Abstract
Soil salinity is among the abiotic stressors that threaten agriculture the most, and purslane (Portulaca oleracea L.) is a dicot species adapted to inland salt desert and saline habitats that hyper accumulates salt and has high phytoremediation potential. Many researchers consider purslane a suitable model species to study the mechanisms of plant tolerance to drought and salt stresses. Here, a robust salinity stress protocol was developed and used to characterize the morphophysiological responses of young purslane plants to salinity stress; then, leaf tissue underwent characterization by distinct omics platforms to gain further insights into its response to very high salinity stress. The salinity stress protocol did generate different levels of stress by gradients of electrical conductivity at field capacity and water potential in the saturation extract of the substrate, and the morphological parameters indicated three distinct stress levels. As expected from a halophyte species, these plants remained alive under very high levels of salinity stress, showing salt crystal-like structures constituted mainly by Na+, Cl−, and K+ on and around closed stomata. A comprehensive and large-scale metabolome and transcriptome single and integrated analyses were then employed using leaf samples. The multi-omics integration (MOI) system analysis led to a data-set of 51 metabolic pathways with at least one enzyme and one metabolite differentially expressed due to salinity stress. These data sets (of genes and metabolites) are valuable for future studies aimed to deepen our knowledge on the mechanisms behind the high tolerance of this species to salinity stress. In conclusion, besides showing that this species applies salt exclusion already in young plants to support very high levels of salinity stress, the initial analysis of metabolites and transcripts data sets already give some insights into other salt tolerance mechanisms used by this species to support high levels of salinity stress.
Supplementary Information
The online version contains supplementary material available at 10.1007/s43657-022-00061-2.
Keywords: Purslane, RNA-Seq, Chemometrics, High-resolution mass spectrometry, Abiotic stress, Multi-omics integration
Introduction
There are many abiotic stressors affecting the plant life cycle, and interfering with growth and productivity (Sunkar et al. 2007). Soil salinity is among the abiotic stressors that threaten agriculture the most, and it is a growing problem in several parts of the world, predominantly in arid and semi-arid regions (Allbed and Kumar 2013), resulting in a considerable restriction on crop productivity (Mahajan and Tuteja 2005). Approximately 20% of the agricultural land in the world has saline or sodic soils, and 25–30% of the irrigated land area is affected by salt (Shahid et al. 2018). Soil salinity spreads over 100 countries (Kumari et al. 2015; Zhang et al. 2012), resulting in an annual cost of 27.3 billion US dollars due to the loss of agricultural production (Qadir et al. 2014).
Approximately 99% of all plant species are glycophytes or salt-sensitive, including all major crops. Halophytes account for the remaining 1% and can complete their life cycle in an environment where the salt concentration is > 200 mM NaCl or > 20 dS m−1 (Flowers & Colmer 2008; Shabala and Mackay 2011). Purslane (Portulaca oleracea L.) is the most well-known and studied species of this single genus from the Portulacaceae family. It is a dicot species adapted to inland salt desert and saline habitats—xerohalophyte, and it is a salt hyperaccumulator plant with a high phytoremediation potential (Devi et al. 2016; Santos et al. 2016; Ozturk et al. 2020). It also produces many bioactive allelopathic compounds—such as growth regulators and natural herbicides, which makes it suitable to be used as an allelopathic plant (El-Shora and El-Gawad 2015; Petropoulos et al. 2016).
Petropoulos et al. (2015), after studying the chemical composition and yield of six genotypes of common purslane, reported that the yield (fresh weight) was affected by genotype, with the highest yield of the tested genotypes being of 33 tons/hectare, and the lowest one being 11.5 tons/hectare, with an average of about 22.5 tons/hectare among those genotypes. On the top of this high biomass productivity seen under a controlled agricultural environment, portulaca is known as a plant with a nutritional quality higher than many other leafy vegetables, as it possesses a large spectrum of pharmacological properties, such as neuroprotective, antimicrobial, antidiabetic, antioxidant, anti-inflammatory, antiulcerogenic, and anticancer activities; and flavonoids, alkaloids, polysaccharides, fatty acids, terpenoids, sterols, proteins, vitamins, and minerals (Zhou et al. 2015; Ozturk et al. 2020).
Due to its well-known tolerance to several abiotic stresses, as well as short growth cycle—completes its life cycle in 2–4 months, Borsai et al. (2018) presented the idea of using P. oleracea as a suitable model to study the mechanisms of plant tolerance to drought and salt stresses. Furthermore, it is a C4 plant that can develop the Crassulacean acid metabolism (CAM) when subjected to water stress and short photoperiod, making the idea of using it as a model plant even more interesting (D’andrea et al. 2014; Koch and Kennedy 1980).
Studies conducted by different researchers have shown that there are differences between the P. oleracea genotypes in terms of defense mechanisms against high salinity (Ozturk et al. 2020). Some studies have suggested an initial view of some of these mechanisms triggered in purslane plants under saline stress, such as the ability to avoid chlorine ion toxicity, the activation of the ethylene signaling pathway, the carriers' ability to discriminate cations, the increased antioxidant activity, the synthesis of osmoprotectant proline, and the synthesis of several other metabolites involved with many biochemical pathways (Sdouga et al. 2019; Xing et al. 2019, 2020; Zaman et al. 2020).
Metabolomics and transcriptomics are techniques intensively used to assist in the systemic understanding of plant responses to salinity, envisioning the use of genetic engineering in sensitive plants of economic importance. The transcriptome and metabolome are the complete set of RNA and metabolites (primary and secondary), respectively, produced under specific circumstances or in a cell, tissue, organ, or an entire organism, in a given moment of its development (Villate et al. 2021). In recent years, due to the technological advances and cost reduction achieved with the RNA-seq technique, as well as mass spectrometry and NMR spectroscopy, we have witnessed an explosion in the amount of transcriptome and metabolome data generated and made public (Lowe et al. 2017; Villate et al. 2021).
Salinity stress is known to alter the metabolome and transcriptome profile of several plant species (Tada et al. 2019; Arif et al. 2020; Wang et al. 2021). There are many reports on the tolerance of purslane plants to salt stress, but not many reporting the use of the so-called multiomics integration (MOI) strategy (Cavill et al. 2016; Jamil et al. 2020). Xing et al. (2020), which performed an integrative analysis of the transcriptome and metabolome profiles of purslane, showed that resisting saline stress reduces the expression levels of chloride channel proteins to avoid the toxicity of chloride, activates the signaling pathway of ethylene, and accumulates pyrophosphate and unsaturated fatty acid to regulate energy supply and minimize oxidative effects on cell structure.
Building up a robust multiomics database on the response of purslane to salt stress, and the subsequent study of it via an MOI analysis can create the basis for a future system biology approach to decode the genetics behind its tolerance to salinity stress. A first step in pursuing this road is to set up and validate a bioassay to evaluate its morphophysiological changes due to this stress. Then, this bioassay will be applied to generate distinct plant material that will undergo comprehensive and large-scale analysis by means of distinct omics platforms. This study reports on a robust salinity stress protocol and the characterization of the morphophysiological responses of young purslane plants to salinity stress using such protocol. Furthermore, it reports metabolome and transcriptome single and integrated analyses that produced data sets and insights into the salt tolerance shown by this halophyte species.
Materials and Methods
Plant Material, Growth Conditions, Experimental Design and Saline Stress
Seeds of the B1 accession of purslane, from the Purslane Collection at Embrapa Agroenergia, were disinfected by soaking in 2% sodium hypochlorite and Tween® 20 for 5 min, under slow agitation, then washed with sterile water and dried on sterile filter paper. After seeded on a culture medium (MS 1/2 strength, Phytagel 0.2%, and pH 5.8) (Murashige and Skoog 1962), the seeds were kept for germination in a growth chamber Conviron mod. Adaptis 1000TC (Controlled Environments Ltd, Winnipeg, Canada) at 150 µmol m2 s−1 of light and 30 °C. After 13 days, plantlets were individually transferred to 200 mL plastic cups containing 100 g of sterilized substrate [clay soil, vermiculite, and a commercial substrate (Bioplant®), 2:1:1 (v:v:v) ratio], and transferred to another Conviron® growth chamber mod. PGW40 at 25 ± 2 °C, 500 ± 20 µmol m−2 s−1 of light, 65 ± 5% air relative humidity, and photoperiod of 16/8 h (light/dark), and kept there until the end of the experiments. The plants were allowed to acclimatize for three days before starting the experiment.
Young purslane plants (13 days after sowing, plus three days of acclimatization in the growth chamber) were submitted to and kept under stress for five days. The treatments used consisted of control (no added salt) and four salinity levels (0.5, 1.0, 1.5, and 2.0 g of NaCl per 100 g of substrate), with ten replicates per treatment in a completely randomized design. NaCl was dissolved in deionized water to salinize the substrate. The amount of deionized water used corresponded to the difference between the amount of water previously present in the substrate and the amount of water necessary for the substrate to reach field capacity. Applying the right amount of water, up to field capacity, was a mean of ensuring no leakage of the solution out of the pot and no loss of Na+ or Cl−. The water lost by evapotranspiration was replaced with deionized water in a daily basis, and the electric conductivity at field capacity (ECfc) and water potential in the substrate solution measured daily, for all replicates.
Phenomic Data
The parameters of leaf gas exchange [net CO2 assimilation rate (A), transpiration rate (E), stomatal conductance to water vapor (gs), and intercellular CO2 concentration (Ci)] were measured by a portable infrared gas analyzer LI-COR Mod. 6400XT (LI-COR, Lincoln, NE, USA) equipped with a measuring chamber (2 × 3 cm) with artificial light system LI-COR Mod. 6400-02B. The extracted data were provided by the OPEN software version 6.3. Measurements were performed daily, between 9 and 11 am, on the first fully expanded leaf. The block temperature was 25 °C, PAR was 1500 µmol m2 s−1, the relative humidity of the air inside the measuring chamber was kept between 50 and 60%, the airflow index was 400 µmol s−1, and the CO2 concentration was 400 ppm in the reference cell, using the CO2 mixer model 6400-01.
The parameters evaluated using the chlorophyll fluorescence technique (Saturation Pulse Method) via a Walz image fluorimeter model IMAGING-PAM Maxi version (Heinz Walz GmbH, Effeltrich, Germany), controlled by the ImaginWin software version 2.40b, were: Fm, Fo, Y(II), Fv/Fm, Y(NPQ), and Y(NO). The IMAG-MAX/L LED lighting head and the CCD camera were mounted on a 15 mm-diameter metal bar on the optional support at a standard distance of 18.5 cm for all plants. We used the following settings: measurement light = 1, saturation pulse = 10 (2800 µmol m2 s−1), pulse amplification factor = 1, damping = 2, amplification factor in red = 25, Fm factor = 1.055, Factor = 0.999, and actinic light = 280 µmol m−2 s−1. Plants were kept in the dark for 30 min before measurement, performed in the dark, and on the same leaf used for gas exchange measurements. The induction curve approach was used with a 40 s delay from the initial saturation pulse until the start of the actinic illumination, and, from then on, a saturation pulse was emitted every 20 s until completing 315 s. After measuring the initial parameters, the software calculated all derivatives parameters. For this, an area of interest (AOI) that did not include the midrib was demarcated on the leaf surface.
We used a hyperspectral camera Resonon Mod Pika XC (Resonon, Bozeman, Massachusetts, USA), controlled by software Spectronon version 2.1, to obtain the spectral signature of the samples and estimate the levels of pigments. The calculated parameters were: chlorophyll index [CI = (R660 − R930) × R930] (Gitelson et al. 2005), photochemical reflectance index [PRI = (R531 − R570)/(R531 + R570)] (Gamon et al. 1992), and carotenoid index [CRI = (R510) − (R550) x R800] (Gitelson et al. 2002). The system consisted of a hyperspectral camera, a linear translation phase, and a fixed lighting system in the assembly tower. Hyperspectral images generated by maintaining the same distance for all plants and the configurations applied according to the manufacturer’s recommendations. Five regions of interest for the average reflectance spectrum were marked on each plant.
Scanning Electron Microscopy Analysis
We collected samples of purslane leaves and immediately immersed them in liquid nitrogen and then stored them at − 80 °C until lyophilization in a freeze dryer LIOTOP® model K120 for 48 h. Lyophilized samples were stored in a desiccator until use. Leaf samples were coated with a gold layer, 12.3 mm thick, using a Quorum Technologies® model Q 150 T ES with the QT GOLD program. High-resolution images of the regions of interest were obtained employing a scanning electron microscope (SEM) with detectors of energy dispersive spectroscopy (EDS). The composition of the sample was identified, qualitatively, in specific points of the image.
Metabolomics Analysis
Leaves from all treatments—five replicates per treatment—were collected at one and four days after treatment (DAT), immediately immersed in liquid nitrogen, and then stored at − 80 °C until extraction of metabolites.
The solvents methanol-grade UHPLC, acetonitrile-grade LC–MS, formic acid-grade LC–MS and sodium hydroxide ACS-grade LC–MS were from Sigma-Aldrich (St. Louis, MO, USA); and the water treated in a Milli-Q system (Millipore, Bedford, MA, USA).
Based on previous works, the analytical protocol employed was optimized for a fast and efficient metabolite extraction method. Aliquots of 50 mg of grounded sample were transferred to 2 mL microtubes, then 1 mL of a solvent mixture (1:3 (v:v) methanol: methyl tert-butyl ether) was added, and left for homogenization at 4 °C on an orbital shaker for 10 min, followed by an ultrasound treatment in an ice bath for another 10 min. Next, 500 μL of a 1:3 (v:v) methanol: water mixture was added to each microtube before centrifugation (12,000 rpm, 4 °C for 5 min) to promote a phase separation. After centrifugation, three phases were generated: an upper nonpolar, a lower polar, and a protein pellet. The apolar and polar fractions were transferred separately to 1.5 mL microtubes and vacuum dried in a Speed vac overnight at room temperature (Centrivap, Labconco, Kansa, MO, USA).
After resuspending the dry polar fraction by adding 500 μL of 1:3 (v:v) methanol:water mixture, samples were transferred to vials and analyzed by UHPLC–MS/MS. We used a UHPLC chromatographic system (Nexera X2, Shimadzu Corporation, Japan) equipped with an Acquity UPLC HSS T3 (1.8 μm, 2.1 × 150 mm) reverse phase column (Waters Technologies, Milford, MA), maintained at 35 °C. A polar mobile phase was used, where solvent A was 0.1% (v/v) formic acid in water and solvent B was 0.1% (v/v) formic acid in acetonitrile/methanol (70/30, v/v). The gradient elution used, with a flow rate of 0.4 mL min−1, was the following: isocratic from 0 to 1 min (0% B), linear gradient from 1 to 3 min (5% B), from 3 to 10 min (50% B), and 10 to 13 min (100% B), isocratic from 13 to 15 min (100% B), followed by re-balancing in the initial conditions for 5 min. The rate of acquisition spectra was 3.00 Hz, monitoring a mass range from m/z 70 to 1200 (polar fraction) and m/z 300 to 1600 (lipidic fraction).
A high-resolution mass spectrometer was used after the UHPLC separation (MaXis 4G Q-TOF MS, Bruker Daltonics, Germany) using electrospray source in positive (ESI(+)-MS) and negative modes (ESI(−)-MS). The settings used on the MS method were: final plate offset, 500 V; capillary voltage, 3800 V; nebulizer pressure, 4 bar; dry gas flow, 9 L min−1, dry temperature, 200 °C. The rate of acquisition spectra was 3.00 Hz. A sodium formate solution (10 mM HCOONa solution in 50/50 v/v isopropanol/water containing 0.2% formic acid) was injected directly through a six-way valve at the beginning of each chromatographic run for external calibration. Ampicillin ([M + H] 350.11867 and [M–H] 348.10288) was added to each sample and was used as an internal standard for peak normalization.
Tandem mass spectrometry (MS/MS) parameters have been adjusted to improve mass fragmentation, with collision energy ranging from 20 to 50 eV, using a step method. Precursor ions were acquired using the 3.0 s cycle time. The general AutoMS settings were mass range, m/z 70–1000 (polar fraction) and m/z 300–1600 (lipidic fraction); spectrum rate, 3 Hz; ionic, positive polarity; pre-pulse storage, 8 μs; funnel 1 RF, 250.0 Vpp. The UHPLC–MS and UHPLC–MS/MS data were acquired by HyStar Application version 3.2 (BrukerDatonics, Germany).
The raw data from UHPLC–MS were exported as mzMXL files, using DataAnalysis 4.2 software (Bruker Daltonics, Germany) and pre-processed using XCMS Online (Gowda et al. 2014; Tautenhahn et al. 2012), for peak detection, retention time correction and alignment of the metabolites detected in the UHPLC–MS analysis. Peak detection was performed using centWave peak detection (∆m/z = 10 ppm; minimum peak width, 5 s; maximum peak width, 20 s) and mzwid = 0.015, minfrac = 0.5, bw = 5 for alignment of retention time. The unpaired parametric t test (Welch t test) was used for statistical analysis.
The processed data (csv file) were exported to MetaboAnalyst 5.0, and submitted to analysis in the Statistical Analysis module (Chong et al. 2019; Chong and Xia 2020). Before the chemometric analysis, all data variables from the polar fraction were normalized by internal standard (ampicillin-rT = 7.9 min; [M + H], m/z = 350.11867, [M-H], m/z = 348.10288); and, all data variables from the lipidic fraction were normalized by internal standard (1,2-diheptadecanoyl-sn-glycero-3-phosphocholine = 4.85 min; [M + H] + m/z = 762.60125). All three sets of data were scaled using the pareto method.
The differentially expressed peaks (DEP) were selected according to the following criteria: variable importance in projection—VIP values ≥ 0.99, obtained from the PLS-DA model; adjusted p value (FDR – False Discovery Rate) ≤ 0.05, of the Welch t test; and Log2 (FC – Fold Change) ≠ 1. The selected DEPs were then submitted to analysis in the MS Peaks to Pathway module (Chong et al. 2019; Chong and Xia 2020) and analyzed using the following parameters: molecular weight tolerance of 5 ppm; mixed ion mode; joint analysis using the mummichog algorithm (Li et al. 2013) with a p value cutoff of 1.0×10–5 and Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005) algorithms, and the latest KEGG version of the Arabidopsis thaliana pathway library.
In the case of a DEP with two or more matched forms (isotopes) and later a matched compound with two or more DEPs, the initial criterion of metabolite selection applied was the mass difference comparing to the metabolite database for choosing the smallest one. The second criterion was the adduct study of each candidate back in its mass spectra. Then, we applied the formula and exact mass data from KEGG; and, finally, performed the putative annotation of the metabolites of interest, with one or two candidates on each detected ion.
The KEGG IDs of the matched compounds were then submitted to pathway analysis (integrating enrichment analysis and pathway topology analysis) and visualization in the Pathway Analysis module (Chong et al. 2019; Chong and Xia 2020) and analyzed using the Hypergeometric Test and the latest KEGG version of the A. thaliana pathway library.
Transcriptomics Analysis
Based on the results of the morphophysiological characterization, we selected the following treatments for transcriptomics analysis: control and stressed plants—0.0 and 1.5 g of NaCl per 100 g of substrate, respectively, at one and four days after treatment (DAT). Leaves for transcriptomics analysis were collected from five replicates, immediately immersed in liquid nitrogen, and then stored at − 80 °C until RNA extraction. Three replicates per treatment were randomly selected for total RNA extraction, library preparation and sequencing.
Total RNA was isolated from purslane shoots using the Qiagen RNeasy® Plant Mini kit (QIAGEN, CA, USA) following the manufacturer’s protocol. RNA quantity was measured using a Nanodrop Qubit 2.0 Fluorometer (Life Technologies, CA, USA), and RNA quality was evaluated with an Agilent Bioanalyzer Model 2100 (Agilent Technologies, Palo Alto, CA, USA). Samples were subjected to RNA-Seq using an Illumina HiSeq platform at the GenOne Company (Rio de Janeiro, Brazil), using the paired-end strategy. The raw sequence data (24 fastq files) have been uploaded in the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information under the BioProject PRJNA575830 and BioSample SAMN12911623 (Portulaca oleracea_B1—TaxID: 46,147).
All the transcriptomics analysis was performed with OmicsBox version 1.3 (OmicsBox 2019). We used FastQC (Andrews 2010) and Trimmomatic (Bolger et al. 2014) to perform the quality control, to filter reads and remove low-quality bases. The minimum average quality of reads kept was 30, and the minimum length of reads was 75. The default parameters from OmicsBox version 1.3 were used to create a “de novo” transcriptome without a reference genome through the software Trinity version 2.8.5 (Grabherr et al. 2011) and Bowtie2 version 2.3.5.1 (Langmead and Salzberg 2012). The RNA-Seq data were aligned to the reference transcriptome using default parameters from OmicsBox version 1.3 through software STAR (Dobin et al. 2013); and we used the default parameters from OmicsBox version 1.3 through HTSeq version 0.9.0 (Anders et al. 2015) to quantify the expression at gene or transcript level.
The pairwise differential expression analysis between different experimental conditions was performed through edgeR version 3.28.0 (Robinson et al. 2010), applying a simple design and an exact statistical test, without the use of filter for low counts genes. The functional analysis of the differentially expressed genes (DEGs) was performed combining differential expression results with functional annotations from the high-throughput functional annotation and data mining pipeline in OmicsBox version 1.3 (Götz et al. 2008).
Integratomics Analysis
Previously to the integration of metabolome and transcriptome data, a fasta file containing all 97,613 ORFs from the reference transcriptome was submitted to analysis in the GhostKOALA annotation tool for K number assignment of KEGG genes (Kanehisa et al. 2016).
Omics Fusion (Brink et al. 2016), the web platform for integrative analysis of Omics data (https://fusion.cebitec.uni-bielefeld.de/), was employed for carrying out the integrative analysis of transcripts and metabolites. The input data used were the Log2 (FC) data of differentially expressed transcripts and metabolites. First, to check the data distribution, we used the Data Overview module and then the Scatter Plot one for the correlation analysis between the two sets of data—a pairwise combination of the different scenarios evaluated.
For subsequent analysis, we used the modules KEGG feature distribution and Map data on the KEGG pathway. The former module employed to verify which metabolic pathways had more transcripts and metabolites differentially expressed, and the latter to map these data differentially expressed in the metabolic paths in question. For the KEGG feature distribution module, we applied the joint analysis of transcripts and metabolites with a threshold of 8, and for the Map data on the KEGG pathways, the organism code bvg (Beta vulgares—sugar beet) was used for mapping.
Results
Morphophysiological Responses of Young Purslane Plants to High Salinity Stress
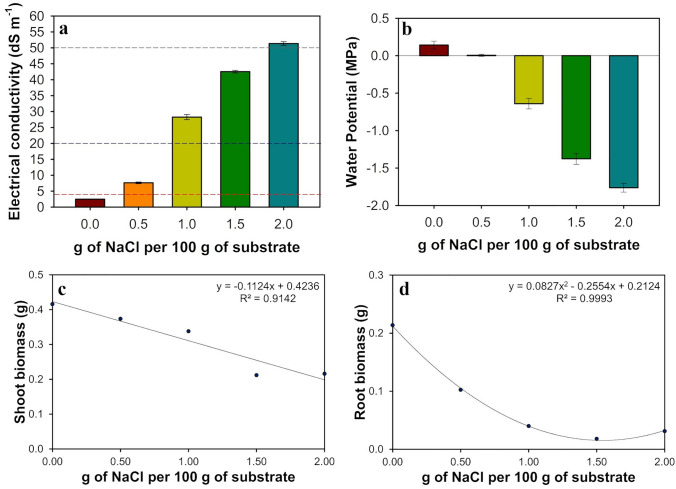
As the amount of NaCl added to the substrate rose, it led to a dose-dependent rise in electrical conductivity at field capacity (ECfc) (Fig. 1a) and a fall in water potential (Fig. 1b) of the saturation extract. As the amount of NaCl in the substrate increased, the shoot (Fig. 1c) and root (Fig. 1d) biomass of young purslane plants decreased about 50% and 90%, respectively, in the stress treatments with ≥ 1.5 g of NaCl/100 g of the substrate. The visual aspect of the purslane plants observed at the end of the experiment was recorded (Fig. 2). Compared to the control, changes in appearance and color of the leaves and stem were evident only in the stress treatments with ≥ 1.0 g of NaCl/100 g of the substrate. In summary, these are pieces of evidence that these plants were under stress.
Fig. 1.
Electrical conductivity at field capacity (a) and water potential (b) of the substrate used for growing purslane plants to which different levels of NaCl have been added. Biomass accumulation in shoots (c) and roots (d) of young purslane plants grown for five days under different concentrations of NaCl. The values represent the average of five replicates, and the bars represent the standard error of the mean. Red dashed line: EC = 4 dS m−1 (above that level is considered saline soil); Blue dashed line: EC = 20 dS m−1 (plant completing its life cycle above this EC is considered as halophyte); and Gray dashed line: EC = 50 dS m−1 (seawater salinity)
Fig. 2.
Young purslane plants grown for five days under different concentrations of NaCl
One day after adding NaCl to the substrate, one already can see a reduction in the rates of CO2 assimilation (A), transpiration (E), and stomatal conductance to water vapor (gs), which correlated with the amount of NaCl used (Fig. 3a, b, c). Those variables remained reasonably constant throughout the experiment. On the other hand, there was an increase in intercellular CO2 concentration (Ci) in the two treatments with the highest amount of NaCl (Fig. 3d). The reductions in A and E were proportional to gs at the intermediate salt stress levels, indicating a stomatal limitation to gas exchange. The increase in Ci at the highest levels of salt stress pointed to a difficulty in CO2 assimilation, which intensified over time.
Fig. 3.
Leaf gas exchange in young purslane plants grown under increasing concentrations of NaCl in the substrate. a Net CO2 assimilation rate (A); b stomatal conductance rate to water vapor (gs); c transpiration rate (E); d intercellular CO2 concentration (Ci). The values represent the average of five replicates, and the bars represent the standard error of the mean
The chlorophyll fluorescence variables remained relatively stable in control plants throughout the experiment (Supplementary Fig. S1). However, changes occurred as the saline concentration in the substrate increased. In general, from the addition of 1.0 g of NaCl to the substrate, the leaves of purslane plants began to show a reduction in the maximum fluorescence in the dark (Fm), the effective quantum yield of photosystem II [Y(II)], and maximum quantum yield of photosystem II (Fv/Fm). On the other hand, there was an increase in initial fluorescence in the dark (Fo), in the quantum yield of regulated [Y(NPQ)], and unregulated [Y(NO)] dissipation of the energy of the light.
Regarding the pigment content, as the period of stress went on, there was a downward trend in the chlorophyll and carotenoid indices in a dose-dependent manner (Fig. 4a and b). At the same time, PRI values were also reduced in a dose-dependent way, indicating that the plants kept the xanthophylls cycle functioning, despite the degradation of photosynthetic pigments (Fig. 4c). In general, as higher was the NaCl level applied, the lower the values of chlorophyll, carotenoid, and photochemical reflectance indexes were on day 5.
Fig. 4.
a Chlorophyll index (CI), b carotenoid index (CRI) and c photochemical reflectance index (PRI) for control and salinity stressed young purslane plants (0.0, 0.5, 1.0, 1.5, 2.0 g NaCl/100 g of the substrate) throughout the period of five days of stress. The values represent an average of five replicates, and bars represent the standard error of the mean
Taken together, the morphological parameters clearly indicated that there are three distinct stress levels: level 1 (Control and 0.5 g of NaCl), level 2, (1.0 g of NaCl), and level 3 (1.5 g and 2.0 g of NaCl).
Appearance of Salt Crystals on the Edges of Purslane Leaves
As some white crystals appeared on the leaves margin in some young purslane plants under saline stress (data not shown), leaf samples from control and stressed plants underwent microscopy analysis. Scanning electron microscopy (SEM) images showed the formation of wrinkles on the leaf surface, probably due to dehydration (Supplementary Fig. S2). In general, the stomata on the leaves of salt-stressed purslane plants started to close already in the lowest NaCl level used and were completely closed on the highest ones. It was possible to see the white crystals surrounding the stomata and on top of it (Supplementary Fig. S2).
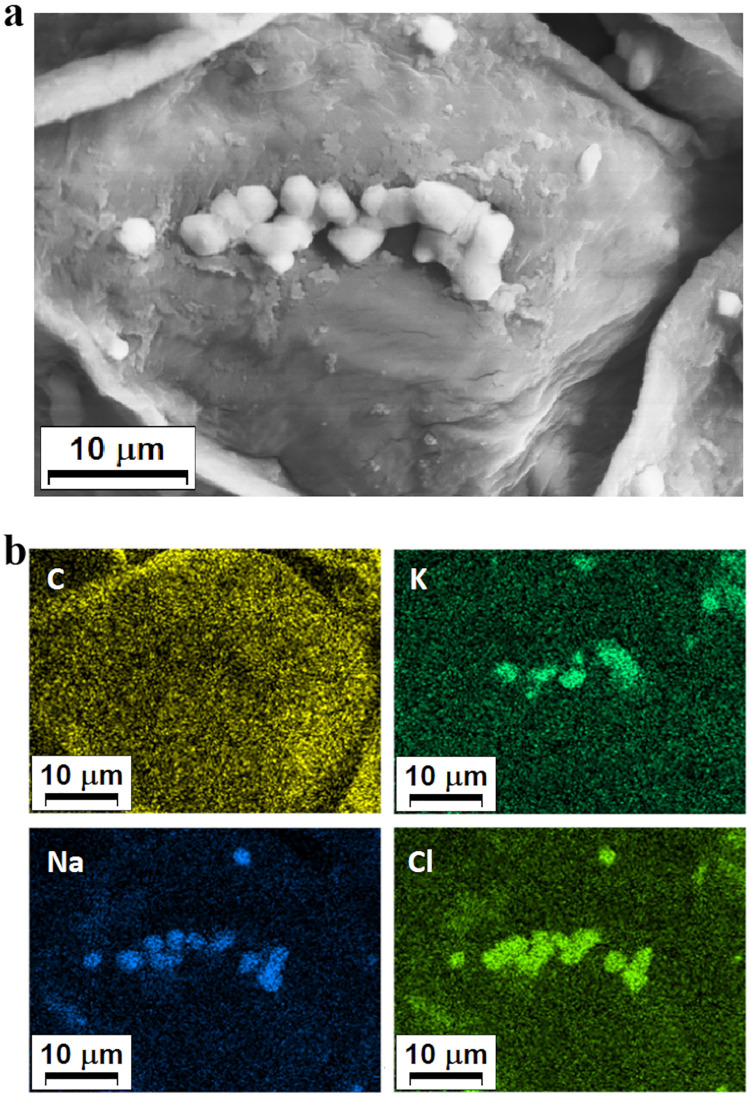
To identify the chemical composition of these crystals, we obtained SEM images with detectors of energy dispersive spectroscopy (EDS) (Fig. 5a, b). The compositional map allowed identifying the crystals' main constituents as Na+, Cl−, and K+ (Fig. 5b). These results indicate that P. oleracea has a mechanism of salt exclusion operating on the leaves, which has its role in salt tolerance.
Fig. 5.
Compositional map of the elements and image obtained by scanning electron microscope (SEM) with detectors of energy dispersive spectroscopy (EDS) of a stoma on the leaf of a salt-stressed purslane plant. a Formation of salt crystals above and around a closed stoma in salt-stressed plants. b Compositional map of the elements showing that the crystals are made of Cl−, K+, and Na+
Purslane Metabolome Under Salinity Stress—Single Analysis
When performing PLS-DA (partial least squares discriminant analysis) permutations tests using all treatments data-set (control and stressed plants at 1 and 4 DAT), applying permutation number = 2000, the model used was validated. When evaluated by the group separation distance, the probability of creating the model by chance was less than 0.0005, independent of the fraction—polar-positive, polar-negative, and lipidic-positive; the evaluation by prediction accuracy also showed a probability less than 0.0005, independent of the fraction. The cross-validation analysis determines the optimal number of components necessary to build the PLS-DA model by measuring three criteria. These criteria are R2 (the sum of squares captured by the model), Q2 (the predictive ability of the model estimated by cross-validation), and Accuracy (Chong et al. 2019). When choosing the three-component model based on Q2, the value obtained for the all treatments data-set was 89.98%, 80.55%, and 85.82%, for the polar-positive, polar-negative, and lipidic-positive fraction, respectively (data not shown).
After organizing the data set as follows, they underwent analysis in the statistics module: age effect—AE (samples from the control plants at 1 and 4 days after treatment—DAT); short-term stress—STS (control vs. stressed plants at 1 DAT); long-term stress 1—LTS1 (control vs. stressed plants at 4 DAT); and long-term stress 2—LTS2 (stressed plants at 1 and 4 DAT). Each data set had three biological replicates per treatment.
The AE data-set contained 1003, 2008, and 1113 peaks, respectively, in the polar-positive, polar-negative, and lipidic-positive fractions (Table 1). All but two peaks did not show a difference in expression between 1 and 4 DAT, showing that there was no age effect modifying the metabolome profile of purslane. As already stated in Materials and Methods, a differentially expressed peak (DEP) is a peak with a VIP ≥ 0.99, a FDR ≤ 0.05, and Log2 (FC) > 0 (upregulated) or Log2 (FC) < 0 (downregulated).
Table 1.
Differentially expressed peaks and genes in the leaves of young purslane plants submitted to salinity stress in four distinct scenarios: age effect—AE (control plants at 1 and 4 days under salinity stress—DAT); short-term stress—STS (control vs stress plants at 1 DAT); long-term stress 1—LTS1 (control vs stress plants at 4 DAT); and long-term stress 2—LTS2 (stressed plants at 1 and 4 DAT)
| Metabolomics | # of Peaks | Up | Down | Same |
|---|---|---|---|---|
| Age effect—AE | Control Plants at 04 DAT vs. Control Plants at 01 DAT | |||
| Polar-Positive | 1003 (100%) | 0 (0.0%) | 0 (0.0%) | 1003 (100%) |
| Polar-Negative | 2008 (100%) | 0 (0.0%) | 2 (0.1%) | 2006 (99.9%) |
| Lipidic-Positive | 1113 (100%) | 0 (0.0%) | 0 (0.0%) | 1113 (100%) |
| Short-term Stress—STS | Stressed Plants at 01 DAT vs. Control Plants at 01 DAT | |||
| Polar-Positive | 1034 (100%) | 141 (13.64%) | 18 (1.74%) | 875 (84.62%) |
| Polar-Negative | 2046 (100%) | 198 (9.67%) | 52 (2.54%) | 1797 (87.79%) |
| Lipidic-Positive | 1114 (100%) | 10 (0.90%) | 4 (0.36%) | 1100 (98.74%) |
| Long-term Stress 1—LTS1 | Stressed Plants at 04 DAT vs. Control Plants at 04 DAT | |||
| Polar-Positive | 1034 (100%) | 46 (4.45%) | 33 (3.19%) | 955 (92.36%) |
| Polar-Negative | 2046 (100%) | 182 (8.89%) | 67 (3.27%) | 1798 (87.84%) |
| Lipidic-Positive | 1114 (100%) | 5 (0.45%) | 10 (0.90%) | 1099 (98.65%) |
| Long-term stress 2—LTS2 | Stressed plants at 04 DAT vs. Stressed plants at 01 DAT | |||
| Polar-Positive | 1034 (100%) | 2 (0.19%) | 53 (5.13%) | 979 (94.68%) |
| Polar-Negative | 2046 (100%) | 15 (0.73%) | 127 (6.21%) | 1904 (93.06%) |
| Lipidic-Positive | 1114 (100%) | 4 (0.36%) | 123 (11.04%) | 987 (88.60%) |
| Transcriptomics | # of Genes | Up | Down | Same |
|---|---|---|---|---|
| Age Effect – AE | Control Plants at 04 DAT vs. Control Plants at 01 DAT | |||
| Reference Transcriptome | 29,737 (100%) | 3512 (11.81%) | 2868 (9.64%) | 23,358 (78.55%) |
| Short-term Stress—STS | Stressed Plants at 01 DAT vs. Control Plants at 01 DAT | |||
| Reference Transcriptome | 29,737 (100%) | 8430 (28.35%) | 8280 (27.84%) | 13,027 (43.81%) |
| Long-term Stress 1—LTS1 | Stressed Plants at 04 DAT vs. Control Plants at 04 DAT | |||
| Reference Transcriptome | 29,737 (100%) | 11,005 (37.01%) | 11,550 (38.84%) | 7182 (24,15%) |
| Long-term Stress 2—LTS2 | Stressed Plants at 04 DAT vs. Stressed Plants at 01 DAT | |||
| Reference Transcriptome | 29,737 (100%) | 8693 (29.23%) | 8994 (30.25%) | 12,050 (40.52%) |
The differentially expressed peaks are those with a variable importance in projection (VIP) value ≥ 0.99, obtained from the PLS-DA model; adjusted p value (FDR) ≤ 0.05, of the Welch t test; and Log2 (FC) ≠ 1 (FC fold change). Differentially expressed transcripts are those with a FDR ≤ 0.05, and Log2 (FC) ≥ 1 (up-regulated) or Log2 (FC) ≤ -1 (down-regulated)
The STS data-set was employed to evaluate how distinct are the metabolome profiles of the control and stressed plants at 1 DAT. The samples applied contained 1034, 2046, and 1114 peaks, respectively (Table 1). On average, 90.38% of the peaks did not show a difference in expression between control and stressed plants at 1 DAT, while 141, 198, and 10 peaks upregulated, and 18, 52, and 4 downregulated, respectively (Table 1). The LTS1 data-set was employed to evaluate how distinct are the metabolome profiles of the control and stressed plants at 4 DAT. The samples applied contained 1034, 2046, and 1114 peaks, respectively. On average, 92.95% of the peaks did not show a difference in expression between control and stressed plants at 4 DAT, while 46, 182, and 5 peaks upregulated, and 33, 67, and 10 downregulated, respectively. The LTS2 data-set was employed to evaluate how distinct are the metabolome profiles of stressed plants at 1 and 4 DAT. The samples applied contained 1034, 2046, and 1114 peaks, respectively. On average, 92.11% of the peaks did not show a difference in expression, while 2, 15, and 4 peaks upregulated, and 53, 127, and 123 downregulated, respectively. The results from STS, LTS1, and LTS2, taken together, showed that less than 10% of the peaks did differentially express; they are the ones to be used to identify the differentially expressed metabolites and the most affected pathways to which they belong.
The metabolome profiles obtained from the analysis of the STS, LTS1, and LTS2 data-sets in the Statistical Analysis module of MetaboAnalyst 5.0 revealed 748 DEPs—those peaks differentially expressed in STS, and those in LTS2 but not in STS, plus those in LTS1 but not in STS or LTS2. These DEPs underwent functional interpretation via analysis in the MS Peaks to Pathway module. After applying the initial criteria of metabolite selection, 109 differentially accumulated metabolites (DEMs) (Supplementary Table S1) were submitted to the pathway topology analysis module, resulting in a list of 63 ranked pathways (Supplementary Fig. S3). The nicotinate and nicotinamide metabolism, C5-branched dibasic acid metabolism, and Phenylpropanoid biosynthesis pathways, with an FDR (false discovery rate) of 0.014831, 0.017755, and 0.041788, came out at the top of this rank, respectively; and an impact of 0.4, 1.0, and 0.2, respectively. These three pathways had 20 DEMs with the highest level of significance in the set of 109 matched metabolites submitted to analysis; being six out of its 13 metabolites in the nicotinate and nicotinamide metabolism pathway, four out of its six in the C5-branched dibasic acid metabolism, and 10 out of its 46 in Phenylpropanoid biosynthesis (Supplementary Table S1).
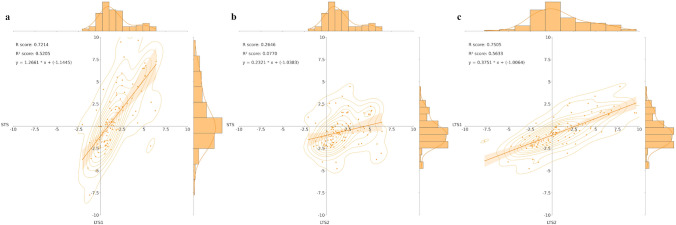
All the 109 DEMs underwent correlation analysis through pairwise comparison of the STS, LTS1, and LTS2 scenarios, using Log2(FC) values. The correlation analysis revealed strong positive correlations when comparing STS vs. LTS1 and LTS1 vs. LTS2 (Fig. 6a, c) and a weak positive correlation in STS vs. LTS2 (Fig. 6b). As STS compares control and stressed plants at 1 DAT, and LTS2 compares at 4 DAT, this weak positive correlation implies that the behavior seen for most of the 109 DEMs at short-term stress does not repeat in the long-term. Meanwhile, the behavior seen in most of the 109 DEMs in STS repeats itself in LTS1; the same is true when comparing LTS1 and LTS2. When comparing STS and LTS1, the Log2(FC) value at LTS1 is already the result of the differential expressed changes seen at 1 and 4 DAT; but when comparing STS and LTS2, one must combine their Log2(FC) values to reflect the final change in expression level. When comparing LTS1 and LTS2, the Log2(FC) value at the former reflects the change in expression level due to the stress.
Fig. 6.
Histogram and correlation analysis of the Log2 (Fold Change) of 109 differentially expressed metabolites by pairwise comparison of three scenarios: short-term stress—STS (control vs stress plants at 1 DAT); long-term stress 1—LTS1 (control vs stress plants at 4 DAT); and long-term stress 2—LTS2 (stressed plants at 1 and 4 DAT). a STS vs LTS1; b STS vs LTS2; and c LTS1 vs LTS2
Taken together, the results from this large-scale metabolome single analysis reveal three pathways affected by salt stress in the leaves of young purslane plants, as well as several metabolites from these and other pathways that should be the focus of further characterization of the role of metabolites in the tolerance of this halophyte species to salt stress.
Purslane Transcriptome Under Salinity Stress—Single Analysis
The raw sequence data generated in this study have been uploaded in the Sequence Read Archive (SRA) database of the National Center for Biotechnology Information under Portulaca oleracea_B1—BioProject number PRJNA575830, BioSample SAMN12911623. A total of 296,493,969 high-quality-pairs of reads—with an average quality of reads ≥ 30 and the minimum length of 75 nucleotides—remained after pre-processing the raw sequence data (data not shown). The assembly of the Reference Transcriptome (RT), the mapping, counting, and differential expression analysis used these high-quality sequences. The RT assembled has 97,613 ORFs, 49,412 complete ORFs, a 42.09% GC content, and an N50 equal to 1493 bases per transcript. The differential expression analysis was performed to measure a possible AE, as well as to measure STS, LTS1, and LTS2; and applied the following criteria: CPM filter equals to 1.0, number of samples reaching CPM filter equals to 3, normalization method TMM (Trimmed mean of M values). Differentially expressed genes (DEGs) are those with a FDR < 0.05, and FC > 1 (upregulated) or FC < 1 (downregulated).
Differential expression analysis of the AE data-set revealed 3512 (11.81%) upregulated and 2868 (9.64%) downregulated genes in the control plants at 4 DAT when comparing to 1 DAT (Table 1). In the STS scenario, a total of 8430 (28.35%) upregulated and 8280 (27.84%) downregulated in the stressed plants comparing to the control plants at 1 DAT. In the LTS1 scenario, a total of 11,005 (37.01%) upregulated and 11,550 (38.84%) downregulated in the stressed plants comparing to the control plants at 4 DAT. At last, in the LTS2 scenario, 8693 (29.23%) upregulated and 8994 (30.25%) downregulated in the stressed plants at 4 DAT comparing to the stressed ones at 1 DAT.
The genes submitted to differential expression analysis in all four scenarios (AE, STS, LTS1, and LTS2) were separated accordingly to the combined profiles in 15 groups (Fig. 7). A total of 442 genes (1,49%) were upregulated in all four scenarios (Fig. 7a), while 2755 (9,26%) did not differentially expressed in all scenarios (Fig. 7b), and 524 (1,76%) were downregulated in all scenarios (Fig. 7c). When considering those genes that up- or downregulated twice in STS and LTS2, but not in AE, a total of 3053 (10,27%) and 2864 (9,63%) genes were selected, respectively. No genes were found up- or downregulated only in AE and STS, or in AE and LTS1, but not in the other scenarios.
Fig. 7.
Venn Diagram of genes from Portulaca oleracea differentially expressed in the leaves of young purslane plants submitted to salt stress, under four distinct scenarios: age effect—AE (control plants at 1 and 4 days under salinity stress—DAT); short-term stress—STS (control vs stress plants at 1 DAT); long-term stress 1—LTS1 (control vs stress plants at 4 DAT); and long-term stress 2—LTS2 (stressed plants at 1 and 4 DAT). a upregulated genes, b non-differentially expressed genes, and c downregulated genes
All 29,737 genes submitted to differential expression analysis underwent correlation analysis through pairwise comparison of the AE, STS, LTS1, and LTS2, using Log2(FC) values. Only the genes differentially expressed in STS underwent correlation analysis against AE, LTS1, and LTS2; the same is true for LTS1 against AE and LTS2, and LTS2 against AE (Fig. 8). The correlation analysis revealed weak positive correlations in AE vs. STS, AE vs. LTS2, and STS vs. LTS2 (Fig. 8a, c, and e), weak negative correlation in AE vs. LTS1 (Fig. 8b), and strong positive correlations in STS vs. LTS1 and LTS1 vs. LTS2 (Fig. 8d and f).
Fig. 8.
Histogram and correlation analysis of the Log2 (Fold Change) of differentially expressed genes by pairwise comparison of four scenarios: age effect—AE (control plants at 1 and 4 days under salinity stress—DAT); short-term stress—STS (control vs stress plants at 1 DAT); long-term stress 1—LTS1 (control vs stress plants at 4 DAT); and long-term stress 2—LTS2 (stressed plants at 1 and 4 DAT). a STS vs AE, b LTS1 vs AE, c LTS2 vs AE, d STS vs LTS1, e STS vs LTS2, f LTS1 vs LTS2
A weak positive or negative correlation implies that the behavior seen for most of the genes in one scenario does not repeat the second scenario evaluated. Even in such a case, it is possible to identify the gene(s) with the behavior of interest for further studies. As an example, one can see in the fourth quadrant in Fig. 8b the existence of a group of genes that the level of upregulation in AE is the same as downregulation in LTS1; meaning that the saline stress affected the expression of these genes in a way that let their expression level to what it was in the control plants at 1 DAT. Another example is in Fig. 8c, on the line separating the first and the second quadrants, near the Log2(FC) value of 10, where genes not differentially expressed in AE experienced an increase in expression of about 1000-fold between 1 and 4 DAT.
In the case of the strong positive correlation seen when comparing STS and LTS1, and LTS1 and LTS2, the behavior seen for most of the genes in one scenario does repeat itself in the second scenario evaluated. When comparing STS and LTS1, the Log2(FC) value at LTS1 is already the result of the differential expressed changes seen at 1 and 4 DAT; but when comparing LTS1 and LTS2, it is the Log2(FC) value at LTS1 that reflects the final change in expression level due to the stress. One can infer that genes upregulated twice—in STS and LTS2—are among the most important to further characterization regarding their role in the response of this halophyte species to salinity stress. Even though there is a weak positive correlation when comparing STS and LTS2, it was possible to identify several genes upregulating twice (Fig. 8e).
The results from this large-scale transcriptome single analysis revealed many genes differentially expressed in all four scenarios, and also allowed the identification of specific behavior mode as a result of the correlation studies. As expected, for some their expression increased and many others decreased. These results allow the selection for functional annotation of genes affected only early in the stress—1 DAT, as well as those affected only latter—4 DAT, or in both. Taken together, these results give us several opportunities to select and further characterize salt-responsive genes aiming either the identification of candidate genes for salt tolerance or the selection of promoter sequence for biotechnological application—salt stress-dependent expression of genes of interest.
Purslane Transcriptome and Metabolome Under Salinity Stress—Integrative Analysis
The annotation of the fasta file containing all 97,613 ORFs from the purslane RT in GhostKOALA, against the genus_prokaryotes + family_eukaryotes databases, resulted in 38,700 entries annotated under functional categories (data not shown). The KEGG Mapper reconstruction analysis allowed the identification of 1,802 enzymes from eudicots (data not shown).
Out of the 415,379 isoform features from the purslane RT, 72,830 remained for differential expression analysis, after applying the following criteria: CPM filter equals to 1.0, number of samples reaching CPM filter equals to 3, normalization method TMM (Trimmed mean of M values). Out of these 72,830 isoform features, 23,834 did not differentially expressed in any of the four scenarios tested (AE, STS, LTS1, and LTS2), and 48,996 did in at least one of these scenarios (data not shown). Differentially expressed isoforms (DEIs) are those with a FDR ≤ 0.05, and FC > 1 (upregulated) or FC < 1 (downregulated). A search for the 1,802 enzymes among the 48,996 DEIs led to a list of 5,883 DEIs with KO number and E.C. number.
Only two out the four scenarios studied underwent transcriptome and metabolome integration analysis: STS and LTS1. To do that, the list of 109 DEMs was first filtered to select only those differentially expressed in STS or LTS1; and the same was done with the list of 5883 DEIs. The filtered lists from DEMs and DEIs were then combined for the integration analysis. In the STS scenario, the glycerophospholipid metabolism pathway came first in the rank of combined enzymes and compounds occurrence, with six enzymes and five metabolites; while in the LTS1 one, the Purine metabolism pathway came first with four enzymes and seven metabolites (Table 2). When considering both scenarios studied, the glycerophospholipid metabolism pathway comes first as the most affected pathway in young purslane plants under high salinity stress, followed by Cysteine and methionine metabolism (Table 2).
Table 2.
List of metabolic pathways in the leaves of young purslane plants affected by salinity stress, obtained after metabolome and transcriptome integration using the Omics Fusion platform.
| Pathway | Pathway id | STS* | LTS1 | ||||
|---|---|---|---|---|---|---|---|
| MOI | Enzymes | Metabolites | MOI | Enzymes | Metabolites | ||
| Glycerophospholipid metabolism | 564 | 11 | 6 | 5 | 8 | 6 | 2 |
| Cysteine and methionine metabolism | 270 | 9 | 3 | 6 | 9 | 3 | 6 |
| Glycine, serine and threonine metabolism | 260 | 8 | 7 | 1 | 9 | 6 | 3 |
| Purine metabolism | 230 | 6 | 4 | 2 | 11 | 4 | 7 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 400 | 9 | 4 | 5 | 7 | 4 | 3 |
| Arginine and proline metabolism | 330 | 7 | 2 | 5 | 6 | 2 | 4 |
| Aminoacyl-tRNA biosynthesis | 970 | 6 | 1 | 5 | 7 | 1 | 6 |
| Tryptophan metabolism | 380 | 5 | 4 | 1 | 7 | 4 | 3 |
| Methane metabolism | 680 | 6 | 1 | 5 | 6 | 1 | 5 |
| Lysine degradation | 310 | 6 | 4 | 2 | 5 | 4 | 1 |
| Terpenoid backbone biosynthesis | 900 | 5 | 4 | 1 | 6 | 4 | 2 |
| Glyoxylate and dicarboxylate metabolism | 630 | 6 | 2 | 4 | 5 | 2 | 3 |
| Inositol phosphate metabolism | 562 | 5 | 4 | 1 | 5 | 4 | 1 |
| Carbon fixation pathways in prokaryotes | 720 | 6 | 1 | 5 | 4 | 2 | 2 |
| Ether lipid metabolism | 565 | 5 | 3 | 2 | 4 | 3 | 1 |
| Glutathione metabolism | 480 | 6 | 2 | 4 | 3 | 2 | 1 |
| Nicotinate and nicotinamide metabolism | 760 | 4 | 2 | 2 | 5 | 2 | 3 |
| Pyrimidine metabolism | 240 | 3 | 2 | 1 | 6 | 3 | 3 |
| Tyrosine metabolism | 350 | 5 | 1 | 4 | 4 | 1 | 3 |
| Ascorbate and aldarate metabolism | 53 | 5 | 2 | 3 | 3 | 2 | 1 |
| Thiamine metabolism | 730 | 5 | 2 | 3 | 3 | 2 | 1 |
| beta-Alanine metabolism | 410 | 4 | 2 | 2 | 4 | 2 | 2 |
| C5-Branched dibasic acid metabolism | 660 | 5 | 1 | 4 | 3 | 1 | 2 |
| Cyanoamino acid metabolism | 460 | 4 | 1 | 3 | 4 | 2 | 2 |
| Sulfur metabolism | 920 | 3 | 1 | 2 | 5 | 2 | 3 |
| Arginine biosynthesis | 220 | 4 | 1 | 3 | 3 | 2 | 1 |
| Chloroalkane and chloroalkene degradation | 625 | 3 | 2 | 1 | 4 | 2 | 2 |
| Butanoate metabolism | 650 | 4 | 1 | 3 | 2 | 1 | 1 |
| Glycolysis / Gluconeogenesis | 10 | 3 | 2 | 1 | 3 | 2 | 1 |
| Pentose phosphate pathway | 30 | 3 | 2 | 1 | 3 | 2 | 1 |
| Pyruvate metabolism | 620 | 4 | 1 | 3 | 2 | 1 | 1 |
| Carbon fixation in photosynthetic organisms | 710 | 3 | 1 | 2 | 3 | 1 | 2 |
| Histidine metabolism | 340 | 3 | 1 | 2 | 2 | 1 | 1 |
| Carbapenem biosynthesis | 332 | 2 | 1 | 1 | 2 | 1 | 1 |
| Indole alkaloid biosynthesis | 901 | 2 | 1 | 1 | 2 | 1 | 1 |
| Alanine, aspartate and glutamate metabolism | 250 | 2 | 2 | 4 | NA | NA | NA |
| Amino sugar and nucleotide sugar metabolism | 520 | 1 | 1 | 2 | NA | NA | NA |
| Fructose and mannose metabolism | 51 | 1 | 1 | 2 | NA | NA | NA |
| Glycerolipid metabolism | 561 | 3 | 1 | 4 | NA | NA | NA |
| Lysine biosynthesis | 300 | 1 | 1 | 2 | NA | NA | NA |
| Sphingolipid metabolism | 600 | 2 | 1 | 3 | NA | NA | NA |
| Starch and sucrose metabolism | 500 | 2 | 1 | 3 | NA | NA | NA |
| Steroid biosynthesis | 100 | 2 | 1 | 3 | NA | NA | NA |
| Valine, leucine and isoleucine biosynthesis | 290 | 1 | 1 | 2 | NA | NA | NA |
| Caffeine metabolism | 232 | NA | NA | NA | 1 | 2 | 3 |
| Citrate cycle (TCA cycle) | 20 | NA | NA | NA | 1 | 1 | 2 |
| Pantothenate and CoA biosynthesis | 770 | NA | NA | NA | 2 | 1 | 3 |
| Porphyrin and chlorophyll metabolism | 860 | NA | NA | NA | 4 | 1 | 5 |
| Propanoate metabolism | 640 | NA | NA | NA | 3 | 1 | 4 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 130 | NA | NA | NA | 2 | 1 | 3 |
| Zeatin biosynthesis | 908 | NA | NA | NA | 1 | 1 | 2 |
STS short-term stress—(control vs stress plants at 1 DAT); LTS1 long-term stress 1 (control vs stress plants at 4 DAT); MOI multi-omics integration system; NA not applicable
*STS (short-term stress), LTS (long-term stress), MOI (multi-omics integration), NA (not applicable)
The five DEMs from the glycerophospholipid metabolism pathway were: glycerylphosphorylethanolamine (C01233), glycerol 3-phosphate (C00093), O-phosphoethanolamine (C00346), glycerophosphocholine (C00670), and phosphodimethylethanolamine (C13482). The overexpression of the OsDRAP1 gene in rice not only resulted in salt tolerance, but also elicited the accumulation of 38 secondary metabolites, including C01233 (Wang et al. 2021), which is also associated with osmotic stress tolerance (Avelange-Macherel et al. 2006).
Discussion
When adding NaCl to a solution or wet substrate, this salt dissociates into its component ions, Na+ and Cl−, enabling the conduction of electric current (Visconti and de Paz 2016). The higher the concentration of such ions in the solution, the greater its EC (Polle and Chen 2015; Rhoades et al. 1999), as seen in Fig. 1a. On the other hand, the dissociation of the component ions of the salt in solution reduces the osmotic potential (Fig. 1b), as water molecules are needed to dissolve the ions (Cordeiro 2001; Ramoliya et al. 2004). The increase in EC and the reduction in water potential affect plant metabolism (Ali et al. 2019; Maksimovic and Ilin 2012; Munns 2002; Parida and Das 2005; Duarte and Souza 2016).
The drop of the leaf gas exchange rates is usually a plant response to salt, and its intensity depends on the level of plant tolerance to salinity stress (Everard et al. 1994; Gale 1975; Koyro 2006; Parihar et al. 2015; Alam et al. 2015; Xing et al. 2019). At the lowest doses of salt, the drop in gas exchange rates for young purslane plants seems to be due to stomatal closure, as net CO2 assimilation, transpiration rate, and stomatal conductance fell in the same proportion (Fig. 3). That is basically due to the reduction of water potential, which makes it difficult for the plants to absorb water and results in partial or total closure of stomata, depending on the severity of the salt stress (Flowers et al. 2015; Gale 1975; Qiu et al. 2003). At higher doses of NaCl, as the intercellular CO2 concentration increased, the activities of CO2-fixing enzymes probably decreased. Such increase did intensify over time (Fig. 3d). Similar results have been reported previously in purslane (Alam et al. 2015; Xing et al. 2019). According to Van Zelm et al. (2020), this finding suggests that there may also be an ionic effect, or at least a stomatal closure-independent effect of sodium on photosynthesis.
In purslane plants under saline stress of approximately 8.0 dS m−1, gas exchange rates did not differ much from the control plants, mainly at the end of the stress period (Fig. 3). These plants managed to lose only a small percentage (13.27%) of shoot biomass (Fig. 1c), while they lost almost 50% of the root biomass (Fig. 1d). Under saline stress of ≥ 20 dS m−1, the stomatal closure increased. As for the intercellular CO2 concentration, the results suggest that the mesophyll’s resistance to carboxylation occurred from the 1.5 g NaCl level (Fig. 3d). In summary, the changes in gas exchange rates at salt stress up to 50 dS m−1 resulted in smaller purslane plants that did not die (Fig. 2).
In general, the aerial part of the plant is more affected than roots by salt stress (Acosta-Motos et al. 2017; Munns and Tester 2008). In some cases, biomass production is stimulated by salinity depending on the salt concentration, as in Atriplex nummularia L. at 300 mM but inhibited at 600 mM NaCl (de Araújo et al. 2006). In Chloris gayana and Salvadora persica, root growth was more affected than shoot (Céccoli et al. 2011; Rao et al. 2004), as observed in young purslane plants (Fig. 1). Due to their direct contact with the soil, roots are considered the first damage sites (Rewald et al. 2013). In another study, purslane plants showed reductions in the aerial part and the roots as a function of salinity (150 mM and 200 mM NaCl) (Xing et al. 2019).
Recently, Xing et al. (2019) reported that, under saline stress between 100 and 200 mM NaCl, purslane plants significantly decreased the net photosynthetic rate, increased the intercellular concentration of CO2, the content of malondialdehyde, the production rate of O2−, and the activity of the enzymes SOD, POD, and CAT. Such results suggest the inhibition of photosynthesis and the occurrence of oxidative stress. Considering the changes in the net photosynthetic rate and the intercellular concentration of CO2, our results correlate to the one found by Xing et al. (2019), even though the NaCl concentrations and plant ages used were different.
Chlorophyll fluorescence variables showed a differential effect of the level of saline stress on the photochemical apparatus (Supplementary Fig. S1). The addition of salt to the substrate, regardless of the dose, caused damage to the chloroplast membrane system, which became more pronounced throughout the stress period, as can be inferred from the increase in Fo, the drop in Fm, and, consequently, in Fv/Fm. The electron flow in the Z scheme of photosynthesis fell over time. After five days, such drop was directly proportional to the level of NaCl added to the substrate, as can be inferred from the Y(II) data. The light energy not used to flow the electrons in the Z scheme was directed towards the generation of heat, as seen in Y(NPQ), but up to a limit of saline stress. This limit was up to 1.0 g of NaCl in the substrate, whose fluorescence emission in the light, despite the initial increase, was practically the same as the control at the end of the stress period (Supplementary Fig. S1). Purslane plants continued to increase the emission of fluorescence in the light with the increase of the level of salt in the substrate, as seen in Y(NO), meaning that plants no longer had the means to regulate the extinction of fluorescence by their photochemical apparatus.
The drop in the levels of CI and CRI (Fig. 4) was an expected result for purslane plants under salt stress, as recurrently reported for several plant species, such as Salicornia persica and S. europaea (Aghaleh et al. 2009), Ricinus communis L. (Li et al. 2010) and Cakile maritime (Megdiche et al. 2008). In the case of purslane, the effect was observed especially under saline stress of ≥ 20 dS m−1, and more sharply at higher levels of NaCl. At the end of the stress period, the decrease in PRI was proportional to the salt stress (Fig. 4c), indicating that the plants probably kept the xanthophyll cycle in total operation at all salt levels. The activation of the xanthophyll cycle is a response observed in some halophyte species under salt stress (Qiu et al. 2003; Rabhi et al. 2012) and is related to the protection of the photosynthetic apparatus against photoinhibition damage (Qiu et al. 2003).
The effect of salt stress on biomass accumulation in purslane plants was most likely related to the osmotic effect, which resulted in stomatal closure and restricted the entry of CO2 into the leaf mesophyll. So much so that the drop in biomass accumulation was proportional to the decrease in net CO2 assimilation rates. Evidently, throughout stress, additional mechanisms contributed to a more pronounced effect of saline stress in the roots than in the aerial part. The reduction of biomass due to high salinity is a usual response from several species such as Atriplex griffithii var. stocksii (Khan et al. 2000b), Suaeda fruticosa L. (Khan et al. 2000a), Mentha piperita L. (Khorasaninejad et al. 2010), and even for Portulaca oleracea (Alam et al. 2014, 2015).
Some purslane plants under salt stress showed structures resembling salt crystals on the leaf surface, which was confirmed to be constituted mainly of Na+, Cl− and K+ (Fig. 5). The ability to exclude salt thought roots and leaves are present in several halophyte species (Rozema et al. 1981; Ramadan 1998; Flowers et al. 2010; Yuan et al. 2016). Such ability represents a self-regulating behavior, and the secretion can occur through epidermal pores and glands located in roots, shoots, and leaves. The intracellular transport mechanisms are responsible for moving excess salt from the surface cells to the outside of leaves or stems, and as the water evaporates, it is possible to observe salt crystals (Arora and Rao 2017). In the present study, the salt crystal-like structures seen on and around closed stomata had their composition mapped, indicating that sodium, chlorine, and potassium ions were excluded probably through salt glands and accumulated on the leaf surface.
The single-metabolomic analysis done in the present study led to the identification of three pathways, that is, Nicotinate and nicotinamide metabolism, C5-Branched dibasic acid metabolism, and Phenylpropanoid biosynthesis. Together, these three pathways had 20 differentially expressed metabolites. He et al. (2021) and Singh et al. (2022) have also reported the links between the Nicotinate and nicotinamide metabolism pathway and salt tolerance in Chinese cabbages and mustard, respectively. A MOI analysis by Shu et al. (2022) and a proteomics analysis by Yan et al. (2022) did show C5-Branched dibasic acid metabolism pathway as one of the most significantly enriched in Brassica napus L. and Reaumuria soongorica during salt stress, respectively. In the case of Phenylpropanoid biosynthesis, Carvalho e Silva et al. (2021) and Cuong et al. (2020) have reported direct effect of salinity stress in this pathway in gliricidia and wheat, respectively.
The (single) analysis of the changes in the transcriptome profile of young purslane plants in response to saline stress led to the identification of several different groups of salt-responsive genes, accordingly to the response seen in four distinct scenarios analyzed. This set of genes is a database valuable for further studies aiming the selection and deep structural/functional characterization of genes that may play a role in purslane high tolerance to salinity stress.
This study did not attempt to connect conclusions from these two single-omic analyses. It has focused on getting insights into candidate genes and metabolites for further characterization via structural/functional characterization of genes and proteins or targeted metabolomics. The strategy applied in this study to perform this element-based correlation analysis, comparing different scenarios in a pairwise manner, turned out to be a powerful tool to select candidate salt-responsive genes for future work aiming for functional genomics studies.
The conceptual integration strategy analyses the different omics data-sets separately (single analysis), and the connection of the resulting conclusions results arbitrarily without further analysis of the data-sets (Cavill et al. 2016; Rai et al. 2017; Jamil et al. 2020). This approach to connect conclusions from the single-omic analysis can produce valuable insights; however, it may miss reproducible associations when multiple omics data-sets are analyzed together (Cavill et al. 2016; Rai et al. 2017). Jamil et al. (2020) state that this approach should not be considered a part of the MOI approach.
The multi-omics integration (MOI) system presented by Jamill et al. (2020) has classified the integration strategies in three levels with increasing degrees of complexity: element- (level 1), pathway- (level 2), and mathematical-based approach (level 3). Here, a level 2 approach integrated transcriptome and metabolome data-sets resulted from single–omics analysis. The differently expressed enzymes selected from the transcriptomics data-set are the central factor needed to perform the level 2 integration strategy in the Omics Fusion (Brink et al. 2016). They allow to link genes and metabolites from a metabolic pathway and consequently point out the ones most affected by the salt stress.
The MOI (transcriptome and metabolome) analysis of the STS scenario led to 44 pathways having differentially expressed transcripts and metabolites, and 42 pathways in the LTS1 scenario. When combining the results of the MOI analysis—STS and LTS1, it showed that 35 were common to both scenarios, while nine were present only in STS, and seven only in LTS1 (Table 2). The glycerophospholipid metabolism pathway was the one with the highest number of combined enzymes and metabolites differentially expressed in both scenarios, followed by the cysteine and methionine metabolism (Table 2). Previously, these pathways were linked to plant response to abiotic stress, including salt stress (Hou et al. 2016; Zhang et al. 2017; Sui et al. 2017; Xia et al. 2019; He and Ding 2020; Han and Yang 2021), and the overexpression of genes from these pathways have shown to be effective in conferring tolerance to salt stress (Sui et al. 2017; Ma et al. 2017; Han and Yang 2021).
Taken together, the results from the MOI analysis reveals a set of pathways with enzymes and metabolites differentially expressed due to salinity stress at 1 DAT (STS) and 4 DAT (LTS), which is also a database valuable for new studies aiming to further characterize the mechanisms behind the strong tolerance seen in young purslane plants to saline stress. This present study did not explore the changes in the metabolome and transcriptome profiles in the roots of young purslane plants. The main reason for that was the small amount of root tissue available at the end of the experiments (Fig. 1d). We are performing subsequent studies where new omics data—transcriptomic, metabolomic, ionomic, and proteomic—are being gathered from adult purslane plants under salinity stress to circumvent this problem seen in young purslane plants (Rodrigues-Neto et al. unpublished).
Conclusion
In conclusion, the protocol developed in this study for assessing the morphophysiological responses of young purslane (P. oleracea) plants to saline stress successfully revealed different stress levels by gradients of electrical conductivity and water potential in the saturation extract of the substrate, according to the added NaCl. As expected from a halophyte species, young plants remained alive under very high levels of salinity stress (> 20 dS m−1). The salt crystal-like structures seen on and around closed stomata on the leaves of these plants are constituted mainly by Na+, Cl−, and K+, indicating that this species has a mechanism of salt exclusion operating on the leaves, which has its role in salt tolerance. By organizing and analyzing the metabolome and transcriptome data-sets under distinct scenarios (AE, STS, LTS1, and LTS2), it was possible to identify metabolic pathways, metabolites, and genes/proteins most affected at an early and a late moment of the salt stress on the leaves. The correlation analysis strategy applied produced a list of salt-responsive metabolites and genes of high value for future studies aiming at prospecting genes conferring high tolerance to salinity stress, as well as promoter sequences from salt-responsive genes. Finally, the MOI strategy applied in this study led to a group of 51 pathways that had at least one enzyme and one metabolite differentially expressed due to salinity stress, where the Glycerophospholipid metabolism pathway was the most affected one when combining STS and LTS1 scenarios. This pathway list is a valuable tool for future target metabolomics and transcriptomics studies that aim to deepen our knowledge on the mechanisms behind the high tolerance of P. oleracea to salinity stress.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 Representative images (1) of the mean of variables derived from the chlorophyll fluorescence technique (saturation pulse method) in purslane plants after 1 and 4 days of submission to increasing levels of NaCl in the growing substrate. The images of Fo, Fv/Fm, and Fm' were obtained in plants adapted to the dark for 30 min, while the images of Y(II), Y(NO) and Y(NPQ) were captured after 5 min of actinic illumination at 280 μmol of light m-2 s-1. The values of the chlorophyll fluorescence parameters in the images can be compared with the color scale in the right bar. Changes over time in chlorophyll fluorescence parameters (2) for control and salinity stressed young purslane plants. The values represent an average of five replicates, and bars represent the standard error of the mean. (a) F - fluorescence intensity; (b) Fo - minimum fluorescence yield on dark-adapted plants; (c) Fm - maximum fluorescence yield on the dark-adapted plants; (d) Fv/Fm - maximum PSII quantum yield; (e) qP - proportion of open PSII; (f) qL - indicator of the PSII redox state; (g) Y(II) - PSII effective quantum yield; (h) qN - non-photochemical quenching that requires measurement of Fo; (i) NPQ - non‐photochemical quenching; (j) Y(NPQ) - regulated energy dissipation quantum yield; (k) Y(NO) - unregulated energy dissipation quantum yield; and (l) ETR - electron transport rate. The images of Fo, Fv/Fm, and Fm were obtained in plants adapted to the dark, while the images of Y(II), Y(NO) and Y(NPQ) were captured after 5 min of actinic illumination at 280 µmol of light/m2/s (JPG 693 kb)
Supplementary file2 Scanning electron microscope (SEM) images of the leaf surface of purslane plants. (a) Control; (b) stressed plant (2.0 g NaCl) showing wrinkle due to dehydration; (c) degree of stomata opening in control compared to (d) stressed (0.5 g NaCl). Formation of salt crystals around (e) and above (f) a closed stomata in stressed plants (JPG 684 kb)
Supplementary file3 Summary of Pathway Analysis using the Pathway Topology Analysis modules of MetaboAnalyst 5.0. The metabolome view resulted from the analysis in the Pathway Topology Analysis module using the Hypergeometric test, the relative betweenness centrality node importance measure, and the latest KEGG version of the A. thaliana pathway library. The three pathways with an FDR (False Discovery Rate) lower than 0.05 are indicated by name: The Nicotinate and nicotinamide metabolism, C5-Branched dibasic acid metabolism, and Phenylpropanoid biosynthesis pathways (JPG 141 kb)
Supplementary file4 List of differentially expressed peaks (m.z) resulted from the Pathway Analysis using the MS Peaks to Pathway module of MetaboAnalyst 5.0. Data-set showing the pathway code, KEGG id of the matched compound, matched form, mass difference, name of the compound, correlation, t.score, p.value, FDR (False Discovery Rate), fold change - FC, Log2 (FC), and profile, in each one of the three scenarios evaluated: short-term stress – STS (control vs stress plants at 1 DAT); long-term stress 1 – LTS1 (control vs stress plants at 4 DAT); and long-term stress 2 – LTS2 (stressed plants at 1 and 4 DAT) (ODS 37 kb)
Acknowledgements
The authors acknowledge funding to V.N.B.S., T.L.C.S., T.M.M.F. and J.C.R.N. by the Coordination for the Improvement of Higher Education Personnel (CAPES), via the Graduate Program in Plant Biotechnology at the Federal University of Lavras (UFLA) and the Graduate Program in Chemistry at the Federal University of Goiás (UFG).
Abbreviations
- AE
Age effect
- STS
Short-term stress
- LTS
Long-term stress
- FDR
False discovery rate
- FC
Fold change
- GSEA
Gene set enrichment analysis
- HRMS
High-resolution mass spectrometry
- KEGG
Kyoto encyclopedia of genes and genomes
- MS
Mass spectrometry
- PLS-DA
Partial least squares discriminant analysis
- UHPLC
Ultra-high performance liquid chromatography
- VIP
Variable importance in projection
- RT
Reference transcriptome
Authors’ Contributions
CAFS and MTSJr designed the research; VNBS, TLCS, TMMF, APL and JAAR performed the experiments; VNBS, TLCS, JCRN, PVA, LFV, CAFS and MTSJr analyzed the data; PVA, LFV, CAFS and MTSJr drafted the manuscript. CAFS and MTSJr have revised the manuscript. All authors read and approved the final manuscript.
Funding
The authors disclose receipt of the following financial support for the research, authorship, and/or publication of this article: the grant (01.13.0315.00—DendePalm Project) for this study was awarded by the Brazilian Research and Innovation Agency (FINEP). The authors confirm that the funder had no influence over the study design, the content of article, or selection of this journal.
Data Availability
The data-sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
References
- Acosta-Motos JR, Ortuño MF, Bernal-Vicente A, Diaz-Vivancos P, Sanchez-Blanco MJ, Hernandez JA. Plant responses to salt stress: adaptive mechanisms. Agronomy. 2017;7(1):18. doi: 10.3390/agronomy7010018. [DOI] [Google Scholar]
- Aghaleh M, Niknam V, Ebrahimzadeh H, Razavi K. Salt stress effects on growth, pigments, proteins and lipid peroxidation in Salicornia persica and S. europaea. Biologia Plantarum. 2009;53(2):243–248. doi: 10.1007/s10535-009-0046-7. [DOI] [Google Scholar]
- Alam M, Juraimi AS, Rafii M, Abdul Hamid A, Aslani F. Screening of purslane (Portulaca oleracea L.) accessions for high salt tolerance. Sci World J. 2014 doi: 10.1155/2014/627916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam M, Juraimi AS, Rafii M, Abdul Hamid A. Effect of salinity on biomass yield and physiological and stem-root anatomical characteristics of purslane (Portulaca oleracea L.) accessions. BioMed Res Int. 2015 doi: 10.1155/2015/105695. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ali A, Maggio A, Bressan RA, Yun D-J. Role and functional differences of HKT1-type transporters in plants under salt stress. Int J Mol Sci. 2019;20(5):1059. doi: 10.3390/ijms20051059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allbed A, Kumar L. Soil salinity mapping and monitoring in arid and semi-arid regions using remote sensing technology: a review. Adv Remote Sens. 2013 doi: 10.4236/ars.2013.24040. [DOI] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. In. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 15 Dec 2021
- Arif Y, Singh P, Siddiqui H, Bajguz A, Hayat S. Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Biochem. 2020;156:64–77. doi: 10.1016/j.plaphy.2020.08.042. [DOI] [PubMed] [Google Scholar]
- Arora S, Rao GG (2017) Bio-amelioration of salt-affected soils through halophyte plant species. In: Arora S, Singh A, Singh Y (eds) Bioremediation of salt affected soils: an Indian perspective. Springer, Cham. 10.1007/978-3-319-48257-6_4
- Avelange-Macherel MH, Ly-Vu B, Delaunay J, Richomme P, Leprince O. NMR metabolite profiling analysis reveals changes in phospholipid metabolism associated with the re-establishment of desiccation tolerance upon osmotic stress in germinated radicles of cucumber. Plant Cell Environ. 2006;29(4):471–482. doi: 10.1111/j.1365-3040.2005.01424.x. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsai O, Al Hassan M, Boscaiu M, Sestras RE, Vicente O. The genus as a suitable model to study the mechanisms of plant tolerance to drought and salinity. The EuroBiotech Journal. 2018;2(2):104–113. doi: 10.2478/ebtj-2018-0014. [DOI] [Google Scholar]
- Brink BG, Seidel A, Kleinbölting N, Nattkemper TW, Albaum SP. Omics fusion–a platform for integrative analysis of omics data. J Integr Bioinform. 2016;13(4):43–46. doi: 10.1515/jib-2016-296. [DOI] [PubMed] [Google Scholar]
- Carvalho da Silva TL, Belo Silva VN, Braga ÍdO, et al. Integration of metabolomics and transcriptomics data to further characterize Gliricidia sepium (Jacq.) Kunth under high salinity stress. Plant Genom. 2022;15(1):e20182. doi: 10.1002/tpg2.20182. [DOI] [PubMed] [Google Scholar]
- Cavill R, Jennen D, Kleinjans J, Briedé JJ. Transcriptomic and metabolomic data integration. Brief Bioinform. 2016;17(5):891–901. doi: 10.1093/bib/bbv090. [DOI] [PubMed] [Google Scholar]
- Céccoli G, Ramos JC, Ortega LI, Acosta JM, Perreta MG (2011) Salinity induced anatomical and morphological changes in Chloris gayana Kunth roots. BIOCELL 35(1):9–17. https://www.techscience.com/biocell/v35n1/37228. Accessed 15 Dec 2021 [PubMed]
- Chong J, Xia J. Using MetaboAnalyst 4.0 for metabolomics data analysis, interpretation, and integration with other omics data. Comput Methods Data Anal Metabol. 2020 doi: 10.1007/978-1-0716-0239-3_17. [DOI] [PubMed] [Google Scholar]
- Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protocols Bioinform. 2019;68(1):e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Cordeiro GG (2001) Salinidade em areas irrigadas. Embrapa Semiárido-Artigo em periódico indexado (ALICE). https://ainfo.cnptia.embrapa.br/digital/bitstream/item/143307/1/ID-31379.pdf. Accessed 15 Dec 2021
- Cuong DM, Kwon S-J, Nguyen BV, Chun SW, Kim JK, Park SU. Effect of salinity stress on phenylpropanoid genes expression and related gene expression in wheat sprout. Agronomy. 2020;10(3):390. doi: 10.3390/agronomy10030390. [DOI] [Google Scholar]
- D’andrea RM, Andreo CS, Lara MV. Deciphering the mechanisms involved in Portulaca oleracea (C4) response to drought: metabolic changes including crassulacean acid-like metabolism induction and reversal upon re-watering. Physiol Plant. 2014;152(3):414–430. doi: 10.1111/ppl.12194. [DOI] [PubMed] [Google Scholar]
- de Araújo SA, Silveira JA, Almeida TD, Rocha I, Morais DL, Viégas RA. Salinity tolerance of halophyte Atriplex nummularia L. grown under increasing NaCl levels. Revista Brasileira de Engenharia Agrícola e Ambiental. 2006;10(4):848–854. doi: 10.1590/S1415-43662006000400010. [DOI] [Google Scholar]
- Devi S, Nandwal A, Angrish R, Arya S, Kumar N, Sharma S. Phytoremediation potential of some halophytic species for soil salinity. Int J Phytorem. 2016;18(7):693–696. doi: 10.1080/15226514.2015.1131229. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte HHF, Souza ERd. Soil water potentials and Capsicum annuum L. under salinity. Revista Brasileira de Ciência do solo. 2016;40:e0150220. doi: 10.1590/18069657rbcs20150220. [DOI] [Google Scholar]
- El-Shora HM, El-Gawad AMA. Response of Cicer arietinum to allelopathic effect of Portulaca oleracea root extract. Phyton (Horn) 2015;55(2):215–232. doi: 10.12905/0380.phyton55(2)2015-0215. [DOI] [Google Scholar]
- Everard JD, Gucci R, Kann SC, Flore JA, Loescher WH. Gas exchange and carbon partitioning in the leaves of celery (Apium graveolens L.) at various levels of root zone salinity. Plant Physiol. 1994;106(1):281–292. doi: 10.1104/pp.106.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. Salinity tolerance in halophytes. New Phytol. 2008;179(4):945–963. doi: 10.1111/j.1469-8137.2008.02531.x. [DOI] [PubMed] [Google Scholar]
- Flowers TJ, Galal HK, Bromham L. Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol. 2010;37(7):604–612. doi: 10.1071/FP09269. [DOI] [Google Scholar]
- Flowers TJ, Munns R, Colmer TD. Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Ann Bot. 2015;115(3):419–431. doi: 10.1093/aob/mcu217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale J. Water balance and gas exchange of plants under saline conditions. Plants Saline Environ. 1975 doi: 10.1007/978-3-642-80929-3_11. [DOI] [Google Scholar]
- Gamon JA, Peñuelas J, Field CB. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sensing Environ. 1992;41:35–44. doi: 10.1016/0034-4257(92)90059-S. [DOI] [Google Scholar]
- Gitelson A, Kaufman Y, Stark R, Rundquist D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens Environ. 2002;80:76–87. doi: 10.1016/S0034-4257(01)00289-9. [DOI] [Google Scholar]
- Gitelson A, Viña A, Ciganda V, Rundquist D, Arkebauer T. Remote estimation of canopy chlorophyll in crops. Geophys Res Lett. 2005 doi: 10.1029/2005GL022688. [DOI] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda H, Ivanisevic J, Johnson CH, Kurczy ME, Benton HP, Rinehart D, Nguyen T, Ray J, Kuehl J, Arevalo B. Interactive XCMS Online: simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal Chem. 2014;86(14):6931–6939. doi: 10.1021/ac500734c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang Y. Phospholipids in Salt Stress Response. Plants. 2021;10(10):2204. doi: 10.3390/plants10102204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Ding N-Z. Plant unsaturated fatty acids: multiple roles in stress response. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.562785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Li J, Shi L, Zhao Q, Wu Z, Zeng S, Li Y, Zhang W, Wang F-D, Zhang S. Exogenous metabolites spray, which identified from metabolomics analysis and transcriptomic analysis, can improve salt tolerance of Chinese cabbages (Brassica rapa L. ssp pekinensis) J Plant Interact. 2021;16(1):452–461. doi: 10.1080/17429145.2021.1969457. [DOI] [Google Scholar]
- Hou Q, Ufer G, Bartels D. Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 2016;39(5):1029–1048. doi: 10.1111/pce.12666. [DOI] [PubMed] [Google Scholar]
- Jamil IN, Remali J, Azizan KA, Nor Muhammad NA, Arita M, Goh H-H, Aizat WM. Systematic multi-omics integration (MOI) approach in plant systems biology. Front Plant Sci. 2020;11:944. doi: 10.3389/fpls.2020.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428(4):726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Khan MA, Ungar IA, Showalter AM. The effect of salinity on the growth, water status, and ion content of a leaf succulent perennial halophyte, Suaeda fruticosa (L.) Forssk. J Arid Environ. 2000;45(1):73–84. doi: 10.1006/jare.1999.0617. [DOI] [Google Scholar]
- Khan MA, Ungar IA, Showalter AM. Effects of salinity on growth, water relations and ion accumulation of the subtropical perennial halophyte, Atriplex griffithii var. stocksii. Annal Bot. 2000;85(2):225–232. doi: 10.1006/anbo.1999.1022. [DOI] [Google Scholar]
- Khorasaninejad S, Mousavi A, Soltanloo H, Hemmati K, Khalighi A. The effect of salinity stress on growth parameters, essential oil yield and constituent of peppermint (Mentha piperita L.) World Appl Sci J. 2010;11:1403–1407. [Google Scholar]
- Khorasaninejad S, Mousavi A, Soltanloo H, Hemmati K, Khalighi A. The effect of drought stress on growth parameters, essential oil yield and constituent of Peppermint (Mentha piperita L.) J Med Plants Res. 2011;5(22):5360–5365. doi: 10.5897/JMPR.9001196. [DOI] [Google Scholar]
- Koch K, Kennedy RA. Characteristics of crassulacean acid metabolism in the succulent C4 dicot Portulaca oleracea L. Plant Physiol. 1980;65(2):193–197. doi: 10.1104/pp.65.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyro H-W. Effect of salinity on growth, photosynthesis, water relations and solute composition of the potential cash crop halophyte Plantago coronopus (L.) Environ Exp Bot. 2006;56(2):136–146. doi: 10.1016/j.envexpbot.2005.02.001. [DOI] [Google Scholar]
- Kumari A, Das P, Parida AK, Agarwal PK. Proteomics, metabolomics, and ionomics perspectives of salinity tolerance in halophytes. Front Plant Sci. 2015;6:537. doi: 10.3389/fpls.2015.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wan S, Zhou J, Yang Z, Qin P. Leaf chlorophyll fluorescence, hyperspectral reflectance, pigments content, malondialdehyde and proline accumulation responses of castor bean (Ricinus communis L.) seedlings to salt stress levels. Ind Crops Prod. 2010;31(1):13–19. doi: 10.1016/j.indcrop.2009.07.015. [DOI] [Google Scholar]
- Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol. 2013;9(7):e1003123. doi: 10.1371/journal.pcbi.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol. 2017;13(5):e1005457. doi: 10.1371/journal.pcbi.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Wang Y, Gu D, Nan J, Chen S, Li H. Overexpression of S-adenosyl-l-methionine synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. Int J Mol Sci. 2017;18(4):847. doi: 10.3390/ijms18040847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444(2):139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Maksimovic I, Ilin Z. Effects of salinity on vegetable growth and nutrients uptake. Irrig Syst Pract Challenging Environ. 2012 doi: 10.5772/29976. [DOI] [Google Scholar]
- Megdiche W, Hessini K, Gharbi F, Jaleel CA, Ksouri R, Abdelly C. Photosynthesis and photosystem 2 efficiency of two salt-adapted halophytic seashore Cakile maritima ecotypes. Photosynthetica. 2008;46(3):410–419. doi: 10.1007/s11099-008-0073-1. [DOI] [Google Scholar]
- Munns R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002;25(2):239–250. doi: 10.1046/j.0016-8025.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- OmicsBox (2019) Bioinformatics made easy https://www.biobam.com/omicsbox/. Accessed 15 Dec 2021
- Ozturk M, Altay V, Guvensen A (2020) Portulaca oleracea: a vegetable from saline habitats. In: Handbook of halophytes. Springer, Switzerland. 10.1007/978-3-030-17854-3_96-1
- Parida AK, Das AB. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60(3):324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Parihar P, Singh S, Singh R, Singh VP, Prasad SM. Effect of salinity stress on plants and its tolerance strategies: a review. Environ Sci Pollut Res. 2015;22(6):4056–4075. doi: 10.1007/s11356-014-3739-1. [DOI] [PubMed] [Google Scholar]
- Petropoulos SΑ, Karkanis A, Fernandes Â, Barros L, Ferreira IC, Ntatsi G, Petrotos K, Lykas C, Khah E. Chemical composition and yield of six genotypes of common purslane (Portulaca oleracea L): an alternative source of omega-3 fatty acids. Plant Foods Human Nutr. 2015;70(4):420–426. doi: 10.1007/s11130-015-0511-8. [DOI] [PubMed] [Google Scholar]
- Petropoulos S, Karkanis A, Martins N, Ferreira IC. Phytochemical composition and bioactive compounds of common purslane (Portulaca oleracea L.) as affected by crop management practices. Trends Food Sci Technol. 2016;55:1–10. doi: 10.1016/j.tifs.2016.06.010. [DOI] [Google Scholar]
- Polle A, Chen S. On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 2015;38(9):1794–1816. doi: 10.1111/pce.12440. [DOI] [PubMed] [Google Scholar]
- Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD. Economics of salt-induced land degradation and restoration. Nat Resour Forum. 2014;38(4):282–295. doi: 10.1111/1477-8947.12054. [DOI] [Google Scholar]
- Qiu N, Lu Q, Lu C. Photosynthesis, photosystem II efficiency and the xanthophyll cycle in the salt-adapted halophyte Atriplex centralasiatica. New Phytol. 2003;159(2):479–486. doi: 10.1046/j.1469-8137.2003.00825.x. [DOI] [PubMed] [Google Scholar]
- Rabhi M, Castagna A, Remorini D, Scattino C, Smaoui A, Ranieri A, Abdelly C. Photosynthetic responses to salinity in two obligate halophytes: Sesuvium portulacastrum and Tecticornia indica. S Afr J Bot. 2012;79:39–47. doi: 10.1016/j.sajb.2011.11.007. [DOI] [Google Scholar]
- Rai A, Saito K, Yamazaki M. Integrated omics analysis of specialized metabolism in medicinal plants. Plant J. 2017;90(4):764–787. doi: 10.1111/tpj.13485. [DOI] [PubMed] [Google Scholar]
- Ramadan T. Ecophysiology of salt excretion in the xero-halophyte Reaumuria hirtella. New Phytol. 1998;139(2):273–281. doi: 10.1046/j.1469-8137.1998.00159.x. [DOI] [Google Scholar]
- Ramoliya P, Patel H, Pandey A. Effect of salinization of soil on growth and macro-and micro-nutrient accumulation in seedlings of Salvadora persica (Salvadoraceae) For Ecol Manage. 2004;202(1–3):181–193. doi: 10.1016/j.foreco.2004.07.020. [DOI] [Google Scholar]
- Rao GG, Nayak A, Chinchmalatpure AR, Nath A, Babu VR. Growth and yield of Salvadora persica, a facultative halophyte grown on saline black soil (Vertic Haplustept) Arid Land Res Manag. 2004;18(1):51–61. doi: 10.1080/15324980490245013. [DOI] [Google Scholar]
- Rewald B, Shelef O, Ephrath JE, Rachmilevitch S (2013) Adaptive plasticity of salt-stressed root systems. In: Ahmad P, Azooz M, Prasad M (eds) Ecophysiology and responses of plants under salt stress, Springer, New York. 10.1007/978-1-4614-4747-4_6
- Rhoades J, Chanduvi F, Lesch S (1999) Soil salinity assessment: methods and interpretation of electrical conductivity measurements. FAO Irrigation and Drainage Paper 57. www.fao.org/3/x2002e/x2002e.pdf
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema J, Gude H, Pollak G (1981) An ecophysiological study of the salt secretion of four halophytes. New Phytologist 89(2):201–217. https://www.jstor.org/stable/2432058. Accessed 15 Dec 2021
- Santos J, Al-Azzawi M, Aronson J, Flowers TJ. eHALOPH a database of salt-tolerant plants: helping put halophytes to work. Plant Cell Physiol. 2016;57(1):e10–e10. doi: 10.1093/pcp/pcv155. [DOI] [PubMed] [Google Scholar]
- Sdouga D, Amor FB, Ghribi S, Kabtni S, Tebini M, Branca F, Trifi-Farah N, Marghali S. An insight from tolerance to salinity stress in halophyte Portulaca oleracea L.: physio-morphological, biochemical and molecular responses. Ecotoxicol Environ Saf. 2019;172:45–52. doi: 10.1016/j.ecoenv.2018.12.082. [DOI] [PubMed] [Google Scholar]
- Shabala S, Mackay A. Ion transport in halophytes. Adv Bot Res. 2011;57:151–199. doi: 10.1016/B978-0-12-387692-8.00005-9. [DOI] [Google Scholar]
- Shahid SA, Zaman M, Heng L (2018) Soil salinity: historical perspectives and a world overview of the problem. In: Guideline for salinity assessment, mitigation and adaptation using nuclear and related techniques, Springer, Cham. 10.1007/978-3-319-96190-3_2
- Shu J, Ma X, Ma H, et al. Transcriptomic, proteomic, metabolomic, and functional genomic approaches of Brassica napus L. during salt stress. PLoS One. 2022;17(3):e0262587. doi: 10.1371/journal.pone.0262587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Singh V, Dutt V, Walia N, Kumawat G, Jakhar ML, Yadava DK, Sharma PC. Insights into salt tolerance of mustard (Brassica juncea L. Czern & Coss): a metabolomics perspective. Environ Exp Bot. 2022;194:104760. doi: 10.1016/j.envexpbot.2021.104760. [DOI] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui N, Tian S, Wang W, Wang M, Fan H. Overexpression of glycerol-3-phosphate acyltransferase from Suaeda salsa improves salt tolerance in Arabidopsis. Front Plant Sci. 2017;8:1337. doi: 10.3389/fpls.2017.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Tada Y, Kawano R, Komatsubara S, Nishimura H, Katsuhara M, Ozaki S, Terashima S, Yano K, Endo C, Sato M. Functional screening of salt tolerance genes from a halophyte Sporobolus virginicus and transcriptomic and metabolomic analysis of salt tolerant plants expressing glycine-rich RNA-binding protein. Plant Sci. 2019;278:54–63. doi: 10.1016/j.plantsci.2018.10.019. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS online: a web-based platform to process untargeted metabolomic data. Anal Chem. 2012;84(11):5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71:403–433. doi: 10.1146/annurev-arplant-050718-100005. [DOI] [PubMed] [Google Scholar]
- Villate A, San Nicolas M, Gallastegi M, Aulas P-A, Olivares M, Usobiaga A, Etxebarria N, Aizpurua-Olaizola O. Metabolomics as a prediction tool for plants performance under environmental stress. Plant Sci. 2021;303:110789. doi: 10.1016/j.plantsci.2020.110789. [DOI] [PubMed] [Google Scholar]
- Visconti F, de Paz JM. Electrical conductivity measurements in agriculture: the assessment of soil salinity. New Trends Develop Metrol. 2016;1:99–126. doi: 10.1016/j.plantsci.2020.110789. [DOI] [Google Scholar]
- Wang Y, Huang L, Du F, Wang J, Zhao X, Li Z, Wang W, Xu J, Fu B. Comparative transcriptome and metabolome profiling reveal molecular mechanisms underlying OsDRAP1-mediated salt tolerance in rice. Sci Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-84638-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z, Zhou X, Li J, Li L, Ma Y, Wu Y, Huang Z, Li X, Xu P, Xue M. Multiple-omics techniques reveal the role of glycerophospholipid metabolic pathway in the response of Saccharomyces cerevisiae against hypoxic stress. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.01398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J-C, Dong J, Wang M-W, Liu C, Zhao B-Q, Wen Z-G, Zhu X-M, Ding H-R, Zhao X-H, Hong L-Z. Effects of NaCl stress on growth of Portulaca oleracea and underlying mechanisms. Braz J Bot. 2019;42(2):217–226. doi: 10.1007/s40415-019-00526-1. [DOI] [Google Scholar]
- Xing J, Zhao B, Dong J, Liu C, Wen Z, Zhu X, Ding H, He T, Yang H, Wang M. Transcriptome and metabolome profiles revealed molecular mechanisms underlying tolerance of Portulaca oleracea to saline stress. Russ J Plant Physiol. 2020;67(1):146–152. doi: 10.1134/S1021443720010240. [DOI] [Google Scholar]
- Yan S, Chong P, Zhao M, Liu H. Physiological response and proteomics analysis of Reaumuria soongorica under salt stress. Sci Rep. 2022;12(1):2539. doi: 10.1038/s41598-022-06502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Lyu M-JA, Leng B-Y, Zhu X-G, Wang B-S. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol Biol. 2016;91(3):241–256. doi: 10.1007/s11103-016-0460-0. [DOI] [PubMed] [Google Scholar]
- Zaman S, Bilal M, Du H, Che S. Morphophysiological and comparative metabolic profiling of purslane genotypes (Portulaca oleracea L.) under salt stress. BioMed Res Int. 2020 doi: 10.1155/2020/4827045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Han B, Wang T, Chen S, Li H, Zhang Y, Dai S. Mechanisms of plant salt response: insights from proteomics. J Proteome Res. 2012;11(1):49–67. doi: 10.1021/pr200861w. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Mao C, Shi Z, Kou X. The amino acid metabolic and carbohydrate metabolic pathway play important roles during salt-stress response in tomato. Front Plant Sci. 2017;8:1231. doi: 10.3389/fpls.2017.01231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y-X, Xin H-L, Rahman K, Wang S-J, Peng C, Zhang H. Portulaca oleracea L.: a review of phytochemistry and pharmacological effects. BioMed Res Int. 2015 doi: 10.1155/2015/925631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 Representative images (1) of the mean of variables derived from the chlorophyll fluorescence technique (saturation pulse method) in purslane plants after 1 and 4 days of submission to increasing levels of NaCl in the growing substrate. The images of Fo, Fv/Fm, and Fm' were obtained in plants adapted to the dark for 30 min, while the images of Y(II), Y(NO) and Y(NPQ) were captured after 5 min of actinic illumination at 280 μmol of light m-2 s-1. The values of the chlorophyll fluorescence parameters in the images can be compared with the color scale in the right bar. Changes over time in chlorophyll fluorescence parameters (2) for control and salinity stressed young purslane plants. The values represent an average of five replicates, and bars represent the standard error of the mean. (a) F - fluorescence intensity; (b) Fo - minimum fluorescence yield on dark-adapted plants; (c) Fm - maximum fluorescence yield on the dark-adapted plants; (d) Fv/Fm - maximum PSII quantum yield; (e) qP - proportion of open PSII; (f) qL - indicator of the PSII redox state; (g) Y(II) - PSII effective quantum yield; (h) qN - non-photochemical quenching that requires measurement of Fo; (i) NPQ - non‐photochemical quenching; (j) Y(NPQ) - regulated energy dissipation quantum yield; (k) Y(NO) - unregulated energy dissipation quantum yield; and (l) ETR - electron transport rate. The images of Fo, Fv/Fm, and Fm were obtained in plants adapted to the dark, while the images of Y(II), Y(NO) and Y(NPQ) were captured after 5 min of actinic illumination at 280 µmol of light/m2/s (JPG 693 kb)
Supplementary file2 Scanning electron microscope (SEM) images of the leaf surface of purslane plants. (a) Control; (b) stressed plant (2.0 g NaCl) showing wrinkle due to dehydration; (c) degree of stomata opening in control compared to (d) stressed (0.5 g NaCl). Formation of salt crystals around (e) and above (f) a closed stomata in stressed plants (JPG 684 kb)
Supplementary file3 Summary of Pathway Analysis using the Pathway Topology Analysis modules of MetaboAnalyst 5.0. The metabolome view resulted from the analysis in the Pathway Topology Analysis module using the Hypergeometric test, the relative betweenness centrality node importance measure, and the latest KEGG version of the A. thaliana pathway library. The three pathways with an FDR (False Discovery Rate) lower than 0.05 are indicated by name: The Nicotinate and nicotinamide metabolism, C5-Branched dibasic acid metabolism, and Phenylpropanoid biosynthesis pathways (JPG 141 kb)
Supplementary file4 List of differentially expressed peaks (m.z) resulted from the Pathway Analysis using the MS Peaks to Pathway module of MetaboAnalyst 5.0. Data-set showing the pathway code, KEGG id of the matched compound, matched form, mass difference, name of the compound, correlation, t.score, p.value, FDR (False Discovery Rate), fold change - FC, Log2 (FC), and profile, in each one of the three scenarios evaluated: short-term stress – STS (control vs stress plants at 1 DAT); long-term stress 1 – LTS1 (control vs stress plants at 4 DAT); and long-term stress 2 – LTS2 (stressed plants at 1 and 4 DAT) (ODS 37 kb)
Data Availability Statement
The data-sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Not applicable.