Abstract
Insulin secretion from pancreatic beta cells is crucial for maintaining glucose homeostasis. The murine insulinoma derived MIN6 cell line is commonly used as a model for insulin secretion studies. However, its glucose responsiveness wanes with passaging, and insulin secretion is traditionally measured by expensive and time-consuming RIA or ELISA. We have developed a MIN6 subclone (MIN6-6) that allows for high throughput assay of insulin secretion in both population and single cells. In addition, MIN6-6 also expresses Cas9, permitting genome wide CRISPR screen of insulin secretion using a pooled sgRNA library. Here we provide methods for assaying insulin secretion both in bulk and in single cells in MIN6-6 cells, as well as for CRISPR screen of insulin secretion.
-
•
A highly glucose responsive beta cell reporter line (MIN6-6) with multiple engineered functionalities.
-
•
Allows for CRISPR/Cas9 mutagenesis, quantification of bulk insulin secretion by a straightforward nanoLuc assay and visualization of intracellular insulin granules.
-
•
Allows for en masse quantification of insulin granule exocytosis in individual cells under multiple conditions.
Keywords: SNAP-tag, Phogrin, FACS, Pancreatic beta cells, GSIS, CRISPR screen
Method name: Insulin secretion assays in a MIN6 reporter cell line.
Graphical abstract
Specifications table
| Subject area: | |
| More specific subject area: | Insulin secretion and cell biology |
| Name of your method: | Insulin secretion assays in a MIN6 reporter cell line |
| Name and reference of original method: | N/A |
| Resource availability: | N/A |
Method details
Background
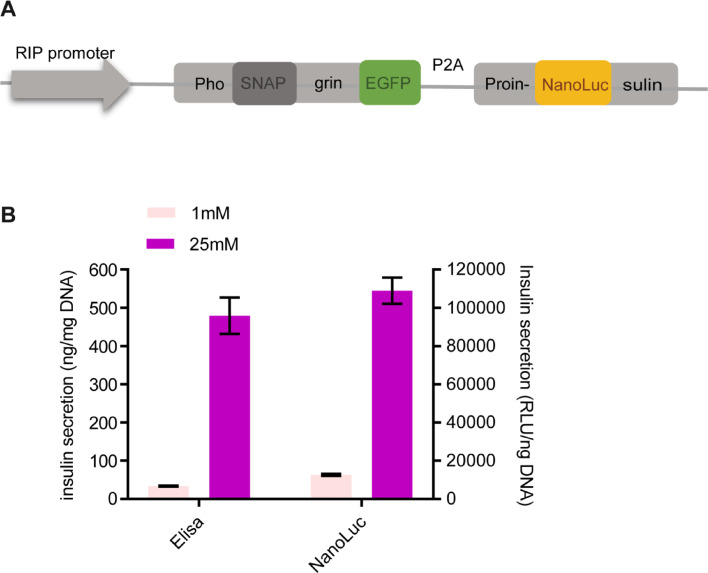
Insulin secretion from pancreatic beta cells is the main controller of glucose homeostasis and energy metabolism [1]. Glucose is the most potent stimulus of insulin secretion. Glucose stimulated insulin secretion (GSIS) has long been a focus of diabetes research in past 6 decades [2]. However, much is still unknown about the regulation of insulin secretion, especially during fasting [3]. MIN6 cell derived from mouse insulinoma is commonly used as a model of beta cells [4]. Currently, RIA or ELISA is the method to assay insulin secretion in bulk. But these methods are time-consuming and expensive. Imaging by total internal reflection fluorescence (TIRF) microscopy is commonly used for assaying insulin secretion in single cells. However, TIRF imaging is low throughput. To facilitate insulin secretion assay in bulk and in single cells, we have recently established a MIN6 subclone (MIN6-6) that expresses a multifunctional reporter and SpCas9 (Fig. 1A) [5]. The multifunctional reporter consists of 6 components. The full length phogrin, a dense core granule specific transmembrane protein, targets its fusion partners to the insulin granule [6,7]. The SNAP tag inserted in the lumenal domain of phogrin covalently self-labels with its substrates [8]. EGFP tagged at the C-terminus of phogrin labels insulin granules. P2A allows translation of two proteins from one transcript [9]. Finally, the preproinsulin directs nanoluciferase (NLuc), which is inserted in the middle of the C-peptide, to insulin granules [10]. Thus, in MIN6-6 cells, NLuc is co-secreted with insulin and serves as an insulin surrogate and SNAP is only accessible to cell impermeant substrates during exocytosis. As a result, insulin secretion can be easily measured by the a NLuc assay and insulin granule exocytosis in individual cells can be quantified using cell impermeant SNAP substrates. Furthermore, the subclone was transduced by a lentivirus that expresses SpCas9-2A-blasticidin [11], enabling blasticidin S resistance and CRISPR/Cas9 mutagenesis in the presence of sgRNA. We have validated all the designed functionalities in MIN6-6. Here we give an overview on using MIN6-6 for the three engineered applications.
Fig. 1.
(A) Schematic of the reporter construct (Created with BioRender.com). (B) Comparison of insulin secretion measured by an ELISA kit and the NanoGlo kit. The same samples were measured with both the insulin ELISA kit and the NanoGlo Luciferase kit.
Assay bulk GSIS using NLuc
MIN6-6 cells express a modified preproinsulin in which the C-peptide is replaced by NLuc. Similar preproinsulin insulin expression construct with Gaussia luciferase has been successfully used as a surrogate for insulin secretion [10,12]. In unmodified MIN6 cells and other beta cell lines, the conditioned medium collected from cell culture is commonly used for insulin radioimmunoassay or enzyme-linked immunosorbent assay. In MIN6-6 cells, the conditioned medium can also be used for NLuc assay. Conditioned medium from cells grown in a 96 or 384 well plate is sufficient due to the high sensitivity of NLuc assay [12].
We performed the following experiments to validate that the NLuc assay is a good surrogate for the traditional ELISA assay. MIN6-6 cells were seeded in a 12-well plate and allowed to grow to 80–90% confluency. The cells were then starved for one hour in 1 mL KRB (114 mM NaCl, 5 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 20 mM HEPES, 2.5 mM CaCl2, 25.5 mM NaHCO3 and 0.2% BSA) containing 1 mM glucose (low glucose KRB). They were then cultured with freshly prepared low glucose KRB or high glucose KRB (25 mM glucose) for another hour. The conditioned buffer was collected from each well and centrifuged at 2000 rpm for 5 min to obtain supernatants. The amount of NLuc in the supernatants was then measured using Promega Nano‐Glo® Luciferase Assay System (Promega). The amount of insulin in the supernants was measured using a mouse insulin ELISA kit (Mercodia) following manufacturer's instruction. Each sample was measured in triplicates. The cells in each well were lysed with RIPA buffer (R0278, Sigma) followed by centrifugation as above to obtain supernatants. The amount of insulin and nanoluciferase was determined as above after dilution. At the end of the assay, the reactions were read in a BioTek Synergy™ H4 Hybrid Microplate Reader. The content of NLuc and insulin in conditioned buffer was normalized to the amount of total NLuc or insulin in each sample. We found that normalization with total DNA content led to similar results. We prefer normalization with DNA content as it can be determined faster and more economically. We measured DNA in the cell lysate using the AccuBlue® dsDNA Quantitation kit (Biotium) following manufacturer's instruction. At least three sets of independently generated samples were analyzed under each condition. Typically, with 1 hour incubation, the ratio of the normalized content in high glucose to low glucose buffer, or GSIS index, was 10 using NLuc assay, or 11 using insulin ELISA assay (Fig. 1B). The NLuc assay is much faster as it only takes 30 min to complete, compared to 3 h using the ELISA assay. Therefore, NLuc secretion assay is a good surrogate for insulin secretion assay.
Assay single cell GSIS using SNAP lableing
MIN6-6 cells also enables high throughput quantification of insulin granules exocytosis in single cells. The lumenal SNAP can only be labeled by impermeant substrates when insulin granules are fused with the plasma membrane, that is, during exocytosis. As multiple spectrally distinct SNAP substrates are commercially available, it enables quantification of exocytosis in the same cell under multiple conditions (Fig. 1A). We have shown that the labeling can be quantified by either fluorescent microscopy or flow cytometry.
To quantify exocytosis using imaging, MIN6-6 cells were seeded on a coverslip. The assay started with starving the cells in DMEM with 10% FBS and 1mM glucose (low glucose medium) for four hours. Then, the cells were incubated with prewarmed low glucose medium containing 5 µM nonfluorescent SNAP-Surface® Block for 15 min to block any surface SNAP. After washing out the Block with low glucose medium, the cells were cultured in low glucose medium containing 5 µM SNAP-Surface® Alexa Fluor® 546 for 20 min. After washing out the substrate, the cells were then cultured in high glucose medium (DMEM with 10% FBS, 25 mM glucose) containing 5 µM SNAP-Surface® Alexa Fluor® 647 for 20 min. After washing out the substrate, cells were fixed in 4% paraformaldhyde (PFA), mounted on slides with AquaMount (Thermo Scientific), and imaged with Zeiss LSM880 laser scanning spectral confocal microscope (Carl Zeiss). All fluorescence are in intracellular vesicles, likely due to recyling of phogrin [13,14]. There were overtly more vesicles labeled by the substrate in the high glucose medium than by the substrate in low glucose medium (Fig. 2A). To deteremine the specificity of the labeling, we labeled parental MIN6 cells and MIN6-6 cells with low glucose and high glucose labeling medium separately for 20 min. There was little SNAP labeling in the parental cells in ether condition (Fig. 2B), indicating labeling specificity. The signal from each SNAP label can be quantified. These results demonstrate glucose-stimulated labeling of SNAP in MIN6-6 cells.
Fig. 2.
(A) Confocal image of MIN6-6 cells sequentially stained with low glucose medium (1 mM) containing the SNAP surface dye Alexa Fluor 546 and high glucose medium (25 mM) with the SNAP surface dye Alexa Fluor 647. Phogrin-EGFP was also imaged. Scale bar, 25 µm. (B) Confocal image of parental MIN6 cells and MIN6-6 cells after 20 min incubation in the low glucose labeling medium or the high glucose labeling medium. Phogrin-EGFP was also imaged. Scale bar, 25 µm.
To quantify exocytosis by flow cytometry, the MIN6-6 cells were pre-seeded in a 6-well plate. Once reaching about 75% confluency, the cells were starved, quenched, and sequentially labelled in low and high glucose medium as above. As controls, we also incubated the cells in high glucose labeling medium containing 300 µM diazoxide or in glucose free labeling medium containing 40 mM KCl in the second labeling step. Diazoxide is a KATP channel opener that prevents glucose-triggered membrane deploarization. In contrast, KCl induces membrane depolarization independent of glucose. The cells were washed twice with fresh DMEM after each labeling. After washing twice in cold PBS, the cells were dissociated by 0.25% Trypsin with EDTA (Gibco), and fixed in 4% PFA-PBS for 15 min at room temperature with intermittently vortexing. The cells were then washed twice with cold PBS and resuspended in PBS + 1% BSA. Fluorescence intensity of GFP, Alexa Fluor 546 and Alexa Fluor 647 of each cell was determined using a flow cytometer such as a 5-laser BD LSRII. The data were analyzed using FlowJo software (BD Biosciences). The signal intensity ratio of Alexa Fluor 647/546 in each cell is thus a measure of a cell's GSIS index. As expected, diazoxide left shifted the distribution of GSIS index while KCl right shifted the distribution (Fig. 2C). These results confirm that MIN6-6 cells allows for high throughput assay of insulin secretion in single cells.
CRISPR screen for GSIS
MIN6-6 cells also express high levels of SpCas9. With the high throughput insulin secretion assay in single cells, it is possible to identify mediators of various insulin secretagogues. As a proof of principle, we conducted a CRISPR/Cas9 screen using the MIN6-6 cell line to uncover genes that regulate the glucose induction index. We used a commercially available pooled library (Brie) that expresses approximately 80,000 sgRNAs targeting close to 20,000 protein coding genes to transduce approximately 150 million MIN6-6 cells at a low multiplicity of infection (0.3–0.4) [15]. The sgRNAs-expressing lentiviral vector also expresses the puromycin N-acetyltransferase that confers puromycin resistance. Transduced cells were selected and expanded in puromycin-supplemented medium over 14 days to allow time for gene-editing and depletion of the target proteins before experiments. After selection, the transduced cells were starved, quenched, and sequentially labeled at low and high glucose conditions with different impermeant fluorescent SNAP substrates, and prepared for flow cytometry as above. We collected the top 5% and bottome 5% according to their Alexa Fluor 647 / Alexa Fluor 546 ratio using a 5-laser BD FACSAria III. Approximately 10 million cells were recovered for each population. Sorted cells were then pelleted by centrifugation, and the cell pellet was frozen at -80 °C before genomic DNA isolation. Approximately 80 million unsorted cells (1000× coverage per library element) were saved as a reference. Genomic DNA was extracted using NK cell lysis buffer (50 mM Tris-HCL, 50 mM EDTA, 1% SDS, pH = 8.0) from sorted or unsorted cells. The genomic DNA were processed for sequencing of sgRNAs. Readcount tables and gene enrichment analysis were performed using the MAGeCK algorithm (https://sourceforge.net/projects/mageck/) [16]. Fig. 3 shows the workflow and the identified candidates of the screen. These data confirm that the multifunctional MIN6-6 cell line is suitable for high-throughput CRISPR screen and can be potentially used in more screens using other modulators of glucose secretion.
Fig. 3.
The workflow of the screen. A genome-wide CRISPR sgRNA lentiviral library (78,637 gRNAs) was used to infect SpCas9-expressing MIN6-6 cells at low MOI (0.3–0.4). After selection, most cells should express only one sgRNA. CRISPR/Cas9-edited cells were sequentially labeled with Alexa Fluor 546 in 1 mM glucose and Alexa Fluor 647 in 25 mM glucose. They were then subjected to FACS. Top and bottom 5% cells were sorted as high and low glucose induction index according to their Alexa Fluor 546 / Alexa Fluor 647 ratio. Sorted cells were lysed for genomic DNA extraction, sgRNA amplification and next generation sequencing. A volcano plot demonstrating the FDR and fold change-based ranking of genes identified in the cells with high glucose induction index.
Summary
We have established a highly glucose responsive beta cell reporter line (MIN6-6) with multiple engineered functionalities. These functionalities were intended to allow for CRISPR/Cas9 mutagenesis, quantification of bulk insulin secretion by a straightforward NLuc assay, and en masse quantification of glucose-stimulated insulin granule exocytosis in single cells. We have validated these functionalities and conducted a genome-wide screen for genes affecting glucose stimulation index. We identified several candidate genes involved in this process. Thus, this MIN6-6 line is a useful tool for studying insulin secretion and identifying chemical and genetic modulators of insulin secretion using high throughput screens.
CRediT authorship contribution statement
Liu Yang: Writing – original draft, Methodology, Software. Wenbiao Chen: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by a Vanderbilt Discovery Grant and National Institutes of Health grants, R01DK117147.
Data availability
No data was used for the research described in the article.
References
- 1.Ferrannini E., Pilo A. Pattern of insulin delivery after intravenous glucose injection in man and its relation to plasma glucose disappearance. J. Clin. Investig. 1979;64(1):243–254. doi: 10.1172/JCI109445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rorsman P., Ashcroft F.M. Pancreatic beta-Cell electrical activity and insulin secretion: of mice and men. Physiol. Rev. 2018;98(1):117–214. doi: 10.1152/physrev.00008.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corkey B.E., Deeney J.T., Merrins M.J. What regulates basal insulin secretion and causes hyperinsulinemia? Diabetes. 2021;70(10):2174–2182. doi: 10.2337/dbi21-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127(1):126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 5.Yang L., Fye M.A., Yang B. Genome-wide CRISPR screen identified a role for commander complex mediated ITGB1 recycling in basal insulin secretion. Mol. Metab. 2022 doi: 10.1016/j.molmet.2022.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pouli A.E., Emmanouilidou E., Zhao C., Wasmeier C., Hutton J.C., Rutter G.A. Secretory-granule dynamics visualized in vivo with a phogrin-green fluorescent protein chimaera. Biochem. J. 1998;333(1):193–199. doi: 10.1042/bj3330193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taraska J.W., Perrais D., Ohara-Imaizumi M., Nagamatsu S., Almers W. Secretory granules are recaptured largely intact after stimulated exocytosis in cultured endocrine cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100(4):2070–2075. doi: 10.1073/pnas.0337526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keppler A., Gendreizig S., Gronemeyer T., Pick H., Vogel H., Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21(1):86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 9.Szymczak A.L., Workman C.J., Wang Y., Vignali K.M., Dilioglou S., Vanin E.F., et al. Correction of multi-gene deficiency in vivo using a single 'self-cleaving' 2A peptide-based retroviral vector. Nat. Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 10.Burns S.M., Vetere A., Walpita D., Dancik V., Khodier C., Perez J., et al. High-throughput luminescent reporter of insulin secretion for discovering regulators of pancreatic beta-cell function. Cell Metab. 2015;21(1):126–137. doi: 10.1016/j.cmet.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Hart T., Chandrashekhar M., Aregger M., Steinhart Z, Brown K.R., MacLeod G., et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 12.Kalwat M.A., Wichaidit C., Nava Garcia A.Y., McCoy M.K., McGlynn K., Hwang I.H., et al. Insulin promoter-driven Gaussia luciferase-based insulin secretion biosensor assay for discovery of beta-cell glucose-sensing pathways. ACS Sens. 2016;1(10):1208–1212. doi: 10.1021/acssensors.6b00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vo Y.P., Hutton J.C., Angleson J.K. Recycling of the dense-core vesicle membrane protein phogrin in MIN6 beta-cells. Biochem. Biophys. Res. Commun. 2004;324(3):1004–1010. doi: 10.1016/j.bbrc.2004.09.147. [DOI] [PubMed] [Google Scholar]
- 14.Tomas A., Yermen B., Regazzi R., Pessin J.E., Halban P.A. Regulation of insulin secretion by phosphatidylinositol-4,5-bisphosphate. Traffic. 2010;11(1):123–137. doi: 10.1111/j.1600-0854.2009.00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34(2):184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W., Xu H., Xiao T., Cong L., Love M.I., Zhang F., et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15(12):554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.