Abstract
CRISPR-Cas defense systems have been naturally coopted for guide RNA-directed transposition by Tn7 family bacterial transposons. We find cyanobacterial genomes are rich in Tn7-like elements, including most of the known guide RNA-directed transposons, the type V-K, I-B1, and I-B2 CRISPR-Cas based systems. We discovered and characterized an example of a type I-D CRISPR-Cas system which was naturally coopted for guide RNA-directed transposition. Multiple novel adaptations were found specific to the I-D subtype, including natural inactivation of the Cas10 nuclease. The type I-D CRISPR-Cas transposition system showed flexibility in guide RNA length requirements and could be engineered to function with ribozyme-based self-processing guide RNAs removing the requirement for Cas6 in the heterologous system. The type I-D CRISPR-Cas transposon also has naturally fused transposase proteins that are functional for cut-and-paste transposition. Multiple attributes of the type I-D system offer unique possibilities for future work in gene editing. Our bioinformatic analysis also revealed a broader understanding of the evolution of Tn7-like elements. Extensive swapping of targeting systems was identified among Tn7-like elements in cyanobacteria and multiple examples of convergent evolution, including systems targeting integration into genes required for natural transformation.
Graphical Abstract
Graphical Abstract.
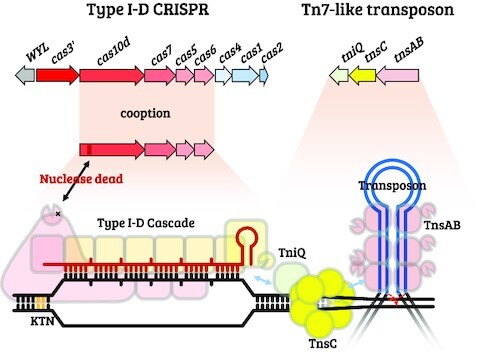
Cooption of a type I-D CRISPR-Cas system for RNA-guided transposition was discovered in cyanobacteria and characterized. CRISPR-Cas cooption occurred with loss of Cas10d nuclease activity and spacer acquisition.
INTRODUCTION
CRISPR-associated transposons (CASTs) are naturally occurring mobile elements that offer exciting possibilities for gene editing due to their capacity to direct a single cargo DNA insertion at a guide RNA-programmed position in one orientation (1). DNA cargo insertion by a transposition mechanism circumvents the need for host DNA double-strand break repair for integrating DNA cargo as found with canonical CRISPR-Cas systems. All the CAST systems that have been characterized are Tn7-like systems with a core set of transposition genes that coopted CRISPR-Cas domain proteins from independent subtypes (2–5). Here, we find that cyanobacteria are a reservoir of diverse Tn7-like elements, including multiple examples of transposon targeting systems formed by convergent evolution. Among the new configurations of Tn7-like elements, we discovered and characterized a new family of CAST elements formed by the cooption of a type I-D CRISPR-Cas system.
Prototypic Tn7 and Tn7-like elements are defined by the control they display over target site selection with two pathways, one targeting a conserved chromosomal site and a second that preferentially targets mobile genetic elements facilitating cell-to-cell transmission (6). Tn7 has a heteromeric transposase formed by TnsA and TnsB. All Tn7-like elements possess an RNaseH family transposase TnsB that allows breakage at the ends of the element and joining to a new target DNA (7,8). Most possess a TnsA nuclease subunit that cleaves the non-transferred strand allowing the element to move by a cut-and-paste mechanism (9). One of the hallmarks of these elements is transposase activity is latent until it is signaled by the identification of a special target DNA. Signaling between the transposase and target site selecting proteins occurs via an AAA+ protein called TnsC (10,11). In one Tn7 targeting pathway, transposition is directed into a single conserved attachment site (att site or attTn7) in the chromosome downstream of the essential glmS gene recognized using a TniQ-family protein, TnsD. In a second targeting pathway, Tn7 preferentially activates transposition directed into mobile plasmids by recognizing special replication features using a second type of protein, TnsE (12,13). Diverse Tn7-like elements have been identified that appear to use one or multiple TniQ family proteins to establish dual targeting pathways (1,2,14–16).
CRISPR-Cas systems have been captured by Tn7-like elements on four separate occasions that have been functionally characterized in a heterologous host. The mechanism used to recognize a fixed insertion site in the chromosome versus sites on mobile elements capable of cell-to-cell transfer differs across the CAST elements. Type I-B1 and I-B2 CAST manage two pathways with separate TniQ family proteins, a TnsD-like protein for targeting a fixed chromosomal attachment (att) site and a second smaller TniQ for interfacing with a coopted CRISPR-Cas system (1,2). A similar configuration with two TniQ proteins is found with a branch of the type I-F3 CAST elements (5), which was also shown to function in a heterologous host as two pathways (17). Alternatively, type V-K CAST and most type I-F3 CAST elements only have one TniQ, which associates with the CRISPR effector, and instead use specialized types of guide RNAs to allow for two transposition pathways (2,5,18).
We find that cyanobacterial genomes are rich in diverse Tn7-like elements showing multiple examples of convergent evolution for target site selection pathways. In addition to having three of the four proven CAST systems, we discover cooption in cyanobacteria of the type I-D CRISPR-Cas system. Similar to previous examples, cooption occurred with loss of spacer acquisition function, but specific to I-D style systems, cooption required both natural inactivation of Cas10d nuclease activity and loss of the central helicase Cas3’ normally used for target degradation. Characterization of the type I-D CAST from Myxacorys californica WJT36-NPBG1 reveals cut and paste transposition with naturally fused TnsA and TnsB proteins and indicates a new mechanism of cooption, not involving the Cas6 mechanism found essential with the type I-F3 elements. Among the novel features of the I-D CAST system is the capacity to use variable length guide RNAs which we show are amenable to guide auto-maturation via ribozymes allowing independence from the steps normally required from Cas6 in most other type I CRISPR-Cas systems. Our bioinformatic analysis also revealed a broader understanding of the evolution of Tn7-like elements, including multiple new examples of convergent evolution, which we show predominate in Tn7-like elements.
MATERIALS AND METHODS
Growth conditions
Escherichia coli strains (Table 1) were grown in lysogeny broth (LB) or on LB agar supplemented with the following concentrations of antibiotics when appropriate: 100 μg/ml carbenicillin, 10 μg/ml gentamicin, 30 μg/ml chloramphenicol, 8 μg/ml tetracycline, 50 μg/ml kanamycin, 50 μg/ml spectinomycin, 20 μg/ml nalidixic acid, 100 μg/ml rifampicin, 50 μg/ml X-gal.
Table 1.
Strains used in the study
| Strain | Genotype | Source, reference |
|---|---|---|
| BW27783 | F−, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, Δ(araH-araF)570(::FRT), ΔaraEp-532::FRT, φPcp8-araE535, rph-1, Δ(rhaD-rhaB)568, hsdR514 | (19) |
| BW20767 | F−, RP4-2(Km::Tn7,Tc::Mu-1), ΔuidA3::pir+, leu-163::IS10, recA1, creC510, hsdR17, endA1, thi | (20) |
| CW51 | F−, ara−, arg-, Δ(lac-pro)XIII, nalR, rifR, recA56 | (13) |
| PO677 | BW27783 attTn7::miniTn7-miniMcCAST(KanR) | This work |
| PO788 | PO677 pOPO717 (McCAST Cascade operon and lacZ spacer 5 under arapBAD control), pOPO636 (TnsABCQ under lac control) | This work |
| PO619 | BW27783 lacZ+ | This work |
| PO704 | BW20767 pOPO701(donor plasmid with mini-transposon of McCAST and an R6K origin of replication) | This work |
Strain and plasmid construction
Strain PO677 was constructed with a mini McCAST element in the chromosome at the neutral attTn7 position within a mini Tn7 element as described previously (5,21). A Lac+ derivative of BW27783, PO619, was constructed by using P1 transduction to move the wild type lac allele from wild type E. coli K-12 (CGSC#: 4401) (22). Strain PO704 was used for delivery of a conditional replicon and oriT (RP4) containing pOPO701 vector with the mini McCAST element from the Pir+ donor strain BW20767 which encodes the RP4 conjugation machinery (20). Standard molecular cloning techniques were used to make the vectors described in supplementary Table S2 according to the vendor instructions. The biomass of Myxacorys californica WJT36-NPBG1 was kindly donated by Dr. Nicole Pietrasiak. The genomic DNA was extracted with DNeasy PowerLyzer Microbial Kit (QIAGEN) as described before (23).
Bioinformatics
Annotated protein fasta files, genomic sequences, and feature tables of cyanobacteria were downloaded from National Center for Biotechnology Information (NCBI) FTP site. In total, there were 2,163 genomes for analysis. Profile HMMs associated with TnsA (PF08722, PF08721), TnsB (PF00665), TnsC (PF11426, PF05621), TniQ (PF06527) downloaded from the European Bioinformatics Institute (EMBL-EBI) Pfam database, were used for detecting homologs with hmmsearch (HMMER3). Candidate proteins were grouped into tnsBC operons, and each operon was then grouped with its neighboring tnsA and tnsQ into one transposon functional unit. The tnsA and tniQ adjacent to more than one tnsBC operon are allocated to the closest one. Only those with at least one tnsA or tnsQ are collected. The dataset of Tn7-like elements with TnsAB fusion generated during this study is available in Supplementary Table S1. The TnsB and TniQ proteins were aligned with MUSCLE (24). Similarity trees were made with FastTree using WAG evolutionary model and the discrete gamma model with 20 rate categories as previously described (1). The visualization of the trees and coloring was done with iTOL (Interactive Tree Of Life) (25).
Mate-out transposition assay
The frequency of transposition was monitored in a large pool of independent transformants, as described previously (5). Briefly, vectors encoding the core transposase genes (TnsABCQ/TnsABC with lactose induction) and target selection genes (Cascade operon, crRNA/TnsD with arabinose induction) were co-transformed into cells (BW27783 background) carrying an F plasmid derivative with the target sequence and the mini-McCAST element (Kanamycin resistance gene flanked by left and right McCAST transposon ends) on a donor plasmid. Plates were grown overnight, and hundreds of transformants were washed off the plate in LB media, pelleted, washed twice with M9 minimal media, and finally resuspended to O.D. 0.6 in M9 minimal media supplemented with 0.2% w/v maltose, required antibiotics, 0.2% w/v arabinose, and 0.1 mM IPTG for induction. After 18 h of incubation with shaking at 30°C, 0.5 ml of the donor cells was spun down, washed twice with LB, and resuspended into 0.5 ml LB supplemented with 0.2% w/v glucose for recovery with shaking at 37°C for 30 min. To monitor transposition from the donor plasmid into the F plasmid target, donor cells were then mixed with mid-log recipient cells (CW51) in LB supplemented with 0.2% w/v glucose at a ratio of 1:5 donor:recipient and incubated with gentle agitation for 90 minutes at 37°C to allow mating. After incubation, cultures were vortexed, placed on ice, then serially diluted in LB 0.2% w/v glucose and plated on LB supplemented with required antibiotics for selecting CW51 recipient cells for transconjugants 20 μg/ml nalidixic acid, 100 μg/ml rifampicin, 50 μg/ml spectinomycin, 50 μg/ml X-gal, with or without 50 μg/ml kanamycin to sample the entire transconjugant population or select for transposition respectively. Plates were incubated at 37°C for 24 h before colonies were counted. For testing the effects of expressing additional Cas11d and Cas7d, pOPO808 or empty vector control pBBR-GenR-ara was co-transformed with the other transposition gene expression vectors, with 10 μg/ml gentamycin supplemented into LB agar and induction M9 minimal media in the following step.
Mapping insertions
To confirm the target site duplication expected with transposition, transposon junctions from insertions in the lacZ gene (guided by lacZ spacer 1) were amplified by colony PCR with primer pairs JEP2257 + JEP2901 and JEP1597 + JEP2903 (Table 2) and subjected to Sanger DNA sequencing. Illumina sequencing was used to map the total insertions from F plasmids from transconjugants. Transconjugants were collected, and F plasmid DNA was isolated using the ZR BAC DNA Miniprep Kit. Insertions were mapped with BBtools (BBMap – Bushnell B. – sourceforge.net/projects/bbmap/).
Table 2.
Oligonucleotide primers used in this work
| Name | Primer and description | Sequence |
|---|---|---|
| JEP2257 | Amplify left end junction | 5′-CCGCGCTGTACTGGAGGCTGAAGTT-3′ |
| JEP2901 | Amplify left end junction | 5′-TTGGTCTCTTCAGCTCCTCATGTAAAAGTGTCTTCAAA-3′ |
| JEP1597 | Amplify right end junction | 5′-CAGCGACCAGATGATCAC-3′ |
| JEP2903 | Amplify right end junction | 5′-TTGGTCTCTCCAATTACCAGCACCATGATCTTTATAA-3′ |
| JEP3375 | Making PAM library | 5′-GTTGCTCTTCAAGAGTTGCCCGGCGCTCT |
| CCGGCTGCCCGGCTTCCATTCAGGTCGAG-3′ | ||
| JEP3376 | Making PAM library | 5′-GTTGCTCTTCATCTGGCTCACAGTACGCG |
| TAGTGCNNNNTGCAGAATCCCTGCTTCGT-3′ |
PAM screening
A PAM library was constructed by PCR amplification of plasmid pBBR-GenR with JEP3375 + JEP3376, subsequent digestion with SapI and self-ligation. The plasmid PAM library was transformed into DH5α, pooled, and plasmid isolated for PAM screening. To screen PAM preference, the PAM library was electroporated into the PO788 (BW27783 with vectors carrying the transposition genes) and plated on LB agar supplemented with the appropriate antibiotics, and 0.1mM IPTG, and 0.2% w/v arabinose for induction. After 17 h of incubation at 37°C, the colonies were scrapped from the plates, and the plasmids extracted then retransformed into DH5α with electroporation for selecting those with insertions on LB agar supplemented with 50 μg/ml kanamycin and 10 μg/ml gentamycin. Each step of the process was repeated to ensure a library coverage greater than 80X. The plasmids with transposon insertions and the original PAM library were sent to Illumina sequencing for comparing their PAM compositions.
Testing the TnsA activity with mate-in transposition assay
To monitor whether the TnsAB fusion protein of McCAST moves by cut-and-paste transposition or forms cointegrates, we monitored vector backbone integration genetically following a mate-in transposition assay with an appropriate control. Briefly, a donor plasmid carrying a mini-McCAST element and TetR marker on its backbone (pOPO701) was delivered by conjugation into recipient cells where the donor plasmid cannot replicate. Transposition by simple insertion or cointegrate formation could be assessed by monitoring whether the backbone TetR marker was retained after transposition in recipient cells. The recipient strain PO619 (Escherichia coli BW27783 lacZ+) was freshly transformed with vectors carrying transposition genes. Overnight cultures of the transformed recipient strain were diluted 50 times into induction media (LB, 0.1mM IPTG, 0.2% (w/v) arabinose, required antibiotics), and grown to mid-log phase. In parallel, an overnight culture of the donor strain PO704 (BW20767 carrying pOPO701) was diluted 25 times into LB with appropriate antibiotics and grown to mid-log phase. The cultures of donor and recipient strains were spun down, washed with LB twice, and resuspended to O.D.600 = 10. The donor was then mixed with recipients in a ratio 1:5, 20 μl of each mixture was spotted on LB plate supplemented with 0.1 mM IPTG and 0.2% (w/v) arabinose. Conjugal mating was conducted at 30°C for 2 h. After mating, each spot was washed up with 3 ml LB medium, serial diluted, and plated on LB plates supplemented with appropriate antibiotics and X-gal. One hundred fifty white colonies (presumably on-lacZ transposition) were purified onto a fresh plate, then streaked on LB agar supplemented with tetracycline to test for cointegration of donor plasmid backbone. As a control, the experiment was repeated with different combinations of vectors carrying transposition genes (TnsABC + TniQ with or without a TnsA active site mutation, Cascade operon with and without target spacer) transformed into the recipient strain as described in the text.
Quantification and statistical analysis
Statistical details are listed in figure legends. When stated, experiments were performed with three biological replicates (n = 3).
RESULTS
Diverse configurations of Tn7-like elements are found in cyanobacteria
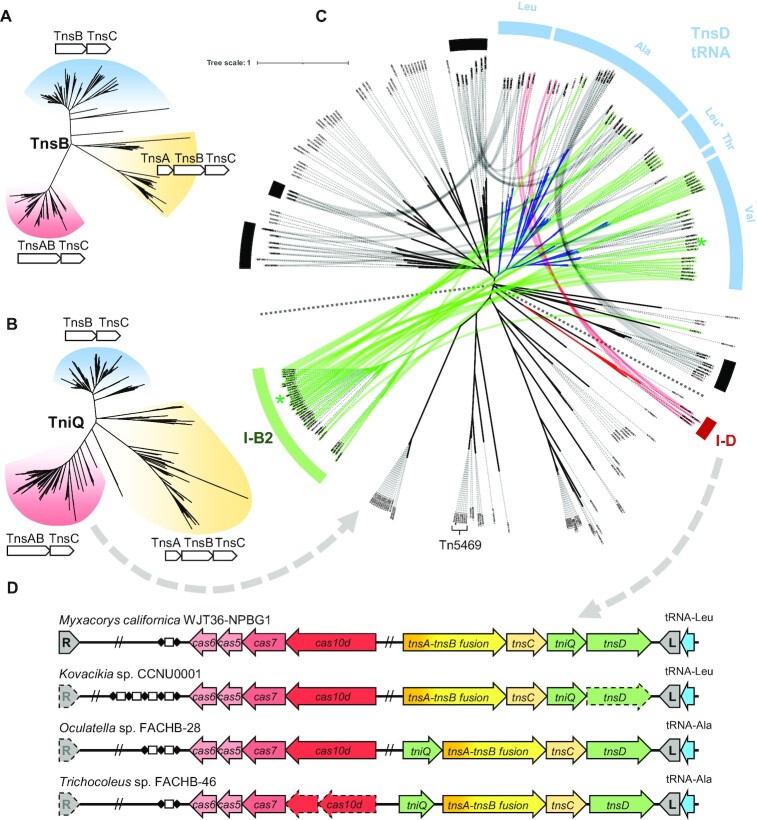
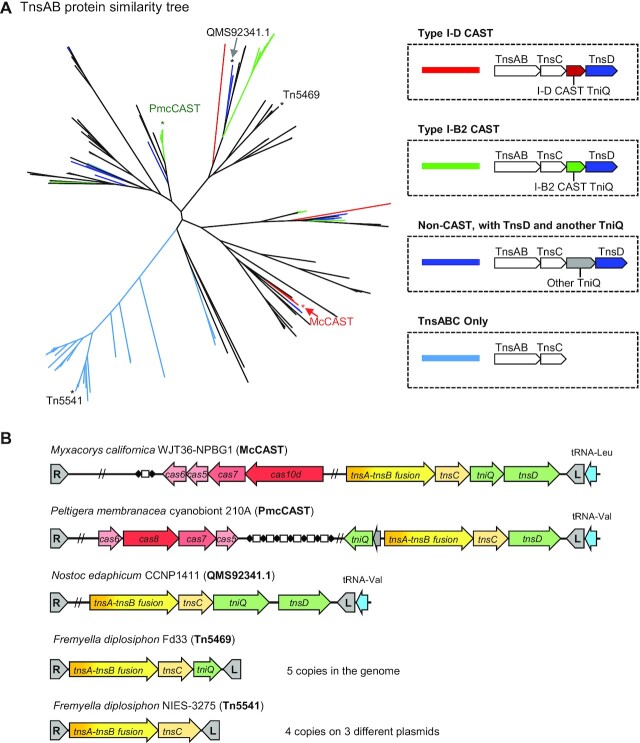
We surveyed 2,163 annotated cyanobacterial genomes on NCBI for Tn7-like transposons, defined as transposons with TnsB and TnsC and encoding either TnsA or TniQ family proteins, and found >800 Tn7-like transposons. Similarity trees of TnsB and TniQ subdivided candidate Tn7-like elements based on basic transposase architecture, elements without TnsA (i.e. only the TnsB transposase in addition to TnsC and TniQ), elements with separate TnsA and TnsB transposase proteins, or derivatives with TnsA and TnsB fused as the transposase (Supplementary Table S1)(Figure 1A, B). Different types of CAST elements are found across all three branches of transposons and distinguished by transposase architecture. The clade that lacked a tnsA gene is predominated by type V-K CAST systems, elements with a separate tnsA gene include type I-B1 CAST elements, and the clade with the tnsAB fused transposase includes type I-B2 CAST elements (1,2,15,18). It is unclear why so much of the known CAST diversity is found in cyanobacteria, but further examination here indicates additional novelty not realized in the previously published studies (see below).
Figure 1.
Bioinformatic analysis reveals a novel family of CAST. (A) TnsB similarity tree of Tn7-like transposons in cyanobacteria. (B) TniQ similarity tree of Tn7-like transposons in cyanobacteria. (C) TniQ similarity tree of Tn7-like transposons with TnsAB fusion in cyanobacteria. The dashed line separates the tree into two parts, the top is mostly large TniQ (>450 a.a.), and the lower half is mostly small TniQ (<350 a.a.). TniQ proteins encoded in the same transposon are connected with curved lines. The tRNA-targeting TnsD are indicated in blue with the specific tRNA indicated, tRNA-Leu* contains a group I intron. The type I-B1 CAST TniQ are indicated in green and type I-D CAST TniQ are indicated in red. The TniQ proteins of PmcCAST are marked with a green asterisk. Another four prominent tRNA-targeting TnsD-associated secondary TniQ groups are marked with black bars. (D) The gene configuration of four putative type I-D CAST are indicated, cargo genes are not shown for simplicity. Dashed outline means the transposon end cannot be found or the gene is a pseudogene. L: transposon left end; R: transposon right end.
To better understand TniQ diversity and CAST pathway acquisition, we primarily focus on the clade with the fused TnsAB transposase in this study (Figure 1C). Most transposons in the TnsAB clade found in tRNA attachment sites that based on similarity trees are likely recognized by a TnsD-like protein with an N-terminal TniQ domain (PF06527) and a C-terminal DNA binding domain (Figure 1C). These elements often also encoded a second TniQ protein. Based on the known behavior of Tn7-like elements to typically have a second pathway that targets mobile plasmids, we examined the TniQ branch positions in elements encoding two TniQ proteins. We hypothesized that if the TnsD-like protein is for targeting transposition into the tRNA gene attachment site, the second TniQ encoded in the element is likely adapted for a targeting pathway facilitating the horizontal transfer of the element. This analysis revealed six prominent TniQ branches as putatively adapted as an alternative targeting pathway based on forming independently branching phylogenetic groups (Marked with black, green, and red bars in Figure 1C) (See discussion). Two of the TniQ branches identified using this analysis consisted of proteins lacking C-terminal DNA binding domains, a feature common among known CAST systems (marked with green and red bars in Figure 1C). One such branch includes the recently validated type I-B2 CRISPR-coopting TniQ (2) (green bar in Figure 1C); however, a second branch within this group of tRNA targeting elements was a group with a distinct branch of small TniQ family proteins (red bar in Figure 1C). Of further interest, instead of possessing a type I-B2 CRISPR-Cas system, this small group associated with type I-D CRISPR-Cas systems suggests a new example of CRISPR-Cas cooption. The type I-D CRISPR-Cas associated transposons are closely related to type I-B2 PmcCAST in the core Tns proteins (∼48% a.a. sequence identity of concatenated TnsABCD). Multiple features of the associated type I-D CRISPR-Cas suggested that the system had been coopted for RNA-guided transposition.
Canonical type I-D CRISPR-Cas systems shares features common to both type I and type III CRISPR systems (26–29). Like other type I CRISPR-Cas systems, I-D systems have the signature Cas3 protein, but the Cas3 functional domains are separated in these systems as the Cas3′ protein and a Cas3″ functional domain is part of the Cas10 protein (30). Cas3′ contains the helicase domain for unwinding dsDNA allowing processive cleavage over long distances. The Cas3″ HD nuclease domain is part of the large subunit Cas10 protein, a protein typically associated with type III CRISPR-Cas systems. In addition, the Cas7 of type I-D CRISPR has a separate nuclease activity, enabling its Cascade complex to cut the target ssDNA strand at 6nt intervals, much like how type III CRISPR-Cas Cascade cut target RNA (26). Examining the architecture of the transposon-associated type I-D systems indicated they lack the cas3′ gene required for processive DNA cleavage found in canonical type I-D systems (Figure 1D and Supplementary Figure S1A, B), reminiscent of the loss of Cas3 in type I-F3 systems (Supplementary Figure S1A, B) (1). In addition, the transposon associated type I-D CRISPR systems maintain short CRISPR arrays and lack the spacer acquisition genes cas1, 2, 4 found in the canonical I-D system (31,32), which are convergent features shared by all known CAST families (Supplementary Figure S1A). Interestingly, closer examination of the Cas10d HD nuclease domain in the transposons reveals a change from the conserved HD residue that is normally required to coordinate a metal essential for nuclease activity (Supplementary Figure S1A–C), whose importance was confirmed experimentally and structurally (29,33). This loss of active-site residues is reminiscent of nuclease-inactivating mutations in the Cas12k proteins in the type V-K CAST systems (15). All observed features strongly support these transposons represent a novel family of CAST.
McCAST is a type I-D CRISPR-guided transposon
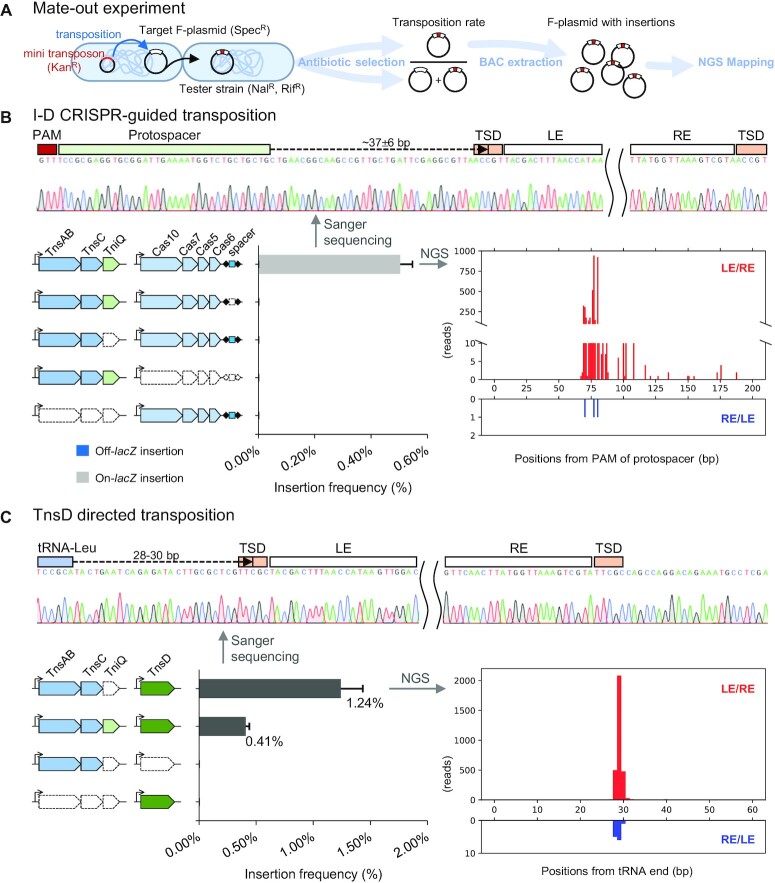
We selected the type I-D CAST from Myxacorys californica WJT36-NPBG1 (McCAST) for experimental validation in a heterologous E. coli host. McCAST is the only type I-D CAST where both ends of the element could be identified along with the characteristic target site duplication indicating transposition was used for the integration of the element. Additionally, all the CRISPR-associated and transposition genes were present and are not pseudogenes in this element (Figure 1D). As in previous validation work, RNA-guided transposition was tested in the heterologous E. coli host using a mate-out assay (5). In our assay, a mini-McCAST transposon with the cis-acting transposon ends flanking an antibiotic resistance determinant was situated on a donor plasmid and a lacZ gene maintained on an F plasmid derivative as a transposition target (Figure 2A). The cas and transposase genes were expressed from separate plasmids. The native single spacer array downstream of the cas operon was replaced with restriction sites for cloning and expressing candidate spacers. A spacer targeting the F plasmid-encoded lacZ gene was used for the transposition assay, and we used a GTT protospacer adjacent motif (PAM) known to be favored in many type I-D CRISPR systems (28,29,32,34). After inducing expression of the system, RNA-guided transposition events were detected and quantified by using conjugation to transfer F plasmids into a tester strain. Transposition assays indicated that the McCAST type I-D CRISPR-Cas was capable of guide RNA programmable transposition (Figure 2B). RNA-guided transposition only occurs when the lacZ targeting spacer and the Cascade and TnsABCQ proteins are expressed. In the assay, on-target and off-target transposition events are roughly estimated with LacZ activity (i.e. blue/white screen with X-Gal). Greater than 99% of the insertions render the F-plasmid LacZ− indicating a high level of guide RNA targeting. RNA-guided transposition was further verified by Sanger sequencing, showing the 5bp target site duplication at the transposon ends (Figure 2B). NGS mapping of F plasmids targets showed that the insertions are concentrated 75 ± 6 bp downstream from the GTT PAM. Deep sequencing also allowed us to visualize a small fraction of insertions trailing downstream from the preferred site, something not observed with other CAST subtypes. Consistent with other Tn7-related transposons, insertion events also show the expected orientation bias, with >99% of insertions having transposon left end adjacent to the target sites.
Figure 2.
In vivo transposition assay of the type I-D CRISPR-guided pathway and TnsD mediated tRNA-targeting with McCAST in E. coli (A) Cartoon representation of the mate-out assay strategy (plasmids expressing transposon and Cas function omitted for clarity). (B) Left: Frequency of McCAST transposition into F-lacZ with different genetic backgrounds. In the diagram, trials of the experiment missing genes or the spacer are shown as white with dashed outlines. Data are shown as mean + SD, n = 3. No off-lacZ insertions were detected, <0.1%. Top: Sanger sequencing of an on-target insertion. (TSD: target site duplication, LE: left end, RE: right end) Right: Type I-D CRISPR-guided insertion distribution revealed through deep sequencing. Red bars are insertions with their left ends proximal to the lacZ protospacer; blue bars are insertions with right ends proximal to the lacZ protospacer (Note differences in scale). (C) Left: Frequency of McCAST transposition into F-tRNA-Leu gene with different genetic backgrounds. Top: Sanger sequencing of an on-target insertion. Right: Insertion distribution of TnsD-guided transposition revealed through deep sequencing.
The second, larger TniQ (TnsD) was predicted to target transposition into the tRNA-Leu attachment site in M. californica WJT36-NPBG1 based on the informatics analysis presented above. To confirm this prediction, we constructed a target F-plasmid carrying a tRNA-Leu gene from M. californica WJT36-NPBG1 and a vector carrying the tnsD gene. We found that the TnsD protein can indeed direct insertions downstream of the tRNA-Leu gene at the position found natively in the M. californica genome (Figure 2C). This pathway requires only TnsABC and TnsD, consistent with a previous study on the PmcCAST tRNA targeting pathway (2). Similar to PmcCAST, we found that the expression of TniQ reduces the efficiency of the tRNA-targeting pathway, indicating the two TniQ family proteins may interfere with each other (2). Compared to the RNA-guided transposition events, the TnsD-guided insertions are more precise; almost all insertions are at 29 ± 1 bp after the target tRNA-Leu, similar to PmcCAST.
The type I-D McCAST system shows features of other guide RNA-directed transposition systems. As found with both canonical and CAST CRISPR-Cas systems, activity can vary between protospacers, even when they all contain the correct PAM. Transposition rates varied greatly when eight spacers in lacZ were randomly selected and tested, all with the predicted GTT PAM (Supplementary Figure S2A). These differences cannot be explained by which strand of DNA was targeted (Supplementary Figure S2A). The distribution of insertions fell within the range of 75 ± 6 bp downstream of the start of GTT PAM and almost all in a single orientation (Supplementary Figure S2B). The experiment confirmed the programmability of McCAST, even though the efficiencies are highly guide RNA-dependent.
To explore any differences from other CAST systems and canonical I-D CRISPR-Cas systems, we also examined mismatch tolerance. Previous structural work with type I systems indicates that every sixth position in the R-loop is flipped out and does not contribute to the specificity of the protospacer (35–38). Consistent with this idea, we found that a spacer with mismatches at every sixth position showed no reduction in transposition efficiency (Supplementary Figure S2C). Mismatches were not tolerated in the seed region and seed proximal region of the spacer. Consecutive 5 bp mismatches at any of the seed-proximal five Cas7 binding sites impairs transposition as much as the scrambled spacer control. Only mismatches at the most distal region where the most distal Cas7 is expected (31–35 bp) showed substantial transposition compared to controls (Supplementary Figure S2C).
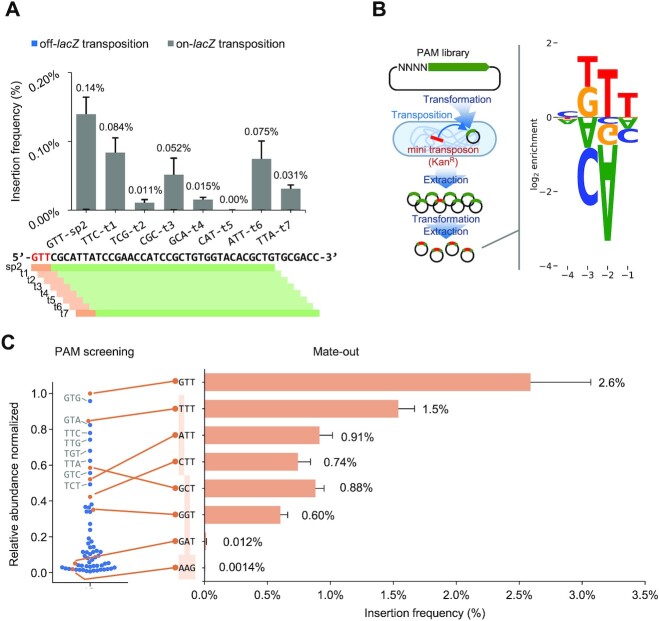
The type I-D McCAST element shows the PAM preference found with canonical I-D elements
Canonical I-F1 systems strongly prefer a CC PAM, while diverse type I-F3 CAST show high levels of PAM promiscuity (17,39) and in one case, an element (Tn7479) lacks any PAM requirement (39). To get more information on the sequence requirements of the type I-D CAST system, we monitored transposition frequency and targeting when we tiled crRNA downstream relative to lacZ spacer 2. The tiling spacer experiment showed that most spacers with non-GTN PAM on their targets allow low but detectable levels of guide RNA-directed transposition (Figure 3A). We conducted a PAM screen to investigate the PAM requirement of the type I-D McCAST system. A PAM library was made on a target plasmid with the most efficient protospacer we identified and used as a transposition target in vivo (Supplementary Figure S2A). Plasmids in the library with preferred PAMs should be over-represented as targets in a population of cells capable of McCAST transposition, and anti-PAMs should be underrepresented following deep sequencing of the population. PAM enrichment is measured by comparing the sequencing results of the PAM library before and after the screen. The type I-D McCAST showed no clear nucleotide preference at –4 position, while there was a clear G/T, T, T bias across the –3 through –1 positions (Figure 3B). There were clear sequences that were also biased against, suggesting anti-PAMs exist in the type I-D CAST system (Figure 3B). By plotting the normalized relative abundances of PAM sequences after screening as a swarm plot, the PAM requirement of McCAST aligns with the general GTN PAM of type I-D CRISPR systems, but in addition to GTN, TTN are also among the top performing PAMs (Figure 3C). In contrast, the NAN PAMs are all disfavored by McCAST; considering the type I-D CRISPR repeat also has an A at its –2 position, it can serve as an anti-PAM signal to reduce self-targeting. Direct testing of selected PAMs in the mate-out transposition assay confirmed the PAM screening results. Although our study revealed an unusual preference toward TTN PAM and showed that some other PAMs can support a modest level of transposition (Figure 3B, C), McCAST does not have PAM promiscuity as observed in many type I-F3 CAST elements (17,40).
Figure 3.
PAM preference of McCAST. (A) Spacers are tiled along lacZ gene in a 1-bp increment from lacZ spacer 2. The transposition efficiency of each spacer is determined with the mate-out assay. The three nucleotide PAM of each spacer is labeled. Data is shown as mean + SD. The off-lacZ transposition rates are too low to be visible in the bar chart. The position of each tiled spacer is illustrated below; green bars indicate protospacers, and orange bars indicate PAM. (B) The PAM screening process is illustrated on the left. The enrichment of PAM is determined with deep sequencing the library before and after selection, log2 scale enrichment of nucleotides at each position is shown on the right. (C) The relative abundance of PAMs normalized by the most abundant PAM are plotted on a swarm plot on the left. The PAMs with different nucleotides at the -4 position showed no clear preference at the position. To confirm the PAM screening results, eight different F plasmids carrying a lacZ fragment with different PAMs were constructed and tested for transposition efficiency with the mate-out assay with results indicated in the bar graph. Data are shown as mean + SD. The off-target rates are not measured in this experiment.
Extended spacers are functional for type I-D McCAST transposition
The CRISPR surveillance complexes of Class I CRISPR systems comprise multiple proteins and a crRNA; oligomerization of Cas7 family proteins on the RNA scaffold forms the backbone, while other proteins cap the ends. In many type I CRISPR-Cascades, Cas11 (small subunit) forms part of the complex on the guide RNA along with the Cas7 filament, similar to type III CRISPR-Cascades (41). In type I-A, I-E, the small subunit is encoded in a separate gene (41,42); while in type I-B, I-C, I-D, the small subunit is encoded within the large subunit gene (Cas8/Cas10) (28) (Supplementary Figure S3A). Many type I CRISPR-Cas systems can accommodate non-standard length spacers with changes in the oligomeric state of Cas7. For examples, the type I-E CRISPR-Cas from E. coli, the type I-Fv2 CRISPR-Cas from Shewanella putrefaciens CN-32, and the type I-F1 CRISPR-Cas from Aggregatibacter actinomycetemcomitans D7S-1 are all functional with extended spacers (43–45). Previous work on type I-F3 CAST Tn6677 found that shortening or extending a spacer greatly reduces the activity of RNA-guided transposition (4).
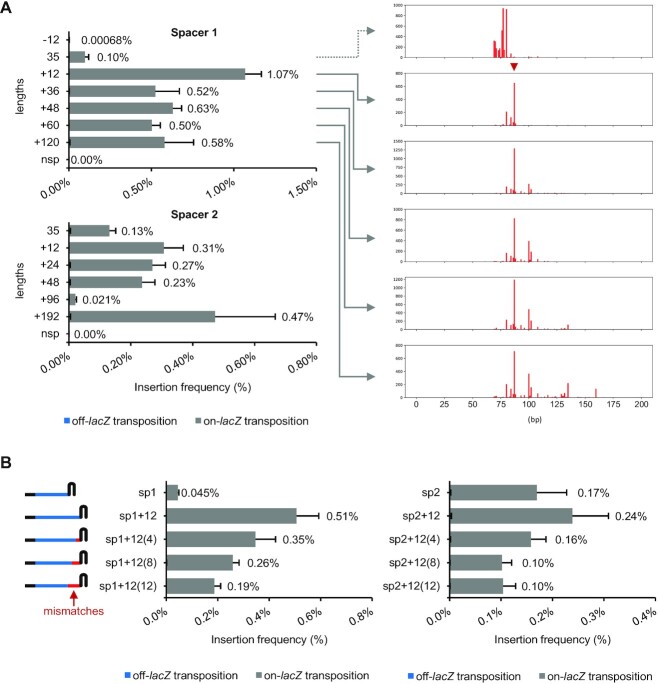
We tested for changes in functionality with changes in guide RNA length in the type I-D McCAST system. While shortening the spacer by 12 bp greatly diminished transposition, extended spacers were functional and generally showed a higher frequency of transposition (Figure 4, Supplementary Figure S4). Mapping insertions using NGS revealed that with the extended spacers, the resulting transposition events shift further downstream. Interestingly, while a portion of the transposition events could be shifted to increasing distances from the PAM with longer spacers, a prominent hotspot of insertions was fixed at ∼75 bp from the PAM. Longer guides revealed additional hotspots at ∼100 and ∼135 bp with this spacer. One possible explanation is that the crRNA of type I-D CRISPR-Cas may be a mixed population with different lengths. A heterogeneous mix of type I-D CRISPR crRNAs was found when transcripts from a native host were examined with high-throughput transcriptome analysis; the less abundant transcripts differ in length by about 6 nt intervals, suggesting the trimming and natural variation in the number of Cas7 (46). Recent structural studies on purified type I-D Cascade also observed the heterogeneity of the length of Cas7 filament (28,29). The pre-crRNA in type I and type III CRISPR-Cas systems is processed from the transcript by a Cas6 family endonuclease (Cas5 for type I-C) into functional guide RNAs. In some CRISPR-Cas families (type I-C, I-E, I-F), the nuclease remains part of the Cascade complexes. However, in other CRISPR-Cas subtypes, the Cas6 endonuclease dissociates (type III, I-A, I-B), and the crRNA is further processed at the 3′-end by an unresolved factor such as a host nuclease(s) (47), usually resulting in a heterogeneous population of crRNAs. Previous work showed the importance of the Cas11 subunit encoded withing the Cas10 gene (28). We overexpressed the Cas7 and Cas11 proteins (see Materials and methods) under the hypothesis that more of these proteins could be needed with extended spacer to coat the longer guide RNAs, but overexpression of these components modestly reduced the frequency of transposition and did not alter the distribution of insertions (Supplementary Figure S3).
Figure 4.
Impact of extended spacers on McCAST transposition and the resulting insertion distributions. (A) The lacZ spacers 1 and 2 with altered lengths were tested for transposition using the mate-out assay; the results are shown on the left as mean + SD. Spacers with native length are labeled with 35 (bp), and spacers with altered lengths are labeled with the number of nucleotides increased or decreased. nsp: the negative control without the spacer. Transposition events with selected spacers were mapped with deep sequencing as indicated. Note that the insertion profile data in the top panel in part A is the same data as in Figure 2B for comparison (indicated with a dotted line). (B) The effect of having mismatches on the distal part of extended spacers was tested with the mate-out assay and shown as mean + SD. The numbers of mismatches are labeled in parentheses. In all mate-out assays, n = 3. The off-lacZ bar is not visible because it is less than one percent of the total.
Extended spacers were also tested for their mismatch tolerance at the PAM distal extension. For CRISPR-Cas that were shown to be able to accommodate extended spacers, the type I-E CRISPR-Cas from E. coli was found to be susceptible to mismatches at the extension (43); on the contrary, the type I-F CRISPR-Cas from A. actinomycetemcomitans D7S-1 was found to be functional as long as its Cascade can form R-loop longer than 32 bp starting from 5′-end of spacer (45). An intermediate phenotype was found when we examined the type I-D McCAST system (Figure 4B, Supplementary Figure S4). The results differed slightly depending on the initial activity of the spacer chosen. Generally, increasing the length of the mismatched segment at the distal end modestly reduced transposition. Nonetheless, extended spacers are functional in McCAST.
The type I-D McCAST system can be engineered for simplified guide RNA maturation and independence from Cas6
Studies suggest that the Cas6 endonuclease may not be functionally essential in canonical type I-D CRISPR-Cas effectors. For example, the type I-D CRISPR-Cas from Sulfolobus islandicus LAL14/1 was shown to be functional in vitro without Cas6. Moreover, type I-D crRNAs extracted from S. islandicus LAL14/1 and Synechocystis sp. PCC6803 were truncated at the 3′-end, no longer maintaining the repeat sequence required for Cas6 binding (26,46). However, type I-D Cascade from Synechocystis sp. PCC6803 when expressed in E. coli showed Cas6 co-purified with full-length crRNA with the same stoichiometry as the complex (28).
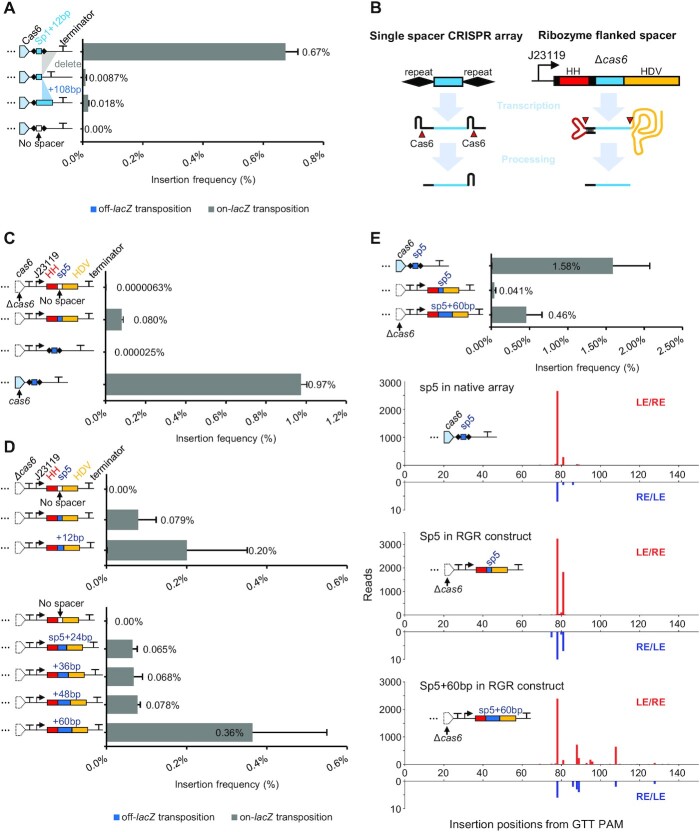
As the first test for Cas6d dispensability for guide RNA-directed transposition in the I-D McCAST system, we removed the downstream repeat normally required for Cas6d processing and binding at the 3′ end of the guide RNA complex. Removing the 3′ repeat drastically reduced transposition, but on-target transposition events were still detected, implying Cas6d activity was not essential (Figure 5A). Constructs with an extended spacer did not compensate for the loss of Cas6d processing (Figure 5A). To directly test if Cas6d was an essential component of the effector complex involved in type I-D McCAST transposition, we deleted cas6 gene and set up a ribozyme-catalyzed system for guide RNA production. In this synthetic construct, a constitutive heterologous J23119 promoter drives the expression of a guide RNA that functions as a self-processed ribozyme guide ribozyme (RGR) construct (Figure 5B). The RGR construct was initially developed to overcome the limitations of gRNA processing in non-native settings. Processing occurs via hammerhead and the hepatitis delta virus (HDV) ribozymes self-cleaving at the 5′ and 3′ of crRNA, respectively, thereby removing the need for native Cas6 processing activity (48). This construct allowed us to directly test Cas6d dependence and provided another mechanism of altering the length of guide RNAs. Guide RNAs produced in the systems were functional for guide RNA-directed transposition in the absence of Cas6d, while the same spacer cannot guide transposition without Cas6d in the context of a normal array (Figure 5C). Transposition rates varied with guide RNA length with the auto-processed RGR construct (Figure 5D), but the same general profile of insertions was found with the Cas6-processed and auto-processed RGR constructs (Figure 5E). Guides engineered to be +60 in length showed the same hotspot as the 35 base spacer, although a portion of the insertions extended from the hotspot with the extended construct (Figure 5E). While these results confirm that Cas6d is not essential for guide RNA-directed transposition with the type I-D CAST system, we cannot rule out a contributory role for Cas6 beyond processing. Optimization will need to be explored to allow the auto-processed system to match the frequency of transposition found with the native system. Guide RNA flexibility will be a promising parameter to explore in future work adapting this CAST system to heterologous hosts across all three domains (see Discussion).
Figure 5.
Examining the requirement of Cas6d for RNA-guided transposition. (A) Transposition efficiency of different array variants determined by the mate-out, data shown as mean + SD. Array structures are illustrated on the left. From top to bottom are lacZ spacer 1 with additional 12 nucleotides flanked by native repeats, the same spacer with downstream repeat removed, lacZ spacer 1 increased by 120 nucleotides with downstream repeat removed, PaqCI entry sites flanked by native repeats without target spacer. (B) Schematic of the crRNA processing by Cas6d and crRNA processing by ribozyme-guide-ribozyme (RGR) construct. HH: hammerhead ribozyme, HDV: Hepatitis delta virus ribozyme. (C) The frequency of transposition found with lacZ spacer 5 in the RGR construct or normal array or normal array construct without cas6. RGR construct without spacer is used as the control of lacZ spacer 5 in RGR construct. (D) Transposition efficiencies of lacZ spacer 5 with different lengths in RGR construct were determined with the mate-out assay and shown as mean + SD. RGR construct were tested at various lengths with PaqCI entry sites is used as no spacer control. The experiments are in two panels because they are done at two different times. (E) Comparing transposition rates of lacZ spacer 5 in a normal array construct with lacZ spacer 5 (and its extended version increased by 60 nucleotides) in the RGR construct. The frequency of transposition was determined with the mate-out assay and shown in mean + SD. The resulting insertions were mapped with deep sequencing. Note that the scales of reads of two insertion orientations are different. In all mate-out assays, n = 3. The off-lacZ bar is not visible because it is less than one percent of the total.
The type I-D McCAST element moves by cut-and-paste transposition and has regulatory characteristics similar to other CAST systems
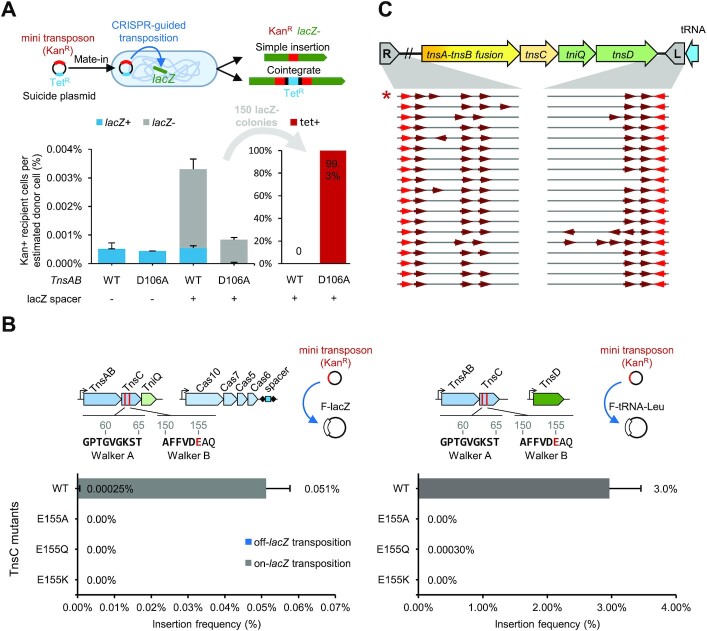
The entire tRNA-targeting branch where the McCAST and PmcCAST elements belong have TnsA and TnsB as a single polypeptide (Figure 1). In spite of having the TnsA nuclease domain, in a previous study, PmcCAST was found to form cointegrates at roughly the rate as found with TnsA-free type V-K CAST, suggesting its TnsAB fusion protein lacked the expected TnsA nuclease activity (2). To investigate the TnsA activity of McCAST, which is a close relative of PmcCAST (TnsAB a.a. identity 54%), we developed a transposition assay to measure the cointegrate rate with McCAST transposition. The assay utilizes a mate-in strategy to deliver a conditional donor plasmid into host cells where plasmid replication is not maintained. The use of the mate-in assay with a conditional plasmid helps guard against potential toxicity that could result from integrating a second origin of DNA replication into the chromosome, something that could favor confounding RecA-mediated cointegrate resolution. As in the mate-out transposition assays described above, transposition of the mini-McCAST element was directed to protospacers in the lacZ locus to estimate successful guide RNA-targeted transposition on agar selection plates containing X-Gal. Targeted transposition required the lacZ spacer in the assay (Figure 6A). The incidence of cointegrates could be monitored phenotypically because the conditional vector backbone used in this assay encoded resistance to tetracycline (TetR). On-target transposition events in the lacZ gene (white colonies on X-gal) were screened for TetR which indicated that none of the transposition events with the type I-D CAST system were cointegrates (0/150) (Figure 6A). As a control for the assay, we also tested an active site mutant predicted to inactive the nuclease activity in the TnsA active site and result in cointegrates (9). In the TnsAB(D106A) mutant, nearly all the transposition events were stable cointegrates (149/150), indicating these could be readily detected in the assay when present (Figure 6A). These experiments indicate that the TnsAB fusion found in the McCAST element has a functional TnsA nuclease activity catalyzing transposition via a cut-and-paste mechanism in the heterologous E. coli host.
Figure 6.
Characteristics of McCAST. (A) Examining TnsA activity with the mate-in assay. The experimental procedure is illustrated. The mini-McCAST element was encoded on a conditionally replicative plasmid that is transferred into recipient cells expressing RNA-guided transposition machinery via conjugation from donor cells. Four kinds of recipient cells are used, those with and without lacZ spacer 1 and having TnsAB(D106A) mutation or TnsAB wild type (WT). The amount of recipient cell colonies carrying mini-McCAST marker per donor cell (%) are shown on the left bar chart. LacZ + colonies are indicated in blue, while LacZ- colonies are indicated in white. White colonies were only found when the lacZ spacer was expressed, supporting RNA-guided transposition into the lacZ gene. Cointegrate formation was judged by testing for the plasmid backbone marker allowing TetR (n = 150), supporting cut-and-paste transposition in the wild-type configuration and coinetegrate formation in the TnsA(D106A) nuclease mutant (Right). (B) Different Walker B motif mutants are tested for their ability to support transposition. The Walker motif sequences and their positions are indicated. The key glutamate residue (E155) required for ATP hydrolysis is marked in red. All E155 mutants inactivate both transposition pathways, suggesting that ATP hydrolysis is required for transpositions. In all mate-out assays, n = 3. The off-lacZ bar is not visible because it is less than one percent of the total. (C) TnsAB binding sites arrangement on the ends of tRNA-targeting transposons with TnsAB fusion in cyanobacteria. Only examples where both ends and the expected target site duplication could be identified are included. Identical sequences were removed. The TnsAB binding site arrangement is unique among known Tn7-like elements. The asterisk marks the McCAST.
The core machinery of Tn7-like transposons is composed of a transposase TnsB and an AAA+ ATPase regulator protein, TnsC. TnsC forms the functional connection between the transposase and the target site selection proteins, playing roles in transposase activation and target immunity (49,50). Recent structural studies showed that in the type V-K ShCAST system TnsC directly interacts with target selection protein TniQ, and its ATPase activity is essential for transposition (11). In prototypic Tn7, ATPase activity of TnsC is also required for targeted transposition. While mutating the TnsC Walker B motif in type I-F3 CAST and V-K CAST systems abolishes transposition (4,11), inactivating Tn7 TnsC ATPase by mutating its Walker B motif resulted in unregulated random transposition (10). We tested different Walker B mutations of McCAST TnsC and found that the predicted loss of ATPase activity impairs both RNA-guided and TnsD-guided transposition pathways (Figure 6B). It is unclear what accounts for the different effects of Walker B mutations across Tn7 family elements, but it may suggest different recruitment mechanisms have evolved for signaling successful target site capture to the transposase.
The TGT/ACA end sequence is not universally conserved in Tn7-like transposons
The ends of Tn7-like family transposons have multiple TnsB binding sites set in an asymmetric arrangement that allows control over insertion orientation (51). The distribution of TnsAB binding sites differed from most other Tn7-like element families; TnsAB binding sites are found in both orientations in the left end instead of a single orientation as found in other elements (Figure 6C). Most Tn7-like transposons experimentally investigated thus far are bounded by 5′-TGT/ACA-3′. McCAST, however, is bounded by 5′-TAC/GTA-3′. Changing the McCAST ends to 5′-TGT/ACA-3′ had a modest effect on transposition frequency and showed no increase in off-site targeting outside lacZ (Supplementary Figure S5A), consistent with the idea that the end sequence requirement is not as strict as originally assumed (1,16). Examples of type I-F3 CASTs that are bounded by 5′-TGA/TCA-3′ were also recently found to be functional in E. coli (17). With these observations, we searched the transposon ends of Tn7-like transposons with tnsAB fusion and identifiable target-site-duplication in cyanobacteria with loosened criteria and found that almost 20% of transposons do not have 5′-TGT/ACA-3′ ends (Supplementary Figure S5B). Although the mechanism behind the conserveness of 5′-TGT/ACA-3′ is yet to be understood, the 5′-TGT/ACA-3′ is not universally conserved.
Extensive targeting flexibility and evidence of convergent evolution with Tn7-like elements in cyanobacteria
The Cas-coopting TniQ of type I-B2 and I-D CAST systems form their own phylogenetic clades indicating a single origin for each of these groups (Figure 1C, marked with green and red bars) (1,2). However, within the TnsAB elements examined in this study, we find that type I-B2 and type I-D CAST do not cluster into distinct branches but scatter across branches of the TnsAB similarity tree (Figure 7A). This indicates that there are horizontal exchanges of type I-B2 and I-D systems between elements. Within the TnsAB Tn7-like elements, we also found Tn5469, an element previously identified as a spontaneous insertion mutagen in cyanobacteria (52). The Tn7-like Tn5469 element has no cargo and only one TniQ and was identified in a screen for spontaneous inactivation of a gene (53), consistent with the idea that the element inserts without targeting an attachment site of a specific DNA sequence. The even simpler Tn5541 Tn7-like element with a TnsAB fusion and TnsC but lacking TniQ and cargo also likely lacks dual targeting pathway choice. Interestingly, the Tn5541 branch of elements has an extra extension at the C-terminal of its TnsC and only appears to be on plasmids in the sequenced representatives suggesting a novel type of targeting preference may be found with these elements found in cyanobacteria. Further work will be needed to better understand if the level of modularity we find is driven solely by selection or characteristics of the underlying components in these closely related systems that show >50% transposase identity.
Figure 7.
Diversity and evolutionary flexibility of Tn7-like transposons with TnsAB fusion in cyanobacteria (A) The unrooted TnsAB protein similarity tree of Tn7-like transposons in cyanobacteria. The branches that belong to transposons with a putative tRNA-targeting TnsD and an additional TniQ protein are colored based on the putative functions of the second TniQ. The legend indicates coopting type I-B2 Cas: green, coopting type I-D Cas: red, others: blue. The transposons without TniQ are colored light blue. Previously described transposons and McCAST are marked with an asterisk and labeled. (B) The gene arrangements of labeled transposons are illustrated.
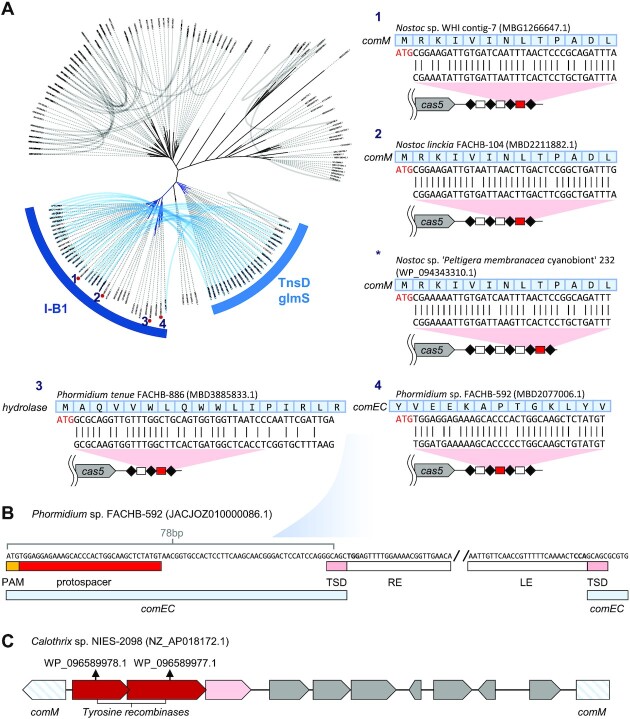
Our bioinformatic analysis indicates that convergent evolution is a repeating theme with Tn7-like elements. Convergent evolution has repeatedly selected diverse tRNA genes as targets by guide RNAs or as fixed sites directly recognized by a DNA binding domain (Figure 1C, Supplementary Table S1) (2,18). We also found examples of convergent evolution with Tn7-like elements acquiring targeting pathways directed at attachment sites where insertion inactivates genes responsible for natural transformation (genetic competency) (Figure 8). Applying the same analysis we used to discover the type I-D CAST systems, we identified multiple cases where the type I-B1 CASTs use the guide RNA system to target an attachment site in the chromosome. Of particular interest, we note multiple examples where candidate competence (com) genes are targets for guide RNA-directed transposition. These cases are reminiscent of the Tn6022 elements that utilize an attachment site that inactivates the comM gene (54). We find multiple examples where the final guide RNA encoded in the CRISPR array also targets the comM gene and in another case where the comEC gene is targeted (Figure 8A, B). Interestingly, as part of our analysis, we also identified a different kind of mobile element, a tyrosine recombinase-based integrating element that also targets the comM gene (Figure 8C). Presumably, there is an advantage for mobile elements to inactivate competence - possibly to prevent recombination from crossing-out the element—but whatever the advantage, it has evolved independently on multiple occasions.
Figure 8.
Convergent evolution observed in type I-B1 CASTs. (A) TniQ protein similarity tree of Tn7-like transposons with a separate TnsA. TniQ proteins encoded in the same transposon are connected with curved lines. The type I-B1 CASTs are indicated in blue. The putative type I-B1 CRISPR coopting TniQ and glmS targeting TnsD are labeled. Some transposons with only type I-B1 CRISPR coopting TniQ are found to carry a chromosome attachment site targeting spacer (att-spacer) similar to results found with type I-F3 and type V-K CASTs. The att-spacers and protospacers of the transposons are illustrated, their positions on the tree are marked with numbers (*: the transposon is not on the tree because its tnsB is a pseudogene). All contained an ATG PAM as expected with type I-B CRISPR-Cas systems. Multiple transposons target comM, one targets comEC, and another targets an unknown hydrolase. (B) The comEC targeting transposon ends and protospacer sequence are illustrated. The transposon is also predicted to have non-TGT/ACA ends. (C) A mobile genetic element with tyrosine recombinases also targets comM from the indicated GenBank accession.
DISCUSSION
Tn7-like elements are abundant in cyanobacterial genomes, including most subtypes that are capable of RNA-guided transposition. Here we discovered and characterized a novel cooption of a type I-D CRISPR-Cas system for RNA-guided transposition. Interestingly, the mechanism used for coopting the CRISPR-Cas system is distinct from the other well-studied examples. The major interface between the TniQ protein and I-F3 Cascade is via Cas6f (55) while in the I-D McCAST system described here, the Cas6d protein is not essential for guide RNA-directed transposition. Both the type I-F3 and I-D systems show a low level of off-site targeting and tight orientation control. Unlike the I-F3 CAST elements, the I-D McCAST element maintains a PAM preference found in the canonical CRISPR-Cas system where it was likely derived and we could identify a robust anti-PAM property with the I-D system. Maintaining the PAM system is advantageous for limiting targeting into the CRISPR array, an issue found with the type I-F3 systems that show extensive PAM ambiguity (17,56). Confirming cut-and-paste transposition with the type I-D system also increases the potential utility of the McCAST system for future work. Flexibility in accommodating a variety of guide RNA lengths and independence from Cas6 through guides that are auto-processed with ribozymes should further simplify adapting the I-D system to new heterologous hosts. The ability to extend the guide RNA may also allow future adaptations, such as tethers that could be appended to the PAM distal region of the guide.
It is unclear why most of the examples of cooption allowing guide RNA-directed transposition are found in cyanobacteria. There may be special unknown benefits to the CAST lifestyle in cyanobacteria, but diverse Tn7-like elements in general are common supporting the idea that tight control of transposase activity likely paved the way for evolving numerous new targeting mechanisms (54). Canonical type I-D and I-B systems are common in cyanobacteria, which may help explain why members of the I-D and I-B groups were coopted. No known canonical type V-K CRISPR-Cas system has been identified, and it is likely that the adaptation system from canonical type I-D systems is used for spacer acquisition with type V-K CAST (18).
The type I-D CRISPR-Cas system shows features suggesting a more recent CRISPR-Cas cooption event than other systems. The type I-F3 CAST systems are more diverged from the canonical I-F1 systems than is found with the type I-D CAST and canonical systems (1,57). Maintenance of a robust PAM system with type I-D CAST is also consistent with the idea that cooption was more recent. Consistent with a recent capture with the I-D system, we find a canonical system that is 56% identical to McCAST with its central Cas10d protein (MBD1847458.1) and also a type I-D CAST element that appears to maintain the Cas1,2,4 adaptation system (Cyanothece sp. PCC 7425, accession number NC_011884).
Convergent evolution is a repeating theme with Tn7-like elements. The capture of the type I-D CRISPR-Cas system represents the fifth experimentally validated independent cooption event with guide RNA-directed transposition systems. Multiple groups of Tn7-like elements converged on the strategy of using separate TniQ family proteins, including the type I-B1 and I-B2 systems and the type I-D system described here (Supplementary Figure S6). In another example of Tn7-like elements using separate TniQ family proteins to allow two transposition pathways, the I-F3 TniQ-Cascade system was coopted by a family of Tn7-like elements with a TniQ targeting an attachment site downstream of parE (1,5,17). The type I-F3 and V-K CAST systems converged on the strategy where separate classes of guide RNA evolved to allow a targeting system that recognizes a conserved attachment site in the chromosome and a separate series of guides that targets mobile elements capable of cell-to-cell transfer. Interestingly, our analysis here indicates that the I-B1 family has undergone a similar transition in re-evolving dual pathways using different guide RNAs with a single TniQ protein (Figure 8 and Supplementary Figure S6). Of further interest, Tn7-like elements have converged on the programmed targeting of the comM gene. Our work here provides multiple new lines of support for the control of host gene uptake systems with the lifestyle of integrating mobile genetic elements. Bacteriophage integration into the comK gene of Listeria monocytogenes and integration of Tn6022 into the comM of Acinetobacter baumannii both compromise natural competence (58,59). A recent preprint also suggests the ϕOXC141 prophage downregulates host natural competence (60).
Bioinformatics analysis of Tn7-like elements continues to reveal interesting new modalities (1,14–16,18). Expanding on our previous assumptions with these elements and mining larger data sources should continue to provide additional insights and design possibilities with Tn7-like elements. Our results here indicate that carefully focused searches in specific groups of prokaryotes will also be important for finding rare types of Tn7-like elements and other mobile genetic elements that do not follow the same rules gleaned from earlier studies.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Elizabeth Kellogg, Michael Petassi, and Zach Barth for their comments on the manuscript. We thank Nicole Pietrasiak for providing the strain used in this work from the Terrestrial Cyanobacterial Culture Collection (NuMex TCCC).
Contributor Information
Shan-Chi Hsieh, Department of Microbiology, Cornell University, Ithaca, NY 14853 USA.
Joseph E Peters, Department of Microbiology, Cornell University, Ithaca, NY 14853 USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01GM129118, R21AI148941]. Funding for open access charge: NIH.
Conflict of interest statement. The Peters lab has corporate funding for research that is not directly related to the work in this publication. Cornell University has filed a provisional patent application on this work with SCH and JEP as inventors
REFERENCES
- 1. Peters J.E., Makarova K.S., Shmakov S., Koonin E.V.. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saito M., Ladha A., Strecker J., Faure G., Neumann E., Altae-Tran H., Macrae R.K., Zhang F.. Dual modes of CRISPR-associated transposon homing. Cell. 2021; 184:2441–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Strecker J., Ladha A., Gardner Z., Schmid-Burgk J.L., Makarova K.S., Koonin E.V., Zhang F.. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019; 365:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Klompe S.E., Vo P.L.H., Halpin-Healy T.S., Sternberg S.H.. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature. 2019; 571:219–225. [DOI] [PubMed] [Google Scholar]
- 5. Petassi M.T., Hsieh S.C., Peters J.E.. Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cell. 2020; 183:1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peters J.E. Tn7. Microbiol. Spectr. 2014; 2: 10.1128/microbiolspec.MDNA3-0010-2014. [DOI] [PubMed] [Google Scholar]
- 7. Bainton R., Gamas P., Craig N.L.. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991; 65:805–816. [DOI] [PubMed] [Google Scholar]
- 8. Sarnovsky R., May E.W., Craig N.L.. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 1996; 15:6348–6361. [PMC free article] [PubMed] [Google Scholar]
- 9. May E.W., Craig N.L.. Switching from cut-and-paste to replicative Tn7 transposition. Science. 1996; 272:401–404. [DOI] [PubMed] [Google Scholar]
- 10. Shen Y., Gomez-Blanco J., Petassi M.T., Peters J.E., Ortega J., Guarne A.. Structural basis for DNA targeting by the Tn7 transposon. Nat. Struct. Mol. Biol. 2022; 29:143–151. [DOI] [PubMed] [Google Scholar]
- 11. Park J.U., Tsai A.W., Mehrotra E., Petassi M.T., Hsieh S.C., Ke A., Peters J.E., Kellogg E.H.. Structural basis for target site selection in RNA-guided DNA transposition systems. Science. 2021; 373:768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parks A.R., Li Z., Shi Q., Owens R.M., Jin M.M., Peters J.E.. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell. 2009; 138:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waddell C.S., Craig N.L.. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 1988; 2:137–149. [DOI] [PubMed] [Google Scholar]
- 14. Benler S., Faure G., Altae-Tran H., Shmakov S., Zheng F., Koonin E.. Cargo genes of Tn7-Like transposons comprise an enormous diversity of defense systems, mobile genetic elements, and antibiotic resistance genes. Mbio. 2021; 12:e0293821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faure G., Shmakov S.A., Yan W.X., Cheng D.R., Scott D.A., Peters J.E., Makarova K.S., Koonin E.V.. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat. Rev. Microbiol. 2019; 17:513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rybarski J.R., Hu K., Hill A.M., Wilke C.O., Finkelstein I.J.. Metagenomic discovery of CRISPR-associated transposons. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2112279118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klompe S.E., Jaber N., Beh L.Y., Mohabir J.T., Bernheim A., Sternberg S.H.. Evolutionary and mechanistic diversity of type I-F CRISPR-associated transposons. Mol. Cell. 2022; 82:616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh S.-C., Peters J.E.. Tn7-CRISPR-Cas12K elements manage pathway choice using truncated repeat-spacer units to target tRNA attachment sites. 2021; bioRxiv doi:06 February 2021, preprint: not peer reviewed 10.1101/2021.02.06.429022. [DOI]
- 19. Khlebnikov A., Datsenko K.A., Skaug T., Wanner B.L., Keasling J.D.. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology (Reading). 2001; 147:3241–3247. [DOI] [PubMed] [Google Scholar]
- 20. Metcalf W.W., Jiang W., Daniels L.L., Kim S.K., Haldimann A., Wanner B.L.. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996; 35:1–13. [DOI] [PubMed] [Google Scholar]
- 21. Sibley M.H., Raleigh E.A.. A versatile element for gene addition in bacterial chromosomes. Nucleic Acids Res. 2012; 40:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peters J.E. Methods for General and Molecular Microbiology. 2014; American Society of Microbiology; 735–755. [Google Scholar]
- 23. Ward R.D., Stajich J.E., Johansen J.R., Huntemann M., Clum A., Foster B., Foster B., Roux S., Palaniappan K., Varghese N.et al.. Metagenome sequencing to explore phylogenomics of terrestrial cyanobacteria. Microbiol. Resour. Announc. 2021; 10:e0025821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Letunic I., Bork P.. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021; 49:W293–W296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lin J., Fuglsang A., Kjeldsen A.L., Sun K., Bhoobalan-Chitty Y., Peng X.. DNA targeting by subtype I-D CRISPR-Cas shows type I and type III features. Nucleic Acids Res. 2020; 48:10470–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F.et al.. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011; 9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McBride T.M., Schwartz E.A., Kumar A., Taylor D.W., Fineran P.C., Fagerlund R.D.. Diverse CRISPR-Cas complexes require independent translation of small and large subunits from a single gene. Mol. Cell. 2020; 80:971–979. [DOI] [PubMed] [Google Scholar]
- 29. Schwartz E.A., McBride T.M., Bravo J.P.K., Wrapp D., Fineran P.C., Fagerlund R.D., Taylor D.W.. Structural rearrangements allow nucleic acid discrimination by type I-D cascade. Nat. Commun. 2022; 13:2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., Abudayyeh O.O., Gootenberg J.S., Makarova K.S., Wolf Y.I.et al.. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017; 15:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kieper S.N., Almendros C., Haagsma A.C., Barendregt A., Heck A.J.R., Brouns S.J.J.. Cas4-Cas1 Is a protospacer adjacent motif-processing factor mediating half-Site spacer integration during CRISPR adaptation. CRISPR J. 2021; 4:536–548. [DOI] [PubMed] [Google Scholar]
- 32. Kieper S.N., Almendros C., Behler J., McKenzie R.E., Nobrega F.L., Haagsma A.C., Vink J.N.A., Hess W.R., Brouns S.J.J.. Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 2018; 22:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Osakabe K., Wada N., Murakami E., Miyashita N., Osakabe Y.. Genome editing in mammalian cells using the CRISPR type I-D nuclease. Nucleic Acids Res. 2021; 49:6347–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osakabe K., Wada N., Miyaji T., Murakami E., Marui K., Ueta R., Hashimoto R., Abe-Hara C., Kong B., Yano K.et al.. Genome editing in plants using CRISPR type I-D nuclease. Commun. Biol. 2020; 3:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson R.N., Golden S.M., van Erp P.B., Carter J., Westra E.R., Brouns S.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B.. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014; 345:1473–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pausch P., Muller-Esparza H., Gleditzsch D., Altegoer F., Randau L., Bange G.. Structural variation of type I-F CRISPR RNA guided DNA surveillance. Mol. Cell. 2017; 67:622–632. [DOI] [PubMed] [Google Scholar]
- 37. Mulepati S., Heroux A., Bailey S.. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014; 345:1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo T.W., Bartesaghi A., Yang H., Falconieri V., Rao P., Merk A., Eng E.T., Raczkowski A.M., Fox T., Earl L.A.et al.. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell. 2017; 171:414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roberts A., Nethery M.A., Barrangou R.. Functional characterization of diverse type I-F CRISPR-associated transposons. Nucleic Acids Res. 2022; 50:11670–11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang S., Zhang Y., Xu J., Zhang J., Zhang J., Yang J., Jiang Y., Yang S.. Orthogonal CRISPR-associated transposases for parallel and multiplexed chromosomal integration. Nucleic Acids Res. 2021; 49:10192–10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Venclovas C. Structure of Csm2 elucidates the relationship between small subunits of CRISPR-Cas effector complexes. FEBS Lett. 2016; 590:1521–1529. [DOI] [PubMed] [Google Scholar]
- 42. Xiao Y., Luo M., Hayes R.P., Kim J., Ng S., Ding F., Liao M., Ke A.. Structure basis for directional R-loop formation and substrate handover mechanisms in type I CRISPR-Cas system. Cell. 2017; 170:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luo M.L., Jackson R.N., Denny S.R., Tokmina-Lukaszewska M., Maksimchuk K.R., Lin W., Bothner B., Wiedenheft B., Beisel C.L.. The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 2016; 44:7385–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gleditzsch D., Muller-Esparza H., Pausch P., Sharma K., Dwarakanath S., Urlaub H., Bange G., Randau L.. Modulating the Cascade architecture of a minimal type I-F CRISPR-Cas system. Nucleic Acids Res. 2016; 44:5872–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tuminauskaite D., Norkunaite D., Fiodorovaite M., Tumas S., Songailiene I., Tamulaitiene G., Sinkunas T.. DNA interference is controlled by R-loop length in a type I-F1 CRISPR-Cas system. BMC Biol. 2020; 18:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scholz I., Lange S.J., Hein S., Hess W.R., Backofen R.. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013; 8:e56470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Behler J., Sharma K., Reimann V., Wilde A., Urlaub H., Hess W.R.. The host-encoded rnase E endonuclease as the crRNA maturation enzyme in a CRISPR-Cas subtype III-Bv system. Nat. Microbiol. 2018; 3:367–377. [DOI] [PubMed] [Google Scholar]
- 48. Gao Y., Zhao Y.. Self-processing of ribozyme-flanked rnas into guide rnas in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant. Biol. 2014; 56:343–349. [DOI] [PubMed] [Google Scholar]
- 49. Stellwagen A.E., Craig N.L.. Mobile DNA elements: controlling transposition with ATP-dependent molecular switches. Trends Biochem. Sci. 1998; 23:486–490. [DOI] [PubMed] [Google Scholar]
- 50. Li Z., Craig N.L., Peters J.E.. Roberts A.P., Mullany P.. Bacterial Integrative Mobile Genetic Elements. 2013; Landes Bioscience; 1–32. [Google Scholar]
- 51. Arciszewska L.K., Craig N.L.. Interaction of the Tn7-encoded transposition protein TnsB with the ends of the transposon. Nucleic Acids Res. 1991; 19:5021–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schaefer M.R., Kahn K.. Cyanobacterial transposons Tn5469 and Tn5541 represent a novel noncomposite transposon family. J. Bacteriol. 1998; 180:6059–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kahn K., Schaefer M.R.. Characterization of transposon Tn5469 from the cyanobacterium Fremyelladiplosiphon. J. Bacteriol. 1995; 177:7026–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peters J.E., Fricker A.D., Kapili B.J., Petassi M.T.. Heteromeric transposase elements: generators of genomic islands across diverse bacteria. Mol. Microbiol. 2014; 93:1084–1092. [DOI] [PubMed] [Google Scholar]
- 55. Halpin-Healy T.S., Klompe S.E., Sternberg S.H., Fernandez I.S.. Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system. Nature. 2020; 577:271–274. [DOI] [PubMed] [Google Scholar]
- 56. Vo P.L.H., Ronda C., Klompe S.E., Chen E.E., Acree C., Wang H.H., Sternberg S.H.. CRISPR RNA-guided integrases for high-efficiency, multiplexed bacterial genome engineering. Nat. Biotechnol. 2021; 39:480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P.et al.. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rabinovich L., Sigal N., Borovok I., Nir-Paz R., Herskovits A.A.. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012; 150:792–802. [DOI] [PubMed] [Google Scholar]
- 59. Godeux A.S., Svedholm E., Lupo A., Haenni M., Venner S., Laaberki M.H., Charpentier X.. Scarless removal of large resistance island AbaR results in antibiotic susceptibility and increased natural transformability in Acinetobacterbaumannii. Antimicrob Agents Chemother. 2020; 64:e00951-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwun M.J., Ion A.V., Cheng H.-C., D’Aeth J.C., Dougan S., Oggioni M.R., Goulding D.A., Bentley S.D., Croucher N.J. Post-vaccine epidemiology of serotype 3 pneumococci identifies transformation inhibition through prophage-driven alteration of a non-coding RNA. 2022; bioRxiv doi:21 September 2022, preprint: not peer reviewed 10.1101/2022.09.21.508813. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.