Abstract
We motivate and present biVI, which combines the variational autoencoder framework of scVI with biophysically motivated, bivariate models for nascent and mature RNA distributions. While previous approaches to integrate bimodal data via the variational autoencoder framework ignore the causal relationship between measurements, biVI models the biophysical processes that give rise to observations. We demonstrate through simulated benchmarking that biVI captures cell type structure in a low-dimensional space and accurately recapitulates parameter values and copy number distributions. On biological data, biVI provides a scalable route for identifying the biophysical mechanisms underlying gene expression. This analytical approach outlines a generalizable strategy for treating multimodal datasets generated by high-throughput, single-cell genomic assays.
Advances in experimental methods for single-cell RNA sequencing (scRNA-seq) allow for the simultaneous quantification of multiple cellular species, such as nascent and mature transcriptomes [1,2], surface [3–5] and nuclear [6] proteomes, and chromatin accessibility [7,8]. While these rich datasets have the potential to enable unprecedented insight into cell type and state in development and disease, joint analyses of distinct modalities remain challenging. We show that principled biophysical “integration” of multimodal datasets can be achieved through parameterization of interpretable mechanistic models [9], scalable to measurements made for thousands of genes in tens of thousands of cells [10].
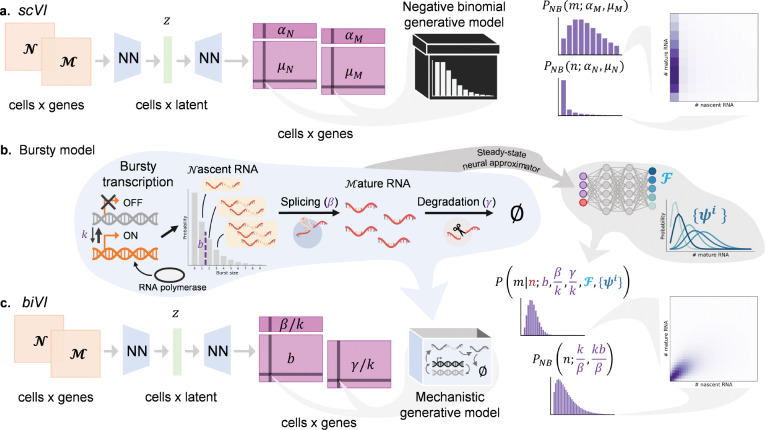
Recent approaches to integrate and reduce the dimensionality of multimodal single-cell genomics data have leveraged advances in machine learning [11–13]. For example, the popular tool scVI is a variational autoencoder (VAE) that uses neural networks to encode scRNA-seq counts to a low-dimensional representation. This is decoded by another neural network to a set of cell- and gene- specific parameters for conditional likelihood distributions of observed counts. These distributions are chosen post hoc to be consistent with the discrete, over-dispersed nature of scRNA-seq counts, but can be derived from biophysical models (Section S1). Extensions of scVI to bimodal data have been attempted for protein [11] and chromatin measurements [14] by jointly encoding data modalities to a single latent space, then employing two decoding networks to produce parameters for independent conditional likelihoods specific to each datatype. Nascent and mature transcripts, available by realigning existing scRNA-seq reads [1,2], could be similarly treated (Figure 1a). However, using independent conditional likelihoods for bimodal measurements derived from the same gene ignores the inherent causality between observations and has no biophysical basis: the generative model is merely part of a neural “black box” used to summarize data.
Figure 1:
biVI reinterprets and extends scVI to infer biophysical parameters. a. scVI can take in concatenated nascent and mature RNA count matrices, encode each cell to a low-dimensional space , and learn per-cell parameters and and per-gene parameters and for independent nascent and mature count distributions. This approach is not motivated by any specific biophysical model. b. A schematic of the telegraph model of transcription: a gene locus has the on rate , the off rate , and the RNA polymerase binding rate . Nascent RNA molecules are produced in geometrically distributed bursts with mean , which are spliced at a constant rate and degraded at a constant rate . Although there is no closed-form solution, this model’s steady-state distribution can be approximated by a pre-trained neural network and a set of basis functions . c. biVI can take in nascent and mature count matrices, produce a low-dimensional representation for each cell, and output per-cell parameters and , as well as the per-gene parameters , for a mechanistically motivated joint distribution of nascent and mature counts.
Nevertheless, good causal model candidates are available: for example, Figure 1b illustrates the extensively validated [15–17] bursty model of transcription. Nascent RNA molecules are produced in geometrically distributed bursts with mean at constant rate and spliced at rate to produce mature molecules, which are degraded with constant rate . While the joint steady-state distribution induced by the bursty model is analytically intractable [18], we have previously shown that it can be approximated by a set of basis functions with neural-network learned weights [19].
We introduce biVI, a strategy that adapts scVI to work with well-characterized stochastic models of transcription. First, we propose several models, formalized by chemical master equations (CMEs), that could give rise to bivariate count distributions for nascent and mature transcripts. We then use the bivariate, CME-derived distribution as the conditional data likelihood distribution for nascent and mature counts (Figure 1c). The inferred conditional likelihood parameters thus have biophysical interpretations as part of a mechanistic model of transcriptional dynamics. Although we focus on the bursty model, biVI implements the closed-form constitutive and extrinsic noise models previously discussed in the literature [9,20,21] (derivations in Section S1 and diagrams in Figures S1 and S2).
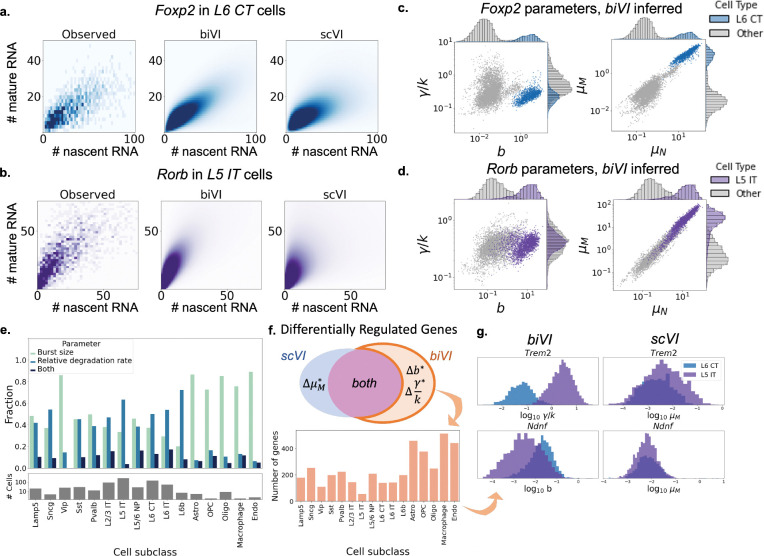
After using simulations to show that biVI models, when compared to scVI, better recapitulate ground-truth distributions and achieve similar clustering of cell types’ latent representations (Figures S3, S4, S5, and Section S6.7), we applied biVI and scVI to experimental data [22] (Section 2.6) from mouse brain tissue [22]. As shown in Figure 2a–b, biVI recapitulates empirical distribution shapes better than scVI (Section 3) while allowing for interpretation of cell-specific parameters to determine how genes are regulated (Section S8). For example, in Figure 2c–d, we illustrate that the upregulation of markers Foxp2 and Rorb can be ascribed to an increase in burst size. We generalize this approach in Figure 2e, which shows the fraction of identified genes in each cell subclass that exhibited significant differences in burst size, relative degradation rate, or both (Section 2.8). Interesting trends across cell subclasses begin to emerge: neuronal cells appear to regulate gene expression via a mix of regulatory strategies, while non-neuronal cells seem to preferentially modulate burst size.
Figure 2:
biVI successfully fits single-cell neuron data and suggests the biophysical basis for expression differences. a.-b. Observed, scVI, and biVI reconstructed distributions of Foxp2, a marker gene for L6 CT (layer 6 corticothalamic) cells, and Rorb, a marker gene for L5 IT (layer 5 intratelencephalic) cells, restricted to respective cell type. c.-d. Cell-specific parameters inferred for Foxp2 and Rorb demonstrate identifiable differences in means and parameters in the marked cell types. e. Cell subclasses show different modulation patterns, with especially pronounced distinctions in non-neuronal cells (top: fractions of genes exhibiting differences in each parameter; bottom: number of cells in each subclass). f. biVI allows the identification of cells which exhibit differences in burst size or relative degradation rate, without necessarily demonstrating differences in mature mean expression. Hundreds of genes demonstrate this modulation behavior, with variation across cell subclasses. g. Histograms of biVI parameters and scVI mature means for two genes that exhibit parameter modulation without identifiable mature mean modulation. Trem2 (top) shows differences in the degradation rate in L5 IT cells, whereas Ndnf (bottom) shows differences in burst size in L6 CT cells.
Finally, biVI can identify distributional differences which do not result in mean expression changes (Section 2.8, Figure 2g–h). For some cell subclasses, there were several hundred such genes, interesting targets for follow-up experimental investigation. For example, the gene Ndnf, which codes for the neuron derived neurotrophic factor NDNF, demonstrated a statistically significant difference in the biVI inferred burst size, but not scVI inferred mature mean, in the neuronal subclass L6 CT (Figure 2i, top row). NDNF promotes the growth, migration, and survival of neurons [23]; characterizing its regulatory patterns could help elucidate its role in neuronal maintenance. As another example, the relative degradation rate of the gene coding for the triggering receptor expressed on myeloid cells-2 (TREM2), variants of which are strongly associated with increased risk of Alzheimer’s disease [24], was found to be greater in the neuronal L5 IT subclass than in other subclasses (Figure 2i, bottom row). While known to be highly expressed in microglia [24], understanding its modulation in other cell subclasses could yield a better understanding of its cell type specific effects on the development of Alzheimer’s disease. Such mechanistic description provides a framework for characterizing the connection between a gene’s role and a cell’s regulatory strategies beyond a mere change in mean expression [25,26].
We have demonstrated that bivariate distributions arising from mechanistic models can be used in variational autoencoders for principled integration of unspliced and spliced RNA-seq data. This improves model interpretability: conditional parameter estimates give insight into the mechanisms of gene regulation that result in differences in expression. While we impose biophysical constraints on species’ conditional joint distributions, orthogonal improvements in interpretability can be made by changing the decoder architecture. biVI models can be instantiated with single-layer linear decoders [27] to directly link latent variables with gene mean parameters via layer weights (Section S9 and Figure S9).
Relaxing assumptions and modeling more molecular modalities (e.g., protein counts and chromatin accessibility) are natural extensions. As single-cell technologies evolve to provide larger-scale, more precise measurements of biomolecules, we anticipate that our approach can be applied and extended for a more comprehensive picture of biophysical processes in living cells.
2. Methods
In order to extend the scVI method to work with multimodal molecule count data in a way that is coherent with biology, we define bivariate likelihood functions that (i) encode a specific, precedented mechanistic model of transcriptional regulation and (ii) are admissible under the assumptions made in the standard scVI pipeline. On a high level, our method entails the following steps:
Choose one of the scVI univariate generative models (Section 2.2), including the functional form of its likelihood and any assumptions about its distributional parameters.
Identify a one-species chemical master equation (CME) that produces this distribution as its steady state, and translate assumptions about distributional parameters into assumptions about the biophysical quantities that parameterize the CME (Section 2.3). The one-species system and its assumptions will typically not be uniquely determined.
Identify a two-species CME and derive assumptions about parameter values consistent with the one-species system (Section 2.3). There will typically be multiple ways to preserve the assumptions but only a single CME.
Modify the autoencoder architecture to output the variables that parameterize the CME solution under the foregoing assumptions, and use this solution as the generative model (Section 2.5).
2.1. Statistical preliminaries
We use the standard parameterization of the Poisson distribution:
| (1) |
We use the shape-mean parameterization of the univariate negative binomial distribution:
| (2) |
We use mean parameterization of the geometric distribution on :
| (3) |
2.2. scVI models
A brief summary of the generative process of the standard, univariate scVI pipeline is useful to contextualize the options and constraints of the bivariate model. In the Bayesian model, each cell has some posterior probability over a low-dimensional space and can be represented as a sample from that posterior. scVI uses the “decoder” neural network to map from realizations to quantities , which describe the compositional abundance of gene in cell as a function of , such that . Furthermore, a cell-specific “size factor” is sampled from a lognormal distribution parameterized by either fit or plug-in estimates of mean and variance such that the mean expression of a gene in a given cell is .
The univariate workflow provides the options of three discrete generative models: Poisson with mean , negative binomial with mean and gene-specific dispersion parameter , and zero-inflated negative binomial, with an additional Bernoulli mixture parameter. We report the master equation models consistent with the first two generative laws below, and discuss a potential basis for and reservations about the zero-inflated model in Section S1.4.
Due to the intractability of the posterior probability , scVI uses variational inference to infer an approximate posterior , which is in form a multivariate Gaussian. Models are trained via stochastic optimization of the Evidence Lower Bound, or ELBO, which minimizes the Kullback-Leibler divergence between the approximate posterior and a prior and maximizes the expectation value of conditional likelihood over the approximate posterior. The Gaussian form of the approximate posterior makes possible a reparameterization trick to calculate gradients of the ELBO over expectation estimates made by Monte Carlo sampling from the approximate posterior. Further, the encoding network amortizes inference by learning a map between data to parameters of the approximate posterior [12,28].
2.3. Master equation models
The one-species CMEs encode reaction schema of the following type:
| (4) |
where is a generic transcript species used to instantiate a univariate scVI generative model, is the transcript’s Markovian degradation rate, and the specific dynamics of the transcription process (first arrow) are deliberately left unspecified for now. Such systems induce univariate probability laws of the form .
The two-species CMEs encode reaction schema of the following type:
| (5) |
where denotes a nascent species, denotes a mature species, and denotes the nascent species’ Markovian conversion rate. Such systems induce bivariate probability laws of the form . We typically identify the nascent species with unspliced transcripts and the mature species with spliced transcripts. We use the nascent/mature nomenclature to simplify notation and emphasize that this identification is natural for scRNA-seq data, but not mandatory in general.
Formalizing a model in terms of the CME requires specifying the precise mechanistic meaning of and . Previous reports equivocate regarding the latter [11], appealing either to cell-wide effects on the biology (in the spirit of [20,21]) or technical variability in the sequencing process (in the spirit of [29]). For completeness, we treat both cases.
Below, we present the theoretical results, including the biophysical models, the functional forms of bivariate distributions consistent with the standard scVI models, and the consequences of introducing further assumptions. The full derivations are given in Section S1.
2.3.1. Constitutive: The Poisson model and its mechanistic basis
The Poisson generative model can be recapitulated by the following schema:
| (6) |
where is a constant transcription rate. This process converges to the bivariate Poisson stationary distribution, with the following likelihood:
| (7) |
where and . If we suppose each gene’s and are constant across cell types, the likelihoods involve a single compositional parameter , such that
| (8) |
where is a gene-specific parameter that can be fit or naïvely estimated by the ratio of the unspliced and spliced averages. On the other hand, if the downstream processes’ kinetics can also change between cell types, we must use two compositional parameters:
| (9) |
We refer to this model as “Poisson,” reflecting its functional form, or “constitutive,” reflecting its biophysical basis.
2.3.2. Extrinsic: The negative binomial model and a possible mixture basis
The negative binomial generative model can be recapitulated by the following schema:
| (10) |
where is the transcription rate, a realization of , a gamma random variable with shape , scale , and mean . This process converges to the bivariate negative binomial (BVNB) stationary distribution, with the following likelihood:
| (11) |
where and . If we suppose that cell type differences only involve changes in the transcription rate scaling factor , with constant and , the likelihoods involve a single compositional parameter . The mean parameters are identical to Equation 8, with an analogous parameter , as well as a gene-specific shape parameter . On the other hand, if the downstream processes’ kinetics can also change between cell types, we must use two compositional parameters, as in Equation 9.
We refer to this model as “extrinsic” to reflect its biophysical basis in extrinsically stochastic rates of transcriptional initiation.
2.3.3. Bursty: The negative binomial model and a possible bursty basis
The negative binomial generative model may be recapitulated by the alternative schema [18]:
| (12) |
where is the burst frequency and is a geometric random variable with mean (Equation 3). This system converges to the following stationary distribution:
| (13) |
where and is arbitrarily set to for simplicity.
Although the nascent marginal is known to be negative binomial, the joint and conditional distributions are not available in closed form. For a given set of parameters, the joint distribution can be approximated over a finite microstate domain , with total state space size . This approach is occasionally useful, if intensive, for evaluating the likelihoods of many independent and identically distributed samples. The numerical procedure entails using quadrature to calculate values of the generating function on the complex unit sphere, then performing a Fourier inversion to obtain a probability distribution [18]. However, this strategy is inefficient in the variational autoencoder framework, where each observation is associated with a distinct set of parameters. Furthermore, it is incompatible with automatic differentiation.
In [19], we demonstrated that the numerical approach can be simplified by approximating with a learned mixture of negative binomial distributions: the weights are given by the outputs of a neural network, whereas the negative binomial bases are constructed analytically. The neural network is trained on the outputs of the generating function procedure. Although the generative model does not have a simple closed-form expression, it is represented by a partially neural, pre-trained function that is a priori compatible with the VAE.
If we suppose cell type differences only involve changes in the burst size , with constant and , we use Equation 13 to evaluate likelihoods. These likelihoods involve a single compositional parameter , with mean parameters identical to Equation 8, with an analogous parameter , as well as a gene-specific shape parameter . On the other hand, if kinetics of the degradation process can also change between cell types, we must use two compositional parameters, as in Equation 9. There is no admissible way to allow modulation in the burst frequency.
We refer to this model as “bursty,” reflecting its biophysical basis.
2.4. biVI bursty generative model
Following the notation of scVI [28], biVI’s generative process for the bursty hypothesis models expression values of and of nascent and mature counts, respectively, in cell c as:
| (14) |
with a standard, multivariate normal prior on the latent space vector. Here, are by default observed mean and variance in log-sequencing depth (‘log-library size’ in scVI) across a cell’s batch, although they can be learned. Further, as in scVI, is neural network that produces fraction of sequencing depth parameters for nascent and mature counts. The sum of nascent and mature fractions is constrained to be 1 over a cell c by a softmax applied to the network output: , where is the number of genes. is a network parameter jointly optimized across all cells during the variational inference procedure. To recover biophysical parameters, is arbitrarily set to . Burst size and relative degradation rate can be recovered according to the following conversions:
| (15) |
We further set with no loss of generality at steady-state. Generative processes for constitutive and extrinsic noise models are discussed in Sections S2 and S3.
2.5. biVI modifications to scVI
Our code is built upon scVI version 0.18.0 [30]; the following outlines the modifications we made for biVI. The scVI framework already supports the constitutive model. By setting conditional likelihood to “poisson,” no modification of scVI architecture is necessary. The conditional data likelihood distribution is the product of two Poisson distributions (Equation 7). Explicitly, unspliced and spliced count matrices can be concatenated along the cell axis to produce a matrix of shape by , where is the number of cells and the number of genes. scVI will then produce Poisson mean parameters for the two Poisson distributions of Equation 7.
For the extrinsic and bursty models, mean parameters for nascent and mature counts, and , and a single shape parameter are necessary. The default scVI architecture returns two independent parameters for nascent and mature counts of the same gene. biVI thus modifies the scVI architecture to update vectors rather than , where is the number of genes. For the extrinsic model, the conditional data likelihood distribution is set to the extrinsic likelihood (Equation 11). For the bursty model, the conditional data likelihood distribution is set to the bursty likelihood (Equation 13). These models also intake concatenated unspliced and spliced matrices of shape by .
2.6. Preprocessing Allen data
Raw 10x v3 single-cell data were originally generated by the Allen Institute for Brain Science [22]. The raw reads in FASTQ format [31] and cluster metadata [32] were obtained from the NeMO Archive. We selected mouse library B08 (donor ID 457911) for analysis.
To obtain spliced and unspliced counts, we first obtained the pre-built mm10 mouse genome released by 10x Genomics (https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest, version 2020-A). We used kallisto|bustools 0.26.0 [2] to build an intronic/exonic reference (kb ref with the option --lamanno). Next, we pseudoaligned the reads to this reference (kb count with the option --lamanno) to produce unspliced and spliced count matrices. We used the outputs produced by the standard bustools filter. This filter was relatively permissive: all (8,424) barcodes given cell type annotations in the Allen metadata were present in the output count matrix (10,975 barcodes).
Based on previous clustering results, we selected cells that were given cell type annotations, and omitted “low quality” or “doublet” barcodes [22], for a total of 6,418 cells. Although any choice to retain or omit cells from analysis is arbitrary, our work models the generating process that produced cells’ nascent and mature counts by presupposing each barcode corresponds to a single cell. Therefore, we propose that cells identified as low-quality (empty cells) or as doublets (two cells measured in one observation) [22] have a fundamentally different data-generating process than individual single cells, and therefore remove them before fitting VAE models. However, we stress that the stochastic nature of transcription and sequencing, the intrinsic uncertainties associated with read alignment, and the numerical compromises made in clustering large datasets mean that previous annotations are not “perfect,” merely a reasonable starting point for comparing alternative methods.
We used Scanpy [33] to restrict our analysis to the most variable genes, which presumably reflect the cell type signatures of interest. The spliced count matrix for the 6,418 retained cells was normalized to sum to 10,000 counts per cell, then transformed with log1p. The top 2,000 most highly variable genes were identified using scanpy.pp.highly_variable_genes on spliced matrices with minimum mean of 0.0125, maximum mean of 3, and minimum dispersion of 0.5 [33]. Spliced and unspliced matrices were subset to include only the 2,000 identified highly variable genes, then concatenated along the cell axis in the order unspliced, spliced to produce a count matrix of size 6,418 by 4,000.
2.7. Fitting Allen data
We applied biVI with the three generative models (bursty, constitutive, and extrinsic) and scVI with negative binomial likelihoods to the concatenated unspliced and spliced count matrix obtained by the filtering procedures outlined above. We made the key assumption that unspliced and spliced counts could be treated as the nascent and mature species of the bursty generative model (see discussion in Section S5). 4,622 cells were used for training with 513 validation cells, and 1,283 cells were held out for testing performance. All models were trained for 400 epochs with a learning rate of 0.001. Encoders and decoder consisted of 3 layers of 128 nodes, and each model employed a latent dimension of 10.
2.8. Bayes factor hypothesis testing for differential expression
After fitting the VAE models, we sought to identify meaningful statistical di↵erences that distinguish cell types. We excluded cell subclasses “L6 IT Car3,” “L5 ET,” “VLMC,” and “SMC” from this analysis, as they contained fewer than ten annotated cells and may require more sophisticated statistical models to account for small sample sizes. The following analysis thus considers 6,398 cells in 16 unique subclasses. We only computed di↵erential expression metrics under the bursty model.
Differential parameter values were tested for each assigned subclass label (as annotated in [22]) versus all others using a Bayes factor hypothesis test following [11]. We reproduce Equations (18) – (21) of [11] below for clarity.
Estimating differential values of any parameter of gene in cells and can be done according to the following Bayesian framework. First, as in Equation (18) of [11], the log fold change (LFC) of between two cells and can be calculated as follows:
| (16) |
Then, as in Equation (19) of [11], the probability that the magnitude of the LFC is greater than some effect threshold can be found by evaluating over the posterior distributions of each cell:
| (17) |
where, in practice, the integral is approximated with many Monte Carlo samples from the two cells’ posteriors. Two hypotheses are tested: , or that the magnitude of the LFC is greater than or equal to threshold , and , or the null hypothesis that the magnitude of the LFC is less than . A Bayes factor for gene between cells and is calculated to compare the two hypotheses, as in Equation (20) of [11]:
| (18) |
Extending this to test differential expression between two groups of cells and amounts to “aggregating the posterior,” as in Equation (21) of [11], or evaluating the same over
| (19) |
In other words, a random sample can be be taken from the approximate posterior of any cell belonging to group and decoded to produce parameter ; likewise a random sample can be taken from the approximate posterior of any cell belonging to group and decoded to produce parameter . The LFC between the two parameters can then be calculated. Repeating this for many Monte Carlo samples over the aggregate posteriors allows estimation of the Bayes factor between two groups.
For the results shown in Figure 1, we used cutoffs of , or a magnitude LFC of ≥ 2, and a Bayes factor threshold of 1.5. The Bayes factors were calculated on normalized burst size and means for biVI, i.e., the fractional inferred burst size or inferred means (before scaling by sampled sequencing depth for that cell), and normalized means for scVI. This controlled for differences in parameters due to sequencing depth that were not biologically meaningful. Relative degradation rate is independent of sequencing depth: hypothesis tests were performed directly on inferred relative degradation rates. While batch identity can also be integrated over to compare groups of cells from different batches, our analysis did not require this as all cells were from the same batch.
3. Reconstructing gene distributions
Let be mechanistic model parameters for gene in cell type . While parameters for a given gene are identical across all cells in a specific cell type, biVI and scVI infer unique parameters for every cell and gene: , where indexes over cells and indexes over genes. To reconstruct distributions for a given gene in a specific cell type , we sample once from the posterior distribution of each cell to obtain point-estimates of conditional parameters , where conditional refers to a single sampling from a cell’s posterior, or a particular realization of . We then average over the cell-specific conditional probabilities for the gene to produce a cell type marginal distribution:
| (20) |
where is the total number of cells in cell type , and indexes over all cells in that cell type. This identity follows immediately from defining the cell type’s distribution as the mixture of the distributions of its constituent cells. In the case of biVI, we plug in Equation 7, 11, or 13 for . In the case of scVI, we use a product of two independent negative binomial laws:
| (21) |
where and are cell- and gene-specific, whereas and are fit separately and take on different values (Section 2.5). For simplicity, this comparison omits uncertainty associated with , which is formally inherited from the uncertainty in the latent representation for each cell .
Thus, Equation 21 is an approximation to the posterior predictive distribution, or marginal distribution of data given the approximated posterior, if we assume Monte Carlo sampling from the approximate posterior distributions of cells within that cell type as a reasonable proxy for sampling from the cell type’s posterior distribution. The posterior predictive, or marginal, distribution is:
where is the approximate posterior. We further note that conditional data likelihood and the marginal distribution are not necessarily of the same form (for example, if the conditional data likelihood distribution is negative binomial, the marginal distribution of genes is not necessarily negative binomial).
Supplementary Material
5. Acknowledgments
M.C., G.G., T.C., and L.P. were partially funded by NIH 5UM1HG012077–02 and NIH U19MH114830. Y.C. was partially funded by T32 GM007377. G.G. thanks Drs. Ido Golding and Heng Xu for the inspiration leading to the explanatory model for the zero-inflated negative binomial distribution in Section S1.4. The RNA illustrations used in Figures 1, S1, and S2 were derived from the DNA Twemoji by Twitter, Inc., used under the CC-BY 4.0 license. We thank the Caltech Bioinformatics Resource Center for GPU resources that helped in performing the analyses.
4. Data availability
Simulated datasets, simulated parameters used to generate them, and Allen dataset B08 and its associated metadata are available in the Zenodo package 7497222. All analysis scripts and notebooks are available at https://github.com/pachterlab/CGCCP_2023. The repository also contains a Google Colaboratory demonstration notebook applying the methods to a small human blood cell dataset.
References
- [1].La Manno Gioele, Soldatov Ruslan, Zeisel Amit, Braun Emelie, Hochgerner Hannah, Petukhov Viktor, Lidschreiber Katja, Kastriti Maria E., Lönnerberg Peter, Furlan Alessandro, Fan Jean, Borm Lars E., Liu Zehua, van Bruggen David, Guo Jimin, He Xiaoling, Barker Roger, Sundström Erik, Castelo-Branco Gonçalo, Cramer Patrick, Adameyko Igor, Linnarsson Sten, and Kharchenko Peter V.. RNA velocity of single cells. Nature, 560(7719):494–498, August 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Melsted Páll, Booeshaghi A. Sina, Liu Lauren, Gao Fan, Lu Lambda, Min Kyung Hoi, da Veiga Beltrame Eduardo, Hjörleifsson Kristján Eldjárn, Gehring Jase, and Pachter Lior. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nature Biotechnology, 39(7):813–818, July 2021. [DOI] [PubMed] [Google Scholar]
- [3].Peterson Vanessa M, Zhang Kelvin Xi, Kumar Namit, Wong Jerelyn, Li Lixia, Wilson Douglas C, Moore Renee, McClanahan Terrill K, Sadekova Svetlana, and Klappenbach Joel A. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology, 35(10):936–939, October 2017. [DOI] [PubMed] [Google Scholar]
- [4].Mimitou Eleni P., Cheng Anthony, Montalbano Antonino, Hao Stephanie, Stoeckius Marlon, Legut Mateusz, Roush Timothy, Herrera Alberto, Papalexi Efthymia, Ouyang Zhengqing, Satija Rahul, Sanjana Neville E., Koralov Sergei B., and Smibert Peter. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nature Methods, 16(5):409–412, May 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Stoeckius Marlon, Hafemeister Christoph, Stephenson William, Houck-Loomis Brian, Chattopadhyay Pratip K, Swerdlow Harold, Satija Rahul, and Smibert Peter. Simultaneous epitope and transcriptome measurement in single cells. Nature Methods, 14(9):865–868, September 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chung Hattie, Parkhurst Christopher N., Magee Emma M., Phillips Devan, Habibi Ehsan, Chen Fei, Yeung Bertrand Z., Waldman Julia, Artis David, and Regev Aviv. Joint single-cell measurements of nuclear proteins and RNA in vivo. Nature Methods, 18(10):1204–1212, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Reyes M., Billman K., Hacohen N., and Blainey P.C.. Simultaneous profiling of gene expression and chromatin accessibility in single cells. Advanced Biosystems, 3,11, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].De Rop Florian, Ismail Joy N, González-Blas Carmen Bravo, Hulselmans Gert J, Flerin Christopher Campbell, Janssens Jasper, Theunis Koen, Christiaens Valerie M, Wouters Jasper, Marcassa Gabriele, de Wit Joris, Poovathingal Suresh, and Aerts Stein. HyDrop enables droplet based single-cell ATAC-seq and single-cell RNA-seq using dissolvable hydrogel beads. eLife, 11:e73971, February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gorin Gennady, Vastola John J., Fang Meichen, and Pachter Lior. Interpretable and tractable models of transcriptional noise for the rational design of single-molecule quantification experiments. Nature Communications, 13(1):7620, December 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Svensson Valentine, Vento-Tormo Roser, and Teichmann Sarah A. Exponential scaling of single-cell RNA-seq in the past decade. Nature Protocols, 13(4):599–604, April 2018. [DOI] [PubMed] [Google Scholar]
- [11].Gayoso Adam, Steier Zöe, Lopez Romain, Regier Jeffrey, Nazor Kristopher L., Streets Aaron, and Yosef Nir. Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nature Methods, 18(3):272–282, March 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gayoso Adam, Lopez Romain, Xing Galen, Boyeau Pierre, Wu Katherine, Jayasuriya Michael, Melhman Edouard, Langevin Maxime, Liu Yining, Samaran Jules, Misrachi Gabriel, Nazaret Achille, Clivio Oscar, Xu Chenling, Ashuach Tal, Lotfollahi Mohammad, Svensson Valentine, da Veiga Beltrame Eduardo, Talavera-López Carlos, Pachter Lior, Theis Fabian J., Streets Aaron, Jordan Michael I., Regier Jeffrey, and Yosef Nir. scvi-tools: a library for deep probabilistic analysis of single-cell omics data. Preprint, bioRxiv: 2021.04.28.441833, April 2021. [Google Scholar]
- [13].Lin Xiang, Tian Tian, Wei Zhi, and Hakonarson Hakon. Clustering of single-cell multi-omics data with a multimodal deep learning method. Nature Communications, 13(1):7705, December 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ashuach Tal, Reidenbach Daniel A., Gayoso Adam, and Yosef Nir. PeakVI: A deep generative model for single-cell chromatin accessibility analysis. Cell Reports Methods, 2(3):100182, March 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Raj Arjun, Charles S Peskin Daniel Tranchina, Vargas Diana Y, and Tyagi Sanjay. Stochastic mRNA Synthesis in Mammalian Cells. PLoS Biology, 4(10):e309, September 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dar R. D., Razooky B. S., Singh A., Trimeloni T. V., McCollum J. M., Cox C. D., Simpson M. L., and Weinberger L. S.. Transcriptional burst frequency and burst size are equally modulated across the human genome. Proceedings of the National Academy of Sciences, 109(43):17454–17459, October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanchez A. and Golding I.. Genetic Determinants and Cellular Constraints in Noisy Gene Expression. Science, 342(6163):1188–1193, December 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Singh Abhyudai and Bokes Pavol. Consequences of mRNA Transport on Stochastic Variability in Protein Levels. Biophysical Journal, 103(5):1087–1096, September 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gorin Gennady, Carilli Maria, Chari Tara, and Pachter Lior. Spectral neural approximations for models of transcriptional dynamics. Preprint, bioRxiv: 2022.06.16.496448, June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ham Lucy, Brackston Rowan D., and Stumpf Michael P. H.. Extrinsic Noise and Heavy-Tailed Laws in Gene Expression. Physical Review Letters, 124(10):108101, March 2020. [DOI] [PubMed] [Google Scholar]
- [21].Elowitz Michael B, Levine Arnold J, Siggia Eric D, and Swain Peter S. Stochastic Gene Expression in a Single Cell. Science, 297(5584):1183–1186, 2002. [DOI] [PubMed] [Google Scholar]
- [22].Yao Zizhen, Liu Hanqing, Xie Fangming, Fischer Stephan, Adkins Ricky S., Aldridge Andrew I., Ament Seth A., Bartlett Anna, Behrens M. Margarita, Van den Berge Koen, Bertagnolli Darren, de Bézieux Hector Roux, Tommaso Biancalani, Booeshaghi A. Sina, Corrada Bravo Héctor, Casper Tamara, Colantuoni Carlo, Crabtree Jonathan, Creasy Heather, Crichton Kirsten, Crow Megan, Dee Nick, Dougherty Elizabeth L., Doyle Wayne I., Dudoit Sandrine, Fang Rongxin, Felix Victor, Fong Olivia, Giglio Michelle, Goldy Jeff, Hawrylycz Mike, Herb Brian R., Hertzano Ronna, Hou Xiaomeng, Hu Qiwen, Kancherla Jayaram, Kroll Matthew, Lathia Kanan, Yang Eric Li Jacinta D. Lucero, Luo Chongyuan, Mahurkar Anup, McMillen Delissa, Nadaf Naeem M., Nery Joseph R., Nguyen Thuc Nghi, Niu Sheng-Yong, Ntranos Vasilis, Orvis Joshua, Osteen Julia K., Pham Thanh, Pinto-Duarte Antonio, Poirion Olivier, Preissl Sebastian, Purdom Elizabeth, Rimorin Christine, Risso Davide, Rivkin Angeline C., Smith Kimberly, Street Kelly, Sulc Josef, Svensson Valentine, Tieu Michael, Torkelson Amy, Tung Herman, Vaishnav Eeshit Dhaval, Vanderburg Charles R., van Velthoven Cindy, Wang Xinxin, White Owen R., Huang Z. Josh, Kharchenko Peter V., Pachter Lior, Ngai John, Regev Aviv, Tasic Bosiljka, Welch Joshua D., Gillis Jesse, Macosko Evan Z., Ren Bing, Ecker Joseph R., Zeng Hongkui, and Mukamel Eran A.. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature, 598(7879):103–110, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kuang XL., Zhao XM., Xu HF., Shi YY., Deng JB., and Sun GT.. Spatio-temporal expression of a novel neuron-derived neurotrophic factor (ndnf) in mouse brains during development. BMC Neurosci, 11, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ulland T. K. and Colonna M.. Trem2 — a key player in microglial biology and alzheimer disease. Nature Reviews Neurology, 14:667–675, 2018. [DOI] [PubMed] [Google Scholar]
- [25].Munsky Brian, Li Guoliang, Fox Zachary R., Shepherd Douglas P., and Neuert Gregor. Distribution shapes govern the discovery of predictive models for gene regulation. Proceedings of the National Academy of Sciences, 115(29):7533–7538, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Munsky Brian, Trinh Brooke, and Khammash Mustafa. Listening to the noise: random fluctuations reveal gene network parameters. Molecular Systems Biology, 5:318, October 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Svensson Valentine, Gayoso Adam, Yosef Nir, and Pachter Lior. Interpretable factor models of single-cell RNA-seq via variational autoencoders. Bioinformatics, 36(11):3418–3421, June 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lopez Romain, Regier Jeffrey, Cole Michael B., Jordan Michael I., and Yosef Nir. Deep generative modeling for single-cell transcriptomics. Nature Methods, 15(12):1053–1058, December 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Jingshu, Huang Mo, Torre Eduardo, Dueck Hannah, Shaffer Sydney, Murray John, Raj Arjun, Li Mingyao, and Zhang Nancy R.. Gene expression distribution deconvolution in single-cell RNA sequencing. Proceedings of the National Academy of Sciences, 115(28):E6437–E6446, July 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gayoso Adam, Lopez Romain, Xing Galen, Boyeau Pierre, Amiri Valeh Valiollah Pour, Hong Justin, Wu Katherine, Jayasuriya Michael, Mehlman Edouard, Langevin Maxime, Liu Yining, Samaran Jules, Misrachi Gabriel, Nazaret Achille, Clivio Oscar, Xu Chenling, Ashuach Tal, Gabitto Mariano, Lotfollahi Mohammad, Svensson Valentine, da Veiga Beltrame Eduardo, Kleshchevnikov Vitalii, Talavera-López Carlos, Pachter Lior, Theis Fabian J., Streets Aaron, Jordan Michael I., Regier Jeffrey, and Yosef Nir. A Python library for probabilistic analysis of single-cell omics data. Nature Biotechnology, February 2022. [DOI] [PubMed] [Google Scholar]
- [31].Allen Institute for Brain Science. FASTQ files for Allen v3 mouse MOp samples, February 2020. [Google Scholar]
- [32].Allen Institute for Brain Science. Analyses for Allen v3 mouse MOp samples, February 2020. [Google Scholar]
- [33].Alexander Wolf F., Philipp Angerer, and Fabian J. Theis. SCANPY: large-scale single-cell gene expression data analysis. Genome Biology, 19(1):15, December 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jahnke Tobias and Huisinga Wilhelm. Solving the chemical master equation for monomolecular reaction systems analytically. Journal of Mathematical Biology, 54:1–26, September 2006. [DOI] [PubMed] [Google Scholar]
- [35].Perez-Carrasco Ruben, Beentjes Casper, and Grima Ramon. Effects of cell cycle variability on lineage and population measurements of messenger RNA abundance. Journal of The Royal Society Interface, 17(168):20200360, July 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gorin Gennady and Pachter Lior. Length biases in single-cell RNA sequencing of pre-mRNA. Biophysical Reports, 3(1):100097, March 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gorin Gennady and Pachter Lior. Monod: mechanistic analysis of single-cell RNA sequencing count data. Preprint, bioRxiv: 2022.06.11.495771, June 2022. [Google Scholar]
- [38].Gorin Gennady and Pachter Lior. Intrinsic and extrinsic noise are distinguishable in a synthesis – export – degradation model of mRNA production. Preprint, bioRxiv: 2020.09.25.312868, September 2020. [Google Scholar]
- [39].Jiang Ruochen, Sun Tianyi, Song Dongyuan, and Li Jingyi Jessica. Statistics or biology: the zero-inflation controversy about scRNA-seq data. Genome Biology, 23:31, January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Svensson Valentine. Droplet scRNA-seq is not zero-inflated. Nature Biotechnology, 38(2):147–150, February 2020. [DOI] [PubMed] [Google Scholar]
- [41].Jia Chen. Kinetic Foundation of the Zero-Inflated Negative Binomial Model for Single-Cell RNA Sequencing Data. SIAM Journal on Applied Mathematics, 80(3):1336–1355, January 2020. [Google Scholar]
- [42].Xu Heng, Sepúlveda Leonardo A, Figard Lauren, Sokac Anna Marie, and Golding Ido. Combining protein and mRNA quantification to decipher transcriptional regulation. Nature Methods, 12(8):739–742, August 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gorin Gennady and Pachter Lior. Modeling bursty transcription and splicing with the chemical master equation. Biophysical Journal, 121(6):1056–1069, February 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rodriguez Joseph and Larson Daniel R.. Transcription in Living Cells: Molecular Mechanisms of Bursting. Annual Review of Biochemistry, 89(1):189–212, June 2020. [DOI] [PubMed] [Google Scholar]
- [45].Xu Heng, Skinner Samuel O., Sokac Anna Marie, and Golding Ido. Stochastic Kinetics of Nascent RNA. Physical Review Letters, 117(12):128101, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Choubey Sandeep, Kondev Jane, and Sanchez Alvaro. Deciphering Transcriptional Dynamics In Vivo by Counting Nascent RNA Molecules. PLOS Computational Biology, 11(11):e1004345, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Choubey Sandeep. Nascent RNA kinetics: Transient and steady state behavior of models of transcription. Physical Review E, 97(2):022402, 2018. [DOI] [PubMed] [Google Scholar]
- [48].Gómez-Schiavon Mariana, Chen Liang-Fu, West Anne E., and Buchler Nicolas E.. BayFish: Bayesian inference of transcription dynamics from population snapshots of single-molecule RNA FISH in single cells. Genome Biology, 18(1):164, December 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wang Mengyu, Zhang Jing, Xu Heng, and Golding Ido. Measuring transcription at a single gene copy reveals hidden drivers of bacterial individuality. Nature Microbiology, 4:2118–2127, September 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zenklusen Daniel, Larson Daniel R, and Singer Robert H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Structural & Molecular Biology, 15(12):1263–1271, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Senecal Adrien, Munsky Brian, Proux Florence, Ly Nathalie, Braye Floriane E., Zimmer Christophe, Mueller Florian, and Darzacq Xavier. Transcription Factors Modulate c-Fos Transcriptional Bursts. Cell Reports, 8(1):75–83, July 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Halpern Keren Bahar, Tanami Sivan, Landen Shanie, Chapal Michal, Szlak Liran, Hutzler Anat, Nizhberg Anna, and Itzkovitz Shalev. Bursty Gene Expression in the Intact Mammalian Liver. Molecular Cell, 58(1):147–156, April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Skinner Samuel O, Xu Heng, Nagarkar-Jaiswal Sonal, Freire Pablo R, Zwaka Thomas P, and Golding Ido. Single-cell analysis of transcription kinetics across the cell cycle. eLife, 5:e12175, January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Shah Sheel, Takei Yodai, Zhou Wen, Lubeck Eric, Yun Jina, Eng Chee-Huat Linus, Koulena Noushin, Cronin Christopher, Karp Christoph, Liaw Eric J., Amin Mina, and Cai Long. Dynamics and Spatial Genomics of the Nascent Transcriptome by Intron seqFISH. Cell, 174(2):363–376.e16, July 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wan Yihan, Anastasakis Dimitrios G., Rodriguez Joseph, Palangat Murali, Gudla Prabhakar, Zaki George, Tandon Mayank, Pegoraro Gianluca, Chow Carson C., Hafner Markus, and Larson Daniel R.. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell, 184(11):2878–2895.e20, May 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wold Barbara and Myers Richard M. Sequence census methods for functional genomics. Nature Methods, 5(1):19–21, January 2008. [DOI] [PubMed] [Google Scholar]
- [57].Reimer Kirsten A., Mimoso Claudia A., Adelman Karen, and Neugebauer Karla M.. Co-transcriptional splicing regulates 3’ end cleavage during mammalian erythropoiesis. Molecular Cell, 81(5):998–1012.e7, March 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Drexler Heather L., Choquet Karine, and Churchman L. Stirling. Splicing Kinetics and Co-ordination Revealed by Direct Nascent RNA Sequencing through Nanopores. Molecular Cell, 77(5):985–998.e8, March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zeisel A., Kostler W. J., Molotski N., Tsai J. M., Krauthgamer R., Jacob-Hirsch J., Rechavi G., Soen Y., Jung S., Yarden Y., and Domany E.. Coupled pre-mRNA and mRNA dynamics unveil operational strategies underlying transcriptional responses to stimuli. Molecular Systems Biology, 7(1):529–529, September 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pimentel Harold, Conboy John G., and Pachter Lior. Keep Me Around: Intron Retention Detection and Analysis. Preprint, arXiv: 1510.00696, October 2015. [Google Scholar]
- [61].Pimentel Harold, Parra Marilyn, Gee Sherry L., Mohandas Narla, Pachter Lior, and Conboy John G.. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Research, 44(2):838–851, January 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gorin Gennady, Fang Meichen, Chari Tara, and Pachter Lior. RNA velocity unraveled. PLOS Computational Biology, 18(9):e1010492, September 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hjörleifsson Kristján Eldjárn, Sullivan Delaney K., Holley Guillaume, Melsted Páll, and Pachter Lior. Accurate quantification of single-nucleus and single-cell RNA-seq transcripts. Preprint, bioRxiv: 2022.12.02.518832, December 2022. [Google Scholar]
- [64].Soneson Charlotte, Srivastava Avi, Patro Rob, and Stadler Michael B.. Preprocessing choices affect RNA velocity results for droplet scRNA-seq data. PLOS Computational Biology, 17(1):e1008585, January 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mazille Maxime, Buczak Katarzyna, Scheiffele Peter, and Mauger Oriane. Stimulus-specific remodeling of the neuronal transcriptome through nuclear intron-retaining transcripts. The EMBO Journal, 41(21):e110192, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sina Booeshaghi A., Yao Zizhen, van Velthoven Cindy, Smith Kimberly, Tasic Bosiljka, Zeng Hongkui, and Pachter Lior. Isoform cell-type specificity in the mouse primary motor cortex. Nature, 598(7879):195–199, October 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kessler O, Jiang Y, and Chasin L A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Molecular and Cellular Biology, 13(10):6211–6222, October 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Allison Coté Chris Coté, Bayatpour Sareh, Drexler Heather L, Alexander Katherine A, Chen Fei, Wassie Asmamaw T, Boyden Edward S, Berger Shelley, Churchman L Stirling, and Raj Arjun. pre-mRNA spatial distributions suggest that splicing can occur post-transcriptionally. Preprint, bioRxiv: 2020.04.06.028092, June 2021. [Google Scholar]
- [69].Gorin Gennady, Yoshida Shawn, and Pachter Lior. Transient and delay chemical master equations. Preprint, bioRxiv: 2022.10.17.512599, October 2022. [Google Scholar]
- [70].Cao Zhixing and Grima Ramon. Analytical distributions for detailed models of stochastic gene expression in eukaryotic cells. Proceedings of the National Academy of Sciences, 117(9):4682–4692, March 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jiang Qingchao, Fu Xiaoming, Yan Shifu, Li Runlai, Du Wenli, Cao Zhixing, Qian Feng, and Grima Ramon. Neural network aided approximation and parameter inference of non-Markovian models of gene expression. Nature Communications, 12(1):2618, December 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Filatova Tatiana, Popović Nikola, and Grima Ramon. Modulation of nuclear and cytoplasmic mRNA fluctuations by time-dependent stimuli: Analytical distributions. Mathematical Biosciences, 347:108828, May 2022. [DOI] [PubMed] [Google Scholar]
- [73].Hansen Maike M.K., Desai Ravi V., Simpson Michael L., and Weinberger Leor S.. Cytoplasmic Amplification of Transcriptional Noise Generates Substantial Cell-to-Cell Variability. Cell Systems, 7(4):384–397.e6, October 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Battich Nico, Stoeger Thomas, and Pelkmans Lucas. Control of Transcript Variability in Single Mammalian Cells. Cell, 163(7):1596–1610, December 2015. [DOI] [PubMed] [Google Scholar]
- [75].Gorin Gennady and Pachter Lior. Special function methods for bursty models of transcription. Physical Review E, 102(2):022409, August 2020. [DOI] [PubMed] [Google Scholar]
- [76].Fu Xiaoming, Patel Heta P, Coppola Stefano, Xu Libin, Cao Zhixing, Lenstra Tineke L, and Grima Ramon. Quantifying how post-transcriptional noise and gene copy number variation bias transcriptional parameter inference from mRNA distributions. eLife, 11:e82493, October 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fu Xiaoming, Patel Heta P., Coppola Stefano, Xu Libin, Cao Zhixing, Lenstra Tineke L., and Grima Ramon. Accurate inference of stochastic gene expression from nascent transcript heterogeneity. Preprint, bioRxiv: 2021.11.09.467882, November 2021. [Google Scholar]
- [78].Pedregosa Fabian, Varoquaux Gael, Gramfort Alexandre, Michel Vincent, Thirion Bertrand, Grisel Olivier, Blondel Mathieu, Prettenhofer Peter, Weiss Ron, Dubourg Vincent, Vanderplas Jake, Passos Alexandre, and Cournapeau David. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research, 12:2825–2830, October 2011. [Google Scholar]
- [79].The Gene Ontology Consortium, Carbon Seth, Douglass Eric, Good Benjamin M, Unni Deepak R, Harris Nomi L, Mungall Christopher J, Basu Siddartha, Chisholm Rex L, Dodson Robert J, Hartline Eric, Fey Petra, Thomas Paul D, Albou Laurent-Philippe, Ebert Dustin, Kesling Michael J, Mi Huaiyu, Muruganujan Anushya, Huang Xiaosong, Mushayahama Tremayne, LaBonte Sandra A, Siegele Deborah A, Antonazzo Giulia, Attrill Helen, Nick H Brown Phani Garapati, Steven J Marygold Vitor Trovisco, Gil dos Santos Kathleen Falls, Tabone Christopher, Zhou Pinglei, Goodman Joshua L, Strelets Victor B, Thurmond Jim, Garmiri Penelope, Ishtiaq Rizwan, Rodríguez-López Milagros, Acencio Marcio L, Kuiper Martin, Astrid Lægreid Colin Logie, Lovering Ruth C, Kramarz Barbara, Saverimuttu Shirin C C, Pinheiro Sandra M, Gunn Heather, Su Renzhi, Thurlow Katherine E, Chibucos Marcus, Giglio Michelle, Nadendla Suvarna, Munro James, Jackson Rebecca, Duesbury Margaret J, Del-Toro Noemi, Meldal Birgit H M, Paneerselvam Kalpana, Perfetto Livia, Porras Pablo, Orchard Sandra, Shrivastava Anjali, Chang Hsin-Yu, Finn Robert Daniel, Mitchell Alexander Lawson, Rawlings Neil David, Richardson Lorna, Sangrador-Vegas Amaia, Blake Judith A, Christie Karen R, Dolan Mary E, Drabkin Harold J, Hill David P, Ni Li, Sitnikov Dmitry M, Harris Midori A, Oliver Stephen G, Rutherford Kim, Wood Valerie, Hayles Jaqueline, Bähler Jürg, Bolton Elizabeth R, De Pons Jeffery L, Dwinell Melinda R, Hayman G Thomas, Kaldunski Mary L, Kwitek Anne E, Laulederkind Stanley J F, Plasterer Cody, Tutaj Marek A, Vedi Mahima, Wang Shur-Jen, D’Eustachio Peter, Matthews Lisa, Balhoff James P, Aleksander Suzi A, Alexander Michael J, Cherry J Michael, Engel Stacia R, Gondwe Felix, Karra Kalpana, Miyasato Stuart R, Nash Robert S, Simison Matt, Skrzypek Marek S, Weng Shuai, Wong Edith D, Feuermann Marc, Gaudet Pascale, Morgat Anne, Bakker Erica, Tanya Z Berardini Leonore Reiser, Subramaniam Shabari, Huala Eva, Cecilia N Arighi Andrea Auchincloss, Axelsen Kristian, Argoud-Puy Ghislaine, Bateman Alex, Blatter Marie-Claude, Boutet Emmanuel, Bowler Emily, Breuza Lionel, Bridge Alan, Britto Ramona, Bye-A-Jee Hema, Casas Cristina Casals, Coudert Elisabeth, Denny Paul, Estreicher Anne, Famiglietti Maria Livia, Georghiou George, Gos Arnaud, Gruaz-Gumowski Nadine, Hatton-Ellis Emma, Hulo Chantal, Ignatchenko Alexandr, Jungo Florence, Laiho Kati, Le Mercier Philippe, Lieberherr Damien, Lock Antonia, Lussi Yvonne, MacDougall Alistair, Magrane Michele, Martin Maria J, Masson Patrick, Natale Darren A, Hyka-Nouspikel Nevila, Orchard Sandra, Pedruzzi Ivo, Pourcel Lucille, Poux Sylvain, Pundir Sangya, Rivoire Catherine, Speretta Elena, Sundaram Shyamala, Tyagi Nidhi, Warner Kate, Zaru Rossana, Wu Cathy H, Diehl Alexander D, Chan Juancarlos N, Grove Christian, Lee Raymond Y N, Muller Hans-Michael, Raciti Daniela, Van Auken Kimberly, Sternberg Paul W, Berriman Matthew, Paulini Michael, Howe Kevin, Gao Sibyl, Wright Adam, Stein Lincoln, Douglas G Howe Sabrina Toro, Westerfield Monte, Jaiswal Pankaj, Cooper Laurel, and Elser Justin. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Research, 49(D1):D325–D334, January 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Simulated datasets, simulated parameters used to generate them, and Allen dataset B08 and its associated metadata are available in the Zenodo package 7497222. All analysis scripts and notebooks are available at https://github.com/pachterlab/CGCCP_2023. The repository also contains a Google Colaboratory demonstration notebook applying the methods to a small human blood cell dataset.