Abstract
In recent years, the increased demand for and regional variability of available water resources, along with sustainable water supply planning, have driven interest in the reuse of produced water. Reusing produced water can provide important economic, social, and environmental benefits, particularly in water-scarce regions. Therefore, efficient wastewater treatment is a crucial step prior to reuse to meet the requirements for use within the oil and gas industry or by external users. Bioremediation using microalgae has received increased interest as a method for produced water treatment for removing not only major contaminants such as nitrogen and phosphorus, but also heavy metals and hydrocarbons. Some research publications reported nearly 100% removal of total hydrocarbons, total nitrogen, ammonium nitrogen, and iron when using microalgae to treat produced water. Enhancing microalgal removal efficiency as well as growth rate, in the presence of such relevant contaminants is of great interest to many industries to further optimize the process. One novel approach to further enhancing algal capabilities and phytoremediation of wastewater is genetic modification. A comprehensive description of using genetically engineered microalgae for wastewater bioremediation is discussed in this review. This article also reviews random and targeted mutations as a method to alter microalgal traits to produce strains capable of tolerating various stressors related to wastewater. Other methods of genetic engineering are discussed, with sympathy for CRISPR/Cas9 technology. This is accompanied by the opportunities, as well as the challenges of using genetically engineered microalgae for this purpose.
Keywords: bioremediation, CRISPR/cas9, genetic engineering, microalgae, produced wastewater
1 Introduction
Microalgae are photosynthetic microscopic organisms, either prokaryotic or eukaryotic that could live in all bodies of water and utilize sunlight and carbon dioxide (CO2) as their sole energy and carbon sources to produce organic compounds via photosynthesis (Zhu et al., 2016; Deviram et al., 2020). This, together with their fast growth rates, and ability to produce high value metabolites of interest for many industrial applications, make microalgae an attractive subject for researchers in the field of sustainable production (Saadaoui et al., 2019; Arias et al., 2020; Rasheed et al., 2020; Saadaoui et al., 2019; Veiga et al., 2020; Saadaoui et al., 2019; Varaprasad et al., 2021; Bounnit et al., 2022; Bello et al., 2022). Furthermore, microalgae have also been found to be able to efficiently remove contaminants from various types of wastewater effluents, including pharmaceutical (Singh et al., 2020), agricultural (Leite et al., 2019), and industrial (Al-Jabri et al., 2021). In the tertiary treatment stage, they can eliminate macro-nutrients from the water mainly nitrogen and phosphorus, as well as heavy metals (Molazadeh et al., 2019). Even in particularly contaminated wastewaters, such as produced water from the oil and gas industry, microalgae have demonstrated the ability to degrade specific components as a treatment step toward clean water (Graham et al., 2017; Rahman et al., 2020; Alsarayreh et al., 2022). Overall, cultivation of microalgae on different types of wastewaters is considered a sustainable technology for bioremediation due to its capacity to thrive by consuming present contaminants (Mehariya et al., 2021). Non-etheless, due to immature technologies, instabilities of wastewater components, and required improvements of bioremediation efficiencies, microalgae are not yet widely applied for wastewater treatment (Feng et al., 2020; Li et al., 2022).
Various methods can be applied to enhance the bioremediation efficiencies of microalgae, including advanced cultivation systems, consortiums, and genetic modification. Microalgal cells are transformable; their genomes can be redesigned to include a desired feature by employing the proper delivery system to introduce DNA for transformation (Gimpel et al., 2015). Transformation of different microalgal species using multiple genetic engineering tools has been successfully applied to enhance metabolite production for biofuels (Radakovits et al., 2010) as well as for other products (Ibuot et al., 2017; Rahman et al., 2020). Genetic engineering is a powerful tool to enhance the ability of many microorganisms’ to bioremediate wastewater. For example (Huang et al., 2015), inserted the arsenite S-adenosylmethionine methyltransferase gene obtained from the red microalgae Cyanidioschyzon merolae into Bacillus subtilis. The transformed Bacillus was able to methylate the inorganic arsenic. Such studies provide proof for concept of using transformed microbes for treating different kinds of contaminants.
This review provides the status of microalgal-based bioremediation of produced water, as well as the most recent microalgal applicable genetic engineering tools: zinc finger nucleases (ZFNs), Transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats (CRISPR/Cas9). Previous reviews of this field have particularly focused on multiple wastewater treatment such as agricultural or municipal, or on genetic engineering as a tool to enhance efficiency the efficiency of microalgae to serve different purposes. This review is the first of its kind to develop a critical overview and explore about the importance of genetic engineering in enhancing algae phytoremediation efficiency. Combining the two topics allows us a forward look at genetic engineering strategies to enhance microalgal efficiency in phytoremediation of produced wastewater.
2 Microalgae: Promising alternative for produced water bioremediation
Produced water (PW) is the highest volume of liquid waste generated and discharged during the production of oil and gas. The worldwide volume of produced water generated is approximately 1.3 times that of hydrocarbon production (Gray, 2020). PW varies in composition and volume from one formation to another, and predominant constituents include total dissolved solids (TDS), such as natural salts and minerals, as well as dissolved and volatile organic compounds, oil and grease, heavy metals, dissolved gases, bacteria, naturally occurring radioactive materials (NORM, radionuclides such as radium), and the additives used in hydrocarbon production (Al-Ghouti et al., 2019). In recent years, the increased demand for and regional variability of available water resources, along with sustainable water supply planning, have driven interest in the reuse of produced water. As freshwater supplies become scarcer, produced water can be a crucial source of water after suitable treatment. There has been an increased focus on reclaiming, reusing, and recycling water that is usually wasted to meet the communities’ needs for freshwater sources (Gray, 2020).
In recent times, a greater focus has appeared on using biological systems, including microalgae, for treating produced wastewater effluents (Graham et al., 2017; Rahman et al., 2020; Alsarayreh et al., 2022). Application of microalgae in wastewater treatment even shows competitive advantages over other treatment methods to improve water quality, due to its high treatment efficiency as well as carbon capture and biomass valorization opportunities (Molazadeh et al., 2019; Leng et al., 2020; Alsarayreh et al., 2022).
It is not surprising that algae have a high potential for PW treatment. For decades, researchers have studied their general wastewater treatment capabilities, optimizing their treatment efficiencies and growth on different types of wastewaters. Salgueiro et al. (2016) for example demonstrated that Chlorella vulgaris was able to remove 99.2% of phosphorus from artificial wastewater containing glucose, ammonium chloride, urea, monopotassium phosphate and reduce the chemical oxygen demand (COD) by 71.1%. Likewise (El-Kassas and Mohamed, 2014), showed that Chlorella vulgaris was able to treat textile wastewater, with COD and color removal percentages of 69.9% and 76.32%, respectively. Furthermore (Wang et al., 2019), investigated the use of the marine diatom Phaeodactylum tricornutum to treat municipal wastewater mixed with seawater. The results revealed that the diatom was able to remove up to 89.9% of COD, 86.7% of Total Nitrogen (TN), 84.2% of ammonium, and 97.0% of total phosphorus. Moreover, the strains were able to produce a high amount of lipids, which could potentially be applied for biodiesel production (Jayakumar et al., 2021). Produced water, on the other hand, can be more challenging, due to potential toxicity and the presence of heavy metals, hydrocarbons, surfactants, and anti-corrosives. Non-etheless, various studies have shown that bioremediation of PW using microalgae has shown a high potential. For example (Das et al., 2019), demonstrated the ability of Chlorella sp. (QUCCCM10) to bioremediate pretreated PW, after pH adjustment and removal of suspended matter. The microalga was found to be able to reduce the concentration of various elements, such as arsenic, cadmium, iron, nickel, and potassium, as well as remove 92% and 94.2% of TN and Total Organic Carbon (TOC), respectively. In a more recent study (Rahman et al., 2021), showed that Galdieria sulphuraria was able to grow in PW concentrations of up to 50%, with biomass productivities of up to .72 g L−1·d−1, and 99.6% and 74.2% nitrogen and phosphorus removal rates, respectively, without the addition of extra micronutrients. Similarly (Ammar et al., 2018), inoculated two marine microalgae species, Nannochloropsis oculata and Isochrysis galbana, with different concentrations of PW obtained from an oil field located in Iraq. Although higher PW loadings were found to have a negative impact on biomass productivities; successive adaptation biomass yields of .31 g L−1 were still achieved for both strains at 50% PW loading. Optimal contaminant removal however occurred at lower PW loadings of 10% and 25%, at which Nannochloropsis oculata was able to remove up to 89% and 81% oil content and 90% and 72% COD, respectively.
The ability of microalgae to remove pollutants from wastewater is due to different mechanisms they can perform. First, microalgae have the ability to take advantage of mixotrophic modes of nutrition. Which means it can switch their metabolism from using only CO2 as a carbon source to using organic matter based on its availability in the growth medium (Alalawy et al., 2019). (Devi et al., 2022) cultivated Scenedesmus sp. DDVG strain in municipal wastewater under mixotrophic condition to test the strain availability to survive and remove major contaminants form the water. The results showed that Scenedesmus efficiently removed ≈75% of COD and ≈100% of total nitrogen with a biomass of nearly 3.4 gL−1 after 10 days of cultivation.
Microalgal removal of organic compounds from the wastewater can be credited to biodegradation and biosorption processes. Biodegradation is known to be the most effective method by which microalgae use different enzymes to eliminate organic micropollutants from the aqueous environment (Nguyen et al., 2021). Biosorption is defined as the physical-chemical process in which substances are removed from solution using biological matter. Due to the negative charge of the cell wall in microalgae, cationic pollutants can efficiently adhere to the surface. However, it is less efficient in term of pollutants removal compared to biodegradation (Nguyen et al., 2021). Biodegradation and biosorption of different contaminants including organic compounds are extensively studied by Pathak et al. (2018).
Microalgae have enormous potential not only for bioremediation of produced water, but also for biomass reutilization in a variety of applications. Algae cultivated in wastewater are rich source of primary and secondary metabolites that could benefit in producing many valuable bioproducts such as biofuels, biofertilizers, and feed supplements (Shahid et al., 2020). Combining algae-based wastewater treatment with producing high-value compounds will greatly reduce the economic cost of the process. For instance (Japar et al., 2021), successfully increased the biomass production and lipid content of Chlorella vulgaris and Chlorella sorokiniana UKM3 grown in industrial wastewater by acclimatization process. . Since wastewater can be used as a sustainable growth medium, a variety of wastewater types have been proposed to increase algal biomass (Srimongkol et al., 2022). In this context, biomass generated from produced water treatment can be a promising source for valuable metabolite production for numerous applications. In Table 1, more detailed information is shown on these and other recent studies investigating algal-mediated PW treatment, including the different strains, experimental and cultivation conditions applied.
TABLE 1.
Selected recent literature on algal bioremediation of PW.
| Strain | Wastewater type | Cultivation conditions | Highest biomass yield (g·L−1) | Pollutants removed | Highest removal efficiency (%) | Reference | |
|---|---|---|---|---|---|---|---|
| 1 | Chlorella sp | produced water from a Qatari local petroleum company | In a temperature-controlled room, a glass bottle was agitated with compressed air and illuminated with an light intensity of 600 µmol photons·m−2·s−1 | 1.72 | TOC | 73 | Das et al. (2019) |
| total Nitrogen | 92 | ||||||
| 2 | Chlorella vulgaris | Produced water from an oil and gas facility in United States. | Tissue Culture Roller Drum Apparatus inside an incubator with a constant level of CO2 of 2%–3% (v/v), a temperature of 28°C with 16:8 h light:dark cycle and an illumination of ∼4000 lux | 3.1 ± 0.5 | total Nitrogen | 100 | Rahman et al. (2021) |
| phosphorus | ≈74.2 | ||||||
| 3 | Chlorella vulgaris | PW from dumping site generated by oil wells in Colombia | fluorescent light at an irradiance of 36.8 ± 4.2 μmol photons m−2 s−1 at the surface of the culture medium, temperature at 20 °C and permanent aeration supplied by a blower | — | Total hydrocarbons | ≈100 | Calderón-delgado et al. (2019) |
| 4 | Chlorella pyrenoidosa | PW from oilfield in Algeria | outdoor, under sunlight radiation, using an open system sited in the desert area in the winter season. The temperatures fluctuated from 26 to 31°C during the day | 1.15 | COD | 89.67% | Rahmani et al. (2022) |
| Ammonium nitrogen | 100% | ||||||
| total Nitrogen | 57.14%, | ||||||
| total Phosphorus | 75.51% | ||||||
| Copper | 73.39 | ||||||
| Lead | 72.80 | ||||||
| Cadmium | 48.42 | ||||||
| 5 | Nannochloropsis oculata | Produced water from oil field in Iraq | Florescence light (2000 lux) at and a light photoperiod of 18:6 h light:dark, 25°C±1°C, continuous filtered air at a constant flow rate via two aquarium air pumps | 1.13 | Oil | 66.5 | Ammar et al. (2018) |
| COD | 54 | ||||||
| 6 | Nannochloropsis oculata | Produced water from a TOTAL operating site in France | 14/10 h light/dark periods, by LED lamps, temperature at (21 ± 1°C), autotrophic conditions with air. CO2 was added in pulse, 5 s each 20–40 min and pH between 7.5 and 9 | — | Ammonium nitrogen | ≈100 | Parsy et al. (2020) |
| COD | 70 | ||||||
| Iron | 100 | ||||||
| 7 | Nannochloropsis oculata | Produced water from an oil field in Brazil | Aerated photobioreactors (3 L min−1), cold white LED lamps with light intensity of 57 μmol m−2·s−1, photoperiod of 12:12 h. Temperature controlled at 21°C±.9°C. The pH was fixed at 7 | — | PAHs | 94 | Marques et al. (2021) |
| NAP | 96 | ||||||
| APT | 95 | ||||||
| FLU | 91 | ||||||
| PHE | 83 | ||||||
| BbF | 95 | ||||||
| DA | 90 | ||||||
| BaP | 95 | ||||||
| Iron | 96.80 | ||||||
| 8 | Galdieria sulphuraria | Produced water from an oil and gas facility in United States. | Tissue Culture Roller Drum Apparatus inside an incubator CO2 level was kept constant at 2%–3% (v/v), temperature 42°C with 24 h of continuous illumination (∼4000 lux) | 5.12 ± .28 | total Nitrogen | ≈100 | Rahman et al. (2021) |
| 9 | Isochrysis galbana | Produced water from oil field in Iraq | Florescence light (2000 lux), and a photoperiod of 18:6 h light: dark, 25°C±1°C, continuous filtered air at a constant flow rate via two aquarium air pumps | 1.01 | Oil | 68 | Ammar et al. (2018) |
| COD | 56 | ||||||
| 10 | Dunaliella tertiolecta | Produced water from an oil production facility in the Permian Basin of southeast New Mexico | Temperature controlled at 24°C in a growth chamber with fluorescent illumination of 100 μmol photons m−2 s−1. agitation was set at at 140 rpm with a 16-h light/8-h dark cycle | ≈0.3 | nitrate | ≈99.6 | Hopkins et al. (2019) |
| Phosphate | ≈99.6 |
PAHs, polycyclic aromatic hydrocarbons; NAP, naphthalene; AP, acenaphthylene; FLU, fluorene; PHE, phenanthrene; BbF, benzo(b)fluoranthene; DA, dibenzo (a, h) anthracene; BaP, benzo(a)pyrene.
To summarize, utilizing microalgae for PW bioremediation can be advantageous and efficient owing to i) its capability of removing heavy metals, hydrocarbons, and other pollutants, ii) its ability to produce high-value bioproducts such as biofuels (Cho et al., 2011), iii) its ability to reduce the need for fresh water and nutrients for algae cultivation (Rahman et al., 2020; Ahmad et al., 2021) and iv) its potential for carbon capture and utilization (CCU) from industrial point-sources (Kalra et al., 2021). Non-etheless, some obstacles and complications still arise when applying this technology, which will need to be mitigated for it to be applied on a large scale. Such concerns include the requirement for long residence times for efficient contaminant removal, the risk of contamination, the existence of contaminants that may inhibit the algae growth, and the cost of nutrients added and for biomass harvesting (Ahmad et al., 2021; Watanabe and Isdepsky, 2021).
3 Genetic engineering as promising tool to enhance bioremediation efficiencies
A possible option to improve the application of algae in industrial processes, such as PW treatment, could be strain improvement through genetic engineering (Ahmad et al., 2022). Even though advanced tools for genetic engineering have emerged at a great pace, they remain underutilized for microalgae as compared to other microorganisms. This is despite the demonstrated benefits of improving yields and overcoming challenges of high production costs (Bajhaiya et al., 2017; Kumar et al., 2020). In this section, we go over the different types of genetic modification which can be applied to algae, as well as how they can be used to improve PW bioremediation efforts.
3.1 Random mutagenesis
One of the powerful trait alteration tools for microalgae is random mutagenesis (Manandhar-Shrestha and Hildebrand, 2013; Arora et al., 2020). Obtaining mutants through random mutagenesis is accomplished through various methods, ranging from chemical, nuclear irradiation, plasmas, and ultraviolet (UV) mutagenesis. This, combined with a strong selective selection pressure, has proven to be a reliable strategy for producing strains with improved stress tolerances, resistance to contaminants, increased productivities (Cabanelas et al., 2016), but also higher metabolite production rates (Lai et al., 2004; Cordero et al., 2011; Doan and Obbard, 2012). Recently (Qi et al., 2018), developed a mutant of Scenedesmus obliquus for the purpose of increasing the CO2 bio-fixation and enhancing biomass production under elevated CO2 levels. The mutants with genetic stability as well as potential for increased CO2 tolerance were found through UV mutagenesis, as well as applying a low pH as a selective pressure. The authors found that the mutant strain was able to achieve higher biomass productivities and light conversion efficiencies under elevated CO2 concentrations compared to the parent strains, as well as contain 37% and 25% higher carbohydrate and lipid contents, respectively. Applying radon mutagenesis to algae strains such as those investigated by (Ammar et al., 2018), followed by exposure to high PW loadings, could potentially result in strains’ abilities to bioremediate PW without dilution, increasing its industrial and economic feasibility as a treatment option. Nevertheless, although interesting, the technology also exhibits some weaknesses, such as the fact that mutants may lose the mutation of interest within a number of generations, thus losing their enhanced trait, as well as the fact that experiments can have a long time span (Arora and Philippidis, 2021), making their application cumbersome.
3.2 Targeted genetic engineering
Besides random mutagenesis, using targeted genetic modification methods for heterologous expression of foreign genes is very promising for enhancing specific algae traits. Recently, genetic engineering has gained momentum because new and strong genetic tools are progressively offered and redesigning and improving metabolic pathways reveal new opportunities for the industrial development of microalgae (Ng et al., 2017). In the past decade, gene-editing tools such as (TALEN), (ZFN) (CRISPR/Cas9) technologies have emerged as the most popular recombinant DNA technologies that have been applied on a variety of different microorganisms, including microalgae (Fajardo et al., 2020; Kumar et al., 2020).
3.2.1 Zinc-finger nucleases (ZFN)
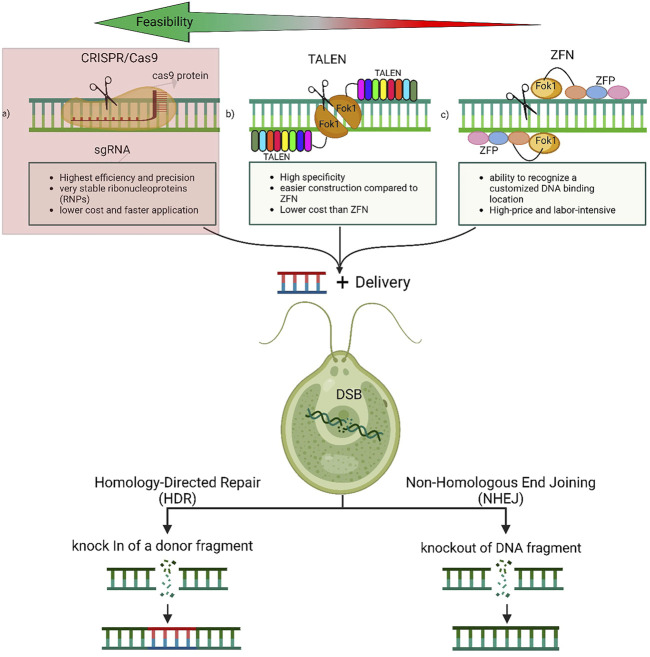
(Nain et al., 2010) describe zinc-finger nucleases (ZFNs) as a powerful tool that reshapes the boundaries of biological research. It is composed of programmable modules that bind to a specific DNA sequence. Excitingly, ZFN enables a wide range of genetic modifications by allowing DNA double-strand breaks that stimulate error repairs at specific genomic sites (Jabalameli et al., 2015; Kanchiswamy et al., 2015). The specificity of ZFNs arises from their versatility and ability to recognize a customized DNA binding location (Gaj et al., 2013) (Figure 2C). ZFNs have been used multiple times for chosen modifications of microalgal genomic DNA. It works by creating a cleavage site where insertions or deletions (INDELs) can take place (Jeon et al., 2017). ZFN technology was successfully applied to the model microalga Chlamydomonas reinhardtii in 2013 (Sizova et al., 2013). The ZFNs were created to target the COP3 gene using paromomycin-resistance as a marker activity. Furthermore, this work proves that transient ZFN expression is not toxic for the cells as it results in stable transformed colonies. Similarly (Greiner et al., 2017), used ZFNs to reliably edit genes by homologous recombination in multiple strains of Chlamydomonas, including the wild type. This work also reported that promising results were achieved when the ZFN protocol was changed to replace glass beads with electroporation. Regardless of the numerous benefits of editing DNA with ZFNs, some issues may arise when it is applied. For example, there are limited sites to be targeted for nuclease selection. Also, there is a potential that double strand breaks may occur at an off-target site (Gupta and Musunuru, 2014). A simplified workflow of ZFN process is illustrated in (Figure 1).
FIGURE 2.
Genetic engineering tools-induced genome editing in Microalgae. The double-stranded breaks (DSBs) introduced at the target site by CRISPR/Cas or TALEN or ZFN complexes stimulates the endogenous DNA repair machineries, non-homologous end joining (NHEJ) in the absence of the donor template or the homology-directed repair (HDR) in presence of the donor template. The NHEJ is generally associated with the introduction of insertions and/or deletions (indels) of varying lengths at the DSB site, often leading to the disruption of the reading frame of the target gene. The HDR pathway results in a precise insertion or deletion at the DSB site by homologous recombination. The preferred tool is the CRISPR/Cas9 which is highlighted in red as it is very accurate, easy, and fast.
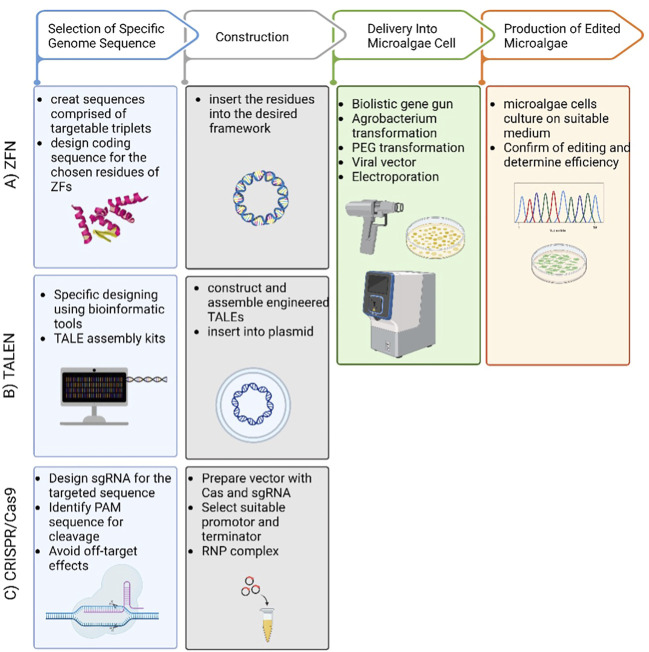
FIGURE 1.
Simplified workflow for ZFN, TALEN and CRISPR/Cas9 mediated microalgae genome editing. (A) ZFN, (B) TALEN and (C) CRISPR/Cas9. The 3 tools have 4 steps (1) using bioinformatic tools to identify the targeted sequence and design accordingly, (2) construct the delivery method by choosing the vector, (3) select the delivery method to insert the plasmid into the microalgae cell, (4) subculture the microalgae on a selective medium and determine the efficiency of gene editing.
3.2.2 Transcription activator-like effector nucleases (TALEN)
Transcription activator-like effector nucleases (TALEN) are one of the first developed molecular editing tools (Zhang et al., 2016). TALEs are proteins that exist in nature in Xanthomonas bacteria (Malzahn et al., 2017), and are distinguished by their ability to recognize and bind to single base pairs of a DNA sequence (Gaj et al., 2013). When TALE proteins fuse with FokI nuclease, a double strand break is induced in the DNA, which enables knocking out genes or introducing mutations (Jeon et al., 2017) (Figure 1B). TALEN has many advantages, most importantly: i) it can be engineered to target a specific site in the genome; ii) it is much easier to design compared to ZFNs; iii) it is available commercially, as well as a TALEN-based library has been constructed; iv) it is not limited by the length of sequence it can bind to; v) fewer obstacles appear when selecting the binding site (Gupta and Musunuru, 2014). On the other hand, some constraints must be taken into consideration when applying TALEN. As such, compared to ZFNs, it appears to be much larger in size, knowing that the large size makes it less specific. Also, the larger the TALENs, the harder it becomes to deliver them to cells. Moreover, the host plasmid vector of the TALEN sequence tends to rearrange after transduction (Kumar et al., 2020). Several studies have reported the modification of the genomes of different microalgae. TALENs were used to enhance lipid metabolic pathways in the genome of the diatom Phaeodactylum tricornutum, according to (Daboussi et al., 2014) and (Hao et al., 2018). Likewise (Takahashi et al., 2018), enhanced the lipid content in the green microalga Coccomyxa sp. Another example of successful utilization of the platinum TALENs is efficiently mutating the nitrate reductase and acyltransferase genes in Nannochloropsis oceanica (Kurita et al., 2020). The process of applying TALEN in microalgae is shown in (Figure 1).
3.2.3 CRISPR/Cas9
Due to its simplicity and versatility, CRISPR/cas9-mediated genome-editing is one of the most promising novel techniques for gene editing and has been shown to be successful in a variety of living organisms (Bortesi and Fischer, 2015; Xu M. et al., 2020). It offers an excellent time and labor efficient system (Doudna and Charpentier, 2014; Zhang et al., 2020) and multiple mitigation strategies succeeded in significantly reducing off-target effects (Han et al., 2020). Moreover, implementing a plasmid-free CRISPR-Cas9 system has resulted in very stable ribonucleoproteins (RNPs) in the studied cells (Song et al., 2019). To use genetically manipulated algae on an industrial scale, transformed cells need to demonstrate a stable gene expression on long term. Therefore, large scale algal cultivation require fully integrated gene cassettes within the genome the transformed strain (Patel et al., 2019). Many cases of CRISPR/Cas9 genetically manipulated microalgae have reported stable mutants, which may alleviate the problem of unsettled mutations (Nymark et al., 2016; Slattery et al., 2018). ((Nymark et al., 2016; Slattery et al., 2018). The mechanism of designing the CRISPR/Cas9 system to manipulate microalgae genome is expressed in (Figure 1) while its structure is shown in (Figure 2A)
The first successful application of CRISPR/Cas9 in microalgae was demonstrated in 2014, by (Jiang et al., 2014) on Chlamydomonas reinhardtii, although improvements were needed to reduce the cytotoxic effect of Cas9 on the algae strain (Baek et al., 2016; Shin et al., 2016). Since then, Chlamydomonas transformation using CRISPR system was accomplished multiple times, e.g., for increasing lipids and pigments content (Song et al., 2022), increasing triacylglycerol productivity (Lee et al., 2022); lipid accumulation (Nguyen T. H. T. et al., 2020); and for understanding CO2 sequestration mechanism (Asadian et al., 2022).
Due to its role in biofuel production; several studies have been conducted on algal cells for increasing lipid content through CRISPR/Cas9 genetic modification (D’Alessandro and Antoniosi Filho, 2016; Shokravi et al., 2020). For instance, a recent study by (Lin and Ng, 2020) investigated the improvement of the lipid content of Chlorella vulgaris, through targeted editing of the omega-3 fatty acid desaturase gene (fad3). Results showed that the genetically modified strain was able to reach up to 46% higher lipid content and 20% higher biomass concentrations, as compared to the wild type of strain. Other studies suggested that knocking out genes involved in the process of fatty acid degradation could induce increased lipid yields in different microalgal species (Nguyen A. D. et al., 2020; Chang et al., 2020). Several other studies were also successful in implementing CRISPR/Cas9 to increase the production of different carotenoids in Dunaliella salina (Hu et al., 2021); and in Chlamydomonas reinhardtii (Baek et al., 2016), to improve the thermal tolerance of Tetraselmis suecica (Xu J. et al., 2020), and to investigate gene function in Phaeodactylum tricornutum (Hao et al., 2022; Llavero-Pasquina et al., 2022). Overall, CRISPR/Cas9 has been successfully applied on many other algae species to improve their biomass productivities, tolerances to abiotic stressors, or increase lipid content, or that of other biomolecules and value-added products (Kumar et al., 2020; Jeon et al., 2021; Wang et al., 2021).
In summary, microalgae are emerging and thriving as sustainable base for biotechnology in general and produced wastewater treatment. Yet, it is not currently viable on the industrial scale due to many obstacles. Therefore, it is considered practical to use genetic engineering to enable the use of microalgae on a bigger scale. Furthermore, genome editing techniques may be used to further our understanding of the mechanisms behind the genes that enable microalgae to survive in such harmful environments. ZFNs, TALENs, and CRISPR/Cas9 were successfully implemented in enhancing various applications of microalgae including production of pharmaceutical products lipid, carotenoids, and protein (Grama et al., 2022). Several reviews summarized the major differences between these technologies stating the advantages and disadvantages of each individually (Jeon et al., 2017; Liu et al., 2022). These new approaches are of great interest to optimize specific traits of microalgae to boost their effectiveness in phytoremediation.
3.3 Applying CRISPR/Cas9 for improving PW treatment: Target genes
Although the application of CRISPR/Cas9 to enhance many microalgal traits, productivities, and production of primary metabolites has been increasing, there are very few studies that report the use of CRISPR/Cas9 to enhance microalgal applications in wastewater treatment (Patel et al., 2019; Feng et al., 2020). In general, there are a plethora of candidate genes that can alter metabolic pathways in favor of bioremediation of wastewaters, with the potential to improve bioremediation and biomass production (Balzano et al., 2020). To enhance microalgal PW treatment through genetic engineering, two strategies can be applied: (a) improving strain tolerance and degradability of certain pollutants, such as hydrocarbons or heavy metals, or (b) expressing and producing of degradation aiding molecules, such as surfactants or antifouling ingredients (Feng et al., 2020). In both cases, the first step is the identification of target genes for transformation into selected microalgae strains.
3.3.1 Tolerance and accumulation of heavy metals
Various wastewaters contain hazardous and toxic pollutants that pose a significant environmental risk, including heavy metals (Gray, 1998). Furthermore, the removal of heavy metals from wastewater is a serious issue and can be challenging in many cases (Kaur and Roy, 2021). However, numerous algal strains have been found to be capable of sequestering metals through the use of extracellular polysaccharides (EPS) and intracellular polyphosphates, which chelate metal ions (Opeolu et al., 2010). Additionally, the potential use of genetically engineered microalgae for metal bioremediation is paving the way for more evaluations and selections of novel genes that are involved in metal accumulation (Cheng et al., 2019).
The most common genetic manipulation techniques for algal-based metal recovery are overexpression of genes and introducing exogenous DNA by constructing transgenic algal strains (Cheng et al., 2019). Interestingly, it is worth mentioning that numerous authors have discussed and studied the introduction of foreign DNA fragments to different organisms for the purpose of increasing their heavy metal tolerance, such as plants and bacteria; only few have highlighted this genetic manipulation strategy for microalgae. One such example in bacteria comes from (Sriprang et al., 2003), who introduced genes for phytochelating synthase (PCSAt) in Mesorhizobium huakuiiwhich. PCSAt is a protease-like enzyme that catalyzes the synthesis of peptides, which in turn chelate metals (Rigouin et al., 2013). It was reported that the transformation resulted in a transgenic strain that accumulated Cd+2 9–19-fold more than the strain that did not contain the PCSAt. In another study, ACC deaminase and iaaM genes were introduced into the Petunia hybrida Vilm plant via agrobacterium-mediated transformation, and the transgenic plants were continuously treated with Copper (II) sulfate CuSO4 and Cobalt (II) chloride CoCl2 to test for heavy metal tolerance (Zhang et al., 2008). The authors planted the transgenic Petunia in heavily heavy metal contaminated soil and found that the mutant plant had double the growth of the wild-type plant. Also, they grew bigger, healthier, and faster in the heavy metal contaminated soil, most likely related to the increased tolerance to cobalt that resulted from the co-expression of both introduced genes. Other genes that could potentially improve heavy metal tolerance and accumulation were identified by (Peng et al., 2020) in the plant Kandelia obovate. This study showed that KoCBF1 and KoCBF3 genes were highly expressed in the presence of lead (Pb (NO3)2), implying that they are involved in growth and heavy metal accumulation. One of the few examples found in literature demonstrated that wild-type C. reinhardtii could not survive in the presence of Cadmium (Cd) in the cultivation media, whereas a transgenic strain with high expression level of the CrMTP4 gene was able to grow well under the metal stress (Ibuot et al., 2017).
3.3.2 Hydrocarbon degradation
It is well known that the composition of wastewater, including PW, varies greatly based on its origin, also applying to the level of TOC, including hydrocarbons. As an example, TOC in PW collected from a petroleum industrial site in Qatar was found to be 720.33 mg L−1 (Das et al., 2019), however, values of up to 2430 mg L−1 have also been reported (Shaikh et al., 2020).
Microbial degradation of hydrocarbons is a common phenomenon, although its applications in bioremediation are still limited (Ławniczak et al., 2020). These microorganisms can however be the source of relevant genes that are in control of hydrocarbon degradation and organic carbon digestion which could be applied in microalgae. For example (Luo et al., 2015), constructed the pCom8 vector to express alkane hydroxylase in Escherichia. coli (E. coli) DH5α, after which it was inoculated in diesel containing media to induce gene expression and perform biodegradation assays. Furthermore, applying a consortium of Acinetobacter and the transgenic E. coli strain improved diesel biodegradation by up to 49% compared to the control. Another study conducted by (Kang et al., 2017) cloned a two-component flavin-diffusible monooxygenase gene (cph) from Arthrobacter chlorophenolicus for enzyme overexpression. This enzyme is responsible for the degradation and removal of 4-chlorophenol (4-CP) and transformed strains could remove up to 82.7% of 4-CP from the media.
Besides the introduction of genes involved in hydrocarbon degradation, increasing biosurfactant production to aid degradation can be of great interest (Ochsner et al., 1994). Several microalgae and cyanobacteria were tested for their capability of producing biosurfactant. Dunaliella salina and Porphyridium cruentum, two marine microalgal species that produce extracellular polymeric substances that can be used as emulsifiers to metabolize oil hydrocarbons (Sukla et al., 2019). Also (Radmann et al., 2015), stated that Arthrospira sp. and other cyanobacteria and microalgae can produce biosurfactant as a by-product in the presence of certain organic carbon. In this context, genetic engineering can be helpful to increase the potential of microalgae to secrete biosurfactant. Numerous research efforts were made to understand their production on the molecular level, and the different genes involved in enhancing it were discovered. For instance, the Emt1, Mmc1, Mac1, Mac2 genes in Ustilago maydis were studied for their association with the expression of Mannosylerythritol Lipids (MELs), which are class of biosurfactant (Markande et al., 2021). These lipids are also expressed in different microalgae (Luca et al., 2021). Moreover, using wastewater as a substrate for Pseudozyma tsukubaensis made it possible to successfully increase the production of MELs (Andrade et al., 2017).An overview of these and other reported genes that influence heavy metal and hydrocarbon degradation is given in Table 2. Using gene-editing technologies like CRISPR to apply such genes to microalgae could potentially increase the feasibility of using it for produced water treatment.
TABLE 2.
Candidate genes for heavy metal tolerance and accumulation and hydrocarbon degradation.
| Gene | Plasmid | Promotor | Application | Reference |
|---|---|---|---|---|
| PCS At | pBBR1MCS-2 pMP220 | nifH | Accumulation of Cd+2 | Sriprang et al. (2003) |
| KoCBF3 | Accumulation of Pb | Peng et al. (2020) | ||
| ACC deaminase | pBI-iaaM/ACC | CaMV 35S | Accumulation of copper and cobalt | Zhang et al. (2008) |
| iaaM | pBI-iaaM | GRP | Accumulation of copper and cobalt | Zhang et al. (2008) |
| CrMTP4 | CrMTP4gDNA-pH2GW7 | Not mentioned | Increase Mn and Cd content in the cell | Ibuot et al. (2017) |
| alkane hydroxylase (alkB) | pCom8 | Not mentioned | Degrading diesel oil | Luo et al. (2015) |
| cph | pET-24a | Not mentioned | Removal of 4-chlorophenol | Kang et al. (2017) |
| mat1 | pET15b | — | Biosurfactant | Hewald et al. (2006) |
In summation, microalgae can ideally contribute to the bioremediation of heavily contaminated water with heavy metals or organic pollutants. Owing to its fast adaptability to use these contaminants to thrive. Additionally, the role of genetic engineering to develop and understand the mechanism in which microalgae consume such pollutants cannot be ignored. Thus, to overcome the growing environmental threat of produced water on the sustainable development; an increased focused research is acquired to strengthen the large-scale use of genetically engineered microalgae and to fill the knowledge gap in this field.
3.4 Challenges and limitations of genetic engineering for enhancing bioremediation efficiencies
Many water treatment methods depend on using microbes that degrade pollutants. Recently, there has been a great focus on genetically modifying such microorganisms to augment their ability for bioremediation. Nevertheless, there are limitations and challenges linked to redesign microorganisms’ DNA related to gene expression efficiency and the stability of introduced genes according to (Fajardo et al., 2020) and (Tran and Kaldenhoff, 2020); the success of genetic transformation is highly dependent on the species selection. Some diatoms and microalgal species are known for their low stability after nuclear transformation such as Thalassiosira weissflogii, Ulva lactuca, and Gracilaria changii (Fajardo et al., 2020). Another major concern is the presence of off-target effects of CRISPR/Cas9(Zhang et al., 2015). Additionally, genome-editing techniques can be very expensive and involve complex procedures with some technical challenges, such as TALEN (Khan, 2019). Furthermore, in order to manipulate the genome of microalgae to improve its removal of a specific contaminant, such as heavy metals, a complete understanding of the cells' metabolism and structure during metal stress is required to ensure the maximum effectiveness of those engineered cells for bioremediation (Ranjbar and Malcata, 2022).
Other limitations of using genetically enhanced microalgae on a large-scale, include environmental concerns and issues related to public health (Janssen and Stucki, 2020; Kumar et al., 2020). Thus, the environmental impact assessments and other assessments that comprise biosafety must be performed prior to using a genetically modified organism (GMO) (Sayler and Ripp, 2000). Nevertheless, several successful studies were conducted about releasing transgenic organisms into the environment. For instance, the field trials of genetically engineered mosquitos in North America (Neuhaus, 2018), and the release of Pseudomonas fluorescens HK44 in a controlled field in the US (Ripp et al., 2000). Also (De Leij et al., 1995), spread transgenic Pseudomonas fluorescens in a wheat field in 1995. Using GMOs in bioremediation poses a risk of horizontal gene transfer as well as regulatory and ecological complications (Singh et al., 2011).
Thus, even though this technology has the potential for future use, its sustainability and large-scale use are still in question. Optimization for a more sustainable use on a large scale should be considered. Finally, microalgae have great potential for many biotechnological applications, but to date, the development and improvement of industrial production using CRISPR/Cas9 or other biotechnologies has yet to be conducted and tested for feasible and satisfying outcomes.
4 Conclusion and recommendations
This review discussed the potential of using the genetic engineering tools ZFNs, TALENs, and CRISPR/Cas9 to manipulate the genome of microalgae. It is believed that they have a great opportunity to improve the tolerance of microalgae to toxic mediums like the produced water. As well as to enhance its ability to remove existing contaminants. Deeper investigation of these technologies is crucial the potential genes and understand their mechanism and function for bioremediation. However, CRISPR/Cas9, the latest and most promising tool for genome modification is believed to hold applicational advantages over the other technologies as it offers more stability to the introduced gene; and it consumes less time and effort with fewer technical issues.
Therefore, it is highly recommended to extend the research on CRISPR/Cas9 application to serve the purpose of bioremediation of produced water using microalgae given that they are efficient and economically feasible candidate to make the PW reusable and lower its negative impact on the environment. Finally, more research effort is required to overcome the problems that arise when CRISPR/Cas9 is being applied such as the off-target effects.
Funding Statement
This work was supported by ExxonMobil funding (QUEX-CAS-EMRQ-20/21). The findings herein reflect the work and are solely the responsibility of the authors.
Author contributions
AH: Writing-original draft preparation, writing- reviewing and editing. IS: Supervision, conceptualization, writing-reviewing and editing; KS: Writing- reviewing and editing, project administration; SA-M and TD: Writing- reviewing and editing. MA and SS: Conceptualization and writing- reviewing and editing. HA-J: Conceptualization, writing- reviewing and editing, and funding acquisition.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CCU, carbon capture and utilization; Cd, Cadmium; CO2, carbon dioxide; CoCl2, Cobalt (II) chloride; COD, chemical oxygen demand; CRISPR, clustered regularly interspaced short palindromic repeats; CuSO4, Copper (II) sulfate; EPS, extracellular polysaccharides; GMO, genetically modified organism; INDELs, insertions or deletions; NORM, naturally occurring radioactive material; PW, Produced water; RNPs, ribonucleoproteins; TALEN, Transcription activator-like effector nuclease; TDS, total dissolved solids; TN, Total Nitrogen; TOC, Total Organic Carbon; UV, ultraviolet; ZFN, Zinc finger nuclease.
References
- Ahmad A., Banat F., Alsafar H., Hasan S. W. (2022). Algae biotechnology for industrial wastewater treatment, bioenergy production, and high-value bioproducts. Sci. Total Environ. 806, 150585. 10.1016/j.scitotenv.2021.150585 [DOI] [PubMed] [Google Scholar]
- Ahmad I., Abdullah N., Koji I., Yuzir A., Mohamad S. E. (2021). Potential of microalgae in bioremediation of wastewater. Bull. Chem. React. Eng. 16, 413–429. 10.9767/bcrec.16.2.10616.413-429 [DOI] [Google Scholar]
- Ahmad I., Abdullah N., Koji I., Yuzir A., Mohamad S. E. (2021). Potential of microalgae in bioremediation of wastewater. Bull. Chem. React. Eng. Catal. 16, 413–429. 10.9767/bcrec.16.2.10616.413-429 [DOI] [Google Scholar]
- Al-Ghouti M. A., Al-Kaabi M. A., Ashfaq M. Y., Da’na D. A. (2019). Produced water characteristics, treatment and reuse: A review. J. Water Process Eng. 28, 222–239. 10.1016/j.jwpe.2019.02.001 [DOI] [Google Scholar]
- Al-Jabri H., Das P., Khan S., Thaher M., Abdulquadir M. (2021). Treatment of wastewaters by microalgae and the potential applications of the produced biomass—A review. WaterSwitzerl. 13, 27. 10.3390/w13010027 [DOI] [Google Scholar]
- Alalawy A. I. A., Sh Alabdraba W. M., Omer E. A. (2019). Nutrients and organic matters removal of ospitals wastewater by microalgae. J. Phys. Conf. Ser. 1294, 072002. 10.1088/1742-6596/1294/7/072002 [DOI] [Google Scholar]
- Alsarayreh M., Almomani F., Khraisheh M., Nasser M. S., Soliman Y. (2022). Biological-based produced water treatment using microalgae: Challenges and efficiency. Sustainability 14 (1), 499. 10.3390/su14010499 [DOI] [Google Scholar]
- Ammar S. H., Khadim H. J., Isam A. (2018). Cultivation of Nannochloropsis oculata and Isochrysis galbana microalgae in produced water for bioremediation and biomass production. Environ. Technol. Innovation 10, 132–142. 10.1016/j.eti.2018.02.002 [DOI] [Google Scholar]
- Andrade C. J., de B, L. M. de A., Rocco S. A., Sforça M. L., Pastore G. M., Jauregi P. (2017). A novel approach for the production and purification of mannosylerythritol lipids (MEL) by Pseudozyma tsukubaensis using cassava wastewater as substrate. Sep. Purif. Technol. 180, 157–167. 10.1016/j.seppur.2017.02.045 [DOI] [Google Scholar]
- Arias D. M., Uggetti E., García J. (2020). Assessing the potential of soil cyanobacteria for simultaneous wastewater treatment and carbohydrate-enriched biomass production. Algal Res. 51, 102042. 10.1016/j.algal.2020.102042 [DOI] [Google Scholar]
- Arora N., Philippidis G. P. (2021). Microalgae strain improvement strategies: Random mutagenesis and adaptive laboratory evolution. Trends Plant Sci. 26, 1199–1200. 10.1016/j.tplants.2021.06.005 [DOI] [PubMed] [Google Scholar]
- Arora N., Yen H.-W., Philippidis G. P. (2020). Harnessing the power of mutagenesis and adaptive laboratory evolution for high lipid production by oleaginous microalgae and yeasts. Sustainability 12, 5125. 10.3390/su12125125 [DOI] [Google Scholar]
- Asadian M., Saadati M., Bajestani F. B., Beardall J., Abdolahadi F., Mahdinezhad N. (2022). Knockout of Cia5 gene using CRISPR/Cas9 technique in Chlamydomonas reinhardtii and evaluating CO2 sequestration in control and mutant isolates. J. Genet. 101, 6. 10.1007/s12041-021-01350-x [DOI] [PubMed] [Google Scholar]
- Baek K., Kim D. H., Jeong J., Sim S. J., Melis A., Kim J.-S., et al. (2016). DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci. Rep. 6, 30620. 10.1038/srep30620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajhaiya A. K., Ziehe Moreira J., Pittman J. K. (2017). Transcriptional engineering of microalgae: Prospects for high-value chemicals. Trends Biotechnol. 35, 95–99. 10.1016/j.tibtech.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Balzano S., Sardo A., Blasio M., Chahine T. B., Dell’Anno F., Sansone C., et al. (2020). Microalgal metallothioneins and phytochelatins and their potential use in bioremediation. Front. Microbiol. 11, 517. 10.3389/fmicb.2020.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortesi L., Fischer R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. 10.1016/j.biotechadv.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Cabanelas I. T. D., van der Zwart M., Kleinegris D. M. M., Wijffels R. H., Barbosa M. J. (2016). Sorting cells of the microalga Chlorococcum littorale with increased triacylglycerol productivity. Biotechnol. Biofuels 9, 183. 10.1186/s13068-016-0595-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-delgado I. C., Mora-solarte D. A., Velasco-Santamaría Y. M. (2019). Physiological and enzymatic responses of Chlorella vulgaris exposed to produced water and its potential for bioremediation. Environ. Monit. Assess. 191, 399. 10.1007/s10661-019-7519-8 [DOI] [PubMed] [Google Scholar]
- Chang K. S., Kim J., Park H., Hong S. J., Lee C. G., Jin E. S. (2020). Enhanced lipid productivity in AGP knockout marine microalga Tetraselmis sp. using a DNA-free CRISPR-Cas9 RNP method. Bioresour. Technol. 303, 122932. 10.1016/j.biortech.2020.122932 [DOI] [PubMed] [Google Scholar]
- Cheng S. Y., Show P., Lau F., Chang J., Ling T. C. (2019). New prospects for modified algae in heavy metal adsorption. Trends Biotechnol. 37, 1255–1268. 10.1016/j.tibtech.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Cho S., Luong T. T., Lee D., Oh Y. K., Lee T. (2011). Reuse of effluent water from a municipal wastewater treatment plant in microalgae cultivation for biofuel production. Bioresour. Technol. 102, 8639–8645. 10.1016/j.biortech.2011.03.037 [DOI] [PubMed] [Google Scholar]
- Cordero B. F., Obraztsova I., Couso I., Leon R., Vargas M. A., Rodriguez H. (2011). Enhancement of lutein production in Chlorella sorokiniana (chorophyta) by improvement of culture conditions and random mutagenesis. Mar. Drugs 9, 1607–1624. 10.3390/md9091607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F., Leduc S., Maréchal A., Dubois G., Guyot V., Perez-Michaut C., et al. (2014). Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 5, 3831–3837. 10.1038/ncomms4831 [DOI] [PubMed] [Google Scholar]
- D’Alessandro E. B., Antoniosi Filho N. R. (2016). Concepts and studies on lipid and pigments of microalgae: A review. Renew. Sustain. Energy Rev. 58, 832–841. 10.1016/j.rser.2015.12.162 [DOI] [Google Scholar]
- Das P., AbdulQuadir M., Thaher M., Khan S., Chaudhary A. K., Alghasal G., et al. (2019). Microalgal bioremediation of petroleum-derived low salinity and low pH produced water. J. Appl. Phycol. 31, 435–444. 10.1007/s10811-018-1571-6 [DOI] [Google Scholar]
- De Leij F. A. A. M., Sutton E. J., Whipps J. M., Fenlon J. S., Lynch J. M. (1995). Field release of a genetically modified Pseudomonas fluorescens on wheat: Establishment, survival and dissemination. Bio/Technology 13, 1488–1492. 10.1038/nbt1295-1488 [DOI] [Google Scholar]
- Devi N. D., Sun X., Ding L., Goud V. V., Hu B. (2022). Mixotrophic growth regime of novel strain Scenedesmus sp. DDVG I in municipal wastewater for concomitant bioremediation and valorization of biomass. J. Clean. Prod. 365, 132834. 10.1016/j.jclepro.2022.132834 [DOI] [Google Scholar]
- Deviram G., Mathimani T., Anto S., Shan T., Arul D., Pugazhendhi A. (2020). Applications of microalgal and cyanobacterial biomass on a way to safe, cleaner and a sustainable environment. J. Clean. Prod. 253, 119770. 10.1016/j.jclepro.2019.119770 [DOI] [Google Scholar]
- Doan T. T. Y., Obbard J. P. (2012). Enhanced intracellular lipid in Nannochloropsis sp. via random mutagenesis and flow cytometric cell sorting. Algal Res. 1, 17–21. 10.1016/j.algal.2012.03.001 [DOI] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- El-Kassas H. Y., Mohamed L. A. (2014). Bioremediation of the textile waste effluent by Chlorella vulgaris. Egypt. J. Aquatic Res. 40, 301–308. 10.1016/j.ejar.2014.08.003 [DOI] [Google Scholar]
- Fajardo C., De Donato M., Carrasco R., Martínez-Rodríguez G., Mancera J. M., Fernández-Acero F. J. (2020). Advances and challenges in genetic engineering of microalgae. Rev. Aquac. 12, 365–381. 10.1111/raq.12322 [DOI] [Google Scholar]
- Feng S., Hu L., Zhang Q., Zhang F., Du J., Liang G., et al. (2020). CRISPR/Cas technology promotes the various application of Dunaliella salina system. Appl. Microbiol. Biotechnol. 104, 8621–8630. 10.1007/s00253-020-10892-6 [DOI] [PubMed] [Google Scholar]
- Gaj T., Gersbach C. A., Barbas C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. 10.1016/j.tibtech.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimpel J. A., Henríquez V., Mayfield S. P. (2015). In metabolic engineering of eukaryotic microalgae: Potential and challenges come with great diversity. Front. Microbiol. 6, 1376. 10.3389/fmicb.2015.01376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grama S. B., Liu Z., Li J. (2022). Emerging trends in genetic engineering of microalgae for commercial applications. Mar. Drugs 20, 285. 10.3390/md20050285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. (2020). “Reuse of produced water in the oil and gas industry,” in SPE International Conference and Exhibition on Health, Safety, Environment and Sustainability SPE-199498-MSy, England, July 20 2020. [Google Scholar]
- Gray S. N. (1998). Fungi as potential bioremediation agents in soil contaminated with heavy or radioactive metals. Biochem. Soc. Trans. 26, 666–670. 10.1042/bst0260666 [DOI] [PubMed] [Google Scholar]
- Greiner A., Kelterborn S., Evers H., Kreimer G., Sizova I., Hegemann P. (2017). Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29, 2498–2518. 10.1105/tpc.17.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. M., Musunuru K. (2014). Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-cas9. J. Clin. Investigation 124, 4154–4161. 10.1172/JCI72992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H. A., Pang J. K. S., Soh B. S. (2020). Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 98, 615–632. 10.1007/s00109-020-01893-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Chen W., Amato A., Jouhet J., Maréchal E., Moog D., et al. (2022). Multiplexed CRISPR/Cas9 editing of the long‐chain acyl‐CoA synthetase family in the diatom Phaeodactylum tricornutum reveals that mitochondrial ptACSL3 is involved in the synthesis of storage lipids. New Phytol. 233, 1797–1812. 10.1111/nph.17911 [DOI] [PubMed] [Google Scholar]
- Hao X., Luo L., Jouhet J., Rébeillé F., Maréchal E., Hu H., et al. (2018). Enhanced triacylglycerol production in the diatom Phaeodactylum tricornutum by inactivation of a Hotdog-fold thioesterase gene using TALEN-based targeted mutagenesis. Biotechnol. Biofuels 11, 312–319. 10.1186/s13068-018-1309-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewald S., Linne U., Scherer M., Marahiel M. A., Bo M., Bölker M. (2006). Identification of a gene cluster for biosynthesis of mannosylerythritol lipids in the basidiomycetous fungus Ustilago maydis. Appl. Environ. Microbiol. 72, 5469–5477. 10.1128/AEM.00506-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins T. C., Graham E. J. S., Schuler A. J. (2019). Biomass and lipid productivity of Dunaliella tertiolecta in a produced water-based medium over a range of salinities. J. Appl. Phycol. 31, 3349–3358. 10.1007/s10811-019-01836-3 [DOI] [Google Scholar]
- Hu L., Feng S., Liang G., Du J., Li A., Niu C. (2021). CRISPR/Cas9-induced β-carotene hydroxylase mutation in Dunaliella salina CCAP19/18. Amb. Express 11, 83. 10.1186/s13568-021-01242-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Chen C., Shen Q., Rosen B. P., Zhao F.-J. (2015). Genetically engineering Bacillus subtilis with a heat-resistant arsenite methyltransferase for bioremediation of arsenic-contaminated organic waste. Appl. Environ. Microbiol. 81, 6718–6724. 10.1128/AEM.01535-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibuot A., Dean A. P., McIntosh O. A., Pittman J. K. (2017). Metal bioremediation by CrMTP4 over-expressing Chlamydomonas reinhardtii in comparison to natural wastewater-tolerant microalgae strains. Algal Res. 24, 89–96. 10.1016/j.algal.2017.03.002 [DOI] [Google Scholar]
- Jabalameli H. R., Zahednasab H., Karimi-Moghaddam A., Jabalameli M. R. (2015). Zinc finger nuclease technology: Advances and obstacles in modelling and treating genetic disorders. Gene 558, 1–5. 10.1016/j.gene.2014.12.044 [DOI] [PubMed] [Google Scholar]
- Janssen D. B., Stucki G. (2020). Perspectives of genetically engineered microbes for groundwater bioremediation. Environ. Sci. Process. Impacts 22, 487–499. 10.1039/c9em00601j [DOI] [PubMed] [Google Scholar]
- Japar A. S., Takriff M. S., Mohd Yasin N. H. (2021). Microalgae acclimatization in industrial wastewater and its effect on growth and primary metabolite composition. Algal Res. 53, 102163. 10.1016/j.algal.2020.102163 [DOI] [Google Scholar]
- Jayakumar S., Bhuyar P., Pugazhendhi A., Rahim M. H. A., Maniam G. P., Govindan N. (2021). Effects of light intensity and nutrients on the lipid content of marine microalga (diatom) Amphiprora sp. for promising biodiesel production. Sci. Total Environ. 768, 145471. 10.1016/j.scitotenv.2021.145471 [DOI] [PubMed] [Google Scholar]
- Jeon M. S., Han S.-I., Jeon M., Choi Y.-E. (2021). Enhancement of phycoerythrin productivity in Porphyridium purpureum using the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 ribonucleoprotein system. Bioresour. Technol. 330, 124974. 10.1016/j.biortech.2021.124974 [DOI] [PubMed] [Google Scholar]
- Jeon S., Lim J. M., Lee H. G., Shin S. E., Kang N. K., Park Y. Il, et al. (2017). Current status and perspectives of genome editing technology for microalgae. Biotechnol. Biofuels 10, 267. 10.1186/s13068-017-0957-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Brueggeman A. J., Horken K. M., Plucinak T. M., Weeks D. P. (2014). Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 13, 1465–1469. 10.1128/EC.00213-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra R., Gaur S., Goel M. (2021). Microalgae bioremediation: A perspective towards wastewater treatment along with industrial carotenoids production. J. Water Process Eng. 40, 101794. 10.1016/j.jwpe.2020.101794 [DOI] [Google Scholar]
- Kang C., Yang J. W., Cho W., Kwak S., Park S., Lim Y., et al. (2017). Oxidative biodegradation of 4-chlorophenol by using recombinant monooxygenase cloned and overexpressed from Arthrobacter chlorophenolicus A6. Bioresour. Technol. 240, 123–129. 10.1016/j.biortech.2017.03.078 [DOI] [PubMed] [Google Scholar]
- Kaur S., Roy A. (2021). Bioremediation of heavy metals from wastewater using nanomaterials. Environ. Dev. Sustain. 23, 9617–9640. 10.1007/s10668-020-01078-1 [DOI] [Google Scholar]
- Khan S. H. (2019). Genome-editing technologies: Concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther. - Nucleic Acids 16, 326–334. 10.1016/j.omtn.2019.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G., Shekh A., Jakhu S., Sharma Y., Kapoor R. (2020). Bioengineering of microalgae: Recent advances, perspectives, and regulatory challenges for industrial application. Front. Bioeng. Biotechnol. 8, 914. 10.3389/fbioe.2020.00914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T., Moroi K., Iwai M., Okazaki K., Shimizu S., Nomura S., et al. (2020). Efficient and multiplexable genome editing using Platinum TALENs in oleaginous microalga, Nannochloropsis oceanica NIES-2145. Genes Cells 25, 695–702. 10.1111/gtc.12805 [DOI] [PubMed] [Google Scholar]
- Lai Y.-P., Huang J., Wang L.-F., Jun Li Z.-R. W., Wu Z. R. (2004). A new approach to random mutagenesis in vitro . Biotechnol. Bioeng. 86, 622–627. 10.1002/bit.20066 [DOI] [PubMed] [Google Scholar]
- Ławniczak Ł., Woźniak‐Karczewska M., Loibner A. P., Heipieper H. J., Chrzanowski Ł. (2020). Microbial degradation of hydrocarbons—basic principles for bioremediation: A review. Molecules 25 (4), 856. 10.3390/molecules25040856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Rudolph P., Miller S. M., Li Y. (2022). Genetic compensation of triacylglycerol biosynthesis in the green microalga Chlamydomonas reinhardtii . Plant J. 111, 1069–1080. 10.1111/tpj.15874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite L. de S., Hoffmann M. T., Daniel L. A. (2019). Microalgae cultivation for municipal and piggery wastewater treatment in Brazil. J. Water Process Eng. 31, 100821–100827. 10.1016/j.jwpe.2019.100821 [DOI] [Google Scholar]
- Leng L., Wei L., Xiong Q., Xu S., Li W., Lv S., et al. (2020). Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere 238, 124680. 10.1016/j.chemosphere.2019.124680 [DOI] [PubMed] [Google Scholar]
- Li M., Zamyadi A., Zhang W., Dumée L. F., Gao L. (2022). Algae-based water treatment: A promising and sustainable approach. J. Water Process Eng. 46, 102630. 10.1016/j.jwpe.2022.102630 [DOI] [Google Scholar]
- Lin W. R., Ng I. S. (2020). Development of CRISPR/Cas9 system in Chlorella vulgaris FSP-E to enhance lipid accumulation. Enzyme Microb. Technol. 133, 109458. 10.1016/j.enzmictec.2019.109458 [DOI] [PubMed] [Google Scholar]
- Liu K., Chen R., Yang R., Chen Y., Zhu C., Tang Y., et al. (2022). “Genome editing approaches applied to microalgae-based fuels,” in 3rd generation biofuels (Netherlands: Elsevier; ), 47–64. 10.1016/B978-0-323-90971-6.00013-9 [DOI] [Google Scholar]
- Llavero‐Pasquina M., Geisler K., Holzer A., Mehrshahi P., Mendoza‐Ochoa G. I., Newsad S. A., et al. (2022). Thiamine metabolism genes in diatoms are not regulated by thiamine despite the presence of predicted riboswitches. New Phytol. 235, 1853–1867. 10.1111/nph.18296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca M. D., Pappalardo I., Limongi A. R., Viviano E., Radice R. P., Todisco S., et al. (2021). Lipids from microalgae for cosmetic applications. Cosmetics 8, 52. 10.3390/cosmetics8020052 [DOI] [Google Scholar]
- Luo Q., He Y., Hou D. Y., Zhang J. G., Shen X. R. (2015). GPo1 <italic>alkB</italic> gene expression for improvement of the degradation of diesel oil by a bacterial consortium. Braz. J. Microbiol. 46, 649–657. 10.1590/S1517-838246320120226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzahn A., Lowder L., Qi Y. (2017). Plant genome editing with TALEN and CRISPR. Cell & Biosci. 7, 21. 10.1186/s13578-017-0148-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manandhar-Shrestha K., Hildebrand M. (2013). Development of flow cytometric procedures for the efficient isolation of improved lipid accumulation mutants in a Chlorella sp. microalga. J. Appl. Phycol. 25, 1643–1651. 10.1007/s10811-013-0021-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markande A. R., Patel D., Varjani S. (2021). A review on biosurfactants: Properties, applications and current developments. Bioresour. Technol. 330, 124963. 10.1016/j.biortech.2021.124963 [DOI] [PubMed] [Google Scholar]
- Marques I. M., Oliveira A. C. V., de Oliveira O. M. C., Sales E. A., Moreira Í. T. A. (2021). A photobioreactor using Nannochloropsis oculata marine microalgae for removal of polycyclic aromatic hydrocarbons and sorption of metals in produced water. Chemosphere 281, 130775. 10.1016/j.chemosphere.2021.130775 [DOI] [PubMed] [Google Scholar]
- Mehariya S., Goswami R. K., Verma P., Lavecchia R., Zuorro A. (2021). Integrated approach for wastewater treatment and biofuel production in microalgae biorefineries. Energies 14, 2282. 10.3390/en14082282 [DOI] [Google Scholar]
- Molazadeh M., Ahmadzadeh H., Pourianfar H. R., Lyon S., Rampelotto P. H. (2019). The use of microalgae for coupling wastewater treatment with CO2 biofixation. Front. Bioeng. Biotechnol. 7, 42. 10.3389/fbioe.2019.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamangala Kanchiswamy C., Sargent D. J., Velasco R., Maffei M. E., Malnoy M. (2015). Looking forward to genetically edited fruit crops. Trends Biotechnol. 33, 62–64. 10.1016/j.tibtech.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Nain V., Sahi S., Verma A. (2010). CPP-ZFN: A potential DNA-targeting anti-malarial drug. Malar. J. 9, 258. 10.1186/1475-2875-9-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus C. P. (2018). Community engagement and field trials of genetically modified insects and animals. Hastings Cent. Rep. 48, 25–36. 10.1002/hast.808 [DOI] [PubMed] [Google Scholar]
- Ng I., Tan S., Kao P., Chang Y., Chang J. (2017). Recent developments on genetic engineering of microalgae for biofuels and bio-based chemicals. biotechnology 12, 1600644. 10.1002/biot.201600644 [DOI] [PubMed] [Google Scholar]
- Nguyen A. D., Kim D., Lee E. Y. (2020a). Unlocking the biosynthesis of sesquiterpenoids from methane via the methylerythritol phosphate pathway in methanotrophic bacteria, using α-humulene as a model compound. Metab. Eng. 61, 69–78. 10.1016/j.ymben.2020.04.011 [DOI] [PubMed] [Google Scholar]
- Nguyen H. T., Yoon Y., Ngo H. H., Jang A. (2021). The application of microalgae in removing organic micropollutants in wastewater. Crit. Rev. Environ. Sci. Technol. 51, 1187–1220. 10.1080/10643389.2020.1753633 [DOI] [Google Scholar]
- Nguyen T. H. T., Park S., Jeong J., Shin Y. S., Sim S. J., Jin E. (2020b). Enhancing lipid productivity by modulating lipid catabolism using the CRISPR-Cas9 system in Chlamydomonas. J. Appl. Phycol. 32, 2829–2840. 10.1007/s10811-020-02172-7 [DOI] [Google Scholar]
- Nymark M., Sharma A. K., Sparstad T., Bones A. M., Winge P. (2016). A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 6, 24951. 10.1038/srep24951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner U., Koch A., Fiechter A., Reiser J. (1994). Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa . J. Bacteriol. 176, 2044–2054. 10.1128/jb.176.7.2044-2054.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opeolu B. O., Bamgbose O., Arowolo T. A., Adetunji M. T. (2010). Utilization of biomaterials as adsorbents for heavy metals removal from aqueous matrices. Sci. Res. Essays 5, 1780–1787. 10.5897/SRE.9000983 [DOI] [Google Scholar]
- Parsy A., Sambusiti C., Baldoni-andrey P., Elan T., Périé F. (2020). Cultivation of Nannochloropsis oculata in saline oil & gas wastewater supplemented with anaerobic digestion effluent as nutrient source. Algal Res. 50, 101966. 10.1016/j.algal.2020.101966 [DOI] [Google Scholar]
- Patel V. K., Soni N., Prasad V., Sapre A., Dasgupta S., Bhadra B. (2019). CRISPR–Cas9 system for genome engineering of photosynthetic microalgae. Mol. Biotechnol. 61, 541–561. 10.1007/s12033-019-00185-3 [DOI] [PubMed] [Google Scholar]
- Pathak B., Gupta S., Verma R. (2018). “Biosorption and biodegradation of polycyclic aromatic hydrocarbons (PAHs) by microalgae,” in Green Adsorbents for pollutant removal environmental chemistry for a sustainable world. Editors Crini G., Lichtfouse E. (Cham: Springer International Publishing; ), 215–247. 10.1007/978-3-319-92111-2_7 [DOI] [Google Scholar]
- Peng Y. L., Wang Y. S., Fei J., Sun C. C. (2020). Isolation and expression analysis of two novel C-repeat binding factor (CBF) genes involved in plant growth and abiotic stress response in mangrove Kandelia obovata. Ecotoxicol. Lond. Engl. 29, 718–725. 10.1007/s10646-020-02219-y [DOI] [PubMed] [Google Scholar]
- Qi F., Wu D., Mu R., Zhang S., Xu X. (2018). Characterization of a microalgal UV mutant for CO 2 biofixation and biomass production. BioMed Res. Int. 2018, 1–8. 10.1155/2018/4375170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovits R., Jinkerson R. E., Darzins A., Posewitz M. C. (2010). Genetic engineering of algae for enhanced biofuel production. Eukaryot. Cell 9, 486–501. 10.1128/EC.00364-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmann E. M., Morais E. G. De, Oliveira C. F., Jorge A. V. C., Alberto J., Costa V. (2015). Microalgae cultivation for biosurfactant production. Afr. J. Microbiol. Res. 9, 2283–2289. 10.5897/AJMR2015.7634 [DOI] [Google Scholar]
- Rahman A., Agrawal S., Nawaz T., Pan S., Selvaratnam T. (2020). A review of algae-based produced water treatment for biomass and biofuel production. WaterSwitzerl. 12 (9), 2351. 10.3390/W12092351 [DOI] [Google Scholar]
- Rahman A., Pan S., Houston C., Selvaratnam T. (2021). Evaluation of galdieria sulphuraria and chlorella vulgaris for the bioremediation of produced water. WaterSwitzerl. 13 (9), 1183. 10.3390/w13091183 [DOI] [Google Scholar]
- Rahmani A., Zerrouki D., Tabchouche A., Djafer L. (2022). Oilfield - produced water as a medium for the growth of Chlorella pyrenoidosa outdoor in an arid region. Environ. Sci. Pollut. Res. 29, 87509–87518. 10.1007/s11356-022-21916-1 [DOI] [PubMed] [Google Scholar]
- Ranjbar S., Malcata F. X. (2022). Is genetic engineering a route to enhance microalgae-mediated bioremediation of heavy metal-containing effluents? Molecules 27, 1473. 10.3390/molecules27051473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigouin C., Nylin E., Cogswell A. A., Schaumlöffel D., Dobritzsch D., Williams D. L. (2013). Towards an understanding of the function of the phytochelatin synthase of schistosoma mansoni. PLoS Neglected Trop. Dis. 7, e2037. 10.1371/journal.pntd.0002037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripp S., Nivens D. E., Ahn Y., Werner C., Jarrell J., Easter J. P., et al. (2000). Controlled field release of a bioluminescent genetically engineered microorganism for bioremediation process monitoring and control. Environ. Sci. Technol. 34, 846–853. 10.1021/es9908319 [DOI] [Google Scholar]
- Saadaoui I., Sedky R., Rasheed R., Bounnit T., Almahmoud A., Elshekh A., et al. (2019). Assessment of the algae-based biofertilizer influence on date palm (Phoenix dactylifera L.) cultivation. J. Appl. Phycol. 31, 457–463. 10.1007/s10811-018-1539-6 [DOI] [Google Scholar]
- Salgueiro J. L., Pérez L., Maceiras R., Sánchez A., Cancela A. (2016). Bioremediation of wastewater using chlorella vulgaris microalgae: Phosphorus and organic matter. Int. J. Environ. Res. 10, 465–470. 10.22059/ijer.2016.58766 [DOI] [Google Scholar]
- Sayler G. S., Ripp S. (2000). Field applications of genetically engineered microorganisms for bioremediation processes. Curr. Opin. Biotechnol. 11, 286–289. 10.1016/S0958-1669(00)00097-5 [DOI] [PubMed] [Google Scholar]
- Shahid A., Malik S., Zhu H., Xu J., Nawaz M. Z., Nawaz S., et al. (2020). Cultivating microalgae in wastewater for biomass production, pollutant removal, and atmospheric carbon mitigation; a review. Sci. Total Environ. 704, 135303. 10.1016/j.scitotenv.2019.135303 [DOI] [PubMed] [Google Scholar]
- Shaikh S. S., Abu-Dieyeh M. H., Al Naemi F. A., Ahmed T., Al-Ghouti M. A. (2020). Environmental impact of utilization of “produced water” from oil and gas operations in turfgrass systems. Sci. Rep. 10 (1), 15051. 10.1038/s41598-020-72069-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S.-E., Lim J.-M., Koh H. G., Kim E. K., Kang N. K., Jeon S., et al. (2016). CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci. Rep. 6, 27810. 10.1038/srep27810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokravi Z., Shokravi H., Chyuan O. H., Lau W. J., Koloor S. S. R., Petrů M., et al. (2020). Improving ‘lipid productivity’ in microalgae by bilateral enhancement of biomass and lipid contents: A review. Sustain. Switz. 12 (21), 9083. 10.3390/su12219083 [DOI] [Google Scholar]
- Singh A., Ummalyma S. B., Sahoo D. (2020). Bioremediation and biomass production of microalgae cultivation in river water contaminated with pharmaceutical effluent. Bioresour. Technol. 307, 123233. 10.1016/j.biortech.2020.123233 [DOI] [PubMed] [Google Scholar]
- Singh J. S., Abhilash P. C., Singh H. B., Singh R. P., Singh D. P. (2011). Genetically engineered bacteria: An emerging tool for environmental remediation and future research perspectives. Gene 480, 1–9. 10.1016/j.gene.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Sizova I., Greiner A., Awasthi M., Kateriya S., Hegemann P. (2013). Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 73, 873–882. 10.1111/tpj.12066 [DOI] [PubMed] [Google Scholar]
- Slattery S. S., Diamond A., Wang H., Therrien J. A., Lant J. T., Jazey T., et al. (2018). An expanded plasmid-based genetic toolbox enables Cas9 genome editing and stable maintenance of synthetic pathways in Phaeodactylum tricornutum . ACS Synth. Biol. 7, 328–338. 10.1021/acssynbio.7b00191 [DOI] [PubMed] [Google Scholar]
- Song I., Kim S., Kim J., Oh H., Jang J., Jeong S. J., et al. (2022). Macular pigment-enriched oil production from genome-edited microalgae. Microb. Cell Fact. 21, 27. 10.1186/s12934-021-01736-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., Zhai Q., Sun L., Huang E., Zhang Y., Zhu Y., et al. (2019). CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 103, 6919–6932. 10.1007/s00253-019-10007-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srimongkol P., Sangtanoo P., Songserm P., Watsuntorn W., Karnchanatat A. (2022). Microalgae-based wastewater treatment for developing economic and environmental sustainability: Current status and future prospects. Front. Bioeng. Biotechnol. 10, 904046. 10.3389/fbioe.2022.904046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriprang R., Hayashi M., Ono H., Takagi M., Hirata K., Murooka Y. (2003). Enhanced accumulation of Cd2+ by a Mesorhizobium sp. transformed with a gene from Arabidopsis thaliana coding for phytochelatin synthase. Appl. Environ. Microbiol. 69, 1791–1796. 10.1128/AEM.69.3.1791-1796.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukla L. B., Subudhi E., Pradhan D. (2019). The role of microalgae in wastewater treatment. Germany: Springer. [Google Scholar]
- Sullivan Graham E. J., Dean C. A., Yoshida T. M., Twary S. N., Teshima M., Alvarez M. A., et al. (2017). Oil and gas produced water as a growth medium for microalgae cultivation: A review and feasibility analysis. Algal Res. 24, 492–504. 10.1016/j.algal.2017.01.009 [DOI] [Google Scholar]
- Takahashi K., Ide Y., Hayakawa J., Yoshimitsu Y., Fukuhara I., Abe J., et al. (2018). Lipid productivity in TALEN-induced starchless mutants of the unicellular green alga Coccomyxa sp. strain Obi. Algal Res. 32, 300–307. 10.1016/j.algal.2018.04.020 [DOI] [Google Scholar]
- Tran N. T., Kaldenhoff R. (2020). Achievements and challenges of genetic engineering of the model green alga Chlamydomonas reinhardtii. Algal Res. 50, 101986. 10.1016/j.algal.2020.101986 [DOI] [Google Scholar]
- Varaprasad D., Narasimham D., Paramesh K., Sudha N. R., Himabindu Y., Keerthi Kumari M., et al. (2021). Improvement of ethanol production using green alga Chlorococcum minutum. Environ. Technol. 42, 1383–1391. 10.1080/09593330.2019.1669719 [DOI] [PubMed] [Google Scholar]
- Veiga M. C., Fontoura M. M., de Oliveira M. G., Costa J. A. V., Santos L. O. (2020). Magnetic fields: Biomass potential of spirulina sp. for food supplement. Bioprocess Biosyst. Eng. 43, 1231–1240. 10.1007/s00449-020-02318-4 [DOI] [PubMed] [Google Scholar]
- Wang Q., Gong Y., He Y., Xin Y., Lv N., Du X., et al. (2021). Genome engineering of Nannochloropsis with hundred‐kilobase fragment deletions by Cas9 cleavages. Plant J. 106, 1148–1162. 10.1111/tpj.15227 [DOI] [PubMed] [Google Scholar]
- Wang X.-W., Huang L., Ji P.-Y., Chen C.-P., Li X.-S., Gao Y.-H., et al. (2019). Using a mixture of wastewater and seawater as the growth medium for wastewater treatment and lipid production by the marine diatom Phaeodactylum tricornutum. Bioresour. Technol. 289, 121681. 10.1016/j.biortech.2019.121681 [DOI] [PubMed] [Google Scholar]
- Watanabe M. M., Isdepsky A. (2021). Biocrude oil production by integrating microalgae polyculture and wastewater treatment: Novel proposal on the use of deep water-depth polyculture of mixotrophic microalgae. Energies 14, 6992. 10.3390/en14216992 [DOI] [Google Scholar]
- Xu J., Soni V., Chopra M., Chan O. (2020a). Genetic modification of the HSP90 gene using CRISPR-cas9 to enhance thermotolerance in T. Suecica. Undergrad. Res. Nat. Clin. Sci. Technol. (URNCST) J. 4, 1–6. 10.26685/urncst.178 [DOI] [Google Scholar]
- Xu M., Weng Q., Ji J. (2020b). Applications and advances of CRISPR/Cas9 in animal cancer model. Briefings Funct. Genomics 19, 235–241. 10.1093/bfgp/elaa002 [DOI] [PubMed] [Google Scholar]
- Zhang S., Guo F., Yan W., Dai Z., Dong W., Zhou J., et al. (2020). Recent advances of CRISPR/Cas9-Based genetic engineering and transcriptional regulation in industrial biology. Front. Bioeng. Biotechnol. 7, 459. 10.3389/fbioe.2019.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-H., Tee L. Y., Wang X.-G., Huang Q.-S., Yang S.-H. (2015). Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 4, e264. 10.1038/mtna.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang H., Zhang B., Lin S. (2016). TALEN- and CRISPR-enhanced DNA homologous recombination for gene editing in zebrafish. Methods Cell Biol. 135, 107–120. 10.1016/bs.mcb.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Zhao L., Wang Y., Yang B., Chen S. (2008). Enhancement of heavy metal accumulation by tissue specific co-expression of iaaM and ACC deaminase genes in plants. Chemosphere 72, 564–571. 10.1016/j.chemosphere.2008.03.043 [DOI] [PubMed] [Google Scholar]
- Zhu L. D., Li Z. H., Hiltunen E. (2016). Strategies for lipid production improvement in microalgae as a biodiesel feedstock. BioMed Res. Int. 2016, 1–8. 10.1155/2016/8792548 [DOI] [PMC free article] [PubMed] [Google Scholar]