Abstract
Background & Aims
Fibrosis is a common complication of inflammatory bowel diseases (IBDs). The pregnane X receptor (PXR) (encoded by NR1I2) suppresses intestinal inflammation and has been shown to influence liver fibrosis. In the intestine, PXR signaling is influenced by microbiota-derived indole-3-propionic acid (IPA). Here, we sought to assess the role of the PXR in regulating intestinal inflammation and fibrosis.
Methods
Intestinal inflammation was induced using dextran sulfate sodium (DSS). Fibrosis was assessed in wild-type (WT), Nr1i2-/-, epithelial-specific Nr1i2-/-, and fibroblast-specific Nr1i2-/- mice. Immune cell influx was quantified by flow cytometry and cytokines by Luminex. Myofibroblasts isolated from WT and Nr1i2-/- mice were stimulated with cytomix or lipopolysaccharide, and mediator production was assessed by quantitative polymerase chain reaction and Luminex.
Results
After recovery from DSS-induced colitis, WT mice exhibited fibrosis, a response that was exacerbated in Nr1i2-/- mice. This was correlated with greater neutrophil infiltration and innate cytokine production. Deletion of the PXR in fibroblasts, but not the epithelium, recapitulated this phenotype. Inflammation and fibrosis were reduced by IPA administration, whereas depletion of the microbiota exaggerated intestinal fibrosis. Nr1i2-deficient myofibroblasts were hyperresponsive to stimulation, producing increased levels of inflammatory mediators compared with WT cells. In biopsies from patients with active Crohn’s disease (CD) and ulcerative colitis (UC), expression of NR1I2 was reduced, correlating with increased expression of fibrotic and innate immune genes. Finally, both CD and UC patients exhibited reduced levels of fecal IPA.
Conclusions
These data highlight a role for IPA and its interactions with the PXR in regulating the mesenchyme and the development of inflammation and fibrosis, suggesting microbiota metabolites may be a vital determinant in the progression of fibrotic complications in IBD.
Keywords: Intestinal Fibrosis, Pregnane X Receptor, Microbiota, Fibroblasts
Abbreviations used in this paper: ABX, antibiotics; αSMA, alpha smooth muscle actin; CD, Crohn’s disease; Col1a1, collagen, type I, alpha 1; Col1a2, collagen, type I, alpha 2; Col3a1, collagen, type 3, alpha 1; CSF2, colony stimulating factor 2; CSF3, colony stimulating factor 3; CXCL1, C-X-C Motif chemokine ligand 1; CXCL2, C-X-C Motif chemokine ligand 2; CXCL8, C-X-C Motif chemokine ligand 8; CXCL9, C-X-C Motif chemokine ligand 9; Cyp3a11, cytochrome P450, family 3, subfamily a, polypeptide 11; DSS, dextran sulfate sodium; G-CSF, granulocyte colony-stimulating factor; GF, germ-free; GM-CSF, granulocyte-macrophage colony-stimulating factor; 4-OHT, 4-hydroxytamoxifen; IBD, inflammatory bowel disease; IFNγ, interferon gamma; IL1b, interleukin 1 beta; IL5, interleukin 5; IPA, indole-3-propionic acid; LP, lamina propria; LPS, lipopolysaccharide; MMP, matrix metalloproteinase; NF-κB, nuclear factor kappa B; NR1I2, nuclear receptor subfamily 1 group I member 2; PBS, phosphate-buffered saline; PCN, pregnenolone 16α-carbonitrile; PXR, pregnane X receptor; SPF, specific pathogen–free; TGFβ, transforming growth factor beta; TNFα, tumor necrosis factor alpha; UC, ulcerative colitis; WT, wild-type
Summary.
PXR signaling in intestinal fibroblasts restrains their inflammatory response and dampens intestinal fibrosis. Luminal sensing of the microbiota-derived metabolites IPA via PXR reduces inflammation and fibrosis, highlighting the impact of the microbiota in the regulation of the intestinal mesenchyme and points to an additional cellular target to treat intestinal inflammation and fibrosis patients with IBD.
Intestinal fibrosis is a frequent occurrence in patients with the inflammatory bowel diseases (IBDs), Crohn’s disease (CD) and ulcerative colitis (UC).1 In CD, fibrotic processes contribute to intestinal strictures and obstruction in 20%–50% of patients over their lifetime,2,3 with many requiring surgery to remove obstructed areas of the intestine. However, surgical removal of the affected segment does not prevent the recurrence of fibrotic events that still occur at a high frequency in CD patients after initial surgery.4,5 Although IBD with ileal involvement is most frequently associated with fibrotic complications, intestinal fibrosis has also been observed in CD of the colon and in UC. Indeed, reports suggest that fibrosis can occur in up to 11% of patients with chronic and progressive UC and can result in significant colonic shortening.6 The pathogenesis of fibrosis is incompletely understood and often perceived as an irreversible process. Despite significant advances in the treatment of IBDs, current therapies do not efficiently block fibrosis and stricture formation.
Chronic inflammation is a significant contributor to intestinal fibrosis by fueling defective responses of mesenchymal cells (eg, fibroblasts) in the intestinal lamina propria (LP). Aberrantly activated fibroblasts, also termed myofibroblasts, are found in inflamed and fibrotic lesions of IBD patients where they express increased levels of smooth muscle actin (αSMA) and other fibrogenic factors, including collagens and matrix metalloproteinases (MMPs).1,7 Myofibroblasts also actively participate in inflammatory responses and are equipped with various receptors, including pattern recognition and cytokine receptors, that can be activated by different stimuli, including components of luminal bacteria such as lipopolysaccharide (LPS) and cytokines released into the local milieu.8,9 By producing cytokines and chemokines through different intracellular signaling pathways, myofibroblasts can provide recruitment, survival, and retention signals for leukocytes to propel the inflammatory cascade.10, 11, 12 In IBD, subsets of stromal cells, including fibroblasts and myofibroblasts, have been observed to respond excessively to inflammatory signals in diseased lesions, contributing to immune cell crosstalk and inflammatory reactions that increase disease severity and support fibrotic processes.11, 12, 13, 14
Immune homeostasis at mucosal surfaces is significantly shaped by the microbiota and microbiota-produced metabolites. To date, several classes of gut microbiota-derived metabolites have been identified that can modulate the immune response and the pathogenesis of IBD,15 including metabolites of tryptophan that can balance mucosal reactivity and prevent inflammation in the intestine.16 Indole-3-propionic acid (IPA) is one such metabolite that is produced by tryptophan metabolizing Clostridia commensals in the intestine17, 18, 19 and can influence mucosal barrier function and inflammation through its interactions with the pregnane X receptor (PXR) (encoded by nuclear receptor subfamily 1 group I member 2 [NR1I2]).20, 21, 22 Activity of the PXR, a master regulator of xenobiotic metabolism, not only influences inflammation but can also regulate fibrotic processes in the liver23,24 and skin25; however, the role of the PXR in intestinal fibrosis has not been explored. Because of this question and the ability of the PXR to sense microbiota-derived metabolites in the gut, we sought to test the hypothesis that the activity of the bacterial metabolite IPA through its interactions with the PXR controls fibrotic responses in the intestine.
Here, we show for the first time that PXR signaling is important for proper resolution of inflammation after colonic injury, and that defective PXR signaling can prolong intestinal inflammation and result in exaggerated fibrosis. This occurs, in part, through the ability of intestinal PXR signaling to restrain the mesenchymal compartment of the intestine from overactive immune responses. Importantly, administration of IPA dampens the development of fibrosis in the healing colon through clearing inflammation and supressing fibrosis promoting interactions between the mesenchyme and leukocytes. In humans with IBD, expression of the PXR and its target genes in the intestinal tissue is significantly suppressed, resulting in heightened inflammatory and fibrotic responses. Furthermore, patients with CD and UC have decreased levels of fecal IPA, a metabolite that is able to suppress inflammatory responses in stimulated fibroblasts.
Results
Loss of PXR Results in Protracted Intestinal Inflammation and Fibrosis
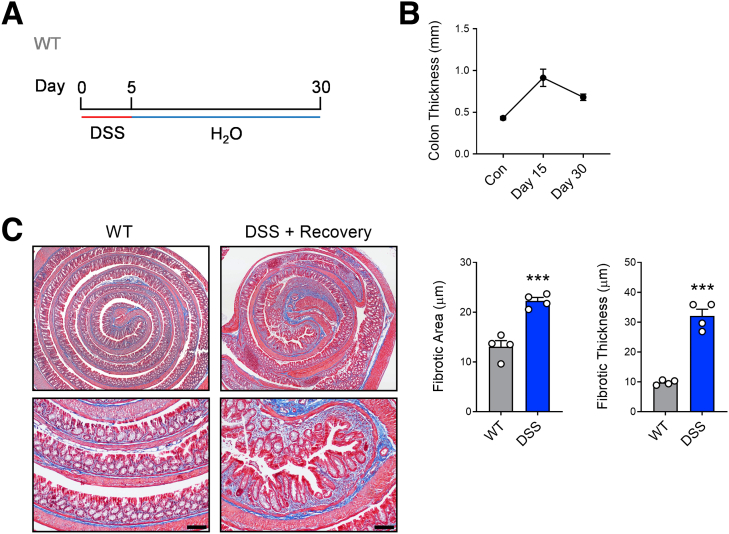
Dextran sulfate sodium (DSS) is a commonly used model of intestinal injury that triggers inflammation and prompts structural remodelling and fibrotic changes in the mouse colon that model human IBD after only a short, acute course.26 To further examine fibrotic processes in our studies, we used a DSS-induced injury-healing model where mice were administered 3% DSS for 5 days to trigger acute injury and inflammation and then switched to normal drinking water and allowed to heal for an additional 25 days (30 days total; Figure 1A). This protocol resulted in gross colonic thickening and increased fibrotic remodeling in the submucosa as indicated by staining with Masson’s trichrome (Figure 2).
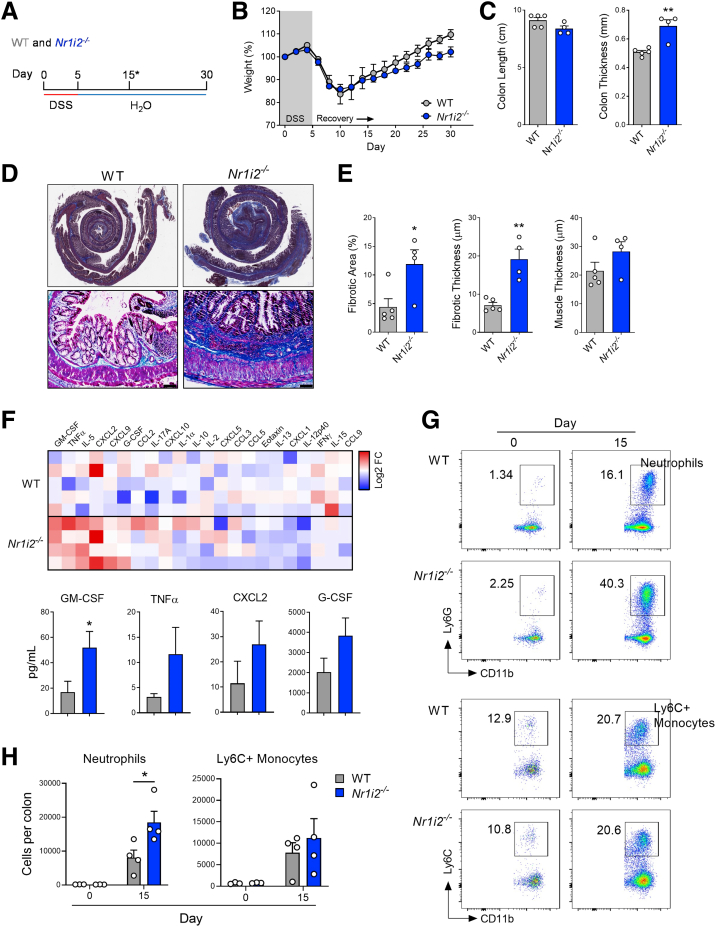
Figure 1.
Loss of Nr1i2 (the gene encoding the pregnane X receptor) exacerbates colonic fibrosis and is accompanied by elevated GM-CSF levels and neutrophilic inflammation after injury. (A) WT(Nr1i2+/+) and Nr1i2-/- mice were administered DSS for 5 days to induce colonic injury and inflammation and then switched to regular drinking water and allowed to heal for an additional 25 days (∗indicates the different analysis time points). (B) Weight recovery and (C) gross colon length and thickness in WT(Nr1i2+/+) and Nr1i2-/- mice 25 days after withdrawal of DSS. (D) Representative images of Masson’s trichrome staining to indicate fibrosis in the colon of WT(Nr1i2+/+) and Nr1i2-/- mice (scale bar = 100 μm). (F) Heatmap of cytokines and chemokines detected in the serum 25 days after withdrawal of DSS (each tile represents an individual animal) and absolute levels of the most differentially produced cytokines in the serum between WT(Nr1i2+/+) and Nr1i2-/- mice (n = 4–5 per group). (G) Representative FACS plots of CD11b+Ly6G+ neutrophils and CD11b+Ly6CHi monocytes in the healing colonic lamina propria of WT(Nr1i2+/+) and Nr1i2-/- mice 10 days after withdrawal of DSS (n = 3 per water groups, n = 4 per DSS groups). Data are presented as mean ± standard error of the mean. Student t test (C, E, and F) and one-way analysis of variance with Tukey post hoc test (H). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Figure 2.
Assessment of fibrotic remodeling in the colon after recovering from DSS-induced injury. (A) WT C57BL/6 mice were administered DSS (3%) for 5 days to induce colonic injury and inflammation and then switched to regular drinking water and allowed to heal for an additional 25 days. (B) Gross colon thickness was measured in mice without DSS and 10 and 25 days after withdrawal of DSS (after 5-day course). (C) Representative images of Masson’s trichrome staining (scale bar = 100 μm) and associated quantification of total fibrotic area and fibrotic thickness in the colon of control mice and mice administered DSS after healing for 25 days (n = 4 per group). Data are presented as mean ± standard error of the mean. Student t test (C). ∗∗∗P < .001.
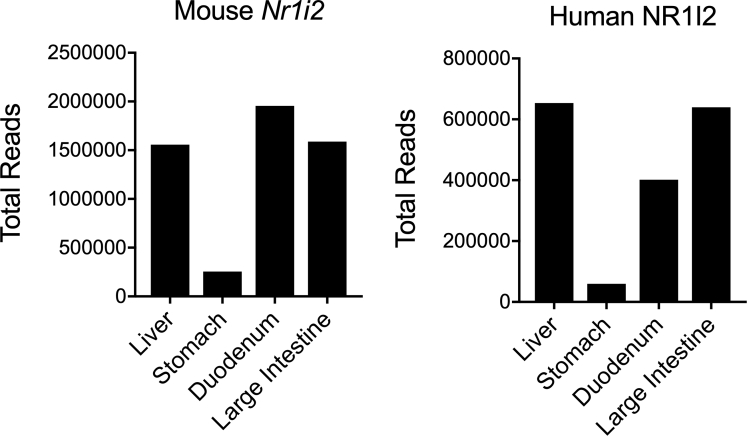
We next used this model to investigate the role of the PXR in influencing intestinal fibrosis because Nr1i2/N1I2 (the gene encoding the PXR) is highly expressed in the colon of both mice and humans (Figure 3). After the induction of colitis, there was a precipitous drop in body weight that did not differ between Nr1i2-/- mice and their wild-type (WT) littermate counterparts. However, once mice began to regain body weight, WT mice gained significantly more weight than Nr1i2-/- mice, suggesting poorer recovery in the absence of the PXR (Figure 1B). Macroscopic examination of the colon in these mice after 25 days of healing did not reveal a difference in colon length between WT and Nr1i2-/- mice; however, there was significantly greater colonic thickening in Nr1i2-/- mice (Figure 1C). Masson’s trichrome stained colonic sections from each strain of mice showed fibrotic changes, with areas of the colonic submucosa occupied by connective tissue; however, this fibrotic layer of connective tissue underlying the mucosa occupied significantly more area and was significantly thicker in Nr1i2-/- mice than in WT littermate counterparts (Figure 1D). Together these results suggest a role for PXR signaling in promoting proper recovery and preventing the development of fibrosis after colonic injury.
Figure 3.
NR1I2 expression in mouse and human colon. Total read counts for Nr1i2 mRNA in different tissues from mice and humans. Mouse counts were obtained from online deposited data at BioProject PRJNA66167 Human counts were obtained from online deposited data at BioProject: PRJEB4337.
Intestinal Inflammation in the Colon of Nr1i2-/- Mice Is Accompanied by Prolonged Innate, Neutrophilic Inflammation
To assess whether there were differences in the immune responses between WT and Nr1i2-/- mice that could explain differences in fibrotic responses in the healing colon, we first examined serum cytokines. The heatmap in Figure 1F shows serum levels of different cytokines between WT and Nr1i2-/- mice after healing (25 days after stopping DSS). Of the mediators assessed, the most differing were granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), tumor necrosis factor alpha (TNFα), interleukin 5 (IL5), C-X-C Motif chemokine ligand 9 (CXCL9), and C-X-C Motif chemokine ligand 2 (CXCL2); each showed greater protein levels in Nr1i2-/- mice compared with their WT littermate counterparts. When examining the serum concentration of each mediator, we observed a significant difference in GM-CSF and IL5, along with observable, but not statistically significant, differences in protein levels of G-CSF, CXCL2, and TNFα. In fibrotic lesions, GM-CSF influences the abundance of leukocytes including neutrophils and monocytes,10,27 and thus, we next examined the influx of these cells into the colonic LP of WT and Nr1i2-/- mice during healing.
We chose to examine the colonic LP at day 15 after the induction of DSS injury to assess a time point with established immune cell influx that was beyond the acute inflammatory phase, but when the colon was in an actively healing state.28, 29, 30 Cells were first fractioned on the basis of the expression of cell surface markers into CD45+ myeloid cells that lacked the expression of MHCII (CD45+MHCII-) (Figure 4). From this population of non–antigen-presenting myeloid cells, we further isolated CD11b+ cells, which were then fractioned into Ly6G+ neutrophils (CD45+MHCII-CD11b+Ly6G+) and Ly6CHi inflammatory monocytes (CD45+MHCII-CD11b+Ly6CHi).31,32 Compared with their naive mice, both DSS-treated WT and Nr1i2-/- mice had a significant increase in Ly6G+ neutrophils and Ly6CHi inflammatory monocytes into the colonic LP on day 15 after the induction of colonic injury. Importantly, when compared with WT mice after DSS, Nr1i2-/- mice had a significantly higher percentage and number of neutrophils in the colonic LP (Figure 1G). However, there was no difference in the influx of Ly6CHi inflammatory monocytes between WT and Nr1i2-/- mice. Together these data suggest a more prolonged inflammatory response in Nr1i2-/- mice after DSS-induced injury, with elevated levels of innate cytokines including GM-CSF and a greater number of recruited neutrophils.
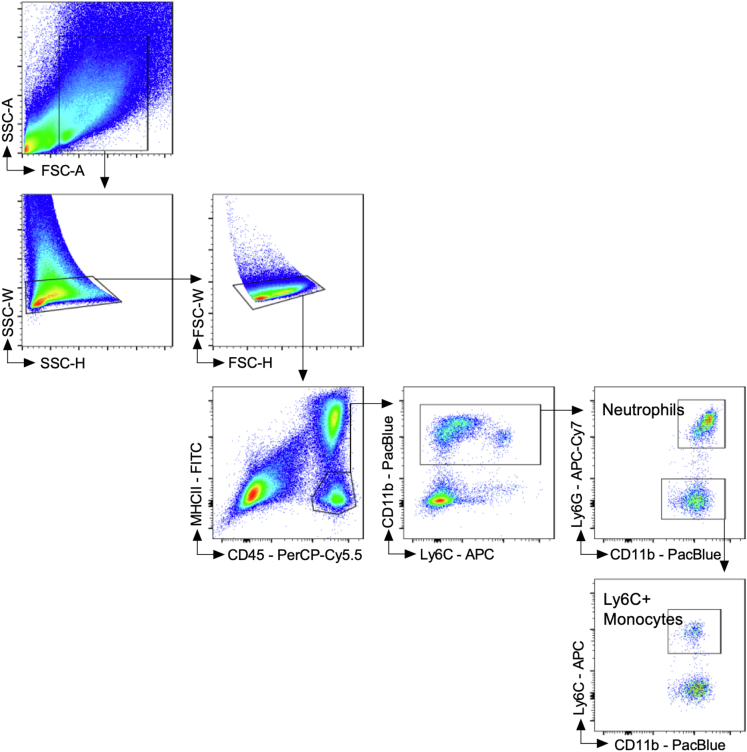
Figure 4.
Gating strategy for immune cells infiltrating the colonic lamina propria. Cells from the digested colonic LP were first discriminated as singlets based on side scatter (SSC-W vs SSC-H) and forward scatter (FSC-W vs FSC-H). Single cells were then fractioned on the basis of the expression of cell surface markers CD45 and MHCII, with CD45+MHCII- being non–antigen-presenting myeloid cells. From this population, MHCII- myeloid cells were further separated into CD11b+ cells, which were then fractioned into Ly6G+ neutrophils (CD45+MHCII-CD11b+Ly6G+). Remaining CD11b+ cells that were Ly6G- were assessed for Ly6C expression with inflammatory monocytes being Ly6CHi (CD45+MHCII-CD11b+Ly6CHi). The example used for gating is from WT C57Bl/6 mouse after 5 days of 3% DSS, followed by a switch to regular drinking water and healing for 10 days (15 days total). The fluorochrome for each cell surface marker is indicated, and each clone used is outlined in the Methods. Data were acquired using the BD FACS Canto cytometer.
Pharmacologic Activation of the PXR Suppresses Intestinal Fibrosis After Injury
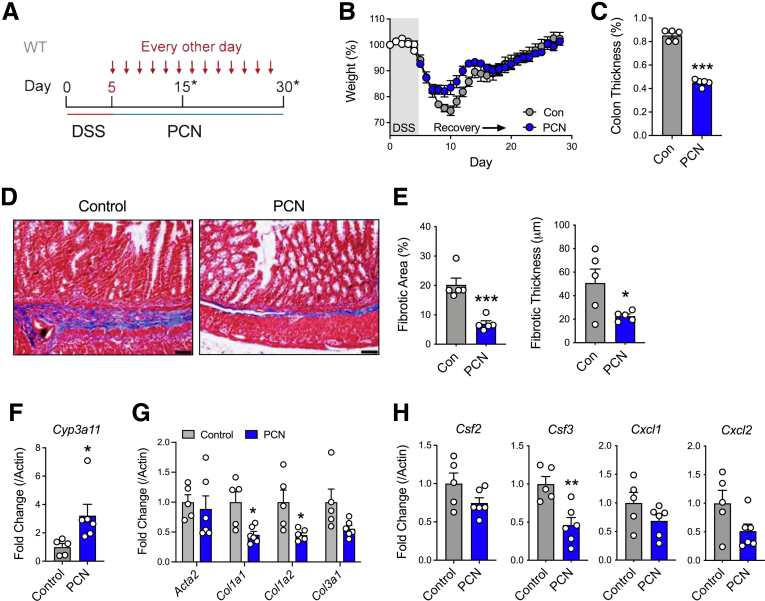
Because of the role of the PXR in regulating the fibrotic response, we tested the ability of PXR activation to mitigate inflammation and fibrosis in the healing colon after DSS-induced injury. To activate the PXR in vivo, we used the selective mouse agonist pregnenolone 16α-carbonitrile (PCN) (administered for the duration of the healing process of 25 days; Figure 5A). Following this protocol, we observed that PCN treatment decreased gross colon thickness, when compared with vehicle-treated mice (Figure 5C). In line with decreased colon thickness, we also observed significantly decreased fibrotic area and thickness in the submucosal area in the colon of PCN-treated mice (Figure 5D and E). In colonic tissue, PCN significantly increased expression of the PXR target gene cytochrome P450, family 3, subfamily a, polypeptide 11 (Cyp3a11), confirming in vivo activation of the PXR (Figure 5F). Activation of the PXR in the healing colon was accompanied by decreased expression of collagen, type I, alpha 1 (Col1a1) and collagen, type I, alpha 2 (Col1a2), as well as an apparent decrease in collagen, type I, alpha 3 (Col3a1) expression that did not reach statistical significance (Figure 5G). Compared with the control group, PCN treatment also significantly decreased colonic mRNA expression of colony stimulating factor 3 (Csf3) and decreased colony stimulating factor 2 (Csf2) and the neutrophil chemokines C-X-C Motif chemokine ligand 1 (Cxcl1) and C-X-C Motif chemokine ligand 2 (Cxcl2), although these were not statically significant (Figure 5H). These data suggest that pharmacologic activation of the PXR can prevent fibrosis development in the healing colon, while modulating some of the mediators that are associated with exaggerated fibrogenesis in Nr1i2-/- mice.
Figure 5.
Pharmacologic activation of the PXR attenuates the development of fibrosis after colonic injury. (A) WT mice were administered DSS for 5 days to induce colonic injury and then switched to regular drinking water and injected interperitoneally with the mouse-specific PXR agonist pregnenolone-15-alpha carbonitrile (PCN) every other day for 25 days. (B) Weight recovery and (C) gross colonic length and thickness in WT(Nr1i2+/+) and Nr1i2-/- mice 25 days after withdrawal of DSS. (D) Representative images of Masson’s trichrome staining and (E) quantification of fibrosis in the colon after injury and subsequent treatment with PCN or vehicle (n = 5–6 per group; scale bar = 100 μm). The expression of (F) the PXR target gene Cyp3a11, (G) fibrotic genes Acta2, Col1a1, Col1a2, and Col3a1, and (H) innate immune genes Csf2, Csf3, Cxcl1, and Cxcl2 in the healing colon of mice (10 days withdrawal of DSS; n = 5–6 per group). Data are represented as mean ± standard error of the mean. Student t test (C, E, and F) and one-way analysis of variance with Tukey post hoc test (H). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Exaggerated Inflammation and Fibrosis in Nr1i2-/- Mice Originates From the Mesenchymal Compartment
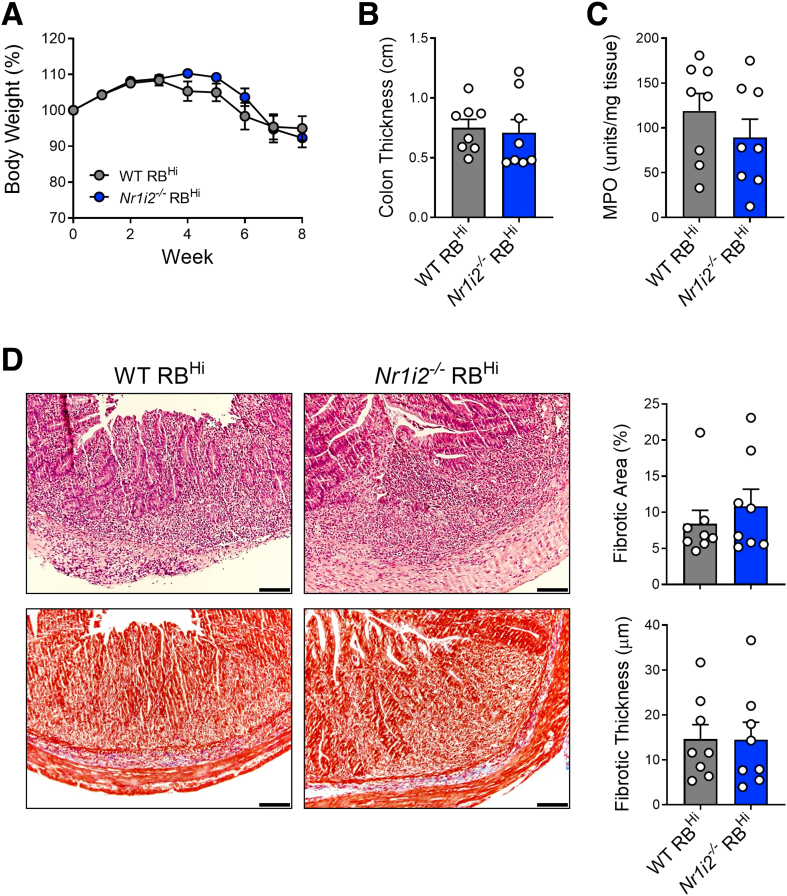
Next, we aimed to identify the cellular compartment responsible for prolonged inflammation and exaggerated fibrosis in Nr1i2-/- mice. Because the PXR has been implicated in T-cell driven fibrosis,25 we used a model of T-cell driven colitis where naive CD4+CD25-RBhi T cells were sorted from WT and Nr1i2-/- mice and transferred into lymphocyte-deficient Rag1-/- mice. Eight weeks after the transfer of either WT and Nr1i2-/- naive CD4+CD25-RBhi T cells, mice displayed significant disease as evidenced by weight loss (∼15%) and colonic inflammation upon histologic examination; however, there was no difference between the inflammation score or macroscopic thickness depth of colon. Furthermore, the degree of fibrosis detected with Masson’s trichrome stain was no different between both groups of mice (Figure 6).
Figure 6.
Loss of the PXR in T cells does not increase the severity of disease or intestinal fibrosis in a chronic model of T-cell driven colitis. (A) Weight loss over the course of 8 weeks after transfer of sorted naive CD4+CD25-CD45RbHi T cells from WT(Nr1i2+/+) and Nr1i2-/- mice into C57BL/6 Rag1-/- mice via intraperitoneal injection. (B) Gross colon thickness and (C) myeloperoxidase (MPO) activity in the colon of Rag1-/- mice 8 weeks after injection of WT or Nr1i2-/- CD4+CD25-CD45RbHi T cells. (D) Masson’s trichrome staining of colonic sections with associated quantification of total fibrotic area and fibrotic thickness in Rag1-/- mice 8 weeks after transfer of WT(Nr1i2+/+) and Nr1i2-/- RbHi naive T cells. (n = 8 per group; scale bar = 100 μm). Data are presented as mean ± standard error of the mean. Student t test.
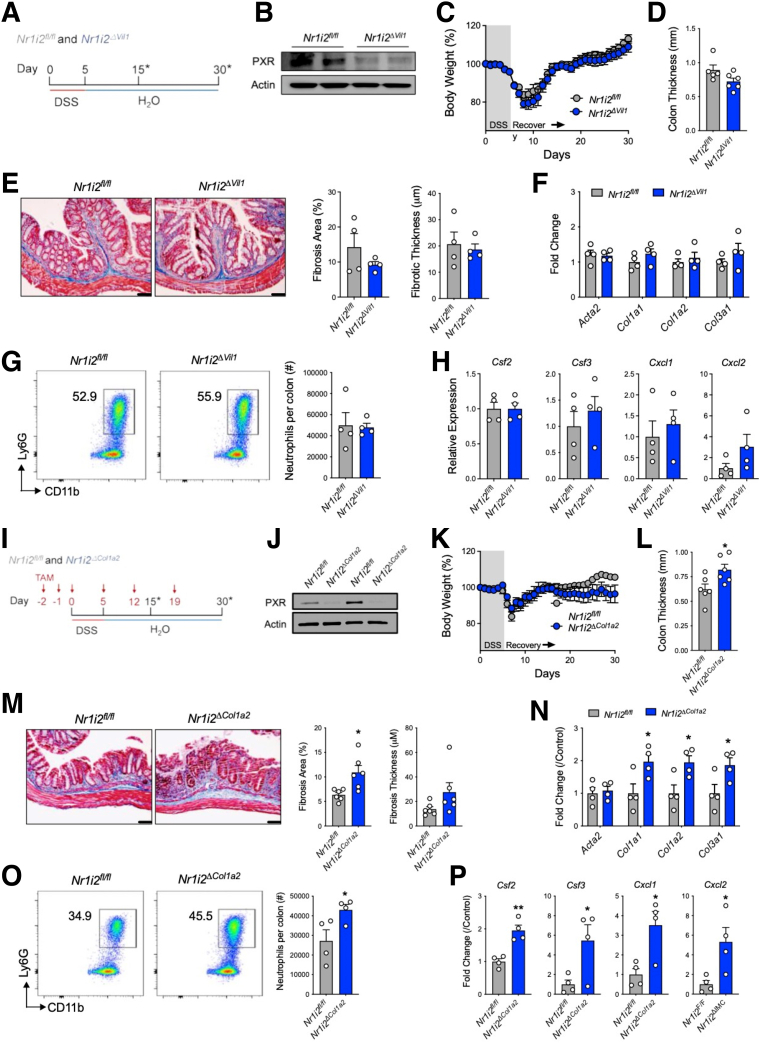
To assess the contribution of the PXR in the epithelium, we used Villin (Vil1)-Cre mice that restrict spontaneous cre-mediated recombination to all cells of epithelial origin. These mice were crossed to Nr1i2-floxed mice to delete Nr1i2 in intestinal epithelial cells (termed Nr1i2ΔVil1) and administered DSS for 5 days and then switched to normal water and allowed to heal for an additional 25 days (Figure 7A). Throughout the course of disease, we did not observe significant differences in weight loss, and after healing there was no difference in gross colon thickness between Nr1i2ΔVil1 mice and their cre-negative Nr1i2-floxed (Nr1i2fl/fl) counterparts (Figure 7C and D). Examination of Masson’s trichrome stained colon sections did not reveal significant difference in collagen deposition (Figure 7E), which coincided with no observable differences in the expression of collagen genes (Figure 7F). In addition, the number of neutrophils recruited into the healing colon of Nr1i2fl/fl and Nr1i2ΔVil1 mice did not differ (Figure 7G), and the expression of innate cytokines or chemokines did not, except for increased but non-significant expression of colonic Cxcl2 (Figure 7H).
Figure 7.
Deletion of Nr1i2 in the mesenchymal compartment, but not epithelial compartment, enhances fibrotic responses in the colon after injury. (A) Experimental outline of Vil1-Cre:Nr1i2fl/fl (Nr1i2ΔVil1) and Nr1i2fl/fl (Cre-) control mice administered DSS for 5 days and then allowed to heal for 25 days (∗indicates the different analysis time points). (B) Western blot analysis of PXR protein in freshly isolated colonic intestinal epithelial cells. (C) Weight recovery and (D) gross colonic length and thickness in Nr1i2ΔVil1and Nr1i2fl/f mice 25 days after withdrawal of DSS. (E) Representative images of Masson’s trichrome staining and quantification of fibrosis in the colon after injury in Nr1i2ΔVil1and Nr1i2fl/fl control mice (n = 4 per group). The expression of (F) fibrotic genes Acta2, Col1a1, Col1a2, and Col3a1 and (G) colonic lamina propria neutrophil infiltration and (H) innate immune gene expression in the healing colonic lamina propria of Nr1i2ΔVil1 and Nr1i2fl/fl mice 10 days after withdrawal of DSS (n = 4 per group). (I) Experimental outline of tamoxifen administration to Col1a2-CreERT:Nr1i2fl/fl (Nr1i2ΔCol1) and Nr1i2fl/fl (Cre-) control mice that were then given DSS for 5 days and then allowed to heal for 25 days while receiving tamoxifen throughout experiment as indicated. (J) Western blot analysis of PXR protein in freshly isolated colonic fibroblasts. (K) Weight recovery and (L) gross colonic length and thickness in Nr1i2ΔCol1a2 and Nr1i2fl/f mice 25 days after withdrawal of DSS. (M) Representative images of Masson’s trichrome staining and quantification of fibrosis in the colon after injury in Nr1i2ΔCol1a2 and Nr1i2fl/fl control mice (n = 6 per group). The expression of (N) fibrotic genes Acta2, Col1a1, Col1a2, and Col3a1 and (O) colonic lamina propria neutrophil infiltration and (P) innate immune gene expression in the healing colonic lamina propria of Nr1i2ΔCol1a2 and Nr1i2fl/fl mice 10 days after withdrawal of DSS (n = 4 per group). Data are presented as mean ± standard error of the mean. Student t test (C–G, K–O). ∗P < .05,∗∗P < .01,∗∗∗P < .001.
Because of our results in Nr1i2ΔVil1 mice, we next targeted the mesenchymal compartment by using tamoxifen-inducible Col1a2Cre-ERT2 mice that restrict cre-mediated recombination to Col1a2-expressing cells in the intestine.33 Col1a2 is expressed in cells in the mesenchymal compartment including the mesenchymal stem cell niche,34,35 and thus our model targeted the mesenchymal compartment. To examine the effect of PXR deficiency in the mesenchymal compartment during intestinal healing, we crossed conditional Nr1i2fl/fl mice with Col1a2Cre-ERT2 mice (termed Nr1i2ΔCol1a2). These mice were administered tamoxifen on days –2, –1, and day 0 when DSS administration was started. Mice also received injections of tamoxifen on day 5 of DSS when mice were switched to normal drinking water and subsequent tamoxifen injections weekly during the healing phase until day 30 (Figure 7I). Western blot analysis of freshly isolated fibroblasts from tamoxifen-treated Nr1i2ΔCol1a2 mice confirmed knockdown of PXR (Figure 7J). Weight loss during the induction of injury with DSS did not differ between Nr1i2fl/fl and Nr1i2ΔCol1a2 mice; however, Nr1i2ΔCol1a2 exhibited a tendency toward attenuated weight gain when allowed to heal for 25 days compared with Nr1i2fl/fl mice (Figure 7K). After healing, the colon of Nr1i2ΔCol1a2 mice was significantly thicker than Nr1i2fl/fl control mice, and examination of Masson’s trichrome stained colon sections showed significantly larger areas of collagen deposition in Nr1i2ΔCol1a2 mice (Figure 7M). Increased levels of fibrosis in Nr1i2ΔCol1a2 mice coincided with significantly increased expression of Col1a1, Col1a2, and Col3a1 (Figure 7N) and a higher number of neutrophils infiltrating the colon (Figure 7O). In addition, the expression of the innate cytokines Csf2, Csf3, Cxcl1, and Cxcl2 were significantly higher in the healing colon of Nr1i2ΔCol1a2 than in counterpart Nr1i2fl/fl mice (Figure 7P). Together, these data suggest that deletion of Nr1i2 in the mesenchymal compartment recapitulates the phenotype of increased inflammation and fibrosis observed in Nr1i2-/- mice, and that PXR signaling in mesenchymal cells may contribute to returning the colon to homeostasis in part by preventing the development of fibrosis after injury.
Intestinal PXR Signaling Restrains the Proinflammatory Potential of Myofibroblasts
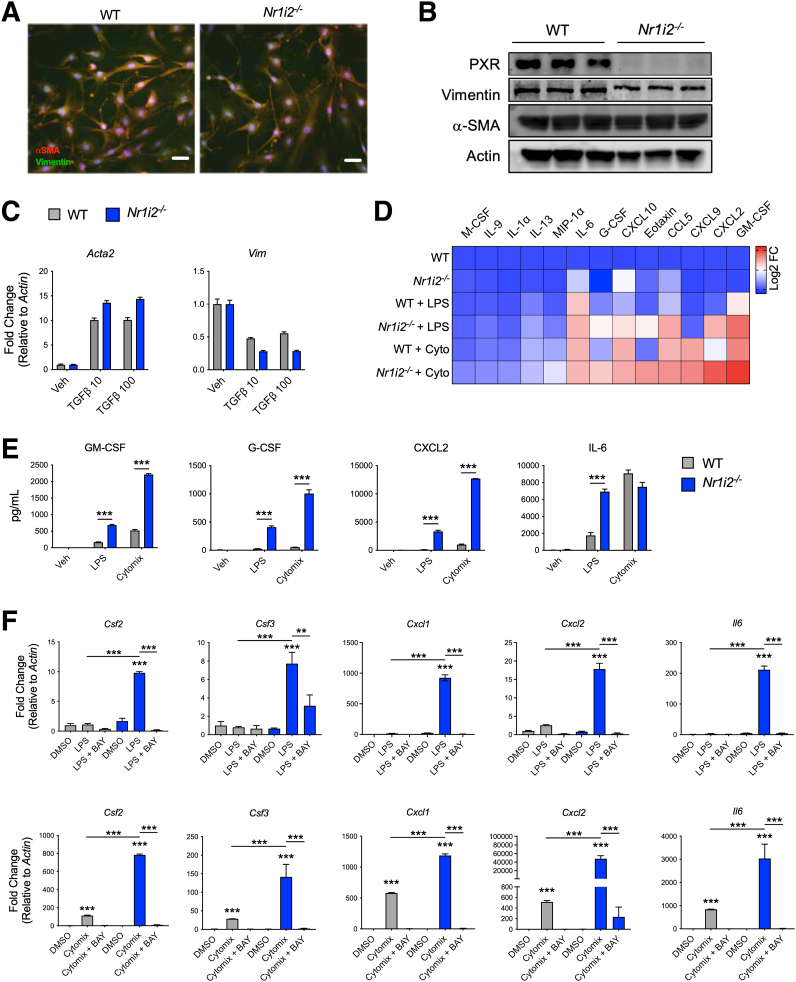
To further understand how loss of the PXR in the mesenchymal compartment drives fibrotic responses, myofibroblasts, a major cell type of the mesenchyme, were isolated from the colon of WT and Nr1i2-/- mice. An initial analysis of colonic myofibroblasts isolated from WT and Nr1i2-/- mice showed no difference morphology (Figure 8A) and expressed similar protein levels of the myofibroblast markers αSMA and vimentin (Figure 8B). Transforming growth factor beta 1 (TGFβ1) is a central regulator of fibroblast activation in different fibrotic diseases36; therefore, we first focused on responses of myofibroblasts in this context. After 24 hours of TGFβ1 stimulation, expression of the activation markers Acta2 (αSMA) and Vim were increased in both WT and Nr1i2-/- myofibroblasts, but there was no observable difference between the 2 groups of cells, suggesting that TGFβ1 responsiveness does not play a strong role in driven fibrotic responses in the absence of the PXR.
Figure 8.
Loss of intestinal PXR signaling imparts increased inflammatory reactivity in intestinal myofibroblasts. (A) Staining of αSMA and vimentin in primary myofibroblasts isolated from the colon of WT(Nr1i2+/+) and Nr1i2-/- mice to show fibroblastic morphology (scale bar = 250 μm). (B) Western blots demonstrating protein levels of PXR, αSMA, and vimentin in myofibroblasts isolated from the colon of WT(Nr1i2+/+) and Nr1i2-/- mice. (C) Responsiveness of colonic myofibroblasts from WT(Nr1i2+/+) and Nr1i2-/- mice to TGFβ (reduced serum conditions for 24 hours and then stimulated with TGFβ1 for 24 hours) measured by mRNA expression for Acta2 (αSMA) and Vim (n = 3 per group). (D) Heatmap showing the most robustly up-regulated cytokines in cell supernatants from colonic myofibroblastss isolated from WT(Nr1i2+/+) and Nr1i2-/- mice after stimulation with LPS or cytomix (TNFα, IL1β, and IFNγ) for 24 hours (each tile is the average of 4 replicates). (E) Quantification of total cytokine levels in respective myofibroblasts groups after 24 hours of stimulation with cytomix or LPS (n = 4 per group). (F) Gene expression of different cytokines in colonic myofibroblasts isolated from WT(Nr1i2+/+) and Nr1i2-/- mice and pretreated with vehicle (DMSO) or the NF-κB kinase inhibitor BAY 11-7082 (BAY) for 4 hours and then stimulated with LPS or cytomix for additional 12 hours (n = 3 per group). Data are presented as mean ± standard error of the mean. One-way analysis of variance with Tukey post hoc test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Because of the observed differences in cytokine responses in the colons of WT and Nr1i2-/- mice, we next assessed the inflammatory potential of intestinal myofibroblasts. Both cytokines and bacterial products, 2 prominent inflammatory stimuli found in the damaged intestine, can trigger cytokine and chemokines production from myofibroblasts.9,37 Thus, we stimulated primary intestinal myofibroblasts from the colon of WT and Nr1i2-/- mice with LPS or cytomix (combination of TNFα, IL1β, and IFNγ) and assessed the culture supernatants for inflammatory mediator production. The heatmap in Figure 8D shows that LPS triggered increased expression of various cytokines/chemokines, but these responses were exaggerated in Nr1i2-/- fibroblasts. Similar results were also observed with cytomix; however, the magnitude of difference between WT and Nr1i2-/- myofibroblasts was even more evident. The cytokines/chemokines that were most highly produced by myofibroblasts and that differed between WT and Nr1i2-/- cells included GM-CSF, CXCL2, MIG, Eotaxin, IL6, IL9, and G-CSF, respectively (Figure 8D). The concentration of cytokines of interest including GM-CSF, G-CSF, and CXCL2 were all produced to much larger degree in Nr1i2-/- myofibroblasts compared with WT cells, when triggered by the same stimuli (LPS or cytomix; Figure 8E). Interestingly, IL6 production was not different between strains after cytomix stimulation, but it was significantly increased in Nr1i2-/- myofibroblasts compared with WT cells after stimulation with LPS (Figure 8E).
Hyperactivity in Myofibroblasts Is Attenuated by Nuclear Factor Kappa B Inhibition
Various reports have described a reciprocal interaction between the PXR and nuclear factor kappa B (NF-κB) that dampens inflammatory signaling.38 Thus, PXR activation can decrease NF-κB–dependent cytokine production, whereas deletion of PXR leads to exaggerated NF-κB activity, increased inflammatory gene expression, and tissue inflammation. To this end, we sought to determine whether inhibition of NF-κB signaling could normalize the hyperinflammatory phenotype observed in Nr1i2-/- myofibroblasts. To test this, WT and Nr1i2-/- myofibroblasts were pretreated with the NF-κB inhibitor BAY 11-7082 for 6 hours, followed by stimulation with LPS or cytomix with or without BAY 11-7082 for 12 hours. As with protein expression, mRNA expression for the inflammatory cytokines Csf2, Csf3, Cxcl1, Cxcl2, and Il6 was induced in myofibroblasts by LPS and cytomix, but to a much greater extent in Nr1i2-/- myofibroblasts than in WT cells. The hyperresponsiveness of Nr1i2-/- myofibroblasts was almost completely abolished by pretreating cells with the selective NF-κB inhibitor BAY 11-7082 (Figure 8F). These data suggest that the loss of PXR signaling leads to unrestrained NF-κB activity and enhanced cytokine expression and production from myofibroblasts.
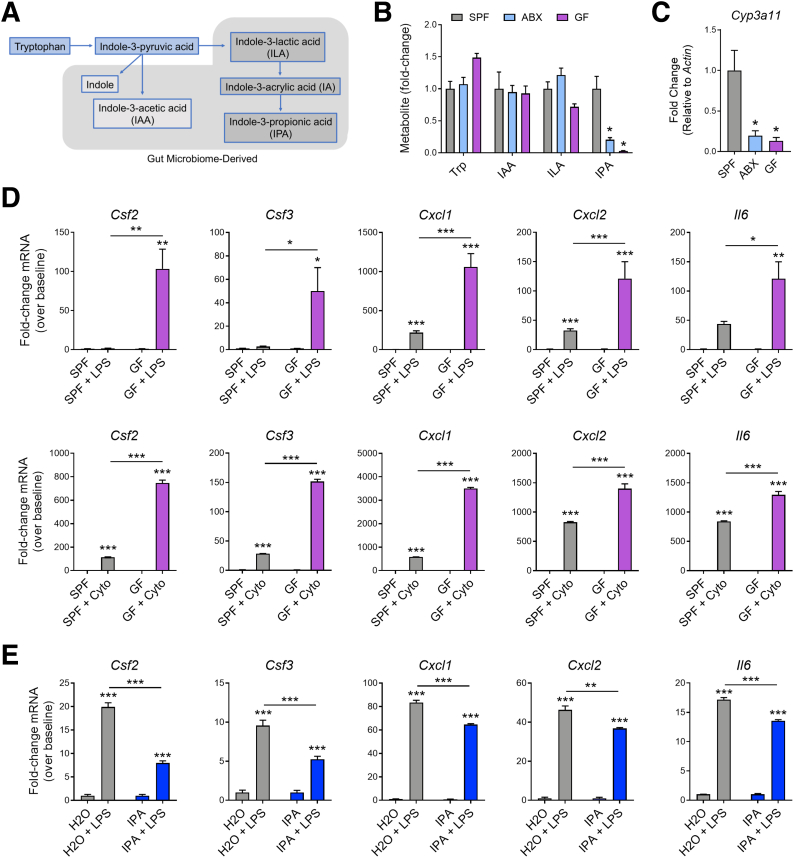
The Microbiota Regulates Systemic IPA Levels and the Proinflammatory Capacity of Intestinal Fibroblasts
The commensal microbiota is a major contributor to systemic IPA levels in the host through catabolic pathways that metabolize tryptophan into bioactive indole metabolites (Figure 9A). In our studies, antibiotic (ABX)-treated and germ-free (GF) mice had significantly depleted levels of IPA compared with specific pathogen–free (SPF) controls (∼80% IPA reduction in ABX-treated mice and ∼97% reduction in GF mice; Figure 9B). Levels of tryptophan, as well as the other tryptophan metabolites indole-3-acetic acid and indole-3-lactic acid, in the serum did not significantly change with ABX treatment or in GF mice (Figure 9B). Further examination also revealed greatly reduced expression of the PXR target gene Cyp3a11 in the colons of ABX-treated and GF mice, which is consistent with previous findings in other parts of the gastrointestinal tract22 (Figure 9C). Remarkably, in a similar fashion to Nr1i2-/- mice, myofibroblasts isolated from the colon of GF mice had a much greater amplitude of inflammatory induction compared with cells isolated from SPF mice. To this extent, compared with baseline, stimulation with LPS or cytomix resulted in significantly greater expression of Csf2, Csf3, Cxcl1, Cxcl2, and Il6 in GF colonic myofibroblasts compared with SPF control cells (Figure 9D).
Figure 9.
The microbiome regulates systemic levels of indole-3-propionic acid (IPA), colonic PXR signaling, and responsiveness of myofibroblasts to inflammatory stimuli. (A) Schematic showing the conversion pathway of tryptophan into different bioactive indole metabolites including those that require the gut microbiota for conversion. (B) Serum levels of tryptophan (trp) and the tryptophan metabolites indole-3-acetic acid (IAA), indole-3-lactic acid (ILA), and indole-3-propionic acid (IPA) in specific pathogen–free (SPF), antibiotic-administered (ABX), and germ-free (GF) mice (expressed as fold-change relative to SPF control; n = 4–5 per group). (C) Expression of the PXR target gene Cyp3a11 as measured by quantitative polymerase chain reaction in the colon of SPF, ABX, and GF mice (n = 4–8 per group). (D) Gene expression of inflammatory mediators in myofibroblasts isolated from the colon of SPF and GF mice and stimulated with LPS or cytomix (cyto) for 12 hours (n = 3 per group). (E) Gene expression of inflammatory mediators in myofibroblasts isolated from the colon of WT mice administered normal drinking water or drinking water supplemented with IPA (pH matched) for 2 weeks and then stimulated with LPS for 12 hours (n = 3 per group). One-way analysis of variance with Tukey post hoc test (B–E). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Because of the hyperresponsiveness of myofibroblasts isolated from GF mice, we next tested whether IPA itself could modulate inflammatory mediator production. We administered normal drinking water or water supplemented with IPA (250 mg/L; pH-matched) to mice for 2 weeks. Compared with cells isolated from mice on normal drinking water, colonic myofibroblasts isolated from mice on IPA-supplemented water had significantly lower expression of mRNA for Csf2, Csf3, Cxcl1, Cxcl2, and Il6 after stimulation with LPS (Figure 9E).
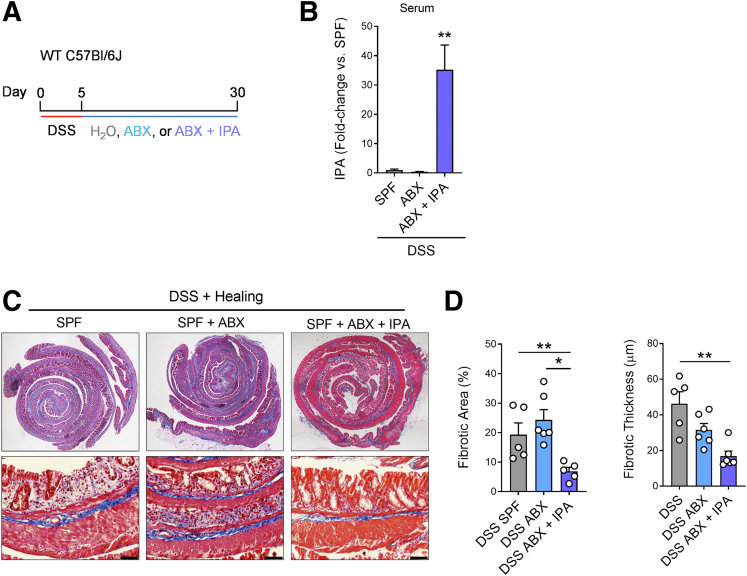
To capture the fibrotic manifestations of these changes to myofibroblasts in an in vivo setting and assess whether IPA was involved in modulating fibrotic processes in the colon, after a 5-day course of DSS to prompt colonic injury, mice were administered normal drinking water, ABX water, or ABX water supplemented with IPA while healing for 25 days (Figure 10A). This protocol significantly increased the circulating levels of IPA when co-administered in the drinking water to ABX-treated mice (Figure 10B). IPA supplementation in ABX-treated mice prevented the development of fibrosis in the colons of mice, which had significantly lower fibrotic area and thinner fibrotic lesions compared with SPF mice and mice administered ABX during healing (Figure 10C). To our surprise, SPF and ABX-treated mice did not differ in the severity of colonic fibrosis. However, we observed that the pattern of fibrosis in the healed colon of ABX-treated mice appeared different than in SPF mice, with a thinner epithelial lining that was predominantly composed of fibrotic tissue (Figure 10C). Thus, it appears that in the absence of luminal microbes to potentiate inflammation, ABX-induced bacterial depletion after intestinal injury creates an environment that still lends to fibrosis development. Importantly, IPA administration alone was enough to prevent fibrosis in this setting (Figure 10C and D).
Figure 10.
The effect of microbiota depletion on the development of fibrosis in the intestine after injury. (A) Experimental outline demonstrating the course of DSS (3%) administration for 5 days, followed by a switch to H2O, H2O supplemented with antibiotic cocktail (ABX), or H2O supplemented ABX and indole-3-propionic acid (IPA) (∗indicate the different analysis time points). (B) Circulating serum levels of IPA in mice after oral administration of IPA (expressed as fold-change compared with WT control mice; n = 3–6 per group). (C) Representative Masson’s trichrome staining of the colon with (D) associated analysis of microscopic fibrotic area and fibrotic thickness in mice receiving H2O or IPA water for 25 days after induction of colitis with DSS. (n = 5–6 per group; scale bar = 200 mm). Data are presented as mean ± standard error of the mean. One-way analysis of variance with Tukey post hoc test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
The Microbial Metabolite IPA Dampens Inflammation and Intestinal Fibrosis After Injury
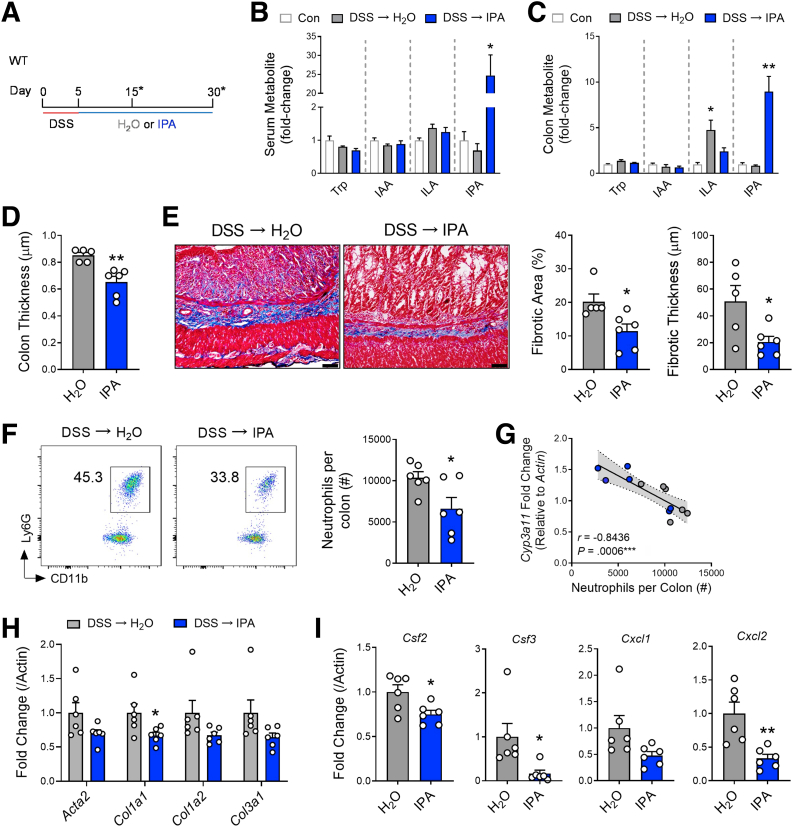
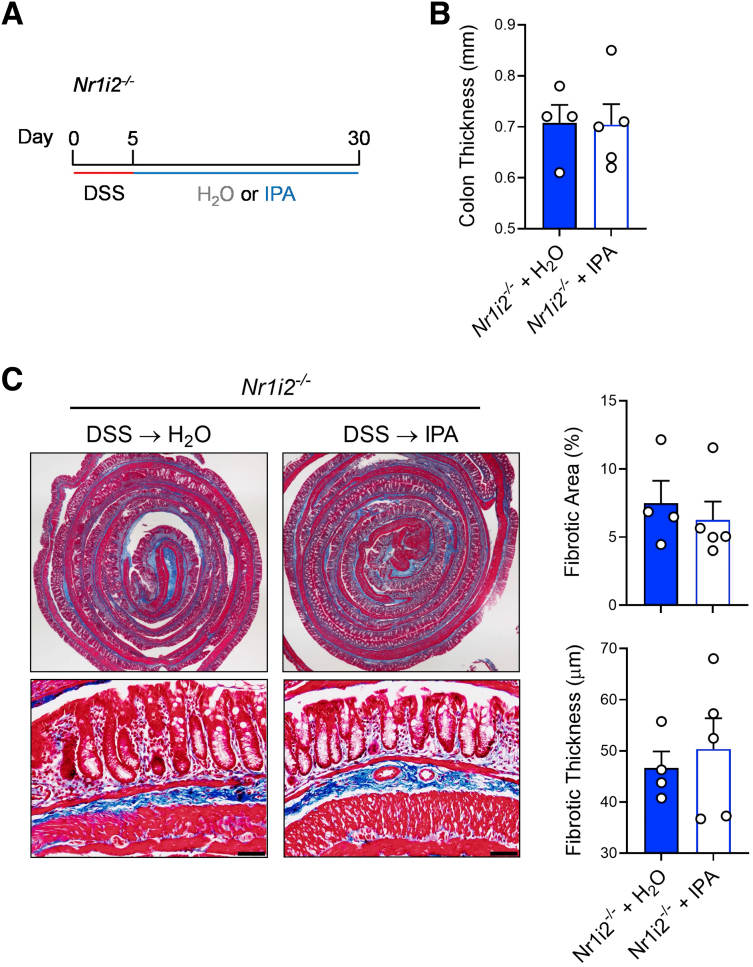
Next, we tested the ability of IPA to influence inflammation and fibrosis in the healing colons of SPF mice after DSS-induced injury (Figure 11A). Administration of IPA (250 mg/L) in the drinking water of mice after 5 days of DSS significantly increased serum levels of IPA and, importantly, increased the levels of IPA in colonic tissue (Figure 11B and C). IPA administration did not affect the serum or colonic levels of tryptophan or the tryptophan metabolites in the same catabolic pathways as IPA (indole-3-acetic acid and indole-3-lactic acid). Interestingly, indole-3-lactic acid was increased in colonic tissue as DSS-induced injury healed, but this was blunted after IPA administration. When compared with SPF mice that received normal, pH-matched drinking water while recovering from DSS-induced injury, SPF mice that received IPA had significantly reduced colonic thickness (Figure 11D). Further histologic examination showed that IPA administration significantly reduced total fibrotic area and fibrotic thickness in the colon, when compared with SPF mice receiving normal drinking water (Figure 11E). Notably, IPA failed to reduce gross colon thickness, as well as total fibrotic area and fibrotic thickness, when administered to Nr1i2-/- mice (Figure 12).
Figure 11.
Oral administration of microbial metabolite indole-3-propionic acid (IPA) prevents development of inflammation and fibrosis in the colon after DSS-induced injury. (A) Experimental outline demonstrating the course of DSS (3%) administration for 5 days, followed by a switch to either H2O or IPA dissolved in pH matched H2O (∗indicates the different analysis time points). (B and C) Circulating serum and colonic levels of different tryptophan metabolites (trp, tryptophan; IAA, indole-3-acetic acid; ILA, indole-3-lactic acid; IPA, indole-3-propionic acid) in mice after oral administration of IPA (expressed as fold-change compared with WT control mice; n = 3–6 per group). (D) Macroscopic colon thickness and (E) representative Masson’s trichrome staining of the colon with associated analysis of microscopic fibrotic area and fibrotic thickness in mice receiving H2O or IPA water for 25 days after induction of colitis with DSS (n = 5–6 per group; scale bar = 100 μm). (F) Representative FACs plots with associated numbers of neutrophils infiltrating into the lamina propria of the healing colon (day 15) of mice receiving either H2O or IPA water (n = 6 per group). (G) Colonic gene expression of the PXR target gene Cyp3a11 correlated to the number of neutrophils infiltrating the colon of mice and colonic gene expression of (H) fibrotic factors and (I) inflammatory cytokines in the healing colon of mice receiving either H2O or IPA water. Data are presented as mean ± standard error of the mean. Student t test (D–F, H, and I) and one-way analysis of variance with Tukey post hoc test (B and C). ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
Figure 12.
IPA administration fails to prevent development of fibrosis in Nr1i2-/-mice. (A) Experimental outline demonstrating the length of DSS administration, followed by a switch to either H2O or IPA dissolved in H2O (pH matched) during healing for 25 days (30 days total). (B) Gross colon thickness and (C) representative Masson’s trichrome staining in the colon with associated analysis of microscopic fibrotic area and fibrotic thickness (n = 4–5 per group; scale bar = 100 μm). Data are presented as mean ± standard error of the mean. Student t test.
When assessing inflammation, we observed that IPA significantly reduced the number of infiltrating neutrophils when compared with mice receiving normal drinking water (Figure 11F). Importantly, neutrophil numbers in the healing colon negatively correlated with colonic mRNA expression of the PXR target gene Cyp3a11 (Figure 11G) and coincided with significantly reduced expression of Col1a1 and a trend toward lower levels of Col1a2 and Col3a1 expression (Figure 11H). IPA also reduced the expression of inflammatory genes Csf2, Csf3, and the neutrophil chemokines Cxcl1 and Cxcl2 (Figure 11I). Together, these data highlight the ability of orally administered IPA to dampen fibrotic and inflammatory responses after colonic injury.
PXR Signaling Alleviates Proteolytic Activity in the Colon After Injury
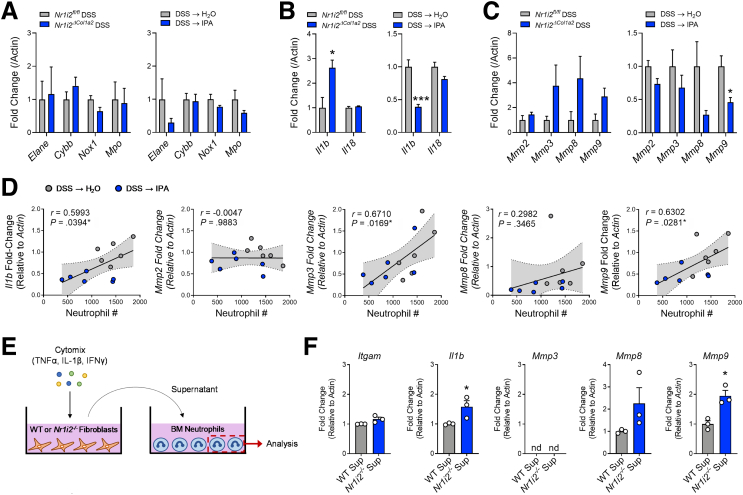
Next, we sought to determine how the loss of PXR signaling and neutrophilic inflammation could culminate in fibrotic activity in the colon. We first focused on common neutrophil antimicrobial enzymes known to lend to tissue destruction and fibrosis.37,39 Examination of mRNA expression for Elane (neutrophil elastase), Cybb (NADPH oxidase 2), Nox1 (NADPH oxidase 1), and Mpo (myeloperoxidase) found no difference in transcript levels in the healing colon between mice with mesenchymal specific deletion of Nr1i2 (Nr1i2ΔCol1a2) and their control counterparts (Nr1i2fl/fl; Figure 13A). Furthermore, IPA administration had no effect on these transcripts (Figure 13A). We next assessed IL1-related cytokines, which are linked to neutrophil function and the promotion of fibrotic responses.37 Colonic mRNA levels of Il1b and Il18 were significantly higher when Nr1i2 was deleted in the mesenchymal compartment (Figure 13B). IPA administration also significantly reduced colonic expression of Il1b.
Figure 13.
Defective responses in myofibroblasts isolated from the colon of Nr1i2-/-mice impart greater proteolytic activity of neutrophils. (A) Expression of genes related to neutrophil antimicrobial function. (B) IL1 family cytokines and (C) matrix metalloproteinases (MMPs) in the healing colon of mice with deletion of Nr1i2 in the mesenchymal compartment (Nr1i2fl/fl vs Nr1i2ΔCol1a2) or mice administered IPA after induction of colonic injury (n = 4 and n = 6 per group per respective experiment). (D) Pearson’s correlation of colonic expression of Il1b and MMP genes against the level of neutrophil infiltration into the colon of mice administered either H2O or IPA water after induction of colonic injury. (E) Experimental schematic and (F) gene expression in bone marrow–derived neutrophils treated with supernatant from cytokine-stimulated colonic myofibroblasts isolated from WT or Nr1i2-/ mice (n = 3 per group). Data are presented as mean ± standard error of the mean. Student t test (A–C, F). ∗P < .05, ∗∗P < .01, ∗∗∗P < .005; nd = no detection.
Finally, we examined the expression of different MMP genes, because these enzymes can be produced by neutrophils and fibroblasts40, 41, 42 and have a role in directly impacting connective tissue organization and fibrotic processes in the intestine.43 The expression of Mmp2, Mmp3, Mmp8, and Mmp9 is reported to be profibrotic in the intestine and elevated in IBD.41,42 Of these MMPs, transcripts for Mmp3, Mmp8, and Mmp9 were elevated in the healing colon of mice with mesenchymal specific deletion of Nr1i2 (Nr1i2ΔCol1a2) and compared with their control counterparts (Nr1i2fl/fl; Figure 13C). Furthermore, IPA administration reduced the expression of MMPs, with a significant decrease in Mmp9. When correlated to neutrophil numbers in the healing colon, transcripts showing significant positive correlation (Pearson’s) were Mmp3, Mmp9, and Il1b in that respective order (Figure 13D). These data are in line with previous reports indicating that neutrophils directly produce Il1b, MMP8, and MMP9.41,42
The Ability of Myofibroblasts to Impart Proteolytic Activity on Neutrophils Is Heightened by Loss of PXR Signaling
To address whether the overexuberant responses observed in Nr1i2-/- myofibroblasts could influence the proteolytic behavior of neutrophils, we treated freshly isolated bone marrow–derived neutrophils from WT mice with the supernatant from WT or Nr1i2-/- colonic myofibroblasts after stimulation with cytomix (IL1β, IFNγ, TNFα; Figure 13E). When comparing the supernatant from WT and Nr1i2-/- myofibroblasts, there was no difference in the ability to alter expression of the adhesion molecule CD11b (Igtam); however, supernatant from Nr1i2-/- cells significantly increased the expression of Il1b in bone marrow neutrophils. Importantly, although Mmp3 expression was not detected in naive bone marrow neutrophils before or after treatment, the supernatant from Nr1i2-/- myofibroblasts increased the expression of Mmp8 and Mmp9 in bone marrow neutrophils to a greater extent than WT supernatant (Figure 13F). These results suggest that the overexuberant responses of myofibroblasts in the absence of PXR signaling impart greater pro-fibrotic activity in neutrophils and provide a link between increased neutrophil influx and enhanced fibrotic remodeling in the absence of the PXR.
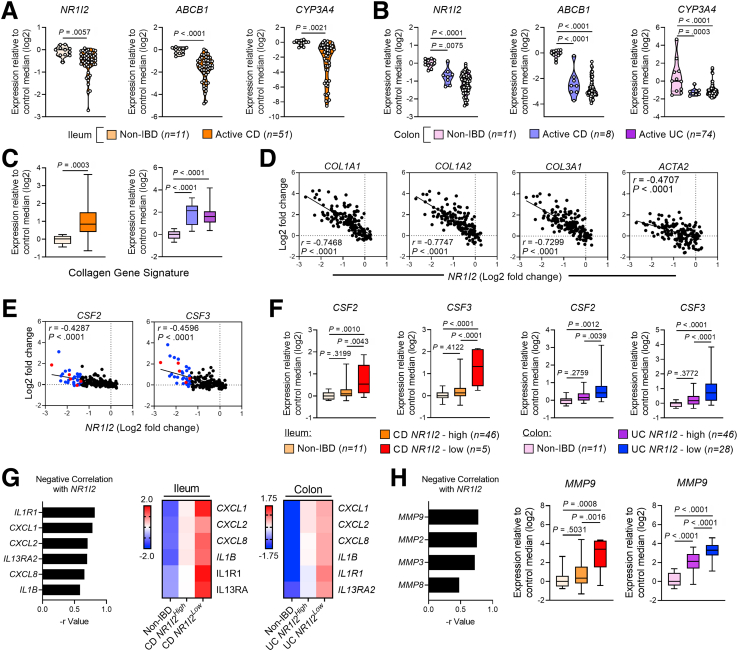
PXR Signaling Signature Is Suppressed in IBD and Correlates With Fibrotic and Inflammatory Gene Expression
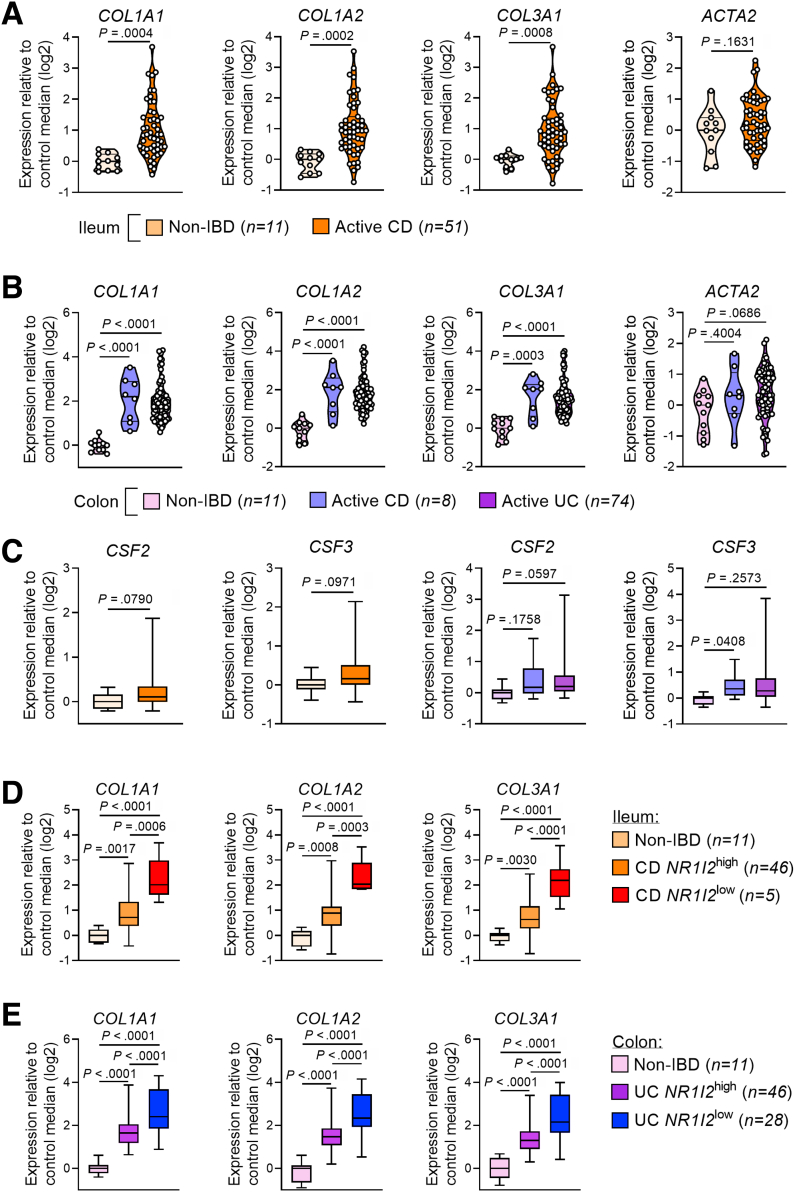
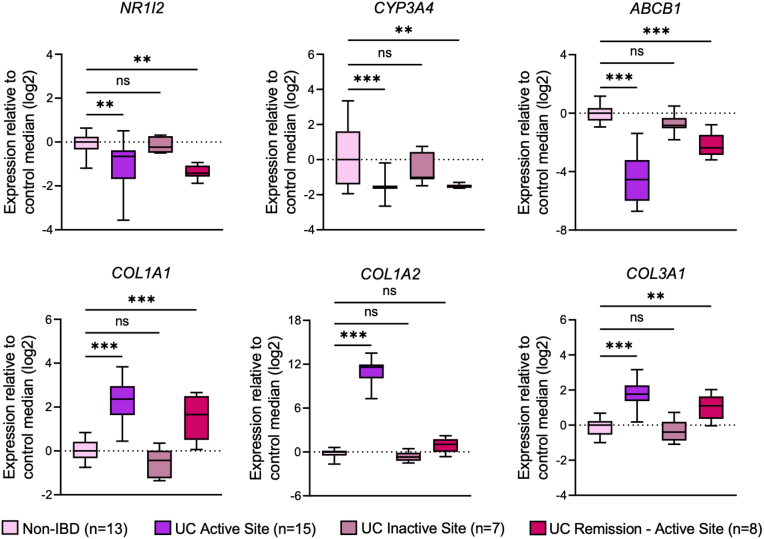
To examine the relationship between PXR signaling genes and the fibrotic and inflammatory responses in human IBD, we examined mRNA expression profiles from 2 cohorts of patients with clinically active ileal CD (GSE75214; n = 51 with CD compared with n = 11 non-IBD controls) and active colonic CD and UC (GSE59071; n = 8 with colonic CD, n = 74 with UC, both compared with n = 11 non-IBD controls). In both cohorts, patients with active disease had significantly lower mRNA expression of NR1I2 (PXR) and significantly lower expression of the PXR target genes ABCB1 and CYP3A4 (Figure 14A and B). Biopsies from both cohorts also had significantly higher levels of mRNA for COL1A1, COL1A2, and COL1A3, which together (average of log2-fold changes) we refer to as the collagen signature (Figure 14C, Figure 15A and B). Each of these collagen genes negatively correlated with the expression of NR1I2 across both patient cohorts (Figure 14D), whereas ACTA2 (αSMA), which was not significantly up-regulated in either cohort of IBD patients, showed a much weaker correlation with NR1I2 expression (Figure 14D, Figure 15A and B). Interestingly, the suppression of NR1I2 expression and downstream PXR target genes CYP3A4 and ABCB1 remained evident in biopsies isolated from UC patients in remission (Figure 16). This was associated with increased expression of both COL1A1 and COL3A1 (Figure 16), suggesting the interplay between the aberrant PXR expression/function and fibrosis may remain active even in the context of endoscopic remission.
Figure 14.
Low levels of NR1I2 expression are associated with increased inflammatory and fibrotic gene expression in patients with active Crohn’s disease (CD) and active ulcerative colitis (UC). Data were derived from Gene Expression Omnibus (GEO) datasets GSE57945 (ileum; n = 11 controls and n = 51 with active CD) and GSE59071 (colon; n = 11 controls, n = 8 with active CD, n = 74 with active UC). (A) Expression of NR1I2 (PXR) and associated target genes, ABCB1 and CYP3A4 in (A) the ileum of non-IBD and active CD patients (GSE57945) and (B) in the colon of non-IBD, active CD, and active UC patients (GSE59071). (C) Expression of a fibrotic gene signature in the ileum of CD patients compared with non-IBD ileum controls and in the colon of UC and CD patients compared with non-IBD colon controls. The fibrotic gene signature was an average of the Log2 expression values of COL1A1, COL1A2, and COL3A1. Spearman correlation of (D) fibrotic genes and (E) CSF2 and CSF3 mRNA expression against NR1I2 expression in pooled IBD cohorts (n = 155, all samples across both GSE57945 and GSE59071 datasets; color dots are representative of subjects with lowest NR1I2 expression: red = CD, blue = UC). (G) Spearman correlation of inflammatory genes with NR1I2 and heatmaps of each inflammatory gene in biopsies when each patient subset is classified as expressing high or low mRNA for NR1I2 (heatmap for each condition depicts z-score of each group). (H) Spearman correlation of proteolytic MMP enzyme expression with NR1I2 and expression of MMP9 when each patient subset is classified as expressing high or low NR1I2. Violin plots display all data points per group, with quartiles depicted by thin black line and median depicted by solid black line. Box-and-whisker plots display quartiles and range, with median depicted by solid black line. Student t test (A and C) and one-way analysis of variance with Tukey multiple-comparison test (B, C, F, and H). ∗P < .05, ∗∗P < .01, ∗∗∗P < .005.
Figure 15.
Expression levels of fibrotic genes and inflammatory cytokines in biopsies from patients with active CD and active UC. Data were derived from Gene Expression Omnibus (GEO) datasets GSE57945 (n = 42 controls and n = 162 with CD) and GSE59071 (n = 11 controls and n = 74 with UC). Expression of ACTA2, COL1A1, COL1A2, and COL3A1 in (A) the ileum of non-IBD and active CD patients (GSE57945) and (B) the colon of non-IBD, active CD, and active UC patients (GSE59071). (C) Expression of CSF2 and CSF3 in the ileum of CD patients compared with non-IBD ileum controls and in the colon of UC and CD patients compared with non-IBD colon controls. (D and E) Expression of COL1A1, COL1A2, and COL3A1in biopsies from CD and UC patients with high and low expression profiles for NR1I2 compared with non-IBD control patients. (E) CSF2 and CSF3 mRNA expression against NR1I2 expression in pooled IBD cohorts (n = 155, all samples across both GSE57945 and GSE59071 datasets. Student t test (A and C) and one-way analysis of variance with Tukey multiple-comparison test (B and C). ∗P < .05, ∗∗P < .01, ∗∗∗P < .005.
Figure 16.
Expression levels of NR1I2, PXR target genes, and fibrotic genes in biopsies from patients with active UC or in remission. Data were derived from Gene Expression Omnibus (GEO) dataset GSE59071 (n = 11 controls and n = 74 with UC). Expression of NR1I2, CYP3A4, ABCB1, COL1A1, COL1A2, and COL3A1 in colonic biopsies from non-IBD, UC active sites, UC inactive sites, and UC remission (formerly active sites). One-way analysis of variance with Tukey multiple-comparison test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005, ns, not significant.
While examining the inflammatory response in these cohorts, we focused on CSF2 and CSF3 mRNA levels, because we found these genes to be linked to PXR signaling during inflammation and fibrosis in mice. Expression of CSF2 mRNA was only mildly up-regulated in the affected ileum of CD patients and the colon of UC patients, while not significantly different in the affected colon of CD patients compared with non-IBD controls (Figure 15C). CSF3 mRNA was not significantly up-regulated in either set of CD patients (ileum or colon) and only mildly elevated in the colon of UC patients (Figure 15C). Furthermore, CSF2 and CSF3 from these IBD cohorts did not strongly correlate with NR1I2 expression (Figure 14E). However, we did identify a portion of patients with elevated levels of CSF2 and CSF3. Upon further examination, we found a gradient of NR1I2 expression across patients and within both CD and UC cohorts. Thus, next we classified patients with disease into 2 groups, those with higher levels of NR1I2 expression (named NR1I2high) closer to control levels and patients with low levels of NR1I2 expression (named NR1I2low). In both the CD ileum and UC colon cohorts, NR1I2low patients had greater than 60% reduction in NR1I2 mRNA expression. When examining mRNA expression in these 2 groups, across both IBD cohorts, we observed increased expression of CSF2 and CSF3 in patients with the lowest expression of NR1I2 (NR1I2low) when compared with NR1I2high patients and non-IBD controls (Figure 14F). Biopsies from CD and UC NR1I2low patients also had significantly elevated mRNA levels of COL1A1, COL1A2, and COL1A3 compared with the biopsies from CD and UC NR1I2high patients and non-IBD controls (Figure 15D).
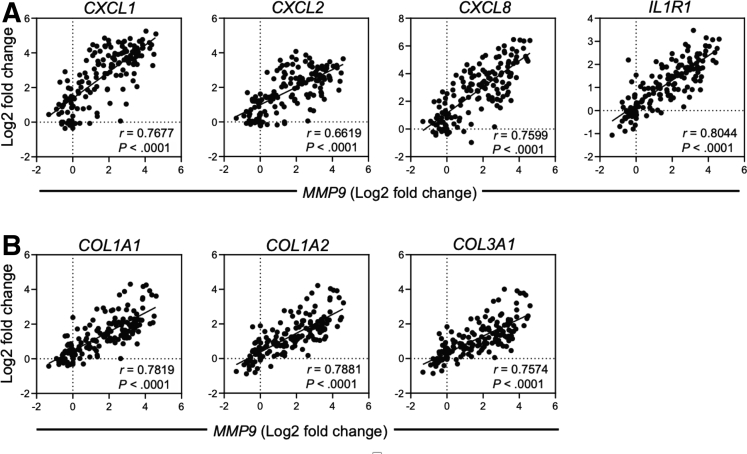
We next examined the expression of other chemokines and cytokines highlighted in this current study along with other signature genes enriched in inflammatory fibroblasts in human IBD.13 Across all patients, there was a significant negative correlation between NR1I2 and the expression of inflammatory genes, with IL1R1 being the most highly correlated, followed by CXCL1 and CXCL8 (Figure 14G). The heatmaps also show higher expression of all these inflammatory markers in the NR1I2low patient subsets in both cohorts compared with NR1I2high patients and non-IBD controls (Figure 14G).
Finally, examination of MMP expression across both patient cohorts found that mRNA expression for MMP9 was most negatively correlated with NR1I2 expression (Figure 14H). Expression of MMP9 in patients across both cohorts most strongly correlated with IL1R1 mRNA expression, as well as the neutrophil-attracting mediators CXCL1, CXCL2, and CXCL8 (Figure 17). Across all patients, a strong correlation between MMP9 and each of the fibrotic genes COL1A1, COL1A2, and COL3A1 was also observed. Finally, MMP9 mRNA expression was highest in both CD and UC NR1I2low patients, with levels significantly higher than NR1I2high patients and non-IBD controls (Figure 14H). Together these results highlight an association between decreased NR1I2 gene expression in IBD patients, the increased expression of different mediators across tightly correlated fibrotic processes (collagen and MMP expression), and innate inflammatory responses.
Figure 17.
Intestinal MMP9 expression positively correlates with innate cytokines expression and collagen gene expression in patients with active CD and active UC. Spearman correlation of (A) inflammatory mediator and (B) collagen mRNA expression against MMP9 expression in pooled IBD cohorts (n = 73, all samples across both GSE57945 and GSE59071 cohorts).
Fecal Levels of IPA Are Lower in IBD
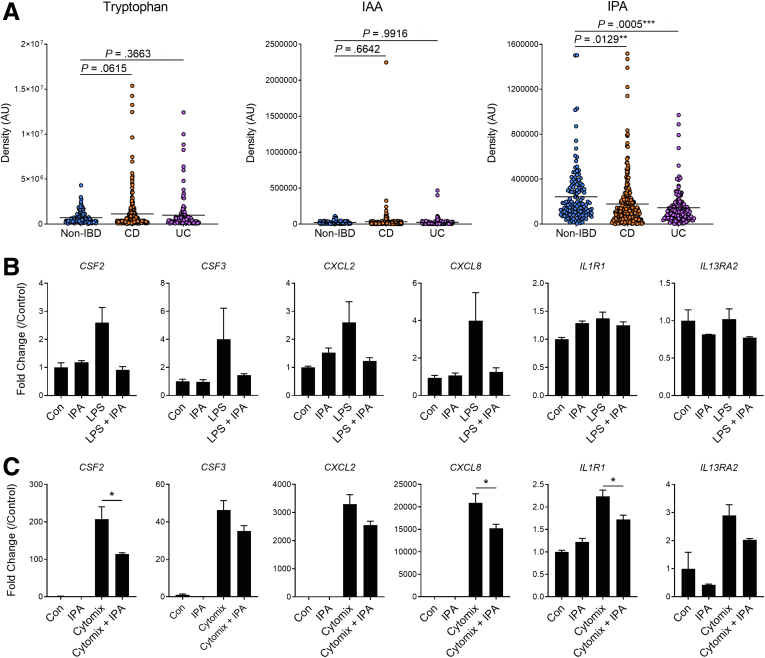
We next examined the relationship between tryptophan metabolites, including IPA, and human IBD. To do this, we reanalyzed the fecal metabolome dataset from non-IBD and IBD patients from 5 different academic hospitals across the United States that were profiled (via liquid chromatography with tandem mass spectrometry) as part of the Human Microbiome Project 2.44 When compared with fecal samples from non-IBD control patients (n = 135), samples from patients with CD (n = 265) and UC (n = 146) did not display significant differences in levels of tryptophan or levels of the microbiota-derived tryptophan metabolite indole-3-acetic acid in the feces. However, fecal samples from both CD and UC patients had significantly reduced levels of IPA (Figure 18A). During sample collection for the Human Microbiome Project 2, disease activity was not taken into consideration at the time of fecal sample collection, which led to the inability to confirm some previous associations with IBD patient data (ie, microbial diversity), which makes it more remarkable that decreased levels of IPA in CD and UC patient held true in our reanalysis.
Figure 18.
Levels of the microbiota-derived metabolite IPA are decreased in fecal samples from patients with Crohn’s disease (CD) and ulcerative colitis (UC). (A) Analysis of fecal metabolomic profiles from non-IBD, CD, and UC patients as part of the Human Microbiome Project 2 (HMP2) deposited on the NIH Metabolomics Workbench under the Project ID PR000639 (n = 135 non-IBD, n = 265 CD, n = 146 UC; IAA, indole-3-acetic acid; IPA, indole-3-propionic acid). Expression of inflammatory genes in human CCD-18Co cells pretreated with IPA (100 mmol/L) for 24 hours before adding (B) LPS or (C) cytomix (human recombinant TNF-α, Il-1β, and IFN-γ) for 12 hours (n = 3 per group). Data are presented as mean ± standard error of the mean. One-way analysis of variance with Tukey post hoc test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
IPA Can Decrease the Proinflammatory Activity of Human Intestinal Myofibroblasts
To test the influence of IPA levels on human mesenchymal cells, intestinal myofibroblasts were pretreated with IPA (100 μmol/L) for 24 hours and then stimulated with LPS or human cytomix (human IL1β, TNFα, and IFNγ) for 12 hours. In these cells, LPS treatment induced the expression of mRNA for CSF2, CSF3, CXCL2, and CXCL8. The expression of these cytokines was dampened by IPA pretreatment, which reduced expression of CSF2, CSF3, CXCL2, and CXCL8 (Figure 18B). To note, LPS treatment did not change the expression of IL1R1 or IL13RA2, 2 cytokine receptors previously demonstrated to mark inflammatory myofibroblasts in human IBD.12,13 IPA pretreatment suppressed the expression of cytokines and chemokines produced in response to human cytomix, significantly reducing the expression of CSF2 and C-X-C Motif chemokine ligand 8 (CXCL8). In addition, IPA also decreased the expression of IL1R1 but had no influence on the expression of IL13RA2 (Figure 18C). Together these results suggest that IPA can suppress the inflammatory capacity of the human intestinal mesenchyme, and thus, lower levels of luminal IPA in IBD patients may create an environment wherein the intestinal mesenchyme is unrestrained and becomes more reactive to inflammatory or injurious insults.
Discussion
Intestinal fibrosis in IBD is a common consequence of chronic inflammation that delays wound healing and interrupts proper tissue remodeling. These processes are tightly regulated by the intestinal mesenchyme that attempt to return tissue to homeostasis after inflammation and injury but can quickly become defective when noxious stimuli are not promptly removed or when inflammation becomes protracted. However, the mesenchymal compartment is not just a passive player ensuring proper tissue remodeling, but instead it is a vital contributor to the development of inflammation through its ability to promote inflammatory responses. In IBD, the intestinal mesenchymal niche can become dysregulated with the emergence of “activated” or “inflammatory” fibroblasts, traditionally termed myofibroblasts11,13 that can fuel inflammation and subsequent fibrogenesis. Inflammatory function of the mesenchyme may be one of the more important contributors to the development of fibrosis, because one of the most salient features of fibrostenotic lesions from IBD patients is consistently elevated levels of innate immune cytokines including IL1β,45 IL6,45 CXCL1,45 and CXCL8.46 Serum levels of anti-CSF2 antibodies, resulting from prolonged intestinal levels of CSF2, in combination with serum COL3A1 have also been used to predict the development of stricturing disease in CD.47 In line with this function of the mesenchymal compartment, our current work shows a role for intestinal PXR function in suppressing the inflammatory and fibrotic capacity of the intestinal mesenchymal compartment. Importantly, inflammation and fibrosis in the healing colon can be dampened by the microbiota-derived metabolite IPA, highlighting a novel circuit in which the microbiota can modulate the function of the mesenchyme in dictating inflammation and fibrosis.
Fibroblasts, the most abundant component of the mesenchyme, express various receptors including TLR1-9 and NOD-like receptors 1 and 2 and can be activated by the corresponding ligands to produce chemokines and cytokines in a NF-κB–dependent manner.9,48,49 Overexuberance of myofibroblasts in this context can contribute to chronic inflammation by sustaining leukocytes infiltration and participating in complex cellular crosstalk with immune cells, epithelial cells, and cells within the smooth muscle layers of the intestine.11 The importance of inflammatory myofibroblasts in IBD have been solidified by studies using small cytoplasmic RNA sequencing to uncover subsets of inflammation-associated myofibroblasts in IBD lesions that are enriched for inflammatory cytokines including Il1B and the neutrophil chemokines CXCL1, CXCL2, CXCL5, and CXCL8.12,13,50 These cells also expressed increased levels of Il1R1 and TNFR and were poised for activation by IL1β- and TNF-producing immune cell subsets to perpetuate inflammatory gene enrichment. One study showed that myofibroblasts were also enriched for genes participating in NF-κB signaling and matrix remodeling.50 More robust contributions of these myofibroblasts to cellular networks in IBD lesions were associated with failure of anti-TNF therapy,12,14 which is indicative of more aggressive disease and a direct predictor of fibrostenotic disease.51 In the current study, our analysis of deposited datasets from actively inflamed biopsies from CD and UC patients showed increased expression of many of the cytokines and receptors reported to be up-regulated in myofibroblasts (including CXCL2, CXCL8, and IL1R1), and importantly, these inflammatory genes negatively correlated with the expression of NR1I2, the gene that encodes PXR, across both datasets. In addition, reduced NR1I2 expression in IBD also correlated with increased levels of fibrotic genes including COL1A1, COL1A2, COL3A1, and MMPs. Using a human mesenchymal cell line, we also demonstrated that IPA was able to supress the expression of CXCL8 and IL1R1, as well as other cytokines, triggered by inflammatory stimuli. Furthermore, activating the PXR in animal models of intestinal healing was able to supress the expression of many of these cytokines, especially neutrophil-attracting Cxcl1 and Cxcl2, while also supressing the expression of collagen and MMP genes.
Many reports have shown a protective role for PXR signaling in experimental and human IBD,52, 53, 54, 55, 56 with suppression of NF-κB and the subsequent dampening of chemokine and cytokine expression being a common mechanism.55 Although the interplay between inflammation and PXR function/expression has been well-studied, it is still not completely understood whether IBD risk variants in the NR1I2 gene are the driving mechanism conferring susceptibility, or whether inflammation-associated suppression of PXR expression may also be involved in intestinal disease pathogenesis. Our current data suggest that PXR signaling in the injured colon provides protection, in part, by shaping the inflammatory capacity of the intestinal mesenchyme and suppressing mesenchymal cells from overactive NF-κB signalling. In the context of NF-κB interactions, the relevant cytokines in both mice and humans include neutrophil-attracting chemokines, which we showed to be enriched in the colon of Nr1i2-/- mice after injury and, importantly, was accompanied by increased infiltration of neutrophils compared with WT mice. How the PXR influences NF-κB signaling to modulate the expression of inflammatory mediators is still not completely resolved, but it is thought to occur through direct interactions at the receptor level or through downstream transcriptional events.38 PXR-mediated suppression of TLR4 signaling may also play a role in protecting the intestine, dampening the inflammatory capacity of the mesenchyme, and preventing immune cell infiltration into the intestine.22,32,57 Because of the ability of the PXR to dampen the fibrotic potential of hepatic stellate cells, while also potentially influencing epithelial-mesenchymal transition,23 the anti-fibrotic effects of PXR signaling may also be occurring through mechanisms that remain to be studied in the context of intestinal injury. Furthermore, pharmacologic activation of PXR signaling in the intestinal epithelium can also supress inflammation20, 21, 22; however, in the current study, deletion of unliganded PXR in the epithelium did not have a significant effect on the inflammatory response or fibrosis observed after colonic injury. We did note an increase in Cxcl2 expression in the healing colon of mice depleted of PXR in the epithelium, raising the possibility of cross-compartmental contributions and crosstalk between epithelial cells and fibroblasts in PXR-mediated control of inflammation and fibrosis. The intimate links between the epithelium and mesenchyme highlight an intriguing balance in the control of mucosal reactivity because of the proximity of the microbiota and its ability to produce IPA, and our current data and others17 showing that increasing luminal IPA levels can increase serum and tissue levels of IPA.
Although alterations in the composition and functional output of the intestinal microbiota have been associated with IBD pathogenesis, how the microbiota influences IBD-related intestinal fibrosis is less well-understood. Current evidence suggests a complex interplay between the microbiota and the mesenchymal compartment that can influence fibrotic processes both directly, where microbes act on mesenchymal cells to dictate fibrotic responses, and indirectly by tailoring the inflammatory potential of myofibroblasts. Indeed, several candidate bacteria have been identified that, in culture conditions, can promote or prevent migration and collagen expression in myofibroblasts.58 Conversely, our findings and others59 suggest that the interactions between microbial metabolites and specific receptors can play an important role in supressing fibrotic events in the intestine. Consistent with our findings, harboring a more complex microbial community in the gut limited inflammation and fibrosis in a liver injury model, potentially through the provision of more complex and protective microbial metabolites.60 These interactions may be particularly important in the mesenchyme, and our current data suggest that myofibroblasts isolated from GF mice are more sensitive to inflammatory stimuli than those from colonized, SPF mice. It is possible that the microbiota can program immunosuppression or hyporesponsiveness of the intestinal mesenchyme in the same way resident intestinal macrophages are programmed by the microbiota and its metabolites to become hyporesponsive to the constant antigen exposure in the gut lumen.61, 62, 63 This is a salient point because of the finding that levels of fecal IPA are decreased in CD and UC patients.44 Another study examining indole-metabolite levels in IBD patients reported no difference in IPA levels; however, samples were collected by aspirating the colon after patients had performed bowel cleanses in preparation for colonoscopy.64 Lower levels of luminal IPA levels after bowel cleanse, which drastically decreases microbiota load,65 lends to the importance of luminal microbes in contributing to levels of IPA and other indole metabolites. In the context of our current data, these human findings raise the possibility that the mesenchymal compartment in IBD patients may be prone to inflammatory overactivity and the promotion of fibrotic events.
The intimate relationship between the intestinal mucosa and the diverse range of community members of the microbiota can also complicate the picture of fibrosis. In a mouse model of intestinal inflammation, the microbiota was required to drive fibrosis,58 whereas in CD patients, antibody responses to microbiota-derived antigens can predict progression toward stricturing disease.66,67 Our current data and others show that the mesenchyme function can be influenced by microbe and microbe-derived products (such as LPS) that breach the intestinal epithelium and can contribute to fibrosis development in the intestine.7,37,43,68 Indeed, during inflammation and injury, mesenchymal cells, including myofibroblasts, are major contributors to various cytokines including GM-CSF (Csf2) and G-CSF (Csf3).69,70 Both GM-CSF and G-CSF are recognized as myeloid growth factors and aid in neutrophil recruitment and promote the survival of neutrophils once in the tissue. In other organ systems, injury-induced GM-CSF and G-CSF production from mesenchymal cells can recruit neutrophils through enhanced chemokine expression, creating a tissue environment that favors tissue destruction and fibrosis.10,71,72 In the current study, primary colonic myofibroblasts from mice produced significant amounts of GM-CSF and G-CSF that were robustly elevated in the absence of the PXR after stimulation with cytokines or LPS. Furthermore, Csf2 and Csf3 expression in the healing colon were elevated in the absence of PXR signaling in the mesenchymal compartment of mice, which was accompanied by increased expression of neutrophil chemokines and the influx of neutrophils into the LP. The oversupply of leukocytes, including neutrophils, promoted by GM-CSF and G-CSF and other downstream mediators (Cxcl1, Cxcl2), can promote fibrosis in different organs through a variety of mechanisms.10,42,71,72
After their recruitment to sites of injury, neutrophils release various mediators, including reactive oxygen species, neutrophil elastase, and myeloperoxidase, directed at controlling the invasion of bacteria across the epithelium. Although highly effective at killing bacteria, these molecules often cause bystander damage if not tightly controlled.42 To curtail inappropriate tissue damage, neutrophils are typically short-lived, and their apoptosis can initiate the resolution processes. However, failure to execute resolution can lead to prolonged neutrophil activation and chronic inflammation and contribute to propagation of intestinal fibrosis. This cycle has been observed in both human IBD and mouse models of intestinal injury, where neutrophil influx can create a “vicious cycle” of tissue degradation and further leukocyte infiltration that can persist in the absence of proper inflammatory resolution.42 In the current study, although we observed increased neutrophil infiltration into the protracted inflammatory environment of the injured colon of mice lacking the PXR, we did not observe increased expression of neutrophil enzymes such as Elane and Mpo. Instead, we observed increased expression of Mmp8 and Mmp9, two enzymes that are produced by neutrophils in inflamed intestinal tissue41,42 and are involved in the degradation and turnover of the extracellular matrix. In human IBD, robustly increased expression of MMPs, including MMP9, has been observed in strictured and ulcerated intestinal tissue, whereas polymorphonuclear neutrophils isolated from the blood of IBD patients had greater MMP proteolytic capacity compared with control patients.40,41,43 We demonstrated that myofibroblasts from PXR-deficient mice are capable of programming neutrophils to express higher levels of MMP9. IPA administration was also able to suppress the expression of MMP9 that correlated with neutrophil tissue levels and corresponded with levels of decreased fibrosis. On the basis of our current data, the ability of the PXR to shape mesenchymal function may serve to prevent inflammation and subsequent neutrophil infiltration from potentiating the vicious cycle of leukocyte-driven tissue proteolysis and damage that has been demonstrated in both IBD tissues and mouse models.
Ultimately, the integration of signals from the microbiota by the mesenchyme appears to be complex, and with our current data, the composition of the microbiota and its resulting metabolites may be an important determinant in fibrosis development. Several receptors that sense microbes and microbe-derived metabolites likely contribute to the overall microenvironment in the intestine that can regulate the underlying processes that promote fibrosis. In particular, receptors that can modulate inflammatory processes to prevent overexuberant inflammation, while also targeting the mesenchymal compartment to promote proper remodeling and healing responses, may be rational targets to treat inflammation-driven fibrosis. To this end, recently developed microbial metabolite mimetics, including analogs of IPA,73 may prove beneficial in controlling the immune reactivity of the mesenchyme and combating IBD-related-fibrosis.
Methods
Ethics Approval and Consent to Participate
All human data presented in this submission were reanalyses of publicly available datasets. All animal experiments were approved by the University of Calgary Health Sciences Animal Care Committee (AC19-0053; AC19-0105; AC20-0138) and were conducted in compliance with the guidelines established by the Canadian Council for Animal Care.
Animals
Male Nr1i2-/- and counterpart WT mice were obtained from Taconic Labs (Albany, NY). Villin-cre (B6.Cg-Tg(Vil1-cre)1000Gum/J), Col1a2-creERT2 (Tg(Col1a2-cre/ERT,-ALPP)7Cpd/J), and Rag1-/- mice on the C57BL/6 background were purchased from The Jackson Laboratory (Bar Harbor, ME). Floxed-Nr1i2 mice were derived at the Center for Genome Engineering at the University of Calgary. To generate the floxed-Nr1i2 mouse, we used an embryonic stem cell targeting vector produced by the International Knockout Mouse Consortium (project 25663). All mice including cre-negative littermate controls were crossed on a C57Bl/6J background for at least 4 generations, and in all experiments, littermate controls were used. To confirm the targeted deletion of Nr1i2 in intestinal epithelial cells within the Nr1i2DVil1 line, the expression of the PXR protein was assessed in freshly isolated colonic intestinal epithelial cells via Western blot. For induction of cre in male Col1a2-creERT2-Nr1i2 mice, 1 mg of 4-hydroxytamoxifen (4-OHT) was injected intraperitoneally. On the day of each injection, 4-OHT was dissolved in ethanol and then diluted in sterile corn oil so that each mouse received 1 mg of 4-OHT in a 200 mL injection volume. Mice were administered 3 consecutive doses of 4-OHT before commencing administration of DSS (3% w/v). Mice received weekly injections of 4-OHT starting 7 days after the first injection of the 3 initial injections. To confirm knockdown of Nr1i2 after 4-OHT treatment, the expression of the PXR protein was assessed in freshly isolated fibroblasts via Western blot. GF C57BL/6 mice were bred and maintained in flexible film isolators at the International Microbiome Centre, University of Calgary, Canada. GF status was confirmed by culture-dependent and -independent methods and were independently confirmed to be pathogen-free. All other mice were housed in Tecniplast ventilated cage systems (standard 12-hour light/dark cycle), with wood shaving-based bedding, free access to food and autoclaved water, and housed with no more than 4 animals per cage. All mice were randomly allocated to cages designated for specific treatment groups by vivarium staff upon delivery from commercial sources or when transferred from the breeding barrier unit into the animal housing room. All mice purchased from vendors or transferred from our breeding barrier unit were acclimatized in the animal housing room for 7 days before commencing experiments. Male mice of 8–12 weeks of age were used for all experiments. At the end of each experiment, mice were anesthetized with isoflurane (5%) and euthanized by cervical dislocation. All experiments were approved by the University of Calgary Health Sciences Animal Care Committee and were conducted in compliance with Canadian Council for Animal Care guidelines.
Reagents
DSS (colitis grade; molecular weight, 36,00–50,00 Da; cat#: SKU 02160110) and PCN (cat#: 156362) were purchased from MPBio (Santa Ana, CA). IPA (cat#: 57400), ampicillin (cat#: A9518), neomycin (cat#: N1876), metronidazole (cat#: M3761), vancomycin (cat#: V8138), and collagenase (Type IV, cat#: C5138) were purchased from Sigma-Aldrich (St Louis, MO). Human intestinal myofibroblasts (cat#: CC2902), SmGM basal media (cat#: CC3181), and SmGM-2 supplements (cat#: CC4149) were purchased from Lonza (Basel, Switzerland). BAY 11-7082 (cat#: 10010266) and 4-OHT (cat#:14854) were purchased from Cayman Chemicals (Ann Arbor, MI). Cytokines (carrier-free) that made up cytomix (recombinant mouse and human versions of TNFα [cat#: mouse-410-MT, cat#: human-210-TA], IL1β (cat#: mouse-401-ML, cat#: human-201-LB), and IFNγ (cat#: mouse-485-MI, cat#: human-285-IF) were purchased from R&D Systems (Minneapolis, MN). LPS (ultrapure, cat#: tlrl-3pelps) was purchased from InVivogen (San Diego, CA). DNase I (cat#: 10104159001) was purchased from Roche (Basel, Switzerland). The following antibodies were purchased from eBioscience (Thermo Fisher Scientific, Waltham, MA): MHCII (I-A/I-E)-FITC (M5/114.15.2, cat #11-5321-82) and Ly6C-APC (HK1.4; cat#: 17-5932-80) and BioLegend (San Diego, CA): CD11b - Pacific Blue (M1/70, cat#: 101224), Ly6G-APC-Cy7 (1A8, cat#: 127624), CD45-PerCP-Cy5.5 (30-F11, cat#: 127624103132), and Fc-Block TruStain FcX (anti-mouse CD16/32, 93, cat#: 101319).
Induction and Assessment of Intestinal Damage and Fibrosis
Intestinal injury was induced using 3% DSS for 5 days after which mice were switched to normal drinking water to recover for the indicated times. At the end of the defined recovery period, mice were anesthetized with isoflurane (5%), euthanized by cervical dislocation, and tissues removed for outcome measures. Colon length was measured with a ruler, and gross colon thickness was measured using digital calipers. Colon sections were fixed in 4% formalin for 24 hours and then embedded in paraffin, sectioned, and stained with Masson’s trichrome by Calgary Lab Services. Chronic T-cell colitis was induced using CD4+CD25-CD45RbHi T-cell sorted (BD FACS Aria) from the spleens of WT or Nr1i2-/- C57BL/6J mice. After sorting, 5 × 105 naive CD4+CD25-CD45RbHi T cells in sterile phosphate-buffered saline (PBS) were injected intraperitoneally into Rag1-/- mice. The level of fibrosis in Masson’s trichrome stained tissue sections was determined on blinded slides using ImageJ (National Institutes of Health, Bethesda, MD). The total area occupied by blue fibrotic staining was expressed as a percentage of the total section area. For fibrotic thickness, the average of 15 measurements across the thickest fibrotic areas of each section was taken.
Intervention Strategies in Mice
After a 5-day course of DSS and the switch to normal drinking H2O, PCN (25 mg/kg dissolved in sterile corn oil; intraperitoneal route; Sigma-Aldrich) was administered to mice every other day for the course of the experiment. IPA was administered in the drinking water at a dose of 250 mg/L. In the DSS studies, IPA water was administered on day 5 after DSS when animals were switched to either water supplemented with IPA or pH-matched water. For colonic fibroblasts studies animals were administered IPA for 2 weeks before isolation.
Depletion of Intestinal Commensal Bacteria
Gut microbiota were depleted using a method modified from a previously described protocol.74 Mice were provided with ampicillin (1 g/L), vancomycin (0.5 g/L), neomycin (1 g/L), and metronidazole (1 g/L) in drinking water. In serum metabolites studies, WT (Nr1i2+/+) mice were administered antibiotics for 2 weeks before sample collection. For intestinal damage studies, water containing antibiotics was administered on day 5 after DSS when animals were switched to normal drinking water, antibiotic water, or antibiotic water supplemented with IPA (250 mg/L).
Quantification of Cytokines/Chemokines
Cytokine levels were assessed in serum and cell culture supernatants using the Mouse Cytokine/Chemokine 32-Plex Discovery Assay (Eve Technologies, Calgary, AB, Canada). Briefly, multiplex immunoassay was used to simultaneously detect 32 inflammatory biomarkers in a single microwell/sample. For serum samples, blood was collected from mice via cardiac puncture in heparinized syringes and immediately spun down for 10 minutes at 5000 rpm, and serum supernatants were snap frozen. Cell culture supernatants were collected directly from culture plates and immediately snap frozen. All samples were diluted 2-fold in sterile PBS before shipment on dry ice.
Lamina Propria Cell Isolation and Flow Cytometry
LP cells were isolated as previously described by Denning et al75 with modifications. Briefly, the large intestine was opened longitudinally, washed of fecal contents, and cut into pieces 0.5 cm in length. To remove epithelial cells colon tissue was subjected to 2 sequential 20-minute incubations in 30 mL RPMI with 5% fetal bovine serum and 2 mmol/L EDTA at 37°C with agitation (250 rpm). After each incubation, medium containing epithelial cells and debris was discarded. The remaining colon tissue was minced and incubated for 20 minutes in RPMI with 5% fetal bovine serum, 1 mg/mL collagenase IV (Sigma-Aldrich), and 40 U/ml DNase I (Roche) at 37°C with agitation (200 rpm). Cell suspensions were collected and passed through a 100-μm strainer and pelleted by centrifugation at 300g. Cell surface staining was performed using antibodies purchased from eBioscience (Thermo Fisher Scientific): MHCII (M5/114.15.2) and Ly6C (HK1.4) and BioLegend: CD11b (M1/70), Ly6G (1A8), and CD45 (30-F11). Fc receptors were blocked with the antibody anti-FcγRIII/II.75 Samples were run on the BD FACS Canto and analyzed using FlowJo (gating strategy in Figure 4).
Primary Myofibroblast Isolation and Cell Culture
Primary myofibroblasts were isolated from the full colon of WT and Nr1i2-/- mice. Briefly, colonic tissue was collected, manually minced into 1- to 2-mm fragments, and washed in ice-cold PBS (Ca2+ and Mg2+ free). The tissue was washed with Hank’s buffered salt solution (Ca2+ and Mg2+ free) containing 5 mmol/L EDTA, rinsed in Hank’s buffered salt solution (with Ca2+ and Mg2+), and then digested with 250 U/mL of collagenase IV (Sigma-Aldrich) for 90 minutes at 37°C. The digested suspension was neutralized with Dulbecco modified Eagle medium F/12 Ham medium containing 10% fetal bovine serum, L-glutamine, antibiotic-antimycotic, sodium pyruvate, nonessential amino acids, and ciprofloxacin and subsequently passed through a 70-μm strainer. The mixture was centrifuged at 1000 rpm for 10 minutes and resuspended in fresh medium, and a differential adherence approach was used to selectively separate myofibroblasts from smooth muscle cells as previously described.76 The cell mixture was placed in prewarmed culture flasks for 20 minutes, where myofibroblasts adhered, and the non-adherent suspension was removed and discarded. After 3 weeks in culture, fibroblasts were cultured in Dulbecco modified Eagle medium F/12 Ham medium containing L-glutamine, antibiotic-antimycotic, and supplement pack (C-39363; PromoCell, Heidelberg, Germany). WT and Nr1i2-/- myofibroblasts were propagated and treated between passages 3 and 20, where N = a separate passage of cells. For in vivo experiments comparing colonic myofibroblasts isolated from water-treated and IPA-treated mice, as well as GF mice, cells were plated and treated immediately after isolation and for subsequent passages.
Culture of Human Myofibroblasts
Commercially sourced human intestinal myofibroblasts (cat#: CC2902; Lonza) were grown in SmGM basal medium (cat#: CC3181; Lonza) with SmGM-2 supplements (cat#: CC4149; Lonza). All in vitro experiments were performed on cells between passages 3 and 10, where N = a separate passage of cells.
In Vitro Treatments
To test the responses of fibroblasts to TGFβ, cells were first transferred to reduced serum medium (Opti-MEM; Thermo Fisher) for 24 hours and then treated with 10 ng/mL or 100 ng/mL of TGFβ1 in Opti-MEM for 24 hours. To test the response to myofibroblasts to inflammatory stimuli, cells were treated with LPS (10 ng/mL) or cytomix (TNFα, IL1β, and IFNγ at 10 ng/mL each) in OptiMEM. Cells were treated for 24 hours to assess the release of cytokines into cell supernatants or 12 hours to assess gene expression. Treatments were initiated when cells reached 85%–90% confluency. To inhibit NF-κB activity, myofibroblasts were pretreated with BAY-11-7085 (20 mmol/L) for 6 hours and then treated with cytomix or LPS in the presence of BAY-11-7085 for 12 hours. In human studies, cells were pretreated with IPA (100 mmol/L) for 24 hours and then stimulated with LPS or human cytomix at a concentration of 10 pg/mL for 12 hours.
Measurement of Metabolite Levels
Serum was first extracted in a 50:50 mix of methanol and molecular grade water. Colon samples were also extracted in a 50:50 methanol:water mix by homogenization (50 mg/mL) using a Bullet Blender (Next Advance, Raymertown, NY), with each sample containing a 5-mm stainless steel bead (Qiagen, Hilden, Germany; #69989). The samples were then spun down (speed) and stored at –80° until being run. Samples were further extracted with 1 mL of water (containing 10 mg/mL butyric acid_D7, 10 mg/mL indole_D7, 10 mg/mL IPA_D2, and 10 nmol 2-chlorophenylalanine as internal standards). The samples were vortexed for 2 minutes and sonicated for 5 minutes. The samples were centrifuged after keeping the sample on ice for 15 minutes. A volume of 500 mL of supernatants was transferred into a glass tube, add 100 mL of internal standards solution. After adding 300 mL of water, 500 mL of propanol: pyridine (v:v, 3:2), and 100 mL of propyl chloroformate, the reaction was performed in sonication for 1 minute. A volume of 300 mL of hexane was added and vortexed for 2 minutes. The samples were then centrifuged for 5 minutes at 1200 rpm. The upper hexane layer was pipetted into a GC sampling vial after passing through the sodium sulfate (anhydrous) to remove excess water. One mL of extract was injected into Agilent CG-MS system with full scan mode (Agilent, Santa Clara, CA). Separation was performed with a DB-1701 column. Helium was used as carrier gas at a consistent flow of 1 mL/min. The oven program was the following: started at 40°C for 2 minutes, 5°C /min to 90°C, 10°C/min to 270°C, and keep for 6 minutes. ChemStation was used for data analysis. The GC-MS analysis was conducted using our adapted propyl chloroformate derivatization method in an untargeted profiling mode. This mode was selected because various metabolites were also being screened. A selected ion mode for indole (m/z = 117) and indole propionic acid (m/z = 231) was selected to calculate the concentrations of indole and IPA in the sample.
Neutrophil Isolation
Bone marrow was collected in sterile conditions from the tibia and femurs of WT mice by centrifugation, after which red blood cells were lysed using Red Blood Cell Lysis Buffer (Sigma-Aldrich). Neutrophils were isolated from resulting bone marrow using the Neutrophil Isolation kit from Miltenyi Biotech (cat#: 130-097-658; Bergisch Gladbach, Germany), according to the manufacturer’s instructions. Five × 105 cells per well were plated (24-well plates) in RPMI supplemented with 10% fetal bovine serum. Supernatant from WT or Nr1i2-/- fibroblasts after treatment with cytomix were incubated with freshly isolated neutrophils for 8 hours and then washed in PBS before isolation of RNA using Trizol (TRI Reagent; Millipore Sigma, Burlington, MA).
Western Blotting
Western blots were performed on normalized protein extracts from primary mouse fibroblast cultures. Membranes were probed with the appropriate primary antibody (anti-αSMA Thermo Fisher Scientific #701457; anti-vimentin, Santa Cruz Biotechnology [Dallas, TX] #sc-6260; β-actin, Santa Cruz Biotechnology, #sc-47778; anti-PXR [H-160], Santa Cruz Biotechnology #sc-25381) and corresponding horseradish peroxidase-conjugated secondary antibody. Membranes were imaged using the MicroChemi Bio-Imaging system.
Real-Time Polymerase Chain Reaction
For in vitro studies, RNA was extracted from cells using Trizol (TRI Reagent; Millipore Sigma) per the manufacturer’s instructions. For in vivo studies, 6 cm of distal colonic tissue was isolated and stored in RNAlater (Thermo Fisher Scientific, Mississauga, Canada) at –20°C. Tissue was homogenized by Bullet Blender (Next Advance), with each sample containing a 5-mm stainless steel bead (Qiagen #69989). RNA was extracted using the RNeasy Mini Kit (Qiagen #74106) per the manufacturer’s instructions. Total RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative polymerase chain reaction was conducted on reactions containing PerfeCTa SYBR Green FastMix (Quantabio, Beverly, MA), cDNA, and validated primers from Qiagen. Quantitative polymerase chain reaction was conducted on reactions containing PerfeCTa SYBR Green FastMix (Quantabio), cDNA, and validated mouse and human primers. The following mouse primers were used: Acta2 (PPM04483A), Col1a1(PPM03845F), Col1a2 (PPM04448F), Col3a1(PPM04784B), Csf2 (PPM02990F), Csf3 (PPM02989B), Cxcl1 (PPM03058C), Cxcl2 (PPM02969F), Cyp3a11 (PPM03917F), Il6 (PPM03015A), Il1b (PPM03109F), Il18 (PPM03112B), Itgam (PPM03671F), Mmp2 (PPM03642C), Mmp3 (PPM03673A), Mmp8 (PPM03610C), Mmp9 (PPM03661C), Mpo (PPM06205A), Nox1 (PPM34199A), Elane (PPM03805E), Cybb (PPM32951A), and Vim (PPM04780B). The following human primers were used: CSF2 (PPH00576C), CSF3 (PPH00723B), CXCL1 (PPH00696C), CXCL2 (PPH00552F), CXCL8 (PPH00568A), IL1R1 (PPH00274A), and IL13RA1 (PPH01257C). In all samples, β-actin (ACTB, mouse: PPM02945B, human: PPH00073G) was used as the endogenous control. Threshold cycle (Ct) values were obtained from the amplification plots and used to calculate fold-change using the ΔΔCt method.
Immunofluorescence
Colonic myofibroblasts were isolated from WT and Nr1i2-/- mice and seeded onto 8-well chamber slides at 37,500 cells per well. Cells were briefly washed with PBS and fixed with 10% formalin for 30 minutes at room temperature. Cells were then permeabilized with 0.5% Triton X-100 for 10 minutes, washed with PBS, and blocked for 1 hour in PBS containing 10% normal goat serum at room temperature. This was followed by incubation with primary antibodies for αSMA (3 μg/mL; cat# 701457; Thermo Fisher Scientific) and vimentin (1:200; Cat# sc-6260; Santa Cruz Biotechnology) overnight at 4°C. The following day, cells were washed with PBS-T and incubated with the appropriate Alexa Fluor conjugated secondary antibodies for 1 hour and subsequently stained with Hoechst for 30 minutes to label nuclei.
Microarray Data
In the present study, the IBD datasets GSE75214 and GSE59071 were acquired as raw data Cel files from Gene Expression Omnibus repository. Cel files were processed through the Transcriptome Analysis Console (Thermo Fisher Scientific) microarray analysis platform that uses RMA correction and the Limma R package to derive corrected intensity values. Corrected intensity values were transformed to Log2 values, and relative expression differences were calculated to the median of the control group.
Metabolomics Data
Metabolomics data were obtained from the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center website via the Metabolomics Workbench (http://www.metabolomicsworkbench.org) under the Project ID PR000639. Metadata were matched to each coded sample in the compiled metabolomic data.
Statistical Analysis
Data were expressed as mean ± standard error of the mean. Unpaired Student t test was used to compare between 2 groups. One-way analysis of variance was used to compare more than 2 groups, followed by Tukey post hoc test. A P value <.05 was considered statistically significant. All tests were two-tailed. All statistical analyses were performed using GraphPad Prism v8.0 software (GraphPad Software Inc, La Jolla, CA). All authors had access to the study data and analysis approaches and reviewed and approved the final manuscript.
Acknowledgments
CRediT Authorship Contributions
Kyle Flannigan (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Writing – original draft: Equal; Writing – review & editing: Equal)
Kristoff Nieves (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Holly Szczepanski (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Alex Serra (Investigation: Supporting; performed protein isolation and western blots: Supporting)
Joshua W. Lee (Investigation: Supporting; performed protein isolation and western blots: Supporting)
Laurie Alston (Investigation: Supporting; Methodology: Supporting; Project administration: Supporting; Writing – review & editing: Supporting)
Hena Ramay (Data curation: Lead; Formal analysis: Lead; Methodology: Supporting)
Sridhar Mani (Conceptualization: Supporting; Resources: Equal; Writing – review & editing: Equal)
Simon Andrew Hirota, PhD (Conceptualization: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding SAH’s salary is supported by the Canadian Institutes of Health Research (CIHR) Canada Research Chair program (Tier II CRC in Host-Microbe Interactions and Chronic Disease); SAH’s laboratory is supported by an infrastructure grant provided by the Canadian Foundation for Innovation John R. Evans Leaders Fund; operating funds from CIHR (SAH; #376341) and Crohn’s & Colitis Canada (SAH); and CA 222469 and Department of Defense Partnering PI (W81XWH-17-1-0479; PR160167) (SM).
References
- 1.Rieder F., Fiocchi C., Rogler G. Mechanisms, management, and treatment of fibrosis in patients with inflammatory bowel diseases. Gastroenterology. 2017;152:340–350 e346. doi: 10.1053/j.gastro.2016.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rieder F. Managing intestinal fibrosis in patients with inflammatory bowel disease. Gastroenterol Hepatol. 2018;14:120–122. [PMC free article] [PubMed] [Google Scholar]
- 3.Thia K.T., Sandborn W.J., Harmsen W.S., Zinsmeister A.R., Loftus E.V., Jr. Risk factors associated with progression to intestinal complications of Crohn’s disease in a population-based cohort. Gastroenterology. 2010;139:1147–1155. doi: 10.1053/j.gastro.2010.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rutgeerts P., Geboes K., Vantrappen G., Beyls J., Kerremans R., Hiele M. Predictability of the postoperative course of Crohn’s disease. Gastroenterology. 1990;99:956–963. doi: 10.1016/0016-5085(90)90613-6. [DOI] [PubMed] [Google Scholar]
- 5.Yaffe B.H., Korelitz B.I. Prognosis for nonoperative management of small-bowel obstruction in Crohn’s disease. J Clin Gastroenterol. 1983;5:211–215. doi: 10.1097/00004836-198306000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Gordon I.O., Agrawal N., Willis E., Goldblum J.R., Lopez R., Allende D., Liu X., Patil D.Y., Yerian L., El-Khider F., Fiocchi C., Rieder F. Fibrosis in ulcerative colitis is directly linked to severity and chronicity of mucosal inflammation. Aliment Pharmacol Ther. 2018;47:922–939. doi: 10.1111/apt.14526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawrance I.C., Maxwell L., Doe W. Altered response of intestinal mucosal fibroblasts to profibrogenic cytokines in inflammatory bowel disease. Inflamm Bowel Dis. 2001;7:226–236. doi: 10.1097/00054725-200108000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Rieder F. The gut microbiome in intestinal fibrosis: environmental protector or provocateur? Sci Transl Med. 2013;5:190ps110. doi: 10.1126/scitranslmed.3004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otte J.M., Rosenberg I.M., Podolsky D.K. Intestinal myofibroblasts in innate immune responses of the intestine. Gastroenterology. 2003;124:1866–1878. doi: 10.1016/s0016-5085(03)00403-7. [DOI] [PubMed] [Google Scholar]
- 10.Anzai A., Choi J.L., He S., Fenn A.M., Nairz M., Rattik S., McAlpine C.S., Mindur J.E., Chan C.T., Iwamoto Y., Tricot B., Wojtkiewicz G.R., Weissleder R., Libby P., Nahrendorf M., Stone J.R., Becher B., Swirski F.K. The infarcted myocardium solicits GM-CSF for the detrimental oversupply of inflammatory leukocytes. J Exp Med. 2017;214:3293–3310. doi: 10.1084/jem.20170689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinchen J., Chen H.H., Parikh K., Antanaviciute A., Jagielowicz M., Fawkner-Corbett D., Ashley N., Cubitt L., Mellado-Gomez E., Attar M., Sharma E., Wills Q., Bowden R., Richter F.C., Ahern D., Puri K.D., Henault J., Gervais F., Koohy H., Simmons A. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–386 e317. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L.S., Nayar S., Greenstein A.J., Dubinsky M., Walker L., Leader A., Fine J.S., Whitehurst C.E., Mbow M.L., Kugathasan S., Denson L.A., Hyams J.S., Friedman J.R., Desai P.T., Ko H.M., Laface I., Akturk G., Schadt E.E., Salmon H., Gnjatic S., Rahman A.H., Merad M., Cho J.H., Kenigsberg E. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508 e1420. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J., Sud M., Andrews E., Velonias G., Haber A.L., Jagadeesh K., Vickovic S., Yao J., Stevens C., Dionne D., Nguyen L.T., Villani A.C., Hofree M., Creasey E.A., Huang H., Rozenblatt-Rosen O., Garber J.J., Khalili H., Desch A.N., Daly M.J., Ananthakrishnan A.N., Shalek A.K., Xavier R.J., Regev A. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730 e722. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.West N.R., Hegazy A.N., Owens B.M.J., Bullers S.J., Linggi B., Buonocore S., Coccia M., Gortz D., This S., Stockenhuber K., Pott J., Friedrich M., Ryzhakov G., Baribaud F., Brodmerkel C., Cieluch C., Rahman N., Muller-Newen G., Owens R.J., Kuhl A.A., Maloy K.J., Plevy S.E., Oxford I.B.D.C.I., Keshav S., Travis S.P.L., Powrie F. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 16.Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F., Carvalho A., Puccetti P., Romani L. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Dodd D., Spitzer M.H., Van Treuren W., Merrill B.D., Hryckowian A.J., Higginbottom S.K., Le A., Cowan T.M., Nolan G.P., Fischbach M.A., Sonnenburg J.L. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature. 2017;551:648–652. doi: 10.1038/nature24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsden S.R., Hilton M.G., Waller J.M. The end products of the metabolism of aromatic amino acids by Clostridia. Arch Microbiol. 1976;107:283–288. doi: 10.1007/BF00425340. [DOI] [PubMed] [Google Scholar]
- 19.Wikoff W.R., Anfora A.T., Liu J., Schultz P.G., Lesley S.A., Peters E.C., Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dvorak Z., Kopp F., Costello C.M., Kemp J.S., Li H., Vrzalova A., Stepankova M., Bartonkova I., Jiskrova E., Poulikova K., Vyhlidalova B., Nordstroem L.U., Karunaratne C.V., Ranhotra H.S., Mun K.S., Naren A.P., Murray I.A., Perdew G.H., Brtko J., Toporova L., Schon A., Wallace W.G., Walton W.G., Redinbo M.R., Sun K., Beck A., Kortagere S., Neary M.C., Chandran A., Vishveshwara S., Cavalluzzi M.M., Lentini G., Cui J.Y., Gu H., March J.C., Chatterjee S., Matson A., Wright D., Flannigan K.L., Hirota S.A., Sartor R.B., Mani S. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Mol Med. 2020 doi: 10.15252/emmm.201911621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garg A., Zhao A., Erickson S.L., Mukherjee S., Lau A.J., Alston L., Chang T.K., Mani S., Hirota S.A. Pregnane X receptor activation attenuates inflammation-associated intestinal epithelial barrier dysfunction by inhibiting cytokine-induced myosin light-chain kinase expression and c-Jun N-terminal kinase 1/2 activation. J Pharmacol Exp Ther. 2016;359:91–101. doi: 10.1124/jpet.116.234096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A.P., Qiu Z., Maher L., Redinbo M.R., Phillips R.S., Fleet J.C., Kortagere S., Mukherjee P., Fasano A., Le Ven J., Nicholson J.K., Dumas M.E., Khanna K.M., Mani S. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haughton E.L., Tucker S.J., Marek C.J., Durward E., Leel V., Bascal Z., Monaghan T., Koruth M., Collie-Duguid E., Mann D.A., Trim J.E., Wright M.C. Pregnane X receptor activators inhibit human hepatic stellate cell transdifferentiation in vitro. Gastroenterology. 2006;131:194–209. doi: 10.1053/j.gastro.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Marek C.J., Tucker S.J., Konstantinou D.K., Elrick L.J., Haefner D., Sigalas C., Murray G.I., Goodwin B., Wright M.C. Pregnenolone-16alpha-carbonitrile inhibits rodent liver fibrogenesis via PXR (pregnane X receptor)-dependent and PXR-independent mechanisms. Biochem J. 2005;387:601–608. doi: 10.1042/BJ20041598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyer C., Skapenko A., Distler A., Dees C., Reichert H., Munoz L., Leipe J., Schulze-Koops H., Distler O., Schett G., Distler J.H. Activation of pregnane X receptor inhibits experimental dermal fibrosis. Ann Rheum Dis. 2013;72:621–625. doi: 10.1136/annrheumdis-2012-202476. [DOI] [PubMed] [Google Scholar]
- 26.Melgar S., Karlsson A., Michaelsson E. Acute colitis induced by dextran sulfate sodium progresses to chronicity in C57BL/6 but not in BALB/c mice: correlation between symptoms and inflammation. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1328–G1338. doi: 10.1152/ajpgi.00467.2004. [DOI] [PubMed] [Google Scholar]
- 27.Xing Z., Ohkawara Y., Jordana M., Graham F., Gauldie J. Transfer of granulocyte-macrophage colony-stimulating factor gene to rat lung induces eosinophilia, monocytosis, and fibrotic reactions. J Clin Invest. 1995;97:1102–1110. doi: 10.1172/JCI118503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bento A.F., Leite D.F., Marcon R., Claudino R.F., Dutra R.C., Cola M., Martini A.C., Calixto J.B. Evaluation of chemical mediators and cellular response during acute and chronic gut inflammatory response induced by dextran sodium sulfate in mice. Biochem Pharmacol. 2012;84:1459–1469. doi: 10.1016/j.bcp.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Choi J.S., Kim K.H., Lau L.F. The matricellular protein CCN1 promotes mucosal healing in murine colitis through IL-6. Mucosal Immunol. 2015;8:1285–1296. doi: 10.1038/mi.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopetuso L.R., De Salvo C., Pastorelli L., Rana N., Senkfor H.N., Petito V., Di Martino L., Scaldaferri F., Gasbarrini A., Cominelli F., Abbott D.W., Goodman W.A., Pizarro T.T. IL-33 promotes recovery from acute colitis by inducing miR-320 to stimulate epithelial restitution and repair. Proc Natl Acad Sci U S A. 2018;115:E9362–E9370. doi: 10.1073/pnas.1803613115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones G.R., Bain C.C., Fenton T.M., Kelly A., Brown S.L., Ivens A.C., Travis M.A., Cook P.C., MacDonald A.S. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol. 2018;9:2764. doi: 10.3389/fimmu.2018.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erickson S.L., Alston L., Nieves K., Chang T.K.H., Mani S., Flannigan K.L., Hirota S.A. The xenobiotic sensing pregnane X receptor regulates tissue damage and inflammation triggered by C difficile toxins. FASEB J. 2020;34:2198–2212. doi: 10.1096/fj.201902083RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng B., Zhang Z., Black C.M., de Crombrugghe B., Denton C.P. Ligand-dependent genetic recombination in fibroblasts : a potentially powerful technique for investigating gene function in fibrosis. Am J Pathol. 2002;160:1609–1617. doi: 10.1016/S0002-9440(10)61108-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Florin L., Alter H., Grone H.J., Szabowski A., Schutz G., Angel P. Cre recombinase-mediated gene targeting of mesenchymal cells. Genesis. 2004;38:139–144. doi: 10.1002/gene.20004. [DOI] [PubMed] [Google Scholar]
- 35.Ponticos M., Abraham D., Alexakis C., Lu Q.L., Black C., Partridge T., Bou-Gharios G. Col1a2 enhancer regulates collagen activity during development and in adult tissue repair. Matrix Biol. 2004;22:619–628. doi: 10.1016/j.matbio.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Biernacka A., Dobaczewski M., Frangogiannis N.G. TGF-beta signaling in fibrosis. Growth Factors. 2011;29:196–202. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrance I.C., Rogler G., Bamias G., Breynaert C., Florholmen J., Pellino G., Reif S., Speca S., Latella G. Cellular and molecular mediators of intestinal fibrosis. J Crohns Colitis. 2017;11:1491–1503. doi: 10.1016/j.crohns.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie W., Tian Y. Xenobiotic receptor meets NF-kappaB, a collision in the small bowel. Cell Metab. 2006;4:177–178. doi: 10.1016/j.cmet.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Wera O., Lancellotti P., Oury C. The dual role of neutrophils in inflammatory bowel diseases. J Clin Med. 2016;5:118. doi: 10.3390/jcm5120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bailey C.J., Hembry R.M., Alexander A., Irving M.H., Grant M.E., Shuttleworth C.A. Distribution of the matrix metalloproteinases stromelysin, gelatinases A and B, and collagenase in Crohn’s disease and normal intestine. J Clin Pathol. 1994;47:113–116. doi: 10.1136/jcp.47.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baugh M.D., Perry M.J., Hollander A.P., Davies D.R., Cross S.S., Lobo A.J., Taylor C.J., Evans G.S. Matrix metalloproteinase levels are elevated in inflammatory bowel disease. Gastroenterology. 1999;117:814–822. doi: 10.1016/s0016-5085(99)70339-2. [DOI] [PubMed] [Google Scholar]
- 42.Koelink P.J., Overbeek S.A., Braber S., Morgan M.E., Henricks P.A., Abdul Roda M., Verspaget H.W., Wolfkamp S.C., te Velde A.A., Jones C.W., Jackson P.L., Blalock J.E., Sparidans R.W., Kruijtzer J.A., Garssen J., Folkerts G., Kraneveld A.D. Collagen degradation and neutrophilic infiltration: a vicious circle in inflammatory bowel disease. Gut. 2014;63:578–587. doi: 10.1136/gutjnl-2012-303252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogler G., Hausmann M. Factors promoting development of fibrosis in Crohn’s disease. Front Med (Lausanne) 2017;4:96. doi: 10.3389/fmed.2017.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lloyd-Price J., Arze C., Ananthakrishnan A.N., Schirmer M., Avila-Pacheco J., Poon T.W., Andrews E., Ajami N.J., Bonham K.S., Brislawn C.J., Casero D., Courtney H., Gonzalez A., Graeber T.G., Hall A.B., Lake K., Landers C.J., Mallick H., Plichta D.R., Prasad M., Rahnavard G., Sauk J., Shungin D., Vazquez-Baeza Y., White R.A., 3rd, Investigators I., Braun J., Denson L.A., Jansson J.K., Knight R., Kugathasan S., McGovern D.P.B., Petrosino J.F., Stappenbeck T.S., Winter H.S., Clish C.B., Franzosa E.A., Vlamakis H., Xavier R.J., Huttenhower C. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheibe K., Kersten C., Schmied A., Vieth M., Primbs T., Carle B., Knieling F., Claussen J., Klimowicz A.C., Zheng J., Baum P., Meyer S., Schurmann S., Friedrich O., Waldner M.J., Rath T., Wirtz S., Kollias G., Ekici A.B., Atreya R., Raymond E.L., Mbow M.L., Neurath M.F., Neufert C. Inhibiting interleukin 36 receptor signaling reduces fibrosis in mice with chronic intestinal inflammation. Gastroenterology. 2019;156:1082–1097 e1011. doi: 10.1053/j.gastro.2018.11.029. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto T., Umegae S., Kitagawa T., Matsumoto K. Postoperative change of mucosal inflammation at strictureplasty segment in Crohn’s disease: cytokine production and endoscopic and histologic findings. Dis Colon Rectum. 2005;48:749–757. doi: 10.1007/s10350-004-0826-2. [DOI] [PubMed] [Google Scholar]
- 47.Ballengee C.R., Stidham R.W., Liu C., Kim M.O., Prince J., Mondal K., Baldassano R., Dubinsky M., Markowitz J., Leleiko N., Hyams J., Denson L., Kugathasan S. Association between plasma level of collagen type III alpha 1 chain and development of strictures in pediatric patients with Crohn’s disease. Clin Gastroenterol Hepatol. 2019;17:1799–1806. doi: 10.1016/j.cgh.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burke J.P., Cunningham M.F., Watson R.W., Docherty N.G., Coffey J.C., O’Connell P.R. Bacterial lipopolysaccharide promotes profibrotic activation of intestinal fibroblasts. Br J Surg. 2010;97:1126–1134. doi: 10.1002/bjs.7045. [DOI] [PubMed] [Google Scholar]
- 49.Miyazaki H., Kobayashi R., Ishikawa H., Awano N., Yamagoe S., Miyazaki Y., Matsumoto T. Activation of COL1A2 promoter in human fibroblasts by Escherichia coli. FEMS Immunol Med Microbiol. 2012;65:481–487. doi: 10.1111/j.1574-695X.2012.00979.x. [DOI] [PubMed] [Google Scholar]
- 50.Huang B., Chen Z., Geng L., Wang J., Liang H., Cao Y., Chen H., Huang W., Su M., Wang H., Xu Y., Liu Y., Lu B., Xian H., Li H., Li H., Ren L., Xie J., Ye L., Wang H., Zhao J., Chen P., Zhang L., Zhao S., Zhang T., Xu B., Che D., Si W., Gu X., Zeng L., Wang Y., Li D., Zhan Y., Delfouneso D., Lew A.M., Cui J., Tang W.H., Zhang Y., Gong S., Bai F., Yang M., Zhang Y. Mucosal profiling of pediatric-onset colitis and IBD reveals common pathogenics and therapeutic pathways. Cell. 2019;179:1160–1176 e1124. doi: 10.1016/j.cell.2019.10.027. [DOI] [PubMed] [Google Scholar]
- 51.de Bruyn J.R., Becker M.A., Steenkamer J., Wildenberg M.E., Meijer S.L., Buskens C.J., Bemelman W.A., Lowenberg M., Ponsioen C.Y., van den Brink G.R., D’Haens G.R. Intestinal fibrosis is associated with lack of response to infliximab therapy in Crohn’s disease. PLoS One. 2018;13 doi: 10.1371/journal.pone.0190999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deuring J.J., Li M., Cao W., Chen S., Wang W., de Haar C., van der Woude C.J., Peppelenbosch M. Pregnane X receptor activation constrains mucosal NF-kappaB activity in active inflammatory bowel disease. PLoS One. 2019;14 doi: 10.1371/journal.pone.0221924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prantera C., Lochs H., Grimaldi M., Danese S., Scribano M.L., Gionchetti P., Retic Study G. Rifaximin-extended intestinal release induces remission in patients with moderately active Crohn’s disease. Gastroenterology. 2012;142:473–481 e474. doi: 10.1053/j.gastro.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 54.Terc J., Hansen A., Alston L., Hirota S.A. Pregnane X receptor agonists enhance intestinal epithelial wound healing and repair of the intestinal barrier following the induction of experimental colitis. Eur J Pharm Sci. 2014;55:12–19. doi: 10.1016/j.ejps.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 55.Ren Y., Yue B., Ren G., Yu Z., Luo X., Sun A., Zhang J., Han M., Wang Z., Dou W. Activation of PXR by alantolactone ameliorates DSS-induced experimental colitis via suppressing NF-kappaB signaling pathway. Sci Rep. 2019;9 doi: 10.1038/s41598-019-53305-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shah Y.M., Ma X., Morimura K., Kim I., Gonzalez F.J. Pregnane X receptor activation ameliorates DSS-induced inflammatory bowel disease via inhibition of NF-kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1114–G1122. doi: 10.1152/ajpgi.00528.2006. [DOI] [PubMed] [Google Scholar]
- 57.Huang K., Mukherjee S., DesMarais V., Albanese J.M., Rafti E., Draghi Ii A., Maher L.A., Khanna K.M., Mani S., Matson A.P. Targeting the PXR-TLR4 signaling pathway to reduce intestinal inflammation in an experimental model of necrotizing enterocolitis. Pediatr Res. 2018;83:1031–1040. doi: 10.1038/pr.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacob N., Jacobs J.P., Kumagai K., Ha C.W.Y., Kanazawa Y., Lagishetty V., Altmayer K., Hamill A.M., Von Arx A., Sartor R.B., Devkota S., Braun J., Michelsen K.S., Targan S.R., Shih D.Q. Inflammation-independent TL1A-mediated intestinal fibrosis is dependent on the gut microbiome. Mucosal Immunol. 2018;11:1466–1476. doi: 10.1038/s41385-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monteleone I., Zorzi F., Marafini I., Di Fusco D., Dinallo V., Caruso R., Izzo R., Franze E., Colantoni A., Pallone F., Monteleone G. Aryl hydrocarbon receptor-driven signals inhibit collagen synthesis in the gut. Eur J Immunol. 2016;46:1047–1057. doi: 10.1002/eji.201445228. [DOI] [PubMed] [Google Scholar]
- 60.Moghadamrad S., Hassan M., McCoy K.D., Kirundi J., Kellmann P., De Gottardi A. Attenuated fibrosis in specific pathogen-free microbiota in experimental cholestasis- and toxin-induced liver injury. FASEB J. 2019;33:12464–12476. doi: 10.1096/fj.201901113R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang P.V., Hao L., Offermanns S., Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith P.D., Smythies L.E., Shen R., Greenwell-Wild T., Gliozzi M., Wahl S.M. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 2011;4:31–42. doi: 10.1038/mi.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ueda Y., Kayama H., Jeon S.G., Kusu T., Isaka Y., Rakugi H., Yamamoto M., Takeda K. Commensal microbiota induce LPS hyporesponsiveness in colonic macrophages via the production of IL-10. Int Immunol. 2010;22:953–962. doi: 10.1093/intimm/dxq449. [DOI] [PubMed] [Google Scholar]
- 64.Li H., Illes P., Karunaratne C.V., Nordstrom L.U., Luo X., Yang A., Qiu Y., Kurland I.J., Lukin D.J., Chen W., Jiskrova E., Krasulova K., Pecinkova P., DesMarais V.M., Liu Q., Albanese J.M., Akki A., Longo M., Coffin B., Dou W., Mani S., Dvorak Z. Deciphering structural bases of intestinal and hepatic selectivity in targeting pregnane X receptor with indole-based microbial mimics. Bioorg Chem. 2021;109 doi: 10.1016/j.bioorg.2021.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jalanka J., Salonen A., Salojarvi J., Ritari J., Immonen O., Marciani L., Gowland P., Hoad C., Garsed K., Lam C., Palva A., Spiller R.C., de Vos W.M. Effects of bowel cleansing on the intestinal microbiota. Gut. 2015;64:1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 66.Mow W.S., Vasiliauskas E.A., Lin Y.C., Fleshner P.R., Papadakis K.A., Taylor K.D., Landers C.J., Abreu-Martin M.T., Rotter J.I., Yang H., Targan S.R. Association of antibody responses to microbial antigens and complications of small bowel Crohn’s disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 67.Schoepfer A.M., Schaffer T., Mueller S., Flogerzi B., Vassella E., Seibold-Schmid B., Seibold F. Phenotypic associations of Crohn’s disease with antibodies to flagellins A4-Fla2 and Fla-X, ASCA, p-ANCA, PAB, and NOD2 mutations in a Swiss cohort. Inflamm Bowel Dis. 2009;15:1358–1367. doi: 10.1002/ibd.20892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Tol E.A., Holt L., Li F.L., Kong F.M., Rippe R., Yamauchi M., Pucilowska J., Lund P.K., Sartor R.B. Bacterial cell wall polymers promote intestinal fibrosis by direct stimulation of myofibroblasts. Am J Physiol. 1999;277:G245–G255. doi: 10.1152/ajpgi.1999.277.1.G245. [DOI] [PubMed] [Google Scholar]
- 69.Becher B., Tugues S., Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity. 2016;45:963–973. doi: 10.1016/j.immuni.2016.10.026. [DOI] [PubMed] [Google Scholar]
- 70.Panopoulos A.D., Watowich S.S. Granulocyte colony-stimulating factor: molecular mechanisms of action during steady state and 'emergency' hematopoiesis. Cytokine. 2008;42:277–288. doi: 10.1016/j.cyto.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng Z., Ou L., Liu Y., Liu X., Li F., Sun B., Che Y., Kong D., Yu Y., Steinhoff G. Granulocyte colony-stimulating factor exacerbates cardiac fibrosis after myocardial infarction in a rat model of permanent occlusion. Cardiovasc Res. 2008;80:425–434. doi: 10.1093/cvr/cvn202. [DOI] [PubMed] [Google Scholar]
- 72.Tsantikos E., Lau M., Castelino C.M., Maxwell M.J., Passey S.L., Hansen M.J., McGregor N.E., Sims N.A., Steinfort D.P., Irving L.B., Anderson G.P., Hibbs M.L. Granulocyte-CSF links destructive inflammation and comorbidities in obstructive lung disease. J Clin Invest. 2018;128:2406–2418. doi: 10.1172/JCI98224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dvořák Z., Kopp F., Costello C.M., Kemp J.S., Li H., Vrzalova A., Stepankova M., Bartonkova I., Jiskrova E., Poulikova K., Vyhlidalova B., Norstroem L.U., Karunaratne C., Ranhotra H., Mun K.S., Naren A.P., Murray I., Perdew G.H., Brtko J., Toporova L., Schon A., Wallace B., Walton W.G., Redinbo M.R., Sun K., Beck A., Kortagere S., Neary M.C., Chandran A., Vishveshwara S., Cavalluzzi M.M., Lentini G., Cui J.Y., Gu H., March J.C., Chaterjee S., Matson A., Wright D., Flannigan K.L., Hirota S.A., Mani S. Targeting the pregnane X receptor using microbial metabolite mimicry. EMBO Molecular Medicine. 2020 doi: 10.15252/emmm.201911621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 75.Denning T.L., Wang Y.C., Patel S.R., Williams I.R., Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 76.Batista Lobo S., Denyer M., Britland S., Javid F.A. Development of an intestinal cell culture model to obtain smooth muscle cells and myenteric neurones. J Anat. 2007;211:819–829. doi: 10.1111/j.1469-7580.2007.00820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]