Abstract
Measles virus (MeV), which is usually non-neurotropic, sometimes persists in the brain and causes subacute sclerosing panencephalitis (SSPE) several years after acute infection, serving as a model for persistent viral infections. The persisting MeVs have hyperfusogenic mutant fusion (F) proteins that likely enable cell-cell fusion at synapses and “en bloc transmission” between neurons. We here show that during persistence, F protein fusogenicity is generally enhanced by cumulative mutations, yet mutations paradoxically reducing the fusogenicity may be selected alongside the wild-type (non-neurotropic) MeV genome. A mutant F protein having SSPE-derived substitutions exhibits lower fusogenicity than the hyperfusogenic F protein containing some of those substitutions, but by the wild-type F protein coexpression, the fusogenicity of the former F protein is enhanced, while that of the latter is nearly abolished. These findings advance the understanding of the long-term process of MeV neuropathogenicity and provide critical insight into the genotype-phenotype relationships of en bloc transmitted viruses.
The concept of “en bloc transmission” advances understanding of the evolutionary process of measles virus neuropathogenicity.
INTRODUCTION
RNA virus populations consist of large numbers of variant genomes because of the high error rates of viral RNA-dependent RNA polymerases (1, 2). Individual mutants in the viral populations sometimes do not act independently but rather functionally interact with each other, which may determine the biological behavior of virus populations (1, 2). Positive and negative interactions are observed among variants in virus populations (3). These internal interactions presuppose the cotransmission of multiple viral genomes to the same cell, which could be mediated by “collective infectious units” that simultaneously transmit groups of viral genomes (“en bloc transmission”) (3–7). En bloc transmission occurs via polyploidy virions, virion aggregation, nonviral-specific structures, and direct cell-to-cell transmission (3).
Measles is an acute viral respiratory illness with high fever, cough, coryza, conjunctivitis, and a pathognomonic enanthema (Koplik spots), followed by a maculopapular rash (8, 9). Measles virus (MeV), the causative agent of measles, is an enveloped RNA virus that belongs to the genus Morbillivirus, the subfamily Orthoparamyxovirinae, and the family Paramyxoviridae. MeV may persist in the brain, causing a fatal progressive neurological disorder, subacute sclerosing panencephalitis (SSPE), several years after acute infection (9–11). The incidence of SSPE is estimated to be approximately 4 to 11 cases per 100,000 cases of measles.
MeV has two kinds of glycoproteins on its envelope, the hemagglutinin (H) and the fusion (F) proteins (10). The H protein binds to receptors on the human cells and subsequently triggers the conformational changes of the trimeric F protein from the prefusion form to the postfusion form, leading to virus-to-cell or cell-to-cell membrane fusion. While the wild-type (WT) F protein cannot induce membrane fusion in neurons lacking MeV receptors [signaling lymphocytic activation molecule family member 1 (SLAMF1) and nectin-4], hyperfusogenic mutant F proteins from SSPE patient–derived MeV strains can do so by using cell adhesion molecule 1 (CADM1) and CADM2 as “cis-acting receptors” (host factors triggering fusion in cis) (12–14). The H protein interacts with CADM1 and CADM2 in cis on the same membrane, triggering the conformational changes of the hyperfusogenic mutant F proteins but not of the WT F protein.
In the brain, multiple MeV genomes are likely transmitted transsynaptically between neurons through cell-cell fusion (15, 16), and thus, the ancestral WT and progeny mutant genomes could be cotransmitted to the same cells, where both the WT F and mutant F proteins are then coexpressed (fig. S1). How in neurons SSPE-derived mutant F proteins interact with the WT F protein remains to be investigated.
The F protein T461I substitution found in multiple SSPE isolates [5 of the 18 SSPE-derived F proteins in the National Center for Biotechnology Information (NCBI) protein database have this substitution] (fig. S2) has been extensively studied (17–22). It confers enhanced fusogenicity (the ability to fuse two distinct lipid bilayers such as cell membranes) on the F protein. Unlike the WT MeV, the recombinant MeV having the F(T461I) protein efficiently spreads in neurons and exhibits neurovirulence in mice and suckling hamsters (18, 19, 21). However, the F proteins of SSPE MeV isolates having the T461I substitution also accumulate other mutations, which have not been functionally examined. All SSPE isolates studied have many mutations in the F protein (11).
In this study, we show that cumulative mutations found in MeV isolates from patients with SSPE could up- or down-regulate the F protein fusogenicity. By using the plasmid-mediated fusion assay and recombinant MeVs, we also demonstrate positive and negative interactions between different F proteins, which could determine the adaptability of MeV populations to persistence in the brain. These results not only provide critical insight into the evolutionary pathway to MeV neuropathogenicity but also indicate that for en bloc transmitted viruses, one cannot fully understand the effect of a mutation without considering interactions between different viral genomes.
RESULTS
Accumulations of multiple hyperfusogenic mutations in the MeV F protein dynamically change its fusogenicity
In the NCBI protein database, we found five SSPE-derived MeV strains that encode F proteins with the T461I substitution (a substitution that confers enhanced fusogenicity on the F protein) (fig. S2). Amino acid alignments revealed that these SSPE-derived F proteins have multiple additional substitutions at different locations (marked in red) as well as T461I (marked in purple) (fig. S3). Among them, our literature search of relevant publications (17–19, 22–25) identified several substitutions at the positions where amino acid substitutions can cause enhanced fusogenicity (Fig. 1A). The F proteins of the patient A (26–28), patient B (26–28), and OSA-3/Bs/B (29) strains have potentially hyperfusogenic mutations N462S/N465S (18), G264E (22), and F375S/N465K (22), respectively, in addition to T461I (Fig. 1A). The phenotypes of F proteins having those mutations (N462S/N465S, G264E, or F375S/N465K) were evaluated by the fusion assay. Briefly, expression plasmids respectively encoding the WT H protein, WT F or one of the mutant F proteins, enhanced green fluorescence protein (EGFP), and SLAMF1 were transfected into 293FT cells, and the cells were observed 24 hours after transfection under a fluorescence microscope. The results confirmed that those mutations in the F gene enhance membrane fusion triggered by SLAMF1 (fig. S4A). The results of the quantitative fusion assay [the dual-split protein (DSP) assay] (13, 30–32) were also consistent with the observations with the fusion assay (fig. S4B).
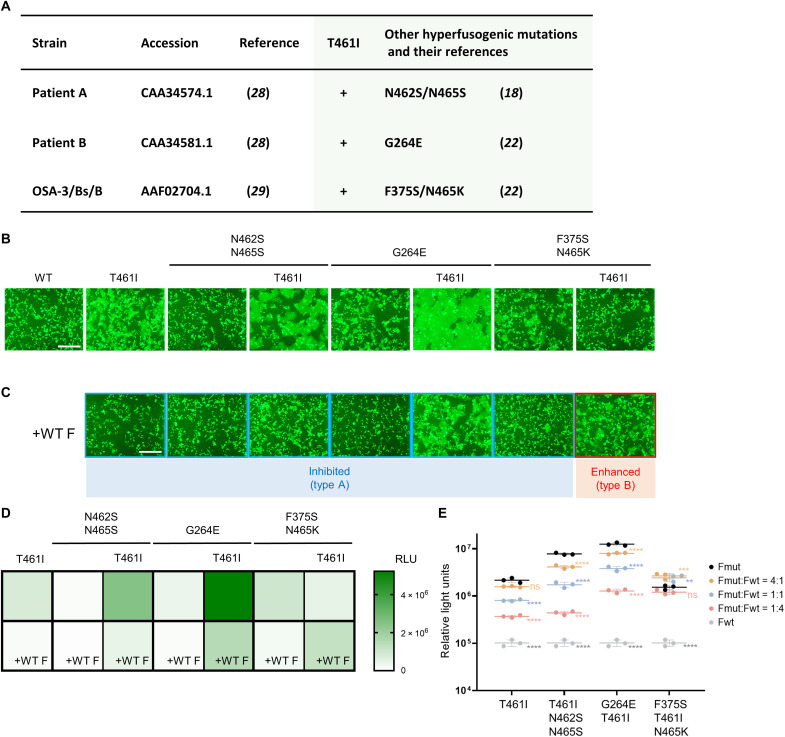
Fig. 1. Influence on the F fusogenicity of cumulative mutations and the WT F protein coexpression.
(A) The MeV isolates from patients with SSPE who have the T461I substitution and other hyperfusogenic mutations in the F protein. Their strain names, accession numbers (GenBank) and references, other hyperfusogenic mutations, and references for the mutations are shown. (B and C) The WT H protein, MeV F [WT F, F(T461I), F(N462S/N465S), F(T461I/N462S/N465S), F(G264E), F(G264E/T461I), F(F375S/N465K), or F(F375S/T461I/N465K)], CADM1, and EGFP were expressed in 293FT cells without (B) or with (C) the WT F protein. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bars, 500 μm. For type A and type B, refer to the main text and fig. S8. (D and E) The WT H protein, MeV F [F(T461I), F(N462S/N465S), F(T461I/N462S/N465S), F(G264E), F(G264E/T461I), F(F375S/N465K), or F(F375S/T461I/N465K)], and CADM1 were expressed in mixed 293FT/DSP1 and 293FT/DSP2 cells without or with the WT F protein. The ratio of each mutant F protein to the WT F protein was 1:1 (D) or 4:1, 1:1, or 1:4 (E). The Renilla luciferase activity in the transfected cells was analyzed 24 hours after transfection. RLU, relative light units. Each data point represents one biological replicate (N = 3). Error bars indicate SDs. Significance of the difference in the luciferase activity (as compared with that obtained by the expression of the corresponding mutant F protein only) was analyzed by two-way ANOVA: ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
We then investigated the CADM1-dependent fusogenicities of the F proteins having these hyperfusogenic mutations with or without T461I by the fusion assay to determine how cumulative mutations affect the F protein function. Expression plasmids respectively encoding the WT H protein, WT F or one of the mutant F proteins, EGFP, and CADM1 were transfected into 293FT cells, and the cells were observed 24 hours after transfection under a fluorescence microscope. Syncytia were detected with the F(T461I), F(G264E), or F(F375S/N465K) protein (Fig. 1B) but not evident with the WT F or F(N462S/N465S) protein. Combinatorial substitutions with T461I resulted in different functional impacts on their fusogenicities. The F(T461I/N462S/N465S) and F(G264E/T461I) proteins exhibited enhanced fusogenicities as compared with the F(T461I) protein, while the F(F375S/T461I/N465K) protein exhibited lower fusogenicity than the F(T461I) or F(F375S/N465K) protein. The surface expression levels of mutant F proteins used here were investigated by the surface biotinylation assay (fig. S5), and there was no correlation between the surface expression levels and the F fusogenicities.
These results indicate that accumulation of hyperfusogenic mutations in the F protein could up- or down-regulate the fusogenicity. However, because neuropathogenic MeVs presumably spread between neurons through cell-cell membrane fusion, it is difficult to understand why a less neurotropic MeV with lower fusogenicity could be selected in the brain. To explain this discrepancy, we turned to the concept of en bloc transmission (collective infectious units) and investigated possible functional interactions between different F proteins encoded in different viral genomes in neurons.
Coexpression of the WT protein influences the fusogenicities of mutant MeV F proteins
Because MeVs are transmitted en bloc between neurons, different MeV genomes encoding the WT and mutant F proteins are likely to coexist in neurons (fig. S1). We therefore evaluated the influence of the WT F protein coexpression on the fusogenicities of mutant MeV F proteins by the fusion assay. When the WT F protein was coexpressed, the levels of syncytium formation induced by all mutant F proteins examined, except for the F(F375S/T461I/N465K) protein, were inhibited as compared with those induced without the WT F protein (Fig. 1C). In contrast, the F(F375S/T461I/N465K) protein exhibited enhanced fusogenicity when coexpressed with the WT F protein (Fig. 1C).
The levels of membrane fusion were also evaluated quantitatively by the DSP assay (Fig. 1D) (13, 30–32). The F(T461I/N462S/N465S) and F(G264E/T461I) proteins exhibited enhanced fusogenicities as compared with the F(T461I) protein, yet their fusogenicities were decreased when the WT F protein was coexpressed. Conversely, the F(F375S/T461I/N465K) protein exhibited lower fusogenicity than the F(T461I) or F(F375S/N465K) protein, but its fusogenicity was increased when the WT F protein was coexpressed. These results were consistent with the observations with the fusion assay (Fig. 1, B and C). Thus, the data show that the interaction between the mutant and WT F proteins could make the effect of the mutation(s) on F protein fusogenicity greatly altered.
Next, the ratios of the plasmid encoding the influenza virus hemagglutinin (HA)–tagged WT F protein to those encoding the respective FLAG-tagged mutant F proteins were changed in the DSP assay (Fig. 1E). The amounts of plasmids used for the transfection correlated well with the expression levels of the corresponding F proteins (fig. S6). As the amount of the plasmid encoding the WT F protein was relatively increased, the fusogenicities of the F(T461I), F(T461I/N462S/N465S), and F(G264E/T461I) proteins became gradually decreased, but that of the F(F375S/T461I/N465K) protein was first increased (when the mutant was in excess) and then decreased. When the WT F protein was expressed in any proportion tested, the fusogenicities of the F(T461I) protein were lower than those of the F(F375S/T461I/N465K) [P < 0.05, two-way analysis of variance (ANOVA)]. Furthermore, when the ratio of the plasmid encoding each mutant F protein to that encoding the WT F protein was 1:4, the fusogenicity of the F(F375S/T461I/N465K) protein was comparable to that of the F(G264E/T461I) protein (P = 0.98, two-way ANOVA) and higher than those of the F(T461I) and F(T461I/N462S/N465K) proteins (P < 0.05, two-way ANOVA). These data suggest that the F(F375S/T461I/N465K) protein may have an advantage for the MeV genome transmission between neurons through membrane fusion over the other mutant F proteins including the F(T461I) protein in the presence of the WT F protein.
Hetero-oligomer formations with the WT F protein may influence the fusogenicities of mutant F proteins
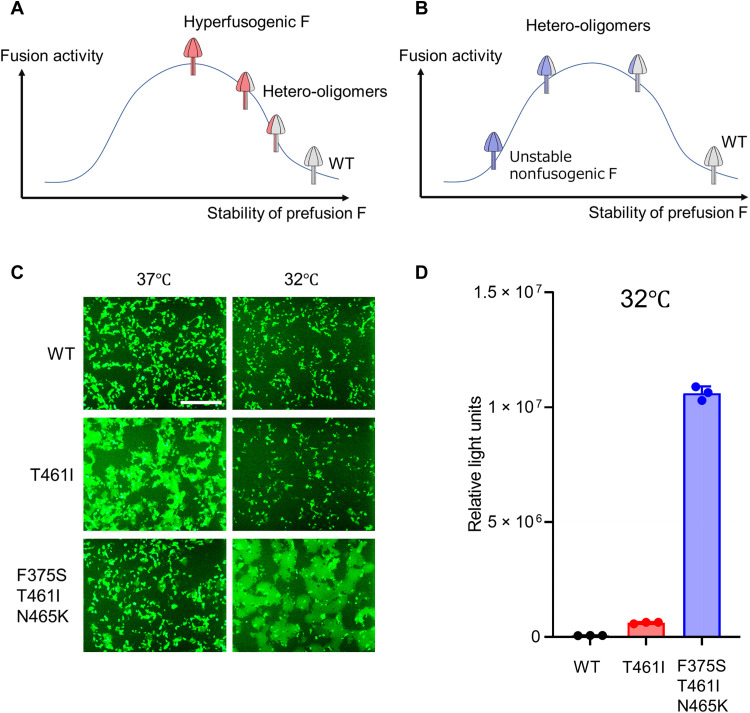
The prefusion form of the MeV F protein forms a trimer (33). A possible mechanism by which the coexpression of the WT F protein affects the function of mutant F proteins is hetero-oligomer (heterotrimer) formation between the WT and mutant F proteins, as we reported previously (23). In that study, we suggested that the conformational stability must be appropriate for the prefusion form of the F protein to exhibit fusion activity upon receptor binding by the H protein. Under certain conditions, the WT F protein (prefusion form) may be too stable to be triggered. Some mutations in the F protein may decrease the conformational stability of the prefusion form (11, 19, 25, 33). When the mutant F protein has reduced stability, its conformational changes could occur easily, exhibiting hyperfusogenicity (Fig. 2A). However, if its stability is too low, the mutant F protein may undergo inappropriate folding before receptor binding, precluding fusion triggering (Fig. 2B). Hetero-oligomer between the WT F protein and a given mutant F protein may have the intermediate stability, exhibiting increased or decreased fusogenicity, depending on the stability of that mutant F protein (Fig. 2, A and B).
Fig. 2. The fusogenicities of MeV F proteins at a low temperature.
(A and B) These figures depict the possible relationship between the stability of the F protein prefusion form and its fusion activity. The cases of ordinary hyperfusogenic F proteins (A) and unstable nonfusogenic proteins (B) are shown, respectively. (C) The WT H protein, MeV F [WT F, F(T461I), or F(F375S/T461I/N465K)], CADM1, and EGFP were expressed in 293FT cells at 37° or 32°C. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm. (D) The WT H protein, MeV F [WT F, F(T461I), or F(F375S/T461I/N465K)], and CADM1 were expressed in mixed 293FT/DSP1 and 293FT/DSP2 cells at 32°C. The Renilla luciferase activity in the transfected cells was analyzed 24 hours after transfection. Each data point represents one biological replicate (N = 3). Error bars indicate SDs.
To examine whether this model can explain our findings, the fusogenicities of the WT F, F(T461I), and F(F375S/T461I/N465K) proteins were evaluated by the fusion assay at 32° and 37°C. At the lower temperature (which is expected to increase the F protein stability) (11, 19, 23, 25), the fusogenicity of the F(T461I) protein was decreased, while that of the F(F375S/T461I/N465K) protein was increased (Fig. 2C). The DSP assay also revealed that at 32°C, the fusogenicity of the F(F375S/T461I/N465K) protein was substantially higher than that of the F(T461I) protein (Fig. 2D). The enhanced fusogenicity of the F(F375S/T461I/N465K) protein was presumably caused by its increased stability at 32°C. Thus, the F(F375S/T461I/N465K) protein likely corresponds to “unstable nonfusogenic F” in Fig. 2B and exhibits increased fusogenicity in the copresence of the WT F protein.
Inclusion of the WT genome influences the neurotropism of recombinant MeVs bearing mutant F proteins
We then generated the recombinant MeVs respectively encoding the WT F, F(T461I), and F(F375S/T461I/N465K) proteins and examined their phenotypes. While the recombinant MeV encoding the WT F protein and the red fluorescent mCherry protein [MV323-mCherry (WT)] and that encoding the F(T461I) protein and the green fluorescent Venus protein [MV323-Venus-F(T461I)] were successfully recovered, the virus encoding the F(F375S/T461I/N465K) protein and the Venus protein [MV323-Venus-F(F375S/T461I/N465K)] did not produce infectious particles (Table 1). We think that the stability of the F(F375S/T461I/N465K) protein is too low to keep virus particle infectivity.
Table 1. Recovery of mixed recombinant viruses.
| Viral genome 1 | Viral genome 2 | Infectious virus |
|---|---|---|
| – | MV323-mCherry (WT) | + |
| MV323-Venus-F(T461I) | – | + |
| MV323-Venus-F(F375S/T461I/N465K) | – | – |
| MV323-Venus-F(T461I) | MV323-mCherry (WT) | + |
| MV323-Venus-F(F375S/T461I/N465K) | MV323-mCherry (WT) | + |
Furthermore, both of the full-length genome plasmids of MV323-mCherry (WT) and MV323-Venus-F(T461I) or MV323-Venus-F(F375S/T461I/N465K) were mixed and used to generate recombinant viruses. MeV is known to have the ability to include several genomes in one viral particle, called polyploidy (34). In this protocol, the mixed genome virus of MV323-mCherry (WT) and MV323-Venus-F(F375S/T461I/N465K) as well as that of MV323-mCherry (WT) and MV323-Venus-F(T461I) produced infectious particles (Table 1). These virus particles could contain both or either of the MV323-mCherry genome and the MV323-Venus-F(T461I) genome or the MV323-Venus-F(F375S/T461I/N465K) genome (Fig. 3).
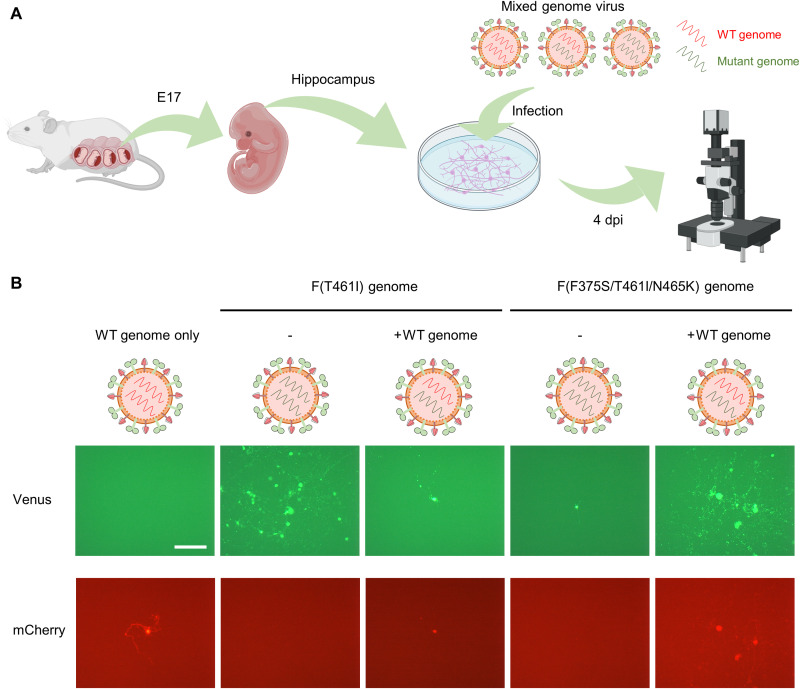
Fig. 3. Infection of mouse primary neurons with mixed genome MeVs.
(A) Schematic diagram of the experimental procedures for the isolation of mouse hippocampus primary neurons and infection with mixed genome viruses. Created with BioRender.com. E17, embryonic day 17. (B) Mouse primary neurons were infected with the MV323-mCherry (WT) or one of mixed genome viruses [MV323-mCherry + MV323-Venus-F(T461I) and MV323-mCherry + MV323-Venus-F(F375S/T461I/N465K)] at an MOI of 0.0001, and the cells were observed 4 days post infection (dpi) under a fluorescence microscope. Scale bar, 200 μm.
We have previously shown that hyperfusogenic MeVs can spread in mouse primary neurons using mouse CADM1 and CADM2 (13). Mouse primary hippocampus neurons were isolated from mouse embryos and infected with these mixed genome viruses (Fig. 3A). The cells were observed 4 days after infection under a fluorescence microscope (Fig. 3B). The MV323-Venus-F(T461I) genome alone spread in neurons, but the spread was restricted when the MV323-mCherry (WT) genome coexisted. In contrast, the MV323-Venus-F(F375S/T461I/N465K) genome spread efficiently in neurons only when the MV323-mCherry (WT) genome was copresent. There were the “no-spread” and “spread” types of infected spots on mouse primary neurons after MeV infection (fig. S7A). When mouse primary neurons were infected with the mixed genome virus of MV323-mCherry (WT) and MV323-Venus-F(T461I), the percentage of the spread type in infected spots expressing the mutant genome was much lower in the presence of the WT genome than that in its absence (fig. S7B). In contrast, when the cells were infected with the mixed genome virus of MV323-mCherry (WT) and MV323-Venus-F(F375S/T461I/N465K), the percentage of the spread type in infected spots expressing the mutant genome was 100% in the presence of the WT genome (~20% in its absence) (fig. S7B).
Thus, the WT MeV genome can be transmitted together with other mutant MeV genomes, influencing the ability of MeV to spread in neurons. These results are consistent with the observations with the fusion assay (Fig. 1, B and C).
F proteins of SSPE MeV strains have cumulative mutations, which make them highly fusogenic
In addition to the substitutions G264E/T461I and F375S/T461I/N465K, other various substitutions are found in the patient B strain and the OSA-3/Bs/B strain, respectively (fig. S3). To investigate whether these other mutations affect the F protein function, we respectively evaluated the F protein fusogenicities of the patient B strain (Fig. 4) and the OSA-3/Bs/B strain (Fig. 5) by the fusion assay.
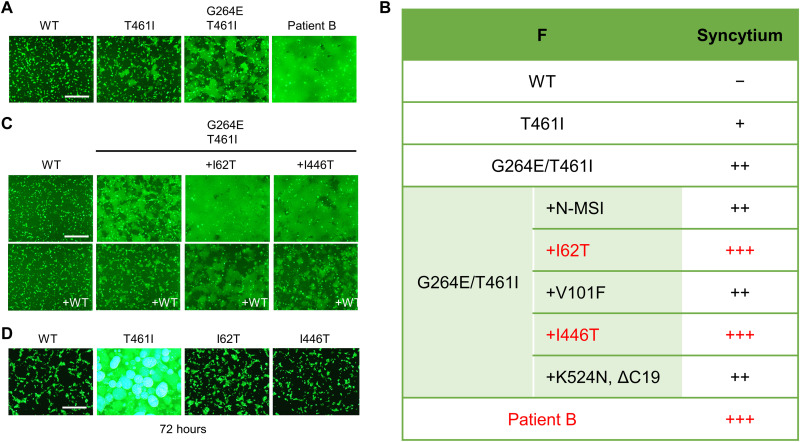
Fig. 4. The F fusogenicity of the patient B strain.
(A) The WT H protein, MeV F [WT F, F(T461I), F(G264E/T461I), and Patient-B-F], CADM1, and EGFP were expressed in 293FT cells. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm. (B) The F mutations that the patient B strain has and the fusogenicities of F proteins having certain combinations of the mutations are shown. N-MSI indicates three amino acids (methionine, serine, and isoleucine) added at the N terminus. ΔC19 indicates a 19–amino acid deletion at the C terminus. (C) The WT H protein, MeV F [WT F, F(G264E/T461I), F(I62T/G264E/T461I), or F(G264E/T461I/I446T)], CADM1, and EGFP were expressed without or with the WT F protein in 293FT cells. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm. (D) The WT H protein, MeV F [WT F, F(T461I), F(I62T), or F(I446T)], CADM1, and EGFP were expressed in 293FT cells. The cells were observed 72 hours after transfection under a fluorescence microscope. Scale bar, 500 μm.
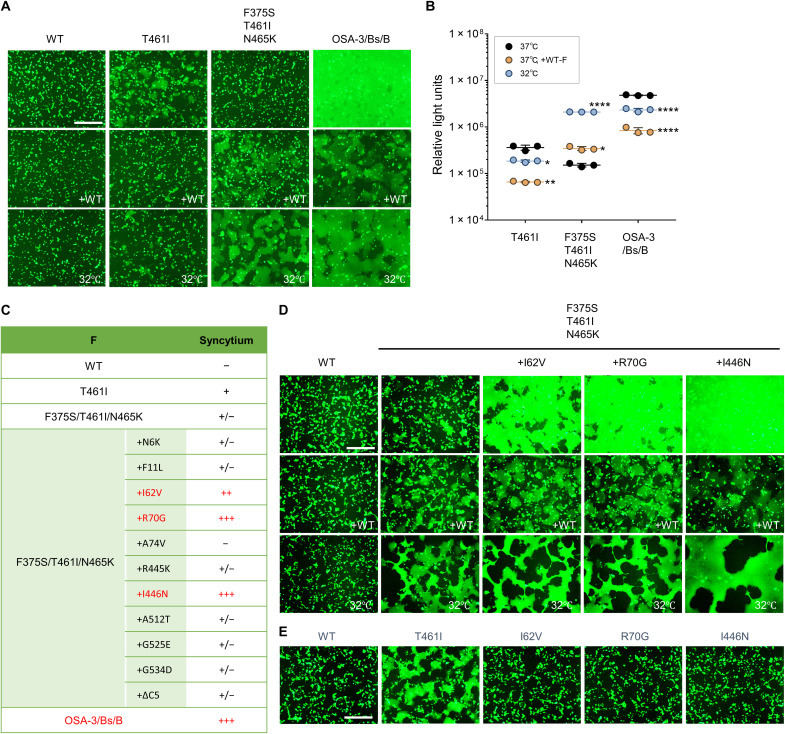
Fig. 5. The F fusogenicity of the OSA-3/Bs/B strain.
(A) The WT H protein, MeV F [WT F, F(T461I), F(F375S/T461I/N465K), and OSA-3/Bs/B-F], CADM1, and EGFP were expressed without or with the WT F protein in 293FT cells at 37° or 32°C. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm. (B) The WT H protein, MeV F [F(T461I), F(F375S/T461I/N465K), and OSA-3/Bs/B-F], and CADM1 were expressed in mixed 293FT/DSP1 and 293FT/DSP2 cells without or with the WT F protein at 37° or 32°C. The Renilla luciferase activity in the transfected cells was analyzed 24 hours after transfection. Each data point represents one biological replicate (N = 3). Error bars indicate SDs. Significance of the difference in the luciferase activity (as compared with that at 37°C) was analyzed by two-way ANOVA: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (C) The F mutations that the OSA-3/Bs/B strain has and the fusogenicities of F proteins having certain combinations of the mutations are shown. ΔC5 indicates a 5–amino acid deletion at the C terminus. (D) The WT H protein, MeV F [WT F, F(F375S/T461I/N465K), F(I62V/F375S/T461I/N465K), F(R70G/F375S/T461I/N465K), or F(F375S/T461I/N465K/I446N)], CADM1, and EGFP were expressed without or with the WT F protein in 293FT cells at 37° or 32°C. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm. (E) The WT H protein, MeV F [WT F, F(T461I), F(I62V), F(R70G), or F(I446N)], CADM1, and EGFP were expressed in 293FT cells. The cells were observed 24 hours after transfection under a fluorescence microscope. Scale bar, 500 μm.
The F protein of the patient B strain (Patient-B-F) had highly enhanced fusogenicity as compared with the F(G264E/T461I) protein (Fig. 4A). Among multiple mutations found in the Patient-B-F protein (fig. S3), the additional substitution I62T or I446T further increased the fusogenicity of the F(G264E/T461I) protein (Fig. 4, B and C). The fusogenicities of these highly fusogenic F proteins were reduced by the WT F protein coexpression (Fig. 4C). Notably, the substitution I62T or I446T alone did not enhance the WT F protein fusogenicity substantially even 72 hours after transfection (Fig. 4D).
The F protein of the OSA-3/Bs/B strain (OSA-3/Bs/B-F) was also highly fusogenic (Fig. 5A). The fusogenicity of the OSA-3/Bs/B-F protein, unlike that of the F(F375S/T461I/N465K) protein, was reduced by the WT F protein coexpression or at 32°C (Fig. 5A). These observations were also confirmed by the DSP assay (Fig. 5B). Among mutations present in the OSA-3/Bs/B-F protein (fig. S3), the additional substitution I62V, R70G, or I446N strongly increased the fusogenicity of the F(F375S/T461I/N465K) protein, and the fusogenicities of these mutant F proteins were reduced by the WT F protein coexpression or at 32°C (Fig. 5, C and D). The substitution I62V, R70G, or I446N alone did not enhance the fusogenicity of the WT F protein (Fig. 5E). These data indicate that the Patient-B-F protein and the OSA-3/Bs/B-F protein have acquired high fusogenicities through other halfway mutant F proteins by accumulating multiple mutations step by step. The data also indicate that fusogenicities of SSPE-derived mutant F proteins are generally reduced in the copresence of the WT F protein, although some exception such as the F(F375S/T461I/N465K) protein could occur.
DISCUSSION
RNA virus populations generally evolve rapidly under selection pressures, because of the high error rates of viral RNA polymerase (1, 2). This viral evolvability is known to cause some of the difficulties in controlling RNA virus infections, such as drug resistance, immune evasion, and expansion of the tropism and host range (1). Furthermore, the error-prone nature of the RNA genome replication leads to virus populations containing various variants (“quasispecies”) (1, 2, 35). Virus populations are not the mere mixture of independently acting variants. Rather, diverse heterogeneous variants sometimes interact positively or negatively to form the “community,” which determines the biological behavior of the populations (1, 2, 35, 36). These interactions within virus populations could be mediated by en bloc transmission that simultaneously transmits groups of viral genomes as a collective infectious unit (4, 5, 35, 37). Several mechanisms have been reported that produce collective infectious units in various species of viruses, such as polyploidy virions (23, 34, 38, 39), virion aggregation (40, 41), nonviral-specific structures (42–48), and direct cell-to-cell transmission (49–56). Studies suggest that collective infectious units contribute to the maintenance of viral genetic diversity and the evolution of “social-like” virus-virus interactions (4, 6). It is reported that MeV spread also relies on en bloc transmission through its polyploidy virion and intercellular pores in well-differentiated primary human airway epithelia (23, 34, 52, 57).
In this study, we show that mutations accumulated in SSPE-derived MeV F genes dynamically change the F protein fusogenicity. The cumulative mutations generally increase the fusogenicity (type A) but sometimes decrease it (type B) (Fig. 1 and fig. S8A). High levels of F protein fusogenicity achieved by cumulative mutations are probably required for efficient viral transmission between neurons to sustain persistent infection. Therefore, the observation that the F(F375S/T461I/N465K) protein exhibits lower fusogenicity than the F(T461I) or F(F375S/N465K) protein was puzzling because it was difficult to explain the selection of mutants having lower fusogenicity and thus the ability to spread in the brain.
By taking en bloc transmission into account, we could solve this paradox. MeV is thought to spread in the brain through cell-cell fusion at synapses, where multiple viral genomes are transmitted simultaneously (15, 16). In these circumstances, the ancestral WT genome and progeny mutant genomes may coexist in neurons, at least in the early phase of MeV persistence in the brain (fig. S1). The coexisting WT F protein, in many cases, down-regulates the fusogenicities of mutant F proteins but may enhance those of others (Fig. 1, C to E). Thus, in the presence of ancestral genomes, both type A and type B mutants likely increase their fusogenicities step by step by accumulating mutations (fig. S8B). This model may also explain the reason why it takes a long time (usually several years) to develop SSPE after acute infection (9, 10). Because the copresence of the WT MeV genome generally interferes with the fusogenicities of mutant F proteins, many additional mutations may be required for persisting mutant viruses to overcome the interference and gain enough fusogenicity to spread between neurons efficiently. Thus, the two related concepts, en bloc transmission and interactions between different viral genomes, can explain the long-term process of SSPE and the emergence of the type B mutants in SSPE-derived MeV isolates. A future challenge would be to determine, using animal models in vivo, how the WT MeV evolves in the brain to gain neurotropism and how en bloc transmission of the viral genomes contributes to that process.
Our results also show that effects of mutations differ depending on the context. For instance, the introduction of the T461I substitution increases the fusogenicity of the WT F, F(N462S/N465S), or F(G264E) protein but decreases that of the F(F375S/N465K) protein (Fig. 1B). Furthermore, the introduction of the substitution I62T, I446T, I62V, R70G, or I446N does not affect the fusogenicity of the WT F protein, but I62T or I446T substantially enhances that of the F(G246E/T461I) protein, and I62V, R70G, or I446N increases that of the F(F375S/T461I/N465K) protein (Figs. 4, C and D, and 5, D and E). These data indicate that the order of mutations introduced is also critical in determining F protein fusogenicity. In addition to hyperfusogenic mutations in the F gene, MeV isolates from patients with SSPE contain other characteristic changes in their genomes (10), such as adenine-to-guanine or uracil-to-cytosine biased hypermutations causing defects in the matrix (M) gene (27, 58) and mutations causing the elongation or shortening of the cytoplasmic tail of the F protein (29, 59). The M genes of the patient A, patient B, and OSA-3 strains have biased hypermutations (27, 60). It is possible that these changes are also selectively advantageous for MeV persistence in the brain. However, recent studies reported that recombinant MeVs lacking the M protein or the cytoplasmic tail of the F protein did not induce syncytia in SLAMF1- and nectin-4–negative cells (18), while SSPE-derived hyperfusogenic mutations in the F protein enable MeV spread between neurons (11, 17–19, 21, 24, 25). More studies are needed to fully elucidate the effect of the mutations affecting the M protein or the cytoplasmic tail of the F protein on MeV persistence in the brain. Most of the hyperfusogenic mutations in the ectodomain of the F protein analyzed in this study are caused by adenine-to-guanine or uracil-to-cytosine nucleotide substitutions (table S1), suggesting the involvement of biased hypermutations.
Whether mutations in the H gene also contribute to the MeV neurotropism is not known. Although the H gene of certain SSPE isolates is not a major determinant of neurovirulence (17, 24, 25), the H mutations could enhance the interaction with CADM1 and CADM2 or allow MeV to use additional receptors. This is an important issue for future studies.
Thus, this study provides important insights into the evolutionary process of MeV neuropathogenicity and genotype-phenotype relationships of oligomeric viral fusion proteins in viral evolution. The concept of en bloc transmission could unveil positive and negative interactions among different fusion proteins in other enveloped viruses. In addition, we suggest that the interpretation of mutations found in en bloc transmitted viruses requires careful evaluations, because the effects of mutations depend on interactions between different viral genomes.
MATERIALS AND METHODS
Cells
293FT cells (R70007, Invitrogen) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Fujifilm, Wako Pure Chemical Corporation, Japan) supplemented with 10% fetal bovine serum (FBS). 293FT cells stably expressing DSP1 and DSP2 (30–32), provided by Z. Matsuda, the University of Tokyo, were maintained in DMEM supplemented with 10% FBS and puromycin (1 μg/ml; InvivoGen, San Diego, CA). The 293FT cell line is a highly transfectable clone derived from human embryonal kidney cells transformed with the SV40 large T antigen. We previously established the method to evaluate the fusion triggering activity of host molecules using 293FT cells, which lack the known MeV receptors SLAMF1 and nectin-4 (12, 13). Mouse primary neurons were isolated from the hippocampus of the C57BL/6 mouse at embryonic day 17 and cultured according to the protocol described previously (61).
Plasmids
The eukaryotic expression vector pCA7 (62) is a derivative of pCAGGS (63). The pCA7 plasmids expressing the MeV H protein (the Ichinose-B (IC-B) strain), MeV F protein (the IC-B strain), and human CADM1 were described previously (13, 18, 19, 64). For Western blot and coimmunoprecipitation analysis, the HA tag (YPYDVPDYA) and FLAG tag (DYKDDDDK) sequences were fused to the C termini of the WT and mutant F proteins.
Fusion assay
293FT cells cultured in 24-well plates were transfected with different combinations of pCA7 plasmids respectively expressing the WT H protein, MeV F protein, EGFP, and SLAMF1 or CADM1 using Lipofectamine LTX (Thermo Fisher Scientific). The induction of cell-cell fusion was evaluated 24 hours after transfection by fluorescence microscopy. For quantification of cell-cell fusion, 293FT/DSP1 and 293FT/DSP2 cells were cocultured in 24-well plates and then transfected with pCA7 plasmids respectively expressing the WT H protein, MeV F protein, and SLAMF1 or CADM1. The Renilla luciferase activity in the transfected cells was analyzed 24 hours after transfection using a Renilla luciferase assay system (Promega, Madison, WI). Chemiluminescence was measured using a Mithras LB940 plate reader (Berthold Technologies, Pforzheim, Germany). In these assays, the cells were incubated after transfection usually at 37°C for the standard assay or at the lower temperature of 32°C, which is expected to increase the stability of the F protein. All measurements were taken from distinct samples.
Cell surface biotinylation assay
Following procedures previously described in (14), 293FT cells cultured on 12-well plates were transfected with 0.5 μg of pCA7 encoding one of FLAG-tagged F proteins with 0.5 μg of pCA7 encoding E-cadherin (a cell surface protein control) using Lipofectamine LTX. At 24 hours after transfection, cells were washed with phosphate-buffered saline (PBS) and then incubated with 200 μl of the biotin reagent solution [2 mM EZ-Link N-hydroxysulfosuccinimide (sulfo-NHS)–biotin (Thermo Fisher Scientific) in PBS] for 30 min at 4°C. After being washed three times with PBS containing 50 mM ammonium chloride for quenching, the cells were lysed in 200 μl of the immunoprecipitation (IP) lysis buffer (Thermo Fisher Scientific) containing the protease inhibitor cocktail (Sigma-Aldrich). The lysates were cleared by centrifugation for 30 min at 17,360g and 4°C. Then, 50 μl of each supernatant was mixed with an equal volume of 2× SDS loading buffer [125 mM tris-HCl (pH 6.8), 10% 2-mercaptoethanol, 4% SDS, 0.1% bromophenol blue, and 20% glycerol], boiled for 5 min, and stored at −20°C as the cell lysate samples. The rest of the supernatant was incubated with avidin-agarose beads (A-9207, Sigma-Aldrich) for 3 hours at 4°C. The samples were centrifuged and washed three times with IP lysis buffer. Pellets were mixed with 30 μl of 2× SDS loading buffer, boiled for 5 min, and stored at −20°C as biotinylated cell surface protein samples.
Western blotting
Following procedures previously described in (14), proteins in samples were separated by SDS–polyacrylamide gel electrophoresis and then blotted onto polyvinylidene difluoride membranes (Hybond-P, Amersham Biosciences). The membranes were incubated with primary antibodies (Abs) for 1 hour. Rabbit polyclonal Abs against HA tag (GTX225044, GeneTex) and FLAG tag (F1804, Sigma-Aldrich) and mouse monoclonal Abs against FLAG tag (F1804, Sigma-Aldrich), E-cadherin (clone SHE78-7, TaKaRa Bio), and β-actin (clone BA3R, BioVision) were used. The membranes were washed with tris-buffered saline containing 0.05% Tween 20 (TBS-T) and incubated with horseradish peroxidase–conjugated goat anti-rabbit or mouse immunoglobulin G (Bio-Rad) for 1 hour at room temperature. After being washed with TBS-T, the membranes were treated with an ECL Plus reagent (Amersham Biosciences), and chemiluminescent signals were detected and imaged using a VersaDoc 5000 imager (Bio-Rad).
Virus preparation
Recombinant MeVs encoding mCherry and the WT F protein or Venus and one of the mutant F proteins [F(T461I) or F(F375S/T461I/N465K)] were recovered as described previously (65). Vero cells stably expressing the human SLAMF1 (Vero/hSLAM cells) cultured in a 10-cm dish were infected with each recombinant MeV at a multiplicity of infection (MOI) of 0.001. The cells were harvested and then lysed by three freeze-thaw cycles. Lysates were centrifuged at 2500g for 10 min at 4°C. Supernatants were collected and stored at −80°C.
Measurements and statistical tests
All measurements were taken from distinct samples. All statistical tests were performed using GraphPad Prism 9 software [two-way ANOVA (Dunnett’s multiple comparisons test)].
Animal experiments
All animal experiments were performed in accordance with procedures approved by the Animal Care and Use Committee of Kyushu University (permit nos. A19-383-0 and A21-044-0) and were in compliance with all relevant ethical regulations.
Acknowledgments
We thank Z. Matsuda, the University of Tokyo, for providing the DSP assay system and A. Leeks and S. Watanabe for invaluable comments. We also appreciate the technical assistance from The Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences.
Funding: This work was supported by JSPS KAKENHI grant numbers JP20K07527 (to Y.S.) and JP20H00507 (to Y.Y.), Takeda Science Foundation (to Y.S.), The Chemo-Sero-Therapeutic Research Institute (to Y.S.), JSPS Core-to-Core Program A grant number JPJSCCA20190008 (to T.H.), and AMED grant number JP22wm0325002h (to T.H.).
Author contributions: Conceptualization: Y.S. Methodology: Y.S., Y.H., and R.T. Investigation: Y.S., H.H., Y.H., and R.T. Formal analysis: Y.S., H.H., Y.H., R.T., T.S., T.H., and Y.Y. Visualization: Y.S. and H.H. Funding acquisition: Y.S., Y.Y., and T.H. Project administration: Y.S. Writing—original draft: Y.S. and Y.Y. Writing—review and editing: Y.S., H.H., Y.H., R.T., T.S., T.H., and Y.Y.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The 293FT/DSP1 and 293FT/DSP2 cell lines can be provided by Z. Matsuda, the University of Tokyo, pending scientific review and a completed material transfer agreement. Requests for the cell lines should be submitted to Z. Matsuda, the Institute of Medical Science, the University of Tokyo (zmatsuda@ims.u-tokyo.ac.jp).
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
REFERENCES AND NOTES
- 1.E. Domingo, J. Sheldon, C. Perales, Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76, 159–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R. Andino, E. Domingo, Viral quasispecies. Virology 479-480, 46–51 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Y. Shirogane, S. Watanabe, Y. Yanagi, Cooperation between different variants: A unique potential for virus evolution. Virus Res. 264, 68–73 (2019). [DOI] [PubMed] [Google Scholar]
- 4.R. Sanjuán, Collective infectious units in viruses. Trends Microbiol. 25, 402–412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Altan-Bonnet, C. Perales, E. Domingo, Extracellular vesicles: Vehicles of en bloc viral transmission. Virus Res. 265, 143–149 (2019). [DOI] [PubMed] [Google Scholar]
- 6.S. L. Díaz-Muñoz, R. Sanjuán, S. West, Sociovirology: Conflict, cooperation, and communication among viruses. Cell Host Microbe 22, 437–441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Leeks, S. A. West, M. Ghoul, The evolution of cheating in viruses. Nat. Commun. 12, 6928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Coughlin, A. Beck, B. Bankamp, P. Rota, M. M. Coughlin, A. S. Beck, B. Bankamp, P. A. Rota, Perspective on global measles epidemiology and control and the role of novel vaccination strategies. Viruses 9, 11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. Mekki, B. Eley, D. Hardie, J. M. Wilmshurst, Subacute sclerosing panencephalitis: Clinical phenotype, epidemiology, and preventive interventions. Dev. Med. Child Neurol. 61, 1139–1144 (2019). [DOI] [PubMed] [Google Scholar]
- 10.D. E. Griffin, Measles virus, in Fields Virology, D. M. Knipe, P. M. Howley, Eds. (Lippincott Williams & Wilkins, ed. 6, 2013), pp. 1042–1069. [Google Scholar]
- 11.S. Watanabe, Y. Shirogane, Y. Sato, T. Hashiguchi, Y. Yanagi, New insights into measles virus brain infections. Trends Microbiol. 27, 164–175 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Y. Shirogane, T. Hashiguchi, Y. Yanagi, Weak cis and trans interactions of the hemagglutinin with receptors trigger fusion proteins of neuropathogenic measles virus isolates. J. Virol. 94, e01727–e01719 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Y. Shirogane, R. Takemoto, T. Suzuki, T. Kameda, K. Nakashima, T. Hashiguchi, Y. Yanagi, CADM1 and CADM2 trigger neuropathogenic measles virus-mediated membrane fusion by acting in cis. J. Virol. 95, e00528–e00521 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R. Takemoto, T. Suzuki, T. Hashiguchi, Y. Yanagi, Y. Shirogane, Short-stalk isoforms of CADM1 and CADM2 trigger neuropathogenic measles virus-mediated membrane fusion by interacting with the viral hemagglutinin. J. Virol. 96, e01949–e01921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Y. Iwasaki, H. Koprowski, Cell to cell transmission of virus in the central nervous system. I. Subacute sclerosing panencephalitis. Lab. Invest. 31, 187–196 (1974). [PubMed] [Google Scholar]
- 16.D. M. P. Lawrence, C. E. Patterson, T. L. Gales, J. L. D’Orazio, M. M. Vaughn, G. F. Rall, Measles virus spread between neurons requires cell contact but not CD46 expression, syncytium formation, or extracellular virus production. J. Virol. 74, 1908–1918 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Ayata, K. Takeuchi, M. Takeda, S. Ohgimoto, S. Kato, L. B. Sharma, M. Tanaka, M. Kuwamura, H. Ishida, H. Ogura, The F gene of the Osaka-2 strain of measles virus derived from a case of subacute sclerosing panencephalitis is a major determinant of neurovirulence. J. Virol. 84, 11189–11199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Watanabe, Y. Shirogane, S. O. Suzuki, S. Ikegame, R. Koga, Y. Yanagi, Mutant fusion proteins with enhanced fusion activity promote measles virus spread in human neuronal cells and brains of suckling hamsters. J. Virol. 87, 2648–2659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Watanabe, S. Ohno, Y. Shirogane, S. O. Suzuki, R. Koga, Y. Yanagi, Measles virus mutants possessing the fusion protein with enhanced fusion activity spread effectively in neuronal cells, but not in other cells, without causing strong cytopathology. J. Virol. 89, 2710–2717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.E. M. Jurgens, C. Mathieu, L. M. Palermo, D. Hardie, B. Horvat, A. Moscona, M. Porotto, Measles fusion machinery is dysregulated in neuropathogenic variants. mBio 6, e02528–e02514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Y. Sato, S. Watanabe, Y. Fukuda, T. Hashiguchi, Y. Yanagi, S. Ohno, Cell-to-cell measles virus spread between human neurons is dependent on hemagglutinin and hyperfusogenic fusion protein. J. Virol. 92, e02166–e02117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. Ikegame, T. Hashiguchi, C.-T. Hung, K. Dobrindt, K. J. Brennand, M. Takeda, B. Lee, Fitness selection of hyperfusogenic measles virus F proteins associated with neuropathogenic phenotypes. Proc. Natl. Acad. Sci. 118, e2026027118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Y. Shirogane, S. Watanabe, Y. Yanagi, Cooperation between different RNA virus genomes produces a new phenotype. Nat. Commun. 3, 1235 (2012). [DOI] [PubMed] [Google Scholar]
- 24.M. Ayata, M. Tanaka, K. Kameoka, M. Kuwamura, K. Takeuchi, M. Takeda, K. Kanou, H. Ogura, Amino acid substitutions in the heptad repeat A and C regions of the F protein responsible for neurovirulence of measles virus Osaka-1 strain from a patient with subacute sclerosing panencephalitis. Virology 487, 141–149 (2016). [DOI] [PubMed] [Google Scholar]
- 25.F. Angius, H. Smuts, K. Rybkina, D. Stelitano, B. Eley, J. Wilmshurst, M. Ferren, A. Lalande, C. Mathieu, A. Moscona, B. Horvat, T. Hashiguchi, M. Porotto, D. Hardie, Analysis of a subacute sclerosing panencephalitis genotype B3 virus from the 2009-2010 South African measles epidemic shows that hyperfusogenic F proteins contribute to measles virus infection in the brain. J. Virol. 93, e01700–e01718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. Baczko, U. G. Liebert, M. Billeter, R. Cattaneo, H. Budka, V. ter Meulen, Expression of defective measles virus genes in brain tissues of patients with subacute sclerosing panencephalitis. J. Virol. 59, 472–478 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.R. Cattaneo, A. Schmid, D. Eschle, K. Baczko, V. ter Meulen, M. A. Billeter, Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55, 255–265 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R. Cattaneo, A. Schmid, P. Spielhofer, K. Kaelin, K. Baczko, V. ter Meulen, J. Pardowitz, S. Flanagan, B. K. Rima, S. A. Udem, M. A. Billeter, Mutated and hypermutated genes of persistent measles viruses which caused lethal human brain diseases. Virology 173, 415–425 (1989). [DOI] [PubMed] [Google Scholar]
- 29.X. Ning, M. Ayata, M. Kimura, K. Komase, K. Furukawa, T. Seto, N. Ito, M. Shingai, I. Matsunaga, T. Yamano, H. Ogura, Alterations and diversity in the cytoplasmic tail of the fusion protein of subacute sclerosing panencephalitis virus strains isolated in Osaka, Japan. Virus Res. 86, 123–131 (2002). [DOI] [PubMed] [Google Scholar]
- 30.N. Kondo, K. Miyauchi, F. Meng, A. Iwamoto, Z. Matsuda, Conformational changes of the HIV-1 envelope protein during membrane fusion are inhibited by the replacement of its membrane-spanning domain. J. Biol. Chem. 285, 14681–14688 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.H. Ishikawa, F. Meng, N. Kondo, A. Iwamoto, Z. Matsuda, Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng. Des. Sel. 25, 813–820 (2012). [DOI] [PubMed] [Google Scholar]
- 32.H. Wang, X. Li, S. Nakane, S. Liu, H. Ishikawa, A. Iwamoto, Z. Matsuda, Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLOS ONE 9, e96790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Hashiguchi, Y. Fukuda, R. Matsuoka, D. Kuroda, M. Kubota, Y. Shirogane, S. Watanabe, K. Tsumoto, D. Kohda, R. K. Plemper, Y. Yanagi, Structures of the prefusion form of measles virus fusion protein in complex with inhibitors. Proc. Natl. Acad. Sci. U.S.A. 115, 2496–2501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.M. Rager, S. Vongpunsawad, W. P. Duprex, R. Cattaneo, Polyploid measles virus with hexameric genome length. EMBO J. 21, 2364–2372 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.L. J. González Aparicio, C. B. López, S. A. Felt, A virus is a community: Diversity within negative-sense RNA virus populations. Microbiol. Mol. Biol. Rev. 86, e0008621 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. E. Jones, V. le Sage, S. S. Lakdawala, Viral and host heterogeneity and their effects on the viral life cycle. Nat. Rev. Microbiol. 19, 272–282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Y. Shirogane, S. Watanabe, Y. Yanagi, Cooperation: Another mechanism of viral evolution. Trends Microbiol. 21, 320–324 (2013). [DOI] [PubMed] [Google Scholar]
- 38.D. Luque, G. Rivas, C. Alfonso, J. L. Carrascosa, J. F. Rodríguez, J. R. Castón, Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc. Natl. Acad. Sci. U.S.A. 106, 2148–2152 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D. R. Beniac, P. L. Melito, S. L. Devarennes, S. L. Hiebert, M. J. Rabb, L. L. Lamboo, S. M. Jones, T. F. Booth, The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLOS ONE 7, e29608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. Combe, R. Garijo, R. Geller, J. M. Cuevas, R. Sanjuán, Single-cell analysis of RNA virus infection identifies multiple genetically diverse viral genomes within single infectious units. Cell Host Microbe 18, 424–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. M. Cuevas, M. Durán-Moreno, R. Sanjuán, Multi-virion infectious units arise from free viral particles in an enveloped virus. Nat. Microbiol. 2, 17078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.J. Slack, B. M. Arif, The baculoviruses occlusion-derived virus: Virion structure and function. Adv Virus Res. 69, 99–165 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Y.-H. Chen, W. Du, M. C. Hagemeijer, P. M. Takvorian, C. Pau, A. Cali, C. A. Brantner, E. S. Stempinski, P. S. Connelly, H.-C. Ma, P. Jiang, E. Wimmer, G. Altan-Bonnet, N. Altan-Bonnet, Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Z. Feng, L. Hensley, K. L. McKnight, F. Hu, V. Madden, L. Ping, S.-H. Jeong, C. Walker, R. E. Lanford, S. M. Lemon, A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.S. Nagashima, S. Jirintai, M. Takahashi, T. Kobayashi, Tanggis, T. Nishizawa, T. Kouki, T. Yashiro, H. Okamoto, Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J. Gen. Virol. 95, 2166–2175 (2014). [DOI] [PubMed] [Google Scholar]
- 46.R. J. Owens, C. Limn, P. Roy, Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J. Virol. 78, 6649–6656 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. M. Robinson, G. Tsueng, J. Sin, V. Mangale, S. Rahawi, L. L. McIntyre, W. Williams, N. Kha, C. Cruz, B. M. Hancock, D. P. Nguyen, M. R. Sayen, B. J. Hilton, K. S. Doran, A. M. Segall, R. Wolkowicz, C. T. Cornell, J. L. Whitton, R. A. Gottlieb, R. Feuer, Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLOS Pathog. 10, e1004045 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.C. M. Robinson, P. R. Jesudhasan, J. K. Pfeiffer, Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe 15, 36–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.L. M. Agosto, P. D. Uchil, W. Mothes, HIV cell-to-cell transmission: Effects on pathogenesis and antiretroviral therapy. Trends Microbiol. 23, 289–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. Jolly, K. Kashefi, M. Hollinshead, Q. J. Sattentau, HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J Exp Med. 199, 283–293 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.N. M. Sherer, M. J. Lehmann, L. F. Jimenez-Soto, C. Horensavitz, M. Pypaert, W. Mothes, Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat. Cell Biol. 9, 310–315 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.N. Cifuentes-Muñoz, R. E. Dutch, R. Cattaneo, Direct cell-to-cell transmission of respiratory viruses: The fast lanes. PLOS Pathog. 14, e1007015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.F. el Najjar, N. Cifuentes-Muñoz, J. Chen, H. Zhu, U. J. Buchholz, C. L. Moncman, R. E. Dutch, Human metapneumovirus induces reorganization of the actin cytoskeleton for direct cell-to-cell spread. PLOS Pathog. 12, e1005922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. Mateo, A. Generous, P. L. Sinn, R. Cattaneo, Connections matter—How viruses use cell–cell adhesion components. J. Cell Sci. 128, 431–439 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Mehedi, T. McCarty, S. E. Martin, C. le Nouën, E. Buehler, Y.-C. Chen, M. Smelkinson, S. Ganesan, E. R. Fischer, L. G. Brock, B. Liang, S. Munir, P. L. Collins, U. J. Buchholz, Actin-related protein 2 (ARP2) and virus-induced filopodia facilitate human respiratory syncytial virus spread. PLOS Pathog. 12, e1006062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.K. L. Roberts, B. Manicassamy, R. A. Lamb, Influenza a virus uses intercellular connections to spread to neighboring cells. J. Virol. 89, 1537–1549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R. Cattaneo, R. C. Donohue, A. R. Generous, C. K. Navaratnarajah, C. K. Pfaller, Stronger together: Multi-genome transmission of measles virus. Virus Res. 265, 74–79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.K. Baczko, J. Lampe, U. G. Liebert, U. Brinckmann, V. ter Meulen, I. Pardowitz, H. Budka, S. L. Cosby, S. Isserte, B. K. Rima, Clonal expansion of hypermutated measles virus in a SSPE brain. Virology 197, 188–195 (1993). [DOI] [PubMed] [Google Scholar]
- 59.A. Schmid, P. Spielhofer, R. Cattaneo, K. Baczko, V. ter Meulen, M. A. Billeter, Subacute sclerosing panencephalitis is typically characterized by alterations in the fusion protein cytoplasmic domain of the persisting measles virus. Virology 188, 910–915 (1992). [DOI] [PubMed] [Google Scholar]
- 60.M. Ayata, T. Kimoto, K. Hayashi, T. Seto, R. Murata, H. Ogura, Nucleotide sequences of the matrix protein gene of subacute sclerosing panencephalitis viruses compared with local contemporary isolates from patients with acute measles. Virus Res. 54, 107–115 (1998). [DOI] [PubMed] [Google Scholar]
- 61.H. Nakashima, K. Tsujimura, K. Irie, M. Ishizu, M. Pan, T. Kameda, K. Nakashima, Canonical TGF-β signaling negatively regulates neuronal morphogenesis through TGIF/Smad complex-mediated CRMP2 suppression. J. Neurosci. 38, 4791–4810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.M. Takeda, S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, Y. Yanagi, Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 79, 14346–14354 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.H. Niwa, K. Yamamura, J. Miyazaki, Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 64.M. Tahara, M. Takeda, Y. Shirogane, T. Hashiguchi, S. Ohno, Y. Yanagi, Measles virus infects both polarized epithelial and immune cells by using distinctive receptor-binding sites on its hemagglutinin. J. Virol. 82, 4630–4637 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.F. Seki, K. Yamada, Y. Nakatsu, K. Okamura, Y. Yanagi, T. Nakayama, K. Komase, M. Takeda, The SI strain of measles virus derived from a patient with subacute sclerosing panencephalitis possesses typical genome alterations and unique amino acid changes that modulate receptor specificity and reduce membrane fusion activity. J. Virol. 85, 11871–11882 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1