Abstract
In this retrospective cohort study of 193 consecutive subjects with primary mitochondrial disease (PMD) seen at the Children’s Hospital of Philadelphia Mitochondrial Medicine Frontier Program, we assessed prevalence, severity, and time of onset of sensorineural hearing loss (SNHL) for PMD of different genetic etiology. Subjects were grouped by genetic diagnosis: mitochondrial DNA (mtDNA) pathogenic variants, single large-scale mtDNA deletions (SLSMD), or nuclear DNA (nDNA) pathogenic variants. SNHL was audiometrically confirmed in 27% of PMD subjects (20% in mtDNA pathogenic variants, 58% in SLSMD and 25% in nDNA pathogenic variants). SLSMD had the highest odds ratio for SNHL. SNHL onset was post-lingual in 79% of cases, and interestingly in all cases with mtDNA pathogenic variants and SLSMD, significantly different from PMD due to nDNA variants. Onset during school age dominated. Regular audiologic assessment is important for PMD patients, and PMD of mtDNA etiology should be considered as a differential diagnosis in pediatric patients and young adults with post-lingual SNHL onset, particularly in the setting of multi-system clinical involvement. Pathogenic mtDNA variants and SLSMD are less likely etiologies in subjects with congenital hearing loss.
Keywords: Mitochondrial disease, primary mitochondrial disease, mitochondrial DNA, sensorineural hearing loss, mitochondria, audiology
Introduction
Patients with primary mitochondrial diseases (PMD) commonly develop bilateral symmetric1 sensorineural hearing loss (SNHL)2,3, likely due to energetic failure in hair cells or stria vascularis in the cochlea4,5. Mitochondria are subcellular organelles providing most of the adenosine triphosphate (ATP) required for cellular homeostasis, generated through oxidative phosphorylation within the respiratory chain. Mitochondrial failure disrupts cellular function and leads to clinical symptoms in potentially any organ6. Tissues and organs with high energy requirements, such as muscles, the brain, and specialized sensory cells in the retina and cochlea are particularly susceptible to mitochondrial dysfunction7. The mitochondria contain mitochondrial DNA (mtDNA), which is a double stranded DNA chromosome with 16,569 base pairs that harbors 37 genes8. Cells may contain a mix of wild-type and mutated mtDNA genomes, a phenomenon known as heteroplasmy, where pathogenic variants may cause symptoms only over a certain threshold. As most mitochondrial proteins are encoded by nuclear DNA (nDNA) and imported into mitochondria, pathogenic variants in nDNA genes may also directly impair mitochondrial function. Thus, pathogenic variants in either mtDNA or nDNA can underlie primary mitochondrial disease (PMD)9,10,11.
Hearing loss (HL) is commonly classified based on pure tone average (PTA), which is the average of the threshold in decibel (dB) for a number of frequencies (often 4) where a subject repeatedly can identify a tone. HL can be either conductive due to failure in the outer or middle ear function or sensorineural, where the inner ear or hearing nerve is damaged. HL caused by mitochondrial disease is typically sensorineural5.
SNHL in PMD varies from mild to profound and can have either cochlear or retrocochlear origin1,12,13, but cochlear origin dominates1,2,4. SNHL is often seen as part of a multi-systemic mitochondrial disorder, although SNHL may be the only or predominant symptom, and an audiological assessment is recommended to be included in the standard care of PMD patients14. The most common isolated SNHL due to primary mitochondrial dysfunction is caused by the pathogenic mtDNA variant m.1555A>G in the 12S mitochondrial ribosomal RNA, which causes a maternally inherited deafness of varying degree and conveys a high susceptibility for aminoglycoside ototoxicity15-17.
In this retrospective cohort study, we sought to describe the prevalence, severity and age of onset of SNHL in a consecutive large cohort of subjects diagnosed with molecularly confirmed PMD at a single US center and characterize the susceptibility to SNHL in specific PMD cohorts based on genetic etiology.
Methods
Study design and patient population
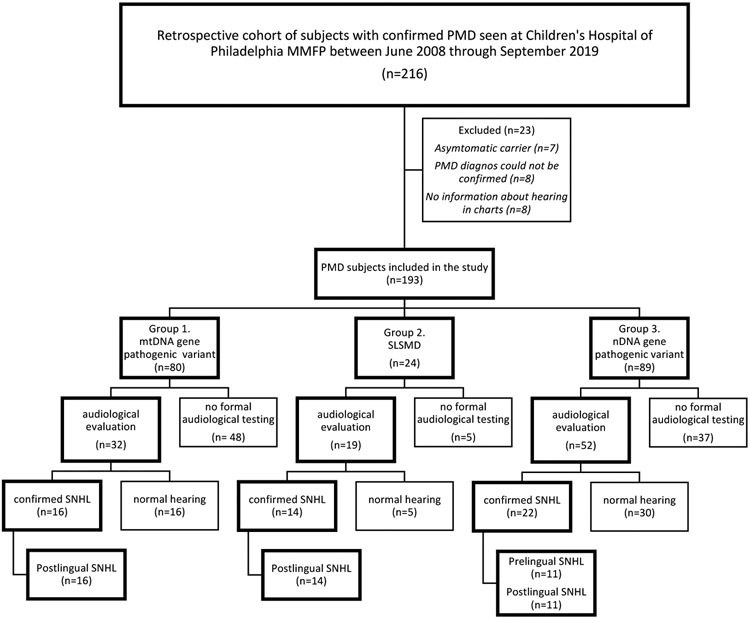
The study, approved by the Institutional Review Board of the Children’s Hospital of Philadelphia (CHOP, IRB #08-006177, PI: M.J. Falk), was an observational retrospective cohort study of all subjects with genetically confirmed PMD evaluated at the CHOP Mitochondrial Medicine Frontier Program (MMFP) between June 2008 to September 2019. The study sample was consecutive and all subjects with molecularly confirmed disease, information about hearing in the medical records and clinical symptoms of PMD were included, to reflect the actual prevalence of SNHL in an unselected clinical patient population. Medical records were reviewed for 216 subjects with confirmed pathogenic variants in either nuclear or mtDNA genes. Twenty-three cases were excluded from further analysis based on absence of clinical symptoms (asymptomatic carriers of pathogenic variants) or insufficient information available regarding genetics and/or hearing function at time of review (Figure 1). Study start was defined as the time of birth for each subject. Data about age, gender, survival, degree of hearing difficulties, onset of hearing concern and genetic etiologies were extracted from the electronic medical records (EMR). Time of SNHL onset was divided into standard developmental age groups (prelingual child < 2 years, preschool child 2-5 years, school-age child 6-12 years, adolescent 13-19 years, young adult 20-40 years, and middle age to elderly >40 years). For subjects deceased at follow-up, age at time of death was used as the follow-up time and for demographic description. The manuscript was prepared according to the EQUATOR network STROBE guidelines for cohort studies18.
Figure 1.
Retrospective cohort study of sensorineural hearing loss in patients with primary mitochondrial disease. Flow-chart of PMD subject enrollment by genetic subgroup etiology, hearing testing performed and degree of SNHL. MMFP, Mitochondrial Medicine Frontier Program; PMD, Primary Mitochondrial Disease; SLSMD, Single large-scale mtDNA deletion; SNHL, Sensorineural hearing loss,
Genetic etiologies
Genetic diagnoses were determined as part of the routine MMFP clinical and/or research evaluations of each subject. No additional genetic testing was performed for the purpose of this retrospective study. All subjects had undergone genetic testing, which reduce exposure bias. Subjects were divided into three groups based on their genotype: (1) mtDNA gene pathogenic variants, (2) single large-scale mtDNA deletions (SLSMD), and (3) nDNA gene pathogenic variants. This grouping was used as exposure in the regression analysis. Throughout this manuscript the term mtDNA gene pathogenic variants refer to pathogenic point mutations or other single-nucleotide pathogenic variants but do not encompass SLSMD.
Hearing function
Hearing function was classified as: normal hearing (≤20 dB), mild HL (21-40 dB), moderate HL (41-60 dB), and severe to profound HL (≥61 dB) on pure tone average (PTA, a mean of the hearing thresholds at a set of specified frequencies, usually 0.5, 1, 2 and 4 kHz) based on available EMR data for 103 of the 193 total subjects. No additional assessment of hearing was performed specifically for this study. SNHL was verified by review of audiological test reports available in the CHOP EMR either performed at CHOP or provided by another care giver. Objective information verifying SNHL was required for a subject to be considered as having SNHL, otherwise normal hearing was assumed to avoid over-reporting and subjective bias. Age of onset of hearing difficulties was noted for each subject, which was based on the earlier time point when self-reported (not guardian or relative) difficulties preceded formal audiologic testing.
Statistical analysis
Data were presented as count and percentage for categorical variables, and median and range for continuous variables. Odds ratios (ORs) with 95% confidence interval (CI) for SNHL according to genetic etiology was determined using multivariable logistic regression, controlling for age and sex, with the nDNA pathogenic variant group as the reference. The occurrence of SNHL and frequency of post- and prelingual onset in each genetic etiology group was compared using Fisher’s exact test across the three groups, and then pairwise if a significant difference was detected. Heteroplasmy levels for subjects with and without hearing loss were compared using students t-test. A sensitivity analysis was conducted by repeating the analyses on patients with formal hearing test performed (n=103). Statistical analyses were performed using R 3.6.1 (www.r-project.org) and Prism GraphPad 9.2.0 (LaJolla, CA, USA). Figures were generated using Prism GraphPad 9.2.0.
Results
Demographics
Medical records were reviewed for 216 subjects with confirmed pathogenic variants in either nuclear or mtDNA genes evaluated at the CHOP Mitochondrial Medicine Frontier Program (MMFP) between June 2008 to September 2019, and 193 subjects were eligible for analysis in the study (Figure 1). Subjects were divided into three groups based on their genotype: (1) mtDNA gene pathogenic variants, (2) single large-scale mtDNA deletions (SLSMD), and (3) nDNA gene pathogenic variants.
The demographics of the study cohort is summarized in Table 1. The study population included 102 females and 91 males. Median age was 14 years (2 days – 78 years), which also is the median follow-up time. The median age of cases in the nDNA group was 8 years as compared to 15 and 19 in the SLSMD group and mtDNA group respectively. Pathogenic mtDNA gene pathogenic variants were confirmed in 80 subjects, including 79 point mutations and 1 single base insertion. Twenty-four subjects had a SLSMD. Pathogenic nDNA gene variants were identified in 89 PMD subjects. 22 subjects were deceased during the study period, of which 18 died before 5 years of age.
Table 1.
Demographics of subjects in PMD cohort by genetic subgroup etiology.
| mtDNA gene pathogenic variant, n=80 |
SLSMD, n=24 |
nDNA gene pathogenic variant, n=89 |
Total, n=193 |
|
|---|---|---|---|---|
| Median age, year (range) | 19 (2-78) | 15 (0–57) | 8 (0–65) | 14 (0-78) |
| Sex | ||||
| Male, n (%) | 36 (45) | 13 (54) | 42 (47) | 91 (47) |
| Female, n (%) | 44 (55) | 11 (46) | 47 (53) | 102 (53) |
| Alive at follow-up, n (%) | ||||
| Yes | 78 (97.5) | 22 (92) | 71 (80) | 171 (89) |
| No | 2 (2.5) | 2 (8) | 18 (20) | 22 (11) |
| Audiological assessment performed | ||||
| Yes, n (%) | 32 (40) | 19 (79) | 52 (58) | 103 (53) |
| No, n (%) | 48 (60) | 5 (21) | 37 (42) | 90 (47) |
| SNHL, n (%) | 16 (20) | 14 (58) | 22 (25) | 52 (27) |
PMD, primary mitochondrial disease; SLSMD, Single large-scale mtDNA deletion; SNHL, Sensorineural hearing loss
PMD genetic etiology subgroups and hearing function
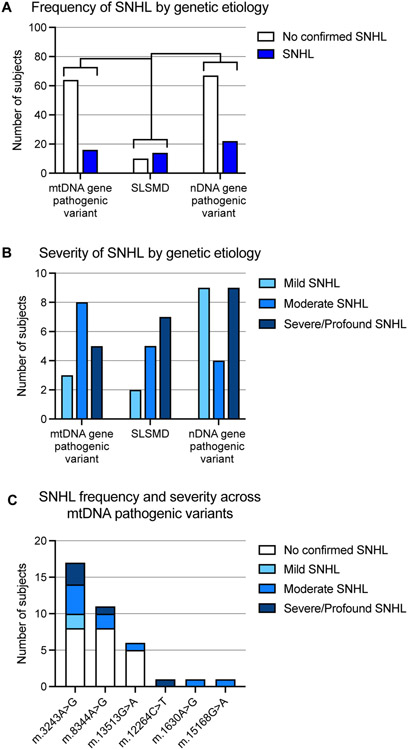
The overall frequency of confirmed SNHL, across the full range from mild to profound, in the PMD study population was 27% (n=52/193), with onset observed at all age groups and among all three genetic etiology groups. Formal audiological tests were performed in 53% (n=103/193) of all PMD cases in this cohort. The audiological test reports were available in 50 cases and in another 53 cases, formal audiological testing had been performed with results available in the EMR. Among the 90 cases without formal audiological assessment, five subjects had subjective hearing complaints, but without objective verification and were hence considered as having normal hearing. Consequently, there may be unrecognized cases of SNHL (particularly isolated high frequency SNHL), and the results reported here should be considered the minimal prevalence of SNHL in this PMD subject cohort. SNHL was present in 20% (n=16/80) of individuals with mtDNA genes pathogenic variant, 58% (n=14/24) of individuals with SLSMD, and 25% (n=22/89) of individuals with nDNA gene pathogenic variants, significantly more common in the SLSMD group compared to the other subgroups (p-value <0.01 and <0.001 respectively, Figure 2A). Among the 52 PMD subjects with SNHL, 40% (n=21/52) had severe to profound SNHL, 33% (n=17/52) had moderate SNHL, and 27% (n=14/52) had mild SNHL (Figure 2B).
Figure 2.
SNHL occurrence and severity in PMD subjects by genetic subgroup etiology. (A) SNHL frequency and (B) SNHL severity are shown among 193 PMD subjects grouped by genetic etiology. (C) SNHL in PMD subjects with mtDNA gene pathogenic variants. Only mtDNA variants identified in individuals with audiological confirmed SNHL are shown. SNHL, sensorineural hearing loss.; PMD, Primary Mitochondrial Disease; **p-value <0.01; *** p-value <0.001
Using multivariable logistic regression with genetic etiology subgroup, age and sex as predictors and the presence of SNHL outcome, an association was found between occurrence of SNHL and genetic etiology subgroup. SLSMD subjects were more than 4 times more likely to have SNHL than were the nDNA gene mutation subjects (Table 2). For individuals with mtDNA variants, no significant difference was seen compared to nDNA variants. Advancing age was found to increase the likeliness of having SNHL. In the sensitivity analysis which only included subjects who had undergone formal testing (n=103) results were similar to the primary analyses. SLSMD subjects were significantly more likely to have SNHL compared to subjects with nDNA pathogenic variants (OR 3.833, 95% CI 1.100 to 15.26, p=0.042).
Table 2.
Multiple logistic regression for presence of SNHL in PMD subjects
| Odds Ratio | p-value | 95% Confidence interval | |
|---|---|---|---|
| Intercept | 0.27 | 0.0001 | 0.13:051 |
| Sex (female ref) | 0.51 | 0.063 | 0.25:1.03 |
| Age (years) | 1.03 | 0.0017 | 1.01:1.05 |
| Genetic subgroup | |||
| nDNA variant | ref | ref | |
| mtDNA variant | 0.50 | 0.089 | 0.21:1.10 |
| SLSMD | 4.29 | 0.0040 | 1.61:11.91 |
PMD, primary mitochondrial disease, SLSMD, Single large-scale mtDNA deletion; SNHL, Sensorineural hearing loss
SNHL characteristics in PMD subjects with mtDNA gene pathogenic variants.
In 80 subjects with mtDNA gene pathogenic variants, 16 had SNHL to variable degree (Figure 2B). This group had widely diverse clinical features ranging from mild neurological symptoms to classical mitochondrial disease syndromes including Leigh syndrome, MERFF (myoclonic epilepsy with ragged red fibers) syndrome, and MELAS (mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes) syndrome. Many mtDNA gene pathogenic variants in this cohort were seen in only a single individual. Among the mtDNA cohort with SNHL, 9 of the 16 (56%) subjects had the common m.3243A>G pathogenic variant, 3 subjects (19%) (unique kindreds) had the m.8344A>G pathogenic variant, and 4 subjects had m.13513G>A, m.12264C>T, m.1630A>G and m.15168G>A, respectively (Figure 2C). Most subjects had moderate to severe SNHL (Figure 2B). Of note, SNHL was not seen in any of the 9 subjects in an extended kindred with the m.3288A>G variant, nor in any of the other 16 subjects with other mtDNA pathogenic variants in whom audiologic testing had been performed. One subject in this group, with a MERRF syndrome phenotype due to the MT-TK m.8344A>G variant received a cochlear implant. SNHL was first noted at 5 years of age and progressed until she received bilateral cochlear implant at 7 years.
SNHL characteristics in PMD subjects with SLSMD.
Subjects with SLSMD had the highest percentage of SNHL (58%, n=14/24, Figure 2A), and the greatest percentage of individuals with severe to profound SNHL (50%, n=7/14). Five (36%) of the remaining SLSMD subjects had moderate SNHL, and 2 (14%) had mild SNHL (Figure 2B). All 14 of the SLSMD subjects with SNHL had a Kearns-Sayre syndrome (KSS) spectrum phenotype19. Four of the SLSMD subjects received a cochlear implant, 3 of whom received unilateral implants at 9, 18 and 25 years of age. The fourth KSS subject was first implanted at age 11 years and then received a contralateral implant at 12 years of age. The size and specific location of the mtDNA deletion was unavailable in 4 cases and the deletion was confirmed only by southern blot (with multiple mtDNA duplications also seen) in 1 case. For the remaining 19 cases, the size range of the mtDNA deletion was 2.7–9.7 kB, with heteroplasmy levels from 9%. Two cases had the typical 4.977 kb deletion, while a 7.4 kb deletion was identified in 3 cases.
SNHL characteristics in PMD subjects with nDNA gene pathogenic variants.
Among the 89 subjects with PMD due to nDNA gene pathogenic variants, 22 subjects (25%) had some degree of SNHL (Figure 2A,B). The nuclear gene etiologies seen in 9 individuals with severe to profound SNHL were RRM2B (n=3), RMND1 (n=2), NDUFS4 (n=1), OPA1 (n=1), POLG (n=1), and SLC25A46 (n=1). Six of these 9 (67%) PMD subjects died during childhood, four of whom died in the first 6 months of life. Nuclear gene etiologies in 4 subjects with moderate SNHL included ELAC2 (n=1), ERCC6 (n=1), FBXL4 (n=1), and TWINKLE (n=1). Nuclear gene etiologies in 9 individuals with mild SNHL included AFG3L2 (n=1), FBXL4 (n=1), MFN2 (n=2), MPV17 (n=1), MRPL3 (n=1), NDUFS8 (n=1), PEPCK (n=1), and POLG (n=1). SNHL was not reported in subjects with pathogenic variants in SURF1 (6 cases) and TRMU (4 cases). In general, MMFP clinical team advised avoidance of aminoglycoside antibiotics in PMD cases whose gene etiologies were reported with known hypersensitivity to these antibiotics, such as in subjects with variants in TRMU20. One subject in this group, with epilepsy and cardiomyopathy due to a variant in RMND1 with mtDNA depletion, received cochlear implants; SNHL was detected at newborn hearing screening with bilateral cochlear implants performed at 2 years of age.
Prelingual or postlingual SNHL onset in PMD subgroups by age and genetic etiology.
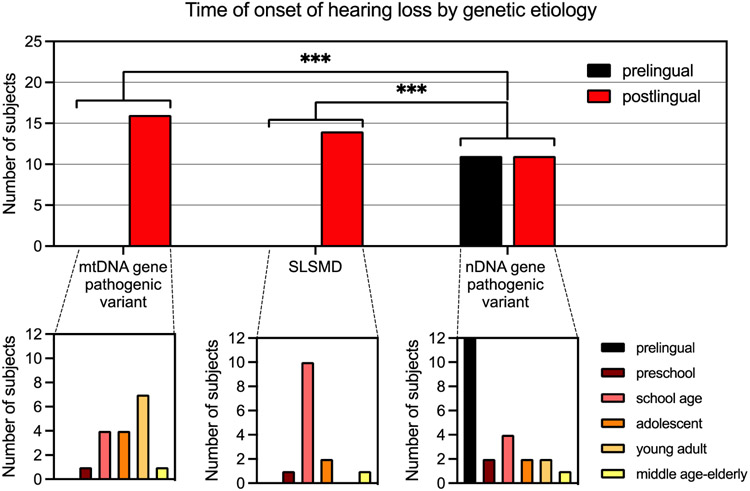
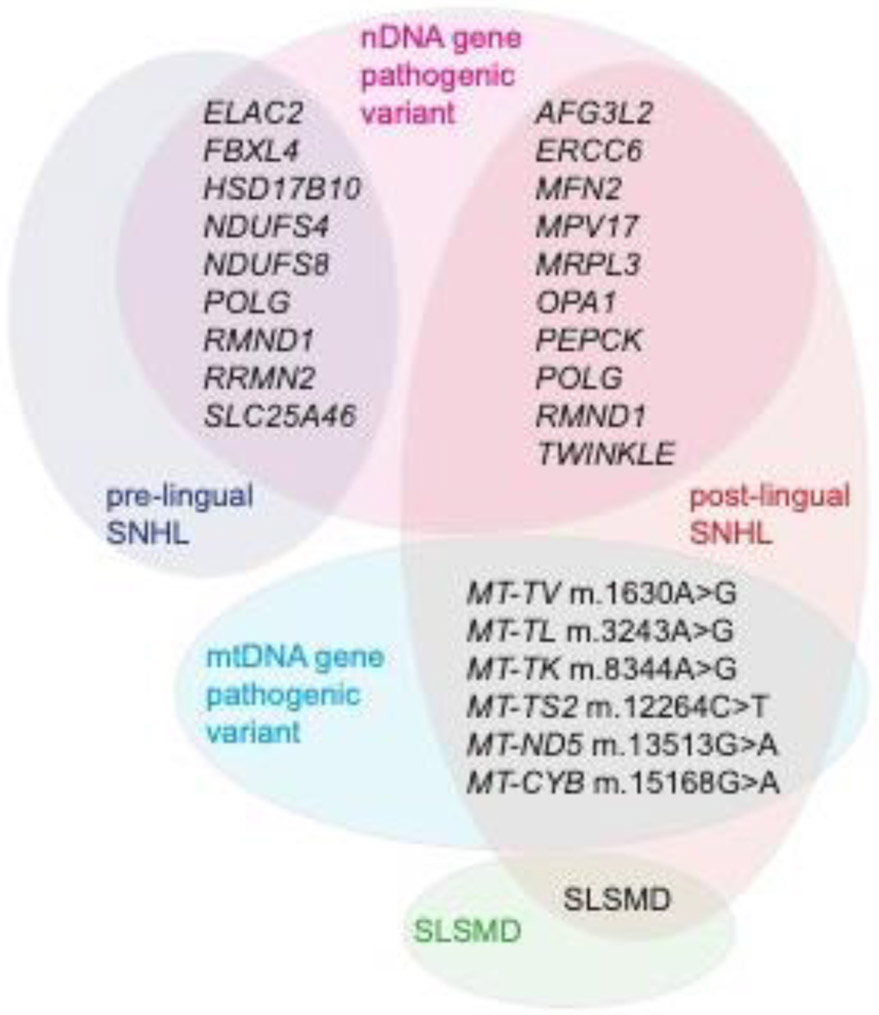
SNHL onset was prelingual in 11 cases, 7 (58%) of whom had severe or profound SNHL. Interestingly, all subjects with prelingual SNHL had nDNA gene pathogenic variants. The causal PMD genes in subjects with prelingual severe to profound SNHL were RRM2B (n=3), POLG (n=1), SLC25A46 (n=1), RMND1 (n=1) and NDUFS4 (n=1). In subjects with prelingual moderate SNHL, causal PMD genes were FBXL4 (n=1) and ELAC2 (n=1). In subjects with prelingual mild SNHL, causal PMD genes were FBXL4 (n=1) and NDUFS8 (n=1). The mtDNA gene pathogenic variant and SLSMD groups had no cases of prelingual SNHL, and consequently differed statistically significantly from the nDNA group (p<0.001) Postlingual SNHL most commonly had onset during school-age (n=18) but occurred in PMD subjects across the spectrum from preschool to elderly adult. 100% of subjects with SNHL due to either mtDNA gene pathogenic variants or SLSMD had postlingual onset of SNHL. In the SLSMD group, 71% (n=10/14) first reported SNHL symptoms during school age (Figure 3 and 5).
Figure 3.
PMD cohort age group at SNHL onset. SNHL age of onset was determined based on first EMR mention of hearing concerns occurring in individuals subsequently diagnosed by formal audiologic evaluation. Age groupings were based on standard development trajectory categorization, including prelingual (before development of spoken language, <2 years), preschool (2-5 years), school-age child (6-12 years), adolescent (13-19 years), young adult (20-40 years), and middle age to elderly adult (>40 years). PMD, Primary mitochondrial disease; SNHL, Sensorineural hearing loss; SLSMD, Single large-scale mtDNA deletions; EMR, Electronic medical records. *** p-value <0.001
Figure 5.
Schematic depiction of pathogenic nuclear genes and mitochondrial (mt) DNA variants in subjects with pre- and post-lingual onset, respectively, of SNHL. PMD, Primary Mitochondrial Disease; SLSMD, Single large-scale mtDNA deletion; SNHL, Sensorineural hearing loss
Relationship between mtDNA heteroplasmy, SNHL and age of onset.
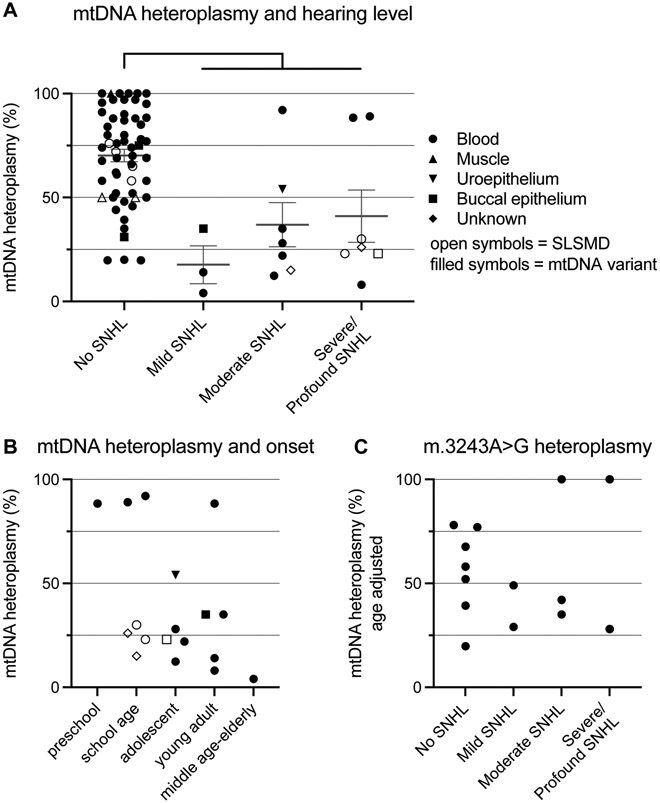
Data for heteroplasmy was available for 79 out of 104 subjects with mtDNA-related disease (mtDNA pathogenic variants or SLSMD) from either blood, muscle, urinary tract, or buccal epithelium (in 7 subjects the tissue of origin could not be established from medical records). Heteroplasmy data were available from blood in 64 subjects, from buccal epithelium in 14 subjects, from urinary tract epithelium in 12 subjects and from muscle in 7 subjects. There was a statistically significant difference in mtDNA heteroplasmy (any tissue) between cases with any degree of SNHL compared to individuals without hearing concerns, with lower heteroplasmy levels in subjects with SNHL (Figure 4A). Heteroplasmy levels in subjects with m.3243A>G (age-adjusted blood data due to known relationship to age in blood21), and in relation to age of onset of SNHL was plotted for visualization purposes but not statistically analyzed due to the limited data available (Figure 4B,C).
Figure 4.
Mitochondrial (mt) DNA heteroplasmy and SNHL. (A) All cases with any mtDNA related disease and mtDNA heteroplasmy data available for any tissue. Student t-test for mtDNA heteroplasmy levels between subjects with no hearing concern and any SNHL, **** p < 0.0001. B) mtDNA heteroplasmy in relationship to hearing loss onset (no age-adjustment) (C) Newcastle age-adjusted blood mtDNA heteroplasmy in relationship to hearing concerns in subjects with m3243A>G. For panel B and C, data was plotted for visualization purposes, and no statistical analysis was performed due to limited data. One data point per subject per panel, all data from latest sampling time point and priority was given to blood data (heteroplasmy levels available in 64 subjects). For panel A and B, other tissues plotted when data from blood was unavailable. mtDNA, mitochondrial DNA; SNHL, Sensorineural hearing loss; SLSMD, Single large-scale mtDNA deletions;
Discussion
Hearing loss in mitochondrial disease
In this single-site retrospective cohort study, we describe the prevalence, severity, age of onset, and genetic etiology of SNHL across a large consecutive cohort of individuals with molecularly confirmed PMD. We specifically show that the minimum prevalence of SNHL in this cohort of 193 molecularly confirmed PMD subjects is 27% (half of the 103 PMD subjects in whom audiologic testing had been performed), largely consistent with previous reports1,22-24. Interestingly, SNHL had particularly high prevalence among SLSMD cases, being significantly higher than in mtDNA and nDNA pathogenic variant subjects. Strikingly, subjects with mtDNA pathogenic variants (both point mutations and SLSMD) presented with postlingual SNHL onset as children, teenagers, or young adults, with none having SNHL documented at the time of newborn screening. This is previously not systematically described. Studies of genetic testing in large cohorts of hearing loss patients typically omit analyses of mtDNA25-28, and hence onset patterns for mtDNA-related hearing loss are largely undescribed. In contrast, prelingual onset of SNHL occurred in half of PMD subjects with SNHL who had causal nDNA gene etiologies. Cochlear implants were placed in only a minority (12%) of the PMD subjects with SNHL.
PMD remains classified as a rare disease, with overall reported prevalence of at least 1 in 4,300 across all ages 29. To our knowledge, we report here the largest cohort of molecularly confirmed PMD subjects in which a retrospective, systematic assessment of hearing function has been performed. The cohort size enabled comparative analysis of SNHL occurrence and likelihood based on major genetic subgroup etiology. However, as PMD is heterogenous in clinical presentation and genetic background, the number of cases for each specific genetic alteration remained generally inadequate to power statistical analyses at the individual gene or variant level.
Van Kempen et al recently assessed hearing in-depth in a relatively large cohort of PMD patients and concluded that largely, SNHL was more severe in high frequencies, symmetrical between ears and of cochlear origin, as shown by maintained speech perception, although potential retrocochlear involvement could be seen in certain patient groups (primarily MELAS, but based on n=2) as demonstrated by prolonged ABR latencies1. Auditory neuropathy, where speech perception is worse than expected from degree of hearing loss in PTA, has previously been shown to occur in mitochondrial disease (reviewed in30), but was hence not present in the van Kempen study, and no auditory neuropathy cases were identified from the medical records in this study. However, in more severe cases of PMD, where the clinical picture is dominated by other signs and symptoms, auditory neuropathy may go unnoticed.
Prevalence of hearing loss in mitochondrial disease in genetic subgroups
The m.3243A>G pathogenic variant in mtDNA has been extensively associated with SNHL, with the known “maternally-inherited diabetes and deafness” (MIDD)31 phenotype. In our study, more than half of subjects (53%, n=9/17) with the m.3243A>G variant had SNHL, generally moderate to severe. This overall prevalence of SNHL in m.3243A>G subjects is largely consistent with other reported studies. The prevalence of SNHL was 58% (n=73/126) in an Italian m.3243A>G cohort 23, 76% (n=25/33) in a Czech cohort 24, and 67% (n= 92/138) in a North American cohort 22. Furthermore, 31% of MERRF subjects with m.8344A>G (12/39) had impaired hearing in this Italian cohort, similar to 27 % of m.8344A>G subjects in our study 23. In the North American Mitochondrial Disease Consortium (NAMDC) registry of PMD patients, SNHL was reported in 18% (n=46/252) of participants with any nDNA gene pathogenic disorder, 39% (n=160/440) of participants with any mtDNA pathogenic variant, 67% (n=92/138) of participants with the m.3243A>G pathogenic variant specifically, and 27% (n=18/67) of participants with SLSMD. The total prevalence of SNHL in the NAMDC registry for SLSMD was lower than the 58% that we report here. Since mitochondrial disease have a broad spectrum of symptoms and signs involving failure of vital function, symptoms such as hearing loss may be overlooked in the setting of severe disease and this discrepancy could potentially be due to underreporting. It should be noted that many of the PMD cases in this current study also are included in the NAMDC registry 22.
Hearing loss and mtDNA heteroplasmy
Hearing loss in mitochondrial disease is thought to be due to failure of energy-intense cochlear hair cells or supportive cells, and it would be reasonable to assume that increasing mutational load would lead to worse hearing. This relationship has previously been demonstrated for muscle heteroplasmy in m.3443A>G, but not for blood2. In this study, increasing levels of heteroplasmy of pathogenic mtDNA variants or SLSMDs could not be shown to lead to more severe hearing loss. To the contrary, in subjects with hearing loss, lower heteroplasmy levels compared to subjects without hearing concerns was noted. There are several potential confounding factors, and the results should be interpreted cautiously. In addition to missing data for heteroplasmy levels in 25 of 104 individuals with mtDNA-related disease there was an inconsistency in sampled tissue, and heteroplasmy levels can vary considerably between tissues. It is to our knowledge not concluded what peripheral tissue best reflects the mtDNA heteroplasmy levels of the inner ear, or neuronal tissue. Heteroplasmy levels may change with age, which specifically has been shown for m.3243A>G in blood21. Age-adjusted m.3243A>G heteroplasmy levels did however not show any visual relationship to degree of hearing loss (muscle heteroplasmy data was only available for one subject) (Fig 4C). Further, there is a considerable risk of selection bias, as investigation of hearing function may be unprioritized in subjects with severe clinical disease (and potentially high heteroplasmic load). Data were insufficient to statistically analyze mtDNA heteroplasmy in relation to age of onset of hearing loss, but it can be noted that most cases with low heteroplasmy levels had a relatively later onset (Fig 4B). Here, no adjustment for age was performed though, as this data encompass all mtDNA variants in the material, and from any tissue. As no cases with pathogenic mtDNA variants or SLSMD had prelingual onset, we speculate that mtDNA variants do not cause inner ear damage until the individual is subject to the strain and stress of extrauterine life, with sound/noise exposure and mitochondrial reactive oxygen species (ROS) generation.
Clinical consequences
Based on the findings in this study, we argue that genetic diagnostic evaluations for SNHL routinely should include analysis of both nDNA and mtDNA genomes32, and that disease related to mtDNA specifically should be considered in unexplained, postlingual onset SNHL33. For adults in particular, the presence of diabetes mellitus in combination with SNHL and a positive maternal family history of SNHL should raise concern for the common m.3243A>G pathogenic variant in mtDNA. Whole exome sequencing25,34, may at some centers and/or diagnostic laboratories include mtDNA genome sequencing for both pathogenic variants and copy number alterations, if requested by the ordering physician.
Future prospective audiologic characterization of PMD subjects at regular intervals by a standardized battery of audiologic tests would be valuable to determine the true prevalence of SNHL across PMD genotypes and phenotypes, ages, severity, progression, and response to hearing aid and/or cochlear implant intervention in the PMD population.
Limitations
Despite being encouraged to undergo yearly audiologic assessment, only 52% of subjects had undergone formal audiology testing with results available. As subjects with subjective hearing concerns were not included in this analysis the prevalence in this study of mild or high-frequency SNHL may be underestimated, and hence the true prevalence of hearing loss in this population could be higher. No specific audiological testing was performed for this study, and hence there is a difference in which specific testing each subject has undergone. The strength of this study is rather the analysis of a large consecutive unselected clinical sample. Further, genetic testing was completed as part of clinical diagnostic evaluations rather than specifically for research purposes in this study cohort, where choice of tissue and methodology varied over time and between individuals.
Conclusions
SNHL is a common PMD symptom, present in 27% of 193 PMD subjects, and in this study more prevalent in SLSMD cases compared to other genetic etiologies. While prelingual onset SNHL occurs in nDNA subjects, in any mtDNA variant (mtDNA gene pathogenic variant or SLSMD) onset is invariably postlingual. Prospective genomic analysis in larger cohorts of individuals with prelingual and postlingual hearing loss respectively are needed to confirm this observation. Individuals with PMD are at high risk for developing SNHL and should undergo regular hearing tests despite having passed their newborn hearing screen. As PMD may be an under-recognized cause of SNHL in the wider population, increased awareness is needed among audiology professionals to consider PMD of mtDNA etiology in cases of postlingual SNHL6,14,35.
Acknowledgements
The authors would like to thank the many PMD subjects who participated in this study, as well as Anna Åkesson, Clinical Studies Sweden – Forum South, Skåne University Hospital, Lund, Sweden for statistical support.
Funding
This work was funded by the National Institutes of Health (R03-DK082521 and R35-GM134863 to MJF), Children’s Hospital of Philadelphia Mitochondrial Medicine Frontier Program, The Swedish Hearing Research Foundation, Regional and Departmental grants for Skane University Hospital, The Swedish Royal Physiographic Society, The Magnus Bergvall Foundation, Fredrik and Ingrid Thuring’s Foundation, The Lars Hierta Memorial Foundation, The Acta Oto-Laryngologica Foundation and The Swedish Society of Medicine. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations or the National Institutes of Health. No funding body had influence over the execution of the study or the writing of the manuscript.
List of abbreviations
- CHOP
Children’s Hospital of Philadelphia
- CI
Confidence interval
- dB
decibel
- EMR
Electronic medical records
- HL
Hearing loss
- KSS
Kearns-Sayre syndrome
- MELAS
Mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes
- MERFF
Myoclonic epilepsy with ragged red fibers
- MMFP
Mitochondrial Medicine Frontier Program
- mtDNA
Mitochondrial DNA
- NAMDC
North American Mitochondrial Disease Consortium
- OR
Odds ratio
- PTA
Pure tone average
- PMD
Primary mitochondrial disease
- ROS
Reactive oxygen species
- SLSMD
Single large-scale mtDNA deletions
- SNHL
Sensorineural hearing loss
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of the Children’s Hospital of Philadelphia (IRB #08-006177, PI: M.J. Falk) and all participants, or their legal guardian as appropriate, provided informed consent for their participation.
Consent for publication
All included subjects, or their legal guardian as appropriate provided consent to the study including publication of the data.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
According to the ICMJE uniform disclosure form the authors have the following to declare: no support, economic or otherwise, from any organisation for the submitted work; MJF is inventor on International Patent Application No. PCT/US19/39631 entitled, “Compositions and Methods for Treatment of Mitochondrial Respiratory Chain Dysfunction and Other Mitochondrial Disorders and Methods for Identifying Efficacious Agents for the Alleviating Symptoms of the Same,” filed in the Name of The Children’s Hospital of Philadelphia on 6/27/2019. M.J.F. is a scientific advisory board member with equity interest in RiboNova, Inc., and scientific board member as paid consultant with Khondrion and with Larimar Therapeutics. M.J.F. has previously been or is currently engaged with several companies involved in mitochondrial disease therapeutic preclinical and/or clinical stage development as a paid consultant (Astellas [formerly Mitobridge] Pharma Inc., Casma Therapeutics, Cyclerion Therapeutics, Epirium Bio, HealthCap, Imel Therapeutics, Minovia Therapeutics, NeuroVive Pharmaceutical, Reneo Pharmaceuticals, Stealth BioTherapeutics, and Zogenix, Inc.) and/or a sponsored research collaborator (AADI Therapeutics, Astellas, Cardero Therapeutics, Cyclerion Therapeutics, Epirium Bio, Imel Therapeutics, Khondrion, Minovia Therapeutics Inc., Mission Therapeutics, NeuroVive Pharmaceutical, PTC Therapeutics, Raptor Therapeutics, REATA Inc., Reneo Therapeutics, RiboNova Inc., Standigm Therapeutics, and Stealth BioTherapeutics). RG is a consultant with Minovia Therapeutics. ZZC is a consultant with Reneo Pharmaceuticals. JKE has had intermittent salary support from, and equity interest in, Abliva (formerly Neurovive Pharmaceutical), a company active in the field of mitochondrial medicine. The remaining authors have no competing interest to report.
References
- 1.van Kempen CMA, Beynon AJ, Smits JJ & Janssen MCH A retrospective cohort study exploring the association between different mitochondrial diseases and hearing loss. Mol Genet Metab 135, 333–341, doi: 10.1016/j.ymgme.2022.02.003 (2022). [DOI] [PubMed] [Google Scholar]
- 2.Chinnery PF et al. The spectrum of hearing loss due to mitochondrial DNA defects. Brain 123 (Pt 1), 82–92, doi: 10.1093/brain/123.1.82 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Scarpelli M et al. Mitochondrial Sensorineural Hearing Loss: A Retrospective Study and a Description of Cochlear Implantation in a MELAS Patient. Genetics research international 2012, 287432, doi: 10.1155/2012/287432 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kullar PJ et al. Both mitochondrial DNA and mitonuclear gene mutations cause hearing loss through cochlear dysfunction. Brain 139, e33, doi: 10.1093/brain/aww051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forli F et al. Mitochondrial syndromic sensorineural hearing loss. Bioscience reports 27, 113–123, doi: 10.1007/s10540-007-9040-5 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Muraresku CC, McCormick EM & Falk MJ Mitochondrial Disease: Advances in clinical diagnosis, management, therapeutic development, and preventative strategies. Curr Genet Med Rep 6, 62–72, doi: 10.1007/s40142-018-0138-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas RH et al. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics 120, 1326–1333, doi: 10.1542/peds.2007-0391 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Greaves LC, Reeve AK, Taylor RW & Turnbull DM Mitochondrial DNA and disease. J Pathol 226, 274–286, doi: 10.1002/path.3028 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Angelini C, Bello L, Spinazzi M & Ferrati C Mitochondrial disorders of the nuclear genome. Acta Myol 28, 16–23 (2009). [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart JB & Chinnery PF The dynamics of mitochondrial DNA heteroplasmy: implications for human health and disease. Nat Rev Genet 16, 530–542, doi: 10.1038/nrg3966 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Picard M & Hirano M Disentangling (Epi)Genetic and Environmental Contributions to the Mitochondrial 3243A>G Mutation Phenotype: Phenotypic Destiny in Mitochondrial Disease? JAMA Neurol 73, 923–925, doi: 10.1001/jamaneurol.2016.1676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sue CM et al. Cochlear origin of hearing loss in MELAS syndrome. Ann Neurol 43, 350–359, doi: 10.1002/ana.410430313 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Cacace AT & Pinheiro JM The mitochondrial connection in auditory neuropathy. Audiol Neurootol 16, 398–413, doi: 10.1159/000323276 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Parikh S et al. Patient care standards for primary mitochondrial disease: a consensus statement from the Mitochondrial Medicine Society. Genet Med 19, doi: 10.1038/gim.2017.107 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaheel H et al. Frequency of mitochondrial m.1555A > G mutation in Syrian patients with non-syndromic hearing impairment. BMC Ear Nose Throat Disord 18, 7, doi: 10.1186/s12901-018-0055-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yelverton JC et al. The clinical and audiologic features of hearing loss due to mitochondrial mutations. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery 148, 1017–1022, doi: 10.1177/0194599813482705 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Bai YH, Ren CC, Gong XR & Meng LP A maternal hereditary deafness pedigree of the A1555G mitochondrial mutation, causing aminoglycoside ototoxicity predisposition. J Laryngol Otol 122, 1037–1041, doi: 10.1017/S0022215107001648 (2008). [DOI] [PubMed] [Google Scholar]
- 18.von Elm E et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet 370, 1453–1457, doi: 10.1016/s0140-6736(07)61602-x (2007). [DOI] [PubMed] [Google Scholar]
- 19.Goldstein A & Falk MJ in GeneReviews((R)) (eds Adam MP et al. ) (1993). [Google Scholar]
- 20.He Z et al. Reduced TRMU expression increases the sensitivity of hair-cell-like HEI-OC-1 cells to neomycin damage in vitro. Sci Rep 6, 29621, doi: 10.1038/srep29621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grady JP et al. mtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol Med 10, doi: 10.15252/emmm.201708262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barca E et al. Mitochondrial diseases in North America: An analysis of the NAMDC Registry. Neurol Genet 6, e402, doi: 10.1212/NXG.0000000000000402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancuso M et al. The m.3243A>G mitochondrial DNA mutation and related phenotypes. A matter of gender? Journal of neurology 261, 504–510, doi: 10.1007/s00415-013-7225-3 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Dvorakova V et al. The phenotypic spectrum of fifty Czech m.3243A>G carriers. Mol Genet Metab 118, 288–295, doi: 10.1016/j.ymgme.2016.06.003 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Downie L et al. Exome sequencing in infants with congenital hearing impairment: a population-based cohort study. Eur J Hum Genet 28, 587–596, doi: 10.1038/s41431-019-0553-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehta D et al. Outcomes of evaluation and testing of 660 individuals with hearing loss in a pediatric genetics of hearing loss clinic. Am J Med Genet A 170, 2523–2530, doi: 10.1002/ajmg.a.37855 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Nishio SY & Usami S Deafness gene variations in a 1120 nonsyndromic hearing loss cohort: molecular epidemiology and deafness mutation spectrum of patients in Japan. Ann Otol Rhinol Laryngol 124 Suppl 1, 49S–60S, doi: 10.1177/0003489415575059 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Sloan-Heggen CM et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet 135, 441–450, doi: 10.1007/s00439-016-1648-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman GS et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol 77, 753–759, doi: 10.1002/ana.24362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santarelli R Information from cochlear potentials and genetic mutations helps localize the lesion site in auditory neuropathy. Genome Med 2, 91, doi: 10.1186/gm212 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Hattab AW, Almannai M & Scaglia F in GeneReviews(®) (eds Adam MP et al. ) (University of Washington, Seattle: Copyright © 1993-2020, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., 1993). [Google Scholar]
- 32.McCormick EM, Muraresku CC & Falk MJ Mitochondrial Genomics: A complex field now coming of age. Curr Genet Med Rep 6, 52–61, doi: 10.1007/s40142-018-0137-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen L, McCormick EM, Muraresku CC, Falk MJ & Gai X Clinical Bioinformatics in Precise Diagnosis of Mitochondrial Disease. Clin Lab Med 40, 149–161, doi: 10.1016/j.cll.2020.02.002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shearer AE & Smith RJ Massively Parallel Sequencing for Genetic Diagnosis of Hearing Loss: The New Standard of Care. Otolaryngology--head and neck surgery : official journal of American Academy of Otolaryngology-Head and Neck Surgery 153, 175–182, doi: 10.1177/0194599815591156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barcelos I, Shadiack E, Ganetzky RD & Falk MJ Mitochondrial medicine therapies: rationale, evidence, and dosing guidelines. Curr Opin Pediatr 32, 707–718, doi: 10.1097/mop.0000000000000954 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]