Abstract
GeneMark-ETP is a computational tool developed to find genes in eukaryotic genomes in consistency with genomic-, transcriptomic- and protein-derived evidence. Developed earlier GeneMark-ET and GeneMark-EP+ used a single type of external to genome data, either fragments of transcripts (short RNA reads) or with homologous protein sequences, respectively. Both the transcript- and protein-derived evidence have uneven distribution across a genome. Therefore, GeneMark-ETP is using an approach dictated by the availability of comprehensive but non-uniform data. It finds the genomic loci where extrinsic data are sufficient for gene identification with ‘high confidence’ and then proceeds with finding of the remaining genes across the whole genome. The initial parameters of the algorithm statistical model, generalized HMM, are estimated on the training set composed of high confidence genes. These model parameters are iteratively re-estimated in the cycles of gene prediction and parameter estimation until reaching convergence and making the final prediction of the whole complement of genes. Since the difficulty of gene prediction task ramps up significantly in large plant and animal genomes, the focus of the new development was on large eukaryotic genomes. The GeneMark-ETP performance was favorably compared with the ones of GeneMark-ET, GeneMark-EP+, BRAKER1, and BRAKER2, the methods using a single type of extrinsic evidence. A comparison was also made with TSEBRA, a tool constructing an optimal combination of gene predictions made by BRAKER1 and BRAKER2, thus utilizing both transcript- and protein-derived evidence.
Introduction
Computational methods of gene identification in novel eukaryotic genomes could use both intrinsic and extrinsic evidence. The intrinsic evidence, the species-specific patterns of nucleotide ordering, could be used as a solo source in a self-training gene finder delivering sufficiently accurate predictions of protein-coding genes in fungal and protist genomes (Lomsadze et al. 2005; Ter-Hovhannisyan et al. 2008). Addition of the extrinsic evidence, however, was indispensable for accurate gene prediction in large eukaryotic genomes that carry long non-coding regions, leaving an ample space for false positive predictions (Guigo et al. 2006; Coghlan et al. 2008; Goodswen et al. 2012; Mudge and Harrow 2016; Scalzitti et al. 2020). Strictly extrinsic evidence-based approaches, using either a space of proteins, exonerate (Slater and Birney 2005), GenomeThreader (Gremme et al. 2005), or ProSplign (Kiryutin et al. 2007), or a space of transcripts, StringTie (Pertea et al. 2015; Kovaka et al. 2019), PsiCLASS (Song et al. 2019), and Cufflinks (Trapnell et al. 2010), were shown to be efficient computational tools, though limited to finding subsets of the whole gene complement. The protein-based evidence helps identify genes whose protein products show detectable similarity to cross-species orthologs. The transcripts-based evidence is useful for finding genes with sufficiently high expression. Attempts to combine the two sets of thus predicted genes were made. For instance, GeMoMa (Keilwagen et al. 2018) delivered quite accurate gene predictions for species having well-annotated genomes of the close relatives.
In absence of extrinsic evidence, an ab initio gene finding algorithm relies on features such as k-mer frequency patterns, splice site motifs, intron/exon length distributions, etc. embedded in a HMM type model (e.g., Genie (Kulp et al. 1996), GENSCAN (Burge and Karlin 1997), GeneID (Parra et al. 2000), SNAP (Korf 2004), AUGUSTUS (Stanke and Waack 2003), GeneMark-ES (Lomsadze et al. 2005)). The ab initio methods were observed to be less accurate for the large eukaryotic genomes that carry long non-coding regions, leaving an ample space for false positive predictions (Guigo et al. 2006; Coghlan et al. 2008; Goodswen et al. 2012; Scalzitti et al. 2020). With the advent of massive sequencing of large plant and animal genomes, it was realized that the addition of extrinsic evidence is necessary for accurate genome annotation. We have participated in the research efforts aimed on developing gene finders relying on both intrinsic evidence as well as extrinsic evidence and produced AUGUSTUS (Stanke et al. 2008), GeneMark-ET (Lomsadze et al. 2014) and BRAKER1 (Hoff et al. 2016) the tools integrating genomic and transcript data, as well as AUGUSTUS-PPX (Keller et al. 2011), GeneMark-EP+ (Bruna et al. 2020) and BRAKER2 (Bruna et al. 2021) the tools integrating genomic and protein data. The next step in this process is developing a tool integrating all the three sources of evidence.
The majority of the existing tools relying on the three sources of evidence work as combiners, e.g., FINDER (Banerjee et al. 2021), LoReAn (Cook et al. 2019), GAAP (Kong et al. 2019), IPred (Zickmann and Renard 2015) Evigan (Liu et al. 2008), EVidenceModeler (Haas et al. 2008), JIGSAW (Allen and Salzberg 2005), Combiner (Allen et al. 2004), or GAZE (Howe et al. 2002). A combiner first generates independent sets of genome-wide gene predictions: ab initio-, transcriptomic- and mapped proteins-based. Next, at a post-processing step, these sets are combined into a final set of predictions.
An alternative approach would integrate the three sources of evidence upon a prediction of each gene. In a self-training algorithm — one working without an expert-defined training set — the integration is included into the cycles of iterative model training and gene prediction.
Here we introduce GeneMark-ETP, a new computational tool integrating genomic, transcriptomic, and protein information throughout all the stages of the algorithm’s training and gene prediction. This integration is facilitated upon gene prediction in long transcripts assembled from RNA-Seq reads and supported by verification of the consistency of protein and transcript information. The estimation of parameters of the statistical models (generalized hidden Markov models, GHMM) used in GeneMark-ETP is done by unsupervised training. Protein based evidence, producing hints to locations of introns and exons in genomic DNA, is generated by using homologous proteins of any evolutionary distance, including remote homologs. Accurate accounting for DNA sequence repeats plays a significant role as well.
Tests of the GeneMark-ETP performance were done on both compact and large, GC-homogeneous and GC-heterogeneous eukaryotic genomes. The results were compared with performances of GeneMark-ET, GeneMark-EP+ as well as their virtual combination. The performance of GeneMark-ETP was also compared with the performances of the pipelines BRAKER1 and BRAKER2 as well as with TSEBRA (Gabriel et al. 2021), a recently developed tool combining the BRAKER1 and BRAKER2 predictions.
Results
Assessment of the GeneMark-ETP prediction accuracy
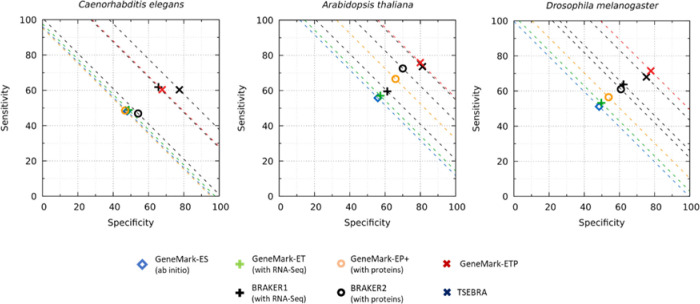
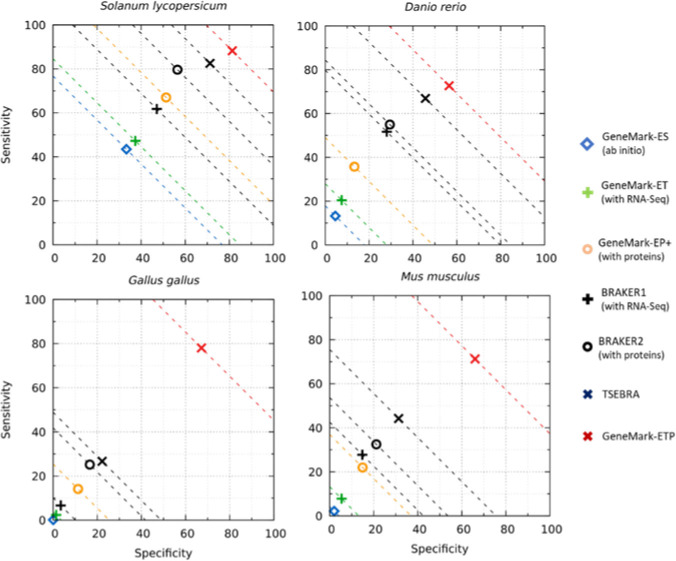
The gene prediction accuracy of GeneMark-ETP (see the algorithm diagram in Fig. 1) was assessed for seven genomes representing diverse genomic organizations and taxonomic clades: Arabidopsis thaliana, Caenorhabditis elegans, Drosophila melanogaster, Solanum lycopersicum, Danio rerio, Gallus gallus and Mus musculus. For the three shorter (compact) genomes A. thaliana, C. elegans, D. melanogaster we accepted genome annotation as the ground truth. For the four large genomes, S. lycopersicum, D. rerio, G. gallus and M. musculus estimations of the gene prediction sensitivity (Sn) were computed for genes present in both NCBI and Ensembl annotations (see Methods) while the gene prediction specificity (Sp) was computed in comparison with the union of the NCBI and Ensemble annotations. We have observed a significant increase in accuracy in comparison with both GeneMark-ET and GeneMark-EP+. Moreover, the improvement was reached also in comparison with both BRAKER1 and BRAKER2 (Figs. 2, 3, S1; Tables S1, S2). The most notable improvements occurred in large genomes, especially the GC-heterogeneous ones. For the groups of compact, large homogeneous, and large heterogeneous genomes, the GeneMark-ETP gene level F1 values increased on average over GeneMark-EP+ by 14.2%, 33.9%, and 55.7%, in each of the above-mentioned groups, respectively (Table S1), while the exon level F1 values for the same three groups of genomes increased on average by 5.4%, 15.2%, and 42.8%, respectively (Table S1).
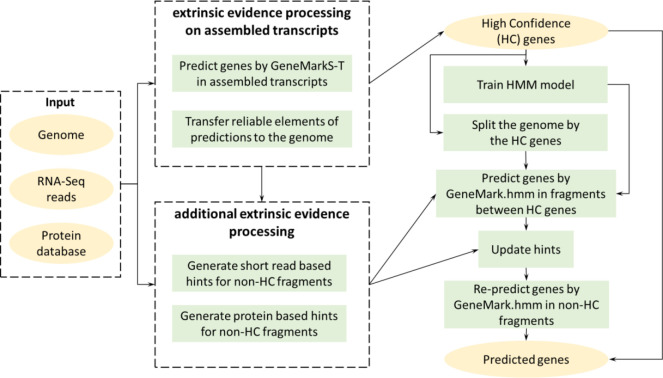
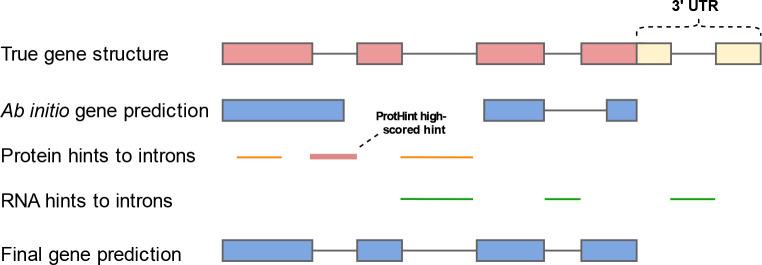
Figure 1.
High-level diagram of the GeneMark-ETP algorithm
Figure 2.
Gene level Sensitivity (Sn) and Specificity (Sp) of GeneMark-ETP for the three compact genomes. The dashed lines correspond to constant levels of (Sn+Sp)/2. The species-specific protein databases used for derivation of protein-based evidence did not include proteins originated from the species from the same taxonomic order. Sn = Tp/(Tp+Fn) and Sp = Tp/(Tp+Fp) where Tp, Fp and Fn are the numbers of true positive, false positive and false negative gene predictions, respectively.
Figure 3.
Gene level Sn and Sp of GeneMark-ETP for the four larger genomes. All other specifications are the same as in Fig. 2.
In comparison with TSEBRA (Figs. 2, 3; Table S2), the average gene level F1 values changed, respectively, by −1.0%, 8.2%, and 39.0% (Table S2) while the average exon F1 values by 0.7%, 1.3%, and 18.6% (Table S2).
For both compact and large genomes, the accuracy of GeneMark-ETP was observed to be significantly higher than the accuracy of the virtual combinations of the sets of gene predictions made by GeneMark-ET and GeneMark-EP+ separately, the union, the intersection or the ‘best’ combination (Figs. 4, S2, Methods Section).
Figure 4:
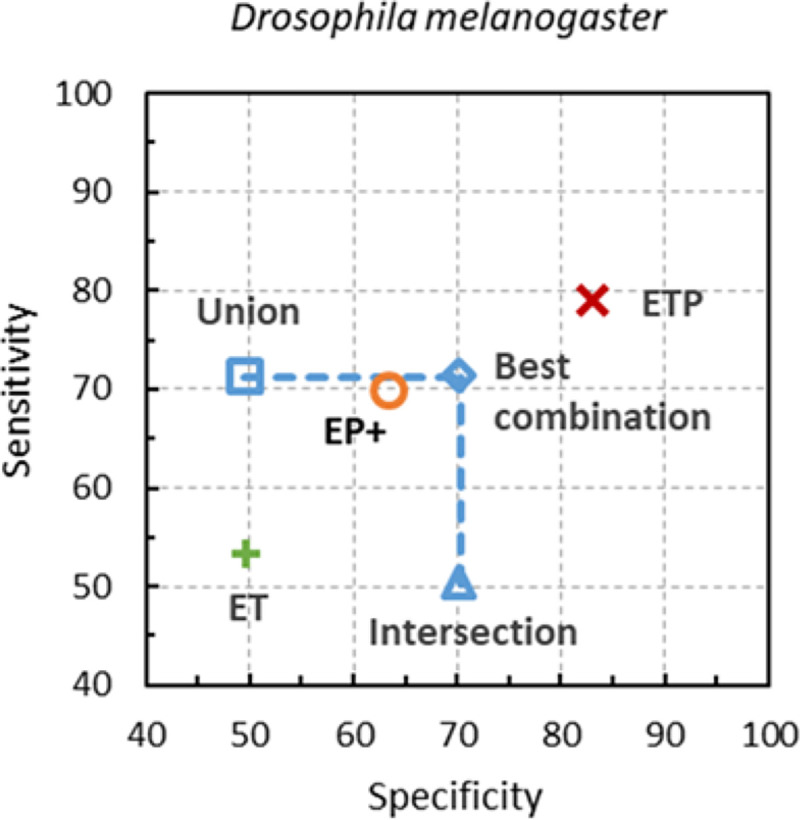
Gene-level Sn and Sp of the artificial combinations of GeneMark-ET and GeneMark-EP+ gene predictions made in genome of D. melanogaster are shown along with the Sn and Sp of GeneMark-ETP. Proteins of the same species were excluded from the reference protein database.
Refinement of the gene predictions in assembled transcripts
To produce a set of high-confidence genes we used information inferred from transcripts and proteins. Gene predictions in assembled transcripts were made by GeneMarkS-T (Tang et al. 2015). Some of these predictions were refined. If an alignment of the predicted protein to a protein in a database indicated a better support to an alternative protein start, the initial prediction was altered (Methods Section 2). We found that for each of the seven species, and for the two types of the protein database made for each species, having either the smaller or of the larger size (Tables 1, S3), this correction increased the gene-level specificity, on average, by 25 percentage points, reaching values higher than 90%.
Table 1.
The gene-level Sn and Sp values of all the GeneMarkS-T gene predictions in the assembled transcripts and for those GeneMarkS-T predictions that were selected as high-confidence (HC) genes. The Sn and Sp values in the second column are shown for these HC genes. databases Proteins from the species of the same taxonomic order were excluded from the corresponding PD0 protein databases.
| Species/ Accuracy | GeneMarkS-T predictions | Selected HC genes | |
|---|---|---|---|
|
| |||
| C. elegans | Sn | 46.8 | 35.7 |
| Sp | 63.4 | 88.4 | |
|
| |||
| A. thaliana | Sn | 51.2 | 56.7 |
| Sp | 79.9 | 97.3 | |
|
| |||
| D. melanogaster | Sn | 59.6 | 55.0 |
| Sp | 81.8 | 94.7 | |
|
| |||
| S. lycopersicum | Sn | 67.8 | 74.9 |
| Sp | 77.8 | 95.2 | |
|
| |||
| D. rerio | Sn | 59.6 | 67.0 |
| Sp | 59.9 | 88.5 | |
|
| |||
| G. gallus | Sn | 49.6 | 74.4 |
| Sp | 47.0 | 89.1 | |
|
| |||
| M. musculus | Sn | 49.6 | 63.5 |
| Sp | 63.2 | 93.2 | |
When the smaller protein databases were used (with proteins of the same taxonomic order excluded from the species-specific reference database - , see Materials) we observed a noticeable increase in gene prediction sensitivity for five of the seven tested genomes (Table S3). For the larger databases, (with only the proteins of the same species excluded from , the increase was observed for all seven genomes (Tables 1, S3). This increase was largely due to the introduction of the refinement of the gene prediction in transcripts. For example, in the case of D. rerio, 2,750 out of 22,979 genes predicted in transcripts were initially classified as 5’ partial by GeneMarkS-T (Table S4). Comparison with annotation revealed 1,349 truly 5’ partial predictions and 1,401 those that contained true complete gene inside. The refinement changed longer partial gene prediction to shorter complete gene prediction for 1,152 of the 1,349 predictions (reaching 82% sensitivity in this set). At the same time, 107 genes from the 1,349 true partial genes (8% error rate) were incorrectly shortened. The results of this type of analysis for all seven genomes are shown in Table S4.
Analysis of the balance of extrinsic and intrinsic evidence
For each of the seven genomes we divided the whole complements of predicted genes into four categories by the type of extrinsic support: fully extrinsic: all elements of the exon-intron structure were supported by significant (high scoring) extrinsic evidence; partially extrinsic: some elements of the exon-intron structure were determined due to significant extrinsic evidence while other were predicted ab initio; ab initio anchored which meant that the whole gene was predicted ab initio, while a match to a low scoring extrinsic evidence for some gene elements was detected a posteriori; ab initio unsupported: none of the gene elements predicted ab initio were supported by any extrinsic evidence even a posteriori.
We observed that the reliability of gene predictions could be reduced significantly upon the decrease of the level of extrinsic support. Particularly, in the three largest genomes, the predictions in the ab initio unsupported category had gene level Sp values below 1.5% (Table 2) and exon level Sp below 3% (Table S5). When we removed such gene predictions from the reported lists of predicted genes, we found that in these four genomes, the gene-level Sp increased, on average, by 21% while Sn decreased by 0.3% (Table S6). For the three smaller genomes, such a pruning would increase on average the gene level Sp by 3.7% with decrease of Sn by 1.7%.
Table 2.
Distribution of predicted genes among the four categories of extrinsic support along with average Sp values (gene level) for each category. Descriptions of the smaller and larger species-specific protein databases are given in Materials.
| Species | Types of evidence for a predicted gene | Smaller protein DB | Larger protein DB | ||
|---|---|---|---|---|---|
|
| |||||
| # of genes | Specificity, % | # of genes | Specificity, % | ||
|
| |||||
| C. elegans | Fully extrinsic | 7,676 | 88.9 | 10,778 | 91.6 |
| Partially extrinsic | 4,804 | 56.4 | 5,417 | 54.4 | |
| Ab initio anchored | 4,020 | 54.7 | 1,548 | 45.2 | |
| Ab initio unsupported | 1,298 | 24.9 | 778 | 18.0 | |
|
| |||||
| A. thaliana | Fully extrinsic | 16,445 | 97.2 | 18,083 | 97.5 |
| Partially extrinsic | 4,825 | 64.4 | 5,807 | 55.7 | |
| Ab initio anchored | 1,794 | 50.2 | 1,360 | 30.1 | |
| Ab initio unsupported | 2,964 | 27.9 | 1,128 | 9.4 | |
|
| |||||
| D. melanogaster | Fully extrinsic | 8,059 | 95.1 | 9,952 | 96.8 |
| Partially extrinsic | 2,328 | 49.3 | 2,751 | 44.9 | |
| Ab initio anchored | 1,043 | 57.1 | 165 | 44.9 | |
| Ab initio unsupported | 1,369 | 41.6 | 377 | 15.9 | |
|
| |||||
| S. lycopersicum | Fully extrinsic | 17,639 | 95.2 | 18,420 | 95.0 |
| Partially extrinsic | 5,174 | 47.3 | 5,813 | 44.3 | |
| Ab initio anchored | 1,577 | 38.4 | 1,484 | 29.7 | |
| Ab initio unsupported | 4,714 | 14.8 | 3,703 | 9.2 | |
|
| |||||
| D. rerio | Fully extrinsic | 15,691 | 89.8 | 15,501 | 92.6 |
| Partially extrinsic | 10,905 | 16.6 | 11,769 | 16.6 | |
| Ab initio anchored | 1,973 | 11.4 | 1,663 | 7.3 | |
| Ab initio unsupported | 12,534 | 0.8 | 11,879 | 0.3 | |
|
| |||||
| G. gallus | Fully extrinsic | 11,856 | 89.3 | 11,547 | 89.9 |
| Partially extrinsic | 4,857 | 19.6 | 5,337 | 20.1 | |
| Ab initio anchored | 527 | 8.9 | 579 | 7.1 | |
| Ab initio unsupported | 11,332 | 0.4 | 11,352 | 0.3 | |
|
| |||||
| M. musculus | Fully extrinsic | 13,556 | 94.6 | 13,769 | 96.2 |
| Partially extrinsic | 7,376 | 20.6 | 7,606 | 19.6 | |
| Ab initio anchored | 957 | 10.1 | 1,155 | 7.3 | |
| Ab initio unsupported | 20,711 | 1.2 | 19,666 | 0.5 | |
Gene prediction accuracy by MAKER2 and comparison with ETP
Another gene prediction pipeline that uses the three data sources, genomic, transcript and protein data, and that could be compared with GeneMark-ETP is MAKER2. To make these comparisons we used the genomes of D. melanogaster, D. rerio and M. musculus along with the RNA-seq and protein data sets being the same for both tools (see details in the Supplementary materials).
The MAKER execution requires training of the three gene finders AUGUSTUS, GeneMark.hmm and SNAP. We wanted to generate MAKER2 gene predictions with accuracy that would be considered as an upper bound. Therefore, instead of de novo training we used models for AUGUSTUS and SNAP that were generated by supervised training on GenBank annotated genes or models provided by the gene finder code developers (available in the software distribution). Both MAKER2 and GeneMark-ETP use the GeneMark.hmm gene finder. MAKER2 uses the version of GeneMark.hmm self-trained by GeneMark-ES, thus, having no use of external evidence either in model training or in gene prediction. To improve the GeneMark.hmm performance in MAKER2 we used the models trained on high confidence genes in GeneMark-ETP. The gene predictions were compared to the existing genome annotations.
We have observed that for all the species the GeneMark-ETP accuracy was higher than the one of MAKER2 (Table 3, S11). The GeneMark-ETP predictions of protein-coding exons had F1 values higher than ones of MAKER2 by approximately 10 points. The difference in the F1 values was more pronounced at gene and transcript level, with GeneMark-ETP F1 values exceeding the ones of MAKER2 by 20 and more points.
Table 3.
Performance of MAKER2 and GeneMark-ETP pipelines on the three model organisms.
| D. melanogaster | D. rerio | M. musculus | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| MAKER2 | GeneMark-ETP | MAKER2 | GeneMark-ETP | MAKER2 | GeneMark-ETP | ||
|
| |||||||
| Exon | Sn | 75.2 | 80.7 | 83.3 | 93.9 | 79.2 | 91.7 |
| Sp | 74.0 | 91.4 | 79.2 | 84.9 | 77.4 | 87.9 | |
| F1 | 74.6 | 85.7 | 81.2 | 89.2 | 78.3 | 89.8 | |
| Gene | Sn | 38.3 | 79.0 | 47.7 | 73.5 | 41.6 | 73.1 |
| Sp | 51.7 | 83.0 | 37.6 | 56.2 | 34.8 | 59.7 | |
| F1 | 44.0 | 81.0 | 42.0 | 63.7 | 37.9 | 65.7 | |
| Transcript | Sn | 60.2 | 54.8 | 42.8 | 68.5 | 33.4 | 61.9 |
| Sp | 55.3 | 79.4 | 35.1 | 55.8 | 32.2 | 61.9 | |
| F1 | 57.7 | 64.8 | 38.5 | 61.5 | 32.8 | 61.9 | |
Notably, for a GC heterogeneous genome of M. musculus, GeneMark-ETP uses the GC specific HMM models, while in MAKER2 the GC specific models are used in AUGUSTUS and not in SNAP or GeneMark.hmm. This can also lead to reduced accuracy by MAKER2 on M. musculus genomes.
Discussion
The purpose of GeneMark-ETP was to generate gene predictions in a eukaryotic genome in consistency with the genomic sequence patterns, protein-coding region determinants elucidated from transcripts, as well as homologous proteins footprints. Solving this task required training of the two GHMM models – one for the assembled transcripts and one for genomic DNA, mapping of the genes predicted in transcripts to genome and finding a set of proteins homologous to a not yet fully predicted gene. All these steps have led to integration of the three layers of information for each genomic locus.
One of the principal differences with the earlier developed tools, GeneMark-ES, GeneMark-ET and GeneMark-EP+ was in the method of training of the genomic GHMM model. In all the just cited tools the gene prediction process started with the heuristic model with parameters determined based on functions approximating dependence of the k-mer frequencies on genome GC content. In GeneMark-ETP we start the process of genomic GHMM training with a model derived from a set of the HC gene loci identified by integration of genomic, transcriptomic and protein evidence. In experiments with well-studied genomes the numbers of HC genes were frequently so large that thus derived initial GHHM model would not significantly change in the further training iterations.
In what follows we discuss the algorithmic steps that make GeneMark-ETP different from other tools.
Identification of a set of genes predicted with high confidence.
The accuracy of gene prediction in assembled transcripts is affected by assembly errors. However, the task of generating complete assembled transcripts from short RMA-Seq reads has presented a well-known challenge (Steijger et al. 2013). New tools, such as StringTie2 (Kovaka et al. 2019), were demonstrated to have a significantly improved performance.
Gene prediction in an assembled transcript was done by GeneMarkS-T (Tang et al., 2015). Predicted proteins were searched against a protein database. The proteins found in the similarity searches could fully support the predicted gene, thus making the prediction more confident. Predicted 5’ partial genes were further refined (see Methods). The resulting set of genes was named high-confidence (HC) genes. In our test, the HC genes had on average significantly better match to the ‘true’ genes than the set of genes derived from the initial GeneMarkS-T gene predictions (Tables 1, S3). Thus, the set of HC genes was identified by using genomic, transcriptomic, and protein data simultaneously.
Identification of a set of genes predicted with the least confidence.
In all the seven genomes, we saw that genome-specific proportions of genes predicted with full and partial extrinsic support went down with the increase in the genome size (Table 2). For example, the percentage of genes predicted with extrinsic support diminished, from 96% for D. melanogaster to 51% for M. musculus (the numbers a given for the case of using larger reference databases).
At the same time, the increase in genome size was accompanied by the increase in the proportion of the genes predicted ab initio (Table 2). For instance, the percentage of genes predicted ab initio were 18.8% and 4.1% for D. melanogaster and 51.0% and 49.3% for M. musculus for smaller and the larger protein databases, respectfully.
Importantly, the fraction of false positives among the ab initio predictions grew even faster with the genome size increase and led to a significant drop in Specificity (Tables 2, S6). The fast growth in the rate of false positive predictions could be caused by a combination of several factors; an increase in the average length of intron and intergenic regions; increased frequency of pseudogenes; increase in the size of populations of transposable elements (repeats), etc. Notably, in the large genomes, the percentage of false positive predictions was in the range observed in our experiments with gene prediction in simulated non-coding regions (data not shown).
Analysis of the results showed that the gene level specificity of ab initio gene predictions could drop significantly, reaching below 10% for ab initio unsupported predictions (the fourth category in Table 2). We observed that such genes comprised more than 10% of the total number of predictions in genomes larger than 300 Mbp (Figs. S3, S4). At the same time, in all the genomes under consideration, the fraction of predictions supported extrinsically (fully or partially) was above 50%. Therefore, we came up with the following empirical rule. For genomes larger than 300 Mbp where a 10% threshold on the fraction of ab initio predictions was exceeded, it was reasonable to eliminate gene predictions that fell into the category ab initio unsupported. For such genomes, all the genes remaining in the final list of predictions would have at least one element of the exon-intron structure supported by extrinsic evidence which was either used in the prediction or detected a posteriori (the first three categories in Table 2).
Transition to the GC-content-specific models
For gene prediction in GC-heterogeneous genomes and transcripts, we used several GC-content-specific sets of the GHMM parameters. This diversification led to the improvement in gene prediction accuracy. The resulting performance was certainly better than the ones of GeneMark-ET or GeneMark-EP+, the tools that used a single model designed for an average genomic GC composition (Fig. 2).
The “GC-heterogeneous” mode could be used for any genome. However, if a genome is rather a true GC-homogeneous one, the use of this mode would increase runtime and slightly decrease the accuracy, due to splitting the overall training set into smaller subsets. Therefore, the degree of GC-heterogeneity is assessed at a pre-processing step, and the “GC-heterogeneous” mode is used if needed.
Processing of repetitive elements
Transposable elements (TEs), particularly families of retrotransposons with thousands of copies of very similar TE sequences, occupy substantial portions of eukaryotic genomes. Errors in gene prediction may happen in presence of repetitive elements with composition similar to protein-coding genes (Yandell and Ence 2012; Torresen et al. 2019). Getting information on the repeat locations, e.g., predicted de novo, helps to reduce the errors. However, some of the predicted repeat sequences may overlap with the protein-coding genes of the host (Bayer et al. 2018).
One could hard-mask all repeats longer than a chosen threshold (Lomsadze et al. 2014; Bruna et al. 2020). Such an approach carries disadvantages: (i) repeats shorter than would not be masked and (ii) protein-coding exons overlapped by the masked repeats could be difficult to find.
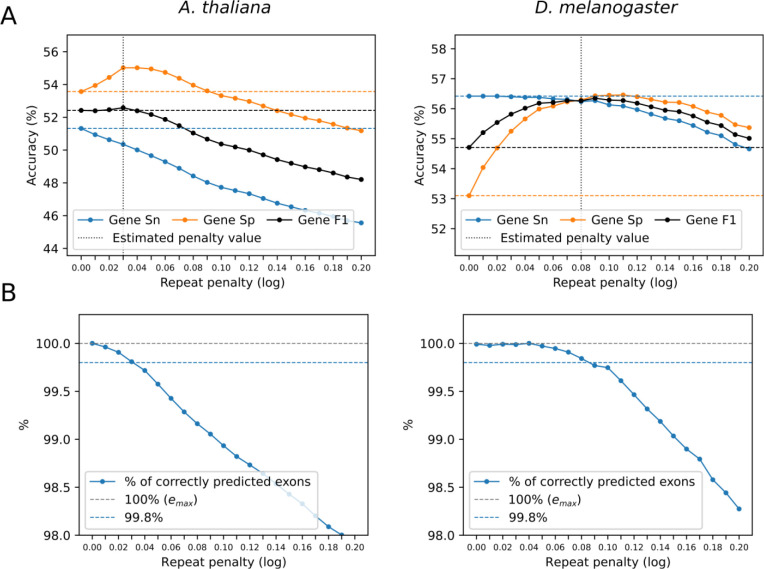
To deal with this issue, the authors of AUGUSTUS have introduced a repeat length-dependent penalty function used in the GHMM Viterbi algorithm (Stanke et al. 2008). A single parameter of this function, q, had the same value for all species. We have shown that in GeneMark-ETP use of a species-specific parameter q produced even better results (Fig. 4, see also Fig. S5, Table S7). The species-specific q value was determined after identification of the HC genes (see Methods). We have observed that the suggested approach was robust with respect to the size of the sample of the HC genes (data not shown).
Robustness of GeneMark-ETP
GeneMark-ETP iterates over training and prediction steps until convergence is reached between two consecutive prediction steps (Fig S6). Add more here about robustness of training from amount of RNA data.
We have observed that if more than 4,000 HC genes were found in the initial step of identifying HC genes supported by transcript and protein data, then, the further iterative training did not lead to significant improvement of the prediction accuracy. Such an outcome was likely due to reaching stationary values of the parameter estimates.
As could be expected, the GeneMark-ETP accuracy was less affected by the change in size of the protein database in comparison with GeneMark-EP+ that was using protein data only. For example, for D. melanogaster, when a larger protein database (proteins of the same species excluded from ) was changed to a smaller database (proteins of the same order excluded from ), the gene level F1 of GeneMark-ETP decreased by 6.4% (Table S1) while the F1 of GeneMark-EP+ decreased by 11.4%. Certainly, the use of the HC genes derived from GeneMarkS-T predictions that did not have full-length protein support did contribute into this effect (Methods Section).
While the increase in the volume of proteins from the closely related species should, generally, lead to increase in gene prediction accuracy, the accuracy of GeneMark-ETP (similarly to GeneMark-EP+) did not critically depend on presence of such proteins in the reference database.
Comparison with other computational tools
GeneMark-ET, GeneMark-EP+, and their virtual combination
GeneMark-ETP performed better than either GeneMark-ET or GeneMark-EP+ in all the tests (see Results). Since both GeneMark-ET and GeneMark-EP+, use only a single source of extrinsic evidence, this result should have been expected.
The virtual tool considered here made an artificial combination of the sets of genes predicted separately by GeneMark-ET and GeneMark-EP+ (Method Section). The largest sensitivity of such a tool could be achieved by the union of the two sets while the largest specificity could be achieved if the intersection of the two sets is used. Implementation of the best-balanced combination would require either a removal of false positives from the union set, or an addition of true positives to the intersection set. When a gene finder is running on a novel genome information on true and false positives is not immediately available. Nevertheless, even if this ideal correction would be made, the accuracy of the best virtual tool would still fall below the accuracy of GeneMark-ETP (Figs 3, S2).
BRAKER1, BRAKER2 and TSEBRA
Earlier developed pipelines—BRAKER1 (Hoff et al. 2016), combining AUGUSTUS and GeneMark-ET, using transcripts as a source of extrinsic evidence, and BRAKER2 (Bruna et al. 2021), combining AUGUSTUS and GeneMark-EP+ supported by cross-species protein data are frequently used tools. We have shown that GeneMark-ETP gene prediction accuracy was higher than either BRAKER1 or BRAKER2, especially for large genomes (Figs. 1, 2). Again, this result could be expected due to the use of twice as many types of extrinsic information in GeneMark-ETP. The recently developed TSEBRA applies well designed rules to select a subset of all predictions made by either BRAKER1 or BRAKER2 and, thus, achieves higher accuracy than any of the BRAKERs (Gabriel et al. 2021). It was shown that TSEBRA performed better than EVidenceModeler, one of the best combiners, as well.
In our tests, it was demonstrated that GeneMark-ETP achieved higher accuracy than TSEBRA in large genomes (Fig. 2), particularly in the GC-heterogeneous ones (G. gallus, D. rerio) where BRAKER1 and BRAKER2 use single statistical models tuned up for “average GC” in each genome. Nevertheless, GeneMark-ETP reached higher than TSEBRA prediction accuracy in the GC-homogeneous genomes of S. lycopersicum and D. rerio. Therefore, there should be yet another factor beyond the training of the GC diversified the statistical models. Such an additional source of accuracy improvement is, arguably, use of hints that integrate both assembled transcript and protein information. The accuracy advantage of GeneMark-ETP was much smaller in the group of compact genomes (Fig. 1), with TSEBRA achieving higher accuracy than GeneMark-ETP in the case of C. elegans.
All over, the new tool integrated transcriptomic and protein evidence of presence of protein-coding function in genomic sequences into hints used consistently at all stages of the algorithm training and gene prediction. We argue that such an approach is more effective than combining the predictions made with a particular single type of extrinsic evidence in a “post-processing” step.
Materials
For the assessment of the GeneMark-ETP gene prediction accuracy, we selected seven genomes from diverse eukaryotic clades (Tables 4, S8). The group included relatively short GC-homogeneous genomes of the well-studied model organisms: A. thaliana, C. elegans, and D. melanogaster. The group also included larger genomes, both GC-homogenous (S. lycopersicum, D. rerio) and GC-heterogeneous (G. gallus, M. musculus). In all the genomic datasets, contigs with no chromosome or organelle assignment were excluded from the analysis.
Table 4.
Genomes used for the assessment of the GeneMark-ETP gene prediction accuracy. For the larger genomes, the numbers in parentheses characterize selected subsets of genes presumed to be more reliably annotated. To compute the numbers of introns per gene we used averages among annotated alternative transcripts.
| Species | Genome length (Mb) | Reference annotation statistics |
|||||
|---|---|---|---|---|---|---|---|
| # of protein-coding genes | # of transcripts | # of introns per gene | |||||
|
| |||||||
| C. elegans (roundworm) | 100 | 19,969 | 28,544 | 4.8 | |||
| A. thaliana (thale cress) | 119 | 27,445 | 40,828 | 4.0 | |||
| D. melanogaster (fruit fly) | 138 | 13,951 | 22,395 | 2.8 | |||
|
| |||||||
| S. lycopersicum (tomato) | 807 | 25,158 | (15,138) | 31,911 | (15,150) | 4.4 | (4.3) |
| D. rerio (zebrafish) | 1,345 | 25,611 | (17,894) | 42,934 | (19,978) | 8.4 | (8.4) |
|
| |||||||
| G. gallus (chicken) | 1,050 | 17,279 | (10,736) | 38,534 | (12,733) | 9.0 | (9.2) |
| M. musculus (mouse) | 2,723 | 22,405 | (16,531) | 58,318 | (20,708) | 6.0 | (8.6) |
To generate the reference sets of proteins used as a source of extrinsic evidence we used the OrthoDB v10.1 protein database (Kriventseva et al. 2019); for more details see (Bruna et al. 2020; Bruna et al. 2021). For each of the seven species, we built an initial species-specific protein database () containing all proteins from the largest clade considered for the given species (Table S9). Also, for each given species, we created two smaller reference databases by removing from PD0 either all proteins of this species itself and its strains, or proteins from all the species that belonged to the same taxonomic order. These, the larger and the smaller databases, were devised to mimic practical scenarios when a species in question would have either a larger or a smaller set of proteins from close relatives present in the reference database. All over, the numbers of proteins in the databases used in the computations ranged from 2.6 to 8.3 million (Table S9).
Transcript datasets, such as the sets of Illumina paired reads, were selected from the NCBI SRA database. The read length varied between 75 to 151 nt; the total volume of RNA-Seq collections varied from 9 Gb for D. melanogaster to 83 Gb for M. musculus (Table S10).
Methods
Outline of the GeneMark-ETP algorithmic steps
In GeneMark-ES, -ET, -EP+, iterative unsupervised training was used to estimate the parameters of the GHHM models (Lomsadze et al. 2005; Lomsadze et al. 2014; Bruna et al. 2020). The iterative cycles of model training and gene prediction resulted in getting a final set of model parameters employed in the prediction of the final set of genes. Since GeneMark-ETP relies on a larger set of extrinsic data, the training procedure was significantly modified.
Another difference with the previous developments is that GeneMark-ETP predicts genes both in genomic DNA as well as in assembled transcripts. Gene prediction in transcripts is done by a self-training tool GeneMarkS-T with GHMM designed for sequences with intron-less genes (Tang et al. 2015). On the other hand, gene prediction in eukaryotic DNA sequences requires GHMM with an exon-intron model (Lomsadze et al. 2005).
At the start of a genome analysis, GeneMark-ETP generates a set of high-confidence (HC) genes (Fig. S7). GeneMarkS-T plays a central role in this step. Next, the parameters of the ‘eukaryotic’ GHMM are estimated, and the Viterbi algorithm is used to predict genes in the regions between the HC genes. If the set of HC genes is not large enough, the initial parameters of the GHMM model are further refined by self-training in the genomic regions situated between HC genes. The use of the transcript and protein evidence continues in all the steps (Fig. 5).
Figure 4.
A. Dependence of the accuracy measures on the repeat penalty parameter q observed for genomes of A. thaliana and D. melanogaster. B. Dependence of % of correctly predicted exons of the HC genes (Sn) on the repeat penalty parameter q for the same genomes as in A (see Methods).
Selection of a set of genes predicted with high confidence.
1. Gene prediction in assembled transcripts
Reads of each RNA-Seq library used in the input for GeneMark-ETP are splice-aligned to the genome by HISAT2 (Kim et al. 2019) and assembled into a set of transcripts by StringTie2 (Kovaka et al. 2019). Since StringTie2 assembles transcripts from reads mapped to genome, the information on intron positions is carried on to define the exon/intron structure of the gene predicted by GeneMarkS-T. All the sets of transcripts are merged into the final non-redundant transcriptome where the low-abundance transcripts are filtered out (Kovaka et al. 2019).
Refinement of the GeneMarkS-T predictions based on protein information.
A gene predicted by GeneMarkS-T is assumed to be 5’ partial if it starts from the first nucleotide of a transcript. An incorrectly predicted 5’ partial gene could have a complete true gene inside (Fig. S8). To determine if an ATG is a true start codon, the two alternative protein translations are used as queries by DIAMOND (Buchfink et al. 2015) in searches against a reference protein database. If among the hits exists at least one target that is i/ common for both queries and ii/ shows better support for the prediction starting at the start of the transcript, the 5’ partial gene is predicted. Otherwise, the shorter sequence is selected as the predicted complete gene. Note, that if the sets of targets do not overlap (those with 25 best scores from each DIAMOND search), the 5’ partial prediction is selected.
The details of the protein similarity based assessment of which of the alternative queries is better supported by a common target is described in Section 1.1 of Supplementary Materials. The 5’ partial prediction (the longer protein query) is chosen if inequality (S1) is fulfilled for at least one common target, otherwise, the shorter protein query (complete gene) is selected (Fig. S9). We have observed that GeneMarkS-T makes very few errors when predicts a start codon within a transcript.
2. High-confidence genes
Complete genes with full protein support
A gene predicted by GeneMarkS-T is said to have full protein support if there is a protein in a database who’s significant BLASTp alignment to the predicted protein satisfies condition (S2) described in Section 1.4 of Supplementary Materials. To find a target satisfying condition (S2), we examine 25 top-scoring alignments of the query to the target proteins. If such a target exists, the query—a 5’ complete gene with full protein support—is classified as a high-confidence gene.
A 5’ complete gene predicted in a transcript may not make the “longest ORF” with respect to the predicted 3’end of the gene, though it was observed that most annotated eukaryotic genes do make the longest ORFs. To correct possible underprediction, both the original prediction and its extension to the “longest ORF” are checked by condition (S2) and, if fulfilled, one of them or even both are classified as HC (alternative) isoforms.
5’-partial genes with full protein support
A 5’ partial gene (see Fig. S10) can be classified as a high-confidence gene if the C-terminal of its protein translation is supported by at least one protein alignment. If the best-scoring protein alignment does not cover the 5’ partial protein from the start (see Fig. S11 where ), the 5’ partial gene is shortened to the first in-frame ATG codon covered by the protein alignment.
Any gene predicted as 3’ partial (unambiguously defined by the lack of a stop codon) is not considered as a candidate for an HC gene.
Genes predicted ab initio.
Complete GeneMarkS-T gene predictions that either have no significant BLASTp hits in the protein database or do not satisfy condition (S2), for an alignment of the predicted protein and any of its best targets in the protein database, still could make high-confidence genes. To be qualified as such, all of the following conditions have to be satisfied: (i) a length of protein-coding region is longer than 299 , (ii) an in-frame stop codon triplet is present in the 5’ UTR, (iii) the GeneMarkS-T log-odds score is ≥ 50 and (iv) the gene structure mapped to genomic DNA does not create any conflict with ProtHint hints (see Section 2 of Supplementary Methods for more details). A single HC isoform (the one with the longest protein-coding region) is selected per locus, where several isoforms are predicted based on multiple transcript assemblies.
High-confidence alternative isoforms
Alternative isoforms of the same gene may belong to the set of HC genes. Selection of HC alternative isoforms is done as follows.
Let be a set of all complete isoforms of gene and is a set of all its partial isoforms. Each isoform is assigned a score -- the bitscore of its best hit to a protein in the reference protein database.
We compute the maximum score of all the complete isoforms for each gene (Eq. 1).
| (1) |
The score of an isoform selected as HC complete isoform must satisfy inequality (2).
| (2) |
For the partial alternative isoforms, we have
| (3) |
If this score is larger than , the partial transcript with this largest score is selected as the partial HC isoform. Moreover, all the complete HC isoforms are removed in this case. Otherwise, if , is lower than , then only the complete isoforms are retained.
3. The GHHM model training
Single step model training
A set of predicted HC genes is used for the initial and often final GHHM parameter estimation. First, the set of HC genes is checked for possible redundancy. In the loci with several complete HC isoforms the isoform with the longest protein-coding region is selected. Next, we determine the GC content distribution of the selected HC genes and if more than 80% of them are contained in a 10% wide GC content interval, the genome is characterized as GC homogeneous, else as GC heterogeneous.
In the GC homogeneous case, the loci of all the selected HC genes are used for the estimation of parameters of a GHHM model (Fig. S6). The GHMM model parameter estimation is done by training on the set of the HC loci, the sequences containing these HC genes with 1,000 margins. An iterative extended training of the GHMM parameters is done similar to the approach described earlier (Lomsadze et al. 2014; Bruna et al. 2020; Lomsadze et al., 2005).
In the GC heterogeneous case, the sequence set of HC loci is split into the three GC bins: low, medium, and high. The borders of the medium GC bin with a fixed width (9% by default) are selected within the GC one dimensional range to include the largest possible number of the HC loci. Setting up these boundaries automatically determines the borders of the low and medium GC bins. The sets of the HC loci contained in each GC bin are used for the training of the three GC-specific GHHM models. Iterations of extended training are done in each bin.
Notably, gene prediction in transcripts by GeneMarkS-T is made with a set of the GC-specific statistical models derived as described in Tang et al, 2015.
Extended GHMM model training
The logic of extended model training is similar but not identical to iterative training used in GeneMark-ET and GeneMark-EP+ (Lomsadze et al. 2014; Bruna et al. 2020).
At the initialization of iterations for genomes with homogeneous GC content, the GHMM model parameters are derived from the sequences of the HC loci contrary to the use of the cruder heuristic model in GeneMark-ET and GeneMark-EP+. The gene prediction is then made only in the genomic sequences situated between HC genes, the HC-intermediate regions. These predictions, serving as ProtHint gene seeds, are translated and used as queries for a protein database search initiating the full run of ProtHint (Bruna et al. 2020). Hints created by ProtHint are combined with hints from RNA-seq alignment to generate a set of high confidence hints in HC-intermediate regions. These hints are enforced in the following steps of iterative training of GHMM parameters and predictions by Viterbi algorithm. Iteration convergence criteria is defined as percent identity between two consecutive gene sets. We used exon identity level 99% as stopping criteria and genes predicted along pure ab initio path were excluded from calculation of convergence criteria.
Three iterations were the maximum number of iterations observed in our experiments. Most frequently GeneMark-ETP was converging on the second iteration.
An important trait of the GHMM training process implemented in GeneMark-ET and GeneMark-EP+ algorithms, was step by step unfreezing of the subsets of the GHMM model parameters. For instance, the Markov chain transition probabilities and durations of functional regions, i.e., intron, exon etc., were fixed during the initial iterations while the values of emission probabilities were free to change. In the later iterations all the parameters were made free. Such gradual unfreezing of the parameters was shown to be unnecessary for GeneMark-ETP. All the GHMM parameters are estimated simultaneously. We attribute this streamlining of the training process to availability of the more accurate initial parameters of GHMM derived from the sequences of HC loci.
In GC heterogeneous genomes the extended GHHM training worked as follows. First, GeneMark-ETP calculated the GC content of each HC-intermediate region and assigned the regions to the corresponding GC bins. The parameters of the initial GC specific GHHM model were trained on the thus selected sets of sequences of HC loci. Subsequently, a GC specific model was used for gene prediction in the HC-intermediate regions of a corresponding GC bin. From this point on, the extended training on the HC-intermediate regions from a particular GC bin was essentially made in the same way as described above for the GC homogeneous case. The iterative training and the final gene prediction step were executed using the GC-specific models updated from iteration to iteration.
Accounting for repeats
Repeat identification and masking.
To identify repetitive sequences, we used RepeatModeler2 (Flynn et al. 2020) and RepeatMasker (www.repeatmasker.org). Repeat libraries were generated de novo using RepeatModeler2. Repeat sequences—interspersed and tandem repeats—were then identified and soft-masked by RepeatMasker.
Selection of the species-specific repeat penalty parameter
To account for an overlap of a protein-coding region with a repetitive sequence, the GeneMark-ETP algorithm changes the probability (likelihood) of a sequence in such an overlap by formula (4) with penalty parameter (n is the length of an overlap):
| (4) |
GeneMark-ETP estimates species-specific parameter for each genome. The goal is to find the value minimizing disruptions to correct gene predictions. The estimation step is made after the first iteration of the GeneMark-ETP model training; this, we have full GHMM model, a set of the HC genes, and the coordinates of the repeats identified in genomic DNA prior to the first iteration. The Viterbi algorithm is then run several times (with different values) in an ab initio mode to predict genes in the soft-masked genomic sequences containing the HC genes (Fig. 4). In each run we compute gene level F1 value of the gene prediction accuracy determined on the test set made from the HC genes. Then we identify the value for which F1 would reach maximum (Fig. 4A). Thus, determined best value was good approximation of the one determined on a test set derived with “full” knowledge of genome annotation (Table S7). Moreover, we found that selecting the value by using the exon level Sn was a more robust method in comparison of using the gene level F1. Technically, we would compute the best by maximizing the number of correctly predicted exons in the HC genes, Such value would be larger than or equal to 1 (Fig. 4B). We found that value at which exons were correctly predicted was a good estimate of the best value q (marked in panel A of Fig. 4). To reduce the running time of the search for the best repeat penalty parameter, we used simulated annealing (Kirkpatrick et al. 1983).
Gene prediction in the HC intermediate regions
Integration of intrinsic and extrinsic evidence
The models trained on the set of the HC loci are used in GeneMark.hmm to create initial gene predictions in the HC-intermediate segments (Fig. S12). These gene predictions can be refined by incorporation of the protein and transcript evidence. This task is solved as follows. The initial gene predictions are used in ProtHint to generate protein-based hints (Bruna et al., 2020). On the other hand, we have a set of genes predicted by GeneMarkS-T in transcripts that were mapped to genome by HISAT2. The mapping that falls into the HC intermediate regions constitute transcript based evidence for the HC intermediate regions. The whole set of hints is then ready for enforcement in a run of the Viterbi algorithm for GHMM (Fig. 6). To reiterate, we have the following categories of hints: 1/ RNA-Seq and ProtHint-derived hints that agree with each other; 2/ solely high score ProtHint hints; 3/ solely RNA-Seq-based intron hints, if they overlap but do not coincide with the ab initio predicted introns; the requirement of the overlap filters out introns mapped from expressed lncRNA; 4/ exons of partial HC genes; partial HC genes are determined at the stage of HC gene identification (Methods Section 2.2). Note that category 1–3 may not necessarily point to elements of the same gene (the RNA-Seq mapped introns or the ProtHint introns). Hints of category 4 should belong to the same gene.
Figure 6.
Integration of extrinsic evidence into the GeneMark-ETP gene predictions in HC-intermediate segments. The figure shows that a low score extrinsic evidence not corroborated by other extrinsic evidence or by ab initio gene prediction is ignored. The low score evidence is shown by thin lines.
The genes predicted in the HC-intermediate segments along with the full set of the HC genes constitute the final set of genes predicted by GeneMark-ETP.
While we did not make experiments with long RNA reads, we could argue that if the high-quality long reads or their assemblies are available, GeneMarkS-T could be run on the long reads to predict intron-less genes that in turn would be mapped to the genome, e.g. with Minimap2 (Li 2018). Thus, the GeneMark-ETP run could be implemented with this type of data.
Filtering out unsupported ab initio predictions
The genes predicted in the HC-intermediate segments could be split into two non-overlapping sets: evidence-supported predictions and pure ab initio predictions (see Discussion and Table 2). The evidence-supported genes must have at least one element of the gene structure supported externally. We have observed that in larger genomes the fraction of correct predictions among unsupported ab initio predictions were sharply decreasing with the genome size (see Sp values in Table 2). For each genome, the lists of genes predicted by GeneMark-ETP contain either the full set of gene predictions or the set of genes remaining after filtering out completely unsupported ab initio predictions. The reported here algorithm accuracy for the larger genomes (longer than 300 Mbp) was computed for the reduced output.
The accuracy assessment of GeneMark-ETP
Selection of gene sets with reliable annotation
Since annotations of well-studied genomes of A. thaliana, C. elegans, and D. melanogaster have been updated multiple times, we considered these complete annotations as “gold standards”, against which the gene prediction accuracy parameters was determined. Arguably, the reference annotations of the other four genomes have been less trustworthy. Therefore, to assess the sensitivity parameters we selected genes with identical annotations in two different sources, e.g., in the NCBI and the Ensembl records (Table S8). On the other hand, the values of prediction specificity for these genomes were defined by comparison with the union of genes annotated by either NCBI or Ensemble or by both.
Description of the sets of genes used for accuracy assessment is given in Table 2. In all the tests, regions of annotated pseudogenes were excluded from consideration.
A virtual combination of GeneMark-ET and GeneMark-EP+ predictions
We compared the accuracy of GeneMark-ETP with the accuracy delivered by a “virtual” tool which output was made of a combination of genes predicted by GeneMark-ET and GeneMark-EP+. Predictions made by GeneMark-ET and GeneMark-EP+ could be combined in two simple ways: by making either union U or intersection I. The intersection contained only genes with identical gene structures. The set U presents the most comprehensive set of predicted genes, while the set I, arguably, presents the most reliable predictions. The sensitivity of U genes is designated as Sn and the specificity of I genes is designated as Sp. Now, if one can reduce set U by taking away only the incorrect predictions, the point in Fig. 3 will move horizontally. If one can add to the set I only correct predictions the point in Fig. 3 will move up vertically. The crossing of the two lines at the point characterizes the accuracy of the virtual tool, implementing the best version of the virtual combiner approach.
Running BRAKER1, BRAKER2, and TSEBRA
To make comparisons with the transcript-supported BRAKER1 (Hoff et al. 2016) and protein-supported BRAKER2 (Bruna et al. 2021) we have run BRAKER1 and BRAKER2, respectively, with the same RNA-Seq libraries and protein databases, as the ones used in experiments with GeneMark-ETP. Also, we ran TSEBRA (Gabriel et al. 2021) that generated a set of genes supported by both RNA-Seq and proteins. TSEBRA selects a subset of the union of gene predictions made by BRAKER1 and BRAKER2. TSEBRA was shown to achieve higher accuracy than (i) either BRAKER1 or BRAKER2 running alone, as well as (ii) EVidenceModeler (Haas et al. 2008), one of the frequently used combiner tools.
Summary
A new eukaryotic gene prediction software tool, GeneMark-ETP was shown to generate better—and in the case of large genomes significantly more accurate—eukaryotic gene predictions in comparison with the earlier developed tools. The algorithm constructs a genomic parse into coding and non-coding regions supported by the combined evidence extracted from genomic, transcriptomic, and protein sequences. Integration of the intrinsic and extrinsic data is consistently implemented through the major steps of the algorithm: the GHMM models training and gene prediction. The margin of the prediction accuracy improvement does grow with the increase of the genome complexity from relatively compact genomes to large, GC-heterogeneous genomes. All over, we believe that we managed to demonstrate the advantage of the simultaneous integration of several sources of evidence into gene prediction over a post-processing-style integration combining several separate streams of gene predictions, each with its own type of extrinsic evidence.
Supplementary Material
Funding
National Institutes of Health [GM128145 to M.B., in part]. Funding for the open access charge: National Institutes of Health [GM128145].
Footnotes
Conflict of interest statement. None declared.
Supplementary materials
URL to be determined (a file is submitted along with the main text)
Availability
GeneMark-ETP is available on GitHub at https://github.com/gatech-genemark/GeneMark-ETP.git and http://topaz.gatech.edu/GeneMark/license_download.cgi. All scripts and data used to generate figures and tables in this manuscript are available at https://github.com/gatech-genemark/GeneMark-ETP-exp. The runtime of GeneMark-ETP depends linearly on the genome size and is comparable to the one of GeneMark-EP+. For example, on a machine with 64 CPU cores, GeneMark-ETP runs on genomes of D. melanogaster, D. rerio, and M. musculus for 1.0, 4.5, and 6.5 hours, respectively.
References
- Allen JE, Pertea M, Salzberg SL. 2004. Computational gene prediction using multiple sources of evidence. Genome Research 14: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JE, Salzberg SL. 2005. JIGSAW: integration of multiple sources of evidence for gene prediction. Bioinformatics 21: 3596–3603. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Bhandary P, Woodhouse M, Sen TZ, Wise RP, Andorf CM. 2021. FINDER: an automated software package to annotate eukaryotic genes from RNA-Seq data and associated protein sequences. BMC Bioinformatics 22: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer PE, Edwards D, Batley J. 2018. Bias in resistance gene prediction due to repeat masking. Nat Plants 4: 762–765. [DOI] [PubMed] [Google Scholar]
- Bruna T, Hoff KJ, Lomsadze A, Stanke M, Borodovsky M. 2021. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom Bioinform 3: lqaa108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna T, Lomsadze A, Borodovsky M. 2020. GeneMark-EP+: eukaryotic gene prediction with self-training in the space of genes and proteins. NAR Genom Bioinform 2: lqaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nature Methods 12: 59–60. [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S. 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Fiedler TJ, McKay SJ, Flicek P, Harris TW, Blasiar D, n GC, Stein LD. 2008. nGASP--the nematode genome annotation assessment project. BMC Bioinformatics 9: 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Valle-Inclan JE, Pajoro A, Rovenich H, Thomma B, Faino L. 2019. Long-Read Annotation: Automated Eukaryotic Genome Annotation Based on Long-Read cDNA Sequencing. Plant Physiol 179: 38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A 117: 9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel L, Hoff KJ, Bruna T, Borodovsky M, Stanke M. 2021. TSEBRA: transcript selector for BRAKER. Bmc Bioinformatics 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodswen SJ, Kennedy PJ, Ellis JT. 2012. Evaluating high-throughput ab initio gene finders to discover proteins encoded in eukaryotic pathogen genomes missed by laboratory techniques. PLoS One 7: e50609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gremme G, Brendel V, Sparks ME, Kurtz S. 2005. Engineering a software tool for gene structure prediction in higher organisms. Inform Software Tech 47: 965–978. [Google Scholar]
- Guigo R, Flicek P, Abril JF, Reymond A, Lagarde J, Denoeud F, Antonarakis S, Ashburner M, Bajic VB, Birney E et al. 2006. EGASP: the human ENCODE Genome Annotation Assessment Project. Genome Biol 7 Suppl 1: S2 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. 2008. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 9: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. 2016. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics 32: 767–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe KL, Chothia T, Durbin R. 2002. GAZE: A generic framework for the integration of gene-prediction data by dynamic programming. Genome Research 12: 1418–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keilwagen J, Hartung F, Paulini M, Twardziok SO, Grau J. 2018. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinformatics 19: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27: 757–763. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD, Vecchi MP. 1983. Optimization by Simulated Annealing. Science 220: 671–680. [DOI] [PubMed] [Google Scholar]
- Kiryutin B, Souvorov A, Tatusova T. 2007. Prosplign: protein to genomic alignment tool. In 11th Annual International Conference in Research in Computational Molecular Biology, San Francisco, USA. [Google Scholar]
- Kong J, Huh S, Won JI, Yoon J, Kim B, Kim K. 2019. GAAP: A Genome Assembly + Annotation Pipeline. Biomed Res Int 2019: 4767354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovaka S, Zimin AV, Pertea GM, Razaghi R, Salzberg SL, Pertea M. 2019. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol 20: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208. [DOI] [PubMed] [Google Scholar]
- Kriventseva EV, Kuznetsov D, Tegenfeldt F, Manni M, Dias R, Simao FA, Zdobnov EM. 2019. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Research 47: D807–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH. 1996. A generalized hidden Markov model for the recognition of human genes in DNA. Proc Int Conf Intell Syst Mol Biol 4: 134–142. [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Mackey AJ, Roos DS, Pereira FC. 2008. Evigan: a hidden variable model for integrating gene evidence for eukaryotic gene prediction. Bioinformatics 24: 597–605. [DOI] [PubMed] [Google Scholar]
- Lomsadze A, Burns PD, Borodovsky M. 2014. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res 42: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. 2005. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res 33: 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge JM, Harrow J. 2016. The state of play in higher eukaryote gene annotation. Nat Rev Genet 17: 758–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigo R. 2000. GeneID in Drosophila. Genome Res 10: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalzitti N, Jeannin-Girardon A, Collet P, Poch O, Thompson JD. 2020. A benchmark study of ab initio gene prediction methods in diverse eukaryotic organisms. BMC Genomics 21: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Sabunciyan S, Yang G, Florea L. 2019. A multi-sample approach increases the accuracy of transcript assembly. Nat Commun 10: 5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Diekhans M, Baertsch R, Haussler D. 2008. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24: 637–644. [DOI] [PubMed] [Google Scholar]
- Stanke M, Waack S. 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19 Suppl 2: ii215–225. [DOI] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engstrom PG, Kokocinski F, Consortium R, Hubbard TJ, Guigo R, Harrow J, Bertone P. 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Lomsadze A, Borodovsky M. 2015. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res 43: e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18: 1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresen OK, Star B, Mier P, Andrade-Navarro MA, Bateman A, Jarnot P, Gruca A, Grynberg M, Kajava AV, Promponas VJ et al. 2019. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res 47: 10994–11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Ence D. 2012. A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet 13: 329–342. [DOI] [PubMed] [Google Scholar]
- Zickmann F, Renard BY. 2015. IPred - integrating ab initio and evidence based gene predictions to improve prediction accuracy. BMC Genomics 16: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GeneMark-ETP is available on GitHub at https://github.com/gatech-genemark/GeneMark-ETP.git and http://topaz.gatech.edu/GeneMark/license_download.cgi. All scripts and data used to generate figures and tables in this manuscript are available at https://github.com/gatech-genemark/GeneMark-ETP-exp. The runtime of GeneMark-ETP depends linearly on the genome size and is comparable to the one of GeneMark-EP+. For example, on a machine with 64 CPU cores, GeneMark-ETP runs on genomes of D. melanogaster, D. rerio, and M. musculus for 1.0, 4.5, and 6.5 hours, respectively.