Abstract
Aims: Vascular smooth muscle cells are key participants in atherosclerosis. Circular RNA hsa_circ_0000345 (circ_0000345) and miR-647 are related to oxygenized low-density lipoprotein (ox-LDL)-induced arterial smooth muscle cell (ASMC) dysregulation. However, the relationship between circ_0000345 and miR-647 in ox-LDL-induced ASMC dysregulation is unclear.
Methods: Relative levels of circ_0000345, miR-647, and PAP-associated domain containing 5 (PAPD5) mRNA in AS patient’s serum and ox-LDL-induced ASMCs were detected via RT-qPCR. Gain-of-function experiments were utilized to analyze the effects of circ_0000345 upregulation on ox-LDL-induced cell proliferation, migration, invasion, and inflammatory response in ASMCs. The relationship between circ_0000345 or PAPD5 and miR-647 was validated by dual-luciferase reporter and RNA immunoprecipitation assays.
Results: Circ_0000345 and PAPD5 were lowly expressed in AS patient’s serum and ox-LDL-induced ASMCs, while miR-647 expression had an opposing trend. Mechanistically, circ_0000345 was verified as a miR-647 sponge, and miR-647 overexpression impaired the inhibitory effects of circ_0000345 upregulation on ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response. Further experiments demonstrated that PAPD5 was a miR-647 target, and circ_0000345 adsorbed miR-647 to mediate PAPD5 expression. Also, PAPD5 inhibition relieved miR-647 silencing-mediated suppression on ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response.
Conclusions: Circ_0000345 elevated PAPD5 expression via acting as a miR-647 sponge, resulting in alleviating ox-LDL-induced ASMC dysregulation. The study highlighted the critical role of circ_0000345 in AS.
Keywords: ox-LDL, AS, circ_0000345, miR-647, PAPD5, ASMC
Introduction
Atherosclerosis (AS), whose pathological basis is lipid metabolism disorder, can lead to coronary heart disease, cerebral infarction, and peripheral vascular disease 1 , 2) . Vascular smooth muscle cells (VSMCs), an important part of the blood vessel wall, have the potential for self-renewal and differentiation 3) . The abnormal proliferation of VSMCs located in the arterial intima (ASMCs) leads to the thickening of the aortic intima, which plays an important role in AS pathogenesis 4) . Low-density lipoprotein (LDL) is an independent risk factor for AS 5) , and oxidized LDL (ox-LDL) can promote the growth, migration, and differentiation of ASMCs, leading to ASMC dysfunction 6 , 7) . Thus, exploring the underlying mechanism of ASMC dysfunction mediated by ox-LDL is essential to understand AS pathogenesis.
Circular RNAs (circRNAs) are covalently circularized RNA moieties that are generated by a noncanonical splicing event. Thanks to their lack of free 5′ and 3′ ends, circRNAs are resistant to exonuclease activity 8) . Furthermore, circRNAs have the characteristics of cell type and tissue type-specific expression, which prompts people to pay attention to the potential functions of circRNAs 9) . Growing up evidence has shown the vital roles of circRNAs in various diseases, including cardiovascular disease 10) . For instance, circ-Ube3a upregulation causes a more severe myocardial fibrosis after acute myocardial infarction 11) . Another example is that circ-ACSL1 aggravates myocarditis by upregulating MAPK14 12) . Researchers have shown the important roles of circRNAs in AS. For example, circ-0030042 exerts a protecting effect on atherosclerotic plaque stability by targeting eIF4A3 13) . Furthermore, circ-0068087 14) and circ-USP36 15) mediate ox-LDL-induced endothelial cell dysfunction, whereas circ-GNAQ plays an inhibiting impact on endothelial cell senescence 16) . Hsa_circ_0000345 (circ_0000345), derived from the RSF1 gene, has been uncovered to take part in endothelial cell damage under ox-LDL stimulation 17 , 18) . Presently, the potential mechanism of circ_0000345-mediated ox-LDL-induced ASMC dysregulation must be further explored.
One of the important functions of circRNAs is to regulate gene expression by acting as a sponge for microRNAs (miRNAs), which play important role in human diseases 19 - 21) . For example, circ-HIPK3 elevates Rac1 expression through adsorbing miR-93-5p, resulting in aggravating myocardial infarction-induced cardiac dysfunction 22) . Recently, regulation of miRNAs was extensively studied for their roles in biological processes 23) . miR-647 exerts a tumor-inhibiting role in glioma 24) , prostate cancer 25) , gastric cancer 26) , and cervical cancer 26) . However, miR-647 has been uncovered to promote ox-LDL-induced ASMC dysregulation 27) . The online prediction tool (Circular RNA Interactome) shows the possible function of circ_0000345 as a sponge for miR-647. Nonetheless, the relationship between circ_0000345 and miR-647 in ox-LDL-induced ASMC dysregulation is indistinct.
Here, we investigated the relationship between circ_0000345 and miR-647 in ox-LDL-induced ASMC dysregulation. Mechanistic investigations revealed that ox-LDL-mediated circ_0000345 downregulation facilitated ASMC proliferation, migration, invasion, and inflammatory response via decreasing PAP-associated domain containing 5 (PAPD5) expression through functioning as a miR-647 sponge.
Materials and Methods
Ethics Statement
Thirty-five patients diagnosed with AS at the Fourth Affiliated Hospital of Harbin Medical University were recruited in the present research, and 15 normal subjects were utilized as controls. Assessment of AS and vascular stenosis was performed via cerebrovascular magnetic resonance angiography. Patients with cerebral AS and vascular stenosis ≥ 50% were included in the AS group. All the participants gave their written informed consent before they entered the study. The serum samples of all participants were used according to the Declaration of Helsinki under a license approved by the hospital ethics committee of the Fourth Affiliated Hospital of Harbin Medical University.
Cell Culture and Treatment
Human ASMCs (ZQ0491, Zhong Qiao Xin Zhou Biotechnology Co., Ltd., Shanghai, China) were cultured in the F-12K complete medium (ZQ-507) in an incubator with 5% carbon dioxide and 20% oxygen at 37℃. The cells were treated with ox-LDL (50 µg/mL) (Sigma-Aldrich, St. Louis, MO, USA) for 24 h to induce the pathological state of AS, with ox-LDL solvent PBS as a control.
Transient Transfection
The FuGENE® HD Transfection Reagent (Promega, Madison, WI, USA) was used to transfect ASMCs with the circ_0000345 overexpression plasmid, which was produced by inserting its full-length sequence into the pCD-ciR vector (Geneseed, Guangzhou, China). The Lipofectamine RNAiMAX Reagent (Thermo, Waltham, MA, USA) was utilized to transfect ASMCs with the synthesized oligonucleotides si-NC, si-circ_0000345, si-PAPD5, miR-647 inhibitor (anti-miR-647), anti-miR-NC, miR-647 mimic, and miR-NC.
RNA Isolation and RT-qPCR
Total RNA was isolated from serum samples and cells using the Total RNA Extraction Kit (Solarbio, Beijing, China) as per the manufacturer’s instructions. The quality and quantity of the extracted RNA were determined by measuring the absorbance at 260 nm/280 nm (A260/A280) using a NanoDrop spectrophotometer (Thermo). RNA integrity was verified using 1% agarose gel electrophoresis in TAE buffer. For RNase R digestion, total RNA from ASMCs was incubated with 3U/µg RNase R (Epicentre Technologies, WI, USA) at 37℃ for 30 min, and RNase R solvent DEPC-treated water served as a control. Fractionation of cytoplasmic and nuclear fractions from ASMCs was performed using Active Motif’s Nuclear Extract Kit (Carlsbad, CA, USA). Generation of cDNA was conducted using Hifair® Ⅱ 1st Strand cDNA Synthesis Kit (Yesen, Shanghai, China) or miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Aarhus, Denmark) along with 1 µg total RNA. Amplifications were done using the Lightcycler 480 (Roche, Basel, Switzerland) in triplicate with a Hieff® qPCR SYBR Green Master Mix (Yesen) and specific primers ( Table 1 ) . All genes were normalized to the housekeeping gene GAPDH or U6 and the results were expressed as relative fold change (equation 2−ΔΔCt).
Table 1. Primer sequences used for RT-qPCR.
| Genes | Primer sequences (5’-3’) |
|---|---|
| circ_0000345 | Forward (F): 5’-ATGTGGCCTTCCAAACCATCC-3’ |
| Reverse (R): 5’-AGAACATTGGCTGTAGAACGG-3’ | |
| PAPD5 | F: 5’-CTCCATGCGGCCTCGTC-3’ |
| R: 5’-TCGTCTCTAGGGGCAGGAAG-3’ | |
| miR-647 | F: 5’-GCCGAGGTGGCTGCACTCACT-3’ |
| R: 5’-AGCAGGGAGATAACGGACTGAC-3’ | |
| GAPDH | F: 5’-GACAGTCAGCCGCATCTTCT-3’ |
| R: 5’-GCGCCCAATACGACCAAATC-3’ | |
| U6 | F: 5’-CTCGCTTCGGCAGCACA-3’ |
| R: 5’-AACGCTTCACGAATTTGCGT-3’ | |
| RSF1 | F: 5’-TACTCCACAGTTGATGAAGGCA-3’ |
| R: 5’-CATCTCCCATGCCCAGGTACT-3’ |
MTT Assay
The MTT Cell Proliferation Assay Kit (Yesen) was utilized as per the manufacturer’s instructions. Approximately 1×104 cells were allowed to grow for 48 h at 96-well plates and then incubated with the MTT working solution (10 µL, 5 mg/mL) for 3 h. After removing the solution from each well, the cells were incubated with the MTT solubilization buffer (100 µL). Four hours later, the absorbance of the solution was measured using a microplate reader (Bio-Tek, Winooski, VT, USA) at 570 nm.
Western Blotting
Whole-cell extract from ASMCs was prepared using RIPA buffer containing protease and phosphatase inhibitor (Thermo). Protein concentration was determined with a bicinchoninic acid kit (Beyotime, Nantong, China). Protein samples were loaded, separated on 8%–10% SDS-PAGE, blotted on immuno-blot PVDF membranes, blocked in 5% skim milk, followed by incubation with primary antibodies against PCNA (ab265609, 1:1000, Abcam, Cambridge, MA, USA), Cleaved caspase-3 (ab32042, 1:500, Abcam), PAPD5 (ab231266, 1:1000, Abcam), and GAPDH (ab8245, 1:5000, Abcam). Horseradish peroxidase-conjugated goat antirabbit IgG H&L was utilized as a secondary antibody. Protein signals were detected using chemiluminescence (Thermo).
Flow Cytometry Assay
Apoptotic cells were detected using the Annexin V-FITC/PI Apoptosis Detection Kit (Yesen) following the manufacturer’s instructions. The trypsinized ASMCs were collected and resuspended in 1×Binding Buffer (100 µL), followed by staining with Annexin V-FITC and PI. A flow cytometry (BD, San Jose, CA, USA) was utilized for the analysis of the samples after reacting for 15 min in the dark.
Wound Healing Assay
A linear scratch was made using a sterile pipet tip after growing to 90%–100% confluence. Pictures were taken of the same area of the scratch after 0 and 24 h of incubation at 37℃. The remaining area of the wound was judged by Image software (Media Cybernetics, Silver Spring, MD, USA).
Transwell Invasion Assay
Cell invasion ability was analyzed using 24-well transwell chambers (Corning, NY, USA) with Matrigel (Corning). In short, 1×104 ASMCs in F-12K medium-serum-free (200 µL) were added to the upper chamber, and 600 µL of F-12K complete medium was added to the bottom chamber. The cells were allowed to grow for 24 h, and the cells passed the membranes were fixed with 4% paraformaldehyde, followed by staining with 0.5% crystal violet (Yesen). The number of invading cells was counted under a microscope (Olympus, Tokyo, Japan) in five randomly chosen fields.
Enzyme-Linked Immunosorbent Assay (ELISA)
The release of proinflammatory cytokines TNF-α and IL-6 from ASMCs was detected using a microplate reader (Bio-Tek) with human TNF-α (E-EL-H0109c, Elabscience, Wuhan, China)/IL-6 (kt34251, Moshak Biological Technology Co., Wuhan, China) ELISA Kit.
Dual-Luciferase Reporter Assay
Sequences of WT-circ_0000345, MUT-circ_0000345, WT-PAPD5 3′UTR, and MUT-PAPD5 3′UTR were ligated into the pGL3-Basic vector (Promega), respectively. ASMCs were cotransfected with a recombinant luciferase plasmid, pGL3-Basic vector, and pRL-TK (Promega) and miR-647 mimic/miR-NC. Forty-eight hours later, the lysates of transfected ASMCs were analyzed on a varioskan™ LUX microplate reader (Thermo) with a dual-luciferase reporter assay kit (Promega).
RNA Immunoprecipitation (RIP)
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was used for RIP analysis with an antibody against Ago2/IgG. Co-precipitated RNA complex was isolated after digestion with proteinase K (Yesen). The RNA complex was purified with the RNeasy kit (Qiagen, Valencia, CA, USA) and subjected to RT-qPCR analysis.
Statistical Analysis
Experimental results, which from three biological replicates, were analyzed with GraphPad Prism software (Version 6.01, GraphPad Software, San Diego, California, USA) and expressed as mean±standard deviation. Significance was evaluated by Student’s t-test (for two groups) or ANOVA (for multiple groups). P<0.05 was defined as significant.
Results
Expression of miR-647 and circ_0000345 Had a Negative Correlation in the AS Patient’s Serum
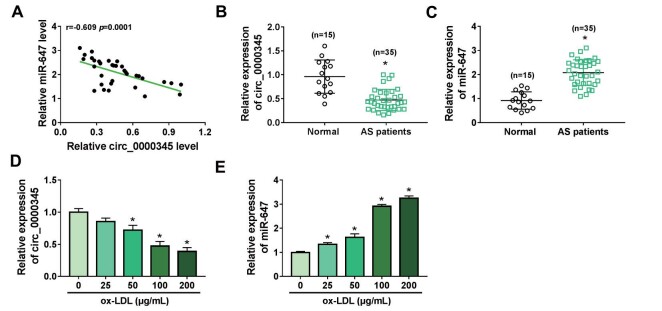
By consulting the references, we found that circ_0000345 17) and miR-647 play opposite roles in AS pathogenesis 27) . We attempted to investigate the association between circ_0000345 and miR-647 in AS pathogenesis. RT-qPCR showed an obvious decrease in circ_0000345 expression and a conspicuous elevation in miR-647 expression in the AS patient’s serum, and their expression had a negative correlation ( Fig.1A-C ) . Also, ox-LDL stimulation decreased circ_0000345 expression in ASMCs in a dose-dependent manner ( Fig.1D ) . By contrast, miR-647 expression was gradually increased in ox-LDL-stimulated ASMCs with the increase of ox-LDL dose ( Fig.1E ) . Together, these findings manifested that the negative correlation between circ_0000345 and miR-647 in the AS patient’s serum might be related to AS pathogenesis.
Fig.1. Expression of miR-647 and circ_0000345 was negatively correlated in the AS patient’s serum.
(A) Pearson’s correlation coefficient analysis of the correlation between miR-647 and circ_0000345 in the AS patient’s serum. (B and C) Relative levels of circ_0000345 and miR-647 in the AS patient’s serum. (D and E) Relative expression levels of circ_0000345 and miR-647 in ASMCs stimulated by different doses of ox-LDL. *P<0.05.
Characteristics of circ_0000345 in ASMCs
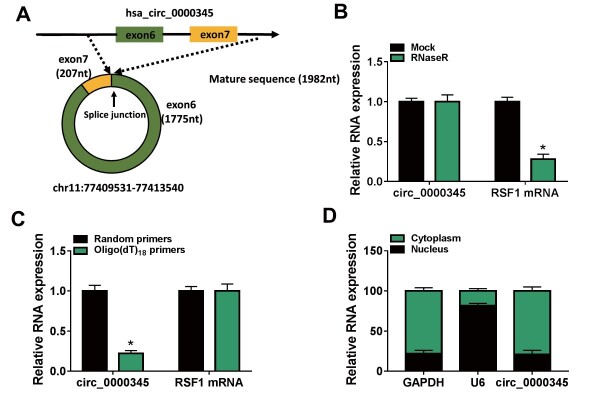
We then explored the characteristics of circ_0000345 in ASMCs. circ_0000345, located on chr11: 77409531-77413540, is derived from exons 6 and 7 of the RSF1 gene ( Fig.2A ) . Because RNase R preferentially degrades linear RNA sequences, we utilized this enzyme to digest ASMCs-derived total RNA. Data in Fig.2B exhibited the resistance of circ_0000345 to RNase R. Also, the expression of circ_0000345 using random primers for reverse transcription was significantly higher than that using oligo (dT)18 primers ( Fig.2C ) . RT-qPCR analysis of nuclear and cytoplasmic RNA showed that circ_0000345 was preferentially expressed in the cytoplasm of ASMCs ( Fig.2D ) . These results manifested that circ_0000345 was a stable circRNA that was mainly localized in the cytoplasm of ASMCs.
Fig.2. The characteristics of circ_0000345 in ASMCs.
(A) Schematic illustration showing the circularization of RSF1 exons 6 and 7 to form circ_0000345. (B) RT-qPCR analysis of circ_0000345 and RSF1 mRNA in ASMCs-derived total RNA pre-treated with or without RNase R. (C) Relative expression levels of circ_0000345 and RSF1 mRNA after reverse transcription with random primers or oligo (dT)18 primers. (D) RT-qPCR analysis of circ_0000345 in ASMCs-derived nuclear RNA and cytoplasmic RNA. *P<0.05.
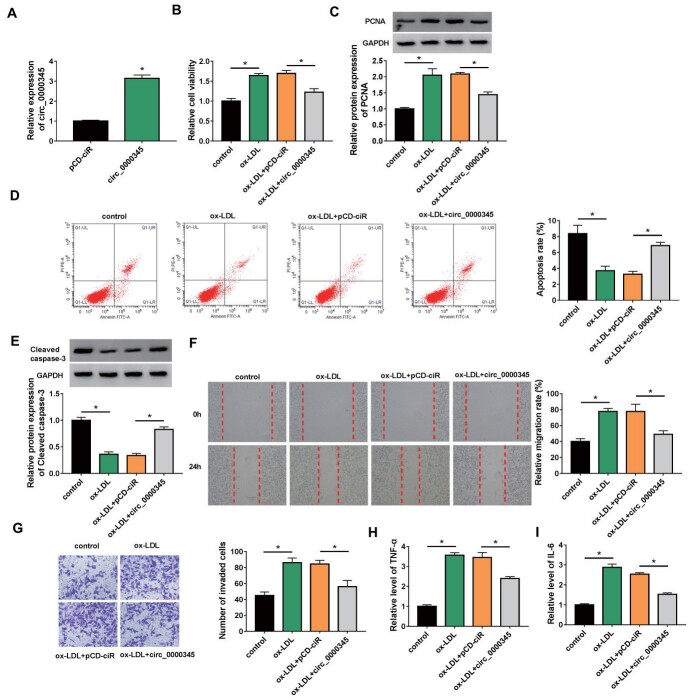
Circ_0000345 Overexpression Weakened ox-LDL-Induced ASMC Proliferation, Migration, Invasion, and Inflammatory Response
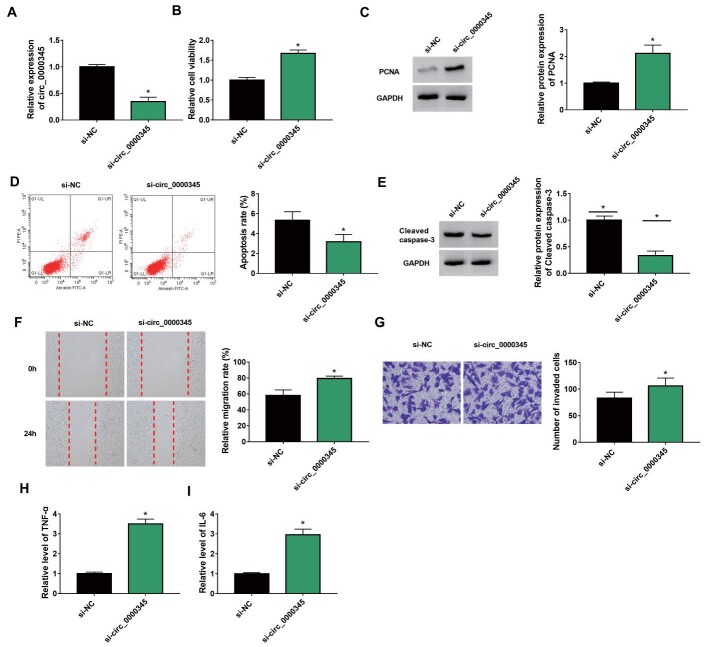
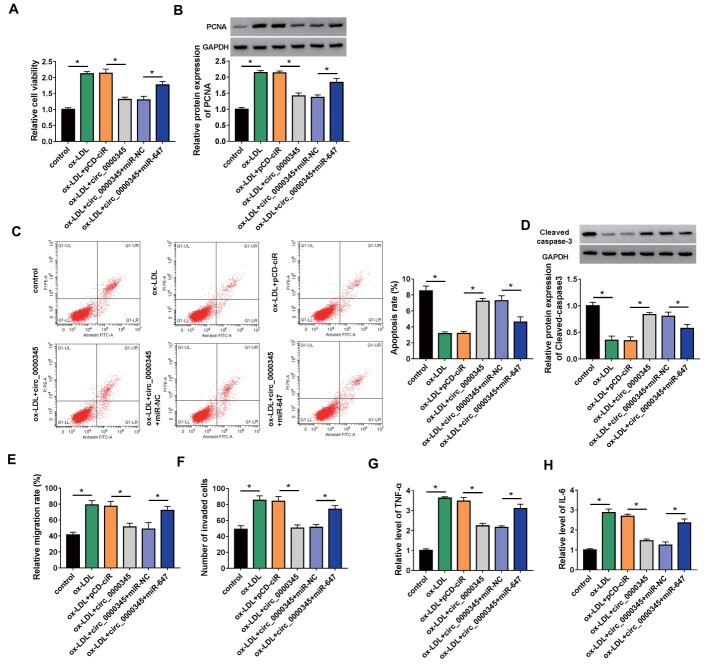
We then tried to investigate whether circ_0000345 was involved in AS pathogenesis. The circ_0000345 overexpression plasmid was constructed and its transfection efficiency was shown in Fig.3A . Furthermore, ox-LDL stimulation elevated cell viability and PCNA protein levels in ASMCs, whereas these changes were weakened by circ_0000345 overexpression ( Fig.3B and C ) . Also, introduction of circ_0000345 reversed the decrease in apoptosis and Cleaved caspase-3 protein levels in ASMCs mediated by ox-LDL stimulation ( Fig.3D and 3E ) . Additionally, the elevated migration and invasion capacities of ASMCs caused by ox-LDL stimulation were overturned after circ_0000345 introduction ( Fig.3F and G ) . Moreover, the increased release of TNF-α and IL-6 in ox-LDL-stimulated ASMCs was impaired by circ_0000345 upregulation ( Fig.3H and I ) . The effects of circ_0000345 silencing on ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response were investigated. The results showed that transfection with si-circ_0000345 decreased circ_0000345 expression in ox-LDL-induced ASMCs ( Supplementary Fig.1A ) . Moreover, circ_0000345 silencing elevated cell viability and PCNA protein levels in ox-LDL-induced ASMCs, implying that circ_0000345 knockdown promoted ox-LDL-induced ASMC proliferation ( Supplementary Fig.1B and C ) . Also, circ_0000345 downregulation reduced cell apoptosis and Cleaved caspase-3 protein levels in ASMCs under ox-LDL stimulation, suggesting that circ_0000345 downregulation inhibited ox-LDL-induced ASMC apoptosis ( Supplementary Fig.1D and E ) . We also observed a marked increase in cell migration and invasion in ox-LDL-induced ASMCs ( Supplementary Fig.1F and G ) . Additionally, the release of TNF-α and IL-6 in ox-LDL-induced ASMCs was elevated after circ_0000345 inhibition ( Supplementary Fig.1H and I ) . Together, these results manifested that circ_0000345 impaired ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response.
Fig.3. Effects of circ_0000345 overexpression on ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response.
(A) RT-qPCR verifying the transfection efficiency of the circ_0000345 overexpression plasmid. (B–I) ASMCs with or without ox-LDL stimulation and circ_0000345/pCD-ciR-transfected ASMCs with ox-LDL stimulation were used. (B) MTT assays showing the viability of ASMCs. (C) Western blotting analysis showing the relative levels of PCNA protein in ASMCs. (D) Flow cytometry assays exhibiting the apoptosis of ASMCs. (E) Western blotting analysis showing the relative levels of Cleaved caspase-3 in ASMCs. (F and G) Wound healing and transwell invasion assays exhibiting the migration and invasion of ASMCs. (H and I) The release of proinflammatory cytokines TNF-α and IL-6 in ASMCs. *P<0.05.
Supplementary Fig.1. Effects of circ_0000345 silencing on cell proliferation, migration, invasion, and inflammatory response in ox-LDL-stimulated ASMCs.
(A-I) ASMCs were transfected with si-NC or si-circ_0000345 and then treated with ox-LDL. (A) Relative expression of circ_0000345 in the above treated ASMCs. (B) The viability of the above treated ASMCs was determined by MTT assay. (C) Relative protein levels of PCNA in the above treated ASMCs were detected by western blotting. (D) The apoptosis of the above treated ASMCs was detected via flow cytometry assay. (E) PCNA protein levels in the above treated ASMCs were measured via western blotting. (F and G) Cell migration and invasion in the above treated ASMCs were evaluated by wound healing and transwell invasion assays. (H and I) ELISA was used to measure the release of TNF-α and IL-6 in the above treated ASMCs. *P<0.05.
Function of circ_0000345 as a miR-647 Sponge
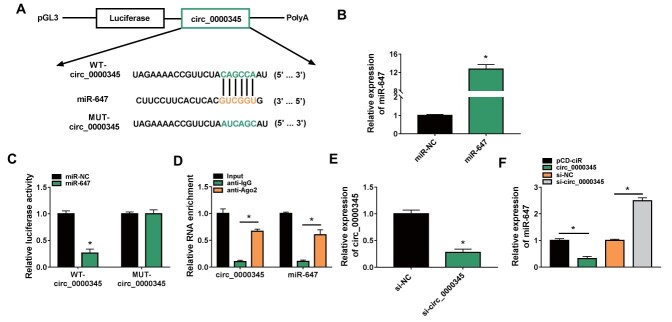
Because circ_0000345 was preferentially located in the cytoplasm, we inferred that circ_0000345 might function as a miRNA sponge. After Circular RNA Interactome prediction, six miRNAs (miR-942 28) , miR-147 29) , miR-647 27) , miR-636 30) , miR-515-5p 31) , and miR-153-3p 32) ) related to AS were selected for further analyzed. We observed that circ_0000345 overexpression repressed miR-647, miR-515-5p, and miR-153-3p expression levels, especially miR-647 ( Supplementary Fig.2 ) . We then constructed WT-circ_0000345 and MUT-circ_0000345 luciferase plasmids, and Fig.4A shows the binding sites between circ_0000345 and miR-647. To verify this prediction, we transfected miR-647 mimic to ASMCs to overexpress miR-647 ( Fig.4B ) . And miR-647 overexpression curbed the luciferase activity of the WT-circ_0000345 luciferase plasmid instead of the MUT-circ_0000345 luciferase plasmid ( Fig.4C ) . Moreover, miR-647 and circ_0000345 were markedly enriched in the RNA complex precipitated by the antibody against Ago2 ( Fig.4D ) . A siRNA targeting circ_0000345 was utilized to interference circ_0000345 in ASMCs ( Fig.4E ) . As expected, miR-647 expression was downregulated in circ_0000345-overexpressed ASMCs and upregulated in circ_0000345-inhibited ASMCs ( Fig.4F ) . Collectively, circ_0000345 functioned as a miR-647 sponge.
Supplementary Fig.2.
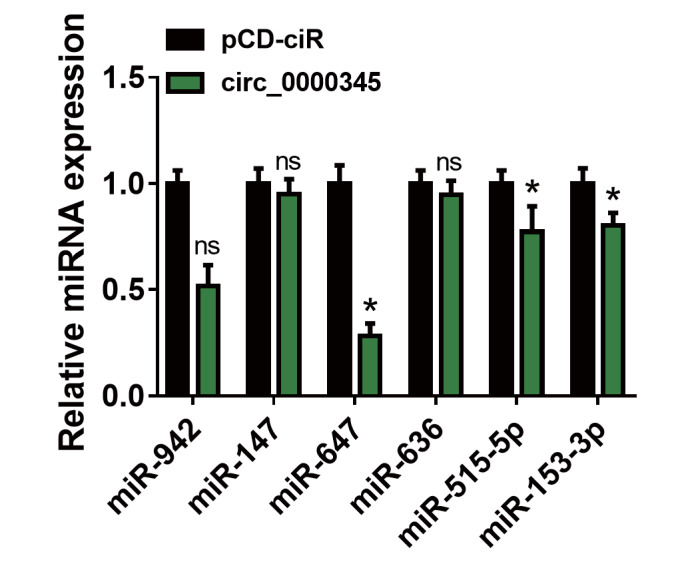
Effect of circ_0000345 overexpression on the expression levels of 6 miRNAs (miR-942, miR-147, miR-647, miR-636, miR-515-5p, and miR-153-3p) in ASMCs was assessed by RT-qPCR.
*P<0.05.
Fig.4. Circ_0000345 acted as a miR-647 sponge.
(A) The WT-circ_0000345 and MUT-circ_0000345 sequences were respectively inserted into the pGL3 plasmid. (B) RT-qPCR showing the transfection efficiency of miR-647 mimic. (C and D) Dual-luciferase reporter and RIP assays were used to validate the relationship between circ_0000345 and miR-647. (E) RT-qPCR showing the interference efficiency of si-circ_0000345. (F) Impacts of circ_0000345 inhibition and overexpression on miR-647 expression. *P<0.05.
Circ_0000345/miR-647 Axis Participated in ox-LDL-Induced ASMC Proliferation, Migration, Invasion, and Inflammatory Response
We then tried to investigate whether ox-LDL induced ASMC proliferation, migration, invasion, and inflammatory response through the circ_0000345/miR-647 axis. The results showed that exogenetic expression of miR-647 weakened circ_0000345 overexpression-mediated inhibition on viability and PCNA protein levels in ox-LDL-stimulated ASMCs ( Fig.5A and B ) . Furthermore, the elevation in apoptosis and Cleaved caspase-3 protein levels in ox-LDL-stimulated ASMCs mediated by circ_0000345 overexpression were whittled by the introduction of miR-647 mimic ( Fig.5C and D ) . Additionally, upregulated miR-647 impaired the inhibitory effects of circ_0000345 overexpression on cell migration, invasion, and inflammatory response in ASMCs under ox-LDL stimulation ( Fig.5E-H ) . In summary, these findings suggested that ox-LDL induced ASMC proliferation, migration, invasion, and inflammatory response via regulation of the circ_0000345/miR-647 axis.
Fig.5. miR-647 impaired circ_0000345 overexpression-mediated effects on cell proliferation, migration, invasion, and inflammatory response in ox-LDL-stimulated ASMCs.
(A–H) ASMCs with or without ox-LDL stimulation and circ_0000345/pCD-ciR/circ_0000345+miR-NC/circ_0000345+miR-647-transfected ASMCs with ox-LDL stimulation were used. (A) MTT assays analysis of the viability of ASMCs. (B) Western blotting detection of the relative levels of PCNA protein in ASMCs. (C) Flow cytometry assays evaluation of the apoptosis of ASMCs. (D) Western blotting investigation of the relative levels of Cleaved caspase-3 protein in ASMCs. (E and F) Wound healing and transwell invasion assays evaluation of the migration and invasion of ASMCs. (G and H) ELISA measurement of proinflammatory cytokines TNF-α and IL-6 in ASMCs. *P<0.05.
PAPD5 was Validated as a miR-647 Target
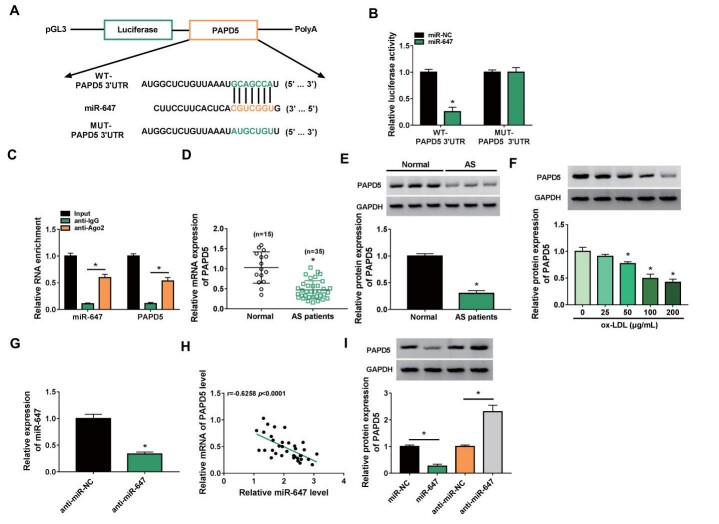
Considering that circRNA can regulate gene expression via sequestering miRNA, we further investigated the target of miR-647. TargetScan predicted that PAPD5 might be a potential target for miR-647. The pGL3 plasmid containing WT-PAPD5 3′UTR or MUT-PAPD5 3′UTR was constructed ( Fig.6A ) . Also, there was a prominent decrease in the luciferase activity of the pGL3 plasmid containing WT-PAPD5 3′UTR in presence of miR-647 mimic ( Fig.6B ) . Moreover, the levels of miR-647 and PAPD5 mRNA were higher in the RNA complex derived from the anti-Ago2 group ( Fig.6C ) . We also observed a significant decrease in PAPD5 mRNA and protein levels in the AS patient’s serum ( Fig.6D and E ) . Analogously, PAPD5 protein levels were decreased in ox-LDL-stimulated ASMCs with the increase in ox-LDL dose ( Fig.6F ) . We also used a miR-647 inhibitor to knockdown miR-647 in ASMCs ( Fig.6G ) . Also, the expression of miR-647 and PAPD5 mRNA was negatively correlated in the AS patient’s serum ( Fig.6H ) . Expectedly, miR-647 mimic repressed PAPD5 protein levels in ASMCs, but the miR-647 inhibitor played an opposing effect ( Fig.6I ) . These findings suggested that PAPD5 was a downstream target for miR-647.
Fig.6. PAPD5 acted as a target for miR-647.
(A) The pGL3 plasmid containing WT-PAPD5 3′UTR or MUT-PAPD5 3′UTR was constructed, respectively. (B and C) The targeting relationship between miR-647 and PAPD5 was demonstrated by dual-luciferase reporter and RIP assays. (D and E) Relative levels of PAPD5 mRNA and protein in the AS patient’s serum. (F) Relative levels of PAPD5 protein in ASMCs stimulated by different doses of ox-LDL. (G) RT-qPCR showing the interference efficiency of miR-647 inhibitor. (H) Pearson’s correlation coefficient analysis of the correlation between miR-647 and PAPD5 mRNA in the AS patient’s serum. (I) Effects of miR-647 inhibition and overexpression on PAPD5 protein levels in ASMCs. *P<0.05.
Inhibition of miR-647 Whittled ox-LDL-Induced ASMC Proliferation, Migration, Invasion, and Inflammatory Response via Upregulating PAPD5
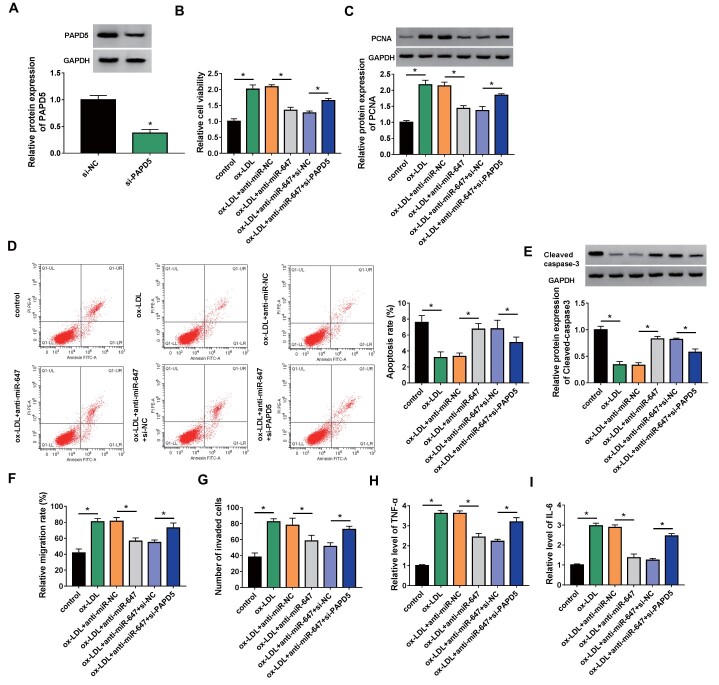
Whether miR-647 mediated ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response via PAPD5 had been further explored. The transfection efficiency of si-PAPD5 was verified and displayed in Fig.7A . Moreover, inhibition of miR-647 weakened the elevation in viability and PCNA protein levels and the reduction in apoptosis and Cleaved caspase-3 protein levels in ASMCs caused by ox-LDL stimulation, but these effects mediated by miR-647 silencing were reversed after PAPD5 knockdown ( Fig.7B-E ) . Additionally, miR-647 knockdown reversed the promoting effects of ox-LDL on ASMC migration, invasion, and inflammatory response, whereas the introduction of si-PAPD5 offset the impacts caused by miR-647 silencing ( Fig.7F-I ) . Collectively, these findings manifested that ox-LDL induced ASMC proliferation, migration, invasion, and inflammatory response via the miR-647/PAPD5 axis.
Fig.7. PAPD5 silencing impaired miR-647 knockdown-mediated effects on ASMC proliferation, migration, invasion, and inflammatory response under ox-LDL stimulation.
(A) Western blotting showing the interference efficiency of si-PAPD5. (B–I) ASMCs with or without ox-LDL stimulation and anti-miR-NC/anti-miR-647/anti-miR-647+si-NC/anti-miR-647+si-PAPD5-transfected ASMCs with ox-LDL stimulation were used. (B) The viability of ASMCs was evaluated. (B) Relative levels of PCNA protein in ASMCs were detected. (C) The apoptosis of ASMCs was detected. (D) Relative levels of Cleaved caspase-3 protein in ASMCs were analyzed. (E and F) The migration and invasion of ASMCs were determined. (G and H) The release of proinflammatory cytokines TNF-α and IL-6 in ASMCs was measured. *P<0.05.
Circ_0000345 Sponged miR-647 to Modulate PAPD5 Expression
Subsequently, we analyzed the involvement between circ_0000345 and PAPD5. Pearson’s correlation coefficient analysis showed a positive correlation between circ_0000345 and PAPD5 in the AS patient’s serum ( Fig.8A ) . Furthermore, circ_0000345 overexpression partly reversed the decreased expression of PAPD5 in ox-LDL-induced ASMCs, but this change mediated by circ_0000345 overexpression was weakened after miR-647 upregulation ( Fig.8B and C ) . Together, these results manifested that circ_0000345 adsorbed miR-647 to regulate PAPD5 expression in ASMCs with ox-LDL stimulation.
Fig.8. Circ_0000345 regulated PAPD5 expression via adsorbing miR-647.
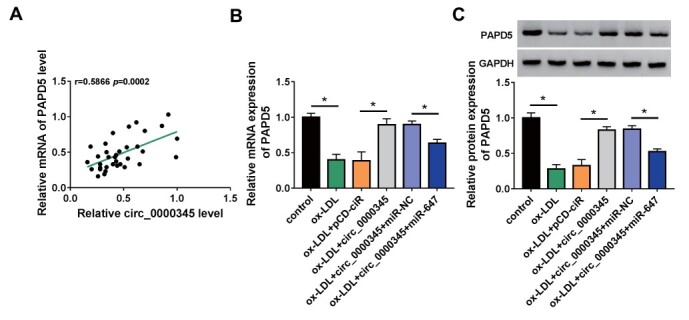
(A) The correlation between circ_0000345 and PAPD5 in the AS patient’s serum. (B and C) Relative levels of PAPD5 protein in ASMCs with or without ox-LDL stimulation and circ_0000345/pCD-ciR/circ_0000345+miR-NC/circ_0000345+miR-647-transfected ASMCs with ox-LDL stimulation were detected. *P<0.05.
Discussion
Insights into the roles of circRNAs in human diseases have made circRNAs attractive tools and targets for novel therapeutic approaches. Accumulated evidence highlights the important function of circRNAs as key regulators in cardiovascular disease 33) . There is sufficient evidence to support that abnormal circRNA expression may be involved in AS pathogenesis 34) . In the present study, we revealed a mechanism by which ox-LDL-mediated circ_0000345 downregulation facilitated ASMC proliferation, migration, invasion, and inflammatory response via the miR-647/PAPD5 axis.
Our results were the same as the previous reports in that circ_0000345 had a lower level in the AS patient’s serum than the control group 17 , 18) . Researchers revealed that circ_0000345 expression was decreased in ox-LDL-induced endothelial cells, and circ_0000345 could weaken ox-LDL-induced endothelial cell damage through miR-758/CCND2 17) and miR-129-5p/TET2 axes 18) . Our results showed a lower circ_0000345 level in ox-LDL-stimulated ASMCs, and circ_0000345 overexpression attenuated ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response. The above evidence uncovered that circ_0000345 could combat ox-LDL-induced ASMC dysregulation and endothelial cell damage.
The canonical regulation of circRNAs in the ceRNA mechanism acts as a miRNA sponge to adsorb miRNA and reduce its abundance 35) . Our results exhibited a negative correlation between circ_0000345 and miR-647 expression in the AS patient’s serum. Also, the function of circ_0000345 as a miR-647 sponge was validated using luciferase reporter and RIP assays. A previous study reported that miR-647 activated the PI3K/AKT pathway via targeting PTEN, resulting in promoting ASMC migration and proliferation 27) . Herein, miR-647 was upregulated in the AS patient’s serum and ox-LDL-induced ASMCs, and miR-647 upregulation whittled the inhibitory effects of circ_0000345 overexpression on ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response. Hence, we inferred that circ_0000345 relieved ox-LDL-induced ASMC proliferation, migration, invasion, and inflammatory response through adsorbing miR-647.
PAPD5, a noncanonical polymerase, can oligoadenylate and destabilize the RNA component of telomerase 36) . PAPD5 can facilitate cellular senescence by regulating the translation of p53 37) . Also, PAPD5 mediates the degradation of cancer miR-21, which is disrupted in other proliferative diseases 38) . Additionally, CASC2 relieved ox-LDL-induced ASMC migration and invasion via upregulating PAPD5 via sponging miR-532-3p 39) . Our data also exhibited an overt decrease in PAPD5 expression in the AS patient’s serum and ox-LDL-induced ASMCs. Also, PAPD5 was verified as a miR-647 target, and PAPD5 silencing partly reversed miR-647 knockdown-mediated repression on ox-LDL-stimulated ASMC proliferation, migration, invasion, and inflammatory response. Of note, circ_0000345 acted as a miR-647 sponge and elevated PAPD5 expression via sponging miR-647 in ox-LDL-stimulated ASMCs. Therefore, we concluded that circ_0000345 decreased ox-LDL-induced proliferation, migration, invasion, and inflammatory response in ASMCs by regulating the miR-647/PAPD5 axis. Unfortunately, we did not collect samples on atherosclerotic plaques, so this study only provided serum levels of circ_0000345, miR-647, and PAPD5. Nevertheless, the levels of circ_0000345, miR-647, and PAPD5 in atherosclerotic plaque samples require further analysis in the future.
In summary, our study verified the downregulation of circ_0000345 in the AS patient’s serum and ox-LDL-stimulated ASMCs. Moreover, our findings identified the important role of circ_0000345/miR-647/PAPD5 axis on ox-LDL-induced ASMC dysregulation, providing new insight into AS pathogenesis.
Acknowledgement
None.
Disclosure of Interest
The authors declare that they have no conflicts of interest.
Funding
None.
References
- 1).Falk E. Pathogenesis of atherosclerosis. J Am Coll Cardiol, 2006; 47: C7-12 [DOI] [PubMed] [Google Scholar]
- 2).Libby P, Bornfeldt KE, Tall AR. Atherosclerosis: Successes, Surprises, and Future Challenges. Circ Res, 2016; 118: 531-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3).Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res, 2016; 118: 692-702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Allahverdian S, Chaabane C, Boukais K, Francis GA, Bochaton-Piallat ML. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res, 2018; 114: 540-550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Nicholls SJ, Tuzcu EM, Nissen SE. Atherosclerosis regression: is low-density lipoprotein or high-density lipoprotein the answer? Curr Atheroscler Rep, 2007; 9: 266-273 [DOI] [PubMed] [Google Scholar]
- 6).Li R, Jiang Q, Zheng Y. Circ_0002984 induces proliferation, migration and inflammation response of VSMCs induced by ox-LDL through miR326/VAMP3 axis in atherosclerosis. J Cell Mol Med, 2021; 25: 8028-8038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Guo X, Liu Y, Zheng X, Han Y, Cheng J. HOTTIP knockdown inhibits cell proliferation and migration via regulating miR-490-3p/HMGB1 axis and PI3K-AKT signaling pathway in ox-LDL-induced VSMCs. Life Sci, 2020; 248: 117445 [DOI] [PubMed] [Google Scholar]
- 8).Kristensen LS, Andersen MS, Stagsted L, Ebbesen KK, Kj Em S J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet, 2019; 20: 675-691 [DOI] [PubMed] [Google Scholar]
- 9).Chen LL. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol, 2020; 21: 475-490 [DOI] [PubMed] [Google Scholar]
- 10).M-Ashraf, Altesha, any Tf, Afaan, Khan, Kexiang, Liu, Xiufen, Zheng. Circular RNA in cardiovascular disease. J Cell Physiol, 2019; 234: 5588-5600 [DOI] [PubMed] [Google Scholar]
- 11).Wang Y, Li C, Zhao R, Qiu Z, Shen C, Wang Z, Liu W, Zhang W, Ge J, Shi B. CircUbe3a from M2 macrophage-derived small extracellular vesicles mediates myocardial fibrosis after acute myocardial infarction. Theranostics, 2021; 11: 6315-6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Zhang L, Han B, Liu H, Wang J, Feng X, Sun W, Cai D, Jia H, Jiang D. Circular RNA circACSL1 aggravated myocardial inflammation and myocardial injury by sponging miR-8055 and regulating MAPK14 expression. Cell Death & Disease, 2021; 12: 487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Yu F, Zhang Y, Wang Z, Gong W, Zhang C. Hsa_circ_0030042 regulates abnormal autophagy and protects atherosclerotic plaque stability by targeting eIF4A3. Theranostics, 2021; 11: 5404-5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14).Li S, Huang T, Qin L, Yin L. Circ_0068087 Silencing Ameliorates Oxidized Low-Density Lipoprotein-Induced Dysfunction in Vascular Endothelial Cells Depending on miR-186-5p-Mediated Regulation of Roundabout Guidance Receptor 1. Front Cardiovasc Med, 2021; 8: 650374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Huang J-g, Tang X, Wang J-j, Liu J, Chen P, Sun Y. A circular RNA, circUSP36, accelerates endothelial cell dysfunction in atherosclerosis by adsorbing miR-637 to enhance WNT4 expression. Bioengineered, 2021; 12: 6759-6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Wu WP, Zhou MY, Liu DL, Min X, Shao T, Xu ZY, Jing X, Cai MY, Xu S, Liang X, Mo M, Liu X, Xiong XD. circGNAQ, a circular RNA enriched in vascular endothelium, inhibits endothelial cell senescence and atherosclerosis progression. Bioengineered, 2021; 26: 374-387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Wei Z, Ran H, Yang C. CircRSF1 contributes to endothelial cell growth, migration and tube formation under ox-LDL stress through regulating miR-758/CCND2 axis. Life Sci, 2020; 259: 118241 [DOI] [PubMed] [Google Scholar]
- 18).Tiliwaldi H, Tursun A, Tohti A, Mamatzunun M, Wu Z. Circ_0000345 Protects Endothelial Cells From Oxidized Low-Density Lipoprotein-Induced Injury by miR-129-5p/Ten-Eleven Translocation Axis. J Cardiovasc Pharmacol, 2021; 77: 603-613 [DOI] [PubMed] [Google Scholar]
- 19).Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet, 2015; 52: 710-718 [DOI] [PubMed] [Google Scholar]
- 20).Yang B, Wang X, Ying C, Peng F, Xu M, Chen F, Cai B. Long Noncoding RNA SNHG16 Facilitates Abdominal Aortic Aneurysm Progression through the miR-106b-5p/STAT3 Feedback Loop. J Atheroscler Thromb, 2021; 28: 66-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Liao B, Zhou MX, Zhou FK, Luo XM, Zhong SX, Zhou YF, Qin YS, Li PP, Qin C. Exosome-Derived MiRNAs as Biomarkers of the Development and Progression of Intracranial Aneurysms. J Atheroscler Thromb, 2020; 27: 545-610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Wu Y, Wu M, Yang J, Li Y, Peng W, Wu M, Yu C, Fang M. Silencing CircHIPK3 Sponges miR-93-5p to Inhibit the Activation of Rac1/PI3K/AKT Pathway and Improves Myocardial Infarction-Induced Cardiac Dysfunction. Front Cardiovasc Med, 2021; 8: 645378 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23).Vishnoi A, Rani S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol Biol, 2017; 1509: 1-10 [DOI] [PubMed] [Google Scholar]
- 24).Qin K, Tian G, Chen G, Zhou D, Tang K. miR‐647 inhibits glioma cell proliferation, colony formation and invasion by regulating HOXA9. The Journal of Gene Medicine, 2020; 22: [DOI] [PubMed] [Google Scholar]
- 25).Chen W, Cen S, Zhou X, Yang T, Wu K, Zou L, Luo J, Li C, Lv D, Mao X. Circular RNA CircNOLC1, Upregulated by NF-KappaB, Promotes the Progression of Prostate Cancer via miR-647/PAQR4 Axis. J Gene Med, 2020; 8: 624764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang Q, Li W, Jiang Y, Zhuang B, Liu H, Shen Z, Wang D, Yan L, Zhang L, Zhou H, Hu Y, Deng H, Liu H, Li G, Qi X. MicroRNA-647 Targets SRF-MYH9 Axis to Suppress Invasion and Metastasis of Gastric Cancer. Theranostics, 2017; 7: 3338-3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Xu CX, Xu L, Peng FZ, Cai YL, Wang YG. MiR-647 promotes proliferation and migration of ox-LDL-treated vascular smooth muscle cells through regulating PTEN/PI3K/AKT pathway. Eur Rev Med Pharmacol Sci, 2019; 23: 7110-7119 [DOI] [PubMed] [Google Scholar]
- 28).Hua Z, Ma K, Liu S, Yue Y, Cao H, Li Z. LncRNA ZEB1-AS1 facilitates ox-LDL-induced damage of HCtAEC cells and the oxidative stress and inflammatory events of THP-1 cells via miR-942/HMGB1 signaling. Life Sci, 2020; 247: 117334 [DOI] [PubMed] [Google Scholar]
- 29).Li L, He M, Zhou L, Miao X, Wu F, Huang S, Dai X, Wang T, Wu T. A solute carrier family 22 member 3 variant rs3088442 G→A associated with coronary heart disease inhibits lipopolysaccharide-induced inflammatory response. J Biol Chem, 2015; 290: 5328-5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Zhang LL. CircRNA-PTPRA promoted the progression of atherosclerosis through sponging with miR-636 and upregulating the transcription factor SP1. Eur Rev Med Pharmacol Sci. 2020; 24: 12437-12449 [DOI] [PubMed] [Google Scholar]
- 31).Xie Q, Li F, Shen K, Luo C, Song G. LOXL1-AS1/miR-515-5p/STAT3 Positive Feedback Loop Facilitates Cell Proliferation and Migration in Atherosclerosis. J Cardiovasc Pharmacol, 2020; 76: 151-158 [DOI] [PubMed] [Google Scholar]
- 32).Guo J, Li J, Zhang J, Guo X, Liu H, Li P, Zhang Y, Lin C, Fan Z. LncRNA PVT1 knockdown alleviated ox-LDL-induced vascular endothelial cell injury and atherosclerosis by miR-153-3p/GRB2 axis via ERK/p38 pathway. Nutr Metab Cardiovasc Dis, 2021; 31: 3508-3521 [DOI] [PubMed] [Google Scholar]
- 33).Tang Y, Bao J, Hu J, Liu L, Xu D. Circular RNA in cardiovascular disease: Expression, mechanisms and clinical prospects. J Cell Mol Med, 2021; 25: 1817-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Cao Q, Guo Z, Du S, Ling H, Song C. Circular RNAs in the pathogenesis of atherosclerosis. Life Sci, 2020; 255: 117837 [DOI] [PubMed] [Google Scholar]
- 35).Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Molecular cancer, 2018; 17: 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Rammelt C, Bilen B, Zavolan M, Keller W. PAPD5, a noncanonical poly(A) polymerase with an unusual RNA-binding motif. Rna, 2011; 17: 1737-1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Burns DM, D’Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature, 2011; 473: 105-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Søkilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, Horie-Inoue K, Ando Y, Burroughs AM, Sasaki C, Suzuki C, Sakai M, Aoki S, Ogawa A, Hasegawa A, Lizio M, Kaida K, Teusink B, Carninci P, Suzuki H, Inoue S, Gunaratne PH, Rovira C, Hayashizaki Y, de Hoon MJ. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc Natl Acad Sci U S A, 2014; 111: 11467-11472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Wang C, Zhao J, Nan X, Guo Z, Huang S, Wang X, Sun F, Ma S. Long noncoding RNA CASC2 inhibits ox-LDL-mediated vascular smooth muscle cells proliferation and migration via the regulation of miR-532-3p/PAPD5. Mol Med, 2020; 26: 74 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]