uniformly sampled patches was further merged, turning the patch-level prediction back to patient level with more clinical significance. In studies using dimension transforming, Nakao, et al. [38] used maximum intensity projection (MIP) to generate nine MIP images from a cube VOI and concatenated them as 2D input, while Jerman, et al. [61] computed 2D intravascular distances to obtain an intensity map from 3D DSA and achieved 100% sensitivity. However, producing such intensity maps requires considerable computing time. For 2D raw data, Podgorsak, et al. [62] directly used a CNN to detect and segment intracranial aneurysms in DSA, and the extracted angiographic parametric imaging radiomic features showed fewer features than human user results. Similar to 3D patch splitting, both Rahmany, et al. [63] and Ueda, et al. [64] extracted 2D patches (window) from raw images and trained on some well-known networks, including pretrained Inception-V3 [65] and untrained ResNet-18 [66] separately. However, these patch-based studies classified patients at the patch level and lacked specific statements about patient-level decisions. In addition, the DL backbones adopted in these papers were less innovative and neglected to integrate some specific medical image characteristics. In addition to spatial information captured from DSA images, Jin, et al. [67] proposed a bidirectional convolutional LSTM module at each level of the U-shape network to depict the contrast medium flow change across the DSA 2D frames, which incorporated both spatial and temporal features of images, successfully detecting 316 out of 354 (89.3%) aneurysms in an independent testing set. However, inputting the whole DSA series in one direction may lose information and increase the interference from the background; thus, Duan, et al. [68] designed a two-stage detection network using information on both frontal and lateral 2D DSA sequences. Specifically, the region localization stage (RLS) located the posterior communicating artery (PCoA) region from the raw images since PCoA has the aneurysm recurrence rate of second place, and the second stage identified intracranial aneurysm from the output of RLS. In addition to the direct detection of aneurysms [69-72], other computer-aided classifications related to aneurysms also adopted CNN or RNN to meet objectives such as distinguishing aneurysmal subarachnoid hemorrhages from other intracranial hemorrhage subtypes [73-75].
Compared with aneurysms, very few studies on AVM detection or segmentation have been published, even though AVMs, as abnormal vascular masses caused by direct communication between cerebral arteries and veins or the absence of capillary structures, are not hard to detect. Lahmiri, et al. applied detrended fluctuation analysis (DFA) to AVM detection based on SVM [76] and made some improvements in another paper [77]. Both approaches split the original MRI into right and left hemispheres and converted each hemisphere to a 1D signal to save time, followed by feature extraction on two 1D signals and final classification. The main difference between these two methods is that the work in [76] assumed at least one brain hemisphere is healthy and performed classification on each side independently, while the improved method [77] incorporated features from both sides. Babin, et al. [78, 79] presented an algorithm to use anatomical vessel differences and in homogeneities in the distribution of pixel gray values to detect and extract AVMs from 3D-CTA images. It used a novel skeletonization method to segment blood vessels first and extracted AVM through density calculation due to its high density of small intertwined vessels. However, this method is not applicable enough to segment the AVM nidus and vessel simultaneously. To handle this problem, Lian, et al. [80] combined the global thresholding technique to distinguish the large nidus and local iterative thresholding technique to identify tiny vessel structures on DSA. For the detection and segmentation of AVMs after radiosurgery, such as gamma knife, fuzzy c-means clustering on T2-weighted MR images [81, 82] and 3D V-net with deep supervision on CT [83] were also investigated.
3.3.1.2. Computer-aided Prediction
Detecting intracranial aneurysms and AVM from imaging scans is a fundamental but essential step in the prevention of potential bleeding, especially for nontraumatic subarachnoid hemorrhage (SAH) since the majority of nontraumatic SAH results from aneurysm rupture and is associated with a high mortality and morbidity rate. Specifically, the initial bleeding of ruptured aneurysms is fatal in 10-20% of instances, and despite improvements in patient management, the incidence of SAH has not declined over time, and the morbidity rate is still reported to be between 25% and 50% in patients surviving aneurysm ruptures [84-86]. Numerous studies have analyzed and revealed the correlation between aneurysm rupture and some risk factors, such as aneurysm size [87], aspect ratio [88], and sex [89]. Nevertheless, rupture risk prediction is still challenging due to the complicated relationship between multiple parameters and aneurysms. Manual calculation was widely used for feature extraction in [90-92]. For example, Liu, et al. [91] took seventeen factors, including thirteen aneurysm morphological parameters, two demographic factors, hypertension and smoking histories, as input into an MLP classifier, reaching an accuracy of 94.8%. However, it is noteworthy that the adaptive synthetic (ADASYN) sampling approach [93] adopted in this paper to solve class imbalance was challenged by de Jong, et al. [94] since ADASYN seemed to perform worse with increasingly imbalanced datasets [95] and the mismatch between the actual population and the population with synthetically generated subjects remains unknown. Therefore, caution should be taken when selecting proper implementations to improve imbalanced data for use in ANNs. In addition to several morphological and clinical features, both Chen, et al. [90] and Shi, et al. [96] employed hemodynamic characteristics (e.g., wall shear stress, oscillatory shear index), which is one of the pathogeneses of aneurysms, to compare the performance of different classifiers. Instead of demanding aneurysm delineation from clinicians to extract features, as in the aforementioned studies, some papers [97, 98] segmented aneurysms automatically first, followed by feature extraction and classification, which saved considerable labor. For deep learning applications, Kim, et al. [99] developed a CNN model to assess rupture risk based on images in six directions from 3D-DSA and performed well even on small-sized intracranial aneurysms. However, it is not a fully automatic classifier because it applies user-selected ROIs around aneurysms in every direction. In addition to rupture or stability prediction [100], Paliwal, et al. [101] modeled flow diverter deployment using a fast virtual stenting algorithm and hemodynamics using image-based computational fluid dynamics, which was used for further feature calculation to predict the treatment outcome by flow diverters. de Toledo, et al. [102] and Xia, et al. [103] adopted machine learning techniques to predict the outcome of aneurysmal SAH and the outcome after rupture of anterior communicating artery aneurysm, respectively.
Different from morphological, geometric or texture features extracted from medical images, Hong, et al. [104] utilized quantitative DSA (QDSA) features, which are salient features from the time-density curve (TDC) of a selected ROI, to reflect the hemodynamics of the cerebrovasculature and predict the rupture of the AVM nidus. Favorable performance on various classifiers suggested the robustness of this feature set. The outcome after surgery was also discussed in [105, 106] with some conventional algorithms.
3.3.1.3. Computer-aided Treatment
The complexity of the disease hinders AI-based methods from making progress in CVDs, especially in fields requiring knowledge from multiple disciplines. Computer-aided treatment in CVDs is a field that may need to integrate segmentation, classification, and hardware implementation. Hence, not only for aneurysms or AVMs but also for other CVDs, computer-aided treatment is a new application, lacking enough specific studies. A study that aimed to segment and track catheters on 2D X-ray sequences during endovascular aneurysm repair was proposed by Zhou, et al. [107]. An encoder-decoder architecture combining the advantages of a recurrent convolutional neural network (RCNN), pretrained components of MobileNetV2 and a gradient harmonizing mechanism (GHM) was developed, in which RCNN extracts temporal information and a GHM addresses problematic class imbalance and misclassified examples. The proposed approach obtained a sensitivity of 95.8% and provided great convenience for the subsequent delivery of guidewires, stents and other interventional instruments. Notably, network backbones, such as ResNet-50 and VGG16, actually slightly outperformed the proposed MobileNetV2 in ablation experiments in terms of precision and F1-score, but the processing speed was remarkably improved with MobileNetV2, showing a reasonable tradeoff between result accuracy and performance speed. Although there is little published literature for reference about auxiliary treatment, some novel conceptions of treatment in hemorrhagic CVDs have been proposed, including automatic selection of interventional materials and real-time guidance in surgery, presenting bright prospects in this area.
3.3.2. Ischemic Cerebrovascular Diseases
Compared with the hemorrhagic type, fewer studies have investigated AI-based applications of ischemic CVDs. One of the reasons is that there is a more complex interaction among different kinds of factors related to ischemic CVDs. Taking AS, for example, hemodynamic factors (e.g., wall shear stress), biological factors (e.g., the biological reaction of the arterial wall), and clinical factors (e.g., cholesterol, hypertension) play important roles in analyzing AS than morphological or gray-level characteristics, which are easier to quantify and widely used in aneurysm detection. Similarly, it is difficult to study MMD comprehensively without considering multiple varieties. To the best of our knowledge, few essays refer to the computer-intervened analysis of ischemic CVDs such as AS and MMD, and most models do not take medical images as input. Gao, et al. [108] introduced an approach to determine the optimal classification function and its membership function by using fuzzy SVM (SFVM) since the scientific diagnosis of AS lacks strict uniform standards and often contains much fuzzy information. Four clinical indicators, including age, body mass index, cholesterol and triglycerides, were selected as the most influential and independent features from fifty cases and were used to build the optimal function, attaining an average error of 11.2% during the testing phase. Similarly, Terrada, et al. [109] also excluded imaging features and utilized only several clinical factors to diagnose AS with ANN and KNN. Additionally, based on a fuzzy algorithm, Ding, et al. [110] extracted features from the time and frequency domains of pulse signals, which presented various changes with the increasing degree of AS. Then, the fuzzy clustering method was used for feature selection, followed by fuzzy classification with three selected features. The photoplethysmogram (PPG) signal was regarded as raw data in [111], and three types of parameters consisting of a waveform in the time domain, harmonic amplitudes in the frequency domain and wavelet energy were calculated from the signal. An SVM model was finally established on the selected features through a genetic algorithm and achieved an accuracy of 98% on the test set.
For MMD, even though medical images have been utilized for relevant applications, they have appeared in very few studies. Kim, et al. [112] initially proposed a deep learning model comprising six convolutional layers, several pooling layers and two fully connected layers to detect MMD from plain skull images. A total of 753 subjects were enrolled for validation with a final AUC of 91%, and both the sensitivity and specificity were 84%. Although there were some limitations, such as using an uncommon modality, causing potential bias by selecting trauma patients rather than normal subjects as the control group, as the first milestone to cover MMD detection based on AI, this study still paved the way for AI-based application on MMD with various other medical imaging modalities. For example, Akiyama, et al. [113] applied a pretrained VGG16 network to differentiate MMD patients with AS patients or normal controls on T2-weighted MR images, and the areas of interest of MMD and AS were visualized by the Grad-CAM technique. To automatically detect MMD on DSA and reduce clinicians’ workloads, Hu, et al. [114] fused spatial and temporal information from DSA images by combining a traditional CNN with a bidirectional convolutional gated recurrent unit for better spatiotemporal feature learning and achieved a high accuracy of 97.88%. To further predict the hemorrhagic risk of MMD, Lei, et al. [115] first employed a CNN to recognize MMD on DSA and then combined a multiview CNN with a squeeze-excitation block to assess the hemorrhagic risk. Their team also investigated the recognition of vascular cognitive impairment (VCI) in adult patients with MMD based on a dynamic resting-state functional connectivity (FC) network [116]. Specifically, they extracted features from constructed low-order and high-order FC networks, followed by sparse representation-based feature selection and final classification, revealing some possible VCI-related brain regions. In addition, real-time and intelligent image fusion in operation navigation is also a popular topic in terms of computer-aided treatment, especially for artery stenting or bypass surgery in ischemic CVDs.
Given the late development of CVDs and few studies in its ischemic subtype, some AI-based applications in cardiovascular diseases are introduced here due to the great similarity between cerebrovascular and cardiovascular diseases. As one of the major causes of cardiovascular diseases, coronary atherosclerosis is an inflammatory disease in which scleroses and obstructions flow through arterial blood vessels, and the presence of coronary artery calcium (CAC) assessed via CT is a direct marker of coronary atherosclerosis. Isgum, et al. [117] first applied ML techniques to coronary calcium detection and scoring from ungated, noncontrast chest CTs. Their following study [118] used a probabilistic coronary calcium map to infer the positions of coronary calcifications before feature extraction and classification. More recent ML approaches have enabled reliable and rapid automated Agatston score measurements from dedicated noncontrast calcium scoring CT scans [119, 120]. A two-stage DL framework was also utilized in several studies [121-125] for coronary calcification detection. For example, Lessmann, et al. [121] used three independently trained CNNs to estimate a bounding box around the heart in axial, coronal and sagittal slices in the first stage and then fed 2D patches from three orthogonal planes into three concurrent CNNs for final detection.
The detection and quantification of coronary stenosis is perhaps the most important clinical application of coronary medical imaging. Kelm, et al. [126] used an ML algorithm to automatically identify, grade and classify coronary stenosis caused by both calcified and noncalcified plaques on 229 CTA volumes. Similarly, Zreik, et al. [127] were the first to train a 3D-CNN paired with an RNN to accurately detect plaque, determine its composition, and classify coronary stenosis as obstructive or nonobstructive. More CTA-based methods, such as temporal constraint CNN [128] and DL radiomics [129], achieved reliable performance as well. In addition, studies on other modalities, such as intravascular optical coherence tomography (IVOCT) [130, 131] and single-photon emission computed tomography (SPECT) myocardial perfusion imaging (MPI) [132, 133], also showed promising results in detecting plaque or coronary artery disease with ML and DL techniques. Since segmentation can be regarded as high-level detection, multiple atherosclerosis-related segmentation methods have been proposed. Specifically, to segment four plaque tissues, Olender, M.L. et al. [134] presented a novel CNN-based domain enriched method that classifies arterial tissue imaged through IVUS pixelwise, while Shen, et al. [135] designed a multitask learning CNN combining three advanced multitask mechanisms. Other ML and DL studies on carotid vessel segmentation also obtained good performance [136-138].
The hemodynamic importance of given stenosis can also be estimated through AI approaches, especially when calculating fractional flow reserve (FFR), which is a functional index quantifying the severity of coronary artery lesions. For instance, a novel DL algorithm to calculate noninvasive FFR demonstrated similar accuracy and faster execution times compared to computationally expensive 3D flow simulations with invasive FFR reference [139] and was further validated in a multicenter study [140]. An unsupervised DL technique was also proposed in [141].
With regard to computer-aided prediction and treatment, AI has been used for ischemia or outcome prediction and image acquisition. Studies [142, 143] showed that combining clinical variables and quantitative plaque metrics on CTA improved the prediction of ischemia using ML. Two DL architectures, including traditional CNN and LSTM, were used for obstructive case prediction [144] and cardiovascular disease risk factor prediction [145], respectively. Moreover, ensemble-based classification approaches, such as LogitBoost, decision tree, and XGBoost, have been adopted in predicting the risk for all-cause mortality [146], myocardial infarction cardiac death [147], calcification [148] and early revascularization [149]. Regarding image acquisition, AI has also been applied to real-time detection and suppression of imaging artifacts using random forest [150] and residual-based U-net [151] architectures.
4. CURRENT CHALLENGES
Despite the recently reported successes of AI in medical imaging and its enormous potential in the clinical application in CVDs, multiple challenges need to be considered when building a technical model and giving the clinical interpretations discussed in the following subsections.
4.1. Questionable Reproducibility
In most AI-based applications of medical imaging, the reproducibility of the proposed method can be limited to variability across raw data processing, feature extraction or selection schemes, choice of classifier and so on. This phenomenon is more obvious in the majority of existing CVD-related literature, as there is considerable manual intervention in their proposed frameworks. For example, since there has not been a standard or commonly used approach for feature extraction in CVDs such as radiomics in tumors [152, 153], some features were designed for a specific problem on a specific dataset by humans [76], making it difficult to conduct reliable replication by other studies. Additionally, the nascent development of AI in CVDs has led to the wide implementation of rule-based methods, such as empirically setting thresholds, which also belong to manual intervention and improve poor reproducibility to a certain extent. To address this problem, on the one hand, some standard feature sets can be established through numerous studies across datasets, covering abundant features in morphology, hemodynamics and so on. On the other hand, the deep learning technique is a promising tool for automatically learning features and the underlying pattern, which means it does not require any prior knowledge provided by humans; thus, it can lower manual intervention and increase the reproducibility of a model.
4.2. Overfitting
Overfitting refers to a model that models the training data too well, resulting in good classification performance on the training data but poor performance on independent testing data [154]. This can be caused by utilizing models with a large number of features from a small sample size or complex models with many parameters [155, 156]. For CVDs, related medical imaging datasets have small sample sizes in general, which indicates that the corresponding model is susceptible to overfitting. It is noteworthy that the majority of the surveyed studies in this review did not have abundant data at the patient level, and even though some of them regarded scanning slices or sliding patches as input, the number of medical imaging samples was still far smaller than the number of natural images. Specifically, for models with DL algorithms, millions of auto extracted features and parameters also increase the probability of overfitting.
As a result, k-fold CV and regularization are routinely used to alleviate overfitting in CVD modeling. For instance, CV quantitatively validates the performance of the model using various groups of test subjects in a statistically robust manner, and regularization, such as the L2-norm, can smooth the decision boundary by restraining the parameter values. However, these strategies may not work effectively on DL models, and from the fundamental point of view, exploiting large quantities of training data is one of the key methods to address overfitting. Therefore, collecting numerous data from multiple centers and applying appropriate data augmentation strategies such as geometric transformation or generative adversarial network (GAN) are general methods for achieving this goal. In addition, some DL tactics, such as global average pooling (GAP) [157] and multitasking [158], are also thought to be useful for overfitting suppression.
4.3. Modality-specific Limitation
To enhance the performance of AI-based applications in CVDs, the aforementioned perspectives should be considered, and integrating complementary information from different imaging modalities is helpful. Typically, the existing CVD-related literature has utilized a single medical imaging modality to build the model, and even if multimodality has been mentioned in a few studies [35, 49], multimodal data were only used to test the generalization capability of their method across modalities without any fusion. Compared with the lower popularity in CVDs, multimodal techniques have been widely adopted in tumors [159, 160] and mental illness [161, 162] to overcome modality-specific limitations with complementary and reliable information, achieving higher accuracy than using a single modality.
Thus, the multimodal idea is expected to be extended to medical imaging in CVDs, as this kind of complex disease requires more comprehensive information. Taking MMD as an example, in addition to common modalities such as DSA and CTA, which reflect clear structural information of vessels, T2-weighted magnetic resonance imaging (T2W MRI) is also useful for identifying occlusive lesions around the circle of Willis and dilated moyamoya vessels [163, 164], and patterns of cerebral hemodynamics and metabolism can also be presented on positron emission computed tomography (PET) by measuring cerebral blood flow (CBF) [165] and cerebral perfusion pressure (CPP) [166]. Such rich information provided by multimodal imaging suggests that modality fusion methods combined with AI-based techniques point to a promising direction for improving CVD applications.
4.4. Challenge on Prospective Research
As mentioned previously, it is more significant to achieve computer-aided prediction and treatment than detection in CVDs, especially predicting the potential risk of stroke caused by CVDs. However, only a few studies covered shallow research in this field, making prospective research full of challenges and opportunities. For instance, although it is clear that SAH due to aneurysm rupture is one of the leading reasons for hemorrhagic stroke, only some shape information, such as the size of the aneurysm, has been used for rupture prediction in many studies based on the assumption of invariant size before and after rupture. Ischemic stroke may be caused by a transient ischemic attack (TIA) from MMD or occlusion from AS, but their risk prediction at present mainly depends on rating scales [167] involving age, duration of symptoms, diabetes and so on, lacking features extracted directly from medical images. With the power of AI algorithms, particularly DL methods, some latent and invisible features can be discovered from images as discriminative markers for prediction. In addition to stroke risk prediction, assistant decision-making, such as predicting the need for stenting, is a potential area for prospective research. Therefore, proposing and developing prospective research on CVD medical images will be a challenging but highly meaningful field.
5. FUTURE DIRECTIONS
Considering the current diagnosis and treatment situation in CVDs and the challenges mentioned above, some future directions are presented in the following section, providing various opportunities in this field.
5.1. Integration of Multitype Information
Medical imaging is not an isolated measure of disease, and integrating multitype information covering static and dynamic fields contributes to the high accuracy of computer-aided detection, prediction and treatment in CVDs. In detail, multimodal imaging affords complementary information through the intrinsic attributes of different imaging techniques. Therefore, extracting features from different perspectives can further diversify the information, which includes static morphological or textural features and hemodynamic features. For example, several studies have proposed that combining shape-based morphologic metrics with hemodynamic factors can improve the rupture risk assessment of aneurysms [168]. In addition, the patient’s clinical background, such as age, sex and medical history, also plays an important role because the performance of CVDs usually varies according to these factors. In addition, since AI is well suited to integrating parallel information streams, expanding the sources of information and integrating them properly can lead to a more comprehensive understanding of CVDs.
5.2. Validation on Multicenter Datasets
Instead of multicenter studies, most of the existing models in CVDs were established on single-center datasets, lacking reliability and robustness to a certain extent. On the one hand, a multicenter study can provide more data, leading to better model performance in the training phase. On the other hand, using multicenter data is more convincing when verifying the generalization and robustness of the model in the validation phase compared with single-center data. However, it is necessary to develop advanced normalization techniques to efficiently handle images acquired from multiple centers since they are cross regional, cross parametric, and even cross racial, and image differences caused by these reasons may be feature differences caused by the pathological characteristics. For routine imaging modalities in CVDs, it seems that image reconstruction in terms of intensity [169] and thickness [170] can serve as an effective approach to ensure the uniformity of datasets.
5.3. Improvement of Deep Learning
It has been proven in many tasks that DL methods have outperformed traditional machine learning algorithms in recent years, but there are two unresolved limitations: the demand for tremendous data and the lack of clinical interpretability. For the data problem, not only should the aforementioned multicenter validation and data normalization be considered, but some semisupervised and unsupervised techniques, such as GAN, are also required because large but unlabeled datasets are easier to collect [171]. Clinical interpretability mainly results from the highly automatic and black box-like characteristics of the DL model, limiting the ability to trace back successive weight layers to the original vascular data. Although the current state of DL research has prioritized performance gains over interpretability and transparency, attaining insight into the internal working mechanism and the extracted features is crucial for clinical explanatory ability. Currently, class activation maps (CAMs) [172, 173] demonstrate which areas of the image significantly participate in the classification, and feature visualization, such as heat map output from the convolutional layer or deconvolution method [174], are common tools for a clinical explanation. Other methods such as special attention maps have also been employed with DL [175, 176] in tumor-related studies.
CONCLUSION
AI-based algorithms on medical imaging have been prevalent in the past few years, but they are still a new field in CVDs due to the complexity of the diseases. Through a systematic review protocol, this paper not only elaborated on most of the recent work in three main applications, computer-aided detection, prediction and treatment of four representative diseases, including aneurysm, AVM, MMD and AS but also introduced the characteristics of algorithms adopted in the literature. Subsequently, the existing challenges and future directions were discussed in the paper, indicating the great potential in this field. Since this paper excluded a large number of search entries when selecting studies, there may be some related literature omitted, which is a limitation of this paper. But in order to achieve a scientific and convincing study in AI applications on CVDs, both model innovation and big data validation should be taken into account in the future. With the development of computational techniques and the further improvement of clinical systems in CVDs, it is believed that AI-based algorithms will shed light on the medical imaging analysis of CVDs.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
This work is granted by the National Natural Science Foundation of China (NSFC) (81801155 and 81771237), the New Technology Projects of Shanghai Science and Technology Innovation Action Plan (18511102800); and the Shanghai Municipal Science and Technology Major Project and ZJLab (2018SHZDZX01).
CONFLICT OF INTEREST
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- 1.Jauch E.C., Saver J.L., Adams H.P., Jr, Bruno A., Connors J.J., Demaerschalk B.M., Khatri P., McMullan P.W., Jr, Qureshi A.I., Rosenfield K., Scott P.A., Summers D.R., Wang D.Z., Wintermark M., Yonas H. Guidelines for the early management of patients with acute ischemic stroke: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2013;44(3):870–947. doi: 10.1161/STR.0b013e318284056a. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones D., Adams R.J., Brown T.M., Carnethon M., Dai S., De Simone G., Ferguson T.B., Ford E., Furie K., Gillespie C., Go A., Greenlund K., Haase N., Hailpern S., Ho P.M., Howard V., Kissela B., Kittner S., Lackland D., Lisabeth L., Marelli A., McDer-mott M.M., Meigs J., Mozaffarian D., Mussolino M., Nichol G., Roger V.L., Rosamond W., Sacco R., Sorlie P., Roger V.L., Thom T., Wasserthiel-Smoller S., Wong N.D., Wylie-Rosett J. Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121(7):e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 3.Roy K., Chaudhury S.S., Burman M., Ganguly A., Dutta C., Banik S., Banik R. A Comparative study of Lung Cancer detection using supervised neural network.; 2019 International Conference on Opto-Electronics and Applied Optics; Kolkata, India. March 18-20, 2019; pp. 1–5. [DOI] [Google Scholar]
- 4.Vas M., Dessai A. Lung cancer detection system using lung CT image processing.; 3rd International Conference on Computing, Commu-nication, Control and Automation; Pune, India. August 17-18, 2017; pp. 1–5. [DOI] [Google Scholar]
- 5.Moradi P., Jamzad M. Detecting lung cancer lesions in CT images using 3D convolutional neural networks.; 4th International Conference on Pattern Recognition and Image Analysis; Tehran, Iran. March 06-07, 2019; pp. 114–118. [DOI] [Google Scholar]
- 6.Wang D., Khosla A., Gargeya R., Irshad H., Beck A.H. Deep learning for identifying metastatic breast cancer. 2016.
- 7.Lin H., Chen H., Dou Q., Wang L., Heng P.A. ScanNet: A fast and dense scanning framework for metastastic breast cancer detection from whole-slide image.; 2018 IEEE Winter Conference on Applications of Computer Vision; Lake Tahoe, NV, USA. March 12-15, 2018; pp. 539–546. [DOI] [Google Scholar]
- 8.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A., Allison T., Arnaout O., Abbosh C., Dunn I.F., Mak R.H., Tamimi R.M., Tempany C.M., Swanton C., Hoffmann U., Schwartz L.H., Gillies R.J., Huang R.Y., Aerts H.J.W.L. Artificial in-telligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019;69(2):127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen W., Zhou M., Yang F., Yang C., Tian J. Multi-scale convolutional neural networks for lung nodule classification.; 24th Interna-tional Conference on Information processing in medical imaging; June 28- July 3, 2015; pp. 588–599. [DOI] [PubMed] [Google Scholar]
- 10.Wang Q., Zheng Y., Yang G., Jin W., Chen X., Yin Y. Multiscale rotation-invariant convolutional neural networks for lung texture classification. IEEE J. Biomed. Health Inform. 2018;22(1):184–195. doi: 10.1109/JBHI.2017.2685586. [DOI] [PubMed] [Google Scholar]
- 11.Jiang H., Ma H., Qian W., Gao M., Li Y., Hongyang Jiang. He Ma; Wei Qian; Mengdi Gao; Yan Li, An automatic detection system of lung nodule based on multigroup patch-based deep learning network. IEEE J. Biomed. Health Inform. 2018;22(4):1227–1237. doi: 10.1109/JBHI.2017.2725903. [DOI] [PubMed] [Google Scholar]
- 12.Li Z., Wang Y., Yu J., Guo Y., Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci. Rep. 2017;7(1):5467. doi: 10.1038/s41598-017-05848-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonekura A., Kawanaka H., Prasath V.B.S., Aronow B.J., Takase H. Improving the generalization of disease stage classification with deep CNN for glioma histopathological images.; 2017 IEEE International Conference on Bioinformatics and Biomedicine; Kansas City, USA. November 13-16, 2017; pp. 1222–1226. [DOI] [Google Scholar]
- 14.González S.R., Zemmoura I., Tauber C. Deep convolutional neural network to predict 1p19q co-deletion and IDH1 mutation status from MRI in low grade Gliomas.; 10th International Conference on Pattern Recognition Systems; Tours, France. 2019. [DOI] [Google Scholar]
- 15.Li Z., Wang Y., Yu J., Shi Z., Guo Y., Chen L., Mao Y. Low-grade glioma segmentation based on CNN with fully connected CRF. J. Healthc. Eng. 2017;2017:, 9283480. doi: 10.1155/2017/9283480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouridsen K., Thurner P., Zaharchuk G. Artificial intelligence applications in stroke. Stroke. 2020;51(8):2573–2579. doi: 10.1161/STROKEAHA.119.027479. [DOI] [PubMed] [Google Scholar]
- 17.Cover T.M., Hart P.E. Nearest neighbor pattern classification. IEEE Trans. Inf. Theory. 1967;13(1):21–27. doi: 10.1109/TIT.1967.1053964. [DOI] [Google Scholar]
- 18.Fisher R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936;7:179–188. doi: 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- 19.Burges C.J.C. A tutorial on Support Vector Machines for pattern recognition. Data Min. Knowl. Discov. 1998;2(2):121–167. doi: 10.1023/A:1009715923555. [DOI] [Google Scholar]
- 20.Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 21.Werbos P.J. Back propagation through time: what it does and how to do it. Proc. IEEE. 1990;78(10):1550–1560. doi: 10.1109/5.58337. [DOI] [Google Scholar]
- 22.Hartigan J.A., Wong M.A. Algorithm AS 136: A K-Means Clustering Algorithm. Appl. Stat. 1979;28(1):100–108. doi: 10.2307/2346830. [DOI] [Google Scholar]
- 23.Bezdek J., Jm K., Krisnapuram R., Pal N. Fuzzy models and algorithms for pattern recognition and image processing. Mass.: Kluwer Academic Publishers; 1999. [DOI] [Google Scholar]
- 24.Lecun Y., Bottou L., Bengio Y., Haffner P. Gradient-based learning applied to document recognition. Proc. IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 25.Ghosal P., Nandanwar L., Kanchan S., Bhadra A., Nandi D. Brain tumor classification using ResNet-101 based squeeze and excitation deep neural network.; 2019 Second International Conference on Advanced Computational and Communication Paradigms; Gangtok, India. February 25-28, 2019; pp. 1–6. [DOI] [Google Scholar]
- 26.Ronneberger O., Fischer P., Brox T. U-Net: Convolutional networks for biomedical image segmentation.; 18th International Conference on Medical Image Computing and Computer-Assisted Intervention; Munich, Germany. October 5-9, 2015; pp. 234–241. [DOI] [Google Scholar]
- 27.Sak H.i., Senior A., Rao K., Beaufays F.o. Fast and accurate recurrent neural network acoustic models for speech recognition.; 16th Annual Conference of the International-Speech-Communi-cation-Association; Dresden, Germany. September 6-10, 2015. [Google Scholar]
- 28.Adel H., Ngoc Thang V., Kraus F., Schlippe T., Li H., Schultz T. Recurrent neural netowrk language modeling for code switching con-versational speech.; 2013 IEEE International Conference on Acoustics, Speech Speech and Signal Processing; Beijing, China. July 6-10, 2013; pp. 8411–8415. [DOI] [Google Scholar]
- 29.Hochreiter S., Schmidhuber J. LSTM can solve hard long time lag problems.; 10th Annual Conference on Neural Information Processing Systems; Denver, USA. December 2-5, 1997; pp. 473–479. [Google Scholar]
- 30.A study of cross-validation and bootstrap for accuracy estimation and model selection. 14th International Joint Conference on Artificial Intelligence, 1995; Morgan Kaufmann: San Francisco, USA, 1995. 1995. p. 1137-1143.
- 31.Bandos A.I., Rockette H.E., Gur D. Subject-centered free-response ROC (FROC) analysis. Med. Phys. 2013;40(5):, 051706. doi: 10.1118/1.4799843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pham D.L., Xu C., Prince J.L. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 33.Uchiyama Y., Ando H., Yokoyama R., Hara T., Fujita H., Iwama T. Computer-aided diagnosis scheme for detection of unruptured intracranial aneurysms in MR angiography. 27th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Shanghai, China, January 17-18, 2006:pp. 3031- 3034. doi: 10.1109/IEMBS.2005.1617113. [DOI] [PubMed] [Google Scholar]
- 34.Arimura H., Li Q., Korogi Y., Hirai T., Katsuragawa S., Yamashita Y., Tsuchiya K., Doi K. Computerized detection of intracranial aneurysms for three-dimensional MR angiography: feature extraction of small protrusions based on a shape-based difference image tech-nique. Med. Phys. 2006;33(2):394–401. doi: 10.1118/1.2163389. [DOI] [PubMed] [Google Scholar]
- 35.Lauric A., Miller E., Frisken S., Malek A.M. Automated detection of intracranial aneurysms based on parent vessel 3D analysis. Med. Image Anal. 2010;14(2):149–159. doi: 10.1016/j.media.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Blezek D.J., Cheng L.T.E., Ryan W.J., Kallmes D.F., Erickson B.J. Computer-aided detection of intracranial aneurysms in MR angiography. J. Digit. Imaging. 2011;24(1):86–95. doi: 10.1007/s10278-009-9254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macía I., Graña M., Maiora J., Paloc C., de Blas M. Detection of type II endoleaks in abdominal aortic aneurysms after endovascular repair. Comput. Biol. Med. 2011;41(10):871–880. doi: 10.1016/j.compbiomed.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Nakao T., Hanaoka S., Nomura Y., Sato I., Nemoto M., Miki S., Maeda E., Yoshikawa T., Hayashi N., Abe O. Deep neural net-work-based computer-assisted detection of cerebral aneurysms in MR angiography. J. Magn. Reson. Imaging. 2018;47(4):948–953. doi: 10.1002/jmri.25842. [DOI] [PubMed] [Google Scholar]
- 39.Malik K.M., Anjum S.M., Soltanian-Zadeh H., Malik H., Malik G.J.I.A. ISADAQ: A framework for intracranial saccular aneurysm detection and quantification using morphological analysis of cerebral angiograms. IEEE Access. 2018;6:7970–7986. doi: 10.1109/ACCESS.2018.2799307. [DOI] [Google Scholar]
- 40.Rahmany I., Arfaoui B., Khlifa N., Megdiche H. Cerebral aneurysm computer-aided detection system by combing MSER, SURF and SIFT descriptors.; 5th International Conference on Control, Decision and Information Technologies; Thessaloniki, Greece. April 10-13, 2018; pp. 1122–1127. [DOI] [Google Scholar]
- 41.Chandra A., Mondal S. One novel algorithm for the detection of Cerebral Aneurysm using morphological filtering.; 2014 International Conference on Communications and Signal Processing; Melmaruvathur, India. April 3-5, 2014; pp. 137–141. [Google Scholar]
- 42.Sulayman N., Al-Mawaldi M., Kanafani Q. Semi-automatic detection and segmentation algorithm of saccular aneurysms in 2D cerebral DSA images. Egypt. J. Radiol. Nucl. Med. 2016;47(3):859–865. doi: 10.1016/j.ejrnm.2016.03.016. [DOI] [Google Scholar]
- 43.Otsu N. Threshold selection method from gray-level histogram. IEEE Trans. Syst. Man Cybern. 1979;9(1):62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 44.Kohout J., Chiarini A., Clapworthy G.J., Klajnšek G. Aneurysm identification by analysis of the blood-vessel skeleton. Comput. Methods Programs Biomed. 2013;109(1):32–47. doi: 10.1016/j.cmpb.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Suniaga S., Werner R., Kemmling A., Groth M., Forkert N.D. Computer-aided detection of aneurysms in 3D time-of-flight MRA da-taset.; 3rd International Workshop on Machine Learning in Medical Imaging; Nice, France. 2012. [DOI] [Google Scholar]
- 46.Jerman T., Pernus F., Likar B., Spiclin Z. Computer-aided detection and quantification of intracranial aneurysms.; 18th International Conference on Medical Image Computing and Computer-Assisted Intervention; Munich, Germany. October 5-9, 2015; pp. 3–10. [DOI] [Google Scholar]
- 47.Hentschke C.M., Beuing O., Nickl R., Toennies K.D. Automatic cerebral aneurysm detection in multimodal angiographic images.; 2011 IEEE Nuclear Science Symposium and Medical Imaging Conference; Valencia, Spain. October 23-29, 2011; pp. 3116–3120. [DOI] [Google Scholar]
- 48.Hentschke C.M., Tonnies K.D., Beuing O., Nickl R. A new feature for automatic aneurysm detection. 9th IEEE International Symposium on Biomedical Imaging, Barcelona, Spain May 2-5,; 2012. p. pp. 800-803. [DOI] [Google Scholar]
- 49.Hentschke C.M., Beuing O., Paukisch H., Scherlach C., Skalej M., Tönnies K.D. A system to detect cerebral aneurysms in multimodal-ity angiographic data sets. Med. Phys. 2014;41(9):, 091904. doi: 10.1118/1.4890775. [DOI] [PubMed] [Google Scholar]
- 50.Arimura H., Li Q., Korogi Y., Hirai T., Yamashita Y., Katsuragawa S., Ikeda R., Doi K. CAD scheme for detection of intracranial aneurysms in MRA based on 3D analysis of vessel skeletons and enhanced aneurysms. Medical Imaging 2005 Conference, San Diego, USA February 15-17; 2005. p. pp. 967-974. [Google Scholar]
- 51.Frangi A.F., Niessen W.J., Vincken K.L., Viergever M.A. Multiscale vessel enhancement filtering.; 1st International Conference on Med-ical Image Computing and Computer-Assisted Intervention; Cambridge, MA., USA. October 11-13, 1998; pp. 130–137. [Google Scholar]
- 52.Hanaoka S., Nomura Y., Nemoto M., Miki S., Yoshikawa T., Hayashi N., Ohtomo K., Masutani Y., Shimizu A. HoTPiG: A novel geometrical feature for vessel morphometry and its application to cerebral aneurysm detection 18th International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany October 5-9,; 2015. p. pp. 103-110. [DOI] [Google Scholar]
- 53.Rahmany I., Khlifa N. Detection of intracranial aneurysm in angiographic images using fuzzy approaches. 1st International Image Processing, Applications and Systems Conference, Sfax, Tunisia Novmber 5-7; 2014. [DOI] [Google Scholar]
- 54.Geng C., Xia W., Huang L., Yang L.Q., Li Y.X., Dai Y.K., Geng D.Y. Automated computer-assisted detection system for cerebral aneurysms in time-of-flight magnetic resonance angiography using fully convolutional network. Biomed. Eng. Online. 2020;19(1):38. doi: 10.1186/s12938-020-00770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park A., Chute C., Rajpurkar P., Lou J., Ball R.L., Shpanskaya K., Jabarkheel R., Kim L.H., McKenna E., Tseng J., Ni J., Wishah F., Wittber F., Hong D.S., Wilson T.J., Halabi S., Basu S., Patel B.N., Lungren M.P., Ng A.Y., Yeom K.W. Deep learning-assisted di-agnosis of cerebral aneurysms using the HeadXNet model. JAMA Netw. Open. 2019;2(6):, e195600. doi: 10.1001/jamanetworkopen.2019.5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sichtermann T., Faron A., Sijben R., Teichert N., Freiherr J., Wiesmann M. Deep learning-based detection of intracranial aneurysms in 3D TOF-MRA. AJNR Am. J. Neuroradiol. 2019;40(1):25–32. doi: 10.3174/ajnr.A5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shahzad R., Pennig L., Goertz L., Thiele F., Kabbasch C., Schlamann M., Krischek B., Maintz D., Perkuhn M., Borggrefe J. Fully automated detection and segmentation of intracranial aneurysms in subarachnoid hemorrhage on CTA using deep learning. Sci. Rep. 2020;10(1):21799. doi: 10.1038/s41598-020-78384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Z., Miao C., Schoepf U.J., Savage R.H., Dargis D.M., Pan C., Chai X., Li X.L., Xia S., Zhang X., Gu Y., Zhang Y., Hu B., Xu W., Zhou C., Luo S., Wang H., Mao L., Liang K., Wen L., Zhou L., Yu Y., Lu G.M., Zhang L.J. A clinically applicable deep-learning model for detecting intracranial aneurysm in computed tomography angiography images. Nat. Commun. 2020;11(1):6090. doi: 10.1038/s41467-020-19527-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joo B., Ahn S.S., Yoon P.H., Bae S., Sohn B., Lee Y.E., Bae J.H., Park M.S., Choi H.S., Lee S.K. A deep learning algorithm may automate intracranial aneurysm detection on MR angiography with high diagnostic performance. Eur. Radiol. 2020;30(11):5785–5793. doi: 10.1007/s00330-020-06966-8. [DOI] [PubMed] [Google Scholar]
- 60.Dai X.L., Huang L.X., Qian Y., Xia S., Chong W., Liu J.J., Di Ieva A., Hou X.X., Ou C.B. Deep learning for automated cerebral aneu-rysm detection on computed tomography images. Int. J. CARS. 2020;15(4):715–723. doi: 10.1007/s11548-020-02121-2. [DOI] [PubMed] [Google Scholar]
- 61.Jerman T., Pernus F., Likar B., Spiclin Z. Aneurysm detection in 3D cerebral angiograms based on intra-vascular distance mapping and convolutional neural networks.; 14th IEEE International Symposium on Biomedical Imaging; Melbourne, Australia. April 18-21, 2017; pp. 612–615. [DOI] [Google Scholar]
- 62.Podgorsak A.R., Rava R.A., Shiraz Bhurwani M.M., Chandra A.R., Davies J.M., Siddiqui A.H., Ionita C.N. Automatic radiomic feature extraction using deep learning for angiographic parametric imaging of intracranial aneurysms. J. Neurointerv. Surg. 2020;12(4):417–421. doi: 10.1136/neurintsurg-2019-015214. [DOI] [PubMed] [Google Scholar]
- 63.Rahmany I., Guetari R., Khlifa N. A fully automatic based deep learning approach for aneurysm detection in DSA images.; 3rd IEEE International Conference on Image Processing, Applications and Systems; Sophia Antipolis, France. December 12-14, 2018; pp. 303–307. [DOI] [Google Scholar]
- 64.Ueda D., Yamamoto A., Nishimori M., Shimono T., Doishita S., Shimazaki A., Katayama Y., Fukumoto S., Choppin A., Shimahara Y., Miki Y. Deep learning for MR angiography: automated detection of cerebral aneurysms. Radiology. 2019;290(1):187–194. doi: 10.1148/radiol.2018180901. [DOI] [PubMed] [Google Scholar]
- 65.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the Inception Architecture for Computer Vision. 29th IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, USA June 27-30, 2016:pp. 2818-2826. doi: 10.1109/CVPR.2016.308. [DOI] [Google Scholar]
- 66.He K.M., Zhang X.Y., Ren S.Q., Sun J. Deep residual learning for image recognition.; 29th IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, USA. June 27-30, 2016; pp. 770–778. [Google Scholar]
- 67.Jin H., Geng J., Yin Y., Hu M., Yang G., Xiang S., Zhai X., Ji Z., Fan X., Hu P., He C., Qin L., Zhang H. Fully automated intracra-nial aneurysm detection and segmentation from digital subtraction angiography series using an end-to-end spatiotemporal deep neural network. J. Neurointerv. Surg. 2020;12(10):1023–1027. doi: 10.1136/neurintsurg-2020-015824. [DOI] [PubMed] [Google Scholar]
- 68.Duan H.H., Huang Y.Z., Liu L.X., Dai H.M., Chen L.Y., Zhou L.X. Automatic detection on intracranial aneurysm from digital subtrac-tion angiography with cascade convolutional neural networks. Biomed. Eng. Online. 2019;18(1):110. doi: 10.1186/s12938-019-0726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stember J.N., Chang P., Stember D.M., Liu M., Grinband J., Filippi C.G., Meyers P., Jambawalikar S. Convolutional neural networks for the detection and measurement of cerebral aneurysms on magnetic resonance angiography. J. Digit. Imaging. 2019;32(5):808–815. doi: 10.1007/s10278-018-0162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng Y., Liu X., Xiao N., Li Y., Jiang Y., Feng J., Guo S. Automatic diagnosis based on spatial information fusion feature for intra-cranial aneurysm. IEEE Trans. Med. Imaging. 2020;39(5):1448–1458. doi: 10.1109/TMI.2019.2951439. [DOI] [PubMed] [Google Scholar]
- 71.Hahn S., Morris C.S., Bertges D.J., Wshah S. Deep learning for recognition of endoleak after endovascular abdominal aortic aneurysm repair.; 16th IEEE International Symposium on Biomedical Imaging; Venice, Italy. April 8-11, 2019; pp. 759–763. [DOI] [Google Scholar]
- 72.Yang X., Xia D., Kin T., Igarashi T. IntrA: 3D intracranial aneurysm dataset for deep learning.; 2020 IEEE Conference on Computer Vision and Pattern Recognition; June 14-19, 2020; pp. 2653–2663. [Google Scholar]
- 73.Ye H., Gao F., Yin Y., Guo D., Zhao P., Lu Y., Wang X., Bai J., Cao K., Song Q., Zhang H., Chen W., Guo X., Xia J. Precise di-agnosis of intracranial hemorrhage and subtypes using a three-dimensional joint convolutional and recurrent neural network. Eur. Radiol. 2019;29(11):6191–6201. doi: 10.1007/s00330-019-06163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danilov G., Kotik K., Negreeva A., Tsukanova T., Shifrin M., Zakharova N., Batalov A., Pronin I., Potapov A. Classification of intracranial hemorrhage subtypes using deep learning on CT scans. Stud. Health Technol. Inform. 2020;272:370–373. doi: 10.3233/SHTI200572. [DOI] [PubMed] [Google Scholar]
- 75.Cho J., Park K.S., Karki M., Lee E., Ko S., Kim J.K., Lee D., Choe J., Son J., Kim M., Lee S., Lee J., Yoon C., Park S. Improving sensitivity on identification and delineation of intracranial hemorrhage lesion using cascaded deep learning models. J. Digit. Imaging. 2019;32(3):450–461. doi: 10.1007/s10278-018-00172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lahmiri S., Boukadoum M., Ieva A.D. Detrended fluctuation analysis of brain hemisphere magnetic resonnance images to detect cere-bral arteriovenous malformations.; 2014 IEEE International Symposium on Circuits and Systems; Melbourne, Australia. June 1-5, 2014; pp. 2409–2412. [DOI] [Google Scholar]
- 77.Lahmiri S., Boukadoum M., Di Ieva A. Fractal-Based Arteriovenous Malformations Detection in Brain Magnetic Resonance Images.; 12th IEEE International New Circuits and Systems Conference; Trois-Rivieres. June 22-25, 2014; pp. 21–24. [DOI] [Google Scholar]
- 78.Babin D., Spyrantis M., Pizurica A., Philips W. Skeleton calculation for automatic extraction of arteriovenous malformation in 3-D CTA images.; 11th IEEE International Symposium on Biomedical Imaging; Beijing, China. April 29- May 2, 2014; pp. 425–428. [DOI] [Google Scholar]
- 79.Babin D., Spyrantis M., Pizurica A., Philips W., Velicki L., Zlokolica V. Pixel profiling for extraction of arteriovenous malformation in 3-D CTA images.; 56th ELMAR International Symposium; Zadar, Croatia. September 10-12, 2014; pp. 183–186. [DOI] [Google Scholar]
- 80.Lian Y.X., Wang Y.Y., Yu J.H., Guo Y., Chen L. Segmentation of Arteriovenous Malformations Nidus and Vessel in Digital Subtrac-tion Angiography Images Based on an Iterative Thresholding Method.; 8th International Conference on Biomedical Engineering and In-formatics; Shenyang, China. October 14-16, 2015; pp. 111–115. [DOI] [Google Scholar]
- 81.Peng S.J., Lee C.C., Wu H.M., Lin C.J., Shiau C.Y., Guo W.Y., Pan D.H.C., Liu K.D., Chung W.Y., Yang H.C. Fully automated tis-sue segmentation of the prescription isodose region delineated through the Gamma knife plan for cerebral arteriovenous malformation (AVM) using fuzzy C-means (FCM) clustering. Neuroimage Clin. 2019;21:, 101608. doi: 10.1016/j.nicl.2018.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee C.C., Yang H.C., Lin C.J., Chen C.J., Wu H.M., Shiau C.Y., Guo W.Y., Hung-Chi Pan D., Liu K.D., Chung W.Y., Peng S.J. In-tervening nidal brain parenchyma and rsk of radiation-induced changes after radiosurgery for brain arteriovenous malformation: A study using an unsupervised machine learning algorithm. World Neurosurg. 2019;125:e132–e138. doi: 10.1016/j.wneu.2018.12.220. [DOI] [PubMed] [Google Scholar]
- 83.Wang T.H., Lei Y., Tian S.B., Jiang X.J., Zhou J., Liu T., Dresser S., Curran W.J., Shu H.K., Yang X.F. Learning-based automatic segmentation of arteriovenous malformations on contrast CT images in brain stereotactic radiosurgery. Med. Phys. 2019;46(7):3133–3141. doi: 10.1002/mp.13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wardlaw J.M., White P.M. The detection and management of unruptured intracranial aneurysms. Brain. 2000;123(Pt 2):205–221. doi: 10.1093/brain/123.2.205. [DOI] [PubMed] [Google Scholar]
- 85.Juvela S. Treatment options of unruptured intracranial aneurysms. Stroke. 2004;35(2):372–374. doi: 10.1161/01.STR.0000115299.02909.68. [DOI] [PubMed] [Google Scholar]
- 86.Suarez J.I., Tarr R.W., Selman W.R.J.S. Aneurysmal subarachnoid hemorrhage. N. Engl. J. Med. 2006;354(4):387–396. doi: 10.1056/NEJMra052732. [DOI] [PubMed] [Google Scholar]
- 87.Morita A., Kirino T., Hashi K., Aoki N., Fukuhara S., Hashimoto N., Nakayama T., Sakai M., Teramoto A., Tominari S., Yoshimo-to T., Investigators U.J. The natural course of unruptured cerebral aneurysms in a Japanese cohort. N. Engl. J. Med. 2012;366(26):2474–2482. doi: 10.1056/NEJMoa1113260. [DOI] [PubMed] [Google Scholar]
- 88.Ujiie H., Tamano Y., Sasaki K., Hori T. Is the aspect ratio a reliable index for predicting the rupture of a saccular aneurysm? Neurosurgery. 2001;48(3):495–502. doi: 10.1097/00006123-200103000-00007. [DOI] [PubMed] [Google Scholar]
- 89.Kongable G.L., Lanzino G., Germanson T.P., Truskowski L.L., Alves W.M., Torner J.C., Kassell N.F. Gender-related differences in aneurysmal subarachnoid hemorrhage. J. Neurosurg. 1996;84(1):43–48. doi: 10.3171/jns.1996.84.1.0043. [DOI] [PubMed] [Google Scholar]
- 90.Chen G., Lu M., Shi Z., Xia S., Ren Y., Liu Z., Liu X., Li Z., Mao L., Li X.L., Zhang B., Zhang L.J., Lu G.M. Development and validation of machine learning prediction model based on computed tomography angiography-derived hemodynamics for rupture status of intracranial aneurysms: a Chinese multicenter study. Eur. Radiol. 2020;30(9):5170–5182. doi: 10.1007/s00330-020-06886-7. [DOI] [PubMed] [Google Scholar]
- 91.Liu J., Chen Y., Lan L., Lin B., Chen W., Wang M., Li R., Yang Y., Zhao B., Hu Z., Duan Y. Prediction of rupture risk in anterior communicating artery aneurysms with a feed-forward artificial neural network. Eur. Radiol. 2018;28(8):3268–3275. doi: 10.1007/s00330-017-5300-3. [DOI] [PubMed] [Google Scholar]
- 92.Ou C.B., Liu J.H., Qian Y., Chong W., Zhang X., Liu W.C., Su H., Zhang N., Zhang J., Duan C., He X. Rupture risk assessment for cerebral aneurysm using interpretable machine learning on multidimensional data. Front. Neurol. 2020;11:, 570181. doi: 10.3389/fneur.2020.570181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He H., Bai Y., Garcia E.A., Li S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. 2008 IEEE International Joint Conference on Neural Networks, Hong Kong, China. 2008:pp. 1322-1328. [Google Scholar]
- 94.de Jong G.A., Aquarius R. Use of artificial neural networks to predict anterior communicating artery aneurysm rupture: possible method-ological considerations. Eur. Radiol. 2019;29(5):2724–2726. doi: 10.1007/s00330-018-5794-3. [DOI] [PubMed] [Google Scholar]
- 95.Tang B., He H. KernelADASYN: Kernel based adaptive synthetic data generation for imbalanced learning.; IEEE Congress on Evolution-ary Computation; Sendai, Japan. May 25-28 2015; pp. 664–671. [DOI] [Google Scholar]
- 96.Shi Z., Chen G.Z., Mao L., Li X.L., Zhou C.S., Xia S., Zhang Y.X., Zhang B., Hu B., Lu G.M., Zhang L.J. Machine Learning-Based Prediction of Small Intracranial Aneurysm Rupture Status Using CTA-Derived Hemodynamics: A Multicenter Study. AJNR Am. J. Neuroradiol. 2021;42(4):648–654. doi: 10.3174/ajnr.A7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lauric A., Miller E.L., Baharoglu M.I., Malek A.M. Rupture status discrimination in intracranial aneurysms using the centroid-radii model. IEEE Trans. Biomed. Eng. 2011;58(10):2895–2903. doi: 10.1109/TBME.2011.2162410. [DOI] [PubMed] [Google Scholar]
- 98.Niemann U., Berg P., Niemann A., Beuing O., Preim B., Spiliopoulou M., Saalfeld S. Rupture status classification of intracranial aneu-rysms using morphological parameters.; 31st IEEE International Symposium on Computer-Based Medical Systems; Karlstad. June 18-21, 2018; pp. 48–53. [DOI] [Google Scholar]
- 99.Kim H.C., Rhim J.K., Ahn J.H., Park J.J., Moon J.U., Hong E.P., Kim M.R., Kim S.G., Lee S.H., Jeong J.H., Choi S.W., Jeon J.P. Machine learning application for rupture risk assessment in small-sized intracranial aneurysm. J. Clin. Med. 2019;8(5):, E683. doi: 10.3390/jcm8050683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Q., Jiang P., Jiang Y., Ge H., Li S., Jin H., Li Y. Prediction of aneurysm stability using a machine learning model based on PyRa-diomics-derived morphological features. Stroke. 2019;50(9):2314–2321. doi: 10.1161/STROKEAHA.119.025777. [DOI] [PubMed] [Google Scholar]
- 101.Paliwal N., Jaiswal P., Tutino V.M., Shallwani H., Davies J.M., Siddiqui A.H., Rai R., Meng H. Outcome prediction of intracranial aneurysm treatment by flow diverters using machine learning. Neurosurg. Focus. 2018;45(5):, E7. doi: 10.3171/2018.8.FOCUS18332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.de Toledo P., Rios P.M., Ledezma A., Sanchis A., Alen J.F., Lagares A. Predicting the outcome of patients with subarachnoid hemor-rhage using machine learning techniques. IEEE Trans. Inf. Technol. Biomed. 2009;13(5):794–801. doi: 10.1109/TITB.2009.2020434. [DOI] [PubMed] [Google Scholar]
- 103.Xia N.Z., Chen J., Zhan C.Y., Jia X.F., Xiang Y.L., Chen Y.C., Duan Y.X., Lan L., Lin B.L., Chen C., Zhao B., Chen X.Y., Yang Y.J., Liu J. Prediction of clinical outcome at discharge after rupture of anterior communicating artery aneurysm using the Random Forest Technique. Front. Neurol. 2020;11:, 538052. doi: 10.3389/fneur.2020.538052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong J.S., Lin C.J., Lin Y.H., Lee C.C., Yang H.C., Meng L.H., Lin T.M., Hu Y.S., Guo W.Y., Chu W.F., Wu Y.T. Machine learning application with quantitative digital subtraction angiography for detection of hemorrhagic brain arteriovenous malformations. IEEE Access. 2020;8:204573–204584. doi: 10.1109/ACCESS.2020.3036692. [DOI] [Google Scholar]
- 105.Asadi H., Kok H.K., Looby S., Brennan P., O’Hare A., Thornton J. Outcomes and complications after endovascular treatment of brain arteriovenous malformations: A prognostication attempt using artificial intelligence. World Neurosurg. 2016;96:562–569.e1. doi: 10.1016/j.wneu.2016.09.086. [DOI] [PubMed] [Google Scholar]
- 106.Oermann E.K., Rubinsteyn A., Ding D., Mascitelli J., Starke R.M., Bederson J.B., Kano H., Lunsford L.D., Sheehan J.P., Hammer-bacher J., Kondziolka D. Using a machine learning approach to predict outcomes after radiosurgery for cerebral arteriovenous malfor-mations. Sci. Rep. 2016;6:21161. doi: 10.1038/srep21161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhou Y.J., Xie X.L., Hou Z.G., Bian G.B., Zhou X.H.F.R.R-N.E.T. Fast recurrent residual networks for real-time catheter segmentation and tracking in endovascular aneurysm repair.; 17th IEEE International Symposium on Biomedical Imaging; April 3-7, 2020; pp. 961–964. [DOI] [Google Scholar]
- 108.Gao W., Zhang J. Estimation of arteriosclerosis based on fuzzy support vector machine.; 1st International Symposium on Computer Net-work and Multimedia Technology; Wuhan, China. January 18-20, 2009; pp. 1–4. [DOI] [Google Scholar]
- 109.Terrada O., Cherradi B., Raihani A., Bouattane O. Atherosclerosis disease prediction using supervised machine learning techniques.; 1st International Conference on Innovative Research in Applied Science, Engineering and Technology; Meknes, Morocco. April 16-19, 2020; pp. 1–5. [Google Scholar]
- 110.Ding L., Zhou R., Liu G. Study on the classification algorithm of degree of arteriosclerosis based on fuzzy pattern recognition.; Interna-tional Conference on Image Processing and Pattern Recognition in Industrial Engineering; Xian, China. August 7-8, 2010; [DOI] [Google Scholar]
- 111.Zhang Y., Pan J. Arteriosclerosis diagnosis based on support vector machine.; 5th International Conference on Information Science and Control Engineering; Zhengzhou, China. July 20-22, 2018; pp. 138–142. [Google Scholar]
- 112.Kim T., Heo J., Jang D-K., Sunwoo L., Kim J., Lee K.J., Kang S-H., Park S.J., Kwon O.K., Oh C.W. Machine learning for detecting moyamoya disease in plain skull radiography using a convolutional neural network. EBioMedicine. 2019;40:636–642. doi: 10.1016/j.ebiom.2018.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Akiyama Y., Mikami T., Mikuni N. Deep learning-based approach for the diagnosis of moyamoya disease. J. Stroke Cerebrovasc. Dis. 2020;29(12):, 105322. doi: 10.1016/j.jstrokecerebrovasdis.2020.105322. [DOI] [PubMed] [Google Scholar]
- 114.Hu T., Lei Y., Su J., Yang H., Ni W., Gao C., Yu J., Wang Y., Gu Y. Learning spatiotemporal features of DSA using 3D CNN and BiConvGRU for moyamoya disease detection. Int. J. Neurosci. 2021:1–14. doi: 10.1080/00207454.2021.1929214. [DOI] [PubMed] [Google Scholar]
- 115.Lei Y., Zhang X., Ni W., Yang H., Su J.B., Xu B., Chen L., Yu J.H., Gu Y.X., Mao Y. Recognition of moyamoya disease and its hemorrhagic risk using deep learning algorithms: sourced from retrospective studies. Neural Regen. Res. 2021;16(5):830–835. doi: 10.4103/1673-5374.297085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lei Y., Chen X., Su J.B., Zhang X., Yang H., Gao X.J., Ni W., Chen L., Yu J.H., Gu Y.X., Mao Y. Recognition of cognitive impair-ment in adult moyamoya disease: A classifier based on high-order resting-state functional connectivity network. Front. Neural Circuits. 2020;14:, 603208. doi: 10.3389/fncir.2020.603208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Isgum I., Rutten A., Prokop M., van Ginneken B. Detection of coronary calcifications from computed tomography scans for automated risk assessment of coronary artery disease. Med. Phys. 2007;34(4):1450–1461. doi: 10.1118/1.2710548. [DOI] [PubMed] [Google Scholar]
- 118.Isgum I., Prokop M., Niemeijer M., Viergever M.A., van Ginneken B. Automatic coronary calcium scoring in low-dose chest computed tomography. IEEE Trans. Med. Imaging. 2012;31(12):2322–2334. doi: 10.1109/TMI.2012.2216889. [DOI] [PubMed] [Google Scholar]
- 119.Martin S.S., van Assen M., Rapaka S., Hudson H.T., Jr, Fischer A.M., Varga-Szemes A., Sahbaee P., Schwemmer C., Gulsun M.A., Cimen S., Sharma P., Vogl T.J., Schoepf U.J. Evaluation of a deep learning-based automated CT coronary artery calcium scoring algo-rithm. JACC Cardiovasc. Imaging. 2020;13(2 Pt 1):524–526. doi: 10.1016/j.jcmg.2019.09.015. [DOI] [PubMed] [Google Scholar]
- 120.Wolterink J.M., Leiner T., Takx R.A.P., Viergever M.A., Isgum I. Automatic coronary calcium scoring in non-contrast-enhanced ECG-triggered cardiac CT with ambiguity detection. IEEE Trans. Med. Imaging. 2015;34(9):1867–1878. doi: 10.1109/TMI.2015.2412651. [DOI] [PubMed] [Google Scholar]
- 121.Lessmann N., Isgum I., Setio A.A.A., de Vos B.D., Ciompi F., de Jong P.A., Oudkerk M., Mali W.P.T.M., Viergever M.A., van Ginneken B. Deep convolutional neural networks for automatic coronary calcium scoring in a screening study with low-dose chest CT. Medical Imaging 2016: Computer-Aided Diagnosis, San Diego, United States February 28-March. 2016.
- 122.Lessmann N., van Ginneken B., Zreik M., de Jong P.A., de Vos B.D., Viergever M.A., Isgum I. Automatic calcium scoring in low-dose chest CT using deep neural networks with dilated convolutions. IEEE Trans. Med. Imaging. 2018;37(2):615–625. doi: 10.1109/TMI.2017.2769839. [DOI] [PubMed] [Google Scholar]
- 123.Li Y.C., Shen T.Y., Chen C.C., Chang W.T., Lee P.Y., Huang C.C. Automatic detection of atherosclerotic plaque and calcification from intravascular ultrasound images by using deep convolutional neural networks. IEEE Trans. Ultrason. Ferroelectr. Freq. Control, 2021. [DOI] [PubMed]
- 124.Wolterink J.M., Leiner T., de Vos B.D., van Hamersvelt R.W., Viergever M.A., Išgum I. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med. Image Anal. 2016;34:123–136. doi: 10.1016/j.media.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 125.Zhang N., Yang G., Zhang W., Wang W., Zhou Z., Zhang H., Xu L., Chen Y. Fully automatic framework for comprehensive coronary artery calcium scores analysis on non-contrast cardiac-gated CT scan: Total and vessel-specific quantifications. Eur. J. Radiol. 2021;134:, 109420. doi: 10.1016/j.ejrad.2020.109420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kelm B.M., Mittal S., Zheng Y., Tsymbal A., Bernhardt D., Vega-Higuera F., Zhou S.K., Meer P., Comaniciu D. Detection, grading and classification of coronary stenoses in computed tomography angiography.; 14th International Conference on Medical Image Compu-ting and Computer-Assisted Intervention; Toronto, Canada. September 18-22, 2011; p. 25. [DOI] [PubMed] [Google Scholar]
- 127.Zreik M., van Hamersvelt R.W., Wolterink J.M., Leiner T., Viergever M.A., Isgum I. A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans. Med. Imaging. 2019;38(7):1588–1598. doi: 10.1109/TMI.2018.2883807. [DOI] [PubMed] [Google Scholar]
- 128.Wu W., Zhang J., Xie H., Zhao Y., Zhang S., Gu L. Automatic detection of coronary artery stenosis by convolutional neural network with temporal constraint. Comput. Biol. Med. 2020;118:, 103657. doi: 10.1016/j.compbiomed.2020.103657. [DOI] [PubMed] [Google Scholar]
- 129.Denzinger F., Wels M., Ravikumar N., Breininger K., Reidelshoefer A., Eckert J., Suehling M., Schmermund A., Maier A. Coronary artery plaque characterization from CCTA scans using deep learning and radiomics.; 22nd International Conference on Medical Image Computing and Computer-Assisted Intervention; Shenzhen, China. October 13-17, 2019; pp. 593–601. [DOI] [Google Scholar]
- 130.Gessert N., Lutz M., Heyder M., Latus S., Leistner D.M., Abdelwahed Y.S., Schlaefer A. Automatic plaque detection in IVOCT pull-backs using convolutional neural networks. IEEE Trans. Med. Imaging. 2019;38(2):426–434. doi: 10.1109/TMI.2018.2865659. [DOI] [PubMed] [Google Scholar]
- 131.Oliveira D.A.B., Macedo M.M.G., Nicz P., Campos C., Lemos P., Gutierrez M.A. Coronary calcification identification in Optical Coherence Tomography using convolutional neural networks. SPIE Conference on Medical Imaging - Biomedical Applications in Molecular, Structural, and Functional Imaging, Houston, USA February 11-13, 2018 doi: 10.1117/12.2293753. [DOI] [Google Scholar]
- 132.Arsanjani R., Xu Y., Dey D., Vahistha V., Shalev A., Nakanishi R., Hayes S., Fish M., Berman D., Germano G., Slomka P.J. Im-proved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J. Nucl. Cardiol. 2013;20(4):553–562. doi: 10.1007/s12350-013-9706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Arsanjani R., Xu Y., Hayes S.W., Fish M., Lemley M., Jr, Gerlach J., Dorbala S., Berman D.S., Germano G., Slomka P. Comparison of fully automated computer analysis and visual scoring for detection of coronary artery disease from myocardial perfusion SPECT in a large population. J. Nucl. Med. 2013;54(2):221–228. doi: 10.2967/jnumed.112.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Olender M.L., Athanasiou L.S., Michalis L.K., Fotiadis D.I., Edelman E.R. A domain enriched deep learning approach to classify ath-erosclerosis using intravascular ultrasound imaging. IEEE J. Sel. Top. Signal Process. 2020;14(6):1210–1220. doi: 10.1109/JSTSP.2020.3002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shen H., Zhang W., Wang H., Ding G., Xie J. NDDR-LCS: A multi-task learning method for classification of carotid plaques.; 2020 IEEE International Conference on Image Processing; Abu Dhabi, United Arab Emirates. October 25-28, 2020; pp. 2461–2465. [DOI] [Google Scholar]
- 136.Jiang M., Spence J.D., Chiu B. Segmentation of 3D ultrasound carotid vessel wall using U-Net and segmentation average network. 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Montreal, Canada July 20-24, 2020:pp. 2043-2046. doi: 10.1109/EMBC44109.2020.9175975. [DOI] [PubMed] [Google Scholar]
- 137.Tsakanikas V.D., Siogkas P.K., Mantzaris M.D., Potsika V.T., Kigka V.I., Exarchos T.P., Koncar I.B., Jovanovic M., Vujcic A., Ducic S., Pelisek J., Fotiadis D.I. A deep learning oriented method for automated 3D reconstruction of carotid arterial trees from MR imaging. 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Montreal, Canada July 20-24, 2020:pp. 2408-2411. doi: 10.1109/EMBC44109.2020.9176532. [DOI] [PubMed] [Google Scholar]
- 138.Zheng Y., Loziczonek M., Georgescu B., Zhou S.K., Vega-Higuera F., Comaniciu D. Machine learning based vesselness measurement for coronary artery segmentation in cardiac CT volumes. Conference on Medical Imaging 2011 - Image Processing, Lake Buena Vista, FL, USA February 14-16, 2011 doi: 10.1117/12.877233. [DOI] [Google Scholar]
- 139.Itu L., Rapaka S., Passerini T., Georgescu B., Schwemmer C., Schoebinger M., Flohr T., Sharma P., Comaniciu D. A machine-learning approach for computation of fractional flow reserve from coronary computed tomography. J. Appl. Physiol. 2016;121(1):42–52. doi: 10.1152/japplphysiol.00752.2015. [DOI] [PubMed] [Google Scholar]
- 140.Coenen A., Kim Y-H., Kruk M., Tesche C., De Geer J., Kurata A., Lubbers M.L., Daemen J., Itu L., Rapaka S., Sharma P., Schwemmer C., Persson A., Schoepf U.J., Kepka C., Hyun Yang D., Nieman K. Diagnostic accuracy of a machine-learning approach to coronary computed tomographic angiography-based fractional flow reserve result from the MACHINE consortium. Circ. Cardiovasc. Imaging. 2018;11(6):, e007217. doi: 10.1161/CIRCIMAGING.117.007217. [DOI] [PubMed] [Google Scholar]
- 141.Zreik M., van Hamersvelt R.W., Khalili N., Wolterink J.M., Voskuil M., Viergever M.A., Leiner T., Isgum I. Deep learning analysis of coronary arteries in cardiac CT angiography for detection of patients requiring invasive coronary angiography. IEEE Trans. Med. Imaging. 2020;39(5):1545–1557. doi: 10.1109/TMI.2019.2953054. [DOI] [PubMed] [Google Scholar]
- 142.Dey D., Gaur S., Ovrehus K.A., Slomka P.J., Betancur J., Goeller M., Hell M.M., Gransar H., Berman D.S., Achenbach S., Botker H.E., Jensen J.M., Lassen J.F., Norgaard B.L. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angi-ography using machine learning: a multicentre study. Eur. Radiol. 2018;28(6):2655–2664. doi: 10.1007/s00330-017-5223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang S., Koo B-K., Hoshino M., Lee J.M., Murai T., Park J., Zhang J., Hwang D., Shin E-S., Doh J-H., Nam C-W., Wang J., Chen S., Tanaka N., Matsuo H., Akasaka T., Choi G., Petersen K., Chang H-J., Kakuta T., Narula J. CT angiographic and plaque predictors of functionally significant coronary disease and outcome using machine learning. JACC Cardiovasc. Imaging. 2021;14(3):629–641. doi: 10.1016/j.jcmg.2020.08.025. [DOI] [PubMed] [Google Scholar]
- 144.Betancur J., Commandeur F., Motlagh M., Sharir T., Einstein A.J., Bokhari S., Fish M.B., Ruddy T.D., Kaufmann P., Sinusas A.J., Miller E.J., Bateman T.M., Dorbala S., Di Carli M., Germano G., Otaki Y., Tamarappoo B.K., Dey D., Berman D.S., Slomka P.J. Deep learning for prediction of obstructive disease from fast myocardial perfusion SPECT a multicenter study. JACC Cardiovasc. Imaging. 2018;11(11):1654–1663. doi: 10.1016/j.jcmg.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Islam M.D.S., Umran H.M., Umran S.M., Karim M. Intelligent Healthcare Platform: Cardiovascular Disease Risk Factors Prediction Using Attention Module Based LSTM.; 2nd International Conference on Artificial Intelligence and Big Data; Chengdu, China. May 25-28, 2019; pp. 167–175. [DOI] [Google Scholar]
- 146.Motwani M., Dey D., Berman D.S., Germano G., Achenbach S., Al-Mallah M.H., Andreini D., Budoff M.J., Cademartiri F., Callister T.Q., Chang H-J., Chinnaiyan K., Chow B.J.W., Cury R.C., Delago A., Gomez M., Gransar H., Hadamitzky M., Hausleiter J., Hin-doyan N., Feuchtner G., Kaufmann P.A., Kim Y-J., Leipsic J., Lin F.Y., Maffei E., Marques H., Pontone G., Raff G., Rubinshtein R., Shaw L.J., Stehli J., Villines T.C., Dunning A., Min J.K., Slomka P.J. Machine learning for prediction of all-cause mortality in pa-tients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur. Heart J. 2017;38(7):500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Commandeur F., Slomka P.J., Goeller M., Chen X., Cadet S., Razipour A., McElhinney P., Gransar H., Cantu S., Miller R.J.H., Rozanski A., Achenbach S., Tamarappoo B.K., Berman D.S., Dey D. Machine learning to predict the long-term risk of myocardial in-farction and cardiac death based on clinical risk, coronary calcium, and epicardial adipose tissue: a prospective study. Cardiovasc. Res. 2020;116(14):2216–2225. doi: 10.1093/cvr/cvz321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee J., Kam H.J., Kim H-Y., Yoo S., Woo K-G., Choi Y-H., Park J.E., Cho S.J. 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Osaka, Japan July 3-7. 2013. Prediction of 4-year risk for Coronary Artery Calcification using Ensemble-based Classification. pp. 3210–3213. [DOI] [PubMed] [Google Scholar]
- 149.Arsanjani R., Dey D., Khachatryan T., Shalev A., Hayes S.W., Fish M., Nakanishi R., Germano G., Berman D.S., Slomka P. Predic-tion of revascularization after myocardial perfusion SPECT by machine learning in a large population. J. Nucl. Cardiol. 2015;22(5):877–884. doi: 10.1007/s12350-014-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lorch B., Vaillant G., Baumgartner C., Bai W., Rueckert D., Maier A. Automated detection of motion artefacts in MR imaging using decision forests. J. Med. Eng. 2017;2017:, 4501647. doi: 10.1155/2017/4501647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hauptmann A., Arridge S., Lucka F., Muthurangu V., Steeden J.A. Real-time cardiovascular MR with spatio-temporal artifact suppres-sion using deep learning-proof of concept in congenital heart disease. Magn. Reson. Med. 2019;81(2):1143–1156. doi: 10.1002/mrm.27480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J-C., Pieper S., Aerts H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Narang S., Lehrer M., Yang D., Lee J., Rao A. Radiomics in glioblastoma: current status, challenges and potential opportunities. Transl. Cancer Res. 2016;5(4):383–397. doi: 10.21037/tcr.2016.06.31. [DOI] [Google Scholar]
- 154.Pereira F., Mitchell T., Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. Neuroimage. 2009;45(1) Suppl.:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Franke K., Ziegler G., Klöppel S., Gaser C., Alzheimers Dis Neuroimaging I. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50(3):883–892. doi: 10.1016/j.neuroimage.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 156.Frost C., Kallis C., Chu C., Draganski B., Scahill R.I., Rohrer J.D., Fox N.C., Ashburner J., Frackowiak R.S.J. Reply: A plea for con-fidence intervals and consideration of generalizability in diagnostic studies. Brain. 2009;132(Pt 4):, e103. doi: 10.1093/brain/awn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin M., Chen Q., Yan S. Network In Network.; 2014 International Conference on Learning Representations; Banff, Canada. 2013. [Google Scholar]
- 158.Zhang Y., Yang Q. A Survey on Multi-Task Learning. 2017. https://arxiv.org/pdf/1707.08114.pdf
- 159.Chaddad A., Desrosiers C., Abdulkarim B., Niazi T. Predicting the gene status and survival outcome of lower grade glioma patients with multimodal MRI features. IEEE Access. 2019;7:75976–75984. doi: 10.1109/ACCESS.2019.2920396. [DOI] [Google Scholar]
- 160.Peng Y., Bi L., Guo Y., Feng D., Fulham M., Kim J. Deep multi-modality collaborative learning for distant metastases predication in PET-CT soft-tissue sarcoma studies. 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Berlin July 23-27. 2019:3685–3688. doi: 10.1109/EMBC.2019.8857666. [DOI] [PubMed] [Google Scholar]
- 161.Liu S., Liu S., Cai W., Che H., Pujol S., Kikinis R., Feng D., Fulham M.J. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans. Biomed. Eng. 2015;62(4):1132–1140. doi: 10.1109/TBME.2014.2372011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Calhoun V.D., Sui J. Multimodal fusion of brain imaging data: A key to finding the missing link(s) in complex mental illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2016;1(3):230–244. doi: 10.1016/j.bpsc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yamada I., Suzuki S., Matsushima Y. Moyamoya disease: diagnostic accuracy of MRI. Neuroradiology. 1995;37(5):356–361. doi: 10.1007/BF00588011. [DOI] [PubMed] [Google Scholar]
- 164.Harada A., Fujii Y., Yoneoka Y., Takeuchi S., Tanaka R., Nakada T. High-field magnetic resonance imaging in patients with mo-yamoya disease. J. Neurosurg. 2001;94(2):233–237. doi: 10.3171/jns.2001.94.2.0233. [DOI] [PubMed] [Google Scholar]
- 165.Ogawa A., Yoshimoto T., Suzuki J., Sakurai Y. Cerebral blood flow in moyamoya disease. Part 1: Correlation with age and regional distribution. Acta Neurochir. (Wien) 1990;105(1-2):30–34. doi: 10.1007/BF01664854. [DOI] [PubMed] [Google Scholar]
- 166.Kuroda S., Houkin K., Kamiyama H., Abe H., Mitsumori K. Regional cerebral hemodynamics in childhood moyamoya disease. Childs Nerv. Syst. 1995;11(10):584–590. doi: 10.1007/BF00300997. [DOI] [PubMed] [Google Scholar]
- 167.Galvin R., Geraghty C., Motterlini N., Dimitrov B.D., Fahey T. Prognostic value of the ABCD² clinical prediction rule: a systematic review and meta-analysis. Fam. Pract. 2011;28(4):366–376. doi: 10.1093/fampra/cmr008. [DOI] [PubMed] [Google Scholar]
- 168.Xiang J., Natarajan S.K., Tremmel M., Ma D., Mocco J., Hopkins L.N., Siddiqui A.H., Levy E.I., Meng H. Hemodynamic-morphologic discriminants for intracranial aneurysm rupture. Stroke. 2011;42(1):144–152. doi: 10.1161/STROKEAHA.110.592923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gao Y., Liu Y., Wang Y., Shi Z., Yu J. A universal intensity standardization method based on a many-to-one weak-paired cycle genera-tive adversarial network for magnetic resonance images. IEEE Trans. Med. Imaging. 2019;38(9):2059–2069. doi: 10.1109/TMI.2019.2894692. [DOI] [PubMed] [Google Scholar]
- 170.Gu J., Li Z., Wang Y., Yang H., Qiao Z., Yu J. Deep generative adversarial networks for thin-section infant image reconstruction. IEEE Access. 2019;7:68290–68304. doi: 10.1109/ACCESS.2019.2918926. [DOI] [Google Scholar]
- 171.Esteva A., Robicquet A., Ramsundar B., Kuleshov V., DePristo M., Chou K., Cui C., Corrado G., Thrun S., Dean J. A guide to deep learning in healthcare. Nat. Med. 2019;25(1):24–29. doi: 10.1038/s41591-018-0316-z. [DOI] [PubMed] [Google Scholar]
- 172.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Learning deep features for discriminative localization.; 2016 IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, USA. June 27-30, 2016; pp. 2921–2929. [Google Scholar]
- 173.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Grad-CAM: Visual explanations from deep networks via gradi-ent-based localization.; 2017 IEEE International Conference on Computer Vision; Venice, Italy. October 22-29, 2017; pp. 618–626. [DOI] [Google Scholar]
- 174.Zeiler M.D., Fergus R. Visualizing and understanding convolutional networks.; 13th European Conference on Computer Vision; Zurich, Switzerland. September 6-12, 2014; pp. 818–833. [Google Scholar]
- 175.Broen M.P.G., Smits M., Wijnenga M.M.J., Dubbink H.J., Anten M.H.M.E., Schijns O.E.M.G., Beckervordersandforth J., Postma A.A., van den Bent M.J. The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro-oncol. 2018;20(10):1393–1399. doi: 10.1093/neuonc/noy048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Banerjee S., Mitra S., Shankar B.U., Hayashi Y. A novel GBM saliency detection model using multi-channel MRI. PLoS One. 2016;11(1):, e0146388. doi: 10.1371/journal.pone.0146388. [DOI] [PMC free article] [PubMed] [Google Scholar]


