Figure 7. Monomeric actin directly binds RPA and actin polymerization is required for efficient RPA deposition at perturbed replication forks.
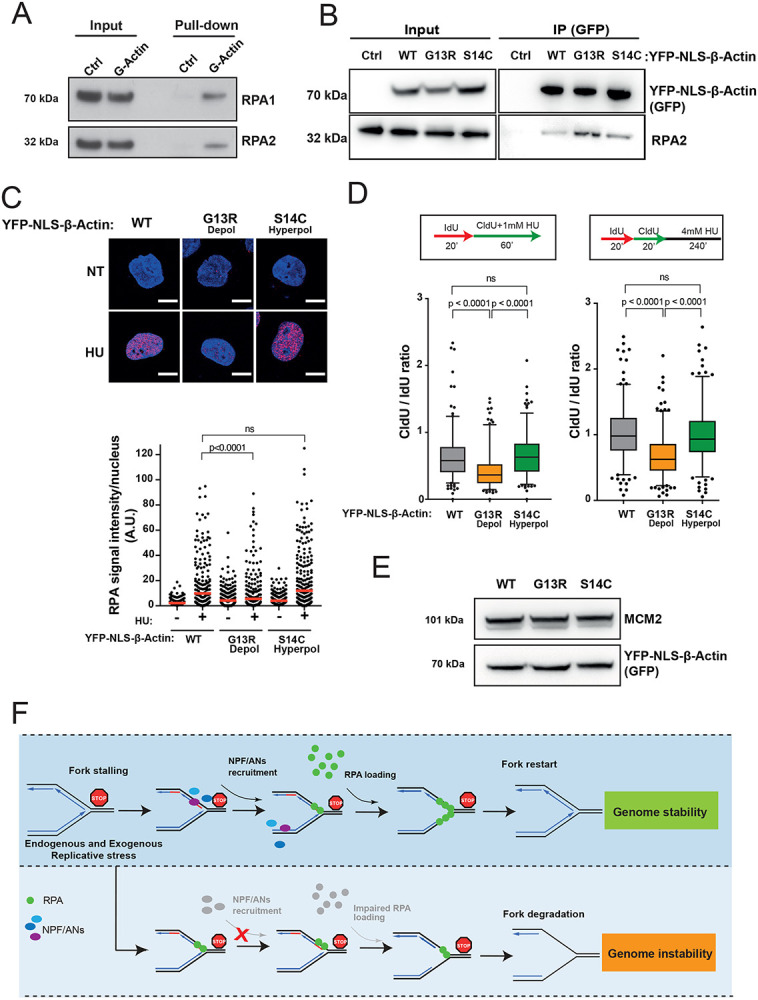
A. RPA directly binds G-actin. WB of RPA in vitro pull down with G-actin conjugated sepharose beads. Ctrl, control.
B. WB of immunoprecipitation (IP) experiments in HEK293TN cells expressing YFP-NLS-β-actin WT, G13R or S14C mutant.
C. Representative images and dot plot of RPA signal intensity in HeLa cells expressing YFP-NLS-β-actin WT, G13R or S14C mutant upon 1mM HU treatment for 1h. Dot plots represent data pooled from three independent experiments. Statistical significance was determined using the Mann-Whitney test. ns, nonsignificant.
D. Box plots of CldU/IdU tract ratios of HeLa cells expressing YFP-NLS-β-actin WT, G13R or S14C mutant proteins treated with 1mM HU during CldU labelling pulse (left panel) or CldU/IdU pulse labelling was followed by treatment with 4mM HU for 4h (right panel). Whiskers indicate 5-95 percentile. Box plots represent data pooled from two independent experiments. Statistical significance was determined using the Mann-Whitney test. ns, nonsignificant.
E. Representative WB showing levels of expression of YFP-NLS-β-actin WT, G13R or S14C in HeLa cells from C and D.
F. Proposed model for the role of NPF/ANs in the replicative stress response. Replicative stress leads to fork stalling and generation of ssDNA, subsequently the association of actin nucleators (ANs) and nucleation promoting factors (NPFs) with sites of ongoing replication facilitates RPA deposition, either directly (via WASp) or indirectly (via actin polymerization), in order to protect ssDNA generated at perturbed forks. This allows for an efficient fork restart and promotes genome stability. Depletion of NPF/ANs and/or interference with actin polymerization impairs RPA loading/stability on ssDNA leading to extensive nascent strand degradation and ultimately genome instability.
