Abstract
Mycobacterium marinum, a relatively rapid-growing fish and human pathogen, has become an important model for the investigation of mycobacterial pathogenesis. M. marinum is closely related to the Mycobacterium tuberculosis complex and causes a disease in fish and amphibians with pathology similar to tuberculosis. We have developed an in vitro model for the study of M. marinum virulence mechanisms using the carp monocytic cell line CLC (carp leukocyte culture). We found that fish monocytes can differentiate between pathogenic and nonpathogenic mycobacterial species. Interestingly, M. marinum enters fish monocytes at a 40- to 60-fold-higher rate than Mycobacterium smegmatis. In addition, M. marinum survives and replicates in fish monocytes while M. smegmatis is killed. We also found that M. marinum inhibits lysosomal fusion in fish monocytes, indicating that these cells may be used to dissect the mechanisms of intracellular trafficking in mycobacteria. We conclude from these observations that monocytic cells from fish, a natural host for M. marinum, provide an extremely valuable model for the identification and characterization of mycobacterial virulence determinants in the laboratory.
Tuberculosis, caused by Mycobacterium tuberculosis, is currently the number one cause of death worldwide from a single infectious agent (11, 28). Genetic analysis of the molecular mechanisms of pathogenesis by M. tuberculosis is ongoing, but the few putative virulence determinants that have been identified are not well understood and specific inactivation of many of these genes by allelic exchange has not been accomplished. The fact that few specific mutations have been constructed in M. tuberculosis may be at least partially due to the fact that homologous recombination is more efficient in rapidly growing mycobacterial species (49, 50). Due to the low growth rate of M. tuberculosis, difficulty of manipulation, risk to research personnel, and cost of running a biosafety level three facility, model systems that would allow a better understanding of the causes of tuberculosis are of great interest (8, 12). Recently, it has become apparent that Mycobacterium marinum has all of the necessary characteristics of an ideal model organism for genetic analysis of M. tuberculosis pathogenesis. M. marinum has a generation time of 4 h compared to 20 h for M. tuberculosis (17, 34), it is a biosafety level two organism, human disease caused by M. marinum presents almost exclusively as lesions on the extremities (4, 18, 20), and construction of specific mutations by homologous recombination is relatively straightforward (52, 53). For these reasons, there is heightened interest in M. marinum as a model for the study of mycobacterial pathogenesis (6, 51, 54, 73).
Since M. marinum is a natural pathogen of poikilothermic organisms (17), the fish (70) and frog (54) animal models should offer the opportunity to closely approximate natural mycobacterial infections in the laboratory. M. marinum is predominantly an aquatic organism that causes systemic tuberculous infections in fish and amphibians (17, 27, 46). This bacterium was first isolated from dying saltwater fish in 1926 (3) and later identified as a human pathogen (40, 47). Humans are infected by exposure to contaminated water or infected fish (34, 38). The primary reason that M. marinum only causes lesions on the extremities in humans is thought to be its optimal growth temperature of 25 to 35°C (17). Despite its preference for lower temperatures, the histopathology of M. marinum infections in both fish and humans is characterized by granuloma formation (20, 74) with similarities to those seen in human tuberculosis (20, 32). Furthermore, M. marinum is very closely related to the M. tuberculosis complex by 16S rRNA and DNA-DNA homology (58, 71). These characteristics make M. marinum a facile and relevant model for mycobacterial pathogenesis.
It has been shown that M. marinum can persist (7) and replicate (44, 51) in murine macrophages, as well as in a number of epithelial cell lines (62, 64), while the nonpathogenic mycobacterial species Mycobacterium smegmatis cannot (10, 51). Most of these experiments, however, were carried out at suboptimal growth temperatures for the host cells which might affect bacterium-host cell interactions. In addition to the above cell lines, environmental protozoa have been used as in vitro models for M. marinum (14). Although protozoa grow well at the optimum temperatures for M. marinum studies, a close association between M. marinum and protozoa in the environment has not been well established. Since fish are natural hosts for M. marinum, fish monocytes should allow us to examine host-pathogen interactions in cells from a natural host at the proper temperature.
In the present study, we characterize a carp monocytic cell line known as CLC (carp leukocyte culture), established from the common carp Cyprinus carpio (30). This cell line has an optimum growth temperature of 28°C, well within the preferred temperature range for M. marinum. We found that M. marinum enters efficiently and replicates in CLC cells while the nonpathogenic species, M. smegmatis, enters poorly and is killed intracellularly. M. marinum is also able to block lysosomal fusion in CLC cells, which makes them a suitable model for examination of intracellular trafficking in mycobacteria. This is the first report of an in vitro model for the study of M. marinum virulence using monocytic cells obtained from fish, a natural host. This model is convenient, easily reproducible, sensitive, and likely to be important for the identification and characterization of mycobacterial virulence determinants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. marinum strain M, a clinical isolate obtained from the skin of a patient (51), was used in this study. M. marinum was grown at 33°C in 7H9 broth (Difco, Detroit, Mich.) supplemented with 0.5% glycerol, 10% albumin-dextrose complex, and 0.25% Tween 80 for 7 to 10 days. M. smegmatis, strain mc2155 (66) cultures were grown in the same manner but only for 3 days at 37°C. The number of viable bacteria was determined for each assay using the LIVE-DEAD assay (Molecular Probes, Eugene, Oreg.) and by plating dilutions for CFU on 7H9 agar (Difco). All inocula used were >99% viable.
Cell lines and culture conditions.
The mouse macrophage cell line J774A.1 (ATCC TIB67) was maintained at 37°C and 5% CO2 in high glucose Dulbecco's modified eagle medium (Dulbecco's MEM) (Gibco, Bethesda, Md.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco) and 2 mM l-glutamine. The adherent carp monocytic/macrophage cell line CLC (European Collection of Cell Cultures no. 95070628) was maintained at 28°C and 5% CO2 using high glucose MEM (Gibco) supplemented with 10% essential amino acids (Gibco), 10% heat-inactivated fetal bovine serum (Gibco), and 2 mM l-glutamine.
Adherence and entry assays.
Entry and adherence assays were carried out with 24-well tissue culture plates (Costar) as described previously (15, 16). J774A.1 cells were seeded at a density of 106 cells per well 18 to 24 h prior to use. CLC cells were seeded in the same manner, at a density of 5 × 105 cells per well. The medium was replaced before use, and bacteria were added to achieve a multiplicity of infection (MOI) of 10. The infection was allowed to proceed for 30 min at 28°C for CLC and 33°C for J774A.1. The cells were washed twice with phosphate-buffered saline (PBS) and incubated in fresh medium plus amikacin (200 μg/ml; Sigma Chemicals, St. Louis, Mo.) for 2 h. The cells were then washed once with PBS and lysed using 1 ml of 0.1% Triton X-100 (Sigma) for 10 min. Dilutions were plated to determine the number of intracellular CFU. Adherence assays were carried out in a similar manner except that bacteria were added to the cells and immediately washed five times with PBS prior to lysis. Adherence assays using fixed cells were carried out as previously described (15). Cells were fixed in 3.7% formaldehyde for 10 min at room temperature and washed three times with PBS prior to addition of bacteria. Bacteria were coincubated with fixed cells for 30 min, and then the cells were washed and lysed and the number of CFU was determined. Triton X-100 had no effect on the viability of M. marinum and M. smegmatis, and all strains used displayed comparable rates of killing by amikacin.
Intracellular growth assays.
Intracellular growth assays were carried out in a similar manner to entry assays, but after amikacin treatment, fresh medium plus amikacin (30 μg/ml) was added. The cells were incubated at the appropriate temperature and then lysed and plated as described above at different time points. Survival is expressed as the percentage of CFU present at each time point (Tx) compared to the number present at time zero (T0, or 2.5 h), i.e., % survival = (CFU at Tx/CFU at T0) × 100.
Microscopy.
For acid-fast stains, CLC and J774A.1 cells were seeded as described above on coverslips in 24-well tissue culture plates. Bacteria were added to achieve an MOI of 10. The infection was allowed to proceed for 5 min at 28 or 33°C, after which the cells were washed two times with PBS and the media were replaced with fresh medium plus amikacin (30 μg/ml). This time point represents time zero. Cells were reincubated at 28 or 33°C, and the medium was removed at appropriate time points. The cells were then fixed with methanol, washed once with PBS, and stained by the Ziehl-Neelsen technique (37), using carbol-fuchsin and malachite green (Sigma). Cells were examined using a Nikon TE300 light microscope with differential interface contrast optics.
Transmission electron microscopy was used to examine the ultrastructure and trafficking of M. marinum and M. smegmatis vacuoles in CLC cells. CLC monocytes were infected at an MOI of 10 for 5 min at 28°C, washed two times with PBS, and incubated at 28°C in MEM plus amikacin (30 μg/ml). After incubation for various times, the cells were suspended in medium with a rubber policeman, pelleted by centrifugation for 2 min at 740 × g at 25°C, fixed, and prepared for electron microscopy as previously described (14). The samples were suspended in 2% glutaraldehyde plus 1% OsO4 for 2 h and postfixed with 0.5% uranyl acetate overnight at 4°C. The cells were embedded and sectioned as described previously (14). In order to quantitate the frequency of lysosomal fusion with mycobacterial vacuoles in CLC cells, they were prelabeled with thorium dioxide (24 μg/ml; Polyscience) for 18 to 24 h prior to infection as described previously (14). The cells were then fixed and prepared for electron microscopy.
Statistical analyses.
Data presented are the means and standard deviations of triplicate samples from a representative experiment. All experiments were repeated at least three times, unless otherwise noted. Significance was determined by analysis of variance using the Student t test. P values of <0.05 were considered significant.
RESULTS
Fish monocytes are bactericidal for nonpathogenic mycobacteria.
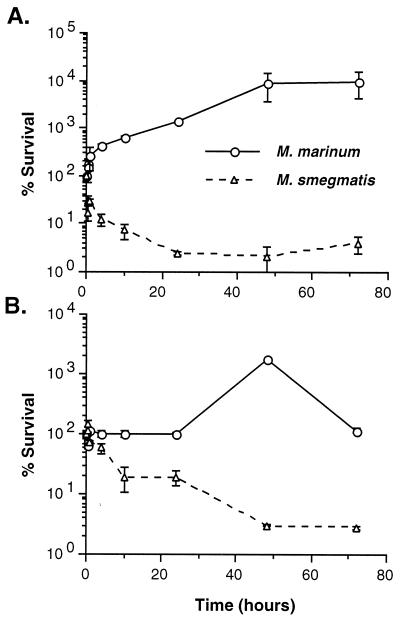
We examined the ability of pathogenic (M. marinum) and nonpathogenic (M. smegmatis) mycobacteria to survive and replicate in the fish monocytic cell line, CLC. As shown in Fig. 1A, we found that M. marinum replicates well in fish monocytes. M. marinum CFU increase 10-fold within 24 h. In contrast, over 90% of the M. smegmatis organisms are killed within the first 24 h. These data are comparable to those obtained with J774A.1 murine macrophages (Fig. 1B), a cell line that has been widely used as an in vitro model for mycobacterial infections (43, 55, 56, 76). As expected, M. marinum replicates in J774A.1 macrophages, while M. smegmatis is killed. Though M. smegmatis has a generation time (1.1 h) that is 3.65-fold that of M. marinum (3.8 h) in broth media, M. smegmatis has a negative generation time in both cell lines while M. marinum doubles in ∼4.9 h within CLC cells. Interestingly, there is a greater difference between the growth of M. marinum and M. smegmatis in fish monocytes than in murine macrophages. This difference is about twofold at 4 h and increases throughout the experiment. These data suggest either that M. marinum is better adapted to parasitize fish monocytes than murine macrophages or that fish monocytes are more bactericidal than murine macrophages for nonpathogenic mycobacteria or that a combination of these factors is responsible.
FIG. 1.
Survival and replication of M. marinum and M. smegmatis in the fish monocytic cell line CLC (A) and the murine macrophage cell line J774A.1 (B). The survival rate was arbitrarily set to 100% at time zero for each strain. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate from a representative experiment.
Ultrastructure of the mycobacterial vacuoles in fish monocytes.
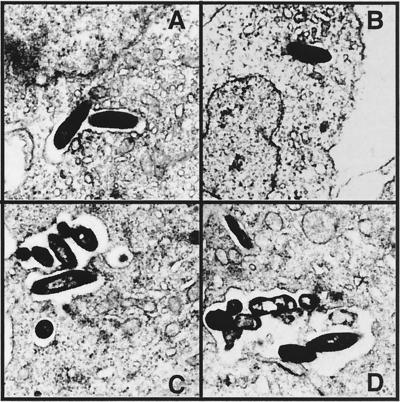
The ultrastructure of the mycobacterial vacuole in fish monocytes was monitored for 2 h postinfection by transmission electron microscopy. We found that both M. marinum and M. smegmatis reside within vacuoles that conform to the shape of the bacteria, which are surrounded by a typical electron-transparent zone (Fig. 2). Quantitation of infected cells by electron microscopy (Table 1) indicates that there is a significant difference (P < 0.05) between the number of cells infected with M. marinum and M. smegmatis, the number of vacuoles per infected cell (P < 0.03), and the number of bacteria per vacuole (P < 0.01).
FIG. 2.
Transmission electron micrographs of M. marinum (C and D) and M. smegmatis (A and B) at 1 h (A and C) and 2 h (B and D) postinfection of the fish monocytic cell line CLC. Magnification, ×10,000.
TABLE 1.
Mycobacterial infections in fish monocytes
| Time (min) after entry | % Infected cellsa
|
No. of vacuoles/cellb
|
No. of bacteria/vacuolec
|
% Fusiond
|
||||
|---|---|---|---|---|---|---|---|---|
| M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | |
| 60 | 25 ± 1.0 | 9 ± 1.0 | 1.31 ± 0.01 | 1.10 ± 0.04 | 1.56 ± 0.12 | 1.00 ± 0.00 | 18 ± 5 | 75 ± 8 |
| 120 | 30 ± 2.0 | 5 ± 1.0 | 1.39 ± 0.01 | 1.19 ± 0.01 | 1.91 ± 0.09 | 1.01 ± 0.01 | 23 ± 6 | 88 ± 53 |
Percentage of cells containing at least one bacterium. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
Number of bacterial vacuoles in each cell. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
Number of bacteria in each vacuole. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
Percentage of lysosomal fusion, calculated by counting bacterial vacuoles containing thorium. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
M. marinum inhibits lysosomal fusion in fish monocytes.
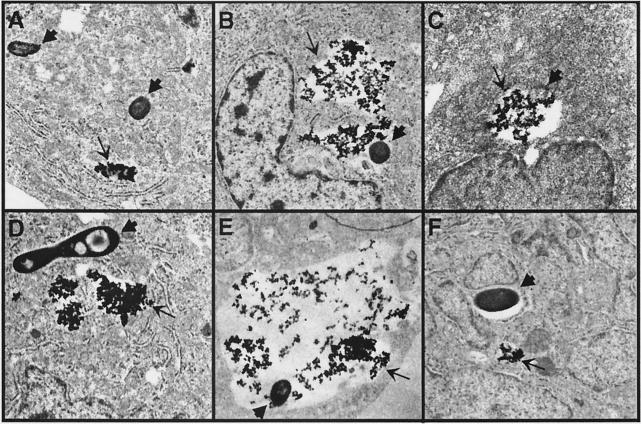
The ability of M. marinum to replicate, while M. smegmatis is killed, in fish monocytes, may be at least partially due to the ability of the former to prevent fusion of the bacterial vacuoles with lysosomes. In order to examine this possibility, we quantitated the frequencies of lysosomal fusion with the M. marinum and M. smegmatis vacuoles in CLC cells (Fig. 3). Fusion of M. smegmatis vacuoles with lysosomes is greater than 75% by 1 h (Table 1), compared to 18% for M. marinum. M. marinum vacuoles display significantly lower levels of fusion with lysosomes than M. smegmatis at all time points (P < 0.01). At 2 h postinfection it was difficult to find intact M. smegmatis in infected cells, as most bacteria were in the process of being degraded (Fig. 3C). On the other hand, at the same time point most bacteria were found in unfused vacuoles in cells infected with M. marinum (Fig. 3F). These data suggest that the survival of M. marinum in CLC cells is at least partially due to the prevention of lysosomal fusion, similar to previous observations of murine macrophages (7).
FIG. 3.
Transmission electron micrographs of fused (B, C, and E) and nonfused (A, D, and F) M. marinum (D to F) and M. smegmatis (A to C) vacuoles with thorium dioxide-labeled lysosomes, at 1 h (A, B, D, and E) and 2 h (C and F) postinfection of the fish monocytic cell line CLC. Thick arrows point to bacteria or bacterial debris (C), while thin arrows point to thorium dioxide-containing vacuoles. (C) M. smegmatis bacterium in the process of being degraded. Magnification, ×10,000.
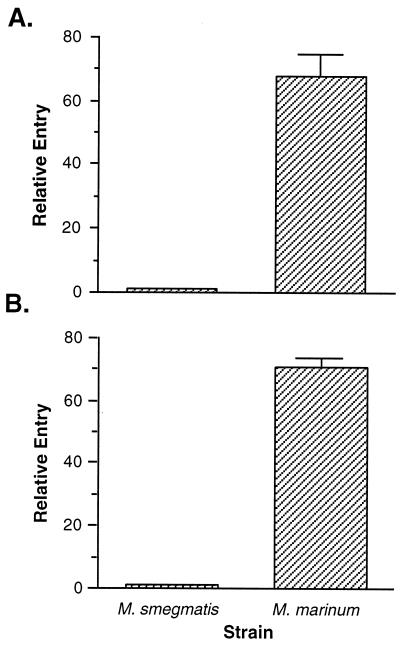
Entry of M. marinum into fish monocytes.
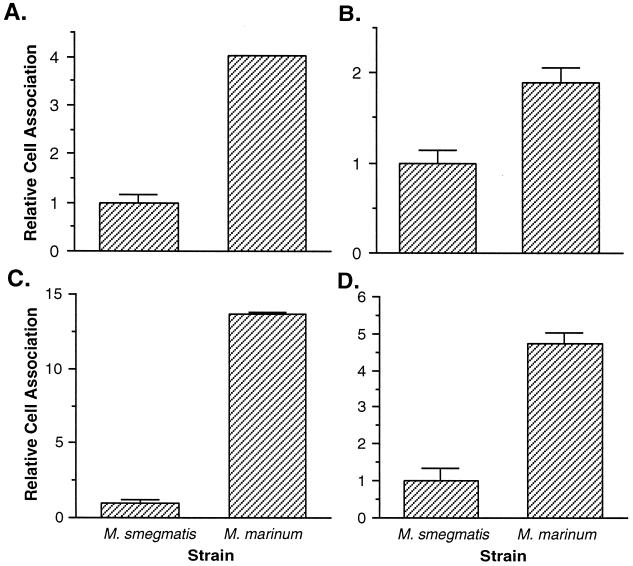
Since entry into human and murine monocytes is thought to play an important role in pathogenesis (29), we wished to characterize the entry of mycobacteria into CLC cells. The ability of M. marinum to enter CLC cells was compared to that of M. smegmatis at 30 min postinfection. As shown in Fig. 4A, we found a significant difference in the entry of M. marinum into CLC cells relative to M. smegmatis (P < 0.01). M. marinum enters fish monocytes at a 60-fold-higher rate than M. smegmatis. Similar results were obtained in murine macrophages (Fig. 4B). These data were confirmed by staining infected monolayers by the Ziehl-Neelsen technique and counting acid-fast bacilli within infected cells. Typically, M. marinum-infected CLC and J774A.1 cells contain more bacilli than those infected with M. smegmatis (data not shown). Quantitation of the number of acid-fast bacilli within infected CLC monocytes (Table 2) indicates that M. marinum infected cells at a higher rate than M. smegmatis (P < 0.01). In addition, more vacuoles per cell (P < 0.01) and more bacteria per vacuole (P < 0.03) are observed with M. marinum than M. smegmatis. By microscopy, M. marinum enters fish monocytes at 27- and 40-fold-higher levels than M. smegmatis after 1 and 2 h, respectively. Similar results were obtained with the murine macrophage cell line J774A.1 (Table 3). The results obtained by microscopy are comparable to those obtained by standard invasion assays. Since acid-fast stains do not differentiate between viable and nonviable bacteria, the differences in entry between M. marinum and M. smegmatis are not solely due to differences in intracellular survival.
FIG. 4.
Entry of M. marinum into the fish monocytic cell line CLC (A) and the murine macrophage cell line J774A.1 (B) relative to M. smegmatis at 30 min postinfection. Entry of M. smegmatis was arbitrarily set at 1. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate from a representative experiment.
TABLE 2.
Entry into fish monocytes
| Time (min) after entry | % Infected cellsa
|
No. of vacuoles/cellb
|
No. of bacteria/vacuolec
|
Infectivity indexd
|
||||
|---|---|---|---|---|---|---|---|---|
| M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | |
| 60 | 17.0 ± 0.22 | 5.5 ± 0.12 | 1.30 ± 0.02 | 1.08 ± 0.00 | 8.35 ± 0.15 | 1.14 ± 0.03 | 27.25 ± 0.22 | 1.0 ± 0.10 |
| 120 | 20.7 ± 0.12 | 5.9 ± 0.11 | 1.72 ± 0.04 | 1.14 ± 0.10 | 8.89 ± 0.09 | 1.19 ± 0.04 | 39.56 ± 0.12 | 1.0 ± 0.10 |
Percentage of cells containing at least one bacterium in a field. Results are the means ± standard deviations for three counts of 25 fields.
Number of bacterial vacuoles in each cell. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
Number of bacteria in each vacuole. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
The infectivity index is calculated as follows: (% infected cells × number of vacuoles per cell × number of bacteria per vacuole for M. marinum)/(% infected cells × number of vacuoles per cell × number of bacteria per vacuole for M. smegmatis).
TABLE 3.
Entry into murine macrophages
| Time (min) after entry | % Infected cellsa
|
No. of vacuoles/cellb
|
No. of bacteria/vacuolec
|
Infectivity indexd
|
||||
|---|---|---|---|---|---|---|---|---|
| M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | M. marinum | M. smegmatis | |
| 60 | 17.4 ± 0.91 | 7.0 ± 0.21 | 1.96 ± 0.04 | 1.18 ± 0.02 | 6.23 ± 0.17 | 1.32 ± 0.05 | 19.49 ± 0.90 | 1.0 ± 0.90 |
| 120 | 24.3 ± 0.20 | 9.1 ± 0.60 | 1.97 ± 0.10 | 1.14 ± 0.20 | 9.85 ± 0.29 | 1.12 ± 0.12 | 40.58 ± 0.29 | 1.0 ± 0.60 |
Percentage of cells containing at least one bacterium in a field. Results are the means ± standard deviations for three counts of 25 fields.
Number of bacterial vacuoles in each cell. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
Number of bacteria in each vacuole. Results are the means ± standard deviations for two counts of 50 cells in different sections of the same preparation.
The infectivity index is calculated as follows: (% infected cells × number of vacuoles per cell × number of bacteria per vacuole for M. marinum)/(% infected cells × number of vacuoles per cell × number of bacteria per vacuole for M. smegmatis).
Adherence of M. marinum to fish monocytes.
To determine whether the differences we observed in entry are at least partially due to differences in adherence, we compared the levels of adherence of M. marinum and M. smegmatis to CLC cells. M. marinum displays a fourfold-higher rate of adherence to fish monocytes than does M. smegmatis (P < 0.01) (Fig. 5A). Similar but somewhat greater differences between these strains are observed with J774A.1 murine macrophages (P < 0.01) (Fig. 5C). We repeated the adherence assays with formaldehyde-fixed cells to prevent the possibility that the effects on adherence are due to uptake or intracellular survival. Adherence assays in fixed CLC (Fig. 5B) and J774A.1 (Fig. 5D) cells show significant (P < 0.01) but smaller differences in adherence of M. marinum and M. smegmatis than those seen with live cells. These data suggest that fish monocytes are a useful in vitro model for examination of M. marinum mechanisms of adherence, entry, trafficking, and intracellular replication.
FIG. 5.
Adherence of M. marinum to the fish monocytic cell line CLC (A and B) and the murine macrophage cell line J774A.1 (C and D) relative to M. smegmatis. Assays were carried out in live (A and C) and formaldehyde-fixed (B and D) cells. Adherence of M. smegmatis was arbitrarily set to 1. Data points and error bars represent the means and standard deviations, respectively, of assays done in triplicate from a representative experiment.
DISCUSSION
We have developed an in vitro model for the study of mycobacterial virulence using M. marinum and fish monocytes. An important feature of this model is that it provides the opportunity to study the pathogenesis of a mycobacterial species in cells from a natural host. Since M. marinum infects more than 150 species of salt- and freshwater fish and replicates in monocytic cells (3, 27, 74), results obtained in this in vitro model should closely resemble those during natural infections. Though murine macrophages have been used extensively as in vitro models for mycobacterial infections, differences exist between their microbicidal mechanisms and those of human macrophages (25, 26, 33, 59, 60). Mice are also naturally resistant to tuberculosis (48), raising the issue of differences in host-pathogen interactions between this model and humans. Human cell lines are available for mycobacterial pathogenesis studies; however, no fully differentiated adherent human macrophage cell line exists. In order to study mycobacterial host cell interactions in adherent macrophages, human monocytes must be treated with phorbol esters (41, 61, 72), lipopolysaccharide (1, 45), or interferon gamma plus tumor necrosis factor alpha (1, 45). Such treatments can complicate analysis of data obtained, since they have pleiotropic effects on macrophages (31, 39, 69, 77). Thus, the use of fish monocytes, in combination with the previously developed systems, should improve our ability to evaluate potential mycobacterial virulence determinants.
Fish monocytes are capable of differentiating between pathogenic and nonpathogenic mycobacteria. There is a much greater difference between the growth of M. marinum and M. smegmatis in fish monocytes than there is between these pathogens in murine macrophages. In support of the high bactericidal ability of fish monocytes, we have also found that they are able to kill an M. marinum oxyR mutant (E. Pagán-Ramos and V. Deretic, unpublished data) that persists in murine macrophages (S. El-Etr and J. Cirillo, unpublished data). This suggests that fish monocytes are more sensitive to subtle differences in virulence within the same species. Generally, all mycobacterial species studied are internalized by macrophages and epithelial cells (36, 62–64). However, only pathogenic species survive and replicate efficiently in mammalian cells (62–64). Like other pathogenic mycobacteria, M. marinum survives and replicates in macrophages and epithelial cells (44, 51, 62, 64). Previous studies of these cells were conducted between 28 and 33°C, temperatures permissive to the growth of M. marinum. The bactericidal ability of J774A.1 macrophages does not appear impaired at 33°C, as judged by their ability to kill nonpathogenic mycobacteria. However, we cannot exclude the possibility that the lower temperature is at least partially responsible for the smaller differences we observed in the intracellular replication of pathogenic and nonpathogenic mycobacteria in murine macrophages.
The ultrastructural characteristics of the M. marinum vacuole in fish monocytes are similar to those previously seen in murine (51) and freshly explanted trout (13) macrophages. As seen in other pathogenic mycobacterial species (19, 23, 67, 75), M. marinum can circumvent the host endocytic pathway blocking fusion of its phagosome with lysosomes (7, 13) and reside in vacuoles that do not acidify (7). Similarly, M. marinum multiplies in typical vacuoles and blocks phagosome-lysosome fusion within fish monocytes. M. smegmatis, on the other hand, is not able to prevent the fusion process, which may contribute to its inability to survive. Results obtained with fish monocytes, therefore, mimic the observations made with M. tuberculosis and M. avium in mammalian macrophages. This observation makes CLC cells suitable for dissection of the mechanisms used by mycobacteria to prevent lysosomal fusion. Though specific lysosomal markers for fish monocytes are not available, lysosomes can be labeled with ferritin (2) or thorium dioxide. Frequencies of lysosomal fusion can also be assessed by assaying for acid phosphatase (57) and fluid phase endosomal tracers like Texas red-ovalbumin (68), which should function regardless of the cell type.
Since M. smegmatis is less efficient than M. avium at entering the laryngeal epithelial cell line HEp-2 (10), it is not surprising that similar results were obtained in our studies. This indicates that fish monocytes differentiate between the entry and adherence mechanisms of pathogenic and nonpathogenic mycobacteria. Comparable levels of entry were obtained with fish monocytes and murine macrophages, suggesting that similar mechanisms are used by mycobacteria to enter both cell types. This is further supported by studies implicating complement and fibronectin receptors in entry of mycobacteria into murine macrophages (29) and by data showing that entry of M. marinum into rainbow trout macrophages is enhanced in the presence of complement (13). The difference between M. marinum and M. smegmatis in adherence to murine macrophages is higher than to fish monocytes. Since both cell types differentiate equally well between the entry of these species, this observation suggests that fish monocytes primarily differentiate pathogenic and nonpathogenic mycobacteria at a step in the entry process subsequent to adherence: signal transduction, phagocytosis, or initial intracellular survival.
Improved animal models would greatly facilitate studies focusing on mycobacterial pathogenesis and vaccines (9, 35). Rabbits (24) and guinea pigs (5, 65) usually develop an acute disease, resulting in death within a few weeks, while many mouse strains are naturally resistant to tuberculosis and do not develop typical granulomas (21, 48). A number of animal models have been described for M. marinum infections (22, 54, 70). In addition, the technology for production of zebra fish germ line chimeras from embryo cell cultures has recently become available (42). Since fish are natural hosts for M. marinum, it is now possible to produce specific transgenic zebra fish to study the genetic basis of host susceptibility and resistance to M. marinum disease. Thus, the animal models available for M. marinum pathogenesis studies are plentiful, are relevant to natural infections, and should allow genetic analysis of the host components involved.
Fish monocytes provide a valuable tool for screening M. marinum strains prior to conducting virulence studies with animals. In addition, they allow pathogenesis studies in a relevant in vitro model at the proper temperature. The CLC fish monocytic cell line used in the present study displays all of the necessary characteristics for examination of the molecular mechanisms of mycobacterial host-pathogen interactions. This model system should allow rigorous analysis of the genetic basis of M. marinum pathogenesis and the identification of novel mycobacterial virulence determinants. However, although M. marinum is closely related to M. tuberculosis, it is likely that some differences in their mechanisms of pathogenesis exist. Thus, it is necessary that, once potential virulence determinants are identified and characterized, their role be confirmed in M. tuberculosis itself. Even with this caveat, the multiple advantages offered by the CLC monocytic cell line suggest that pathogenesis research in M. marinum will yield vast benefits in the coming years.
ACKNOWLEDGMENTS
We thank Tom Bargar and Kit Lee for assistance with electron microscopy.
This study was supported by Nebraska Agricultural Experimental Station grant NEB-14-077 from the United States Department of Agriculture.
Footnotes
Journal no. 13365 of the Nebraska Agricultural Experimental Station.
REFERENCES
- 1.Adams D O, Hamilton T A. The cell biology of macrophage activation. Annu Rev Immunol. 1984;2:283–318. doi: 10.1146/annurev.iy.02.040184.001435. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J A, Hart P D A. Response of cultured macrophages to Mycobacterium tuberculosis with observations on fusion of lysosomes with phagosomes. J Exp Med. 1971;134:713–740. doi: 10.1084/jem.134.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson J D. Spontaneous tuberculosis in salt water fish. J Infect Dis. 1926;39:315–320. [Google Scholar]
- 4.Bailey J P, Jr, Stevens S J, Bell W M, Mealing H G, Jr, Loebl D H, Cook E H. Mycobacterium marinum infection. A fishy story. JAMA. 1982;247:1314. [PubMed] [Google Scholar]
- 5.Balasubramanian V, Wiegeshaus E H, Smith D W. Mycobacterial infection in guinea pigs. Immunobiology. 1994;191:395–401. doi: 10.1016/S0171-2985(11)80445-6. [DOI] [PubMed] [Google Scholar]
- 6.Barker L P, Brooks D M, Small P L. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol Microbiol. 1998;29:1167–1177. doi: 10.1046/j.1365-2958.1998.00996.x. [DOI] [PubMed] [Google Scholar]
- 7.Barker L P, George K M, Falkow S, Small P L C. Differential trafficking of live and dead Mycobacterium marinum organisms in macrophages. Infect Immun. 1997;65:1497–1504. doi: 10.1128/iai.65.4.1497-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barkley W E, Kubica G P. Biological safety in the experimental tuberculosis laboratory. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 61–71. [Google Scholar]
- 9.Barletta R G, Donis R O, Chacon O, Shams H, Cirillo J D. Vaccines against intracellular pathogens. Subcell Biochem. 2000;33:559–599. doi: 10.1007/978-1-4757-4580-1_22. [DOI] [PubMed] [Google Scholar]
- 10.Bermudez L E, Shelton K, Young L S. Comparison of the ability of Mycobacterium avium, M. smegmatis and M. tuberculosis to invade and replicate within HEp-2 epithelial cells. Tuber Lung Dis. 1995;76:240–247. doi: 10.1016/s0962-8479(05)80012-7. [DOI] [PubMed] [Google Scholar]
- 11.Bloom B R, Murray C J L. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control. Guidelines for preventing the transmission of tuberculosis in health-care settings, with special focus on HIV-related issues. Morb Mortal Wkly Rep. 1990;39:19. [PubMed] [Google Scholar]
- 13.Chen S C, Adams A, Thompson K D, Richards R H. Electron microscope studies of the in vitro phagocytosis of Mycobacterium spp. by rainbow trout Oncorhynchus mykiss head kidney macrophages. Dis Aquat Organ. 1998;32:99–110. doi: 10.3354/dao032099. [DOI] [PubMed] [Google Scholar]
- 14.Cirillo J D, Falkow S, Tompkins L S, Bermudez L E. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cirillo S L, Bermudez L E, El-Etr S H, Duhamel G E, Cirillo J D. Legionella pneumophila entry gene rtxA is involved in virulence. Infect Immun. 2001;69:508–517. doi: 10.1128/IAI.69.1.508-517.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cirillo S L, Lum J, Cirillo J D. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology. 2000;146:1345–1359. doi: 10.1099/00221287-146-6-1345. [DOI] [PubMed] [Google Scholar]
- 17.Clark H F, Shepard C C. Effect of environmental temperatures on infection with Mycobacterium marinum (balnei) of mice and a number of poikilothermic species. J Bacteriol. 1963;86:1057–1069. doi: 10.1128/jb.86.5.1057-1069.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark R B, Spector H, Friedman D M, Oldrati K J, Young C L, Nelson S C. Osteomyelitis and synovitis produced by Mycobacterium marinum in a fisherman. J Clin Microbiol. 1990;28:2570–2572. doi: 10.1128/jcm.28.11.2570-2572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemens D L, Horwitz M A. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J Exp Med. 1995;181:257–270. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins C H, Grange J M, Noble W C, Yates M D. Mycobacterium marinum infections in man. J Hyg. 1985;94:135–149. doi: 10.1017/s0022172400061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins F M, Montalbine V. Distribution of mycobacteria grown in vivo in the organs of intravenously infected mice. Am Rev Respir Dis. 1976;113:281–286. doi: 10.1164/arrd.1976.113.3.281. [DOI] [PubMed] [Google Scholar]
- 22.Collins F M, Montalbine V, Morrison N E. Growth and immunogenicity of photochromogenic strains of mycobacteria in the foot-pads of normal mice. Infect Immun. 1975;11:1079–1087. doi: 10.1128/iai.11.5.1079-1087.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crowle A, Dahl R, Ross E, May M H. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis or Mycobacterium avium in cultured human macrophages are not acidic. Infect Immun. 1991;59:1823–1827. doi: 10.1128/iai.59.5.1823-1831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dannenberg A M., Jr . Rabbit model of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 149–156. [Google Scholar]
- 25.Denis M. Interferon-gamma-treated murine macrophages inhibit growth of tubercle bacilli via the generation of reactive nitrogen intermediates. Cell Immunol. 1991;132:150–157. doi: 10.1016/0008-8749(91)90014-3. [DOI] [PubMed] [Google Scholar]
- 26.Denis M. Killing of Mycobacterium tuberculosis within human monocytes: activation by cytokines and calcitriol. Clin Exp Immunol. 1991;84:200–206. doi: 10.1111/j.1365-2249.1991.tb08149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dulin M P. A review of tuberculosis (mycobacteriosis) in fish. Vet Med Small Anim Clin. 1979;74:731–735. [PubMed] [Google Scholar]
- 28.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 29.El-Etr S H, Cirillo J D. Entry mechanisms of mycobacteria. Front Biosci. 2001;6:737–747. doi: 10.2741/sahar. [DOI] [PubMed] [Google Scholar]
- 30.Faisal M, Ahne W. A cell line (CLC) of adherent peripheral blood mononuclear leucocytes of normal common carp Cyprinus carpio. Dev Comp Immunol. 1990;14:255–260. doi: 10.1016/0145-305x(90)90097-x. [DOI] [PubMed] [Google Scholar]
- 31.Fan X D, Goldberg M, Bloom B R. Interferon-gamma-induced transcriptional activation is mediated by protein kinase C. Proc Natl Acad Sci USA. 1988;85:5122–5125. doi: 10.1073/pnas.85.14.5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenton M J, Vermeulen M W. Immunopathology of tuberculosis: roles of macrophages and monocytes. Infect Immun. 1996;64:683–690. doi: 10.1128/iai.64.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flesch I E, Kaufmann S H. Activation of tuberculostatic macrophage functions by gamma interferon, interleukin-4, and tumor necrosis factor. Infect Immun. 1990;58:2675–2677. doi: 10.1128/iai.58.8.2675-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gluckman S J. Mycobacterium marinum. Clin Dermatol. 1995;13:273–276. doi: 10.1016/0738-081x(95)00023-9. [DOI] [PubMed] [Google Scholar]
- 35.Griffin J F, Mackintosh C G, Buchan G S. Animal models of protective immunity in tuberculosis to evaluate candidate vaccines. Trends Microbiol. 1995;3:418–424. doi: 10.1016/s0966-842x(00)88994-5. [DOI] [PubMed] [Google Scholar]
- 36.Hart P D A, Armstrong J A, Brown C A, Draper P. Ultrastructural study of the behavior of macrophages toward parasitic mycobacteria. Infect Immun. 1972;5:803–807. doi: 10.1128/iai.5.5.803-807.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heifets L B, Good R C. Current laboratory methods for the diagnosis of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 85–110. [Google Scholar]
- 38.Huminer D, Pitlik S D, Block C, Kaufman L, Amit S, Rosenfeld J B. Aquarium-borne Mycobacterium marinum skin infection. Report of a case and review of the literature. Arch Dermatol. 1986;122:698–703. [PubMed] [Google Scholar]
- 39.Koide Y, Yoshida A. The signal transduction mechanism responsible for gamma interferon-induced indoleamine 2,3-dioxygenase gene expression. Infect Immun. 1994;62:948–955. doi: 10.1128/iai.62.3.948-955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Linell F, Norden A. Mycobacterium balnei: a new acid-fast bacillus occurring in swimming pools and capable of producing skin lesions in humans. Acta Tuberc Scand. 1954;33(Suppl.):1–84. [PubMed] [Google Scholar]
- 41.Lotem J, Sachs L. Regulation of normal differentiation in mouse and human myeloid leukemic cells by phorbol esters and the mechanism of tumor promotion. Proc Natl Acad Sci USA. 1979;76:5158–5162. doi: 10.1073/pnas.76.10.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma C, Fan L, Ganassin R, Bols N, Collodi P. Production of zebrafish germ-line chimeras from embryo cell cultures. Proc Natl Acad Sci USA. 2001;98:2461–2466. doi: 10.1073/pnas.041449398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehta P K, King C H, White E H, Murtagh J J, Jr, Quinn F D. Comparison of in vitro models for the study of Mycobacterium tuberculosis invasion and intracellular replication. Infect Immun. 1996;64:2673–2679. doi: 10.1128/iai.64.7.2673-2679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mor N. Multiplication of Mycobacterium marinum within phagolysosomes of murine macrophages. Infect Immun. 1985;48:850–852. doi: 10.1128/iai.48.3.850-852.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrison D C, Rudbach J A. Endotoxin-cell-membrane interactions leading to transmembrane signaling. Contemp Top Mol Immunol. 1981;8:187–218. doi: 10.1007/978-1-4684-3917-5_6. [DOI] [PubMed] [Google Scholar]
- 46.Nigrelli R F, Vogel H. Spontaneous tuberculosis in fishes and in other cold-blooded vertebrates with special reference to Mycobacterium fortuitum Cruz from fish and human lesions. Zoologica. 1963;48:131–144. [Google Scholar]
- 47.Norden A, Linell E. A new type of pathogenic mycobacterium. Nature. 1951;168:826. doi: 10.1038/168826a0. [DOI] [PubMed] [Google Scholar]
- 48.Orme I M, Collins F M. Mouse model of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 113–134. [Google Scholar]
- 49.Pavelka M S, Jr, Jacobs W R., Jr Comparison of the construction of unmarked deletion mutations in Mycobacterium smegmatis, Mycobacterium bovis bacillus Calmette-Guérin, and Mycobacterium tuberculosis H37Rv by allelic exchange. J Bacteriol. 1999;181:4780–4789. doi: 10.1128/jb.181.16.4780-4789.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelicic V, Jackson M, Reyrat J M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramakrishnan L, Falkow S. Mycobacterium marinum persists in cultured mammalian cells in a temperature-restricted fashion. Infect Immun. 1994;62:3222–3229. doi: 10.1128/iai.62.8.3222-3229.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramakrishnan L, Federspiel N A, Falkow S. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 53.Ramakrishnan L, Tran H T, Federspiel N A, Falkow S. A crtB homolog essential for photochromogenicity in Mycobacterium marinum: isolation, characterization and gene disruption via homologous recombination. J Bacteriol. 1997;179:5862–5868. doi: 10.1128/jb.179.18.5862-5868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakrishnan L, Valdivia R H, McKerrow J H, Falkow S. Mycobacterium marinum causes both long-term subclinical infection and acute disease in the leopard frog (Rana pipiens) Infect Immun. 1997;65:767–773. doi: 10.1128/iai.65.2.767-773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rastogi N, Blom-Potar M C. A comparative study on the activation of J-774 macrophage-like cells by gamma-interferon, 1,25-dihydroxyvitamin D3 and lipopeptide RP-56142: ability to kill intracellularly multiplying Mycobacterium tuberculosis and Mycobacterium avium. Zentbl Bakteriol. 1990;273:344–361. doi: 10.1016/s0934-8840(11)80438-x. [DOI] [PubMed] [Google Scholar]
- 56.Rastogi N, Bloom-Potar M C, David H L. Comparative intracellular growth of difficult-to-grow and other mycobacteria in a macrophage cell line. Acta Leprol. 1989;7(Suppl. 1):156–159. [PubMed] [Google Scholar]
- 57.Robinson J M. Improved localization of intracellular sites of phosphatases using cerium and cell permeabilization. J Histochem Cytochem. 1985;33:749–754. doi: 10.1177/33.8.2991362. [DOI] [PubMed] [Google Scholar]
- 58.Rogall T, Wolters J, Flohr T, Böttger E. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 59.Rook G A, Steele J, Ainsworth M, Champion B R. Activation of macrophages to inhibit proliferation of Mycobacterium tuberculosis: comparison of the effects of recombinant gamma-interferon on human monocytes and murine peritoneal macrophages. Immunology. 1986;59:333–338. [PMC free article] [PubMed] [Google Scholar]
- 60.Rook G A, Steele J, Fraher L, Barker S, Karmali R, O'Riordan J, Stanford J. Vitamin D3, gamma interferon, and control of proliferation of Mycobacterium tuberculosis by human monocytes. Immunology. 1986;57:159–163. [PMC free article] [PubMed] [Google Scholar]
- 61.Rovera G, Santoli D, Damsky C. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc Natl Acad Sci USA. 1979;76:2779–2783. doi: 10.1073/pnas.76.6.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shepard C C. A comparison of the growth of selected mycobacteria in HeLa, monkey kidney and human amnion cells in tissue culture. J Exp Med. 1958;107:237–250. doi: 10.1084/jem.107.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shepard C C. Growth characteristics in HeLa cells of the rapidly growing acid fast bacteria, Mycobacterium fortuitum, Mycobacterium phlei, and Mycobacterium smegmatis. J Bacteriol. 1957;73:722–726. doi: 10.1128/jb.73.6.722-726.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shepard C C. Growth characteristics of tubercle bacilli and certain other mycobacteria in HeLa cells. J Exp Med. 1957;105:39–48. doi: 10.1084/jem.105.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith D W, Balasubramanian V, Wiegeshaus E. A guinea pig model of experimental airborne tuberculosis for evaluation of the response to chemotherapy: the effect on bacilli in the initial phase of treatment. Tubercle. 1991;72:223–231. doi: 10.1016/0041-3879(91)90013-i. [DOI] [PubMed] [Google Scholar]
- 66.Snapper S B, Melton R E, Mustafa S, Kieser T, Jacobs W R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 67.Sturgill-Koszycki S, Schlesinger P H, Chakraborty P, Haddix P L, Collins H L, Fok A K, Allen R D, Gluck S L, Heuser J, Russell D G. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science. 1994;263:678–681. doi: 10.1126/science.8303277. [DOI] [PubMed] [Google Scholar]
- 68.Sturgill-Koszycki S, Swanson M S. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192:1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takashiba S, Van Dyke T E, Amar S, Murayama Y, Soskolne A W, Shapira L. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor κB. Infect Immun. 1999;67:5573–5578. doi: 10.1128/iai.67.11.5573-5578.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talaat A M, Reimschuessel R, Wasserman S S, Trucksis M. Goldfish, Carassius auratus, a novel animal model for the study of Mycobacterium marinum pathogenesis. Infect Immun. 1998;66:2938–2942. doi: 10.1128/iai.66.6.2938-2942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tonjum T, Welty D B, Jantzen E, Small P L. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J Clin Microbiol. 1998;36:918–925. doi: 10.1128/jcm.36.4.918-925.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Totterman T H, Nilsson K, Sundstrom C. Phorbol ester-induced differentiation of chronic lymphocytic leukaemia cells. Nature. 1980;288:176–178. doi: 10.1038/288176a0. [DOI] [PubMed] [Google Scholar]
- 73.Trucksis M. Fishing for mycobacterial virulence genes: a promising animal model. ASM News. 2000;66:668–671. [Google Scholar]
- 74.van Diujn C., Jr Tuberculosis in fish. J Small Anim Pract. 1981;22:391–411. doi: 10.1111/j.1748-5827.1981.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 75.Xu S, Cooper A, Sturgill-Koszycki S, van Heyningen T, Chatterjee D, Orme I, Allen P, Russell D G. Intracellular trafficking in Mycobacterium tuberculosis and Mycobacterium avium-infected macrophages. J Immunol. 1994;153:2568–2578. [PubMed] [Google Scholar]
- 76.Yajko D M, Nassos P S, Sanders C A, Hadley W K. Killing by antimycobacterial agents of AIDS-derived strains of Mycobacterium avium complex inside cells of the mouse macrophage cell line J774. Am Rev Respir Dis. 1989;140:1198–1203. doi: 10.1164/ajrccm/140.5.1198. [DOI] [PubMed] [Google Scholar]
- 77.Yoshida A, Koide Y, Uchijima M, Yoshida T O. IFN-gamma induces IL-12 mRNA expression by a murine macrophage cell line, J774. Biochem Biophys Res Commun. 1994;198:857–861. doi: 10.1006/bbrc.1994.1122. [DOI] [PubMed] [Google Scholar]