Abstract
Previously, we have shown that Helicobacter pylori can spontaneously and reversibly change its membrane lipid composition, producing variants with low or high content of lysophospholipids. The “lyso” variant contains a high percentage of lysophospholipids, adheres better to epithelial cells, and releases more proteins such as urease and VacA, compared to the “normal” variant, which has a low content of lysophospholipids. Prolonged growth of the normal variant at pH 3.5, but not under neutral conditions, leads to enrichment of lyso variant colonies, suggesting that the colony switch is relevant to acid adaptation. In this study we show that the change in membrane lipid composition is due to phase variation in the pldA gene. A change in the (C) tract length of this gene results in reversible frameshifts, translation of a full-length or truncated pldA, and the production of active or inactive outer membrane phospholipase A (OMPLA). The role of OMPLA in determining the colony morphology was confirmed by the construction of an OMPLA-negative mutant. Furthermore, variants with an active OMPLA were able to survive acidic conditions better than variants with the inactive form. This explains why the lyso variant is selected at low pH. Our studies demonstrate that phase variation in the pldA gene, resulting in an active form of OMPLA, is important for survival under acidic conditions. We also demonstrated the active OMPLA genotype in fresh isolates of H. pylori from patients referred to gastroscopy for dyspepsia.
Helicobacter pylori is one of the most common human pathogens. Infections with H. pylori are associated with chronic gastritis and its progression to gastric and duodenal ulcers. Infection with H. pylori is also a major risk for developing gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (16). Virulence factors such as urease, flagella, and adhesins appear to be necessary for the development of disease, but many elements of pathogenesis still remain unclear at the molecular level (31).
H. pylori colonizes the gastric surface of the human stomach. Colonization of the surface of the mucosa requires not only survival but also growth under acidic conditions. The metabolism of the organism must therefore be adapted to allow this to occur. McGowan et al. have found a gene (wbcJ) whose expression was induced after exposure to acidic pH (28). The wbcJ-negative mutant failed to express O antigen and was sensitive to acid stress. The authors of that study concluded that H. pylori might alter its lipopolysaccharide (LPS) structure in response to acidic pH (28). This suggests that changing the membrane composition is an adaptation facilitating H. pylori colonization of the acidic gastric environment.
We have previously reported that H. pylori can spontaneously and reversibly change its membrane lipid composition (11). The switch is associated with a dramatic change in the membrane phospholipids: from <2% lysophospholipids in one variant (“normal” [L] variant) to >50% lysophospholipids for the other variant (“lyso” [S] variant) (44). The lyso variant is more hemolytic, releases more urease and VacA, and adheres better to epithelial cells in vitro. Furthermore, prolonged growth at low pH results in an almost complete conversion to the lyso variant (11).
The appearance of lysophospholipids in bacteria is normally caused by the action of outer membrane phospholipase A (OMPLA) on the phospholipids (13, 38). OMPLA catalyzes the hydrolysis of acyl ester bonds in phospholipids to yield lysophospholipids and free fatty acids (Fig. 1). In growing bacterial cells, the OMPLA enzyme appears to be dormant (9), and the actual concentrations of lysophospholipids are usually very low (54). Regarding the phospholipid pattern in the lyso (S) variant compared to that in the normal (L) variant (44), we hypothesized that the L-to-S transition might be due to the activation of a PLA.
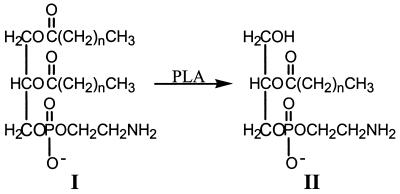
FIG. 1.
Structures of phospholipids. I, phosphatidylethanolamine; II, lysophosphatidylethanolamine made by the action of PLA.
Examination of the H. pylori 26695 and J99 genome sequences (1, 45) revealed the presence of a putative protein with homology to Escherichia coli OMPLA. Analysis of the reported DNA sequence revealed a homopolymer (C) tract in the HP0499 gene. Homopolymeric tracts enable phase variation in the following manner. During replication, nucleotide repeats may spontaneously become 1 bp shorter or longer through DNA slippage (55). This slippage results in shifts in the open reading frame leading to on-off switching of the associated gene products at the translational level. A phase variation in the C tract of the pldA gene could result in variable PLA activity. DNA slippage in poly(C) tracts causes phase variation in Helicobacter pylori α1,2- and α1,3-fucosyl transferases (2, 52). Meningococci are another example of microorganisms that use a reversible on-off change in the expression state of surface-associated components to choose between alternative lifestyles: commensal or pathogenic (10). However, a similar biological significant role of phase variation in H. pylori has not yet been found.
In this study we demonstrate that the reversible change in membrane lipid composition is due to phase variation in the HP0499 gene (pldA). Growth at low pH results in a selection of the best-adapted (S) phenotype and an almost complete conversion of normal (L) variant (C7, OMPLA “off”) to the lyso (S) variant (C8, OMPLA “on”). Consequently, this gene might contribute to survival of H. pylori at low pH.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
H. pylori was routinely cultured on 5% sheep blood agar plates for 3 days at 37°C in a microaerobic atmosphere with 5% O2, 10% CO2, and 85% N2. When appropriate, the blood agar plates were supplemented with kanamycin (30 mg/liter). For growth at acidic pH, the pH of the blood base was adjusted to the desired pH with HCl directly after sterilization and addition of blood.
As described earlier (11), isolate 17B/RH of H. pylori, a representative isolate displaying colony variation, was obtained from a patient with non-ulcer dyspepsia and maintained at −70°C. The 17B/RH formed two distinct colony forms, i.e., L and S variants. The parent L colony variant was designated 17L. Cultivation of 17L on blood plates at pH 7.4 resulted in spontaneous switches to S variant colonies designated 17S. A backswitch variant from 17S to L morphology was designated 17Lr. A S colony variant (17Si) was produced by growing 17L for more than five passages on blood agar plates at pH 5 and restreaking the bacteria on blood agar plates at pH 7.4 (11). Restreaking at pH 7.4 was necessary to observe the change in colony morphology.
As control strains, three isolates from dyspeptic patients were used: 12/LH, 23/LH, and 25/LH. To avoid selection of a special variant on the culture medium, only bacteria from the first passage on blood agar plates were used for DNA isolation.
E. coli strains DH5α and 1793 were used as hosts for recombinant plasmids. E. coli strains were grown either in liquid or on solid Luria-Bertani (LB) medium. For selection, E. coli media were supplemented with either chloramphenicol (20 mg/liter) or kanamycin (50 mg/liter).
AFLP analysis.
The 17L and 17S variants originate from a clinical isolate. To confirm on the genetic level that S and L variants are clonally related, amplified fragment length polymorphism (AFLP) analysis was performed on the variants. The bacteria were grown for 3 days, and the DNA was isolated by the method of Boom et al. (6). The AFLP analysis was carried out as described by van Doorn et al. (50).
Extraction and TLC of phospholipids.
Lipid extracts of the bacterial variants were prepared by standard methods (23). Briefly, ca. 100 colonies were suspended in chloroform-methanol (2:1), the nonlipid material was removed by centrifugation, and the supernatant containing the lipids was washed once with water. The chloroform phase was used for thin-layer chromatography (TLC) analysis. TLC was performed on silica gel plates (Merck, Darmstadt, Germany) developed in chloroform-methanol-water (75:22:3). Relative amounts of phosphatidylethanolamine and lysophosphatidylethanolamine were estimated by staining with ninhydrin (23).
ELISA.
For enzyme-linked immunosorbent assays (ELISAs), polystyrene 96-well microtiter plates were coated at 7.5 × 106 CFU/ml with bacteria washed in phosphate-buffered saline (PBS), and the bacteria were tested for reactivity with monoclonal antibodies (MAbs) at 1 μg/ml as described previously (3). The MAbs used in this study are shown in Table 1.
TABLE 1.
MAbs used in this study
| MAb | Specificity | Isotypea | Sourceb | Reference(s) |
|---|---|---|---|---|
| 1E52 | Ley | IgM | R. Negrini | 32, 34 |
| 54.1F6A | Lex | IgM | G. J. van Dam | 47 |
| 4D2 | H type 1 | IgM | R. Negrini | 32, 34 |
| 6A5 | Core | IgG | P. Doig | 14 |
| 3H11 | Urease | IgM | R. Negrini | 33 |
IgM, immunoglobulin M; IgG, immunoglobulin G.
R. Negrini, Biotechnology Laboratory, General Hospital, Brescia, Italy; G. J. van Dam, Department of Parasitology, University of Leiden, Leiden, The Netherlands; P. Doig, Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia, Canada.
SDS-PAGE.
The size distribution of LPS molecules of 17L, 17S, and 17Lr was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining. Bacterial cells were first digested with proteinase K before fractionation by Tricine-SDS-PAGE (15% gel). Subsequently, the LPS was detected by silver staining (26, 49).
Phospholipase enzyme activity assay.
The various bacterial variants were harvested from blood agar plates in PBS and washed once. H. pylori cells of a 10-ml suspension with optical density at 650 nm (OD650) of 1 were collected by centrifugation and resuspended in 1 ml of lysis buffer (50 mM Tris, 2 mM EDTA [pH 8.3]). Cells were lysed at 0°C by brief sonication. Whole cells were removed by centrifugation for 20 s at 20,800 × g, and the cell envelopes were collected by centrifugation of the supernatant at 20,800 × g for 30 min. The pellet was resuspended in 500 μl of dilution buffer (25 mM Tris, 2 mM EDTA, 3.5 mM Triton X-100 [pH 8.3]). The substrate, [1-14C]palmitoyl-2-laurylcarbamoyl-oxy-sn-glycero-3-phosphocholine (specific activity, 200 dpm/nmol), was prepared by acylation of the corresponding lyso phospholipid via standard procedures (19). The radioactive substrate has a nonhydrolyzable carbamoyl bond rather than an ester bond on the sn-2 position, resulting in an exclusive measurement of PLA1 activity. To determine the optimal conditions for activity measurements, incubations without or with calcium at different pHs were performed. As a result, the OMPLA1 activity of the variants was assayed in a total volume of 500 μl of incubation buffer (50 mM Tris, 10 mM CaCl2, 0.4 mM Triton X-100 [pH 7.0]) containing 100 nmol of substrate together with 50 or 100 μl of cell envelope fraction. After 20 h of incubation at 37°C, the liberated 14C-labeled palmitic acid was extracted from the reaction mixture by a modified Dole extraction procedure (48), and radioactivity was determined in disintegrations per minute (dpm) by liquid scintillation spectrometry. The amount of total protein in the cell envelope fractions was determined as described previously (7). The activity was expressed as the dpm per microgram of total cell envelope protein.
DNA sequence determination.
The HP0499 genes from H. pylori strains 17L, 17S, 17Lr, and 17Si and the control strains 12/LH, 23/LH, and 25/LH were amplified by PCR, with PLA-F (TGTCCAATTCTTGGTATCTC) as the forward primer and PLA-R (ATGCGATAGGTATAGCCTAA) as the reverse primer, at an annealing temperature of 55°C. The 800-bp pldA fragments of 17L, 17S, and 17Lr were ligated in a pGEM-T vector (Promega, Madison, Wis.) and transformed into E. coli DH5α. The resulting plasmids were isolated by using QIAprep Spin Miniprep Kit (Qiagen GmbH, Hilden, Germany) and applied as templates in a sequencing reaction with the Thermo-Sequenase premixed cycle sequence kit (Amersham, Buckinghamshire, England) by using the standard M13 forward or reverse primers labeled with Texas red. Sequencing was performed on an Amersham Vistra 725 Sequencer. The 800-bp pldA PCR products of 17L, 17S, 17Si, 12/LH, 23/LH, and 25/LH were also sequenced directly by using ABI PRISM BigDye Terminator sequencing kit (PE Applied Biosystems, Foster City, Calif.) and the PCR primers. The sequencing products were analyzed by ABI PRISM 310 Genetic Analyzer (PE Applied Biosystems).
Creation of a pldA mutant.
The 800-bp 17S pldA PCR product was cut out from the pGEM-T vector by SacII and SpeI (New England Biolabs Inc., Beverly, Mass.) and ligated in a pBCα3 vector containing a kanamycin cassette (5), also cut with SacII and SpeI. The recombinant plasmid was transformed into E. coli ER1793 with selection on LB kanamycin plates. Uncut plasmid was naturally transformed into 17S with selection on blood agar kanamycin plates (53).
Survival of the variants at pH 3.5.
Aliquots of ca. 108 CFU of 17L and 17S were spread onto blood agar plates of pH 3.5 and incubated under microaerobic conditions at 37°C for 16 h. The bacteria were harvested from the plates and suspended in 100 μl of PBS. Aliquots (90 μl) at dilutions of 100, 10−1, and 10−2 were plated onto blood agar plates at pH 7.4. After 5 days of microaerobic incubation, the colonies were counted. As a control, the suspensions of 17L and 17S were plated directly onto blood agar plates at pH 7.4.
Surface urease expression of the variants at pH 5.
We have previously shown that 17S grown at pH 7 releases large amounts of urease into the media, whereas 17L at this pH retains most of the urease intracellularly (11). The release of urease at pH 5 was compared by colony blotting with anti-urease MAb 3H11 by the method described by Appelmelk et al. (3). Serial dilutions of 17L and 17S were plated onto blood agar plates at pH 5 and 7.4 and incubated under microaerobic conditions at 37°C for 3 days to yield single colonies. Colonies were transferred to a nitrocellulose filter and baked. The washed and blocked filters were incubated overnight with anti-urease MAb 3H11, diluted in blocking buffer-PBS with 0.05% Tween 80 (1:1) at a concentration of 1 μg/ml. The blots were then incubated with goat anti-mouse immunoglobulin M-peroxidase and developed as previously described (49). Subsequently, the blots of 17L and 17S were compared and scanned. The scanned blots were analyzed by using the quantitation software Quantity One (Bio-Rad, Hercules, Calif.).
Statistical methods.
Statistical comparisons of means of data were made by one-way analysis of variance by using the SPSS (Chicago, Ill.) Data Analysis program (SPSS for Windows). The level of significance was P = 0.01.
RESULTS
Characterization of variants.
AFLP data for the variants are shown in Fig. 2. Variants 17L and 17S display a 96% similarity confirming that these colony variants originate from one H. pylori clone. All colony variants are closely related to each other. Strain NCTC 11637 was used as a control and showed a similarity of ca. 40% to all variants.
FIG. 2.
AFLP pattern of H. pylori phase variants compared to type strain NCTC 11637, confirming that the variants originate from one H. pylori clone.
Analysis of LPS.
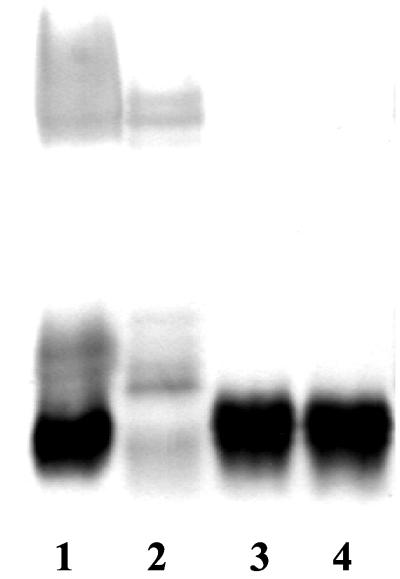
Examination of proteinase K-treated cells of the variants by ELISA was performed to assess possible differences in O-antigen expression. 17L was strongly positive for Ley and H type 1 expression; in contrast, 17S was negative for both of these determinants (Table 2). Analysis by SDS-PAGE confirmed that 17L displayed a smooth-type profile, whereas 17S displayed a rough-type profile in which the O-polysaccharide side chains were absent (Fig. 3). The backswitch variant (17Lr) continued to express the same serotype as 17S, indicating that the morpholgical change cannot be explained by an LPS variation. Cultivation in liquid media did not affect the LPS phenotype or serotype (data not shown).
TABLE 2.
ELISA reactivity of H. pylori phase variants with LPS MAbsa
| Phase variant | ELISA reactivitya with:
|
|||
|---|---|---|---|---|
| Anti-Ley | Anti-Lex | Anti-H type 1 | Anti-core | |
| 17L | +++ | + | ++ | − |
| 17Lr | − | − | − | +++ |
| 17S | − | − | − | +++ |
| 17Si | − | − | − | +++ |
Scores (OD492): −, <0.3; +, 0.3 to <1.3; ++, 1.3 to <2.3; +++, ≥2.3.
FIG. 3.
SDS-PAGE and silver staining of H. pylori phase variants. Lane 1, NCTC 11637; lane 2, 17L; lane 3, 17S; lane 4, 17Lr.
OMPLA activity.
In order to test the hypothesis that the differences in phospholipid composition of the variants were caused by the activity of an OMPLA, the variants were subjected to a phospholipase assay. No phospholipase activity was detected with whole cells of both S and L variants, indicating that the substrate was unable to enter the cell envelope and that PLA is not exposed. When the cells were sonicated, the H. pylori OMPLA activity could be detected only in the presence of Ca2+. Further measurements indicate that the OMPLA-catalyzed hydrolysis exhibited a pH optimum at ca. pH 7 (data not shown). The OMPLA activity of 17S (5.81 dpm/μg of protein, standard deviation [SD] = 0.481), a variant with a high content of lysophospholipid, is ca. 11 times higher than those of 17L (0.71 dpm/μg of protein, SD = 0.205) and 17Lr (0.42 dpm/μg of protein, SD = 0.140), variants with a low content of lysophospholipids. Statistical analysis showed that the differences in PLA activity between 17L or 17Lr and 17S was significant (P = 0.005) The activity of 17S was fivefold lower compared to E. coli (data not shown). The activity data confirmed our hypothesis that OMPLA is involved in the variation of membrane phospholipid composition.
Sequencing of pldA gene.
To determine whether the colony phase variation is due to C-tract length changes in HP0499, the pldA genes from 17L, 17S, 17Lr, and 17Si were sequenced. We found that 17L and 17Lr (i.e., normal variants) have a C7 tract that would lead to a truncated PLA. Variants 17S and 17Si (i.e., lyso variants) have a C8 tract that leads to a full-length and thus active PLA (Fig. 4). Sequencing of 17L and 17S were performed directly on the PCR product and after cloning into a p-GEMT vector; both methods yielded identical results.
FIG. 4.
Partial sequences. (A) Partial sequence of the pldA gene from H. pylori phase variants 17L/17Sr and 17S/Si. The change in ORF caused by the slipped-strand mispairing results in the translation of a truncated or full-length pldA. (B) Partial sequence of the pldA gene from H. pylori isolates 12/LH, 23/LH, and 25/LH compared to the colony phase variant 17S, showing that the gene is “on” in all isolates.
The pldA genes from the control strains 12/LH, 23/LH, and 25/LH were also sequenced and found to be “on” (i.e., C8 tract) (Fig. 4).
Construction of a pldA mutant.
To establish the role of OMPLA in colony morphology variation, HP0499 was mutated in 17S. The mutant (17SpldA) expressed the L-variant phenotype and did not produce detectable amounts of lysophospholipids. The PLA-negative mutant was also tested for phospholipase activity, which was found to be close to background values. Statistical analysis showed that the differences in PLA activity between 17S (5.81 dpm/μg of protein, SD = 0.481) and 17S pldA mutant (0.36 dpm/μg of protein, SD = 0.106) was significant (P = 0.005). This proves that pldA is responsible for the colony phase variation.
Survival of the variants at low pH.
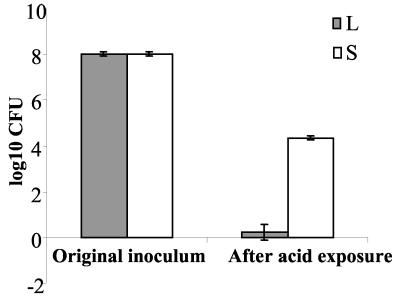
In order to explain the phase shift from 17L to 17Si on acid medium, the survival of the variants at low pH was studied. Both 17L and 17S cultivated at pH 7.4 gave good growth with the expected colony morphology. The L variant survived and grew at pH 5. However, no living bacteria from the aliquot of 108 L-variant bacteria exposed to pH 3.5 and replated at pH 7.4 were observed. The S variant, on the other hand, could survive an acid shock at pH 3.5 (Fig. 5).
FIG. 5.
Survival of H. pylori phase variants after one passage on blood agar plates at pH 3.5 incubated under microaerobic conditions at 37°C for 16 h. The bacteria were harvested at pH 3.5, plated onto blood agar plates (pH 7.4), and then counted after reincubation.
Expression of urease at low pH on the surface of the phase variants.
We finally studied the presence of colony-associated urease of the S and L variants at pH 5 semiquantitatively. The colonies from L and S variants grown at pH 5 were identical in size. Both variants reacted with the anti-urease MAb, but the reaction was 6.5 times stronger with the S variant than with the L variant. Statistical analysis showed that the differences in intensity (int) between 17S (258 int/mm2, SD = 13.6) and 17L (40 int/mm2, SD = 17.7) was significant (P = 0.005).
DISCUSSION
Genomic variation has been shown to be an important event in the interaction between bacteria and their hosts. This has been demonstrated in several pathogenic systems, including flagellar phase shift in Salmonella spp. and pilin variation in Neisseria gonorrhoeae (30). In Shigella sonnei the classically described variation from a smooth to a rough colony type is associated with a loss of virulence (39). The rough colony type is correlated with the loss of both the virulence-associated plasmid and the LPS side chains.
In the present study we have described a genomic variation in H. pylori that results in a major change in the bacterial phospholipid profile. The described phase shift, from L to S, occurs when the OMPLA becomes expressed in its active form. A change from C7 (17L and 17Lr) to C8 (17S and 17Si) in the C tract of HP0499 causes a shift in the reading frame of the gene and results in the production of a full-length and active OMPLA in 17S (Fig. 4). This was confirmed by measurement of OMPLA enzyme activity and phospholipid analysis. It has already been shown by Dorrell et al. (15) that insertional mutagenesis of pldA had no polar effects. The gene order in our strain is the same as in the one studied by Dorrell et al. (15; data not shown). As far as we are aware, phase variation on the translational level through C tracts has never been shown to have polar effects.
The human stomach is an environment of fluctuating acidity. The median luminal pH is ca. 1.4, but there are periods when this falls to ca. 1.0 or increases to as high as 6.0 (37). The C8 variant, with its rather uncommon phospholipid cell wall composition, seems to be better adapted to an acidic environment than is the C7 variant. To determine whether the C8 variant could be found in vivo, fresh H. pylori isolates from three consecutive patients referred to gastroscopy for dyspepsia were examined. Sequencing of the pldA gene from these isolates revealed C8 in the C tract of HP0499. This finding indicates that the lyso variant is not uncommon in vivo. However, the relationship between in vivo acid variation, clinical status (ulcer disease/nonulcer disease), and the state of the OMPLA requires a carefully designed epidemiological study.
OMPLA is widespread among gram-negative bacteria, and the amino acid sequence is rather well conserved (8). Crystal structures of the OMPLA enzyme isolated from E. coli have shown that OMPLA is a serine hydrolase with a unique Asn156-His142-Ser144 catalytic triad (13). The C tract of H. pylori pldA is located in the region between nucleotides encoding for Ser171 and amino acid 221 (H. pylori numbering corresponding to E. coli Ser144 and Asn156) (8). Thus, the length of the C tract decides whether or not the third residue of the catalytic triad will be formed. Interestingly, the studied 17RH strain has a glutamine in position 221 and not an asparagine. Mutation studies in E. coli have confirmed that Asp156, the third residue of the catalytic triad, is more tolerant to replacements than the essential residues Ser144 and His142, since an Asn156Gln mutant showed residual activity of ca. 3% (24). The protein sequence of our studied strain is similar to that reported for Hp26695 (45), whereas the optimal Asn221 was found in J99 (1). We observed that the OMPLA activity of 17S was fivefold lower compared to E. coli, and this finding can probably be explained by the glutamine in position 221. This suggests that other H. pylori strains might have a higher OMPLA activity than the strains studied here.
The presence of proteins homologous to OMPLA in gram-negative bacteria indicates that its function must be important for the bacteria (8). However, a clear physiological role for OMPLA has not been reported to date. The presence of OMPLA in membranes of other bacterial species is usually not associated with any detectable hydrolysis of the membrane lipids (9). Even recombinant strains that overproduce OMPLA do not show any phenotypic deviation relative to normal producing strains (21). E. coli OMPLA enzymatic activity is observed only after perturbation of the membrane by, for example, heat shock of the cells or by phage-induced lysis (9). The only report on phenotypic changes in relation to PLA describes envC mutation in E. coli, which leads to the modification of the outer membrane composition and thereby activation of OMPLA (43). Here we propose that H. pylori OMPLA is responsible for the degradation of membrane phospholipids observed in 17S and probably also for the increased hemolytic activity by release of lysophospholipids and fatty acids (18). The fatty acids and lysophospholipids destabilize the membrane bilayer and may thus facilitate the release of proteins (42). Our data suggest that PLA at pH 5 is involved in the process of urease release. An active OMPLA will degrade the bacterial cell wall, resulting either in autolysis or, less dramatically, in production of a leaky membrane, both circumstances culminating in the release of urease.
The change from L to S colonies seems to be independent of the LPS change. Bertram-Drogatz et al. (4) described a similar phenotypic variation in H. pylori, which these authors explained as a smooth-rough transition in the LPS. Phase variation in H. pylori LPS has been previously reported (3). An on-off switching of glucosyltransferase resulted in variants with smooth or rough LPS. The 17L variant had a smooth LPS and the 17S had a rough LPS, whereas after the backswitch from S to L (Lr) the rough LPS was retained. We do not understand yet why the switch from L to S morphology is always followed by a shortening of the LPS, but from our results we conclude that the observed difference in colony morphology cannot be explained by an LPS variation. It has been suggested that the LPS O antigen may provide a permeability barrier, reducing proton influx and thereby helping to maintain a periplasmic and intracellular pH within a nonlethal range (28). Hynes et al., on the other hand, have studied the influence of LPS composition on outer membrane permeability and found that it is the phosphorylation in the lipid A part of the LPS that contributes most to the permeability barrier (22). In contrast to McGowan et al. (28), our data show that variants with only core LPS can live through periods of acid exposure, indicating that survival at low pH is accomplished by other factors than LPS.
Urease activity is one of the most important factors for the acid tolerance of H. pylori (46). It has previously been postulated that surface urease might locally elevate the pH of the gastric microenvironment of H. pylori to allow survival at gastric pH (35). Krishnamurthy et al. have shown that H. pylori containing only cytoplasmic urease is susceptible to acid and is thus dependent on surface-localized urease for survival at low pH (25). Phadnis and coworkers have proposed a mechanism whereby urease is released as a result of autolysis of a fraction of bacteria and becomes adsorbed to the surface of the remaining intact bacteria (17, 35). Other authors claim that specific and selective mechanisms, rather than autolysis, are involved in the secretion of H. pylori proteins (12, 36, 51). Scott et al. proposed, on the other hand, that internal rather than external urease activity is important for acid protection of the organism and even for growth in acidic environments (40). We have previously reported that the switch from L to S type colony morphology can be induced by cultivating 17L at low pH (11). Since the phase variation is caused by a spontaneous DNA slippage which is most likely random, this event can only be explained by a mechanism of selection at low pH (20). We found that H. pylori OMPLA has its pH optimum at 7.0. Both L and S variants were able to grow at pH 5. But minimal amounts of lysophospholipids were produced, confirming that the complete and active OMPLA of the S variant had a reduced activity at this pH. However, the release of urease from the S variant at pH 5 was still much more pronounced than from the L variant. The local neutralization by urease is likely to be sufficient to give optimal conditions for PLA and thus restoring the normal activities of the S variant. We found that L variants survive and grow at pH 5, but not at pH 3.5; at this low pH, only a few colonies were formed, all with the S-colony form. This indicates that only the spontaneously formed S variants have sufficient extracellular urease to survive at this low pH.
PLA has been proposed as a possible virulence factor for H. pylori (27, 29, 41). PLA is suggested to play a part in lipid mucus degradation and the hydrolysis of the gastric mucosal phospholipids, causing mucosal damage (41). One could also envisage the involvement of OMPLA in pathogenesis by the degradation of the host cell. Dorrell et al. have reported that mice inoculated with an H. pylori pldA mutant showed no colonization at either 2 or 8 weeks postinfection, suggesting that the OMPLA is required for growth of the bacteria in their natural environment (15). Thus, the data presented here may explain how phase variation in pldA contributes to acid adaptation.
ACKNOWLEDGMENTS
We thank A. J. Aarsman (Utrecht University, Utrecht, The Netherlands) for performing the PLA activity assay; R. Negrini (Brescia Hospital, Brescia, Italy) for providing MAbs; and J. J. Maaskant, T. Verboom, and J. G. Kusters (Vrije Universiteit, Amsterdam, The Netherlands) for theoretical and practical support.
REFERENCES
- 1.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, de Jonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Appelmelk B J, Martin S L, Monteiro M, Clayton C A, McColm A A, Zheng P, Verboom T, Maaskant J J, van den Eijnden D H, Wirth H-P, Hokke C H, Perry M B, Vandenbroucke-Grauls C M J E, Kusters J G. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lenghts of poly(C) tracts in α3-fucosyltransferase genes. Infect Immun. 1999;67:5361–5366. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appelmelk B J, Shiberu B, Trinks C, Tapsi N, Zheng P Y, Verboom T, Maaskant J, Hokke C H, Schiphorst W E, Blanchard D, Simoons-Smit I M, van den Eijnden D H, Vandenbroucke-Grauls C M. Phase variation in Helicobacter pylori lipopolysaccharide. Infect Immun. 1998;66:70–76. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertram-Drogatz P A, Sobek-Klocke I, Möller C, Wingbermühle D, Beil W, Sewing K-F, Manns M P, Wagner S. Growth characteristics and influence of antibiotics on rough/smooth phenotypic variants of Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1999;18:490–495. doi: 10.1007/s100960050329. [DOI] [PubMed] [Google Scholar]
- 5.Bijlsma J J, Vandenbroucke-Grauls C M, Phadnis S H, Kusters J G. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect Immun. 1999;67:2433–2440. doi: 10.1128/iai.67.5.2433-2440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom R, Sol C J A, Salimans M M M, Jansen C L, van Dillen P M E W, van den Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;260:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brok R G, Boots A P, Dekker N, Verheij H M, Tommassen J. Sequence comparison of outer membrane phospholipases A: implications for structure and for the catalytic mechanism. Res Microbiol. 1998;149:703–710. doi: 10.1016/s0923-2508(99)80017-5. [DOI] [PubMed] [Google Scholar]
- 9.Brok R G, Dekker N, Gerrits N, Verheij H M, Tommassen J. A conserved histidine residue of Escherichia coli outer-membrane phospholipase A is important for activity. Eur J Biochem. 1995;234:934–938. doi: 10.1111/j.1432-1033.1995.934_a.x. [DOI] [PubMed] [Google Scholar]
- 10.Bucci C, Lavitola A, Salvatore P, Del Giudice L, Massardo D R, Bruni C B, Alifano P. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol Cell. 1999;3:435–445. doi: 10.1016/s1097-2765(00)80471-2. [DOI] [PubMed] [Google Scholar]
- 11.Bukholm G, Tannæs T, Nedenskov P, Esbensen Y, Grav H J, Hovig T, Ariansen S, Guldvog I. Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand J Gastroenterol. 1997;32:445–454. doi: 10.3109/00365529709025079. [DOI] [PubMed] [Google Scholar]
- 12.Cao P, McClain M S, Forsyth M H, Cover T L. Extracellular release of antigenic proteins by Helicobacter pylori. Infect Immun. 1998;66:2984–2986. doi: 10.1128/iai.66.6.2984-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dekker N. Outer membrane phospholipase A: known structure, unknown biological function. Mol Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 14.Doig P, Trust T J. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect Immun. 1994;62:4526–4533. doi: 10.1128/iai.62.10.4526-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorrell N, Martino M C, Stabler R A, Ward S J, Zhang Z W, McColm A A, Farthing M J G, Wren B W. Characterization of Helicobacter pylori pldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology. 1999;117:1098–1104. doi: 10.1016/s0016-5085(99)70394-x. [DOI] [PubMed] [Google Scholar]
- 16.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dunn B E, Vakil N B, Schneider B G, Miller M M, Zitzer J B, Peutz T, Phadnis S H. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun. 1997;65:1181–1188. doi: 10.1128/iai.65.4.1181-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant K A, Belandia I U, Dekker N, Richardson P T, Park S F. Molecular characterization of pldA, the structural gene for a phospholipase A from Campylobacter coli, and its contribution to cell-associated hemolysis. Infect Immun. 1997;65:1172–1180. doi: 10.1128/iai.65.4.1172-1180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta C M, Bali A. Carbamyl analogs of phosphatidylcholines: synthesis, interaction with phospholipases, and permeability behavior of their liposomes. Biochim Biophys Acta. 1981;663:506–515. doi: 10.1016/0005-2760(81)90178-8. [DOI] [PubMed] [Google Scholar]
- 20.Henderson I R, Owens P, Nataro J P. Molecular switches: the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 21.Homma H, Kobayashi T, Chiba N, Karasawa K, Mizushima H, Kudo I, Inoue K, Ikeda H, Sekiguchi M, Nojima S. The DNA sequence encoding pldA gene, the structural gene for detergent-resistant phospholipase A of E. coli. J Biochem. 1984;96:1655–1664. doi: 10.1093/oxfordjournals.jbchem.a134997. [DOI] [PubMed] [Google Scholar]
- 22.Hynes S O, Helander I, Lepisto P, Moran A P. Outer membrane permeability of Helicobacter pylori as assessed with a novel fluorescent probe. Gut. 2000;47(Suppl. I):A3. [Google Scholar]
- 23.Kates M. Laboratory techniques in biochemistry and molecular biology. 3:2. Amsterdam, The Netherlands: Elsevier; 1986. Techniques of lipidology: isolation, analysis and identification of lipids; pp. 396–404. [Google Scholar]
- 24.Kingma R L, Fragiathaki M, Snijder H J, Dijkstra B W, Verheij H M, Dekker N, Egmond M R. Unusual catalytic triad of Escherichia coli outer membrane phospholipase A. Biochemistry. 2000;39:10017–10022. doi: 10.1021/bi000786d. [DOI] [PubMed] [Google Scholar]
- 25.Krishnamurthy P, Parlow M, Zitzer J B, Vakil N B, Mobley H L, Levy M, Phadnis S H, Dunn B E. Helicobacter pylori containing only cytoplasmic urease is susceptible to acid. Infect Immun. 1998;66:5060–5066. doi: 10.1128/iai.66.11.5060-5066.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:280–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Marshall B J. Virulence and pathogenicity of Helicobacter pylori. J Gastroenterol Hepatol. 1991;6:121–124. doi: 10.1111/j.1440-1746.1991.tb01450.x. [DOI] [PubMed] [Google Scholar]
- 28.McGowan C C, Necheva A, Thompson S A, Cover T L, Blaser M J. Acid-induced expression of an LPS-associated gene in Helicobacter pylori. Mol Microbiol. 1998;30:19–31. doi: 10.1046/j.1365-2958.1998.t01-1-01079.x. [DOI] [PubMed] [Google Scholar]
- 29.Mégraud F. Pathogenic diversity of Helicobacter pylori. J Gastroenterol. 1997;32:278–281. doi: 10.1007/BF02936383. [DOI] [PubMed] [Google Scholar]
- 30.Meyer T F, Gibbs C P, Haas R. Variation and control of protein expression in Neisseria. Annu Rev Microbiol. 1990;44:451–477. doi: 10.1146/annurev.mi.44.100190.002315. [DOI] [PubMed] [Google Scholar]
- 31.Moran A P, Wadstrom T. Pathogenesis of Helicobacter pylori. Curr Opin Gastroenterol. 1998;14:S9–S14. [Google Scholar]
- 32.Negrini R, Lisato I, Zanella S, Cavazzini S, Gullini S, Villanacci V, Poiesi C, Albertini A, Ghielmi S. Helicobacter pylori infection induces antibodies cross-reacting with human gastric mucosa. Gastroenterology. 1991;101:437–445. doi: 10.1016/0016-5085(91)90023-e. [DOI] [PubMed] [Google Scholar]
- 33.Negrini R, Lisato L, Cavazzini L, Maini P, Gullini S, Basso O, Lanza G, Garofalo M, Nenci I. Monoclonal antibodies for specific immunoperoxidase detection of Campylobacter pylori. Gastroenterology. 1989;96(Pt. 1):414–420. doi: 10.1016/0016-5085(89)91565-5. [DOI] [PubMed] [Google Scholar]
- 34.Negrini R, Savio A, Poiesi C, Appelmelk B J, Buffoli F, Paterlini A, Cesari P, Graffeo M, Vaira D, Franzin G. Antigenic mimicry between Helicobacter pylori and gastric mucosa in the pathogenesis of body atrophic gastritis. Gastroenterology. 1996;111:655–665. doi: 10.1053/gast.1996.v111.pm8780570. [DOI] [PubMed] [Google Scholar]
- 35.Phadnis S H, Parlow M H, Levy M, Ilver D, Caulkins C M, Connors J B, Dunn B E. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect Immun. 1996;64:905–912. doi: 10.1128/iai.64.3.905-912.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rektorschek M, Buhmann A, Weeks D, Schwan D, Bensch K W, Eskandari S, Scott D, Sachs G, Melchers K. Acid resistance of Helicobacter pylori depends on the ureI membrane protein and an inner membrane proton barrier. Mol Microbiol. 2000;36:141–152. doi: 10.1046/j.1365-2958.2000.01835.x. [DOI] [PubMed] [Google Scholar]
- 37.Rektorschek M, Weeks D, Sachs G, Melchers K. Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology. 1998;115:628–641. doi: 10.1016/s0016-5085(98)70142-8. [DOI] [PubMed] [Google Scholar]
- 38.Rock C O, Jackowski S, Cronan J E., Jr . Lipid metabolism in prokaryotes. In: Vance D E, Vance J E, editors. Biochemistry of lipids, lipoproteins and membranes. New York, N.Y: Elsevier Science Publishing; 1996. pp. 35–74. [Google Scholar]
- 39.Sansonetti P J, Kopecko D J, Formal S B. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott D R, Weeks D L, Hong C, Postius S, Melchers K, Sachs G. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology. 1998;114:58–70. doi: 10.1016/s0016-5085(98)70633-x. [DOI] [PubMed] [Google Scholar]
- 41.Slomiany B L, Slomiany A. Mechanism of Helicobacter pylori pathogenesis: focus on mucus. J Clin Gastroenterol. 1992;14(Suppl. 1):S114–S121. [PubMed] [Google Scholar]
- 42.Snijder H J, Ubarretxena-Belandia I, Blaauw M, Kalk K H, Verheij H M, Egmond M R, Dekker N, Dijkstra B W. Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature. 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 43.Starkova Z, Thomas P, Starka J. Morphological mutants of Escherichia coli: nature of the permeability barrier in mon and envC cells. Ann Microbiol. 1978;129:265–284. [PubMed] [Google Scholar]
- 44.Tannæs T, Grav H J, Bukholm G. Lipid profiles of Helicobacter pylori colony variants. APMIS. 2000;108:349–356. doi: 10.1034/j.1600-0463.2000.d01-67.x. [DOI] [PubMed] [Google Scholar]
- 45.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 46.Tsuda M, Karita M, Morshed M G, Okita K, Nakazawa T. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Dam G. Circulating gut-associated antigens of Schistosoma mansoni: biological, immunological and molecular aspects. Ph.D. thesis. Leiden, The Netherlands: University of Leiden; 1995. [Google Scholar]
- 48.van den Bosch H, Aarsman A J, van Deenen L L M. Isolation and properties of a phospholipase A1 activity from beef pancreas. Biochim Biophys Acta. 1974;348:197–209. doi: 10.1016/0005-2760(74)90231-8. [DOI] [PubMed] [Google Scholar]
- 49.van der Meer N M, Appelmelk B J, Verweij-van Vught A M, Nimmich W, Kosma P, Thijs L G, de Graaff J, MacLaren D M. Binding studies of a monoclonal antibody specific for 3-deoxy-d-manno-octulosonic acid (KDO) with a panel of Klebsiella pneumoniae lipopolysaccharides representing all the O serotypes. Infect Immun. 1994;62:1052–1057. doi: 10.1128/iai.62.3.1052-1057.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Doorn N E, Namavar F, Sparrius M, Stoof J, van Rees E P, van Doorn L J, Vandenbroucke-Grauls C M. Helicobacter pylori-associated gastritis in mice is host and strain specific. Infect Immun. 1999;67:3040–3046. doi: 10.1128/iai.67.6.3040-3046.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanet A, Labigne A. Evidence for specific secretion rather than autolysis in release of some Helicobacter pylori proteins. Infect Immun. 1998;66:1023–1027. doi: 10.1128/iai.66.3.1023-1027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang G, Rasko D A, Sherburne R, Taylor D E. Molecular genetic basis for the variable expression of Lewis Y antigen in Helicobacter pylori: analysis of the alpha (1,2) fucosyltransferase gene. Mol Microbiol. 1999;31:1265–1274. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;139:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 54.Weltzien H U. Cytolytic and membrane-perturbing properties of lysophosphatidylcholine. Biochim Biophys Acta. 1979;559:259–287. doi: 10.1016/0304-4157(79)90004-2. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q L, Gotschlich E C. Variation of gonococcal lipooligosaccharide structure is due to alterations in poly-G tracts in lgt genes encoding glycosyl transferases. J Exp Med. 1996;183:323–327. doi: 10.1084/jem.183.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]