Abstract
Expression of superoxide dismutases (FeSOD and MnSOD) and catalases by laboratory strains of Pseudomonas aeruginosa is modulated by exogenous factors. Whether clinical isolates behave similarly and whether antioxidant enzyme expression influences P. aeruginosa virulence remain unclear. Fifty-seven P. aeruginosa blood culture isolates, plus seven pairs of blood and local-site isolates, were examined for FeSOD, MnSOD, and catalase production in vitro. Under iron-replete growth conditions FeSOD and catalase activities were maximized. MnSOD was not detected. FeSOD and catalase activity decreased under iron-limited growth conditions, whereas MnSOD activity appeared. SOD and catalase activity did not change with site of isolation or by patient. MnSOD could not be expressed by one isolate due to a missense mutation in sodA that produced a premature stop codon. Eleven percent of the isolates expressed a novel, rapidly migrating MnSOD that was associated with missense mutations in the normal stop codon of sodA. We conclude that clinical P. aeruginosa isolates vary little in FeSOD and catalase expression. Some strains produce a newly described MnSOD variant, whereas one is deficient in MnSOD production. The absence of MnSOD expression in a P. aeruginosa strain causing invasive human disease indicates that MnSOD is probably not essential for P. aeruginosa virulence.
Pseudomonas aeruginosa is an important cause of nosocomial pneumonia, burn wound infections, and urinary tract infections, some of which lead to bacteremia and septic shock (19). As a by-product of its aerobic metabolism, as well as through interaction with host phagocytic cells, P. aeruginosa is exposed to endogenous and exogenous fluxes of cytotoxic oxidants such as superoxide (O2⋅−) and hydrogen peroxide (H2O2) (7). Like many other bacteria, P. aeruginosa possess antioxidant enzymes such as the superoxide dismutases (SOD) and catalase, which catabolize O2⋅− and H2O2, respectively (4, 7, 12, 18). P. aeruginosa expresses two forms of SOD, one in which Mn is present at the active site (MnSOD) and another in which iron serves this function (FeSOD) (7, 12, 18). Recent studies have shown that a variety of exogenous factors modulate expression of the SODs and catalase in P. aeruginosa (4, 7, 9, 17). For example, in the presence of relatively high concentrations of extracellular iron, FeSOD is preferentially expressed, whereas under iron-limited conditions, expression of FeSOD decreases and MnSOD is produced (7).
With limited exceptions, information regarding the nature and regulation of antioxidant enzymes in P. aeruginosa has been derived from studies of laboratory strains. There have not been extensive studies of these enzymes in clinical isolates of this organism. This raises the possibility that levels of one or more or these enzymes could differ between isolates causing local infection and those leading to septicemia. In addition, regulation of these enzymes could differ or novel forms of one or more of the enzymes could exist among P. aeruginosa strains causing human infection.
Given these possibilities, we examined the expression of FeSOD, MnSOD, and catalase in a group of P. aeruginosa strains isolated from the urinary tracts, respiratory tracts, and blood of infected patients. Comparisons were made among isolates from different patients as well as between those isolated from a localized site of infection and from the blood of the same patient. In the process, we identified a potentially novel form of MnSOD in some of the clinical isolates.
(This work was presented in abstract form at the Pseudomonas '99: Biotechnology and Pathogenesis meeting of the American Society for Microbiology, Maui, Hawaii, 4 September 1999.)
MATERIALS AND METHODS
Organisms studied.
Clinical strains of P. aeruginosa were selected in a random fashion from banked isolates in the collection of the Clinical Microbiology Laboratory of the University of Iowa Hospitals and Clinics. These consisted of 57 bloodstream isolates from separate patients as well as pairs of blood and local-site (urine [4 patients], respiratory tract [2 patients], and tissue biopsy specimen [1 patient]) isolates from 7 patients. P. aeruginosa strain PAO1 (ATCC 15692; American Type Culture Collection, Manassas, Va.) was used as a reference strain.
Culture conditions.
Each isolate was passed on tryptic soy agar plates at 37°C. Colonies were then transferred from the plate to one of two broth media, tryptic soy broth (TSB) or succinate medium, for growth under iron-rich or iron-depleted conditions, respectively (6, 7). Organisms were inoculated into the broth and grown to mid-logarithmic phase, as determined by monitoring of the optical density at 600 nm, 3 to 4 h for TSB and overnight for succinate medium. Then organisms were pelleted by centrifugation (at 2,500 × g for 10 min). The supernatant was discarded, and the bacterial pellet was then suspended in 50 mM potassium phosphate (KPi) (pH 7.8) (1). Bacteria were lysed by a combination of sonication (30 s, three times) and two freeze-thaw cycles using either liquid N2 or a −80°C freezer (minimum cycle, 30 min). The sample was centrifuged at 13,000 × g for 10 min to remove cellular debris. The supernatant was then removed and frozen at −80°C until needed. These frozen samples were not subjected to more than three to four additional freeze-thaw cycles so as to maintain antioxidant enzyme activity.
Staining for antioxidant enzyme activity using native-protein polyacrylamide gel electrophoresis (PAGE).
Proteins contained in bacterial cell extracts were separated by electrophoresis on 8% (for catalase) and 12% (for SOD) nondenaturing polyacrylamide minigels. Prior to application of the sample, a current was applied to each gel in the presence of Tris (187.5 mM) and EDTA (1 mM) to remove oxidants (e.g., ammonium persulfate) which might alter enzyme activity. Bacterial extracts were then separated by electrophoresis in the presence of Tris (50 mM), glycine (300 mM), and EDTA (1.8 mM) at a constant current (40 mA) for 2 to 3 h. Gel lanes were loaded with 7.5 × 109 and 4.5 × 108 bacterium equivalents for SOD and catalase determinations, respectively. SOD activity was then determined as previously described (21). Briefly, following electrophoresis, gels were placed in the dark and soaked in a SOD staining solution containing riboflavin (0.028 mM), nitroblue tetrazolium (0.25 mM), EDTA (1 mM), and N,N,N′,N′-tetramethylethylenediamine (TEMED) (28 mM) in 50 mM KPi (1, 2). After 45 min of incubation, the staining solution was removed and replaced with 50 mM KPi. Gels were then exposed to light. Areas of SOD activity appeared as achromatic bands on a blue background (1). Each gel contained purified Escherichia coli MnSOD or FeSOD as a positive control. For analysis of catalase, following electrophoresis, gels were extensively rinsed with double-distilled water. After they were soaked for 10 min in 0.003% H2O2, a staining solution consisting of 2% potassium ferricyanide and 2% ferric chloride was added (21). Areas of catalase activity appeared as clear (negative-staining) bands on a blue-green background (21). The enzyme activities of the samples were quantitated and compared by gel spectrometry for both SOD and catalase using a Shimadzu (Kyoto, Japan) model CS-9000 densitometer system. Note that the E. coli MnSOD, FeSOD, and catalase standards were included to demonstrate the specificity of the activity determinations and are not present to confirm the electrophoretic mobility of a given activity.
DNA sequence analysis.
DNA sequences comprising the sodA gene, which encodes MnSOD, were determined as previously described (12) by PCR on double-stranded DNA derived from individual P. aeruginosa strains.
Removal and replacement of metal at the active site of MnSOD.
Metal was removed from, and restored to, the active site of MnSOD as previously described (3). Briefly, bacteria were grown in iron-limited succinate medium for 72 h at 37°C. Each culture (approximately 30 ml) was centrifuged at 2,000 × g for 30 min. Bacterial pellets were resuspended in 5 mM sodium phosphate at a concentration of approximately 6 × 1011 organisms/ml and then sonicated on ice. Samples were then centrifuged at 16,000 × g for 15 min. Supernatants were decanted, and the pellet was discarded. At this point samples could be stored at −70°C with no noticeable loss of enzymatic activity. Samples were then divided into aliquots, with one aliquot saved at each step of the following procedure. Samples were dialyzed overnight at 4°C against a solution of 5 mM Tris HCl (pH 3.8) containing 20 mM 8-hydroxyquinoline, 2.5 mM guanidine HCl, and 0.1 mM EDTA. This removed the metal from the active site of MnSOD. To restore activity, samples were dialyzed overnight at 4°C following two changes of 5 mM Tris HCl (pH 7.8) containing 0.1 mM MnCl2. This was followed by dialysis overnight with three solution exchanges against 5 mM Tris HCl (pH 7.8) containing 0.1 mM EDTA. The resulting samples were then assayed for SOD activity using the native gel electrophoresis system described above.
Ribotyping.
Ribotyping was performed on clinical P. aeruginosa isolates using the RiboPrinter Microbial Characterization System (Qualicon, Wilmington, Del.) according to the manufacturer's protocol and as described by Hollis et al. (13). In brief, colonies were streaked for growth on brain heart infusion agar plates. Cells were suspended in lysis buffer, lysing enzymes were added, and the tubes were placed in the RiboPrinter system. Within the Riboprinter, cells were lysed and the DNA was digested with PvuII restriction enzyme. The restriction fragments were then separated by electrophoresis and transferred to nylon membranes. A chemiluminescent-labeled nucleic acid probe containing the rRNA operon (rrnB) from E. coli was hybridized to the DNA on the membrane. The chemiluminescent patterns were electronically imaged and analyzed using the RiboPrinter Microbial Characterization System computer. Assignment to a particular ribogroup was based on differences in both band position and the signal intensity of each band. Isolates were considered to represent different ribotypes if the coefficient of similarity was <0.9 (13).
Guidelines.
All work was conducted in accordance with the guidelines for the conduct of clinical research of the United States Department of Health and Human Services and the institutional policies of the Department of Veterans Affairs and the University of Iowa.
RESULTS
Expression of antioxidant enzymes by isolates grown in iron-replete medium.
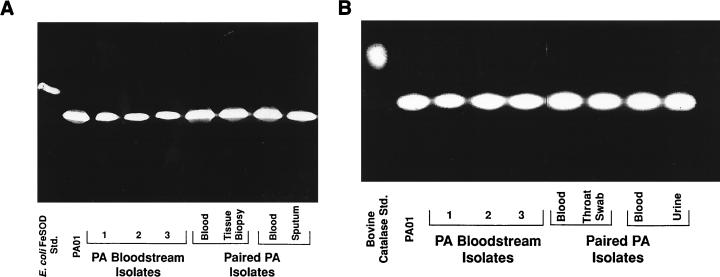
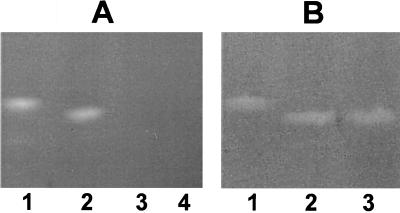
Consistent with previous work using laboratory strains (7), when patient-derived P. aeruginosa isolates were grown in TSB, which provides ample amounts of exogenous iron, and subjected to native gel electrophoresis, a single band of SOD activity (71 isolates examined) or catalase activity (KatA [41 isolates examined]) was observed by enzymatic analysis (Fig. 1A and B, respectively). This SOD activity reflected the presence of FeSOD rather than MnSOD, as demonstrated by both its electrophoretic mobility and its ability to be inhibited by H2O2 (data not shown) (7, 24). No difference in FeSOD or catalase activity was seen between isolates from different patients or from different sites of infection in the same or different patients (Fig. 1).
FIG. 1.
Native (nondenaturing) PAGE analysis demonstrating SOD and catalase activities and migration patterns for P. aeruginosa (PA) strains grown to log phase in TSB. Shown are SOD activities (A) and catalase activities (B) of the PAO1 laboratory strain, three separate bloodstream isolates, and paired local and bloodstream isolates taken from the same patient. Also shown on the far left of each gel is a positive control: E. coli FeSOD (A) or bovine catalase (B). Results are representative of those obtained with all of the clinical isolates examined (71 for SOD and 41 for catalase). Std., standard.
Expression of antioxidant enzymes in organisms grown under iron-limited conditions.
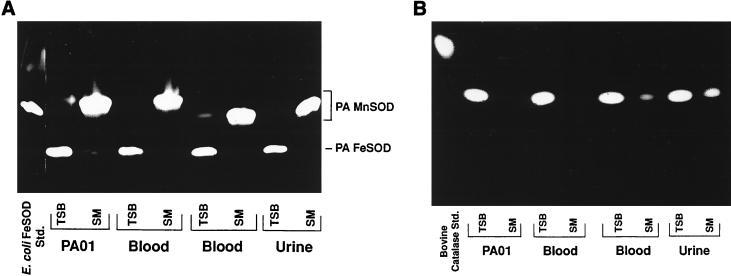
Consistent with previous results with other strains (7, 18), growth of each of the 71 clinical isolates in succinate-based (iron-limited) medium yielded a near-100% decrease in expression of FeSOD (Fig. 2A) and a 36 to 56% decrease in catalase activity (Fig. 2B). The loss of FeSOD activity was accompanied by the appearance of a new band of SOD activity which migrated into the gel more slowly and which paralleled a band known to represent MnSOD in strain PAO1 (Fig. 2A). These bands were resistant to inactivation by H2O2 (data not shown), confirming that they were due to MnSOD activity (7).
FIG. 2.
Native (nondenaturing) PAGE analysis of SOD and catalase activities and migration patterns for P. aeruginosa (PA) strains grown to log phase in TSB compared to those for strains grown in succinate medium (SM). Shown are SOD activities (A) and catalase activities (B) of the PAO1 laboratory strain, two distinct bloodstream isolates, and a urine isolate. Also shown on the far left of each gel is a positive control: E. coli FeSOD (A) and bovine catalase (B). Note that the MnSOD activity of the second blood isolate runs lower than those of the other three P. aeruginosa strains. Std., standard.
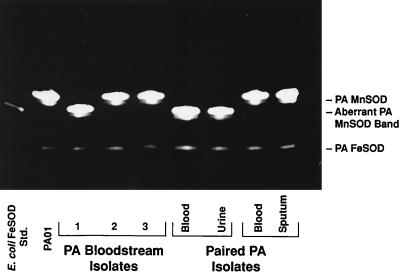
Surprisingly, five of the P. aeruginosa isolates from the 63 individual patients exhibited MnSOD activity which consistently migrated into the gel slightly faster than those of the other isolates (Fig. 3). The higher band was not observed, and this activity was again resistant to inactivation by H2O2 and NaCN (data not shown). Regardless of whether the lower or higher migrating band of MnSOD was expressed, the magnitudes of MnSOD activity among the isolates appeared similar. In addition to the strains expressing the aberrant form of MnSOD, one isolate failed to demonstrate any evidence of MnSOD activity in spite of the near-disappearance of FeSOD (data not shown).
FIG. 3.
Native (nondenaturing) PAGE analysis of SOD activities of P. aeruginosa (PA) strains grown to log phase in succinate medium. Shown are results with the PAO1 strain, three distinct blood culture isolates, and two groups of paired blood and local tissue isolates. Also shown on the far left is SOD activity resulting from the presence of E. coli FeSOD, which was included as a positive control. Std., standard.
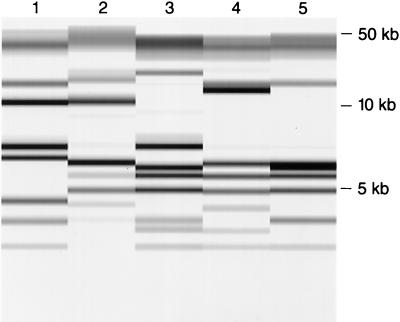
No epidemiologic link among the isolates expressing the atypical MnSOD could be ascertained. Ribotyping analysis revealed them to be different P. aeruginosa strains (Fig. 4).
FIG. 4.
Normalized PvuII riboprint patterns of the five isolates of P. aeruginosa demonstrating aberrant MnSOD expression. These isolates were ribotyped with the RiboPrinter Microbial Characterization System (Qualicon). Relative molecular sizes are given on the right.
Aberrant MnSOD enzyme expression does not reflect alternate metal insertion at the active site.
MnSOD is a homodimer. In E. coli (3), P. aeruginosa (7), and Staphylococcus aureus (5), Fe can be inserted in place of Mn into the active site of one or both of the enzyme monomers. Substitution of Fe is associated with a loss of enzyme activity of that component of the holoenzyme, i.e., homodimers with one Fe atom and one Mn atom have 50% SOD activity, and if both monomers have Fe in place of Mn, the enzyme has no activity (3, 5, 7). Relevant to the aberrant MnSOD we observed in some of the P. aeruginosa strains, it had previously been shown that in E. coli when one of the two Mn atoms are replaced with Fe, the enzyme migrates further into native gels than the fully Mn loaded enzyme (3). This suggested the possibility that the aberrant MnSOD we observed in the P. aeruginosa strains was due to the unique expression of an Fe-Mn hybrid enzyme. In order to address this possibility, we employed a previously described technique in which metals could be removed from and then selectively reinserted into the active site of the MnSOD protein (3, 5, 7). We reasoned that if the aberrant enzyme reflected an Fe-Mn hybrid, if we removed the metals from the active site and then reloaded the protein under conditions in which only Mn was present, the migration pattern of the enzyme would return to that of the wild type. As shown in Fig. 5A, we were able to remove the metals and then restore Mn to the active site of the enzyme with a return of activity. However, this had no effect on the migration pattern of the aberrant form relative to that of the wild type (Fig. 5B). Thus, the aberrant form of MnSOD does not appear to be due to formation of an Fe-Mn hybrid.
FIG. 5.
(A) Native (nondenaturing) PAGE analysis of SOD activities of succinate-grown strain PAO1 and a clinical isolate that expresses the atypical MnSOD activity before (lanes 1 and 2, respectively) and after (lanes 3 and 4, respectively) the samples were subjected to a procedure that removes the metal from the active site of MnSOD. The procedure led to the expected loss of SOD activity in both strains. (B) Native (nondenaturing) PAGE analysis of SOD activity after Mn reloading. Lanes 1 and 3 , SOD activities of the samples from panel A, lanes 3 and 4, which were incubated with Mn in such a way as to reload the active site of the protein with Mn. Activity was restored with restitution of the migration pattern that existed prior to removal of metal and reloading. The migration of the atypical form of MnSOD of the clinical isolate was identical to that exhibited prior to the procedure (lane 2).
Analysis of the sodA genes of bacteria with altered MnSOD electrophoretic profiles.
The altered migration pattern and/or absence of expression of MnSOD in some of the P. aeruginosa isolates suggested that there could be mutations within the sodA locus in these strains. To test this hypothesis, the sodA gene and a region immediately downstream were amplified by PCR and sequenced on both strands. Four of the five strains expressing the aberrant MnSOD form and the one strain unable to express MnSOD were available for analysis. Surprisingly, all strains possessed substitution mutations, the most significant of which were T-to-G and G-to-A missense mutations within the stop codon (Fig. 6). This would allow transcription to proceed and eventually terminate with another stop codon (TAG) 18 codons downstream. Thus, all of the strains that express the aberrant form of MnSOD would predictably have MnSOD isozymes with molecular sizes ∼5 kDa larger than the wild-type MnSOD dimer (∼45 kDa). The additional amino acids could alter the secondary structure of the MnSOD monomer, possibly contributing to the different electrophoretic migration pattern observed when it assembles as the normal dimeric form. In the case of the non-MnSOD-expressing strain, a missense mutation was also detected at position 85; this mutation results in the generation of a premature stop codon (Fig. 6), presumably explaining the inability of the strain to produce a functional MnSOD.
FIG. 6.
DNA and amino acid sequences of the wild-type P. aeruginosa PAO1 sodA gene (www.pseudomonas.com) (12), of four clinical strains possessing MnSOD isozymes with altered electrophoretic profiles (sequences 1, 2, 4, and 5), and of one clinical strain that was not able to produce MnSOD at all (sequence 3). Base substitutions are depicted in boldface, with the base change and strain indicated above. In sequence 3 the base substitution at position 85 results in the conversion of a tryptophan to a stop codon. In each of the five mutant strains, there is a base substitution in the sequence coding for the normal stop codon that converts it to an amino acid. The sequence extended beyond the normal sodA stop codon (star) is shown so as to indicate the next stop codon in the DNA sequence of each strain.
DISCUSSION
The present work confirms that most clinical isolates express similar amounts of FeSOD, MnSOD, and catalase relative to a laboratory control strain (PAO1) grown under the same in vitro conditions. As expected from earlier data (7, 10, 11, 18), FeSOD and catalase enzymatic activities were maximized under iron-replete growth conditions. Activities for both enzymes decreased when strains were grown in iron-limited succinate medium, whereas MnSOD activity now appeared. These effects appear to be the result of the interaction of iron with regulatory elements which modulate expression of the genes encoding MnSOD (sodA), FeSOD (sodB), and catalase (katA and katB), as well as the lack of iron available to insert in the active site of FeSOD and protoporphyrin IX of the catalase heme group (8, 10). Preliminary immunoblot analysis of several clinical P. aeruginosa isolates grown in TSB versus succinate suggests a decrease in immunoreactive catalase activity in succinate-grown organisms.
No difference in the magnitude of SOD or catalase activity was seen when clinical isolates were grouped according to site of isolation, nor were there differences between patients. Thus, we conclude that there are no significant differences in antioxidant enzyme expression among P. aeruginosa clinical isolates grown in vitro. This is not unexpected, since neutrophils, which serve as a major mechanism of host defense against P. aeruginosa, do not appear to rely on oxygen-dependent killing as their principal means for eradicating P. aeruginosa (14–16, 20). Neutrophils from individuals with chronic granulomatous disease of childhood, which are unable to generate O2⋅− and H2O2, kill P. aeruginosa as effectively as those from normal individuals (14). Such patients do not suffer from recurrent infections with P. aeruginosa (23).
An interesting and unexpected result of our study is that 11% of the clinical isolates we examined express a form of MnSOD which migrates more rapidly in a native gel system than the MnSOD of the other clinical isolates and the standard laboratory strain (PAO1) that we examined. This alternate form of MnSOD does not decrease the virulence of the organism, as each P. aeruginosa strain expressing this MnSOD form was isolated from a clinically significant site of human infection, in most cases blood. We also found a blood culture isolate that was unable to produce MnSOD. A bronchopulmonary isolate lacking the ability to produce MnSOD has been reported previously (18). The fact that our strain was isolated from blood provides strong evidence that the ability to produce MnSOD is not required for virulence (22). This is consistent with previous work indicating that FeSOD is more important than MnSOD in protecting P. aeruginosa from oxidative stress (10).
In the four strains expressing the atypical form of MnSOD that were available for analysis, we identified missense mutations within the stop codon of the sodA locus. This would lead to the production of a protein approximately 5 kDa greater than wild-type MnSOD, since transcription would continue until the next stop codon was encountered 18 codons downstream. How production of a larger-than-normal MnSOD monomer leads to the formation of an active dimeric protein that migrates faster on a nondenaturing “activity” gel is unclear. The exact mechanism responsible for the migration pattern of the alternative MnSOD form requires additional analysis but likely relates to conformational or charge density changes of the dimer that in turn lead to its altered migration under nondenaturing gel conditions.
In summary, clinical isolates of P. aeruginosa differ little from standard laboratory strains previously studied in their production of FeSOD, MnSOD, and catalase as well as in the regulation of their expression. The exception to this finding is the identification in a number of the clinical strains of an MnSOD variant that is associated with point mutations in the normal stop codon of the sodA locus. The similarity in SOD and catalase activity levels among the various isolates and the ability to cause invasive disease in the absence of the genetic capacity to produce MnSOD suggest that antioxidant enzyme differences do not contribute to the relative virulence of P. aeruginosa strains.
ACKNOWLEDGMENTS
This work was supported by grants from the Department of Veterans Affairs (to B.E.B. and M.L.M.) and Public Health Service awards RO1AI34954 (to B.E.B.), T32AI07343 to (R.A.M.), and RO1AI40541 (to D.J.H.) and was performed in part during the tenure of B.E.B. as an Established Investigator of the American Heart Association.
We thank Richard Hollis for performing the ribotype analyses and Michael Hayek for technical assistance.
REFERENCES
- 1.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to polyacrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 2.Benov L T, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 3.Beyer W F, Jr, Fridovich I. In vivo competition between iron and manganese for occupancy of the active site region of the manganese-superoxide dismutase of Escherichia coli. J Biol Chem. 1991;266:303–308. [PubMed] [Google Scholar]
- 4.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox C D. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassett D J, Charniga L, Bean K, Ohman D E, Cohen M S. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassett D J, Howell M L, Ochsner U A, Vasil M L, Johnson Z, Dean G E. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol. 1997;179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassett D J, Ma J F, Elkins J G, McDermott T R, Ochsner U A, West S E H, Huang C T, Fredericks J, Burnett S, Stewart P S, McFeters G, Passador L, Iglewski B H. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol Microbiol. 1999;34:1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x. [DOI] [PubMed] [Google Scholar]
- 10.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassett D J, Sokol P A, Howell M L, Ma J F, Schweizer H T, Ochsner U, Vasil M L. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J Bacteriol. 1996;178:3996–4003. doi: 10.1128/jb.178.14.3996-4003.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hassett D J, Woodruff W A, Wozniak D J, Vasil M L, Cohen M S, Ohman D E. Cloning and characterization of the Pseudomonas aeruginosa sodA and sodB genes encoding manganese- and iron-cofactored superoxide dismutase: demonstration of increased manganese superoxide dismutase activity in alginate-producing bacteria. J Bacteriol. 1993;175:7658–7665. doi: 10.1128/jb.175.23.7658-7665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollis R J, Bruce J L, Fritschel S J, Pfaller M A. Comparative evaluation of an automated ribotyping instrument versus pulse-field gel electrophoresis for epidemiological investigation of clinical isolates of bacteria. Diagn Microbiol Infect Dis. 1999;34:1–6. doi: 10.1016/s0732-8893(99)00033-4. [DOI] [PubMed] [Google Scholar]
- 14.Holmes-Gray B. Metabolic stimulation and bactericidal function of polymorphonuclear leukocytes. J Reticuloendothel Soc. 1977;22:87–88. [Google Scholar]
- 15.Hovde C J, Gray B H. Characterization of a protein from normal human polymorphonuclear leukocytes with bactericidal activity against Pseudomonas aeruginosa. Infect Immun. 1986;54:142–148. doi: 10.1128/iai.54.1.142-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizgerd J P, Brain J D. Reactive oxygen species in the killing of Pseudomonas aeruginosa by human leukocytes. Curr Microbiol. 1995;31:124–128. doi: 10.1007/BF00294288. [DOI] [PubMed] [Google Scholar]
- 17.Ochsner U A, Vasil M L, Alsabbagh E, Parvatiyar K, Hassett D J. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polack B, Dacheux D, Delic-Attree I, Toussaint B, Vignais P M. Role of manganese superoxide dismutase in a mucoid isolate of Pseudomonas aeruginosa: adaptation to oxidative stress. Infect Immun. 1996;64:2216–2219. doi: 10.1128/iai.64.6.2216-2219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollack M. Pseudomonas aeruginosa. In: Mandell G L, Bennett J E, Dolin R, editors. Principles and practice of infectious diseases. 5th ed. New York, N.Y: Churchill Livingstone; 2000. pp. 2310–2335. [Google Scholar]
- 20.Siefferman C M, Regelmann W E, Gray B H. Pseudomonas aeruginosa variants isolated from patients with cystic fibrosis are killed by a bactericidal protein from human polymorphonuclear leukocytes. Infect Immun. 1991;59:2152–2157. doi: 10.1128/iai.59.6.2152-2157.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Elwell J H, Oberley L W. A simultaneous visualization of the antioxidant enzymes glutathione peroxidase and catalase on polyacrylamide gels. Free Radic Res Commun. 1988;5:67–75. doi: 10.3109/10715768809066913. [DOI] [PubMed] [Google Scholar]
- 22.Toussaint B, Delic-Attree I, Vignais P M. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun. 1993;196:416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- 23.Winkelstein J A, Marino M C, Johnston R B, Jr, Boyle J, Curnutte J, Gallin J I, Malech H L, Holland S M, Ochs H, Quie P, Buckley R H, Foster C B, Chanock S J, Dickler H. Chronic granulomatous disease—report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Yamakura F, Suzuki K. Inactivation of Pseudomonas iron-superoxide dismutase by hydrogen peroxide. Biochim Biophys Acta. 1986;874:23–29. doi: 10.1016/0167-4838(86)90097-x. [DOI] [PubMed] [Google Scholar]