Abstract
Chlorambucil (CLB) belongs to the class of nitrogen mustards (NMs), which are highly reactive bifunctional alkylating agents and were the first chemotherapeutic agents developed. They form DNA interstrand crosslinks (ICLs), which cause a blockage of DNA strand separation, inhibiting essential processes in DNA metabolism like replication and transcription. In fast replicating cells, e.g., tumor cells, this can induce cell death. The upregulation of ICL repair is thought to be a key factor for the resistance of tumor cells to ICL-inducing cytostatic agents including NMs. To monitor induction and repair of CLB-induced ICLs, we adjusted the automated reversed fluorometric analysis of alkaline DNA unwinding assay (rFADU) for the detection of ICLs in adherent cells. For the detection of monoalkylated DNA bases we established an LC-MS/MS method. We performed a comparative analysis of adduct formation and removal in five human cell lines and in peripheral blood mononuclear cells (PBMCs) after treatment with CLB. Dose-dependent increases in adduct formation were observed, and suitable treatment concentrations were identified for each cell line, which were then used for monitoring the kinetics of adduct formation. We observed significant differences in the repair kinetics of the cell lines tested. For example, in A2780 cells, hTERT immortalized VH10 cells, and in PBMCs a time-dependent repair of the two main monoalkylated DNA-adducts was confirmed. Regarding ICLs, repair was observed in all cell systems except for PBMCs. In conclusion, LC-MS/MS analyses combined with the rFADU technique are powerful tools to study the molecular mechanisms of NM-induced DNA damage and repair. By applying these methods to a spectrum of human cell systems of different origin and transformation status, we obtained insight into the cell-type specific repair of different CLB-induced DNA lesions, which may help identify novel resistance mechanisms of tumors and define molecular targets for therapeutic interventions.
Abbreviations: NM, Nitrogen mustard; CLB, chlorambucil; PBMCs, peripheral blood mononuclear cells; rFADU, reverse fluorometric analysis of alkaline DNA unwinding; ICL, interstrand crosslink; MS, mass spectrometry; NER, nucleotide excision repair; BER, base excision repair; dG, 2'-deoxyguanosine; SD, standard deviation; PI, propidium iodide; hTERT, human telomerase reverse transcriptase; RPE-1, human retinal pigment epithelial; VH10, human foreskin fibroblasts
Keywords: Nitrogen mustard, Chlorambucil, Interstrand crosslink, Monoalkylated DNA adducts, DNA repair kinetics, Mass spectrometry
1. Introduction
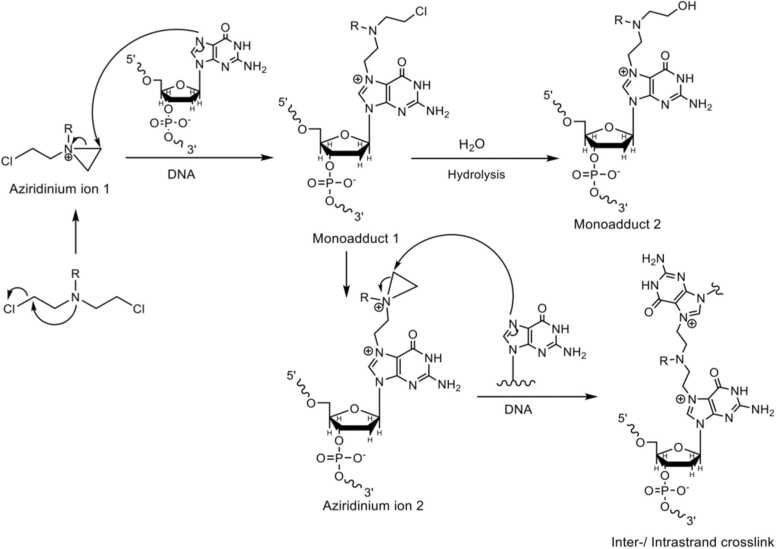
NMs are highly reactive bifunctional alkylating agents and represent the oldest chemotherapeutic drugs still in clinical use [1], [2], [3], [4], [5], [6], [7]. One prominent example is the drug Leukeran, with the nitrogen mustard CLB as its active component [8]. CLB is mostly used in the chemotherapeutic treatment of chronic lymphocytic leukemia, non-Hodgkin's lymphoma, and Hodgkin's disease [9]. Because of the sustained importance of these mustards as anticancer agents, their mechanism of action and the adducts formed have been studied extensively [10], [11], [12], [13], [14]. This class of cytostatic agents reacts via an intramolecular cyclization reaction of the SN1 type, thereby forming aziridinium ions [4], [15], [16]. Due to the high reactivity of these ions, they react immediately with nucleophilic groups of, e.g., water, DNA, RNA, lipids, or proteins. When reacting with nucleobases in DNA, a monoadduct is formed (Fig. 1A, monoadduct 1). Gruppi et al. showed that in a cellular context, the hydrolysis of this monoadduct leads to the main product of the reaction, i.e. the corresponding alcohol derivative (Fig. 1A, monoadduct 2) [17]. If no solvolysis takes place a second aziridinium ion is formed, which likewise can react with one of the above-mentioned nucleophiles. As a consequence, unreactive monoadducts, as well as DNA, RNA, or protein crosslinks are formed [17], [18]. In Fig. 1A, the formation of a DNA N7-guanine crosslink is depicted. The most abundant lesions induced by NMs like CLB are monoadducts at N3-adenine and N7-guanine as well as inter- and intrastrand crosslinks, e.g., bis-N7-guanine crosslinks at 5’-GNC-3’ sequences; crosslinks between N7 of guanine and N3 of adenine; or bis-N3-adenine adducts (see Fig. 1B) [17], [19], [20], [21], [22]. To a lesser extent, alkylations at N3 of cytosine and N1 of adenine were also found [20].
Fig. 1.
General reaction scheme of a NM with guanine and the major DNA lesions induced. From [19] with some modifications.
Even though the N7-guanine and N3-adenine monoadducts are the most abundant products of the reaction with NMs like CLB, there is a lot of evidence that the small amount of ICLs that is formed (1–5%) is the main contributing factor in the cytostatic effect of these drugs [20], [23], [24], [25]. In line with this finding is the fact that the half-mustards, carrying only one reactive center, are much less toxic than their bifunctional counterparts [26], [27]. One explanation for this effect is that, compared to monoadducts, the repair of an ICL is a complex task for the DNA repair machinery of the cell. Several different repair pathways have to be orchestrated to remove an ICL, and the presence (S-Phase) or absence (G1-Phase) of an intact sister chromatid also plays an important role regarding the repair factors to be recruited [28], [29]. Unrepaired ICLs lead to a complete block of the separation of the complementary DNA strands and therefore inhibit several essential processes such as DNA replication and transcription [28], [30]. If the replication fork stalls at an ICL, it is processed into a double-strand break (DSB), which in turn stops cell cycle progression [28], [31], [32], [33]. Another consequence of ICLs is that DNA-protein interactions (e.g., transcription factors binding to DNA) at the affected sites of the DNA are no longer possible. Especially in fast replicating cells such as tumor cells, this will lead to apoptosis or, if this is no longer possible, to mitotic catastrophe due to DNA damage accumulation after NM treatment [23], [34]. The presence of as few as 20–40 unrepaired ICLs is thought to be lethal for a mammalian cell [23], [35]. The correlation between the number of DNA alkylation lesions in cancer cells and the response to the chemotherapeutic treatment is the reason why DNA adducts are thought to be predictive markers in precision medicine. However, due to the low abundance of these lesions in the cell (∼ 0.01–10 adducts per 108 nucleobases), only a very limited number of methods is available for the detection of the adducts [36]. In previous studies the repair of site-specific ICLs in plasmids was mostly monitored via the quantification of the PCR products of the plasmids [37], [38], [39] or via the ability of restriction enzymes to cut at specific sites [40], [41], [42]. For quantification of the number of ICLs formed after treatment with crosslinking agents in the DNA of cellular samples, the modified alkaline comet assay was performed [43], [44]. However, the inter-experiment variability of this assay is high due to standardization issues and the problem with objective sample evaluation [45], [46]. A more reliable method is mass spectrometry (MS), which is frequently used in the detection and quantification of DNA adducts. MS-based methods were also used to identify and quantify ICLs after treatment with crosslinking agents [35], [47], [48], [49], [50], [51], [52]. Even though this method is the gold standard in DNA adductomics, it has a clear disadvantage regarding the detection of ICLs: An ICL is not distinguishable from an intrastrand crosslink if only the nucleobase adduct is detected. Since an intrastrand crosslink is nothing more than a very bulky monoadduct, the cellular response is very different from that of an ICL. Therefore, it has to be ascertained beforehand, e.g., by synthesizing a site-specific ICL, that it is indeed an ICL that is being monitored and not an intrastrand crosslink. In previous studies, our research group was able to implement the automated rFADU method for the detection of ICLs in PBMCs after treatment with a crosslinking agent and, due to the automation of the pipetting process, we could overcome standardization and identification issues of the manually performed FADU assay [46].
As the ICLs formed after treatment with crosslinking agents are thought to be the main cytotoxic lesion, only a very limited number of studies was focused on the aspects of the formation and repair of the monoadducts after treatment with NMs, even though effective repair of monoadducts may lead to a reduced formation of ICLs and therefore to a poor outcome of chemotherapy. In general, bulky DNA adducts involving only one strand are repaired via the nucleotide excision repair (NER) pathway. However, regarding the removal of the DNA-adducts formed after treatment with NMs, there is evidence for either the involvement of the NER pathway [24], [53], [54] or the base excision repair (BER) pathway [55], [56], [57]. The proteins/genes involved in the repair of melphalan-induced monoadducts have been reviewed recently, and an involvement of both the BER and the NER pathway seems possible [58]. In a study performed by Vassilis et al., the repair of the monoadducts as well as the ICLs formed after in-vitro treatment with melphalan were monitored. The authors were able to show a reduction in both adduct types after several hours, but the method used did not allow structural analysis and the different adducts could not be distinguished [59].
To fully elucidate the cellular effects and the repair of the CLB monoadducts in comparison to the ICLs, we developed a mass spectrometric platform to detect the N3-adenine and N7-guanine alkylation monoadducts formed after treatment with CLB and adapted the automated rFADU assay to simultaneously detect ICLs in the adherent cell lines used. A combined analysis of the MS data and the rFADU data revealed a dose-dependent increase in adduct formation in different human cell lines and in PBMCs after treatment with CLB. A suitable treatment concentration was identified for each cell system and used for the subsequent experiments, where the removal of adducts formed was monitored. It was important to simultaneously monitor cell proliferation and cell death in order to exclude that the apparent decrease in adduct levels was the consequence of preferential death of cells carrying a high adduct burden, or else, preferential proliferation of cells with low adduct burden, rather than active DNA repair. We discovered significant differences in the repair capacity of the cell lines tested as well as in the kinetics of the removal of the various adducts.
2. Materials and methods
2.1. Chemicals
CLB with a purity of at least 98% (tested via HPLC analysis), as well as LC-MS grade ammonium acetate were purchased from Sigma-Aldrich (Merck). LC-MS grade formic acid was purchased from Honeywell and LC-MS grade acetonitrile from Carl Roth.
2.2. Cell culture
U2OS [60], HaCaT [61] and hTERT immortalized VH10 cells [62] were cultured in DMEM (Gibco) supplemented with 10% fetal calf serum (Biochrom), 100 µg/ml streptomycin, and 100 U/ml penicillin (Gibco) at 37 °C and 5% CO2 in a humidified incubator. A2780 cells (ATCC) were cultured in RPMI medium (Gibco) with the same supplements. PBMCs were cultured in TexMACS medium (MACS Miltenyi Biotec) under the same conditions. For the cultivation of hTERT immortalized RPE-1 cells [63] a 1:1 mixture of DMEM (Gibco) and Ham’s F12 medium (Gibco) supplemented with 10% fetal calf serum (Biochrom), 100 µg/ml streptomycin, and 100 U/ml penicillin (Gibco) was used. The cells were washed with PBS (Biochrome). Trypsin-EDTA (0.05%; Thermo Scientific) was used to detach the cells. hTERT immortalized podocytes (Evercyte) were cultured in a 1:1 mixture of DMEM (Gibco) and Ham’s F12 (Gibco) supplemented with 1% fetal calf serum (Biochrom), 1% ITS (Insulin, Transferrin, Selenium; Gibco), 1% GlutaMAX (Gibco), 100 µg/ml streptomycin, and 100 U/ml penicillin (Gibco). Podocytes were washed with PBS (Biochrome) and detached with accutase (Sigma-Aldrich; Merck). Culture dishes used for podocytes were coated with human collagen type 1 (Sigma-Aldrich; Merck). The absence of mycoplasma contamination was confirmed using mycoplasmacheck from Eurofins.
2.3. Isolation of PBMCs
Eighty to 90 ml blood from healthy volunteers was collected in S-monovette 10 ml 9 NC (Sarstedt). The collection of the blood samples was approved by the Ethics Committee of the University of Konstanz. Whole blood was centrifuged at 300 ×g for 10 min at room temperature without brake. The plasma was discarded and the hematocrit pellet was resuspended with pre-warmed PBS to 50 ml. 15 ml Biocoll (Biochrome) was carefully overlaid with 25 ml of the hematocrit suspension and centrifuged at 900 ×g for 15 min at room temperature without brake. The PBMC fraction was collected and ice-cold PBS was added until a volume of 50 ml was reached. After centrifugation (300 ×g, 10 min, 4 °C), the cells were resuspended in PBS and counted by using a CASY cell counter (model TT; Schärfe-System). Two million cells were transferred into a new reaction tube and PBS was added until a volume of 1 ml was reached. This cell suspension was treated with CLB.
2.4. CLB treatment of cells
Prior to each experiment, predilutions of CLB in EtOH/HCl (95%/0.5%; v/v) were freshly prepared. For the final treatment solution, the respective volume of the predilution was added to the pre-warmed PBS in a ratio of 1:1000. The cells were washed with PBS and the treatment solution was added. The cells were incubated with the CLB solution for 1 h at 37 °C in a humidified incubator. Then, the treatment solution was removed, the cells were washed with pre-warmed PBS and medium was added for further culturing, or alternatively the cells were detached with trypsin. The harvested cells were washed with ice-cold PBS and the pellets were either frozen in liquid nitrogen or directly used for the rFADU assay. The frozen pellets were stored at − 80 °C until further use.
2.5. Cell lysis and DNA extraction
The extraction of the DNA of all cell systems used in this study was performed with the Quick-DNA Miniprep Plus Kit from Zymo Research according to the manufacturer’s manual. Briefly, cell pellets were thawed and the cell ghosts were resuspended in 200 µl PBS. 200 µl cell buffer (red) and 20 µl Proteinase K were added. The samples were vortexed and incubated for 10 min at 55 °C. 420 µl genomic binding buffer was added to each sample. After vortexing, the samples were transferred to a Zymo Spin IIC-XL column and centrifuged. Following three washing steps with 400 µl DNA pre-wash buffer as well as 700 µl and 200 µl g-DNA wash buffer, the DNA was eluted from the column with 50 µl pre-warmed elution buffer (70 °C) after 5 min incubation. This step was repeated and the purified DNA was then enzymatically digested.
The DNA of each sample was digested as described in Zubel et al. (2018) [64]. Briefly, 2 U benzonase (Sigma-Aldrich) and 3.2 U DNase I (Roche) were added to each sample in a Tris-HCl (630 mM)/MgCl2 (63 mM) buffer with a pH of 8 (final concentration Tris/MgCl2: 10.3 mM/1 mM). After thorough mixing, the samples were incubated at 37 °C for 2 h. Then, 5 U alkaline phosphatase (Sigma-Aldrich) and 2.5 U phosphodiesterase (Qiagen) were added to the sample in a NaOAc buffer (final concentration: 12.4 mM, pH 7.8). After incubation of the samples at 37 °C for 2 h, the samples were filtered using the NanoSep Omega 10 K centrifugal devices (Pall) at 17,000 ×g for 10 min. Finally, the samples were thermally hydrolyzed by incubation at 70 °C for 1 h.
2.6. LC-MS/MS analysis
To detect the analytes in the samples prepared, an ACQUITY UPLC H-Class coupled to a XEVO TQ-S triple quadrupole mass spectrometer (Waters) was used. The autosampler was cooled to 15 °C, and separation of the sample mixture was performed using a BEH C18 column (130 Å; 1.7 µm; 2.1 ×50 mm; Waters) heated to 30 °C with a flow of 0.353 ml/min. Milli-Q water was used as mobile phase A and LC-MS grade acetonitrile (Carl Roth) as mobile phase B. Both mobile phases were supplemented with 0.01% formic acid (Fluka). To separate the sample mixture, a gradient with the starting conditions of 100% A was used. The starting conditions were held constant until 4.7 min. Then, B was increased to 15% until a runtime of 5.7 min. A further increase of B to 30% followed. These conditions were held constant until a runtime of 6.6 min. Finally, the gradient ended with the starting conditions (100% A) held constant until 9.5 min
An ESI source in the positive ion mode was used for the ionization. The capillary voltage was set to 0.9 kV, the cone voltage to 50 V, the source offset to 50 V, whereas the desolvation temperature was set to 500 °C. The source was heated to 100 °C and the desolvation gas flow was set to 1000 l/h, whereas the cone gas flow was set to 150 l/h. Finally, the nebulizer gas flow was set to 7 bar and the collision gas flow to 0.15 ml/min. The analyzer resolution used was the quantitative mass resolution.
The monoalkylated DNA adducts of adenine (N3-CLB-Ade) and guanine (N7-CLB-Gua) formed after the treatment with CLB were measured, and the background signal was subtracted in the calculation. A cone voltage of 6 V and a collision voltage of 10 V were used for the MS measurement. The dwell time was set to 43 ms. The fragmentation pattern [M+H]+ of the two analytes was the following: N3-CLB-Ade 385.2 > 250.1 and N7-CLB-Gua 401.2 > 250.1.
2.7. Synthesis and purification of the external and internal MS standards
Synthesis and purification of the analytical standards were based on a method established by Zubel et al. (2019), with some modifications [64]. Briefly, solutions of 2'-deoxyguanosine (dG) (Sigma-Aldrich, 800 µg/ml) and 13C–15N-labeled dG (Euriso-Top, 1 mg/ml) were prepared using Na2HPO4 buffer (Sigma-Aldrich, 50 mM, pH 7) as diluent. The nucleoside solutions were treated with 5 mM CLB for 4 h at room temperature and stored at − 20 °C for 12 h. To remove the deoxyribose, thermal hydrolysis at 90 °C was performed for 1 h. For purification purposes, a Discovery DSC-18 SPE tube (5 mg, Sigma-Aldrich) was used. First, the column was equilibrated with 8 ml 100% methanol followed by 8 ml 1% methanol (v/v). The column was washed with 30 ml 1% methanol (v/v), 30 ml 5% methanol (v/v), 30 ml 10% methanol (v/v), and 15% methanol (v/v) after the sample was loaded. To elute the N7-CLB-Gua, 16 ml of 20% methanol (v/v) and 8 ml of 80% methanol (v/v) was used. Subsequently, two regeneration steps with 8 ml methanol were performed. Collected fractions were analyzed using the established UPLC-MS/MS method (chapter 2.6), and those containing the desired N7-CLB-Gua were pooled and concentrated using a vacuum concentrator (Vacusafe comfort, Integra Biosciences). For the final purification step, a 2695 Alliance Separation module (Waters) along with a Synergi 4 µm Fusion-RP column (80 Å, 250 ×10 mm, Phenomenex) with a flow of 3.3 ml/min was used. The gradient started with 100% solvent A (2 mM ammonium acetate buffer, pH 3.0). Solvent B (acetonitrile with 0.1% acetic acid) was increased in the first 5 min to 7.5%. Until 20 min, solvent B was further increased to 15%. Finally, the level of solvent B was increased to 30% until 25 min and held constant for another 25 min. After collecting the purified N7-CLB-Gua, the standard was concentrated again and stored at − 20 °C until further usage. Verification of the standard was achieved by a product ion scan using a high-end XEVO TQ-S triple quadrupole mass spectrometer (Waters) with a collision energy of 10 V and a cone voltage of 6 V.
2.8. Automated rFADU assay
The rFADU assay was performed as described in Debiak et al. (2011), which is based on Moreno-Villanueva et al. (2009) [46], [65]. Briefly, 106 cells were resuspended in 1 ml ice-cold suspension buffer (10 mM Na3PO4; 0.25 mM meso-inositol; 1 mM MgCl2; pH 7.4) and 100 µl of the cell suspension was pipetted into 2-ml reaction tubes (Sarstedt). The cells were kept on ice and were X-irradiated with a dose of 25 Gy. All of the following pipetting steps were performed by the liquid handling device Genesis RSP 100 (Tecan AG), which was pre-cooled to − 5 °C. 500 µl ice-cold suspension buffer was added to the samples, and 70 µl of this suspension were transferred into a 96-well plate in triplicates. To lyse the cells, 70 µl of lysis buffer (9 M urea; 10 mM NaOH; 2.5 mM 1,2-cyclohexanedinitrilotetraacetic acid; 0.1% SDS) was added (speed: 150 µl/s) and the cell suspension was incubated for 12 min at 0 °C. Then, 70 µl ice-cold alkaline buffer (42.5% lysis buffer; 0.2 M NaOH) was added at 10 µl/s, and the samples were incubated for 60 min at 30 °C. To stop the alkaline unwinding process, 140 µl neutralization solution (1 M glucose; 14 mM β-mercaptoethanol) was added (speed: 200 µl/s) and the samples were incubated for 30 min at 22 °C. To detect the total amount of DNA present in each sample, control samples were first incubated with neutralization buffer and then with alkaline buffer. For the detection of dsDNA, 156 µl of a SybrGreen/H2O solution (1:8333; v/v; Thermo Fisher Scientific) was added to each sample. After mixing, the fluorescence was measured using a Tecan plate reader (Tecan) with an excitation wavelength of 485 nm and an emission wavelength of 535 nm. The following samples were analyzed: P0: untreated sample (with alkaline unwinding); Px: treated sample (with alkaline unwinding); T0: untreated sample (without unwinding); and Tx: treated sample (without unwinding). The amount of dsDNA remaining after the alkaline unwinding step, which is directly proportional to the amount of ICLs, was calculated as described in Debiak et al. (2011) [46]:
2.9. Annexin V/propidium iodide analysis
One day prior to the experiment, the cells were seeded in 6-well plates (Corning) such that on the treatment day 75% confluence was reached. In particular, the following cell numbers were seeded: 4 × 105 (U2OS, A2780), 6 × 105 (HaCaT), 2 × 105 (RPE-1), 3.3 × 105 (VH10), 7 × 105 (Podocytes). Regarding the PBMCs, 2 × 106 cells were used for the treatment after the isolation from whole blood. The cells were treated as indicated in the respective figure. 24 h after the treatment, medium was removed and cells were washed with PBS. Cells were detached with trypsin and the cell suspension was added to the collected medium and PBS. After centrifugation at 300 ×g for 10 min, the pellet was resuspended in 5 ml ice-cold PBS and the cell number was determined by using a CASY cell counter. The volume containing 2 × 105 cells was transferred into a new reaction tube and the cells were pelleted at 4 °C. Subsequently, the cells were resuspended in 100 µl Annexin V binding buffer (10 mM HEPES, pH 7.4; 140 mM NaCl; 2.5 mM CaCl2) and stained with one drop of the Annexin V APC ready flow conjugate (Thermo Fischer Scientific) as well as 5 µl propidium iodide (PI) (1 mg/ml; Sigma-Aldrich). After incubation of the cells in the staining solution for 15 min in the dark, 400 µl Annexin V binding buffer were added and the samples were kept on ice until analysis with a FACSLyric instrument (BD Sciences). 10,000 events per condition were analyzed. The amount of viable, apoptotic, and dead cells was calculated using the FlowJo software (version 10.7.1; Becton Dickinson & Company).
2.10. Carboxyfluorescein succinimidyl ester assay
Cells were detached and washed with PBS twice. The cell number was determined using a CASY cell counter and the cell suspension was then centrifuged for 10 min at 400 ×g. For each condition 2 × 105 cells were either stained with 10 µM carboxyfluorescein succinimidyl ester (CFSE) staining solution (Abcam) in PBS or left unstained (untreated sample). For the staining procedure, 1 ml staining solution per 106 cells consisting of a 1:1 mixture of 20 µM CFSE solution and PBS was used. The pellet was resuspended in the staining solution. After incubation of the cells with the staining solution for 10 min at 37 °C, uptake of the staining solution was quenched with 30 ml ice-cold medium. To remove the remaining CFSE, the cells were washed with PBS. The pellet was resuspended in pre-warmed medium and 2 × 105 cells per well were seeded in 6-well plates. 24 h later, the cells were treated with CLB as described in chapter 2.3. After the treatment samples were incubated for the time points indicated. Subsequently, cells were washed and detached from the plates. To stop proliferation, the cells were washed with ice-cold PBS and centrifuged at 300×g for 12 min at 4 °C. The pellet was resuspended in 500 µl FACS buffer (PBS; 10% FCS; 10 mM EDTA), then 5 µl PI (1 mg/ml; Sigma-Aldrich) was added. After incubation in the dark for 15 min, the samples were analyzed with a FACSLyric instrument (BD Sciences). 10,000 events per condition were analyzed. The amount of viable and proliferating cells was calculated using the FlowJo software (version 10.7.1; Becton Dickinson & Company). To account for the loss of dead cells, which were excluded from the analysis, a correction factor for each sample was used to normalize the number of proliferating cells. The correction factor used was the percentage of PI negative cells.
2.11. Statistics
Statistical analysis was performed using the GraphPad Prism software (GraphPad software Inc.) version 6.07. For the analysis of the experiments, one-way ANOVA with Dunnett’s post-test was conducted. P-values are reported as follows: *p˂0.05, * *p˂0.01, * **p˂0.001, and * ** *p˂0.0001.
3. Results
3.1. Development of a mass spectrometric platform to measure CLB-induced DNA adducts
Calf thymus DNA treated with CLB was used as a model system for the development of a mass spectrometric platform to measure the major DNA lesions induced by this NM. To obtain the desired nucleobase adducts (Fig. 2B), the CLB-treated DNA was enzymatically hydrolyzed using benzonase and DNase I as well as phosphodiesterase I and phosphatase. After the completion of the enzymatic hydrolysis, the sugar moiety of the nucleosides that had formed was removed by thermal hydrolysis. A gradient for the separation of the two adducts of interest, N3-CLB-Ade and N7-CLB-Gua, via UPLC was successfully established. With this gradient, separation of the two adducts could be achieved with a retention time of 5.5 – 5.7 min for N3-CLB-Ade and 6.2 – 6.4 min for N7-CLB-Gua (Fig. 2A).
Fig. 2.
LC-MS chromatograms of the main DNA adducts formed after treatment with CLB. A. Calf thymus DNA was treated with 500 µM CLB for 4 h at room temperature and the DNA monoadducts formed (N3-CLB-Ade and N7-CLB-Gua) as well as their non-reacted counterparts (dA and dG) were detected using UPLC-MS/MS. The adducts were separated on a BEH C18 column using a water-acetonitrile gradient. Shown is a representative chromatogram. B. Structural formulae of the two monoadducts N3-CLB-Ade (m/z 385.19) and N7-CLB-Gua (m/z 411.20). The arrow indicates the fragmentation point after CID in the MS of the two adducts and the mass-to-charge ratio of the detected CLB-derived fragment (m/z 250.14).
Unlabeled standards were successfully synthesized using nucleosides, which were treated in a similar manner to the calf thymus DNA, except for the enzymatic hydrolysis and a harsher thermal hydrolysis at 90 °C for 1 h. The standards were purified via C18-based solid-phase extraction and subsequent separation via HPLC. For verification of the synthesized standards, product ion scans were performed (Suppl. Figs.1 and 2). For the synthesis of the labeled N7-CLB-Gua standards, the same procedure was conducted with 13C, 14N-labeled nucleosides. The verification via product ion scan is shown in Suppl. Fig. 3. In all three product ion scans conducted, the respective precursor ion (labeled N7-CLB-Gua m/z 411; unlabeled N7-CLB-Gua m/z 401; unlabeled N3-CLB-Ade m/z 385) as well as the CLB fragment formed after the fragmentation of the precursor ion with a m/z of approx. 250 could be detected.
Fig. 3.
Method development for the detection of ICLs in adherent cells using the rFADU assay. HaCaT cells were treated for 1 h at 37 °C with HN2 solutions (A) or CLB solutions (B) at the concentrations indicated. After treatment, the cells were X-irradiated on ice with 25 Gy. The amount of ICLs was determined via the rFADU assay. Each column represents the mean of 3 independent experiments ± standard deviation (SD). Statistical analysis was performed using one-way ANOVA test with the Dunnett’s post-test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001. (C) Collagen-coating test experiment with RPE-1 cells treated with CLB. RPE-1 cells were treated with the doses of CLB indicated above, and the induction of the ICLs was measured via the rFADU assay. The treatment was performed with cells growing on culture dishes, which were coated with collagen or not coated. Three independent experiments were performed and the mean ± the SD is shown. Statistical analysis was performed using the one-way ANOVA test with the Dunnett’s post-test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
A calibration curve of the unlabeled N7-CLB-Gua standard was recorded (Suppl. Fig. 4). To do this, the sample was spiked with the purified isotopically labeled N7-CLB-Gua standard to account for sample losses during the injection and ionization process. The detection range was linear for up to 1000 fmol of N7-CLB-Gua.
Fig. 4.
Cellular effects of CLB treatment in the human ovarian carcinoma cell line A2780. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. A2780 cells were treated with CLB for 1 h at 37 °C and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 4 independent experiments, except 500 µM CLB (n = 3). Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C, and ICLs were monitored using the rFADU assay. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Time-course analysis of CLB-induced DNA ICLs. Cells were treated with 50 µM CLB for 1 h at 37 °C, and ICLs were monitored using the reverse automated FADU assay. Treated cells were allowed to recover for the time points indicated, up to 51 h. Data represents the mean ± SD from 4 independent experiments, except 28 h and 32.5 h (n = 3). Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (E) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 100 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 30 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments, except 3 h and 21.5 h (n = 3). Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation in C. and E. (F) Cytotoxicity as well as proliferation status of cells after the treatment with the indicated treatment concentrations of CLB were monitored as a function of time. A2780 cells were stained with 10 µM CFSE and after 24 h, the cells were treated for 1 h with 50 µM (△) or 100 µM of CLB (▲). Treated cells were allowed to recover for the time points indicated, and the samples were additionally stained with PI before flow cytometric analysis. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
For the subsequent dose-response and time-course experiments to be performed by MS, the specific focus was on the relative changes, including time-course analyses. Therefore, the synthesized standards were only used for setting up the mass spectrometric detection method and for the identification of the adducts of interest. To ensure the suitability of this approach, U2OS cells were treated with increasing concentrations of CLB (0 – 1000 µM), and the N7-CLB-Gua adducts formed were measured with or without the synthesized standards (Suppl. Fig. 5). As no significant difference between the two measurement options was observed, the above mentioned strategy was implemented.
Fig. 5.
Cellular effects of CLB treatment in the human osteosarcoma U2OS cell line. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. U2OS cells were treated with CLB for 1 h at 37 °C and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C and ICLs were monitored using the reverse automated FADU assay. Data represents the mean ± SD from 4 independent experiments, except for 2.5 µM and 5 µM (n = 3). Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Time-course analysis of CLB-induced DNA ICLs. Cells were treated with 100 µM CLB for 1 h at 37 °C and ICLs were monitored using the rFADU assay. Treated cells were allowed to recover for the time points indicated, up to 51 h. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (E) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 200 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 30 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation in C. and E. (F) Cytotoxicity as well as proliferation status of cells after the treatment with the indicated treatment concentrations of CLB were monitored as a function of time. U2OS cells were stained with 10 µM CFSE and after 24 h, the cells were treated for 1 h with 100 µM (△) or 200 µM of CLB (▲). Treated cells were allowed to recover for the time points indicated, and the samples were additionally stained with PI before the flow cytometric analysis. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
3.2. Development of an rFADU method to measure CLB-induced DNA ICLs
We have previously described an automated version of the rFADU method to measure the amount of ICLs formed in suspension cells after treatment with crosslinking agents [46]. To enable detection of ICLs in adherent cells, several adjustments had to be made, and the suitability of this method had to be verified. To do this, HaCaT cells were treated with increasing concentrations of HN2 and CLB, both being crosslinking agents of the NM family, and subjected to the sample preparation protocol described in the Materials and Methods section. A statistically significant and dose-dependent increase in the formation of ICLs was visible for both crosslinking agents, with HN2 showing twice the level of ICLs as compared to CLB (Fig. 3, A & B). To ensure that this method can also be used for cells growing on plates coated with collagen, RPE-1 cells were seeded on coated and on non-coated plates. After the treatment with CLB, ICLs were detected and no difference between the two conditions was observed (Fig. 3C). This result suggests that the established rFADU protocol is indeed suitable for the detection of ICLs in adherent cells.
3.3. Cellular impact of the treatment with CLB on different target cell systems
The repair capacity of each cell type may vary vastly from one another and is highly dependent on the type of the DNA lesion that should be removed. Especially in the field of chemotherapy, the question of whether or how fast a DNA lesion is repaired is crucial. An enhanced or efficient repair of the DNA lesions formed after treatment with a chemotherapeutic agent is a characteristic feature of a chemotherapy resistant tumor [66]. To gain insights into the cellular impact of the treatment of different human cells with the chemotherapeutic agent CLB, including the DNA repair capacity, the following endpoints were systematically examined in all biological systems investigated in this study: Viability, proliferation, the amount of monoalkylated as well as bisalkylated DNA adducts, and the removal of these lesions over time.
3.3.1. Cancer cells
3.3.1.1. A2780 cells
For the analysis of the DNA adducts, a treatment window had to be determined. Analysis of the kinetics of the removal of these lesions is only possible with a sublethal treatment. Therefore, the human ovarian carcinoma cell line A2780 was treated with various concentrations of CLB, and the viability of the cells was determined (Fig. 4A). Annexin V/PI staining revealed a significant reduction in viable cells, starting at a concentration of 100 µM CLB.
For the detection of ICLs, the automated rFADU assay was used. To determine the sensitivity of this method, A2780 cells were treated with concentrations of CLB as indicated, and the fluorescence intensity, which is proportional to the amount of ICLs, was detected (Fig. 4B). A dose-dependent increase in the number ICLs could be observed. Starting at a concentration of 50 µM CLB, the ICLs could readily be detected. In combination with the results of the Annexin V/PI staining, 50 µM was chosen as the concentration of CLB, which was high enough to induce a significantly increased adduct load as well as being sublethal. Based on these results, the cells were treated with 50 µM CLB for the subsequent time-course experiments, where the kinetics of the formation and removal of the CLB-induced ICLs was monitored (Fig. 4C). After an initial increase of ICLs in the first 6 h, which was also observed in earlier studies, the number of ICLs was significantly reduced to approx. 65% of the maximum value after 22.5 h [46], [67]. Over the course of 51 h, the number of ICLs was further reduced by 10%. All in all, a reduction of this DNA adduct of 45% could be observed in this cancer cell line.
ICLs, the most toxic lesion induced by CLB, are not the most abundant ones. ICLs only make up 1–5% of the lesions, whereas the monoalkylated adenine and guanine bases N7-CLB-Gua and N3-CLB-Ade are formed in a much higher frequency [19], [20], [23]. For the detection of these two monoadducts, the MS method we have developed was used. As for the determination of its sensitivity and the detection of the formation and removal of these lesions in A2780 cells, absolute quantification was not considered necessary to gain the desired insights into the repair capacity status line. Therefore, the synthesized standards were not used for these experiments. The sensitivity of the MS method was determined for the A2780 cells by performing a dose-response experiment with increasing concentrations of CLB (Fig. 4D). At a concentration range of 50–100 µM CLB, a significant increase in the signal intensity could be observed. Throughout the concentration series, a clear-cut dose-dependent increase in the signal for both monoadducts was visible. Taken together, a suitable treatment concentration for the time-course experiment, where the formation and removal of the two mostly formed monoadducts N7-CLB-Gua and N3-CLB-Ade was to be monitored, was 100 µM of CLB. After treatment with this concentration, an initial increase in both adducts could be observed until 3 h post-treatment (Fig. 4E). This increase was followed by a rapid decrease for both adducts resulting in a significantly reduced signal at 21.5 h (N3-CLB-Ade 40%; N7-CLB-Gua 58%). Interestingly, the reduction of the N7-CLB-Gua signal was in general slower than that of the N3-CLB-Ade adduct. In comparison to the ICLs formed in A2780 cells after treatment with CLB, the removal of the monoadducts was significantly faster. After 51 h, a reduction in adducts of 48% (N7-CLB-Gua) or 65% (N3-CLB-Ade) of the maximum signal was observed.
To exclude the possibility of a pseudo-removal of the bis- or the monoadducts due to a dilution effect by preferential cell proliferation of those cells with the least DNA adducts formed after the treatment, the proliferation status of CLB-treated cells was assessed by CFSE proliferation assay. For each cell line tested, the assay settings were verified for its suitability. A representative image of the gating strategy (6 A), the resulting histograms of untreated (6B), and of the treated cells (6 C) as well as an image of the analysis using the proliferation tool of FlowJo is shown in Suppl. Fig. 6. To ensure that the treatment with the fluorescent dye CFSE has no effect on the formation of both monoadducts, a control experiment was performed where the cells were treated with either CLB or CFSE in combination with CLB (Suppl. Fig. 7). As no significant difference between the two treatment options was visible, the pre-treatment with CFSE does not seem to have any effect on the formation of DNA monoadducts after the treatment with CLB. To verify the removal of the CLB-DNA adducts over a time-course of 30 h (monoadducts) or 51 h (ICLs), A2780 cells were treated with the same concentrations of CLB as in the performed time-course experiments (Fig. 4C & E). To exclude a bias of the data due to a hypothetical preferential cell death, the survival rate of these cells was monitored simultaneously. With this experiment, we wanted to see if the observed reduction in DNA adducts was indeed due to the actual repair of these lesions. Until 6 h post-treatment, the viability increased for both CLB concentrations (Fig. 4F). For the 50 µM treatment concentration, the survival rate was stagnant up to 32.5 h, but after 51 h the survival rate dropped to 84%. For the higher treatment concentration, a significant drop in the viability could be detected after 32.5 h. At 51 h, the viability was decreased to 78%. Simultaneously, the division rate increased to 8% (100 µM) or 4% (50 µM) at 32.5 h (Fig. 4F). After this slight increase in cells, which underwent one proliferation cycle, the division rate either dropped (100 µM) or increased (50 µM) again to 6%, respectively.
Fig. 6.
Cellular effects of CLB treatment in the hTERT immortalized RPE-1 cell line. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. RPE-1 cells were treated with CLB for 1 h at 37 °C and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 4 independent experiments, except for 200 µM H2O2 (n = 3). Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C and ICLs were monitored using the reverse automated FADU assay. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Time-course analysis of CLB-induced DNA ICLs. Cells were treated with 200 µM CLB for 1 h at 37 °C and ICLs were monitored using the rFADU assay. Treated cells were allowed to recover for the time points indicated, up to 51 h. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 5 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (E) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 300 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 30 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation in C. and E. (F) Cytotoxicity as well as proliferation status of cells after the treatment with the indicated treatment concentrations of CLB were monitored as a function of time. RPE-1 cells were stained with 10 µM CFSE and after 24 h, the cells were treated for 1 h with 200 µM (△) or 300 µM of CLB (▲). Treated cells were allowed to recover for the time points indicated and the samples were additionally stained with PI before the flow cytometric analysis. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
Fig. 7.
Cellular effects of CLB treatment in the hTERT immortalized VH10 cell line. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. VH10 cells were treated with CLB for 1 h at 37 °C, and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C and ICLs were monitored using the rFADU assay. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Time-course analysis of CLB-induced DNA ICLs. Cells were treated with 100 µM CLB for 1 h at 37 °C and ICLs were monitored using the rFADU assay. Treated cells were allowed to recover for the time points indicated, up to 51 h. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (E) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 400 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 30 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation in C. and E. (F) Cytotoxicity as well as proliferation status of cells after the treatment with the indicated treatment concentrations of CLB were monitored as a function of time. VH10 cells were stained with 10 µM CFSE and after 24 h, the cells were treated for 1 h with 100 µM (△) or 400 µM of CLB (▲). Treated cells were allowed to recover for the time points indicated, and the samples were additionally stained with PI before the flow cytometric analysis. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
3.3.1.2. U2OS cells
To investigate if the DNA repair of CLB-induced lesions is tissue or cell type specific, further cell lines were tested including the p53 wild-type U2OS cell line, which was isolated from the biopsy of a mesenchymal tumor [68]. In contrast to the A2780 cells, the susceptibility of the U2OS cells towards the treatment with CLB and H2O2, which was detected via the Annexin V/PI staining, was far less pronounced (Fig. 5A). No apoptosis could be induced, even at 500 µM H2O2. A significant drop in the viability could only be detected at concentration of 600 µM CLB or higher.
The sensitivity of the rFADU assay for the detection of ICLs in U2OS cells was determined with a dose-response experiment, where the cells were treated with increasing doses of CLB and the increase in ICLs was monitored (Fig. 5B). The three highest concentrations 100 µM, 200 µM, and 500 µM CLB induced the formation of ICLs significantly. Based on the results obtained with this cell line, 100 µM CLB was chosen as the treatment concentration for time-course experiments. In these experiments, an increase in the number of ICLs directly after the treatment with CLB was observed (Fig. 5C). This increase was visible until 4 h post-treatment, where the maximum signal was reached. Only at 32.5 h or later, a significant reduction of the ICLs to 78% was visible. The signal further decreased to 56%, indicating that after 51 h only approx. half of the ICLs were removed.
The sensitivity of the MS method was assessed by the measurement of U2OS cell-derived samples treated with increasing concentrations of CLB. A clear-cut dose-dependent increase in the number of N3-CLB-Ade and N7-CLB-Gua adducts could be observed. The lowest concentration where both adducts could be reliably detected was 200 µM of CLB (Fig. 5D). As U2OS cells only showed a reduction of viable cells after a treatment concentration of 600 µM CLB, 200 µM CLB was used as a treatment concentration for monitoring the removal of the monoadducts over time. Compared to the results obtained with the A2780 cells, the difference between the N3-CLB-Ade and N7-CLB-Gua adduct was even more pronounced (Fig. 5E). Over the course of 30 h no adduct removal could be observed for the N7-CLB-Gua adduct, whereas the N3-CLB-Ade adduct was reduced to 36% of the maximum value after 21.5 h. In the last 8.5 h nearly no change in the adduct level of N3-CLB-Ade could be detected.
A presumed dilutive effect of preferential proliferation of cells with low adduct burden on the removal of CLB-induced adducts was analyzed in the U2OS cells with the CFSE proliferation assay using the respective treatment concentrations for the time-course experiments (100 µM ICLs; 200 µM monoadducts). Furthermore, the influence of a possible preferential cell death on the adduct removal was assessed simultaneously with the PI cell death staining. A significant reduction in the survival rate to 85% for both treatment concentrations could only be observed at 32.5 h (Fig. 5F). After 51 h, the survival rate was reduced to 83% (100 µM CLB) or 75% (200 µM CLB). Simultaneously, the percentage of divided cells was significantly increased at 32.5 h to approx. 2.5% for the treatment concentration of 200 µM or 4% for the treatment with 100 µM CLB (Fig. 5F). Following this slight increase, the number of cells, which underwent one proliferation cycle increased further to 9% (200 µM CLB) or 16% (100 µM CLB).
3.3.2. hTERT immortalized cell systems
3.3.2.1. RPE-1 cells
The usage of cancer cell lines is very common in the DNA damage response field due to their cheap and easy handling. These cell lines, however, have the inherent disadvantage of deficiencies or mutations of genes, which may be crucial for the response to DNA damaging agents [69]. For this reason, we wished to compare the repair capacity of the cancer cell lines used with cell lines that had been immortalized via the overexpression of the human telomerase reverse transcriptase (hTERT). Previous studies confirmed that the overexpression of hTERT led to the immortalization of the cell system without causing changes which are usually associated with cancer [70], [71]. The first hTERT immortalized cell system tested was the human retinal pigment epithelial (RPE-1) cell line. The cytotoxicity induced by the treatment of these cells with CLB was detected using the Annexin V/PI staining (Fig. 6A). A significant increase in dead cells was only observed at the two highest concentrations of 1 mM and 2 mM CLB.
In line with the low cytotoxicity displayed by RPE-1 cells after the treatment with CLB, the sensitivity of the rFADU for the detection of the CLB-induced ICLs was in the upper treatment range. Only at treatment concentrations of 200 µM and 500 µM CLB significant increases in ICLs were observable with this method (Fig. 6B). Therefore, the cells were treated with 200 µM CLB for the subsequent time-course experiment. An increase in ICLs was detected in the first 4–6 h post-treatment (Fig. 6C). The ICL signal did not change significantly in the time period between 6 h and 32.5 h. The removal of the ICLs was only detectable after 49.5 h and 51 h, where the signal was reduced to approx. 70% of the maximum signal.
For the detection of N7-CLB-Gua and N3-CLB-Ade, the same phenomenon as for the rFADU dose-response experiment was observed. Only the two highest concentrations (200 µM and 500 µM) in the dose-response experiment showed a significant increase in both monoadducts (Fig. 6D). However, the signal increase for the N7-CLB-Gua at a treatment concentration of 200 µM CLB was only marginal. Furthermore, concentrations of CLB up to 1 mM were sublethal for this cell line. Therefore, 300 µM of CLB was used for the time-series experiment. Due to the high SD displayed by the RPE-1 cells throughout the performed MS experiments, no significant decrease in the signal for both monoadducts could be observed over the course of 30 h. However, a downward trend of the N7-CLB-Gua and the N3-CLB-Ade signal was detected (Fig. 6E).
The survival of these cells after the treatment with 200 µM and 300 µM CLB was monitored for 51 h to exclude adduct loss due to excessive cell death. After an initial increase of viable cells post-treatment, a slight reduction was visible for both concentrations (Fig. 6F). The initial percentage of viable cells was the lowest throughout the experiment with 87% (200 µM) and 85% (300 µM). 51 h post-treatment, the percentage of viable cells was at 91% for the cells treated with 200 µM CLB and at 88% after the treatment with 300 µM CLB. The proliferative capacity of the RPE-1 cells after the treatment with both concentrations was only significantly increased 51 h post-treatment (Fig. 6F). Interestingly, the cells treated with 300 µM CLB had a higher percentage of divided cells with 6.5% in comparison to the cells treated with 200 µM CLB, where after 51 h only 4.4% of all cells had completed one cycle of proliferation.
3.3.2.2. VH10 cells
Due to the low susceptibility and the minor response towards the treatment of the immortalized RPE-1 cells, nonmalignant hTERT immortalized human foreskin fibroblasts (VH10) were also used to identify the cellular response towards the treatment with CLB. The Annexin V/PI analysis revealed a similar trend in the VH10 cells as in the RPE-1 cells (Fig. 7A). No decrease in the number of viable cells was observed for the cells treated with 2.5 µM to 1 mM of CLB or 200 µM H2O2. Only cells treated with 2 mM CLB showed a significantly increased percentage of dead cells.
The rFADU dose-response experiment performed with VH10 cells showed that there was a linear increase in ICLs with increasing treatment concentrations of CLB (Fig. 7B). This linear increase was observed starting at a concentration of 50 µM CLB. The first significant increase in ICLs was detected at a concentration of 100 µM CLB. This concentration was used for the time-course experiment. After an initial increase in the first 6 h, no significant decrease of ICLs was detected except for the 51 h time point (Fig. 7C). 51 h post-treatment, only 30% of the ICLs formed were removed. This outcome was similar to the RPE-1 cells.
VH10 cells were treated with the indicated concentrations of CLB, and N7-CLB-Gua and N3-CLB-Ade were detected via LC-MS/MS (Fig. 7D). This dose-response experiment was performed to detect the treatment concentration, which induces a significantly increased amount of adduct for the analysis of both monoadducts in VH10 cells. As for the ICLs, a linear increase was observed for N7-CLB-Gua and N3-CLB-Ade with increasing concentrations of CLB. Only for 200 µM and 500 µM treated samples a significant increase in both adducts was detected. As the signal increase for 200 µM CLB was minimal and concentrations up to 2 mM CLB were sublethal for the VH10 cells, 400 µM CLB was chosen as a treatment concentration for the subsequent kinetic experiment. In the first 1.5 h post-treatment, an increase in both adducts was observed (Fig. 7E). Starting at the 8 h time point, a significant decrease of N3-CLB-Ade to 52% of the maximum signal was detected. The removal of N7-CLB-Gua was slower than the removal of N3-CLB-Ade. Only at 21.5 h post-treatment a significant decrease of the guanine adduct to 52% was observed. A reduction to 34% could be detected for the N3-CLB-Ade adduct at the same time-point. No further decrease in both monoadducts could be detected in the time span from 21.5 h to 30 h.
No significant decrease in the viability of VH10 cells in the course of 51 h was detected after the treatment with 100 µM CLB (Fig. 7F). When treated with 400 µM CLB, however, a slight decrease to 87% viable cells at 51 h post-treatment was observed. With the CFSE proliferation assay, a slight increase in the percentage of divided cells could be observed for both treatment concentrations. Starting at 24 h, the increase was significant for the treatment concentration of 400 µM CLB. For cells treated with 100 µM CLB, an increase in the number of cells that underwent one cycle of proliferation was detected at 32.5 h post-treatment. At the 51 h time point, cells for both conditions reached a division rate of 5.3%.
3.3.2.3. Podocytes
Podocytes are highly specialized cells of the kidney, and a recent study showed that a defect in NER causes glomerulosclerosis [72]. These findings indicate the presence of an intact repair machinery of this cell type. To analyze the repair capacity of these cells after the treatment with CLB, a suitable treatment concentration had to be determined. Therefore, the viability of hTERT immortalized podocytes after the treatment with increasing concentrations of CLB was detected using the Annexin V/PI cell death staining. A dose dependent decrease of the viability of these cells was detected, but this decrease was only significant for the treatment with 1 mM and 2 mM CLB (Fig. 8A). However, in comparison with the other two hTERT immortalized cell lines used in this study (RPE-1 and VH10), a clear-cut dose-response after the treatment with CLB could be observed.
Fig. 8.
Cellular effects of CLB treatment in the hTERT immortalized podocytes. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. Podocytes were treated with CLB for 1 h at 37 °C, and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C and ICLs were monitored using the reverse automated FADU assay. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Time-course analysis of CLB-induced DNA ICLs. Cells were treated with 50 µM CLB for 1 h at 37 °C, and ICLs were monitored using the rFADU assay. Treated cells were allowed to recover for the time points indicated, up to 51 h. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 5 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (E) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 200 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 30 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation in C. and E. (F) Cytotoxicity as well as proliferation status of cells after the treatment with the indicated treatment concentrations of CLB were monitored as a function of time. Podocytes were stained with 10 µM CFSE and after 24 h, the cells were treated for 1 h with 50 µM (△) or 200 µM of CLB (▲). Treated cells were allowed to recover for the time points indicated and the samples were additionally stained with PI before the flow cytometric analysis. Data represents the mean ± SD from 3 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
For the detection of the sensitivity of the rFADU, podocytes were treated with increasing concentrations of CLB and the amount of ICLs was monitored. Starting at a concentration of 50 µM, a clear-cut linear dose-dependent and significant increase in ICLs was detected (Fig. 8B). As at 50 µM CLB no detectable increase in cell death, but a significant increase in ICLs with the rFADU method was observed, this concentration was used for the subsequent time-course experiment. In the first 6 h post-treatment an increase in ICLs was detected, similar to the other two hTERT immortalized cell lines tested (Fig. 8C). However, already after 22.5 h a significant decrease in ICLs was observed. The decrease of ICLs, beginning 6 h post-treatment, was nearly linear, and over the course of 51 h the number of ICLs was reduced to 51% of the maximum detected signal.
To be able to measure the removal of N3-CLB-Ade and N7-CLB-Gua over time, the sensitivity of the MS method for the podocytes had to be determined. Thus, an experiment was performed where cells were treated with increasing concentrations of CLB. At a concentration range of 100–200 µM, a significant increase in the signal intensity could be observed (Fig. 8D). Throughout the concentration series, a clear dose-dependent increase in the signal for both monoadducts was visible. Taken together, a suitable sublethal treatment concentration for the time-course experiment, where the formation and removal of both monoadducts could be monitored, was 200 µM of CLB. During the first 3 h post-treatment, an upward trend for N3-CLB-Ade and N7-CLB-Gua was observed (Fig. 8E). After this initial increase, the signal of both monoadducts dropped simultaneously to 65% of the maximum detected signal. Between 8 h and 30 h post-treatment, N3-CLB-Ade decreased linearly even further to 28%, whereas no significant removal of N7-CLB-Gua was visible.
To avoid a bias of the time-course data due to preferential cell death of the cells most affected or proliferation of the cells least affected by the treatment with CLB, the survival as well as the division rate was monitored over a time span of 51 h. An overall increase in viability of the podocytes was observed after the treatment with 50 µM and 200 µM over the course of 51 h and no cell death could be detected (Fig. 8F). Furthermore, when treated with 200 µM CLB, no increase in divided cells was observed. For the podocytes treated with 50 µM CLB however, a significant increase of the percentage of divided cells to 2.8% was detected 24 h post-treatment. A linear increase for these specific conditions was identified. 51 h post-treatment, the division rate reached 15%.
3.3.2.4. PBMCs
To exclude an influence of the hTERT immortalization on the repair capacity of the cell systems tested, human PBMCs, which are primary cells with intact repair genes, were analyzed. PBMCs are a perfect model system for the analysis of the DNA damage repair capacity, especially because in previous studies it was shown that PBMCs are able to repair the guanine and adenine adducts formed after the treatment with CEES, which is a monofunctional analog of sulfur mustard [64]. To test whether this is also the case for the adducts formed after the treatment with the bifunctional agent CLB, the susceptibility of the PBMCs to treatment with increasing concentrations of CLB was analyzed via the flow cytometry-based Annexin V/PI assay. Compared to the other cell systems tested in this study, PBMCs were by far the ones most affected by the treatment. A clear-cut dose-dependent increase of dead cells was visible and starting at a concentration of only 50 µM a significant decrease in viable cells was observed (Fig. 9A).
Fig. 9.
Cellular effects of CLB treatment in human PBMCs. (A) Cytotoxicity of the indicated doses of CLB was detected using the annexin V/PI staining and flow cytometry. PBMCs were treated with CLB for 1 h at 37 °C and the cell death status was analyzed 24 h later. ‘Dead’ refers to annexin V/PI positive cells, ‘apoptotic’ to annexin V positive/PI negative cells and ‘viable’ to annexin V/PI negative cells. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using two-way-ANOVA followed by a post hoc Dunnett‘s test. (B) Dose-dependent induction of ICLs after the treatment with CLB. Cells were treated with the indicated doses of CLB for 1 h at 37 °C and ICLs were monitored using the rFADU assay. Data represents the mean ± SD from 5 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (C) Dose-response relationship of the treatment with CLB and the amount of monoalkylated DNA adducts. Cells were treated with the indicated doses of CLB for 1 h at 37 °C. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 6 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. (D) Time-course analysis of CLB-induced monoalkylated adducts in DNA (N7-CLB-Gua and N3-CLB-Ade). Cells were treated with 10 µM CLB for 1 h at 37 °C and were allowed to recover for the time points indicated, up to 50 h. The amount of N7-CLB-Gua and N3-CLB-Ade was monitored using MS. Data represents the mean ± SD from 4 independent experiments. Statistical analysis was performed using one-way-ANOVA followed by a post hoc Dunnett‘s test. For clarity, the display of the statistically significant differences of data points was omitted in the phase of adduct formation. *p˂0.05, * *p˂ 0.01, * **p˂ 0.001, * ** *p˂ 0.0001.
In view of to the high sensitivity of the PBMCs to the treatment with CLB, only concentrations up to 50 µM were tested for the detection of the sensitivity of the rFADU assay. In this sublethal treatment range no increase in ICLs was observed (Fig. 9B). Therefore, the experiment was repeated with concentrations up to 500 µM CLB. With this wider range of treatment concentrations, a significant increase in ICLs in PBMCs could be detected starting at a concentration of 100 µM CLB (Suppl. Fig. 8). Unfortunately, the percentage of viable cells at 100 µM CLB was reduced to < 50%. For this reason, no suitable treatment concentration for the monitoring of the removal of ICLs over time was found for this cell type.
Regarding the sensitivity of the MS method, however, we were able to detect a significant increase in N3-CLB-Ade and N7-CLB-Gua for a concentration as low as 10 µM CLB (Fig. 9C). As no toxicity for this concentration was observed in the Annexin V/PI experiment, the subsequent time-course experiment was conducted with 10 µM CLB as treatment concentration. After an initial increase of both adducts in the first 3 h post-treatment, N7-CLB-Gua did not significantly decrease until the last time point of 50 h (56%), whereas for N3-CLB-Ade a first drop to 51% was detected as soon as 19 h post-treatment (Fig. 9D). The downwards trend continued for the N3-CLB-Ade adduct and at 50 h post-treatment a reduction to 25% was observed.
4. Discussion
4.1. Direct analysis of DNA damage
DNA is the carrier of the genetic code. Therefore, alterations or damage of the DNA has serious implications. DNA lesions are the driver of somatic mutagenesis as well as carcinogenesis [73], [74], [75], but also of cell death or cellular senescence [76], [77] if the affected cell is not able to repair the lesions or if the damage load is too high. This concept is used in cancer chemotherapy. Fast replicating cancer cells are prone to the cytotoxic effects of radiotherapy or of chemotherapeutic agents that induce ICLs. The induced DNA lesions lead to the rapid death of the cancer cells. In normal cells the DNA repair machinery is responsible for preventing mutagenesis and ultimately cancer formation by removal of altered DNA. Here, this is a highly desirable event. However, in the context of chemotherapy a highly active and efficient DNA repair can reduce the efficacy of the treatment [78], [79], [80], [81]. Therefore, genetic polymorphisms in DNA repair genes, which were proven to affect the DNA repair efficacy [82], [83], may determine the success of the chemotherapy. For this reason, the quantification of DNA adducts as biomarkers of exposure is a highly researched topic. Most studies focusing on the quantification and repair of adducts induced by crosslinking agents, such as CLB, were only detecting ICLs and were performed using indirect detection methods. With these methods, it was not the adduct itself that was detected, but rather the decrease in comet tail moment (modified alkaline comet assay [84], [85], [86]), the absence of a PCR product or the reactivation of reporter genes (host cell reactivation assay) of plasmids bearing a site-specific ICL [37], [87], [88], the formation of repair intermediates (e.g. DNA strand breaks) [89], or simply the sensitization of knockout cells to crosslinking agents [90], [91], [92]. Even methods that actually detect the specific adduct vary vastly in their sensitivity and accuracy, e.g. detection via antibodies in comparison to detection via MS. The detection using immunoassays is cheap and quick, but is often lacking specificity, whereas MS is considered the gold standard for adduct quantification [93], [94], [95]. But also the usage of MS in the quantification of crosslink adducts has its drawbacks. Due to the identical chemical structure and mass of ICLs and intrastrand crosslinks, no distinction between the two adducts can be made. However, ICLs are not the only and by far not the most abundant adducts formed upon mustard treatment. Only a limited number of studies have focused on the monoalkylated DNA lesions, and therefore not much is known about their contribution to the cellular consequences of the treatment [58], [59], [96]. To allow for the simultaneous analysis of both mono- as well as bis-adducts, we developed in this study a dual method for the detection of the monoalkylated DNA adducts as well as the ICLs formed after treatment with CLB. Our strategy comprises (i) an MS method for the detection of the main monoalkylated DNA adducts and (ii) the automated rFADU assay that was developed in our lab for the direct and reliable detection of ICLs in the same samples.
To enable quantification of the main DNA monoadducts formed after the treatment with CLB, isotopically-labeled (N7-CLB-Gua) as well as unlabeled standards (N7-CLB-Gua, N3-CLB-Ade) have been synthesized, characterized via product ion scan, and are now available for further studies. As biomarkers of exposure, the adducts formed can provide further insights into the cellular response of different cell types of various backgrounds (malignant or non-malignant) and are therefore important for predicting the treatment outcome of cancer patients receiving CLB as chemotherapeutic agent. These standards were used to successfully establish an MS-based detection method. Up to now, it is still a matter of debate which repair pathway is responsible for the repair of the main monoadducts formed after treatment with CLB. Therefore, a suitable, selective, and specific method was successfully established to detect differences between cell systems regarding the kinetics of formation and removal of the aforementioned adducts.
As the monoalkylated DNA adducts are the ones mostly formed (∼95%), but not the most cytotoxic lesions [23], [24], [97], a detection method for the simultaneous monitoring of the ICLs was established by adjusting the automated rFADU method for the usage of adherent cells. This method proved to be highly sensitive for the detection of ICLs formed in human cell lines after the treatment with NMs. Based on these results, a statistically significant ICL-induction was detectable with treatment concentrations as low as 2.5 µM (HN2) and 100 µM (CLB), respectively, in HaCaT cells. Using this method, we were able to circumvent the problem of distinguishing ICLs from intrastrand crosslinks using an MS-based assay, because the detection of ICLs with the rFADU method relies solely on the fluorescent dye SYBR® Green, which binds preferably to double-stranded DNA and therefore to sequences that flank ICLs [98]. With these methods at hand, we were able to detect, with the highest precision and specificity, the main DNA-CLB adducts in different cellular systems.
4.2. Cytotoxicity of CLB exposure
To exclude that a reduction of the adducts in the time-course experiments was due to preferential death of the cells in the population that are the most affected by the treatment, a sublethal treatment concentration for each cell type was determined. In our toxicity studies a clear trend regarding the sensitivity towards the treatment with CLB was visible. Of the six cell systems tested, the cytotoxicity increased in ascending order: RPE-1 < VH10 < podocytes < U2OS < A2780 < PBMC. Interestingly, the hTERT immortalized cell lines (RPE-1, VH10 and podocytes) showed a reduced sensitivity towards the treatment with CLB in comparison to the cancer cell lines (U2OS and A2780). The toxicity of a compound, besides the inherent toxic potential of the compound itself, depends highly on its toxicokinetics. As CLB enters the cells via passive diffusion, the cellular uptake of the substance is not responsible for the observed difference [99]. Crosslinking agents are supposed to be most toxic for fast replicating (cancer) cells, because of the DNA ICLs formed, which are an insurmountable obstacle for the replication machinery of the cell [100]. However, the doubling times of the cells investigated are in a similar range, except for the hTERT immortalized podocytes. According to the literature, the doubling times of hTERT immortalized RPE-1 cells vary from 14 to 24 h [101], whereas U2OS cells have a doubling time of approx. 22 h [102], and A2780 cells of 25 h [103]. The hTERT immortalized podocytes however, have a doubling time of 50 h [104]. Even though podocytes are the cells with the slowest proliferation rate, they were not the ones least affected by the treatment. Furthermore, PBMCs, which are generally resting, show the highest cytotoxicity after the treatment with CLB. Given the fact that the two smallest cell types of the tested cells were the most affected ones, and that CLB enters the cell via passive diffusion, the high sensitivity of A2780 cells and PBMCs towards the treatment with CLB might be due to the higher accessibility of the nucleus and therefore the DNA. All NMs are highly reactive and form adducts as soon as a nucleophile attacks the electrophilic intermediate (aziridinium ion) [4]. Therefore, more unreacted CLB can enter the nucleus if the compound has less encounters with proteins, etc in the cytosol, and more DNA adducts are formed as compared to cells with a larger cytosol and an increased size.
4.3. Appropriate range of CLB concentrations
For analyzing the kinetics of the formation and removal of the main adducts N7-CLB-Gua, N3-CLB-Ade and the ICLs formed in human cell lines and PBMCs, which were included because the DNA repair machinery might be altered in immortalized cell lines, the dose-response experiments performed were essential to determine a suitable treatment concentration. The chosen treatment concentration had not only to be sublethal, but had to induce a detectable amount of adducts. As it is not known how many adducts are formed in the different cell types after treatment with a specific concentration of CLB, a concentration was selected that induced a significantly increased amount of adducts in comparison to the solvent control. For each cell type such a concentration could be determined except for the detection of ICLs in PBMCs. No sublethal concentration was detected that induced a sufficient amount of ICLs. It has to be noted that for the monoadduct detection, A2780 cells and PBMCs were the cell types where a lower concentration was sufficient to induce a level of adducts high enough to be detected via the MS in comparison to the other cell lines. This finding correlates with the detected higher sensitivity of these cell types in comparison to the hTERT immortalized cell lines and U2OS cells tested. Furthermore, nearly no difference could be detected in the induction of both monoadducts for each treatment concentration.
4.4. Time course of DNA damage formation and removal
With suitable treatment concentrations at hand, time-course experiments were performed. We were able to detect cell specific differences in the removal of ICLs in the five cell lines tested. The time-course analyses revealed a rapid increase in ICLs for all cell lines in the first 3–6 h after treatment, which is in line with earlier findings from Souliotis et al. [59]. The increase of the adducts for such a prolonged time might be due to the slower second reaction after the formation of a monoadduct [105]. Furthermore, as CLB is a lipophilic compound [99], the excess CLB might be stored in lipophilic compartments of the cell and released at later time-point. Subsequent to this initial increase, a significant reduction of the ICLs was confirmed in all cell types in the course of 51 h. However, in the hTERT immortalized VH10 and RPE-1 cells this reduction was very minor and only detectable after 51 h or 49.5 h. The third cell line immortalized with hTERT on the other hand, showed a clear and steady decrease in ICLs beginning at 21.5 h. Therefore, the ICL repair pathway seems to be more active in the podocytes compared to the RPE-1 and VH10 cells. As podocytes have very long doubling times compared to the other cell lines [104], some ICLs might also undergo repair via a replication-independent mechanism. The removal of ICLs in U2OS cells set in at a later time point compared to A2780 cells, which is also a cancer cell line. The ultimate reduction of the level of ICLs was, however, nearly identical. Taken together, the removal of ICLs was most efficient in the cancer cell lines U2OS and A2780. This phenomenon reinforces the hypothesis that DNA repair might be one factor causing the resistance of tumors to chemotherapeutic agents [66]. Interestingly, the hTERT immortalized podocytes also proved to be a suitable model system for researching the repair of ICL, as a clear removal of ICLs over time was detectable. However, a potential confounding effect of adduct dilution by cell proliferation has to be considered as a possible reason for the observed adduct reduction.
The time-course analysis of the main monoadducts N7-CLB-Gua and N3-CLB-Ade in different cellular systems after treatment with sublethal concentrations of CLB revealed consistent differences in the removal of the N7-CLB-Gua adduct in comparison to the N3-CLB-Ade adduct. In all cell systems, except for the A2780 cells, the levels of both adducts were very similar in the first 3 h. However, after the initial increase, the reduction of the N7-CLB-Gua signal was detectably slower than the reduction of the N3-CLB-Ade adduct. We had observed a similar effect in a previous study, where sulfur mustard, which is a structural analogue of HN2, was used to treat PBMCs [64]. Therefore, this phenomenon seems to be highly conserved throughout the class of sulfur mustards as well as NMs. Since both monoadducts are thought to be repaired via the same repair pathway, either the NER [54] or the BER pathway [53], [56], a slower repair of one of the adducts seems not to be responsible for this difference. Furthermore, a preferential depurination of the adenine adduct and therefore a faster adduct removal can also be excluded, as guanine is known to have a 1.5 higher depurination rate than adenine [106], [107]. One reason, however, could be faster recognition of the N3-CLB-Ade adduct, but this hypothesis has to be further examined in future studies. In comparison to ICLs formed after the treatment with CLB, the removal of the monoadducts was significantly faster in most cell systems tested (< 30 h), which was expected as ICL repair is far more complex and more factors are involved in this repair pathway than in the NER or the BER. Beside the preferential removal of N3-CLB-Ade, cell specific differences in removal of both adducts were detected. In RPE-1 cells, U2OS cells and in podocytes, no significant removal of the N7-CLB-Gua adduct was detected, whereas a reduction of the adduct was observed in PBMCs, but only after 50 h of incubation. Also in VH10 and A2780 cells adduct removal was observed. Interestingly, in all cell systems except for the RPE-1 cells, a significant reduction of the N3-CLB-Ade level was detected. Even though RPE-1 cells are the cells with slowest removal rate of CLB-induced ICLs or monoadducts, the displayed cytotoxicity after the treatment was the lowest out of all cell systems in this study. Furthermore, in A2780 cells, which were showing the highest removal rates of all adducts, the treatment induced a high level of toxicity. Actually, with a more effective repair, the toxicity should be reduced in comparison to cell lines not showing any repair. In this study however, we had a different outcome. One reason for the observed toxicity might be an error-prone repair of the adducts, which would lead to a higher toxicity of the treatment in cells with a higher rate of adduct repair. As the repair of the CLB-induced monoadducts is still not fully understood, more studies have to be conducted in order to prove this hypothesis.
4.5. Preferential cell death and cell proliferation as possible confounding factors
The detection of the removal of DNA adducts in a cellular system can have three different underlying mechanisms: First, the preferential death of the cells with the most adducts. Second, the dilution of the adduct levels by the preferential proliferation of cells with a very low number of adducts [108], and third, genuine repair. By using only sublethal concentrations in this study, the removal of adducts due to selective cell death could be excluded. Furthermore, the effect of a dilutive cell proliferation was excluded in PBMCs as they are non-proliferative cells. In this cell system, the removal of the monoaddcuts detected indeed accounts for genuine repair. In the case of the other cell types tested, the dilution effect of proliferation had to be excluded as the tested cells were all immortalized. The results of the proliferation assay (CFSE dilution assay) indicated only a low division rate in all cell lines. When compared to the level of adduct removal neither a reduction in survival rate, nor an increase in cells after division could account for the detected removal. We were therefore able to successfully detect genuine DNA repair of ICLs in the cancer cell lines A2780, U2OS, and in the hTERT immortalized RPE-1 cells, VH10 cells and podocytes as well as the repair of the main monoadducts (N7-CLB-Gua, N3-CLB-Ade) in A2780, VH10 cells and PBMCs. In U2OS cells and in podocytes, no repair of N7-CLB-Gua, but only the repair of N3-CLB-Ade was observed, whereas in RPE-1 cells no monoadduct repair was detected. Taken together, we were able to detect cell specific differences in the repair of the main adducts formed after treatment with CLB. Furthermore, the repair process was very slow, which led to a long adduct persistence.
5. Conclusion
In conclusion, we have demonstrated that upon CLB treatment, the cytotoxicity profiles reveal notable differences between the six cell systems tested. Furthermore, both main monofunctional adducts formed (N7-CLB-Gua and N3-CLB-Ade) were readily detected by MS at subtoxic concentrations, whereas ICLs were detected in all cell systems at subtoxic concentrations except for PBMCs. In almost all cell systems studied, a time-dependent decrease of the two monoadducts and the ICLs could be detected. In addition, we have shown that the decrease over time not only differed between the two monoadducts N7-CLB-Gua and N3-CLB-Ade, but was also dependent on the cell system tested. Taken together, these methods are powerful tools to study the toxicokinetics, toxicodynamics, and molecular mechanisms of NM-induced DNA damage and repair. Based on this methodological platform, providing specific and precise readouts, genetic studies are now possible to identify molecular targets for therapeutic interventions in CLB-resistant tumors. Such genetic studies will include a screening of the DNA repair factors responsible for the removal of the DNA lesions induced by CLB using siRNA-induced knockdown or CRISPR/Cas9 mediated gene disruption. For the identification of, e.g., the repair pathway(s) responsible for the removal of the monoadducts, genes encoding proteins in the NER pathway or in the BER pathway will be preferred targets. The genetically modified cells will then be treated with optimal concentrations of CLB (as reported in the present paper), and any changes in the repair kinetics will be detected using the mass spectrometric platform we established. When the relevant repair factors are identified, the expression of the respective genes of the cell lines showing differences in the repair kinetics of the adducts will be analyzed. If it is possible to confirm differences in the expression level of these genes, clinical samples, e.g., tumor biopsies, can then be analyzed for the gene expression of the same factors in order to verify if CLB resistance can be correlated with DNA repair gene expression in primary human tumor cells. If the latter were the case it would represent a first step towards unravelling the resistance mechanism of the respective tumor.
Funding
SK was supported by a fellowship of the Avicenna-Studienwerk. The Avicenna-Studienwerk had no involvement in the design and the execution of this study.
Author contributions
SK, AB, and AM: Conceptualization; SK, MM, NP, JW, CS, and NS: Formal analysis; SK and AB: Funding acquisition; SK, MM, NP, JW, CS, NS, and GS: Investigation, SK: Methodology; SK and AB: Project administration; AB: Resources; AB and AM: Supervision; SK and AB: Validation; SK and JW: Visualization; SK: Writing – original draft; SK, AB and AM: Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Dr. Annette Sommershof, Head of the FlowKon Facility (flow cytometry) of the University of Konstanz, for her kind assistance in the establishment of the FACS-based assays. We also thank Dr. Tabea Zubel for her instructions with regards to operating the mass spectrometer. Furthermore, we thank Professor Andreas Marx and his group for infrastructural support and helpful discussions on the project. We would also like to acknowledge the joint efforts with the research group of Professor Daniel Dietrich in characterizing the DNA repair capabilities of the PODO-TERT256 cells.
Handling Editor: L.H. Lash
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.toxrep.2023.01.010.
Appendix A. Supplementary material
Supplementary material.
.
Data Availability
Data will be made available on request.
References
- 1.Gilman A. The initial clinical trial of nitrogen mustard. Am. J. Surg. 1963;105:574–578. doi: 10.1016/0002-9610(63)90232-0. [DOI] [PubMed] [Google Scholar]
- 2.Gilman A., Philips F.S. The biological actions and therapeutic applications of the b-chloroethyl amines and sulfides. Science. 1946;103:409–436. doi: 10.1126/science.103.2675.409. [DOI] [PubMed] [Google Scholar]
- 3.Inturi S., Tewari-Singh N., Agarwal C., White C.W., Agarwal R. Activation of DNA damage repair pathways in response to nitrogen mustard-induced DNA damage and toxicity in skin keratinocytes. Mutat. Res. 2014;763–764:53–63. doi: 10.1016/j.mrfmmm.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kundu G.C., Schullek J.R., Wilson I.B. The alkylating properties of chlorambucil. Pharmacol. Biochem. Behav. 1994;49:621–624. doi: 10.1016/0091-3057(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 5.Dogliotti I., Ragaini S., Vassallo F., Boccellato E., de Luca G., Perutelli F., Boccomini C., Clerico M., Botto B., Grimaldi D., Orsucci L., Ferrero S., Vitale C., Ferrero D., Coscia M., Cavallo F. Real life use of bendamustine in elderly patients with lymphoid neoplasia. J. Pers. Med. 2021;11:249. doi: 10.3390/jpm11040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou D., Xu W., Ma H., Zhao C., Hu Y., Zhao Y., Wu D., Zhao X., He Y., Yan J., Wang C., Meng F., Jin J., Zhang X., Yu K., Hu J., Lv Y. Bendamustine versus chlorambucil in treatment of chronic lymphocytic leukaemia in China: a randomized, open-label, parallel-controlled, phase III clinical trial. Invest N. Drugs. 2022:1–12. doi: 10.1007/s10637-021-01206-2. [DOI] [PubMed] [Google Scholar]
- 7.Chanan-Khan A., Cramer P., Demirkan F., Fraser G., Silva R.S., Grosicki S., Pristupa A., Janssens A., Mayer J., Bartlett N.L., Dilhuydy M.-S., Pylypenko H., Loscertales J., Avigdor A., Rule S., Villa D., Samoilova O., Panagiotidis P., Goy A., Mato A., Pavlovsky M.A., Karlsson C., Mahler M., Salman M., Sun S., Phelps C., Balasubramanian S., Howes A., Hallek M. Ibrutinib combined with bendamustine and rituximab compared with placebo, bendamustine, and rituximab for previously treated chronic lymphocytic leukaemia or small lymphocytic lymphoma (HELIOS): a randomised, double-blind, phase 3 study. Lancet Oncol. 2016;17:200–211. doi: 10.1016/S1470-2045(15)00465-9. [DOI] [PubMed] [Google Scholar]
- 8.Tessler H.H. In: Intraocular Inflammation. Zierhut M., Pavesio C., Ohno S., Orefice F., Rao N.A., editors. Springer Berlin Heidelberg; Berlin, Heidelberg: 2016. Chlorambucil; pp. 315–321. [Google Scholar]
- 9.NICOLLE A., PROCTOR S.J., SUMMERFIELD G.P. High dose chlorambucil in the treatment of lymphoid malignancies. Leuk. Lymphoma. 2004;45:271–275. doi: 10.1080/10428190310001595704. [DOI] [PubMed] [Google Scholar]
- 10.Kallama S., Hemminki K. Alkylation of guanosine by phosphoramide mustard, chloromethine hydrochloride and chlorambucil. Acta Pharmacol. Et. Toxicol. 1984;54:214–220. doi: 10.1111/j.1600-0773.1984.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 11.Tilby M.J., McCartney H., Gould K.A., O'Hare C.C., Hartley J.A., Hall A.G., Golding B.T., Lawley P.D. A monofunctional derivative of melphalan: preparation, DNA alkylation products, and determination of the specificity of monoclonal antibodies that recognize melphalan-DNA adducts. Chem. Res. Toxicol. 1998;11:1162–1168. doi: 10.1021/tx980129a. [DOI] [PubMed] [Google Scholar]
- 12.Edler M., Jakubowski N., Linscheid M. Quantitative determination of melphalan DNA adducts using HPLC - inductively coupled mass spectrometry. J. Mass Spectrom. 2006;41:507–516. doi: 10.1002/jms.1009. [DOI] [PubMed] [Google Scholar]
- 13.Mohamed D., Mowaka S., Thomale J., Linscheid M.W. Chlorambucil-adducts in DNA analyzed at the oligonucleotide level using HPLC-ESI MS. Chem. Res. Toxicol. 2009;22:1435–1446. doi: 10.1021/tx900123r. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed D., Linscheid M. Separation and identification of trinucleotide-melphalan adducts from enzymatically digested DNA using HPLC-ESI-MS. Anal. Bioanal. Chem. 2008;392:805–817. doi: 10.1007/s00216-008-2236-0. [DOI] [PubMed] [Google Scholar]
- 15.E.S. Greenwald, Cancer chemotherapy, William Heinemann Medical Books Ltd, London, 1967.
- 16.Shukla P.K., Mishra P.C., Suhai S. Reactions of DNA bases with the anti-cancer nitrogen mustard mechlorethamine: a quantum chemical study. Chem. Phys. Lett. 2007;449:323–328. doi: 10.1016/j.cplett.2007.10.072. [DOI] [Google Scholar]
- 17.Gruppi F., Hejazi L., Christov P.P., Krishnamachari S., Turesky R.J., Rizzo C.J. Characterization of nitrogen mustard formamidopyrimidine adduct formation of bis(2-chloroethyl)ethylamine with calf thymus DNA and a human mammary cancer cell line. Chem. Res. Toxicol. 2015;28:1850–1860. doi: 10.1021/acs.chemrestox.5b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng J., Ye F., Dan G., Zhao Y., Zhao J., Zou Z. Formation and degradation of nitrogen mustard-induced MGMT-DNA crosslinking in 16HBE cells. Toxicology. 2017;389:67–73. doi: 10.1016/j.tox.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Haapala E., Hakala K., Jokipelto E., Vilpo J., Hovinen J. Reactions of N,N-bis(2-chloroethyl)-p-aminophenylbutyric acid (chlorambucil) with 2'-deoxyguanosine. Chem. Res. Toxicol. 2001;14:988–995. doi: 10.1021/tx000249u. [DOI] [PubMed] [Google Scholar]
- 20.Florea-Wang D., Pawlowicz A.J., Sinkkonen J., Kronberg L., Vilpo J., Hovinen J. Reactions of 4-Bis(2-chloroethyl)aminobenzenebutanoic acid (chlorambucil) with DNA. Chem. Biodivers. 2009;6:1002–1013. doi: 10.1002/cbdv.200800327. [DOI] [PubMed] [Google Scholar]
- 21.Povirk L.F., Shuker D.E. DNA damage and mutagenesis induced by nitrogen mustards. Mutat. Res. /Rev. Genet. Toxicol. 1994;318:205–226. doi: 10.1016/0165-1110(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 22.Remias M.G., Lee C.S., Haworth I.S. Molecular dynamics simulations of chlorambucil/DNA adducts. A structural basis for the 5'-GNC interstrand DNA crosslink formed by nitrogen mustards. J. Biomol. Struct. Dyn. 1995;12:911–936. doi: 10.1080/07391102.1995.10508784. [DOI] [PubMed] [Google Scholar]
- 23.Schärer O.D. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. ChemBioChem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 24.Wijen J.P., Nivard M.J., Vogel E.W. The in vivo genetic activity profile of the monofunctional nitrogen mustard 2-chloroethylamine differs drastically from its bifunctional counterpart mechlorethamine. Carcinogenesis. 2000;21:1859–1867. doi: 10.1093/carcin/21.10.1859. [DOI] [PubMed] [Google Scholar]
- 25.Gould K.A., Nixon C., Tilby M.J. p53 elevation in relation to levels and cytotoxicity of mono- and bifunctional melphalan-DNA adducts. Mol. Pharmacol. 2004;66:1301–1309. doi: 10.1124/mol.104.000596. [DOI] [PubMed] [Google Scholar]
- 26.Yaghi B.M., Turner P.M., Denny W.A., Turner P.R., O'Connor C.J., Ferguson L.R. Comparative mutational spectra of the nitrogen mustard chlorambucil and its half-mustard analogue in Chinese hamster AS52 cells. Mutat. Res. 1998;401:153–164. doi: 10.1016/S0027-5107(98)00005-0. [DOI] [PubMed] [Google Scholar]
- 27.Gould K.A., Nixon C., Tilby M.J. p53 elevation in relation to levels and cytotoxicity of mono- and bifunctional melphalan-DNA adducts. Mol. Pharmacol. 2004;66:1301–1309. doi: 10.1124/mol.104.000596. [DOI] [PubMed] [Google Scholar]
- 28.Semlow D.R., Walter J.C. Mechanisms of vertebrate DNA interstrand cross-link repair. Annu. Rev. Biochem. 2021;90:107–135. doi: 10.1146/annurev-biochem-080320-112510. [DOI] [PubMed] [Google Scholar]
- 29.Rogers C.M., Simmons Iii R.H., Fluhler Thornburg G.E., Buehler N.J., Bochman M.L. Fanconi anemia-independent DNA inter-strand crosslink repair in eukaryotes. Prog. Biophys. Mol. Biol. 2020;158:33–46. doi: 10.1016/j.pbiomolbio.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deans A.J., West S.C. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011;11:467–480. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niedernhofer L.J., Odijk H., Budzowska M., van Drunen E., Maas A., Theil A.F., de Wit J., Jaspers N.G.J., Beverloo H.B., Hoeijmakers J.H.J., Kanaar R. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsabar M., Mock C.S., Venkatachalam V., Reyes J., Karhohs K.W., Oliver T.G., Regev A., Jambhekar A., Lahav G. A switch in p53 dynamics marks cells that escape from DSB-induced cell cycle arrest. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vare D., Groth P., Carlsson R., Johansson F., Erixon K., Jenssen D. DNA interstrand crosslinks induce a potent replication block followed by formation and repair of double strand breaks in intact mammalian cells. DNA Repair. 2012;11:976–985. doi: 10.1016/j.dnarep.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Jose Portugal, Sylvia Mansilla, Marc Bataller, Mechanisms of Drug-Induced Mitotic Catastrophe in Cancer Cells, Current Pharmaceutical Design 16 69–78. [DOI] [PubMed]
- 35.Li L., Li S., Sun G., Peng R., Zhao L., Zhong R. Influence of the expression level of O6-alkylguanine-DNA alkyltransferase on the formation of DNA interstrand crosslinks induced by chloroethylnitrosoureas in cells: a quantitation using high-performance liquid chromatography-mass spectrometry. PLOS ONE. 2015;10 doi: 10.1371/journal.pone.0121225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stornetta A., Zimmermann M., Cimino G.D., Henderson P.T., Sturla S.J. DNA adducts from anticancer drugs as candidate predictive markers for precision medicine. Chem. Res. Toxicol. 2017;30:388–409. doi: 10.1021/acs.chemrestox.6b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ben-Yehoyada M., Wang L.C., Kozekov I.D., Rizzo C.J., Gottesman M.E., Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodskinson M.R., Bolner A., Sato K., Kamimae-Lanning A.N., Rooijers K., Witte M., Mahesh M., Silhan J., Petek M., Williams D.M., Kind J., Chin J.W., Patel K.J., Knipscheer P. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature. 2020;579:603–608. doi: 10.1038/s41586-020-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams H.L., Gottesman M.E., Gautier J. Replication-independent repair of DNA interstrand crosslinks. Mol. Cell. 2012;47:140–147. doi: 10.1016/j.molcel.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knipscheer P., Räschle M., Schärer O.D., Walter J.C. Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol. Biol. 2012;920:221–243. doi: 10.1007/978-1-61779-998-3_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu R.A., Semlow D.R., Kamimae-Lanning A.N., Kochenova O.V., Chistol G., Hodskinson M.R., Amunugama R., Sparks J.L., Wang M., Deng L., Mimoso C.A., Low E., Patel K.J., Walter J.C. TRAIP is a master regulator of DNA interstrand crosslink repair. Nature. 2019;567:267–272. doi: 10.1038/s41586-019-1002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato N., Kawasoe Y., Williams H., Coates E., Roy U., Shi Y., Beese L.S., Schärer O.D., Yan H., Gottesman M.E., Takahashi T.S., Gautier J. Sensing and Processing of DNA Interstrand Crosslinks by the Mismatch Repair Pathway. Cell Rep. 2017;21:1375–1385. doi: 10.1016/j.celrep.2017.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Chen Z., Ye Y. SLX4IP acts with SLX4 and XPF-ERCC1 to promote interstrand crosslink repair. Nucleic Acids Res. 2019;47:10181–10201. doi: 10.1093/nar/gkz769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jost P., Svobodova H., Stetina R. Induction and repair of DNA cross-links induced by sulfur mustard in the A-549 cell line followed by a comet assay. Chem. -Biol. Interact. 2015;237:31–37. doi: 10.1016/j.cbi.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Forchhammer L., Bräuner E.V., Folkmann J.K., Danielsen P.H., Nielsen C., Jensen A., Loft S., Friis G., Møller P. Variation in assessment of oxidatively damaged DNA in mononuclear blood cells by the comet assay with visual scoring. Mutagenesis. 2008;23:223–231. doi: 10.1093/mutage/gen006. [DOI] [PubMed] [Google Scholar]
- 46.Dębiak M., Panas A., Steinritz D., Kehe K., Bürkle A. High-throughput analysis of DNA interstrand crosslinks in human peripheral blood mononuclear cells by automated reverse FADU assay. Toxicology. 2011;280:53–60. doi: 10.1016/j.tox.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Chesner L.N., Degner A., Sangaraju D., Yomtoubian S., Wickramaratne S., Malayappan B., Tretyakova N., Campbell C. Cellular repair of DNA-DNA cross-links induced by 1,2,3,4-diepoxybutane. Int. J. Mol. Sci. 2017;18:1086. doi: 10.3390/ijms18051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li L., Zhao L., Zhong R. Quantification of DNA interstrand crosslinks induced by ACNU in NIH/3T3 and L1210 cells using high-performance liquid chromatography/electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2014;28:439–447. doi: 10.1002/rcm.6800. [DOI] [PubMed] [Google Scholar]
- 49.Liu S., Wang Y. A quantitative mass spectrometry-based approach for assessing the repair of 8-methoxypsoralen-induced DNA interstrand cross-links and monoadducts in mammalian cells. Anal. Chem. 2013;85:6732–6739. doi: 10.1021/ac4012232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malayappan B., Johnson L., Nie B., Panchal D., Matter B., Jacobson P., Tretyakova N. Quantitative high-performance liquid chromatography-electrospray ionization tandem mass spectrometry analysis of bis-N7-guanine DNA-DNA cross-links in white blood cells of cancer patients receiving cyclophosphamide therapy. Anal. Chem. 2010;82:3650–3658. doi: 10.1021/ac902923s. [DOI] [PubMed] [Google Scholar]
- 51.Paz M.M., Ladwa S., Champeil E., Liu Y., Rockwell S., Boamah E.K., Bargonetti J., Callahan J., Roach J., Tomasz M. Mapping DNA adducts of mitomycin C and decarbamoyl mitomycin C in cell lines using liquid chromatography/ electrospray tandem mass spectrometry. Chem. Res. Toxicol. 2008;21:2370–2378. doi: 10.1021/tx8002615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H., Cui Z., Hejazi L., Yao L., Walmsley S.J., Rizzo C.J., Turesky R.J. Kinetics of DNA adducts and abasic site formation in tissues of mice treated with a nitrogen mustard. Chem. Res. Toxicol. 2020;33:988–998. doi: 10.1021/acs.chemrestox.0c00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McHugh P.J., Gill R.D., Waters R., Hartley J.A. Excision repair of nitrogen mustard-DNA adducts in Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:3259–3266. doi: 10.1093/nar/27.16.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grant D.F., Bessho T., Reardon J.T. Nucleotide excision repair of melphalan monoadducts. Cancer Res. 1998;58:5196–5200. [PubMed] [Google Scholar]
- 55.Mattes W.B., Lee C.S., Laval J., O'Connor T.R. Excision of DNA adducts of nitrogen mustards by bacterial and mammalian 3-methyladenine-DNA glycosylases. Carcinogenesis. 1996;17:643–648. doi: 10.1093/carcin/17.4.643. [DOI] [PubMed] [Google Scholar]
- 56.Sousa M.M.L., Zub K.A., Aas P.A., Hanssen-Bauer A., Demirovic A., Sarno A., Tian E., Liabakk N.B., Slupphaug G. An inverse switch in DNA base excision and strand break repair contributes to melphalan resistance in multiple myeloma cells. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0055493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Alencar T.A.M., Leitão A.C., Lage C. Nitrogen mustard- and half-mustard-induced damage in Escherichia coli requires different DNA repair pathways. Mutat. Res. 2005;582:105–115. doi: 10.1016/j.mrgentox.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 58.van Kan M., Burns K.E., Helsby N.A. A systematic review of inter-individual differences in the DNA repair processes involved in melphalan monoadduct repair in relation to treatment outcomes. Cancer Chemother. Pharmacol. 2021;88:755–769. doi: 10.1007/s00280-021-04340-z. [DOI] [PubMed] [Google Scholar]
- 59.Souliotis V.L., Dimopoulos M.A., Sfikakis P.P. Gene-specific formation and repair of DNA monoadducts and interstrand cross-links after therapeutic exposure to nitrogen mustards. Clin. Cancer Res. 2003;9:4465–4474. [PubMed] [Google Scholar]
- 60.Ronson G.E., Piberger A.L., Higgs M.R., Olsen A.L., Stewart G.S., McHugh P.J., Petermann E., Lakin N.D. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat. Commun. 2018;9:746. doi: 10.1038/s41467-018-03159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boukamp P., Petrussevska R.T., Breitkreutz D., Hornung J., Markham A., Fusenig N.E. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tomicic M.T., Reischmann P., Rasenberger B., Meise R., Kaina B., Christmann M. Delayed c-Fos activation in human cells triggers XPF induction and an adaptive response to UVC-induced DNA damage and cytotoxicity. Cell. Mol. Life Sci. 2011;68:1785–1798. doi: 10.1007/s00018-010-0546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanzlikova H., Gittens W., Krejcikova K., Zeng Z., Caldecott K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 2017;45:2546–2557. doi: 10.1093/nar/gkw1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zubel T., Hochgesand S., John H., Steinritz D., Schmidt A., Bürkle A., Mangerich A. A mass spectrometric platform for the quantitation of sulfur mustard-induced nucleic acid adducts as mechanistically relevant biomarkers of exposure. Arch. Toxicol. 2019;93:61–79. doi: 10.1007/s00204-018-2324-7. [DOI] [PubMed] [Google Scholar]
- 65.Moreno-Villanueva M., Pfeiffer R., Sindlinger T., Leake A., Müller M., Kirkwood T.B.L., Bürkle A. A modified and automated version of the 'Fluorimetric Detection of Alkaline DNA Unwinding' method to quantify formation and repair of DNA strand breaks. BMC Biotechnol. 2009;9:39. doi: 10.1186/1472-6750-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansoori B., Mohammadi A., Davudian S., Shirjang S., Baradaran B. The different mechanisms of cancer drug resistance: a brief review. Adv. Pharm. Bull. 2017;7:339–348. doi: 10.15171/apb.2017.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauer G.B., Povirk L.F. Specificity and kinetics of interstrand and intrastrand bifunctional alkylation by nitrogen mustards at a G-G-C sequence. Nucleic Acids Res. 1997;25:1211–1218. doi: 10.1093/nar/25.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pontén J., Saksela E. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer. 1967;2:434–447. doi: 10.1002/ijc.2910020505. [DOI] [PubMed] [Google Scholar]
- 69.Squatrito M., Brennan C.W., Helmy K., Huse J.T., Petrini J.H., Holland E.C. Loss of ATM/Chk2/p53 pathway components accelerates tumor development and contributes to radiation resistance in gliomas. Cancer Cell. 2010;18:619–629. doi: 10.1016/j.ccr.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee K.M., Choi K.H., Ouellette M.M. Use of exogenous hTERT to immortalize primary human cells. Cytotechnology. 2004;45:33–38. doi: 10.1007/10.1007/s10616-004-5123-3. https://doi.org/10.1007/10.1007/s10616-004-5123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li N.F., Broad S., Lu Y.J., Yang J.S., Watson R., Hagemann T., Wilbanks G., Jacobs I., Balkwill F., Dafou D., Gayther S.A. Human ovarian surface epithelial cells immortalized with hTERT maintain functional pRb and p53 expression. Cell Prolif. 2007;40:780–794. doi: 10.1111/j.1365-2184.2007.00462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braun F., Blomberg L., Akbar-Haase R., Puelles V.G., Wong M.N., Rahmatollahi M., Fermin D., Slaats G.G., Koehler S., Brinkoetter P.T., Lindenmeyer M.T., Cohen C.D., Kann M., Bloch W., Sampson M.G., Huber T.B., Schermer B., Benzing T., Schumacher B., Kurschat C.E. DNA damage activates mTORC1 signaling in podocytes leading to glomerulosclerosis. bioRxiv. 2020 doi: 10.1101/2020.09.13.295303. 2020.09.13.295303. [DOI] [Google Scholar]
- 73.Ames B.N., Shigenaga M.K., Gold L.S. DNA lesions, inducible DNA repair, and cell division: three key factors in mutagenesis and carcinogenesis. Environ. Health Perspect. 1993;101(Suppl 5):35–44. doi: 10.1289/ehp.93101s535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poetsch A.R. The genomics of oxidative DNA damage, repair, and resulting mutagenesis. Comput. Struct. Biotechnol. J. 2020;18:207–219. doi: 10.1016/j.csbj.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basu A.K., Essigmann J.M. Establishing linkages among DNA damage, mutagenesis, and genetic diseases. Chem. Res. Toxicol. 2022 doi: 10.1021/acs.chemrestox.2c00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Matt S., Hofmann T.G. The DNA damage-induced cell death response: a roadmap to kill cancer cells. Cell. Mol. Life Sci. 2016;73:2829–2850. doi: 10.1007/s00018-016-2130-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodier F., Coppé J.-P., Patil C.K., Hoeijmakers W.A.M., Muñoz D.P., Raza S.R., Freund A., Campeau E., Davalos A.R., Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dai C.-H., Li J., Chen P., Jiang H.-G., Wu M., Chen Y.-C. RNA interferences targeting the Fanconi anemia/BRCA pathway upstream genes reverse cisplatin resistance in drug-resistant lung cancer cells. J. Biomed. Sci. 2015;22:77. doi: 10.1186/s12929-015-0185-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li L., Guan Y., Chen X., Yang J., Cheng Y. DNA repair pathways in cancer therapy and resistance. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.629266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darzynkiewicz Z., Traganos F., Wlodkowic D. Impaired DNA damage response--an Achilles' heel sensitizing cancer to chemotherapy and radiotherapy. Eur. J. Pharmacol. 2009;625:143–150. doi: 10.1016/j.ejphar.2009.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Spanswick V.J., Craddock C., Sekhar M., Mahendra P., Shankaranarayana P., Hughes R.G., Hochhauser D., Hartley J.A. Repair of DNA interstrand crosslinks as a mechanism of clinical resistance to melphalan in multiple myeloma. Blood. 2002;100:224–229. doi: 10.1182/blood.V100.1.224. [DOI] [PubMed] [Google Scholar]
- 82.Vodicka P., Kumar R., Stetina R., Sanyal S., Soucek P., Haufroid V., Dusinska M., Kuricova M., Zamecnikova M., Musak L., Buchancova J., Norppa H., Hirvonen A., Vodickova L., Naccarati A., Matousu Z., Hemminki K. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25:757–763. doi: 10.1093/carcin/bgh064. [DOI] [PubMed] [Google Scholar]
- 83.van Kan M., Burns K.E., Helsby N.A. A systematic review of inter-individual differences in the DNA repair processes involved in melphalan monoadduct repair in relation to treatment outcomes. Cancer Chemother. Pharmacol. 2021;88:755–769. doi: 10.1007/s00280-021-04340-z. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y., Li J., Zhou Y., Li Z., Peng C., Pei H., Zhu W. And-1 coordinates with the FANCM complex to regulate Fanconi Anemia signaling and cisplatin resistance. Cancer Res. 2022 doi: 10.1158/0008-5472.CAN-22-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Spanswick V.J., Hartley J.M., Hartley J.A. Drug-DNA Interaction Protocols. Humana Press; 2010. Measurement of DNA Interstrand Crosslinking in Individual Cells Using the Single Cell Gel Electrophoresis (Comet) Assay; pp. 267–282. [DOI] [PubMed] [Google Scholar]
- 86.Wynne P., Newton C., Ledermann J.A., Olaitan A., Mould T.A., Hartley J.A. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br. J. Cancer. 2007;97:927–933. doi: 10.1038/sj.bjc.6603973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hodskinson M.R., Bolner A., Sato K., Kamimae-Lanning A.N., Rooijers K., Witte M., Mahesh M., Silhan J., Petek M., Williams D.M., Kind J., Chin J.W., Patel K.J., Knipscheer P. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature. 2020;579:603–608. doi: 10.1038/s41586-020-2059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahn B., Kang D., Kim H., Wei Q. Repair of mitomycin C cross-linked DNA in mammalian cells measured by a host cell reactivation assay. Mol. Cells. 2004;18:249–255. [PubMed] [Google Scholar]
- 89.Zhang N., Lu X., Zhang X., Peterson C.A., Legerski R.J. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Mol. Cell. Biol. 2002;22:2388–2397. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bunting S.F., Callén E., Kozak M.L., Kim J.M., Wong N., López-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A., Chen H.-T., Fernandez-Capetillo O., D'Andrea A., Nussenzweig A. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 2012;46:125–135. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takata K., Reh S., Tomida J., Person M.D., Wood R.D. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat. Commun. 2013;4:1–11. doi: 10.1038/ncomms3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feeney L., Muñoz I.M., Lachaud C., Toth R., Appleton P.L., Schindler D., Rouse J. RPA-mediated recruitment of the E3 ligase RFWD3 is vital for interstrand crosslink repair and human health. Mol. Cell. 2017;66:610–621. doi: 10.1016/j.molcel.2017.04.021. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Monien B.H. Advancements of Mass Spectrometry in Biomedical Research. Springer; Cham: 2019. Mass spectrometric DNA adduct quantification by multiple reaction monitoring and its future use for the molecular epidemiology of cancer; pp. 743–751. [Google Scholar]
- 94.Tang Y., Zhang J.-L. Recent developments in DNA adduct analysis using liquid chromatography coupled with mass spectrometry. J. Sep. Sci. 2020;43:31–55. doi: 10.1002/jssc.201900737. [DOI] [PubMed] [Google Scholar]
- 95.Zubel T., Bürkle A., Mangerich A. Mass spectrometric analysis of sulfur mustard-induced biomolecular adducts: are DNA adducts suitable biomarkers of exposure? Toxicol. Lett. 2018;293:21–30. doi: 10.1016/j.toxlet.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 96.Souliotis V.L., Dimopoulos M.A., Episkopou H.G., Kyrtopoulos S.A., Sfikakis P.P. Preferential in vivo DNA repair of melphalan-induced damage in human genes is greatly affected by the local chromatin structure. DNA Repair. 2006;5:972–985. doi: 10.1016/j.dnarep.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 97.Palom Y., Suresh Kumar G., Tang L.-Q., Paz M.M., Musser S.M., Rockwell S., Tomasz M. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbamoyl mitomycin C and mitomycin C. Chem. Res. Toxicol. 2002;15:1398–1406. doi: 10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 98.Dragan A.I., Pavlovic R., McGivney J.B., Casas-Finet J.R., Bishop E.S., Strouse R.J., Schenerman M.A., Geddes C.D. SYBR Green I: fluorescence properties and interaction with DNA. J. Fluor. 2012;22:1189–1199. doi: 10.1007/s10895-012-1059-8. [DOI] [PubMed] [Google Scholar]
- 99.Hill B.T. Studies on the transport and cellular distribution of chlorambucil in the Yoshida ascites sarcoma. Biochem. Pharmacol. 1972;21:495–502. doi: 10.1016/0006-2952(72)90322-X. [DOI] [PubMed] [Google Scholar]
- 100.Brulikova L., Hlavac J., Hradil P. DNA interstrand cross-linking agents and their chemotherapeutic potential. Curr. Med. Chem. 2012;19:364–385. doi: 10.2174/092986712803414295. [DOI] [PubMed] [Google Scholar]
- 101.Uetake Y., Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a "tetraploidy checkpoint". J. Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chung S., Kim S.-H., Seo Y., Kim S.-K., Lee J.Y. Quantitative analysis of cell proliferation by a dye dilution assay: Application to cell lines and cocultures. Cytom. A. 2017;91:704–712. doi: 10.1002/cyto.a.23105. [DOI] [PubMed] [Google Scholar]
- 103.Behrens B.C., Hamilton T.C., Masuda H., Grotzinger K.R., Whang-Peng J., Louie K.G., Knutsen T., McKoy W.M., Young R.C., Ozols R.F. Characterization of a cis-diamminedichloroplatinum(II)-resistant human ovarian cancer cell line and its use in evaluation of platinum analogues. Cancer Res. 1987;47:414–418. [PubMed] [Google Scholar]
- 104.Evercyte GmbH, PODO/TERT256 - Human podocytes hTERT immortalized. https://www.pharmaceutical-networking.com/wp-content/uploads/2017/10/PODO-TERT256-Human-podocytes-hTERT-immortalized.pdf, 2022 (accessed 29 August 2022).
- 105.Neog B., Sinha S., Bhattacharyya P.K. Alkylation of DNA by nitrogen mustards: A DFT study. Comput. Theor. Chem. 2013;1018:19–25. doi: 10.1016/j.comptc.2013.06.002. [DOI] [Google Scholar]
- 106.Suzuki T., Ohsumi S., Makino K. Mechanistic studies on depurination and apurinic site chain breakage in oligodeoxyribonucleotides. Nucleic Acids Res. 1994;22:4997–5003. doi: 10.1093/nar/22.23.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.An R., Jia Y., Wan B., Zhang Y., Dong P., Li J., Liang X. Non-enzymatic depurination of nucleic acids: factors and mechanisms. PLOS ONE. 2014;9 doi: 10.1371/journal.pone.0115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.de Flora S., Izzotti A., Randerath K., Randerath E., Bartsch H., Nair J., Balansky R., van Schooten F., Degan P., Fronza G., Walsh D., Lewtas J. DNA adducts and chronic degenerative diseases. Pathogenetic relevance and implications in preventive medicine. Mutat. Res. /Rev. Genet. Toxicol. 1996;366:197–238. doi: 10.1016/S0165-1110(96)00043-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Data Availability Statement
Data will be made available on request.