Abstract
The functions of proteins depend on their spatial and temporal distributions, which are not directly measured by static protein abundance. Under protein misfolding stress, the unfolded protein response (UPR) pathway remediates proteostasis in part by altering the turnover kinetics and spatial distribution of proteins, yet a global view of these spatiotemporal changes has yet to emerge and it is unknown how they affect different cellular compartments and pathways. Here we describe a mass spectrometry-based proteomics strategy and data analysis pipeline, named Simultaneous Proteome Localization and Turnover (SPLAT), to measure concurrently the changes in protein turnover and subcellular distribution in the same experiment. Investigating two common UPR models of thapsigargin and tunicamycin challenge, we find that the global suppression of protein synthesis during UPR is dependent on subcellular localization, with more severe slowdown in lysosome vs. endoplasmic reticulum (ER) protein turnover. Most candidate translocation events affect pre-existing proteins and likely involve vesicular transport across endomembrane fractions including an expansion of an ER-derived vesicle (ERV) compartment containing RNA binding proteins and stress response proteins. In parallel, we observed specific translocations involving only newly synthesized protein pools that are indicative of endomembrane stalling. The translocation of a subclass of cell surface proteins to the endomembrane including EGFR and ITGAV upon UPR affects only heavy labeled proteins, which suggest their internalization is driven by nascent protein trafficking rather than ligand dependent endocytosis. The approach described here may be broadly useful for inferring the coordinations between spatial and temporal proteome regulations in normal and stressed cells.
Keywords: Unfolded protein response, protein turnover, spatial proteomics, mass spectrometry, stress response, subcellular localization, spatiotemporal
Introduction
Protein turnover is an important cellular process that maintains the quality and quantity of protein pools in homeostasis, and involves fine regulation of the rate of synthesis and degradation of proteins. A close relationship exists between turnover kinetics with the spatial distribution of proteins. Cellular organelles including the cytosol, endoplasmic reticulum (ER), and mitochondria are equipped with distinct quality control and proteolytic mechanisms that maintain protein folding and regulate protein degradation in a localization dependent manner (Lemberg and Strisovsky, 2021; Mårtensson et al., 2019; Tsai et al., 2022). Newly synthesized proteins need to be properly folded and trafficked to their intended subcellular localization through subcellular targeting and sorting mechanisms, whose capacity has to be coordinated to match the rate of protein synthesis (Chartron et al., 2016; Jan et al., 2014; Lakkaraju et al., 2008). A primary subcellular trafficking mechanism of new proteins is the ER vesicular transport and secretory pathway through the endomembrane system, which is a rate-limiting step in the production of membrane and extracellular proteins. A mismatch between temporal synthesis rate and spatial localization capacity can lead to ER stress and subsequently mislocalization of newly synthesized proteins (Hetz et al., 2020).
Disruption of protein turnover and homeostasis is broadly implicated in human diseases including cardiomyopathies, cancer, and neurodegeneration (Hetz et al., 2020; Ren et al., 2021). In stressed cells, the accumulation of misfolded proteins triggers the unfolded protein response (UPR), which involves signaling pathways that suppress protein synthesis and promote protein folding and proteolysis to relieve proteostatic stress. At the same time, UPR activation invokes a spatial transmission of stress status through the translocation of UPR pathway mediators ATF4, ATF6, and XBP1 to the nucleus, the retrotranslocation of misfolded ER proteins to the cytosol for proteasomal clearance under endoplasmic-reticulum-associated protein degradation (ERAD), and the sequestration of RNA and interacting proteins to stress granules. Despite ongoing research, how the cellular proteome reorganizes under proteostatic stress is incompletely understood, and in particular, the full scope of differentially localized proteins in UPR remains to be elucidated.
We wondered whether the spatial and temporal regulations of protein are coordinated under UPR, such that proteins with differential spatial distributions are also more prone to show differential turnover. Advances in mass spectrometry methods have allowed the turnover rate and subcellular localization of proteins to be measured on a large scale. The turnover rate and half-life of proteins can be measured using in vivo stable isotope labeling in cells and in intact animals followed by mass spectrometry measurements of isotope signatures and kinetics modeling to derive rate constants (Claydon and Beynon, 2012; Doherty et al., 2009; Hammond et al., 2022; Schwanhäusser et al., 2011). Quantitative comparison of turnover rates provide a temporal view into proteostatic regulations and can implicate new pathological signatures and pathways over steady-state mRNA and protein levels (Andrews et al., 2022; Lam et al., 2014; Lau et al., 2018). In parallel, spatial proteomics methods have allowed increasing power to discern the subcellular localization of proteins on a large scale (Christopher et al., 2021; Geladaki et al., 2019; Kennedy et al., 2020; Mulvey et al., 2021; Orre et al., 2019). In recent work using a differential solubility fractionation strategy and mass spectrometry, we observed broad substantial rearrangement of proteins across three subcellular fractions in an acute paraquat challenge model of UPR in the mouse heart, consistent with protein translocation being an important layer of proteome regulation under proteostatic stress (Dostal et al., 2020). Nevertheless, an integrated strategy that can simultaneously measure protein turnover kinetics and spatial information has thus far not been realized.
Here we extended protein turnover measurements to include subcellular localization dynamics, by integrating dynamic SILAC labeling with differential ultracentrifugation based spatial proteomics profiling strategies. We describe an experimental strategy and computational analysis pipeline to perform simultaneous proteome localization and turnover (SPLAT) measurements in baseline and stressed cells. SPLAT builds on prior work in protein turnover measurements and subcellular localization profiling, by combining dynamic SILAC isotope labeling, differential ultracentrifugation, isobaric TMT labeling, and kinetic modeling to concurrently measure changes in the turnover dynamics and subcellular distributions of whole cell proteomes under perturbation in a single experiment and infer the mechanism of spatial redistribution events. Applying this method to human AC16 cardiomyocytes under thapsigargin and tunicamycin induced UPR, we delineated prominent changes in the spatial and temporal distributions of proteins on a proteome scale. The inclusion of spatial information of light and heavy SILAC labeled proteins moreover allowed disaggregation of the localization and trafficking of existing and newly synthesized proteins.
Methods
Cell culture, metabolic labeling, UPR induction
AC16 cells procured from Millipore between passage number 11 and 16 were cultured in DMEM/F12 supplemented with 10% FBS and no antibiotics. Cells were maintained at 37°C with 5% CO2 and 10% O2. For isotopic labeling, SILAC DMEM/F12 (Thermo Scientific) deficient in both L-lysine and L-arginine was supplemented with 1% dialyzed FBS and heavy amino acids 13C615N2 L-Lysine-2HCl and 13C615N4 L-Arginine-HCl (Thermo Scientific) at concentrations of 0.499 mM and 0.699 mM, respectively. Light media was switched to heavy media and cells were labeled for 16 hours prior to harvest. UPR was induced with 1 μM thapsigargin (SelleckChem) or 1 μg/mL tunicamycin (Sigma) at the same time as isotopic labeling.
To verify ER stress induction, thioflavin T staining was performed in replicate plates of AC16 cells cultured and treated identically as above. At 80% confluency, the cells were treated with UPR inducing compounds for 16 hours to generate 6 replicates of untreated control, 3 replicates of 1 μg/ml tunicamycin treated cells, and 3 replicates of 1 μM thapsigargin treated wells; thioflavin T (abcam) was added in the final hour of drug treatment by dissolving in 2.5 mL 70% ethanol and dilution in DMEM/F12 media to create a 5 mM stock solution. 2 μL of the stock solution was applied to each well (2 mL final volume) for a final concentration of 5 μM. Cell nuclei were counterstained with NucBlue Hoechst live cell stain (Invitrogen) for 20 minutes prior to imaging. The cells were imaged on an EVOS M5000 microscope (Thermo) using the GFP light cube for thioflavin T (ex/em 470/525 nm) and the DAPI lightcube for NucBlue (ex/em 357/447 nm).
Cell harvest, differential centrifugation, and isobaric labeling
Cell harvest and subcellular fraction was performed based on the LOPIT-DC differential ultracentrifugation protocol as described in Geladaki et al. (Geladaki et al., 2019). Briefly, AC16 cells were harvested with trypsinization, washed 3× with room temperature PBS, and resuspended in a detergent free gentle lysis buffer. 1.5 mL of suspension at a time was lysed using an Isobiotec ball bearing homogenizer with a 16 μM clearance size until ~80% of cell membranes were lysed, as verified with trypan blue (approximately 15 passes through the chamber). Lysates were spun 3 times each in a 4°C swinging bucket centrifuge 200 g, 5 min to remove unlysed cells. The supernatant was retained and used to generate 9 ultracentrifugation pellets using spin parameters shown in Table 1.
Table 1.
Ultracentrifugation speeds and durations used for generation of each pellet.
| Pellet | Speed (g) | Time (min) |
|---|---|---|
| Pellet 1 | 1000 | 10 |
| Pellet 2 | 3000 | 10 |
| Pellet 3 | 5000 | 10 |
| Pellet 4 | 9000 | 15 |
| Pellet 5 | 12000 | 15 |
| Pellet 6 | 15000 | 15 |
| Pellet 7 | 30000 | 20 |
| Pellet 8 | 79000 | 43 |
| Pellet 9 | 120000 | 45 |
The supernatant generated in the final spin was removed and all pellets and the final supernatant were stored at −80°C until proceeding. Supernatant was thawed on ice and precipitated in 3× the volume of cold acetone overnight at −20°C.This was used to generate pellet 10 by centrifuging at 13,000 × g for 10 minutes at 4°C. Excess acetone was removed and the pellet was allowed to dry briefly before resuspension in a resolubilization buffer of 8 M urea, 50 mM HEPES pH 8.5, and 0.5% SDS with 1x Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). The suspension was sonicated in a Biorupter with settings 20× 30s on 30s off at 4°C.
Pellets from the ultracentrifugation fractions 1 to 9 were resuspended in RIPA buffer with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) with sonication in a Biorupter with settings 10× 30s on 30s off at 4°C. Insoluble debris was removed from all samples (1–10) by centrifugation at 14,000xg, 5 minutes. Protein concentration of all samples was measured with Rapid Gold BCA. The samples were digested and isobarically tagged using the iFASP protocol (McDowell et al., 2013). 25 ug protein per sample in 250 uL 8M urea was loaded onto Pierce Protein Concentrators PES, 10K MWCO prewashed with 100 mM TEAB. The samples were again washed with 8 M urea to denature proteins and remove SDS. The samples were washed with 300 uL 100 mM TEAB twice. The samples were then reduced and alkylated with TCEP and CAA for 30 minutes at 37°C in the dark. CAA and TCEP were removed with centrifugation and the samples were washed 3x with 100 mM TEAB. Samples were digested atop the filters overnight at 37°C with mass spectrometry grade trypsin (Promega) at a ratio of 1:50 enzyme:protein.
Briefly, 0.2 mg of TMT-10plex isobaric labels (Thermo Scientific) per differential centrifugation fraction were equilibrated to room temperature and reconstituted in 20 μL LC-MS grade anhydrous acetonitrile. In each experiment, labels were randomly assigned to each fraction with a random number generator to mitigate possible batch effect. Isobaric tags were added to peptides still atop the centrifugation filters and incubated at room temperature for 1 hour with shaking. The reactions were quenched with 1 μL hydroxylamine at room temperature for 30 minutes with shaking. Labeled peptides were eluted from the filters with centrifugation. To further elute labeled peptides 40 μL 50 mM TEAB was added and filters were again centrifuged. All 10 labeled fractions per experiment were combined and mixed well before dividing each experiment into two aliquots. Aliquots were dried with speed-vac and stored at −80°C.
Liquid chromatography and mass spectrometry
One aliquot per experiment was reconstituted in 50 μL 20 mM ammonium formate pH 10 in LCMS grade water (solvent A) for high pH reverse phase liquid chromatography (RPLC) fractionation. The entire sample was injected into a Jupiter 4 μm Proteo 90 Å LC Column of 150 × 1 mm on a Ultimate 3000 HPLC system. The gradient was run with a flow rate of 0.1 mL/min as follows: 0–30 min: 0%–40% Solvent B (20 mM ammonium formate pH 10 in 80% LCMS grade acetonitrile); 30–40 min: 40%−80% Solvent B; 40–50 min: 80% Solvent B. Fractions were collected every minute and pooled into a total of 20 peptide fractions, then dried with speed-vac.
The dried fractions were reconstituted in 10 μL each of pH 2 MS solvent A (0.1% formic acid) and analyzed with LC-MS/MS on a Q-Exactive HF orbitrap mass spectrometer coupled to an LC with electrospray ionization source. Peptides were separated with a PepMap RSLC C18 column 75 μm × 15 cm, 3 μm particle size (ThermoScientific) with a 90 minute gradient from 0 to 100% pH 2 MS solvent B (0.1% formic acid in 80% LCMS grade acetonitrile). Full MS scans were acquired with a 60,000 resolution. A stepped collision energy of 27, 30 and 32 was used and MS2 scans were acquired with a 60,000 resolution and an isolation window of 0.7 m/z.
Mass spectrometry data processing and turnover analysis
Mass spectrometry raw data were converted to mzML format using ThermoRawFileParser v.1.2.0 (Hulstaert et al., 2020) then searched against the UniProt SwissProt human canonical and isoform protein sequence database (retrieved 2022-10-27) using Comet v.2020_01_rev3 (Eng et al., 2015). The fasta database was further appended with contaminant proteins using Philosopher v4.4.0 (total 42,402 forward entries). The search settings were as follows: peptide mass tolerance: 10 ppm; isotope error: 0/1/2/3; number of enzyme termini: 1; allowed missed cleavages: 2; fragment bin tolerance: 0.02; fragment bin offset: 0; variable modifications: TMT-10plex tag +229.1629 for TMT experiments, and lysine + 8.0142, arginine + 10.0083 for all SILAC experiments; fixed modifications: cysteine + 57.0214. The search results were further reranked and filtered using Percolator v3.0 (The et al., 2016) at a 5% FDR. Following database search, the mzML files and Percolator PSMs were input to the SPLAT pipeline. The dynamic SILAC data were analyzed using RIANA v0.7.1 (Hammond et al., 2022) to integrate the peak intensity within a 25 ppm error of the light (+0), heavy (+8, +10), and double K/R (+16, +18, +20) peptide peaks over a 20 second retention time window encompassing the first and last MS2 scan where the peptide is confidently identified. We then calculated the fractional synthesis of all K/R containing peptides as the intensity of the 0th isotopomer peak (m0) over the sum of applicable light and heavy isotopomers (e.g., m0/m0+m8 for a peptide with one lysine). RIANA then performs intensity-weighted least-square curve-fitting using the scipy optimize function to a first-order exponential rise model to find the best-fit peptide turnover rate. Protein turnover rates are calculated as the harmonic mean of peptide turnover rates
Unfractionated protein abundance experiments
To measure the protein abundance changes following thapsigargin and tunicamycin, AC16 cells were cultured as described above without SILAC reagents. At 80% confluency, the cells were treated with UPR inducing compounds (1 μg/ml tunicamycin or 1 μM thapsigargin). The treated wells were harvested at 8 and 16 hour timepoints. Control wells were harvested at 16 hours. For each drug, 3 control wells and 3 replicates per time point were digested, tagged with TMT 10-plex reagents (Thermo), fractionated with RPLC, and analyzed with LC-MS/MS as in the SPLAT experiments. Database search and quantification was performed as above in the absence of variable SILAC modifications.
Spatial proteomics analysis
To analyze the spatial proteomics data in SPLAT, we created a new version of the pyTMT script, which we previously described and performs TMT label quantification for the Comet/Percolator workflow. Two new features were included specifically for the analysis of the spatial proteomics data here. First, we account for isotope impurities in TMT tags which can lead to up to 10% spillover to neighbor channels and decrease quantitative accuracy. Because the TMT data are row normalized in the LOPIT-DC design, we incorporated an automatic non-negative least square correction of isotope contamination of TMT channels based on the batch contamination data sheet, in order to correct for isotope impurity without column normalization and allow individual channel randomization across experiments. Second, we implemented an isoform-aware quantitative rollup of peptide channel intensities into the protein level for the downstream spatial proteomics analysis. Standard protein inference invokes parsimony rules that assign peptides to the protein within a protein group with the highest level of evidence, but razor peptides can conflate spatial information from different proteins with different localization. Here the TMT-quantified peptides are summed into protein groups using a more conservative aggregate method, such that the identified peptides that are assigned to two or more top-level UniProt protein accessions are discarded to avoid confounding of spatial information in the TMT channels. Moreover, protein groups containing two or more proteins belonging to the same top-level UniProt accession are removed from consideration if one of the non-canonical isoforms contain a unique peptide, and are otherwise rolled up to the canonical protein. Protein isoforms are only included in downstream analysis through quantified isoform-unique peptides. Following RIANA and pyTMT processing, the SPLAT pipeline combines the dynamic SILAC and TMT information by peptides and appends a heavy (“_H”) tag to the UniProt accession of all peptides containing dynamic SILAC modifications for separate localization analyses. The data were then used for temporal kinetics summaries and subcellular localization classification.
Subcellular localization classification and translocation predictions were performed using the pRoloc (Gatto et al., 2014) and the BANDLE (Crook et al., 2022) packages in R/Bioconductor. Briefly, the subcellular localization markers were selected from the intersecting proteins with a prior data set generated from human U-2 OS osteosarcoma cells (Geladaki et al., 2019). A subset of the intersecting markers were selected using a random walk algorithm to maximize normalized between-class separation. For differential localization analyses we used the Markov-chain Monte-Carlo (MCMC) and non-parametric model in BANDLE to find unknown protein classification and evaluate differential localization probability. MCMC parameters are 8 chains, 20,000 iterations, and 5,000 burn-in, and 20 thinning; convergence of the Markov chains is assessed visually by rank plots as recommended (Crook et al., 2019). For additional analysis to describe baseline protein classification, a T-augmented Gaussian mixture model with a maximum a posteriori (MAP) method in pRoloc was used.
Data and code availability
Mass spectrometry data have been updated to ProteomeXchange under the accession number PXD038054. Code for the SPLAT pipeline is available at https://github.com/lau-lab/splat.
Results
Simultaneous acquisition of turnover and spatial information using a double labeling strategy
To initiate UPR, we treated human AC16 cardiomyocytes with 1 μM thapsigargin or 1 μg/mL tunicamycin for 16 hours. Both molecules at the dosages used are commonly used to induce ER stress and integrated stress response in cardiomyocytes and other cell types. Thapsigargin is known to induce ER stress through the inhibition of sarco/endoplasmic reticulum Ca2+-ATPase (SERCA) whereas tunicamycin induces proteostatic stress through the inhibition of nascent protein glycosylation.
Thapsigargin and tunicamycin treatment at 16 hours robustly induced ER stress and UPR, as indicated by increased thioflavin T fluorescence (Supplemental Figure S1A) and increased protein abundance of BiP/GRP78 (Thapsigargin: 3.5-fold, adjusted P 6.6e–10; Tunicamycin: 4.5 fold, adjusted P: 5.1e–11) and other stress response proteins including PDIA4 (Thapsigargin: 1.4-fold, adjusted P: 5.5e–5; Tunicamycin 2.1-fold, adjusted P 3.4e–8) and GRP170/HYOU1 (Thapsigargin 1.6-fold, adjusted P: 1.1e–4; Tunicamycin 2.0-fold, adjusted P: 2.9e–6) (Supplemental Figure S1B)
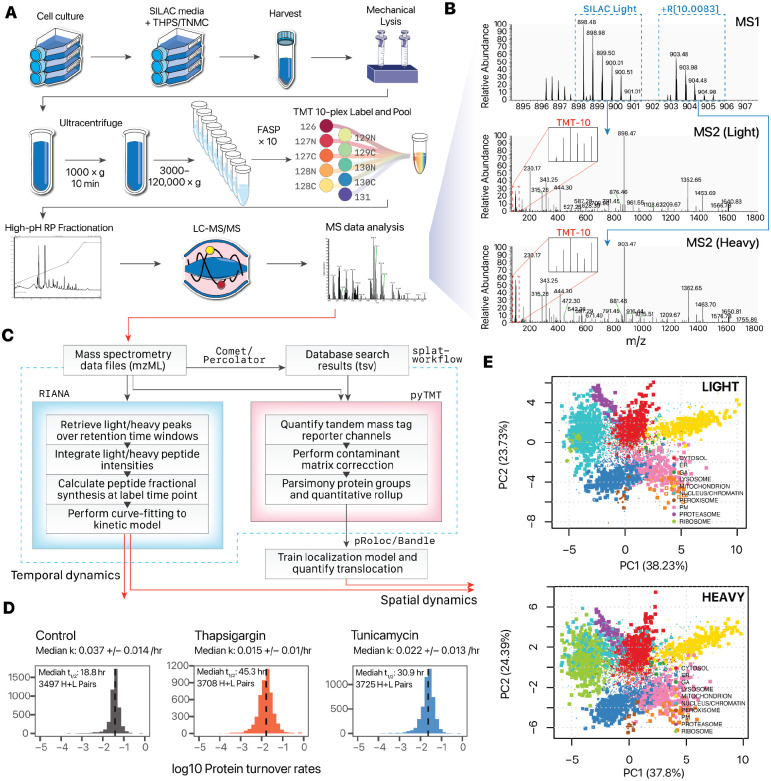
We designed a workflow that combines in vivo metabolic labeling, spatial separation, and isobaric peptide tagging to simultaneously measure new protein synthesis as well as subcellular localization under baseline and perturbation conditions (Figure 1A). To determine the rate of protein turnover during control, thapsigargin, and tunicamycin conditions, a dynamic SILAC strategy was used to measure the rate of appearance of post-labeling synthesized protein. Briefly, cells were pulsed with a lysine and arginine depleted media supplemented with heavy labeled lysine and arginine concurrently with drug treatment to label post-treatment synthesized proteins and derive fractional synthesis rates through kinetic modeling. Upon harvesting, the cells were further fractionated and labeled following a differential centrifugation-based strategy (LOPIT-DC) to identify subcellular protein localization. Briefly, the cells were lysed under gentle conditions and then sequentially pelleted through ultracentrifugation steps, which pellets subcellular fractions based on their sedimentation rate and which is a function of particle mass, shape, and volume. The ultracentrifugation fractions were each subsequently solubilized, and the extracted proteins were digested and further labeled with tandem mass tag (TMT) isobaric stable isotope labels. The acquired mass spectrometry data therefore carries temporal information in the dynamic SILAC tags and spatial information in the TMT channel intensities (Figure 1B). We analyzed the data using a custom computational pipeline comprising database search and post-processing, and quantification for dynamic SILAC and TMT data (Figure 1C). The turnover kinetics information from the dynamic SILAC data is analyzed using a mass spectrometry software tool we developed, RIANA (Hammond et al., 2022), which integrates the areas-under-curve of mass isotopomers from peptides over a specified retention time window, then performs kinetic curve-fitting to a mono-exponential model to measure the fractional synthesis rates (FSR) of each dynamic SILAC-labeled (K and R containing) peptide. To extract the ultracentrifugation fraction quantification information for spatial analysis, we developed a new version of the pyTMT tool (Dostal et al., 2020) to account for spatial proteomics data features (see Methods).
Figure 1.
Overview of the SPLAT strategy. A. Experimental workflow. Human A16 cardiomyocytes were labeled with 13C615N2 L-Lysine and 13C615N4 L-Arginine and treated by thapsigargin or tunicamycin. After 16 hours, cells were harvested and mechanically disrupted, followed by differential ultracentrifugation steps to pellet proteins across cellular compartments. Proteins from the ultracentrifugation fractions were digested and labeled using tandem mass tag (TMT) followed by mass spectrometry. B. Dynamic SILAC labeling allowed differentiation of pre-existing (unlabeled) and post-labeling (heavy lysine or arginine) synthesized peptides at 16 hours. The light and heavy peptides were isolated for fragmentation separately to allow the protein sedimentation profiles containing spatial information to be discerned from TMT channel intensities. C. Computational workflow. Mass spectrometry raw data were converted to mzML format to identify peptides using a database search engine. The mass spectra and identification output were processed using RIANA (left) to quantitate the time dependent change in SILAC labeling intensities and determine the protein half-life, and using pyTMT (right) to extract and correct TMT channel intensities from each light or heavy peptide MS2 spectrum. The TMT data were further processed using pRoloc/Bandle to predict protein subcellular localization via supervised learning. D. Histograms of the determined log10 protein turnover rates in control, thapsigargin, and tunicamycin cells. E. Principal component analysis showing light and heavy protein TMT channel distributions, and the predicted subcellular localization.
In total, we estimated the turnover rates of 3497, 3708, and 3725 light and heavy dynamic SILAC protein pairs with at least 2 peptides in control, thapsigargin, and tunicamycin as 0.037 +/− 0.014 h−1, 0.015 +/− 0.010 h−1, and 0.022 +/− 0.014 h−1 proteins in control, thapsigargin, and tunicamycin treated cells respectively (medians +/− MAD), corresponding to a median fractional synthesis half-life of 18.8, 45.3, and 30.9 hours, respectively (Figure 1D). In parallel, we analyzed the spatial fractionation patterns of the proteins following ultracentrifugation and TMT labeling, and predicted protein localization using a Bayesian TAGM-MAP and TAGM-MCMC classifications as previously described (Crook et al., 2018). The models were trained based on 304 annotated subcellular localization markers for 10 subcellular localizations (32 in the cytosol, 52 in the ER, 7 in the GA, 16 in the lysosome, 65 in the mitochondrion, 38 in the nucleus, 8 in the peroxisome, 17 on the PM, 19 in the proteasome, and 50 in the ribosome). In total, we quantified the ultracentrifugation fraction profiles of 7876, 6867, and 7546 light and heavy proteins in the control, thapsigargin, and tunicamycin samples, respectively, among which 3789, 3253, and 3580 are assigned to a single subcellular localization with 90% or higher confidence (Figure 1E). Thus we were able to acquire proteome-wide spatial and temporal information in matching samples from a single experiment.
Protein turnover changes in unfolded protein response is dependent on cellular localization
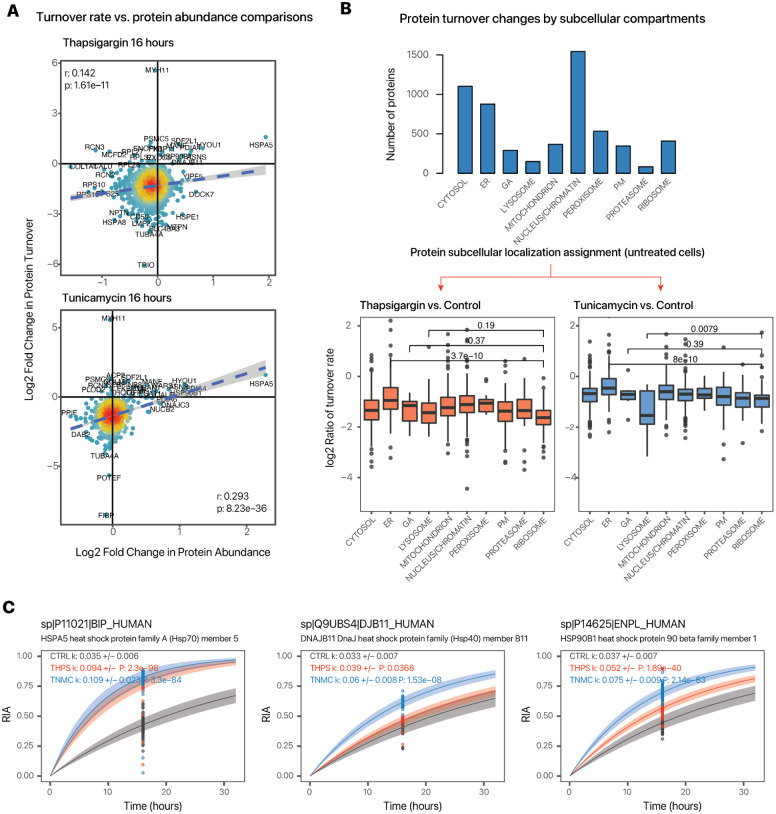
Considering first the turnover kinetics data, we observed a proteome-wide decrease in fractional synthesis rates under both thapsigargin and tunicamycin challenge. Nevertheless, we also observed a wide range of turnover rates for proteins in both the untreated and thapsigargin and tunicamycin treated conditions, suggesting that the cells are actively synthesizing and degrading proteins on top of the passive growth and diffusion from cell doubling (Figure 2A). There was a weak but statistically significant positive correlation between the proteome-wide changes in protein abundance and turnover rates in both models (thapsigargin r: 0.142; tunicamycin r: 0.293). A notable increase in the abundance and turnover rates of BiP/GRP78 (HSPA5) was observed in both models. The observed magnitudes of turnover changes were generally higher than that of abundance changes. There were also notable divergences between turnover and abundance changes, including MYH11 which showed increased turnover rates but not abundance. Of interest, a prior study showed that an overexpression of Myh11 at the transcript level in mouse smooth muscle cells led to ER stress and protein degradation (Kwartler et al., 2014). The turnover data here are consistent with the finding that levels of MYH11 may be regulated primarily by turnover and degradation. Pathway enrichment analyses show that slow and fast turnover proteins are preferentially associated with different cellular functions: whereas slow-turnover proteins are statistically enriched in protein metabolism processes including translation, rRNA processing, and proteolysis; fast turnover proteins are enriched in signaling and regulatory processes including receptor tyrosine kinase signaling, G-protein coupled receptor signaling, and SUMOylation (Supplemental Figure S2).
Figure 2.
Changes in protein temporal kinetics under UPR. A. Scatter plots showing the distribution of protein log2 fold change in abundance (x-axis) and log2 fold change in turnover rates (y-axis) in thapsigargin and tunicamycin treatment. B. Relationship between turnover changes and subcellular localization. Bar charts show the allocation of predicted subcellular location of all measured proteins in normal cells. The box plots show log2 ratio of turnover rate constants (k) between thapsigargin and control (left) cells and between tunicamycin and control (right) cells based on the predicted subcellular location. P values: ANOVA post-hoc between ER, Golgi, lysosome proteins with ribosomal proteins. C. Example of best fit curves in the first-order kinetic model at the protein level between control (gray), thapsigargin (red); and tunicamycin (blue) treated cells, showing an increase in turnover in UPR of several known ER stress response proteins BiP/HSPA5, DJB11, and HSP90B1. Points represent different tryptic peptides.
To interrogate how protein turnover is regulated with regard to protein spatial distributions, we cross-referenced the protein kinetics data to spatial proteomics information, and found that the turnover changes in UPR are stratified across subcellular localizations and moreover the spatial contexts of protein turnover regulation differ by the two UPR models (Figure 2B). Under thapsigargin, proteins that are predicted to co-localize with ribosome markers had the greatest reduction in fractional synthesis rates consistent with a suppression of protein synthesis through ribosome remodelingProteins in the ER by contrast had the least reduction in fractional synthesis rates and also includ. ed 8 out of the 13 proteins with significantly increased turnover (>1.25-fold, P value < 0.01) following thapsigargin, and 13 out of 21 proteins in tunicamycin.
In parallel, tunicamycin treatment led to a lesser suppression of turnover rates than thapsigargin, whereas the greatest reduction in synthesis was observed among proteins classified to be located to the lysosome, a compartment closely linked to glycosylation and recycling of glycans. In both drugs, we observed a number of proteins with increased turnover rates despite the global synthesis decrease and which are concentrated in proteins predicted to be located in the ER (Figure 2C). These include a complement of established ER stress response factors, including most prominently BiP/HSPA5 (3.0-fold increase in thapsigargin; P: 7.6e–63; 2.9-fold increase in tunicamycin, P: 3.1e–84), as well as HSP90B1, PDIA4, HYOU1, MANF, ALDH1L2; as well as some proteins whose function in ER stress/UPR remain incompletely understood. For instance, SDF2L1 (stromal-cell derived factor 2 like 1) is recently described to form a complex with the ER chaperone DNAJB11 to retain it in the ER (Hanafusa et al., 2019). In control cells, we found that the SDF2L1 protein has a basal turnover rate of 0.025/hr; upon thapsigargin treatment, its turnover rate increased to 0.056 /hr (P: 2.1e–5) and in tunicamycin treated cells 0.067 /hr (P: 2.8e–7). DNAJB11 also experienced increased turnover rates (1.2-fold in thapsigargin, P: 3.6e–3; 1.9-fold in tunicamycin, P: 1.3e–10) suggesting both proteins may be preferentially synthesized during UPR upon ER stress.
We also observed different kinetic signatures in select proteins that may reflect the mechanistic differences in the two UPR activators. CALR (calreticulin), a glycan-binding ER chaperone, shows increased turnover in tunicamycin (1.58-fold, P: 1.5e–9) but not thapsigargin (0.95-fold, P: 0.19). RCN3 (reticulocalbin 3) is an ER lumen calcium binding protein that regulates collagen production (Martínez-Martínez et al., 2017) and shows an increased turnover in thapsigargin (1.7-fold, P: 8.4e–7) but not in tunicamycin.
Taken together, the data confirm UPR induction in AC16 cardiomyocyte and corroborate complex protein regulations through temporal kinetics changes. Moreover, such changes are dependent on the UPR inducing agent and the subcellular localization of proteins. Overall, the results suggest an interaction between protein turnover kinetics in UPR and the compartment to which the proteins are localized, and that the incorporation of spatial proteomics data provided further regulatory contexts to kinetics measurements.
Localization of existing protein pools and new protein synthesis
Because each of the SILAC labeled pairs (light and heavy) and their associated TMT profiles were separately quantified by mass spectrometry as independent ions, we reason the SPLAT experimental design will enable additional power to determine cellular localization and changes (translocation) during perturbation in two ways; first, the heavy peptide localization profiles allow additional confirmation of localization classification in the same experiment as technical replicates. On the contrary, if newly synthesized proteins reside in a different cellular locale (e.g., ribosome or ER), the SPLAT design may help improve the resolution of spatial signals by preventing the heavy peptides from otherwise contaminating the spatial signals from the pre-existing protein pool.
We examined whether the light (existing) and heavy (new synthesis) proteins are localized to the identical cellular locales. By training the TAGM-MAP models of light and heavy proteins separately, we found that both existing and post-labeling synthesis proteins supported the construction of subcellular proteome maps with similar localization patterns, which corroborates the overall quality of spatial localization classification as the heavy proteins are never used as markers to train the models. When both light and heavy proteins were classified in the same model, we found that under baseline conditions, 93.3% (1921 out of 2059) proteins for which there is a spatial localization call at ≥90% probability at either or both the light or heavy isotopologue were localized to the same subcellular compartments. When proteins where both the light and heavy labeled isotopologues are confidently localized, 98.7% (1387 out of 1405) are localized to the same compartment. Hence, the spatial information shows good reproducibility across SILAC light and heavy labeled proteins.
Spatial distribution of existing and newly synthesized proteins enables distinction of translocation mechanisms
A change in spatial distribution of proteins following perturbation could be due to distinct localization mechanisms. In the case of a relocation of an existing protein, the protein in question may respond to a signaling cue such as a post-translational modification status. The protein subsequently exits a specific subcellular locale and migrates to a different subcellular location. Alternatively, a protein translocation could also be due to the alternate localization of newly synthesized proteins. The latter is expected to be a feature in UPR as protein synthesis pathways become stalled. The incorporation of dynamic SILAC data in SPLAT distinguishes these two mechanisms by considering whether the light (preexisting) and heavy labeled (post-treatment synthesized) protein pools exhibit a change in localization or only the heavy labeled pool does. In other words, we might anticipate the relocation of existing proteins to be reflected by concurrent differential localization of light and heavy SILAC labeled proteins, assuming translocation occurs at a higher rate than protein synthesis, whereas conversely we may expect only the heavy SILAC labeled protein to display a change in location when there is endomembrane stalling or delayed translocation.
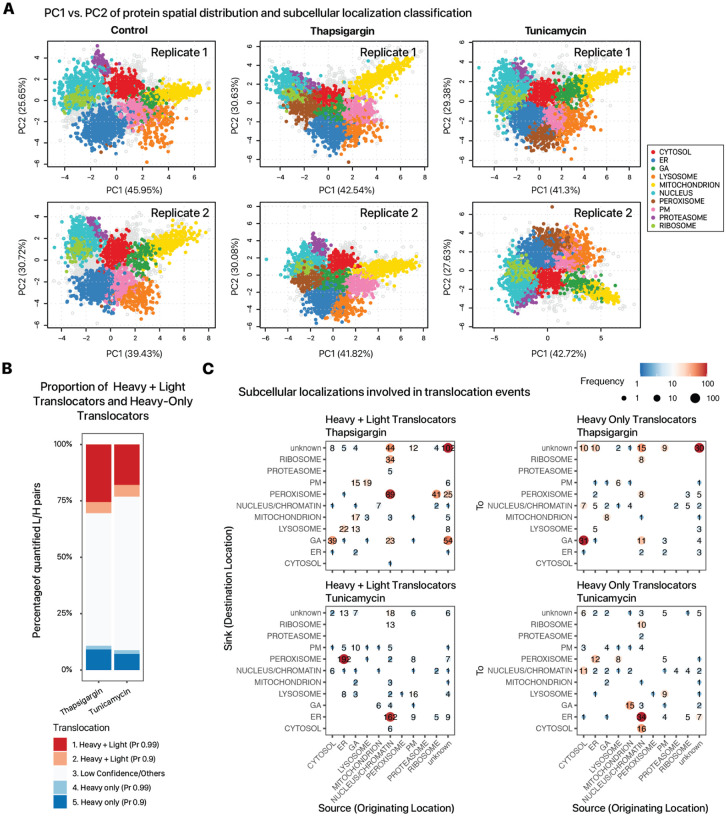
To find instances of relocation of existing protein pools, we therefore analyzed the spatial proteomics data to find proteins where both light (pre-existing) and heavy (newly synthesized) proteins show a change in predicted localization in UPR. To do so, we used a Bayesian statistical model implemented in the BANDLE package to estimate the differential spatial localization of proteins (Figure 3A). In total, we identified 2172 candidate translocators (light or heavy BANDLE probability > 0.99) in thapsigargin and 1946 in tunicamycin, which we prioritized to 1012 and 973 protein pairs in thapsigargin and tunicamycin, respectively, for which the localization of both light and heavy isotopologues were quantified, and for which either or both isotopologues are candidate translocators (Figure 3B). Manual inspection of these results discerned three major categories of differential localization behaviors.
Figure 3.
Protein translocation of light and heavy proteins following thapsigargin and tunicamycin treatment. A. Protein localization classification at 95% confidence in control, thapsigargin, and tunicamycin treated cells across 2 biological replicates. B. Bar plot showing the proportion of detected translocation events by BANDLE where either the heavy and light proteins are predicted to be translocated at 90% and 99% probability (red), vs. where only the heavy protein is predicted to be translocated at 90% and 99% probability (blue) in thapsigargin (left bars) and tunicamycin (right bars) treated cells. C. Density of translocation events where heavy and light proteins translocate (left) vs. only heavy proteins translocate (right) in thapsigargin (upper) and tunicamycin (lower) treated cells. In each plot, the x axis represents the originating subcellular location of a candidate protein translocation event, and the y axis represents the target location. Data point size and color denote the frequency of candidate translocation events in each source-destination location pair.
First, we saw that in both thapsigargin and tunicamycin treated cells, there is a more apparent behavior of proteins classified into a class of peroxisome-cosedimenting fraction (Supplemental Figure S3). Translocation to this fraction involves primarily proteins whose light and heavy isotopologues show differential localization, suggesting vesicular transport that concerns pre-existing proteins (Figure 3C). In mammalian cells, ER and peroxisomes are spatially adjacent; the peroxisome spatial fraction here is distinguished from the ER fraction as it begins to sedimentation at earlier ultracentrifugation steps (F3 and F4) at 5000–9000 × g in the LOPIT-DC protocol, and moreover contains canonical peroxisome markers PEX14 and ACOX1; however, critically we also observer that the majority of proteins categorized into this fraction do not conform to canonical peroxisome functions, and moreover there are known components of the ER-to-Golgi COPII transport vesicles including SEC23/24 in thapsigargin treated samples, and the snare protein SEC22 in tunicamycin. We interpret the expansion of this peroxisome co-sedimenting fraction instead to include also ER-derived transport vesicles (ERVs). We identified previously known translocators including the above mentioned vesicular transport proteins (SEC23A/B, SEC24A/B/C/D, SEC31A); dyneins (DYNC1H1, DYNC1I2, DYNC1LI2); other vesicle proteins (VAPA, VAPBB) toward the peroxisome/ERV fraction, as well as known stress response genes (TMEM33, TOR1A, TOR1B, and UFL1 in tunicamycin). Curiously, the primary origin of the translocation events to this fraction differs by the UPR induction mechanism. Whereas in thapsigargin, there is a greater occurrence of a cluster of nucleic acid binding proteins that change in locations, including DNA repair proteins and RNA binding proteins (CNOT10, CPSF6, CPSF7, EDF1, HNRNPL, HNRNPM, MOV10, RBM10, RBM14, RBMX, SRSF9, TAF15) which indicates a possible egress from the nucleus/ribosome associated fractions toward the ERV enriched fraction (Supplemental Figure S4A). On the contrary, in tunicamycin, there is a preponderance of stress response proteins including UFL1, TOR1A, TOR1B, and TMEM33 that translocate from the main ER fraction to the peroxisome/ERV fraction.
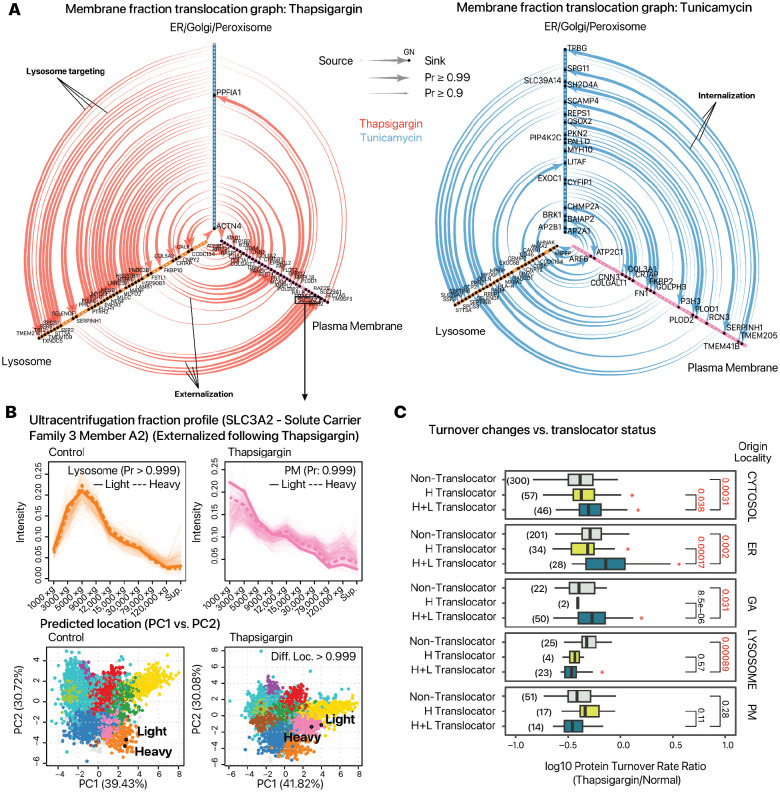
Secondly, in close relation to vesicular remodeling, we observed a complex network of externalization and internalization of proteins from the plasma membrane to the ER/Golgi, lysosome, and plasma membrane axes (Figure 4A). Among the externalized proteins are the dibasic amino acid transporter component SLC3A2, and the histidine/neutral amino acid transporter SLC7A5/LAT1 (Figure 4B). The correlation of these translocations to function is not clear; however, there are close links between ER stress conditions and amino acid biosynthesis; UPR activation is known to induce the biosynthesis of non-essential or partly-essential amino acids despite protein synthesis suppression (Gonen et al., 2019); the recycling of lysosomal lysine and arginine regulates the sensitivity to ER stress (Higuchi-Sanabria et al., 2020); whereas deprivation of amino acids is known to activate downstream pathways of UPR including CHOP in vitro (Harding et al., 2003). Conversely, a number of proteins were internalized from the plasma membrane fraction toward the ER and the Golgi, including several known cell surface transducer such as PDGFRB, PDLIM7, PLCB3, STK10, and VASP in thapsigargin treated cells.
Figure 4.
Membrane trafficking and protein redistribution in UPR. A. The graphs show the detected candidate translocating events across three principal membrane location groups (ER, Golgi, and peroxisome/ERV (ER); lysosome; and plasma membrane (PM)) observed upon thapsigargin (left) and tunicamycin (right) treatment. Arrows of the edge denote the directionality of the candidate translocation events. Example lysosome targeting (ER to Lysosome), externalization (Lysosome to PM), and internalization (PM to ER) events are highlighted. B. The ultracentrifugation profile and predicted location of SLC3A2 in control vs. thapsigargin treated cells. SLC3A2 is localized to the lysosome in control cells and becomes externalized upon thapsigargin. In the line plots (upper), light proteins are represented by the heavy solid lines, heavy proteins by the heavy dashed lines. Transparent lines represent the ultracentrifugation fraction distributions of all proteins classified to the lysosome (orange) and the plasma membrane (PM; pink). In the PCA plots (lower), the black circle denotes the predicted location of light and heavy SLC3A2 in each condition, overlaid on the spatial distribution and predicted subcellular localization of other proteins (colors). C. Translocating proteins originating from the cytosol, ER, and Golgi showed relatively higher turnover rate ratios in thapsigargin vs. normal cells compared to non-translocating proteins in the same compartment, suggesting changes in subcellular localization are coupled to protein kinetics changes. P values: student’s t test. Asterisks: P < 0.05 vs. non-translocators. Parenthesis: number of proteins in each group.
Thirdly, there was also evidence of lysosome targeting from other endomembrane compartments, including IKBIP in thapsigargin and tunicamycin treated cells, and MANF in thapsigargin treated cells. Among these proteins were also extracellular matrix remodeling and collagen modifying enzymes, including P3H1, P4HA1, SERPINH1 in both thapsigargin and tunicamycin; as well as the collagen lysyl hydroxylases PLOD1 and PLOD2 in tunicamycin treated cells. SERPINH1/HSP47 in particular is a collagen chaperone as well as UPR sensor that binds with IRE1alpha which shows a strong translocation from the ER/Golgi to the lysosome fraction under UPR (Supplemental Figure S4B). Interestingly, prior work has found no obvious correlation between the protein levels of collagen modifying enzymes with the observed suppression in collagen synthesis in chondrocytes and fibroblasts under ER stress (Vonk et al., 2010). The results here suggest that correlates with functional decrease may instead reside with a change in the subcellular localization of collagen modifying enzymes in AC16 cells.
Interestingly, translocating proteins (BANDLE differential localization probability ≥ 0.9) originating from the cytosol, ER, and Golgi had relatively higher turnover ratios in thapsigargin treated compared to normal cells than other proteins in the same compartments (Figure 4C). Hence, changes in localization are accompanied by change in turnover relative to the proteome average. We speculate that proteins that change in localization also tend to have higher turnover because these proteins are specifically involved in stress response. Under protein misfolding stress, cells may need to rapidly alter the functions of certain proteins. Here, proteins that change in localization may be those that need to be shuttled to different parts of the cell in order to perform new functions, and the fact that they also have higher turnover may be attributable to their increased activity and importance during this time.
Preferential translocation of newly synthesized protein pools
We next considered another case of interconnected temporal and spatial dynamics, namely whether some localization changes are contingent upon protein pool lifetime, such as where the light (existing) protein does not change in spatial distribution but the heavy (post-labeling synthesis) proteins display differential translocation upon UPR. This experimental design is distinct from time-course experiments where static samples are taken across multiple time points post-synthesis, as it allows the interactions between post-synthesis lifetime of the protein and their spatial localization to be discerned. From the thapsigargin and tunicamycin data, we discerned 192 and 183 candidate proteins each that show temporal dependent spatial distributions, respectively, where the heavy isotopologue shows translocation (99% probability) but the light isotopologue does not (<50% probability), which correspond to ~20% of candidate translocators.
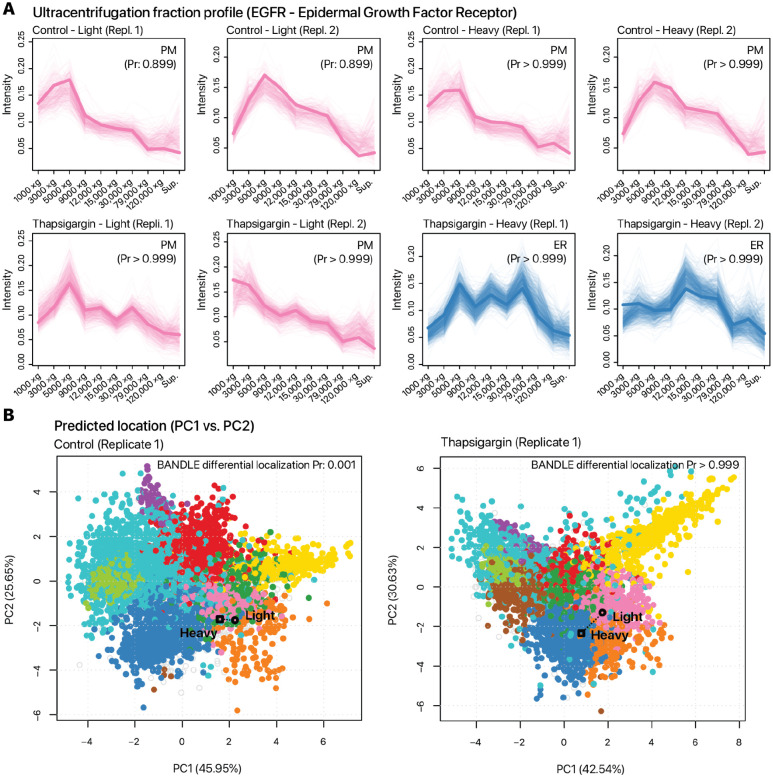
We highlight EGFR1 which shows an ER retention of newly synthesized proteins in both thapsigargin and tunicamycin treatment, and MAN2B1 which shows ER retention in tunicamycin-treated cells. Epidermal growth factor receptor (EGFR/ErbB1/HER1) is a receptor tyrosine kinase with multiple subcellular localizations and signaling roles, and is implicated in cardiomyocyte survival (Lee et al., 2020). Following a variety of stressors, EGFR is known to be inactivated by intracellular trafficking, including being internalized to the early endosome and lysosome following oxidative stress and hypoxia in cancer cells (Tan et al., 2016). In the spatial proteomics data, the spatial distribution of EGFR borders the lysosome and plasma membrane fractions, which we interpret as EGFR having potential multiple pools including a cell surface fraction (Figure 5A). In the thapsigargin and tunicamycin treated cells, the pre-existing (light) EGFR pool shows no signs of translocation (BANDLE differential localization: 0.001) whereas the dynamic SILAC labeled pool (heavy) becomes internalized (BANDLE differential localization: 1.000) (Figure 5B). The data therefore suggests that the internalization of EGFR under thapsigargin and tunicamycin is likely ligand-independent and instead due to endomembrane stalling or redistribution upon new protein synthesis.
Figure 5.
Protein lifetime dependent translocation. EGFR is shown as an example where the heavy SILAC labeled (new synthesis pool) protein shows differential response to UPR. A. Post-labeling synthesized EGFR is differentially distributed and shows ER retention, whereas the preexisting EGFR pool remains to show a likely cell surface localization after thapsigargin or tunicamycin challenge. B. Overlaid comparison of EGFR light and heavy proteins on PC1 vs PC2 of the spatial distribution data in control (left) and thapsigargin treated (right) cells.
We observed other instances of candidate endomembrane stalling events, for instance, cathepsin A (CTSA/PPGB) is localized to the lysosomal fraction in the control cells (light and heavy) but becomes localized to the peroxisome/ERV fraction in the newly synthesized pool upon tunicamycin treatment. Hence whereas the light protein shows no evidence of translocation (BANDLE differential localization < 0.001), the heavy protein likely becomes mislocalized (BANDLE differential localization: 1.000) (Supplemental Figure S5A–B). In thapsigargin-treated cells, newly-synthesized ITGAV (integrin subunit alpha V) shows an internalization from the plasma membrane fraction to the ER fraction (BANDLE differential localization: 1.000) but not the existing light protein pool (BANDLE differential localization: 0.002) (Supplemental Figure S6A–B). With the function of integrins as cell surface receptors that function in intracellular-to-extracellular and retrograde communication, the internalization of a newly synthesized V class integrin subunit would be indicative of a decrease in integrin signaling through spatial regulation whereas the preferential internalization of newly synthesized pools might suggest that this process is ligand independent. Taken together, these and other examples suggest the SPLAT strategy is able to distinguish translocation from new synthesis trafficking changes.
Discussion
Advances in spatial proteomics have opened new avenues to discover the subcellular localization of proteins on a proteome scale. Thus far however, few efforts have linked the spatial dynamics of the proteome to other dynamic proteome parameters, which hinders a multi-dimensional view of protein function (Burnum-Johnson et al., 2022). To our knowledge, this is the first work to incorporate time-resolved stable isotope labeling kinetics to connect proteome-wide protein life time and translocation in stress response. Applying this workflow to human AC16 cardiomyocytes under thapsigargin and tunicamycin induced ER stress and the associated UPR, we captured the coordinated activation of dozens of known ER stress mediators through their increased turnover. At the same time, protein spatial profiles recapitulated known endomembrane remodeling events as well as revealed a large list of over 500 subcellular translocating proteins under thapsigargin and tunicamycin mediated ER stress. At the time of writing, a recent preprint has also reported ~75 translocator protein candidates under acute low-dose thapsigargin (250 nM for 1 hr) in U-2 OS cells, although the experimental approach was more optimized toward mRNA detection (Villanueva et al., 2022). The data here therefore represents one of the more comprehensive lists of protein translocation under ER stress and UPR, and suggest a number of novel salient features including the differential localization of DNA and RNA binding proteins to peroxisome/ER-derived transport fractions, the externalization of amino acid transporters, as well as endomembrane redistribution of extracellular matrix and collagen synthesis proteins. By comparing the independent translocation behaviors of light and heavy SILAC peptides, the reported method can distinguish between two distinct translocation mechanisms, namely the relocation of existing protein molecules and the redirection of newly synthesized proteins, with potentially different mechanistic and translational implications.
It is likely other biologically relevant translocators reside in the data. Upward of 1,000 candidate translocation patterns were detected with significant differences in localization (99% probability) by BANDLE, but many proteins presented a challenge to clear interpretation upon manual inspection. Some fractions including the nucleus/chromatin proteins and the ribosome have similar sedimentation profiles in the LOPIT-DC protocol; and because translocation may be partial stoichiometrically, translocated proteins often have lower confidence in classification of location. Additional analysis may shed light on these proteins.
The current SPLAT experimental design has several limitations. Firstly, although there is no inherent limit to the number of dynamic SILAC labeling time points that can be investigated, here we have chosen one time point (16 hrs post thapsigargin or tunicamycin), which was selected based on the median half-life of proteins in proliferating AC16 cells based on prior experience. Synthesized proteins may have further relocalized following 16 hours, hence acute translocation responses may be missed. The inclusion of earlier time points might provide insight into early translocation events but may be technically challenging, as the acquisition of spatial localization information from the heavy SILAC labeled peptides would be hindered by their low intensity. Secondly, the double labeling design requires independent MS2 acquisition of light and heavy peptides, which can decrease the depth and data completeness of mass spectrometry-based proteomics analysis. Future work may alleviate this limitation by modifying the mass spectrometry acquisition methods to automatically trigger the acquisition of heavy peptides and reduce incomplete light-heavy pairs. Thirdly, the differential ultracentrifugation method employed places a limit on the number of classifiable subcellular localizations here, which are in line with other LOPIT-DC studies, but could not resolve some other fractions, e.g., the nucleus vs. chromatin, 40S vs. 60S ribosomes, and actin-associated fractions. This may be alleviated in future work that couples turnover analysis to gradient-based sedimentation approaches with higher spatial resolution.
Future work may also improve upon current limitations inherent to spatial proteomics strategies, which face challenges in recognizing proteins with multiple localizations or partial translocations. For instance, the multi-functional ERAD protein p97/VCP is known to have multiple subcellular localizations and in our data set is shown to be a potential translocator. However, its precise subcellular translocation profile is difficult to interpret from the TMT data. In the control cells, it is unclassified and likely falls between ER and cytosol in the profile; in thapsigargin treated sample, its location remains unclassified but shares features of ER/Golgi and peroxisome-coseidmenting fractions. Progress in this area may require development of spatial separation methods that combine orthogonal separation principles.
In conclusion, we describe an experimental workflow and data analysis pipeline that integrates dynamic SILAC based proteome turnover analysis with differential ultracentrifugation-based subcellular proteomics to characterize the temporal and spatial aspects of UPR in a single experiment. The simultaneous analysis of spatiotemporal parameters may be useful for understanding of the function and behaviors of proteins inside the cell, and may provide new insight into the mechanisms that regulate protein stability and localization in stress and disease.
Supplementary Material
Acknowledgments
This work was funded in part by NIH awards R01-HL141278 and R01-GM144456 to M.P.L. and NIH awards R03-OD032666, R35-GM146815, and R00-HL144829 to E.L.
References
- Andrews B., Murphy A.E., Stofella M., Maslen S., Almeida-Souza L., Skehel J.M., Skene N.G., Sobott F., Frank R.A.W., 2022. Multidimensional Dynamics of the Proteome in the Neurodegenerative and Aging Mammalian Brain. Mol. Cell. Proteomics 21, 100192. 10.1016/j.mcpro.2021.100192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnum-Johnson K.E., Conrads T.P., Drake R.R., Herr A.E., Iyengar R., Kelly R.T., Lundberg E., MacCoss M.J., Naba A., Nolan G.P., Pevzner P.A., Rodland K.D., Sechi S., Slavov N., Spraggins J.M., Van Eyk J.E., Vidal M., Vogel C., Walt D.R., Kelleher N.L., 2022. New Views of Old Proteins: Clarifying the Enigmatic Proteome. Mol. Cell. Proteomics 21, 100254. 10.1016/j.mcpro.2022.100254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartron J.W., Hunt K.C.L., Frydman J., 2016. Cotranslational signal-independent SRP preloading during membrane targeting. Nature 536, 224–228. 10.1038/nature19309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopher J.A., Stadler C., Martin C.E., Morgenstern M., Pan Y., Betsinger C.N., Rattray D.G., Mahdessian D., Gingras A.-C., Warscheid B., Lehtiö J., Cristea I.M., Foster L.J., Emili A., Lilley K.S., 2021. Subcellular proteomics. Nat. Rev. Methods Primer 1, 32. 10.1038/s43586-021-00029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claydon A.J., Beynon R., 2012. Proteome dynamics: revisiting turnover with a global perspective. Mol. Cell. Proteomics MCP 11, 1551–1565. 10.1074/mcp.O112.022186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook O.M., Breckels L.M., Lilley K.S., Kirk P.D.W., Gatto L., 2019. A Bioconductor workflow for the Bayesian analysis of spatial proteomics. F1000Research 8, 446. 10.12688/f1000research.18636.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook O.M., Davies C.T.R., Breckels L.M., Christopher J.A., Gatto L., Kirk P.D.W., Lilley K.S., 2022. Inferring differential subcellular localisation in comparative spatial proteomics using BANDLE. Nat. Commun. 13, 5948. 10.1038/s41467-022-33570-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook O.M., Mulvey C.M., Kirk P.D.W., Lilley K.S., Gatto L., 2018. A Bayesian mixture modelling approach for spatial proteomics. PLOS Comput. Biol. 14, e1006516. 10.1371/journal.pcbi.1006516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M.K., Hammond D.E., Clague M.J., Gaskell S.J., Beynon R.J., 2009. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112. 10.1021/pr800641v [DOI] [PubMed] [Google Scholar]
- Dostal V., Wood S.D., Thomas C.T., Han Y., Lau E., Lam M.P.Y., 2020. Proteomic signatures of acute oxidative stress response to paraquat in the mouse heart. Sci. Rep. 10, 18440. 10.1038/s41598-020-75505-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J.K., Hoopmann M.R., Jahan T.A., Egertson J.D., Noble W.S., MacCoss M.J., 2015. A deeper look into Comet--implementation and features. J. Am. Soc. Mass Spectrom. 26, 1865–1874. 10.1007/s13361-015-1179-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto L., Breckels L.M., Wieczorek S., Burger T., Lilley K.S., 2014. Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics 30, 1322–1324. 10.1093/bioinformatics/btu013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geladaki A., Kočevar Britovšek N., Breckels L.M., Smith T.S., Vennard O.L., Mulvey C.M., Crook O.M., Gatto L., Lilley K.S., 2019. Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics. Nat. Commun. 10, 331. 10.1038/s41467-018-08191-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen N., Meller A., Sabath N., Shalgi R., 2019. Amino Acid Biosynthesis Regulation during Endoplasmic Reticulum Stress Is Coupled to Protein Expression Demands. iScience 19, 204–213. 10.1016/j.isci.2019.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond D.E., Simpson D.M., Franco C., Wright Muelas M., Waters J., Ludwig R.W., Prescott M.C., Hurst J.L., Beynon R.J., Lau E., 2022. Harmonizing Labeling and Analytical Strategies to Obtain Protein Turnover Rates in Intact Adult Animals. Mol. Cell. Proteomics 21, 100252. 10.1016/j.mcpro.2022.100252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafusa K., Wada I., Hosokawa N., 2019. SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum localization and chaperone activity of ERdj3 protein. J. Biol. Chem. 294, 19335–19348. 10.1074/jbc.RA119.009603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D.F., Bell J.C., Hettmann T., Leiden J.M., Ron D., 2003. An Integrated Stress Response Regulates Amino Acid Metabolism and Resistance to Oxidative Stress. Mol. Cell 11, 619–633. 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- Hetz C., Zhang K., Kaufman R.J., 2020. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 21, 421–438. 10.1038/s41580-020-0250-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi-Sanabria R., Shen K., Kelet N., Frankino P.A., Durieux J., Bar-Ziv R., Sing C.N., Garcia E.J., Homentcovschi S., Sanchez M., Wu R., Tronnes S.U., Joe L., Webster B., Ahilon-Jeronimo A., Monshietehadi S., Dallarda S., Pender C., Pon L.A., Zoncu R., Dillin A., 2020. Lysosomal recycling of amino acids affects ER quality control. Sci. Adv. 6, eaaz9805. 10.1126/sciadv.aaz9805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert N., Shofstahl J., Sachsenberg T., Walzer M., Barsnes H., Martens L., Perez-Riverol Y., 2020. ThermoRawFileParser: Modular, Scalable, and Cross-Platform RAW File Conversion. J. Proteome Res. 19, 537–542. 10.1021/acs.jproteome.9b00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., Weissman J.S., 2014. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science 346, 1257521. 10.1126/science.1257521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.A., Hofstadter W.A., Cristea I.M., 2020. TRANSPIRE: A Computational Pipeline to Elucidate Intracellular Protein Movements from Spatial Proteomics Data Sets. J. Am. Soc. Mass Spectrom. 31, 1422–1439. 10.1021/jasms.0c00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwartler C.S., Chen J., Thakur D., Li S., Baskin K., Wang S., Wang Z.V., Walker L., Hill J.A., Epstein H.F., Taegtmeyer H., Milewicz D.M., 2014. Overexpression of smooth muscle myosin heavy chain leads to activation of the unfolded protein response and autophagic turnover of thick filament-associated proteins in vascular smooth muscle cells. J. Biol. Chem. 289, 14075–14088. 10.1074/jbc.M113.499277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju A.K.K., Mary C., Scherrer A., Johnson A.E., Strub K., 2008. SRP Keeps Polypeptides Translocation-Competent by Slowing Translation to Match Limiting ER-Targeting Sites. Cell 133, 440–451. 10.1016/j.cell.2008.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M.P.Y., Wang D., Lau E., Liem D.A., Kim A.K., Ng D.C.M., Liang X., Bleakley B.J., Liu C., Tabaraki J.D., Cadeiras M., Wang Y., Deng M.C., Ping P., 2014. Protein kinetic signatures of the remodeling heart following isoproterenol stimulation. J. Clin. Invest. 124, 1734–1744. 10.1172/JCI73787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E., Cao Q., Lam M.P.Y., Wang J., Ng D.C.M., Bleakley B.J., Lee J.M., Liem D.A., Wang D., Hermjakob H., Ping P., 2018. Integrated omics dissection of proteome dynamics during cardiac remodeling. Nat. Commun. 9, 120. 10.1038/s41467-017-02467-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Koeppen M., Seo S.-W., Bowser J.L., Yuan X., Li J., Sibilia M., Ambardekar A.V., Zhang X., Eckle T., Yoo S.-H., Eltzschig H.K., 2020. Transcription-independent Induction of ERBB1 through Hypoxia-inducible Factor 2A Provides Cardioprotection during Ischemia and Reperfusion. Anesthesiology 132, 763–780. 10.1097/ALN.0000000000003037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberg M.K., Strisovsky K., 2021. Maintenance of organellar protein homeostasis by ER-associated degradation and related mechanisms. Mol. Cell 81, 2507–2519. 10.1016/j.molcel.2021.05.004 [DOI] [PubMed] [Google Scholar]
- Mårtensson C.U., Priesnitz C., Song J., Ellenrieder L., Doan K.N., Boos F., Floerchinger A., Zufall N., Oeljeklaus S., Warscheid B., Becker T., 2019. Mitochondrial protein translocation-associated degradation. Nature 569, 679–683. 10.1038/s41586-019-1227-y [DOI] [PubMed] [Google Scholar]
- Martínez-Martínez E., Ibarrola J., Fernández-Celis A., Santamaria E., Fernández-Irigoyen J., Rossignol P., Jaisser F., López-Andrés N., 2017. Differential Proteomics Identifies Reticulocalbin-3 as a Novel Negative Mediator of Collagen Production in Human Cardiac Fibroblasts. Sci. Rep. 7, 12192. 10.1038/s41598-017-12305-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell G.S., Gaun A., Steen H., 2013. iFASP: combining isobaric mass tagging with filter-aided sample preparation. J. Proteome Res. 12, 3809–3812. 10.1021/pr400032m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey C.M., Breckels L.M., Crook O.M., Sanders D.J., Ribeiro A.L.R., Geladaki A., Christoforou A., Britovšek N.K., Hurrell T., Deery M.J., Gatto L., Smith A.M., Lilley K.S., 2021. Spatiotemporal proteomic profiling of the pro-inflammatory response to lipopolysaccharide in the THP-1 human leukaemia cell line. Nat. Commun. 12, 5773. 10.1038/s41467-021-26000-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orre L.M., Vesterlund M., Pan Y., Arslan T., Zhu Y., Fernandez Woodbridge A., Frings O., Fredlund E., Lehtiö J., 2019. SubCellBarCode: Proteome-wide Mapping of Protein Localization and Relocalization. Mol. Cell 73, 166–182.e7. 10.1016/j.molcel.2018.11.035 [DOI] [PubMed] [Google Scholar]
- Ren J., Bi Y., Sowers J.R., Hetz C., Zhang Y., 2021. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat. Rev. Cardiol. 18, 499–521. 10.1038/s41569-021-00511-w [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M., 2011. Global quantification of mammalian gene expression control. Nature 473, 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Tan X., Lambert P.F., Rapraeger A.C., Anderson R.A., 2016. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends Cell Biol. 26, 352–366. 10.1016/j.tcb.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The M., MacCoss M.J., Noble W.S., Käll L., 2016. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 27, 1719–1727. 10.1007/s13361-016-1460-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P.-L., Cameron C.J.F., Forni M.F., Wasko R.R., Naughton B.S., Horsley V., Gerstein M.B., Schlieker C., 2022. Dynamic quality control machinery that operates across compartmental borders mediates the degradation of mammalian nuclear membrane proteins. Cell Rep. 41, 111675. 10.1016/j.celrep.2022.111675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva E., Smith T., Pizzinga M., Elzek M., Queiroz R.M.L., Harvey R.F., Breckels L.M., Crook O.M., Monti M., Dezi V., Willis A.E., Lilley K.S., 2022. A system-wide quantitative map of RNA and protein subcellular localisation dynamics (preprint). Cell Biology. 10.1101/2022.01.24.477541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk L.A., Doulabi B.Z., Huang C.-L., Helder M.N., Everts V., Bank R.A., 2010. Endoplasmic reticulum stress inhibits collagen synthesis independent of collagen-modifying enzymes in different chondrocyte populations and dermal fibroblasts. Biochem. Cell Biol. Biochim. Biol. Cell. 88, 539–552. 10.1139/o09-174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been updated to ProteomeXchange under the accession number PXD038054. Code for the SPLAT pipeline is available at https://github.com/lau-lab/splat.