Abstract
Mitochondrial disorders are clinically heterogeneous, resulting from nuclear gene and mitochondrial mutations that disturb the mitochondrial functions and dynamics. There is a lack of evidence linking mtDNA mutations to neurodegenerative disorders, mainly due to the absence of noticeable neuropathological lesions in postmortem samples. This review describes various gene mutations in Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis, multiple sclerosis, and stroke. These abnormalities, including PINK1, Parkin, and SOD1 mutations, seem to reveal mitochondrial dysfunctions due to either mtDNA mutation or deletion, the mechanism of which remains unclear in depth.
Keywords: Alzheimer's disease, amyotrophic lateral sclerosis, mtDNA, multiple sclerosis, neurodegeneration, Parkinson's disease, stroke
1. INTRODUCTION
Mitochondria are known as the powerhouse of cells. The mitochondrial respiratory chain plays a significant role in oxidative phosphorylation and the mitochondrial genes associated with the chain. Mitochondrial DNA (mtDNA), a 16.5 kb circular DNA, is inherited from the mother [1]. Any impairment or damage to mtDNA can shed the possibility of abnormal protein accumulations leading to various mitochondrial diseases. Aging produces functional impairment of the mitochondrial respiratory chain in various tissues, including the central nervous system, leading to abnormalities in oxidative energy production [1-3]. Mitochondria is crucially involved in various intracellular functions, such as biosynthesis of lipids, calcium signaling, and apoptosis [4], and these processes are involved in various neurodegenerative diseases [5, 6]. Mitochondrial dysfunction is now an emerging area of study in aging as well as neurodegeneration. A large proportion of the mitochondrial dysfunction is associated with aging resulting from the mtDNA mutation accumulation due to a constant replication cycle. In individual cells, the mutations happen to be unique and accumulate, leading to cell function impairment [1]. Several copies of mtDNA are initiated. Some of the copies may have mutations known as heteroplasmy, which accumulates during life, suggesting that mtDNA mutations associated with aging dysfunction can be due to an interaction of genetic and acquired mtDNA damage [1, 7].
Neurodegenerative disorders involve diseases that cause neuronal cells' dysfunction, producing devastating effects, and are still a challenging area with limited treatment available. The most common neurodegenerative diseases are Alzheimer's Disease (AD), Parkinson's Disease (PD), Amyotrophic Lateral Sclerosis (ALS), Huntington's Diseases (HD), and Multiple sclerosis (MS), which have various pathophysiological etiology. Recent studies highlight the role of mitochondrial dysfunction in promoting neurodegeneration or other mechanisms, including apoptosis, mitophagy, and autophagy. Mitochondrial dysfunction may occur due to mutation in mitochondrial DNA (mtDNA) and its association with the mutation of various genes contributing to neurodegenerative disorders [8].
2. MITOCHONDRIAL STRUCTURE, PHYSIOLOGY, GENETICS, AND PATHOPHYSIOLOGY
2.1. Mitochondrial Structure and Physiology
Mitochondria are subcellular organelles with a double membrane. They are autonomous, primitive as well as similar to bacteria [9]. More than 90% of cellular energy in the human body is produced by oxidative phosphorylation in the mitochondria [10]. Apart from oxidative phosphorylation [11], mitochondria are involved in other functions such as maintenance of calcium homeostasis and inorganic cofactor iron-sulfur clusters, Reactive Oxygen Species (ROS) generation and signaling, metabolism of lipid in close association with the Endoplasmic Reticulum (ER), and a role in membrane donation whereby dysfunctional/damaged mitochondria will be subjected to cell death or transported to lysosomes. Mitochondria is a double-membrane structure which consists of (i) an outer membrane having lesser protein content and being far more permeable than the inner membrane, (ii) an inner membrane, having a higher percentage of protein content and impermeability to polar molecules, and (iii) a matrix in the form of cristae providing a large surface area for the proton gradient and oxidative phosphorylation maintenance (Fig. 1) [12]. Substances of small molecular weight can permeate the outer membrane. The mitochondrial respiratory chain enzymes are seen in the inner membrane, which is highly folded and segregates the intermembrane space and the mitochondrial matrix. The mitochondrial genome is present in the matrix and the enzymes associated with the tricarboxylic acid cycle and oxidation of fatty acids. The inner mitochondrial membrane consists of cristae to have a high surface area between enzymes of the respiratory chain and biochemical substrates present in the matrix. The mitochondrial inner membrane is almost impermeable except oxygen, water, and carbon dioxide [9]. Mitochondrial oxidative phosphorylation produces ATP from ADP and depends on multi-subunit Complexes I to VI encoded by nuclear and mitochondrial genes except for Complex II, entirely coded by nuclear genes [9].
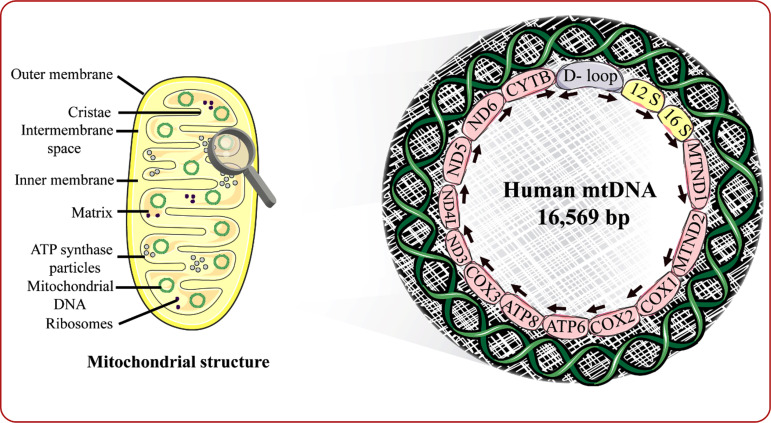
Fig. (1).
Schematic representation of mitochondrial structure and genome. D (Displacement) loop indicates the non-coding region, and the arrows represent the path of transcription of mtDNA; 12S and 16S RNA indicates the two ribosomal RNAs; MTND genes 1 and 2 represent ND genes 1 and 2 in combination with mitochondrial tRNA; COX1, COX2, COX3 represent cytochrome C oxidase subunits 1-3; ND3, ND4L, ND5, ND6 represent the subunits of NADH dehydrogenase, and CYTB represents the gene coding for cytochrome B. (A) mtDNA non-coding region: D loop; (B) Encoding of complex I, III, IV, V subunits. Complex I: MTND1, MTND2, ND3, ND4, ND4L, ND5, ND6. complex III: Cytochrome B. complex IV: COX1, COX2, COX3. complex V: ATPase 6 and ATPase 8.
Mitochondria are found in almost all the cells, including the neuronal cells, and produce necessary proteins for carrying out various physiological effects within the body, including the brain tissue, with mitochondrial DNA (mtDNA). Any dysfunction of mtDNA or aberration in the accumulation of proteins can result in mitochondrial diseases affecting various systems of the body, such as the cardiovascular system, central nervous system, urinary and endocrine, causing neurodegenerative disorders, movement disorders, muscle weakness, cardiovascular diseases, renal dysfunction, and endocrine disorders [13]. Successive oxidoreduction happens, transporting electrons along the respiratory chain to oxygens producing an electrochemical gradient by proton extrusion. This gradient drives the synthesis of Complex VI (ATP synthase). Sometimes mitochondria form a network inside the cells, and there occur a continuous fusion and fission of mitochondria. Mitochondrial disease occurs when the above continuous mitochondrial dynamics are disturbed, and there can be an aggregation of mutations [14].
2.2. Mitochondrial Genome
The mtDNA was discovered in 1963. It is seen in the mitochondrial matrix as supercoiled, double-stranded, and circular molecule. It shows association with proteins producing nucleotide, mtDNA polymerase γ, mtDNA binding helicases, proteins, and mtDNA transcription promoting factors [15,16]. It is seen as multiple copies, usually 2-10 mtDNA in a mitochondrion and thousand to a hundred thousand copies in a cell. The mitochondrial genome has 16,569 base pairs and is the only kind of extrachromosomal DNA seen in humans, as illustrated in Fig. (1).
The mtDNA was first sequenced in 1981 and is known as the Cambridge reference sequence and was late revised to correct the errors due to bovine fragments in that project [17,18]. The mtDNA comprises a heavy H-strand, a light L-strand, and a non-coding control area (D-loop). The H-strand is rich in Guanine, and the L-strand is rich in Cytosine. An individual cell contains about 100-10,000 mitochondria, and every mitochondrion consists of two to ten mtDNA copies [19]. The mitochondrial genome encodes 37 genes; out of 28 are located on the H-strand and 9 located on the L-strand. The mitochondrial genome codes for 13 respiratory chain subunits and the genetic information needed for 22 transfer RNAs (tRNA) and two ribosomal RNAs (rRNA) (12 S rRNA, a small ribosomal subunit, and a 16 S rRNA, a large ribosomal subunit), essential for the intramitochondrial sequence of the proteins [9].
Most of the mitochondrial genes are adjacent and may be separated by one or a couple of base pairs that are non-coding. About 93% of mtDNA encodes proteins. A significant non-coding area is in the displacement loop with the mtDNA replication initiation site [18]. Nuclear genes code major mitochondrial proteins, which form various respiratory chain subunits, and those proteins essential for mtDNA maintenance [9]. The mutations in nuclear genes can affect the proteins associated with the replication and repair of mtDNA, significantly affecting the mitochondrial genome. The mitochondrial genome is vulnerable to various damages, and the rate of mutations is about ten times more than that of the nuclear genes. In mtDNA, the protective histones are absent, and the repair mechanisms are also somewhat limited. The mtDNA is also vulnerable to nucleolytic assaults by free radicals generated in oxidative phosphorylation. The mtDNA is entirely composed of exons and has very little redundancy. A deletion or a point mutation in the mtDNA can easily and quickly translate into a biochemical defect [9].
2.3. Replication, Transcription, and Translation of mtDNA
The mitochondrial non-coding control area, D-loop, is about 1.1 kb and consists of vital sequences for the beginning of replication and transcription. The mtDNA replication is asynchronous, starting from the two points OL and OH [20]. The RNA primer produced from the light chain starts mtDNA replication at heavy chain origin, seen in D-loop. A nuclear gene synthesizes the strand of DNA coded DNA polymerase γ [9]. The light chain origin is a non-coding area where tRNA genes are present. It is exposed while the heavy strand is passing through the area approximately two-thirds on its route around the genome, initiating replicating the L-strand in the opposite route. A strand-coupled model of replication of mtDNA proposes that both strands are simultaneously synthesized [21]. The L-strand synthesis begins soon after heavy strand replication initiation and includes extensive RNA synthesis before the synthesis of DNA. The debate is ongoing regarding the predominant mtDNA replication, and there can be a replication of different forms in various tissues [9]. The mtDNA transcription begins from L and H-strand promoters producing polycistronic transcripts later undergoing processing, generating tRNA, mRNA, and rRNA molecules. The activity needs mitochondrial RNA polymerase TFAM, a transcription activator, and B2 or B1 mitochondrial transcription factors [22]. The mtDNA translation mechanism is not yet completely understood. The control is exerted by proteins coded by nuclear genes, such as two particular mitochondrial initiation factors, 3 elongation factors, and a minimum of one termination release factor [23-26].
2.4. Mutations of mtDNA
The initial pathogenic mutations of mtDNA were elucidated in 1988. Later, more than 300 mutations of mtDNA were discovered (Mitomap) [27]. The mutations can be point mutations or deletions as well as duplications. These mutations can produce a biochemical dysfunction either by protein synthesis if a disruption of rRNA or tRNA sequence occurs or alter respiratory chain function if a gene that codes for the subunit is altered [9].
2.4.1. Germline Variation
The mtDNA tends to undergo mutation. Mutagenesis can produce mutations that can be neutral or pathogenic and can produce primary mitochondrial disease, depending on the kind of mutation [28, 29]. The neutral mutations and the non-pathogenic protein-changing variants can form stable homoplasmic polymorphisms seen in populations segregated by common sequence variation called haplogroups [30, 31]. The initial mtDNA haplogroups were identified in the native Americans and were named by letters A, B, C, and D [32]. All alphabetic letters except O have been utilized with various sub-haplogroups [33]. In neurodegenerative studies, the association with mtDNA haplogroups was studied for AD [34-36], PD [37-39], ALS [40], stroke [41], and Frontotemporal Dementia (FTD) [42].
2.4.2. Somatic mtDNA Mutation
2.4.2.1. Point Mutations
Various intracellular events, including ROS, nucleases, and impulsive hydrolytic processes, can produce mtDNA distraction. The replication of mtDNA showing the nature of a single strand and the absence of histone protein make it vulnerable to damage [43]. The oxidative damage measured by 7,8 dihydro-8-oxo-deoxyguanosine (8-oxo dG) produces G-C to T-A transversion mutation [44]. These are not consistent with transitional mutations frequently occurring in the aged mammalian brain [45]. The mice showing DNA polymerase γ (POLG) mutations can produce transitional germline mutations [46, 47] as well as somatic point mutations similarly seen in the human tissues that are aged [45]. The impairment in replication can be an initial event in the generation of somatic point mutation instead of oxidative damage. The inefficient maintenance of mtDNA can also lead to mtDNA point mutations. The replication errors of mtDNA can lead to mtDNA somatic point mutations [1].
2.4.2.2. The mtDNA Deletions
The mtDNA single-strand breaks can form due to destruction mediated by ROS or aberrant mtDNA replication. These are identified by poly (ADP ribose) Polymerase Proteins (PARP) [48]. The repair utilizes an identical machine and process to base excision repair (BER) [49]. The Double-Strand Breaks (DSB) mechanism is yet to be understood [50]. A DSB misrepair is suspected to be producing deletion in mtDNA and can be observed in human tissues undergone aging [51].
2.4.3. Heteroplasmy and Homoplasmy
Homoplasmy indicates identical mtDNA in a cell. Heteroplasmy indicates a combination of wild-type and mutant mtDNA in a cell. The mtDNA replication and the somatic mutagenesis can facilitate the clonal expansion of the mutations via random drift and selective processes. They can form in different proportions of heteroplasmy within the cells [52]. Next-generation sequencing methods resulted in the identification of universal mtDNA heteroplasmy in tissues [53]. These indicate mutations earlier thought to be somatic and present de novo in the CNS and other tissues can be due to heteroplasmic variant clonal expansion, which could not be identified with other sequencing technologies [7, 53, 54]. In mice with POLG mutation, heteroplasmic variants which are low level and inherited can undergo clonal expansion through generations worsening aging producing neurodevelopmental anomalies [7].
2.4.4. Somatic Mutations in Aged Tissues
High-resolution observation of cytochrome C oxidase (COX) functional impairment was observed [55, 56]. Inside these COX impaired cells, point mutations and mtDNA deletions that are clonally expanded were of high frequency and were believed to be associated with the biochemical deficiency [57]. Accretion of mtDNA mutations reaches a biochemical threshold effect that produces functional impairment of the mitochondrial respiratory chain [57-59]. The wild-type molecules cannot make up for the mutant-type cellular mtDNA. Various research analyzing the mtDNA mutation association with mitochondrial function studied mitochondrial complex activity [60, 44], the expression of the subunit [61, 60], oxidative phosphorylation as well as the synthesis of ATP [62, 63], and clinical phenotype [29]. Studies suggested that a high concentration of heteroplasmy consisting of mtDNA (about 60-90%) is needed for functional impairment within a cell. Heteroplasmy in high concentration is usually tolerated before producing aberration in cellular activity [64]. The mtDNA shows disproportionate transcription and gene mutation of structural complex subunit gives proportionate mutated mRNA [65]. A heteroplasmy of high concentration is also tolerated in tRNA genes before the occurrence of respiratory chain impairment [66]. A high degree of heteroplasmy is required for affecting the protein translation [60]. Even in a lowered subunit expression, the enzymatic activity of mitochondria can continue as normal [60]. Even if there is functional impairment of one complex, the mitochondrial respiratory chain conserves oxidative phosphorylation and synthesis of ATP [62,63] via respiratory chain compositional or organisational alterations [64].
2.4.5. Brain mtDNA Mutations
2.4.5.1. Deletions
In a healthy aged brain, mtDNA4977, a base pair deletion is seen lower than 2.5% [67-71] but is relatively higher than the muscle and the heart [72]. The mtDNA4977 levels can also be associated with brain locations as substantia nigra shows 2.9% (69), frontal cortex 0.2% [69], cerebellum less than 0.001% and temporal cortex 0.0092% [73]. The fetal brain does not show mtDNA deletions, indicating that most mtDNA deletions can be de novo with later clonal expression [67]. It is also believed that a healthy aged brain can have mtDNA deletions which are unique in comparison to those observed in neurodegenerative disorders, such as AD and PD. A recent study of mtDNA deletions suggested that a healthy brain shows about 5% of D-loop removing and 20% of origin of L-strand replication deletions, and those deletions above 20% show 3' breakpoint between 16,000 and 16,100 bases [74]. The D-loop removing mtDNA deletion is not seen in PD. The clear nature of the breakpoints, their concentration mechanism of occurrence, and the mtDNA mutation associations with neurodegenerative diseases need to be comprehensively analyzed in further studies.
2.5. Mitoepigenetics in Neurodegenerative Disorders
Many researchers who have been working on role of mitochondrial impairment in neurodegenerative disorders have come to a conjecture that mitoepigenetics possibly have a role in neurodegeneration [75]. A report by Sharma et al. concludes that mitochondrial dysfunction in the form of altered gene expression and ATP production, resulting from epigenetic changes, can contribute to age-related neurodegenerative disorders, including AD [76]. Few preclinical as well as clinical studies on AD, PD and ALS, revealed the impairment of D-loop methylation level [75].
Cerebellum, as well as Superior and Middle Temporal Gyrus (SMTG) of seven late onset AD patients, showed elevation in 5 hydroxymethylated (5-hmC) cytosine residues in mtDNA, which seemed to be less significant [77]. However, the entorhinal cortex of eight AD patients showed a marked increase in methylated (5-mC) cytosine residues in the D-loop of mtDNA with a higher degree of methylation in the early stages of the disease and a reduction in MT-ND1 methylation levels [78]. Another study conducted on 133 late-onset AD patients blood samples showed a marked decrease in 5-mC in mtDNA D-loop [79]. Amalgamating data obtained from all these studies can benefit the characterisation of various stages of AD by assessing the degree of methylation in mtDNA regions [75]. In a study conducted on ten PD patients, the substantia nigra region showed a decrease in 5-mC levels in mtDNA D-loop [78]. Preclinical studies in spinal cord motor neurons and brain mitochondria, as well as clinical studies in cortical motor neurons, reveals an elevation in DNA methyltransferase 1 (DNMT1) and global 5-mC level in ALS mice and patients, respectively [80]. In contrast to AD, D-loop hypomethylation was observed in pre-symptomatic ALS mice as well as patients. Additionally, an elevation in methylation of the 16S rRNA gene and DNMT3A were observed in the ALS-linked SOD1 mutants indicating the role of the antioxidant enzyme, Superoxide Dismutase 1 (SOD1) counteracting the damage due to oxidative stress [75].
2.6. Role of Sirtuins in Neurodegeneration
Sirtuins (SIRTs), a histone deacetylases (HDACs) member, is known to have an association with aging and neurodegenerative diseases by means of interaction with various Transcription Factors (TFs), poly (ADP-ribose) polymerases (PARPs), and signaling proteins. SIRT1, located mainly in the nucleus, is also found in mitochondria and studies reveal its role in mitochondrial biogenesis, neuroprotective via peroxisome proliferator-activated receptor γ co-activator-1α (PGC-1α), impairment of which can contribute to neurodegenerative disorders such as AD, PD and HD [81]. Mitochondrial SIRTs, SIRT3-5 are mainly found in the mitochondrial matrix and its expression is interdependent. For example, the expression of SIRT5 within mitochondria decreases with an increase in SIRT3 level. SIRT3 can enhance thioredoxin 2, peroxiredoxins, Mn-SOD via FOXO3 and activation of PINK1 potentiates mitophagy and mitochondrial fusion. Apart from influencing antioxidant defenses within mitochondria, SIRTs (SIRT1-7) are also involved in the mitigation of aging by reducing oxidative stress, and restoration of Insulin/IGF Signaling (IIS) for modulating longevity and stress resistance via various signaling pathways, including JNK1, Akt, IIS-mTor signaling, FOXO1, NRF-2, NF-κB, and cross-talk of SIRT-HIF [81].
In AD, it was observed that SIRT1 plays a neuroprotective role by balancing APP processing via NF-κB inhibition and activation of the Notch pathway, thereby contributing to neurogenesis and differentiation. Contrarily an increased SIRT2 level can contribute to the pathophysiology of neurodegenerative disorders, including AD [81]. In vitro studies reveal a proportionate increase of SIRT3,4,5 with Aβ1-42 and inverse correlation of presenilin and APP [82, 83]. The role of SIRTs in coordination with IIS and FOXOs can improve longevity and aging-associated neurodegenerative disorders. However, there is a lacunae of in-depth knowledge of IIS association in AD and need to be further investigated [81]. Analogous to AD, SIRT1 shows neuroprotection in PD and ALS models via PGC-1α activation, and Heat Shock Factor 1 (HSF1) deacetylation, respectively. Studies show that SIRT2 inhibition can improve defective behavioural and neurological characteristics in MPTP-induced aged PD mice models [81, 84].
3. NEURODEGENERATIVE DISORDERS AND THEIR ASSOCIATION WITH MITOCHONDRIAL DYSFUNCTION
3.1. Alzheimer's Disease
AD is the most commonly observed neurodegenerative disorder that progresses in aging [8]. AD's pathophysiology can be explained through various hypotheses such as amyloid, tau, cholinergic, and metal ion hypothesis [85, 86]. The former two explain the aggregation of Aβ and tau (τ) proteins, respectively, which results in neurofibrillary tangles or plaque formation, causing neuronal death. On the other hand, the metal ion hypothesis causes an imbalance in the metal ions that causes an elevation in both aggregations of Aβ and τ protein, leading to plaque formation, all of which contributes to memory impairment. On the other hand, the cholinergic hypothesis emphasizes decreased acetylcholine release capable of memory impairment. All these hypotheses can be interlinked with mitochondrial dysfunction and play either primary secondary role in AD progression. According to a recent article elucidating the molecular mechanism involved in zinc (Zn) neurotoxicity by Narayanan et al., metal ions like Zn can cause cell death either by Aβ-Zn complex formation leading to Aβ induced neurotoxicity or via various mechanisms within the postsynaptic neurons, including τ phosphorylation in serine 214, ROS production within mitochondria, and ERK pathway induced neurofibrillary tangle formation [86]. Genomic studies suggest the role of mutation of various variants or genes, including Amyloid Precursor Protein (APP), apolipoprotein E4 (APOE4), clusterin (CLU), phosphatidylinositol binding clathrin assembly protein (PICALM), presenilin 1 and 2 (PSEN1, PSEN2) genes, SORL1, BIN1, CR1, ABI3, PLCG2, and TREM2 in causation or progress of AD [87, 88].
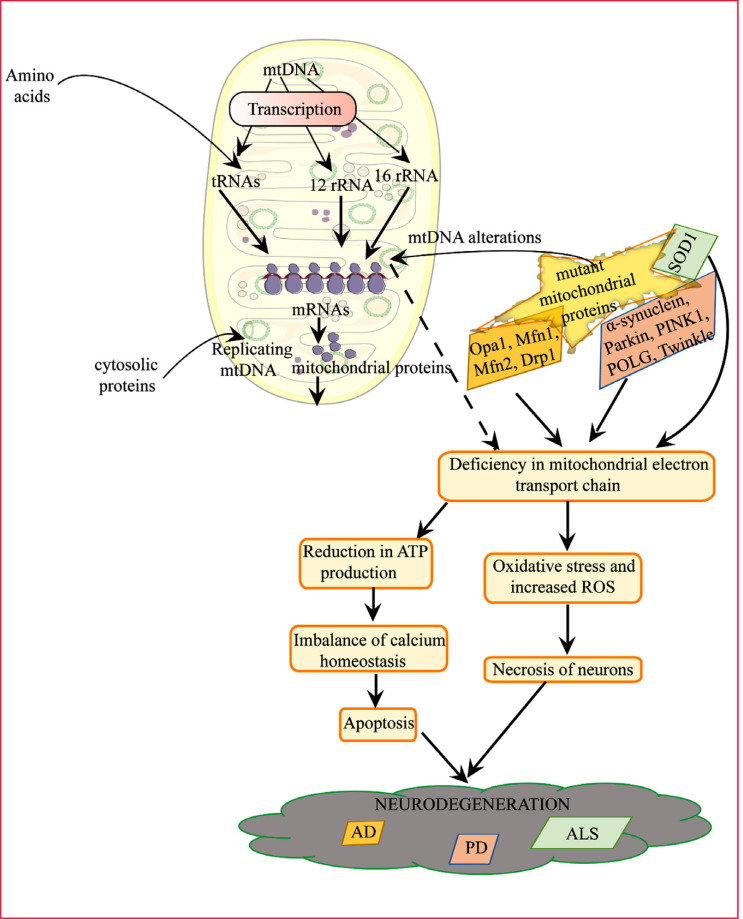
Clinical studies showing the reduction in oxygen and glucose metabolism in AD patients' brains indicate a possible role of mitochondrial impairment in the disease that needs to be investigated at the genetic and epigenetic levels. The link between the variants mentioned above and how they can contribute to the mitochondrial impairment or mtDNA mutations requires more profound study. However, mitochondrial involvement can either be primary or secondary. The exclusive involvement of maternal mtDNA states direct involvement inherited that develops reduced glucose metabolism in AD brain due to oxidative stress and mitochondrial ROS production contributing to amyloid and τ deposition. Secondary involvement is backed up by the evidence stating mitochondrial fission-fusion imbalance due to both downregulations of Opa1, Mfn1, Mfn2, and upregulation of Drp1 proteins mRNA levels. In addition to this, various hypothesis proposing the role of mitochondrial Aβ and τ deposition leading to mitochondrial dysfunction in association with an imbalance in calcium homeostasis and fission-fusion mechanism, respectively.
Mitochondrial dysfunction plays a role in AD pathogenesis (Fig. 2). In AD, mosaic respiratory anomalies are seen in cortical neurons, a scenario identical to PD [89, 90] along with cerebral energy hypometabolism [91, 92]. There is a physical interaction seen between mitochondria and Aβ [93]. Regarding the mitochondrial respiratory chain, mitochondrial complex IV activity has been seen reduced in the AD brain's cortical tissue [94, 95], which can be linked to AD brain hypometabolism [96, 97].
Fig. (2).
Illustration of neurodegenerative diseases in association with mitochondrial dysfunction.
3.1.1. Homplasmic Population Variants
In AD, mtDNA haplogroup research failed to prove miochondrial haplogroups contributing to AD [98]. Epidemiological data showed weak evidence for rare homoplasmic variants contributing to AD development [99].
3.1.2. Heteroplasmic Variants
There is a lack of evidence linking somatic mtDNA mutations with AD. Coskum et al. found many heteroplasmic point mutations in the mtDNA hypervariable [100] but not seen in other AD studies at a high frequency [101] and are found in the population as common homoplasmic variants [102]. Another study exploring the cold gene somatic mutations yielded results comparable to the control group [103]. All these studies suggest the lack of evidence linking mtDNA variations with AD.
3.2. Parkinson's Disease
PD is the second common neurodegenerative disorder that affects the serotonergic, adrenergic, and dopaminergic systems, causing a depletion in the neurotransmitters of the respective systems developing defective motor non-motor coordination [104-106]. Studies suggest the role of mutation of various variants, including α-synuclein, Leucine-rich repeat kinase 2 (LRRK 2), glucocerebrosidase gene (GBA), microtubule-associated protein τ (MAPT), MDR1, Parkin, and PINK1 [87,88]. PD is characterized by motor coordination dysfunction, and evidence claims a striking role of mitochondria/ mtDNA in PD pathogenesis. One aspect is the upregulation of mtDNA deletions/rearrangement, and the other is point mutations comprising both homoplasmic (MT-ND1, MT-ND2) and heteroplasmic (MT-ND5) mutations. Compagnoni et al. mustered various studies and proposed PD's association with genes, POLG, and Twinkle involved in mtDNA maintenance. Among various gene mutations, PINK1 and Parkin play a prominent role in maintaining mitochondrial fission-fusion mechanism show possibilities of its involvement in PD pathology via mitochondrial dysfunction mechanism, which has been mainly confirmed in Drosophila models and rodents [8]. Compagnoni et al. also aggregate information on α-synuclein overexpression in mitochondrial dysfunction in rat models and the deficiency of Complex I though the mechanism remains unclear [8].
Research worldwide proposes the involvement of environmental toxins such as 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), rotenone, paraquat, and maneb in developing PD via mitochondrial dysfunction. MPTP is the by-product of 1-methyl-4-phenyl-4-propionoxypiperidine (Desmethylprodine), an analogue of meperidine and exposure to MPTP was known to develop parkinsonian symptoms, due to which this is accepted as a PD model. MPTP being lipophilic, crosses the blood-brain barrier and, in the presence of monoamine oxidase present in glial cells, gets converted to 1-methyl-4-phenylpyridinium (MPP+). MPP+ via dopamine transporter is then taken into the dopaminergic neurons and gets accumulated in mitochondria, thereby inhibiting the mitochondrial complexes I, III, IV, increasing ROS generation and potentiating oxidative stress [107, 108]. In vitro studies utilising MPP+ reported a decrease in mitochondrial gene expressions and function, in vivo studies additionally report a decrease in tyrosine hydroxylase expression. Sub-toxic dose of MPTP can alter mitochondrial proteins, among which about 270 proteins have specifically altered substantia nigra and striatum, suggesting the association with nigrostriatal pathway [108].
In PD, mtDNA mutations can impair respiratory chain functions, and a mosaic pattern of deficiency is seen in the postmortem PD brain [109]. A study indicated a high number of neurons in substantia nigra of PD patients are COX deficient compared to the control group, and mtDNA deletion appeared more significant in neurons that are COX deficient single-cell real-time PCR. PCR cloning techniques suggested discrete breakpoints showing intracellular clonal expansion as the mtDNA deletion mechanism [3].
In patients with PD, mtDNA deletions can be high all over the brain, and these deletions are produced somatically and undergo clonal expansion leading to COX deficiency. Still, the question of whether mtDNA deletion is linked to PD pathophysiology stays unclear. In substantia nigra, mtDNA deletions can lead to adaptive responses such as raised mtDNA copy number, higher striatal dopamine, and better respiration instead of harmful [110]. There is not much evidence proving mitochondrial respiratory chain deficiency facilitates Lewy body generation, as some studies showed normal RC complex activity in Lewy body-positive cells [111, 112]. The mtDNA point mutations can lead to early propagation of Lewy bodies, raising the chance of early neuronal death, leading to mtDNA's survival.
3.3. Amyotrophic Lateral Sclerosis
This neurological disease causes motor neuron degeneration in the spinal cord, cortex, and brain stem, thereby weakening the muscle tissues and developing spasticity and atrophy [113]. ALS is known to have an association with mutation of variants including annexin A11 (ANXA11), chromosome 21 open reading frame 2 protein (C21orf2), chromosome 9 open reading frame 72 (C9orf72), cyclin F (CCNF), coiled- coil-helix-coiled-coil-helix domain-containing protein 10 (CHCHD10), FUS RNA-binding protein (FUS), matrin 3 (MATR3), NIMA-related kinase 1 (NEK1), profilin 1 (PFN1), superoxide dismutase 1 (SOD1), TAR DNA-binding protein (TARDBP), TANK-binding kinase 1 (TBK1), T cell-restricted intracellular antigen-1 (TIA1), tubulin alpha 4A protein (TUBA4A), valosin containing protein (VCP) [87]. Among various gene mutations, mutant SOD1 that can be found in the intermembrane space, outer mitochondrial membrane, and matrix causes impairment in calcium loading within the mitochondria, and activities of electron transport chain are hindered by the release of cytochrome C and associated apoptosis triggering in addition to mutant SOD1-Bcl-2 complex formation [113, 114].
3.3.1. Homoplasmic Population Variants
There are significantly few biological observations in ALS and sporadic FTD indicating mitochondrial dysfunction compared with AD and PD, where noticeable hypometabolism or dysfunction occurs in the mitochondrial respiratory chain. There is no significant association found linking mitochondrial haplogroups with ALS [40]. Although there is no significant association, some studies indicated rare germline mtDNA variation linked with the disease [115].
3.3.2. Heteroplasmic Variants
Based on a case report, SOD1 mutations as well as frame deletion in mitochondrial complex I have been suggested to have an association with ALS progression [116,117] and rearrangement in the mitochondrial genome at a large scale associated with the ALS variant, Progressive Muscular Atrophy (PMA) [118]. The muscle biopsies on SOD1 mutant showed mtDNA4977 deletion elevation in 3 patients and the highest deletion seen in the severe case [7]. More studies are required to link mutations of mtDNA with monogenic and sporadic ALS [1].
4. PHARMACOLOGICAL APPROACH TO MITOCHONDRIAL DYSFUNCTION IN NEURODEGENERATIVE DISORDERS
Neuroprotection via SIRT1 highlights the opportunity of enhancing the use of SIRT1 activators. One such is resveratrol, a potent activator of SIRT1, which functions against Aβ42 toxicity. Another to be listed is the well-known phytocompound curcumin which naturally activates SIRT1 in addition to antioxidant and antiapoptotic effect in neurons rich in Aβ deposit. Unlike SIRT1, SIRT2 inhibitors hold the chance of promoting neuroprotection in neurodegenerative disorders, with major reports available on PD. Nicotinamide, a SIRT inhibitor showed restoration of AD-associated defects in cognition, phospho-tau (Thr231) reduction, and an increase in acetylated SIRT2 brain substrate, α-tubulin [119]. Underlying mechanisms can be SIRT1 and PARP-1 inhibition associated NAD+ and ATP levels maintenance, p53 inactivation associated antiapoptotic and anti-inflammatory effect mediated by SIRT1, α-secretase activity enhancement, mitochondrial mPTP closure mediated by SIRT3 [120].
PGC-1α enhancement can protect oxidative stress and neuronal death, revealing its potential as a candidate in PD treatment. Several studies have been conducted, including resveratrol induced PGC-1α activation in dopaminergic neurons, PGC-1α delivery via adenoviral vector showed an increase in dopaminergic death possibly due to PGC-1α overexpression causing an increase in ROS productivity and mitochondrial hyperactivity. Ameliorating PD symptoms require the usage of an amalgam of pharmacological agents to develop neuroprotective effects as the drugs currently available have limitations in slowing down disease progression. Modulation of pathways such as PPARγ and PGC-1α simultaneously can enhance neuroprotection. Invitro studies and various PD models reveal the neuroprotective activity of PGC-1α and PPARγ agonists, which opens the door for future therapy [121]. Another cause of nigral degeneration in PD is the decrease in mitochondrial respiratory chain complex 1 activity, which can be enhanced by mitochondrial complexes I, II electron acceptor, coenzyme Q10. Studies show that prolonged coenzyme Q10 administration can enhance the production of dopamine in the presynaptic neurons [122]. Coenzyme Q10 was subjected to a randomized clinical trial and can proceed for further studies in PD [123].
CONCLUSION
Our knowledge regarding somatic mtDNA mutations and germline mtDNA variations in neurodegeneration and aging achieved significant progress in the past decades. Evidence indicates mtDNA point mutations and mtDNA deletions developing with age, which in specific cells can expand to heteroplasmic levels leading to dysfunction of the respiratory chain. Still, the action of lower-level variants on neuronal functions is currently not understood. In PD, the somatic mtDNA mutations and Lewy body pathology and germlines mtDNA variants contributing as a risk factor to the disease need to be explored in detail as the mechanism is not yet understood. The mtDNA mutations and their effects on cell dysfunction are also a promising area of research. The inherited low-level variants in mtDNA mutations are also worth exploring to elucidate the heritability of neuronal degeneration and aging. Various studies point out that various genes are involved in the pathogenesis of neurodegenerative disorders such as ALS, PD, AD, MS, or stroke, mtDNA mutation, and the nature of mutation deletion single mutation remains controversial within species, and mechanisms remain unclear. Hence, in-depth study on the variants and mechanisms needs to be carried out to shed light in this area and propose a targeted therapy or serve the purpose as an adjuvant for the ailment/ alleviation of these diseases.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Keogh M.J., Chinnery P.F. Mitochondrial DNA mutations in neurodegeneration. Biochim. Biophys. Acta. 2015;1847(11):1401–1411. doi: 10.1016/j.bbabio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 2.Trounce I., Byrne E., Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: Possible factor in ageing. Lancet. 1989;1(8639):637–639. doi: 10.1016/S0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- 3.Bender A., Krishnan K.J., Morris C.M., Taylor G.A., Reeve A.K., Perry R.H., Jaros E., Hersheson J.S., Betts J., Klopstock T., Taylor R.W., Turnbull D.M. High levels of mitochondrial DNA deletions in Substantia nigra neurons in aging and Parkinson disease. Nat. Genet. 2006;38(5):515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 4.van der Giezen M., Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6(6):525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celsi F., Pizzo P., Brini M., Leo S., Fotino C., Pinton P., Rizzuto R. Mitochondria, calcium and cell death: A deadly triad in neurodegeneration. Biochim. Biophys. Acta. 2009;1787(5):335–344. doi: 10.1016/j.bbabio.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossy-Wetzel E., Barsoum M.J., Godzik A., Schwarzenbacher R., Lipton S.A. Mitochondrial fission in apoptosis, neurodegeneration and aging. Curr. Opin. Cell Biol. 2003;15(6):706–716. doi: 10.1016/j.ceb.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 7.Ross J.M., Stewart J.B., Hagström E., Brené S., Mourier A., Coppotelli G., Freyer C., Lagouge M., Hoffer B.J., Olson L., Larsson N.G. Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013;501(7467):412–415. doi: 10.1038/nature12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monzio Compagnoni G., Di Fonzo A., Corti S., Comi G.P., Bresolin N., Masliah E. The role of mitochondria in neurodegenerative diseases: the lesson from Alzheimer’s disease and Parkinson’s disease. Mol. Neurobiol. 2020;57(7):2959–2980. doi: 10.1007/s12035-020-01926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull H.E., Lax N.Z., Diodato D., Ansorge O., Turnbull D.M. The mitochondrial brain: From mitochondrial genome to neurodegeneration. Biochim. Biophys. Acta. 2010;1802(1):111–121. doi: 10.1016/j.bbadis.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright A.F., Murphy M.P., Turnbull D.M. Do organellar genomes function as long-term redox damage sensors? Trends Genet. 2009;25(6):253–261. doi: 10.1016/j.tig.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35(Database issue) Suppl. 1:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simcox E.M., Reeve A.K. An introduction to mitochondria, their structure and functions.Mitochondrial Dysfunction in Neurodegenerative Disorders. Springer; 2016. pp. 3–30. [DOI] [Google Scholar]
- 13.Li H., Liu D., Lu J., Bai Y. Physiology and pathophysiology of mitochondrial DNA. Advances in Mitochondrial Medicine. Springer; 2012. pp. 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson G., Amati-Bonneau P., Blakely E.L., Stewart J.D., He L., Schaefer A.M., Griffiths P.G., Ahlqvist K., Suomalainen A., Reynier P., McFarland R., Turnbull D.M., Chinnery P.F., Taylor R.W. Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: A novel disorder of mtDNA maintenance. Brain. 2008;131(Pt 2):329–337. doi: 10.1093/brain/awm272. [DOI] [PubMed] [Google Scholar]
- 15.Bogenhagen D.F. Mitochondrial DNA nucleoid structure. Biochim. Biophys. Acta. 2012;1819(9-10):914–920. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Bobba K.N., Binoy A., Koo S., Nedungadi D., Podder A., Sharma A., Mishra N., Kim J.S., Bhuniya S. Direct readout protonophore induced selective uncoupling and dysfunction of individual mitochondria within cancer cells. Chem. Commun. (Camb.) 2019;55(45):6429–6432. doi: 10.1039/C9CC01483G. [DOI] [PubMed] [Google Scholar]
- 17.Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., Schreier P.H., Smith A.J., Staden R., Young I.G. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 18.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23(2):147–147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 19.Wiesner R.J., Rüegg J.C., Morano I. Counting target molecules by exponential polymerase chain reaction: Copy number of mitochondrial DNA in rat tissues. Biochem. Biophys. Res. Commun. 1992;183(2):553–559. doi: 10.1016/0006-291X(92)90517-O. [DOI] [PubMed] [Google Scholar]
- 20.Clayton D.A. Replication of animal mitochondrial DNA. Cell. 1982;28(4):693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- 21.Holt I.J., Lorimer H.E., Jacobs H.T. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell. 2000;100(5):515–524. doi: 10.1016/S0092-8674(00)80688-1. [DOI] [PubMed] [Google Scholar]
- 22.Scarpulla R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008;88(2):611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 23.Rorbach J., Soleimanpour-Lichaei R., Lightowlers R.N., Chrzanowska-Lightowlers Z.M.A. How do mammalian mitochondria synthesize proteins? Portland Press Ltd.; 2007. [DOI] [PubMed] [Google Scholar]
- 24.Liao H-X., Spremulli L.L. Initiation of protein synthesis in animal mitochondria. Purification and characterization of translational initiation factor 2. J. Biol. Chem. 1991;266(31):20714–20719. doi: 10.1016/S0021-9258(18)54767-0. [DOI] [PubMed] [Google Scholar]
- 25.Schwartzbach C.J., Spremulli L.L. Bovine mitochondrial protein synthesis elongation factors. Identification and initial characterization of an elongation factor Tu-elongation factor Ts complex. J. Biol. Chem. 1989;264(32):19125–19131. doi: 10.1016/S0021-9258(19)47276-1. [DOI] [PubMed] [Google Scholar]
- 26.Soleimanpour-Lichaei H.R., Kühl I., Gaisne M., Passos J.F., Wydro M., Rorbach J., Temperley R., Bonnefoy N., Tate W., Lightowlers R., Chrzanowska-Lightowlers Z. mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol. Cell. 2007;27(5):745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandon M.C., Lott M.T., Nguyen K.C., Spolim S., Navathe S.B., Baldi P., Wallace D.C. MITOMAP: A human mitochondrial genome database--2004 update. Nucleic Acids Res. 2005;33(Database issue) Suppl. 1:D611–D613. doi: 10.1093/nar/gki079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuppen H.A., Blakely E.L., Turnbull D.M., Taylor R.W. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta. 2010;1797(2):113–128. doi: 10.1016/j.bbabio.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 29.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6(5):389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Oven M., Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 2009;30(2):E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 31.Sigurğardóttir S., Helgason A., Gulcher J.R., Stefansson K., Donnelly P. The mutation rate in the human mtDNA control region. Am. J. Hum. Genet. 2000;66(5):1599–1609. doi: 10.1086/302902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torroni A., Schurr T.G., Cabell M.F., Brown M.D., Neel J.V., Larsen M., Smith D.G., Vullo C.M., Wallace D.C. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 1993;53(3):563–590. [PMC free article] [PubMed] [Google Scholar]
- 33.Richards M.B., Macaulay V.A., Bandelt H-J., Sykes B.C. Phylogeography of mitochondrial DNA in western Europe. Ann. Hum. Genet. 1998;62(Pt 3):241–260. doi: 10.1046/j.1469-1809.1998.6230241.x. [DOI] [PubMed] [Google Scholar]
- 34.Mancuso M., Nardini M., Micheli D., Rocchi A., Nesti C., Giglioli N.J., Petrozzi L., Rossi C., Ceravolo R., Bacci A., Choub A., Ricci G., Tognoni G., Manca M.L., Siciliano G., Murri L. Lack of association between mtDNA haplogroups and Alzheimer’s disease in Tuscany. Neurol. Sci. 2007;28(3):142–147. doi: 10.1007/s10072-007-0807-z. [DOI] [PubMed] [Google Scholar]
- 35.Lakatos A., Derbeneva O., Younes D., Keator D., Bakken T., Lvova M., Brandon M., Guffanti G., Reglodi D., Saykin A., Weiner M., Macciardi F., Schork N., Wallace D.C., Potkin S.G. Association between mitochondrial DNA variations and Alzheimer’s disease in the ADNI cohort. Neurobiol. Aging. 2010;31(8):1355–1363. doi: 10.1016/j.neurobiolaging.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridge P.G., Maxwell T.J., Corcoran C.D., Norton M.C., Tschanz J.T., O’Brien E., Kerber R.A., Cawthon R.M., Munger R.G., Kauwe J.S. Mitochondrial genomic analysis of late onset Alzheimer’s disease reveals protective haplogroups H6A1A/H6A1B: The Cache County Study on Memory in Aging. PLoS One. 2012;7(9):e45134. doi: 10.1371/journal.pone.0045134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehta P., Mellick G.D., Rowe D.B., Halliday G.M., Jones M.M., Manwaring N., Vandebona H., Silburn P.A., Wang J.J., Mitchell P., Sue C.M. Mitochondrial DNA haplogroups J and K are not protective for Parkinson’s disease in the Australian community. Mov. Disord. 2009;24(2):290–292. doi: 10.1002/mds.22389. [DOI] [PubMed] [Google Scholar]
- 38.Latsoudis H., Spanaki C., Chlouverakis G., Plaitakis A. Mitochondrial DNA polymorphisms and haplogroups in Parkinson’s disease and control individuals with a similar genetic background. J. Hum. Genet. 2008;53(4):349–356. doi: 10.1007/s10038-008-0259-1. [DOI] [PubMed] [Google Scholar]
- 39.Ghezzi D., Marelli C., Achilli A., Goldwurm S., Pezzoli G., Barone P., Pellecchia M.T., Stanzione P., Brusa L., Bentivoglio A.R., Bonuccelli U., Petrozzi L., Abbruzzese G., Marchese R., Cortelli P., Grimaldi D., Martinelli P., Ferrarese C., Garavaglia B., Sangiorgi S., Carelli V., Torroni A., Albanese A., Zeviani M. Mitochondrial DNA haplogroup K is associated with a lower risk of Parkinson’s disease in Italians. Eur. J. Hum. Genet. 2005;13(6):748–752. doi: 10.1038/sj.ejhg.5201425. [DOI] [PubMed] [Google Scholar]
- 40.Ingram C.J., Weale M.E., Plaster C.A., Morrison K.E., Goodall E.F., Pall H.S., Beck M., Jablonka S., Sendtner M., Fisher E.M., Bradman N., Kasperavičiūtė D. Analysis of European case-control studies suggests that common inherited variation in mitochondrial DNA is not involved in susceptibility to amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2012;13(4):341–346. doi: 10.3109/17482968.2012.654394. [DOI] [PubMed] [Google Scholar]
- 41.Chinnery P.F., Elliott H.R., Syed A., Rothwell P.M. Mitochondrial DNA haplogroups and risk of transient ischaemic attack and ischaemic stroke: A genetic association study. Lancet Neurol. 2010;9(5):498–503. doi: 10.1016/S1474-4422(10)70083-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose G., Longo T., Maletta R., Passarino G., Bruni A.C., De Benedictis G. No evidence of association between frontotemporal dementia and major European mtDNA haplogroups. Eur. J. Neurol. 2008;15(9):1006–1008. doi: 10.1111/j.1468-1331.2008.02222.x. [DOI] [PubMed] [Google Scholar]
- 43.Caldecott K.W. Single-strand break repair and genetic disease. Nat. Rev. Genet. 2008;9(8):619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 44.Spelbrink J.N., Van Oost B.A., Van den Bogert C. The relationship between mitochondrial genotype and mitochondrial phenotype in lymphoblasts with a heteroplasmic mtDNA deletion. Hum. Mol. Genet. 1994;3(11):1989–1997. doi: 10.1093/hmg/3.11.1989. [DOI] [PubMed] [Google Scholar]
- 45.Baines H.L., Stewart J.B., Stamp C., Zupanic A., Kirkwood T.B., Larsson N-G., Turnbull D.M., Greaves L.C. Similar patterns of clonally expanded somatic mtDNA mutations in the colon of heterozygous mtDNA mutator mice and ageing humans. Mech. Ageing Dev. 2014;139:22–30. doi: 10.1016/j.mad.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R., Törnell J., Jacobs H.T., Larsson N.G. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429(6990):417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 47.Stewart J.B., Freyer C., Elson J.L., Wredenberg A., Cansu Z., Trifunovic A., Larsson N.G. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008;6(1):e10. doi: 10.1371/journal.pbio.0060010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi M.N., Carbone M., Mostocotto C., Mancone C., Tripodi M., Maione R., Amati P. Mitochondrial localization of PARP-1 requires interaction with mitofilin and is involved in the maintenance of mitochondrial DNA integrity. J. Biol. Chem. 2009;284(46):31616–31624. doi: 10.1074/jbc.M109.025882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szczesny B., Tann A.W., Longley M.J., Copeland W.C., Mitra S. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008;283(39):26349–26356. doi: 10.1074/jbc.M803491200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleff S., Kemper B., Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11(2):699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krishnan K.J., Reeve A.K., Samuels D.C., Chinnery P.F., Blackwood J.K., Taylor R.W., Wanrooij S., Spelbrink J.N., Lightowlers R.N., Turnbull D.M. What causes mitochondrial DNA deletions in human cells? Nat. Genet. 2008;40(3):275–279. doi: 10.1038/ng.f.94. [DOI] [PubMed] [Google Scholar]
- 52.Elson J.L., Samuels D.C., Turnbull D.M., Chinnery P.F. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 2001;68(3):802–806. doi: 10.1086/318801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Payne B.A., Wilson I.J., Yu-Wai-Man P., Coxhead J., Deehan D., Horvath R., Taylor R.W., Samuels D.C., Santibanez-Koref M., Chinnery P.F. Universal heteroplasmy of human mitochondrial DNA. Hum. Mol. Genet. 2013;22(2):384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khrapko K. The timing of mitochondrial DNA mutations in aging. Nat. Genet. 2011;43(8):726–727. doi: 10.1038/ng.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Müller-Höcker J. Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: An age-related alteration. J. Neurol. Sci. 1990;100(1-2):14–21. doi: 10.1016/0022-510X(90)90006-9. [DOI] [PubMed] [Google Scholar]
- 56.Müller-Höcker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart--an age-related phenomenon. A histochemical ultracytochemical study. Am. J. Pathol. 1989;134(5):1167–1173. [PMC free article] [PubMed] [Google Scholar]
- 57.Fayet G., Jansson M., Sternberg D., Moslemi A-R., Blondy P., Lombès A., Fardeau M., Oldfors A. Ageing muscle: Clonal expansions of mitochondrial DNA point mutations and deletions cause focal impairment of mitochondrial function. Neuromuscul. Disord. 2002;12(5):484–493. doi: 10.1016/S0960-8966(01)00332-7. [DOI] [PubMed] [Google Scholar]
- 58.Nambiar J., Vijayakumar G., Drishya G., Shaji S.K., Pandurangan N., Kumar G.B., Nair B.G. (I-3,II-3)-Biacacetin-mediated cell death involves mitochondria. Mol. Cell. Biochem. 2019;451(1-2):79–90. doi: 10.1007/s11010-018-3395-8. [DOI] [PubMed] [Google Scholar]
- 59.Rossignol R., Malgat M., Mazat J-P., Letellier T. Threshold effect and tissue specificity. Implication for mitochondrial cytopathies. J. Biol. Chem. 1999;274(47):33426–33432. doi: 10.1074/jbc.274.47.33426. [DOI] [PubMed] [Google Scholar]
- 60.Boulet L., Karpati G., Shoubridge E.A. Distribution and threshold expression of the tRNA(Lys) mutation in skeletal muscle of patients with myoclonic epilepsy and ragged-red fibers (MERRF). Am. J. Hum. Genet. 1992;51(6):1187–1200. [PMC free article] [PubMed] [Google Scholar]
- 61.Hayashi J., Ohta S., Kikuchi A., Takemitsu M., Goto Y., Nonaka I. Introduction of disease-related mitochondrial DNA deletions into HeLa cells lacking mitochondrial DNA results in mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA. 1991;88(23):10614–10618. doi: 10.1073/pnas.88.23.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.James A.M., Wei Y.H., Pang C.Y., Murphy M.P. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J. 1996;318(Pt 2):401–407. doi: 10.1042/bj3180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Aurelio M., Pallotti F., Barrientos A., Gajewski C.D., Kwong J.Q., Bruno C., Beal M.F., Manfredi G. In vivo regulation of oxidative phosphorylation in cells harboring a stop-codon mutation in mitochondrial DNA-encoded cytochrome c oxidase subunit I. J. Biol. Chem. 2001;276(50):46925–46932. doi: 10.1074/jbc.M106429200. [DOI] [PubMed] [Google Scholar]
- 64.Rossignol R., Faustin B., Rocher C., Malgat M., Mazat J-P., Letellier T. Mitochondrial threshold effects. Biochem. J. 2003;370(Pt 3):751–762. doi: 10.1042/bj20021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai Y., Shakeley R.M., Attardi G. Tight control of respiration by NADH dehydrogenase ND5 subunit gene expression in mouse mitochondria. Mol. Cell. Biol. 2000;20(3):805–815. doi: 10.1128/MCB.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enriquez J.A., Chomyn A., Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nat. Genet. 1995;10(1):47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 67.Cortopassi G.A., Shibata D., Soong N.W., Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc. Natl. Acad. Sci. USA. 1992;89(16):7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corral-Debrinski M., Shoffner J.M., Lott M.T., Wallace D.C. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat. Res. 1992;275(3-6):169–180. doi: 10.1016/0921-8734(92)90021-G. [DOI] [PubMed] [Google Scholar]
- 69.Meissner C., Bruse P., Mohamed S.A., Schulz A., Warnk H., Storm T., Oehmichen M. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: A useful biomarker or more? Exp. Gerontol. 2008;43(7):645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 70.Corral-Debrinski M., Horton T., Lott M.T., Shoffner J.M., Beal M.F., Wallace D.C. Mitochondrial DNA deletions in human brain: Regional variability and increase with advanced age. Nat. Genet. 1992;2(4):324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 71.Soong N.W., Hinton D.R., Cortopassi G., Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat. Genet. 1992;2(4):318–323. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 72.Cooper J.M., Mann V.M., Schapira A.H.V. Analyses of mitochondrial respiratory chain function and mitochondrial DNA deletion in human skeletal muscle: Effect of ageing. J. Neurol. Sci. 1992;113(1):91–98. doi: 10.1016/0022-510X(92)90270-U. [DOI] [PubMed] [Google Scholar]
- 73.Hamblet N.S., Castora F.J. Elevated levels of the Kearns-Sayre syndrome mitochondrial DNA deletion in temporal cortex of Alzheimer’s patients. Mutat. Res. 1997;379(2):253–262. doi: 10.1016/S0027-5107(97)00158-9. [DOI] [PubMed] [Google Scholar]
- 74.Damas J., Samuels D.C., Carneiro J., Amorim A., Pereira F. Mitochondrial DNA rearrangements in health and disease--a comprehensive study. Hum. Mutat. 2014;35(1):1–14. doi: 10.1002/humu.22452. [DOI] [PubMed] [Google Scholar]
- 75.Coppedè F., Stoccoro A. Mitoepigenetics and neurodegenerative diseases. Front. Endocrinol. (Lausanne) 2019;10:86. doi: 10.3389/fendo.2019.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sharma N., Pasala M.S., Prakash A. Mitochondrial DNA: Epigenetics and environment. Environ. Mol. Mutagen. 2019;60(8):668–682. doi: 10.1002/em.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradley-Whitman M.A., Lovell M.A. Epigenetic changes in the progression of Alzheimer’s disease. Mech. Ageing Dev. 2013;134(10):486–495. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanch M., Mosquera J.L., Ansoleaga B., Ferrer I., Barrachina M. Altered mitochondrial DNA methylation pattern in Alzheimer disease-related pathology and in Parkinson disease. Am. J. Pathol. 2016;186(2):385–397. doi: 10.1016/j.ajpath.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 79.Stoccoro A., Siciliano G., Migliore L., Coppedè F. Decreased methylation of the mitochondrial D-loop region in late-onset Alzheimer’s disease. J. Alzheimers Dis. 2017;59(2):559–564. doi: 10.3233/JAD-170139. [DOI] [PubMed] [Google Scholar]
- 80.Chestnut B.A., Chang Q., Price A., Lesuisse C., Wong M., Martin L.J. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31(46):16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jęśko H., Wencel P., Strosznajder R.P., Strosznajder J.B. Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem. Res. 2017;42(3):876–890. doi: 10.1007/s11064-016-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cieślik M., Czapski G.A., Strosznajder J.B. The molecular mechanism of amyloid β42 peptide toxicity: The role of sphingosine kinase-1 and mitochondrial sirtuins. PLoS One. 2015;10(9):e0137193. doi: 10.1371/journal.pone.0137193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W., Zou Y., Zhang M., Zhao N., Tian Q., Gu M., Liu W., Shi R., Lü Y., Yu W. Mitochondrial Sirt3 expression is decreased in APP/PS1 double transgenic mouse model of Alzheimer’s disease. Neurochem. Res. 2015;40(8):1576–1582. doi: 10.1007/s11064-015-1630-1. [DOI] [PubMed] [Google Scholar]
- 84.Guan Q., Wang M., Chen H., Yang L., Yan Z., Wang X. Aging-related 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced neurochemial and behavioral deficits and redox dysfunction: Improvement by AK-7. Exp. Gerontol. 2016;82:19–29. doi: 10.1016/j.exger.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 85.Harilal S., Jose J., Parambi D.G.T., Kumar R., Mathew G.E., Uddin M.S., Kim H., Mathew B. Advancements in nanotherapeutics for Alzheimer’s disease: Current perspectives. J. Pharm. Pharmacol. 2019;71(9):1370–1383. doi: 10.1111/jphp.13132. [DOI] [PubMed] [Google Scholar]
- 86.Narayanan S.E., Rehuman N.A., Harilal S., Vincent A., Rajamma R.G., Behl T., Uddin M.S., Ashraf G.M., Mathew B. Molecular mechanism of zinc neurotoxicity in Alzheimer’s disease. Environ. Sci. Pollut. Res. Int. 2020;27(35):43542–43552. doi: 10.1007/s11356-020-10477-w. [DOI] [PubMed] [Google Scholar]
- 87.Harilal S., Jose J., Parambi D.G.T., Kumar R., Unnikrishnan M.K., Uddin M.S., Mathew G.E., Pratap R., Marathakam A., Mathew B. Revisiting the blood-brain barrier: A hard nut to crack in the transportation of drug molecules. Brain Res. Bull. 2020;160:121–140. doi: 10.1016/j.brainresbull.2020.03.018. [DOI] [PubMed] [Google Scholar]
- 88.Alexiou A., Nizami B., Khan F.I., Soursou G., Vairaktarakis C., Chatzichronis S., Tsiamis V., Manztavinos V., Yarla N.S., Md Ashraf G. Mitochondrial dynamics and proteins related to neurodegenerative diseases. Curr. Protein Pept. Sci. 2018;19(9):850–857. doi: 10.2174/1389203718666170810150151. [DOI] [PubMed] [Google Scholar]
- 89.Cottrell D.A., Blakely E.L., Johnson M.A., Ince P.G., Turnbull D.M. Mitochondrial enzyme-deficient hippocampal neurons and choroidal cells in AD. Neurology. 2001;57(2):260–264. doi: 10.1212/WNL.57.2.260. [DOI] [PubMed] [Google Scholar]
- 90.Kish S.J., Bergeron C., Rajput A., Dozic S., Mastrogiacomo F., Chang L-J., Wilson J.M., DiStefano L.M., Nobrega J.N. Brain cytochrome oxidase in Alzheimer’s disease. J. Neurochem. 1992;59(2):776–779. doi: 10.1111/j.1471-4159.1992.tb09439.x. [DOI] [PubMed] [Google Scholar]
- 91.de Leon M.J., Ferris S.H., George A.E., Christman D.R., Fowler J.S., Gentes C., Reisberg B., Gee B., Emmerich M., Yonekura Y., Brodie J., Kricheff I.I., Wolf A.P. Positron emission tomographic studies of aging and Alzheimer disease. AJNR Am. J. Neuroradiol. 1983;4(3):568–571. [PMC free article] [PubMed] [Google Scholar]
- 92.Small G.W., Ercoli L.M., Silverman D.H., Huang S-C., Komo S., Bookheimer S.Y., Lavretsky H., Miller K., Siddarth P., Rasgon N.L., Mazziotta J.C., Saxena S., Wu H.M., Mega M.S., Cummings J.L., Saunders A.M., Pericak-Vance M.A., Roses A.D., Barrio J.R., Phelps M.E. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2000;97(11):6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lustbader J.W., Cirilli M., Lin C., Xu H.W., Takuma K., Wang N., Caspersen C., Chen X., Pollak S., Chaney M., Trinchese F., Liu S., Gunn-Moore F., Lue L.F., Walker D.G., Kuppusamy P., Zewier Z.L., Arancio O., Stern D., Yan S.S., Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304(5669):448–452. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 94.Parker W.D., Jr, Parks J., Filley C.M., Kleinschmidt-DeMasters B.K. Electron transport chain defects in Alzheimer’s disease brain. Neurology. 1994;44(6):1090–1096. doi: 10.1212/WNL.44.6.1090. [DOI] [PubMed] [Google Scholar]
- 95.Parker W.D., Jr, Filley C.M., Parks J.K. Cytochrome oxidase deficiency in Alzheimer’s disease. Neurology. 1990;40(8):1302–1303. doi: 10.1212/WNL.40.8.1302. [DOI] [PubMed] [Google Scholar]
- 96.Alexiou A., Soursou G., Chatzichronis S., Gasparatos E., Kamal M.A., Yarla N.S., Perveen A., Barreto G.E., Ashraf G.M. Role of GTPases in the regulation of mitochondrial dynamics in Alzheimer’s disease and CNS-related disorders. Mol. Neurobiol. 2019;56(6):4530–4538. doi: 10.1007/s12035-018-1397-x. [DOI] [PubMed] [Google Scholar]
- 97.Alexiou A., Chatzichronis S., Ashraf G.M. Prediction of Alzheimer’s disease. Diagnosis and Management in Dementia. Elsevier; 2020. pp. 365–378. [DOI] [Google Scholar]
- 98.Hudson G., Sims R., Harold D., Chapman J., Hollingworth P., Gerrish A., Russo G., Hamshere M., Moskvina V., Jones N., Thomas C., Stretton A., Holmans P.A., O’Donovan M.C., Owen M.J., Williams J., Chinnery P.F. No consistent evidence for association between mtDNA variants and Alzheimer disease. Neurology. 2012;78(14):1038–1042. doi: 10.1212/WNL.0b013e31824e8f1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Edland S.D., Silverman J.M., Peskind E.R., Tsuang D., Wijsman E., Morris J.C. Increased risk of dementia in mothers of Alzheimer’s disease cases: Evidence for maternal inheritance. Neurology. 1996;47(1):254–256. doi: 10.1212/WNL.47.1.254. [DOI] [PubMed] [Google Scholar]
- 100.Coskun P.E., Beal M.F., Wallace D.C. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc. Natl. Acad. Sci. USA. 2004;101(29):10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chinnery P.F., Taylor G.A., Howell N., Brown D.T., Parsons T.J., Turnbull D.M. Point mutations of the mtDNA control region in normal and neurodegenerative human brains. Am. J. Hum. Genet. 2001;68(2):529–532. doi: 10.1086/318204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Howell N., Elson J.L., Chinnery P.F., Turnbull D.M. mtDNA mutations and common neurodegenerative disorders. Trends Genet. 2005;21(11):583–586. doi: 10.1016/j.tig.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 103.Lin M.T., Simon D.K., Ahn C.H., Kim L.M., Beal M.F. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer’s disease brain. Hum. Mol. Genet. 2002;11(2):133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 104.Parambi D.G.T., Saleem U., Shah M.A., Anwar F., Ahmad B., Manzar A., Itzaz A., Harilal S., Uddin M.S., Kim H., Mathew B. Exploring the therapeutic potentials of highly selective oxygenated chalcone based MAO-B inhibitors in a haloperidol-induced murine model of Parkinson’s disease. Neurochem. Res. 2020;45(11):2786–2799. doi: 10.1007/s11064-020-03130-y. [DOI] [PubMed] [Google Scholar]
- 105.Palakkathondi A., Oh J.M., Dev S., Rangarajan T.M., Kaipakasseri S., Kavully F.S., Gambacorta N., Nicolotti O., Kim H., Mathew B. (Hetero-)(arylidene)arylhydrazides as Multitarget-Directed Monoamine Oxidase Inhibitors. ACS Comb. Sci. 2020;22(11):592–599. doi: 10.1021/acscombsci.0c00136. [DOI] [PubMed] [Google Scholar]
- 106.Chaudhary S.S., Chaudhary S., Rawat S., Natesan S., Pardeshi T., Alexiou A. Recent developments in the etiology, treatment, and potential therapeutic targets for Parkinson’s disease: A focus on biochemistry.Diagnosis and Management in Parkinson’s Disease. Elsevier; 2020. pp. 73–90. [DOI] [Google Scholar]
- 107.Mapa M.S.T., Le V.Q., Wimalasena K. Characteristics of the mitochondrial and cellular uptake of MPP+, as probed by the fluorescent mimic, 4'I-MPP. PLoS One. 2018;13(8):e0197946. doi: 10.1371/journal.pone.0197946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Subramaniam S.R., Chesselet M-F. Mitochondrial dysfunction and oxidative stress in Parkinson’s disease. Prog. Neurobiol. 2013;106-107:17–32. doi: 10.1016/j.pneurobio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itoh K., Weis S., Mehraein P., Müller-Höcker J. Cytochrome c oxidase defects of the human substantia nigra in normal aging. Neurobiol. Aging. 1996;17(6):843–848. doi: 10.1016/S0197-4580(96)00168-6. [DOI] [PubMed] [Google Scholar]
- 110.Perier C., Bender A., García-Arumí E., Melià M.J., Bové J., Laub C., Klopstock T., Elstner M., Mounsey R.B., Teismann P., Prolla T., Andreu A.L., Vila M. Accumulation of mitochondrial DNA deletions within dopaminergic neurons triggers neuroprotective mechanisms. Brain. 2013;136(Pt 8):2369–2378. doi: 10.1093/brain/awt196. [DOI] [PubMed] [Google Scholar]
- 111.Reeve A.K., Park T-K., Jaros E., Campbell G.R., Lax N.Z., Hepplewhite P.D., Krishnan K.J., Elson J.L., Morris C.M., McKeith I.G., Turnbull D.M. Relationship between mitochondria and α-synuclein: A study of single Substantia nigra neurons. Arch. Neurol. 2012;69(3):385–393. doi: 10.1001/archneurol.2011.2675. [DOI] [PubMed] [Google Scholar]
- 112.Müller S.K., Bender A., Laub C., Högen T., Schlaudraff F., Liss B., Klopstock T., Elstner M. Lewy body pathology is associated with mitochondrial DNA damage in Parkinson’s disease. Neurobiol. Aging. 2013;34(9):2231–2233. doi: 10.1016/j.neurobiolaging.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 113.Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 114.Beal M.F. Mitochondria take center stage in aging and neurodegeneration. Ann. Neurol. 2005;58(4):495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 115.Grazina M., Silva F., Santana I., Santiago B., Mendes C., Simões M., Oliveira M., Cunha L., Oliveira C. Frontotemporal dementia and mitochondrial DNA transitions. Neurobiol. Dis. 2004;15(2):306–311. doi: 10.1016/j.nbd.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 116.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O’Regan J.P., Deng H.X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 117.Comi G.P., Bordoni A., Salani S., Franceschina L., Sciacco M., Prelle A., Fortunato F., Zeviani M., Napoli L., Bresolin N., Moggio M., Ausenda C.D., Taanman J.W., Scarlato G. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann. Neurol. 1998;43(1):110–116. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- 118.Zoccolella S., Artuso L., Capozzo R., Amati A., Guerra F., Simone I., Logroscino G., Petruzzella V. Mitochondrial genome large rearrangements in the skeletal muscle of a patient with PMA. Eur. J. Neurol. 2012;19(7):e63–e64. doi: 10.1111/j.1468-1331.2012.03720.x. [DOI] [PubMed] [Google Scholar]
- 119.Polito L., Biella G., Albani D. Sirtuin modulation as novel neuroprotective strategy for Alzheimer’s disease.Neuroprotection in Alzheimer’s Disease. Elsevier; 2017. pp. 149–173. [DOI] [Google Scholar]
- 120.Hwang E.S. Pharmacological nicotinamide: mechanisms centered around SIRT1 activity. Pharmacoepigenetics. Elsevier; 2019. pp. 781–799. [DOI] [Google Scholar]
- 121.Corona J.C., Duchen M.R. PPARγ and PGC-1α as therapeutic targets in Parkinson’s. Neurochem. Res. 2015;40(2):308–316. doi: 10.1007/s11064-014-1377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Müller T., Büttner T., Gholipour A-F., Kuhn W. Coenzyme Q10 supplementation provides mild symptomatic benefit in patients with Parkinson’s disease. Neurosci. Lett. 2003;341(3):201–204. doi: 10.1016/S0304-3940(03)00185-X. [DOI] [PubMed] [Google Scholar]
- 123.Kieburtz K., Ravina B., Galpern W.R., Tilley B., Shannon K., Tanner C. A randomized clinical trial of coenzyme Q10 and GPI-1485 in early Parkinson disease. Neurology. 2007;68(1):20–28. doi: 10.1212/01.wnl.0000250355.28474.8e. [DOI] [PubMed] [Google Scholar]