Abstract
With advances in force fields and algorithms, robust tools have been developed for molecular simulation of three-dimensional structures of nucleic acids and investigation of aptamer-target interactions. The traditional aptamer discovery technique, Systematic Evolution of Ligands by EXponential enrichment (SELEX), continues to suffer from high investment and low return, while in vitro screening by simulated SELEX remains a challenging task, where more accurate structural modeling and enhanced sampling limit the large-scale application of the method. Here, we proposed a modified aptamer enhanced library design strategy to facilitate the screening of target-binding aptamers. In this strategy, a comprehensive analysis of the original complexes and the target secondary structure were used to construct an enhanced initial library for screening. Our enhanced sequence library design strategy based on the target secondary structure explored a certain sequence space while ensuring the accuracy of the structural conformation and the calculation method. In an enhanced library of only a few dozen sequences, four sequences showed a similar or better binding free energy than the original aptamer, with consistently high binding stability over three rounds of multi-timescale simulations, ranging from − 30.27 to − 32.25 kcal/mol. Consequently, the enhanced library strategy based on the target secondary structure is shown to have very significant potential as a new aptamer design and optimization strategy.
Keywords: Nucleic-acid aptamer, Target secondary structure, Enhanced library, Binding free energy
Graphical Abstract

1. Introduction
Aptamers, single-stranded nucleic acids including DNA and RNA, are capable of folding into a variety of secondary structures and forming unique three-dimensional structures upon recognition and binding to specific target molecules. In comparison with other biomolecules, aptamers have various advantages, including easy in vitro synthesis, high stability, structural accessibility for modification and low cost of detection, which make them useful tools for targeted therapy, detection, biosensor, and cell imaging. Due to the high selectivity and high affinity of the aptamer for the target molecule, it is also termed a “chemical antibody”. A variety of research studies have demonstrated that aptamers are capable of interacting with a variety of targets, including metal ions, amino acids, antibiotics, coenzymes, polypeptides, and fluorophore molecules [1], [2], [3], [4]. Recognition and interaction with different target molecules allow aptamers be applied as biosensors, target recognition and target delivery. For instance, aptamers that specifically recognize the natural EpCAM protein on the surface of live human cancer cells have important application potential in assisting the novel targeted cancer therapy [5]. Fluorescence-activated aptamers with extraordinary potential for live cell imaging, aptamers that activate dye molecular fluorophores, since Jaffrey and colleagues pioneered RNA mimics of GFPs (RMFPs), a series of fluorescent dye molecules and aptamers developed for GFP fluorophores and their derivatives (e.g., Spinach [6], Broccoli [7], Mango [8], Pepper aptamer [9], [10]) were discovered successively and have become effective methods for real-time imaging of cellular RNA and real-time tracking of protein-RNA utility [9]. Aptamers are expected to show their extraordinary potential in a wider range of domains.
The aptamers were usually identified via an in vitro procedure, named Systematic Evolution of Ligands by EXponential enrichment (SELEX) [11]. It involves three stages: (i) a random library containing a large number of random sequences, usually 1013 to 1015 different sequence motifs; (ii) the library is incubated with the target molecules and the unbound nucleic acids are eluted, followed by polymerase chain reaction (PCR) amplification of the bound nucleic acids for the next round of screening; (iii) Sequencing the final enrichment for aptamers with high binding affinity and specificity. SELEX has the potential to obtain any number of nucleic acid aptamers with targeted function and high affinity. It is, however, a time-consuming and low-reward strategy with some shortcomings. The selection of aptamers with acceptable affinity depends on the initial library, and the manually prepared aptamer library often occupies only a small fraction of the total sequence space for aptamers. Furthermore, in the enrichment problem after the screening, there may also be aptamers with a high affinity that are not detected. To cover this gap, several sequence analysis algorithms were developed to identify valid sequences from high-throughput sequencing (HTS) data. These molecular identification algorithms efficiently and accurately identify ligand recognition aptamers by multidimensional evaluation of sequence motif enrichment, aptamer family abundance, and structural stability [12], [13]. Moreover, molecular simulation based approaches [1] have also been reported for the optimal modification design of aptamers, where the binding free energy calculation in combination with the implicit model significantly helped the experimentalists to optimize the minimum useful length of the aptamer [14].
A few computational tools have been developed to facilitate the design and optimization of aptamers. RNA structure prediction and modeling programs such as Mfold [15], RNAfold[16], RNAComposer [17], and SimRNA [18], have achieved high precision in structure prediction, which makes it more feasible to conduct computational simulation studies. Structure-based molecular docking programs such as Glide [19] and Dock 6 [20], have also been specifically optimized for nucleic acids and have better accuracy in scoring algorithms. The key mechanisms by which nucleic acids and proteins interact with drugs have been successfully revealed through molecular simulations on the spatial and temporal scales, providing a theoretical basis for subsequent molecular optimization. For instance, multi-scale simulations successfully reveal the influence of RNA environment on spectral tuning and photoisomerization mechanism in Spinach-DFHBDI [6], contributing new observations to the principle and in the design of other fluorescent RNAs [21]. Yang's group rapidly identified binding surfaces/sites through molecular docking and molecular simulations, combined with mutation experiments to identify crucial residues, allowing significant time savings in the binding site identification and subsequent applications [13].
In this study, we proposed an aptamer design strategy based on the target secondary structure of an existing RNA-ligand complex, taking theophylline (1,3-dimethyl-7H-purine-2,6-dione) aptamer complex as a case study (Fig. 1a, d). This two-step approach began with molecular docking and molecular dynamics (MD) simulation of the original aptamer-ligand complex to identify key bases that are retained in subsequent sequence library design based on the target secondary structure. Subsequently, multiple rounds of virtual screening were performed based on the binding free energy in MD. Exhausting the entire sequence space to find high-affinity aptamers for target compounds is extremely difficult, both in experimental and virtual screening. The original complex-based analysis with a secondary structure-based sequence library design approach allows us to build a more efficient aptamer library. With an enhanced library design, it is easier to search for aptamers with high binding to target compounds than with a completely random sequence library. In this way, we screened four sequences with similar or better binding free energy than the original aptamer in a random batch of 50 enhanced sequences. These results demonstrated that the initial library design based on the target secondary structure would be an effective strategy for aptamer screening and optimization.
Fig. 1.
Secondary and tertiary structures of RNA-theophylline complex. a) Chemical structure of theophylline. b) Minimum free energy secondary structure of aptamer predicted by Mfold. c) The target secondary structure is described based on the tertiary structure determined by NMR. d) Three-dimensional structure of theophylline-aptamer complex. e) Bases around the TEP, and stacking structure of TEP formed by C8-G26-A28. f) RNA-ligand interaction analysis from PLIP.
2. Materials and methods
2.1. Aptamer discovery via enhanced library strategy
For a general aptamer-ligand complex, the secondary structure of the aptamer at minimum free energy is different from the secondary structure when it binds to the target, and the secondary structure when it is stably bound to the target we call the target secondary structure [22] of the aptamer. After binding to the target, the stem or small loop of the aptamer undergoes a few minor structural changes to adapt to the ligand. Therefore, it is important to focus on the target secondary structure of the aptamer-ligand complex after binding to the molecule, and characterizing the secondary structure of the aptamer RNA can also explain the aptamer folding process and explore the RNA fragmentation pattern to some extent [23]. The changes in the secondary structure not only suggest the location of the binding pocket of the ligand but also provide us with a solution for designing new aptamers.
For each aptamer-ligand complex, the minimal secondary structure of the aptamer alone is similar to but different from that of the bound state. In our study, aptamer formed a target secondary structure (Fig. 1c) after binding to theophylline, which was stably bound together by intermolecular forces such as electrostatic attraction and van der Waals forces, showing strong binding energy. From the structural point of view, the local part of the aptamer and the theophylline molecule form a ‘sandwich’ structure after binding, and this structure allows the theophylline to bind stably in the aptamer (Fig. 1d). Thus, for each sequence, there is minimum free energy (MFE, ∆GMFE), and free energy for folding to the target secondary motif (∆Gtarget), the difference between them (∆GMFE - ∆Gtarget) is called ∆∆Ggap. When we use the target secondary structure to design a new aptamer, the difference of ∆∆Ggap will converge to zero, which means that the newly designed aptamer can fold itself from the minimum free energy structure to the desired target secondary structure and bind to theophylline without additional energy loss, which increases the binding stability and conformational stability of both to some extent. In addition, a larger value of ∆∆Ggap makes the aptamer need to pay more energy sacrifice to convert to the target motif of binding to the ligand. It is not only unfavorable to the stability of the complex structure but also has a large conformational change due to its need to undergo a large change in folding, which puts the ligand at a disadvantage at the beginning of binding and is likely to fail to form a conformation that the ligand can recognize. Accordingly, our target structure motif-based design approach allows the designed aptamer to bind to theophylline without the need to undergo large folding and without crossing the energy barrier, and the binding pocket of the aptamer does not change significantly due to the maintenance of the target secondary structure, which facilitates the recognition and entry of the ligand. Thus, we will design new theophylline aptamers based on the target secondary structure.
The aptamer design method based on the target secondary structure is a two-step method to design the initially enhanced library and screen the aptamer (Fig. 2).
Fig. 2.
Overall process of enhanced library design and screening based on target secondary structure. a) Conversion from minimum free energy secondary structure to target secondary structure after recognition of ligand by an aptamer. b) Docking and molecular simulation analysis of the complex structure to identify significant residues. c) Sequence library design based on target secondary structure with retention of important residues. d) X3DNA was used for homology modeling of RNA. e) Multi-round screening based on binding free energy.
2.2. Comprehensive analysis of the original aptamer complex
After determining the aptamer design strategy based on the target secondary structure, we believe that by fully analyzing the interaction between the original aptamer and theophylline, and through the insight of the interaction and binding principle, we can guide the design of the aptamer sequence library. Thus, we performed molecular docking and molecular dynamics simulation analysis on the original aptamer complex, the Protein-Ligand Interaction Profiler (PLIP) [24] online service system was also utilized to analyze the aptamer-ligand interactions as a reference for the results. We clarified which bases play which roles in the interaction with theophylline by processing and summarizing the analysis results.
2.2.1. Molecular docking analysis
The theophylline-aptamer complex structure was obtained from the RCSB (PDB ID:1O15 [25]). In structure preparation, Protein Preparation Wizard in Maestro was used to complete the missing bonds and atoms in the structure and energy minimization of the protein structure under the OPLS4 force field [26]. Theophylline was generated in a variety of 3D structural conformations by LigPrep. The position of the original ligand is the center of the box, the outer box size was set to 14 Å, and other default settings were kept. Selecting the docked RMSD for output. After the docking was completed, the Ligand Interaction Diagram module was used to analyze the docking results. The top-score conformation in the docking results was selected for the initial structure of the MD simulation (Fig. 2b).
2.2.2. Analysis of MD simulations
To elucidate the interaction mechanism of the original theophylline-aptamer complex, we performed molecular dynamics simulations of the original complex using GROMACS 2018.8 [27] in the hope of obtaining the original aptamer binding and other relevant data to facilitate the design of subsequent mutant sequence libraries (Fig. 2b). For the RNA-ligand complex, the AMBER.ff99bsc0 [28] force field was used to simulate the RNA, the ligand was passed through the AnteChamber Python Parser interface [29] to generate a GAFF [30] force field based molecular topology file. Complex structures were placed in cubic boxes with boundaries of 11 Å. A total of 13,151 TIP3P [31] water molecules were placed inside the box, and Na+ and Mg2+ ions [32] were added as counterbalances to keep the whole simulated system electrically neutral.
First, the system undergoes a 50,000-step energy minimization, then the system is gradually heated from 0 to 300 K and subsequently reaches an initial equilibrium of 100 ps at a constant volume. In addition, there are 1 ns at constant temperature and constant pressure in equilibrium to thermostats for two groups with independent stabilization temperatures (RNA-ligand and water-ions groups). All hydrogen-related atoms using the LINCS algorithm [33] to constraint. An electrostatic interaction with long-range distances was handled by the Particle Mesh Ewald summation scheme [34], which used a fourth-order third interpolation method and a 0.12 nm lattice point size. The structural changes in the theophylline-aptamer complex and the reasons for the ligand binding stability were investigated by molecular simulation trajectories. The root-mean-square deviation (RMSD), the hydrogen bond atoms and hydrogen bond [35] occupancy were analyzed during the molecular simulation.
2.2.3. Binding free energy calculation
Binding free energy calculation is a widely used method to assess the binding capacity complexes like RNA-ligand and protein-ligand, etc. The MM-GB/PBSA method, thermodynamic integration, and free energy perturbation are common methods for calculating binding free energies [36], [37]. For the binding free energy of RNA to theophylline, Poisson Boltzmann surface area (MM-PBSA) versus Molecular Mechanics/Generalized Born surface area (MM-GBSA) [38] was figured by MMPBSA.py [39] in AmberTools21 [40]. Molecule-mechanics energies combined with the Poisson–Boltzmann or generalized Born and surface area continuum solvation methods are popular methods for estimating the free energy of binding ligands to macromolecules. Where the meaning of Eq. (1) splits the total binding energy of the complex in the solvent into two parts for calculation: the molecular mechanics term (free energy of binding in vacuum) and the solvation energy. For Eq. (2), the EMM represents the molecular mechanics term in the gas phase, which can be obtained directly by calculating the trajectories from molecular dynamics simulations, where Eele, Evdw, and Eint represent the electrostatic interaction energy, Van der Waals interaction energy, and gas-phase internal energy, respectively.
| Gcomplex, RNA, Ligand= EMM+ Gsol– TS | (1) |
| EMM= Eele+Evdw+Eint | (2) |
| Gsol= Epol+ Enopol= Epol+ ( γSASA+ β) | (3) |
For the solvation energy calculation, it was split into two parts: polar solvation free energy Epol and nonpolar solvation free energy Enopol. In the calculation of nonpolar solvation free energy, we used the estimation of surface accessible area (SASA). The parameter settings for the calculation were adopted according to the system. The surface tension parameters γ and β were 0.00542 and 0.92, respectively, and the salt concentration was kept at 100 mM. The binding free energy calculations were performed by averaging 1000 frames from the second half of the trajectory.
2.3. Multi-round screening based on binding free energy
Simulations of the complex structure at multiple time scales provide a more observable evaluation of the stability and binding affinity of the structure. Accordingly, we proposed to screen the enhanced sequence library in three rounds based on binding free energy (Fig. 2e). The first round of molecular dynamics simulations was conducted over each of the complex structures for 2 ns, and the second half of the 1 ns simulation trajectory was used to calculate binding free energy for 1000 frames at 1 ps intervals. Then, based on the ranking of the binding free energy results calculated by MMPBSA.py, we performed a second round of 30 ns MD simulations for each of the structures that exhibited lower binding energies in the first round of binding free energy calculations (∆GMM-PBSA≤ −25.00 kcal/mol), and took the second half of the 15 ns simulation traces with 15 ps interval output for 1000 frames of the structure for binding free energy calculations. The third round of screening was to perform 100 ns molecular dynamics simulations for the aptamer sequence that still performed well in the binding energy calculation in the second round (∆GMM-PBSA ≤ −25.00 kcal/mol) and to take the 1000 frames of fragment structures output at 50 ps intervals from the latter 50 ns trajectory for the binding free energy calculation.
3. Results
3.1. Comprehensive analysis of the RNA-theophylline complex
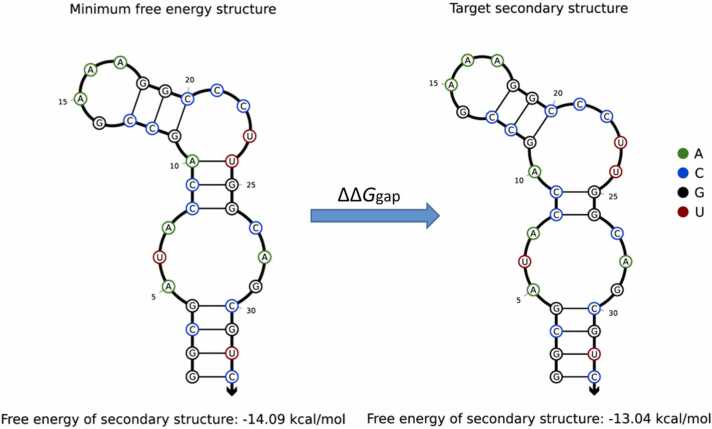
The structure of the theophylline-aptamer complex was obtained from the Protein Data Bank (PDB, http://www.rcsb.org/). Mfold was used to predict the dot-bracket formula of the aptamer sequence at the minimum free energy state as follows: ((((.((((((.))).)))).)))) (Fig. 1b). Target secondary structure dot-bracket formula was obtained using the "3D to (.)" module of RNApdbee 2.0 web server [41] as follows: ((((.((.(((.))).))).)))). It is observed that the secondary structure changes slightly after binding to the theophylline, and a pair of brackets becomes two points, indicating that one pair of bases has lost its pairing. We used Analysis [42] and Utilities in NUPACK web server [43] to calculate that the minimum free energy and the free energy in the target motif state as − 14.09 kcal/mol and − 13.04 kcal/mol respectively at 25 ℃ (Fig. 3). The value of ∆∆Ggap is not too large, which is probably one of the reasons for their stable binding.
Fig. 3.
The energy of the theophylline aptamer at 25 ℃ for the minimum free energy structure and the target secondary structure by NUPACK.
From the interaction relationship after docking (Fig. S1), using glide docking, it is possible to identify the interaction between theophylline and the aptamer in the docking mode where the receptor is rigid and the ligand is flexible, as C22 and U24 form three hydrogen bonds with theophylline, which stabilizes it. Also combined with the protein ligand interaction profiler (PLIP) Web Server, results were consistent with docking (Fig. 1f). The theophylline is just in the same direction with the conjugated aromatic rings in the upper and lower parallel C8 and A28 bases, forming a stable parallel π-stacking interaction. Based on the above hydrogen bonding and π-stacking together, the theophylline binds to the aptamer stably and forms a stable structural conformation. The RMSD value of the docking result is 0.538, which is in general agreement with the result in Li's work [44]. The resultant complexes were calculated using the MM-GBSA module in prime, and the approximate MM-GBSA value obtained was − 32.89 kcal/mol and the gscore was − 8.876 kcal/mol.
On the other hand, from the docking results, the binding site of the theophylline after docking is the same as that of the original complex, and the difference in the structural conformation after docking is also very small, which shows that the "sandwich"-like structure formed by the original binding site and the theophylline molecule is stable for the binding of the theophylline, which is beneficial not only to the entry of theophylline, but also to its binding. In addition, from the interaction relationships, the bases on both sides of the pocket also can stabilize the theophylline well through the joint action of hydrogen bonding and π-stacking, and we can conclude that the original complex structure has good structural stability. Therefore, it also shows the reliability of the method of aptamer design based on the target secondary structure.
The number of hydrogen bonds and RMSD value were calculated for the molecular simulation trajectories (Fig. 4). In the hydrogen bonding analysis, we analyzed the number of hydrogen bonds at 100 ps intervals for the 1000 ns molecular simulation trajectory, the number of hydrogen bonds between the aptamer and theophylline is stable around 3, and the number of hydrogen bonds is more stable after 500 ns in the second half. Compared with the results of docking, it indicates that the theophylline gradually finds a better binding site during the molecular dynamics, forms stable hydrogen bonds with the bases in the binding pocket, and the binding stability is further increased. Hydrogen bonding analysis between acceptor and donor was performed on the trajectories after molecular simulation using gmx_hbdat script, in which we used geometric criterion to determine the presence of hydrogen bonds (Table S1). Among the stability of hydrogen bonds, hydrogen bonds formed by atoms 703 and 701 of C22 base with theophylline molecule are more important in maintaining the stability of the structure, with an occupancy rate of 94.3% and 63.8%, and the atoms in U24 also form stable hydrogen bonds with theophylline. All the above result data indicate that C22 and U24 bases play a great role in maintaining the stability of the theophylline with the aptamer. Even in the molecular dynamics simulation at 1000 ns, it was stable in the binding pocket of the aptamer, and formed a stable number of hydrogen bonds with C22 and U24. Therefore, it should be reserved in the sequence design based on the target secondary structure.
Fig. 4.
Time dependence of Root mean square deviation (a) and hydrogen bonds (b) of the original RNA-ligand during a molecular simulation of 1000 ns.
In addition, MMPBSA.py was used to perform a decomposition residue free energy analysis of the binding free energy, which provides a more intuitive analysis of the binding energy contribution and error bars of the 33 bases of the aptamer. The analysis results showed that most of the bases have a very low binding free energy contribution, the main binding energy contributors were the bases in the binding pocket, and the bases at positions 23 and 26 also have free energy contributions with theophylline (Fig. 5 and Table 1). U23 is close to theophylline and has both van der Waals force and electrostatic interaction force with theophylline in the process of molecular simulation, however, it is relatively weak. G26 shows its very essential role in theophylline binding, which has a strong interaction force with theophylline. In terms of position, the G26 base is on the side of theophylline in position, and it is easy to interact with theophylline in the continuous simulation change. In addition, the parallel G26 bases can just form π-stacking interaction with the conjugation plane of the theophylline, thus G26 plays an equally important role in maintaining the stability of theophylline, and we also reserve it in the subsequent template design. Notably, although bases G25, C27 were also in the close proximity of the binding pocket, they contribute negligibly to the binding free energy (Table S2 and Fig. S2). Weak electrostatic interactions did not overcome the polar solvation free energy, an unfavorable factor, has an adverse effects on binding.
Fig. 5.
Binding free energy of the decomposition residues of the original aptamer.
Table 1.
Energy decomposition analysis of important residues (kcal/mol).
| Residue index | ∆EVDW | ∆EELE | ∆EPOL | ∆ENOPOL | ∆EBIND |
|---|---|---|---|---|---|
| C 8 | -3.92 ± 0.45 | 1.29 ± 0.52 | 0.01 ± 0.48 | -0.28 ± 0.03 | -2.90 ± 0.44 |
| C 22 | -0.47 ± 0.64 | -9.91 ± 1.36 | 7.45 ± 0.59 | -0.05 ± 0.01 | -2.97 ± 0.82 |
| U 23 | -1.42 ± 0.26 | -1.06 ± 0.43 | 1.57 ± 0.33 | -0.01 ± 0.01 | -0.91 ± 0.33 |
| U 24 | -0.76 ± 0.49 | -4.25 ± 0.84 | 3.30 ± 0.39 | -0.06 ± 0.01 | -1.76 ± 0.47 |
| G 26 | -2.72 ± 0.43 | -2.03 ± 0.46 | 2.49 ± 0.43 | -0.15 ± 0.03 | -2.41 ± 0.44 |
| A 28 | -5.15 ± 0.47 | 0.17 ± 0.54 | 1.89 ± 0.45 | -0.40 ± 0.02 | -3.48 ± 0.49 |
∆EELE =Van der Waals energy; ∆EELE = Electrostatic energy; ∆EPOL = Polar solvation energy; ∆ENOPOL = Non-Polar solvation energy; ∆EBIND = Total binding free energy.
According to the above analysis of the interaction between the original aptamer residues and theophylline, when designing new aptamers based on the target secondary structure (Fig. 2b), we chose residues retaining the above five important sites (8, 22, 24, 26, 28).
3.2. Design of enhanced sequence libraries
Our approach is based on the target secondary structure of the original complex and important sites for enhanced sequence libraries design, and in the above study we indicated the dot-bracket notation of the aptamer in the original complex as ((((.((.(((.))).))).)))). The template design for the enhanced sequence library will be guided by the results of the comprehensive analysis above. Based on the results of the above analysis, only five bases at sites 8, 22, 24, 26, and 28 were retained, the rest of the sites we chose as random mutations, but keep the structure of the complex when the mutated sequence binds to theophylline still as the above target structure.
Design of an enhanced sequence library based on the target secondary structure we obtained and the above-mutated sequence template. The RNAinverse Web Server in ViennaRNA Package [16] was used to generate an enhanced aptamer library. By setting the structure and sequence as above, the generated aptamer library can be guaranteed to be the target secondary structure we want to achieve while keeping the above 5 loci unchanged (Fig. 2c). A homology model was performed on the sequences of the enhanced library using X3DNA [45] (Fig. 2d), a modeling approach can ensure that the aptamer and theophylline remain in a “sandwich” structure and that the theophylline has suitable binding sites for the aptamer.
3.3. Binding free energy-based sequence screening
Based on the identified target secondary structure and the design method of the mutant sequence library, we performed binding free energy calculations for the generated 50 aptamer sequences to select the aptamer with well-behaved binding energy (Table S3). In the previous analysis (Fig. 4a), we learned that when molecular simulations were performed on the original complex structure, less than 100 ns of molecular simulations were sufficient to bring the complex to a stable structure when combined with changes in RMSD values. Therefore, when performing binding free energy calculations using MMPBSA.py, we adopted a stepwise approach of gradually increasing the simulation time to explore the binding ability of the complex. Thus, in the first round of simulations, the results obtained from the binding free energy calculations with a sequence structure of 2 ns are shown in Table S4, from the MM-PBSA binding free energy that more than half of the sequences performed well. In the second round of screening, we took the aptamer sequences with ∆GMM-PBSA ≤ −25.00 kcal/mol in the first round for the second round of 30 ns molecular dynamics simulations, and the simulation results are shown in Table S5. For the sequences that performed well in the first round, only about one-third of the aptamers in the second round of screening had binding energies greater than the set threshold. Among the 12 aptamers with underperforming binding energies, 9 aptamers had binding energies between − 25.00 and − 23.00 kcal/mol. This implies that even if they were screened down in the second round of underperformance in binding energy, the reason was not a sudden drop in binding affinity due to excessive conformational changes, but rather they remained at a relatively stable level, showing that the first round of 2 ns simulation was reasonable as a setting for the initial screening. In the second round of screening, we observed that some sequences such as 9, 43, 4, 50, 22, and other aptamers that performed poorly in the first round showed stronger affinity to the target in the second round of 30 ns simulation. That may be due to the longer simulation, some sequences got a more reasonable and stable conformation compared to the first 2 ns simulation. In the third round of 100 ns simulation with binding free energy calculation (Tables 2 and S6), only three sequences exhibit a binding free energy below − 25 kcal/mol and up to 85% of the sequences have well binding free energy performance. In four of these sequences, binding free energies were comparable or exceeded those of the original aptamer, and free energy decomposition analysis of them (Fig. S3) indicated that their interactions with theophylline were usually stronger at two positions (e.g. bases 8 and 26) than the original, leading to enhanced π-π stacking interactions. Based on this stable conformation of base stacking, theophylline binds more strongly to the sequence, resulting in higher binding affinity. In addition, we were surprised to see that although most of the sequences always maintain a high binding energy level, there has been a significant change in the ranking. This phenomenon also suggests to us that to obtain a more accurate binding affinity for a pair of RNA-ligand complexes, simulations on longer time scales are necessary to better reflect the real kinematic conformation and actual performance of the complexes.
Table 2.
Ranking of binding free energy after the third round of 100 ns simulation.
| sequence ID | ∆∆GMM-GBSA (kcal/mol) | ∆∆GMM-PBSA (kcal/mol) |
|---|---|---|
| original | 2.23 | 0.00 |
| sequence 20 | -2.29 | -2.16 |
| sequence 24 | -1.72 | -0.71 |
| sequence 7 | -0.69 | -0.21 |
| sequence 41 | -5.45 | -0.18 |
| sequence 40 | -1.50 | 0.43 |
| sequence 14 | -1.32 | 0.52 |
| sequence 10 | 2.66 | 1.00 |
| sequence 38 | 0.02 | 1.26 |
| sequence 18 | -2.06 | 1.86 |
| sequence 46 | 0.04 | 2.04 |
| sequence 35 | 0.10 | 3.12 |
| sequence 22 | -1.03 | 3.71 |
| sequence 36 | 0.31 | 3.78 |
| sequence 9 | 0.75 | 4.24 |
| sequence 50 | -1.55 | 4.36 |
| sequence 4 | 1.08 | 4.47 |
| sequence 3 | -1.14 | 4.67 |
| sequence 31 | -0.30 | 5.94 |
| sequence 43 | 0.31 | 6.15 |
| sequence 23 | 7.17 | 13.92 |
Finally, based on the above results, we concluded that: in case the binding free energy of the aptamer and theophylline could still be stabilized within a reasonable range of variation with the extension of the molecular simulation time, their gradual discovery of a suitable binding conformation within this simulation time also indicated that they had a relatively stable binding affinity. The reasons if the binding free energy is less or the range of variation is larger after the molecular simulation time is prolonged may be as follows: (1) The molecular simulation time is not enough for the aptamer to find the correct binding mode and conformation with theophylline and form a stable structure, and a longer simulation is needed. (2) This aptamer is not a good candidate sequence to form a stable structure with the theophylline molecule. It was also considered reasonable to set a time for each screening round. The sequences with binding energy below − 25.00 kcal/mol after the first round of screening were also overwhelmingly stabilized below − 25 kcal/mol in the second and third rounds of screening. This indicates that the setting of 2 ns in the first round of screening is reasonable, and the first round of screening has screened out most of the poorly performing aptamers, which also saves our arithmetic resources for the subsequent rounds. In the subsequent 30 ns and 100 ns simulations, we found that the sequence ranking had changed significantly, and most of the sequences gradually found more reasonable binding concepts with the extension of simulation, which also provided a prerequisite for the accuracy of our calculation.
4. Discussion
Our work introduces a strategy that designs aptamers based on the secondary structure of the target and then searches for new aptamers by using an enhanced library of targeted design sequences. This approach encompassed two parts, the first is a comprehensive analysis based on the original complex structure, which requires the application of docking and molecular dynamics simulation programs. In this process, the positions of nucleic acids are determined that are essential to maintaining the stability of aptamers and ligands in subsequent targeting sequence library design processes. Subsequently, the remaining bases were randomly mutated, but keeping the minimum free energy structure of the mutated sequence structure as our previously determined target secondary structure. In the second part, the obtained sequence structures were subjected to three rounds of binding free energy-based screening. The sequences with high binding energy were finally enriched.
Our approach allows the generation of a sequence of aptamers with a specific target secondary structure in the sequence library. This aptamer design strategy further optimizes the initial library, i.e., an enhanced aptamer library is created. Such enhanced sequence libraries were not completely random but had certain characteristics that allowed us to explore other better sites while keeping the good ones. Moreover, since we employ the target secondary structure-based aptamer design method, the minimum free energy structure of the newly designed aptamer is the target secondary structure of the original complex, which theoretically binds to the ligand without excessive energy loss and forms a well-known conformation with a target. This also makes it easier and more reliable to calculate the theoretical affinity between them and screen the sequence library through computational simulations.
Traditional aptamer selection process, SELEX, identification inefficiency and low success rates [13]. Despite iterative improvements, the technology remains an unexplainable black box. That means, the goodness of the sequence library can’t be determined, to maximize the diversity and number of sequences in preparing sequence libraries. In contrast, our strategy involves analyzing the original complex to build the enhanced library: C22 and U24 bases form hydrogen bonds with the theophylline. Among the stacking bases parallel to the theophylline, C8 and A28 further stabilize the theophylline through strong π-π stacking. The decomposition of the binding free energy further demonstrates that the binding energy contribution of the G26 base pair to the theophylline includes both electrostatic and π-stacking interactions. In combination with the structural observation, as a stacking base parallel to the theophylline, its position is just to the sides of the theophylline, allowing electrostatic interactions with the terminal atoms of the theophylline molecule to occur and further stabilize the conformation of the theophylline in the binding pocket. Thus, the target secondary structure-based aptamer optimization and screening strategy are not designed to exhaust the entire sequence space to screen for strongly binding aptamers, but rather a new optimization strategy that analyzes the binding pattern of the original aptamer complexes, and in this way, the targeted design of the enhanced library makes it possible to screen for aptamers that bind strongly to the target with a much higher probability. Compared with the experiment, in an enhanced library of only a few dozen sequences, four sequences showed a similar or better binding free energy than the original aptamer, with consistently high binding stability over three rounds of multi-timescale simulations, ranging from − 30.27 to − 32.25 kcal/mol. Sequence space exhaustive search for random mutations of a sequence to screen for aptamers is feasible [1], nevertheless, this approach assumes that all complexes bind in the same way. which is controversial. Moreover, in contrast to methods that analyze the frequency of aptamer sequence occurrence, secondary structure and enrichment of high-throughput data to identify valid aptamers, our optimization strategy does not require previous motif information [13]. It is suitable for post-SELEX process optimization. By using this enhanced sequence library, we can discover aptamers by screening multiple times under the assumption that the binding conformation is reasonable. As the simulation proceeds at different time scales, the complex structure gradually finds the point with the lowest energy, resulting in more informative calculated results. The enhanced library also allows for a reduction in screening time, which is a clear advantage over traditional experiments. During the screening, when increasing the time scale of the simulation, the majority of sequences found more reasonable and stable binding sites than in the first simulation round, and the binding energy was more stable, showing that this optimization strategy can effectively identify effective aptamers. The effectiveness of this optimization strategy has also been demonstrated in certain research. Yang’s group [10] performed in vitro affinity and other assays by randomly mutating key residues of well-bound aptamers into others, and their results showed that deletion of even just one residue at a key position typically resulted in a substantial decrease in affinity or a dramatic reduction or near disappearance of the fluorescence value of the fluorescent dye ligand at a particular wavelength. In another interesting experiment, they reconstructed the aptamer and transformed several key residues interacting with the ligand from RNA ribonucleic acid to DNA deoxyribonucleic acid and then performed in vitro experiments to verify that both affinity and fluorescence values were dramatically reduced. The above results not only reveal the importance of key residues but also reveal that hydrogen bonding plays a great role in the binding stability of aptamers and ligands.
Following three rounds of multi-timescale simulations and accurate binding free energy calculations, four sequences were enriched that were superior or similar to the original aptamer (Table 2). These results manifest that the aptamer-enhanced library design strategy based is an effective and reasonable optimization tool, and the designed sequences can finally enrich the aptamer with a high binding affinity to the target. It helps us to further clarify the interactions of the complexes and optimize the original aptamers with less time and experimental cost.
However, this aptamer design approach based on the target secondary structure also has drawbacks. Although only five nucleic acid bases are retained for the initial library design, the remaining 28 bases can be mutated at will, the secondary structure richness of the newly generated sequences is still relatively homogeneous, and we have not done enough in exploring a better binding pocket structure for theophylline. The sequence space explored was also limited, and no more sequence space was explored in this study for several reasons. First, exploring more structural space would also lead to too large computational resources, and second, when we choose to explore larger structural sequence space, various new theophylline-aptamer binding patterns would no longer apply to our more accurate screening method based on homology modeling via in silico base mutations and sequence screening based on binding free energy. There is a need to improve the existing calculation methods and incorporate sequence and ligand treatment, such as accurate three-dimensional structure prediction[46], metadynamics of enhanced sampling[47] to obtain the correct binding conformation, to make the results more reliable. Yet, this results in too much computational burden for large-scale screening, as opposed to our approach, which strikes a balance between computational accuracy and large-scale screening. As well as maintaining the rationality of the structural conformation and the accuracy of the computational method during molecular simulation, it also heavily explores the structure within a certain sequence space.
Funding
This study was supported by the National Key Research and Development Program of China (Grant 2019YFA0904800), the National Natural Science Foundation of China (Grants 82173746 and 82104066), Shanghai Frontier Science Research Base of Optogenetic Techniques for Cell Metabolism (Shanghai Municipal Education Commission, Grant 2021 Sci & Tech 03–28), and the 111 Project (Grant BP0719034).
CRediT authorship contribution statement
Long Chen: Conceptualization, Methodology, Formal analysis, Visualization, Writing − original draft. Bibi Zhang: Validation. Zengrui Wu: Validation. Guixia Liu: Validation, Project administration. Weihua Li: Validation, Project administration. Yun Tang: Project administration, Supervision, Funding acquisition, Writing − review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.01.002.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Zhou Q., Xia X., Luo Z., et al. Searching the Sequence Space for Potent Aptamers Using SELEX in Silico. J Chem Theory Comput. 2015;11:5939–5946. doi: 10.1021/acs.jctc.5b00707. [DOI] [PubMed] [Google Scholar]
- 2.Tuma Sabah J., Zulkifli R.M., Shahir S., et al. In vitro selection and characterization of single stranded DNA aptamers for luteolin: A possible recognition tool. Anal Biochem. 2018;549:72–79. doi: 10.1016/j.ab.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Song Y., Song J., Wei X., et al. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Q., Sun X., Xia X., et al. Exploring the mutational robustness of nucleic acids by searching genotype neighborhoods in sequence space. J Phys Chem Lett. 2017;8:407–414. doi: 10.1021/acs.jpclett.6b02769. [DOI] [PubMed] [Google Scholar]
- 5.Song Y., Zhu Z., An Y., et al. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal Chem. 2013;85:4141–4149. doi: 10.1021/ac400366b. [DOI] [PubMed] [Google Scholar]
- 6.Huang H., Suslov N.B., Li N.-S., et al. A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filonov G.S., Moon J.D., Svensen N., et al. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc. 2014;136:16299–16308. doi: 10.1021/ja508478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolgosheina E.V., Jeng S.C.Y., Panchapakesan S.S.S., et al. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 9.Chen X., Zhang D., Su N., et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs. Nat Biotechnol. 2019;37:1287–1293. doi: 10.1038/s41587-019-0249-1. [DOI] [PubMed] [Google Scholar]
- 10.Huang K., Chen X., Li C., et al. Structure-based investigation of fluorogenic Pepper aptamer. Nat Chem Biol. 2021 doi: 10.1038/s41589-021-00884-6. [DOI] [PubMed] [Google Scholar]
- 11.Stoltenburg R., Reinemann C., Strehlitz B. SELEX--a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Iwano N., Adachi T., Aoki K., et al. Generative aptamer discovery using RaptGen. Nat Comput Sci. 2022 doi: 10.1038/s43588-022-00249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J., Zheng Y., Huang M., et al. A sequential multidimensional analysis algorithm for aptamer identification based on structure analysis and machine learning. Anal Chem. 2020;92:3307–3314. doi: 10.1021/acs.analchem.9b05203. [DOI] [PubMed] [Google Scholar]
- 14.Anderson P.C., Mecozzi S. Identification of a 14mer RNA that recognizes and binds flavin mononucleotide with high affinity. Nucleic Acids Res. 2005;33:6992–6999. doi: 10.1093/nar/gki992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz R., Bernhart S.H., Siederdissen C.H.Z., et al. ViennaRNA Package 2.0. Algorithms for Molecular Biology. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antczak M., Popenda M., Zok T., et al. New functionality of RNAComposer: an application to shape the axis of miR160 precursor structure. Acta Biochim Pol. 2016;63:737–744. doi: 10.18388/abp.2016_1329. [DOI] [PubMed] [Google Scholar]
- 18.Boniecki M.J., Lach G., Dawson W.K., et al. SimRNA: a coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkv1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friesner R.A., Banks J.L., Murphy R.B., et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 20.Allen W.J., Balius T.E., Mukherjee S., et al. DOCK 6: Impact of new features and current docking performance. J Comput Chem. 2015;36:1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X., Chung L.W., Li G. Multiscale simulations on spectral tuning and the photoisomerization mechanism in fluorescent RNA Spinach. J Chem Theory Comput. 2016;12:5453–5464. doi: 10.1021/acs.jctc.6b00578. [DOI] [PubMed] [Google Scholar]
- 22.Shao Y., Chan C.Y., Maliyekkel A., et al. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybarczyk A., Jackowiak P., Figlerowicz M., et al. Computational prediction of non-enzymatic RNA degradation patterns. Acta Biochim Pol. 2016:63. doi: 10.18388/abp.2016_1331. [DOI] [PubMed] [Google Scholar]
- 24.Adasme M.F., Linnemann K.L., Bolz S.N., et al. PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Res. 2021;49:W530–W534. doi: 10.1093/nar/gkab294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clore G.M., Kuszewski J. Improving the accuracy of NMR structures of RNA by means of conformational database potentials of mean force as assessed by complete dipolar coupling cross-validation. J Am Chem Soc. 2003;125:1518–1525. doi: 10.1021/ja028383j. [DOI] [PubMed] [Google Scholar]
- 26.Lu C., Wu C., Ghoreishi D., et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J Chem Theory Comput. 2021;17:4291–4300. doi: 10.1021/acs.jctc.1c00302. [DOI] [PubMed] [Google Scholar]
- 27.Mja A., Tm D., Rsb C., et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers - ScienceDirect. SoftwareX. 2015;1–2:19–25. (s) [Google Scholar]
- 28.Pérez A., Marchán I., Svozil D., et al. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α / γ Conformers. Biophys J. 2007:92. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sousa da Silva A.W., Vranken W.F. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J., Wolf R.M., Caldwell J.W., et al. Development and Testing of a General AMBER Force Field. J Comput Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 31.Price D.J., Brooks C.L., 3rd A modified TIP3P water potential for simulation with Ewald summation. J Chem Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 32.Anderson P.C., Mecozzi S. Unusually Short RNA Sequences: Design of a 13-mer RNA that Selectively Binds and Recognizes Theophylline. J Am Chem Soc. 2005;127:5290–5291. doi: 10.1021/ja0432463. [DOI] [PubMed] [Google Scholar]
- 33.Ȧqvist J. Ion-water interaction potentials derived from free energy perturbation simulations. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- 34.Essmann U., Perera L., Berkowitz M.L., et al. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 35.van der Spoel D., van Maaren P.J., Larsson P., et al. Thermodynamics of Hydrogen Bonding in Hydrophilic and Hydrophobic Media. J Phys Chem B. 2006;110:4393–4398. doi: 10.1021/jp0572535. [DOI] [PubMed] [Google Scholar]
- 36.Jorgensen William L. Free energy calculations: a breakthrough for modeling organic chemistry in solution. Acc. chem. res. 1989;22:184–189. [Google Scholar]
- 37.Van G.W.F. The role of computer simulation techniques in protein engineering. Protein Eng. 1988:5–13. doi: 10.1093/protein/2.1.5. [DOI] [PubMed] [Google Scholar]
- 38.Sun H., Duan L., Chen F., et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Phys Chem Chem Phys. 2018;10:1039. doi: 10.1039/c7cp07623a. C1037CP07623A. [DOI] [PubMed] [Google Scholar]
- 39.MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314-3321, Journal of Chemical Theory and Computation. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 40.Case D.A., Iii T., Darden T., et al. The Amber biomolecular simulation programs. J Comput Chem. 2010;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zok T., Antczak M., Zurkowski M., et al. RNApdbee 2.0: multifunctional tool for RNA structure annotation. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fornace M.E., Porubsky N.J., Pierce N.A. A unified dynamic programming framework for the analysis of interacting nucleic acid strands: enhanced models, scalability, and speed. ACS Synthetic Biol. 2020:9. doi: 10.1021/acssynbio.9b00523. [DOI] [PubMed] [Google Scholar]
- 43.Zadeh J.N., Steenberg C.D., Bois J.S., et al. NUPACK: Analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 44.Li Y., Shen J., Sun X., et al. Accuracy assessment of protein-based docking programs against RNA targets. J Chem Inform Modeling. 2010;50:1134–1146. doi: 10.1021/ci9004157. [DOI] [PubMed] [Google Scholar]
- 45.Li S., Olson W.K., Lu X.-J. Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 2019;47:W26–W34. doi: 10.1093/nar/gkz394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watkins A.M., Rangan R., Das R. FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure. 2020;28(963–976) doi: 10.1016/j.str.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dama J.F., Hocky G.M., Sun R., et al. Exploring valleys without climbing every peak: more efficient and forgiving metabasin metadynamics via robust on-the-fly bias domain restriction. J Chem Theory Comput. 2015;11:5638–5650. doi: 10.1021/acs.jctc.5b00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.