Abstract
The elucidation of the genomic sequence of Mycobacterium tuberculosis revealed the presence of a novel multigene family designated PE/PE_PGRS that encodes numerous, highly related proteins of unknown function. In this study, we demonstrate that a transposon insertion in a PE_PGRS gene (1818PE_PGRS) found in Mycobacterium bovis BCG Pasteur, which is the BCG homologue of the M. tuberculosis H37Rv gene Rv1818c, introduces new phenotypic properties to this BCG strain. These properties include dispersed growth in liquid medium and reduced infection of macrophages. Complementation of the 1818PE_PGRS::Tn5367 mutant with the wild-type gene restores both aggregative growth (clumping) in liquid medium and reestablishes infectivity of macrophages to levels equivalent to those for the parent BCG strain. Western blot analysis using antisera raised against the 1818PE_PGRS protein shows that PE_PGRS proteins are found in cell lysates of BCG and M. tuberculosis H37Ra and in the cell wall fraction of M. tuberculosis H37Rv. Moreover, immunofluorescent labeling of mycobacteria indicates that certain PE_PGRS proteins are localized at the cell surface of BCG and M. tuberculosis. Together these results suggest that certain PE_PGRS proteins may be found at the surface of mycobacteria and influence both cell surface interactions among mycobacteria as well as the interactions of mycobacteria with macrophages.
It is well recognized that an important first step in bacterial infection of the host is the manifestation of specific surface components by the pathogen to facilitate its entry of host cells and tissues. Many organisms have evolved novel adhesins that can bind to molecules commonly found on the eukaryotic cell surface, such as integrins (17, 38), glycosaminoglycans (16, 22, 41), or specific sugar residues (8, 10). A number of complex surface structures are also expressed by bacteria to seek out the appropriate host cells; these include the formation of fimbriae (pili) (2) and complex cell wall structures exemplified by the type III secretion systems (14, 19, 26). Following entry into the host, Mycobacterium tuberculosis shows a tropism for macrophages but can also infect epithelial cells (30, 33, 43). M. tuberculosis has also been shown to have ligands that bind to extracellular matrix proteins like fibronectin (1, 37, 47) and proteoglycans (16, 33). Schlesinger et al. (42) have described complement and mannose receptors on macrophages that promote the phagocytosis of mycobacteria. Genetic studies of M. tuberculosis have identified numerous genes, such as mce (3), eis (45), and erp (7), encoding proteins that enhance mycobacterial entry and survival within macrophages. Although progress has been made, the molecular mechanisms of mycobacterial infection of host cells remains unexplained.
Transposon mutagenesis has been successfully used to identify novel genes that encode for bacterial virulence factors and surface components (6, 27). In the past few years, transposon mutagenesis systems specific for mycobacteria have been developed (4, 24, 34) and have been used to generate auxotrophic mutants in mycobacteria (29) as well as identify new virulence factors (7, 20). An insertional mutagenesis strategy, combined with the information available from the sequencing of the M. tuberculosis genome (12), constitutes a powerful approach for characterizing the function of mycobacterial proteins.
In this investigation, we initially performed a genetic screen of Mycobacterium bovis BCG Pasteur mutagenized with Tn5367 in an attempt to identify novel mycobacterial adhesins. Here we show that a transposon inserted into a gene encoding a PE_PGRS protein present in BCG results in a mutant showing dispersed growth in liquid media and impaired ability to enter and/or survive within macrophages. The results indicate that certain PE_PGRS proteins may be localized to the cell surface and influence the interactions of mycobacteria with other cells.
MATERIALS AND METHODS
Microorganisms and growth conditions.
A library of Tn5367 transposon mutants was generated in BCG Pasteur (obtained from the Statens Serum Institut, Copenhagen, Denmark) as described previously (4). Individual colonies from a library of 1,920 independent mutants were propagated in 96-well plates and screened for cells with dispersed growth phenotypes. All mycobacteria were cultured on 7H11 agar (Difco, Detroit, Mich.) or in stationary culture flasks containing 7H9 media supplemented with oleic acid-albumin-dextrose-catalase enrichment (Becton-Dickinson, Cockeysville, Md.), 0.05% Tween 80, and 20 μg of kanamycin per ml or 50 μg of hygromycin per ml when appropriate. For expression of histidine-tagged antigens, the Escherichia coli BL21(DE3)pLysS strain (Invitrogen, San Diego, Calif.) was used for transformation with pET15b expression constructs. The cell wall and culture filtrate preparations from M. tuberculosis H37Rv were obtained from John Belisle under National Institute of Allergy and Infectious Diseases, National Institutes of Health contract NO1-AI-75320.
Determination of location of Tn5367 insertion.
To identify the location of the Tn5367 insertion in the mc21525 mutant, genomic DNA was isolated as described previously (4). A cosmid genomic library was constructed by partially digesting the chromosomal DNA with Sau3A and cloning into the BamHI site of pYUB328. Upon transduction of E. coli, two independent kanamycin-resistant clones were identified from 2,000 independent, ampicillin-resistant transductants. The DNA sequence was determined for each cosmid using primers from Tn5367 (4) and was found to be identical for analogous insertions for each clone. The sequences derived from the Tn5367 junctions were GCCAACGCGGCCGCCGCGG TCCCGACCACGACGG TG T TGGCC GCCGCCGCCGATGAGGTG TCGGCGGCGATGGCGGCAT TG T TC TCCGGACACGCCCAGGCC TATCAGGCGCTGAGCGCCCAGGCGGCGCTGTTTCAC and TGTTTCACGAGCAGT TCG TGCGGGCGC TCACCGCCGGGGCGGGC TCG TATGCGGCCGCCGAGGCCGCCAGCGCGGCCCCGC TAGAGGG TGTGC TCGACGTGATCAACGCCCCCGCCC TGGCGC TGTTGGGGCGCCCAC TGATCGGTAAC, respectively. These sequences were subjected to BLASTN alignment to the sequence database in TubercuList (12). From the alignments it is clear that both sequences match with a member of the PE_PGRS family. However, only one open reading frame displays 100% homology, and it aligns with the sequence of the Rv1818c gene of M. tuberculosis. This analysis shows that Tn5367 has been inserted 219 bp downstream from the start of the BCG homologue of the M. tuberculosis Rv1818c gene.
Construction of vectors and recombinants.
The Rv1818c gene of M. tuberculosis H37Rv was amplified by PCR using the Vent Polymerase (New England Biolabs, Beverly, Mass.), and the 1,500-bp fragment was cloned into pCRBlunt (Invitrogen). The forward primer 5′-ACGTAGCATATGTCATTTGTGGTC ACGATCCCGGAG-3′, containing an NdeI site, and the reverse primer 5′-TAGCGAGGATCCCTACGGTAACCCGTTCATCCC-3′ with a BamHI site were used to amplify the fragment prior to insertion into the pET15b E. coli expression vector. The forward primer 5′-ACGTCCATGGGCTCA TTTGTGGTCACGATCCCGGAG-3′, with an NcoI site, was used with the reverse primer described above to amplify the fragment before insertion into the pMV1-18 vector. A 390-bp fragment containing the hsp60 promoter region from M. tuberculosis (kindly provided by Joseph A. DeVito) was inserted into the multicloning site of pMV206 to produce pMV1-18. The Rv1818c gene was inserted in frame with the promoter region in pMV1-18 to produce pMV1-23, which expresses the 1818PE_PGRS protein in transformed mycobacteria. The mutant BCG, mc21525, was transformed with pMV1-18 and pMV1-23 using standard procedures (4), and transformants were selected in 7H11/oleic acid-albumin-dextrose-catalase agar plates containing 20 μg of kanamycin per ml and 50 μg of hygromycin per ml. Rv1818c was also cloned into the pET15b expression vector (Novagen Inc., Madison, Wis.), fused to a histidine tag, and expressed. Proteins were purified using nickel chromatography and the X-Press system (Invitrogen) (16). Sequence analysis of transposon insertions in mutants was performed as previously described (15, 20).
Macrophage infectivity assay.
J774 macrophage-like cells (American Type Culture Collection, Rockville, Md.) were cultured in RPMI media (GIBCO BRL, Gaithersburg, Md.) supplemented with 10% fetal calf serum (HyClone, Logan, Utah), 2 mM glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 5 × 10−5 M β-mercaptoethanol, and 50 μg of gentamicin (Life Technologies, Grand Island, N.Y.) per ml. Cells were placed in 24-well tissue culture plates (Costar, Corning, N.Y.) 24 h prior to infection with mycobacteria for an estimated cell number of 2 × 105 cells per well. Plates were washed three times with Dulbecco's modified Eagle's medium (DMEM) (Life Technologies), and mycobacteria were added at a multiplicity of infection (MOI) of 1:1 or 10:1 (see figure legends for specific numbers of bacteria added). Mycobacteria were washed two times in attachment media (complete RPMI media with 1% fetal calf serum and without gentamicin), sonicated one time for 10 s, and diluted to the appropriate concentration in attachment media. For some experiments, mycobacteria were directly scraped from 7H11 plates into attachment media, allowed to “settle” as described previously (42), and diluted as described above. This method results in a more dispersed population of mycobacteria being added to the wells containing macrophages and eliminates false determinations due to clumping of bacteria. Aliquots of the final dilution were plated for determination of mycobacteria added at the “0” time point. Following incubation at 37°C in a CO2 atmosphere for 4 h, wells were washed three times with DMEM, and either RPMI media containing gentamicin were added for further incubation of the macrophages or cells were immediately lysed in 0.5% Triton X-100 in phosphate-buffered saline (PBS), pH 7.5. Aliquots of the lysates from each well were diluted in PBS containing 0.04% Tween 80, and numbers of CFU were determined by plating on 7H11 agar containing the appropriate antibiotic for maintaining selective pressure on BCG variants. Additional wells were used to determine the number of macrophages per well following the incubation periods. Electron microscopy indicates that, after washing, the number of mycobacteria determined per well reflects mostly bacteria that have entered the macrophage cells, since very few bacteria are found associated with the cell surface (M. J. Brennan, unpublished observations). The number of bacteria recovered ranges from 5 to 10% of the total number of bacteria added per well.
Mouse bone marrow macrophages (BMMφ) were collected as previously described (39) by flushing the femurs of C57BL6/J mice and then culturing the cells in DMEM containing 10% fetal calf serum (Hy-Clone), 2 mM glutamine, 10 mM HEPES, 0.1 mM nonessential amino acids, 50 μg of gentamicin per ml, and 10% L-929 conditioned media. Invasion assays using BMMφ were performed as described for the J774 cells, except that cells were seeded into wells several days before performing the assay and that the DMEM containing the L-929 conditioned media supplement (39) was used for the assay.
Chromatographic separation of M. tuberculosis proteins.
M. tuberculosis H37Ra were harvested after stationary culture in liquid media, centrifuged, and lysed by sonication as previously described (16). The cell lysate was centrifuged at 15,000 × g, and the supernatant was collected, dialyzed against Tris-buffered saline (TBS) (20 mM Tris-HCl, pH 7.0–150 mM NaCl 150 mM), and used in the following chromatographic steps. For ion-exchange chromatography, the lysate was added to DEAE Sephacel matrix (Pharmacia LKB, Uppsala, Sweden) in TBS buffer at a flow rate of 0.5 ml/min using a Bio-Rad Econo System (Bio-Rad, Hercules, Calif.), and the column was washed extensively using TBS. The bound material was then eluted by a 0.15 to 1 M NaCl gradient in TBS, and selected fractions from the DEAE chromatography were pooled, dialyzed against buffer A [2 M (NH4)2PO4–0.025 M K2HPO4, pH 6.0], and then subjected to hydrophobic interaction chromatography on a phenyl-Sepharose column (Pharmacia). The material was bound to the column in buffer A at a flow rate of 0.5 ml/min, and the column was washed extensively using the same buffer. Three different elution steps were carried out in order to elute and separate the mycobacterial proteins according to their degree of hydrophobicity. First, the column was eluted using a 15-ml gradient of buffer A mixed with buffer B (0.025 M K2HPO4, pH 6.0) followed by a second gradient of buffer B diluted with buffer C (0.025 M K2HPO4, pH 6.0, and 80% [vol/vol] ethylene glycol) and a third gradient of buffer C mixed with buffer D (0.025 M K2HPO4, pH 6.0, and 50% [vol/vol] glycerol). From the 1-ml fractions collected during the chromatography, 0.2 ml was concentrated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using the Nitro Blue Tetrazolium chloride–5-bromo-4-chloro-3-indolylphosphate (BCIP) system (Life Technology, Gaithersburg, Md.) for detection.
Immunoassays.
For immunoblots, SDS-PAGE was performed as described originally by Laemmli (25) and proteins were transferred to nitrocellulose membranes as described by Harlow and Lane (23). Antisera directed against 1818PE_PGRS protein were collected from five C57BL/6 mice following three immunizations with 50 μg each of purified His-tagged recombinant 1818PE_PGRS or from mice immunized with three injections of 100 μg each of DNA vaccine encoding 1818cPE_PGRS fused to a tissue plasminogen activator signal sequence as described elsewhere (16a). Pooled sera were used for incubations with nitrocellulose blots at a dilution of 1:500 to 1:1,500 and were visualized using alkaline phosphatase-conjugated anti-mouse whole molecule immunoglobulin G (Sigma, St. Louis, Mo.) and the NPP/BCIP alkaline phosphatase development system (Life Technology).
An indirect immunofluorescence assay was performed using bacteria cultured for 12 days in liquid media without shaking. Bacteria were washed in PBS, dried onto lysine-coated glass slides, and then fixed with 4% paraformaldehyde. Slides were bathed in PBS before addition of pooled anti-1818PE_PGRS sera or preimmune-phase mouse sera at a dilution of 1:50 for 2 h at room temperature. After washing in PBS, fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin (Zymed Laboratories, Inc., South San Francisco, Calif.) was added at a dilution of 1:50 for 1 h. Slides were viewed using a Nikon Optiphot-2 microscope equipped with a 100X fluorescence objective, and digital images were obtained using a SPOT RT color camera (Diagnostics, Inc., Sterling Heights, Mich.). Fluorescent and phase contrast images were merged using SPOT RT software and posterized using Adobe Photoshop software.
RESULTS
Identification of a BCG transposon mutant that has a dispersed growth phenotype.
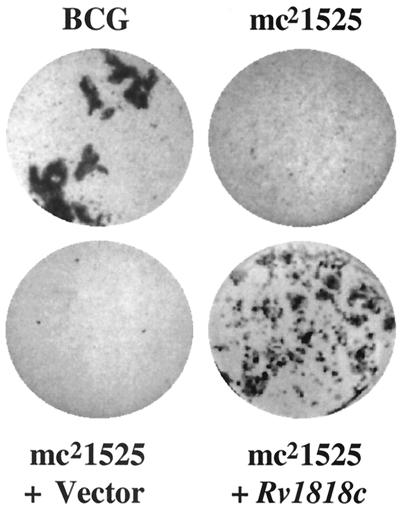
Certain bacterial adhesins including a previously described mycobacterial heparin-binding hemagglutinin (HBHA) induce autoaggregation of bacteria (9, 32, 33). We reasoned that mutants with defective adhesins might not form typical large cell aggregates when grown in liquid media (Fig. 1) but rather would show a dispersed growth phenotype. A library of 1,920 independent transposon mutants of BCG (4) grown in liquid media was screened by visual examination for mutants that show a more dispersed phenotype. Nine potential mutants were selected and rescreened microscopically. One mutant, mc21525, was notably deficient in aggregation (Fig. 1) and was analyzed in more detail. When grown in liquid media, BCG began to show prominent clumping at approximately day 7, while no clumping was observed with mc21525 until day 15. In a number of immunoassay experiments, using a monoclonal antibody specific for HBHA (33) to examine mc21525, it was found that the deficiency in aggregation shown by mc21525 is not due to a lack of expression of HBHA, a mycobacterial protein known to induce autoaggregation (data not shown). This initial observation suggested that the transposon in mc21525 may have interrupted a novel gene that encodes a surface protein that promotes mycobacterial aggregation.
FIG. 1.
Comparison of the phenotypic growth patterns of the BCG Pasteur parent line, the transposon variant mc21525, and mc21525 transformed with the pMV1-18 vector only and with the pMV1-23 vector carrying the Rv1818c gene. Cultures were seeded at a similar cell number into 7H9 broth and incubated at 36°C for 12 days.
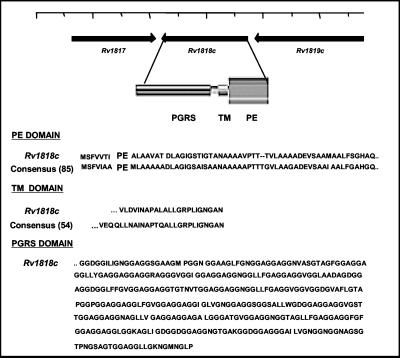
Sequence analysis of the transposon insertion revealed that mc21525 had an insertion of Tn5367 in the BCG equivalent of the M. tuberculosis H37Rv PE_PGRS gene, Rv1818c, at bp 219 of the gene causing a disruption at amino acid 73 of the open reading frame, which encodes a total of 498 amino acids (Fig. 2). For convenience, we have designated this gene 1818PE_PGRS. Analysis of the genes surrounding 1818PE_PGRS in the M. tuberculosis genome suggested that, in M. tuberculosis and most probably in BCG as well, the 1818PE_PGRS gene is an independent transcriptional unit (Fig. 2). Therefore, the dispersed growth phenotype of mc21525 most likely results from a disruption of the 1818PE_PGRS gene.
FIG. 2.
Properties of the PE_PGRS protein encoded by the Rv1818c gene. The N-terminal domain of the 1818PE_PGRS protein has significant homology with the PE domain consensus sequence derived from 85 members of the PE multigene family; it contains a putative transmembrane domain (TM) found in 54 members of the family, and the PGRS domain extending to the C terminus is composed of 21 GGAGGX repeats. The position of the Rv1818c gene in the genome of the M. tuberculosis strain H37Rv (12) indicating the putative direction of transcription is shown.
Complementation of mc21525 with Rv1818c restores the wild-type phenotype.
To confirm that the dispersed growth phenotype in the mutant mc21525 strain results from the inactivation of 1818PE_PGRS, we performed a complementation analysis. Plasmid pMV1-23 containing the Rv1818c open reading frame expressed from an hsp60 promoter was used to transform the mutant BCG strain mc21525. Microscopic analysis of liquid cultures of a number of clone-purified, independent transformants showed a growth phenotype characteristic of the wild-type BCG (Fig. 1). Therefore, complementation of mc21525 with Rv1818c restored the clumping phenotype similar to the in vitro growth of the BCG parent strain.
To verify that the Rv1818c gene is expressed following genetic complementation, we performed Western blot analysis of whole-cell lysates of mc21525 expressing 1818PE_PGRS by using sera from mice immunized with an 1818PE_PGRS DNA vaccine (16a). These sera are not specific for 1818PE_PGRS but cross-react with a number of putative PE_PGRS proteins (Fig. 3B below). As shown in Fig. 3A, lanes 1 and 2, protein bands of about 42 kDa cross-reactive with the anti-1818PE_PGRS sera were observed in all strains of BCG tested by using the anti-1818PE_PGRS sera in immunoblots, including mc21525 (Fig. 3A, lane 1) as well as the BCG-Pasteur parent line (data not shown). On Western blots containing lysates of mc21525 transformed with 1818PE_PGRS (Fig. 3A, lane 2), a new band of about 41 kDa reactive with the anti-1818PE_PGRS sera was observed compared with mc21525 containing the vector only (Fig. 3A, lane 1). These results indicate that the appearance of a new protein band in mc21525 lysates is associated with complementation and restoration of the clumping phenotype.
FIG. 3.
Immunoblot detecting the presence of PE_PGRS proteins in mycobacterial preparations. (A) A 4 to 20% gradient SDS-polyacrylamide gel containing a cell lysate of mc21525 transformed with the vector only (lane 1), a cell lysate of mc21525 transformed with Rv1818c (lane 2), 3 μg of purified His-tagged 1818PE_PGRS protein (lane 3), 10 μg (protein) of a M. tuberculosis H37Rv cell wall fraction (lane 4), and 10 μg (protein) of a M. tuberculosis H37Rv culture filtrate preparation (lane 5) were transferred to nitrocellulose and probed with antisera directed against 1818PE_PGRS protein. The arrow shows the additional reactive band present in the complemented mc21525 strain. (B) Western blot containing proteins from cell lysates of M. tuberculosis H37Ra and partially purified by DEAE chromatography (lane 1) followed by phenyl-Sepharose chromatography and subsequent elution of hydrophobic material (lanes 2 to 6). Aliquots of the eluted fractions were separated by SDS-PAGE and transferred to nitrocellulose membranes, and blots were incubated with sera obtained from mice immunized with a DNA vaccine encoding 1818PE_PGRS. The position of the molecular mass standards is shown.
PE_PGRS proteins are localized to the surface of mycobacteria.
The protein encoded by 1818PE_PGRS is a member of a family of 61 M. tuberculosis genes designated PE_PGRS (12). The N-terminal region of 1818PE_PGRS is highly homologous with most members of the PE_PGRS family as well as the PE family (Fig. 2). The PGRS domain, which extends to the C terminus, contains a large number of Gly-Ala repeats. The PE_PGRS proteins, including 1818PE_PGRS, also contain a putative transmembrane domain, which suggests that they could be interculated into the mycobacterial cell wall. Partial characterization of the antisera raised against 1818PE_PGRS in mice has indicated that the antibodies are directed toward the PGRS domain, which contains the Gly-Ala repeats, and that it cross-reacts with a number of putative PE_PGRS proteins (16a). These anti-1818PE_PGRS sera were used to determine if PE_PGRS proteins are found in cellular fractions of M. tuberculosis. On Western blots, the antisera recognized purified recombinant 1818PE_PGRS (Fig. 3A, lane 3) as well as major protein bands of 42 to 45 kDa and lower-molecular-weight bands in a cell wall preparation of M. tuberculosis H37Rv (Fig. 3A, lane 4). No reactivity was observed with culture filtrate proteins prepared from M. tuberculosis H37Rv (Fig. 3A, lane 5). To further analyze the antisera used in this study, lysates of Mtb H37Ra were concentrated by DEAE ion-exchange chromatography and multiple bands were detected at approximately 40 kDa (Fig. 3B, lane 1). Upon further separation of this material by hydrophobic interaction chromatography, the 1818PE_PGRS antisera detected at least three major bands in Western blots containing fractions eluted according to increasing hydrophobicity (Fig. 3B, lanes 2 to 6). This result suggests that multiple PE_PGRS proteins, immunologically related to 1818PE_PGRS, are expressed by M. tuberculosis in culture, albeit in low amounts. To date, attempts to specifically identify these proteins, including 1818PE_PGRS, by N-terminal amino acid sequencing of the separated immunoreactive protein bands have failed. Together these results suggest that a number of PE_PGRS proteins may be expressed by BCG and M. tuberculosis in culture, some of which may be localized to the cell wall.
Indirect immunofluorescence was also performed using the anti-1818PE_PGRS polyclonal sera and FITC-conjugated anti-mouse sera to determine if PE_PGRS antigens can be visualized on the surface of BCG. A specific, pronounced, punctate-like surface staining of the BCG parent strain was observed (Fig. 4A) compared with control sera (Fig. 4C). The mc21525 strain showed some staining but much less than the parent strain (Fig. 4B). We also observed a very pronounced immunofluorescent staining at the surface of M. tuberculosis strain CDC1551 (data not shown). These findings are consistent with the suggestion that PE_PGRS proteins are found at the cell surface of mycobacteria.
FIG. 4.
Localization of PE_PGRS antigens at the cell surface of BCG by immunofluorescence. The BCG parent strain (A and C) and mc21525 (B) were incubated with anti-1818PE_PGRS sera (A and B) or with preimmune mouse sera (C) followed by FITC-conjugated anti-mouse sera. Arrows designate the pronounced surface staining of clusters of BCG in panel A and the reduced punctate staining observed on mc21525 cells in panel B. Fluorescence was visualized using a Nikon Optiphot-2 microscope with a 100× fluorescent objective. Images were taken using a SPOT RT digital camera and were posterized using Adobe Photoshop software. (×1,000).
The BCG 1818PE_PGRS::Tn5367 mutant mc21525 is deficient in its ability to infect macrophages.
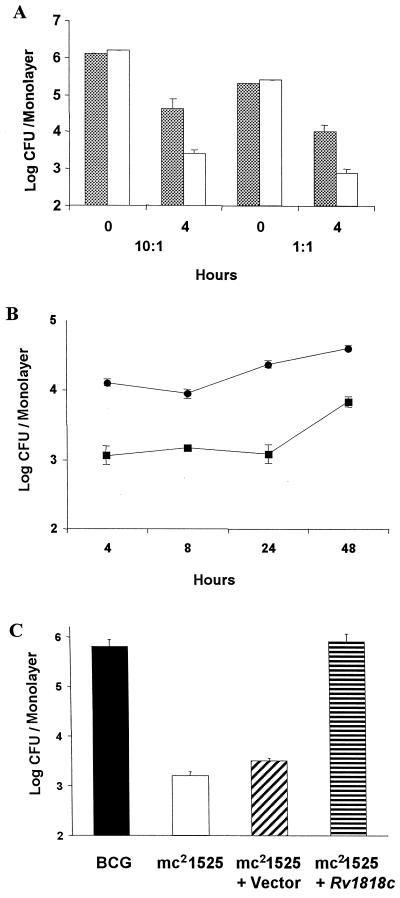
To test whether the mc21525 variant is also attenuated in its interactions with eukaryotic cells, we compared the ability of mc21525 and the BCG parent strain to enter and reside within J774 macrophage-like cells. Mycobacteria from log-phase cultures were incubated at an MOI of 10:1 (approximately 106 mycobacteria per 105 J774 cells) or 1:1 (approximately 105 mycobacteria per 105 J774 cells) with monolayers of J774 cells for 4 h, and then cells were lysed and plated for the determination of colonies. At an MOI of both 10:1 and 1:1, mc21525 infected J774 cells about 1 log less well than did the BCG parent line (Fig. 5A). We then compared the ability of mc21525 and BCG to enter and survive within BMMφ over a 2-day period (Fig. 5B). As observed for the J774 macrophage cell line, there is approximately a 1-log difference in the number of mc21525 organisms found within primary BMMφ cells after 4 h of incubation and this initial reduced level of bacterial infection is maintained over the 2-day period. These results suggest that the alteration of a gene in mc21525 also attenuates the ability of the bacteria to interact with macrophages, although mycobacteria that enter the macrophages can survive for a number of days.
FIG. 5.
Comparison of the infection of J774 cells or mouse BMMφ by BCG Pasteur, the mc21525 mutant, and mc21525 complemented with the Rv1818c gene. (A) The number of parent BCG (solid bars) and mc21525 (clear bars) in log CFU per monolayer of J774 cells is shown at the time added (0) and following a 4-h incubation period. For the experiments shown, approximately 2.5 × 106 mycobacteria per well were added for an MOI of 10:1 and approximately 2 × 105 mycobacteria per well for an MOI of 1:1. Cell counts showed that there were 2.2 × 105 J774 cells per monolayer for the 10:1 experiment and 2.5 × 105 cells per monolayer for the 1:1 experiment. The results are an average of triplicate wells and show that the number of mc21525 associated with J774 cells is significantly less after 4 h than the parent BCG at P < 0.01, as measured by the t test. Three independent experiments gave results like those shown here. (B) Approximately 2.2 × 105 BMMφ cells per monolayer were incubated for 4 h with about 3 × 105 mycobacteria (BCG = 3.2 × 105 per well and mc21525 = 3.7 × 105 per well), and log CFU per monolayer were determined at 4, 8, 24, and 48 h. The results shown are an average of triplicate wells and show that the number of mc21525 organisms (solid squares) associated with BMMφ cells is significantly less after 4 h and throughout the test period than the number of parent BCG organisms (solid circles) as measured by the t test. (C) Approximately 4 × 105 organisms of BCG Pasteur, the mc21525 mutant, mc21525 transformed with the vector only, and mc21525 transformed with the Rv1818c gene were incubated with 2 × 105 J774 cells per monolayer for 4 h, and the numbers of CFU were determined from triplicate wells. The infectivity of J774 cells by mc21525 alone and mc21525 containing only the vector is similar and significantly reduced, as determined by t test measurements, compared to their infectivity by both BCG and mc21525 complemented with Rv1818c. Similar results were obtained in three separate experiments.
Since complementation of mc21525 with the Rv1818c gene restores the clumping phenotype, we also investigated the ability of complementation to augment the ability of the BCG 1818PE_PGRS::Tn5367 mutant to infect J774 cells. As seen in Fig. 5C, while the mc21525 strain containing only the vector showed the same reduced level of entry into J774 cells as mc21525 after 4 h, the number of mc21525 organisms expressing Rv1818c residing within J774 cells was equivalent to that of the BCG parental strain. These results indicate that complementation of the 1818PE_PGRS::Tn5367 mutation with the wild-type gene restores cell surface properties of the bacterium, including the ability of the BCG variant to efficiently infect macrophages.
DISCUSSION
In this investigation, we have used transposon mutagenesis to identify a novel mycobacterial protein that is associated with phenotypic changes in the parent strain that alter the adhesive properties of the mycobacteria. Recently, a similar BCG transposon library was used to identify a mycolic acid cyclopropane synthetase as a virulence factor in M. tuberculosis by screening for variants that are deficient in cord formation (20). For our investigation, we reasoned that a mutant which exhibited a nonaggregative morphology may identify a gene that influences the interaction of mycobacteria with other cells, since there is evidence that adhesins can also induce autoaggregation of bacteria (9, 32). In fact, it has previously been shown that the mycobacterial adhesin HBHA promotes bacterial aggregation (16, 31, 33).
In this study, after screening more than 1,900 BCG transposon insertion mutants, one mutant strain, mc21525, was singled out for demonstrating an obvious difference in clumping morphology in culture compared to the parent strain. Surprisingly, sequence analysis revealed that the Tn5367 transposon had interrupted a gene identical to the Rv1818c gene of M. tuberculosis H37Rv, which is a member of the multigene PE_PGRS family distributed throughout the M. tuberculosis genome (12). As recently shown (11, 12), the PE_PGRS genes are a subgroup of the PE family of genes that are composed of 38 PE and 61 PE_PGRS genes. Both have a highly conserved N-terminal domain of 110 to 130 amino acids containing the PE motif. The protein encoded by the Rv1818c gene of M. tuberculosis is typical of the PE_PGRS family in that (i) it is highly homologous with many of the average-sized PE_PGRS genes; for example, Rv1818c shows about 60% identity with the PE_PGRS gene Rv1756c (12), (ii) the N-terminal PE region of Rv1818c shows a very high degree of homology with the genes in the PE family, including those that encode only the PE domain, and (iii) it is composed of 41% glycine and 20% alanine, and the PGRS domain contains 21 GGAGGX repeats (Fig. 2).
To date, evidence suggests that the occurrence of numerous PE and PE_PGRS genes is restricted to members of the M. tuberculosis complex and a few other mycobacterial species (11, 12, 18, 35) which infect primarily humans and other mammals (13, 40). This implies that the PE multigene family may have a role in mycobacterial infection and survival within host tissues or, as has been suggested, that the numerous PE_PGRS genes could function as a source of antigenic variability for M. tuberculosis in order to evade the host immune response (12, 44). However, very little is known about the function of the proteins encoded by the M. tuberculosis PE_PGRS multigene families. Although it has been demonstrated that some PE_PGRS proteins are preferentially expressed within host cells (36), our experiments associating loss of aggregation and reduced infection of macrophages with a mutation in the 1818PE_PGRS gene indicate that certain PE_PGRS proteins are also expressed in culture. Also, as shown by immunoblots, antisera raised against 1818PE_PGRS protein recognized multiple protein bands at approximately 40 to 45 kDa in strains of M. tuberculosis and M. bovis, which is the predicted molecular size of several PE_PGRS proteins (12) both in a cell wall preparation of M. tuberculosis and in whole-cell lysates of M. tuberculosis and BCG strains. This is consistent with other studies in which it has been demonstrated that the major humoral response to 1818PE_PGRS is directed against the PGRS domain, which contains the Gly-Ala repeats present in most PE_PGRS proteins (16a). Immunofluorescence studies using the anti-1818PE_PGRS sera also indicate that PE_PGRS proteins are located on the cell surface of BCG as well as M. tuberculosis. It is of interest that glycine-rich proteins with some homology with the PGRS region of the PE_PGRS proteins are found in the cell wall of a number of plants (28, 46). Together these data and the fact that there is a putative transmembrane domain predicted in the sequence of PE_PGRS proteins (12) suggest that certain of these proteins may be associated with the cell surface of mycobacteria.
The capacity of mycobacteria such as M. tuberculosis and M. bovis to form cell aggregates (clumps) while growing in liquid culture is altered by the inactivation of the Rv1818c-like gene in mc21525, as is its ability to efficiently interact with macrophages. It is possible that mc21525 demonstrates less infectivity of macrophages because it does not aggregate like the parent strain, therefore resulting in fewer bacteria accumulating within the macrophages. However, we have used a method of preparing the bacteria for the infectivity assay (42) which should minimize the number of aggregated bacteria being added to the macrophages. Utilizing this method we observed that mc21525 enters macrophages about 1 log less well than does the parent strain. The time course studies using primary BMMφ also suggest that it is only the initial adherence event that is affected by the loss of 1818PE_PGRS and that mc21525 organisms are able to survive as well as the parent strain within the macrophages for 48 h, albeit in lesser numbers. However, we cannot rule out the possibility that the reduced numbers of mc21525 organisms observed after 4 h is due to increased sensitivity of the strain to microbicidal activity within the macrophages.
Recent investigations indicate that certain genes in the PE family may be deleted in M. bovis as well as in attenuated BCG strains (5, 21), and there is evidence that there is sequence variation between M. tuberculosis H37Rv and BCG Pasteur in one PE_PGRS gene, Rv0746 (12). However, since there are likely to be numerous PE_PGRS genes in BCG, it is surprising that the disruption of one PE_PGRS gene results in the phenotypic changes noted in this study. There are several possible explanations for the alteration in phenotypic properties of M. bovis BCG observed in these studies which are areas for continued exploration. These include the possibility that 1818PE_PGRS is directly involved in the cell adhesion events because it is a member of the PE_PGRS family that is preferentially localized to the surface of the bacterium or, alternatively, that 1818PE_PGRS is a critical participant in the formation of a complex cell wall structure with adhesive properties, perhaps composed of a number of PE_PGRS proteins. Examples of complex prokaryotic structures such as fimbriae (pili) (2) and the type III secretion structures that are composed of many closely related proteins and promote bacterium-host interactions (14, 19, 26) could serve as models for further study. It is of interest that another PE_PGRS protein encoded by the M. tuberculosis gene Rv1759c has been characterized as a fibronectin-binding protein (1, 18). However, we cannot exclude the possibility that loss of the 1818PE_PGRS indirectly affects the surface properties of the mycobacteria by a polar effect on other proteins or cell wall components which then alters the cell surface properties of the bacterium. Also, the attenuation of 1818PE_PGRS could result in a less viable strain which has phenotypic characteristics like those documented in this study. The direct influence of 1818PE_PGRS on mycobacterial interactions with other cells awaits further experimentation with purified protein and specific antibody probes as well as the construction of M. tuberculosis strains carrying specific gene deletions in the PE_PGRS gene(s).
Ramakrishnan et al. (36) have recently shown that certain members of the PE_PGRS family are expressed within host tissues and macrophages infected with Mycobacterium marinum. In this study, the evidence suggests that certain PE_PGRS proteins may also be expressed by mycobacteria living outside the host cell and may be located at the cell surface where they can influence the interactions of mycobacteria with other mycobacteria and with the macrophage target cell of the host. Thus, the mycobacterial PE_PGRS proteins may have multiple roles in the infectious process which culminates in tuberculosis.
ACKNOWLEDGMENTS
We are grateful to Thames Pickett of CBER/FDA for the anti-1818PE_PGRS mouse polyclonal antisera used in immunofluorescence and to Catherine Bosio of CBER/FDA for advice on isolation of mouse BMMφ.
REFERENCES
- 1.Abou-Zeid C, Garbe T, Lathigra R, Wiker H G, Harboe M, Rook G A, Young D B. Genetic and immunological analysis of Mycobacterium tuberculosis fibronectin-binding proteins. Infect Immun. 1991;59:2712–2718. doi: 10.1128/iai.59.8.2712-2718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham S N, Jonsson A B, Normark S. Fimbriae-mediated host-pathogen cross-talk. Curr Opin Microbiol. 1998;1:75–81. doi: 10.1016/s1369-5274(98)80145-8. [DOI] [PubMed] [Google Scholar]
- 3.Arruda S, Bomfim G, Knights R, Huima-Byron T, Riley L W. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science. 1993;261:1454–1457. doi: 10.1126/science.8367727. [DOI] [PubMed] [Google Scholar]
- 4.Bardarov S, Kriakov J, Carriere C, Yu A, Vaamonde C, McAdam R A, Bloom B R, Hatfull G F, Jacobs W R., Jr Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr M A, Wilson M A, Gill W P, Salamon H, Schoolnik G K, Rane S, Small P M. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- 6.Berg C M, Berg D E, Groisman E A. Transposable elements and the genetic engineering of bacteria. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 879–925. [Google Scholar]
- 7.Berthet F-X, Rauzier J, Lim E M, Philipp W, Gicquel B, Portnoi D. Characterization of the Mycobacterium tuberculosis erp gene encoding a potential cell surface protein with repetitive structures. Microbiology. 1995;141:2123–2130. doi: 10.1099/13500872-141-9-2123. [DOI] [PubMed] [Google Scholar]
- 8.Brennan M J, Hannah J H, Leininger E. Adhesion of Bordetella pertussis to sulfatides and to the GalNAcβ4Gal sequence found in glycosphingolipids. J Biol Chem. 1991;266:18827–18831. [PubMed] [Google Scholar]
- 9.Chiang S L, Taylor R K, Koomey M, Mekalanos J J. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 10.Cisar J O, Sandberg A L, Reddy G P, Abeygunawardana C, Bush C A. Structural and antigenic types of cell wall polysaccharides from viridans group streptococci with receptors for oral actinomyces and streptococcal lectins. Infect Immun. 1997;65:5035–5041. doi: 10.1128/iai.65.12.5035-5041.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole S T. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Lett. 1999;452:7–10. doi: 10.1016/s0014-5793(99)00536-0. [DOI] [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Collins F M. Tuberculosis: the return of an old enemy. Crit Rev Microbiol. 1993;19:1–16. doi: 10.3109/10408419309113520. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis G R, Van Gijsegem F. Assembly and function of type III secretory systems. Annu Rev Microbiol. 2000;54:735–774. doi: 10.1146/annurev.micro.54.1.735. [DOI] [PubMed] [Google Scholar]
- 15.Cox J S, Chen B, McNeil M, Jacobs W R., Jr Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 16.Delogu G, Brennan M J. Functional domains present in the mycobacterial hemagglutinin, HBHA. J Bacteriol. 1999;181:7464–7469. doi: 10.1128/jb.181.24.7464-7469.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Delogu G, Brennan M J. Comparative immune response to PE and PE_PGRS antigens of Mycobacterium tuberculosis. Infect Immun. 2001;69:5606–5611. doi: 10.1128/IAI.69.9.5606-5611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dersch P, Isberg R R. An immunoglobulin superfamily-like domain unique to the Yersinia pseudotuberculosis invasin protein is required for stimulation of bacterial uptake via integrin receptors. Infect Immun. 2000;68:2930–2938. doi: 10.1128/iai.68.5.2930-2938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espitia C, Laclette J P, Mondragon-Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology. 1999;145:3487–3495. doi: 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 19.Galan J E. Molecular genetic bases of Salmonella entry into host cells. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 20.Glickman M S, Cox J S, Jacobs W R., Jr A novel mycolic acid cyclopropane synthetase is required for coding, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. [DOI] [PubMed] [Google Scholar]
- 21.Gordon J E, Brosch R B, Billault A, Farnier T, Eiglmeier K, Cole S T. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 22.Hannah J H, Menozzi F D, Renauld G, Locht C, Brennan M J. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (FHA) and mapping of the heparin-binding domain on FHA. Infect Immun. 1994;62:5010–5019. doi: 10.1128/iai.62.11.5010-5019.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies, a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 24.Kalpana G V, Bloom B R, Jacobs W R., Jr Insertional mutagenesis and illegitimate recombination in mycobacteria. Proc Natl Acad Sci USA. 1991;88:5433–5437. doi: 10.1073/pnas.88.12.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 27.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsui M, Toyosawa I, Fukuda M. Novel N-terminal sequence of a glycine-rich protein in the aleurone layer of soybean seeds. Biosci Biotechnol Biochem. 1994;58:1920–1922. doi: 10.1271/bbb.58.1920. [DOI] [PubMed] [Google Scholar]
- 29.McAdam R A, Weisbrod T R, Martin J, Scuderi J D, Brown A M, Cirillo J D, Bloom B R, Jacobs W R., Jr In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect Immun. 1995;63:1004–1012. doi: 10.1128/iai.63.3.1004-1012.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonough K A, Kress Y. Cytotoxicity for lung epithelial cells is a virulence-associated phenotype of Mycobacterium tuberculosis. Infect Immun. 1995;63:4802–4811. doi: 10.1128/iai.63.12.4802-4811.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menozzi F D, Bischoff R, Fort E, Brennan M J, Locht C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menozzi F D, Boucher P E, Riveau G, Gantiez C, Locht C. Surface-associated filamentous hemagglutinin induces autoagglutination of Bordetella pertussis. Infect Immun. 1994;62:4261–4269. doi: 10.1128/iai.62.10.4261-4269.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menozzi F D, Rouse J H, Alavi M, Laude-Sharp M, Muller J, Bischoff R, Brennan M J, Locht C. Identification of a heparin-binding hemagglutinin present in mycobacteria. J Exp Med. 1996;184:993–1001. doi: 10.1084/jem.184.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelicic V, Jackson M, Reyrat J-M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poulet S, Cole S T. Characterization of the highly abundant polymorphic GC-rich-repetitive sequence (PGRS) present in Mycobacterium tuberculosis. Arch Microbiol. 1995;163:87–95. doi: 10.1007/BF00381781. [DOI] [PubMed] [Google Scholar]
- 36.Ramakrishnan L, Federspiel N A, Falkow S. Granuloma-specific expression of mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science. 2000;288:1436–1439. doi: 10.1126/science.288.5470.1436. [DOI] [PubMed] [Google Scholar]
- 37.Ratliff T L, McCarthy R, Telle W B, Brown E J. Purification of a mycobacterial adhesin for fibronectin. Infect Immun. 1993;61:1889–1894. doi: 10.1128/iai.61.5.1889-1894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Relman D A, Domenighini M, Tuomanen E, Rappouli R, Falkow S. Recognition of a bacterial adhesion by an integrin: macrophage CR3 (alpha M beta 2, CD11b/CD18) binds filamentous hemagglutinin of Bordetella pertussis. Cell. 1990;61:1375–1382. doi: 10.1016/0092-8674(90)90701-f. [DOI] [PubMed] [Google Scholar]
- 39.Rhoades E R, Orme I M. Susceptibility of a panel of virulent strains of Mycobacterium tuberculosis to reactive nitrogen intermediates. Infect Immun. 1997;65:1189–1195. doi: 10.1128/iai.65.4.1189-1195.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rook G A, Bloom B R. Mechanisms of pathogenesis of tuberculosis. In: Bloom B R, editor. Tuberculosis: pathogenesis, protection, and control. Washington, D.C.: ASM Press; 1994. pp. 485–501. [Google Scholar]
- 41.Rostand K S, Esko J D. Microbial adherence to and invasion through proteoglycans. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlesinger L S, Bellinger-Kawahara C G, Payne N R, Horwitz M A. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- 43.Shepard C. Growth characteristics of tubercle bacilli and certain other mycobacteria in HeLa cells. J Exp Med. 1957;105:39–48. doi: 10.1084/jem.105.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tekaia F, Gordon S V, Garnier T, Brosch R, Barrell B G, Cole S T. Analysis of the proteome of Mycobacterium tuberculosis in silico. Tuber Lung Dis. 1999;79:329–342. doi: 10.1054/tuld.1999.0220. [DOI] [PubMed] [Google Scholar]
- 45.Wei J, Dahl J L, Moulder J W, Roberts E A, O'Gaora P, Young D B, Friedman R L. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol. 2000;182:377–384. doi: 10.1128/jb.182.2.377-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ye Z H, Song Y R, Marcus A, Varner J E. Comparative localization of three classes of cell wall proteins. Plant J. 1991;1:175–183. doi: 10.1111/j.1365-313x.1991.00175.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhao W, Schorey J S, Groger R, Allen P M, Brown E J, Ratliff T L. Characterization of the fibronectin binding motif for a unique mycobacterial fibronectin attachment protein, FAP. J Biol Chem. 1999;274:4521–4526. doi: 10.1074/jbc.274.8.4521. [DOI] [PubMed] [Google Scholar]