Abstract
Replication stress is a major source of endogenous DNA damage. Despite the identification of numerous proteins on replication forks to modulate fork or replication machinery activities, it remains unexplored whether noncoding RNAs can localize on stalled forks and play critical regulatory roles. Here, we identify an uncharacterized long noncoding RNA NONMMUT028956 (Lnc956 for short) predominantly expressed in mouse embryonic stem cells. Lnc956 is accumulated on replication forks to prevent fork collapse and preserve genomic stability and is essential for mouse embryogenesis. Mechanistically, it drives assembly of the Lnc956-TRIM28-HSP90B1 complex on stalled forks in an interdependent manner downstream of ataxia telangiectasia and Rad3-related (ATR) signaling. Lnc956-TRIM28-HSP90B1 complex physically associates with minichromosome maintenance proteins 2 (MCM2) to minichromosome maintenance proteins 7 (MCM7) hexamer via TRIM28 and directly regulates the CDC45-MCM-GINS (CMG) helicase retention on chromatin. The regulation of Lnc956-TRIM28-HSP90B1 on CMG retention is mediated by HSP90B1’s chaperoning function. These findings reveal a player that actively regulates replisome retention to prevent fork collapse.
LncRNA Lnc956 resides on replication forks and promotes CMG helicase retention to ensure stem cell genomic stability.
INTRODUCTION
Pluripotent stem cells (PSCs) are capable of self-renewal and differentiation into all cell types in the body and are cellular basis for organism development. Because of their unique functions, maintaining stable genome is fundamental for stem cells. Perturbations on genome stability can cause stem cell apoptosis, impair differentiation potentials, and induce the tumorigenicity (1–3). It has been well recognized that PSCs have superior stable genome over differentiated cells. For instance, mouse embryonic stem cells (ESCs) display 100-fold lower mutation rate than their isogenic mouse embryonic fibroblasts (MEFs) (4). How this is achieved in PSCs remains largely unknown. Understanding the mechanisms can not only provide valuable insights into the developmental failure/defects but also help address the issue of genomic instability seen in cultured human PSCs, which hampers the full applications of PSCs in cell-based regenerative medicine.
Limited studies suggested that PSCs use unique strategies and regulators to efficiently safeguard the genomic stability in a range of cellular activities. For example, a homologous recombination (HR)–based way that involves PSC-specific proteins ZSCAN4 and DCAF11 operates in telomere lengthening (5, 6). In addition, telomere protection in ESCs does not require TRF2, which is instead essential in somatic cells (7, 8). In response to DNA double-strand breaks (DSBs), PSCs prefer to choose the HR-mediated repair pathway (9). What determine this pathway choice remains mysterious, but PSC-specific proteins SALL4 and Filia were identified to play parts in HR repair processes (10, 11). DNA replication stress represents a major source of endogenous DNA damages and imposes big threat to genomic integrity (12). PSCs contain high level of DNA replication stress due to the lack of G1 checkpoint (13), but they are able to efficiently resolve replication stress (14). Although our recent study identified a PSC-specific Filia-Floped protein complex on replication forks as a regulator (14), the underlying mechanisms are complicative and far from clear.
DNA replication is carried out by a large molecular machine called replisome. The core components of replisome include the CDC45-MCM-GINS (CMG) DNA helicase, proliferating cell nuclear antigen (PCNA), and replicative DNA polymerases. In addition, numerous auxiliary factors associate with replisome to modulate its assembly, disassembly, and activities under unperturbed and perturbed conditions in a context-dependent manner (15). CMG helicase, which is composed of CDC45, minichromosome maintenance proteins 2 to 7 (MCM2 to MCM7), and the go-ichi-ni-san (GINS) complex (comprising SLD5, PSF1, PSF2, and PSF3), plays rate-limiting role in DNA replication by unwinding DNA along the leading strand template. Upon replication fork stalling, the intra–S phase replication checkpoint mediated by kinase ataxia telangiectasia and Rad3-related (ATR) is activated, and the local and global responses coordinate to preserve stalled forks and promote fork restart after obstacles are removed. Failure to repair and restore the stalling forks will cause fork collapse, which is defined as the inability to restart forks and characterized by the breakage of DNA into DSBs and the dissociation of replisomes from forks (16–18). Thanks to the innovative methods such as isolation of proteins on nascent DNA (iPOND) and nascent chromatin capture (19, 20), which coupled with mass spectrometry have provided systematic insights into the dynamic protein compositions and regulations of replication forks under normal and stressed conditions (17, 21, 22).
To date, only protein regulators have been reported on replication forks. It is unclear whether noncoding RNAs can reside on replication forks and regulate forks or replisomes. Long noncoding RNAs (lncRNAs) are able to bind DNA, RNAs, or proteins, can drive highly efficient condensation of interacting proteins, and largely enhance the protein functions (23, 24). In light of the knowledge, we wonder whether lncRNAs can reside on replication forks in PSCs and lncRNA-mediated regulation attributes to the efficient replication stress response in PSCs. In this study, we modified iPOND method to isolate fork-associated RNA species and named this method as isolation of RNAs on nascent DNA (iROND). Using iROND coupled with RNA sequencing (RNA-seq), we systematically identified the lncRNA species localized on replication forks of PSCs under normal and replication stress conditions. Further, we characterized in detail one ESC-prevalent lncRNA Lnc956 (NONCODE ID: NONMMUT028956, Lnc956 for short). Our results showed that ATR replication checkpoint induced the assembly of Lnc956-TRIM28-HSP90B1 complex on stalled forks under replication stress. Lnc956-TRIM28-HSP90B1 promotes CMG helicase retention to prevent replication fork collapse. Thus, our study revealed a lncRNA-mediated mechanism in resolving replication stress.
RESULTS
iROND identifies lncRNAs on replication forks
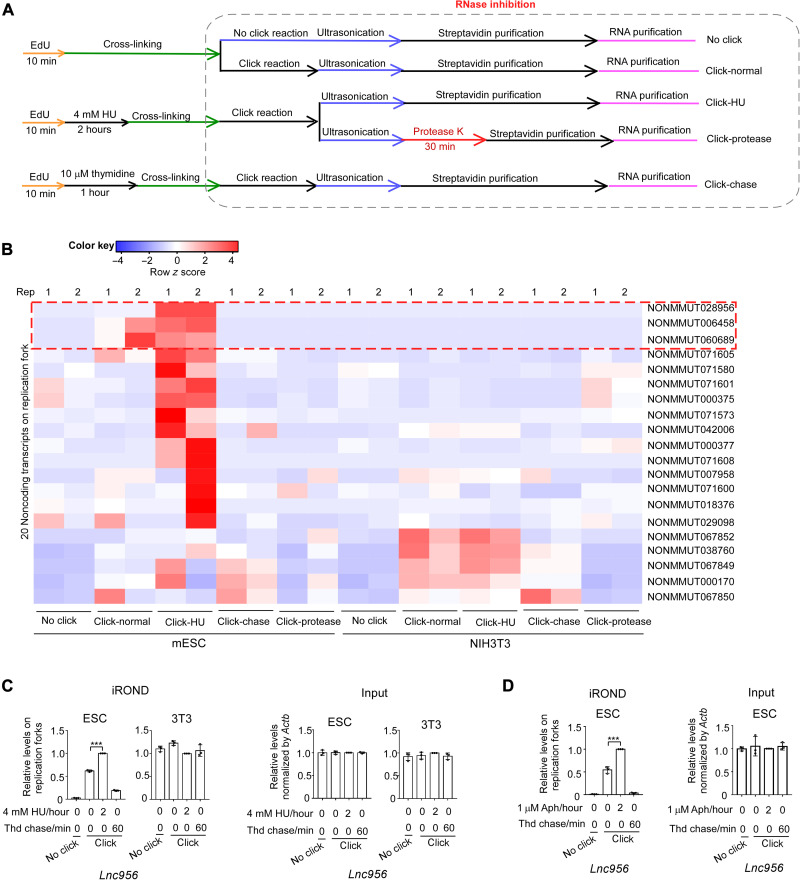
To identify whether there are lncRNAs on replication forks, we adapted iPOND with subtle modification. Briefly, cells were pulse-labeled with 5-ethynyl-2'-deoxyuridine (EdU) for 10 min to trace the active unperturbed replication forks. Cells were then immediately fixed, and the click reaction was carried out. After this step, the ribonuclease (RNase) inhibitors were included in all the following procedures to avoid RNA degradation. RNAs were extracted from purified nascent DNA samples by TRIzol and prepared for RNA-seq. We named the method as iROND. In performing iROND, we set up the following experimental designs (Fig. 1A): (i) no click group, in which cells were cultured in normal condition, and click reaction was omitted in the procedures; (ii) click-normal group, in which cells were cultured in normal condition, and standard iROND was performed; (iii) click-HU group, in which cells were treated with 4 mM hydroxyurea (HU) for 2 hours following EdU pulse label to induce fork stalling; (iv) click-protease group, in which all procedures were same as in group 3 except that cell lysis was treated with protease K (100 μg/ml) for 30 min to degrade proteins; (v) click-chase group, in which cells were chase labeled with thymidine for 1 hour after EdU pulse label. Among these settings, groups 1 and 5 were set as negative controls to evaluate the specific isolation of nascent DNA. The success of nascent DNA isolation was validated by immunoblot examination of isolated protein samples (fig. S1A).
Fig. 1. Identification of lncRNAs on replication forks by iROND.
(A) Scheme of iROND procedures in five groups. (B) Two replicative (rep) iROND samples were prepared from mESCs and NIH3T3 cells under five different conditions described in (A). RNA-seq analyses of iROND samples reproducibly identified a set of lncRNAs highly enriched on unperturbed and/or perturbed replication forks in ESCs and NIH3T3 cells, respectively. Samples without click reaction (no click) or with 10 μM thymidine chase (click-chase) were included as negative control. Protease K treatment (click-protease) disrupted the enrichment of lncRNAs on replication forks. (C and D) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of iROND samples validated the enrichment of Lnc956 on replication forks in ESCs, but not in NIH3T3 cells, and the levels were largely increased by HU (C) or aphidicolin (Aph) (D) treatment. The levels of Lnc956 in inputs were normalized by Actb. Data were shown as means ± SEM, two-tailed Student’s t test. ***P < 0.001. Thd, thymidine chase.
We performed iROND RNA-seq in mouse ESCs (mESCs) and NIH3T3 cells in two replicates. Intriguingly, a small number of lncRNAs, annotated according to the NONCODE database (www.noncode.org/), were reproducibly and specifically detected on replication forks in ESCs and NIH3T3 cells, respectively (fold change ≥ 2 when compared to no click and click-chase negative controls) (Fig. 1B and dataset S1). These lncRNAs were no longer detected when the protease K was included in cell lysis to degrade the proteins. Notably, there was no coding RNA reproducibly detected in replication forks (fig. S1B). These observations suggested that some lncRNAs might reside on replication forks and play roles, and their allocation on replication forks may require proteins. Intriguingly, we found that specific regions were reproducibly detected for each lncRNA in RNA-seq (fig. S1C).
To further confirm the presence of specific lncRNAs on replication forks in mESCs, we chose the top three candidate lncRNAs (NONMMUT060689, NONMMUT028956, and NONMMUT006458), which displayed the specific and highest enrichment on replication forks in click-HU condition compared to no click control, for validation by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of iROND samples. As shown, three lncRNAs were highly enriched on unperturbed (normal culture condition) and stalled (HU or aphidicolin treatment condition) replication forks of ESCs, when compared to no click and 1-hour thymidine chase negative controls (Fig. 1, C and D, and fig. S1, D and E). Notably, the fork-allocated Lnc956 increased about twofold after forks were perturbed (Fig. 1, C and D). As a control, lncRNA Neat1, which localizes on nuclear paraspeckles (25), was not detected on replication forks of ESCs (fig. S1F). In sharp contrast to the observation in ESCs, the three lncRNAs were not detected on replication forks of NIH3T3 cells (Fig. 1C and fig. S1, D and E). These observations supported the presence of specific lncRNAs on replication forks of mESCs.
ESC-prevalent lncRNA Lnc956 is essential for fork restart after replication stress
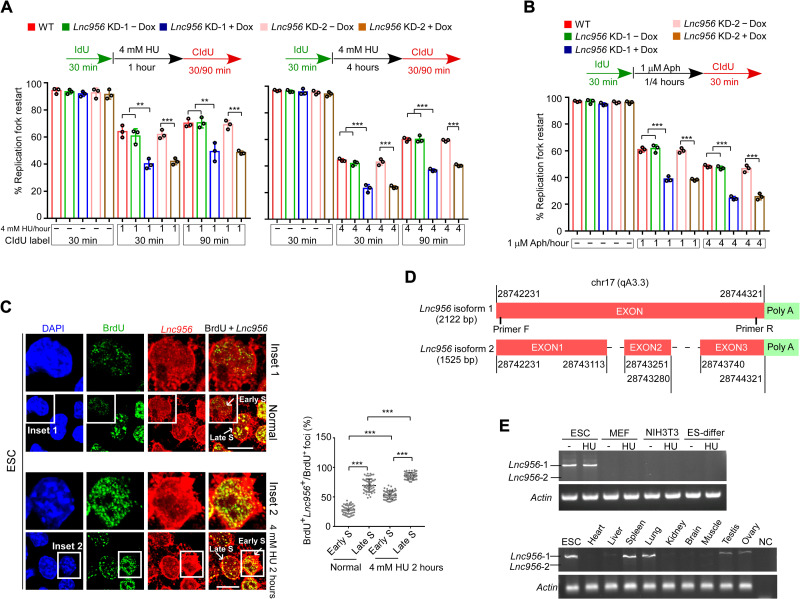
Among the top three replication fork-resided lncRNAs in mESCs, NONMMUT006458 was expressed in both ESCs and differentiated cells, whereas NONMMUT028956 and NONMMUT060689 were prevalently expressed in ESCs when compared to differentiated cells (fig. S2A) or various tissue samples (fig. S2B). Their expression levels in ESCs did not markedly change after HU treatment (fig. S2A). To explore whether the fork-resided lncRNAs have functions on replication forks, we took lncRNAs NONMMUT028956 (Lnc956 for short) and NONMMUT006458 (Lnc458 for short) as examples to examine their functions. Lnc956 was efficiently knocked down (KD) by two independent doxycycline (Dox)–inducible short hairpin RNAs (shRNAs) (fig. S3A), and Lnc458 was KD via two shRNAs (fig. S3B). We then investigated the influence on replication forks under normal and replication stress conditions using DNA fiber assay (26). Lnc956 KD neither affect replication fork speed under normal culture condition (fig. S3C) nor did it cause the nascent DNA degradation under replication stress condition (fig. S3D). However, Lnc956 KD severely impaired the stalled fork restart after HU (Fig. 2A) or aphidicolin treatment (Fig. 2B) for 1 or 4 hours. Intriguingly, Lnc458 KD reduced fork progression rate under normal culture condition (fig. S3E) and mildly compromised stalling fork restart under stress condition (fig. S3F). No nascent DNA degradation was observed in Lnc458 KD ESCs after HU treatment (fig. S3G). Together, these observations suggested that fork-allocated lncRNAs were functional in regulating fork behaviors. In the following studies, we focused on Lnc956 to elucidate its functions and molecular mechanisms.
Fig. 2. ESC-prevalent lncRNA Lnc956 promotes stalled fork restart after replication stress.
(A and B) ESCs were treated with HU (A) for 1 hour (left) or 4 hours (right) or treated with aphidicolin (Aph) for 1 or 4 hours (B). DNA fiber assay revealed that Lnc956 KD impaired the stalled fork restart at all conditions. At least 200 fibers from three independent experiments were analyzed. (C) RNA-FISH revealed the partial colocalization of the Lnc956 with nascent DNA labeled by BrdU for 5 min, indicating the allocation of Lnc956 on replication forks in ESCs (left). ESCs at early and late S phase were indicated. Insets 1 and 2 showed the early S phase cells under normal culture and HU treatment conditions, respectively. Right showed the percentages of Lnc956-containing replication forks in each cell. Fifty cells were analyzed in each group. Scale bars, 10 μm. (D) The information of Lnc956 two isoforms. Red boxes indicate exons. (E) RT-PCR showed the prevalent expression of Lnc956 long isoform in ESCs and other differentiated cells (top) or tissues (bottom). The locations of two primers were shown in (D). ES-differ refers to the ESC-differentiated cells in the presence of retinoid acid. Three independent experiments were repeated with similar results. All data were shown as mean ± SEM from three replications, two-tailed Student’s t test. **P < 0.01 and ***P < 0.001.
To better understand the function of Lnc956, we examined the subcellular localization of Lnc956 by fluorescence in situ hybridization (FISH) combined with 5-bromo-2′-deoxyuridine (BrdU) immunostaining. Nascent DNA fragments were pulse-labeled with BrdU for 5 min. In ESCs, partial BrdU foci showed colocalization with Lnc956 foci. The colocalization rate increased at late S phase and under the replication stress condition (Fig. 2C). In NIH3T3 cells, however, Lnc956 seldomly formed foci and did not show obvious colocalization with BrdU (fig. S4A). We also obtained the full length of Lnc956 by 5′-end and 3′-end RACE (rapid amplification of cDNA ends). Intriguingly, two isoforms were identified (Fig. 2D), with the longer isoform being dominant in both ESCs and tissue samples (Fig. 2E). We found that the sequence targeted by Lnc956 shRNA 2 was missing in short isoform, which failed to be KD by shRNA 2 (fig. S4B). Because short isoform was barely expressed, this did not affect the overall KD efficiency of Lnc956. In the following studies, we only focused on long isoform. To examine whether Lnc956 undergo subcellular translocation upon replication stress in ESCs, we fractionated the cellular compartments into cytoplasm, soluble nucleoplasm, and insoluble chromatin. qRT-PCR analysis revealed a dynamic relocation from cytoplasm to nucleus after HU treatment (fig. S4C). We also investigated the coding ability of several potential coding frames in Lnc956 and failed to detect any translation of protein or peptide (fig. S4D). In addition, Lnc956 is predicted to be partially conserved among mammalian species (fig. S4E). Specifically, mouse and human Lnc956 sequences had around 30% conservation (fig. S4F).
Lnc956 promotes the retention of CMG helicase on chromatin to maintain genomic stability under replication stress
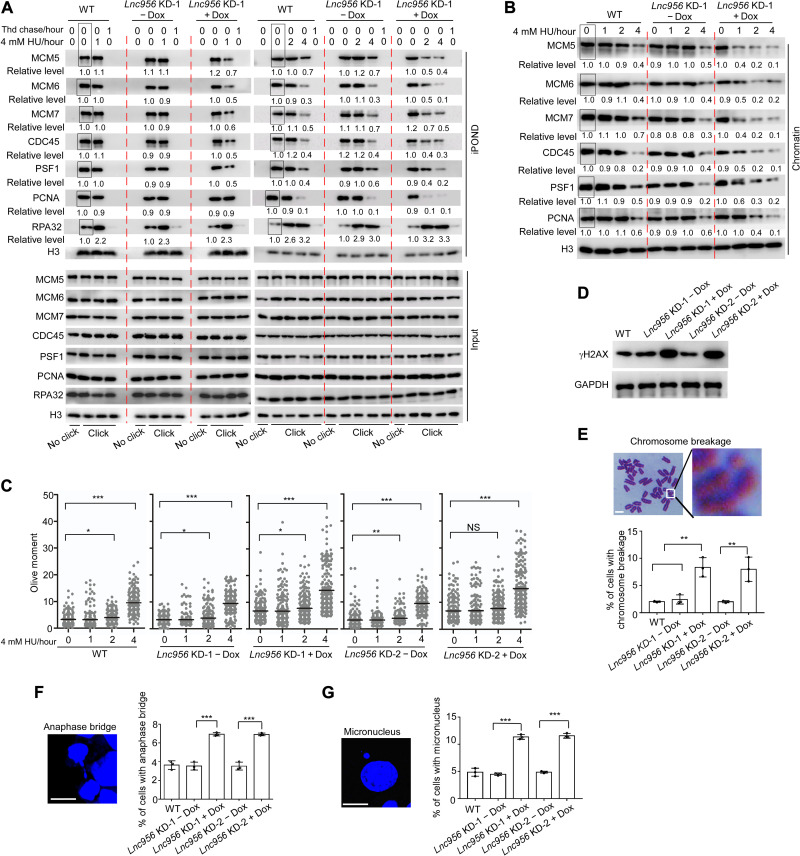
Depletion of Lnc956 compromised the stalled fork restart following replication stress. Failure to restart forks is defined as fork collapse and is associated with replisome dissociation and DNA DSB formation. We went on to examine the influences of Lnc956 depletion on dynamics of the two events after HU treatment. The unperturbed active replication forks were pulse-labeled. Following pulse label, the forks were then isolated by iPOND or were subject to HU treatment to induce fork stalling before isolation. The amounts of replisome core components, including CMG and PCNA, were then evaluated. The levels of these proteins on forks were comparable between Lnc956-proficient and Lnc956-deficient ESCs under unperturbed condition. Upon HU treatment, the CMG components and PCNA in Lnc956-proficient ESCs remained stable on replication forks at the initial 1 and 2 hours of HU treatment but displayed obvious unloading at 4 hours of treatment. Compared to Lnc956-proficient ESCs, the dissociation of CMG components from replication forks was markedly accelerated in Lnc956 KD cells. At 1 hour of HU treatment, CMG components already displayed obvious dissociation in both KD cells. PCNA also dissociated at 2 hour of HU treatment (Fig. 3A and fig. S5A). Further, we isolated chromatin and the chromatin-bound CMG components, and PCNA displayed consistent dynamics (Fig. 3B and fig. S5B). Moreover, recovery for 30 and 90 min following HU treatment (1 or 4 hours) as depicted in Fig. 2A did not change the CMG dissociation pattern (fig. S5, C and D). Thus, Lnc956 KD induced premature unloading of replisome components when replication forks were stalled but had no influence on unperturbed forks. We then examined the dynamics of DNA DSB formation indicative of fork breakage under the same treatment conditions. Compared to Lnc956-proficient ESCs, Lnc956 KD ESCs contained higher basal level of DNA DSBs under normal culture condition, probably due to the elevated fork breakage induced by endogenous replication stress. HU treatment stimulated DSB formation in all ESC groups. However, Lnc956 KD had no influence on the kinetics of DSB formation under stressful condition. Only at 4 hours of HU treatment were the stalled forks broken down to form DSBs in a manner irrelevant to Lnc956 KD (Fig. 3C). Thus, Lnc956 KD induced premature replisome dissociation without accelerating the DNA DSB formation after HU treatment. This observation suggested that Lnc956 might directly regulate the replisome retention on replication forks to prevent fork collapse under stressful conditions.
Fig. 3. Lnc956 ensures the CMG helicase retention on stalled forks to preserve genomic stability.
(A) iPOND showed that Lnc956 KD (KD-1) accelerated the unloading of CMG components (MCM5, MCM6, MCM7, CDC45, and PSF1) from stalled replication forks after HU treatment. The premature dissociation occurred at as early as 1 hour following HU treatment. (B) Lnc956 KD accelerated the dissociation of CMG components from chromatin. In (A) and (B), the relative protein levels were normalized by histone H3, and the levels in samples marked with box were set as 1. (C) DNA DSB was measured by neutral comet assay. Although Lnc956 KD ESCs contained higher basal level of DSBs, the kinetics of HU-induced DSB formation was not affected by Lnc956 KD. At least 200 tails were analyzed in each group. (D) Lnc956 KD cells accumulated more DNA damages than wild-type (WT) ESCs, as indicated by higher level of γH2AX. GAPDH, glyceraldehyde phosphate dehydrogenase. (E) Lnc956 KD mESCs had higher rate of chromosome breakage. At least 50 metaphase spreads in three independent replications were analyzed. (F) Lnc956 KD increased the rate of anaphase bridge. At least 50 anaphase-stage cells from three independent replications were analyzed in each group. (G) Lnc956 KD stimulated micronuclei formation. At least 50 visual fields containing 1000 cells were analyzed in three replications in each group. Experiments were repeated three times with similar results. All data were shown as mean ± SEM, two-tailed Student’s t test. Scale bars, 10 μm. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant.
CMG helicase plays rate-limiting role in DNA replication. Its core component MCM hexamer is loaded on chromatin in G1 phase and cannot be reloaded during S phase. Undesirable dissociation of MCM from chromatin during S phase prevents DNA replication restart, leading to fork collapse and DNA under-replication, which can be manifested by chromosomal breakages, chromosomal bridges during mitosis, and micronuclei formation (27). Lnc956 KD ESCs contained more DNA DSBs under normal culture conditions, as measured by immunoblotting analysis of γH2AX (Fig. 3D). The frequencies of chromosomal breakages (Fig. 3E), mitotic chromosomal bridges (Fig. 3F), and micronuclei formation (Fig. 3G) were significantly elevated in Lnc956 KD ESCs. These defects were further enhanced by HU treatment (fig. S6, A to D).
Loss of Lnc956 causes embryo lethality
DNA replication–associated genomic instability often affects embryogenesis. For instance, MCM4chaos3 mutation destabilizes MCM2-MCM7 hexamer complex and significantly reduces the chromatin-associated hexamers, leading to genomic instability and embryonic lethality (28, 29). On the basis of the genomic instability phenotypes observed in ESCs, we speculated that Lnc956 might play essential roles in embryo development. To test this hypothesis, we went on to generate Lnc956 knockout (KO) mice by CRISPR-Cas9–mediated mutation strategy, which aimed to delete the sequence spanning the 5′ end and 3′ end of Lnc956 (fig. S7A). The Lnc956 sequence did not contain any coding gene. In addition, mouse ENCODE data showed that the H3K27ac and H3K4me1 signal peaks, indicative of potential promoter and enhancer, respectively, overlapped with Lnc956 sequence only in several adult tissues (fig. S7B). Moreover, two nearby genes Fkbp5 and Armc12, which are potentially subject to the regulation of the promoter or enhancer, are dispensable for embryogenesis as demonstrated by gene KO in mice (30, 31). Thus, the Lnc956 KO defects on embryogenesis, if any, could not be attributed to disturbance on other genes.
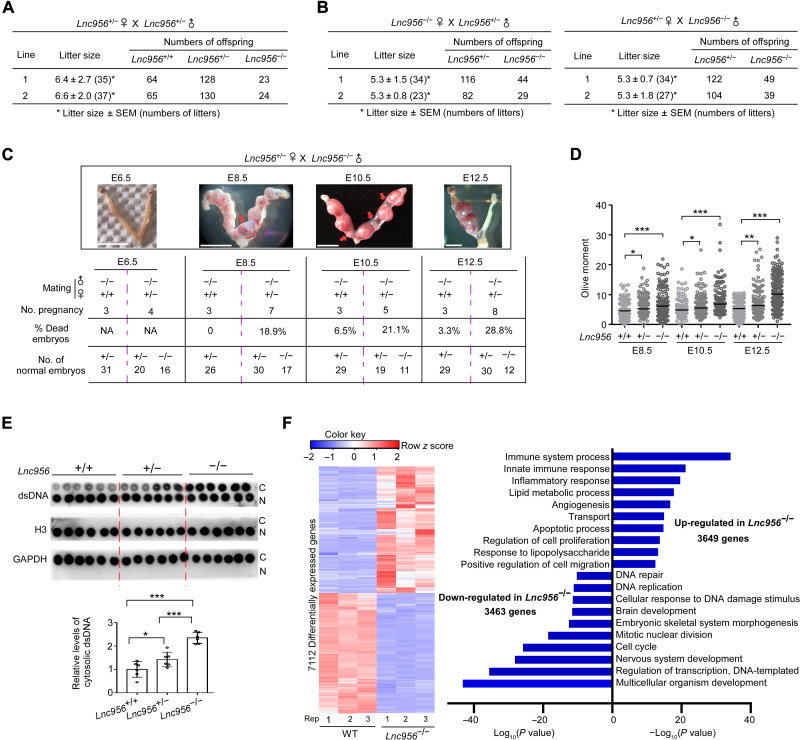
Two KO mouse lines were obtained from the same targeting design. Lnc956 KO mice were validated by Sanger sequencing of amplified DNA fragment (fig. S7C) and were genotyped by PCR examination of tail DNA samples (fig. S7D). Because the two mouse lines harbored similar fragment deletion (fig. S7C), we used line 1 for detailed investigations. No gross abnormality was detected in heterozygous mice. However, crossing of heterozygous mice did not produce offsprings in normal Mendelian ratio, with the homozygotes severely underrepresented (Fig. 4A). Other breeding strategies (Lnc956−/− females × Lnc956+/− males, and Lnc956+/− females × Lnc956−/− males) led to the consistent results (Fig. 4B). Thus, the complete loss of Lnc956 caused embryonic lethality.
Fig. 4. Lnc956 KO causes embryo lethality.
(A and B) Mating experiments with different strategies revealed that complete loss of Lnc956 caused embryonic death. (C) Representative images of uterus collected from Lnc956+/− pregnant females mated with Lnc956−/− males at E6.5, E8.5, E10.5, and E12.5 (top). Red arrows indicated the absorbed dead embryos. Bottom showed the embryo information at each developmental stage. The mating (female Lnc956+/+ × male Lnc956−/−) was used as control. NA, Null acl. Scale bars, 1 cm. (D) Single cells were prepared from embryos. Neutral comet assay showed that loss of one or both allels of Lnc956 induced higher level of DNA DSBs when compared to WT counterparts. At least 200 tails were analyzed in each group. Data were from three independent replications. (E) Dot immunoblotting (top) showed that Lnc956−/− embryonic cells contained higher level of cytosolic dsDNA than Lnc956+/− or Lnc956+/+ embryonic cells. Bottom showed the relative levels of cytosolic dsDNA from three independent replications. N refers to nucleus, and C refers to cytoplasm. (F) Heatmap (left) and gene ontology (GO) enrichments (right) of differentially expressed genes (DEGs) between WT and Lnc956−/− embryos at E12.5. All data were shown as mean ± SEM, two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
We next examined the time window when the embryos die. Lnc956+/− females were mated with Lnc956−/− males, and the blastocysts were collected from uterus. The genotypes of blastocysts displayed normal Mendelian ratio, indicating that Lnc956 KO did not impair the preimplantation development. We then collected after implantation embryos from embryonic day 6.5 (E6.5) to E12.5. Compared to the control (Lnc956+/+ females × Lnc956−/− males), Lnc956+/− females mated with Lnc956−/− males produced more dead or absorbed embryos at all examined developmental stages (Fig. 4C). This observation suggested that Lnc956−/− embryos might die at stages around gastrulation.
To find whether embryo lethality is associated with DNA damage, we collected embryos at E8.5, E10.5, and E12.5. Neutral comet assay showed that the DNA DSB level was increased in Lnc956+/− embryos when compared to Lnc956+/+ embryos and was further elevated in Lnc956−/− embryos at all stages (Fig. 4D). DNA replication–associated genomic instability is usually characterized by the formation of cytosolic DNA fragments, which are able to activate the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) pathway and lethal inflammation (32). We then examined whether Lnc956 KO increased cytosolic DNA fragments and induced inflammation response in embryos. Dot immunoblotting analysis of E12.5 embryos showed that Lnc956−/− embryonic cells contained higher level of cytosolic double-strand DNA (dsDNA) than Lnc956+/− or Lnc956+/+ counterparts (Fig. 4E). Further, we performed RNA-seq to compare the mRNA expressions between wild-type (WT) and Lnc956−/− embryos at E12.5. A total of 7112 differentially expressed genes (DEGs) were identified to show consistent up- or down-regulation between two groups (fold change ≥ 2) (Fig. 4F and dataset S2). Gene ontology (GO) enrichment analyses of these DEGs revealed that innate immune response and inflammation response were the top processes enriched in DEGs up-regulated in Lnc956−/− embryos, whereas processes including cellular response to DNA damage stimulus, DNA replication, and repair were enriched in DEGs down-regulated in Lnc956−/− embryos (Fig. 4F and fig. S8). These lines of evidence suggested that Lnc956 KO evoked severe DNA DSBs and cytosolic dsDNA, which induced innate immune and inflammation responses in postimplantation embryos. These results suggested that Lnc956 KO embryos died of the genomic instability and associated lethal inflammation.
Lnc956-TRIM28-HSP90B1 form complex on stalled replication forks in an interdependent manner
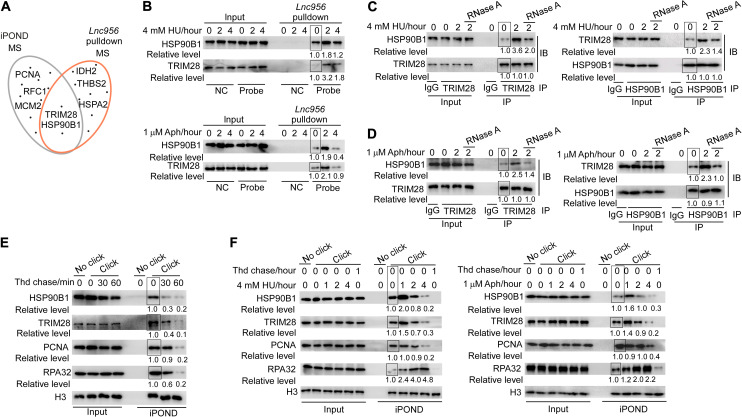
Lnc956 regulates CMG/replisome retention on chromatin to preserve genomic stability upon replication stress. To find out the underlying molecular mechanisms, we first examined the ATR signaling, whose activation failure can induce replisome dissociation (33). Notably, the ATR-CHK1 signaling central to replication checkpoint was normally activated, and the activation level was higher in Lnc956 KD ESCs than in Lnc956 proficient ESCs, probably due to the elevated basal level of DNA replication stress (fig. S9A). This observation indicated that the regulation of Lnc956 on replisome retention was not mediated by ATR signaling. We then performed in vivo RNA pulldown combined with mass spectrometry analysis to identify the potential interaction proteins of Lnc956 (dataset S3). Meanwhile, we also performed iPOND followed by mass spectrometry to find proteins on replication forks of ESCs (dataset S3). By comparing the two sets of protein list, we identified TRIM28 and HSP90B1 (also called GRP94) as overlapped candidates (Fig. 5A) and proposed that Lnc956 might interact with TRIM28 and HSP90B1 on replication forks. To test this hypothesis, we first examined the association of Lnc956 with both proteins. Immunoblotting analysis of in vivo RNA pulldown samples showed that Lnc956 interacted with TRIM28 and HSP90B1, and the association was stimulated by HU or aphidicolin treatment (Fig. 5B). The direct interaction of Lnc956 with TRIM28 and HSP90B1 was further validated by in vitro RNA pulldown assay, and the region involved in binding to TRIM28 and HSP90B1 was mapped to C terminus of Lnc956 (fig. S9, B and C). Intriguingly, TRIM28 and HSP90B1 physically interacted, and their association was robustly enhanced by treatment with HU (Fig. 5C) or aphidicolin (Fig. 5D). Notably, the stress-induced TRIM28-HSP90B1 interaction was repressed by inclusion of RNase A in cell lysis, implicating that RNAs were required to enhance TRIM28-HSP90B1 association (Fig. 5, C and D). We also validated the localization of TRIM28 and HSP90B1 on replication forks by iPOND assay (Fig. 5E). Recruitment of the two proteins to replication forks was dynamically regulated by replication stress in WT ESCs. Initially, HU or aphidicolin treatment stimulated the accumulation of two proteins on replication forks. However, the amount of fork-resided proteins gradually decreased along with the replication stress persistence (Fig. 5F). This dynamics was different from the pattern observed for other components of replisomes including PCNA and RPA32 (Fig. 5F). The decrease could be attributed to the fact that prolonged replication stress continuously induced stalling of newly fired forks (34), and the increasing numbers of stalled forks gradually exhaust the available TRIM28-HSP90B1 pool, leading to the reduced amount of TRIM28-HSP90B1 on each replication fork.
Fig. 5. Lnc956-TRIM28-HSP90B1 associate with each other on replication forks.
(A) Mass spectrometry (MS) analyses identified TRIM28 and HSP90B1 as candidates to reside on replication forks and interact with Lnc956. (B) In vivo RNA pulldown validated the interaction of Lnc956 with HSP90B1 and TRIM28. The interaction was enhanced by HU (top) or aphidicolin (Aph) treatment (bottom). Pulldown assay using sense probe was included as negative control (NC). (C and D) Reciprocal immunoprecipitation (IP) validated the physical interaction of HSP90B1 with TRIM28. The interaction was enhanced by HU (C) or aphidicolin (D) treatment and required the mediation of RNA species. IgG, immunoglobulin G. (E) Immunoblotting (IB) of iPOND samples validated the localization of HSP90B1 and TRIM28 on replication forks of ESCs under normal culture condition. Cells were chased with 10 μM thymidine chase for different times. (F) The accumulation of HSP90B1 and TRIM28 on replication forks was dynamically regulated by HU (left) or aphidicolin treatment (right). The relative protein levels were normalized by input (B to D) or by histone H3 (E and F), and the levels in samples marked with box were set as 1. In (B) to (F), all experiments were repeated three times with similar results.
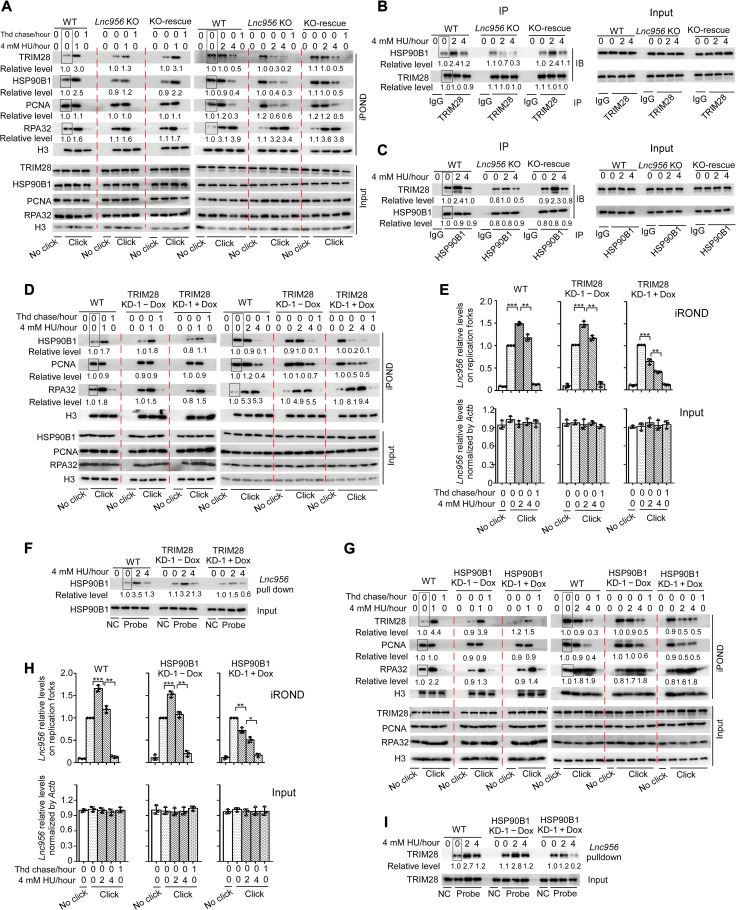
To gain more evidence supporting the physical association of Lnc956-TRIM28-HSP90B1 on replication forks, we examined the interdependence of the three components on replication forks. To this end, we established Lnc956 KO ESCs (fig. S10A), which had normal S phase cell population (fig. S10B). Like Lnc956 KD ESCs, KO ESCs displayed same defects in stalling fork restart and CMG dissociation, which could be fully rescued by reexpression of Lnc956 (fig. S10, C to E). In Lnc956 KO ESCs, the loading of TRIM28 and HSP90B1 on replication forks was not affected under unperturbed condition. However, their recruitment to stalling forks was impaired by Lnc956 KO under HU treatment condition (Fig. 6A). Similarly, Lnc956 KO had no influence on TRIM28-HSP90B1 association under unperturbed condition but compromised their interaction upon replication stress (Fig. 6, B and C). Reexpression of full-length Lnc956 in KO ESCs completely resued the phenotypes (Fig. 6, A to C). However, reexpression of truncate Lnc956 (Δ1500-2122), which did not interact with TRIM28-HSP90B1, had no rescue effect (fig. S11, A to C). Thus, under perturbed condition, the accumulation of TRIM28 and HSP90B1 on stalled forks and their increased association required Lnc956. Next, we efficiently KD TRIM28 by two independent Dox-inducible shRNAs (fig. S12A) and examined its influences on HSP90B1 and Lnc956. Similar to Lnc956 KO, TRIM28 KD compromised the accumulation of HSP90B1 on replication forks only after HU treatment, and no influence was detected under unperturbed condition (Fig. 6D and fig. S12B). TRIM28 KD reduced the cell population at S phase (fig. S12C), which made it difficult to compare the enrichment of Lnc956 on replication forks between TRIM28-proficient and TRIM28-deficient ESCs. We therefore assessed its influence on the dynamics of Lnc956 enrichment on replication forks before and after HU treatment. In TRIM28-proficient ESCs, HU treatment increased the level of Lnc956 on stalled replication forks. However, HU treatment decreased the level of Lnc956 on stalling forks in TRIM28 KD ESCs (Fig. 6E and fig. S12D). Concordantly, the Lnc956-HSP90B1 association was impaired under HU treatment condition (Fig. 6F and fig. S12E). Likewise, HSP90B1 KD (fig. S13A), which did not change the S phase cell population (fig. S13B), exhibited similar effects on TRIM28 and Lnc956 under stressful condition (Fig. 6, G to I, and fig. S13, C to E). Together, these data suggest that replication stress/fork stalling stimulates the accumulation and physical association of Lnc956-TRIM28-HSP90B1 on replication forks. Notably, ATR replication checkpoint regulated this process. Inhibition of ATR activation by specific inhibitor VE-821 (fig. S14A) compromised the accumulation of TRIM28, HSP90B1 (fig. S14B), and Lnc956 (fig. S14C) on stalled replication forks. TRIM28-HSP90B1 association was also reduced by VE-821 (fig. S14, D and E). Although the basal levels of Lnc956, TRIM28, and HSP90B1 were present on unperturbed replication forks, they did not influence each other and therefore did not form an interdependent complex.
Fig. 6. Lnc956-TRIM28-HSP90B1 form an interdependent complex on stalled replication forks.
(A) iPOND showed that Lnc956 KO impaired the recruitment of HSP90B1 and TRIM28 on stalled replication forks at all time points of HU treatment. Reexpression of Lnc956 (KO-rescue) rescued the defects. (B and C) Reciprocal IP showed that Lnc956 KO weakened the TRIM28-HSP90B1 interaction under replication stress. The defect was rescued by reexpression of Lnc956. (D) TRIM28 KD (KD-1) reduced the accumulation of HSP90B1 on stalled replication forks. (E) iROND were performed using same numbers of cells. qRT-PCR analysis of iROND samples showed that Lnc956 levels on replication forks were increased by fork stalling in TRIM28-proficient ESCs, whereas fork-resided Lnc956 was reduced upon fork stalling in TRIM28 KD ESCs. (F) TRIM28 KD compromised the association of HSP90B1 with Lnc956 as revealed by in vivo RNA pulldown. (G to I) Similarly, HSP90B1 KD (KD-1) decreased the accumulation of TRIM28 (G) and Lnc956 (H) on stalled replication forks and impaired the interaction of TRIM28 with Lnc956 (I). In vivo RNA pulldown using sense probe was set as negative control (NC) in (F) and (I). The protein levels were normalized by histone H3 in (A), (D), and (G) and by input in (B), (C), (F), and (I). The relative protein levels in samples marked with box were set as 1. All experiments were repeated three times with consistent results. Data were shown as mean ± SEM, two-tailed Student’s t test. *P < 0.05, **P < 0.01, and ***P < 0.001.
Because Lnc956-TRIM28-HSP90B1 formed complex in an interdependent manner on stalling replication forks, we speculated that loss of each functional component would exhibit similar phenotypes. TRIM28 KD or HSP90B1 KD accelerated the dissociation of replisome from chromatin, as indicated by the reduced levels of CMG components and PCNA on purified replication forks (fig. S15, A and B) or on whole chromatin (fig. S15, C and D). Consistently, stalled fork restart was compromised in TRIM28 KD (fig. S15E) or HSP90B1 KD ESCs (fig. S15F). Notably, Lnc956 KO ESCs expressing the truncate Lnc956 (Δ1500-2122), which did not bind TRIM28-HSP90B1 and had no rescue effect on TRIM28 and HSP90B1 (fig. S11, A to C), displayed same defects in CMG dissociation (fig. S11, A and D) and stalled fork restart (fig. S11E).
Lnc956-TRIM28-HSP90B1 physically interacts with MCM2 to MCM7 hexamer
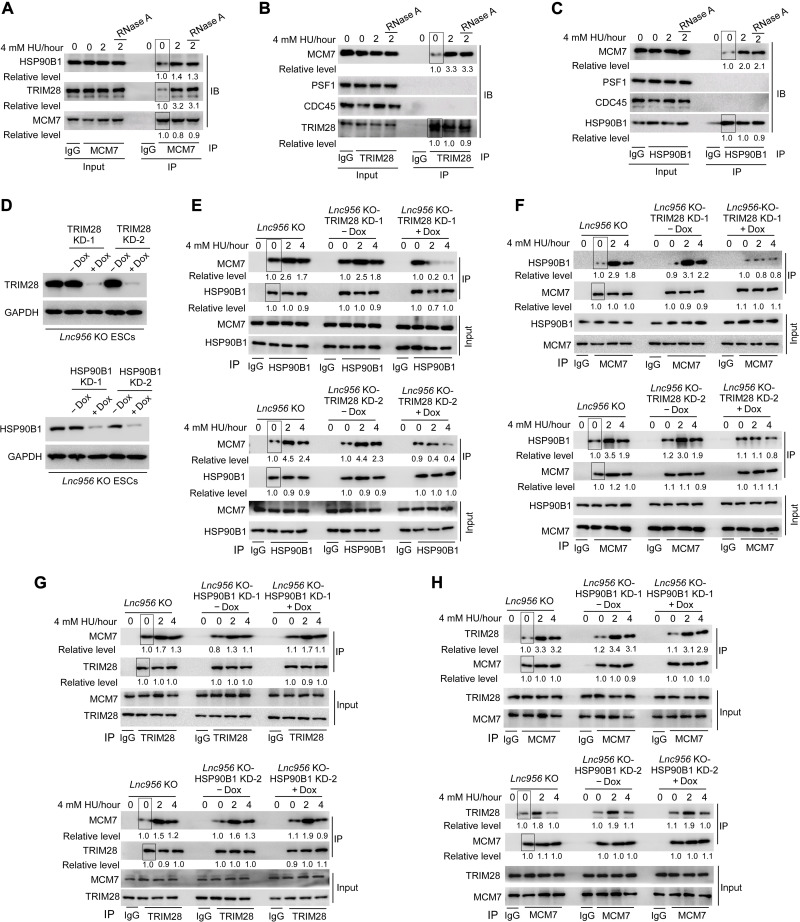
Next, we sought to understand how the Lnc956-TRIM28-HSP90B1 complex regulated CMG/replisome retention on chromatin under replication stress. We first examined whether this complex physically interacted with replisomes. Proteomic analysis of TRIM28 immunoprecipitates identified a long list of potential interaction proteins (dataset S3). HSP90B1, the validated interaction partner, was in the list. Intriguingly, we detected all six components of MCM2 to MCM7 hexamer (MCM2, MCM3, MCM4, MCM5, MCM6, and MCM7), whereas the other components of active CMG complex, CDC45 and GINS, were not detected (dataset S3). This suggested that Lnc956-TRIM28-HSP90B1 might interact with MCM hexamer. We then went on to validate the association between the two complexes. MCM7 could pull down TRIM28 and HSP90B1 (Fig. 7A). Similarly, TRIM28 or HSP90B1 coprecipitated with MCM7 but not CDC45 or GINS (Fig. 7, B and C). In line with the induced association of Lnc956-TRIM28-HSP90B1 under replication stress, Lnc956-TRIM28-HSP90B1-MCM interaction was elevated after HU treatment (Fig. 7, A to C). Thus, Lnc956-TRIM28-HSP90B1 complex physically associated with MCM hexamer upon replication stress.
Fig. 7. Lnc956-TRIM28-HSP90B1 complex physically interacts with MCM hexamer via TRIM28.
(A) MCM7 coimmunoprecipitated with TRIM28 and HSP90B1. The interaction was enhanced by HU treatment and did not rely on RNA species. (B and C) Reciprocally, TRIM28 (B) and HSP90B1 (C) could immunoprecipitate with MCM7 but not PSF1 or CDC45. The interaction was also stimulated by HU treatment and did not rely on RNAs. (D) KD of TRIM28 (top) or HSP90B1 (bottom) in Lnc956 KO mESC using two independent Dox-inducible shRNAs. (E and F) KD of TRIM28 in Lnc956 KO ESCs impaired the association of HSP90B1 with MCM7, as revealed by reciprocal immunoprecipitation. (G and H) However, KD of HSP90B1 in Lnc956 KO ESCs had no influence on the interaction between TRIM28 and MCM7, as shown by reciprocal immunoprecipitation. The relative protein levels were normalized by input, and the levels in samples marked with box were set as 1. All experiments were repeated three times with consistent results.
We then investigated which component of Lnc956-TRIM28-HSP90B1 complex mediated the association. To dissect whether Lnc956 is necessary for interaction with MCM, we treated the cell lysis with RNase A to degrade Lnc956 before conducting immunoprecipitation. The interaction between MCM7-TRIM28 and MCM7-HSP90B1 was not affected by RNase A treatment (Fig. 7, A to C), suggesting that Lnc956 was dispensable for association with MCM. Consistently, in vivo (dataset S3) or in vitro RNA pulldown assay (fig. S16A) did not detect the interaction between Lnc956 and MCM7. To understand the role of TRIM28 and HSP90B1 in mediating the interaction with MCM, we individually depleted TRIM28 or HSP90B1 in Lnc956 KO ESCs (Fig. 7D). Reciprocal immunoprecipitation showed that TRIM28 KD with two independent shRNAs severely impaired the association between MCM7 and HSP90B1 under replication stress condition (Fig. 7, E and F). However, HSP90B1 KD did not affect MCM7-TRIM28 interaction (Fig. 7, G and H). These data together supported that TRIM28 mediated the interaction between Lnc956-TRIM28-HSP90B1 complex and MCM hexamer. A previous study reported the association of TRIM28 (also known as KAP1) with MCM and PCNA at heterochromatin regions (35). We wondered whether the Lnc956-TRIM28-HSP90B1 localized at heterochromatin. Counterstaining of HP1 (marker of heterochromatin) with Lnc956 revealed partial colocalization (fig. S16B). Thus, the function of Lnc956-TRIM28-HSP90B1 was not limited to heterochromatin regions.
HSP90B1 regulates the retention of CMG helicase under replication stress
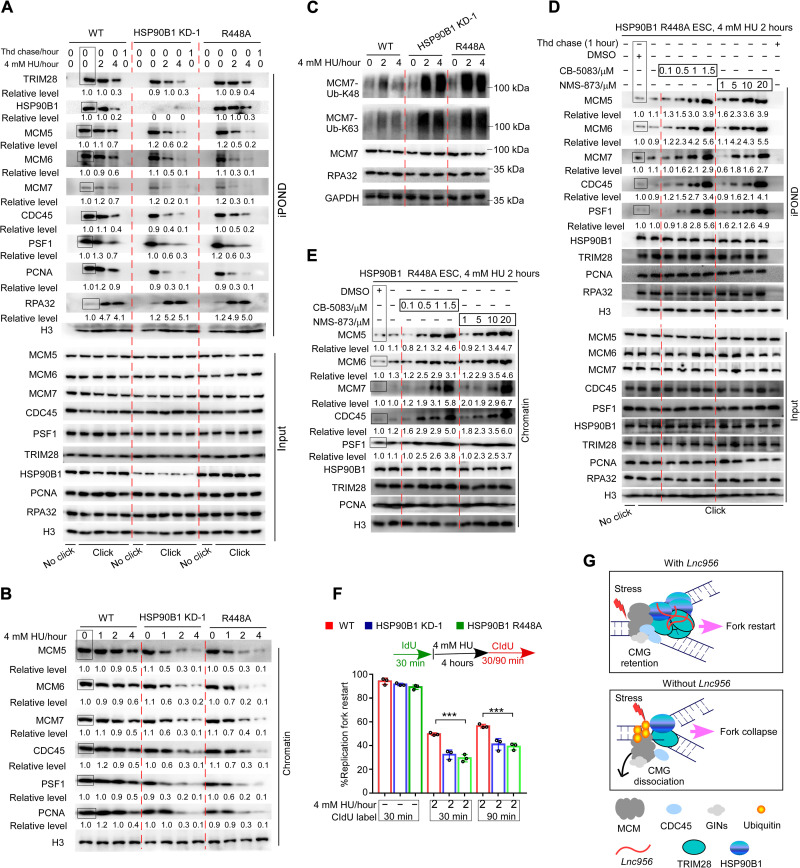
Unloading of CMG from chromatin is regulated by a series of sequential events. Initially, MCM7 in CMG complex undergoes K48 polyubiquitylation. Following ubiquitylation, the CMG complex is extracted from chromatin by segregase P97 (36). The ubiquitylation is prohibited by the DNA structure of active forks during elongation (37). HSP90B1 is a chaperone protein, which has adenosine triphosphatase activity and functions as protein folding reservoir and stabilizes its clients (38–40). Considering that DNA structural change during fork stalling could induce MCM7 ubiquitylation and CMG unloading, we speculated that HSP90B1 may buffer the stress-induced changes and prevent the ubiquitylation reaction and CMG extraction by P97. To test this hypothesis, we mutated Arg448 in HSP90B1 to Alanine (HSP90B1-R448A), which reduces the hydrolysis activity by more than 85% (40), and investigated the influences on CMG retention, MCM7 ubiquitylation, and stalling fork restart. R448A mutation did not impair the recruitment of TRIM28 and HSP90B1 to replication forks (Fig. 8A). However, it accelerated the unloading of CMG components in a manner similar to HSP90B1 KD (Fig. 8, A and B). We then examined the ubiquitylation of MCM7 proteins, which were enriched by immunoprecipitation through MCM7 antibody. MCM7 underwent K48 and K63 ubiquitylation after HU treatment, and the modification level increased with the stress intensity (41, 42). Notably, HSP90B1-R448A mutation, like HSP90B1 KD, sharply increased both types of ubiquitylation levels (Fig. 8C), indicating that HSP90B1 suppressed the MCM7 ubiquitylation. HSP90B1 recruitment to stalling forks was compromised in TRIM28 KD, Lnc956 KO, or truncate Lnc956 (Δ1500-2122)-rescued ESCs. Concordantly, MCM7 ubiquitylation was increased in these ESCs (fig. S16, C and D). These results together support the regulatory role of HSP90B1 on MCM7 ubiquitylation.
Fig. 8. HSP90B1 stabilizes CMG helicase under replication stress.
(A and B) iPOND (A) and chromatin purification (B) analyses showed that HSP90B1 mutation (R448A) did not affect the recruitment of HSP90B1 and TRIM28 to replication forks. However, R448A mutation, similar to HSP90B1 KD, induced the premature dissociation of CMG helicase from stalled replication forks. (C) HSP90B1 R448A mutation robustly increased the K48 and K63 ubiquitylation of MCM7 under the HU treatment condition. (D and E) HSP90B1 R448A mutant ESCs were treated with HU to induce K48 and K63 ubiquitylation of MCM7. Under this condition, blocking P97 activity by CB-5083 or NMS-873 preserved CMG retention on stalled replication forks, as shown by iPOND (D) and chromatin purification analyses (E). (F) HSP90B1 R448A mutation impaired the stalled fork restart. At least 200 fibers from three independent experiments were analyzed. (G) The working model of Lnc956-TRIM28-HSP90B1 complex on replication forks. The relative protein levels were normalized by histone H3 in (A), (B), (D), and (E). The levels in samples marked with box were set as 1. All experiments were repeated three times with similar results. Data were shown as mean ± SEM, two-tailed Student’s t test. ***P < 0.001.
To further clarify the involvement of the ubiquitylation-P97 pathway in the extraction of CMG, we blocked P97 activity in HSP90B1-R448A mutant ESCs and examined whether it could prevent the extraction of CMG. P97 inhibitor NMS-873 or CB-5083 (43, 44) prevented the extraction and increased the retention of CMG components in a dose-dependent manner, as revealed by iPOND (Fig. 8D) and chromatin purification analyses (Fig. 8E). Together, these data suggested that HSP90B1 promoted the retention of CMG helicase probably through blocking the MCM7 ubiquitylation-P97 extraction pathway. CMG helicase cannot be reloaded once unloaded during S phase, and we speculated that HSP90B1-R448A mutation compromised stalled fork restart. HSP90B1-R448A decreased the fork restart rate to a level comparable to that in HSP90B1 KD ESCs (Fig. 8F).
Ectopic expression of Lnc956 enhances CMG/replisome retention in somatic cells
Depletion of Lnc956 in mESCs by KO or shRNA-mediated KD revealed its essential role in promoting the Lnc956-TRIM28-HSP90B1 complex assembly as well as CMG retention on chromatin under replication stress condition. To further illustrate whether Lnc956 is sufficient to drive Lnc956-TRIM28-HSP90B1 association and stimulate CMG retention, we ectopically expressed Lnc956 in NIH3T3 cells (fig. S17A) and investigated the effects. As shown, expression of Lnc956 in NIH3T3 increased the fork-allocated levels of Lnc956 (fig. S17B), TRIM28, and HSP90B1 (fig. S17C). Concordantly, TRIM28-HSP90B1 interaction was enhanced by Lnc956 overexpression (fig. S17D). As a result, Lnc956 expression promoted the CMG retention on chromatin as revealed by iPOND (fig. S17C) and chromatin purification analyses (fig. S17E). The stalling fork restart after HU treatment was also stimulated by overexpression of Lnc956 (fig. S17F). These results supported that introduction of Lnc956 in somatic cells was able to enhance the association and function of Lnc956-TRIM28-HSP90B1 complex on replication forks, albeit with low efficiency when compared to the ESC context. Other additional ESC-specific factors could be required. In summary, we identified an ESC-prevalent lncRNA Lnc956 that localizes on replication forks and potentiates the assembly of Lnc956-TRIM28-HSP90B1 complex under replication stress condition. This complex physically interacts with MCM hexamer via TRIM28 and regulates MCM2 to MCM7 retention on chromatin via HSP90B1 (Fig. 8G). Lnc956-TRIM28-HSP90B1 complex formation significantly increases the local concentrations of TRIM28 and HSP90B1, conferring cells with greater ability to stabilize CMG helicase on chromatin and to ensure genomic stability under replication stress condition.
DISCUSSION
Replication stress is the major source of endogenous DNA damages. Dysregulation of replication stress response can cause variable human diseases ranging from developmental defects to cancers (15, 45). Numerous efforts have been made to understand the local and global cellular responses to stalled forks (17). Owing to the innovative methods such as iPOND, an increasing numbers of proteins were identified on replication forks under normal and/or stressed conditions. These regulatory proteins play critical roles in ensuring fork intact and restart under stressed conditions (17, 46). To our knowledge, it remains unexplored whether there exist noncoding regulatory RNAs on replication forks. LncRNAs are able to enhance the functions of their interacting protein by driving efficient protein condensation (23, 24). Using PSC as a model, we reported the presence of functional lncRNA on stalled replication forks and revealed its critical role in regulating CMG helicase retention. Stable retention of CMG helicase confers PSCs superior abilities to restart stalled forks and preserve genomic stability. In addition, we identified a lncRNA Lnc956 essential for embryogenesis and provided clue to decipher the cause of recurrent pregnancy loss in clinic.
Certain levels of Lnc956, TRIM28, and HSP90B1 were detected on unperturbed replication forks. However, depletion of each component had no influence on the other two components under unperturbed condition, suggesting that Lnc956, TRIM28, and HSP90B1 did not form an interdependent complex on unperturbed forks. Moreover, depletion of each component had no influence on CMG retention on unperturbed forks, implicating that they were dispensable for the unstressed forks. Upon fork stalling, more Lnc956, TRIM28, and HSP90B1 were recruited to forks. In addition, depletion of one component impaired the accumulation as well as association of the other two on stalled forks, indicating that they formed an interdependent complex upon replication stress. This complex preserved the retention of CMG on stalled forks and promoted the stalled fork restart. The assembly of Lnc956-TRIM28-HSP90B1 complex on stalled forks was regulated by replication stress–evoked ATR signaling. ATR kinase can phosphorylate and activate many downstream proteins. Whether TRIM28 or HSP90B1 was ATR target and how ATR signaling cascade stimulates the assembly of this complex warrant future investigation. Previous studies demonstrated that lncRNAs (for instance, NORAD, Xist, and Neat1) could drive the condensation and phase separation of interacting RNA binding proteins (RBPs), thereby drastically increasing the local concentration and probably the activity of the RBPs (24, 47, 48). In the future, it would be intriguing to clarify whether Lnc956 induces the Lnc956-TRIM28-HSP90B1 complex formation through phase separation.
Lnc956-TRIM28-HSP90B1 promotes CMG retention under replication stress. This regulation was direct rather than an indirect consequence of blocking fork breakage. First, CMG helicase unloading induced by Lnc956 depletion occurred independent of the induction of fork breakage and DSB formation. Second, Lnc956-TRIM28-HSP90B1 physically associates with MCM hexamer, providing a basis for direct regulation of Lnc956-TRIM28-HSP90B1 on CMG helicase. Previous studies suggested that the major functions of ATR replication checkpoint were to repair stalled forks, prevent fork cleavage, and modulate replisome function rather than regulate replisome stability (49–51). To date, it still remains unclear whether replisome unloading is simply the outcome of fork collapse or underlies fork collapse (17). The Lnc956-TRIM28-HSP90B1 complex was assembled under the control of ATR signaling, therefore being a component of replication checkpoint. Our work suggested that the ATR replication checkpoint could also actively regulate replisome stability, which underlies fork collapse in certain cellular context. In summary, we identified an lncRNA-based mechanism that is a component of ATR replication checkpoint and directly regulates the retention of CMG to prevent stalled fork collapse. This mechanism plays essential roles in embryogenesis. Future works are warranted to investigate the human Lnc956 expressions and functions and its relevance to human developmental abnormalities.
METHODS
Generation of Lnc956 KO mice
The protocols of gene KO, animal care, and manipulation were approved by the Institutional Animal Care and Use Committee of the Kunming Institute of Zoology, Chinese Academy of Sciences. The Lnc956+/− C57Bl/6J mice were generated by Cyagen Company (www.cyagen.com) using CRISPR-based gene editing technology. The guide RNAs (gRNAs) were designed on the basis of mouse Lnc956 genome region sequence (chr17 qA3.3: 28742231-28744321). Cas9 mRNA and a pair of gRNAs (5′-AGCGGGCAGCGCCCGGGAGGAGG-3′; 5′-TTGGAAGAGTAACACGTTTTGGG-3′) were coinjected into one-cell embryos to generate targeted KO mice. The genome DNA from mouse tails was used for genotype confirmation by PCR amplification and DNA sequence analysis. The PCR program was 94°C for 5 min (94°C for 15 s, 60°C for 30 s, and 72°C for 30 s), 35 cycles, 72°C for 15 min. Genotyping primers were shown in table S1.
Cell culture
The MEFs were obtained from embryos of CD1 mice at E13.5. Mitomycin-treated MEFs were used as coculture feeder cells for derivation and culture of mESCs. WT and Lnc956−/− mESCs were derived from the inner cell mass of E3.5 blastocysts.
The culture of MEFs, NIH3T3, and mESCs were described in detail in our previous papers (14). Briefly, MEFs and NIH3T3 cells were maintained in Dulbecco’s minimum essential medium (DMEM) (Gibco, 11965) with 10% fetal bovine serum (Gibco, 10099141C). mESCs were cultured in DMEM/F12 medium supplemented with 20% KO serum replacement (Gibco, 10828028), 2 mM l-glutamine (Sigma-Aldrich, G8540), 1 mM sodium pyruvate (Gibco, 11360070), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, M7522), 1% nonessential amino acids (Gibco, 11140-035), and mouse leukemia inhibitory factor (1000 U/ml; Millipore, ESG1107). All cells were grown at 37°C and 5% CO2 atmosphere. Regular mycoplasma testing was performed using the LookOut Mycoplasma PCR detection (Sigma-Aldrich, catalog no. MP0035).
Lentivirus-mediated gene manipulation
The shRNA sequences and primers were listed in table S2. Following the manufacturer’s instructions (Open Biosystems, QCHENG BIO, QCP0703), all the shRNA sequences were constructed into pTRIPZ lentivirus tet-on inducible shRNAmir system, except the Lnc458 shRNA sequences that were constructed into PLKO.1. The Lnc956 and Trim28 coding regions (CDS) were inserted into pTOMO–internal ribosomal entry site (IRES)–enhanced green fluorescent protein (EGFP) lentiviral expression vector (Addgene, #26291). Lnc956 ORF1, ORF2, ORF3, and Floped CDS containing Flag-tag were inserted into pcDNA3.1 vector. For the virus package, the shRNAmir or pTOMO-IRES-EGFP lentiviral expression vectors were mixed with packaging plasmids psPAX2 and PMD2.G (shRNAmir or pTOMO-IRES-EGFP: psPAX2: PMD2.G = 2:1:1) and cotransfected into 293T cells. For the groups of shRNAmir-mediated gene KD, 48 hours after lentivirus infection, the puromycin (0.5 μg/ml) was added into culture medium for selection. For the groups of pTOMO-IRES-EGFP–mediated gene expression, GFP-positive cells were sorted by flow cytometry for extended culture and analysis.
Isolation of proteins on nascent DNA
iPOND was performed as described in previous paper (19). To harvest enough S phase cells, NIH3T3 cells were synchronized by 2 mM thymidine (Sigma-Aldrich, T1895) for 15 hours, followed by fresh medium culture for 2.5 hours. For each sample, 108 synchronized NIH3T3 or normal cultured mESCs were incubated with 10 mM EdU (Life Technologies, A10044) for 10 min, followed by phosphate-buffered saline (PBS) wash and HU treatment. Thymidine chase (10 μM) was included as chromatin control. Next, the cells were fixed with 1% formaldehyde (Sigma-Aldrich, F1635) and then quenched with 0.125 M glycine (Sangon Biotech, A100167). The cells were scraped and permeabilized by ice-cold 0.25% Triton X-100/PBS for 30 min at room temperature. After washing with 0.5% bovine serum albumin (BSA)/PBS, the cells were resuspended in click reaction buffer including 10 mM sodium ascorbate, 2 mM CuSO4,10 μM Biotin-azide (Life Technologies, B10184) or dimethyl sulfoxide (negative control) for 1 hour at room temperature. After the click reaction, cells were washed with 0.5% BSA/PBS for three times and resuspended in lysis buffer [50 mM tris-HCl (pH 8.0), 1% SDS, aprotinin (1 μg/ml), and leupeptin (1 μg/ml)] for sonication (30-s pulse/30-s pause, 60 cycles) using the Bioruptor UCD-200 machine. Last, samples were centrifuged at 10,000g for 10 min at 4°C to remove the precipitate. Each sample (volume ratio of 1:1000) was collected as an input sample. The remaining samples were incubated with streptavidin magnetic beads (Millipore, 69203) to purify replicative DNA fragments. The 2× SDS Laemmli sample buffer [0.4 g of SDS, 2 ml of 100% glycerol, 1.25 ml of 1 M tris (pH 6.8), and 0.01 g of Bromophenol blue in 8 ml of H2O] was used to elute nascent DNA binding proteins. The eluted samples were boiled at 100°C for 10 min and used for Western blotting analysis or mass spectrometry screening.
Isolation of RNAs on nascent DNA
To purify potential RNA components on replication forks, we modified the iPOND protocol described above and developed the iROND protocol. The basic principles and general procedures of iROND are the same as iPOND. To avoid RNA degradation, all the buffers and reagents were prepared using diethyl pyrocarbonate–treated double distilled water (ddH2O) and RNase inhibitors (10 U/μl; Invitrogen, EO0382) in the whole procedure, and all the procedures were performed on ice or 4°C whenever possible. Five hundred microliters of TRIzol reagent (Tiangen, DP424) was used for RNA purification of each sample, and RNA samples were used for RNA-seq or qRT-PCR analysis. The quality control, library building, and sequencing were performed by Biolinker Technology (Kunming) Co. Ltd.
Immunoblotting
Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer (Beyotime, P0013J). The protein lysate was electrophoresed into 6 to 10% SDS–polyacrylamide gel electrophoresis gels and transferred to a polyvinylidene difluoride membrane (Roche, 03010040001). After blocking with 5% BSA, the membranes were incubated with primary antibodies overnight at 4°C. After washing with tris buffered saline with 0.05% tween-20 (TBS-T) buffer for three times, the membranes were labeled with the corresponding horseradish peroxidase–conjugated secondary antibodies at room temperature for 1 hour. After washing with TBS-T buffer for three times, images were collected from the ProteinSimple FluorChem system (protein simple, Fluorchem M FM0561) after incubation with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, catalog no. 34580). All antibody information is listed in table S3.
RNA-seq and analysis
The iROND samples and embryonic RNA samples were reverse-transcribed into cDNA libraries using the TruSeq RNA Sample Preparation Kit (Illumina). RNA-seq was performed with an Illumina HiSeq 3000 or HiSeq X Ten platform. The clean reads of mESC iROND samples were processed with TopHat2 and Cufflinks. Noncoding transcripts were annotated with NONCODE database (www.noncode.org/) (version 4.0). The Coding-Non-Coding Index software was used to identify lncRNAs. DEGseq software was used to analyze differentially expressed lncRNAs.
The clean reads of embryonic RNA samples were mapped to the mouse reference genome (MM10) using TopHat2 software, and coded gene expression was calculated using Cufflinks. DEGs were determined by Cuffdiff software. GO was enriched by an online tool (http://geneontology.org/), with default parameters of “gplot” r package to create heatmaps. RNA-seq data have been deposited in Gene Expression Omnibus database (login number: GSE196639).
Real-time qRT-PCR
TRIzol (Tiangen, DP424) was used to extract total RNA from the cells. The quality and concentration of the RNA samples were determined by spectrophotometry (NanoDrop, ND1000). One gram of RNA was subjected to deoxyribonuclease (DNase) treatment and reverse-transcribed into cDNA. The TB Green Premix Ex Taq II kit (Takara, RR820A) and the CFX96TM-Real Time System (Bio-Rad, CFX96Touch) were used for real-time quantitative analysis. All reactions were normalized by Actb. The primers were provided in table S1.
RNA-FISH and immunofluorescence staining
RNA-FISH was performed as described in published paper (52). All operations were performed in RNase-free conditions. The cells were cultured on a glass lid (ZEHAO, 12-545-82) precoated with Matrigel (3 mg/ml). To label replication forks, 10 μM BrdU was added into culture medium for 5 min. Cells were then fixed with 4% paraformaldehyde for 10 min, followed by 0.2% Triton X-100/PBS treatment for 5 min at room temperature. Cells were hybridized with the 2-ng DNA probe (Guangzhou Ribo Biological Co. Ltd.) at 37°C overnight and then washed three times with 4× sodium citrate buffer (SSC), one time with 2× SSC, one time with 1× SSC at 42°C, and one time with PBS at room temperature.
For the colabeling of proteins and BrdU, the slides were refixed with 4% paraformaldehyde for 10 min at room temperature, denatured with 2.5 M HCl for 30 min at room temperature, followed by neutralization with 0.1 M sodium borate for 10 min. Then, the slides were blocked with 2% BSA for 1 hour and incubated with the primary antibodies overnight at 4°C. After washing three times with 0.2% Tween 20/PBS, cells were incubated with fluorescence-conjugated secondary antibodies for 1 hour at room temperature. 4′,6-Diamidino-2-phenylindole (DAPI) was counterstained for 10 min. Slides were sealed with glycerin. Images were captured using a confocal microscope (Olympus, FV1000).
Rapid amplification of cDNA ends
The RACE was performed according to a published protocol (53). Briefly, the template-switching oligonucleotides, oligo-dT primers, and in situ polymerase chain reaction (ISPCR) primers were ordered as described in Smart-seq2. To obtain the 5′ end of Lnc956, gene-specific primer 1 (GSP1) was used for RT, template switching was performed with TSO primers, GSP1 and ISPCR primer were used to amplify the 5′ end by PCR. To obtain the 3′ end of Lnc956, oligo-dT primers was used for RT, and GSP2 and ISPCR primers were used to amplify the 3′ end by PCR. The products were constructed into the pEasy-blunt vector and verified by sequencing. The primers were provided in table S1.
Subcellular fractionation
A total of 2 × 107 cells were lysed in 200 μl of cold cytoplasmic lysis buffer [0.15% NP-40, 10 mM tris (pH 7.5), and 150 mM NaCl] for 5 min, added 500 μl of sucrose buffer [10 mM tris (pH 7.5), 150 mM NaCl, and 24% sucrose], and centrifuged at 13,000g for 10 min, The supernatant was collected as cytoplasmic fraction. The pellet was washed with 200 μl of cytoplasmic lysis buffer without NP-40 and resuspended in 200 μl of ice-cold glycerol buffer [20 mM tris (pH 7.5), 75 mM NaCl, 0.5 mM EDTA, 50% glycerol, and 0.85 mM dithiothreitol (DTT)] and 200 μl of ice-cold nuclei lysis buffer [20 mM Hepes (pH 7.5), 7.5 mM MgCl2, 0.2 mM EDTA, 0.3 M NaCl, 1 M Urea, 1% NP-40, and 1 mM DTT], vortexed, and incubated on ice for 1 min. After centrifuge at 14,000g for 2 min, the supernatant was collected as nucleoplasm fraction. The pellet was resuspended in 50 μl of PBS as chromatin fraction. The separated fractions were mixed with 2× SDS Laemmli sample buffer for immunoblotting or mixed with TRIzol reagent for RNA extraction and qRT-PCR analysis. To avoid RNA degradation, all the buffers and reagents were added with RNase inhibitors (10 U/μl; Invitrogen, EO0382) in the whole procedures.
Protein ubiquitylation assay
A total of 106 cells were harvested in 150 μl of SDS lysis buffer [50 mM tris-HCl (pH 6.8) and 1.5% SDS]. The samples were boiled at 100°C for 15 min. One hundred microliters of protein lysate was diluted with 1.2 ml of embryonic cell lysis solution (EBC)/BSA buffer [50 mM tris-HCl (pH 6.8), 180 mM NaCl, 0.5% CA630, and 0.5% BSA] and incubated with protein G beads (Sigma-Aldrich, 16-662) and MCM7 primary antibody overnight at 4°C with rotation. The beads were collected by centrifugation at 5000g for 3 min at 4°C and washed five times with 1 ml of ice-cold EBC/BSA buffer. Proteins were resuspended with 30 μl of SDS sample loading buffer and analyzed by immunoblotting.
Dot blotting detection of cytosolic dsDNA
E12.5 embryos were dissociated into single cells with collagenase (Sigma-Aldrich, C6745) at 37°C. Cells were used for downstream subcellular fractionation. Two microliters of cytoplasmic fraction was dropped onto nylon membrane (Sigma-Aldrich, 15356) for immunoblotting, and nuclear fractionation was used as control.
DNA fiber assay
DNA fiber analysis was performed as described (14, 26). 5-Iodo-2′-deoxyuridine (IdU; 50 μM; Sigma-Aldrich, I7125) was used to label the cells for 30 min, with or without HU (Selleck, S1896) treatment, and then 250 μM 5-chloro-2′-deoxyuridine (CldU; Sigma-Aldrich, C6891) was used for the second labeling. After labeling, ~3000 cells in 2.5 μl of suspension were dropped onto one end of the glass slide, mixed with 7.5 μl of lysis buffer [50 mM EDTA, 0.5% SDS, and 200 mM tris-HCl (pH 7.5)], and incubated for 8 min at room temperature. Then, the slides were tilted to 15° to spread the DNA fibers along the slide. The slides were then treated with 2.5 M hydrochloric acid, incubated with rat anti-BrdU/CIdU (BU1/75) monoclonal antibody (Novus, NB500-169), and mouse anti-IdU monoclonal antibody (BD, 347580). The secondary antibodies were Alexa Fluor Cy3–conjugated goat anti-rat and Alexa Fluor 488–conjugated goat anti-mouse, respectively. Images were captured using a confocal microscope (Olympus, FV1000). ImageJ software was used to measure the length of DNA fibers, and the formula that 1 μm = 2.59 kb was used to convert the micrometer value to kilobase. New replication origin firing analysis was performed as described (54). Cells were labeled with CIdU for 10 min (without HU) or with 100 μM HU for 20 min.
Neutral comet assay
The neutral comet assay was performed as described (55). Glass slides were coated with 0.8% agarose for 5 s and air-dried. Single-cell suspension (~104 cells/10 μl) was mixed with 70 μl of 0.8% low–melting point agarose (LMP, Sangon Biotech, A600015-0025) kept at 37°C. Cell agarose suspension was spread onto the prepared slides and covered with coverslip. After being kept at 4°C for 10 min, slides were incubated in lysis solution [2.5 M NaCl, 100 mM Na2EDTA, 10 mM tris, 1% N-lauroylsarcosine, and 1% Triton X-100 (pH 9.5)] for 60 min at 4°C in the dark without coverslip and then washed and incubated with cold electrophoresis buffer [300 mM sodium acetate and 100 mM tris (pH 8.3)] for 20 min. Electrophoresis was performed for 30 min at 1 V/cm, 80 mA. Slides were washed with neutralization buffer [0.4 M tris-HCl (pH 7.4)] and fixed in ethanol until air dried. Slides were stained with DAPI (10 ng/ml) and immediately analyzed. Comets were analyzed with Komet 7 comet assay software (Andor Technology). At least 200 cells were analyzed in three independent replications.
Mitotic chromosomal defects analysis
Cells were cultured under normal conditions on Matrigel-coated glass slides (ZEHAO, 12-545-82). When the confluence reached about 70%, cells were fixed with 4% paraformaldehyde for 15 min at room temperature and stained with DAPI. Fifty mitotic cells were analyzed for each sample to detect chromosome bridge and micronuclei. Each experiment was repeated three times independently.
Karyotyping
Cells were cultured until about 70% confluence, treated with KARYOMAX Colcemid (120 ng/ml; Gibco, 15212-012) for 2 hours. Cells were then digested with 0.05% trypsin-EDTA (Invitrogen, 25200072), resuspended with hypotonic buffer [10 mM tris-HCl (pH 7.4), 40 mM glycerol, 20 mM NaCl, 1.0 mM CaCl2, and 0.5 mM MgCl2] for 15 min at 37°C to swell the cells, followed by fixation with methanol/glacial acetic acid (3:1) for 30 min. Cells were dropped onto ice-cold wet glass slides, air-dried, incubated at 37°C for 24 hours, and stained with 3% Giemsa solution (Gibco, 10092013) at pH 6.8 for 10 min. Images were captured using a Leica TCS SP5 confocal microscope system (Leica Microsystems). At least 50 metaphases were analyzed in three independent replications.
In vivo RNA pulldown
The in vivo RNA pulldown was performed as described (52). DNA probes labeled with biotin at 3′ end were purchased from Guangzhou RiboBio Co. Ltd. To perform in vivo RNA pulldown, 108 cells were prepared and cross-linked with 265 nm of ultraviolet light at 400 mJ of energy in ice-cold PBS. Then, the cells were incubated in CSKT buffer with 1 mM phenylmethylsulfonyl fluoride (PMSF; Beyotime, ST505) and SUPERaseIn (Invitrogen, AM2684) at 4°C for 10 min and centrifuged at 1200g for 10 min. The pellet was treated with 3 ml of DNase I buffer [50 mM tris (pH 7.5), 0.1% sodium lauroyl sarcosine, 0.5% NP-40, 1× protease inhibitors, 600 U of RNase-free DNase I, SUPERaseIn, and 10 mM vanadyl ribonucleoside complex] at 37°C, centrifuged, and discarded the precipitation. The supernatant was precleared using 50 μl of M-280 streptavidin Dynabeads (Thermo Fisher Scientific, 00781251) for 20 min at room temperature and incubated with 100 pmol of probes and 160 μl of beads at 65°C for 15 min. After cool down gradually to 37°C, the beads were washed three times with wash buffer 1 [50 mM tris (pH 7.5), 1% SDS, 0.3 M LiCl, 0.5% NP-40, 1 mM PMSF, 1 mM DTT, and 1× protease cocktail inhibitors] at 37°C, incubated with 20 U of DNase I for 10 min at 37°C, and washed twice with wash buffer 1 and once with wash buffer 2 (1% SDS, 5 mM EDTA, 1 mM DTT, 150 mM NaCl, and 1 mM PMSF). The beads were boiled at 100°C in 2× SDS protein loading buffer. The proteins were used for mass spectrometry or Western blot analysis.
In vitro RNA pulldown
The in vitro RNA pulldown was performed as described (56). Lnc956 variants were cloned into p-EASY blunt vector for in vitro transcription. RNAs were transcribed using T7 RNA polymerase (FERMENTAS Inc., EP0111) and labeled with biotin following the instruction of the Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Fisher Scientific, 20163). Fifty picomoles of biotinylated RNAs was used for in vitro RNA pulldown using the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, 20164). The pulldown samples were boiled at 100°C in 2× SDS protein loading buffer and were then used for downstream analysis.
Immunoprecipitation
A total of 107 cells were collected and lysed in RIPA buffer (Beyotime, P0013J) including 1× protease inhibitor (Beyotime, P1006). The lysate was incubated with 40 μl of Protein G Dynabeads (Thermo Fisher Scientific, 88847) and 2 μg antibody at 4°C overnight. After that, the supernatant was removed and the magnetic beads were washed three times. The beads were boiled at 100°C in 2× SDS protein loading buffer for 10 min. The supernatant was used for mass spectrometry analysis or immunoblotting.
Statistical analysis
Data were analyzed using GraphPad Prism 7 (GraphPad Software, La Jolla, CA, USA) through two-tailed Student’s t test. P < 0.05 was considered significant. All data were provided as the mean ± SEM.
Acknowledgments
We thank Y. Ning in our laboratory for deriving Lnc956 KO ESC lines and making the Lnc956 variant constructs.
Funding: This work was supported by National Natural Science Foundation of China (31930027 to P.Z. and 32000422 to W.Z.), National Key Research and Developmental Program of China (2021YFA1102002), Yunnan Fundamental Research Projects (202001AT070140 and 2019FB049), and the exchange program of State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences (GREKF21-14 to Z.C.).
Author contributions: W.Z. and M.T. performed most of the experiments, analyzed the data, and prepared the figures. L.W. analyzed the RNA-seq data and obtained the full-length of Lnc956. H.Z. and J.G. performed the mass spectrometry analysis. Z.C. constructed some expression plasmids. B.Z. developed the iROND method, identified Lnc956, and supervised the figure preparation. P.Z. supervised the study and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data for iROND samples and E12.5 embryos have been deposited in Gene Expression Omnibus database under accession numbers GSE186663 and GSE196639, respectively. The sequence of full-length Lnc956 transcript can be downloaded from the NONCODE database (www.noncode.org/).
Supplementary Materials
This PDF file includes:
Figs. S1 to S17
Tables S1 to S3
Legends for datasets S1 to S3
Other Supplementary Material for this : manuscript includes the following:
Datasets S1 to S3
REFERENCES AND NOTES
- 1.I. Vitale, G. Manic, R. De Maria, G. Kroemer, L. Galluzzi, DNA damage in stem cells. Mol. Cell 66, 306–319 (2017). [DOI] [PubMed] [Google Scholar]
- 2.S. P. Wyles, E. B. Brandt, T. J. Nelson, Stem cells: The pursuit of genomic stability. Int. J. Mol. Sci. 15, 20948–20967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. Behrens, J. M. van Deursen, K. L. Rudolph, B. Schumacher, Impact of genomic damage and ageing on stem cell function. Nat. Cell Biol. 16, 201–207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.E. D. Tichy, P. J. Stambrook, DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 314, 1929–1936 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Zalzman, G. Falco, L. V. Sharova, A. Nishiyama, M. Thomas, S.-L. Lee, C. A. Stagg, H. G. Hoang, H.-T. Yang, F. E. Indig, R. P. Wersto, M. S. Ko, Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R. Le, Y. Huang, Y. Zhang, H. Wang, J. Lin, Y. Dong, Z. Li, M. Guo, X. Kou, Y. Zhao, M. Chen, Q. Zhu, A. Zhao, J. Yin, J. Sun, Z. Su, K. Shi, Y. Gao, J. Chen, W. Liu, L. Kang, Y. Wang, C. Li, X. Liu, R. Gao, H. Wang, Z. Ju, S. Gao, Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell stem cell 28, 732–747.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 7.P. Ruis, D. Van Ly, V. Borel, G. R. Kafer, A. McCarthy, S. Howell, R. Blassberg, A. P. Snijders, J. Briscoe, K. K. Niakan, P. Marzec, A. J. Cesare, S. J. Boulton, TRF2-independent chromosome end protection during pluripotency. Nature 589, 103–109 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Markiewicz-Potoczny, A. Lobanova, A. M. Loeb, O. Kirak, T. Olbrich, S. Ruiz, E. Lazzerini Denchi, TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 589, 110–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.E. D. Tichy, R. Pillai, L. Deng, L. Liang, J. Tischfield, S. J. Schwemberger, G. F. Babcock, P. J. Stambrook, Mouse embryonic stem cells, but not somatic cells, predominantly use homologous recombination to repair double-strand DNA breaks. Stem Cells Dev. 19, 1699–1711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.J. Xiong, D. Todorova, N.-Y. Su, J. Kim, P.-J. Lee, Z. Shen, S. P. Briggs, Y. Xu, Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J. Cell Biol. 208, 513–520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.B. Zhao, W.-D. Zhang, Y.-L. Duan, Y.-Q. Lu, Y.-X. Cun, C.-H. Li, K. Guo, W.-H. Nie, L. Li, R. Zhang, P. Zheng, Filia is an ESC-specific regulator of DNA damage response and safeguards genomic stability. Cell Stem Cell 16, 684–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.M. K. Zeman, K. A. Cimprich, Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.A. K. Ahuja, K. Jodkowska, F. Teloni, A. H. Bizard, R. Zellweger, R. Herrador, S. Ortega, I. D. Hickson, M. Altmeyer, J. Mendez, M. Lopes, A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 7, 10660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.B. Zhao, W. Zhang, Y. Cun, J. Li, Y. Liu, J. Gao, H. Zhu, H. Zhou, R. Zhang, P. Zheng, Mouse embryonic stem cells have increased capacity for replication fork restart driven by the specific Filia-Floped protein complex. Cell Res. 28, 69–89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R. Bellelli, S. J. Boulton, Spotlight on the replisome: Aetiology of DNA replication-associated genetic diseases. Trends Genet. 37, 317–336 (2021). [DOI] [PubMed] [Google Scholar]
- 16.D. Cortez, Preventing replication fork collapse to maintain genome integrity. DNA Repair 32, 149–157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Berti, D. Cortez, M. Lopes, The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 21, 633–651 (2020). [DOI] [PubMed] [Google Scholar]
- 18.H. Techer, S. Koundrioukoff, A. Nicolas, M. Debatisse, The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet. 18, 535–550 (2017). [DOI] [PubMed] [Google Scholar]
- 19.B. M. Sirbu, F. B. Couch, D. Cortez, Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat. Protoc. 7, 594–605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.C. Alabert, J.-C. Bukowski-Wills, S.-B. Lee, G. Kustatscher, K. Nakamura, F. de Lima Alves, P. Menard, J. Mejlvang, J. Rappsilber, A. Groth, Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. Liao, F. Ji, T. Helleday, S. Ying, Mechanisms for stalled replication fork stabilization: New targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 19, e46263 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.K. Rickman, A. Smogorzewska, Advances in understanding DNA processing and protection at stalled replication forks. J. Cell Biol. 218, 1096–1107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Statello, C.-J. Guo, L.-L. Chen, M. Huarte, Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. M. Elguindy, J. T. Mendell, NORAD-induced Pumilio phase separation is required for genome stability. Nature 595, 303–308 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. M. Clemson, J. N. Hutchinson, S. A. Sara, A. W. Ensminger, A. H. Fox, A. Chess, J. B. Lawrence, An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.H. Técher, S. Koundrioukoff, D. Azar, T. Wilhelm, S. Carignon, O. Brison, M. Debatisse, B. Le Tallec, Replication dynamics: Biases and robustness of DNA fiber analysis. J. Mol. Biol. 425, 4845–4855 (2013). [DOI] [PubMed] [Google Scholar]
- 27.M. Petropoulos, S. Champeris Tsaniras, S. Taraviras, Z. Lygerou, Replication licensing aberrations, replication stress, and genomic instability. Trends Biochem. Sci. 44, 752–764 (2019). [DOI] [PubMed] [Google Scholar]
- 28.N. Shima, A. Alcaraz, I. Liachko, T. R. Buske, C. A. Andrews, R. J. Munroe, S. A. Hartford, B. K. Tye, J. C. Schimenti, A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat. Genet. 39, 93–98 (2007). [DOI] [PubMed] [Google Scholar]
- 29.B. S. Heyer, A. MacAuley, O. Behrendtsen, Z. Werb, Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev. 14, 2072–2084 (2000). [PMC free article] [PubMed] [Google Scholar]
- 30.J. C. O’Leary III, S. Dharia, L. J. Blair, S. Brady, A. G. Johnson, M. Peters, J. Cheung-Flynn, M. B. Cox, G. de Erausquin, E. J. Weeber, U. K. Jinwal, C. A. Dickey, A new anti-depressive strategy for the elderly: Ablation of FKBP5/FKBP51. PlOS ONE 6, e24840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K. Shimada, S. Park, H. Miyata, Z. Yu, A. Morohoshi, S. Oura, M. M. Matzuk, M. Ikawa, ARMC12 regulates spatiotemporal mitochondrial dynamics during spermiogenesis and is required for male fertility. Proc. Natl. Acad. Sci. U.S.A. 118, e2018355118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.A. J. McNairn, C.-H. Chuang, J. C. Bloom, M. D. Wallace, J. C. Schimenti, Female-biased embryonic death from inflammation induced by genomic instability. Nature 567, 105–108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. L. Ragland, S. Patel, R. S. Rivard, K. Smith, A. A. Peters, A.-K. Bielinsky, E. J. Brown, RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 27, 2259–2273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.L. I. Toledo, M. Altmeyer, M.-B. Rask, C. Lukas, D. H. Larsen, L. K. Povlsen, S. Bekker-Jensen, N. Mailand, J. Bartek, J. Lukas, ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 (2013). [DOI] [PubMed] [Google Scholar]
- 35.S. M. Jang, A. Kauzlaric, J.-P. Quivy, J. Pontis, B. Rauwel, A. Coluccio, S. Offner, J. Duc, P. Turelli, G. Almouzni, D. Trono, KAP1 facilitates reinstatement of heterochromatin after DNA replication. Nucleic Acids Res. 46, 8788–8802 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.J. M. Dewar, J. C. Walter, Mechanisms of DNA replication termination. Nat. Rev. Mol. Cell Biol. 18, 507–516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.T. D. Deegan, P. P. Mukherjee, R. Fujisawa, C. Polo Rivera, K. Labib, CMG helicase disassembly is controlled by replication fork DNA, replisome components and a ubiquitin threshold. eLife 9, e60371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Taipale, D. F. Jarosz, S. Lindquist, HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 (2010). [DOI] [PubMed] [Google Scholar]
- 39.A. Rohl, J. Rohrberg, J. Buchner, The chaperone Hsp90: Changing partners for demanding clients. Trends Biochem. Sci. 38, 253–262 (2013). [DOI] [PubMed] [Google Scholar]
- 40.D. E. Dollins, J. J. Warren, R. M. Immormino, D. T. Gewirth, Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol. Cell 28, 41–56 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. S. Mirsanaye, D. Typas, N. Mailand, Ubiquitylation at stressed replication forks: Mechanisms and functions. Trends Cell Biol. 31, 584–597 (2021). [DOI] [PubMed] [Google Scholar]
- 42.T. D. Deegan, J. F. X. Diffley, MCM: One ring to rule them all. Curr. Opin. Struct. Biol. 37, 145–151 (2016). [DOI] [PubMed] [Google Scholar]
- 43.D. J. Anderson, R. Le Moigne, S. Djakovic, B. Kumar, J. Rice, S. Wong, J. Wang, B. Yao, E. Valle, S. Kiss von Soly, A. Madriaga, F. Soriano, M.-K. Menon, Z. Y. Wu, M. Kampmann, Y. Chen, J. S. Weissman, B. T. Aftab, F. M. Yakes, L. Shawver, H.-J. Zhou, D. Wustrow, M. Rolfe, Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell 28, 653–665 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Y. Wei, J. I. Toth, G. A. Blanco, A. A. Bobkov, M. D. Petroski, Adapted ATPase domain communication overcomes the cytotoxicity of p97 inhibitors. J. Biol. Chem. 293, 20169–20180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.E. Lecona, O. Fernández-Capetillo, Replication stress and cancer: It takes two to tango. Exp. Cell Res. 329, 26–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.T. Thakar, G.-L. Moldovan, The emerging determinants of replication fork stability. Nucleic Acids Res. 49, 7224–7238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.T. Yamazaki, S. Souquere, T. Chujo, S. Kobelke, Y. S. Chong, A. H. Fox, C. S. Bond, S. Nakagawa, G. Pierron, T. Hirose, Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 48.A. Pandya-Jones, Y. Markaki, J. Serizay, T. Chitiashvili, W. R. Mancia Leon, A. Damianov, C. Chronis, B. Papp, C.-K. Chen, R. McKee, X.-J. Wang, A. Chau, S. Sabri, H. Leonhardt, S. Zheng, M. Guttman, D. L. Black, K. Plath, A protein assembly mediates Xist localization and gene silencing. Nature 587, 145–151 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.H. Dungrawala, K. L. Rose, K. P. Bhat, K. N. Mohni, G. G. Glick, F. B. Couch, D. Cortez, The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell 59, 998–1010 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.G. De Piccoli, Y. Katou, T. Itoh, R. Nakato, K. Shirahige, K. Labib, Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol. Cell 45, 696–704 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Y. Liu, L. Wang, X. Xu, Y. Yuan, B. Zhang, Z. Li, Y. Xie, R. Yan, Z. Zheng, J. Ji, J. M. Murray, A. M. Carr, D. Kong, The intra-S phase checkpoint directly regulates replication elongation to preserve the integrity of stalled replisomes. Proc. Natl. Acad. Sci. U.S.A. 118, e2019183118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.H.-P. Chu, C. Cifuentes-Rojas, B. Kesner, E. Aeby, H.-G. Lee, C. Y. Wei, H. J. Oh, M. Boukhali, W. Haas, J. T. Lee, TERRA RNA antagonizes ATRX and protects telomeres. Cell 170, 86–101.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. Picelli, O. R. Faridani, Å. K. Björklund, G. Winberg, S. Sagasser, R. Sandberg, Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 54.X. Q. Ge, J. Han, E.-C. Cheng, S. Yamaguchi, N. Shima, J.-L. Thomas, H. F. Lin, Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Rep. 5, 185–194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.U. Swain, K. Subba Rao, Study of DNA damage via the comet assay and base excision repair activities in rat brain neurons and astrocytes during aging. Mech. Ageing Dev. 132, 374–381 (2011). [DOI] [PubMed] [Google Scholar]
- 56.P. Wang, Y. Xue, Y. Han, L. Lin, C. Wu, S. Xu, Z. Jiang, J. Xu, Q. Liu, X. Cao, The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S17
Tables S1 to S3
Legends for datasets S1 to S3
Datasets S1 to S3