Abstract
Human genetic variation affects the gut microbiota through a complex combination of environmental and host factors. Here we characterize genetic variations associated with microbial abundances in a single large-scale population-based cohort of 5,959 genotyped individuals with matched gut microbial metagenomes, and dietary and health records (prevalent and follow-up). We identified 567 independent SNP–taxon associations. Variants at the LCT locus associated with Bifidobacterium and other taxa, but they differed according to dairy intake. Furthermore, levels of Faecalicatena lactaris associated with ABO, and suggested preferential utilization of secreted blood antigens as energy source in the gut. Enterococcus faecalis levels associated with variants in the MED13L locus, which has been linked to colorectal cancer. Mendelian randomization analysis indicated a potential causal effect of Morganella on major depressive disorder, consistent with observational incident disease analysis. Overall, we identify and characterize the intricate nature of host–microbiota interactions and their association with disease.
Humans have coevolved with the microbial communities that colonize them, resulting in a complex assembly of thousands of microbial species mutualistically living in their gastrointestinal tract. A fine-tuned interplay between microbial and human physiologies can impact multiple aspects of development and health to the point that dysbiosis is often associated with disease1,2. As such, increasing evidence points to the influence of human genetic variation on the composition and modulation of their gut microbiota.
Past genetic studies have collectively revealed important host– microbe interactions3–13. Previous twin studies detected substantial heritability signal from the presence and abundance of only a few microbial taxa, such as some Firmicutes14, suggesting a strong transientness and variability in gut microbial composition, as well as an important influence from external factors5,14–17. Nonetheless, a well-described association between Bifidobacterium levels and LCT-MCM6, governing the phenotype of lactase persistence throughout adulthood in Europeans, was uncovered in 2015 (ref. 3) and subsequently replicated by later studies5,6,8–11, suggesting a very strong influence of the evolution of dairy diet in modern humans on their gut bacteria. Additionally, genes involved in immune and metabolic processes8 but also disease18 were also associated with gut microbial variation. Despite several promising findings, reproducibility across studies varying in sampling and methods is generally poor, and most previously reported associations lose significance after multiple testing correction19. The individual gut microbiota is largely influenced by environmental variables, mostly diet and medication20–22, which could explain a larger proportion of microbiome variance than identifiable host genetic factors8,9. Biological factors could also influence the cross-study reproducibility of results. Genome-wide association studies (GWASs) would typically not reproducibly identify genetic associations with taxa harbouring microbial functions potentially shared by multiple unrelated species23,24. Indeed, a certain degree of functional redundancy has been observed in human gut microbial communities24, which is believed to play a role in the resistance and resilience to perturbations25–27. However, both assembly and functioning in human gut microbial communities seem to be driven by the presence of a few particular and identifiable keystone taxa28, which exert key ecological and modulatory roles on gut microbial composition independently of their abundance29,30. Such taxa are relatively prevalent across individuals and thought to be part of the human ‘core’ microbiota29,30, which makes them potentially identifiable through GWAS.
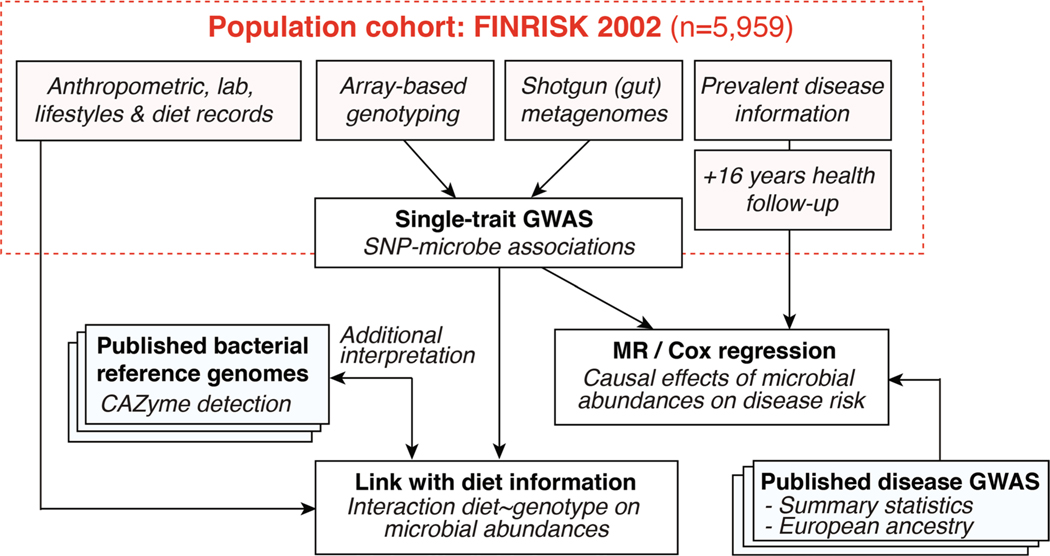
Increasing sample size in studied populations could yield novel and robustly associated results, and alleviate the effect of confounding technical or biological factors. This could be achieved either by performing meta-analyses of GWASs conducted in various populations11, or by using larger cohort datasets. In this study, we used a large single homogenous population cohort with matching human genotypes and shotgun fecal metagenomes (N = 5,959; FINRISK 2002 (FR02)) to identify genome-wide associations between human genotypes and gut microbial abundances (Extended Data Fig. 1). We further leveraged additional and extensive health registry and dietary individual data to investigate the effects of diet and genotype on particular host–microbial associations, and to predict incident disease linked to gut microbial variation.
Results
Genome-wide association analysis of gut microbial taxa.
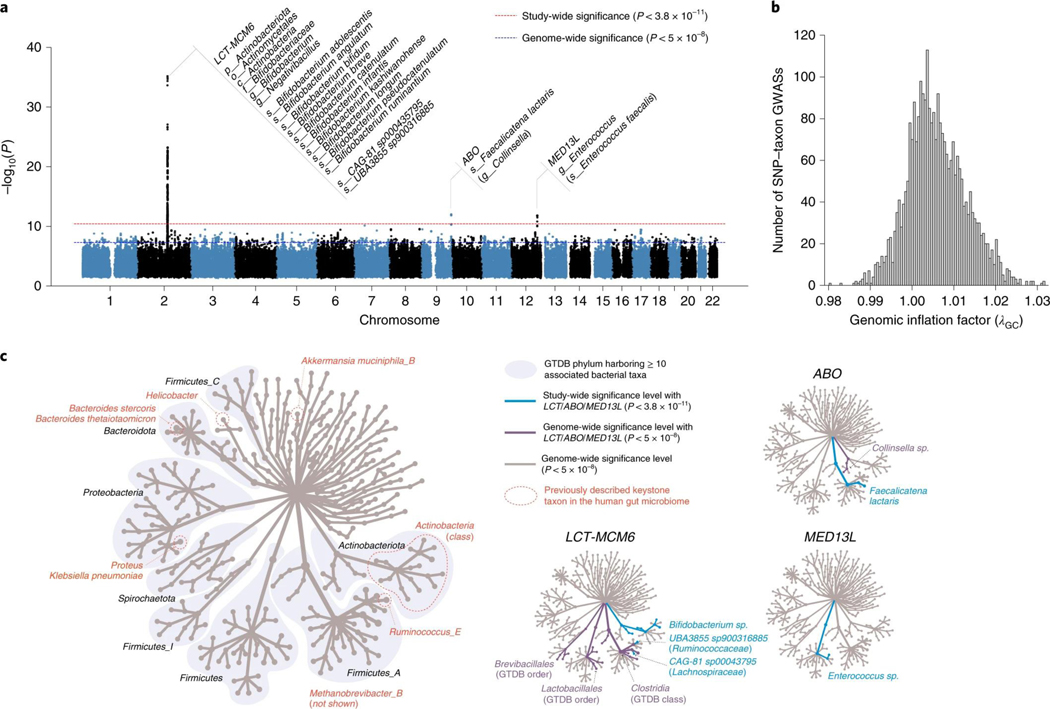
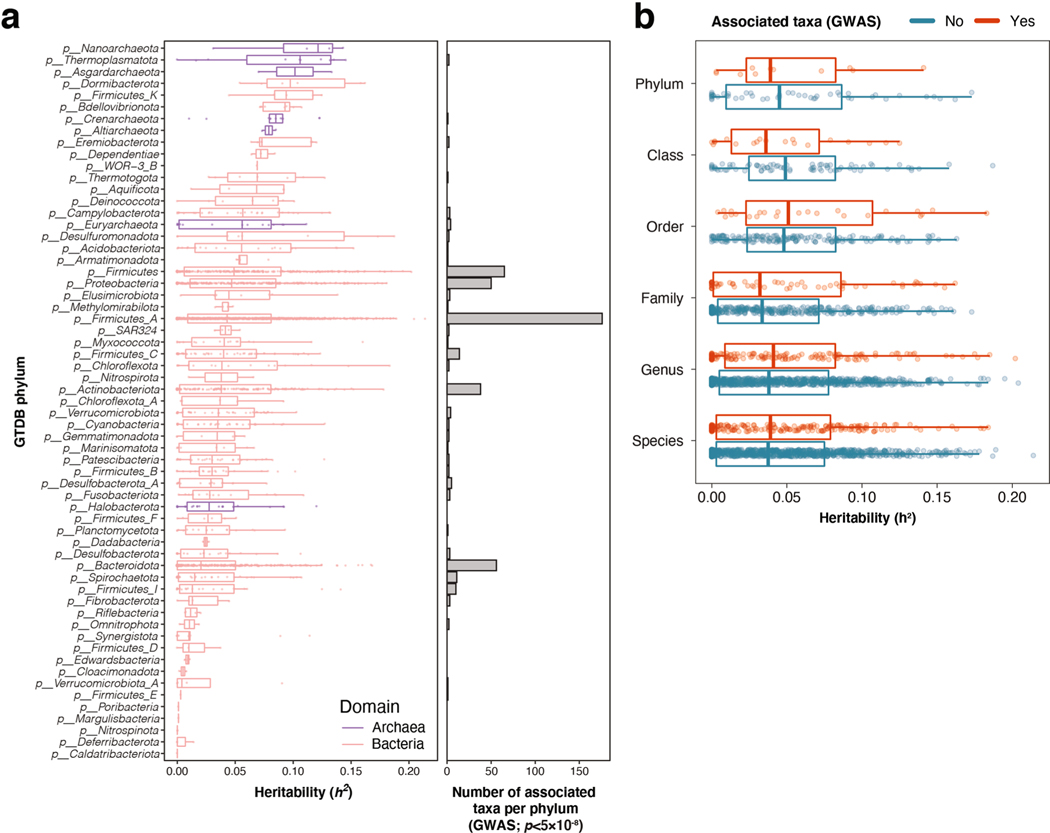
Genome-wide association tests were applied to 2,801 microbial taxa and 7,967,866 human genetic variants from 5,959 individuals enrolled in the FR02 cohort (Supplementary Table 9), which includes all taxa discovered to be prevalent in >25% of the cohort (Methods). Using a genome-wide significance threshold (P < 5.0 × 10−8), a total of 471 distinct Genome Taxonomy Database (GTDB) taxa, which represented 17% of all tested taxa and included 11 phyla, 19 classes, 24 orders, 62 families, 146 genera and 209 species, were found to be associated with at least one genetic variant (Fig. 1 and Supplementary Table 1). Conditional analysis found 567 independent SNP–taxon associations at genome-wide significance in 411 loci (Supplementary Table 1). Heritability across the 2,801 taxa ranged between h2 = 0.001 and 0.214, with the highest values observed for taxa belonging to the Firmicutes and Firmicutes_A GTDB phyla, both of which encompassed half (237 of 471, 50.3%) of all associated taxa with genetic variation (Extended Data Fig. 2). There was no difference in SNP heritability between groups of associated or nonassociated taxa at genome-wide significance (P = 0.23). Adjusting for antibiotic prescription did not change any study-wide significant associations, and only 32 of 567 genome-wide associations moved slightly above P = 5 × 10−8, which is likely by chance given inclusion of any additional covariate (Supplementary Table 10). After adjustment, beta estimates were highly correlated (Pearson r > 0.999).
Fig. 1 |. Genome-wide association of human genetic and gut microbial variations.
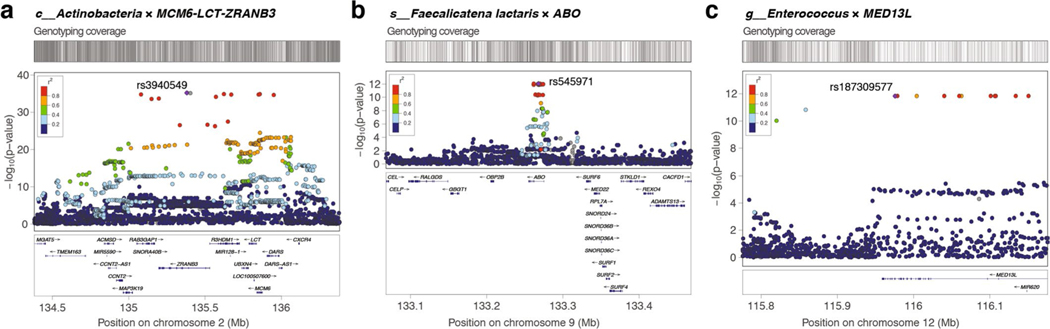
a, Manhattan plot aggregating the top associations with microbial variation. Each SNP was tested against each of the 2,801 taxa and the Manhattan plot shows the lowest resulting P value for each SNP. Loci with associations above study-wide significance level (P < 3.8 × 10−11; red dashed line) are annotated with the human locus name and the corresponding associated microbial taxa. The blue dashed line denotes genome-wide significance level (P < 5 × 10−8). Of all genome-wide significant associations shown on the Manhattan plot, 320 of 567 (56.4%) involved 265 lead SNPs with MAF between 1% and 5%, and 247 of 567 (43.6%) involved 185 lead SNPs with MAF > 5%. P values denote significance of the joint analysis model using GTCA-COJO. b, The distribution of genomic inflation factor (λGC) in 2,801 tested taxa (median(λGC)=1.0051; mean(λGC)=1.0059). c, Tree-based visualization of the taxonomic diversity of genome-wide associated microbial taxa. The central root of the tree represents the Bacteria domain, the first connected node represents phylum, the second connected node class, the third order and the fourth family. Every node represents at least one associated taxon in the GWAS at genome-wide significance level. The three smaller trees on the right highlight all taxonomic groups containing at least one taxon identified as associated with the LCT-MCM6, ABO and MED13L loci (blue edges and nodes denote taxa associated at study-wide significance level and purple edges and nodes denote taxa associated at genome-wide significance level). The main tree is annotated to indicate phyla harboring >10 distinct genome-wide associated taxa, as well as previously described keystone taxa. MAF, minor allele frequency.
Three loci were strongly associated with microbial variation at study-wide significance, as shown on a Manhattan plot showing the lowest resulting P value for each SNP tested against each of the 2,801 taxa (Fig. 1 and Supplementary Table 1). There was no evidence of excess false positive rate in the GWAS (median genomic inflation factor (λGC) = 1.0051) (Fig. 1b and Supplementary Table 9). After conditional analysis, the strongest association by far (P = 5.0 × 10−35) involved members of class Actinobacteria and rs3940549, a variant in the LCT-MCM6-ZRANB3 locus region which is in high linkage disequilibrium (LD) (r2 = 0.87) with the well-described LCT variant rs4988235, causing lactase persistence in adults of European ancestry (Extended Data Fig. 3). In total, 29 taxa were associated with the LCT-MCM6 region, including 18 below study-wide significance (Fig. 1 and Supplementary Table 1). These involved Bifidobacterium-related Actinobacteriota and three taxa from the GTDB Firmicutes_A phylum which included two uncultured species defined from metagenome-assembled reference genomes (UBA3855 sp900316885 and CAG-81 sp000435795). The association of these three Firmicutes_A with LCT was still genome-wide significant after adjusting for Bifidobacterium abundances, as were 11 other taxa associated with the LCT-MCM6 region (Supplementary Table 2). Additionally, the abundance of these LCT-MCM6-associated taxa were not, or very weakly, associated with the Bifidobacterium abundances. A variant in ABO (rs545971), expressing the histo-blood group ABO system transferase, was strongly associated (P = 1.1 × 10−12) with levels of Faecalicatena lactaris. There was evidence for a second independent signal at ABO associated with the Collinsella genus (chr9:133271182; P = 2.5 × 10−8). rs187309577 and rs143507801 in MED13L, expressing the Mediator complex subunit 13L, were found to be associated with genus Enterococcus (P = 1.8 × 10−12) and the Enterococcus faecalis species (P = 7.26 × 10−11), respectively.
Details on the replication of previously reported microbiome GWAS signals in our study are included in the Supplementary Note.
Gut microbial keystone taxa associate with genetic variation.
In total, we identified 31 distinct genetic variants associated (P < 5.0 × 10−8) with 39 microbial taxa related to identified keystone species as listed by Banerjee et al. (2018)28,31, which included the Actinobacteria class, Helicobacter pylori, Bacteroides stercoris, Bacteroides thetaiotaomicron, Ruminococcus bromii, Klebsiella pneumoniae, Proteus mirabilis, Akkermansia muciniphila and the archaeon Methanobrevibacter smithii (Fig. 1c and Supplementary Table 1). Keystone species are defined as members of a microbial community exerting selective modulation and not broad effects on microbiome composition variation. Only one documented keystone species from Banerjee et al., Bacteroides fragilis, was not associated with genetic variation in our study28. Although a lot of computationally identified keystone species remain to be experimentally verified32,33, this observation suggests that they would generally associate with human genetic variation. This would indicate an intimate association with the human gut niche in line with their reported key ecological roles in microbiome modulation and functioning. Our work highlights human genotypes associating with keystone taxa (Supplementary Table 1), which could further improve our understanding of their ecology.
Combined effect of genetics and diet on LCT-associated taxa.
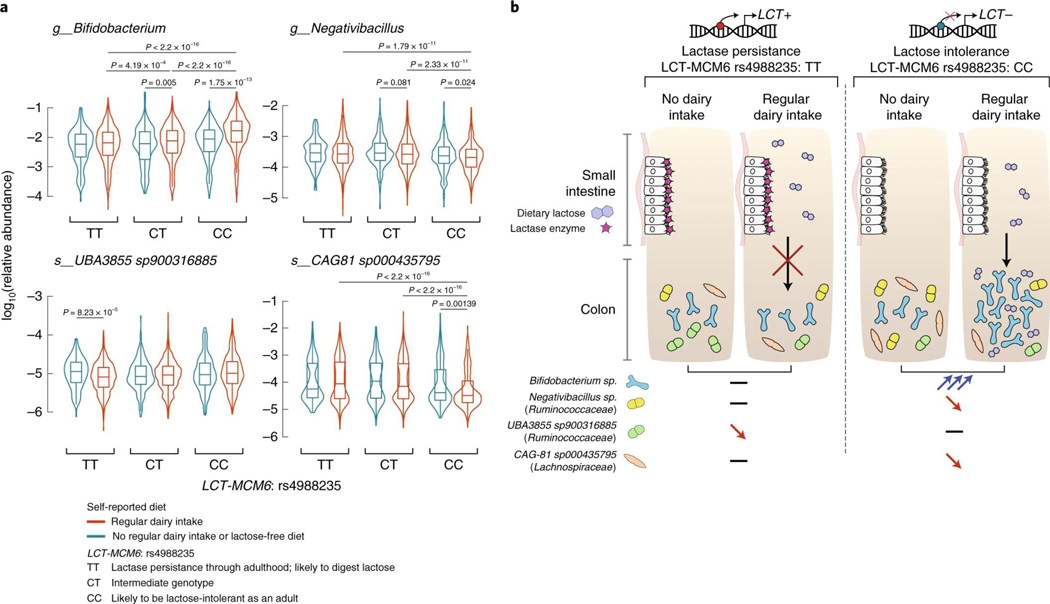
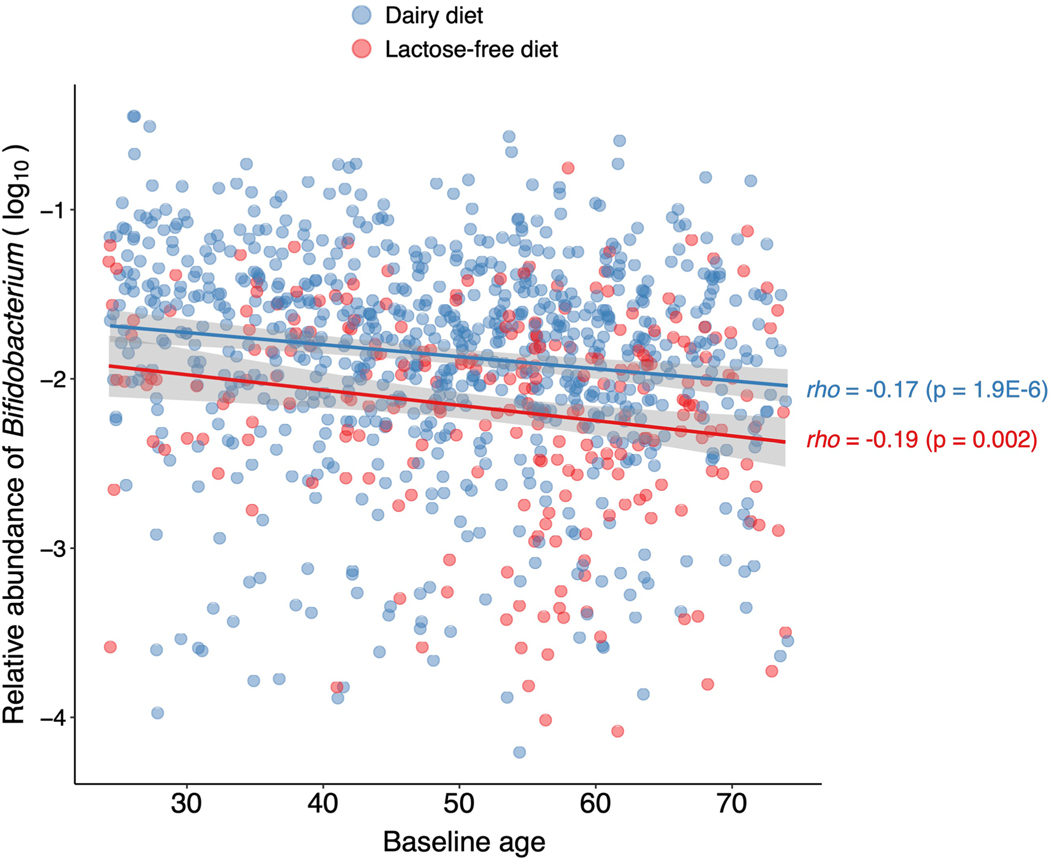
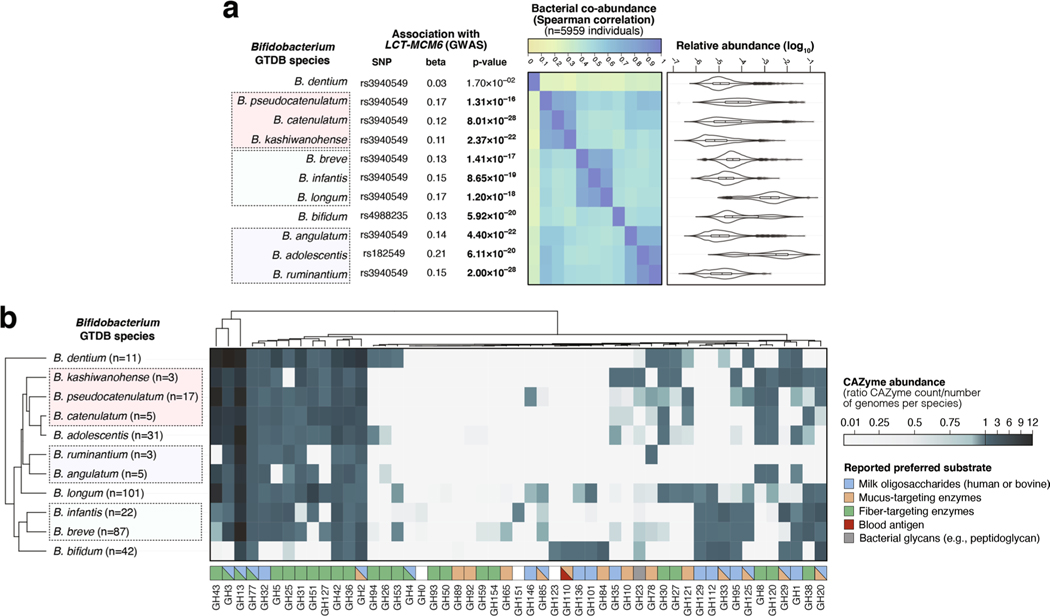
We compared the abundances of four bacterial taxa strongly associated with the LCT locus (Bifidobacterium genus, Negativibacillus genus, UBA3855 sp900316885 and CAG-81 sp000435795) in individuals with different rs4988235 genotypes and dairy diets (Fig. 2a). The abundance of Bifidobacterium in individuals producing lactase through adulthood (rs4988235:TT) was unaffected by dairy intake. However, lactose-intolerant individuals (rs4988235:CC) self-reporting a regular dairy diet had a significant increase in Bifidobacterium abundance (P = 1.75 × 10−13; Wilcoxon rank test). An intermediate genotype (rs4988235:CT) was linked to an intermediate increase in Bifidobacterium abundance (Fig. 2a). This trend did not seem to be affected by age34 (Extended Data Fig. 4). Additionally, we observed a moderate negative correlation between Bifidobacterium abundances and age in rs4988235:CC individuals reporting a regular dairy diet (Spearman’s ⍴ = −0.17, P = 1.9 × 10−6) and in rs4988235:CC individuals reporting a low-lactose or lactose-free diet (⍴ = −0.19, P = 0.002). Furthermore, the Spearman correlation between the Bifidobacterium residual abundance and dairy diet was still significant (⍴ = −0.22, P = 2 × 10−12) in rs4988235:CC individuals. This indicated that the associations with age were consistent in individuals with and without regular dairy intake, and did not confound the association between Bifidobacterium and dairy diet.
Fig. 2 |. Interaction of human genotype, dairy diet and gut bacterial variation with the LCT locus.
a, The four panels present variation in microbial relative abundances (not CLR-transformed) for the four taxa associated at study-wide significance level with the LCT locus at P < 3.8 × 10−11: Bifidobacterium, Negativibacillus, UBA3855 sp900316885 and CAG-81 sp000435795. Abundances are compared across stratified groups of individuals from the FR02 cohort according to LCT-MCM6:rs4988235 genotype and self-reported dietary lactose intake (red, regular dairy diet; blue, lactose-free diet). Sample sizes for groups of individuals self-reporting a regular dairy diet: rs4988235:TT (n = 1,786), CT (n = 2,413), CC (n = 736); self-reporting a nonregular dairy diet or lactose-free diet: TT (n = 150), CT (n = 198), CC (n = 245). All statistical comparisons denote the P values of Wilcoxon rank test on the distributions of untransformed relative abundances. Only significantly different comparisons (P < 0.05) are indicated. For all box plots, the central line, box and whiskers represent the median, interquartile range (IQR) and 1.5 times the IQR, respectively. Violin plots represent the distribution density of the data points.
b, Host genetics and gut microbes interact in the context of dairy intake and lactose intolerance.
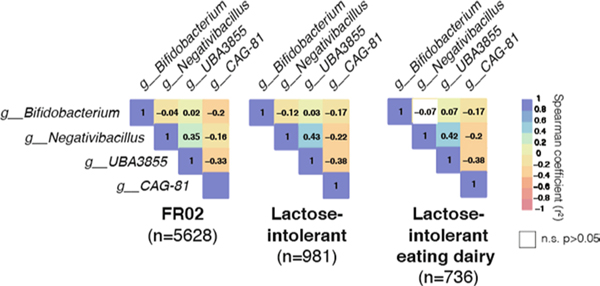
An inverse pattern was observed for the abundance distributions of Negativibacillus and uncultured CAG-81 sp000435795, for which abundances decreased in lactose-intolerant individuals reporting dairy intake, as compared with rs4988235:TT individuals consuming dairy products (Fig. 2a). Levels of UBA3855 sp900316885 were unaffected by a dairy diet in lactose-intolerant individuals but were surprisingly lower in rs4988235:TT individuals who reported dairy intake (P = 8.23 × 10−5). These opposite and contrasting effects of dairy on associated bacterial abundances in lactose-intolerant individuals could reflect competition for lactose in the gut. CAG-81 abundances were the most negatively correlated with those of the other LCT-associated taxa (Extended Data Fig. 5), which suggests that this competition could be strong and prevalent enough to drive coassociation at the LCT locus, possibly mediated by lactose intake (Fig. 2b).
Functional profiling of carbohydrate-active enzymes (CAZymes) in 11 Bifidobacterium species.
Of all 11 Bifidobacterium species prevalent enough in our study population to be included in the GWAS, only Bifidobacterium dentium was not associated with the LCT locus (P = 1.70 × 10−2), nor was it coabundant with any other Bifidobacterium species (Extended Data Fig. 6a). B. dentium has previously been suggested to have different metabolic abilities35. A clustering of CAZyme profiles from reference genomes of all 11 Bifidobacterium species revealed that B. dentium clustered apart from the ten other species, which grouped consistently with their coabundance patterns (Extended Data Fig. 6b). B. dentium harbored more genes encoding CAZyme families with preferred fiber/plant-related substrates (GH94, GH26, GH53) than other Bifidobacterium species, which seemed to harbor more milk oligosaccharide-targeting CAZyme families (GH129, GH112) than B. dentium (Extended Data Fig. 6b), which could relate to the observed association differences. This suggests that bacterial metabolic abilities can be strong drivers of coabundance, and of association with human genetic variation.
Impacts of genotype and fiber intake on ABO-associated taxa.
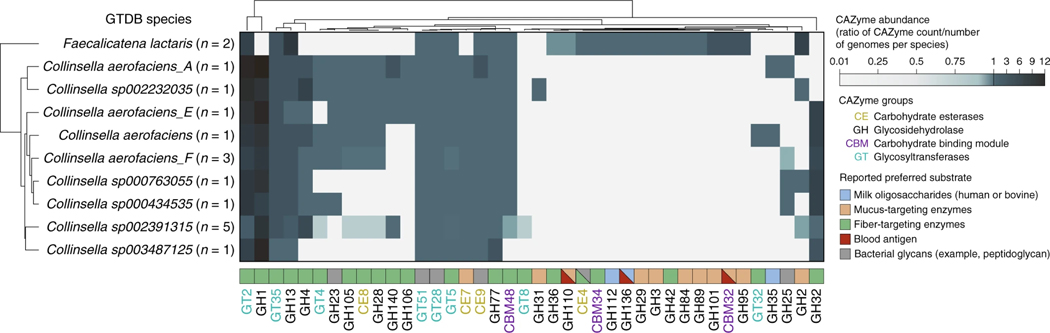
A variety of bacteria metabolize blood antigens, with potential applications in synthetic universal donor blood production36,37. Gut bacteria are particularly exposed to A- and B-antigens in the gut mucosa of secretor individuals38. Our associations of F. lactaris (P = 1.10 × 10−12) and Collinsella (P = 2.59 × 10−8) with ABO suggest a possible metabolic link with blood antigens. A comparison of CAZyme profiles across a set of reference genomes revealed three CAZymes with blood-related activities in F. lactaris (GH110 (ref. 39), GH136 (ref. 40), CBM32 (ref. 41)), but none in any of nine Collinsella species (Fig. 3). More mucus-targeting and fewer fiber-degrading enzymes were found in F. lactaris than Collinsella, suggesting distinct functions in the gut.
Fig. 3 |. Functional profiling of reference genomes from two bacterial taxa associated with the ABO locus.
CAZyme distribution patterns in F. lactaris and Collinsella reference genomes (from the GTDB release 89 index used to classify metagenomes in this study). The heatmap indicates species abundance in corresponding CAZyme families, corresponding to the total count of detected families for each species divided by the number of reference genomes examined for the same species. Values < 1 (white to light blue) indicate that less than one copy per genome of the corresponding CAZyme family was detected for each species; values > 1 (light blue to dark blue) indicate that more than one copy per genome was detected. Preferred substrate groups are based on literature search and descriptions on CAZypedia.org.
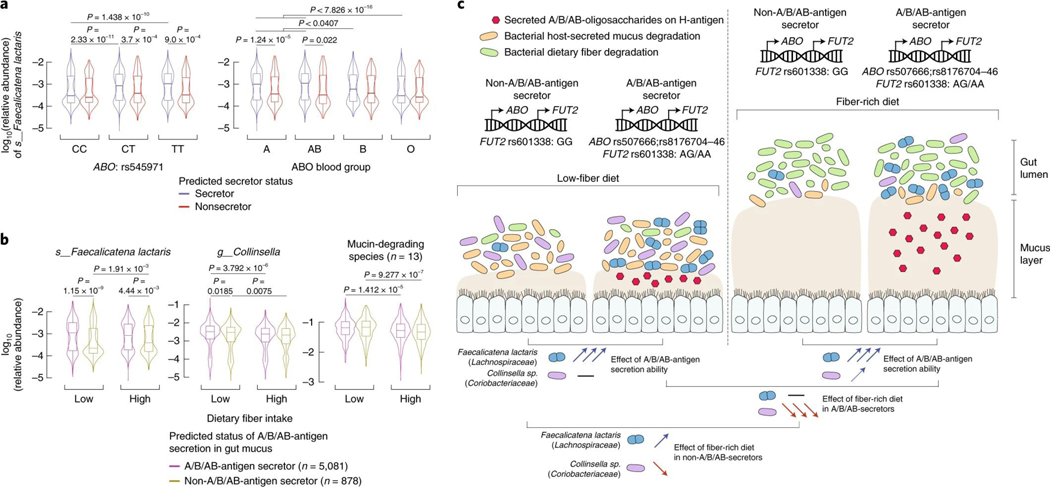
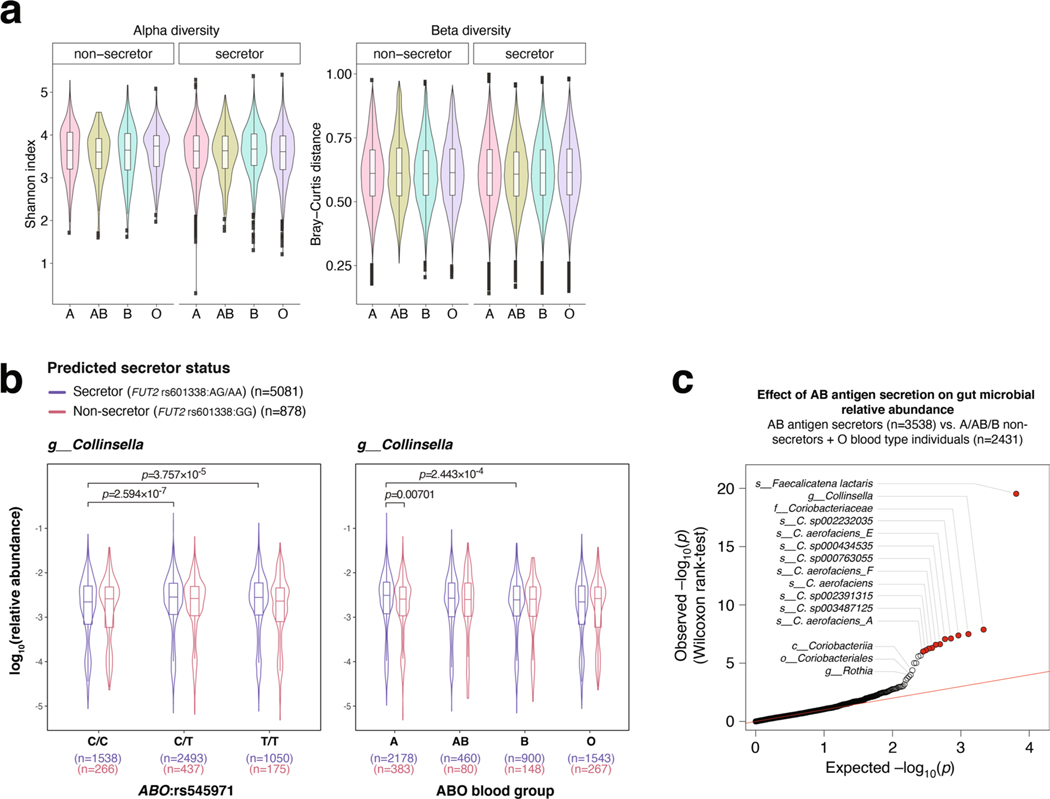
As previously reported4, neither ABO blood types nor secretor status had an impact on alpha- and beta-diversity (Extended Data Fig. 7a). However, we observed that the effects of ABO genotypes on F. lactaris levels, underlying the association, were largely driven by secretor status, with increased abundances in secretor individuals from genotype groups rs545971:CT and rs545971:TT, A and AB blood type groups, but not in rs545971:CC genotype, or B and O blood type individuals (Fig. 4a). Levels in nonsecretors did not vary across ABO genotypes or blood types. Despite a slight increase in blood type A secretors, Collinsella only remained minimally affected by secretor status or blood group (Extended Data Fig. 7b). Taken together, this suggests that the secretion of soluble A- and B-antigens strongly affects F. lactaris in the gut, possibly through reduced opportunity to use them as substrate. Levels of both F. lactaris and Collinsella were significantly higher when individuals were predicted to secrete A-, B- and AB-antigens in their gut mucosa (Extended Data Fig. 7c).
Fig. 4 |. Effects of host genetics and dietary fiber intake on gut abundance variation of two bacterial taxa associated with the ABO locus.
a, ABO-associated F. lactaris relative abundances (not CLR-transformed) are compared across stratified groups of individuals from the FR02 cohort according to (left panel) ABO:rs4988235 genotype and predicted secretor status (blue, secretor status conferred by FUT2 rs601338:AG/AA genotype; red, nonsecretor status conferred by FUT2 rs601338:GG genotype), and (right panel) according to predicted A, AB, B and O blood types, and predicted secretor status. Sample sizes for compared groups: secretor status with rs545971:C/C (n = 1,538), C/T (n = 2,493), T/T (n = 1,050) and blood group A (n = 2,178), AB (n = 460), B (n = 900), O (n = 1,543); nonsecretor status with rs545971:C/C (n = 266), C/T (n = 437), T/T (n = 175) and blood group A (n = 383), AB (n = 80), B (n = 148), O (n = 267). b, ABO-associated F. lactaris and Collinsella sp. relative abundances, as well as compounded abundances from 13 mucin-degrading species from Tailford et al. (2015)43, are compared across stratified groups of individuals from the FR02 cohort according to the predicted A/B/AB-antigen secretion status and dietary fiber intake. Secretion status was defined to segregate individuals. A/B/AB-antigen secretors were defined as secretor individuals from blood types A, AB and B. Non-A/B/AB-antigen secretors were defined as nonsecretor individuals and O-antigen secretors. Fiber intake was compared in individual groups from the top and bottom quartiles of total fiber score (Methods). Sample sizes for compared groups of individuals: A/B/AB-antigen secretors (n = 1,393) following a low-fiber diet (n = 723) or a fiber-rich diet (n = 670), or non-A/B/AB-antigen secretors (n = 952) following a low-fiber diet (n = 490) or a fiber-rich diet (n = 462). All statistical comparisons denote the P values of Wilcoxon rank test on the distributions of untransformed relative abundances. For all box plots (b and c), the central line, box and whiskers represent the median, IQR and 1.5 times the IQR, respectively. Violin plots represent the distribution density of the data points. c, Host genetics and gut microbes interact in the context of fiber intake, secretor status and blood types.
A high-fiber diet is thought to induce a metabolic transition from mucus-degrading to fiber-degrading activities in the colon, as carbohydrates from fiber are more easily metabolized42. The increase in F. lactaris abundances in A/B/AB-secretors (defined as secreting A-, B- and AB-antigens) compared with non-A/B/AB-secretors remained strongly significant irrespective of fiber intake (P = 1.15 × 10−9 in the low-fiber diet group, and P = 4.4 × 10−3 in the high-fiber diet group), suggesting that F. lactaris has a strong affinity for secreted A/B/AB-antigens, does not efficiently degrade dietary fiber or will not easily switch to it as an energy source (Fig. 4b). F. lactaris levels were increased in non-A/B/AB-secretors with a high-fiber diet compared with a low-fiber diet, implying a switch to fiber degradation or interaction with fiber-degrading bacteria (Fig. 4b). Collinsella variation in both A/B/AB-secretors and non-A/B/AB-secretors with high- and low-fiber diets was similar to the compounded abundances of 13 major mucin-degrading species in the human gut43, suggesting a similar ecological response in stark contrast with F. lactaris (Fig. 4b,c).
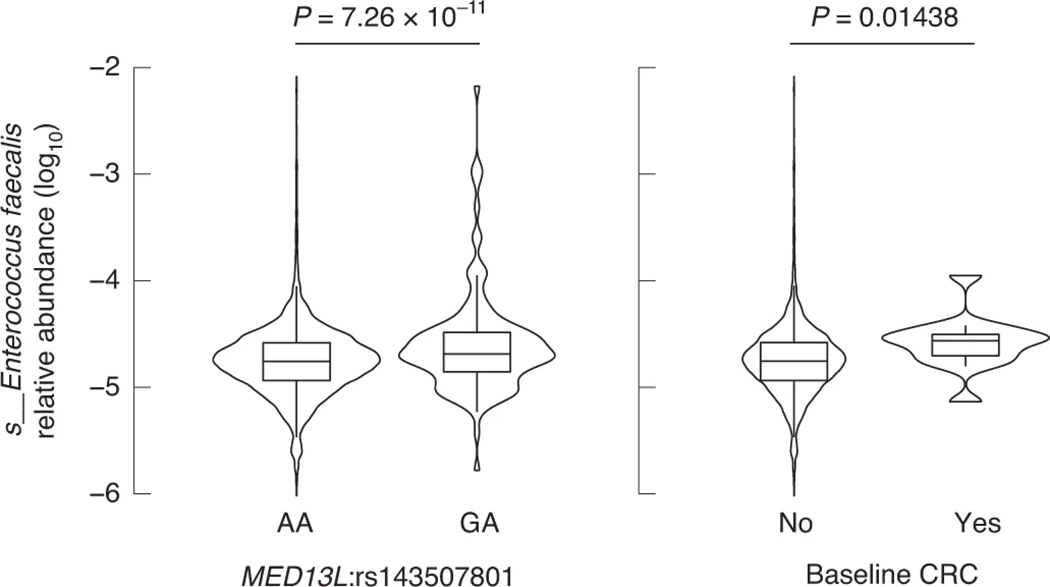
MED13L-associated E. faecalis as a putative link with colorectal cancer (CRC).
The allele frequency of the MED13L rs143507801 variant (A > G), associated with levels of E. faecalis (P = 7.26 × 10−11), was low (minor allele frequency = 0.0111), consistent with reported allele frequencies in the gnomAD database44. In our study population, 131 individuals carried rs143507801:G allele, 130 being heterozygous (GA) and only one being homozygous (GG). We observed that E. faecalis levels were increased in heterozygous rs143507801:GA individuals (Fig. 5). E. faecalis is a gut commensal, but also an opportunist pathogen believed to play a role in CRC development, possibly through direct damaging of colorectal cells45–47. MED13L and MED13 encode for Mediator transcriptional coactivator complex modules associating with RNA polymerase II (ref.48), and as such specifically interact with cyclin-dependent kinase 8 (CDK8) modules described for their oncogenic activation of transcription during colon tumorigenesis49. Consequently, we observed slightly higher levels of E. faecalis (P = 0.014) in 14 individuals enrolled in FR02 with a history of CRC at the time of sampling (Fig. 5). Groups of individuals segregated by allelic variant and CRC status could not be compared robustly due to small sample size. Taken together, these results suggest a possible link between E. faecalis and CRC through the MED13 activation of CDK8 in colorectal tumors, which will need to be investigated further.
Fig. 5 |. Effect of host genetics and prevalent CRC on gut levels of E. faecalis associated with MED13L variation across participants of the FR02 cohort.
Abundances are compared across individuals grouped according to (left panel) MED13L:rs143507801 genotype and (right panel) CRC prevalence according to the Finnish Cancer Registry. The comparison between E. faecalis variation and MED13L:rs143507801 reflects the GWAS results (Supplementary Table 1). The comparison of E. faecalis abundances in individuals with or without a history of CRC at the time of sampling was performed using a Wilcoxon rank test. Sample sizes for compared groups of individuals: rs143507801:A/A (n = 5,825), G/A (n = 130) (note: only 1 of 5,959 individuals in our cohort was G/G); with CRC (n = 14), without a history of CRC at baseline (n = 5,941). For all box plots, the central line, box and whiskers represent the median, IQR and 1.5 times the IQR, respectively. Violin plots represent the distribution density of the data points.
Mendelian randomization (MR) highlights possible causal effect of Morganella on major depressive disorder (MDD).
Interpreting results of causal inference prediction using bacterial information entails particular caution, due to the possibility of multiple and unaccounted confounding factors10, but can be useful to highlight potential focus for future research. Here we predicted 96 causal effects in both microbe-to-disease and disease-to-microbe directions using bidirectional MR. Of these, 34 were from microbial levels as exposure to disease as outcome, with a large proportion of causal effects in psychiatric and neurological diseases (Supplementary Table 5). For example, MR suggested an increased abundance of Faecalicoccus may have a causal effect on anorexia nervosa (odds ratio = 1.8 per s.d. increase in bacterial abundance; 95% confidence interval (95% CI) = 1.3–2.5; P = 2.0 × 10−4, MR method inverse-variance weighted) (Methods). Other examples included increasing abundances of Morganella and Raoultella predicted to have causal effects on MDD (Supplementary Table 5). When MR was performed in the reverse direction, using disease risk as an exposure and microbial level as an outcome, most predicted causal effects involved autoimmune and inflammatory diseases, but the strongest predicted causal effect involved type 2 diabetes (Supplementary Table 6). Doubling the genetic risk of type 2 diabetes (possibly accompanied by external factors such as hypoglycemic medications or metformin intake) was predicted to reduce levels of the uncultured CAG-345 sp000433315 species (Firmicutes phylum) by 0.14 s.d. (standard error = 0.04, P = 3.0 × 10−4, MR method inverse-variance weighted). A few other examples included some degree of literature validation, such as the higher genetic risk for primary sclerosing cholangitis causally impacting levels of the cholesterol-reducing Eubacterium_R coprostanoligenes50. Furthermore, a higher genetic risk for celiac disease was predicted to increase abundances in four species previously reported to be more abundant in patients with celiac disease than in controls51 (Supplementary Table 6). Finally, a higher genetic risk for multiple sclerosis was predicted to cause a reduction in the abundance of Lactobacillus_B ruminis, consistent with the report that Lactobacillus sp. can reduce symptom severity in an animal model of multiple sclerosis52.
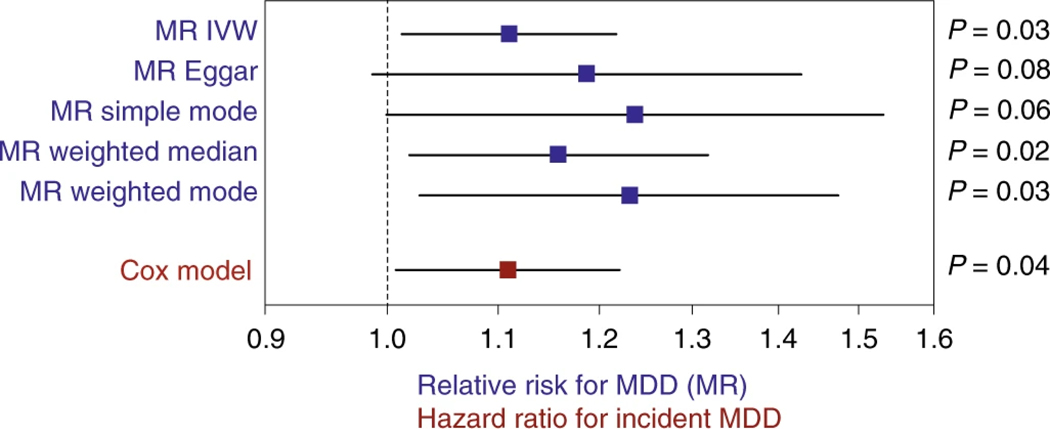
The availability in our study dataset of up to 16 yr of electronic health record follow-up after the initial sampling of the microbiota allowed for observational validation of predicted effects using MR. Of all causal predictions identified using MR, only the effect of Morganella on MDD could be validated by a statistically significant association with incident MDD (n = 181 cases; hazard ratio = 1.11, 95% CI = 1.01–1.22, per s.d. increase of bacterial abundance), after accounting for age, sex and BMI (Fig. 6). In our GWAS, Morganella variation in the study population associated with a variant (rs192436108; P = 6.16 × 10−8) in the PDE1A locus, which has previously been linked to depression53,54 and psychiatric disorders55. We did not find that the development of MDD could be linked to an abnormal incidence of microbiome-related diseases (Supplementary Table 8). Taken together, these predicted links between Morganella and MDD suggest more efforts should be deployed into exploring the possible roles of this bacterium as part of the brain–gut axis metabolic modulation of health.
Fig. 6 |. MR-based causal effects and incident depression analysis link Morganella with MDD.
Forest plot (in blue) representing the magnitude of the effect on MDD risk per 1-s.d. increase in bacterial abundance. MR analysis was carried out with 28 genetic instruments and their effect sizes from FR02 (5,959 samples) and MR-Base summary statistics (173,005 samples). In red is shown the hazard ratio for incident MDD in the FR02 cohort up to 16 yr after baseline sampling, using Cox model (Methods). Error bars represent the 95% CIs. IVW, inverse-variance weighted.
Discussion
Through GWAS and the subsequent investigation of functional and ecological factors contributing to the most robust human–microbe associations, we present a diverse and global picture of human– microbe interactions in a single cohort of ~6,000 European individuals. We find three genetic loci to be strongly associated with gut microbial variation. Two of these loci, LCT and ABO, are well known and very segregated in human populations, possibly explaining why our homogenous European cohort identified them as being associated so strongly. A third more mysterious association with the MED13L locus highlights possible links with cancer while causal inference highlights several diseases as being causally linked to gut microbes.
Lactase persistence, or the continued ability to digest lactose into adulthood, is the most strongly selected single-gene trait over the last 10,000 yr in multiple human populations56, believed to have spread amongst humans with the advent of animal domestication and the culturally transmitted practice of dairying57. In our study, as in previous work3,5,6,10, the association of LCT variants with Actinobacteria, more specifically Bifidobacterium, is by far the most statistically significant, suggesting a profound interaction between Actinobacteria and the human gut, in line with their reported keystone activities29. We reported a strong increase of Bifidobacterium levels in genetically lactose-intolerant people reporting a regular consumption of dairy products8. This increase was not confounded by age in adults, despite Bifidobacterium levels generally decreasing with age in our cohort. While self-reported dietary information is not entirely reliable due to various reasons58,59, our study population was large and the differences were significant enough to consider this a robust observation, which can be explained by the evolutionary adaptation of Bifidobacterium to specifically metabolize human and bovine milk oligosaccharides60. In lactase-deficient adults, consumed lactose is likely to become available for colonic bacteria as an energy source for which to compete. Hints of a possible competitive relationship between Bifidobacterium and Negativibacillus were revealed, which could depend upon lactose intake and should be investigated in functional studies.
Two considerations stem from our findings. First, the genetic determinants of lactose intolerance are known to vary across ethnicity61 and cross-population heterogeneity in the LCT–Bifidobacterium association was recently reported11. As more non-European-centric genetic studies are conducted worldwide11,62,63, examining this combined interaction between dairy diet and Bifidobacterium in different genetic backgrounds could bring new insights. Second, despite recent progresses, lactose intolerance is still largely underdiagnosed, and genetic prediction rates from large population studies exceed lactose intolerance prevalence rates obtained using physical tests61. In our study, we lacked information on lactose malabsorption symptoms in lactose-intolerant individuals reporting a regular dairy diet. Lactose-free (<0.01% lactose content) or low-lactose (<0.1%) dairy products have been available in Finland since 1978 and are popular among people experiencing symptoms of lactose malabsorption. Our data did not allow us to make the distinction between lactose-intolerant individuals aware of their symptoms and consuming low-lactose products as a result, and intolerant individuals unaware of the cause of their symptoms while consuming dairy. The latter would either experience discomfort symptoms without knowingly implicating their lactose intake, or the ability of a higher concentration of Bifidobacterium to degrade lactose in their intestines may alleviate the perceived symptoms of discomfort associated with lactose intolerance, therefore encouraging individuals to continue consuming indigestible lactose asymptomatically64. This possible probiotic effect should be investigated in controlled studies.
The ABO gene expresses a glycosyltransferase in many cell types, which determines the ABO blood group of an individual by modifying the oligosaccharides on cell-surface glycoproteins. A comparison of humans and nonhuman primates has identified ABO (along with the major histocompatibility complex) as harboring ancient multiallelic polymorphisms that are maintained across species65,66. Many infectious diseases such as norovirus infection, bacterial meningitis, malaria, cholera67 or even more recently SARS-CoV-2 (refs. 68,69) are associated with host blood type and secretor status67, suggesting that infection could be a driver of a strong balancing selection that has maintained ABO polymorphisms. Furthermore, blood type variation has been linked to various chronic diseases67, such as heart and vascular diseases, gastric cancers, diabetes, asthma or even dementia67. As many of these chronic diseases are also associated with dysbiosis of the gut microbiota, this prompts an interesting but largely unexplored parallel between gut commensals, blood types and disease38. Our study confirms previous findings4 that secretor status or blood types do not seem to globally affect gut microbial alpha- or beta-diversity. It also confirms reports from two very recent studies: first, a meta-analysis across five German cohorts, using 16S ribosomal RNA sequencing to characterize the gut microbiota, linked Bacteroides and Faecalibacterium to ABO and FUT2 (ref.70). The second study functionally associated bacterial lactose and galactose degradation genes to ABO variation in a cohort of 3,432 Chinese individuals71. Taken together, these findings suggest a broad association of ABO polymorphisms with microbial variation in various human populations.
An important research effort aiming to enzymatically produce synthetic universal donor blood has driven a push for screening a large diversity of CAZymes, including bacteria, revealing substrate affinities for blood antigens across various microbes36,37. Here we highlight F. lactaris (formerly Ruminococcus lactaris) as a mucin-degrading commensal likely able to digest blood antigens through its predicted GH110, GH136 and CBM32 CAZyme family genes39–41. F. lactaris is strongly associated with ABO genetic variation in our European cohort, and is differentially abundant in people according to their predicted gut mucosal secretion of A/B/ AB-antigens. Interestingly, our findings are not consistent with F. lactaris switching to a fiber-degrading activity in individuals reporting a high-fiber diet, unlike other mucin-degrading bacteria in our study and in the literature42 and Collinsella, another ABO-associated taxon. Our work suggests that some gut commensals such as F. lactaris appear to be very efficient and adapted metabolizers of A/B/AB-antigens in the gut, despite their predicted ability to degrade simpler carbohydrates in fiber. This could be an example of ecological niche differentiation in the gut, with impacts on associated F. lactaris microbial communities, to which Collinsella, also associated with ABO, may belong.
Although validation of the association is inconclusive because of the low prevalence of CRC cases and genetic variation in our study population, the association of MED13L rs143507801 variant with E. faecalis suggested a putative link with CRC. It has been shown that MED13 could directly link a CDK8 module to Mediator72,73, which is a CRC oncogene, amplified in colorectal tumors and activating transcription-driving colon tumorigenesis leading to CRC49. This could explain a long-suspected link between E. faecalis and development of CRC after having been found in higher concentrations in patients with CRC than in healthy individuals46,74. The suspected mode of action of E. faecalis on CRC development is currently unclear, but could be linked to extracellular free radical production directly leading to DNA breaks, point mutations and chromosomal instability in colorectal cells47. Although we saw a trend of E. faecalis being increased in abundance in individuals with a history of CRC, and in MED13L variation, more focused work including incident CRC and a larger sample size will be required to precisely pinpoint a link between this bacterium and CRC through the Mediator complex, if any.
Besides suggesting a link between gut microbes and autoimmune and inflammatory diseases, in line with previous studies75, causal inference analysis highlighted a very particular and promising example of interplay between a gut microbe and a complex disease. Among other suggested links with psychiatric diseases, we predicted increasing abundances of Morganella and Klebsiella (ex-Raoultella76,77) to have causal effects on MDD. Members of the Enterobacteriaceae family, such as these two genera, have previously been found in higher levels in patients with MDD78. Although caution is required when interpreting predictions of causality79, increasing evidence suggests that gut microbes are likely to influence host behavior via a systemic modulation of hormones and metabolites along the gut–brain axis80–82. Importantly, our MR-based result was consistent with observed hazards using follow-up observational data up to 16 yr after sampling. This observation supports previous experimental results showing an increase of IgM- and IgA-related immune responses against Morganella-secreted lipopolysaccharide in major depression83. A recent retrospective cohort study performed on 311 individuals including 156 MDD cases highlighted bacterial functions, metabolites and species involved in the interaction between the gut microbiome and MDD84. Although Morganella was not specifically highlighted, levels of several other Enterobacteriales species were found to significantly differ between patients with MDD and healthy controls84. Taken together, our findings highlight the intimate influence of the gut–brain axis on humans, with more mechanistic studies required to untangle and further interpret these predictions.
Our study highlights the benefits of increasing sample size to increase the statistical power for discovery. Although the LCT locus has been reported multiple times to be associated with bacterial taxa, to the best of our knowledge, our work is the first to report study-wide significant associations in a single cohort, at the strongest significance ever reported. The association with Bifidobacterium in our study was even stronger than the recent findings that used integrative data from 18,473 individuals in 28 different cohorts11, emphasizing the importance of standardized methodology and homogeneity in participant ethnicity (especially when studying geographically distributed traits such as lactose intolerance85). ABO allelic variation is also notoriously affected by geography86, which could explain why some meta-analyses in nonhomogenous populations could miss it. Also, metagenomic sequencing with standardized, robust taxonomic definitions87,88 can provide species-level characterization of microbial profiles in the gut of individuals, unlike 16S rRNA-based studies. An example from our work is the observation that B. dentium was prevalent but not associated with the LCT locus similar to all other Bifidobacterium species in the population. Observed differences in CAZymes commonly found in other Bifidobacterium species may explain this difference35. This should be confirmed in future experiments using more deeply sequenced metagenomes unambiguously linking function to particular metagenome-assembled genomes. Furthermore, GTDB taxonomic standardization results in greater taxon granularity, that is, smaller, more discrete clades of similar phylogenetic depth than commonly known lineages or species87,88. In theory, this would increase overall accuracy89, as a weak association with a poorly defined lineage may be caused by a strong association with a well-defined subset of that lineage, defined as a coherent group using GTDB88. Finally, a myriad of microbial taxa that are now solely defined and represented by uncultured metagenome-assembled genomes in the GTDB database were found to be independently associated with various loci. Along with recent reports that the more gut microbiome diversity is explored, the more novel, unknown species are discovered90,91, this suggests that many discoveries are yet to be made in the field of human microbiome studies.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41588-021-00991-z.
Methods
Study population.
The FINRISK study has been extensively described elsewhere92. FINRISK population surveys have been performed every 5 yr since 1972 to monitor trends of cardiovascular and other noncommunicable disease risk factors in the Finnish population92,93. The study population of this study consists of the participants of the FR02 study, including men and women aged between 25 and 74 yr from six geographical areas of Finland92,94,95. The sampling was stratified by sex, region and 10-yr age group so that each stratum had 250 participants. The overall participation rate was 65.5% (n = 8,798). Participants filled out a questionnaire, then participated in a clinical examination carried out by specifically trained nurses and gave a blood sample on which various laboratory measurements were performed. They also received a sampling kit and instructions to donate a stool sample at home and mailed it to the Finnish Institute for Health and Welfare in an overnight mail. The survey was done in Finland during winter months (January to March 2002), with average temperatures well below 0 °C. Special care was additionally taken to ensure that samples did not remain sitting in a post office more than 1 d, or over the weekend. Upon reception at the Finnish Institute for Health and Welfare (THL), samples were immediately frozen to −20 °C and kept unthawed until shipped to the University of California San Diego (USA), where they were processed and sequenced. The use of antibiotics was recorded from participants in the FR02 questionnaire and by linking with prescription registry. In addition, participants in each study site were asked whether they had an acute infection and were, as a general rule, asked to reschedule their examinations and stool sampling if they had.
The follow-up of the cohort took place by record linkage of the study data with the Finnish national electronic health registers (Hospital Discharge Register and Causes of Death Register), which provide in practice 100% coverage of relevant health events in Finnish residents. For present analyses involving follow-up data, we used a follow-up which extended until 31 December 2018.
The study protocol of FR02 was approved by the Coordinating Ethical Committee of the Helsinki and Uusimaa Hospital District (Ref. 558/E3/2001). All participants signed an informed consent. The study was conducted according to the World Medical Association’s Declaration of Helsinki on ethical principles.
Cohort phenotype metadata and specific dietary information.
The phenotype data in this study comprised demographic characteristics, life habits, disease history, clinical measurements, laboratory test results and follow-up electronic health records. More specifically, baseline dietary factors were collected. Details of the method have been described previously93. To broadly assess diet information within the cohort participants, a binary variable was used to indicate whether individuals were self-reporting to follow various possible dietary restrictions. Dietary consumption of specific food product categories was also reported. Habitual diet was assessed using a food propensity questionnaire which contained 42 food items or groups and had choices ranging 1–6 for consumption frequency, ranging from ‘Less than once a month’ to ‘Once a day or more often’. The consumption frequencies were converted to frequencies per month, ranging from 0.5 times per month to 30, 45 or 60 times per month. Food items that are rarely eaten more than once a day were given the value of 30 times per month. Food items that are often eaten multiple times a day such as fresh vegetables, breads, and so on were given a value of 60 times per month. Food items that fall in between these two groups were given 45 points.
Self-reporting of lactose-free diet and dietary fiber consumption.
Allelic distribution at the LCT-MCM6:rs4988235 variant responsible for lactase persistence in Europeans was as follows in our study population: 1,936 (35%) individuals had the T/T allele conferring a lactase persistence phenotype through adulthood, allowing them to digest lactose, while 981 (18%) individuals had the C/C allele conferring lactose intolerance. Most individuals (n = 2,611, 47%) had the intermediate allele, C/T, making them likely to be able to digest lactose. Most individuals reported a regular dairy intake in their diet (n = 5,002, 89%), while 706 (12.5%) individuals reported a regular lactose-free diet.
A total fiber consumption score was calculated from the questionnaires, reflecting the overall consumption of a combination of various fiber sources such as high-fiber bread, vegetables (vegetarian dishes, fresh vegetables, and boiled vegetables and legumes) and fruits, berries and natural juices. The resulting total fiber index values ranged from 9 (low dietary fiber intake) to 48 (high dietary fiber intake), with a median of 33. Comparisons of the effects of low- versus high-fiber diets were made between the 1st (n = 1,213) and 4th (n = 1,132) quartiles of the total fiber index.
Genotyping, imputation and quality control.
The genotyping was performed on Illumina genome-wide SNP arrays (the HumanCoreExome BeadChip, the Human610-Quad BeadChip and the HumanOmniExpress) and has been described previously96. Stringent criteria were applied to remove samples and variants of low quality. Samples with call rate < 95%, sex discrepancies, excess heterozygosity and non-European ancestry were excluded. Variants with call rate < 98%, deviation from Hardy–Weinberg equilibrium (P < 1 × 10−6) and minor allele count < 3 were filtered. Data were prephased by using Eagle2 v.2.3 (ref. 97). Imputation was performed using IMPUTE2 v.2.3.0 (ref. 98) with two Finnish-population-specific reference panels: 2,690 high-coverage whole-genome sequencing and 5,092 whole-exome sequencing samples. To evaluate the imputation quality, we compared the sample allele frequencies with reference populations and examined imputation quality (INFO scores) distributions. Imputed SNPs with INFO > 0.7 were kept for analysis. Postimputation quality control was carried out by using plink v.2.0 (ref. 99). Samples with >10% missing rate were removed. Individuals with extreme height or BMI values were further excluded (31 individuals with height < 1.47 m, 5 with BMI > 50 were removed). Both genotyped and imputed SNPs were kept for analysis if they met the following criteria: call rate > 90%, no significant deviation from Hardy–Weinberg equilibrium (P > 1.0 × 10−6) and minor allele frequency > 1%. SNP filtering was based on all individuals for which genotype information was available (n = 7,280), not on the 5,959 individuals selected subsequently for GWAS after quality control. The postquality control dataset comprised 7,967,866 SNPs.
Metagenomic sequencing from stool samples.
Stool samples were collected by participants and mailed overnight to the Finnish Institute for Health and Welfare for storing at −20 °C; the samples were sequenced at the University of California San Diego in 2017. No special arrangements were made regarding the temperature of the samples when they were shipped from the field clinics to the laboratory in THL but, as the survey was done in Finland during the winter months (January to March 2002), the average temperatures were well below 0 °C. Special care was anyway additionally taken to ensure that samples did not remain sitting in a post office over the weekend. The gut microbiome was characterized by shallow shotgun metagenomics sequencing with Illumina HiSeq 4000 Systems. We successfully performed stool shotgun sequencing in n = 7,231 individuals. The detailed procedures for DNA extraction, library preparation and sequence processing have been previously described95. Adapter and host sequences were removed. To preserve the quality of data while retaining most of the disease cases, samples with a total number of sequenced reads lower than 400,000 were removed.
Taxonomic profiling, quality filtering and data transformation.
Taxonomic profiling of FR02 metagenomes was performed as follows: briefly, raw shotgun metagenomic sequencing reads were mapped using the k-mer-based metagenomic classification tool Centrifuge v.1.0.4 (ref. 100) to an index database custom-built to encompass reference genomes that followed the taxonomic nomenclature introduced and updated in the GTDB release 89 (refs. 87–89). This implies that unless specified otherwise, all taxonomic names in our study refer to their nomenclature in GTDB, which can be related to the original National Center for Biotechnology Information (NCBI) nomenclature using the GTDB database server: https://gtdb.ecogenomic.org/taxon_history/. The same profiling approach has also been used and described in recent studies from our consortium94,95,101. Our study present results involving F. lactaris, which is named differently in NCBI and subsequent GTDB releases. A particular note on the evolution of this nomenclature can be found in the Supplementary Note.
Gut microbial composition was represented as the relative abundance of taxa. For each metagenome at phylum, class, order, family, genus and species levels, the relative abundance of a taxon was computed as the proportion of reads assigned to the clade rooted at this taxon among total classified reads. The relative abundance of a taxon with no reads assigned in a metagenome was considered as zero in the corresponding profile. For the purpose of this association study and because of reduced accuracy and power when considering rare taxa, we focused on common and relatively abundant microbial taxa, defined as prevalent in >25% of studied individuals, and defined with at least ten mapped reads per individual. For the purpose of association, and as previous studies have reported that only some microbial taxa are inheritable102, we also removed taxa with zero SNP heritability. This filtering resulted in a microbial dataset composed of a total of 2,801 taxa, including 59 phyla, 95 classes, 187 orders, 415 families, 922 genera and 1,123 species.
Taxonomic profiles derived from sequencing data are by nature compositional because of an arbitrary total imposed by the instrument103. The compositional data of microbial taxa are not independent and can lead to inappropriate use of linear regression. To overcome this artificial bias, all relative abundance values were transformed by center-log-ratio (CLR)104. More information about data transformation can be found in the Supplementary Note.
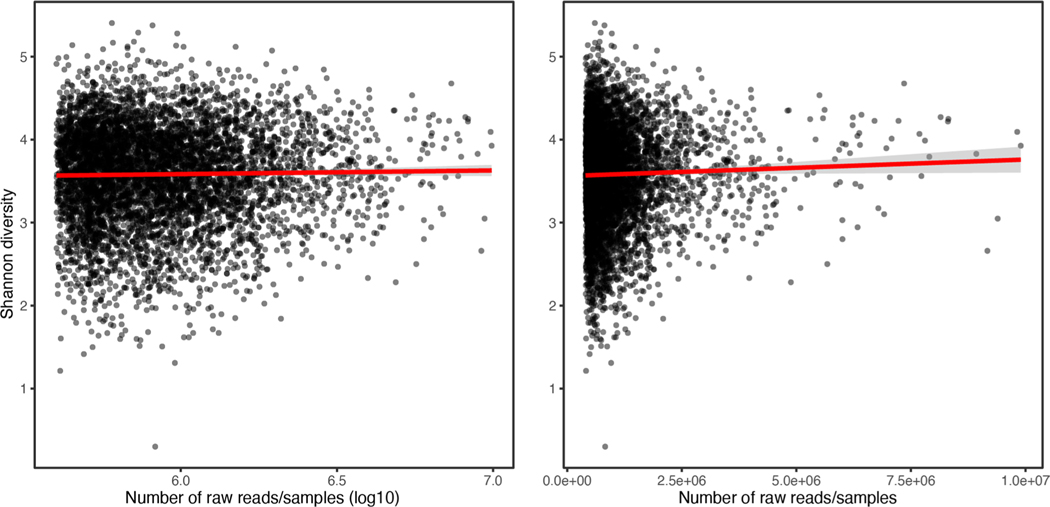
When visually comparing relative abundances in groups of individuals throughout the manuscript, we used untransformed relative abundances, for better interpretability. Alpha- (Shannon index) and beta- (Bray–Curtis distance) diversity were calculated at genus level used functions in the R package vegan v.2.5–6. We did not find a correlation between sequencing depth and Shannon diversity index (Spearman’s ⍴ = −0.001598, P = 0.90) in n = 5,959 samples (Extended Data Fig. 8). Additionally, to define CLR-transformed abundances of higher taxonomic levels than species, we summed the raw abundances of all taxa (for example, species) belonging to a specific higher taxonomic taxon (for example, genus), and then applied a CLR transformation. Additionally, we observed that Eastern and Western Finnish populations did not have different microbiome diversity, despite having overall slightly different lifestyles and mortality rates. To further investigate this, we visualized potential geographical effects using a Principal Coordinates Analysis (PCoA) plot on beta-diversity (Bray–Curtis dissimilarity) from metagenomic profiles of samples used in the GWAS from our study (n = 5,959; Extended Data Fig. 9).
Genome-wide association analysis.
The protocol followed in this study was described elsewhere105. Briefly, a linear mixed model (LMM) implemented in BOLT-LMM v.2.3.2 (ref. 106) was used to search for genome-wide associations accounting for the individual similarity. Since BOLT-LMM only accepts <1 million SNPs in modeling the genetic relationship matrix, SNPs were pruned at the threshold of r2 < 0.1 (plink2 (ref. 99), command–indep-pairwise 1000 80 0.1), resulting in 106,201 independent SNPs. This list of independent SNPs was used to estimate heritability using BOLT-LMM. Additionally, BOLT-LMM automatically performs leave-one-chromosome-out analysis to avoid proximal contamination. Although the LMM accounts for the cryptic relatedness in individuals, there are still large population structures that cannot be addressed. Thus, the top ten genetic principal components (calculated by FlashPCA v.2.0 (ref. 107) based on the pruned SNPs mentioned above) were included as covariates, in addition to age, sex and genotyping batch. We did not adjust for microbiome sequencing batch, as we observed that it had no effect on microbiome composition variation (Extended Data Fig. 9). As no genetic variant was reported to have a large effect size on gut microbiota, statistical estimates were based on infinitesimal model which assumes a small nonzero effect for a large number of genetic variants. To identify independent associations, GCTA-COJO v.1.91.3 (ref. 108) was used to conduct approximate conditional and joint analysis using individual genetic data. Window size was set to 10 megabases (Mb), assuming SNPs on different chromosomes or more than 10-Mb distance apart are uncorrelated. The resulting effect size (beta coefficient) indicated the number of standard deviation changes of a taxon’s CLR-transformed abundance corresponding to one effective allele increase of SNPs. Additionally, for all but two reported SNPs (rs146740485 and rs2797225), the effect allele was the reference allele in the GWAS cohort.
As microbes interact nonindependently with each other in the gut, as part of larger ecological and functional communities, matSpDlite v.1.0 (refs. 109,110) was used to estimate the number of independent tests based on eigenvalue variance—the larger the eigenvalue variance, the smaller the number of effective tests. The number of independent tests was 1,328 for 2,801 tested taxa. We used this information to calculate a Bonferroni-adjusted study-wide significance level for significant associations, which was set to 5 × 10−8/1,328 = 3.8 × 10−11. A genome-wide significance threshold was set as 5 × 10−8. The identified SNPs were annotated using ANNOVAR v.2018Apr16 (ref. 111) and grouped into genetic loci using 200-kilobase windows flanking the top SNPs.
We also examined whether antibiotic prescription before baseline sampling could be an important confounder of results. We obtained individual information on the prescription of any antibiotic up to 1 month before baseline fecal sampling, corresponding to 250 individuals of 5,959 (4.2%). We examined whether individual microbial profiles (via beta-diversity estimates using Bray–Curtis dissimilarity) were broadly affected by recent antibiotic prescription and observed a slight effect along PCoAs with significant variance explained (Extended Data Fig. 9c). After repeating the GWAS for all microbial taxa for which we initially had found at least one significantly associated locus, this time adjusting for previous antibiotic prescription status (‘yes’ versus ‘no’) (Supplementary Table 9), we found that recent antibiotic prescriptions had very minor effects on the GWAS association results. Adjusting for antibiotic prescription did not change any study-wide significant associations and only 32 of 567 genome-wide associations moved slightly above P = 5 × 10−8 (the largest P value was 3.2 × 10−7), which is likely by chance given inclusion of any additional covariate (Supplementary Table 9). In addition, the beta estimates with and without the adjustment of antibiotics usage were highly consistent (Pearson r = 0.9999487).
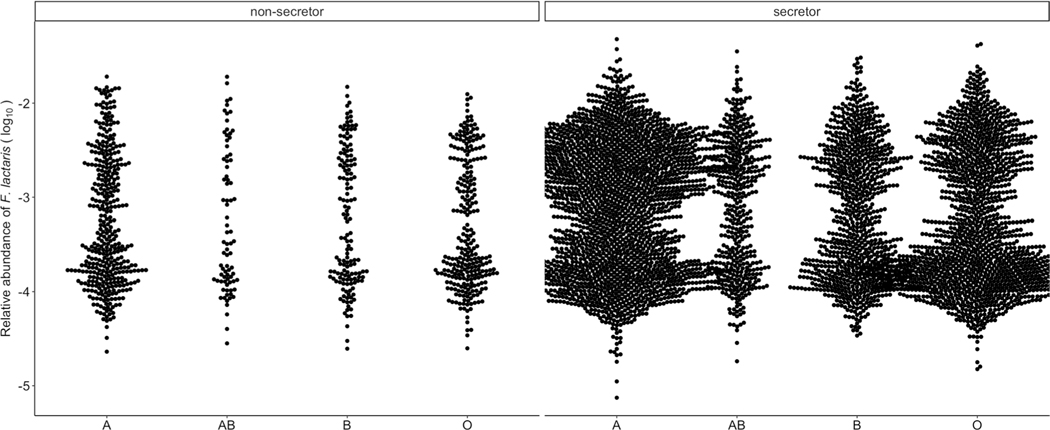
One important association in our study involved F. lactaris abundance and variants in the ABO locus. We observed that the distribution of F. lactaris abundance in our GWAS cohort (n = 5,959) was slightly bimodal (Extended Data Fig. 10). To investigate whether a logistic model gives the same result for this taxon, we arbitrarily coded F. lactaris abundance as ‘1’ if the relative abundance was higher than 5 × 10−4 (n = 2,866), and ‘0’ if smaller (n = 3,093). Akaike information criterion (AIC) value was smaller for logistic than for linear models (AIC = 8,196 versus AIC = 12,463, respectively), and the strongest association was also observed in the same top SNP (rs545971, P = 5.5 × 10−18) as when using linear regression (rs545971, P = 1.1 × 10−12).
Replication of previously reported associations.
To evaluate the reproducibility of our results with previously reported associations, we collected GWAS summary results from eight studies published in peer-reviewed journals at the time of this work3,6–10,102,112. These studies reported associations between 548 SNPs and microbial features. ANNOVAR was used to annotate the reported SNPs to the hg38 human reference genome111 and we used plink2 (ref. 99) to identify a further 15,427 SNPs in high LD (r2 > 0.8, within 5 Mb) with any of these 548 SNPs. To assess replication, we first examined whether previously reported associations could be matched in our results to identical or linked SNPs, with an association below the Bonferroni-corrected suggestive significance threshold, which was set to 0.05/548 = 9.124 × 10−5. More details about the replication methods and the use of the GTDB taxonomic system can be found in the Supplementary Note.
Prediction of ABO blood groups and secretor status.
SNP-based typing of ABO histo-blood group was performed. A combination of four SNPs113 was used for the prediction, and a 98% concordance with phenotypically typed ABO histo-blood group has been reported for this method4. For blood group allele A, the two different types, A1 and A2, were predicted by rs507666 and rs8176704, respectively. Blood group allele B was inferred from rs8176746 and blood group allele O was predicted by rs687289. As the combinations of these SNPs are exclusive, no haplotype information was needed. To validate the accuracy of prediction, we compared it with the prediction using a different combination of SNPs68. The two predictions were highly consistent, with over 99.9% concordance. In addition, the distribution of ABO groups was consistent with the population distribution found in public databases. Secretor status was predicted by the genotype of FUT2 variant rs601338, where AA or AG genotypes are secretors and GG genotypes are nonsecretors. A 100% concordance between the variation in rs601338 and secretor status was reported in a study on Finnish individuals114.
Bidirectional two-sample MR analysis.
Causal relationships between diseases and gut microbiota were investigated at genus and species levels only to maximize interpretability. In total, 213 species and 148 genera associated with at least one variant at genome-wide significant level (P < 1 × 10−8) were included. GWAS summary results were collected for 46 diseases from MR-Base115 (Supplementary Table 4). These included 12 autoimmune or inflammatory diseases, 9 cardiometabolic diseases, 13 psychiatric or neurological diseases, 4 bone diseases and 8 cancers. For diseases with more than one GWAS record, the record with the largest sample size was kept.
Bidirectional causal inference was performed to infer causal effects of microbial abundance variation (exposure) on disease risk (outcome), and of disease (exposure) on microbial abundance levels (outcome). To select the SNP instruments for microbial exposures in our study (Supplementary Table 7), we followed recommendations from a previous study showing that associated SNPs below a significance threshold of P < 1 × 10−5 had the largest explained variance on microbial features116. For each taxon, GCTA-COJO was used to perform a conditional analysis to select independently associated SNPs at P < 1 × 10−5. F statistics were calculated to estimate the strength of instruments for each bacterial exposure, and were found to be >10 for all exposures (Supplementary Table 5). SNP instruments for disease exposures were selected at genome-wide significance threshold (P < 5 × 10−8). Subsequently LD-clumping with a strict threshold (r2 < 0.001 in the 1000 Genomes European data within 10 Mb windows) was conducted to select independent instruments with the lowest P values for taxa and diseases, respectively.
Details about the precise methods used for MR inference can be found in the Supplementary Note.
Cox proportional hazards regression.
Cox proportional hazards regression was conducted to test the association between baseline abundance of gut microbes and incident major depression (16 yr of follow-up, n = 181 incident events). Microbial abundances were CLR-transformed and standardized to zero-mean and unit-variance. The Cox models were stratified by sex and adjusted for age and log-transformed BMI, with time-on-study as the time scale. Participants with prevalent major depression at baseline were excluded. R function coxph() in the R package survival v.3.1–8 was used for this analysis.
Profiling of CAZymes in bacterial genomes.
The standalone run_dbCAN2 v.2.0.11 tool117 (https://github.com/linnabrown/run_dbcan) was used to scan for the presence of CAZyme genes in public assembled bacterial genomes taken from the GTDB release 89 reference. We used a CAZyme reference database taken from the CAZy database118 (31 July 2019 update). In total, we scanned 327 Bifidobacterium sp., 2 F. lactaris and 15 Collinsella sp. reference genomes included in GTDB release 89. Three methods were compared as part of the run_dbCAN2 procedure (HMMER, DIAMOND and Hotpep). We considered it a positive detection result when all three methods agreed on a CAZyme family identification. Identification of preferred reported substrates for the various CAZyme families was done manually from key publications42,119, from literature searches and from the CAZypedia website120. Certain CAZyme families have a broad range of substrates, many of which are still unknown, which results in our reported preferred substrates to be as accurate as possible, but nonexhaustive.
Carbon impact and offsetting.
We used GreenAlgorithms v.1.0 (ref. 121) to estimate that the main computational work in this study had a carbon impact of at least 2,660 kg of CO2 emissions (CO2e), corresponding to 233 tree-years. As a commitment to the reduction of carbon emissions associated with computation in research, we consequently funded planting of 30 trees through a local Australian charity, which across their lifetime will sequester a combined estimated 8,040 kg of CO2e, or three times the amount of CO2e generated by this study.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Complete summary statistics of microbial taxa with genome-wide significant hits are publicly available in the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) from accession GCST90032172 to GCST90032644. The metagenomic data from FINRISK 2002 samples are available from the European Genome- Phenome Archive (study ID: EGAS00001005020). The phenotype data contain sensitive information from healthcare registers and are not publicly available to avoid compromising research participant privacy/consent. They are available through the THL biobank upon submission of a research plan and signing a data transfer agreement (https://thl.fi/en/web/thl-biobank/for-researchers/application-process). Additional databases used in this work include GTDB release 89 (https://gtdb.ecogenomic.org/) and CAZy (last accessed 31 July 2019) (http://www.cazy.org/).
Code availability
Scripts used to analyze nonidentifiable data in this study have been made available on Zenodo (https://doi.org/10.5281/zenodo.5641303).
Extended Data
Extended Data Fig. 1 |.
Study flowchart.
Extended Data Fig. 2 |. Heritability of SNPs associated with microbial taxa.
(a) Associated SNP heritability (h2) for all 2,801 taxa included in the genome-wide association analysis, grouped into their 61 corresponding GTDB phyla, and ordered by median heritability per phylum. Red denotes bacterial phyla, and purple denotes archaeal phyla. The right panel indicates the number of genome-wide significant associated taxa for each phylum. (b) Associated SNP heritability is shown for each associated taxon, grouped by its taxonomic rank. Genome-wide significance was defined as a threshold of p < 5 × 10−8 for all p-values obtained after joint analysis using GTCA-COJO in the GWAS (see Methods). For all box plots (A and B), the central line, box and whiskers represent the median, interquartile range (IQR) and 1.5 times the IQR, respectively.
Extended Data Fig. 3 |. LocusZoom plots for three loci with study-wide significant associations (p < 3.8 × 10−11).
Associations with top taxa are shown. Top SNPs are indicated in purple diamond. Other SNPs are coloured by their linkage disequilibrium (LD) values with the top SNPs. Genes covered by the region are indicated in the bottom and the genotyping coverage is indicated on top of the plot. (A) Associated SNPs at the LCT locus spans over a 2 Mbp genomic region, while they are grouped on a 400 kbp region for both (B) ABO and (C) the MED13L loci.
Extended Data Fig. 4 |. Correlation between individual baseline age and the relative abundance of bacteria from the Bifidobacterium genus in lactose intolerant individuals.
Only genetically lactose intolerance individuals (rs4988235:CC) are shown, and coloured by dietary dairy habits (blue: self-reported regular consumption of dairy, n = 763; and red: self-reported irregular dairy diet or lactose-free diet, n = 253). Best fitted lines and 95% confidence intervals are indicated. Two-sided Spearman correlation coefficients are given.
Extended Data Fig. 5 |. Spearman correlation of relative abundances in 4 taxa associated with the LCT locus.
Abundances of the Bifidobacterium, Negativibacillus, UBA3855 and CAG-81 genera are compared. Abundances in the entire FR02 cohort is compared to those in a subset of genetically lactose-intolerant individuals, and to a subset of genetically lactose-intolerant individuals who reported a regular dairy diet. Coloured boxes denote the strength of correlation (ranging from −1 in red to 1 in dark blue), while a white square denotes a non-significant p-value for the two-sided Spearman correlation (p > 0.05).
Extended Data Fig. 6 |. Co-abundance and carbohydrate-active enzymes (CAZyme) distribution patterns in 11 Bifidobacterium species harboured by > 25% of individuals in the FR02 cohort.
(a) Associations between the LCT-MCM6 locus and 11 Bifidobacterium species; (left) top association results between variation of 11 Bifidobacterium species and the LCT locus, with study-wide significant associations (with p-values from the joint analysis using GTCA-COJO below the p < 3.8 × 10−11 threshold) highlighted in bold; (middle) Two-sided Spearman coefficients calculated on CLR-transformed abundances; (right) relative abundances across the FR02 cohort, ranging from 0 (light green) to 1 (dark blue). (b) CAZyme distribution patterns in 327 previously published reference genomes from 11 Bifidobacterium GTDB species which were included in the GTDB release 89 index used to classify metagenomes in this study. The heatmap indicates abundance of corresponding CAZyme families in species, corresponding to the total count of detected families for each species divided by the number of reference genomes examined for the same species. Values <1 (white to light blue) indicate that less than one copy per genome of the corresponding CAZyme family was detected for each species, values >1 (light blue to dark blue) indicate that more than one copy per genome was detected. Preferred substrate groups are based on literature search and descriptions on CAZypedia.org. For all box plots (A), the central line, box and whiskers represent the median, interquartile range (IQR) and 1.5 times the IQR, respectively. Violin plots represent the distribution density of the data points.
Extended Data Fig. 7 |. Effect of ABO genotypes, blood type and secretor status on microbial diversity and gut levels of ABO-associated taxa.
(a) (left) Alpha diversity represented by Shannon indices; (right) beta diversity, represented by Bray-Curtis distances. Alpha and beta diversity were calculated from individual taxonomic profiles at the genus level. Individuals were segregated according to their predicted blood type and secretor status, both predicted from genotype data. (b) Abundances are compared across stratified groups of individuals from the FR02 cohort according to (left panel): ABO:rs545971 genotype and predicted secretor status (blue: secretor status conferred by FUT2 rs601338:AG/AA genotype; red: non-secretor status conferred by FUT2 rs601338:GG genotype) and (right panel) according to predicted A, AB, B and O blood types, and predicted secretor status. All statistical comparisons denote the p-values of Wilcoxon rank test on the distributions. (c) Effect of AB antigen secretion on gut microbial relative abundance, using the 2,801 taxa considered for GWAS in our study. Taxa with FDR adjusted p value <0.05 are highlighted in red. Red line indicates the expected distribution of p values under the null hypothesis. P values were calculated using Wilcoxon rank test. For all box plots (A and B), the central line, box and whiskers represent the median, interquartile range (IQR) and 1.5 times the IQR, respectively. Violin plots represent the distribution density of the data points.
Extended Data Fig. 8 |. Sequencing depth does not influence alpha diversity.
Alpha-diversity (Shannon index) were computed and plotted against the log10 (left) or the raw (right) number of sequencing reads for each 5,959 individual gut metagenome in this study. No correlation was observed between sequencing depth and Shannon diversity index (two-sided Spearman’s ⍴=−0.001598, p = 0.90). Grey shaded area corresponds to the 95% confidence interval.
Extended Data Fig. 9 |. Effect of geographical region of sampling, microbiome sequencing batch or antibiotic prescription on overall microbiome diversity.
Beta-diversity (Bray Curtis dissimilarity indices) was calculated using the R package vegan, and the 4 top PCoA axes (explaining a combined 25.9% of the total microbiome variation) were plotted against each other, with each individual point labelled according to geographical sampling (panel A), gut metagenomic sequencing batch (panel B), or whether antibiotics were prescribed up to 1 month (n = 250/5959) before baseline sampling.
Extended Data Fig. 10 |. Distribution of F. lactaris relative abundance in groups of individuals with different predicted blood types.
A beeswarm plot is used to visualise the distribution of relative abundances.
Supplementary Material
Acknowledgements
The study protocol of FINRISK 2002 was approved by the Coordinating Ethical Committee of the Helsinki and Uusimaa Hospital District (Ref. 558/E3/2001). All participants signed an informed consent. The study was conducted according to the World Medical Association Declaration of Helsinki on ethical principles. All necessary patient/participant consent has been obtained and the appropriate institutional forms have been archived. We thank all participants of the FINRISK 2002 survey for their contributions to this work. The FINRISK surveys are mainly funded by budgetary funds from the Finnish Institute for Health and Welfare with additional funding from several domestic foundations. Y.Q. was partially supported by The Albert Shimmins Fund (Faculty of Science Postgraduate Writing-Up Award 2020). M.I. was supported by the Munz Chair of Cardiovascular Prediction and Prevention and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). V.S. was supported by the Finnish Foundation for Cardiovascular Research. L.L. was supported by the Academy of Finland (decision 295741) and EU/H2020 (FindingPheno; grant 952914). T.N. was supported by the Emil Aaltonen Foundation, the Finnish Medical Foundation, the Paavo Nurmi Foundation and the Academy of Finland (grant no. 321351). A.S.H. was supported by the Academy of Finland, grant no. 321356. R.L. receives funding support from NIEHS (grant no. 5P42ES010337), NCATS (grant no. 5UL1TR001442), NIDDK (grant nos. U01DK061734, R01DK106419, P30DK120515, R01DK121378, R01DK124318) and DOD PRCRP (grant no. W81XWH-18-2-0026). S.C.R. is funded by a BHF Programme Grant (RG/18/13/33946). This study was supported by the Victorian Government’s Operational Infrastructure Support (OIS) program, and by core funding from the British Heart Foundation (grant no. RG/13/13/30194; RG/18/13/33946) and the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). National Institute for Health Research (Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust) (The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care). This work was supported by Health Data Research UK, which is funded by the UK Medical Research Council, the Engineering and Physical Sciences Research Council, the Economic and Social Research Council, the Department of Health and Social Care (England), the Chief Scientist Office of the Scottish Government Health and Social Care Directorates, the Health and Social Care Research and Development Division (Welsh Government), the Public Health Agency (Northern Ireland), the British Heart Foundation and Wellcome. We thank Dr Annalisa Buniello (EMBL-EBI, Cambridge, UK) for valuable help with GWAS Catalog submissions.
Footnotes
Competing interests
V.S. has consulted for Novo Nordisk and Sanofi and received honoraria from these companies. He also has ongoing research collaboration with Bayer AG, all unrelated to this study. R.L. serves as a consultant or advisory board member for Anylam/Regeneron, Arrowhead Pharmaceuticals, AstraZeneca, Bird Rock Bio, Boehringer Ingelheim, Bristol-Myer Squibb, Celgene, Cirius, CohBar, Conatus, Eli Lilly, Galmed, Gemphire, Gilead, Glympse bio, GNI, GRI Bio, Inipharm, Intercept, Ionis, Janssen Inc., Merck, Metacrine, Inc., NGM Biopharmaceuticals, Novartis, Novo Nordisk, Pfizer, Prometheus, Promethera, Sanofi, Siemens and Viking Therapeutics. In addition, his institution has received grant support from Allergan, Boehringer Ingelheim, Bristol-Myers Squibb, Cirius, Eli Lilly and Company, Galectin Therapeutics, Galmed Pharmaceuticals, GE, Genfit, Gilead, Intercept, Grail, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, NuSirt, Pfizer, pH Pharma, Prometheus and Siemens. He is also cofounder of Liponexus, Inc. The remaining authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41588-021-00991-z.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41588-021-00991-z.
Peer review information Nature Genetics thanks Kenneth Croitoru and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Belizário JE & Napolitano M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Front. Microbiol 6, 1050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levy M, Kolodziejczyk AA, Thaiss CA & Elinav E. Dysbiosis and the immune system. Nat. Rev. Immunol 17, 219–232 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Blekhman R. et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 16, 191 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davenport ER et al. ABO antigen and secretor statuses are not associated with gut microbiota composition in 1,500 twins. BMC Genomics 17, 941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodrich JK et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonder MJ et al. The effect of host genetics on the gut microbiome. Nat. Genet 48, 1407–1412 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Turpin W. et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet 48, 1413–1417 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Wang J. et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet 48, 1396–1406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothschild D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Hughes DA et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol 5, 1079–1087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurilshikov A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet 53, 156–165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolde R. et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med. 10, 6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rühlemann MC et al. Application of the distance-based F test in an mGWAS investigating β diversity of intestinal microbiota identifies variants in SLC9A8 (NHE8) and 3 other loci. Gut Microbes 9, 68–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodrich JK et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie H. et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 3, 572–584.e3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim MY et al. The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut 66, 1031–1038 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Le Roy CI et al. Heritable components of the human fecal microbiome are associated with visceral fat. Gut Microbes 9, 61–67 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodrich JK, Davenport ER, Clark AG & Ley RE The relationship between the human genome and microbiome comes into view. Annu. Rev. Genet 51, 413–433 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurilshikov A, Wijmenga C, Fu J. & Zhernakova A. Host genetics and gut microbiome: challenges and perspectives. Trends Immunol. 38, 633–647 (2017). [DOI] [PubMed] [Google Scholar]
- 20.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falony G. et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 22.Zhernakova A. et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 352, 565–569 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eng A. & Borenstein E. Taxa-function robustness in microbial communities. Microbiome 6, 45 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrer M. et al. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure: metaproteomic insights associated to human obesity. Environ. Microbiol 15, 211–226 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Moya A. & Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 24, 402–413 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Louca S. et al. Function and functional redundancy in microbial systems. Nat. Ecol. Evol 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Louca S. et al. High taxonomic variability despite stable functional structure across microbial communities. Nat. Ecol. Evol 1, 0015 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Banerjee S, Schlaeppi K. & van der Heijden MGA Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol 16, 567–576 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Trosvik P. & de Muinck EJ Ecology of bacteria in the human gastrointestinal tract—identification of keystone and foundation taxa. Microbiome 3, 44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shetty SA, Hugenholtz F, Lahti L, Smidt H. & de Vos WM Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS Microbiol. Rev 41, 182–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chia LW et al. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van Leeuwenhoek 111, 859–873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee S, Schlaeppi K. & van der Heijden MGA Reply to ‘Can we predict microbial keystones?’. Nat. Rev. Microbiol 17, 194 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Röttjers L. & Faust K. Can we predict keystones? Nat. Rev. Microbiol 17, 193 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Kato K. et al. Age-related changes in the composition of gut Bifidobacterium species. Curr. Microbiol 74, 987–995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engevik MA et al. Bifidobacterium dentium fortifies the intestinal mucus layer via autophagy and calcium signaling pathways. mBio 10, e01087–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahfeld P. & Withers SG Toward universal donor blood: enzymatic conversion of A and B to O type. J. Biol. Chem 295, 325–334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu QP et al. Bacterial glycosidases for the production of universal red blood cells. Nat. Biotechnol 25, 454–464 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Arnolds KL, Martin CG & Lozupone CA Blood type and the microbiome—untangling a complex relationship with lessons from pathogens. Curr. Opin. Microbiol 56, 59–66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu QP et al. Identification of a GH110 subfamily of α1,3-galactosidases: novel enzymes for removal of the α3GAL xenotransplantation antigen. J. Biol. Chem 283, 8545–8554 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichler MJ et al. Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat. Commun 11, 3285 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ficko-Blean E. & Boraston AB The interaction of a carbohydrate-binding module from a Clostridium perfringens N-acetyl-β-hexosaminidase with its carbohydrate receptor. J. Biol. Chem 281, 37748–37757 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Desai MS et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tailford LE, Crost EH, Kavanaugh D. & Juge N. Mucin glycan foraging in the human gut microbiome. Front. Genet 6, 81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Genome Aggregation Database Consortium et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amarnani R. & Rapose A. Colon cancer and enterococcus bacteremia co-affection: a dangerous alliance. J. Infect. Public Health 10, 681–684 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Khan Z, Siddiqui N. & Saif MW Enterococcus faecalis infective endocarditis and colorectal carcinoma: case of new association gaining ground. Gastroenterol. Res 11, 238–240 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huycke MM, Abrams V. & Moore DR Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 23, 529–536 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Allen BL & Taatjes DJ The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Firestein R. et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455, 547–551 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li L, Batt SM, Wannemuehler M, Dispirito A. & Beitz DC Effect of feeding of a cholesterol-reducing bacterium, Eubacterium coprostanoligenes, to germ-free mice. Lab. Anim. Sci 48, 253–255 (1998). [PubMed] [Google Scholar]
- 51.Marasco G. et al. Gut microbiota and celiac disease. Dig. Dis. Sci 61, 1461–1472 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Lavasani S. et al. A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS ONE 5, e9009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomita H. et al. G protein-linked signaling pathways in bipolar and major depressive disorders. Front. Genet 4, 297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong M-L et al. Phosphodiesterase genes are associated with susceptibility to major depression and antidepressant treatment response. Proc. Natl Acad. Sci. USA 103, 15124–15129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schork AJ et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat. Neurosci 22, 353–361 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burger J. et al. Low prevalence of lactase persistence in Bronze Age Europe indicates ongoing strong selection over the last 3,000 years. Curr. Biol 10.1016/j.cub.2020.08.033 (2020). [DOI] [PubMed]
- 57.Gerbault P. et al. Evolution of lactase persistence: an example of human niche construction. Philos. Trans. R. Soc. Lond. B Biol. Sci 366, 863–877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hebert JR et al. Social desirability trait influences on self-reported dietary measures among diverse participants in a multicenter multiple risk factor trial. J. Nutr 138, 226S–234S (2008). [DOI] [PubMed] [Google Scholar]
- 59.Schoeller DA How accurate is self-reported dietary energy intake? Nutr. Rev 48, 373–379 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Sakanaka M. et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv 5, eaaw7696 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Storhaug CL, Fosse SK & Fadnes LT Country, regional, and global estimates for lactose malabsorption in adults: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol 2, 738–746 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Liu X. et al. A genome-wide association study for gut metagenome in Chinese adults illuminates complex diseases. Cell Discov. 7, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sirugo G, Williams SM & Tishkoff SA The missing diversity in human genetic studies. Cell 177, 26–31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szilagyi A. Adaptation to lactose in lactase non persistent people: effects on intolerance and the relationship between dairy food consumption and evalution of diseases. Nutrients 7, 6751–6779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ségurel L, Gao Z. & Przeworski M. Ancestry runs deeper than blood: the evolutionary history of ABO points to cryptic variation of functional importance. Bioessays 10.1002/bies.201300030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Segurel L. et al. The ABO blood group is a trans-species polymorphism in primates. Proc. Natl Acad. Sci. USA 109, 18493–18498 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewald DR & Sumner SCJ Blood type biochemistry and human disease. Wiley Interdisp. Rev. Syst. Biol. Med 8, 517–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellinghaus D. et al. Genomewide asociation study of severe Covid-19 with respiratory failure. N. Engl. J. Med 10.1056/NEJMoa2020283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shelton JF et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat. Genet 53, 801–808 (2021). [DOI] [PubMed] [Google Scholar]
- 70.Rühlemann MC et al. Genome-wide association study in 8,956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat. Genet 53, 147–155 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Liu X. et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Preprint at bioRxiv 10.1101/2020.06.30.181438 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Knuesel MT, Meyer KD, Bernecky C. & Taatjes DJ The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23, 439–451 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tsai K-L et al. A conserved Mediator–CDK8 kinase module association regulates Mediator–RNA polymerase II interaction. Nat. Struct. Mol. Biol 20, 611–619 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Almeida C. et al. Differential responses of colorectal cancer cell lines to Enterococcus faecalis’ strains isolated from healthy donors and colorectal cancer patients. J. Clin. Med 8, 388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marchesi JR et al. The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma Y. et al. Proposal for reunification of the genus Raoultella with the genus Klebsiella and reclassification of Raoultella electrica as Klebsiella electrica comb. nov. Res. Microbiol 10.1016/j.resmic.2021.103851 (2021). [DOI] [PubMed] [Google Scholar]
- 77.Wyres KL, Lam MMC & Holt KE Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol 18, 344–359 (2020). [DOI] [PubMed] [Google Scholar]
- 78.Jiang H. et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun 48, 186–194 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Wade KH & Hall LJ Improving causality in microbiome research: can human genetic epidemiology help? Wellcome Open Res. 4, 199 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Foster JA & McVey Neufeld K-A Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 36, 305–312 (2013). [DOI] [PubMed] [Google Scholar]
- 81.Fung TC, Olson CA & Hsiao EY Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci 20, 145–155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Valles-Colomer M. et al. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol 4, 623–632 (2019). [DOI] [PubMed] [Google Scholar]
- 83.Maes M, Kubera M. & Leunis J-C The gut-brain barrier in major depression: intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuro Endocrinol. Lett 29, 117–124 (2008). [PubMed] [Google Scholar]
- 84.Yang J. et al. Landscapes of bacterial and metabolic signatures and their interaction in major depressive disorders. Sci. Adv 6, eaba8555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mattar R, de Campos Mazo DF & Carrilho FJ Lactose intolerance: diagnosis, genetic, and clinical factors. Clin. Exp. Gastroenterol 5, 113–121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bodmer W. Genetic characterization of human populations: from ABO to a genetic map of the British people. Genetics 199, 267–279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Parks DH et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol 36, 996–1004 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Parks DH et al. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol 10.1038/s41587-020-0501-8 (2020). [DOI] [PubMed] [Google Scholar]
- 89.Méric G, Wick RR, Watts SC, Holt KE & Inouye M. Correcting index databases improves metagenomic studies. Preprint at bioRxiv 10.1101/712166 (2019). [DOI] [Google Scholar]
- 90.Pasolli E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Almeida A. et al. A unified catalog of 204,938 reference genomes from the human gut microbiome. Nat. Biotechnol 10.1038/s41587-020-0603-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Borodulin K. et al. Cohort profile: the national FINRISK study. Int. J. Epidemiol 47, 696–696i (2018). [DOI] [PubMed] [Google Scholar]
- 93.Borodulin K. et al. Forty-year trends in cardiovascular risk factors in Finland. Eur. J. Public Health 25, 539–546 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Liu Y. et al. Early prediction of liver disease using conventional risk factors and gut microbiome-augmented gradient boosting. Preprint at medRxiv 10.1101/2020.06.24.20138933 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Salosensaari A. et al. Taxonomic signatures of cause-specific mortality risk in human gut microbiome. Nat. Commun 12, 2671 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.FinnGen et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat. Med 26, 549–557 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Loh P-R et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet 48, 1443–1448 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Howie B, Fuchsberger C, Stephens M, Marchini J. & Abecasis GR Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet 44, 955–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim D, Song L, Breitwieser FP & Salzberg SL Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 26, 1721–1729 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruuskanen MO et al. Links between gut microbiome composition and fatty liver disease in a large population sample. Gut Microbes 13, 1–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Goodrich JK, Davenport ER, Waters JL, Clark AG & Ley RE Cross-species comparisons of host genetic associations with the microbiome. Science 352, 532–535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gloor GB, Macklaim JM, Pawlowsky-Glahn V. & Egozcue JJ Microbiome datasets are compositional: and this is not optional. Front. Microbiol 8, 2224 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aitchison J, Barceló-Vidal C, Martín-Fernández JA & Pawlowsky-Glahn V. Logratio Analysis and Compositional Distance. Math. Geol 32, 271–275 (2000). [Google Scholar]
- 105.Qin Y. et al. Genome-wide association and Mendelian randomization analysis prioritizes bioactive metabolites with putative causal effects on common diseases. Preprint at medRxiv 10.1101/2020.08.01.20166413 (2020). [DOI] [Google Scholar]
- 106.Loh P-R et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abraham G, Qiu Y. & Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics 33, 2776–2778 (2017). [DOI] [PubMed] [Google Scholar]
- 108.Genetic Investigation of ANthropometric Traits (GIANT) Consortium et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet 44, 369–375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li J. & Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 95, 221–227 (2005). [DOI] [PubMed] [Google Scholar]
- 110.Nyholt DR A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet 74, 765–769 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang K, Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Davenport ER et al. Seasonal variation in human gut microbiome composition. PLoS ONE 9, e90731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Paré G. et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 4, e1000118 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wacklin P. et al. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS ONE 6, e20113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hemani G. et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sanna S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet 51, 600–605 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang H. et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cantarel BL et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cantarel BL, Lombard V. & Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS ONE 7, e28742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.The CAZypedia Consortium. Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes. Glycobiology 28, 3–8 (2018). [DOI] [PubMed] [Google Scholar]
- 121.Lannelongue L, Grealey J. & Inouye M. Green algorithms: quantifying the carbon footprint of computation. Adv. Sci 8, 2100707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete summary statistics of microbial taxa with genome-wide significant hits are publicly available in the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/) from accession GCST90032172 to GCST90032644. The metagenomic data from FINRISK 2002 samples are available from the European Genome- Phenome Archive (study ID: EGAS00001005020). The phenotype data contain sensitive information from healthcare registers and are not publicly available to avoid compromising research participant privacy/consent. They are available through the THL biobank upon submission of a research plan and signing a data transfer agreement (https://thl.fi/en/web/thl-biobank/for-researchers/application-process). Additional databases used in this work include GTDB release 89 (https://gtdb.ecogenomic.org/) and CAZy (last accessed 31 July 2019) (http://www.cazy.org/).