Abstract
Background
Patients with COVID-19 are at increased risk of thrombosis, which is associated with altered platelet function and coagulopathy, contributing to excess mortality.
Objectives
To characterize the mechanism of altered platelet function in COVID-19 patients.
Methods
The platelet proteome, platelet functional responses, and platelet-neutrophil aggregates were compared between patients hospitalized with COVID-19 and healthy control subjects using tandem mass tag proteomic analysis, Western blotting, and flow cytometry.
Results
COVID-19 patients showed a different profile of platelet protein expression (858 altered of the 5773 quantified). Levels of COVID-19 plasma markers were enhanced in the platelets of COVID-19 patients. Gene ontology pathway analysis demonstrated that the levels of granule secretory proteins were raised, whereas those of platelet activation proteins, such as the thrombopoietin receptor and protein kinase Cα, were lowered. Basally, platelets of COVID-19 patients showed enhanced phosphatidylserine exposure, with unaltered integrin αIIbβ3 activation and P-selectin expression. Agonist–stimulated integrin αIIbβ3 activation and phosphatidylserine exposure, but not P-selectin expression, were decreased in COVID-19 patients. COVID-19 patients had high levels of platelet-neutrophil aggregates, even under basal conditions, compared to controls. This association was disrupted by blocking P-selectin, demonstrating that platelet P-selectin is critical for the interaction.
Conclusions
Overall, our data suggest the presence of 2 platelet populations in patients with COVID-19: one of circulating platelets with an altered proteome and reduced functional responses and another of P-selectin-expressing neutrophil–associated platelets. Platelet–driven thromboinflammation may therefore be one of the key factors enhancing the risk of thrombosis in COVID-19 patients.
keywords: COVID-19, platelet activation, proteomics, SARS-CoV-2, thromboinflammation
1. Introduction
The COVID-19 pandemic started in China in 2019 and rapidly spread over the world causing more than 6.5 million deaths to date [1]. The disease is caused by a single positive-sense RNA virus, severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2), which can cause a wide variety of clinical syndromes, with most hospitalized patients diagnosed with pneumonitis. During the COVID-19 pandemic, it has become apparent that patients with COVID-19 have enhanced rates of venous and arterial thrombosis, including deep-vein thrombosis, pulmonary embolism, myocardial infarction, and ischemic stroke [[2], [3], [4], [5], [6]]. Circulating platelet microaggregates were detected in COVID-19 patients, showing a strong correlation with COVID-19 severity [7,8]. Postmortem examinations revealed microthrombi in the lung, kidney, liver, heart, and brain, suggesting that COVID-19 can induce systemic thrombosis, thereby contributing to multiorgan failure [[9], [10], [11], [12], [13], [14]]. COVID-19 is also associated with elevated serum coagulation markers, such as fibrinogen and D-dimer, with the latter related to disease severity and poor prognosis [2,15,16]. Thromboprophylaxis, such as administration of heparin, has been shown to improve outcomes in hospitalized COVID-19 patients, but it had limited effect on mortality rates of the subset of patients with severe COVID-19 [[17], [18], [19], [20]]. Thrombosis thus remains a prominent feature of COVID-19 despite thromboprophylaxis, suggesting that the picture is complex, with increased thrombin generation not the only contributing factor.
Major players in thrombosis are platelets, which are blood cells that are not only essential for hemostasis but also contribute to thrombosis when inappropriately activated. Data obtained during the COVID-19 pandemic suggest that platelets become hyperactive, with reports of increased secretion of dense α-granules, increased aggregation, and increased formation of platelet-leukocyte aggregates [[21], [22], [23]]. Platelet transcriptome analysis showed an overrepresentation of pathways involved in antigen presentation and mitochondrial dysfunction in COVID-19 patients, potentially contributing to platelet hyperactivity [21]. However, there are also reports of impaired or reduced platelet functional responses in COVID-19 patients [24,25], suggesting that the platelet response is complex.
The pathogenesis of thrombosis in COVID-19 patients is not completely understood, but it has many hallmarks of thromboinflammation [2,26]. It is likely that SARS-CoV-2 activates endothelial cells via the angiotensin–converting enzyme 2 (ACE2) receptor [27], thereby causing vascular dysfunction [28]. Damaged endothelium leads to activation of the innate immune system mediated through complement, proinflammatory cytokines, tissue factor expression, and neutrophil recruitment [2,[28], [29], [30], [31]]. Together, this results in upregulation of adhesion molecules and enhances the expression and/or release of prothrombotic factors such as neutrophil extracellular traps and von Willebrand factor. The latter will bind and activate platelets, leading not only to aggregate and thrombus formation but also provide feedback to further activation of the innate immune system by the release of cytokines/chemokines and direct platelet-leukocyte interactions [14]. Platelets then amplify thrombin generation through the expression of tissue factor and phosphatidylserine (PS) [2,32], further promoting platelet activation and the cleavage of fibrinogen into the fibrin network required for clot formation.
This intricate network of interactions between platelets, the innate immune system, and the coagulation cascade is the likely factor contributing to thrombosis in COVID-19 patients [26], as well as indirect activation through damaged endothelium. It is also hypothesized that SARS-CoV-2 directly interacts with platelets through receptors, such as TLR7, FcγRIIA, and CD147, and has been evidenced to contribute to platelet hyperactivity and thrombosis as well [[33], [34], [35]]. Platelet expression of ACE2, a receptor involved in SARS-CoV-2 binding and cellular infection, is controversial; although a few studies reported platelet ACE2 expression [36], most of them failed to detect ACE2 by Western blotting, transcriptomics, and proteomics [[37], [38], [39]].
Although the emphasis has been predominantly on coagulopathy and activation of the immune system as important precipitants of thrombosis in patients with COVID-19, it more recently has become apparent that platelets are important, but poorly understood, drivers in this process [40]. In this study, we therefore aimed to assess the effect of COVID-19 on platelet proteome and relate this to platelet functional responses and platelet-neutrophil aggregate formation in patients hospitalized with COVID-19.
2. Methods
2.1. Study population
Twenty adult patients hospitalized with COVID-19 from the DISCOVER study at Southmead Hospital, Bristol, UK, and 19 healthy control adult participants (from the University of Bristol, UK) were enrolled in the study between October 2020 and February 2021 after providing written informed consent. A detailed description of the DISCOVER cohort is available elsewhere [41], but briefly, consecutive patients presenting to hospital with suspected or RT-PCR–confirmed COVID-19 were recruited. Participants were recruited from the COVID-19 ward and therefore exhibited COVID-19 of moderate severity. Demographics, comorbidities, medications, treatment records, and admission results were recorded. Control participants did not have COVID-19 (self-reported) and were matched for blood draw time (within 1 h of each other). Control participants were not recruited if they were pregnant, taking antiplatelet drugs, or had known clotting, bleeding disorders, or a blood–borne disease. Full blood counts were measured in COVID-19 patients and healthy controls using the Sysmex XN-20 hematology analyzer.
2.2. Ethics
Approval to this study was granted by The Yorkshire and The Humber—South Yorkshire Research Ethics Committee (NHS-REC reference 20/YH/0121) and South Central—Hampshire A Research Ethics Committee (NHS-REC reference 20/SC/0222).
2.3. Isolation of platelet-rich plasma and washed platelets
Venous blood was taken by venepuncture into 4.5-mL vacutainers containing 3.2% sodium citrate (1:9 v/v) and centrifuged (180 g, 17 minutes) to produce platelet-rich plasma (PRP). PRP was extracted ensuring that there was sufficient PRP left to not disturb the buffy coat layer. For washed platelets, PRP was acidified with 1:7 v/v acidic citrate dextrose before a second centrifugation (500 g, 10 minutes). Platelets were double washed in modified CGS buffer followed by centrifugation (500 g, 10 minutes). Washed platelets were resuspended in modified HEPES-Tyrode’s buffer. No apyrase was added in the experiments using PRP. See Supplementary Methods for buffer information.
2.4. Preparation of platelet samples for tandem mass tag proteomic analysis
Washed platelets from 8 COVID-19 patients and 7 control subjects were lysed with the radioimmunoprecipitation assay buffer (see Supplementary Methods). Samples were cleared by centrifugation at 4 °C, heated to 55 °C, and stored at −80 °C. In total, 50 μg of the sample was digested with trypsin, labeled with tandem mass tag (TMT) eleven plex reagents, and subjected to TMT analysis as described in the Supplementary Methods section.
2.5. Comparison of platelet proteome in COVID-19 and healthy control subjects
Principal component analysis was performed on the full protein dataset using the R package FactoMineR that identified 1 patient and 1 control outlier. The 2 outliers were removed, and 7 COVID-19 patient samples and 6 control samples were used for further analysis (see Supplementary Methods for more information). The log2 fold change (logFC) was calculated by subtracting the mean abundance of the control subjects from that of the COVID-19 patients. Statistical significance was determined using Welch’s t-tests and then corrected for multiple testing using the Benjamini–Hochberg method. Strength of association was determined by a composite of the logFC in protein levels and the p value.
2.6. Exploring platelet pathway changes using gene ontology pathways
We explored whether plasma or serum proteins reported to be affected by COVID-19 were also affected in platelets [[42], [43], [44]]. To explore the effect that a COVID-19–mediated change in platelet proteins may have on platelet function, we integrated proteomic results with gene ontology (GO) pathway annotations. Proteins involved in GO pathways were extracted from AmiGO2 (http://geneontology.org/). We specifically extracted a list of proteins involved in platelet secretion and platelet activation. The latter was explored using 7 GO pathways (see Supplementary Methods). Following extraction of the GO pathways, deduplication was performed so that each protein was only mentioned once in each list. Proteins involved in the GO pathways were merged into our proteomics results using R, and GO proteins that were associated with COVID-19 according to our TMT analysis (858 proteins in total) were identified.
2.7. Western blotting
Washed platelets were lysed in NuPage LDS sample buffer supplemented with 1:10 v/v dithiothreitol followed by Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) and immunoblotted for the thrombopoietin (TPO) receptor (cMpl), protein kinase Cα (PKCα), RalB, protein kinase A (PKA)-RIIb, interferon–induced transmembrane membrane protein 3 (IFITM3), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as described previously [45]. These proteins were chosen because they are functionally relevant and known to produce clear, individual bands. Samples used for Western blotting were not the same samples used for TMT proteomics analysis.
2.8. Flow cytometry analysis
Washed platelets, PRP, and whole blood were diluted in HEPES-Tyrode’s buffer [2 × 107 washed platelets/mL, PRP (1:40 v/v) and whole blood (1:10 v/v)]. Platelets were incubated with agonists [concentration-response curves for PAR1-AP, ADP, or collagen–related peptide (CRP)] or costimulated with 1 U/mL thrombin and 10 μg/mL CRP for 10 minutes where indicated. To measure integrin αIIbβ3 activation and P-selectin expression, PRP or whole blood was incubated with FITC–conjugated PAC1 or PE–conjugated CD62P antibodies, respectively (1:10 (v/v)). Platelet receptor levels were explored using FITC- or PE–conjugated antibodies in PRP. PS exposure was measured using Annexin V in washed platelets. Samples were fixed with 1xFixLyse (3:1 v/v, whole blood only), followed by a final concentration of 4% PFA (PRP, whole blood) or immediately quenched with HEPES-Tyrode’s buffer containing 2 mM CaCl2 (4:1 v/v, washed platelet PS exposure only). A total of 10 000 platelet events were collected on a BD Accuri C6 Plus flow cytometer (BD Biosciences) in the platelet FSC/SSC gate (washed platelets and PRP) and FSC/SSC gate/CD42b (whole blood). The neutrophil population in whole blood was gated on CD45 and SSC parameters with 1000 neutrophil events captured. Data are expressed as the median fluorescence intensity (MFI) for PAC1 and P-selectin, % of positive platelets (annexin V), or as % of neutrophils associated with platelets (CD41+/CD45+ events).
2.9. Platelet function data
If data met the assumption of normality, as tested by a Shapiro–Wilk test, data were analyzed using parametric tests (unpaired, two-tailed Student’s t-test); otherwise, a nonparametric test was used (Mann–Whitney U-test). Concentration-response curves were fitted using a 4-parameter logistic curve fit. Differences in curve fit were explored by comparing the individual logEC50s and curve maximum values of each group. Moreover, p values are reported throughout, where p < .05 is used as guidance for sufficient evidence to reject the null hypothesis. Analyses were performed using Prism 8 or R version 4.0.2 [46].
3. Results
3.1. Participant characteristics
The mean age of the recruited COVID-19 cohort was 59 years (SD 15.7 years), whereas the healthy control donor cohort recruited were younger, with a mean age of 39 years (SD 13.8) (p = .0002, Table ). Control and COVID-19 groups included a similar proportion of men and women. The mean BMI for COVID-19 patients was 30.6 kg/m2 (SD 4.4 kg/m2) compared with 22.7 (SD 2.8kg/m2) for controls (p < .0001). Patients were supported with oxygen therapy (88%) and additional treatments, including heparin (95%), dexamethasone (89%), and rivaroxaban (6%) (Table).
Table.
Characteristics of study participants.
| Variable | Control group mean (± SD) or % | Control group N | COVID-19 group mean (± SD) or % | COVID-19 group N | p value for difference |
|---|---|---|---|---|---|
| Age (y) | 39.0 (± 13.8) | 18 | 59.1 (± 15.7) | 19 | .0002 |
| Sex | |||||
| Male | 63% | 19 | 68% | 19 | .73a |
| Female | 37% | 32% | |||
| BMI (kg/m2) | 22.7 (± 2.8) | 18 | 30.6 (± 4.4) | 15 | <.0001 |
| Days in hospital (N) | NA | NA | 7.3 (± 20) | ||
| Oxygen required (N) | NA | NA | 88% | 16 | NA |
| Medication | |||||
| Heparinb | NA | NA | 95% | 18 | NA |
| Dexamethasone | 89% | ||||
| Rivaroxaban | 6% |
Chi-squared test.
Patients were treated with heparin from the time of hospitalization.
3.2. Full blood count results
COVID-19 patients had elevated neutrophil counts (6.6 × 109/L ± SD 3.5 vs 2.6 × 109/L ± SD 0.8, p = 6 × 10-5), but a reduced lymphocyte count (1.1 × 109/L ± SD 1 vs 1.8 × 109/L ± SD 0.4, p = .01). Platelet parameters (platelet count, immature platelet fraction, and immature platelet count) were similar across groups (Supplementary Table S1).
3.3. The platelet proteome signature is altered in COVID-19 patients
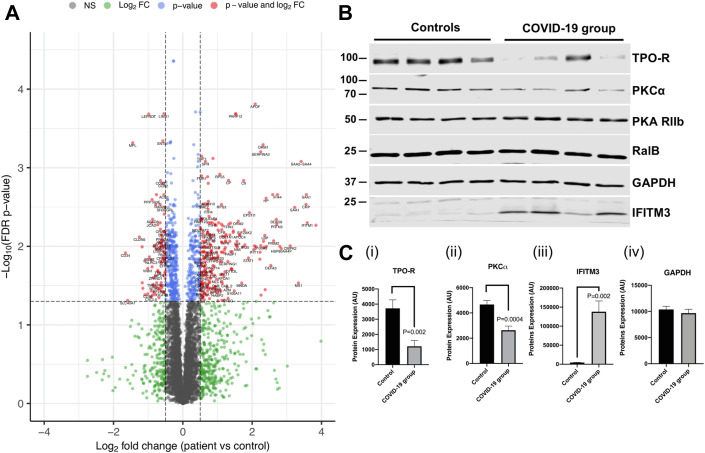
To study whether SARS-CoV-2 viral infection alters the platelet proteome, we performed TMT labeling on lysates of washed platelet samples from 7 COVID-19 patients and 6 healthy controls. After accounting for multiple testing using a false discovery rate (FDR) p value adjustment, 858 proteins of 5773 were altered in abundance in platelet lysates from patients with COVID-19 compared with healthy controls (FDR p < .05, 14.9% of total proteins detected) (Figure 1 A, Supplementary Table S2). A total of 436 proteins were more abundant in platelets from COVID-19 patients compared with controls, and the levels of a total of 422 proteins were reduced in in the platelets of COVID-19 patients compared with control platelets. The levels of C-reactive protein and S100A8/A9 were increased in platelets of COVID-19 patients (logFC 3.55, p = .003 and logFC 2.25, p = 9.8 × 10-3, respectively). We also found weak evidence for an increase in the amount of the antiviral immune protein IFITM3 in platelets from COVID-19 patients compared with healthy controls, with a logFC of 2.02 (p = .096); however, this did not pass the FDR multiple testing threshold. Interestingly, multiple platelet activation signaling proteins were decreased in platelets from COVID-19 patients compared with healthy controls, including the TPO receptor cMpl (logFC −1.45, p = 4.8 × 10-4) and PKCα (logFC −.60, p = .008). Platelet receptors generally showed a downregulation in COVID-19 patients, with a reduction in the levels of the P2Y12 receptor (logFC −0.44, p = .010), PAR-4 (logFC −0.43, p = .007), CD36 (logFC −1.01, p = .040), and GPV (logFC −0.31, p = .042).
Figure 1.
The platelet proteome is altered in patients with COVID-19. (A) Volcano plot of the proteins altered in platelet lysates from patients (N = 7) and controls (N = 6) using tandem mass tag proteomics. Protein changes were analyzed by Welch’s t-test. The proteins that had both a log2 fold change (logFC) greater than ± 0.5 and a false discovery rate–adjusted p value less than .05 are shown in red. Proteins with only a false discovery rate p value less than .05 are shown in blue. The proteins indicated in blue and red make up the 858 proteins that were altered in amount in COVID-19 compared with control patients. Proteins that only had logFC greater than ± 0.5 are shown in green. Proteins that did not reach the p value or logFC threshold are shown in gray. One protein was excluded from the figure (which had a logFC of over −4) to be able to see other data points more clearly. Plot was made using the Enhanced Volcano R package (https://github.com/kevinblighe/EnhancedVolcano). (B) Representative Western blot using platelet lysates from 4 controls and 4 patients for the TPO receptor, PKCα, PKA (control), RalB, GAPDH (control), and IFITM3. Analyses were performed using Student’s t-test. (C) (i) Western blot quantification of the TPO receptor (mean + SEM, N = 9). (ii) Western blot quantification of PKCα (mean + SEM, N = 9). (iii) Western blot quantification of IFITM3 protein expression (mean + SEM, N = 8). (iv) Western blot quantification of GAPDH loading control (mean + SEM, N = 8, p = .58).
To confirm the untargeted TMT proteomic results, we next performed Western blotting analysis on platelet lysates from at least 8 COVID-19 patients and 8 healthy controls. Platelet lysates from the COVID-19 group again showed a reduction in the expression of the TPO receptor cMpl (1211 AU ± SEM 567.4 vs 3719 AU ± SEM 382, p = .002) and PKCα (2629 AU ± SEM 323.5 vs 4688 AU ± SEM 319.2, p = .0004) compared with healthy controls (Figure 1B, 2Ci, and 2Cii). By contrast, PKA expression levels were unaltered, in agreement with our proteomic findings. We also found evidence for an increase in levels of IFITM3 in platelet lysates from COVID-19 patients compared with healthy controls (Figure 1B, 1Ciii), as found in a previous study [21]. Interestingly, proteomic analysis failed to detect any of the SARS-CoV-2 viral proteins reported by Davidson et al. [47] in COVID-19 patient samples, indicating that the virus had not entered and/or replicated in platelets. The receptor for SARS-CoV2, ACE2, was also not detected in any sample. By contrast, CD147 (basigin), a receptor with suggested interaction with SARS-CoV2 [48], was present in both control and COVID-19 patient samples and was unaltered in the COVID-19 group (Supplementary Table S2).
3.4. Enhanced platelet content of COVID-19 plasma biomarkers
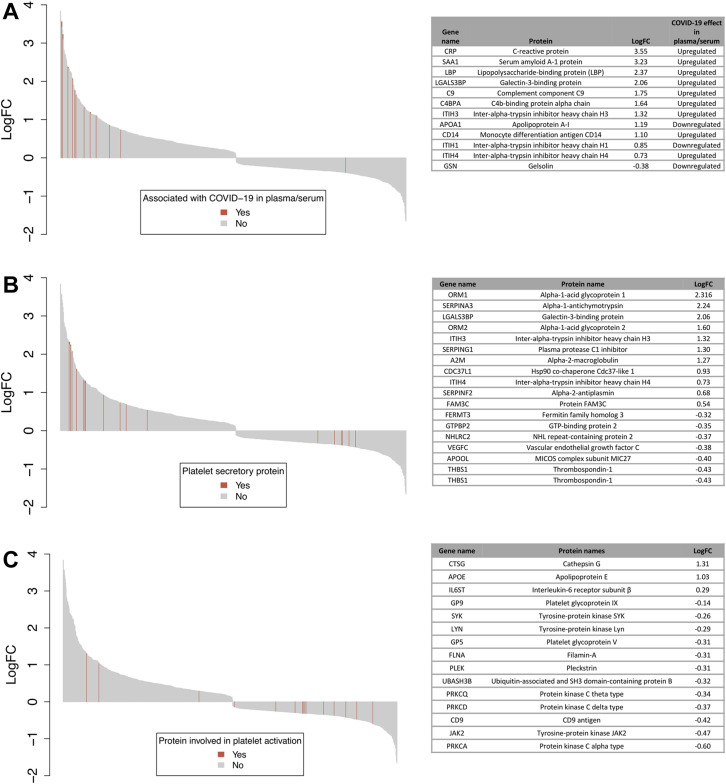
Previous studies have identified multiple plasma or serum proteins associated with COVID-19 [[42], [43], [44]]. Platelets are not only able to release their granule content but can also selectively take up proteins from their environment [49]. We were therefore interested in whether the platelet content of these plasma/serum biomarkers is altered in COVID-19 patients. Using TMT proteomic analysis, we detected 38 proteins that have been demonstrated to be associated with COVID-19 in plasma/serum in at least 2 of 3 studies [[42], [43], [44]]. Of these, 12 proteins were altered in COVID-19 patients’ platelets (Figure 2 A) compared with controls. Interestingly, except for gelsolin, levels of 11 platelet plasma biomarkers were enhanced in COVID-19 platelets, as shown in the waterfall plot in Figure 2A. Of these, the following 4 proteins showed more than a 4-fold increase (logFC > 2) compared with control platelets—serum amyloid A (SAA1), C-reactive protein, lipopolysaccharide binding protein (LBP), and galectin 3-binding protein (G3BP). These results suggest that platelets take up a subset of COVID-19–associated plasma proteins.
Figure 2.
Waterfall plots displaying the log2 fold change of platelet proteins in COVID-19 patients compared with controls. Each protein is represented by a gray bar, and proteins in specific pathways are indicated with red lines. (A) Proteins highlighted in red and listed in the table have been shown to be associated with COVID-19 in plasma or serum in at least 2 studies by Messner et al. [42], Leng et al. [43], or Roh et al. [44]. (B) Proteins highlighted in red and listed in the table have known involvements in gene ontology (GO) pathways α- and dense-granule lumen gene products (GO:0031093 and GO:0031089, respectively). (C) Proteins highlighted in red and listed in the table have known involvement in platelet activation or platelet aggregation GO pathways. Results are plotted using the barplot() function from the “graphics” R package.
3.5. Alterations in proteins involved in platelet secretion pathways in COVID-19 platelets
To investigate whether proteins involved in granule secretion were altered in platelets from COVID-19 patients compared with healthy controls, we compared changes in proteins using GO pathway annotations [50,51]. A total of 70 (68 unique) proteins were involved in the α-granule and dense granule lumen gene product GO pathways and detected in the current study. Of these, 18 (17 unique) proteins were associated with COVID-19, with 11 proteins being increased and 7 proteins decreased in platelets from COVID-19 patients (Figure 2B). Some of the proteins with increased expression in platelets were also enhanced in COVID-19 plasma, such as G3BP, inter-α-trypsin inhibitor heavy chain 3 (ITIH3) and 4 (ITIH4), suggesting that the increase may be due to uptake of proteins that are enriched in the plasma. By contrast, α-granule proteins, such as VEGFC and thrombospondin-1, were reduced in platelets from COVID-19 patients.
3.6. Reduction in proteins involved in platelet activation or aggregation pathways in COVID-19
In total, 37 unique proteins involved in platelet activation or platelet aggregation were detected in the current study. Fifteen of these platelet activation proteins were associated with COVID-19 (Figure 2C). Of these, 12 were reduced in platelets from COVID-19 patients compared with healthy controls, including protein tyrosine kinases (LYN, SYK, and JAK2) and Ser/Thr kinases (PKCα, PKCδ, and PKCξ). Furthermore, the receptor subunits IX and V, which are part of the platelet GPIb-IX-V complex, were also reduced. By contrast, levels of cathepsin G, apolipoprotein E, and interleukin-6 receptor subunit were increased.
3.7. Impaired agonist–induced integrin αIIbβ3 activation in patients with COVID-19
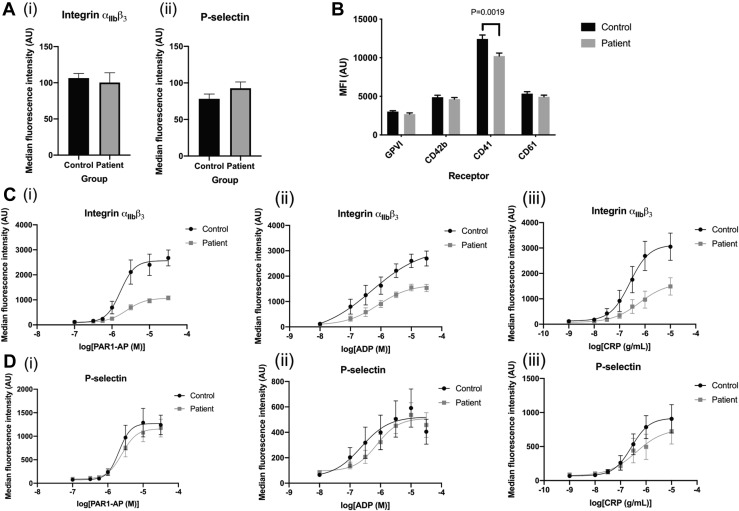
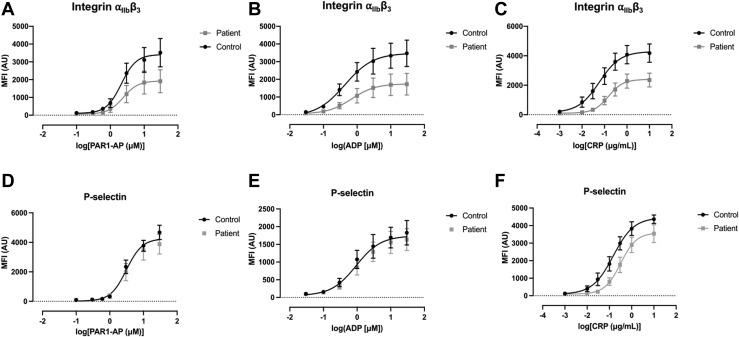
To relate our proteomic findings to platelet functional responses, we performed flow cytometry studies using PRP to explore activation markers and surface receptors in platelets from COVID-19 patients and controls. Our studies focused on activation of integrin αIIbβ3, the receptor responsible for platelet aggregation, and the α-granule marker P-selectin, which is expressed on the surface membrane upon α-granule secretion. Under basal conditions, there was no difference in the activation of integrin αIIbβ3 or surface P-selectin expression between circulating COVID-19 and healthy control platelets (Figure 3 A). Platelets from COVID-19 patients showed a small reduction in basal levels of the αIIb integrin (CD41) but had similar surface receptor levels for GPVI, GPIbα (CD42b) and β3 integrin (CD61) (Figure 3B). We next performed concentration-response experiments for PAR1-AP, ADP, and CRP to evaluate changes in integrin αIIbβ3 activation and P-selectin expression. Our data demonstrate that agonist–induced integrin αIIbβ3 activation is impaired in platelets from COVID-19 patients with a reduction of the curve max values, but no change in EC50 values (Figure 3C). In contrast to integrin αIIbβ3 activation, agonist–stimulated P-selectin expression was unchanged in platelets from COVID-19 patients (Figure 3D), demonstrating that platelet α-granule secretion remains intact. Similar findings for integrin αIIbβ3 activation and P-selectin expression were observed for whole blood (Figure 4 ).
Figure 3.
Comparison of integrin αIIbβ3 activation, P-selectin expression, and surface receptor levels. (A) Comparison of surface receptors and platelet activation markers in diluted platelet-rich plasma (PRP) using flow cytometry. Statistical test used is an unpaired t-test unless otherwise indicated. (i) Basal integrin αIIbβ3 activation in diluted PRP detected using FITC-PAC1 antibody in COVID-19 patients compared with controls (N = 9, Mann–Whitney U-test, p = .18). (ii) Basal P-selectin expression in diluted PRP detected using anti-CD62P-PE antibody in COVID-19 samples compared with controls (N = 9, Mann–Whitney U-test, p = .15). (B) Basal surface receptor levels measured in COVID-19 patients and controls: GPVI (N = 17-18, p = .40), CD42b (N = 17-18, p = .47), CD41 (N = 17-18, p = .0019), and CD61 (N = 13, p = .24). (C) Concentration-response curves of integrin αIIbβ3 activation with (i) PAR1-AP (Mann–Whitney U-test comparison of pEC50s, p = .61, comparison of curve max p = .0001), (ii) ADP (comparison of pEC50s, p = .53, curve max comparison using Mann–Whitney U-test, p = .003), and (iii) CRP (comparison of pEC50s, p = .27, comparison of curve max, p = .12). (D) Concentration-response curves of P-selectin expression in response to increasing concentrations of (i) PAR1-AP (comparison of pEC50s, p = .76, Mann–Whitney U-test of comparison of curve max, p = .96), (ii) ADP (Mann–Whitney U-test for comparison of pEC50s, p = .67, Mann–Whitney U-test for comparison of curve max, p = .34), and (iii) CRP (comparison of pEC50s, p = .18, comparison of curve max, p = .93).
Figure 4.
Comparison of integrin αIIbβ3 activation and P-selectin expression in whole blood samples of COVID-19 patients with those of healthy controls. (A–C) PAC-1 binding using flow cytometry in whole blood samples in response to 10-minute stimulation with increasing concentrations of agonist. Differences between COVID-19 patient and healthy control group compared using Student’s t-test. (A) PAR1-AP (N = 5-6, max p = .17, EC50 p = .45). (B) ADP (N = 5-6, max p = .10, EC50 p = .10). (C) CRP (N = 8, max p = .02, EC50 p = .11) (D–F) CD62P binding in whole blood (mean ± SEM, N = 6-8) in response to 10-minute stimulation with increasing concentrations of agonist. Differences between COVID-19 patient and healthy control group compared using Student’s t-test. (D) PAR1-AP (N = 5-6, max p = .35, EC50 p = .93). (E) ADP (N = 5-6, max p = .57, EC50 p = .89). (F) CRP (N = 8, max p = .10, EC50 p = .45).
3.8. Impaired agonist–induced PS exposure in COVID-19
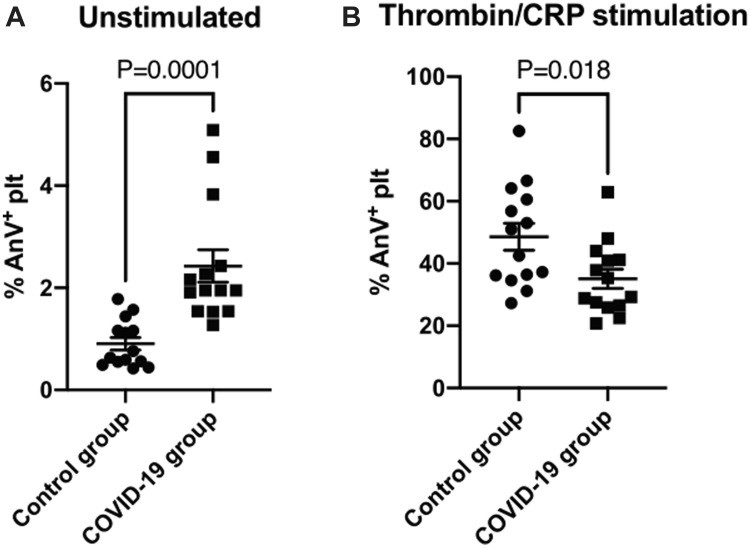
COVID-19 is associated with hypercoagulopathy, and platelets can contribute to coagulation by flipping PS lipids to their outer membranes. We therefore determined platelet procoagulant activity in patients by measuring PS exposure in washed platelets using FITC–tagged Annexin V. Under unstimulated conditions, platelets from COVID-19 patients had a small, elevated level of PS exposure compared with controls (2.43% ± SEM 0.32% vs 0.91% ± SEM 0.12%, p = .0001, Figure 5 A). By contrast, PS exposure following stimulation with both thrombin and CRP was reduced in the COVID-19 group (35.1% ± 3.1% vs 48.6% ± 4.3%, p = .018, Figure 5B).
Figure 5.
Annexin V binding under basal conditions and after dual stimulation with thrombin and CRP. PS exposure measured by Annexin V binding using flow cytometry in washed platelets at 2 × 108/mL under (A) basal conditions (mean ± SEM, N = 14) and (B) upon stimulation with 1 U/mL thrombin and 10 μg/mL CRP (mean ± SEM, N = 14). p values shown for unpaired Student’s t-test.
3.9. Increased platelet-neutrophil interactions in COVID-19
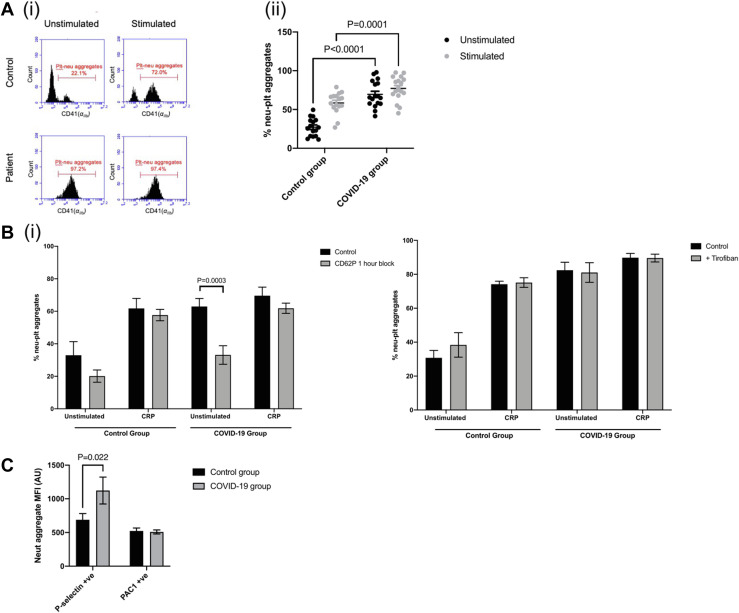
Platelets can contribute to thromboinflammation by interacting with inflammatory cells such as neutrophils. We therefore determined platelet-neutrophil aggregate formation in whole blood from healthy controls and COVID-19 patients using flow cytometry, measuring the platelet marker αIIb (CD41) in the neutrophil gate. Figure 6 A demonstrates that 27% (SEM 3%) of the neutrophils from healthy donors were bound to platelets under basal unstimulated conditions. When control platelets were activated with CRP, 58% (SEM 4%) of the neutrophil population were associated with platelets (Figure 6Ai, ii), demonstrating that platelet activation can drive platelet-neutrophil association. By contrast, platelet-neutrophil aggregate formation was already near maximal under unstimulated conditions in the COVID-19 group (72%, SEM 5%, p < .0001 compared with control unstimulated) (Figure 6A). There was no evidence that the proportion of neutrophils bound to platelets further increased (80%, SEM 4%, p > .05) when stimulated with CRP. To determine the receptors involved in the platelet-neutrophil association, we incubated whole blood for 1 hour in the presence of a P-selectin CD62P blocking antibody and the αIIbβ3 inhibitor tirofiban. Interestingly, the CD62P blocking antibody reduced basal platelet-neutrophil interactions in blood from COVID-19 patients to the level observed in healthy controls (Figure 6Bi). By contrast, the CD62P blocking antibody had no effect on CRP–stimulated platelet-neutrophil interactions in both healthy control and COVID-19 samples, indicating that additional receptor interactions are likely taking place. Incubation with tirofiban did not prevent or reduce platelet-neutrophil aggregate formation in both healthy controls and COVID-19 patients, suggesting that this interaction is not driven by the integrin αIIbβ3 (Figure 6Bi).
Figure 6.
Platelet-neutrophil interactions in patients with COVID-19. (A)(i) An example of platelet-neutrophil aggregate formation in whole blood using flow cytometry. Left hand side shows CD41+ events in gated neutrophils under unstimulated conditions, and right-hand side shows events following stimulation with 5 μg/mL CRP for 15 minutes. (ii) Quantification of the percentage of platelets bound to neutrophils in controls and COVID-19 patients (mean ± SEM, N = 17). Control and COVID-19 patient groups compared using an unpaired Student’s t-test with p values shown where p < .05. (B) p values indicated in the following figures where p < .05. (i) Percentage of platelets bound to neutrophils after 1-hour incubation with CD62P blocking antibody (N = 4-6, mean ± SEM). Control groups and COVID-19 patient groups compared using two-way analysis of variance with multiple comparisons. (ii) Percentage of platelets bound to neutrophils after incubation with tirofiban (mean ± SEM, N = 7). Results analyzed with a mixed effects model with multiple comparisons. (C) P-selectin and PAC-1–positive platelets within gated neutrophils (mean ± SEM, N = 8). Results analyzed with a two-way analysis of variance with multiple comparisons.
4. Discussion
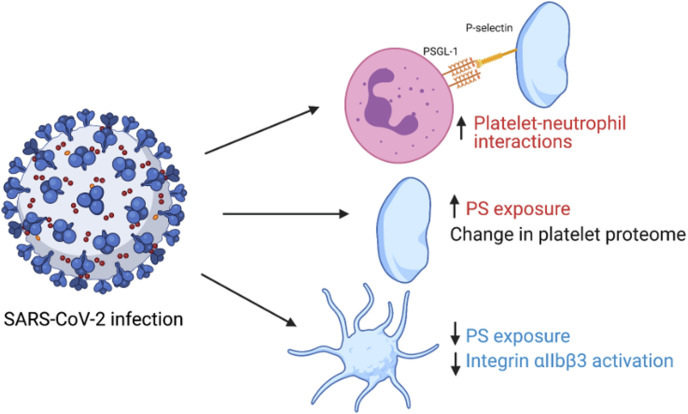
In this study, we assessed the effect of COVID-19 on the platelet proteome and platelet functional responses. We demonstrate that blood from patients hospitalized with COVID-19 contain at least 2 platelet populations—free circulating, functionally defective platelets and neutrophil–associated platelets. Proteomic studies on the free circulating platelets showed increased levels of a subset of COVID-19–associated plasma proteins and reduced levels of intracellular signaling proteins. These platelets had impaired agonist–mediated responses, such as PS exposure and integrin αIIbβ3 activation, whereas P-selectin expression was unaltered. Whole blood analysis indicated that the majority of neutrophils in COVID-19 patients are associated with platelets, an interaction that is dependent on P-selectin. This suggests that platelets drive platelet-neutrophil interaction and immunothrombosis in COVID-19 patients. These main findings are summarized in Figure 7 .
Figure 7.
Graphical summary of study findings. Red indicates an increase in variable in patients with COVID-19 vs healthy controls and blue indicates a decrease. Figure made using biorender.com.
This study showed a large change in the platelet proteome in patients with COVID-19, with strong evidence for altered levels of 858 proteins compared with healthy controls. We replicated some of the proteomic effects of COVID-19 found by Trugilho et al. [52], where 157 of the proteins they deemed to be associated with COVID-19 were also associated in our study. In addition, we found evidence for a decrease in platelet receptors that have not previously been shown to associate with COVID-19 such as P2Y12, PAR-4, CD36, and GPV. These changes are indicative of functionally defective circulating platelets and may contribute to the impairment in integrin αIIbβ3 responses. Another interesting finding was the reduction in levels of the TPO receptor cMpl in platelets from COVID-19 patients. This reduction is potentially because of the enhanced plasma levels of TPO in COVID-19 patients. Increased TPO levels driven, for example, by raised IL-6 levels [21,53], may lead to platelet TPO receptor endocytosis and/or receptor destruction. Conversely, potentially impaired endocytosis of TPO receptors platelets that intrinsically express fewer TPO receptors could also result in an increase in plasma TPO levels. The latter would be consistent with previous platelet transcriptomic findings in COVID-19 patients, reporting a 3-fold reduction in cMpl expression [21], which is in the same order of magnitude as the reduction in TPO receptor protein expression levels. We also found reduced protein levels of the signaling molecule JAK2, which constitutively binds to the cytoplasmic tail of the TPO receptor [54]. Both TPO receptor desensitization and internalization and platelets being produced with fewer TPO receptors/JAK2 may thus contribute to the phenotype. Despite the changes in platelet TPO receptor/JAK2 levels, we and others found that platelet counts are in the normal range in COVID-patients [21,55]. It is possible that this can be explained by enhanced expression levels of downstream signaling molecules, such as STAT1, STAT2, and STAT3 in COVID-19, which may compensate for reduced TPO/JAK2 expression levels through signal amplification. We can however not rule out that any potential changes are masked by platelet association with immune cells or dexamethasone treatment of COVID-19 patients [56]. Further work is required to replicate these findings and explore the signaling consequences.
We also compared our platelet proteomic findings with the 3 recently reported studies on changes in the plasma/serum proteome in COVID-19 patients [[42], [43], [44]]. Using a threshold setting of the protein being detected in 2 of the 3 published studies in plasma/serum, we found a subset of the proteins that were highly enriched in platelets from COVID-19 patients. Proteins that were more than 4-fold enriched in platelets include the following acute phase proteins—C-reactive protein, SAA1, LBP, and G3BP. We also found an enrichment of proteins related to the complement system (complement component C9 and C4b-binding protein α chain), ITIH3 and ITIH4, and the leukocyte marker CD14. Interestingly, the following 2 proteins that were downregulated in COVID-19 plasma were upregulated in platelets: apolipoprotein A-1 and ITIH1. Apolipoprotein A-1 plasma levels are inversely associated with COVID-19 severity, suggesting a protective function [57,58]. For the plasma/serum proteins that were associated with COVID-19 and with COVID-19 in platelets, a transcriptomic study did not find a difference in levels of transcripts for these genes [21], suggesting platelet protein uptake from plasma rather than an alteration in megakaryocyte gene expression being responsible for enhanced levels. One exception is G3BP, whose transcript level is upregulated in platelets from COVID-19 patients [21]. Platelet–derived G3BP may therefore contribute to the observed increase in plasma G3BP levels in COVID-19 patients. Of note, G3BP and its receptor/ligand galectin-3 have been reported to contribute to platelet hyperactivity and venous thrombosis [59,60].
When assessing proteins involved in platelet secretion and/or granule content, we also found proteins that were COVID-19 plasma biomarkers (G3BP, ITIH3, and ITIH4) and acute phase proteins. Interestingly, proteins known to be released upon α-granule secretion, such as thrombospondin-1 and VEGF-C, were reduced in COVID-19 platelets, suggesting that granule markers are reduced, potentially pointing to platelet preactivation/exhaustion. The latter is also reflected by a reduction in a range of signaling proteins detected in platelets from COVID-19 platelets. However, in contrast to previous studies [21,32,61,62], we did not find evidence of platelet preactivation as basal integrin αIIbβ3 and P-selectin expression in platelets from COVID-19 patients were unaltered. The difference with previous studies may be due to the analysis method (we used MFI instead of the percentage of positive platelets as a more indicative measure of the extent of platelet activation and independent of threshold settings), the moderate severity of COVID-19 in our study (Hottz et al. [32] found an increase in P-selectin expression in severe COVID-19 patients only), and/or the high level of platelet-neutrophil interactions, the latter not being captured in the platelet channel.
There was a small increase in PS exposure, although this study may have been underpowered to detect a subtle increase. Although previous studies have suggested platelet hyperactivity in COVID-19 patients [25,32,61,63], we observed that integrin αIIbβ3 activation in response to platelet agonists was impaired, whereas P-selectin expression was unaltered, confirming previous observations in response to collagen [21,25]. Similarly, we found that agonist–stimulated PS exposure was attenuated in the COVID-19 group, in agreement with previous findings [64]. The impairment in platelet function could potentially contribute to bleeding, as has been reported among individuals hospitalized with COVID-19 [65]. Both the proteomics and pathway analysis pointed toward impaired platelet signaling, with a reduction in levels of PKCα and inhibition of Tec kinase and phospholipase C. Impairment in these signaling pathways may underlie the observed reduction in platelet function. Furthermore, a small reduction in levels of CD41 (αIIb) expression in COVID-19 patients may further contribute to the measured impairment in agonist–induced integrin αIIbβ3 activation. Interestingly, as previous studies found normal or enhanced platelet aggregation in COVID-19 patients [21,66], the residual integrin αIIbβ3 activation is likely to be sufficient for aggregation in the COVID-19 group.
The plasma protein S100A8/A9 did not pass our selection criteria of being elevated in 2 of 3 proteomic studies [[42], [43], [44]]; however, several studies have reported increased plasma S100A8/A9 in COVID-19 patients, and plasma levels correlated to poor outcome [[67], [68], [69], [70]]. S100A8/A9 can induce platelet procoagulant responses and promote platelet/neutrophil interaction by enhancing P-selectin levels [69]. Here, we found that S100A8/A9 protein levels were enhanced by more than 4-fold (logFC 2.25) in platelets in COVID-19 patients. Because platelet transcript levels in COVID-19 patients were unchanged [21], this is likely to be the result of enhanced platelet binding and uptake of plasma S100A8/A9, potentially through GPIb/CD36 receptor interaction [69]. The 2-fold reduction in CD36 (logFC −1.01) and the enhanced basal platelet PS exposure in COVID-19 patients detected in our study would be consistent with this.
In whole unstimulated blood, patients with COVID-19 exhibited platelet-neutrophil interactions, which is consistent with previous studies reporting enhanced platelet-leukocyte interactions in COVID-19 [21,25,32]. The majority of neutrophils in COVID-19 patients were associated with platelets. Platelet-neutrophil interactions were found to be dependent on P-selectin and not integrin αIIbβ3. This agrees with our finding that neutrophil–associated platelets were positive for P-selectin but lacked activated integrin αIIbβ3. The latter may be explained by the reversibility of integrin αIIbβ3 activation, where initial platelet activation leads to both P-selectin expression and integrin αIIbβ3 activation, but integrin αIIbβ3 then reverts to a low affinity state [71,72]. However, we cannot rule out that COVID-19 conditions may induce platelet P-selectin expression in the absence of integrin αIIbβ3 activation, such as has recently observed in a small fraction of CRP/PAR-1-activated platelets [73]. The recent study by Colicchia et al. [69] is also of interest because they demonstrated that S100A8/A9 can induce P-selectin expression, although inducing an integrin αIIbβ3 activation state that lacked aggregatory ability and had low-fibrinogen binding capacity.
One mechanism by which COVID-19 may affect platelet function is by direct modulation by SARS-CoV-2 [74]. Most studies were unable to detect platelet expression of the SARS-CoV-2 entry receptor ACE2 at both mRNA and protein levels [21,23], although a few studies reported ACE2-mediated regulation of platelet function [36]. Data from our proteomics study support the lack of ACE2 expression in human platelets. We cannot exclude that there are alternative direct modes of regulation, such as through CD147 (basigin) [48], which we here confirm is expressed on platelets. We did however not find evidence for viral protein expression in platelets from COVID-19 patients, suggesting that SARS-CoV-2 cannot enter and/or replicate in human platelets.
There are a few limitations that are important to note. First, because most patients were on dexamethasone and heparin, we cannot rule out that these medications may contribute to the platelet phenotype we have observed. However, because this was the recommended care for those hospitalized with COVID-19, it was challenging to specifically recruit participants who were not on these medications. Second, the healthy controls were younger than the recruited COVID-19 patients and, on average, had a lower BMI. Therefore, we cannot rule out confounding from variables such as BMI. Finally, the control participants self-reported being negative for SARS-CoV-2. It is possible that control participants could have been carrying the virus and were asymptomatic. However, we would expect that this would only result in smaller differences in platelet function between groups.
Overall, our data suggest the presence of 2 platelet populations in patients with COVID-19. The first is circulating platelets with an altered proteome, increased basal PS exposure, and reduced agonist–induced integrin αIIbβ3 activation. The second platelet population is P-selectin expressing neutrophil–associated platelets. Furthermore, circulating platelets from COVID-19 patients have a unique protein signature, with multiple COVID-19 associated plasma proteins being markedly enhanced. Our data show a complex picture and suggest that platelet–driven thromboinflammation may be one of the key drivers enhancing the risk of thrombosis in COVID-19 patients. The data also point toward potential mechanisms of this effect, which now need to be further characterized.
Acknowledgments
This work was supported by the University of Bristol Elizabeth Blackwell Institute, the University of Bristol Alumni, the Southmead Hospital Charity, the Wellcome Trust (216277/Z/19/Z and 219472/Z/19/Z), and the British Heart Foundation (FS/17/60/33474, RG/15/16/31758, PG/17/62/33190, PG/21/10760, and SP/F/21/150023). L.J.G. is supported by the BHF Accelerator Award AA/18/1/34219. We thank Borko Amulic and Christopher Rice for their helpful discussions over the observed interactions between platelets and neutrophils. This research was funded in whole, or in part, by the Wellcome Trust. For the purpose of Open Access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author contributions
I.H. obtained ethical approval and led the study. L.J.G., C.M.W., and I.H. designed the experiments. F.H., D.A., and A.M. were involved in obtaining patient samples and patient information. L.J.G., C.M.W., J.K., and K.L.B. performed the experiments. P.A.L. and K.J.H. generated the proteomics results. L.J.G., K.L.B., and I.H. analyzed and interpreted the proteomics data. A.D. supported the interpretation of the proteomics results. L.J.G. and I.H. wrote the manuscript. A.W.P. and S.J.M. contributed to discussion. All authors reviewed and/or edited the manuscript.
Declaration of competing interests
There are no competing interests to disclose.
Footnotes
Funding information This work was supported by the University of Bristol Elizabeth Blackwell Institute, the University of Bristol Alumni, the Southmead Hospital Charity, the Wellcome Trust (216277/Z/19/Z and 219472/Z/19/Z), and the British Heart Foundation (FS/17/60/33474, RG/15/16/31758, PG/17/62/33190, PG/21/10760, and SP/F/21/150023). L.J.G. is supported by the BHF Accelerator Award AA/18/1/34219.
Manuscript handled by: M Rondina
Final decision: M Rondina, 19 January 2023
The online version contains supplementary material available at https://doi.org/10.1016/j.jtha.2023.01.018.
Supporting Information
References
- 1.WHO. WHO Coronavirus (COVID-19) Dashboard. 2022.
- 2.McFadyen J.D., Stevens H., Peter K. The emerging threat of (micro)thrombosis in COVID-19 and its therapeutic implications. Circ Res. 2020;127:571–587. doi: 10.1161/CIRCRESAHA.120.317447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klok F.A., Kruip M., van der Meer N.J.M., Arbous M.S., Gommers D., Kant K.M., Kaptein F.H.J., van Paassen J., Stals M.A.M., Huisman M.V., Endeman H. Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res. 2020;191:145–147. doi: 10.1016/j.thromres.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Middeldorp S., Coppens M., van Haaps T.F., Foppen M., Vlaar A.P., Muller M.C.A., Bouman C.C.S., Beenen L.F.M., Kootte R.S., Heijmans J., Smits L.P., Bonta P.I., van Es N. Incidence of venous thromboembolism in hospitalized patients with COVID-19. J Thromb Haemost. 2020;18:1995–2002. doi: 10.1111/jth.14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Violi F., Ceccarelli G., Cangemi R., Cipollone F., D'Ardes D., Oliva A., Pirro M., Rocco M., Alessandri F., D'Ettorre G., Lichtner M., Pignatelli P., Ferro D., Ruberto F., Lip G.Y.H., Pugliese F., Mastroianni C.M. Intensive Care IDC-SGoSU. Arterial and venous thrombosis in coronavirus 2019 disease (Covid-19): relationship with mortality. Intern Emerg Med. 2021;16:1231–1237. doi: 10.1007/s11739-020-02621-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knight R., Walker V., Ip S., Cooper J.A., Bolton T., Keene S., Denholm R., Akbari A., Abbasizanjani H., Torabi F., Omigie E., Hollings S., North T.L., Toms R., Jiang X., Angelantonio E.D., Denaxas S., Thygesen J.H., Tomlinson C., Bray B., et al. Association of COVID-19 with major arterial and venous thrombotic diseases: a population-wide cohort study of 48 million adults in England and Wales. Circulation. 2022;146:892–906. doi: 10.1161/CIRCULATIONAHA.122.060785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishikawa M., Kanno H., Zhou Y., Xiao T.H., Suzuki T., Ibayashi Y., Harmon J., Takizawa S., Hiramatsu K., Nitta N., Kameyama R., Peterson W., Takiguchi J., Shifat E.R.M., Zhuang Y., Yin X., Rubaiyat A.H.M., Deng Y., Zhang H., Miyata S., et al. Massive image-based single-cell profiling reveals high levels of circulating platelet aggregates in patients with COVID-19. Nat Commun. 2021;12:7135. doi: 10.1038/s41467-021-27378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rampotas A., Pavord S. Platelet aggregates, a marker of severe COVID-19 disease. J Clin Pathol. 2021;74:750–751. doi: 10.1136/jclinpath-2020-206933. [DOI] [PubMed] [Google Scholar]
- 9.Rapkiewicz A.V., Mai X., Carsons S.E., Pittaluga S., Kleiner D.E., Berger J.S., Thomas S., Adler N.M., Charytan D.M., Gasmi B., Hochman J.S., Reynolds H.R. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: a case series. EClin Med. 2020;24 doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., Silvestri F., Zacchigna S., Giacca M. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMed. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kondo R., Kawaguchi N., McConnell M.J., Sonzogni A., Licini L., Valle C., Bonaffini P.A., Sironi S., Alessio M.G., Previtali G., Seghezzi M., Zhang X., Sun Z., Utsumi T., Strazzabosco M., Iwakiri Y. Pathological characteristics of liver sinusoidal thrombosis in COVID-19 patients: a series of 43 cases. Hepatol Res. 2021 doi: 10.1111/hepr.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauter J.L., Baine M.K., Butnor K.J., Buonocore D.J., Chang J.C., Jungbluth A.A., Szabolcs M.J., Morjaria S., Mount S.L., Rekhtman N., Selbs E., Sheng Z.M., Xiao Y., Kleiner D.E., Pittaluga S., Taubenberger J.K., Rapkiewicz A.V., Travis W.D. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020;77:915–925. doi: 10.1111/his.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz L.A., Idalsoaga F., Cannistra M., Candia R., Cabrera D., Barrera F., Soza A., Graham R., Riquelme A., Arrese M., Leise M.D., Arab J.P. High prevalence of hepatic steatosis and vascular thrombosis in COVID-19: a systematic review and meta-analysis of autopsy data. World J Gastroenterol. 2020;26:7693–7706. doi: 10.3748/wjg.v26.i48.7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryce C., Grimes Z., Pujadas E., Ahuja S., Beasley M.B., Albrecht R., Hernandez T., Stock A., Zhao Z., AlRasheed M.R., Chen J., Li L., Wang D., Corben A., Haines G.K., 3rd, Westra W.H., Umphlett M., Gordon R.E., Reidy J., Petersen B., et al. Pathophysiology of SARS-CoV-2: the Mount Sinai COVID-19 autopsy experience. Mod Pathol. 2021;34:1456–1467. doi: 10.1038/s41379-021-00793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levi M., Thachil J., Iba T., Levy J.H. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol. 2020;7:e438–e440. doi: 10.1016/S2352-3026(20)30145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leentjens J., van Haaps T.F., Wessels P.F., Schutgens R.E.G., Middeldorp S. COVID-19-associated coagulopathy and antithrombotic agents-lessons after 1 year. Lancet Haematol. 2021;8:e524–e533. doi: 10.1016/S2352-3026(21)00105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hippensteel J.A., LaRiviere W.B., Colbert J.F., Langouët-Astrié C.J., Schmidt E.P. Heparin as a therapy for COVID-19: current evidence and future possibilities. Am J Physiol Lung Cell Mol Physiol. 2020;319:L211–L217. doi: 10.1152/ajplung.00199.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connors J.M., Levy J.H. COVID-19 and its implications for thrombosis and anticoagulation. Blood. 2020;135:2033–2040. doi: 10.1182/blood.2020006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceccato A., Camprubi-Rimblas M., Campana-Duel E., Areny-Balaguero A., Morales-Quinteros L., Artigas A. Anticoagulant treatment in severe ARDS COVID-19 patients. J Clin Med. 2022;11:2695. doi: 10.3390/jcm11102695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manne B.K., Denorme F., Middleton E.A., Portier I., Rowley J.W., Stubben C., Petrey A.C., Tolley N.D., Guo L., Cody M., Weyrich A.S., Yost C.C., Rondina M.T., Campbell R.A. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136:1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Comer S.P., Cullivan S., Szklanna P.B., Weiss L., Cullen S., Kelliher S., Smolenski A., Murphy C., Altaie H., Curran J., O'Reilly K., Cotter A.G., Marsh B., Gaine S., Mallon P., McCullagh B., Moran N., Ní Áinle F., Kevane B., Maguire P.B., et al. COVID-19 induces a hyperactive phenotype in circulating platelets. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaid Y., Puhm F., Allaeys I., Naya A., Oudghiri M., Khalki L., Limami Y., Zaid N., Sadki K., Ben El Haj R., Mahir W., Belayachi L., Belefquih B., Benouda A., Cheikh A., Langlois M.A., Cherrah Y., Flamand L., Guessous F., Boilard E. Platelets can associate with SARS-Cov-2 RNA and are hyperactivated in COVID-19. Circ Res. 2020 doi: 10.1161/CIRCRESAHA.120.317703. 1404–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrottmaier W.C., Pirabe A., Pereyra D., Heber S., Hackl H., Schmuckenschlager A., Brunnthaler L., Santol J., Kammerer K., Oosterlee J., Pawelka E., Treiber S.M., Khan A.O., Pugh M., Traugott M.T., Schorgenhofer C., Seitz T., Karolyi M., Jilma B., Rayes J., et al. Adverse outcome in COVID-19 is associated with an aggravating hypo-responsive platelet phenotype. Front Cardiovasc Med. 2021;8 doi: 10.3389/fcvm.2021.795624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taus F., Salvagno G., Cane S., Fava C., Mazzaferri F., Carrara E., Petrova V., Barouni R.M., Dima F., Dalbeni A., Romano S., Poli G., Benati M., De Nitto S., Mansueto G., Iezzi M., Tacconelli E., Lippi G., Bronte V., Minuz P. Platelets promote thromboinflammation in SARS-CoV-2 pneumonia. Arterioscler Thromb Vasc Biol. 2020;40:2975–2989. doi: 10.1161/ATVBAHA.120.315175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Portier I., Campbell R.A., Denorme F. Mechanisms of immunothrombosis in COVID-19. Curr Opin Hematol. 2021;28:445–453. doi: 10.1097/MOH.0000000000000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pons S., Fodil S., Azoulay E., Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit Care. 2020;24:353. doi: 10.1186/s13054-020-03062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo J., Spittle D.A., Newnham M. COVID-19, immunothrombosis and venous thromboembolism: biological mechanisms. Thorax. 2021:412–420. doi: 10.1136/thoraxjnl-2020-216243. [DOI] [PubMed] [Google Scholar]
- 30.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puhm F., Allaeys I., Lacasse E., Dubuc I., Galipeau Y., Zaid Y., Khalki L., Belleannee C., Durocher Y., Brisson A.R., Wolberg A.S., Langlois M.A., Flamand L., Boilard E. Platelet activation by SARS-CoV-2 implicates the release of active tissue factor by infected cells. Blood Adv. 2022;6:3593–3605. doi: 10.1182/bloodadvances.2022007444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hottz E.D., Azevedo-Quintanilha I.G., Palhinha L., Teixeira L., Barreto E.A., Pao C.R.R., Righy C., Franco S., Souza T.M.L., Kurtz P., Bozza F.A., Bozza P.T. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maugeri N., De Lorenzo R., Clementi N., Antonia Diotti R., Criscuolo E., Godino C., Tresoldi C., Bio Angels For COVID-BioB Study Group. Bonini C., Clementi M., Mancini N., Ciceri F., Rovere-Querini P., Manfredi A.A. Unconventional CD147-dependent platelet activation elicited by SARS-CoV-2 in COVID-19. J Thromb Haemost. 2021:434–448. doi: 10.1111/jth.15575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apostolidis S.A., Sarkar A., Giannini H.M., Goel R.R., Mathew D., Suzuki A., Baxter A.E., Greenplate A.R., Alanio C., Abdel-Hakeem M., Oldridge D.A., Giles J.R., Wu J.E., Chen Z., Huang Y.J., Belman J., Pattekar A., Manne S., Kuthuru O., Dougherty J., et al. Signaling through FcgammaRIIA and the C5a-C5aR pathway mediate platelet hyperactivation in COVID-19. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.834988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fard M.B., Fard S.B., Ramazi S., Atashi A., Eslamifar Z. Thrombosis in COVID-19 infection: role of platelet activation-mediated immunity. Thromb J. 2021;19:59. doi: 10.1186/s12959-021-00311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S., Liu Y., Wang X., Yang L., Li H., Wang Y., Liu M., Zhao X., Xie Y., Yang Y., Zhang S., Fan Z., Dong J., Yuan Z., Ding Z., Zhang Y., Hu L. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J Hematol Oncol. 2020;13:120. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garma L.D., Deng H., Goldschmidt E. Integrated analysis of transcriptomic data reveals the platelet response in COVID-19 disease. Sci Rep. 2022;12:6851. doi: 10.1038/s41598-022-10516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen S., Zhang J., Fang Y., Lu S., Wu J., Zheng X., Deng F. SARS-CoV-2 interacts with platelets and megakaryocytes via ACE2-independent mechanism. J Hematol Oncol. 2021;14:72. doi: 10.1186/s13045-021-01082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T., Yang Y., Li Y., Wang Z., Ma F., Luo R., Xu X., Zhou G., Wang J., Niu J., Lv G., Crispe I.N., Tu Z. Platelets mediate inflammatory monocyte activation by SARS-CoV-2 spike protein. J Clin Invest. 2022:132. doi: 10.1172/JCI150101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebeyer-Masotta M., Eichhorn T., Weiss R., Laukova L., Weber V. Activated platelets and platelet-derived extracellular vesicles mediate COVID-19-associated immunothrombosis. Front Cell Dev Biol. 2022;10 doi: 10.3389/fcell.2022.914891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arnold D.T., Hamilton F.W., Milne A., Morley A.J., Viner J., Attwood M., Noel A., Gunning S., Hatrick J., Hamilton S., Elvers K.T., Hyams C., Bibby A., Moran E., Adamali H.I., Dodd J.W., Maskell N.A., Barratt S.L. Patient outcomes after hospitalisation with COVID-19 and implications for follow-up: results from a prospective UK cohort. Thorax. 2021;76:399–401. doi: 10.1136/thoraxjnl-2020-216086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messner C.B., Demichev V., Wendisch D., Michalick L., White M., Freiwald A., Textoris-Taube K., Vernardis S.I., Egger A.S., Kreidl M., Ludwig D., Kilian C., Agostini F., Zelezniak A., Thibeault C., Pfeiffer M., Hippenstiel S., Hocke A., von Kalle C., Campbell A., et al. Ultra-high-throughput clinical proteomics reveals classifiers of COVID-19 infection. Cell Syst. 2020;11:11–24.e4. doi: 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leng L., Li M., Li W., Mou D., Liu G., Ma J., Zhang S., Li H., Cao R., Zhong W. Sera proteomic features of active and recovered COVID-19 patients: potential diagnostic and prognostic biomarkers. Signal Transduct Target Ther. 2021;6:216. doi: 10.1038/s41392-021-00612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roh J.D., Kitchen R.R., Guseh J.S., McNeill J.N., Aid M., Martinot A.J., Yu A., Platt C., Rhee J., Weber B., Trager L.E., Hastings M.H., Ducat S., Xia P., Castro C., Singh A., Atlason B., Churchill T.W., Di Carli M.F., Ellinor P.T., et al. Plasma proteomics of COVID-19-associated cardiovascular complications: implications for pathophysiology and therapeutics. JACC Basic Transl Sci. 2022;7:425–441. doi: 10.1016/j.jacbts.2022.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sledz K.M., Moore S.F., Vijayaragavan V., Mallah S., Goudswaard L.J., Williams C.M., Hunter R.W., Hers I. Redundant role of ASK1-mediated p38MAPK activation in human platelet function. Cell Signal. 2020;68 doi: 10.1016/j.cellsig.2020.109528. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2019. https://www.R-project.org/ [accessed October 18, 2022].
- 47.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K., Chen W., Zhang Z., Deng Y., Lian J.Q., Du P., Wei D., Zhang Y., Sun X.X., Gong L., Yang X., He L., Zhang L., Yang Z., Geng J.J., Chen R., Zhang H., Wang B., Zhu Y.M., Nan G., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee M., Whiteheart S.W. The ins and outs of endocytic trafficking in platelet functions. Curr Opin Hematol. 2017;24:467–474. doi: 10.1097/MOH.0000000000000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G.M., Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Consortium G.O. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trugilho M.R.O., Azevedo-Quintanilha I.G., Gesto J.S.M., Moraes E.C.S., Mandacaru S.C., Campos M.M., Oliveira D.M., Dias S.S.G., Bastos V.A., Santos M.D.M., Carvalho P.C., Valente R.H., Hottz E.D., Bozza F.A., Souza T.M.L., Perales J., Bozza P.T. Platelet proteome reveals features of cell death, antiviral response and viral replication in covid-19. Cell Death Discov. 2022;8:324. doi: 10.1038/s41420-022-01122-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grozovsky R., Giannini S., Falet H., Hoffmeister K.M. Regulating billions of blood platelets: glycans and beyond. Blood. 2015;126:1877–1884. doi: 10.1182/blood-2015-01-569129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varghese L.N., Defour J.P., Pecquet C., Constantinescu S.N. The thrombopoietin receptor: structural basis of traffic and activation by ligand, mutations, agonists, and mutated calreticulin. Front Endocrinol (Lausanne) 2017;8:59. doi: 10.3389/fendo.2017.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boccatonda A., D'Ardes D., Rossi I., Grignaschi A., Lanotte A., Cipollone F., Guagnano M.T., Giostra F. Platelet count in patients with SARS-CoV-2 infection: a prognostic factor in COVID-19. J Clin Med. 2022;11:4112. doi: 10.3390/jcm11144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng Y., Wong R.S., Soo Y.O., Chui C.H., Lau F.Y., Chan N.P., Wong W.S., Cheng G. Initial treatment of immune thrombocytopenic purpura with high-dose dexamethasone. N Engl J Med. 2003;349:831–836. doi: 10.1056/NEJMoa030254. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Z., Yang Y., Fan L., Ye S., Lou K., Hua X., Huang Z., Shi Q., Gao G. Low serum level of apolipoprotein A1 may predict the severity of COVID-19: a retrospective study. J Clin Lab Anal. 2021;35 doi: 10.1002/jcla.23911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulloque-Badaracco J.R., Hernandez-Bustamante E.A., Herrera-Anazco P., Benites-Zapata V.A. Prognostic value of apolipoproteins in COVID-19 patients: a systematic review and meta-analysis. Travel Med Infect Dis. 2021;44 doi: 10.1016/j.tmaid.2021.102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DeRoo E.P., Wrobleski S.K., Shea E.M., Al-Khalil R.K., Hawley A.E., Henke P.K., Myers D.D., Jr., Wakefield T.W., Diaz J.A. The role of galectin-3 and galectin-3-binding protein in venous thrombosis. Blood. 2015;125:1813–1821. doi: 10.1182/blood-2014-04-569939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen Y., Fu W., Zheng Y., Yang J., Liu Y., Qi Z., Wu M., Fan Z., Yin K., Chen Y., Gao W., Ding Z., Dong J., Li Q., Zhang S., Hu L. Galectin 3 enhances platelet aggregation and thrombosis via Dectin-1 activation: a translational study. Eur Heart J. 2022;43:3556–3574. doi: 10.1093/eurheartj/ehac034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bongiovanni D., Klug M., Lazareva O., Weidlich S., Biasi M., Ursu S., Warth S., Buske C., Lukas M., Spinner C.D., Scheidt M.V., Condorelli G., Baumbach J., Laugwitz K.L., List M., Bernlochner I. SARS-CoV-2 infection is associated with a pro-thrombotic platelet phenotype. Cell Death Dis. 2021;12:50. doi: 10.1038/s41419-020-03333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Léopold V., Pereverzeva L., Schuurman A.R., Reijnders T.D.Y., Saris A., de Brabander J., van Linge C.C.A., Douma R.A., Chouchane O., Nieuwland R., Wiersinga W.J., van 't Veer C., van der Poll T. Platelets are hyperactivated but show reduced glycoprotein VI reactivity in COVID-19 patients. Thromb Haemost. 2021;121:1258–1262. doi: 10.1055/a-1347-5555. [DOI] [PubMed] [Google Scholar]
- 63.Zhang S., Liu Y., Wang X., Yang L., Li H., Wang Y., Liu M., Zhao X., Xie Y., Yang Y., Fan Z., Dong J., Yuan Z., Ding Z., Zhang Y., Hu L. SARS-CoV-2 binds platelet ACE2 to enhance thrombosis in COVID-19. J Hematol Oncol. 2020;13:120. doi: 10.1186/s13045-020-00954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denorme F., Manne B.K., Portier I., Petrey A.C., Middleton E.A., Kile B.T., Rondina M.T., Campbell R.A. COVID-19 patients exhibit reduced procoagulant platelet responses. J Thromb Haemost. 2020;18:3067–3073. doi: 10.1111/jth.15107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Samkari H., Karp Leaf R.S., Dzik W.H., Carlson J.C.T., Fogerty A.E., Waheed A., Goodarzi K., Bendapudi P.K., Bornikova L., Gupta S., Leaf D.E., Kuter D.J., Rosovsky R.P. COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood. 2020;136:489–500. doi: 10.1182/blood.2020006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaid Y., Guessous F., Puhm F., Elhamdani W., Chentoufi L., Morris A.C., Cheikh A., Jalali F., Boilard E., Flamand L. Platelet reactivity to thrombin differs between patients with COVID-19 and those with ARDS unrelated to COVID-19. Blood Adv. 2021;5:635–639. doi: 10.1182/bloodadvances.2020003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mahler M., Meroni P.L., Infantino M., Buhler K.A., Fritzler M.J. Circulating calprotectin as a biomarker of COVID-19 severity. Expert Rev Clin Immunol. 2021;17:431–443. doi: 10.1080/1744666X.2021.1905526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo Q., Zhao Y., Li J., Liu J., Yang X., Guo X., Kuang M., Xia H., Zhang Z., Cao L., Luo Y., Bao L., Wang X., Wei X., Deng W., Wang N., Chen L., Chen J., Zhu H., Gao R., et al. Induction of alarmin S100A8/A9 mediates activation of aberrant neutrophils in the pathogenesis of COVID-19. Cell Host Microbe. 2021;29:222–235 e4. doi: 10.1016/j.chom.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colicchia M., Schrottmaier W.C., Perrella G., Reyat J.S., Begum J., Slater A., Price J., Clark J.C., Zhi Z., Simpson M., Bourne J., Poulter N.S., Khan A.O., Nicolson P.L.R., Pugh M.R., Harrison P., Iqbal A.J., Rainger G.E., Watson S.P., Thomas M.R., et al. S100A8/A9 drives the formation of procoagulant platelets through GPIbα. Blood. The Journal of the American Society of Hematology. 2022;140:2626–2643. doi: 10.1182/blood.2021014966. [DOI] [PubMed] [Google Scholar]
- 70.Shi H., Zuo Y., Yalavarthi S., Gockman K., Zuo M., Madison J.A., Blair C., Woodward W., Lezak S.P., Lugogo N.L., Woods R.J., Lood C., Knight J.S., Kanthi Y. Neutrophil calprotectin identifies severe pulmonary disease in COVID-19. J Leukoc Biol. 2021;109:67–72. doi: 10.1002/JLB.3COVCRA0720-359R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Willigen G., Hers I., Gorter G., Akkerman J.W. Exposure of ligand-binding sites on platelet integrin alpha IIB/beta 3 by phosphorylation of the beta 3 subunit. Biochem J. 1996;314:769–779. [PMC free article] [PubMed] [Google Scholar]
- 72.van Willigen G., Akkerman J.W. Protein kinase C and cyclic AMP regulate reversible exposure of binding sites for fibrinogen on the glycoprotein IIB-IIIA complex of human platelets. Biochem J. 1991;273:115–120. doi: 10.1042/bj2730115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Veninga A., Baaten C., De Simone I., Tullemans B.M.E., Kuijpers M.J.E., Heemskerk J.W.M., van der Meijden P.E.J. Effects of platelet agonists and priming on the formation of platelet populations. Thromb Haemost. 2022;122:726–738. doi: 10.1055/s-0041-1735972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaid Y., Guessous F. The ongoing enigma of SARS-CoV-2 and platelet interaction. Res Pract Thromb Haemost. 2022;6 doi: 10.1002/rth2.12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.