Abstract
Background
In coeliac disease (CoD), the role of B-cells has mainly been considered to be production of antibodies. The functional role of B-cells has not been analysed extensively in CoD.
Methods
We conducted a study to characterize gene expression in B-cells from children developing CoD early in life using samples collected before and at the diagnosis of the disease. Blood samples were collected from children at risk at 12, 18, 24 and 36 months of age. RNA from peripheral blood CD19+ cells was sequenced and differential gene expression was analysed using R package Limma.
Findings
Overall, we found one gene, HNRNPL, modestly downregulated in all patients (logFC −0·7; q = 0·09), and several others downregulated in those diagnosed with CoD already by the age of 2 years.
Interpretation
The data highlight the role of B-cells in CoD development. The role of HNRPL in suppressing enteroviral replication suggests that the predisposing factor for both CoD and enteroviral infections is the low level of HNRNPL expression.
Funding
EU FP7 grant no. 202063, EU Regional Developmental Fund and research grant PRG712, The Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research (SyMMyS) 2012–2017, grant no. 250114) and, AoF Personalized Medicine Program (grant no. 292482), AoF grants 292335, 294337, 319280, 31444, 319280, 329277, 331790) and grants from the Sigrid Jusélius Foundation (SJF).
Keywords: Coeliac disease, B-cells, High-throughput mRNA sequencing
1. Introduction
Coeliac disease (CoD) is an immune-mediated disorder characterized by leukocyte infiltration and damaged tissue architecture in the small intestine. Dietary gluten is the triggering agent of CoD in genetically predisposed individuals. Genetic risk of CoD is determined by the presence of HLA-DQ2.5 in most and HLA-DQ2.2 or HLA-DQ8 in a minority of cases. Another important factor in CoD is tissue transglutaminase 2 (TG2), which is involved in generating deamidated gliadin. Celiac disease is classically characterized as a T-cell mediated disease and B-cells are mostly related to the production of antibodies against gliadin and TG2. The latter are used as a marker to assess disease activity, as they disappear in response to a gluten-free diet [1].
Beyond the importance in the antibody production, the contribution of B-cells in the immune response extends to non-antibody mediated mechanisms such as cytokine production and antigen presentation [2]. The importance of B-cells in CoD is well described by Iversen and Sollid emphsizing their role in TG2 presentation and autoantibody production [3]. Less is known about peripheral B-cell transcriptome. So far, changes in B-cell gene expression have been reported in patients affected by CoD only in the study by Garber et al. [4]. They showed that the gluten-induced small intestine injury score is inversely correlated with expression levels of a subset of peripheral blood B-cell (but not T-cell) genes in a microarray analysis, suggesting a potential protective role for B-cells. Therefore, the aim of our study was to detect possible changes in gene expression related to non-antigen specific funtions of B-cells, preceding the disease onset, that might shed light on the mechanisms of early pathogenesis.
2. Methods
2.1. Study design
In the present case-control study we set out to investigate changes in B-cell gene expression in genetically predisposed children, prospectively followed from birth until the age of 3 years, who developed CoD during follow-up. CD19+ B-cells were purified from PBMC samples collected at a maximum of four time points (12, 18, 24 and 36 mo) and analysed with RNAseq. The subjects were matched with controls for sex, HLA DR/DQ genotype, country of birth, and date of birth. The Ethics Committee of Hospital District of Helsinki and Uusimaa approved the research project “Pathogenesis of Type 1 Diabetes: DIABIMMUNE” (228/13/03/03/2008).
2.2. Subjects
The study material includes PBMC (peripheral blood mononuclear cells) samples from 12 children with HLA-conferred susceptibility to CoD from the international DIABIMMUNE Study. Six of them developed CoD during the study period (CoD progressors) and six of them were controls and they did not develop any autoantibodies or CoD during first three years of life.
The DIABIMMUNE study cohort and material collection have been described in detail previously [5]. Briefly, 2714 infants from maternity hospitals in Estonia and 3105 in Finland were enrolled. Samples were screened for T1D/CoD associated HLA DR/DQ alleles. HLA-eligible infants were invited for a 3-year follow-up study. Altogether, 258 children in Estonia (61% of eligible) and 305 in Finland (43% of eligible) completed the follow-up period with planned visits at the age of 3, 6, 12, 18, 24, and 36 months (±1 month). At each visit, peripheral blood samples were collected for 1) serum for screening of CoD-associated antibodies (anti-tTG2) and 2) for the isolation of peripheral blood mononuclear cells (PBMCs). When the antibody test indicated CoD, a gastroduodenoscopy and small intestinal biopsy were performed to confirm the diagnosis of CoD.
During the study period of 3 years, nine children were diagnosed with CoD. PBMC samples from six of those children who progressed to CoD (one Estonian and five Finnish children) were available for the current study: two cases were diagnosed at or near 24 months of age (CoD24mo) and four cases at or near 36 months of age (CoD36mo). For every child who progressed to CoD, one control was selected from the DIABIMMUNE cohort matched for the HLA DR/DQ genotype, country of birth, date of birth, and sex. All children in the control group were negative for anti-tTG2 antibodies. In the current study, we retrospectively included PBMC samples from CoD progressors starting from 12 months of age until the diagnosis of CoD (24 months or 36 months of age) and their controls. Samples were collected at a maximum of four time points: 12, 18, 24 and 36 months.
2.3. HLA analysis
The HLA DR/DQ genotypes were determined by a PCR-based lanthanide labeled oligonucleotide hybridization method using time-resolved fluorometry detection. HLA genotyping was described in detail previously [6].
2.4. CoD-associated autoantibodies
Anti-tTG2 IgA antibodies were measured by a fully automated solid-phase fluoroenzyme immunoassay technology (ImmunoCAP EliA, Phadia AB, Uppsala, Sweden) as described earlier [5]. None of the participants had IgA deficiency.
2.5. Diagnosis of CoD
CoD was diagnosed in accordance with the European Society for Paediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN) guidelines: positive IgA-tTG antibodies and a biopsy of the small intestine with an abnormal microscopic finding [7] as described earlier [5]. The histological examination of the small intestine specimens were classified in accordance with the Marsh classification modified by Oberhuber [8].
2.6. Isolation of PBMCs and CD19+ B-cells
PBMCs were isolated by Ficoll density centrifugation and were stored in liquid nitrogen. Cryopreserved PBMCs were thawed in a 37 °C water bath and when the sample started to melt, then warm RPMI medium with supplements (10% AB serum, 2 mM l-glutamine, 100 IU/ml penicillin, 100 IU/ml streptomycin) was added. B-cells were isolated trough positive selection using superparamagnetic polystyrene beads (Dynabeads™ CD19 Pan B, number 11143D, Thermo Fisher Scientific) coated with a monoclonal antibody specific for the CD19 antigen and beads were then removed using DETACHaBEAD® CD19 kit (number 12506D, Thermo Fisher Scientific) according to the manufacturer's instructions. The purity of B-cells was confirmed by the flow cytometry where B-cells were determined by the expression of CD20 and the lack of CD3 and the purity was at least 97%. Purified B-cells were resuspended in RLT Plus buffer (Qiagen) to lyse the cells and protect the RNA from degradation.
2.7. RNA extraction
The total RNA was isolated via AllPrep DNA/RNA/miRNA Universal Kit (Qiagen) according to the manufacturer's instructions. Purified RNA was stored at −80 °C. The quality of the RNA samples was ensured using Agilent Bioanalyzer 2100 and RNA integrity number for all the samples was >9. Qubit® Fluorometric Quantitation (Life Technologies) was used to measure RNA concentration.
2.8. RNA sequencing
The library preparation was started from 5 ng of total RNA. cDNA was generated using SMART-Seq v4 Ultra Low Input RNA Kit. Library preparation and single-end sequencing with 50bp read length were performed by the Finnish Functional Genomics Centre (FFGC) at Turku Bioscience Centre using Illumina HiSeq 3000 sequencer. All sample libraries were pooled and sequencing was done in two lanes. The quality of the sequenced reads was checked using FastQC tool version 0.11.4. The alignment of sequencing reads to the human reference genome (Homo sapiens hg38) was performed with Star version 2.5.2b. The uniquely mapped reads were then associated with known genes according to RefSeq gene annotation and the number of reads associated with each gene was counted using subreads package (v. 1.5.1). RNA seq data were deposited to GEO with accession number GSE185503.
2.9. Statistics
Statistical analysis was carried out with R (v. 3.4.1)/Bioconductor (v. 3.6) where Limma package was used to identify differentially expressed genes (DEGs) between groups. The Benjamini-Hochberg procedure was used to adjust for multiple testing and false discovery rate adjusted p-value (FDR) < 0·05 was set for the selection of DEG. First, all cases were compared with a control group regardless of the age of CoD diagnosis. Next, cases were divided into two groups: 1) children diagnosed with CoD at 24 months of age (CoD24mo); 2) children diagnosed with CoD at 36 months of age (CoD36mo). Further, principal component analysis was used to study RNAseq data. Heatmap of DEGs was created using Morpheus online tool (software.broadinstitute.org/morpheus). Functional enrichment analysis was performed against the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using topGO and GOstats packages in R/Bioconductor.
For analysing the longitudinal profiles of the B-cell gene expression in the samples obtained from the CoD progressors and their matched controls linear mixed-effects (LMEs) models were used. LME incorporated group, age, sex, and group/age interaction as fixed effects and matched case-control pair were used as the random effect.
2.10. Peptide analysis in silico
Peptide binding to HLA alleles was analysed with netMHC-4.0 package [9] in the Tartu University High Performance Computing Center. Peptide structures were visualized using Foldit [10].
3. Results
We analysed RNA-seq data from peripheral blood B-cells from multiple samples of CoD patients and their matched control subjects. Altogether, the differences in gene expression between the CoD and healthy groups were small and none of the genes was significantly differentially expressed after normalization procedures and correction for multiple testing. However, the top scoring gene, HNRNPL (logFC −0·7, q = 0·09) was modestly downregulated in all patients, in all samples except one.
Next, we divided study participants into two groups based on the time of CoD diagnosis and analysed them separately:
-
1)
samples from two cases diagnosed at 24 months of age and their controls (CoD24mo);
-
2)
samples from four cases diagnosed at 36 months of age and their controls (CoD36mo).
The rationale for analyzing the early-onset group separately was the idea that whatever the mechanisms might be, the differences should be more drastic when leading to rapid disease development.
The CoD24mo group included 12 samples (six samples from cases and six from controls) collected at 12mo, 18mo and 24mo. CoD36mo included 22 samples collected at 12mo, 18mo, 24mo and 36mo.
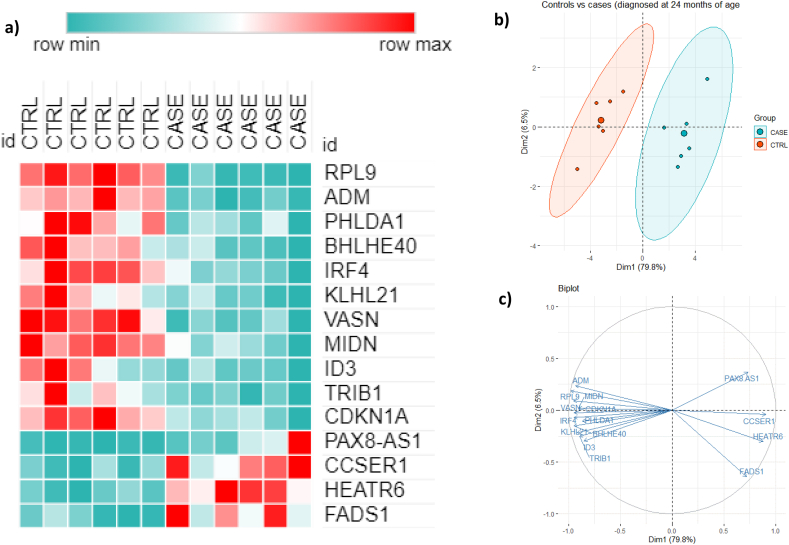
In the CoD24mo group, we identified 275 DEG (93 genes upregulated and 182 genes downregulated in cases compared to controls) and in the CoD36mo group, there were 20 DEG (ten down-regulated and ten up-upregulated genes compared to controls). After correction for multiple testing (FDR ≤0·05), 15 genes remained significantly differentially expressed (four genes were upregulated and 11 were down-regulated compared to controls) in the CoD24mo group (shown in Table 1 and Fig. 1a). These 15 genes were differently expressed at all time points: before and at the time of CoD diagnosis. However, in the CoD36mo group none of the genes remained significant when correction for multiple testing was applied.
Table 1.
Sixteen differently expressed genes between the CoD24mo cases and their controls (DEG ordered as in Fig. 1).
| Gene symbol | Gene name | Mean expression level (CPM) CASES | Mean expression level (CPM) CONTROLS | FC | log FC | p-value | FDR-adjusted p-value |
|---|---|---|---|---|---|---|---|
| RPL9 | ribosomal protein L9 | 14·2 | 110·5 | −8·9 | −3·2 | 6·0E-10 | 0·00001 |
| ADM | adrenomedullin | 2·3 | 12·3 | −6·3 | −2·7 | 3·6E-07 | 0·00210 |
| BHLHE40 | basic helix-loop-helix family member e40 | 103·7 | 313·5 | −3·3 | −1·7 | 4·7E-06 | 0·01343 |
| PHLDA1 | pleckstrin homology like domain family A member 1 | 8·5 | 23·3 | −3·4 | −1·8 | 5·4E-05 | 0·04117 |
| IRF4 | interferon regulatory factor 4 | 240·0 | 600·3 | −2·6 | −1·4 | 1·0E-05 | 0·02049 |
| KLHL21 | kelch like family member 21 | 53·2 | 112·7 | −2·1 | −1·1 | 1·1E-05 | 0·02049 |
| VASN | vasorin | 7·4 | 17·3 | −2·4 | −1·3 | 1·3E-05 | 0·02082 |
| MIDN | midnolin | 101·6 | 225·3 | −2·3 | −1·2 | 2·9E-05 | 0·03494 |
| ID3 | inhibitor of DNA binding 3, HLH protein | 61·2 | 132·2 | −2·1 | −1·1 | 4·4E-05 | 0·03917 |
| TRIB1 | tribbles pseudokinase 1 | 38·7 | 81·9 | −2·1 | −1·1 | 4·4E-05 | 0·03917 |
| CDKN1A | cyclin dependent kinase inhibitor 1A | 1275·4 | 2574·0 | −2·1 | −1·0 | 4·4E-05 | 0·03917 |
| PAX8-AS1 | PAX8 antisense RNA 1 | 13·4 | 2·2 | 5·5 | 2·4 | 4·8E-05 | 0·03494 |
| CCSER1 | coiled-coil serine rich protein 1 | 11·6 | 4·3 | 2·8 | 1·5 | 7·3E-05 | 0·03950 |
| HEATR6 | HEAT repeat containing 6 | 49·8 | 19·6 | 2·5 | 1·3 | 4·4E-06 | 0·04657 |
| FADS1 | fatty acid desaturase 1 | 12·8 | 3·9 | 3·4 | 1·8 | 2·7E-05 | 0·01343 |
Fig. 1.
Heatmap and PCA based on 15 DEG across all three timepoints (12mo, 18mo, 24mo) identified in comparison of cases diagnosed at 24 mo (CoD24mo) and their controls.a: Heatmap, b: PCA plot and c: PCA biplot showing DEG and the direction of change.
Next, we performed a principal component analysis to determine whether the controls and cases in the CoD24mo group cluster separately based on the DEG. In the PCA analysis (Fig. 1b and c), we used 15 DEG (filtered by FDR ≤0·05 and FC ≥ 2) from samples over all time points (12mo–24mo). Our results demonstrate that the cases who were diagnosed at 24mo clearly clustered separately from controls at all time points (before diagnosis and at the time of diagnosis).
Subsequently, 275 significant DEG (filtered by p-value ≤0·05) revealed from the comparison of cases diagnosed at 24mo and their controls were analysed for overrepresentation. Top ten GO terms among each category are shown in Table 2.
Table 2.
Top 10 gene ontology (GO) terms in all three GO domains (biological processes, cellular component and molecular function) calculated for CoD24mo group.
| GO.ID | Term | Annotated | Significant | Expected | P-value | |
|---|---|---|---|---|---|---|
| Biological processes | GO:0051094 | positive regulation of developmental processes | 627 | 41 | 14·86 | 1·90E-09 |
| GO:0051239 | regulation of multicellular organismal processes | 1467 | 68 | 34·78 | 1·20E-08 | |
| GO:0007154 | cell communication | 3247 | 117 | 76·98 | 2·30E-08 | |
| GO:0009605 | response to external stimulus | 1093 | 55 | 25·91 | 3·20E-08 | |
| GO:0045597 | positive regulation of cell differentiation | 465 | 32 | 11·02 | 4·50E-08 | |
| GO:0051240 | positive regulation of multicellular organismal process | 791 | 44 | 18·75 | 6·30E-08 | |
| GO:0023052 | signaling | 3233 | 115 | 76·65 | 7·90E-08 | |
| GO:0009888 | tissue development | 860 | 46 | 20·39 | 9·60E-08 | |
| GO:0050793 | regulation of developmental process | 1293 | 60 | 30·65 | 1·20E-07 | |
| Cellular component | GO:0031225 | anchored component of membrane | 38 | 5 | 0·9 | 1·85E-03 |
| GO:0000788 | nuclear nucleosome | 13 | 3 | 0·31 | 3·10E-03 | |
| GO:0005886 | plasma membrane | 2201 | 69 | 51·88 | 5·01E-03 | |
| GO:0071944 | cell periphery | 2248 | 69 | 52·98 | 8·35E-03 | |
| GO:0031224 | intrinsic component of membrane | 2271 | 69 | 53·53 | 1·06E-02 | |
| GO:0031226 | intrinsic component of plasma membrane | 592 | 23 | 13·95 | 1·24E-02 | |
| GO:0005887 | integral component of plasma membrane | 563 | 22 | 13·27 | 1·35E-02 | |
| GO:0016021 | integral component of membrane | 2236 | 66 | 52·7 | 2·34E-02 | |
| GO:0044459 | plasma membrane part | 1100 | 36 | 25·93 | 2·56E-02 | |
| GO:0009986 | cell surface | 335 | 14 | 7·9 | 2·72E-02 | |
| Molecular function | GO:0001077 | transcriptional activator activity, RNA ··· | 130 | 13 | 3·05 | 1·10E-05 |
| GO:0019955 | cytokine binding | 51 | 8 | 1·2 | 2·20E-05 | |
| GO:0001228 | transcriptional activator activity, RNA ··· | 195 | 15 | 4·57 | 5·30E-05 | |
| GO:0000981 | RNA polymerase II transcription factor a··· | 396 | 23 | 9·29 | 5·40E-05 | |
| GO:0000982 | transcription factor activity, RNA polym··· | 202 | 15 | 4·74 | 7·90E-05 | |
| GO:0003700 | DNA binding transcription factor activit··· | 732 | 34 | 17·17 | 8·80E-05 | |
| GO:0038023 | signaling receptor activity | 320 | 19 | 7·51 | 1·80E-04 | |
| GO:0046983 | protein dimerization activity | 717 | 31 | 16·82 | 6·50E-04 | |
| GO:0060089 | molecular transducer activity | 443 | 22 | 10·39 | 7·10E-04 | |
| GO:0001159 | core promoter proximal region DNA bindin··· | 206 | 13 | 4·83 | 1·11E-03 |
In the analysis above, we searched for the genes, which were differentially expressed at all time points. Next, we used a linear mixed-effects model to assess dynamic changes in B-cell transcriptomics over time.
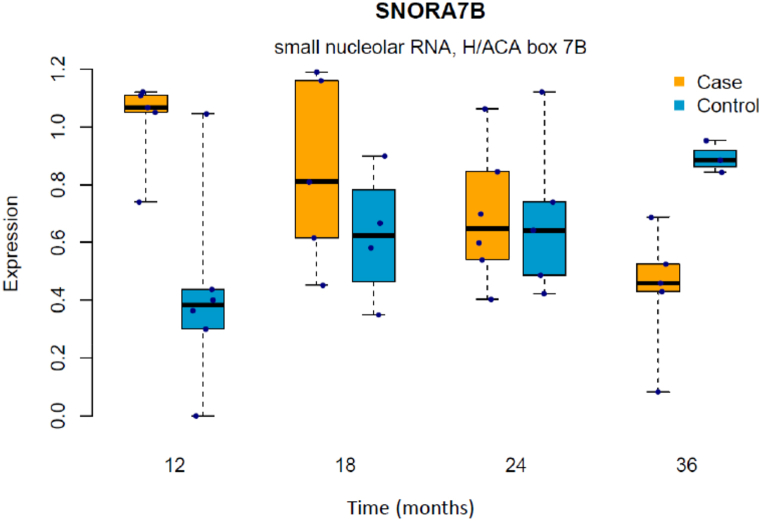
Based on linear mixed-effects model analysis there were 72 significant genes, that are associated with CoD progression (p < 0·01) (Supplementary Table 1). After FDR correction, SNORA7B remained significantly different between cases and controls over time. SNORA7B expression was decreasing with age in cases but showed an opposite trend in controls as SNORA7B level was increasing with age (Fig. 2). Although only SNORA7B remained significant, these 72 genes included four more snoRNAs (SNORD26, SNORA14B, SNORA84, SNORD100).
Fig. 2.
Dynamic changes in SNORA7B expression in B-cells in cases and controls.
When the group-time interaction was analysed, we found 37 genes in cases and controls showing an association with CoD (p < 0·01), but none of them remained significant after FDR (Supplementary Table 1).
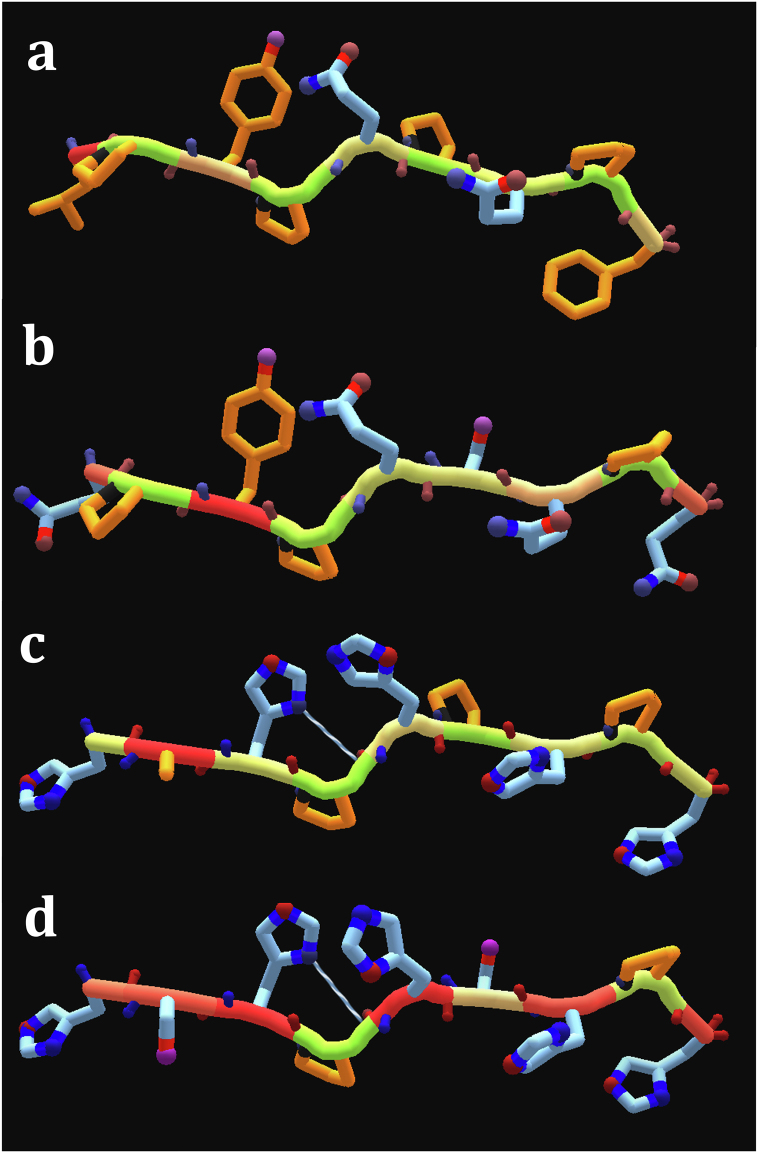
We also studied the differentially expressed proteins for potential MHC-I restricted cross-reactivity with bacterial proteins and gliadin. Extensive epithelium infiltration by CD8+T lymphocytes represents a recurrent histological finding and an important hallmark of the overt Coeliac Disease. A subset of these cells has been identified as reactive to A-gliadin peptide 123–132 (QLIPCMDVVL) in HLA-A*0201-restricted manner [11]. Also, A*0101 and B*0801-restricted CD8+ T-cell responses towards several gliadin peptides have been identified in CoD patients [12]. PHLDA1 amino acid sequence contains a polyglutamine region and a region rich in proline and glutamine, two features shared with gliadin. We carried out a BLAST search for bacterial proteins bearing homology to PHLDA1 proline-rich region. Remarkably, a region rich in proline and histidine displays strong homology to numerous bacterial proteins involved in metal ion binding – including cobalamin biosynthesis protein CbiX, nickel transporter, and urease accessory protein, in Actinobacteria and beta-proteobacteria (Table 3). These motifs are also capable of binding zinc ions. Next, we studied the binding of peptides of the proteins with homologous regions to human HLA-I alleles using netMHC-4.0 package [9] and found several high-affinity homologous peptides with theoretical potential for cross-reactivity. We found a high-affinity HAHPHPHPH peptide (Fig. 3c) presented on HLA-C0303. This peptide is highly similar to HSHPHSHPH of human PHLDA1 (Fig. 3d), and also two HLA-C0303-restricted peptides of gliadin, QPYPQSQPQ (Fig. 3b) and LPYPQPQPF (Fig. 3a), when the position of positive and negative charges is considered. It is possible that the observed low expression of genes is resulting from negative selection of high-expressing cells by cytotoxic T cells, as a consequence of cross-reactivity.
Table 3.
Potentially cross-reactive HLA-C0303-restricted peptides in gliadin, PHLDA1 and CbiX.
| Species | Protein ID | Protein | Position | Peptide | Predicted affinity, nM |
|---|---|---|---|---|---|
| Triticum (wheat) | P18573 | Gliadin | 100–108 | LPYPQPQPF | 1270·38 |
| Triticum (wheat) | P18573 | Gliadin | 112–120 | QPYPQSQPQ | 71·92 |
| Oerskovia sp | WP_157257442.1 | CbiX | 200–208 | HAHPHPHPH | 71·56 |
| H. sapiens | Q8WV24 | PHLDA1 | 359–367 | HSHPHSHPH | 1089·64 |
Fig. 3.
Structural representation of HLA-C0303 restricted peptides. a: Gliadin100-108; b: Gliadin112-120; c: CbiX200-208; d: PHLDA1359-367.
4. Discussion
In this prospective study, we investigated peripheral blood CD19+ B-cell transcriptomics changes in children who developed CoD and in their controls. Most abundantly expressed genes over the whole data set included a number of genes that are involved in the antigen presentation: CD74, HLA-B, HLA-DRA, HLA-DRB1, which is confirming the broader role of B-cells in immune activation in CoD. Furthermore, many of these genes (ACTB, CD69, CD74, DUSP1, EZR, FOS, FTH1, H3F3B, JUNB, JUND, KLF6, NFKBIA, TSC22D3) were recently identified by Snir et al. [13] among top 50 most abundantly expressed non–Ig genes in plasma cells in CoD patients as well as in disease controls. This confirms that there are many similarities in gene expression profiles of plasma cells and other B-cell subsets.
Heterogeneous nuclear ribonucleoprotein L (gene: HNRNPL; protein: HNRPL) is a multifunctional protein that is widely expressed in multiple tissues. The highest levels have, however, been found in the lymph node and bone marrow [14]. HNRPL regulates alternative splicing [15], affecting the expression of genes such as CEACAM1 [16] and CD44 [17]. Importantly, the HNRPL protein binds also viral RNA sequences, such as Foot-and-Mouth Disease Virus (FMDV) Internal Ribosome Entry Site (IRES) and inhibits its replication [18]. FMDV belongs to the Picornaviridae family (genus Aphthovirus) along with the genus Enterovirus. Both families contain the 5’ IRES. In children with CoD, enteroviral infections have been observed to be more frequent compared to controls matched for time of birth, sex, and HLA-DQ genotype, whereas the frequency of adenoviral infections did not differ [19]. In addition to enterocytes, Enteroviruses can infect blood cells, in particular cells with antibody receptors (which includes B-cells), by a process termed Antibody Dependent Enhancement [20]. Also, since the tissue expression pattern of HNRNPL is very broad, it is possible that the reduced expression is also evident in enterocytes, however this hypothesis requires independent verification. It is possible that the predisposing factor for both CoD and enteroviral infections is the low level of HNRNPL expression.
It has been proposed that plasma cells are the most abundant antigen-presenting cells in CoD patients [21]. Other studies have also confirmed the importance of B-cells in CoD [22,23]. We did see a difference in B-cell gene expression between controls and cases who were diagnosed at 24 months of age. When controls and cases diagnosed at 36 months of age were compared, none of the genes remained significantly differently expressed after FDR. This substantial difference between these two groups might be associated with more extensive immune activation and faster disease development in the group of younger children. Among young children B-cells have been shown to undergo extensive changes during a relatively short period of time. Subgroups of memory B-cells and plasma cells expressing different Heavy Chain isotypes peak in abundance between 2 and 4 years of age and decline thereafter [24].
The current study revealed differentially expressed transcription factors (TF) between cases diagnosed with CoD at 24 months of age (CoD24mo group) and controls. The transcription factors displaying reduced expression in CoD are BHLHE40, ID3, and IRF4. It has been shown in mice that the deletion of BHLHE40 results in an abnormal activation of both B-cells and T cells [25]. Also, it has been shown that Bhlhe40 together with Bhlhe41 are regulating the development, self-renewal, surface phenotype and restricted repertoire of B-cell receptors of innate-like B-1a cells [26]. BHLHE40 emerging role in immune reactions and autoimmunity was just recently highlighted in the review by Cook et al., as it is involved in the regulation of various immune cells including B-cells [27].
Another TF downregulated in cases (CoD24mo) was IRF4, which is one of the key players in plasma cell differentiation [1,28]. Further, mice studies have shown the role of IRF4 in the production of IL-10 [29]. It has been reported that the expression of IRF4 is up-regulated in the intestinal mucosa of patients with active CoD [30]. Considering the role of IRF4 in upregulating IL-10 production [30], it may be theorized that its lower basal levels in peripheral blood promote inflammation as is observed in CoD.
ID3 (also known as BHLHB25, HEIR-1, 1R21) is an inhibitor of DNA-binding proteins and it prevents the binding between DNA and Basic helix-loop-helix proteins. ID3 acts as a TF and its expression is dependent on the stage of B-cell development [28]. In mice, Id3 is found to be highly expressed in follicular B-cells and its expression declines in germinal center B-cells [31]. Id3 is also involved in the negative regulation of plasma cell differentiation [32]. Id3 and Bhlhe40 have also been reported to be among key TFs regulating the development of memory CD8+ T cells [33]. Our results demonstrated the downregulation of ID3 in cases diagnosed with CoD at the age of 24 months. This result may indicate that B-cells in those patients are more strongly directed to develop towards plasma cells.
Our results also identified differential expression of adrenomedullin (ADM) being downregulated in cases diagnosed at 24 months’ of age. ADM is a multifunctional peptide belonging to the superfamily of vasoactive peptide hormones [34] and it is expressed in plasma cells [28]. Further, ADM was recently described among other proangiogenic cytokines secreted by novel IgG4+ CD49b + CD73+ B-cell subset [35]. It has been shown that ADM upregulation in PBMCs is a characteristic of several autoimmune conditions such as rheumatoid arthritis, psoriasis, and inflammatory bowel disease [36]. On the contrary, our results showed the down-regulation of ADM in peripheral blood B-cells at the time of diagnosis of CoD and in earlier time-points as well. The discrepancy between our study and previous data may derive from different study material, as we specifically investigated CD19+ B-cells and previous data is obtained from the analysis of PBMC. In accordance with our results, Leonard et al. showed that in the intestinal mucosa of active adult CoD patients ADM is down-regulated [30].
TRIB1 (also known as GIG-2, SKIP1, TRB-1, C8FW) belongs to the Tribbles (TRIB) family of pseudokinases. Under normal physiological conditions, TRIB1 is involved in the regulation of the cell cycle and liver function, immunoglobulin production and hematopoiesis. The dysregulation of pseudokinases plays a role in many diseases, such as numerous cancers [37]. Trib1 overexpression in B-cells is associated with systemic lupus erythematosus [38].
CDKN1A (also known as P21, CIP1, CDKN1, CAP20, MDA-6, CIP1, SDI1) is related to a broad range of different functions including cell cycle arrest, transcriptional regulation, and apoptosis [39]. It was recently shown that CDKN1A is downregulated in intestinal mucosa samples from both celiac patients on a gluten-free diet and gluten challenged patients in comparison to healthy controls [40]. CDKN1A was also observed to be upregulated in untreated CoD patients (but not in patients on a gluten-free diet) in TG-2 specific plasma cells compared to plasma cells specific for deamidated gluten peptides and plasma cells of unknown specificity [41].
PHLDA1, also known as PHRIP, TDAG51, and DT1P1B11 is an evolutionarily conserved proline-histidine rich nuclear protein. It is involved in the regulation of translation. In human T-cells, it has been shown to couple TCR signaling to inhibition of protein biosynthesis in activated cells [42]. In intestinal epithelium, PHLDA1 expression marks the putative epithelial stem cells. In normal epithelium, PHLDA1 is expressed in discrete crypt base cells, while in tumours its function contributes to migration and proliferation [43].
PHLDA1 amino acid sequence contains a polyglutamine region and a region rich in proline and glutamine, two features shared with gliadin and numerous bacterial proteins involved in metal ion binding, including zinc ions.
Zinc deficiency has been associated with CoD, and has been attributed to the malabsorption syndrome characterizing CoD. However, zinc is required for intestinal epithelial barrier maintenance via the regulation of claudin-3 and occludin expression [44]. Therefore, persistent deficiency promotes epithelial damage and creates a vicious cycle maintaining the disease. It can be theorized that a transient episode of enteric colonization with bacteria expressing high levels of metal-binding proteins might initiate metal deficiency, epithelial damage, and cross-reactive CD8 mediated immunity.
KLHL21 (also known as CRL3) is an E3 ubiquitin ligase and a negative regulator of NFkB signaling. KLHL21 specifically binds to the kinase domain of IKKb via its Kelch domains [45].
The role of B-cells in CoD has been highlighted by a recent finding that B-cell depletion can prevent the development of CoD in a mouse model. Lejeune et al. suggested that B-cells may be involved in the amplification of the CD4+ T-cell response to a magnitude sufficient to affect the activation of cytotoxic IELs and the ensuing tissue destruction [46]. Altogether, most of the downregulated genes identified in our study function in various aspects of cell signaling inhibition. Therefore, it appears that the changes in gene expression detected by us represent a state of functional activation for B-cells.
Further, we applied a linear mixed-effects model to study dynamical changes in B-cell gene expression. Our results showed the potential role of small nucleolar RNAs in CoD pathogenesis, since we revealed highly different expression of SNORA7B between controls and children who progressed to CoD. Most interestingly, the expression of SNORA7B showed an opposite trend in these two groups. Small nucleolar RNAs (snoRNAs) represent a class of regulatory RNAs responsible for posttranscriptional maturation of ribosomal RNAs. snoRNAs have been studied in the context of cancer, neurodegenerative diseases and viral infections. However, no specific information on the role of SNORA7B exists [47]. Accordingly, the exact role of snoRNA in the context of B-cells and CoD remains to be elucidated.
One of the strengths of our work is that we specifically studied purified CD19+ B-cells, whereas most previous studies have investigated bulk PBMCs. Garber et al. [4] who also studied B-cell transcriptomics, showed that the extent of mucosal damage correlated with reduced expression of a subset of B-cell genes. The limitation of their study was, that CD3+ T cells and CD19+ B-cells were isolated and their gene profiles were investigated together. To our knowledge, our study is the first specifically investigating changes in CD19+ B-cell gene expression in the context of CoD. Yet, the subpopulation composition of peripheral blood B-cells between and within study groups may affect the results and the changes we observed. Thus, we cannot exclude the possibility that our results may be related to a specific B-cell subset (transitional B-cells, Bregs, MZ B-cells, GC B-cells, etc). This is supported by the recent single-cell transcriptomics study by Horns et al., who showed that BHLHE40, CDKN1A, and MIDN are among genes differently expressed between naive and memory B-cells and/or memory and activated memory B-cells [48]. Further, our study included only children with a narrow age range. It has been also shown that the gene expression profile differs between patients diagnosed in childhood or in adulthood [49].
5. Conclusions
The objective of this prospective study was to investigate peripheral blood B-cell gene expression with the further aim to identify biomarkers differentiating children at risk of developing CoD. We studied B-cell gene expression in genetically predisposed children during the time of development and diagnosis of CoD. We used two different approaches to study changes in B-cell transcriptional profiles in cases and controls. First, we studied genes, which were differentially expressed between cases and controls at all time points. We showed, that there are 16 DEG between cases from the CoD24mo group and their controls. Secondly, we aimed to study dynamic changes in B-cell gene expression over time. With this approach, we found that SNORA7B expression exhibits a reverse trend in cases and controls. In one of the genes expressed at lower levels in CoD we identified C0303-restricted epitopes potentially cross-reactive with gliadin and certain bacterial metal-binding proteins. Double cross-reactivity between bacterial, human, and gliadin sequenes may play a role in the pathogenesis of CoD.
Research in context.
Evidence before this study: The role of B-cells in CoD has been highlighted by a recent finding that B-cell depletion can prevent the development of CoD in a mouse model. Lejeune et al. suggested that B-cells may be involved in the amplification of the CD4+ T-cell response to a magnitude sufficient to affect the activation of cytotoxic lymphocytes and the ensuing tissue destruction. The importance of B-cells in CoD is well described by Iversen and Sollid emphsizing their role in TG2 presentation and autoantibody production. Less is known about peripheral B-cell transcriptome. So far, changes in B-cell gene expression have been reported in patients affected by CoD only in one study. Garber et al. showed that the gluten-induced small intestine injury score is inversely correlated with expression levels of a subset of peripheral blood B-cell (but not T-cell) genes in a microarray analysis, suggesting a potential protective role for B-cells.
Added value of this study: Our study specifically investigates changes in CD19+ B-cell gene expression in the context of CoD. Altogether, the differences in gene expression between the CoD and healthy groups were small and none of the genes was significantly differentially expressed after normalization procedures and correction for multiple testing. However, the top scoring gene, HNRNPL (logFC −0·7, q = 0·09) was modestly downregulated in all patients. In a subgroup of patients who developed CoD at 24 months of age, the set of differentially expressed genes suggests a state of functional activation of B-cells preceding the onset of disease.
Implications of all the available evidence:In children with CoD, enteroviral infections have been observed to be more frequent compared to controls matched for time of birth, sex, and HLA-DQ genotype. It is possible that the predisposing factor for both CoD and enteroviral infections is the low level of HNRNPL expression. The differentially expressed genes between in the group diagnosed with CoD at 24 months of age indicate that a state of functional activation of B-cells precedes the disease onset.
Data sharing
The data collected for the study, including individual participant data and a data dictionary defining each field in the set, have been deposited in NCBI GEO, (Submission GSE185503) and will be made available to others upon publication.
No additional, related documents will be available.
Author contribution statement
Astrid Oras: Performed the experiments; Analysed and interpreted the data; Wrote the paper.
Henna Kallionpää; Tomi Suomi; Satu Koskinen; Asta Laiho; Laura L. Elo: Performed the experiments; Wrote the paper.
Mikael Knip; Riitta Lahesmaa; Raivo Uibo: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Alar Aints: Analysed and interpreted the data; Wrote the paper.
Funding statement
Mikael Knip was supported by FP7 International Cooperation [202063], Academy of Finland [250114].
Raivo Uibo was supported by Eesti Teadusagentuur [PRG712].
Riitta Lahesmaa was supported by Academy of Finland [292482; 292335; 294337; 319280; 31444; 329277; 331790] and grants from the Sigrid Jusélius Foundation (SJF).
Data availability statement
Data associated with this study has been deposited at GEO under the accession number GSE185503.
Declaration of interest's statement
The authors declare no competing interests.
Acknowledgments
We thank DIABIMMUNE consortium for organizing the recruitment of the participants and for the collection of the medical background data from the participants. Also, we would like to thank Mrs. Kristi Alnek, MSc, and Helis Janson, MSc, for performing autoantibody assays and Dr. Oivi Uibo for clinical investigation of the children with CoD. We would like to thank the Finnish Functional Genomics Centre, at Turku Bioscience Centre, University of Turku and Åbo Akademi, supported by Biocenter Finland.
Footnotes
The dataset made available includes deidentified participant data.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.heliyon.2023.e13147.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Du Pré M.F., Sollid L.M. T-cell and B-cell immunity in celiac disease. Best Pract. Res. Clin. Gastroenterol. 2015;29:413–423. doi: 10.1016/j.bpg.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Getahun A., Cambier J.C. Non-antibody-secreting functions of B cells and their contribution to autoimmune disease. Annu. Rev. Cell Dev. Biol. 2019;35:337–356. doi: 10.1146/annurev-cellbio-100617-062518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iversen R., Sollid L.M. Autoimmunity provoked by foreign antigens: in celiac disease, exogenous gluten drives T cell-B cell interactions that cause autoimmunity. Science. 2020;368(80-):132–133. doi: 10.1126/science.aay3037. [DOI] [PubMed] [Google Scholar]
- 4.Garber M.E., Saldanha A., Parker J.S., Jones W.D., Kaukinen K., Laurila K., Lähdeaho M.L., Khatri P., Khosla C., Adelman D.C., Mäki M. A B-cell gene signature correlates with the extent of gluten-induced intestinal injury in celiac disease. CMGH. 2017;4:1–17. doi: 10.1016/j.jcmgh.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simre K., Uibo O., Peet A., Tillmann V., Kool P., Hämäläinen A.M., Härkönen T., Siljander H., Virtanen S.M., Ilonen J., Knip M., Uibo R., Koski M., Ryhänen S., Ormisson A., Ulich V., Kuzmicheva E., Mokurov S., Markova S., Pylova S., Isakova M., Shakurova E., Petrov V., Dorshakova N.V., Karapetyan T., Varlamova T., Kiviniemi M., Alnek K., Janson H., von Mutius E., Weber J., Ahlfors H., Kallionpää H., Laajala E., Lahesmaa R., Lähdesmäki H., Moulder R., Nieminen J., Ruohtula T., Vaarala O., Honkanen H., Hyöty H., Kondrashova A., Oikarinen S., Harmsen H.J.M., De Goffau M.C., Welling G., Alahuhta K. Exploring the risk factors for differences in the cumulative incidence of coeliac disease in two neighboring countries: the prospective DIABIMMUNE study. Dig. Liver Dis. 2016;48:1296–1301. doi: 10.1016/j.dld.2016.06.029. [DOI] [PubMed] [Google Scholar]
- 6.Hermann R., Turpeinen H., Laine A.P., Veijola R., Knip M., Simell O., Sipilä I., Åkerblom H.K., Ilonen J. HLA DR-DQ-encoded genetic determinants of childhood-onset type 1 diabetes in Finland: an analysis of 622 nuclear families. Tissue Antigens. 2003;62:162–169. doi: 10.1034/j.1399-0039.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- 7.Husby S., Koletzko S., Korponay-Szabó I.R., Mearin M.L., Phillips A., Shamir R., Troncone R., Giersiepen K., Branski D., Catassi C., Lelgeman M., Mäki M., Ribes-Koninckx C., Ventura A., Zimmer K.P. European society for pediatric gastroenterology, hepatology, and nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012;54:136–160. doi: 10.1097/MPG.0b013e31821a23d0. [DOI] [PubMed] [Google Scholar]
- 8.Oberhuber G., Granditsch G., Vogelsang H. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur. J. Gastroenterol. Hepatol. 1999;11:1185–1194. doi: 10.1097/00042737-199910000-00019. [DOI] [PubMed] [Google Scholar]
- 9.Andreatta M., Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class i system. Bioinformatics. 2016;32:511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koepnick B., Flatten J., Husain T., Ford A., Silva D.A., Bick M.J., Bauer A., Liu G., Ishida Y., Boykov A., Estep R.D., Kleinfelter S., Nørgård-Solano T., Wei L., Players F., Montelione G.T., DiMaio F., Popović Z., Khatib F., Cooper S., Baker D. De novo protein design by citizen scientists. Nature. 2019;570:390–394. doi: 10.1038/s41586-019-1274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gianfrani C., Troncone R., Mugione P., Cosentini E., De Pascale M., Faruolo C., Senger S., Terrazzano G., Southwood S., Auricchio S., Sette A. Celiac disease association with CD8 + T cell responses: identification of a novel gliadin-derived HLA-A2-restricted epitope. J. Immunol. 2003;170:2719–2726. doi: 10.4049/jimmunol.170.5.2719. [DOI] [PubMed] [Google Scholar]
- 12.Picascia S., Sidney J., Camarca A., Mazzarella G., Giardullo N., Greco L., Auricchio R., Auricchio S., Troncone R., Sette A., Gianfrani C. Gliadin-specific CD8 + T cell responses restricted by HLA class I A*0101 and B*0801 molecules in celiac disease patients. J. Immunol. 2017;198:1838–1845. doi: 10.4049/jimmunol.1601208. [DOI] [PubMed] [Google Scholar]
- 13.Snir O., Kanduri C., Lundin K.E.A., Sandve G.K., Sollid L.M. Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production. United Eur. Gastroenterol. J. 2019;7:1399–1407. doi: 10.1177/2050640619862461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., Asplund A., Sjostedt E., Lundberg E., Szigyarto C.A.K., Skogs M., Ottosson Takanen J., Berling H., Tegel H., Mulder J., Nilsson P., Schwenk J.M., Lindskog C., Danielsson F., Mardinoglu A., Sivertsson A., Von Feilitzen K., Forsberg M., Zwahlen M., Olsson I., Navani S., Huss M., Nielsen J., Ponten F., Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heiner M., Hui J., Schreiner S., Hung L.H., Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7 doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 16.Dery K.J., Gaur S., Gencheva M., Yen Y., Shively J.E., Gaur R.K. Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M. J. Biol. Chem. 2011;286:16039–16051. doi: 10.1074/jbc.M110.204057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh T.J., Cho S., Moon H., Jang H.N., Williams D.R., Jung D.W., Kim I.C., Ghigna C., Biamonti G., Zheng X., Shen H. HnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochim. Biophys. Acta - Gene Regul. Mech. 2015;1849:743–750. doi: 10.1016/j.bbagrm.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Sun C., Liu M., Chang J., Yang D., Zhao B., Wang H., Zhou G., Weng C., Yu L. Heterogeneous nuclear ribonucleoprotein L negatively regulates foot-and-mouth disease Virus replication through inhibition of viral RNA synthesis by interacting with the internal ribosome Entry site in the 5′ untranslated region. J. Virol. 2020;94 doi: 10.1128/jvi.00282-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oikarinen M., Puustinen L., Lehtonen J., Hakola L., Simell S., Toppari J., Ilonen J., Veijola R., Virtanen S.M., Knip M., Hyöty H. Enterovirus infections are associated with the development of celiac disease in a birth cohort study. Front. Immunol. 2021;11:3844. doi: 10.3389/fimmu.2020.604529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S.M., Chen I.C., Su L.Y., Huang K.J., Lei H.Y., Liu C.C. Enterovirus 71 infection of monocytes with antibody-dependent enhancement. Clin. Vaccine Immunol. 2010;17:1517–1523. doi: 10.1128/CVI.00108-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Høydahl L.S., Richter L., Frick R., Snir O., Gunnarsen K.S., Landsverk O.J.B., Iversen R., Jeliazkov J.R., Gray J.J., Bergseng E., Foss S., Qiao S.W., Lundin K.E.A., Jahnsen J., Jahnsen F.L., Sandlie I., Sollid L.M., Løset G.Å. Plasma cells are the most abundant gluten peptide MHC-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology. 2019;156:1428–1439.e10. doi: 10.1053/j.gastro.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sangineto M., Graziano G., D'Amore S., Salvia R., Palasciano G., Sabba C., Vacca M., Cariello M. Identification of peculiar gene expression profile in peripheral blood mononuclear cells (PBMC) of celiac patients on gluten free diet. PLoS One. 2018;13 doi: 10.1371/journal.pone.0197915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlesworth R.P.G., Agnew L.L., Scott D.R., Andronicos N.M. Celiac disease gene expression data can be used to classify biopsies along the Marsh score severity scale. J. Gastroenterol. Hepatol. 2019;34:169–177. doi: 10.1111/jgh.14369. [DOI] [PubMed] [Google Scholar]
- 24.Blanco E., Pérez-Andrés M., Arriba-Méndez S., Contreras-Sanfeliciano T., Criado I., Pelak O., Serra-Caetano A., Romero A., Puig N., Remesal A., Torres Canizales J., López-Granados E., Kalina T., Sousa A.E., van Zelm M., van der Burg M., van Dongen J.J.M., Orfao A. Age-associated distribution of normal B-cell and plasma cell subsets in peripheral blood. J. Allergy Clin. Immunol. 2018;141:2208–2219.e16. doi: 10.1016/j.jaci.2018.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Samten B. Regulation of B-1a cells: another novel function of the basic helix-loop-helix transcriptional regulator BHLHE41, Cell. Mol. Immunol. 2017;14:802–804. doi: 10.1038/cmi.2017.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kreslavsky T., Vilagos B., Tagoh H., Poliakova D.K., Schwickert T.A., Wöhner M., Jaritz M., Weiss S., Taneja R., Rossner M.J., Busslinger M. Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells. Nat. Immunol. 2017;18:442–455. doi: 10.1038/ni.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook M.E., Jarjour N.N., Lin C.C., Edelson B.T. Transcription factor Bhlhe40 in immunity and autoimmunity. Trends Immunol. 2020;41:1023–1036. doi: 10.1016/j.it.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhan F., Tian E., Bumm K., Smith R., Barlogie B., Shaughnessy J. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101:1128–1140. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 29.Baba Y., Saito Y., Kotetsu Y. Heterogeneous subsets of B-lineage regulatory cells (Breg cells) Int. Immunol. 2019;32:155–162. doi: 10.1093/intimm/dxz068. [DOI] [PubMed] [Google Scholar]
- 30.Leonard M.M., Bai Y., Serena G., Nickerson K.P., Camhi S., Sturgeon C., Yan S., Fiorentino M.R., Katz A., Nath B., Richter J., Sleeman M., Gurer C., Fasano A. RNA sequencing of intestinal mucosa reveals novel pathways functionally linked to celiac disease pathogenesis. PLoS One. 2019;14 doi: 10.1371/journal.pone.0215132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S., Miyazaki M., Chandra V., Fisch K.M., Chang A.N., Murre C. Id3 orchestrates germinal center B cell development. Mol. Cell Biol. 2016;36:2543–2552. doi: 10.1128/mcb.00150-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gloury R., Zotos D., Zuidscherwoude M., Masson F., Liao Y., Hasbold J., Corcoran L.M., Hodgkin P.D., Belz G.T., Shi W., Nutt S.L., Tarlinton D.M., Kallies A. Dynamic changes in Id3 and E-protein activity orchestrate germinal center and plasma cell development. J. Exp. Med. 2016;213:1095–1111. doi: 10.1084/jem.20152003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu G., Chen J. A genome-wide regulatory network identifies key transcription factors for memory CD8 + T-cell development. Nat. Commun. 2013;4 doi: 10.1038/ncomms3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schönauer R., Els-Heindl S., Beck-Sickinger A.G. Adrenomedullin – new perspectives of a potent peptide hormone. J. Pept. Sci. 2017;23:472–485. doi: 10.1002/psc.2953. [DOI] [PubMed] [Google Scholar]
- 35.van de Veen W., Globinska A., Jansen K., Straumann A., Kubo T., Verschoor D., Wirz O.F., Castro-Giner F., Tan G., Rückert B., Ochsner U., Herrmann M., Stanić B., van Splunter M., Huntjens D., Wallimann A., Fonseca Guevara R.J., Spits H., Ignatova D., Chang Y.T., Fassnacht C., Guenova E., Flatz L., Akdis C.A., Akdis M. A novel proangiogenic B cell subset is increased in cancer and chronic inflammation. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aaz3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesko B., Poliska S., Szegedi A., Szekanecz Z., Palatka K., Papp M., Nagy L. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med. Genom. 2010;3 doi: 10.1186/1755-8794-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richmond L., Keeshan K. Pseudokinases: a tribble-edged sword. FEBS J. 2020;287:4170–4182. doi: 10.1111/febs.15096. [DOI] [PubMed] [Google Scholar]
- 38.Simoni L., Delgado V., Ruer-Laventie J., Bouis D., Soley A., Heyer V., Robert I., Gies V., Martin T., Korganow A.S., San Martin B.R., Soulas-Sprauel P. Trib1 is overexpressed in systemic lupus erythematosus, while it regulates immunoglobulin production in murine B cells. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair. 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Dotsenko V., Oittinen M., Taavela J., Popp A., Peräaho M., Staff S., Sarin J., Leon F., Isola J., Mäki M., Viiri K. Genome-Wide transcriptomic analysis of intestinal mucosa in celiac disease patients on a gluten-free diet and postgluten challenge. CMGH. 2021;11:13–32. doi: 10.1016/j.jcmgh.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindeman I., Zhou C., Eggesbø L.M., Miao Z., Polak J., Lundin K.E.A., Jahnsen J., Qiao S.W., Iversen R., Sollid L.M. Longevity, clonal relationship, and transcriptional program of celiac disease-specific plasma cells. J. Exp. Med. 2021;218 doi: 10.1084/jem.20200852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinz T., Flindt S., Marx A., Janssen O., Kabelitz D. Inhibition of protein synthesis by the T cell receptor-inducible human TDAG51 gene product. Cell. Signal. 2001;13:345–352. doi: 10.1016/S0898-6568(01)00141-3. [DOI] [PubMed] [Google Scholar]
- 43.Sakthianandeswaren A., Christie M., D'Andreti C., Tsui C., Jorissen R.N., Li S., Fleming N.I., Gibbs P., Lipton L., Malaterre J., Ramsay R.G., Phesse T.J., Ernst M., Jeffery R.E., Poulsom R., Leedham S.J., Segditsas S., Tomlinson I.P.M., Bernhard O.K., Simpson R.J., Walker F., Faux M.C., Church N., Catimel B., Flanagan D.J., Vincan E., Sieber O.M. PHLDA1 expression marks the putative epithelial stem cells and contributes to intestinal tumorigenesis. Cancer Res. 2011;71:3709–3719. doi: 10.1158/0008-5472.CAN-10-2342. [DOI] [PubMed] [Google Scholar]
- 44.Miyoshi Y., Tanabe S., Suzuki T. Cellular zinc is required for intestinal epithelial barrier maintenance via the regulation of claudin-3 and occludin expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311 doi: 10.1152/ajpgi.00405.2015. G105–G116. [DOI] [PubMed] [Google Scholar]
- 45.Mei Z.Z., Chen X.Y., Hu S.W., Wang N., Ou X.L., Wang J., Luo H.H., Liu J., Jiang Y. Kelch-like Protein 21 (KLHL21) targets IκB Kinase-β to regulate nuclear factor κ-light chain enhancer of activated B Cells (NF-κB) signaling negatively. J. Biol. Chem. 2016;291:18176–18189. doi: 10.1074/jbc.M116.715854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lejeune T., Meyer C., Abadie V. B lymphocytes contribute to celiac disease pathogenesis. Gastroenterology. 2021;160:2608–2610.e4. doi: 10.1053/j.gastro.2021.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepanov G.A., Filippova J.A., Komissarov A.B., Kuligina E.V., Richter V.A., Semenov D.V., Tang T.H. Regulatory role of Small nucleolar RNAs in human diseases. BioMed Res. Int. 2015;2015 doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horns F., Dekker C.L., Quake S.R. Memory B cell activation, broad anti-influenza antibodies, and bystander activation revealed by single-cell transcriptomics. Cell Rep. 2020;30:905–913.e6. doi: 10.1016/j.celrep.2019.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pascual V., Medrano L.M., López-Palacios N., Bodas A., Dema B., Fernández-Arquero M., González-Pérez B., Salazar I., Núñez C. Different gene expression signatures in children and adults with celiac disease. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at GEO under the accession number GSE185503.