Abstract
In recent years, N7-methylguanosine (m7G) methylation, originally considered as messenger RNA (mRNA) 5′ caps modifications, has been identified at defined internal positions within multiple types of RNAs, including transfer RNAs, ribosomal RNAs, miRNA, and mRNAs. Scientists have put substantial efforts to discover m7G methyltransferases and methylated sites in RNAs to unveil the essential roles of m7G modifications in the regulation of gene expression and determine the association of m7G dysregulation in various diseases, including neurological disorders. Here, we review recent findings regarding the distribution, abundance, biogenesis, modifiers, and functions of m7G modifications. We also provide an up-to-date summary of m7G detection and profile mapping techniques, databases for validated and predicted m7G RNA sites, and web servers for m7G methylation prediction. Furthermore, we discuss the pathological roles of METTL1/WDR-driven m7G methylation in neurological disorders. Last, we outline a roadmap for future directions and trends of m7G modification research, particularly in the central nervous system.
Key words: MT: RNA/DNA editing, m7G, epitranscriptome, METTL1, WDR4, central nervous system, neurological disorders, neurodevelopment, cancer, stroke
Graphical abstract
m7G modifications are novel regulators of gene expression. Corresponding authors and colleagues detail the biogenesis, modifiers, functions, detection approaches, and high-throughput analyses of m7G modifications, review databases, and webservers for m7G sites, and discuss potential pathological roles of m7G methylation in neurological diseases, pointing out future directions of m7G research.
Introduction
RNA methylation is the most well-characterized type of RNA modifications, which has been discovered for more than six decades.1,2,3 To date, more than 70 types of RNA methylations have been reported, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), N5-methylcytosine (m5C), and N7-methylguanosine (m7G) (Figure 1A).1 Although m6A methylation is the most well-investigated one, other RNA methylations particularly m7G methylation have obtained growing attention in recent years.1,4,5 m7G methylation is a prevalent and evolutionarily conserved RNA modification that was first discovered at the 5′ cap of messenger RNAs (mRNAs).1,6,7 Most eukaryotic mRNAs have a methyl group and a positive charge at the N7 position of the terminal guanosine at its 5′ cap.4 This essential cap modification stabilizes transcripts against exonucleolytic degradation8,9,10 and mediates cap-related biological functions including transcription elongation,11 pre-mRNA splicing,12,13 polyadenylation,14 nuclear export,15 and translation.16
Figure 1.
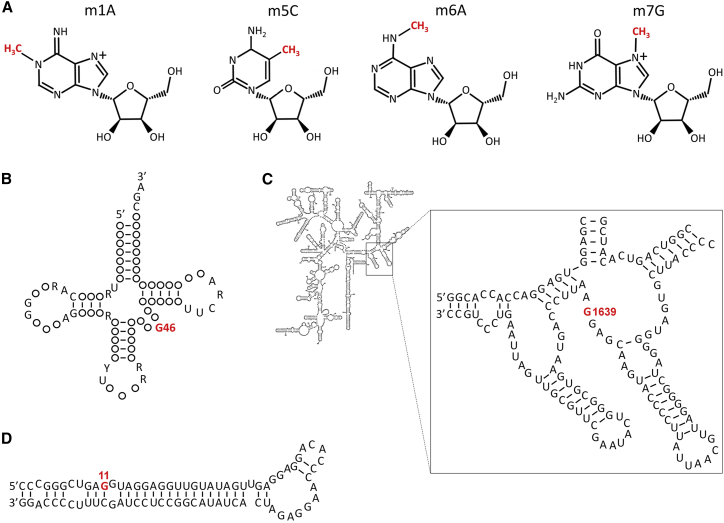
Molecular composition of common RNA methylations and m7G modifications in RNAs
(A) Molecular composition of common RNA methylations including m1A, m5C, m6A, and m7G. (B) Secondary structure of tRNA. m7G modification occurs at nucleotide position 46 in the extra loop. (C) Human small ribosomal subunit rRNA m7G can be methylated in the 18S rRNA 3′ major domain at nucleotide position 1639, which locates at a ridge forming a steric block between the P-site and E-site tRNAs at the back of the small subunit head. (D) Secondary structure of pri-let-7e-5p in which m7G modification occurs at nucleotide position 11.
Besides being a part of the cap structure, m7G methylation has been found at internal positions within mRNAs, transfer RNAs (tRNAs), and ribosomal RNAs (rRNAs).17,18 To date, internal m7G modifications have been demonstrated to play a very important role in many aspects of RNA metabolism including RNA processing, stabilization, maturation, and translation.5,19,20 m7G modifications are dynamically regulated by methyltransferases. The most well-characterized methyltransferase for internal m7G methylation is methyltransferase-like 1 (METTL1), the mammalian ortholog of yeast Trm8.18 METTL1 forms a functional methyltransferase complex with co-factor WD repeat domain 4 (WDR4), the human ortholog of the yeast Trm82.18
Despite recent advances in m7G research, the locations and functions of internal m7G modifications remain largely unknown. Internal m7G modifications have been suggested to be widely associated with the dysregulations of disease-related proteins, implicating internal m7G methylation as a novel epitranscriptome that contributes to multiple neurological disorders.21,22,23,24 In this review, we limit our discussions to internal m7G modifications and hence do not discuss modifications occurring as a part of the mRNA cap structure. We focus on the latest progresses made in internal m7G modification research, provide an up-to-date summary of established databases and web servers for validated and predicted m7G RNA sites, and discuss the association of m7G modifications with neurological disorders. With that, our review may provide insight into the development of m7G-centered diagnostic and prognostic biomarkers, as well as therapeutic strategies in neurological disorders.
The internal m7G methylation in eukaryotes
The internal m7G methylation in eukaryotes is first identified within yeast tRNA.18 tRNA nucleotide position 46 (m7G46) is the most prevalent m7G methylation site (Figure 1B).18,25 m7G46 is found in the variable loop region of a subset of tRNAs, forming a C13-G22-m7G46 base triple interaction in the tRNA three-dimensional core.5 m7G46 has no charge under physiological conditions and is conferred with positive charge via hydrogen bonding to bases G22 and C13.5,26 The tertiary base pair of the m7G46-C13-G22 and the site-specific electrostatic charge of m7G46 methylation stabilize the tRNA three-dimensional core of tRNA.5,26 tRNA m7G methylation may occur at positions other than position 46. For instance, m7G has been found at position 34 in the anticodon of mitochondrial (mt) tRNASer(GCU) of marine invertebrates.27 The D-arm of the mt tRNASer(m7GCU) is responsible for reading all four AGN codons as Ser on account of an unusual secondary structure.28 However, whether m7G34 exists in vertebrates remains to be certified.
m7G methylation is prevalent and conserved to rRNA as well.29,30 m7G modification has been identified in the 18S rRNA 3′ major domain at yeast rRNA nucleotide position 1,575, which is conserved to human small ribosomal subunit rRNA nucleotide position 1,639 (Figure 1C).31,32 m7G1575 methylation is at a ridge forming a steric block between the P-site and E-site tRNAs at the back of the small subunit head.29 The m7G1575 modification can be found throughout eukaryotes but not in bacteria or Archaea.29
Besides, m7G modification is also present in mRNAs. Based on bulk analyses, m7G modification widely exists on mRNAs, and its levels range from approximately 0.02% to 0.05% of all guanosines within mRNAs across several human and mouse cell lines.17,33 To date, more than 44,000 internal mRNA m7G sites within mammalian transcriptomes have been experimentally validated,34 and more than 1,200 disease-associated genetic mutations probably function through regulation of m7G methylation.34,35 Moreover, one methylated guanosine at position 11 (G11) of primary let-7e-5p transcript (pri-let-7e-5p) has been detected using a new protocol to detect m7G within low-abundant RNAs, referred to as borohydride reduction sequencing (BoRed-seq) (Figure 1D).20 G11 belongs to a 16-nt-long G-rich sequence that folds into the alternative Hoogsteen base-paired G-quadruplex structure to inhibit microRNA (miRNA) processing.36 Because G-quadruplexes exist in the precursors of miR-92b and miR-149 as well, m7G modification may occur in other miRNAs.37,38 Notably, the presence of m7G modification on miRNAs and their precursors has been questioned by other studies.39 The re-analysis of liquid chromatography-tandem mass spectrometry data implied that BoRed-seq detected m7G modifications on LSU rRNA rather than let-7e-5p.39 This finding can be supported by the AlkAniline-seq and m7G-MaP-seq results that m7G modifications were detected only in very few nucleotides of mRNAs, and no m7G signals was detected for noncoding RNAs, including miRNAs in eucaryotic cells.33,35,40 These findings imply low frequencies of m7G modifications in mRNAs/miRNAs; however, such results may also be due to technical issues like the incomplete dephosphorylation of m7G-capped mRNAs.33
Hence, m7G modifications have been detected in tRNAs, rRNAs, and maybe mRNAs and miRNAs. With the combination of advanced m7G sequencing at single nucleotide resolution with antibody-based m7G purification approaches, the discovery of more types of RNAs with m7G modifications and more methylation sites is a near possibility.
The functions of internal m7G methylation
Numerous studies have shown distinct functions of internal m7G modifications in various types of RNAs (Figure 2). Among them, tRNA m7G46 modification is the most intensively investigated one. m7G modification is important for the regulation of tRNA expression, structural stabilization, and function. For example, m7G46 modifications are essential for tRNA expression in mouse embryonic stem cells (mESCs), although with uncertified molecular mechanisms.41 m7G46 modification also confers a positive charge to tRNA, facilitating the interaction among G46, G22, and C13 to stabilize the tRNA three-dimensional core of tRNA.41 Moreover, tRNA m7G46 modification enhances translation efficiency of mRNAs in a m7G tRNA-decoded codon (m7G codon)-dependent manner.41,42 tRNA m7G modifications regulate ribosome translocation, whereas the lack of m7G tRNA induces ribosome pausing at m7G codons in the charged tRNA binding sites (A sites).42 In this way, m7G tRNAs are reported to control neural lineage commitment of mESCs by influencing pro-neural gene expression in mESCs.42 Except for developmental regulation, m7G46 modification also participates in the expression regulation of disease-associated genes.42 For instance, m7G modification of Arg-TCT tRNA facilitates translation of AGA codon-enriched mRNAs corresponding with cell growth-related genes, thereby driving oncogenic cell transformation.43
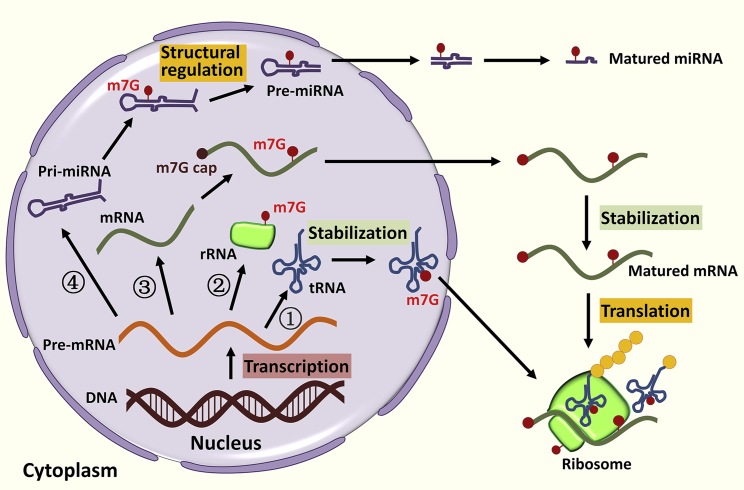
Figure 2.
The functions of m7G methylation in RNA metabolism
m7G methylation is involved in multiple steps of RNA metabolism processes. First, G46 on tRNA can be methylated by METTL1/WDR4 complex and conferred with positive charge via hydrogen bonding to bases G22 and C13, stabilizing the tRNA three-dimensional core of tRNA. Second, rRNA m7G can be methylated by WBSCR22/TRMT112 complex at human small ribosomal subunit rRNA G1639 with uncertified function. Third, mRNA m7G can be methylated by METTL1/WDR4 complex, leading to stabilization and translation of mRNA. Fourth, m7G of pri-let-7e-5p can also be methylated by METTL1/WDR4 complex, which, destabilizes G-quadruplexes and thereby promotes DROSHA-mediated cleavage of primary miRNA transcripts.
The m7G modifications of mRNAs and pri-miRNAs exhibit distinct functions from that of tRNA. m7G methylation stabilizes mRNA and stimulates its translation, as a hallmark of protein expression regulation.19 Zhao et al.19 have reported that m7G methylation of VEGFA transcript directly enhanced mRNA translation. However, whether mRNA m7G methylation modulates mRNA metabolism and translation through the same mechanisms as those of m6A remains to be determined. The m7G modifications of transcripts also play an important role in miRNA processing. Pandolfini et al.20 have claimed that m7G11 of pri-let-7e-5p leads to destabilization of G-quadruplexes, thereby promoting DROSHA-mediated cleavage of primary miRNA transcripts.37,38 Their findings indicate that m7G modifications modulate gene/miRNA expression by fine-tunning the secondary and tertiary structures of the transcripts. However, more rigorous studies are needed to confirm the existence of m7G modifications in mRNAs and miRNAs and clarify their functions.
Although m7G methylation has been reported within eukaryotic rRNAs, its exact role remains vague. Although rRNA m7G1639 modification and the pre-rRNA processing are under the control of the same protein complex WBSCR22/TRMT112, the methylation activities of the complex are not necessarily required for 18S rRNA synthesis, leaving the biological functions of rRNA m7G1639 unclear.29,32 Interestingly, multiple studies have demonstrated altered expression of WBSCR22 and TRMT112 in various pathological conditions, including various cancers44,45 and inflammation.46,47 These findings imply potential contribution of WBSCR22/TRMT112 complex-mediated rRNA m7G modification on human diseases, which needs to be confirmed in future work.
The modifying enzymes of m7G methylation
Although we are far away from clearly understanding the m7G modification processes, multiple enzymes with m7G methyltransferase activities have been identified. Among them, the most well known are the Trm82/Trm8 complex in yeast and its mammalian orthologs METTL1/WDR4 complex.18,48
In 2002, Alexandrov et al.18 identified two yeast open reading frames (ORFs) YDL201w and YDR165w (encoding Trm8 and Trm82, respectively) with tRNA m7G-methyltransferase activity, using an assay of genomic collection of glutathione S-transferase-ORF pools for m7G methyltransferase activity. These two proteins are unrelated and have no homology to each other. The Trm8 protein has an S-adenosyl-L-methionine Rossmann fold methyltransferase and serves as the catalytic subunit of the Trm82/Trm8 complex.49 Trm82 is a member of the WD fold family that adopts a propeller fold and contains seven blades.25,50 Trm82 induces structural rearrangements in Trm8 to influence the catalytic site.50 The noncatalytic subunits of the complex are involved in stabilizing the catalytic subunit or controlling the activity.49,50
Internal m7G methylation in mammals is carried out through the METTL1/WDR4 complex.48 The METTL1 gene, mapped to 12q13, contains seven exons and produces at least three different transcripts because of the differential splicing of two exons.21 The most frequently found METTL1 transcript consists of 1,292 nucleotides and codes for a protein of 276 amino acids.48 METTL1 protein are with high sequence similarities among mouse, Drosophila melanogaster, Arabidopsis thaliana, Caenorhabditis elegans, and yeast (39.8% identity between all six species).48 METTL1 is expressed in various organs and tissues and is inactivated by protein kinase B- and ribosomal S6 kinase-mediated phosphorylation at Ser27.51
Interestingly, m7G modifications in 18S rRNA are not introduced by METTL1/WDR4 complex. Instead, rRNA m7G sites at a ridge forming a steric block between P- and E-site tRNAs are methylated by Bud23/Trm112 methyltransferase complex in yeast and WBSCR22/TRMT112 one in human cells.29,52,53 Bud23/Trm112 complex mediates m7G1575 methylation in the yeast 18S rRNA.31,52 WBSCR22/TRMT112 complex mediates human 18S rRNA m7G1639 methylation.32 Bud23 and WBSCR22 are the catalytic subunits of methyltransferase complexes, and Trm112 and TRMT112 act as coactivators to regulate the metabolic stability of the complexes.29,52 Bud23 and Trm112 interact and form a β-zipper involving main chain atoms, which stabilizes the complex by burying an important hydrophobic surface.53 Moreover, the catalytic activity of these methyltransferases is not required for the pre-18S rRNA processing and the nuclear export of pre-40S ribosomal subunit.29,32,52 Thus, these methyltransferases may regulate rRNA maturation independent of its catalytic function, suggesting the interaction of these methyltransferases to pre-18S rRNA as a conserved quality control mechanism during rRNA maturation.
The distinct m7G methyltransferases for different types of RNAs are highly likely due to their conserved domains.54 Both METTL1 and WBSCR22 contain the conserved domain of the S-adenosyl-Lmethionine (AdoMet) methyltransferase that catalyzes the methylation reaction, however, their binding motifs are slightly different. The AdoMet binding motif of METTL1 is DIGCGYGGLLVELSPLFPDTLILGLEIR,48 and that of WBSCR22 is MAGRALELLYLPENKPCYLLDIGCG.55 METTL1 and WBSCR22 may also form their own special structures. Based on current knowledge on Trm8/Trm82, the major recognition sites of METTL1/WDR4 complex are the D- and T-stem structures of tRNA, and the Py48 sequence in the variable region was required for efficient methylation.56 WBSCR22 and TRMT112 form a heterodimeric methyltransferase complex.29 Although the binding sites or substrates of the WBSCR22/TRMT112 complex remain to be identified, the association of the WBSCR22/TRMT112 complex with pre-ribosomes, an early step occurring on nascent nucleolar transcripts, has been proposed as a key process for rRNA maturation.29
It is worth-noting that there are other methyltransferases for m7G, such as RNMT for m7G cap modification, TrmB family for prokaryotic tRNA m7G46 methylation, and sisomicingentamicin methylase for prokaryotic 16S rRNA m7G1405 methylation.57,58 These m7G cap and prokaryotic enzymes are not summarized here since they are out of the scope of this review.
The detection and high-throughput sequencing technologies of internal m7G methylation
Internal m7G modification can be measured by two different strategies, regular molecular biology experiments and high-throughput sequencing approaches (Table 1). The most classic molecular biology methods to detect m7G methylation are northern blot and immune-dot blot. The m7G modification levels of tRNAs are usually determined by northwestern blot41 and those of rRNAs are examined through primer extension.29,31 The m7G modification of mRNAs and miRNAs can be detected by RNA immune-dot blots using m7G-specific antibodies.20 The m7G modification levels of mRNAs and miRNAs can be further quantified by methylated RNA immunoprecipitation (MeRIP)-qPCR19 and mass spectrometry-based analyses such as liquid chromatography-electrospray ionization-tandem mass spectrometry and differential enzyme treatment coupled with high-resolution mass spectrometry analysis.20 Regular molecular biology experiments are sufficient to examine the m7G methylation at certain positions; however, these methods have a common limitation: they are not able to detect m7G methylation at the single-nucleotide level.
Table 1.
Current approaches for detecting m7G in different types of RNAs
| Technique | Throughput | Quantitative | RNA selection | Detection resolution | References |
|---|---|---|---|---|---|
| Northern blot | low | yes | tRNA | bulk level | Ma et al., 202141 |
| Primer extension | low | no | rRNA | RNA fragment level | Zorbas et al., 201529 |
| Immune-dot blot | low | no | mRNA, pri-miRNA | bulk level | Pandolfini et al., 201920 |
| MeRIP-qPCR | low | yes | mRNA, pri-miRNA | RNA fragment level | Zhao et al., 202119 |
| Liquid chromatography-electrospray ionization-tandem mass spectrometry | low | yes | mRNA | bulk level | Pandolfini et al., 201920 |
| Enzyme treatment coupled with high-resolution mass spectrometry analysis | low | yes | mRNA | bulk level | Pandolfini et al., 201920 |
| MeRIP-seq | high | yes | mRNA | RNA fragment level | Li et al., 202224 |
| m7G-seq | high | yes | tRNA, mRNA | single-nucleotide level | Zhang et al., 202162 |
| AlkAniline-seq | high | yes | tRNA, rRNA | single-nucleotide level | Marchand et al., 201833 |
| TRAC-seq | high | yes | tRNA | single-nucleotide level | Lin et al., 201961 |
| m7G-MaP-seq | high | yes | tRNA, rRNA | single-nucleotide level | Enroth et al., 201935 |
Next-generation sequencing-based technologies has offered an unprecedented opportunity to map the transcriptome-wide atlas of internal m7G modification under various conditions. MeRIP sequencing (MeRIP-seq) is an immunocapturing approach for unbiased transcriptome-wide localization of methylated RNA in high resolution that is widely applied to map the profile of m6A, m7G, m1A, m5C, and other RNA modifications.24,42 However, MeRIP-seq has been restricted because of the requirement of a large amount of total RNAs, the limited reproducibility for the detection of changes in RNA methylation, non-specific signals introduced by antibody pulldown, and the lack of capacity to map m7G down to single-base resolution.17,59,60 Inspiringly, MeRIP-seq with low-input RNA samples can be achieved by optimizing immunoprecipitation conditions and applying a post-amplification rRNA depletion strategy,59 and MeRIP-seq reproducibility can be enhanced significantly via an improved statistical approach implementing R package DESeq2, edgeR, and QNB, that partially overcomes the limitations of m7G-MeRIP-seq.60 Unlike meRIP-seq for the enrichment of methylated RNA sequences, chemically based approaches including m7G sequencing (m7G-seq), alkaline hydrolysis and aniline cleavage sequencing (AlkAniline-seq), tRNA reduction and cleavage sequencing (TRAC-seq), and m7G mutational profiling sequencing (m7G-MaP-seq) can provide nucleotide-level resolution of modification sites and determine the methylation states of highly methylated sites to reveal frequently modified m7G sites.17,33,61,62 These chemically based sequencing techniques take advantage of the unique reactivity of m7G that is reduced and de-purinated to induce structural transformation, which leads to misincorporation during reverse transcription to identify the methylated m7G site. In AlkAniline-seq, RNA fragments cleaved at the N+1 nucleotide to the modification site are produced via a specific sequence of chemical reactions, and selectively converted into sequencing libraries to increase the specificity and the sensitivity of sequencing. Since modified nucleotides are derived from different parental nucleotides, AlkAniline-seq simultaneously detects different RNA modifications such as m7G and m3C, which dramatically expands its applicability and efficiency.33 TRAC-seq is a more specialized chemically based approach to profile m7G modification within tRNAs.42,61 Compared with AlkAniline-seq, TRAC-seq uses an existing strategy that includes small RNA selection, AlkB demethylation, and sodium borohydride reduction to specifically and efficiently profile m7G sites in tRNAs,63 which can be further adapted to chemical cleavage-mediated detection of other RNA modifications.61 In m7G-MaP-seq, m7G-modified positions are converted to abasic sites using sodium borohydride reduction steps similar to TRAC-seq.35 Modified positions are then directly recorded as cDNA mutations through reverse transcription and sequenced. m7G-MaP-seq efficiently detects known m7G modifications in rRNA and identified uncharacterized evolutionarily conserved rRNA modification. However, these chemically based approaches have their limitations. The mild chemical reactions for selective m7G reduction and depurination may fail to achieve quantitative yields. Moreover, only a portion of m7G is converted into the abasic sites and a portion of converted m7G is labeled with biotin hydrazide, leading to decreased accuracy of the modification level measurements.
The databases and web servers for validated and predicted internal m7G methylation
Currently, multiple databases have been established for the annotation of mRNA modifications (Table 2).34 Since internal m7G modification is an emerging research topic, the development of database for m7G is much slower than that for m6A. Inspiringly, Song et al.34 built a platform, namely, m7GDB based on the data generated by high-throughput transcriptome-wide sequencing techniques including m7G-seq, m7G-MeRIP-seq, m7G-miCLIP-seq, and m7G-MaP-seq. m7GDB collects more than 44,000 experimentally validated internal m7G sites within mRNAs, rRNAs, and tRNAs; annotates m7G with the potentially affected post-transcriptional regulations; and demonstrates the pathological roles of internal m7G methylation.
Table 2.
Current platforms/methods/web servers for validated and predicted m7G sites, the impacts of genetic mutations on m7G, and m7G-disease associations
| Platform/model | Functions | RNA types | Developed year | Web server links | References |
|---|---|---|---|---|---|
| m7GDB | m7G database | mRNA, tRNA, rRNA | 2020 | http://www.xjtlu.edu.cn/biologicalsciences/m7ghub | Song et al., 202034 |
| iRNA-m7G | m7G prediction | mRNA | 2019 | http://lin-group.cn/server/iRNA-m7G/home.php | Chen, et al., 201964 |
| m7GFinder | m7G prediction | mRNA | 2020 | http://www.xjtlu.edu.cn/biologicalsciences/m7ghub | Song et al., 202034 |
| m7Gpredictor | m7G prediction | mRNA | 2020 | – | Liu et al., 202068 |
| XG-m7G | m7G prediction | mRNA | 2020 | http://flagship.erc.monash.edu/XG-m7G/ | Bi et al., 201065 |
| m7G-DPP | m7G prediction | mRNA | 2021 | – | Zou and Yin, 202167 |
| m7G-IFL | m7G prediction | mRNA | 2021 | http://server.malab.cn/m7G-IFL/ | Dai et al., 202166 |
| m7GSNPer | Mutation impacts on m7G | mRNA | 2020 | http://www.xjtlu.edu.cn/biologicalsciences/m7ghub | Song et al., 202034 |
| m7GDiseaseDB | m7G-disease association | mRNA | 2020 | http://www.xjtlu.edu.cn/biologicalsciences/m7ghub | Song et al., 202034 |
| m7GDisAI | m7G-disease association | mRNA | 2021 | http://180.208.58.66/m7GDisAI | Ma et al., 202169 |
In addition, various web servers, methods, and models have been built to provide high-accuracy prediction of putative internal m7G sites, such as iRNA-m7G,64 XG-m7G,65 m7G-IFL,66 m7GFinder,34 m7G-DPP,67 and m7GPredictor (Table 2).68 Basically, all computational predictors are built via a machine learning-based strategy. In these predictors, RNA sequences are encoded by different types of features, and the obtained features (e.g., pseudo dinucleotide composition, pseudo k-tuple composition, K monomeric units, K-spaced nucleotide pair frequencies, and nucleotide chemical property) were fed into different classification algorithms such as support vector machine and extreme gradient boosting support vector machine to discriminate m7G from non-m7G. All these predictors achieved decent classification accuracies that range from 86% to 92.5%, determined by performance assessment method with a 10-fold cross-validation. In addition, multiple computational methods have been developed to predict potential disease-associated m7G sites.34,69 For example, m7GSNPer has been developed to evaluate the impact of genetic variants on internal m7G RNA methylation, unraveling the potential functions of genetic mutation via the epitranscriptome regulation.34 Based on m7GSNPer, m7GDiseaseDB is built to provide details of disease-associated genetic variants and their affected m7G sites.34 Another database, m7GDisAI, similarly highlights m7G sites predicted to be associated with diseases with high prediction scores through a heterogeneous network-based m7G-disease associations inference method.69 All these databases and web servers greatly facilitate the investigations that aim to determine the conservation of m7G among species, evaluate the impact of genetic mutations on the m7G methylation states, demonstrate the physiological and pathological roles of currently known and predicted m7G modification, and identify novel internal m7G sites. With fast extension of our knowledge for internal m7G methylation and its regulations, these m7G databases will expand quickly, and these web servers for m7G sites prediction and disease association will acquire greater prediction accuracies in the foreseeable future.
The contribution of m7G methylation deregulation to neurological disorders
To date, studies that focus on the pathological roles of m7G modification deregulation have been initiated and inspiring results have been acquired (Table 3). The first evidence comes from analyses of genome and transcriptome data obtained from patients with neurological disorders and their corresponding animal models.
Table 3.
m7G and its machinery proteins associated with neurological disorders
| Disease | m7G related proteins | m7G RNA types | (Potential) role of m7G | References |
|---|---|---|---|---|
| Microcephalic primordial dwarfism | WDR4 (mutated) | tRNA | Wdr4-mediated tRNA m7G controls neuroectoderm commitment during development | Ma et al., 2021, Shaheen et al., 201569,70 |
| Down syndrome | WDR4 (mutated) | tRNA | Wdr4 maintains cognition function and hippocampal plasticity | Trimouille et al., 2018, Bull, 2017, Michaud et al., 200071,72,73 |
| Ischemic stroke/hypoxia | METTL1 (inhibited) | mRNA | mRNA m7G under hypoxic conditions activates apoptotic and inflammatory pathways | Li, et al., 2022, Androvic et al., 202024,82 |
| Alzheimer’s disease | METTL1 (?) | – | – | Jiang et al., 2020, Srinivasan et al., 202097,98 |
| Multiple sclerosis | METTL1 (mutated) | – | METTL1 regulates T cell and macrophage/microglial activities via vitamin D metabolism modulation | Zhao et al., 2021, Alcina, et al., 2013, Australia, 2009, Hadjigeorgiou et al., 201919,21,108,109 |
m7G methylation deregulation and neurodevelopmental disorders
Microcephalic primordial dwarfism (MPD) is a group of rare single-gene neurological disorders causing extreme reductions in brain and body size from early development onwards. MPD is characterized by pleasant personality, characteristic facial features, severe intrauterine and postnatal growth restriction, and microcephaly. Whole exome sequencing (WES) of two MPD patients in Egypt identified a novel missense mutation in WDR4 as the likely causal variant.70 In human, this WDR4 mutation causes a distinct form of MPD characterized by facial dysmorphism, brain malformation, and severe encephalopathy with seizures.70,71
Down syndrome (DS) is the commonest chromosomal disorder with impaired brain development and function, leading to mild to moderate intellectual disability. DS occurs in approximately 1 of 800 births globally, and more than 200,000 individuals are living with DS in the United States.72 Multiple genes on chromosome 21 and other chromosomes have been identified to contribute to the variation in clinical manifestations.72 In a recent study, WDR4 has been identified as a candidate gene for some of DS phenotypes, including the intellectual disability caused by trisomy of this chromosomal region in human patients.73 Animal study further supports that Wdr4 is one of the candidate genes for DS.74 Wdr4 expression levels are significantly decreased in the hippocampal tissues of DS mice.75 The overexpression of Wdr4 facilitates cognition function and hippocampal plasticity, thereby rescuing DS phenotypes.74 Thus, current evidence indicates the mutations of WDR4 as a key contributor to neurodevelopmental disorders.
Given the importance of METTL1/WDR4 mutations, scientists have investigated the roles of these mutations and consequent deregulation of m7G methylation in neural fate commitment. A study on human induced pluripotent stem cells has demonstrated that METTL1 silencing impairs neurectoderm formation, contributing to the disturbance of neurodevelopment.76 Multiple mechanisms may be involved in METTL1/WDR4 mutations-mediated neurodevelopmental defect. Cells derived from an MPD patient with WDR4 mutations have displayed decreased levels of m7G on tRNA.70 Similarly, Mettl1 deficiency specifically abolishes the enrichment of m7G-modified tRNAs in mESCs.42 The loss of METTL1/WDR4-mediated m7G tRNA methylome decreases the expression of neuroectoderm determinant genes like Otx2, Sox1, and Pax6. It is probably due to the longer coding DNA sequence and untranslated region (UTR) of these genes and the higher frequency of codons decoded by m7G methylated tRNAs that disrupts mESC differentiation to neural lineages.42 Besides, m7G methylation within transcripts such as pri-let-7 also participates in the regulation of neurodevelopment.20 By targeting the 3′UTR of Hmga2, Lin28, Ascl1 transcripts, and let-7 serves as a master miRNA in controlling the timing of neural lineage commitment,77 neurogliogenesis,78 and adult neurogenic niches.79 Disturbed let-7 maturation due to METTL1/WDR4 mutation-mediated m7G methylation deregulation thus may act as a potential mechanism for the pathogenesis of METTL1/WDR4 mutation-associated neurodevelopment including MPD,70,71 though further direct evidences from experimental studies (loss-of-function or gain-of-function studies) are needed to demonstrate this inference.
m7G methylation deregulation and ischemic stroke
Ischemic stroke (IS) is one of the most common causes of mortality and disability worldwide.80 Approximately 26% of the new cases are not able to perform basic daily activities, and more than 50% of patients exhibit hemiparesis and decreased mobility.81 Thus, stroke constitutes a major public health and economic problem of global significance, especially in the era that aging of the population has emerged as a key feature of population development. Neuronal injury and death are the main outcome of IS that result in mortality and disability. The mechanisms of neuronal death after the onset of IS are not yet completely understood. m7G modifications provide a novel perspective to investigate the pathogenesis of stroke. Multiple RNA-seq analyses of the mouse brain tissues with IS have revealed misexpression of Mettl1 mRNA after middle cerebral artery occlusion (MCAo).82 The altered Mettl1 expression may contribute to the pathogenesis of IS via multiple mechanisms. Because the outcome of IS is determined by the severity of hypoxia-related neuronal loss in the affected brain regions to a large degree,80 MeRIP-seq has been performed to compare m7G patterns in zebrafish brain under hypoxic and normoxic conditions, which displayed distinct mRNA m7G methylation signatures.24 The unbiased analyses identified more than 100 up-regulated and 300 down-regulated transcript methylation modification peaks, enriched in the 5′UTRs and start codon regions, in the brain tissue under hypoxic condition.24 These genes that participated in the methylation modification peaks are involved in the regulation of prominent pathways that mediate the hormesis-like response to hypoxia in stroke including Notch signaling.24,80 Notch signaling interacts with nuclear factor-κB,83 hypoxia-inducible factor-1α,84 p53,85 and c-Jun86 to promote neuronal death and neuroinflammation after IS, implying that hypoxia-induced m7G methylation of Notch-related transcripts as a causal factor of IS. Moreover, the expression levels of let-7e-5p, whose transcripts have been identified as a validated target of METTL1, significantly elevated in the serum and cerebral spinal fluid (CSF) collected from IS patients,87 matching with the increased Mettl1 expression in MCAo mouse brains.82 Let-7 has been reported to repress neuroprotection under ischemic conditions by down-regulating insulin-like growth factor-1 and to enhance inflammation via targeting interleukin-10, thereby increasing the risk of IS and worsening the prognosis of IS.88,89,90 Thus, METTL1-mediated pri-let-7 m7G modification may also contribute to the pathogenesis of IS, although again, we are waiting for evidence from gain-of-function or loss-of-function studies to demonstrate this inference.
m7G methylation deregulation and neurodegenerative diseases
Neurodegenerative diseases (NDs) are a group of disorders that include Alzheimer disease (AD), multiple sclerosis (MS), Parkinson disease, Huntington disease, and amyotrophic lateral sclerosis, characterized by progressive dysfunction and loss of selectively vulnerable populations of neurons.91,92 NDs impair learning, memory, cognitive, and motor functions of patients. Unfortunately, none of them is curable currently.91,93 All available treatments are only able to either manage the symptoms or halt disease progression.91,93
AD is the most common age-related ND and the leading cause for dementia in the aged population globally, accounting for 60%–80% of cases.94,95 To date, an estimated 5.5 million individuals suffer from AD in the United States, and the number of patients will increase to 13.8 million by mid-century.95 Moreover, the overall deaths resulting from AD increased 89% between 2000 and 2014.95 However, current treatments including cholinesterase inhibitors and memantine have no effect on the course of illness or the rate of decline,96 suggesting the importance to identify novel targets for AD diagnosis and treatment. Interestingly, virtually all DS patients develop AD in their 30s and at least 70% of adults with DS develop dementia by age 55–60 years, suggesting tight pathologic and mechanistic links between DS and AD.94 Because of the association of METTL1/WDR4 mutation with DS,74,75 the function and regulation of METTL1/WDR4-mediated m7G modifications have emerged as a new perspective to understand the pathogenesis of AD. Based on the information obtained from scREAD, a single-cell RNA-seq database for AD, the expression of Mettl1 transcripts was found to be repressed in excitatory neurons, astrocytes, and microglia in the cortex and hippocampus of AD model mice.97 However, conflicting results have been reported by Srinivasan et al.98 that the expression levels of METTL1 may not be significantly altered in different types of brain cells purified from post-mortem frozen superior frontal gyrus of AD and control patients. Besides, evidence also implicated the association of m7G modification with AD neuroinflammation. CD47, a receptor belonging to the immunoglobulin superfamily, plays an essential role in Aβ-induced activation of mast cells and cytokine release of microglia, thereby stimulating inflammatory responses of immune cells in the brain in the onset and progression of AD.99,100 A recent study reported that CD47 interacts with exportin-1 to modulate the package of m7G-modified miRNAs and mRNAs (e.g., let-7) in the extracellular vesicles (EVs). The Let-7 family acts as a ligand for toll-like receptor 7 on microglia and neurons to trigger intracellular signaling that leads to neuroinflammation and neurodegeneration in the central nervous system (CNS).101 In the CSF of AD patients, high levels of multiple let-7 family members like let-7b and let-7e have been detected, which is likely mediated by EV-dependent miRNA release.102 Given the importance of EV-mediated spreading of AD-related miRNAs and mRNAs within the CNS,103,104 m7G modification could be a novel mechanisms for the selective sorting of neurotoxic and pro-inflammatory RNAs into brain cell-derived EVs, and the deregulation of m7G modification thereby contributes to the pathogenesis of AD.105
MS is a potentially progressive autoimmune disease of the CNS. In MS, autoreactive immune cells induce myelin and axon injury in a progressive manner, leading to sustained motor and sensory function loss.106 Demyelination and neurodegeneration in the entire brain are central pathological features of MS. In the 21st century, the worldwide prevalence of MS keeps elevating and is highest in North America and Europe, which results in huge personal and socioeconomical burdens.107 Recently, METTL1 mutations have been reported in MS by various studies. Tag-single nucleotide polymorphism (SNP) analysis performed by the Australian and New Zealand Multiple Sclerosis Genetics Consortium describes an associated SNP (rs703842) located at the 3′UTR of the METTL1 gene.108 This finding was confirmed by a replication study of genome-wide association studies (GWASs) risk loci in Greek MS patients.109 Similarly, WES and SNP analyses identified a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with MS in the UK and Spain.21,22,23 This locus is required to enhance cytotoxic T lymphocyte antigen-4 expression in effector and regulatory CD4+ T cells, leading to a complex immune dysregulation syndrome in an MS model.110 Furthermore, the summary data-based Mendelian randomization method that integrating data from large scale GWAS and quantitative trait locus studies identified the interaction of METTL1 with METTL21B and TSFM as an important risk factor for MS.111 The literature has implied that an intronic region of the METTL1 gene and that of METTL21B gene function as enhancers of CYP27B1 gene to promote the expression of the latter, thus regulating the inflammatory responses of T cells and macrophages via modulating the mechanistic homeostasis of vitamin D metabolism.112 Notably, multiple studies have demonstrated mutation of METTL1 in MS; however, more investigations are required to clarify the contribution of METTL1 misexpression/mutation to m7G-mediated pathogenesis, accelerating the development of therapeutic strategies for treating MS.
Taken together, pioneer studies have implied that METTL1/WDR4 mutations and misexpressions and their mediated m7G methylation deregulation widely exist in the pathogenesis of various NDs. However, we need to note that most current evidences that support a pathological role of m7G methylation deregulation in NDs are from observational studies. With the help of high-throughput m7G-seq techniques and more exhaustive functional investigations for m7G and its methyltransferases, the m7G signatures of the whole brain and specific brain cell types in NDs can be further unveiled. Interventional studies that provide more direct causal-effect evidences are also in need to demonstrate the pathogenic role of m7G methylation deregulation in NDs, which will fill our knowledge gap.
m7G methylation deregulation and glioma
CNS tumors are relatively rare types of tumor; however, they are a significant cause of cancer morbidity and mortality.113 Malignant CNS tumors are those that can be classified into primary (start within the brain) and secondary tumors (spread from other organisms). Among all types of malignant brain tumors, gliomas, including both low-grade gliomas (LGGs) and glioblastomas (GBMs), are the most common and dismal primary ones in adult.114 According to the standards formulated by the World Health Organization, LGGs are defined as lower stage gliomas (I–III) with good prognosis, while GBMs are classified as the highest grade of glioma (IV) with worse prognosis.115 Currently, the main treatment for gliomas is surgery combined with radiotherapy and chemotherapy. The overall clinical outcome for glioma patients, particularly GBMs, remains poor.116,117 Therefore, it is critical to develop innovative and trustworthy diagnostic and prognostic biomarkers and therapeutic targets to improve the outcomes of patients with gliomas.
The literature has demonstrated the association of dysregulation of m7G RNA modification with various cancers, including lung cancer41 and intrahepatic cholangiocarcinoma.118 Recently, m7G methylation deregulation has been found to be linked with the progression of glioma through bioinformatic analyses. Researchers extracted potential m7G regulators from Gene Set Enrichment Analysis (GSEA; http://www.gsea-msigdb.org/gsea/index.jsp), and analyzed the expression patterns of these genes using transcriptome data from public databases including Cancer Genome Atlas, Gene Expression Omnibus, and Chinese Glioma Genome Atlas.119,120,121,122 These analyses identified various up-regulated m7G regulators (e.g., DCPS, NUDT1, NUDT5, NUDT3) and down-regulated ones (e.g., CYFIP2, LARP1, EIF4G3, NCBP2) in glioma groups versus controls.120,122 Importantly, the expression patterns of m7G RNA modification regulators are strongly correlated with the malignant state of glioma. Moreover, 4 m7G regulators including DCPS, EIF4E1B, NUDT1, and NUDT16L1 displayed distinct expression levels between GBMs and LGGs, suggest their contribution to the malignancy of glioma.121 The prognostic values of differentially expressed m7G methylation regulators in glioma have been further determined by univariate and multivariate Cox regression analyses, and prognostic signatures constructed on m7G regulators have demonstrated outstanding prediction potentials for the overall survival of glioma patients.120,121,122 However, it is worth noting that there are conflicting results obtained in both analyses.120,122 For instance, EIF4E1B and EIF4E3, which were identified as a glioma-enriched genes,122 were reported to be repressed in glioma tissue.120 These contradictory findings are majorly due to the inclusion of different RNA-seq transcriptome data and clinical data and the use of different algorithm in bioinformatic analyses, which is hopeful to be overcome in future comprehensive investigations. More important, we need to be aware that we are in lack of direct causal-effect evidence from experimental studies to support the inference from above findings from observational studies at current stage. Further investigations are needed to demonstrate the regulatory role and underlying mechanism of m7G RNA modification in the pathogenesis and pathologies of glioma. Meanwhile, fastidious studies are necessary to validate the exact roles of these predicted m7G regulators on m7G modification, which may identify novel m7G writers, erasers, and readers in near future.
Besides, 12 m6A/m5C/m1A/m7G-associated long noncoding RNAs (lncRNAs; e.g., AL080276.2, AC092111.1, SOX21-AS1, DNAJC9-AS1, AC025171.1, AL356019.2, AC017104.1, AC099850.3, UNC5B-AS1, C006064.2, AC010319.4, and AC016822.1) have been identified as prognosis-related lncRNAs.119 These lncRNAs are with high values for prediction of overall survival of glioma and evaluation of immunotherapy, ascertained by the receiver operating characteristic and the tumor immune dysfunction and exclusion analyses.119 However, all these studies used cohorts of glioma patients with a distinct genetic background that significantly increase the risk of bias in analyses and failed to explain the underlying mechanisms of differential expression of m7G-associated genes and lncRNAs. Therefore, more original studies, especially ones that specifically examine the m7G signatures of gliomas and BGMs using recently developed m7G high-throughput analyses are urgently needed to clarify the involvement of m7G methylation in glioma.
Conclusions and future perspectives
The rapid development of RNA modification researches has revealed an important role of METTL1/WDR4 complex-mediated m7G methylation in neurodevelopment and neural function. However, we are with limited knowledge of m7G modification regulation and how it influences the CNS. Thus, we prospect for the future development of the directions and trends of m7G methylation, particularly in the CNS.
The first one is to discover novel m7G writers, erasers, and readers. To date, more than 20 m6A modification proteins have been identified, suggesting the complexity of RNA modification regulation mechanisms.1 Current literature has demonstrated METTL1/WDR4 complex as m7G writer for tRNA, mRNA, and miRNA, and Bud23/Trm112 and WBSCR22/TRMT112 complexes as m7G writer for rRNA in eukaryotic cells.29,42,48 However, none of m7G erasers and readers has been reported. Since RNA demethylases may demethylate multiple types of modifications (e.g., FTO demethylates m6A and m3U), it is interesting to investigate that whether the currently known RNA demethylases function as m7G erasers using well established high-performance liquid chromatography-based biochemistry assays.123 Taking a cue from m6A researches, putative m7G readers can be predicted through bioinformatics analyses using published RNA-binding protein cross-linking immunoprecipitation associated to high-throughput sequencing datasets and known m7G modification sites.124,125 m7G reader candidates can be confirmed by mass spectrometry analysis after RNA pull-down using methylated single-stranded RNA bait with the consensus m7G motifs.124 Moreover, GSEA database also identified multiple potential m7G regulators, which helps us to narrow searching scope for novel m7G writers, erasers, and readers.
Second, it is of great importance in scientific and clinical research to identify new m7G modifications in the CNS. Being the most complex organ, the brain consists of many cell types, including neurons, astrocytes, and microglia. More important, these types of cells are with high heterogeneity. We, together with other independent groups, have demonstrated the distinct mRNA m6A signatures of brain tissues126 and specific cell types including microglia46,127 under pathological conditions (e.g., hypoxia, neuroinflammation). Similarly, mRNA m7G profiles are highly likely to be altered during neurodevelopment and the progressions of neurological disorders, which can be addressed by m7G MeRIP-seq and other high-throughput analyses. Following the functional characterization of novel m7G-modified mRNAs, the roles of mRNA m7G modification (de)regulation in CNS development and neurological disorders pathogenesis can be further unveiled.
Third, to unveil the functions of rRNA and tRNA m7G modification in the CNS and other organ systems is necessary to expand our understanding on m7G modification. Although studies have demonstrated the involvement of m7G modification in tRNAs in neurodevelopment, they are just the tip of the iceberg regarding the exact roles of tRNA m7G modification on neurological disorders and the underlying mechanisms. More important, nothing is known regarding the function of rRNA m7G modification in gene expression regulation and diseases. Inspiringly, studies have shown elevated expression levels of WBSCR22 in human glioma cells, leading to enhanced tumor cell survival and a poor prognosis.44 Our recent RNA-seq analyses also suggested significant up-regulation of Trmt112 expression in differentiating neural stem cells,128 and reduction of expression levels of Wbscr22 transcripts in activated microglia.46 These results provide confidence to clarify the contribution of the deregulation of WBSCR22/TRMT112 expression and rRNA m7G modification disorder on brain development and diseases in future studies.
Fourth, the potential applications of m7G modification as diagnostic/prognostic biomarkers and therapeutic targets is also an interesting topic. Multiple studies have revealed a great clinical value of METTL family members in the diagnosis and prognosis of cancer.129 As preliminary investigations have found altered METTL1 expression in activated microglia, it is important to investigate the diagnostic/prognostic values of METTL1/WDR4 for neuroinflammation and related neurological disorders. Investigating the states of m7G modification and the expression levels of METTL1/WDR4 in accessible biofluids, such as urine, blood, or CSF, might be an important first step to demonstrate the feasibility of using m7G profiling as potential biomarkers. We lack such studies, given that the field is still in its very early stages. Yet interestingly, proof-of-concept studies have shown that STM2457, a small molecule inhibitor of METTL3, is with promising anti-tumor effects thanks to the considerable functions of METTL3-mediated m6A methylation in tumorigenesis.130 Given the important findings of METTL1/WDR4 mutation in the pathogenesis of MPD,70,71 DS,74,75 and MS,108,109 it is essential to develop METTL1 agonists/antagonists and examine their therapeutic effects on neurological disorders.
In summary, being a novel RNA modification, internal m7G methylation is widely associated with the modulation of tRNA and rRNA functions, post-transcriptional regulation of mRNAs, and the processing of miRNAs. More importantly, pioneer studies have implied essential pathological roles of m7G modification in NDs. More comprehensive studies that aim to expand our understanding on the roles of m7G methylation in the CNS will shed light on the development of novel strategies for the diagnosis, treatment, and prognosis of neurological disorders.
Acknowledgments
Supported in part by research grants from the National Natural Science Foundation of China (No. 81971145 and No. 82271477 to X.X., No. 91949204 and 81830037 to J.C.Z.). We thank Huiran Wu for proofreading the manuscript.
Author contributions
X.X., J.C.Z. conceived the manuscript. X.X. collected references. X.X. and Y.W. wrote the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no conflict of interests regarding the publication of this paper.
Contributor Information
Xiaohuan Xia, Email: xiaohuan_xia1@163.com.
Jialin C. Zheng, Email: jialinzheng@tongji.edu.cn.
References
- 1.Wiener D., Schwartz S. The epitranscriptome beyond m(6)A. Nat. Rev Genet. 2021;22:119–131. doi: 10.1038/s41576-020-00295-8. [DOI] [PubMed] [Google Scholar]
- 2.Xie S., Chen W., Chen K., Chang Y., Yang F., Lin A., Shu Q., Zhou T., Yan X. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int. 2020;20:585. doi: 10.1186/s12935-020-01679-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler M., Weissmann B., Gutman A.B. Occurrence of methylated purine bases in yeast ribonucleic acid. J. Biol. Chem. 1958;230:717–723. [PubMed] [Google Scholar]
- 4.Furuichi Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2015;91:394–409. doi: 10.2183/pjab.91.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomikawa C. 7-Methylguanosine modifications in transfer RNA (tRNA) Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19124080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramanathan A., Robb G.B., Chan S.H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowling V.H. Regulation of mRNA cap methylation. The Biochemical Journal. 2009;425:295–302. doi: 10.1042/BJ20091352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuichi Y., LaFiandra A., Shatkin A.J. 5'-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 9.Shimotohno K., Kodama Y., Hashimoto J., Miura K.I. Importance of 5'-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc Natl Acad Sci USA. 1977;74:2734–2738. doi: 10.1073/pnas.74.7.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murthy K.G., Park P., Manley J.L. A nuclear micrococcal-sensitive, ATP-dependent exoribonuclease degrades uncapped but not capped RNA substrates. Nucleic Acids Res. 1991;19:2685–2692. doi: 10.1093/nar/19.10.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pei Y., Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 12.Konarska M.M., Padgett R.A., Sharp P.A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984;38:731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- 13.Lindstrom D.L., Squazzo S.L., Muster N., Burckin T.A., Wachter K.C., Emigh C.A., McCleery J.A., Yates J.R., 3rd, Hartzog G.A. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond D.R., Armstrong J., Colman A. The effect of capping and polyadenylation on the stability, movement and translation of synthetic messenger RNAs in Xenopus oocytes. Nucleic Acids Res. 1985;13:7375–7394. doi: 10.1093/nar/13.20.7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis J.D., Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 16.Muthukrishnan S., Both G.W., Furuichi Y., Shatkin A.J. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33–37. doi: 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L.S., Liu C., Ma H., Dai Q., Sun H.L., Luo G., Zhang Z., Zhang L., Hu L., Dong X., et al. Transcriptome-wide mapping of internal N(7)-methylguanosine methylome in mammalian mRNA. Mol. Cell. 2019;74:1304–1316.e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov A., Martzen M.R., Phizicky E.M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y., Kong L., Pei Z., Li F., Li C., Sun X., Shi B., Ge J. m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front. Cell Dev. Biol. 2021;9:642080. doi: 10.3389/fcell.2021.642080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandolfini L., Barbieri I., Bannister A.J., Hendrick A., Andrews B., Webster N., Murat P., Mach P., Brandi R., Robson S.C., et al. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell. Jun 20 2019;74:1278–1290.e9. doi: 10.1016/j.molcel.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alcina A., Fedetz M., Fernandez O., Saiz A., Izquierdo G., Lucas M., Leyva L., Garcia-Leon J.A., Abad-Grau Mdel M., Alloza I., et al. Identification of a functional variant in the KIF5A-CYP27B1-METTL1-FAM119B locus associated with multiple sclerosis. J. Med. Genet. 2013;50:25–33. doi: 10.1136/jmedgenet-2012-101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramagopalan S.V., Dyment D.A., Cader M.Z., Morrison K.M., Disanto G., Morahan J.M., Berlanga-Taylor A.J., Handel A., De Luca G.C., Sadovnick A.D., et al. Rare variants in the CYP27B1 gene are associated with multiple sclerosis. Ann. Neurol. 2011;70:881–886. doi: 10.1002/ana.22678. [DOI] [PubMed] [Google Scholar]
- 23.Pytel V., Matias-Guiu J.A., Torre-Fuentes L., Montero-Escribano P., Maietta P., Botet J., Alvarez S., Gomez-Pinedo U., Matias-Guiu J. Exonic variants of genes related to the vitamin D signaling pathway in the families of familial multiple sclerosis using whole-exome next generation sequencing. Brain Behav. 2019;9:e01272. doi: 10.1002/brb3.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W., Li X., Ma X., Xiao W., Zhang J. Mapping the m1A, m5C, m6A and m7G methylation atlas in zebrafish brain under hypoxic conditions by MeRIP-seq. BMC Genomics. 2022;23:105. doi: 10.1186/s12864-022-08350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov A., Grayhack E.J., Phizicky E.M. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA. 2005;11:821–830. doi: 10.1261/rna.2030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agris P.F., Sierzputowska-Gracz H., Smith C. Transfer RNA contains sites of localized positive charge: carbon NMR studies of [13C]methyl-enriched Escherichia coli and yeast tRNAPhe. Biochemistry. 1986;25:5126–5131. doi: 10.1021/bi00366a022. [DOI] [PubMed] [Google Scholar]
- 27.Matsuyama S., Ueda T., Crain P.F., McCloskey J.A., Watanabe K. A novel wobble rule found in starfish mitochondria. Presence of 7-methylguanosine at the anticodon wobble position expands decoding capability of tRNA. J Biol. Chem. 1998;273:3363–3368. doi: 10.1074/jbc.273.6.3363. [DOI] [PubMed] [Google Scholar]
- 28.Watanabe K., Yokobori S. tRNA modification and genetic code variations in animal mitochondria. J. Nucleic Acids. 2011;2011:623095. doi: 10.4061/2011/623095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zorbas C., Nicolas E., Wacheul L., Huvelle E., Heurgue-Hamard V., Lafontaine D.L. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell. 2015;26:2080–2095. doi: 10.1091/mbc.E15-02-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motorin Y., Helm M. RNA nucleotide methylation: 2021 update. Wiley Interdiscip Rev RNA. 2022;13:e1691. doi: 10.1002/wrna.1691. [DOI] [PubMed] [Google Scholar]
- 31.White J., Li Z., Sardana R., Bujnicki J.M., Marcotte E.M., Johnson A.W. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell Biol. 2008;28:3151–3161. doi: 10.1128/MCB.01674-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haag S., Kretschmer J., Bohnsack M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA. 2015;21:180–187. doi: 10.1261/rna.047910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marchand V., Ayadi L., Ernst F.G.M., Hertler J., Bourguignon-Igel V., Galvanin A., Kotter A., Helm M., Lafontaine D.L.J., Motorin Y. AlkAniline-seq: profiling of m(7) G and m(3) C RNA modifications at single nucleotide resolution. Angew Chem. Int. Ed. Engl. 2018;57:16785–16790. doi: 10.1002/anie.201810946. [DOI] [PubMed] [Google Scholar]
- 34.Song B., Tang Y., Chen K., Wei Z., Rong R., Lu Z., Su J., de Magalhaes J.P., Rigden D.J., Meng J. m7GHub: deciphering the location, regulation and pathogenesis of internal mRNA N7-methylguanosine (m7G) sites in human. Bioinformatics. 2020;36:3528–3536. doi: 10.1093/bioinformatics/btaa178. [DOI] [PubMed] [Google Scholar]
- 35.Enroth C., Poulsen L.D., Iversen S., Kirpekar F., Albrechtsen A., Vinther J. Detection of internal N7-methylguanosine (m7G) RNA modifications by mutational profiling sequencing. Nucleic Acids Res. 2019;47:e126. doi: 10.1093/nar/gkz736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016;13:841–844. doi: 10.1038/nmeth.3965. [DOI] [PubMed] [Google Scholar]
- 37.Mirihana Arachchilage G., Dassanayake A.C., Basu S. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem Biol. 2015;22:262–272. doi: 10.1016/j.chembiol.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 38.Pandey S., Agarwala P., Jayaraj G.G., Gargallo R., Maiti S. The RNA stem-loop to G-quadruplex equilibrium controls mature MicroRNA production inside the cell. Biochemistry. 2015;54:7067–7078. doi: 10.1021/acs.biochem.5b00574. [DOI] [PubMed] [Google Scholar]
- 39.Vinther J. No Evidence for N7-Methylation of Guanosine (m(7)G) in Human let-7e. Mol. Cell. 2020;79:199–200. doi: 10.1016/j.molcel.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Chu J.M., Ye T.T., Ma C.J., Lan M.D., Liu T., Yuan B.F., Feng Y.Q. Existence of internal N7-methylguanosine modification in mRNA determined by differential enzyme treatment coupled with mass spectrometry analysis. ACS Chem. Biol. 2018;13:3243–3250. doi: 10.1021/acschembio.7b00906. [DOI] [PubMed] [Google Scholar]
- 41.Ma J., Han H., Huang Y., Yang C., Zheng S., Cai T., Bi J., Huang X., Liu R., Huang L., et al. METTL1/WDR4-mediated m(7)G tRNA modifications and m(7)G codon usage promote mRNA translation and lung cancer progression. Mol. Ther. 2021;29:3422–3435. doi: 10.1016/j.ymthe.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin S., Liu Q., Lelyveld V.S., Choe J., Szostak J.W., Gregory R.I. Mettl1/Wdr4-Mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell. 2018;71:244–255.e5. doi: 10.1016/j.molcel.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Orellana E.A., Liu Q., Yankova E., Pirouz M., De Braekeleer E., Zhang W., Lim J., Aspris D., Sendinc E., Garyfallos D.A., et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell. 2021;81:3323–3338.e14. doi: 10.1016/j.molcel.2021.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi Y., Liang Z., Guo Y., Chen D., Lu L., Lin J., Qiu S., Wang X., Qiu E., Lin F., et al. WBSCR22 confers cell survival and predicts poor prognosis in glioma. Brain Res Bull. 2020;161:1–12. doi: 10.1016/j.brainresbull.2020.04.024. [DOI] [PubMed] [Google Scholar]
- 45.Khan A.A., Huang H., Zhao Y., Li H., Pan R., Wang S., Liu X. WBSCR22 and TRMT112 synergistically suppress cell proliferation, invasion and tumorigenesis in pancreatic cancer via transcriptional regulation of ISG15. Int J Oncol. 2022;60 doi: 10.3892/ijo.2022.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding L., Wu H., Wang Y., Li Y., Liang Z., Xia X., Zheng J.C. m6A reader Igf2bp1 regulates the inflammatory responses of microglia by stabilizing Gbp11 and cp mRNAs. Front. Immunol. 2022;13:872252. doi: 10.3389/fimmu.2022.872252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jangani M., Poolman T.M., Matthews L., Yang N., Farrow S.N., Berry A., Hanley N., Williamson A.J., Whetton A.D., Donn R., et al. The methyltransferase WBSCR22/Merm1 enhances glucocorticoid receptor function and is regulated in lung inflammation and cancer. J Biol. Chem. 2014;289:8931–8946. doi: 10.1074/jbc.M113.540906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bahr A., Hankeln T., Fiedler T., Hegemann J., Schmidt E.R. Molecular analysis of METTL1, a novel human methyltransferase-like gene with a high degree of phylogenetic conservation. Genomics. 1999;57:424–428. doi: 10.1006/geno.1999.5780. [DOI] [PubMed] [Google Scholar]
- 49.Guy M.P., Phizicky E.M. Two-subunit enzymes involved in eukaryotic post-transcriptional tRNA modification. RNA Biol. 2014;11:1608–1618. doi: 10.1080/15476286.2015.1008360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leulliot N., Chaillet M., Durand D., Ulryck N., Blondeau K., van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure. 2008;16:52–61. doi: 10.1016/j.str.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 51.Cartlidge R.A., Knebel A., Peggie M., Alexandrov A., Phizicky E.M., Cohen P. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J. 2005;24:1696–1705. doi: 10.1038/sj.emboj.7600648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Figaro S., Wacheul L., Schillewaert S., Graille M., Huvelle E., Mongeard R., Zorbas C., Lafontaine D.L., Heurgue-Hamard V. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell Biol. 2012;32:2254–2267. doi: 10.1128/MCB.06623-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Letoquart J., Huvelle E., Wacheul L., Bourgeois G., Zorbas C., Graille M., Heurgue-Hamard V., Lafontaine D.L. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. USA. 2014;111:E5518–E5526. doi: 10.1073/pnas.1413089111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng W., Gao A., Lin H., Zhang W. Novel roles of METTL1/WDR4 in tumor via m(7)G methylation. Mol. Ther Oncolytics. 2022;26:27–34. doi: 10.1016/j.omto.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doll A., Grzeschik K.H. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 2001;95:20–27. doi: 10.1159/000057012. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto K., Toyooka T., Tomikawa C., Ochi A., Takano Y., Takayanagi N., Endo Y., Hori H. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex) FEBS Lett. 2007;581:1599–1604. doi: 10.1016/j.febslet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 57.Husain N., Tkaczuk K.L., Tulsidas S.R., Kaminska K.H., Cubrilo S., Maravic-Vlahovicek G., Bujnicki J.M., Sivaraman J. Structural basis for the methylation of G1405 in 16S rRNA by aminoglycoside resistance methyltransferase Sgm from an antibiotic producer: a diversity of active sites in m7G methyltransferases. Nucleic Acids Res. 2010;38:4120–4132. doi: 10.1093/nar/gkq122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim M., Park S., Lee S.J. Global transcriptional regulator TrmB family members in prokaryotes. J Microbiol. 2016;54:639–645. doi: 10.1007/s12275-016-6362-7. [DOI] [PubMed] [Google Scholar]
- 59.Zeng Y., Wang S., Gao S., Soares F., Ahmed M., Guo H., Wang M., Hua J.T., Guan J., Moran M.F., et al. Refined RIP-seq protocol for epitranscriptome analysis with low input materials. PLoS Biol. 2018;16:e2006092. doi: 10.1371/journal.pbio.2006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McIntyre A.B.R., Gokhale N.S., Cerchietti L., Jaffrey S.R., Horner S.M., Mason C.E. Limits in the detection of m(6)A changes using MeRIP/m(6)A-seq. Sci. Rep. 2020;10:6590. doi: 10.1038/s41598-020-63355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S., Liu Q., Jiang Y.Z., Gregory R.I. Nucleotide resolution profiling of m(7)G tRNA modification by TRAC-Seq. Nat. Protoc. 2019;14:3220–3242. doi: 10.1038/s41596-019-0226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L.S., Liu C., He C. Transcriptome-wide detection of internal N(7)-methylguanosine. Methods Mol. Biol. 2021;2298:97–104. doi: 10.1007/978-1-0716-1374-0_6. [DOI] [PubMed] [Google Scholar]
- 63.Zueva V.S., Mankin A.S., Bogdanov A.A., Baratova L.A. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur J Biochem. 1985;146:679–687. doi: 10.1111/j.1432-1033.1985.tb08704.x. [DOI] [PubMed] [Google Scholar]
- 64.Chen W., Feng P., Song X., Lv H., Lin H. iRNA-m7G: Identifying N(7)-methylguanosine sites by fusing multiple features. Mol. Ther. Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bi Y., Xiang D., Ge Z., Li F., Jia C., Song J. An interpretable prediction model for identifying N(7)-methylguanosine sites based on XGBoost and SHAP. Mol. Ther. Nucleic Acids. 2020;22:362–372. doi: 10.1016/j.omtn.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dai C., Feng P., Cui L., Su R., Chen W., Wei L. Iterative feature representation algorithm to improve the predictive performance of N7-methylguanosine sites. Brief Bioinform. 2021:22. doi: 10.1093/bib/bbaa278. [DOI] [PubMed] [Google Scholar]
- 67.Zou H., Yin Z. m7G-DPP: Identifying N7-methylguanosine sites based on dinucleotide physicochemical properties of RNA. Biophys. Chem. 2021;279:106697. doi: 10.1016/j.bpc.2021.106697. [DOI] [PubMed] [Google Scholar]
- 68.Liu X., Liu Z., Mao X., Li Q.m.7.G.P. An improved machine learning-based model for predicting internal m7G modifications using sequence properties. Anal Biochem. 2020;15:609. doi: 10.1016/j.ab.2020.113905. [DOI] [PubMed] [Google Scholar]
- 69.Ma J., Zhang L., Chen J., Song B., Zang C., Liu H. m(7)GDisAI: N7-methylguanosine (m(7)G) sites and diseases associations inference based on heterogeneous network. BMC Bioinf. 2021;22:152. doi: 10.1186/s12859-021-04007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaheen R., Abdel-Salam G.M., Guy M.P., Alomar R., Abdel-Hamid M.S., Afifi H.H., Ismail S.I., Emam B.A., Phizicky E.M., Alkuraya F.S. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 2015;16:210. doi: 10.1186/s13059-015-0779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trimouille A., Lasseaux E., Barat P., Deiller C., Drunat S., Rooryck C., Arveiler B., Lacombe D. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin Genet. 2018;93:374–377. doi: 10.1111/cge.13074. [DOI] [PubMed] [Google Scholar]
- 72.Bull M.J. Down syndrome. N. Engl. J. Med. 2020;382:2344–2352. doi: 10.1056/NEJMra1706537. [DOI] [PubMed] [Google Scholar]
- 73.Michaud J., Kudoh J., Berry A., Bonne-Tamir B., Lalioti M.D., Rossier C., Shibuya K., Kawasaki K., Asakawa S., Minoshima S., et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics. 2000;68:71–79. doi: 10.1006/geno.2000.6258. [DOI] [PubMed] [Google Scholar]
- 74.Pereira P.L., Magnol L., Sahun I., Brault V., Duchon A., Prandini P., Gruart A., Bizot J.C., Chadefaux-Vekemans B., Deutsch S., et al. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum. Mol. Genet. 2009;18:4756–4769. doi: 10.1093/hmg/ddp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sahun I., Marechal D., Pereira P.L., Nalesso V., Gruart A., Garcia J.M., Antonarakis S.E., Dierssen M., Herault Y. Cognition and hippocampal plasticity in the mouse is altered by monosomy of a genomic region implicated in Down syndrome. Genetics. 2014;197:899–912. doi: 10.1534/genetics.114.165241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deng Y., Zhou Z., Ji W., Lin S., Wang M. METTL1-mediated m(7)G methylation maintains pluripotency in human stem cells and limits mesoderm differentiation and vascular development. Stem Cell Res. Ther. 2020;11:306. doi: 10.1186/s13287-020-01814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rehfeld F., Rohde A.M., Nguyen D.T., Wulczyn F.G. Lin28 and let-7: ancient milestones on the road from pluripotency to neurogenesis. Cell Tissue Res. 2015;359:145–160. doi: 10.1007/s00441-014-1872-2. [DOI] [PubMed] [Google Scholar]
- 78.Xia X., Ahmad I. let-7 microRNA regulates neurogliogenesis in the mammalian retina through Hmga2. Dev. Biol. 2016;410:70–85. doi: 10.1016/j.ydbio.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 79.Nishino J., Kim I., Chada K., Morrison S.J. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arumugam T.V., Baik S.H., Balaganapathy P., Sobey C.G., Mattson M.P., Jo D.G. Notch signaling and neuronal death in stroke. Prog. Neurobiol. 2018;165-167:103–116. doi: 10.1016/j.pneurobio.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Collaborators G.B.D.N. Global, regional, and national burden of neurological disorders, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:459–480. doi: 10.1016/S1474-4422(18)30499-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Androvic P., Kirdajova D., Tureckova J., Zucha D., Rohlova E., Abaffy P., Kriska J., Valny M., Anderova M., Kubista M., et al. Decoding the transcriptional response to ischemic stroke in young and aged mouse brain. Cell Rep. 2020;31:107777. doi: 10.1016/j.celrep.2020.107777. [DOI] [PubMed] [Google Scholar]
- 83.Arumugam T.V., Cheng Y.L., Choi Y., Choi Y.H., Yang S., Yun Y.K., Park J.S., Yang D.K., Thundyil J., Gelderblom M., et al. Evidence that gamma-secretase-mediated Notch signaling induces neuronal cell death via the nuclear factor-kappaB-Bcl-2-interacting mediator of cell death pathway in ischemic stroke. Mol. Pharmacol. 2011;80:23–31. doi: 10.1124/mol.111.071076. [DOI] [PubMed] [Google Scholar]
- 84.Cheng Y.L., Park J.S., Manzanero S., Choi Y., Baik S.H., Okun E., Gelderblom M., Fann D.Y., Magnus T., Launikonis B.S., et al. Evidence that collaboration between HIF-1alpha and Notch-1 promotes neuronal cell death in ischemic stroke. Neurobiol. Dis. 2014;62:286–295. doi: 10.1016/j.nbd.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balaganapathy P., Baik S.H., Mallilankaraman K., Sobey C.G., Jo D.G., Arumugam T.V. Interplay between Notch and p53 promotes neuronal cell death in ischemic stroke. J. Cereb Blood Flow Metab. 2018;38:1781–1795. doi: 10.1177/0271678X17715956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cheng Y.L., Choi Y., Seow W.L., Manzanero S., Sobey C.G., Jo D.G., Arumugam T.V. Evidence that neuronal Notch-1 promotes JNK/c-Jun activation and cell death following ischemic stress. Brain Res. 2014;1586:193–202. doi: 10.1016/j.brainres.2014.08.054. [DOI] [PubMed] [Google Scholar]
- 87.Peng G., Yuan Y., Wu S., He F., Hu Y., Luo B. MicroRNA let-7e Is a Potential Circulating Biomarker of Acute Stage Ischemic Stroke. Transl Stroke Res. 2015;6:437–445. doi: 10.1007/s12975-015-0422-x. [DOI] [PubMed] [Google Scholar]
- 88.Selvamani A., Sathyan P., Miranda R.C., Sohrabji F. An antagomir to microRNA Let7f promotes neuroprotection in an ischemic stroke model. PLoS One. 2012;7:e32662. doi: 10.1371/journal.pone.0032662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang L., Yang J., Xue Q., Yang D., Lu Y., Guang X., Zhang W., Ba R., Zhu H., Ma X. An rs13293512 polymorphism in the promoter of let-7 is associated with a reduced risk of ischemic stroke. J Thromb Thrombolysis. 2016;42:610–615. doi: 10.1007/s11239-016-1400-1. [DOI] [PubMed] [Google Scholar]
- 90.Mueller M., Zhou J., Yang L., Gao Y., Wu F., Schoeberlein A., Surbek D., Barnea E.R., Paidas M., Huang Y. PreImplantation factor promotes neuroprotection by targeting microRNA let-7. Proc. Natl. Acad. Sci. USA. 2014;111:13882–13887. doi: 10.1073/pnas.1411674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kovacs G.G. Concepts and classification of neurodegenerative diseases. Handb. Clin. Neurol. 2017;145:301–307. doi: 10.1016/B978-0-12-802395-2.00021-3. [DOI] [PubMed] [Google Scholar]
- 92.Heemels M.T. Neurodegenerative diseases. Nature. 2016;539:179. doi: 10.1038/539179a. [DOI] [PubMed] [Google Scholar]
- 93.Gupta S.P. Advances in studies on neurodegenerative diseases and their treatments. Curr. Top. Med. Chem. 2020;20:2379. doi: 10.2174/156802662026201013111327. [DOI] [PubMed] [Google Scholar]
- 94.Hartley D., Blumenthal T., Carrillo M., DiPaolo G., Esralew L., Gardiner K., Granholm A.C., Iqbal K., Krams M., Lemere C., et al. Down syndrome and Alzheimer's disease: common pathways, common goals. Alzheimers Dement. 2015;11:700–709. doi: 10.1016/j.jalz.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Weller J., Budson A. Current understanding of alzheimer's disease diagnosis and treatment. F1000Res. 2018;7 doi: 10.12688/f1000research.14506.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mossello E., Ballini E. Management of patients with Alzheimer's disease: pharmacological treatment and quality of life. Ther. Adv. Chronic Dis. 2012;3:183–193. doi: 10.1177/2040622312452387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang J., Wang C., Qi R., Fu H., Ma Q. scREAD: a single-cell RNA-seq database for alzheimer's disease. iScience. 2020;23:101769. doi: 10.1016/j.isci.2020.101769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Srinivasan K., Friedman B.A., Etxeberria A., Huntley M.A., van der Brug M.P., Foreman O., Paw J.S., Modrusan Z., Beach T.G., Serrano G.E., et al. Alzheimer's patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep. 2020;31:107843. doi: 10.1016/j.celrep.2020.107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gheibihayat S.M., Cabezas R., Nikiforov N.G., Jamialahmadi T., Johnston T.P., Sahebkar A. CD47 in the brain and neurodegeneration: an update on the role in neuroinflammatory pathways. Molecules. 2021:26. doi: 10.3390/molecules26133943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Niederhoffer N., Levy R., Sick E., Andre P., Coupin G., Lombard Y., Gies J.P. Amyloid beta peptides trigger CD47-dependent mast cell secretory and phagocytic responses. Int. J. Immunopathol. Pharmacol. 2009;22:473–483. doi: 10.1177/039463200902200224. [DOI] [PubMed] [Google Scholar]
- 101.Lehmann S.M., Kruger C., Park B., Derkow K., Rosenberger K., Baumgart J., Trimbuch T., Eom G., Hinz M., Kaul D., et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 102.Derkow K., Rossling R., Schipke C., Kruger C., Bauer J., Fahling M., Stroux A., Schott E., Ruprecht K., Peters O., et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with Alzheimer's disease. PLoS One. 2018;13:e0200602. doi: 10.1371/journal.pone.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gao G., Zhao S., Xia X., Li C., Li C., Ji C., Sheng S., Tang Y., Zhu J., Wang Y., et al. Glutaminase C regulates microglial activation and pro-inflammatory exosome release: relevance to the pathogenesis of Alzheimer's disease. Front. Cell. Neurosci. 2019;13:264. doi: 10.3389/fncel.2019.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ding L., Yang X., Xia X., Li Y., Wang Y., Li C., Sun Y., Gao G., Zhao S., Sheng S., et al. Exosomes mediate APP dysregulation via APP-miR-185-5p Axis. Front. Cell Dev. Biol. 2022;10:793388. doi: 10.3389/fcell.2022.793388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kaur S., Saldana A.C., Elkahloun A.G., Petersen J.D., Arakelyan A., Singh S.P., Jenkins L.M., Kuo B., Reginauld B., Jordan D.G., et al. CD47 interactions with exportin-1 limit the targeting of m(7)G-modified RNAs to extracellular vesicles. J. Cell Commun Signal. 2022;16:397–419. doi: 10.1007/s12079-021-00646-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cotsapas C., Mitrovic M., Hafler D. Multiple sclerosis. Handb. Clin. Neurol. 2018;148:723–730. doi: 10.1016/B978-0-444-64076-5.00046-6. [DOI] [PubMed] [Google Scholar]
- 107.Belbasis L., Bellou V., Evangelou E., Ioannidis J.P., Tzoulaki I. Environmental risk factors and multiple sclerosis: an umbrella review of systematic reviews and meta-analyses. Lancet Neurol. 2015;14:263–273. doi: 10.1016/S1474-4422(14)70267-4. [DOI] [PubMed] [Google Scholar]
- 108.Australia. New Zealand Multiple Sclerosis Genetics, C Genome-wide association study identifies new multiple sclerosis susceptibility loci on chromosomes 12 and 20. Nat. Genet. 2009;41:824–828. doi: 10.1038/ng.396. [DOI] [PubMed] [Google Scholar]
- 109.Hadjigeorgiou G.M., Kountra P.M., Koutsis G., Tsimourtou V., Siokas V., Dardioti M., Rikos D., Marogianni C., Aloizou A.M., Karadima G., et al. Replication study of GWAS risk loci in Greek multiple sclerosis patients. Neurol. Sci. 2019;40:253–260. doi: 10.1007/s10072-018-3617-6. [DOI] [PubMed] [Google Scholar]
- 110.Spanier J.A., Nashold F.E., Nelson C.D., Praska C.E., Hayes C.E. Vitamin D3-mediated resistance to a multiple sclerosis model disease depends on myeloid cell 1,25-dihydroxyvitamin D3 synthesis and correlates with increased CD4(+) T cell CTLA-4 expression. J. Neuroimmunol. 2020;338:577105. doi: 10.1016/j.jneuroim.2019.577105. [DOI] [PubMed] [Google Scholar]
- 111.Gil-Varea E., Fedetz M., Eixarch H., Spataro N., Villar L.M., Urcelay E., Saiz A., Fernandez O., Leyva L., Ramio-Torrenta L., et al. A new risk variant for multiple sclerosis at 11q23.3 locus is associated with expansion of CXCR5+ circulating regulatory T cells. J. Clin. Med. 2020;9 doi: 10.3390/jcm9030625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meyer M.B., Pike J.W. Mechanistic homeostasis of vitamin D metabolism in the kidney through reciprocal modulation of Cyp27b1 and Cyp24a1 expression. J. Steroid Biochem Mol. Biol. 2020;196:105500. doi: 10.1016/j.jsbmb.2019.105500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McNeill K.A. Epidemiology of brain tumors. Neurol. Clin. 2016;34:981–998. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 114.Schwartzbaum J.A., Fisher J.L., Aldape K.D., Wrensch M. Epidemiology and molecular pathology of glioma. Nat. Clin. Pract. Neurol. 2006;2:494–503. doi: 10.1038/ncpneuro0289. quiz 1 p following 516. [DOI] [PubMed] [Google Scholar]
- 115.Ostrom Q.T., Patil N., Cioffi G., Waite K., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020;22:iv1–iv96. doi: 10.1093/neuonc/noaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tykocki T., Eltayeb M. Ten-year survival in glioblastoma. A systematic review. J. Clin. Neurosci. 2018;54:7–13. doi: 10.1016/j.jocn.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 117.Le Rhun E., Preusser M., Roth P., Reardon D.A., van den Bent M., Wen P., Reifenberger G., Weller M. Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019;80:101896. doi: 10.1016/j.ctrv.2019.101896. [DOI] [PubMed] [Google Scholar]
- 118.Dai Z., Liu H., Liao J., Huang C., Ren X., Zhu W., Zhu S., Peng B., Li S., Lai J., et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell. 2021;81:3339–3355.e8. doi: 10.1016/j.molcel.2021.07.003. [DOI] [PubMed] [Google Scholar]
- 119.Shao D., Li Y., Wu J., Zhang B., Xie S., Zheng X., Jiang Z. An m6A/m5C/m1A/m7G-Related long non-coding RNA signature to predict prognosis and immune features of glioma. Front. Genet. 2022;13:903117. doi: 10.3389/fgene.2022.903117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chen Z., Zhang Z., Ding W., Zhang J.H., Tan Z.L., Mei Y.R., He W., Wang X.J. Expression and potential biomarkers of regulators for M7G RNA modification in gliomas. Front. Neurol. 2022;13:886246. doi: 10.3389/fneur.2022.886246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang Z., Zhong Z., Jiang Z., Chen Z., Chen Y., Xu Y. A novel prognostic 7-methylguanosine signature reflects immune microenvironment and alternative splicing in glioma based on multi-omics analysis. Front. Cell Dev. Biol. 2022;10:902394. doi: 10.3389/fcell.2022.902394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu X., Li C., Wang Z., Zhang Y., Liu S., Chen S., Chen S., Liu W., Liu X. A bioinformatic analysis study of m(7)G regulator-mediated methylation modification patterns and tumor microenvironment infiltration in glioblastoma. BMC Cancer. 2022;22:729. doi: 10.1186/s12885-022-09791-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Malbec L., Zhang T., Chen Y.S., Zhang Y., Sun B.F., Shi B.Y., Zhao Y.L., Yang Y., Yang Y.G. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Research. 2019;29:927–941. doi: 10.1038/s41422-019-0230-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xing L., Cai Y., Yang T., Yu W., Gao M., Chai R., Ding S., Wei J., Pan J., Chen G. Epitranscriptomic m6A regulation following spinal cord injury. J. Neurosci. Res. 2021;99:843–857. doi: 10.1002/jnr.24763. [DOI] [PubMed] [Google Scholar]
- 127.Li Q., Wen S., Ye W., Zhao S., Liu X. The potential roles of m(6)A modification in regulating the inflammatory response in microglia. J. Neuroinflammation. 2021;18:149. doi: 10.1186/s12974-021-02205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xia X., Li C., Wang Y., Deng X., Ma Y., Ding L., Zheng J. Reprogrammed astrocytes display higher neurogenic competence, migration ability and cell death resistance than reprogrammed fibroblasts. Transl. Neurodegener. 2020;9:6. doi: 10.1186/s40035-020-0184-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cai W.Y., Chen X., Chen L.P., Li Q., Du X.J., Zhou Y.Y. Role of differentially expressed genes and long non-coding RNAs in papillary thyroid carcinoma diagnosis, progression, and prognosis. J. Cell Biochem. 2018;119:8249–8259. doi: 10.1002/jcb.26836. [DOI] [PubMed] [Google Scholar]
- 130.Yankova E., Blackaby W., Albertella M., Rak J., De Braekeleer E., Tsagkogeorga G., Pilka E.S., Aspris D., Leggate D., Hendrick A.G., et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature. 2021;593:597–601. doi: 10.1038/s41586-021-03536-w. [DOI] [PMC free article] [PubMed] [Google Scholar]