Abstract
Genome-wide association studies (GWAS) of human complex traits or diseases often implicate genetic loci that span hundreds or thousands of genetic variants, many of which have similar statistical significance. While statistical fine-mapping in individuals of European descent has made important discoveries, cross-population fine-mapping has the potential to improve power and resolution by capitalizing on the genomic diversity across ancestries. Here we present SuSiEx, an accurate and computationally efficient method for cross-population fine-mapping, which builds on the single-population fine-mapping framework, Sum of Single Effects (SuSiE). SuSiEx integrates data from an arbitrary number of ancestries, explicitly models population-specific allele frequencies and LD patterns, accounts for multiple causal variants in a genomic region, and can be applied to GWAS summary statistics when individual-level data is unavailable. We comprehensively evaluated SuSiEx using simulations, a range of quantitative traits measured in both UK Biobank and Taiwan Biobank, and schizophrenia GWAS across East Asian and European ancestries. In all evaluations, SuSiEx fine-mapped more association signals, produced smaller credible sets and higher posterior inclusion probability (PIP) for putative causal variants, and retained population-specific causal variants.
INTRODUCTION
Genome-wide association studies (GWAS) of human complex traits or diseases often implicate genetic loci that span hundreds or thousands of genetic variants, many of which have similar statistical significance. These loci may contain one or a handful of causal variants, while the associations of other variants are driven by their linkage disequilibrium (LD) with the causal variant(s). Statistical fine-mapping refines a GWAS locus to a smaller set of likely causal variants to facilitate interpretation and computational and experimental functional studies. Fine-mapping studies in samples of European ancestry have made important advances, with some disease-associated loci resolved to single-variant resolution1–3. Since non-causal variants tagging causal signals have marginally different effects across populations due to differences in LD patterns, cross-population fine-mapping, which integrates data from multiple populations and capitalizes on the genomic diversity across ancestries (e.g., smaller LD blocks in African populations), holds the promise to further improve fine-mapping resolution.
Cross-population fine-mapping analysis can be broadly classified into three categories, namely the meta-analysis-based approach, the post hoc combining approach, and Bayesian statistical methods (Figure 1). The meta-analysis-based approach applies single-population fine-mapping methods to meta-analyzed GWAS summary statistics and LD matrices, and has been widely used in the field, including in several seminal studies4,5. This approach, however, assumes no heterogeneity in effect sizes and LD patterns across populations, which is often not true and may lead to false positives and miscalibration of the inferred probability of a variant being causal6. The post hoc combining approach analyzes data from each population independently and integrates single-population fine-mapping results post hoc. While conducive to identifying population-specific causal variants7, this approach fails to leverage the increased sample size, potential genetic correlations and LD diversity across populations to facilitate loci discovery and improve fine-mapping resolution, and may be sensitive to the choice of methods that combine population-specific results. Bayesian methods8,9 provide a principled way to fine-map causal variants across populations and have been employed in the analyses of several complex traits or diseases8–12. That said, current cross-population Bayesian fine-mapping methods often suffer from inflated false positive rates, poor computational scalability, and inability to distinguish multiple causal signals in the same genomic locus, impeding their applications to emerging biobank-scale datasets of diverse ancestries.
Figure 1: Overview of fine-mapping methods.
An illustration of the inputs and outputs for single-population and cross-population fine-mapping methods, the latter of which includes meta-analysis-based approaches, post hoc combining approaches, previously published Bayesian fine-mapping methods as well as SuSiEx.
Recently, Wang et al. proposed a single-population fine-mapping method, SUm of SIngle Effects (SuSiE)13, which improved the calibration, computational efficiency and interpretation of statistical fine-mapping. Here, we extend the SuSiE model to a cross-population fine-mapping method, SuSiEx, which integrates multiple population-specific GWAS summary statistics and LD panels to enable more powerful and accurate fine-mapping. We evaluated the calibration, power, resolution and computational scalability of SuSiEx along with alternative fine-mapping methods via extensive simulations. We further used SuSiEx to fine-map 25 quantitative traits shared between the UK Biobank14 and Taiwan Biobank15, and to fine-map schizophrenia genetic risk loci across European and East Asian ancestries.
RESULTS
Overview of SuSiEx
SuSiEx extends the single-population fine-mapping model, SuSiE13, by integrating population-specific GWAS summary statistics and LD reference panels from multiple populations. In SuSiE, the genetic influence on a trait or disease within a genomic locus is modeled as the summation of several distinct effects, each contributed by a single causal variant, which naturally allows for the modeling of multiple association signals and assigns each inferred putative causal variant to a credible set with a posterior inclusion probability (PIP) (Figure 1). Building on this framework, SuSiEx couples each single effect by assuming that the causal variants are shared across populations (i.e., we report a single PIP rather than population-specific PIPs for each variant in a credible set), while allowing them to have varying effect sizes (including null effects) across ancestries. In addition, SuSiEx allows for a variant to be missing in an ancestry (e.g., due to its low allele frequency), in which case the ancestry does not contribute to the PIP estimate, effectively reducing the total sample size. Similar to SuSiE, SuSiEx builds on the Bayesian variable selection in regression16,17 and applies the iterative Bayesian stepwise selection13 to model fitting. Further modeling and computational details for SuSiEx are discussed in Methods.
Compared with the meta-analysis-based fine-mapping approach4,5, SuSiEx explicitly models population-specific GWAS summary statistics and LD patterns (Figure 1; Extended Data Figure 1a), which is expected to improve the fine-mapping resolution and more accurately control the false positive rates, while allowing for heterogeneous effect sizes and retaining population-specific causal variants (Extended Data Figure 1c). Compared with post hoc analysis to combine single-population fine-mapping results7, SuSiEx leverages the sample size, genetic correlation and LD diversity across ancestries to improve the resolution of fine-mapping, especially for loci that are under-powered to fine-map in individual datasets (Figure 1; Extended Data Figure 1b). Compared with other Bayesian cross-population fine-mapping methods such as PAINTOR9,18 and MsCAIVAR8, SuSiEx infers distinct credible sets for each causal signal (Figure 1), facilitating the interpretation of fine-mapping results, and is orders of magnitudes more scalable computationally (discussed later), enabling the analysis of large, complex loci and biobank-scale datasets across many complex traits and diseases.
SuSiEx outperformed single-population and naive cross-population fine-mapping methods in simulations
We conducted a series of simulations to systematically evaluate the performance of SuSiEx. Specifically, we generated simulation data under different numbers of causal variants (ncsl) per locus, genetic correlations across populations (rg) and SNP heritability (h2) (Methods). To examine the impact of these genetic parameters on fine-mapping results, we defined a standard simulation setting with ncsl = 1, rg = 0.7 and h2 = 0.1%, and then varied these parameters to produce a range of local genetic architectures (Supplementary Tables 1 & 2). Given a set of genetic parameters, we further assessed the impact of different population (European - EUR; African - AFR; East Asian - EAS) and discovery sample size combinations (Supplementary Table 3) on fine-mapping results. Throughout the simulation study, in single-population fine-mapping, we analyzed loci that reached genome-wide significance in population-specific GWAS (P<5×10−8); in cross-population fine-mapping, we analyzed loci that reached genome-wide significance in at least one of the population-specific GWAS or in the cross-population fixed-effect meta-analysis. We assessed the performance of different fine-mapping methods using an array of metrics: (i) Coverage/Calibration: the proportion of credible sets that include at least one true causal variant across simulation replicates; (ii) Power: the number of true causal variants identified (i.e., covered by a credible set); (iii) Resolution: the size of credible sets and the number of fine-mapped variants with high confidence (e.g., PIP >95%); (iv) Scalability: the computational cost/feasibility to perform fine-mapping in large genomic loci; (v) Robustness: the proportion of runs in which the fine-mapping algorithm converges and returns sensible results (defined later).
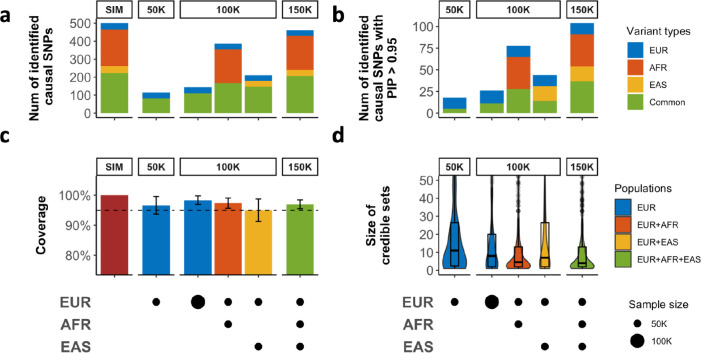
As expected, in the standard simulation setting (Figure 2; Supplementary Figures 1 & 2), compared with single-population fine-mapping even with the same total sample size, integrating data across populations using SuSiEx led to better power (i.e., more true causal variants being identified; Figure 2a), had higher resolution (i.e., smaller credible sets and more causal variants with high PIP; Figure 2b & 2d) and retained population-specific causal variants (Figure 2a & 2b). Meanwhile, SuSiEx had well controlled coverage at 95%, regardless of the populations from which data were combined (Figure 2c). The magnitude of improvements in power and resolution is a result of both the increase in the total sample size and the LD diversity in the discovery samples (Figure 2; Supplementary Table 4). For example, adding 50K EUR individuals to an existing EUR sample of 50K individuals increased the number of identified causal variants with PIP >95% from 18 to 26 and reduced the median size of the credible set from 11 to 8. The yield of causal variants with PIP >95% was much greater (increased from 18 to 78) and the median size of the credible set was much smaller (reduced from 11 to 5) if the added 50K individuals were of AFR instead of EUR ancestry, demonstrating the importance of genetic diversity in cross-population fine-mapping. The inclusion of 50K individuals of EAS ancestry also provided a greater yield of causal variants with PIP >95% (increased from 18 to 44) and smaller credible sets (reduced from 11 to 7) relative to adding 50K EUR samples, although the advantages were less pronounced than when the AFR samples were added, due to the smaller LD blocks in the African ancestries19,20.
Figure 2: The performance of SuSiEx in simulations.
Simulated data were generated under the standard parameter setting (Methods). a, The number of identified true causal variants (true causal variants covered by a credible set) when integrating data from different populations with different sample sizes for fine-mapping. b, The number of true causal variants mapped to PIP >95%. c, The coverage of credible sets (the proportion of credible sets that contain a true causal variant). The dashed line indicates 95% coverage and error bars indicate 95% confidence intervals. d, Distribution of the size of credible sets. The upper and lower bounds of the box indicate the 75th and 25th percentiles, respectively. The middle line in the box indicates the median. In a-d, top labels of each subpanel indicate the total sample size, and the bottom panels indicate the sample size from each population. In a and b, we defined variants with MAF >0.5% only in one population as specific to that population, and all other variants as “shared” (i.e., shared variants across populations).
A widely used approach in recent multi-ancestry genetic studies4 is to apply a single-population fine-mapping method to meta-analyzed GWAS summary statistics and LD matrices (e.g., using a sample size weighted approach). Despite of its convenience, this method can be miscalibrated and does not unleash the full potential of genomic diversity, likely due to its over-simplified modeling of LD across populations, the presence of population-specific variants, and the strong assumption on cross-population effect size heterogeneity in fixed-effect meta-analysis6. We confirmed, using the standard simulation setting, that fine-mapping using meta-analyzed GWAS and sample size weighted LD suffered substantial loss in both power and coverage (Supplementary Figures 3 & 4; Supplementary Table 5). In contrast, SuSiEx, through explicit and flexible modeling of population-specific association statistics and LD, identified many more causal variants (Supplementary Figure 4a) and was well calibrated (Supplementary Figure 4b).
Another recently proposed strategy uses post hoc analysis to combine single-population fine-mapping results, which has been applied to multiple large-scale biobanks with promising biological discoveries7. However, this approach does not make use of subthreshold association signals, and does not leverage LD diversity to improve the resolution of fine-mapping. In simulations, SuSiEx found more true causal variants especially when the GWAS sample size is moderate or small, as expected for current non-EUR GWAS (Supplementary Table 5). For example, when analyzing 50K EUR and 20K AFR individuals under the standard simulation setting, the post hoc approach identified a smaller number of causal variants compared with SuSiEx (159 vs. 175). Although the numbers of true causal variants discovered by both approaches become closer when the GWAS sample sizes become larger, SuSiEx still outperformed post hoc analysis in resolution. In simulations, SuSiEx always identified more true causal variants with high PIP (50% or 95%) than post hoc analysis (Supplementary Figure 5 and Supplementary Table 5). For example, when analyzing 200K EUR and 200K AFR individuals under the standard simulation setting, the post hoc approach identified a smaller number of causal variants with PIP > 95% compared with SuSiEx (140 vs. 161). And the median size of the credible set was 10 vs. 8 when combining data from 50K EUR and 20K AFR individuals for post hoc and SuSiEx respectively, and 4 vs. 2 when analyzing 200K EUR and 200K AFR individuals (Supplementary Table 5).
SuSiEx outperformed existing Bayesian cross-population fine-mapping methods in simulations
We further compared SuSiEx with two published Bayesian cross-population fine-mapping methods, PAINTOR9,18 and MsCAVIAR8, using the standard simulation setting (Supplementary Table 2). We noted that neither of the two methods is capable of analyzing all common variants (MAF >1% in EUR, EAS or AFR) in a 1 Mb locus (6,548 variants per locus on average; Figure 3a, left column). In particular, MsCAVIAR is not computationally scalable and cannot complete analyzing a genetic locus within 24 hours, while PAINTOR always returned unreasonable results, in which the sum of PIP across variants in a genomic locus >5 or <0.1. We note that in the standard simulation setting, the number of true causal variants was set to one in each locus, and thus a sum of PIP >5 or <0.1 appears “unreasonable” and may indicate severe model fitting issues such as failure to converge. We then filtered the discovery summary statistics to fewer variants to enable performance evaluation across methods. Specifically, we created three input datasets with increasingly stringent selection criteria: “p < 0.05”, “top 500” and “top 150”, corresponding to marginal P <0.05, the top 500 and the top 150 most associated variants, respectively. With these filtered input datasets, the “enumerate” mode of PAINTOR, with the number of causal variants set to one (which matched the simulation parameter, and was thus a favorable setting for PAINTOR), still returned unreasonable results (sum of PIP >5 or <0.1) for approximately 25% of the analyses (Figure 3a), while the “MCMC” mode of PAINTOR returned unreasonable results for almost all the analyses, with zero PIP for every variant (Supplementary Table 6). The “enumerate” mode of PAINTOR was also highly sensitive to the parameter “maximum number causal SNPs”, which is typically unknown a priori and difficult to set in practice (Extended Data Figure 2). The other Bayesian fine-mapping method, MsCAIVAR, was only able to analyze the smallest input dataset (“top 150”), as larger dataset took more than 24 hours per locus (Figure 3a), although the results were generally “reasonable” (Extended data Figure 2; Supplementary Table 6).
Figure 3: Comparison of SuSiEx, PAINTOR and MsCAVIAR in simulations.
a, The job completion summary (scalability and robustness) for Bayesian fine-mapping methods using different numbers of input variants. PAINTOR was run using the “enumerate” mode with “-enumerate=1” (which matched the simulation parameter). Unfinished: jobs taking longer than 24 hours wall time. Unreasonable: jobs returning unreasonable results, defined as the sum of PIP across variants in the genomic locus >5 or <0.1 (1 is expected). Successful: jobs completed within 24 hours of wall time and returned reasonable results. b, Number of identified true causal variants with PIP >50% (x-axis) versus the coverage of credible sets (y-axis) for different input datasets and fine-mapping methods. Only simulation runs that were completed within 24 hours and returned reasonable results were included.
For each method, we then focused on simulation runs that returned reasonable PIP estimates. PAINTOR, with the “enumerate” mode and the number of causal variants set to one, had calibrated results at 95% coverage and identified a similar number of high-PIP causal variants to SuSiEx in the EUR-only and EUR + EAS fine-mapping (PIP >50%; Figure 3b). MsCAVIAR, however, identified much fewer causal variants with PIP >50% (Figure 3b). This is because MsCAVIAR tends to return large credible sets containing almost all the variants in the input dataset, each having a small PIP (Supplementary Table 7). SuSiEx outperformed PAINTOR and MsCAVIAR in the number of causal variants identified with PIP >50%, when AFR samples were included in the discovery GWAS (Figure 3b), suggesting that SuSiEx can leverage genomic diversity to fine-map more causal variants with high accuracy. For example, when combining 200K EUR and 200K AFR samples, SuSiEx identified 261 unique causal variants with PIP >50% using the full GWAS summary statistics, comparing with 209 identified by PAINTOR and 7 identified by MsCAIVAR across the four input datasets (Figure 3b; Supplementary Table 7). We note that the coverage for SuSiEx was well calibrated in most settings but dropped below 95% when the top 150 most associated variants were used as input, likely due to information loss from variant filtering. As using the full GWAS summary statistics as input was computationally tractable and yielded optimal results for SuSiEx, we do not consider this a limitation for SuSiEx and do not recommend any prefiltering of variants when using SuSiEx in practice.
SuSiEx is robust to varying cross-population genetic architectures
We further examined the calibration, power and resolution of SuSiEx by varying key parameters in the standard simulation setting. The cross-population genetic correlation (rg) can be less than one for many complex traits and diseases21. SuSiEx accounts for imperfect genetic correlation by allowing for varying genetic effects across populations. Using simulated data with rg of 0.4, 0.7, and 1.0, we confirmed that SuSiEx was robust to a range of rg values, with good calibration and similar power and resolution (Supplementary Figures 6–10; Supplementary Table 8). The local heritability (h2) and the number of causal variants (ncsl) per locus can differ across the genome for a given trait or disease1,22–24. We set the heritability per locus to 0.05%, 0.1%, 0.2%, 0.3%, 0.4% and 0.5%, and for a given per-locus heritability, varied ncsl from 1 to 5 with each genetic effect drawn from a normal distribution (Methods). As expected, SuSiEx performed better when h2 increased (Supplementary Figures 11–15; Supplementary Table 9) and ncsl decreased (Supplementary Figures 16–20; Supplementary Table 10), which corresponds to higher per-variant heritability and thus larger statistical power. Nonetheless, SuSiEx was always well calibrated at 95% coverage (Supplementary Figures 12 & 17), and was able to capture multiple causal variants in the same locus as ncsl increased.
We additionally assessed the robustness of SuSiEx under model misspecifications. SuSiEx assumes that causal variants are shared across populations. While a reasonable assumption for most genetic associations underlying human complex traits and diseases as supported by recent studies25–28, SuSiEx allows for different effect sizes (including null effects) of a causal variant across populations, and thus can accommodate violations of this modeling assumption. We empirically evaluated the robustness of SuSiEx by simulating variants that had non-zero effect sizes in one population but were null in other populations. We found that adding null data had little impact on fine-mapping results (Supplementary Figure 21 and Supplementary Table 11), confirming the robustness of SuSiEx to model misspecifications. Lastly, we note that in-sample LD is preferred in fine-mapping as it matches the correlation pattern between variants in the discovery GWAS sample. Unfortunately, in-sample LD is not always available, especially in large-scale GWAS comprising multiple cohorts. Using an external LD reference panel from a genetically close population can be a pragmatic solution despite its limitations6,29–31. Here, we evaluated the impact of LD mismatch on SuSiEx. Consistent with previous findings, analysis using in-sample LD produced excellent calibration and power, while using external LD led to coverage and power loss as the genetic distance between the external reference panel and the discovery sample increased (Supplementary Figure 4 and Supplementary Table 12).
SuSiEx increased the power and resolution of fine-mapping in biobank analysis
Encouraged by simulation results, we applied SuSiEx to data from the Pan-UKBB project and the Taiwan Biobank (TWB). The Pan-UKBB project is a multi-ancestry resource derived from the UK Biobank (UKBB)14 by analyzing six continental ancestry groups across 7,228 phenotypes. We included summary statistics of EUR and AFR (NEUR up to 419,807; NAFR up to 6,570, Supplementary Table 13) ancestries from Pan-UKBB. We additionally included TWB, one of the largest biomedical databases in East Asia (NEAS = 92,615) with close to 100,000 study samples15,32. We selected 25 quantitative traits shared between Pan-UKBB and TWB (Supplementary Table 13), and defined 13,420 genomic loci that reached genome-wide significance in at least one of the single-population association analysis or the meta-analysis across the three populations (Methods; Supplementary Table 14). We then performed single-population fine-mapping using SuSiE, and cross-population fine-mapping using SuSiEx, combining EUR, AFR and EAS data.
SuSiEx identified 14,400 credible sets across 9,826 loci, while single-population fine-mapping identified 12,784, 48, and 1,475 credible sets for the EUR, AFR and EAS populations, respectively (Supplementary Table 14). Aligning credible sets across analyses (Methods) led to 2,953 (20.5%) credible sets identified by SuSiEx that were not identified by single-population fine-mapping (Supplementary Table 14). Among the 14,400 credible sets, 1,413 (9.8%) credible sets reached genome-wide significance in the meta-analysis but not in any population-specific GWAS (as indexed by the maximum PIP variant), and thus would have been missed if fine-mapping was only conducted in single populations (Supplementary Table 14; Extended Data Figure 3b as an example). In addition to identifying and mapping more genetic associations through integrating data from multiple populations, SuSiEx also improved fine-mapping resolution. Relative to single-population fine-mapping in the EUR population, adding AFR and EAS data increased the average of the maximum PIP for a variant across all aligned credible sets from 0.44 to 0.47 (P = 3.7×10−6; two-sided t test), and reduced the average size of credible sets from 29.4 to 27.2 (P = 0.015; two-sided t test; Figure 4a & 4b; Supplementary Table 15; Extended Data Figure 3a as an example). Additionally, cross-population fine-mapping identified 2,485 putative causal variants with PIP >95% (Figure 4c; Supplementary Table 16), among which 575 were not discovered by any single-population fine-mapping. For example, SuSiEx identified a credible set containing a single variant associated with total bilirubin at PIP >99%, a missense variant of TRIM5 (rs11601507). This credible set failed to reach genome-wide significance in any population and was thus missed in single-population fine-mapping (Figure 5a and Extended Data Figure 4). Similarly, SuSiEx identified a two-variant credible set associated with albumin that failed to reach genome-wide significance in any population (Figure 5b; Extended Data Figure 5). The lead variant in the credible set is an intron variant of ALOX5AP with PIP 97.4%. This variant was fine-mapped to be an eQTL variant regulating the expression of ALOX5AP in whole blood (PIP >99%), artery aorta (PIP = 86.1%) and spleen (PIP = 77.9%) (Figure 5b; Extended Data Figure 5)33. In both examples, SuSiEx identified putative causal variants and resolved a genetic locus to its gene target that would have been missed if only single-population fine-mapping was performed.
Figure 4: Cross-population fine-mapping analysis in biobanks.
a, The distribution of the maximum PIP from all credible sets. b, The distribution of the size of all credible sets. c, The number of variants mapped to PIP >95% for all credible sets. d, The number of variants mapped to PIP >95% in single-credible-set loci. e, The maximum PIP from SuSiEx versus the maximum value of the maximum PIP in the three single-population fine-mapping using SuSiE. Only genomic loci with a single credible set aligned across analyses were included. f and g, The marginal per-allele effect size of the maximum PIP variant in EUR vs. EAS and EUR vs. AFR populations. We included variants in single-credible-set loci with PIP >95% estimated by SuSiEx and minor allele frequencies >5% in all populations. In a-b, red dots indicate the mean, the middle line in the box indicates the median, and the upper and lower bounds of the box indicate the 75th and 25th percentiles, respectively.
Figure 5. SuSiEx identified variants missed in single-population fine-mapping.
Each sub-figure consists of five panels, which are aligned vertically, with the x-axis representing the genomic position. The top three panels visualize GWAS association statistics of the European (Pan-UKBB Europan), African (Pan-UKBB African) and East Asian (Taiwan biobank) populations following the LocusZoom37 style. The second to bottom panel visualizes the fine-mapping results from SuSiEx, which integrated GWAS summary statistics from the three populations. The bottom panel shows gene annotations. For GWAS panels, the left y-axis represents the −log10(p-value) of each SNP. The gray horizontal dash line represents the genome-wide significance threshold (5×10−8). The purple rectangle for each locus represents the lead (most associated) variant. Variants are colored by descending LD with the lead variant (ordered red, orange, green, light blue, and dark blue dots). For fine-mapping panels, different colors were used to distinguish different credible sets. The diamond represents the maximum PIP variant of each credible set. a, Association with total bilirubin on chr11: 5,100,000–5,700,000. b, Association with albumin on chr13: 31,150,000–31,450,000.
Next, we restricted the comparison to loci that were mapped to a single credible set by both single- and cross-population fine-mapping such that our results were not affected by multiple causal variants in LD and the algorithm of credible set alignment. In these single-credible-set loci, SuSiEx continued to outperform single-population fine-mapping in power and resolution, identifying more credible sets with high confidence (best PIP >95%; Figure 4d), and improving the maximum PIP of a credible set in general relative to single-population fine-mapping (P = 6.4e-5; two-sided t test; Figure 4e). In particular, SuSiEx improved the maximum PIP of 30 credible sets from <80% to >95% (Figure 4e; orange and red dots), among which 9 were improved from <50% to >95% (Figure 4e; red dots). We note that the maximum PIP for one credible set dropped substantially, from 99% to 21%, in the cross-population fine-mapping (Figure 4e; blue dot). Further investigation of this locus revealed that the putative causal variant (12-67643414-T-A) is located in a low complexity genomic region, where the quality of variant calling and imputation may be negatively affected34. This variant is also represented in fewer than 50% of individuals in gnomAD v2.1.1 genomes35, and violates Hardy-Weinberg equilibrium.
Biobank analyses further confirmed that SuSiEx can retain population-specific causal variants (Extended Data Figure 3c as an example). Despite a dominating EUR sample size, SuSiEx recaptured 83% of the findings from single-population fine-mapping. A non-trivial proportion of credible sets from single-population fine-mapping that were not captured by SuSiEx may be driven by quality issues, defined as (i) the best PIP variant is in the low complexity region (LCR); (ii) the best PIP variant is in allelic imbalance or violates Hardy Weinberg equilibrium in gnomAD35; or (iii) the best PIP variant is multi-allelic or colocalizes with indels at the same genomic position, which might influence imputation quality. For example, 17.5% (29/166) of the putative causal variants with PIPs dropped by 10–20% in cross-population fine-mapping relative to single-population fine-mapping had quality issues, compared with 41.2% (7/17) of the variants with PIPs dropped by >40% (Extended Data Figure 6). These results suggest that, through the joint modeling of multiple populations and datasets, SuSiEx provides the additional benefit of identifying and removing likely low-quality findings from single-population analyses.
We used Ensembl Variant Effect Predictor (VEP)36 to annotate each variant into high, moderate or low functional impact, as well as modifiers. As the inferred PIPs increased, the proportion of variants with high impact clearly increased (Extended Data Figure 7), suggesting that confidently fine-mapped variants were enriched among mutations of functional importance. In total, we identified 2,286 high or moderate impact variants in 95% credible sets located in 1,630 genes. Among these variants, 425 had a PIP greater than 50% (Supplementary Table 17), and 275 had a PIP greater than 95% (Supplementary Table 18). There were 28 genes containing at least two high/moderate impact SNPs with PIP greater than 95%, while only 23 were detected in the three single-population fine-mapping analyses. In particular, IQGAP2 and PIEZO1 carried 3 missense variants associated with multiple blood biomarkers with PIPs >95%.
Lastly, we compared the per-allele effect sizes of high-confidence putative causal variants (PIP >95% in single- or cross-population fine-mapping) located in single-credible-set loci among EUR, AFR and EAS populations (Figure 4f & 4g). As no secondary association was found in these loci, we used marginal effect sizes in the comparison. Overall, the effect sizes were highly concordant between EUR and EAS populations (r = 0.82) but less consistent between EUR and AFR populations (r = 0.21), likely reflecting the larger uncertainties of the effect size estimates in AFR samples due to the limited GWAS sample size. We suggest the nature and cause of such inconsistency should be subject to a more thorough investigation with expanded non-European resources. At the current state, the imperfect genetic correlations across populations suggested the importance of accounting for variants with varying population-specific effect sizes in fine-mapping models.
SuSiEx identified additional putative causal candidates for schizophrenia
We applied SuSiEx to schizophrenia GWAS summary statistics of EUR (Ncase = 53,251, Ncontrol = 77,127) and EAS (Ncase = 14,004, Ncontrol = 16,757) ancestries from the Psychiatric Genomics Consortium (PGC), and fine-mapped the same 250 autosomal loci in the recent PGC publication4. SuSiEx successfully identified 215 credible sets out of 193 loci (not all loci converged to a credible set, as in all fine-mapping analyses), among which 11 had a SNP with PIP >95% (Figure 6a; Supplementary Tables 19 & 20). As expected, SuSiEx outperformed published PGC fine-mapping results, which applied a single-population fine-mapping method, FINEMAP38, to meta-analyzed GWAS summary statistics and sample size weighted LD4. Specifically, SuSiEx mapped 57% (33 vs. 21) more signals to a single variant with PIP >50% in single-credible-set loci (Figure 6). Most of the SuSiEx-improved credible sets had a marginally genome-wide significant signal (P-value between 5E-8 and 1E-15; Figure 6b & 6c). SuSiEx also produced credible sets for three loci that could not be resolved by FINEMAP in the original analysis. In these loci, FINEMAP inferred five independent credible sets, each containing a single variant that was not statistically significant in the GWAS, likely due to inaccurate reference panel39. Furthermore, SuSiEx substantially increased the resolution of fine-mapping by reducing the average size of credible sets from 87.1 to 60.3 (P = 0.015; paired two-sided t test), and increasing the average of maximum PIP across credible sets from 0.25 to 0.27 (P = 0.012; paired two-sided t test).
Figure 6: Fine-mapping of schizophrenia risk loci across European and East Asian populations.
a, The number of putative causal variants mapped to PIP >50% and >95% by FINEMAP and SuSiEx in single-credible-set loci. b, The maximum PIP for each credible set within single-credible-set loci, estimated by SuSiEx and FINEMAP. c, The difference of the maximum PIP, estimated by SuSiEx and FINEMAP (y-axis), within each single-credible-set locus, plotted against the -log10(p-value) of the most associated variant in the cross-population meta-analysis. In b and c, red dots represent credible sets with a maximum PIP >95% estimated by SuSiEx; orange dots represent credible sets with a maximum PIP >50% estimated by SuSiEx.
DISCUSSION
We presented SuSiEx, a cross-population fine-mapping method which links multiple population-specific sum of single effects (SuSiE) models by assuming the sharing of underlying causal variants. Through flexible and accurate modeling of varying population-specific causal effect sizes and LD patterns, SuSiEx improves the power and resolution of fine-mapping while producing well-calibrated false positive rates and retaining the ability to identify population-specific causal variants. We showed, via comprehensive simulation studies, that SuSiEx is highly computationally efficient, outperforms alternative cross-population fine-mapping methods in calibration, power and resolution, and is robust to model misspecifications. In particular, as the two state-of-the-art Bayesian cross-population fine-mapping methods, PAINTOR is sensitive to the predefined (yet unknown) number of causal variants, while MsCAVIAR is computationally intractable when the total number of input variants is greater than a few hundreds. Moreover, neither method has the capacity to analyze summary statistics from a comprehensive set of common variants in loci greater than 1MB. SuSiEx overcomes these limitations and offers effective and efficient cross-population fine-mapping that can be applied on biobank-scale datasets for the first time.
SuSiEx is designed to flexibly integrate genomic data from multiple populations, where effect sizes and/or LD patterns can be different. For two or more GWAS conducted in independent samples from the same population where effect sizes and LD patterns are highly concordant, we recommend a fixed-effect meta-analysis to combine these GWAS, which is often more statistically powerful than modeling these GWAS separately in SuSiEx without imposing any assumptions on the correlation of SNP effect sizes across samples. A recent study proposed SuSiE-inf40, which incorporates a term of infinitesimal effects in addition to a small number of single-variant causal effects, and showed that the new model can produce more calibrated fine-mapping results. While the calibration of SuSiEx was excellent in simulation studies, expanding the SuSiEx model to include this feature in the future may improve the fine-mapping of complex traits and diseases that have a highly polygenic architecture.
We note that throughout this work we tried to use in-sample LD reference panels for fine-mapping. Mismatch between the LD of the discovery sample and the reference panel may produce spurious credible sets and causal signals, especially in genomic loci that harbor strong association signals. This has been shown in prior work39 and our simulations studies, and is a limitation of all fine-mapping methods. We therefore recommend using in-sample LD for SuSiEx whenever possible, and applying aggressive filtering of low-quality variants and secondary credible sets in complex genomic loci if external LD reference panels have to be used.
There are several limitations of SuSiEx and the present study. First, we restricted our analyses to SNPs to avoid potential strand flippings and alignment errors when analyzing indels across biobanks. This may produce false positives if fine-mapped SNP(s) are proxies for causal indels or structural variations (SV). Second, we did not incorporate functional annotations into SuSiEx. Adding functional priors to the model may improve fine-mapping resolution when multiple variants in strong LD have similar statistical significance, and may aid prioritization of follow-up functional studies. That said, the biology underlying the observed variant-phenotype association may be complex, and the modeling of functional data may be error-prone and inflate false positive rates. Extending the Bayesian framework of SuSiEx to leverage functional or other omics data by introducing a proper prior to the model can be a promising future direction. Third, our cross-population fine-mapping in biobanks had an encouraging but modest improvement over the resolution of credible sets identified by European-only analyses, which was largely due to the limited discovery sample size of the African GWAS. However, we have shown that the largest improvements of SuSiEx come with the most diverse datasets, and thus expect that SuSiEx will become increasingly useful as the scale of genomic research in underrepresented populations continues to expand in global biobanks41 and disease-focused consortia. Lastly, it remains unclear how SuSiEx would perform in admixed samples, in which the local ancestry (and thus the causal variants and their effect sizes) may vary from individual to individual. Developing and evaluating statistical fine-mapping methods in populations with complex genetic ancestries is an important future direction.
In summary, SuSiEx provides robust, accurate and scalable fine-mapping that integrates GWAS summary statistics from diverse populations. Together with the ability to distinguish multiple causal variants within a genomic region, SuSiEx enables the analysis of large, complex genomic loci and aids the interpretation of fine-mapping results. Future work that combines SuSiEx with the rapidly expanding non-European genomic resources may facilitate the discovery of functionally-important disease-causing variants computationally and experimentally.
METHODS
Cross-population Sum of Single Effect (SuSiEx) model
Model description.
We extend the “SUm of SIngle Effects” (SuSiE) regression model to fine-mapping studies across multiple populations:
where for an population s (e.g., European, Asian or African), Ys is a vector of standardized phenotypes (zero mean and unit variance) from Ns individuals, Xs = [xs1, xs2, …, xsM] is an NS × M matrix of standardized genotypes (each column xSj is mean centered and has unit variance) in a genomic region that harbors at least one strong association signal, βs is a vector of SNP effect sizes, and ∊s is a vector of residuals with i.i.d. elements, each following a normal distribution with zero mean and variance . We assume that βs is the sum of L single-effect vectors bsl, l = 1,2, …, L, each has exactly one non-zero element (equals to bsl). The position of the non-zero element is determined by the binary vector γl, which follows a multinomial distribution. π = [π1, π2, …, πM]T is a vector that gives the prior probability of a SNP being causal, and is the prior variance on the effect size bsl of the causal SNP. We note that all populations share the same underlying causal SNPs (γl does not depend on s), but the effect sizes of a causal SNP across populations are allowed to be different (bsl depends on s).
Model fitting.
Assuming and are known, the SuSiEx model can be fitted using a simple extension of the iterative Bayesian stepwise selection (IBSS) algorithm. Specifically, with an initialization of the posterior mean effect size of bsl, denoted as (e.g., for all s and l), the fitting procedure involves iteratively updating bsl, given estimates of other effects bsl′, l′ ≠ l, until convergence:
- Compute residuals:
- Compute the posterior inclusion probabilities (PIPs):
where , , , - Update the posterior distribution for bsl:
where , . - Compute the posterior mean for bsl:
Where αl = [αl1, αl2, …, αlM]T, μl = [μsl1, μsl2, …, μslM]T, and ○ is element-wise multiplication.
Credible sets.
The PIPs αl can be used to compute a level-ρ credible set CS(αl; ρ), which has a probability no less than ρ of containing at least one causal SNP. Specifically, let (i1, i2, …, iM) denote the indices that sort αij in decreasing order, i.e., , and let . Then , where k0 = min{k: Sk ≥ ρ}. When L exceeds the number of detectable effects in the data, some αl become diffuse and the corresponding credible sets will be large, containing many uncorrelated SNPs. Such credible sets have no inferential value and can be discarded if they have purity below a threshold (e.g., 0.5), where purity is defined as the smallest absolute correlation among all pairs of variants within the credible set.
Using GWAS summary statistics.
Let denote the marginal least squares effect size estimate of SNP jin the ethnic group s, and denote the LD matrix for ethnic group s, which can be estimated using an LD reference panel. Note that . Therefore, IBSS can be turned into a summary statistics based algorithm.
The multi-step model fitting approach.
To determine the maximum number of single effects L, we designed a heuristic, multi-step model fitting approach. Specifically, we start with L = 5 and fit the SuSiEx model. If the model does not converge, we sequentially reduce L by 1 until the algorithm converges. If the model converges with L = 5 and returns 5 credible sets, suggesting that more than 5 credible sets may exist, we set L = 10 and rerun the model fitting algorithm. If the model does not converge with L = 10, we sequentially reduce L by 1 until the algorithm converges.
Simulations
Genomic data.
We simulated individual-level genotypes of EUR, EAS and AFR populations using HAPGEN242 with ancestry-matched 1000 Genomes Project (1KG) Phase III43 superpopulation samples as the reference panel. We grouped CEU, IBS, FIN, GBR and TSI into the EUR superpopulation, CDX, CHB, CHS, JPT and KHV into the EAS superpopulation, and ESN, MSL, LWK, GWD and YRI into the AFR superpopulation. To calculate the genetic map (cM) and recombination rate (cM/Mb) for each superpopulation, we downloaded the maps and rates for their constituent subpopulations (Data availability), linearly interpolated the genetic map and recombination rate at each position (Code availability), and averaged the genetic maps and recombination rates across the subpopulations in each superpopulation. We simulated 400,000 EUR samples, 200,000 EAS samples and 200,000 AFR samples, and confirmed that the allele frequencies and LD patterns of the simulated genotypes were highly similar to those of the 1KG reference panels. We randomly selected 100 1MB regions from chromosome 1 (Supplementary Table 1), and filtered for bi-allelic common (MAF >1%) SNPs in at least one of the three superpopulations.
Phenotypic data.
We randomly selected ncsl causal variants within each genomic locus. The allelic effect sizes of each selected causal variant for the EUR, EAS and AFR populations were generated under a multivariate normal distribution N(0, Σ3×3), where Σ3×3 was defined as, Σij = 1, if i = j, and Σij = rg, if i ≠ jwhere rg is the genetic correlation between populations. For each locus, we then generated the phenotype by adding a normally distributed noise term to the genetic component to produce the given per-locus heritability h2.
To assess SuSiEx in a wide range of settings, we generated simulation data with varying genetic correlations (rg), per-locus heritability (h2), and the number of causal variants (ncsl) per locus. We defined a standard simulation setting using ncsl = 1, rg = 0.7 and h2 = 0.1%. We then varied rg (rg = 0.4 and 1.0) to reflect different levels of cross-population genetic correlations, varied h2 (h2 = 0.05%, 0.2%, 0.3%, 0.4% and 0.5%) to reflect different per-locus heritability values, and varied ncsl (ncsl = 2, 3, 4, 5) with h2 = 0.5% to reflect the scenario of multiple causal variants in a genomic locus. To evaluate the robustness of SuSiEx to model misspecification, we simulated 200K EUR and 200K AFR samples with no causal variants, and included these null data in cross-population fine-mapping. For each parameter setting, we replicated the simulation five times for each locus (Supplementary Table 2), producing 500 simulation runs.
Association analysis and LD calculation.
We used the linear regression implemented in PLINK44 to generate GWAS summary statistics, and calculated in-sample LD for each genomic locus. To evaluate the impact of LD mismatch on fine-mapping results, we additionally calculated LD matrices using subpopulation samples within the EUR and AFR superpopulations.
Fine-mapping analysis with SuSiEx, SuSiE, PAINTOR, and MsCAVIAR.
We compared SuSiEx, SuSiE, PAINTOR and MsCAVIAR using the standard simulation setting. SuSiEx and SuSiE were performed and evaluated on additional settings beyond the standard simulations. As PAINTOR and MsCAVIAR are not computationally scalable to full GWAS summary statistics, we restricted the analysis to three filtered sets of variants: “p < 0.05”, “top 500” and “top 150”, corresponding to marginal p-values <0.05, the top 500 and the top 150 most associated variants from GWAS, respectively. PAINTOR provides two model fitting options, “MCMC” and “enumerate”. The “MCMC” mode automatically learns the number of causal variants in a locus while the “enumerate” mode requires pre-setting the maximum number of causal variants. We ran PAINTOR using “-mcmc”, “-enumerate=1”, “-enumerate=2” and “-enumerate=3”. All other parameters were set to default. We set the maximum runtime to 24 hours in our high-performance computing (HPC) system, the maximum memory to 8 GB, and the number of CPUs to one. For SuSiEx, we used the multi-step model fitting approach described above to determine the number of causal variants. Credible sets that did not contain any genome-wide significant variant (marginal P <5E-8) in any single-population GWAS nor cross-population meta-GWAS were filtered out. We ran MsCAVIAR with the default parameters and set the confidence level of credible sets as 0.95.
Biobank analysis
Cohorts.
GWAS summary statistics of 25 quantitative traits, available from both the UK Biobank (UKBB) and Taiwan Biobank (TWB), were used in our biobank fine-mapping analysis (Supplementary Table 13). European (EUR; NEUR up to 419,807) and African (AFR; NAFR up to 6,570) GWAS summary statistics were obtained from the Pan-ancestry genetic analysis of the UK Biobank (Pan-UKBB). East Asian GWAS summary statistics were obtained from the Taiwan Biobank (EAS; NEAS = 92,615).
Loci definition.
We used a 6-way LD clumping-based method to define the genomic loci, using 1KG data as the LD reference for clumping. CEU, GBR, TSI, FIN and IBS were combined as the reference for the EUR population; ESN, GWD, LWK, MSL and YRI were combined as the reference for the AFR population; CHB, CHS, CDX, JPT and KHV were combined as the reference for the EAS population. We extracted all variants with MAF >0.5%, and for each of the 25 traits, performed the LD clumping in the three populations using the corresponding reference panel and PLINK44. To include loci that reached genome-wide significance (P <5E-8) only in the meta-analysis, we further performed clumping for the meta-GWAS across the three populations, using the three reference panels, respectively. For each clumping, we set the p-value threshold of the leading variant as 5e-8 (--clump-p1) and the threshold of the tagging variant as 0.05 (--clump-p2), and set the LD threshold as 0.1 (--clump-r2) and the distance threshold as 250 kb (--clump-kb). We then took the union of the 6-way LD clumping results and extended the boundary of each merged region by 100 kb upstream and downstream. Finally, we merged adjacent loci if the LD (r2) between the leading variants was larger than 0.6 in any LD reference panel.
In-sample LD calculation.
We used the in-sample LD of the three populations in the fine-mapping analysis. We extracted all variants with MAF >0.5% from each population and calculated the LD using PLINK44. Multi-allelic variants and indels were excluded to avoid potential strand flipping and alignment errors.
Fine-mapping.
We applied SuSiEx to the 25 quantitative traits to integrate GWAS summary statistics derived from the three populations. We filtered out credible sets that did not contain any genome-wide significant variant (p <5E-8) in any population-specific GWAS or cross-population meta-GWAS.
Credible set alignment.
To compare the results between single-population and cross-population fine-mapping, we aligned the inferred credible sets across the four sets of analyses using a weighted Jaccard similarity index-based method7. Specifically, for a given pair of overlapping credible sets in a genomic locus, we computed the PIP-weighted Jaccard similarity index, defined as ∑i min(xi, yi)/ ∑i max(xi, yi), where xi and yi are PIP values (or zero if missing) for the same variant i from the two credible sets. Pairs of credible sets with a similarity index greater than 0.1 were aligned. If one credible set can be aligned with multiple credible sets, the set with the highest similarity was selected.
Cross-population fine-mapping in schizophrenia cohorts.
Schizophrenia GWAS summary statistics of European (EUR; Ncase = 53,251, Ncontrol = 77,127) and East Asian (EAS; Ncase = 14,004, Ncontrol = 16,757) ancestries were obtained from the recently published Psychiatric Genomics Consortium (PGC) schizophrenia analysis4. We fine-mapped the same 255 loci defined in the PGC publication. We calculated LD by applying LD-Store v1.139 to each cohort and locus, and then calculated an effective sample size weight LD matrix45 across cohorts for the EUR and EAS populations, respectively (Code availability; LDmergeFM). We applied SuSiEx to integrate EUR and EAS schizophrenia GWAS summary statistics to perform cross-population fine-mapping. Credible set level was set to 99%. Credible sets that did not contain any genome-wide significant variant (marginal P <5E-8) in single-population GWAS or cross-population meta-GWAS were filtered out.
Extended Data
Extended Data Figure 1: Schematic illustration of meta-based, post hoc and SuSiEx fine-mapping methods.
All panels were created following the LocusZoom style17. Variant positions are shown on the x axis. The gold diamond for each locus represents the lead (most associated) variant. The association strengths for other variants are colored by descending degrees of linkage disequilibrium (LD) with the lead variant (ordered red, orange, green, and blue dots). The purple bars represent the posterior inclusion probability (PIP) inferred by fine-mapping methods. The light gray boxes represent the credible set estimated by fine-mapping. a1-a5, Example of a strong causal signal shared across populations. b1-b5, Example of a weak causal signal shared across populations. c1-c5, Example of a population-specific causal signal.
Extended Data Figure 2: Comparison of SuSiEx, PAINTOR and MsCAVIAR in simulations.
a, The job completion summary for the three Bayesian fine-mapping methods using different parameters and input datasets. Red stands for jobs taking longer than 24 hours. Yellow stands for jobs returning unreasonable results, defined as the sum of PIP across variants in the genomic locus >5 or <0.1 (1 is expected). Green stands for jobs that were completed within 24 hours and returned reasonable results. b, Number of identified true causal SNPs with PIP >0.5 (x-axis) versus the coverage of credible sets (y-axis) for different input datasets and fine-mapping methods. Color represents the combination of discovery populations; size of the symbols represents the total discovery sample size, and the shape of the symbols represents different methods and parameters. Only simulation runs that were completed within 24 hours and returned reasonable results were included.
Extended Data Figure 3: Examples of the improvement of SuSiEx over single-population fine-mapping in biobank analysis.
Each of the three sub-figures consists of eight panels, which are aligned vertically, with the x-axis representing the genomic position. The top six panels visualize GWAS association statistics and single-population fine-mapping results of the European (Pan-UKBB Europan), African (Pan-UKBB African) and East Asian (Taiwan biobank) populations. For association statistics, the left y-axis represents the −log10(p-value) of each SNP. The color stands for the descending degrees of LD with the lead SNP (from red, orange to blue). The right y-axis represents the recombination rate in the centimorgan per Megabase. The solid line indicates the population-specific recombination maps obtained from the 1000 Genomes Project. Different colors were used to distinguish different credible sets in the fine-mapping results. The second to bottom panel visualizes results from SuSiEx. The bottom panel shows gene annotations if any. a, Association with albumin on chr8:9,170,000–9,190,000, an example of a strong causal signal shared across populations. b, Association with platelets count on chr12:104,900,000–105,050,000, an example of a weak causal signal shared across populations. c, Association with albumin on chr12:13,100,000–13,400,000, an example of population-specific causal signals.
Extended Data Figure 4: Association with total bilirubin on chr11: 5,100,000–5,700,000.
Panels are aligned vertically, with the x-axis representing the genomic position. The top six panels visualize GWAS association statistics and single-population fine-mapping results of the European (Pan-UKBB Europan), African (Pan-UKBB African) and East Asian (Taiwan biobank) populations following the LocusZoom37 style. The second to bottom panel visualizes the fine-mapping results from SuSiEx, which integrated GWAS summary statistics from the three populations. The bottom panel shows gene annotations. For GWAS panels, the left y-axis represents the −log10(p-value) of each SNP. The gray horizontal dash line represents the genome-wide significance threshold (5×10−8). The purple rectangle for each locus represents the lead (most associated) variant. Variants are colored by descending LD with the lead variant (ordered red, orange, green, light blue, and dark blue dots). For fine-mapping panels, different colors were used to distinguish different credible sets. The diamond represents the maximum PIP variant of each credible set. The left y-axis represents the PIP from fine-mapping and the right y-axis represents the recombination map obtained from the 1000 Genomes Project (for the SuSiEx panel, the average recombination rate across three populations was used).
Extended Data Figure 5: Association with albumin on chr13: 31,150,000–31,450,000.
Panels are aligned vertically, with the x-axis representing the genomic position. The top six panels visualize GWAS association statistics and single-population fine-mapping results of the European (Pan-UKBB Europan), African (Pan-UKBB African) and East Asian (Taiwan biobank) populations following the LocusZoom37 style. The second to bottom panel visualizes the fine-mapping results from SuSiEx, which integrated GWAS summary statistics from the three populations. The bottom panel shows gene annotations. For GWAS panels, the left y-axis represents the −log10(p-value) of each SNP. The gray horizontal dash line represents the genome-wide significance threshold (5×10−8). The purple rectangle for each locus represents the lead (most associated) variant. Variants are colored by descending LD with the lead variant (ordered red, orange, green, light blue, and dark blue dots). For fine-mapping panels, different colors were used to distinguish different credible sets. The diamond represents the maximum PIP variant of each credible set. The left y-axis represents the PIP from fine-mapping and the right y-axis represents the recombination map obtained from the 1000 Genomes Project (for the SuSiEx panel, the average recombination rate across three populations was used).
Extended Data Figure 6: Proportion of variants showing quality issues binned by the drop in PIP from single- to multi-population fine-mapping.
Quality issues were defined as (i) the best PIP variant is in the low complexity region; (ii) the best PIP variant is in allelic imbalance or violates Hardy Weinberg equilibrium in gnomAD33; or (iii) the best PIP variant is multi-allelic or colocalizes with indels at the same genomic position, which might influence imputation quality.
Extended Data Figure 7: Proportion of variants with high/moderate functional impact in cross-population biobank fine-mapping analyses.
The functional impact of each variant was annotated using VEP, with the definition and classification of functional impact obtained from https://useast.ensembl.org/info/genome/variation/prediction/predicted_data.html. The high impact category includes transcript ablation, splice acceptor variants, splice donor variants, etc; moderate impact includes missense variants, protein-altering variants, etc; low impact includes synonymous variants, splice region variants, etc; modifier impact includes introns and intergenic variants among others.
Supplementary Material
ACKNOWLEDGMENTS
UKBB European and African GWAS summary statistics were obtained from the PanUKBB Project. We thank the Schizophrenia Working Group of the Psychiatric Genomic Consortium (PGC) for providing the GWAS summary statistics and in-sample LD for the schizophrenia analysis. H.H. acknowledges supports from National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) K01DK114379 and R01DK129364, National Institute of Mental Health (NIMH) U01MH109539 and R01MH130675, Brain and Behavior Research Foundation Young Investigator Grant (28450), the Zhengxu and Ying He Foundation, and the Stanley Center for Psychiatric Research. T.G. is supported by National Institute on Aging (NIA) R00AG054573 and National Human Genome Research Institute (NHGRI) R56HG012354. Y.F.L. is supported by the National Health Research Institutes (NP-109-PP-09), and the Ministry of Science and Technology (109-2314-B-400-017) of Taiwan.
Footnotes
COMPETING INTERESTS
W.S. and C.S. are employees of Digital Health China Technologies Corp. Ltd.. M.J.D. is a founder of Maze Therapeutics. C.Y.C. is an employee of Biogen. H.H. received consultancy fees from Ono Pharmaceutical and honorarium from Xian Janssen Pharmaceutical.
CODE AVAILABILITY
The code used in this study is available from the following websites:
SuSiEx: https://github.com/getian107/SuSiEx;
PAINTOR: https://github.com/gkichaev/PAINTOR_V3.0;
MsCAVIAR: https://github.com/nlapier2/MsCAVIAR;
HAPGEN2: https://mathgen.stats.ox.ac.uk/genetics_software/hapgen/hapgen2.html;
PLINK1.9: https://www.cog-genomics.org/plink;
LDmergeFM: https://github.com/Pintaius/LDmergeFM
ETHICS
Collection of the UKBB data was approved by the Research Ethics Committee of the UKBB. UKBB individual-level data used in the present work were obtained under application no. 32568. Collection of the TWB data was approved by the Ethics and Governance Council (EGC) of TWB and the Department of Health and Welfare, Taiwan (Wei-Shu-I-Tzu no.1010267471). TWB obtained informed consent from all participants for research use of the collected data. Access to, and use of, TWB data in the present work was approved by the EGC of TWB (approval number: TWBR10907-05) and the Institutional Review Board of National Health Research Institutes, Taiwan (approval number: EC1090402-E).
DATA AVAILABILITY
Publicly available data are available from the following sites: 1KG Phase 3 reference panels: https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html; Genetic map for each subpopulation: https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130507_omni_recombination_rates; PanUKBB summary statistics: https://pan.ukbb.broadinstitute.org/downloads; TWB data used in this study contain protected health information and are thus under controlled access. Application to access such data can be made to the TWB (https://www.twbiobank.org.tw/new_web_en/); PGC schizophrenia GWAS: https://pgc.unc.edu/for-researchers/download-results
REFERENCES
- 1.Huang H. et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maller J. B. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 44, 1294–1301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Identification of multiple risk variants for ankylosing spondylitis through high-density genotyping of immune-related loci. Nat. Genet. 45, 730–738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium, T. S. W. G. of T. P. G., The Schizophrenia Working Group of the Psychiatric Genomics Consortium, Ripke S., Walters J. T. R & O’Donovan M. C Mapping genomic loci prioritises genes and implicates synaptic biology in schizophrenia. Preprint at 10.1101/2020.09.12.20192922. [DOI] [Google Scholar]
- 5.Mahajan A. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 54, 560–572 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanai M. et al. Meta-analysis fine-mapping is often miscalibrated at single-variant resolution. bioRxiv (2022) doi: 10.1101/2022.03.16.22272457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanai M. et al. Insights from complex trait fine-mapping across diverse populations. bioRxiv (2021) doi: 10.1101/2021.09.03.21262975. [DOI] [Google Scholar]
- 8.LaPierre N. et al. Identifying causal variants by fine mapping across multiple studies. PLoS Genet. 17, e1009733 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kichaev G. & Pasaniuc B. Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. Am. J. Hum. Genet. 97, 260–271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wyss A. B. et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat. Commun. 9, 2976 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gharahkhani P. et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 12, 1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson C. C. et al. Fine-mapping, trans-ancestral and genomic analyses identify causal variants, cells, genes and drug targets for type 1 diabetes. Nat. Genet. 53, 962–971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G., Sarkar A., Carbonetto P. & Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. R. Stat. Soc. Series B Stat. Methodol. 82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudlow C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y.-C. A. et al. Taiwan Biobank: A rich biomedical research database of the Taiwanese population. Cell Genomics 2, 100197 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell T. J. & Beauchamp J. J. Bayesian Variable Selection in Linear Regression. J. Am. Stat. Assoc. 83, 1023–1032 (1988). [Google Scholar]
- 17.George McCulloch. Approaches for Bayesian variable selection. Stat. Sin. [Google Scholar]
- 18.Kichaev G. et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 10, e1004722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lonjou C. et al. Linkage disequilibrium in human populations. Proc. Natl. Acad. Sci. U. S. A. 100, 6069–6074 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H. et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat. Commun. 12, 1098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning Z., Pawitan Y. & Shen X. High-definition likelihood inference of genetic correlations across human complex traits. Nat. Genet. 52, 859–864 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Abell N. S. et al. Multiple causal variants underlie genetic associations in humans. Science 375, 1247–1254 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu C.-C., Liu C.-C., Kanekiyo T., Xu H. & Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tehranchi A. et al. Fine-mapping cis-regulatory variants in diverse human populations. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marigorta U. M. & Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet. 9, e1003566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y. R. & Keating B. J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Medicine vol. 6 Preprint at 10.1186/s13073-014-0091-5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam M. et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 51, 1670–1678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulirsch J. C. et al. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nature Genetics vol. 51 683–693 Preprint at 10.1038/s41588-019-0362-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weissbrod O. et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 52, 1355–1363 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ulirsch J. C. & Kanai M. An annotated atlas of causal variants underlying complex traits and gene expression. Under review. [Google Scholar]
- 32.Chen C.-Y. et al. Analysis across Taiwan Biobank, Biobank Japan and UK Biobank identifies hundreds of novel loci for 36 quantitative traits. medRxiv (2021). [DOI] [PubMed] [Google Scholar]
- 33.Wang Q. S. et al. Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs. Nat. Commun. 12, 3394 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karczewski K. J. et al. Author Correction: The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 590, E53 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLaren W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruim R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benner C. et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics 32, 1493–1501 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benner C. et al. Prospects of Fine-Mapping Trait-Associated Genomic Regions by Using Summary Statistics from Genome-wide Association Studies. Am. J. Hum. Genet. 101, 539–551 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui R. et al. Improving fine-mapping by modeling infinitesimal effects. bioRxiv 2022.10.21.513123 (2022) doi:10.1101/2022.10.21.513123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou W. et al. Global Biobank Meta-analysis Initiative: Powering genetic discovery across human disease. Cell Genomics 2, 100192 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Z., Marchini J. & Donnelly P. HAPGEN2: simulation of multiple disease SNPs. Bioinformatics 27, 2304–2305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available data are available from the following sites: 1KG Phase 3 reference panels: https://mathgen.stats.ox.ac.uk/impute/1000GP_Phase3.html; Genetic map for each subpopulation: https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20130507_omni_recombination_rates; PanUKBB summary statistics: https://pan.ukbb.broadinstitute.org/downloads; TWB data used in this study contain protected health information and are thus under controlled access. Application to access such data can be made to the TWB (https://www.twbiobank.org.tw/new_web_en/); PGC schizophrenia GWAS: https://pgc.unc.edu/for-researchers/download-results