Abstract
Introduction
Artificial intelligence based on machine learning has made large advancements in many fields of science and medicine but its impact on pharmacovigilance is yet unclear.
Objective
The present study conducted a scoping review of the use of artificial intelligence based on machine learning to understand how it is used for pharmacovigilance tasks, characterize differences with other fields, and identify opportunities to improve pharmacovigilance through the use of machine learning.
Design
The PubMed, Embase, Web of Science, and IEEE Xplore databases were searched to identify articles pertaining to the use of machine learning in pharmacovigilance published from the year 2000 to September 2021. After manual screening of 7744 abstracts, a total of 393 papers met the inclusion criteria for further analysis. Extraction of key data on study design, data sources, sample size, and machine learning methodology was performed. Studies with the characteristics of good machine learning practice were defined and manual review focused on identifying studies that fulfilled these criteria and results that showed promise.
Results
The majority of studies (53%) were focused on detecting safety signals using traditional statistical methods. Of the studies that used more recent machine learning methods, 61% used off-the-shelf techniques with minor modifications. Temporal analysis revealed that newer methods such as deep learning have shown increased use in recent years. We found only 42 studies (10%) that reflect current best practices and trends in machine learning. In the subset of 154 papers that focused on data intake and ingestion, 30 (19%) were found to incorporate the same best practices.
Conclusion
Advances from artificial intelligence have yet to fully penetrate pharmacovigilance, although recent studies show signs that this may be changing.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40264-022-01176-1.
Key Points
| We reviewed 393 papers in the intersection of pharmacovigilance (PV) and machine learning, and most involved signal detection as opposed to data intake or data analysis. |
| There has been a rapid rise in the use of deep learning in the PV literature, but corresponding dramatic success has not been seen in other fields such as computer vision, natural language processing, and healthcare. |
| There are opportunities to implement machine learning approaches throughout the PV pipeline. |
Introduction
Pharmacovigilance (PV) is fundamentally a data-driven field as it requires the collection, management, and analysis of a large amount of data gathered from a wide range of disparate sources [1]. The primary type of data used in PV are individual case safety reports (ICSRs), which are records of suspected adverse events collected via multiple channels, aggregated and organized into large databases, and constantly monitored to detect safety signals [2]. ICSRs come from a multitude of sources, including chatbot interactions, electronic health records (EHRs), published literature, patient registries, patient support programs, or even directly from patients via social media [3]. Reports are collected worldwide and characterized by heterogeneity in format, language, and unique characteristics of the underlying healthcare systems. Adverse events must be identified and analyzed in order to find potential emerging safety issues in medicines and vaccines.
The central challenge of PV is how to make sense of these large and heterogeneous data to quickly and reliably find the ‘needles in the haystack,’ which are safety signals that require escalation and triage [4]. Given the rise of artificial intelligence (AI) powered by new advancements in machine learning (ML) across many fields of science [5–7] and medicine [8–11] over the last decade [12, 13], many have speculated [14, 15] that these same technologies could be brought to bear on the core problems of PV. The use of these methods for human safety data first appeared in the early 1990s [16] and has steadily increased since the 2000s. The goal of this review is to systematically identify works that use ML, broadly defined, for safety data to characterize the current state of ML in PV, and to provide clarity on ways that recent advances in AI and ML can be translated to improve various components of PV.
Care must be taken when attempting to extrapolate the success of ML in other areas compared with PV since there are specific factors that may account for the recent success of ML that may or may not be present for PV applications [15]. More so than any other ML technique, it has been the rise of ‘deep learning’ methods that have catalyzed the current AI revolution [13]. These methods are scalable and can train on petabytes of data through the use of graphical processing units (GPUs) [17] and continue to improve even when the performance of non-deep learning methods has saturated [18, 19]. In addition to scalability, deep learning’s modular nature brings the added benefit of easily incorporating domain-specific knowledge (often called an inductive bias) to point the model in the direction of good or parsimonious solutions [20]. Although image recognition is not commonly a task in the current PV pipeline, deep learning models known as convolutional neural networks (CNNs) offer a particularly salient example of how potent the combination of large data and domain knowledge can be.
CNNs were introduced in 1988 [21] but it was not until 2010 when datasets [22] with millions of images became available that they began to transform the field of computer vision [23, 24]. Moreover, the convolution operator and the network structure (modeled loosely on the visual cortex [25]) in CNNs are powerful image-specific inductive biases that give the deep learning model a head start when learning a new image recognition task. Without either of these components (large data and the inductive bias of convolution), it is unlikely that deep learning would have caused the computer vision revolution of the 2010s. Indeed, numerous studies have found that without large data and inductive biases, deep learning is often no better than traditional statistical models [26–28]. These lessons have been born out repeatedly in subsequent applications of game playing [29, 30], biology [5, 6], natural language processing (NLP) [31, 32], and image generation [33–35].
Taken as a whole, it is thus reasonable to expect that a field is unlikely to experience a true paradigm shift from the current crop of deep learning-powered AI techniques without having at least some of these prerequisites in place. Despite the widespread interest in AI and its application to safety data [36, 37], including several review articles [14, 15], there are no scoping reviews that critically assess the extent to which PV is poised to be improved by AI under this framework. Previous reviews have focused on specific elements such as NLP techniques for clinical narrative mining in EHRs [38] or in reducing the frequency or impact of adverse events to patients [39]. Our review is unique in that it seeks to fill this gap to provide a clearer understanding of how current AI/ML practices and standards in PV align with the critical factors for success identified in adjacent areas such as biology and medicine.
To be as comprehensive as possible, we take a broad definition of ML for safety data and include traditional signal detection methods such as Bayesian Confidence Propagation Neural Networks (BCPNN) and related techniques [40, 41] given their roots in ML (see the Methods section for the full search details). We surveyed a 21-year period from the year 2000 to September 2021, reflective of the time before and after significant AI breakthroughs in 2012–2015 [23, 42–46], to see what effect, if any, these results had on PV. The scope of this review is limited only to (1) ingestion of safety data from all sources, including the safety data pipeline, social media, EHRs, and scientific literature, followed by (2) the processing and structuring of data, as well as (3) the processes of analyzing, understanding, linking, and disseminating or sharing, the safety data. While ML has made advances in healthcare more broadly [47–51] and ML research in these areas does have the potential to impact PV, we sought to characterize the use of ML that directly analyzed safety data (e.g. social media, forums, or ICSRs such as those in the FDA Adverse Event Reporting System [FAERS]) and excluded studies that performed adjacent kinds of tasks (e.g. ML research on biochemical pathways or meta research on drug safety). Thus, for the purposes of this review, we have chosen to retain our focus by limiting the review to those topics related specifically to the application of ML and human safety data (i.e. work that explicitly analyzes data on suspected adverse events of drugs and vaccines) for data ingestion or analysis.
Methods
Study Design
We queried four databases (PubMed, Embase, Web of Science Core Collection, and IEEE Xplore) for abstracts of full-text research papers containing terms related to ML and PV. The searches were carried out on 9 September 2021 and were limited to articles published in the year 2000 or later. Furthermore, our review was limited to full-text English articles and conference abstracts (non-English papers were excluded). The full list of search terms and the search query used to identify the articles in this review are available as an electronic supplementary file.
We focused our search criteria on ML terms related to disproportionality analysis, common to PV research, as well as modern ML techniques (e.g. deep learning). This allowed us to compare traditional methods of PV alongside cutting-edge ML research. Articles solely focused on rules-based methods or knowledge graph- or ontology-based methods (e.g. Merrill et al. [52, 53]) were excluded from this review since, on their own, these are not direct ML methods per se.
Two independent reviewers determined if an abstract was in the scope of this review. A third reviewer adjudicated conflicts between reviewers or indecision by a reviewer (indicated by a ‘maybe’ vote). For studies that met the inclusion criteria, one reviewer conducted a full-text review to extract data. Analysis of extracted data was performed using the R statistical programming language [54]. Statistical significance for testing for a difference in proportions Chi-square test enabled subgroup analysis of studies that proposed methods for the intake and processing of safety data. This subgroup was enriched for ML models of interest. Topic modeling, described below, enabled an analysis of temporal trends in methods development.
Evaluation Criteria
To understand the extent to which PV studies are amenable to current trends in the broader ML literature, we assessed each paper using the following criteria:
- Task type: We categorized each study into one of three categories reflective of the primary approach: signal detection, data intake, or data analysis.
- Signal detection: Papers that are ‘traditional’ PV analyses that seek to estimate a statistical quantity (e.g. information component, odds ratio, etc.) for signal detection. This category could also include alternative ML methods for signal detection.
- Data intake: Papers that use ML models to process safety data of various kinds for storage in databases or for downstream activities such as signal detection. Examples include adverse event detection, named entity recognition, and other preprocessing activities.
- Data analysis: Papers that leverage safety data but do not fall into either of the previous categories. Examples include clustering of adverse events and topic modeling.
Dataset and dataset size: We collected the name of the dataset and number of data points each study used to train and/or assess its methodology. In the case of multiple reported dataset sizes, we reported the ‘most specific’ number. For example, if a study reported using millions of safety reports from FAERS, but trained and assessed models on a subset of thousands of reports related to acute kidney injury, we went with the smaller number.
- Modeling approach: We identified the primary algorithm or model used in each study in addition to any secondary algorithms or techniques.
- Examples: BCPNNs, reporting odds ratios (ROR), random forests, transformers, etc.
Method novelty: Given the importance of domain adaptation seen in other fields such as computer vision and NLP, we subjectively assessed whether researchers in each study used an ‘off the shelf’ ML algorithm (e.g. random forests, support vector machines [SVMs]) or used a model tailored to the task, or otherwise made non-trivial modifications to an existing algorithm to improve performance (e.g. beyond hyperparameter tuning).
Use of external information or pretrained models: One of the great benefits of current deep learning models is the ability to leverage external data and pretrained models when labeled data are scarce. We checked for additional information as inputs to the model (e.g. incorporation of known adverse effects or molecular structure). We also looked for the use of models that had been trained on other datasets then transported to the PV task at hand.
Reproducibility: We searched for dataset and code availability to indicate whether or not a study was reproducible. We did consider social media data as publicly available, but acknowledge that it may be difficult to exactly reproduce a dataset based on a social media crawl. For code availability, we identified all papers that provided a link to Github or other web-hosted code, or provided supplementary materials with code. We manually assessed this subset of papers for currently available code (e.g. no dead links).
Overall evaluation: We recorded a binary subjective evaluation indicating whether each study was reflective of the best practices in the broader ML literature (e.g. appropriate inductive biases, no obvious test-train leakage, tuning hyperparameters, cross validation). This determination was based on how well each study reflected the other criteria on this list.
Topic Modeling
We trained a structural topic model (Latent Dirichlet Allocation [LDA]) using the ‘stm’ R package [55]. In order to process the documents included in our final screen, we preprocessed the text to remove all non-alpha-numeric text and removed references (e.g. ‘[1]’). We then removed punctuation and limited analysis to words between 2 and 20 characters. For each document, we only considered words that appeared at least 10 times in that respective document. After preprocessing by removing stop words and stemming words, we needed to select the number of topics, K, to instantiate the topic model with. We considered a range of 5–45 topics and chose K = 25 based on log-likelihood on held-out data, exclusivity, and semantic coherence. Finally, we fit the topic model using a semi-collapsed variational expectation-maximization algorithm regressed on the year of publication.
Results
Main Results
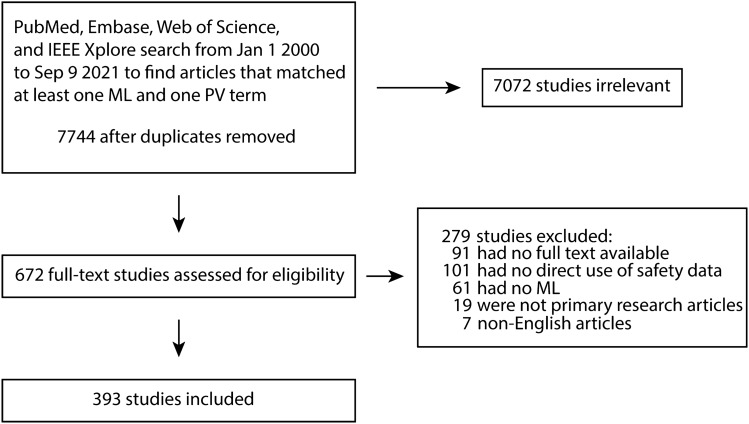
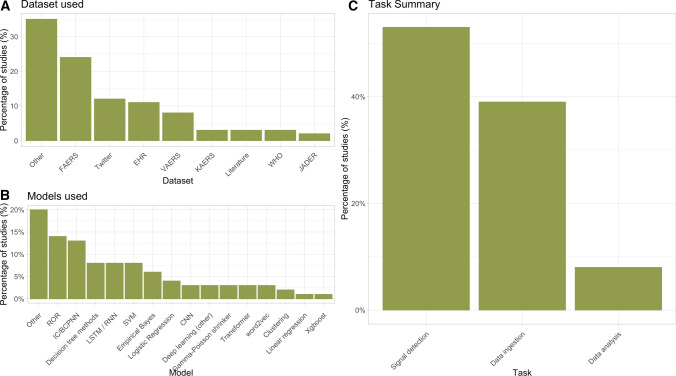
We manually reviewed 7744 unique abstracts that were identified by searching the PubMed, Embase, Web of Science, and IEEE Xplore databases. After manual screening by at least two reviewers, 672 (8.7%) abstracts passed inspection and had their full-text retrieved (Fig. 1). Of these, 279 (41.5%) did not meet the inclusion criteria due to lack of relevance after reviewing the full text, or did not have the full text available, resulting in 393 articles for analysis. Figure 2 displays summary information for the primary datasets and models in addition to the task classification for each study.
Fig. 1.
Summary of inclusion and exclusion process. Articles identified in one of the four databases using keyword and MeSH term searches were manually screened for inclusion. MeSH Medical Subject Heading, ML machine learning, PV pharmacovigilance
Fig. 2.
Summary of datasets, primary algorithms, and task type of the included studies. a Primary dataset used in each study. b Primary analysis method or model used by each study. c Study task classification. EHR electronic health record, FAERS FDA Adverse Event Reporting System, JADER Japanese Adverse Drug Event Report, KAERS Korea Adverse Event Reporting System, VAERS Vaccine Adverse Event Reporting System, WHO World Health Organization, ROR reporting odds ratio, IC information component, BCPNN Bayesian Confidence Propagation Neural Network, LSTM long short-term memory, RNN recurrent neural network, SVM support vector machine, CNN convolutional neural network
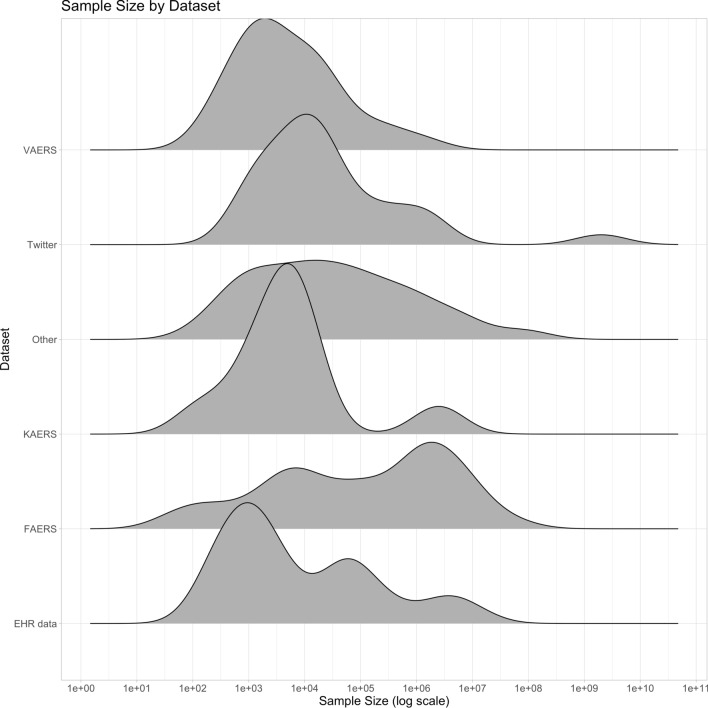
Overall, FAERS was the most popular single database (Fig. 2a) and was used by 24% of the studies. Social media data were used by 12% of studies, while EHR data were used by 11% of studies. Traditional statistical PV methods such as disproportionality scores remain very popular, with 144 (37%) of the included studies using one of them as the primary analysis model. Sample sizes varied greatly across studies and across datasets (Table 1 and Fig. 3), with FAERS being notable for having a mean sample size of 4.3 million and a median sample size of 243,510. Note that the notion of sample size is difficult to compare across data sources since the unit of analysis can be quite different. For example, studies using social media data often reported the number of posts (e.g. Tweets), while EHR studies often reported the number of patients. Ten percent of studies reported no explicit sample size at all.
Table 1.
Summary statistics for types of data utilized and sample sizes used
| Dataset | n | Mean | Median | SD | IQR |
|---|---|---|---|---|---|
| EHR | 43 | 628,859 | 2633 | 2,004,196 | 63,516 |
| FAERS | 93 | 4,306,251 | 243,510 | 14,602,542 | 3,134,268 |
| JADER | 9 | 136,331 | 33,852 | 184,969 | 282,323 |
| KAERS | 12 | 254,293 | 4982 | 789,068 | 6993 |
| Literature | 11 | 17,200 | 3000 | 34,797 | 14,964 |
| Other | 138 | 3,067,200 | 26,508 | 14,025,930 | 259,820 |
| Social media | 47 | 46,685,103 | 14,570 | 304,970,307 | 100,183 |
| VAERS | 30 | 64,363 | 2619 | 234,839 | 13,714 |
| Vigibase | 10 | 365,710 | 11,144 | 941,166 | 14,944 |
SD standard deviation, IQR interquartile range, EHR electronic health record, FAERS FDA Adverse Event Reporting System, JADER Japanese Adverse Drug Event Report, KAERS Korea Adverse Event Reporting System, VAERS Vaccine Adverse Event Reporting System
Fig. 3.
Distribution of sample size for the most popular datasets. EHR electronic health record, FAERS FDA Adverse Event Reporting System, KAERS Korea Adverse Event Reporting System, VAERS Vaccine Adverse Event Reporting System
With respect to method novelty, the vast majority (73%) used ‘off-the-shelf’ methods with little to no problem-specific adaptation or domain knowledge. Method novelty varied with respect to method type; 61% of papers using deep learning or other ML methods had novel adaptations, while only 10% of disproportionality papers did. Similarly, 92% trained a model ‘from scratch’, leaving only 8% of studies that leverage a pretrained model in some capacity, and only 18% explicitly used some kind of external information or data. Six-three percent of the studies used data that were publicly available, while 7% had code that was publicly accessible at some point in time. Our reviewers’ subjective evaluation found that 42 (10%) studies were reflective of modern best practices in ML.
Subgroup Analysis of Data Intake and Pipeline Studies
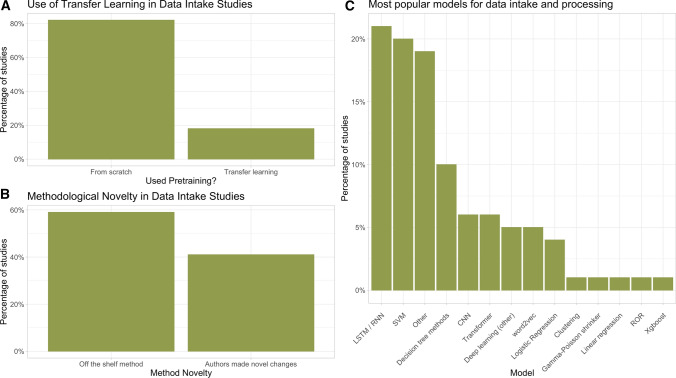
We performed a subgroup analysis of studies that proposed methods for the intake and processing of safety data. The use of transfer learning, methodological novelty, and the types of models used are shown in Fig. 4 and the sample size by dataset is shown in Table 2. Compared with all included studies of the previous section, this category had significantly higher levels of methodological innovation and novelty (40% vs. 27%; p = 0.03) and had higher uses of pretrained models (19% vs. 8%; p = 0.03). We found that 30 (20%) of these studies reflect current best practices, higher than the 10% estimate when considering all included papers.
Fig. 4.
Breakdown of the use of a transfer learning, b methodological novelty, and c popular algorithms for data intake and pre-processing studies. LSTM long short-term memory, RNN recurrent neural network, SVM support vector machine, CNN convolutional neural network, ROR reporting odds ratio
Table 2.
Summary of sample sizes used in intake and processing pipeline studies
| Dataset | n | Mean | Median | SD | IQR |
|---|---|---|---|---|---|
| EHR data | 22 | 327,731 | 1237 | 1,082,972 | 6184 |
| FAERS | 4 | 1,158,504 | 70,357 | 2,223,398 | 1,146,282 |
| Other | 80 | 2,249,619 | 10,296 | 11,094,273 | 267,589 |
| Social media | 26 | 77,012,220 | 13,450 | 392,214,186 | 29,502 |
| VAERS | 3 | 5939 | 6034 | 3380 | 3379 |
SD standard deviation, IQR interquartile range, EHR electronic health record, FAERS FDA Adverse Event Reporting System, VAERS Vaccine Adverse Event Reporting System
Temporal Trends
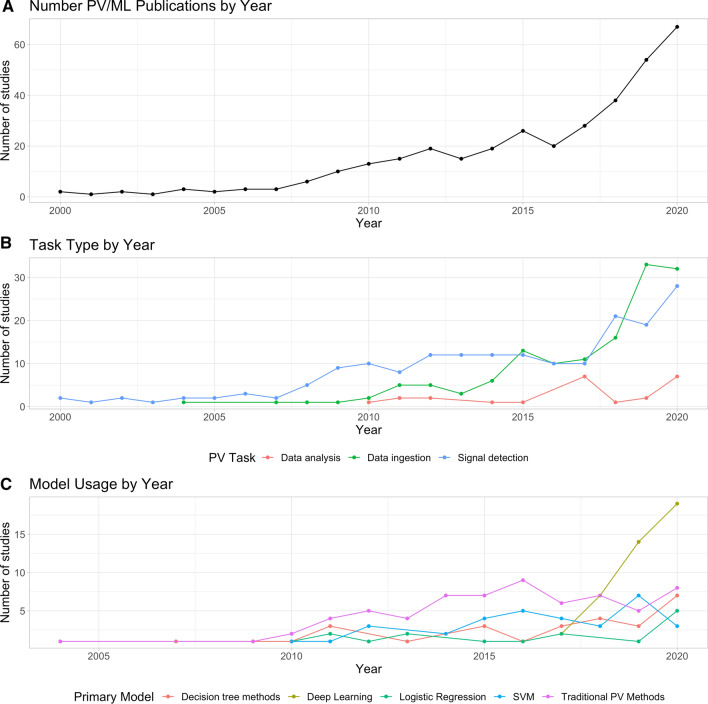
Next, we assessed how some of the patterns from the previous sections varied during the study period. Figure 5 shows trends for the number of publications, task type, and model use for each year in the study period. By and large, the volume of ML-related PV publications went up year-over-year (Fig. 5a). Starting in 2015, the number of studies leveraging ML for data intake (Fig. 5b) markedly increased, which coincided with a rapid increase in the number of studies using deep learning (Fig. 5c), and, by 2020, deep learning was the most popular technique used in the included PV studies. These trends suggest that, especially for data intake studies, ML may be starting to gain traction.
Fig. 5.
Temporal trends in the pharmacovigilance literature. a Total number of publications by year shows an increasing volume of articles that use ML for PV. b The type of task performed by each study. c Trends in usage for several classes of models. ML machine learning, PV pharmacovigilance, SVM support vector machine
Topic Model Analysis
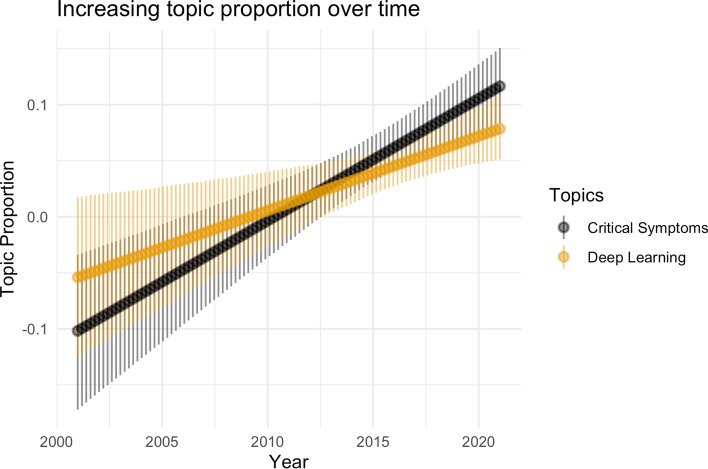
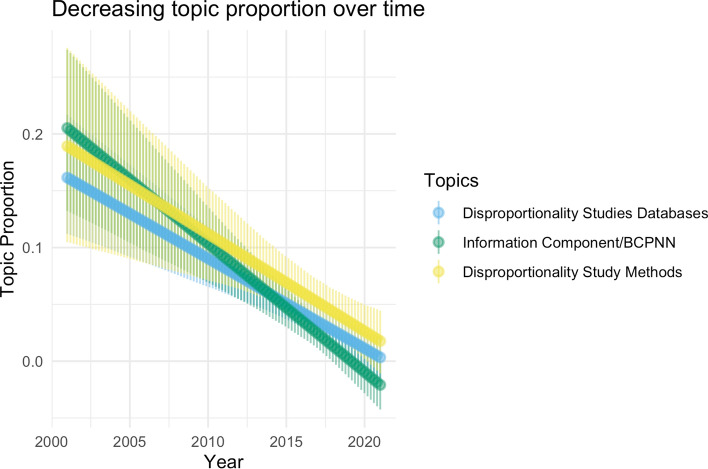
We then performed a topic model analysis of the full text using Latent Dirichlet Allocation (LDA) [56] to see what high-level trends were present. In Table 3, we show four of the most prevalent topics discovered by LDA along with keywords that identify the topic and our subjective assessment of topic focus. We analyzed topic model results using the ‘stm’ and ‘LDAvis’ packages [55, 57]. Figures 6 and 7 show topics whose expected relative proportion increased and decreased, respectively, during the study period. These results align with those from the manual annotation presented in the previous section. Deep learning has seen relative gains in use recently and is likely to see further increases in coming years.
Table 3.
Top four topics discovered by LDA by prevalence
| Topic focus | Keywords | Prevalence (%) |
|---|---|---|
| Deep learning | Embed, LSTM, layer, model | 7.01 |
| Adverse events post-vaccination | Vaccine, VAERS, Guillain–Barré syndrome, report | 6.17 |
| Signal detection | BCPNN, PRR, Gamma Poisson Shrinker, shrinkage, method | 5.87 |
| Information extraction/NLP | Annotate, entity, ADE, sentence | 5.70 |
Topic focus is a subjective assessment of the content based on the most relevant keywords for each topic, as determined by setting the relevancy metric λ to 0.3 in the ‘LDAvis’ package (see electronic supplementary Table S1 for a full list of topics). NLP natural language processing, VAERS Vaccine Adverse Event Reporting System, BCPNN Bayesian Confidence Propagation Neural Network
Fig. 6.
Topics on deep learning and critical systems had their relative expected proportions increase
Fig. 7.
Topics on disproportionality analysis, BCPNN had their relative expected proportions decrease. BCPNN Bayesian Confidence Propagation Neural Network
Discussion
Our scoping review revealed several interesting trends. First and most obvious, traditional signal detection methods in PV (e.g. BCPNN) and data sources remain quite popular and, until very recently, comprised the bulk of signal detection research. That is not to say that the use of these approaches has slowed, but that research development has shifted to other areas of method development. Interest at the intersection of ML and PV is growing; Fig. 5a shows that the number of publications has increased approximately sixfold in the past 10 years alone. Figure 5c indicates that deep learning-based methods have recently eclipsed statistical methods in terms of publication numbers. This may have been enabled by new developments in deep learning for text analysis (e.g. transformers [32]), as initial deep learning progress focused on image recognition and was thus less relevant to PV tasks. Moreover, easy to use frameworks such as Tensorflow and PyTorch have enabled rapid development of ML models. In particular, there has been a rise in the amount of papers focusing on more sophisticated ML techniques. This indicates that the field is shifting towards classification or regression tasks in addition to the more traditional safety signal statistical analyses. Figure 5b shows this fourfold increase in supervised tasks over the last 5 years alone.
In our subgroup analysis of studies that proposed methods for the intake and processing of safety data, we found articles within this category demonstrated higher levels of methodological innovation and novelty (40% vs. 27%; p = 0.03) and made more use of pretrained models (19% vs. 8%; p = 0.03). This could be the result of more freedom to define the task and model when compared with signal detection tasks and the ability to leverage existing pretrained models trained on other types of non-PV text data. Although these crucial ML ingredients were more frequently present in this task, this is lower than what would be expected for other areas where transfer learning is ubiquitous [11, 58].
One limitation of our review is a property of scientific publishing: only novel results are typically published in peer-reviewed journals. For signal detection papers, that means our scoping review has covered both novel methods of signal detection and new drug/adverse event relationships identified by standard methods. In contrast, innovative data intake and analysis methods have been included in our review, but routine use of ML for these parts of PV are missing from the published record. This is partially reflected in our method novelty results; 61% of papers with deep learning or ML made novel changes, while only 10% of disproportionality analyses made novel changes in our reviewers’ estimations. While this bias is unavoidable, we believe that this scoping review likely captures most uses of ML for data ingestion and analysis due to the fact that the rapid rise of ML has been so recent. This bias will affect the ratio between signal detection and other papers, but the conclusions within subgroups should be unaffected.
However, returning to our original framing, it is not yet clear whether PV has assembled the critical mass of ingredients needed to benefit from the recent AI revolution powered primarily by large-scale deep learning methods, although it may be trending in that direction. Figure 3 shows that there is a large amount of human safety data that can serve as fodder for model training. However, most studies are bespoke, one-off exercises that do not introduce methodological novelty beyond what has been published in other areas, and do not use pretrained models or external data despite an increasing use of deep learning methods. In contrast to other areas that have rapid growth and transformation, PV studies still mostly focus on a single task and use a narrow subset of available data, neglecting to use pretrained models or external data. Models that leverage multimodal data (e.g. text and structured data) have been particularly useful in other areas [34, 59, 60]. Another contrast with other areas is the availability of code. Only 7% of PV studies in our review had publicly available code at one point in time (approximately 1% of papers have dead links to code). This is in contrast to 2019 estimates of code availability, where 21% of studies in ML for healthcare more broadly, 39% of computer vision papers, and 48% of NLP papers provided code [61]. Indeed, sharing of code, data, and rapid dissemination of results through preprint servers have accelerated progress in other areas of ML [62–64]. Code-sharing enables others to build on previous work rapidly, which is extremely important when model complexity is high, as can be the case with many complicated deep learning models.
We wish to emphasize that we are not suggesting that PV researchers must follow the deep learning template, nor do we believe that deep learning is the only viable method for PV tasks. However, if PV tasks are to be improved by current approaches based on deep learning, then the criteria of large datasets, the use of pretrained models when appropriate, method novelty, and reproducibility are a reasonable set of requirements. By these criteria, we found that 10% of all studies and 20% of pipeline/intake studies were reflective of current trends based on deep learning. With the increasing use of large datasets and the rise of more modern ML techniques observed in Fig. 5, it is likely safe to project that this percentage will increase over the next several years.
Recommendations
We provide some concrete recommendations that we believe could enhance AI and deep learning applications in PV.
Incorporating domain knowledge: Incorporating domain-specific knowledge biases in ML PV models (e.g. one-dimensional CNNs to detect symptoms next to medications in Tweets or graph neural networks to leverage molecular structure). Known relationships or ontologies that relate symptoms, diseases, and drugs could also be directly incorporated into the model to improve performance [65].
External information and pretrained models: Incorporating external information about mechanisms of action, common adverse effects, or geographic location of reports. This could be used to help triage reports; if multiple reports with the same constellation of symptoms appear for a particular medication that one would not expect (e.g. as encoded by a prior distribution), this would be a clear sign for further investigation. There are numerous pretrained models available for text [66] and molecular data [67] that serve as good foundations for extracting information for PV text and could provide strong prior information when detecting adverse events in case reports.
Methodological innovation available in other areas of ML: Incorporating new advances in ML literature, such as uncertainty quantification [68, 69], federated learning, and fairness ideas. Causal inference [70], an emerging field of epidemiology and computer science, is another promising avenue improving ML PV by incorporating known information about causal relationships directly into the model.
- Data sharing and reproducibility: Common data formats, benchmarks, and code sharing to foster reproducibility. There are several established benchmark datasets that have been used in the literature (n2c2 2018, MADE 1.0, etc.), but there is no equivalent of MNIST or CIFAR-10 for PV that can objectively measure progress on a difficult but standardized task.
- Many efforts (e.g. Lindquist et al. [71], Hochberg et al. [72], Harpaz et al. [73]) towards benchmark tasks have resulted in rich labels for true positive and true negative drug/ADR relationships. However, these datasets do not come with accompanying preprocessed data (e.g. safety reports or social media posts) that would provide an easy-to-use benchmark.
- Few investigations release their data after they scrape a public social media platform or forum. Although in theory one could recreate the authors’ work, it is nearly impossible to capture the exact same posts and process them in the same way.
- Studies have, in general, not published their code or calculations. We appreciate this is more understandable and less problematic for disproportionality analysis with reporting odds ratio, proportional reporting ratio (PRR), or BCPNN, but even for such studies, there is value in code provision to enable reproducibility. For projects that include more complex/modern ML models, public code repositories are lacking. In contrast to PV, this has been an influential factor that has spurred rapid development in the ML community.
Promising Near-Term Applications of Machine Learning in Pharmacovigilance
While our review found that there is still much room for improvement, we wish to offer some near-term tasks that could benefit from the well-executed use of ML today. We offer suggestions for areas across the PV pipeline [74], where ML may have impact in the near- to medium-term, but note that some of these tasks will likely still require substantial effort to achieve.
Translation and multi-language models: Case reports and other safety data can be submitted from anywhere in the world and written in hundreds of different languages. Processing these often necessitates translation into a common language of record before further evaluations can take place. In recent years, ML has become exceedingly good at translation [75, 76], even for low-resource languages that do not have large amounts of training text available [77, 78]. There are even individual models that have been trained on vast amounts of language data and are capable of processing hundreds of languages [79]. Moreover, many of these models are publicly available and could be easily repurposed for the PV intake and processing pipeline. Integrating the translation model directly into the PV pipeline in a trainable way will allow it to adapt the capabilities to a variety of tasks when compared with treating translation as an auxiliary and separate preprocessing task.
Named Entity Recognition (NER): Automatic extraction of key phrases and nouns is a common task in PV data intake and is known in the NLP literature as named entity recognition. There has been rapid progress on this task in other areas of ML [31, 80–82], including scenarios with multiple languages [83, 84], biomedical applications [85–87], and when labeled data are scarce (see the example in Yao et al. [88] detailed in the next section).
Text summarization and generation: Case reports can often contain large volumes of unstructured text that individual case examiners must sift through and synthesize. Abstractive summarization by deep learning has also experienced an impressive leap in capabilities in recent years [89, 90] and thus could easily be applied to the analogous task in PV. Likewise, reports must be generated using codified and structured data, and the generative capabilities of deep learning models could be used for this task.
Causal inference: The critical question of PV is whether a drug is actually causing the adverse events that have been reported in safety reports. Causal inference [91] is a statistical field that provides estimates of treatment effects in real-world data. There has recently been heavy interest in the intersection of causal inference and ML [92]. There is a nearly one-to-one translation of the ideas of causal inference to PV and this could serve as another tool for signal detection and data analysis.
Exemplar Studies
In this section, we wish to highlight several studies that were determined to reflect current ideas and trends in ML to provide good exemplars for how future studies might be conducted. Du et al. [88] provide an example of how an accurate adverse event annotation pipeline can be built, even when there are not large amounts of annotated data, using transfer and self-supervised learning. In the investigation, the authors only had a small set of labeled data in the form of 91 annotated VAERS reports, and the goal was to construct an ML system to automatically extract mentions of named entities (e.g. adverse events, procedures, social circumstances, etc.) from the reports. They accomplished this goal by leveraging a pretrained transformer model known as BioBERT [66] that was fine-tuned on an unannotated set of 43,240 VAERS reports. They show that this approach leads to significantly better performance for this task when compared with traditional NLP methods and when compared against deep learning methods that did not employ transfer learning. Additionally, the annotated dataset they created is publicly available so that others may build on their work [93].
Zhang et al. [94] showed how ML can be useful in complex adverse drug reaction recognition tasks. Adverse drug reactions can be found in all types of media, including scientific literature, EHR data, and Tweets. In these settings, the drug and reaction are not necessarily in the same sentence or near each other in text. Typical ML methods rely on local semantic information (e.g. words in a single sentence) and can struggle in identifying these adverse drug reactions. Zhang et al. leverage a novel mechanism known as multi-hop attention to endow models with the ability to focus across multiple words in a single sentence and between sentences. They used the publicly available benchmarks TwiMed and ADE to assess model performance and to compare with baselines. They demonstrate that their method outperforms well-established ML models such as SVM, CNNs, and LSTMs. Additionally, they show multi-hop attention is superior at identifying adverse drug reactions compared with self-attention and multi-head self-attention, two recent mechanisms found in transformer models. They also performed a comprehensive ablation study to isolate which of their innovations resulted in improved performance.
Finally, Wang et al. [95] demonstrate how ML can assist in determining causation from case reports. The authors utilize causal inference, which is a conceptual framework that, under certain assumptions, allows for estimation of causal effects. This means that one can answer counterfactual or ‘what if’ questions such as ‘what if a patient took medication A rather than medication B?’. They combine causal inference with transformer models that are trained on FAERS safety reports. Wang et al. assess their proposed transformer-causal inference model on two tasks: identifying causes of analgesic-induced acute liver failure and identifying causes of tramadol-related mortalities. Their model is able to recapitulate known risk factors for these adverse events (e.g. acetaminophen consumption for liver failure, and suicidality for tramadol mortalities). Moreover, the model was able to identify potential secondary risk factors that predispose individuals to liver failure. Importantly, Wang et al. published their code and preprocessed data (i.e. FAERS reports). This will enable future researchers to reproduce and extend their work.
Conclusions
We have conducted a scoping review of the use of ML for PV applications. Our aim was to assess the extent to which PV has been or is ready to be improved by current deep learning-based AI techniques. We found that while certain modern practices have begun to appear, many of the primary reasons for the recent success of AI have yet to be translated. We conclude that without certain structural changes, PV is unlikely to experience similar kinds of advancements from current approaches to AI.
Supplementary Information
Below is the link to the electronic supplementary material.
Declarations
Funding
GlaxoSmithKline (GSK) was a sponsor of this project and provided funding to Harvard. No additional external funding was received for the conduct of this study or the preparation of this article.
Conflicts of Interest
Jeffery L. Painter and Andrew Bate were current full-time employees and stockholders of GSK at the time this study was completed. Benjamin Kompa, Joe B. Hakim, Anil Palepu, Kathryn Grace Kompa, Michael Smith, Paul A. Bain, Stephen Woloszynek, and Andrew L. Beam have no conflicts of interest to declare.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data
The datasets generated and/or analyzed during this scoping review are available on Github (https://github.com/beamlab-hsph/drug_safety_scopingreview). All references included in the final analysis are available in the ‘references’ folder in the repo. Additionally, a companion website to our review is available at https://beamlab.shinyapps.io/MLforPVReview.
Code Availability
The code to reproduce our figures and LDA analysis is available at https://github.com/beamlab-hsph/drug_safety_scopingreview
Authors’ Contributions
Conceptualization: AB, ALB. Methodology: BK, JBH, PAB, SW, JLP, AB, ALB. Query formation: BK, JBH, PAB, JLP, AB, ALB. Abstract review: BK, JBH, AP, KGK, MS, SW, ALB. Abstract review conflict adjudication: BK, ALB. Full-text review: BK, JBH, AP, KGK, MS, SW, ALB. Full-text conflict adjudication: BK, ALB. Data extraction: BK, JBH, AP, KGK, MS, ALB. Formal analysis and investigation: BK, SW, ALB. Writing – original draft preparation: BK, ALB. Writing – review and editing: BK, JLP, AB, ALB. Funding acquisition: AB, ALB. Resources: AB, ALB. Supervision: AB, ALB.All authors read and approved the final version.
Footnotes
Link to PRISMA-ScR Checklist: http://prisma-statement.org/Extensions/ScopingReviews.
The original online version of this article was revised due to a retrospective Open Access order.
Change history
2/24/2023
A Correction to this paper has been published: 10.1007/s40264-023-01273-9
References
- 1.Mann RD, Andrews EB. Pharmacovigilance. John Wiley & Sons; 2007.
- 2.Office of the Commissioner. Individual Case Safety Reports. 2019. Available at: https://www.fda.gov/industry/fda-resources-data-standards/individual-case-safety-reports
- 3.Stergiopoulos S, Fehrle M, Caubel P, Tan L, Jebson L. Adverse drug reaction case safety practices in large biopharmaceutical organizations from 2007 to 2017: an industry survey. Pharmaceutical Med. 2019;33(6):499–510. doi: 10.1007/s40290-019-00307-x. [DOI] [PubMed] [Google Scholar]
- 4.Edwards RI. Adverse drug reactions: finding the needle in the haystack. BMJ. 1997;315:500. doi: 10.1136/bmj.315.7107.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stokes JM, Yang K, Swanson K, Jin W, Cubillos-Ruiz A, Donghia NM, et al. A Deep learning approach to antibiotic discovery. Cell. 2020;180:688–702.e13. doi: 10.1016/j.cell.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ML, Doddi A, Royer J, Freschi L, Schito M, Ezewudo M, et al. Beyond multidrug resistance: leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. EBioMedicine. 2019;43:356–369. doi: 10.1016/j.ebiom.2019.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;304:649–656. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 9.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, et al. CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv [cs.CV]. 2017. Available at: http://arxiv.org/abs/1711.05225
- 10.Beam AL, Kohane IS. Big data and machine learning in health care. JAMA. 2018;319:1317–1318. doi: 10.1001/jama.2017.18391. [DOI] [PubMed] [Google Scholar]
- 11.Schmaltz A, Beam AL. Sharpening the resolution on data matters: a brief roadmap for understanding deep learning for medical data. Spine J. 2021;21:1606–1609. doi: 10.1016/j.spinee.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beam AL, Kohane IS. Translating artificial intelligence into clinical care. JAMA. 2016;316(22):2368–2369. doi: 10.1001/jama.2016.17217. [DOI] [PubMed] [Google Scholar]
- 13.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 14.Basile AO, Yahi A, Tatonetti NP. Artificial intelligence for drug toxicity and safety. Trends Pharmacol Sci. 2019;40:624–635. doi: 10.1016/j.tips.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bate A, Hobbiger SF. Artificial intelligence, real-world automation and the safety of medicines. Drug Saf. 2021;44:125–132. doi: 10.1007/s40264-020-01001-7. [DOI] [PubMed] [Google Scholar]
- 16.Alvager T, Smith TJ, Vijai F. Neural-network applications for analysis of adverse drug reactions. Biomed Instrum Technol. 1993;27:408–411. [PubMed] [Google Scholar]
- 17.Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: a system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ′16). 2–4 Nov 2016: Savannah, GA. pp. 265–83. Available at: https://www.usenix.org/conference/osdi16/technical-sessions/presentation/abadi
- 18.Kaplan J, McCandlish S, Henighan T, Brown TB, Chess B, Child R, et al. Scaling laws for neural language models. arXiv [cs.LG]. 2020. Available at: http://arxiv.org/abs/2001.08361
- 19.Whitman N. A bitter lesson. Acad Med. 1999;74(1):1. doi: 10.1097/00001888-199901001-00001. [DOI] [PubMed] [Google Scholar]
- 20.Bronstein MM, Bruna J, LeCun Y, Szlam A, Vandergheynst P. Geometric deep learning: going beyond Euclidean data. IEEE Signal Process Mag. 2017;34:18–42. doi: 10.1109/MSP.2017.2693418. [DOI] [Google Scholar]
- 21.Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86(11):2278–2324. doi: 10.1109/5.726791. [DOI] [Google Scholar]
- 22.Deng J, Dong W, Socher R, Li LJ, Li K. Imagenet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. 2009. pp. 248–155
- 23.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. NIPS. 2012. p. 4. Available at: https://proceedings.neurips.cc/paper/2012/file/c399862d3b9d6b76c8436e924a68c45b-Paper.pdf
- 24.Cireşan D, Meier U, Masci J, Schmidhuber J. A committee of neural networks for traffic sign classification. The 2011 International Joint Conference on Neural Networks; 2011. pp. 1918–21.
- 25.Lindsay GW. Convolutional neural networks as a model of the visual system: past, present, and future. J Cogn Neurosci. 2021;33:2017–2031. doi: 10.1162/jocn_a_01544. [DOI] [PubMed] [Google Scholar]
- 26.Christodoulou E, Ma J, Collins GS, Steyerberg EW, Verbakel JY, Van Calster B. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019;110:12–22. doi: 10.1016/j.jclinepi.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Bellamy D, Celi L, Beam AL. Evaluating progress on machine learning for longitudinal electronic healthcare data. arXiv [cs.LG]. 2020. Available at: http://arxiv.org/abs/2010.01149
- 28.Xiao C, Choi E, Sun J. Opportunities and challenges in developing deep learning models using electronic health records data: a systematic review. J Am Med Inform Assoc. 2018;25:1419–1428. doi: 10.1093/jamia/ocy068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassabis D. AlphaGo: using machine learning to master the ancient game of go. Google blog. 2016. Available at: https://googleblog.blogspot.com/2016/01/alphago-machine-learning-game-go.html
- 30.Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J. Human-level control through deep reinforcement learning. Nature. 2015;518:523–533. doi: 10.1038/nature14236. [DOI] [PubMed] [Google Scholar]
- 31.Brown TB, Mann B, Ryder N, Subbiah M, Kaplan J, Dhariwal P, et al. Language models are few-shot learners. arXiv [cs.CL]. 2020. Available at: http://arxiv.org/abs/2005.14165
- 32.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, et al. Attention is all you need. In: Guyon I, Luxburg UV, Bengio S, Wallach H, Fergus R, Vishwanathan S, et al. (eds). Advances in neural information processing systems 30. Curran Associates, Inc.; 2017. pp. 5998–6008. Available at: http://papers.nips.cc/paper/7181-attention-is-all-you-need.pdf
- 33.Radford A, Kim JW, Hallacy C, Ramesh A, Goh G, Agarwal S, et al. Learning transferable visual models from natural language supervision. arXiv [cs.CV]. 2021. Available at: http://arxiv.org/abs/2103.00020
- 34.Ramesh A, Pavlov M, Goh G, Gray S, Voss C, Radford A, et al. Zero-shot text-to-image generation. arXiv [cs.CV]. 2021. Available at: http://arxiv.org/abs/2102.12092
- 35.Karras T, Aila T, Laine S, Lehtinen J. Progressive growing of GANs for improved quality, stability, and variation. arXiv [cs.NE]. 2017. Available at: http://arxiv.org/abs/1710.10196
- 36.Schmider J, Kumar K, LaForest C, Swankoski B, Naim K, Caubel PM. Innovation in pharmacovigilance: use of artificial intelligence in adverse event case processing. Clin Pharmacol Ther. 2019;105:954–961. doi: 10.1002/cpt.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Surendra E, Garlapati R. Development of pharmacovigilance culture. Int J Pharmacy Res Technol 2020;10:01–4. Available at: http://www.ijccts.org/fulltext/17-1569150791.pdf (Synthesishub-Advanced Scientific Research)
- 38.Luo Y, Thompson WK, Herr TM, Zeng Z, Berendsen MA, Jonnalagadda SR, et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf. 2017;40:1075–1089. doi: 10.1007/s40264-017-0558-6. [DOI] [PubMed] [Google Scholar]
- 39.Syrowatka A, Song W, Amato MG, Foer D, Edrees H, Co Z, et al. Key use cases for artificial intelligence to reduce the frequency of adverse drug events: a scoping review. Lancet Digit Health. 2022;4:e137–e148. doi: 10.1016/S2589-7500(21)00229-6. [DOI] [PubMed] [Google Scholar]
- 40.Bate A. Bayesian confidence propagation neural network. Drug Saf. 2007;30:623–625. doi: 10.2165/00002018-200730070-00011. [DOI] [PubMed] [Google Scholar]
- 41.Szarfman A, Machado SG, O’Neill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA’s spontaneous reports database. Drug Saf. 2002;25:381–392. doi: 10.2165/00002018-200225060-00001. [DOI] [PubMed] [Google Scholar]
- 42.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015. pp. 1–9. Available at: https://www.cv-foundation.org/openaccess/content_cvpr_2015/html/Szegedy_Going_Deeper_With_2015_CVPR_paper.html
- 43.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. arXiv preprint. 2015; Available at: http://arxiv.org/abs/1512.03385
- 44.Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–58. Available at: http://www.jmlr.org/papers/volume15/srivastava14a/srivastava14a.pdf?utm_content=buffer79b43&utm_medium=social&utm_source=twitter.com&utm_campaign=buffer
- 45.Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative Adversarial Nets. In: Ghahramani Z, Welling M, Cortes C, Lawrence N, Weinberger KQ (eds). Advances in neural information processing systems. Curran Associates, Inc.; 2014. Available at: https://proceedings.neurips.cc/paper/2014/file/5ca3e9b122f61f8f06494c97b1afccf3-Paper.pdf
- 46.Ioffe S, Szegedy C. Batch Normalization: Accelerating deep network training by reducing internal covariate shift. In: Bach F, Blei D (eds). Proceedings of the 32nd international conference on machine learning. Lille, France: PMLR; 2015. pp. 448–56. Available at: https://proceedings.mlr.press/v37/ioffe15.html
- 47.Ben-Israel D, Bradley Jacobs W, Casha S, Lang S, Ryu WHA, de Lotbiniere-Bassett M, et al. The impact of machine learning on patient care: a systematic review. Artif Intell Med. 2020;103:101785. doi: 10.1016/j.artmed.2019.101785. [DOI] [PubMed] [Google Scholar]
- 48.Senders JT, Staples PC, Karhade AV, Zaki MM, Gormley WB, Broekman MLD, et al. Machine learning and neurosurgical outcome prediction: a systematic review. World Neurosurg. 2018;109:476–86.e1. doi: 10.1016/j.wneu.2017.09.149. [DOI] [PubMed] [Google Scholar]
- 49.Stafford IS, Kellermann M, Mossotto E, Beattie RM, MacArthur BD, Ennis S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit Med. 2020;3:30. doi: 10.1038/s41746-020-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. bioRxiv. 2018; Available at: http://biorxiv.org/content/early/2018/01/19/142760.abstract [DOI] [PMC free article] [PubMed]
- 51.Rivera SC, Liu X, Chan A-W, Denniston AK, Calvert MJ, SPIRIT-AI and CONSORT-AI Working Group Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. BMJ. 2020;370:m3210. doi: 10.1136/bmj.m3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merrill GH, Ryan PB, Painter JL. Construction and annotation of a UMLS/SNOMED-based drug ontology for observational pharmacovigilance. Methods. 2008; Available from: https://www.academia.edu/4416282/Construction_and_Annotation_of_a_UMLS_SNOMED-based_Drug_Ontology_for_Observational_Pharmacovigilance
- 53.Merrill GH, Ryan PB, Eng M, Painter JL. Using SNOMED to normalize and aggregate drug references in the Safetyworks observational pharmacovigilance project. KR-MED. 2008;126. Available at: https://www.researchgate.net/profile/Christopher-Seebregts/publication/30511023_Integration_of_SNOMED_CT_into_the_OpenMRS_electronic_medical_record_system_framework/links/02bfe50de830584300000000/Integration-of-SNOMED-CT-into-the-OpenMRS-electronic-medical-record-system-framework.pdf#page=131
- 54.Ihaka R, Gentleman R. R: a language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 55.Roberts ME, Stewart BM, Tingley D. stm: R package for Structural Topic Models. 2017. Available at: http://www.structuraltopicmodel.com.
- 56.Jelodar H, Wang Y, Yuan C, Feng X, Jiang X, Li Y, et al. Latent Dirichlet allocation (LDA) and topic modeling: models, applications, a survey. Multimed Tools Appl. 2019;78:15169–15211. doi: 10.1007/s11042-018-6894-4. [DOI] [Google Scholar]
- 57.Sievert C, Shirley K. LDAvis: a method for visualizing and interpreting topics. Proceedings of the workshop on interactive language learning, visualization, and interfaces. 2014. pp. 63–70. Available at: https://aclanthology.org/W14-3110.pdf
- 58.Ribani R, Marengoni M. A survey of transfer learning for convolutional neural networks. In: 2019 32nd SIBGRAPI conference on graphics, patterns and images tutorials (SIBGRAPI-T). 2019. pp. 47–57.
- 59.Zhang Y, Jiang H, Miura Y, Manning CD, Langlotz CP. Contrastive learning of medical visual representations from paired images and text. arXiv [cs.CV]. 2020. Available at: http://arxiv.org/abs/2010.00747
- 60.Azizi S, Mustafa B, Ryan F, Beaver Z, Freyberg J, Deaton J, et al. Big self-supervised models advance medical image classification. arXiv [eess.IV]. 2021. Available at: http://arxiv.org/abs/2101.05224
- 61.McDermott MBA, Wang S, Marinsek N, Ranganath R, Foschini L, Ghassemi M. Reproducibility in machine learning for health research: still a ways to go. Sci Transl Med. 2021 doi: 10.1126/scitranslmed.abb1655. [DOI] [PubMed] [Google Scholar]
- 62.Oakden-Rayner L, Beam AL, Palmer LJ. Medical journals should embrace preprints to address the reproducibility crisis. Int J Epidemiol. 2018;47:1363–1365. doi: 10.1093/ije/dyy105. [DOI] [PubMed] [Google Scholar]
- 63.Beam AL, Manrai AK, Ghassemi M. Challenges to the reproducibility of machine learning models in health care. JAMA. 2020;323(4):305–306. doi: 10.1001/jama.2019.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDermott MBA, Wang S, Marinsek N, Ranganath R, Ghassemi M, Foschini L. Reproducibility in machine learning for health. arXiv [cs.LG]. 2019. Available at: http://arxiv.org/abs/1907.01463 [DOI] [PubMed]
- 65.Choi E, Bahadori MT, Song L, Stewart WF, Sun J. GRAM: graph-based attention model for healthcare representation learning. KDD. 2017;2017:787–795. doi: 10.1145/3097983.3098126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee J, Yoon W, Kim S, Kim D, Kim S, So CH, et al. BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics. 2020;36:1234–1240. doi: 10.1093/bioinformatics/btz682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Honda S, Shi S, Ueda HR. SMILES transformer: pre-trained molecular fingerprint for low data drug discovery. arXiv [cs.LG]. 2019. Available at: http://arxiv.org/abs/1911.04738
- 68.Kompa B, Snoek J, Beam A. Empirical frequentist coverage of deep learning uncertainty quantification procedures. Entropy. 2021;23(12):1608. doi: 10.3390/e23121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kompa B, Snoek J, Beam AL. Second opinion needed: communicating uncertainty in medical machine learning. NPJ Digit Med. 2021;4:4. doi: 10.1038/s41746-020-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hernan MA, Robins JM. Causal inference. Taylor & Francis; 2023. [Google Scholar]
- 71.Lindquist M, Ståhl M, Bate A, Edwards IR, Meyboom RH. A retrospective evaluation of a data mining approach to aid finding new adverse drug reaction signals in the WHO international database. Drug Saf. 2000;23:533–542. doi: 10.2165/00002018-200023060-00004. [DOI] [PubMed] [Google Scholar]
- 72.Hochberg AM, Hauben M, Pearson RK, O’Hara DJ, Reisinger SJ, Goldsmith DI, et al. An evaluation of three signal-detection algorithms using a highly inclusive reference event database. Drug Saf. 2009;32:509–525. doi: 10.2165/00002018-200932060-00007. [DOI] [PubMed] [Google Scholar]
- 73.Harpaz R, Odgers D, Gaskin G, DuMouchel W, Winnenburg R, Bodenreider O, et al. A time-indexed reference standard of adverse drug reactions. Sci Data. 2014;1:140043. doi: 10.1038/sdata.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ghosh R, Kempf D, Pufko A, Barrios Martinez LF, Davis CM, Sethi S. Automation opportunities in pharmacovigilance: an industry survey. Pharmaceut Med. 2020;34:7–18. doi: 10.1007/s40290-019-00320-0. [DOI] [PubMed] [Google Scholar]
- 75.Brants T, Popat AC, Xu P, Och FJ, Dean J. Large language models in machine translation. 2007; Available at: http://research.google/pubs/pub33278.pdf
- 76.Singh SP, Kumar A, Darbari H, Singh L, Rastogi A, Jain S. Machine translation using deep learning: an overview. In: 2017 International Conference on Computer, Communications and Electronics (Comptelix). 2017. pp. 162–7.
- 77.Cui J, Kingsbury B, Ramabhadran B, Saon G, Sercu T, Audhkhasi K, et al. Knowledge distillation across ensembles of multilingual models for low-resource languages. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). 2017. pp. 4825–9.
- 78.Murthy R, Khapra MM, Bhattacharyya P. Improving NER tagging performance in low-resource languages via multilingual learning. In: ACM Transactions on Asian and Low-Resource Language Information Processing. 2019. pp. 1–20.
- 79.Fan A, Bhosale S, Schwenk H, Ma Z, El-Kishky A, Goyal S, et al. Beyond english-centric multilingual machine translation. J Mach Learn Res. 2021;22:1–48. [Google Scholar]
- 80.Li J, Sun A, Han J, Li C. A survey on deep learning for named entity recognition. IEEE Trans Knowl Data Eng. 2020 doi: 10.1109/TKDE.2020.2981314. [DOI] [Google Scholar]
- 81.Arkhipov M, Trofimova M, Kuratov Y, Sorokin A. Tuning multilingual transformers for language-specific named entity recognition. In: Proceedings of the 7th Workshop on Balto-Slavic Natural Language Processing. 2019. pp. 89–93. Available at: https://www.aclweb.org/anthology/W19-3712.pdf
- 82.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv [cs.CL]. 2018. Available at: http://arxiv.org/abs/1810.04805
- 83.Wu S, Song X, Feng Z. MECT: Multi-metadata embedding based cross-transformer for chinese named entity recognition. arXiv [cs.CL]. 2021. Available at: http://arxiv.org/abs/2107.05418
- 84.Arkhipov M, Trofimova M, Kuratov Y, Sorokin A. Tuning multilingual transformers for named entity recognition on slavic languages. BSNLP-2019. 2019;89. Available at: http://aclanthology.lst.uni-saarland.de/W19-37.pdf#page=101
- 85.Wang X, Yang C, Guan R. A comparative study for biomedical named entity recognition. Int J Mach Learn Cybern. 2018;9:373–382. doi: 10.1007/s13042-015-0426-6. [DOI] [Google Scholar]
- 86.Yao L, Liu H, Liu Y, Li X, Anwar MW. Biomedical named entity recognition based on deep neutral network. Int J Hybrid Inf Technol. Global Vision Press; 2015;8:279–88. Available from: http://gvpress.com/journals/IJHIT/vol8_no8/29.pdf
- 87.Habibi M, Weber L, Neves M, Wiegandt DL, Leser U. Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics. 2017;33:i37–48. doi: 10.1093/bioinformatics/btx228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Du J, Xiang Y, Sankaranarayanapillai M, Zhang M, Wang J, Si Y, et al. Extracting postmarketing adverse events from safety reports in the vaccine adverse event reporting system (VAERS) using deep learning. J Am Med Inform Assoc. 2021;28:1393–1400. doi: 10.1093/jamia/ocab014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paulus R, Xiong C, Socher R. A deep reinforced model for abstractive summarization. arXiv [cs.CL]. 2017. Available at: http://arxiv.org/abs/1705.04304
- 90.Gehrmann S, Deng Y, Rush AM. Bottom-up abstractive summarization. arXiv [cs.CL]. 2018. Available at: http://arxiv.org/abs/1808.10792
- 91.Hernán MA, Robins JM. Causal inference. Boca Raton, FL: CRC; 2010. Available at: https://grass.upc.edu/en/seminar/presentation-files/causal-inference/chapters-1-i-2/@@download/file/BookHernanRobinsCap1_2.pdf
- 92.Luo Y, Peng J, Ma J. When causal inference meets deep learning. Nat Mach Intell. 2020;2:426–427. doi: 10.1038/s42256-020-0218-x. [DOI] [Google Scholar]
- 93.Vaccine-AE-recognition. Github [cited 16 Nov 2021]. Available at: https://github.com/UT-Tao-group/Vaccine-AE-recognition
- 94.Zhang T, Lin H, Ren Y, Yang L, Xu B, Yang Z, et al. Adverse drug reaction detection via a multihop self-attention mechanism. BMC Bioinform. 2019;20:479. doi: 10.1186/s12859-019-3053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, Xu X, Tong W, Roberts R, Liu Z. InferBERT: a transformer-based causal inference framework for enhancing pharmacovigilance. Front Artif Intell. 2021;4:659622. doi: 10.3389/frai.2021.659622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.