Abstract
Objectives
A classification system for endocervical adenocarcinoma (ECA) based on high‐risk human papillomavirus (HPV) status has been established; however, the immunohistochemical markers distinguishing HPV‐independent and HPV‐associated ECAs have not been fully described. Here, we aimed to characterize ECA immunopathological features.
Methods
We evaluated the immunohistochemical profile of CLDN18, CDX2, PAX8, p16, p53, and CEA in 60 ECAs comprising 10 HPV‐independent ECAs and 50 HPV‐associated ECAs. Both the membranous and nuclear expression levels of CLDN18 were analyzed.
Results
Membranous CLDN18 (CLDN18 [M]) was found to be expressed in the mucinous epithelium of all HPV‐independent ECAs, including eight gastric‐type ECAs (G‐ECAs), one endometrioid ECA, and one clear cell ECA, but no nuclear CLDN18 (CLDN18 [N]) expression was detected in HPV‐independent ECAs. Among HPV‐associated ECAs, CLDN18 (M) expression levels in intestinal‐type (I‐ECAs) and usual‐type ECAs (U‐ECAs) were significantly different from those in invasive stratified mucin‐producing (iSMILE) carcinomas (p = 0.036). Positive CLDN18 (M) staining was present in 55.6% (5/9) of intestinal‐type and 39.4% (13/33) of usual‐type ECAs and was not present in iSMILE ECAs. Silva pattern C cancers expressed higher levels of CLDN18 (M) than Silva pattern A and B cancers (p = 0.004), whereas the CLDN18 (N) expression levels in cancers showing Silva pattern A were significantly higher than those in cancers exhibiting Silva patterns B and C (p < 0.001).
Conclusion
Membranous CLDN18 is expressed in ECAs and is particularly frequently expressed in HPV‐independent ECAs, and membranous CLDN18 expression has potential as a therapeutic target. Nuclear staining of CLDN18 is a new immunohistochemical marker for diagnosing Silva pattern A HPV‐associated ECAs and is associated with a good prognosis. Further studies should investigate the therapeutic and prognostic significance of membranous and nuclear CLDN18 expression and develop a related test that can be implemented in the clinical evaluation of ECAs.
Keywords: CLDN18, endocervical adenocarcinoma, HPV, immunohistochemistry, membranous expression, nuclear expression
Membranous claudin‐18 expression is expressed in human papillomavirus (HPV)‐independent endocervical adenocarcinomas (ECAs), particularly gastric‐type ECAs. Membranous claudin‐18 expression indicates its potential as a therapeutic target for ECAs. Nuclear staining of claudin‐18 is a new immunohistochemical marker for diagnosing Silva A pattern in HPV‐associated ECAs.
1. INTRODUCTION
Endocervical adenocarcinoma (ECA) accounts for 20%–25% of cervical cancers, and its incidence is increasing relative to that of cervical squamous cell carcinoma (SQCC) and in absolute numbers, 1 , 2 , 3 especially in the era of the HPV vaccine. According to the International Endocervical Adenocarcinoma Criteria and Classification (IECC), it is divided into two categories, HPV‐associated ECA and HPV‐independent ECA. 4 ECAs are considered to be heterogeneous tumors with different etiologies, pathogenesis mechanisms, molecular features, treatment responses, and prognoses. 5 It has been suggested that HPV‐independent ECA is associated with a poorer prognosis than HPV‐associated ECA. 6 , 7 , 8 Among the HPV‐independent ECAs, the gastric type is most common; however, other types (endometrioid adenocarcinoma, clear cell adenocarcinoma, serous adenocarcinoma, and mesonephric carcinoma) are rare. 6
CLDN18 is an adhesion molecule extensively expressed in the cytoplasmic membrane of gastric epithelial cells in normal and gastric cancer tissue. CLDN18 is expressed along the basolateral membrane in all types of differentiated gastric epithelial cells, including surface mucous cells, chief cells, parietal cells, and endocrine cells. 9 Therefore, membranous CLDN18 is considered to be a highly selective immunohistochemical marker to detect gastric differentiation in pancreatobiliary, 10 colorectal, 11 and ovarian mucinous neoplasms. 9 , 12
The CLDN family comprises at least 27 transmembrane proteins that are the main components of tight junctions (TJs). CLDNs generally localize to the apical and basolateral regions of the cell membrane and play an important role in cell–cell adhesion, maintenance of cell polarity and selective paracellular permeability, regulating epithelial‐mesenchymal transformation (EMT), cell proliferation, migration and infiltration in cancer. On the other hand, claudins have one putative or multiple nuclear localization sequences (NLSs) that facilitate transport into the nucleus. Nuclear CLDNs interact with transcriptional regulators that impact gene expression and the regulation of cell apoptosis. 13 Recently, nuclear staining of CLDN18 was shown to be a new immunohistochemical marker for diagnosing intramucosal well‐differentiated gastric adenocarcinoma. 14
A few studies have shown membranous immunohistochemical expression of CLDN18 in endocervical lesions, such as gastric‐type adenocarcinoma in situ, gastric‐type ECAs and nongastric‐type ECAs. 15 , 16 , 17 No relevant studies have investigated CLDN18 and its different subcellular locations in HPV‐independent and HPV‐associated ECAs. Moreover, diagnostic classification of ECAs has been a significant challenge due to their histopathologically similar morphology, especially when only a limited amount of biopsy material is examined, suggesting that a series of immunohistochemical markers is required to improve diagnostic accuracy. 18 Therefore, we conducted the present study to evaluate the membranous and nuclear expression levels of CLDN18 and their association with clinicopathological and immunophenotypic parameters in HPV‐independent and HPV‐associated ECAs.
2. MATERIALS AND METHODS
2.1. Patient selection
This study was performed in accordance with the institutional review board approval from the medical ethical committee of the Affiliated Hospital of Qingdao University. There were 62 patients who had undergone radical hysterectomy or hysterectomy and were histologically diagnosed with ECA at West Coast Hospital and Shinan District Hospital, the Affiliated Hospital of Qingdao University, between 2014 and 2019. All formalin‐fixed paraffin‐embedded (FFPE) tissue specimens were selected from the pathology archives at the Department of Pathology. All whole hematoxylin and eosin (HE)‐stained sections were reviewed by two pathologists, and the histological subtypes were classified according to the 2020 WHO classification of tumors of the uterine cervix and the International Endocervical Adenocarcinoma Criteria and Classification (IECC) system. 16 Two patients with no adequately suitable specimens for immunohistochemical analysis were excluded. There were no patients with mesonephric or serous ECA. The 60 patients (age range, 31–81 years; median age, 51 years) comprised 33 patients with usual‐type endocervical adenocarcinomas (U‐ECAs), nine patients with intestinal‐type endocervical adenocarcinomas (I‐ECAs), eight patients with invasive stratified mucin‐producing carcinomas (iSMILE‐ECAs), eight patients with gastric‐type endocervical adenocarcinomas (G‐ECAs), one patient with clear cell adenocarcinoma and one patient with endometrioid adenocarcinoma. Normal endocervical glands were found in 17 of the 60 patients. Furthermore, 10 normal endocervical glandular mucosae (NEM) samples and 20 cervical SQCC samples were evaluated.
2.2. Immunohistochemistry analysis
Serial sections of 4 mm thickness were taken from representative FFPE tissue blocks, affixed to 3‐aminopropyl triethoxysilane‐coated slides, and air‐dried overnight at 37°C. The sections were then subjected to HE and immunohistochemical staining. Immunohistochemical staining was performed using the following immunoperoxidase polymer methods. After dewaxing and antigen retrieval, endogenous peroxidase activity was blocked with 0.3% hydrogen peroxide for 10 min. After blocking with goat serum, the sections were incubated for 30 min at 37°C with rabbit anti‐CLDN18 (1:500 dilution, clone ab203563; Abcam), rabbit anti‐CDX2 (1:50 dilution, clone A20222; ABclonal Technology Co), rabbit anti‐PAX8 (1:100 dilution, clone bs‐1201R; BIOSS ANTIBODIES), mouse anti‐CEA (1:300 dilution, clone 12‐140‐10; ZSGB‐BIO), mouse anti‐p53 (1:300 dilution, clone DO‐7; ZSGB‐BIO), and mouse anti‐P16 (1:100 dilution; clone MX007, MXB). The slides were visualized with diaminobenzidine (DAB) and counterstained with hematoxylin. Both membranous and nuclear CLDN18 levels were evaluated. The stained slides were scanned with a Pannoramic SCAN slide scanner (3DHISTECH Ltd.) to obtain a whole slide image.
2.3. Immunohistochemical evaluation
CDX2, PAX8, CEA, and CLDN18 expression were assessed using a semiquantitative pathology H‐score, according to staining intensity and percent of corresponding intensity tumor cells. The intensity was scored as follows: No staining as 0, weak staining as 1+, moderate staining as 2+, and strong staining as 3+. In brief, the H‐score was calculated as follows: (Percentage of expression 1+ tumor cells) × 1 + (percentage of expression 2+ tumor cells) × 2 + (percentage of expression 3+ tumor cells) × 3. The score can range from 0 to 300. In this study, a cutoff of 50 was used, and scores were categorized as negative or low (0–50) and positive or high (51–300). 19 , 20 The criterion for subclonal CLDN18 was defined as abnormal staining of more than 12 consecutive cells on the background of abnormal CLDN18 staining. 21 P16 was evaluated as positive with diffuse, block‐like expression, whereas no or patchy expression was negative. 4 P53 expression was considered aberrant when it was overexpressed, completely absent in the nucleus, or expressed in the cytoplasm. 22
2.4. High‐risk HPV DNA assay
For high‐risk HPV testing, each sample was evaluated using the Roche Cobas® 4800 real‐time PCR HPV test. The assay detects 14 high‐risk types, with individual genotyping for HPV16 and HPV18, and the other 12 types were reported by a pooled result. 23
2.5. Silva system
Invasive ECAs were classified based on the pattern of invasion (Pattern A, B or C). 24 Briefly, Pattern A is characterized by well‐demarcated glands without destructive stromal invasion or lymphovascular invasion. Pattern B is characterized by early destructive stromal invasion arising from otherwise well‐demarcated glands and can present with lymphovascular invasion. Pattern C is characterized by extensive and diffuse destructive stromal invasion.
2.6. Statistical analysis
The differences and associations between immunohistochemical results and clinicopathological features were compared using the χ 2 test and Fisher's exact test, with statistical significance at p < 0.05.
3. RESULTS
3.1. CLDN18 expression in ECAs
Nonneoplastic cervical epithelial cells, columnar epithelium and squamous epithelium did not express CLDN18. Cancer cells were considered CLDN18‐positive if membranous or nuclear staining was present. Overall, CLDN18 (M) expression was present in 46.7% (28/60) of cases, 11 with membranous staining and 17 with mixed cytoplasmic/membranous staining. Of note, 14 primary cases (23.3%) demonstrated CLDN18 (N) expression (6 with nuclear expression and 8 with mixed nuclear/cytoplasmic expression) (Figure 1).
FIGURE 1.
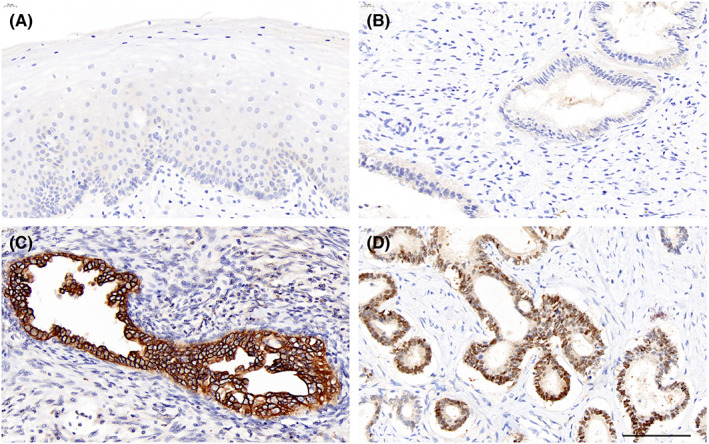
Different subcellular locations of CLDN18 expression in ECAs. (A, B) Nonneoplastic cervical epithelial cells, columnar epithelium and squamous epithelium are negative for CLDN18 expression. (C) ECA sample positive for CLDN18 (M) expression. (D) ECA sample positive for CLDN18 (N) expression (bar = 100 μm). ECA, endocervical adenocarcinoma.
3.2. CLDN18 expression in HPV‐independent ECAs
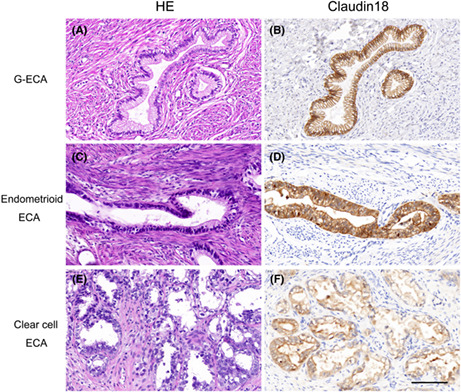
All 8 G‐ECAs were positive for CLDN18 (M) expression in the mucinous epithelium, with seven showing strong staining patterns and one exhibiting moderate staining. One endometrioid ECA and one clear cell ECA were strongly and weakly positive for CLDN18 (M), respectively (Figure 2). However, no CLDN18 (N) expression was detected in HPV‐independent ECAs.
FIGURE 2.
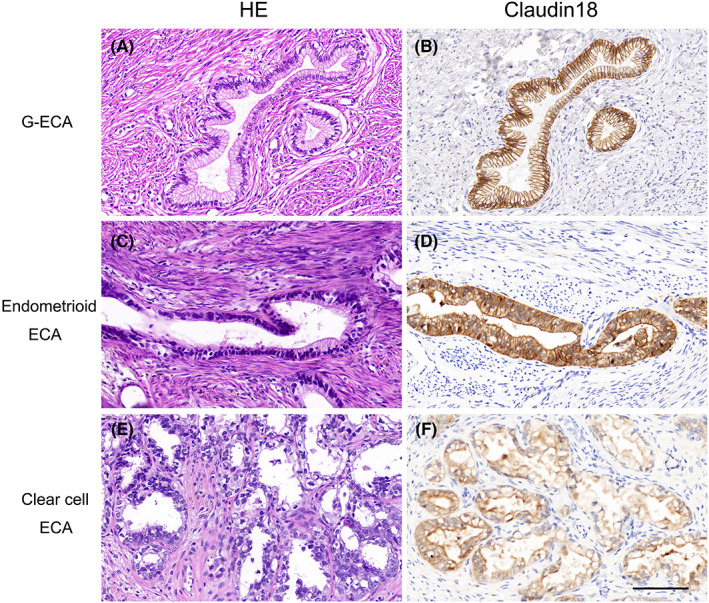
CLDN‐18 expression in HPV‐independent ECAs. (A) Gastric‐type ECA (HE); (B) strong CLDN18 (M) staining. (C) Endometrioid ECA (HE); (D) strong CLDN18 (M) staining. (E) Clear cell ECA (HE); (F) weak CLDN18 (M) staining (bar = 100 μm). ECA, endocervical adenocarcinoma; HE, hematoxylin and eosin; HPV, human papillomavirus.
3.3. CLDN18 expression and clinicopathological associations in HPV‐associated ECAs
In HPV‐associated ECAs, CLDN18 (M) was expressed in the mucinous epithelium of numerous ECAs (18/50 cases, 36.0%). CLDN18 (M) expression positivity showed a statistically significant association with histological type and was significantly higher in intestinal and usual‐type ECAs than in iSMILE‐type ECAs (p = 0.036). Positive CLDN18 (M) staining was present in 5/9 (55.6%) and 13/33 (39.4%) intestinal‐type and usual‐type ECAs, respectively, and was not present in iSMILE‐ECAs. Furthermore, high CLDN18 (M) expression was also associated with the Silva pattern and stromal invasion, and a higher incidence of positive cases was found among the Silva pattern C (p = 0.004) and deep stromal invasion (p = 0.016) groups. CLDN18 (M) expression was not significantly associated with age, grade, stage, lymph node metastases, nerve invasion, or lymphovascular space involvement (LVSI) (Table 1).
TABLE 1.
Clinicopathological features of the considered series according to claudin‐18 (M) and claudin‐18 (N) status in HPV‐associated ECAs
| Characteristic | n | Claudin‐18 (M) n (%) | p | Claudin‐18 (N) n (%) | p |
|---|---|---|---|---|---|
| Age (years) | |||||
| <50 years | 24 | 10 (41.7) | NS | 5 (20.8) | NS |
| ≥50 years | 26 | 8 (30.8) | 9 (34.6) | ||
| Histologic grade | |||||
| G1/2 | 44 | 15 (34.1) | NS | 12 (27.3) | NS |
| G3 | 6 | 3 (50.0) | 2 (33.3) | ||
| Stage | |||||
| I | 40 | 12 (30.0) | NS | 14 (35.0) | NS |
| II | 2 | 2 (100) | 0 (0) | ||
| III | 6 | 2 (33.3) | 0 (0) | ||
| IV | 2 | 2 (100) | 0 (0) | ||
| Lymph node metastases | |||||
| Positive | 6 | 2 (33.3) | NS | 2 (33.3) | NS |
| Negative | 44 | 16 (36.4) | 12 (27.3) | ||
| LVSI | |||||
| Positive | 6 | 2 (33.3) | NS | 4 (66.7) | NS |
| Negative | 44 | 16 (36.4) | 10 (22.7) | ||
| Nerve invasion | |||||
| Positive | 2 | 2 (100) | NS | 0 (0) | NS |
| Negative | 48 | 16 (33.3) | 14 (29.2) | ||
| Stromal invasion | |||||
| ≤1/2 | 29 | 6 (20.7) | 0.016 | 9 (32.1) | NS |
| >1/2 | 21 | 12 (57.1) | 5 (22.7) | ||
| Sillva type | |||||
| A | 14 | 1 (7.1) | 0.004 | 10 (71.4) | <0.001 |
| B | 6 | 1 (16.7) | 2 (33.3) | ||
| C | 30 | 16 (53.3) | 2 (6.7) | ||
| Histologic type | |||||
| I‐EAC | 9 | 5 (55.6) | 0.036 | 0 (0) | NS |
| EAC‐iSMILE | 8 | 0 (0) | 2 (25.0) | ||
| U‐EAC | 33 | 13 (39.4) | 12 (36.4) | ||
Abbreviations: ECA‐iSMILE, invasive stratified mucin‐producing carcinoma; I‐ECA, intestinal‐type ECA; U‐ECA, usual‐type ECA.
Of note, among 50 HPV‐associated ECAs, 14 primary cases (28.0%) demonstrated CLDN18 (N) expression. High CLDN18 (N) expression was associated with the Silva pattern, and a higher incidence of positive cases was found in the Silva pattern A group (p < 0.001). CLDN18 (N) expression was not significantly associated with age, grade, stage, or pathological type. Furthermore, CLDN18 (N) expression was not associated with lymph node metastases, nerve invasion, stromal invasion, or LVSI (Table 1).
Interestingly, six tumors (12.0%) showed concomitant nuclear and membranous CLDN18 expression within subclonal areas. Tumors were considered heterogeneous for CLDN18 if subclonal patterns exhibited different patterns of positive staining with membranous or nuclear CLDN18 in one whole section. Even areas with different staining intensities and distinct subcellular localization of CLDN18 staining were observed within the same tumor focus (Figure 3). Among 50 HPV‐associated ECAs, CLDN18 subclonal expression patterns were seen in two intestinal‐type (2/9, 22.2%) and 4 usual‐type cases (4/33, 12.1%). In the whole tumor sections, all 6 cases showed more than one subclonal area, and in 3/6 cases, subclonality was focal (<10%).
FIGURE 3.
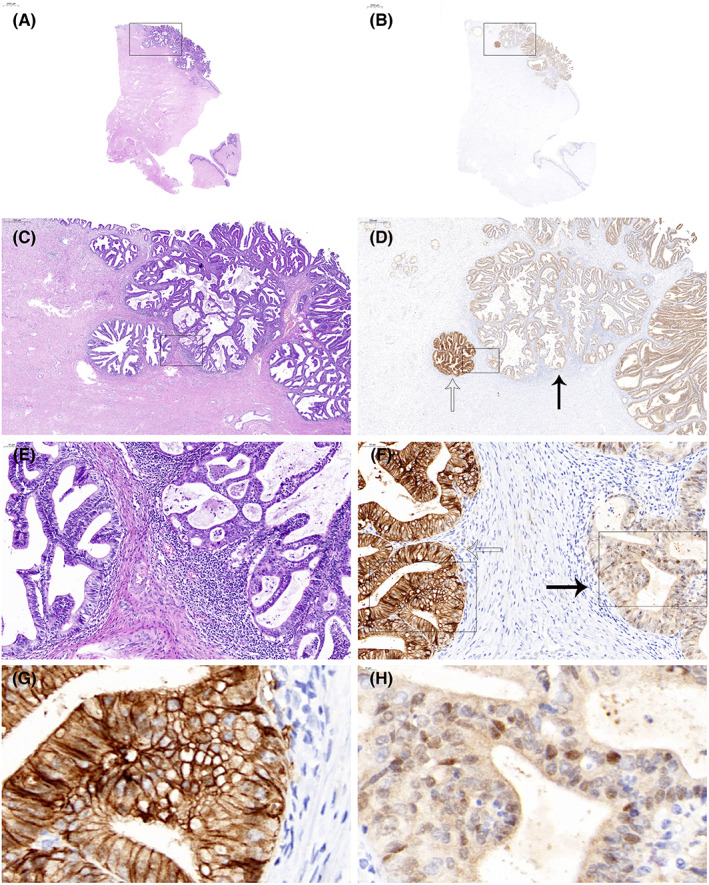
Usual‐type ECA with heterogeneous or subclonal CLDN18 expression. (A, C, E) HE staining at different low‐ and high‐power views. (B) Low‐power view shows CLDN18 expression in the ECA. (D) Areas with different CLDN18 staining intensities were admixed in the same tumor focus (strong expression at the hollow arrow and weak expression at the solid arrow). (F) Areas with different subcellular localizations of CLDN18 expression were admixed in the same tumor focus (region at the hollow arrow is mainly located in the membrane and cytoplasm, and the solid arrow indicates nuclear and cytoplasmic localization). (G) In some areas, CLDN18 expression is strong and located in the cell membrane. (H) In other areas, CLDN18 expression is weak and shows nuclear and cytoplasmic localization. ECA, endocervical adenocarcinoma; HE, hematoxylin and eosin.
3.4. Expression of immunohistochemical markers in different ECA subtypes
Among 60 ECA lesions, 33 (55.0%) were usual‐type, eight (13.3%) were gastric‐type, nine (15.0%) were intestinal‐type, and eight (13.3%) were iSMILE‐type. Figure 4 illustrates the immunohistochemical marker expression of CDX2, PAX8, p16, p53, and CEA, as well as CLDN18 (M) and CLDN18 (N), in different subtypes. As shown in Figure 4, CDX2 was expressed more frequently in intestinal‐type cancers than in gastric‐, usual‐ and iSMILE‐type cancers (p < 0.001). PAX8 expression was significantly lower in gastric‐ and iSMILE‐type cancers than in intestinal‐ and usual‐type cancers (p < 0.001). CLDN18 (M) was expressed in all gastric‐type cancers and tended to be expressed more often in intestinal‐ and usual‐type cancers than in iSMILE‐type cancers (p < 0.001). P16 was expressed in HPV‐associated intestinal‐type, usual‐type, and iSMILE‐type ECAs but was not expressed in HPV‐independent gastric‐type ECAs (p < 0.001). The rates of abnormal P53, CEA, and claudin (N) expression were not significantly different among the four subtypes.
FIGURE 4.
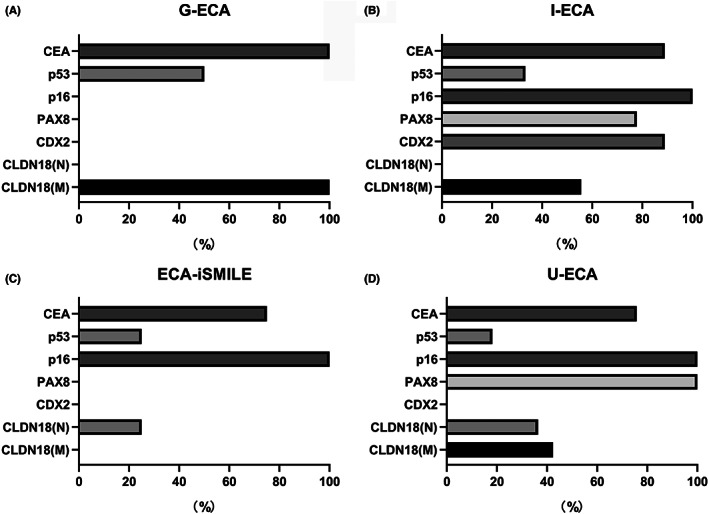
Expression of immunohistochemical markers in different ECA subtypes. Each graph shows the positive or aberrant expression rates of CDX2, PAX8, p16, p53, CEA, CLDN18 (M), and CLDN18 (N). (A) Gastric‐type ECA (n = 8). (B) Intestinal‐type ECA (n = 9). (C) iSMILE‐type ECA (n = 8). (D) Usual‐type ECA (n = 33). ECA, endocervical adenocarcinoma.
Based on the morphological features and results of the immunohistochemical analysis, we combined three phenotypic markers, claudin 18 (M) as gastric differentiation, CDX2 as intestinal differentiation, and PAX8 as Müllerian differentiation. We subclassified the ECAs as follows: Gastric‐type ECA, positive CLDN18 (M), and negative CDX2 expression; intestinal‐type ECA, positive CDX2 and positive PAX8 expression; usual‐type ECA, positive PAX8 and negative CDX2 expression; and iSMILE‐type ECA, negative expression of all three markers (CLDN18 [M], CDX‐2 and PAX8). The representative histological and immunohistochemical features of each ECA subtype are shown in Figure 5.
FIGURE 5.
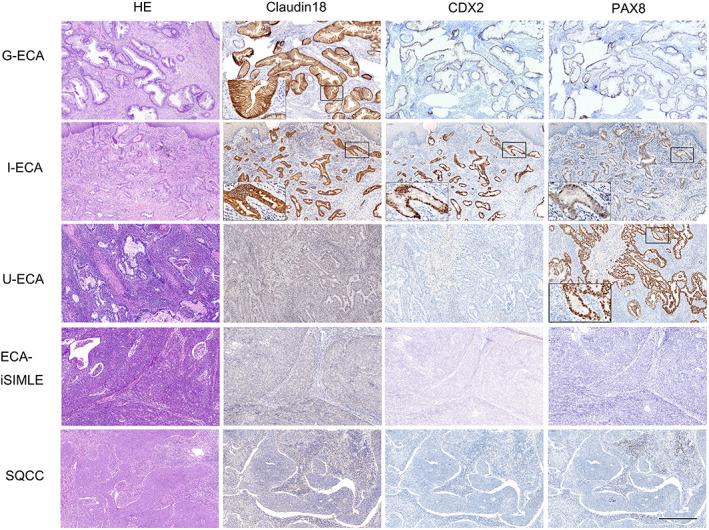
Immunohistochemical expression profiles of CLDN18 (M), CDX2, and PAX8 in G‐ECA, I‐ECA, U‐ECA, ECA‐iSMILE, and SQCC. The tumor cells of the I‐ECA exhibited gastric differentiation characterized by CLDN18 (M)‐positivity. ECA‐iSMILE, invasive stratified mucin‐producing carcinoma; G‐ECA, gastric‐type ECA; I‐ECA, intestinal‐type ECA; SQCC, squamous cell carcinoma (bar = 500 μm); U‐ECA, usual‐type ECA.
3.5. Gastric differentiation in intestinal‐type and usual‐type ECAs
Gastric differentiation characterized by CLDN18 (M) expression was observed in the mucinous epithelium of intestinal‐type and usual‐type ECAs. In a subset of usual‐type ECA cases (13/33, 39.4%), the tumor comprised mucinous epithelium exhibiting an intermediate gastric and usual phenotype and diffuse positive CLDN18 (M) and PAX8 expression. In another subset of intestinal‐type ECA cases (5/9, 55.6%), the tumor comprised mucinous epithelium exhibiting an intermediate gastric and intestinal phenotype and diffuse positive CLDN18 (M) and CDX2 expression. There was no CLDN18 (M) expression in iSMILE ECAs. Representative histological and immunohistochemical features with gastric differentiation in the intestinal subtype are shown in Figure 5.
4. DISCUSSION
In our study, the presence of CLDN18 (M) was documented in HPV‐independent ECA primary cases and intestinal‐type or usual‐type ECA with gastric differentiation. In addition to 10 HPV‐independent ECAs positive for CLDN18 (M) expression, of the 33 usual ECAs, 13 cases (39.4%) showed CLDN18 (M) expression, and CLDN18 (M) expression was present in 5 of 9 I‐ECA cases (55.6%). Similar expression with a gastric differentiation phenotype has been previously reported in a variety of tumors, including pancreatobiliary tumors, 10 , 25 ovarian mucinous tumors, 9 , 12 and endocervical adenocarcinomas. 16 , 17 Moreover, we found a significant association between CLDN18 (M) expression and Silva pattern and stromal invasion level (i.e., cancers with Silva pattern C and deep stromal invasion).
CLDN18 has two isoforms, CLDN18 splice variant 1 (CLDN18.1) and CLDN18 splice variant 1 (CLDN18.2). CLDN18.1 is specific for epithelial cells in normal and cancerous lung tissues, and CLDN18.2 is expressed in normal gastric epithelial cells and in gastric, pancreatic, and esophageal cancer cells. 26 The monoclonal antibody claudiximab (IMAB362, zolbetuximab) targets the membranous CLDN18.2 isoform 27 and induces cancer cell apoptosis and immune effectors by activating antibody‐dependent cellular cytotoxicity (ADCC) and complement‐dependent cytotoxicity (CDC). 28 Currently, several clinical trials have demonstrated improved efficacy among the subgroup of patients with high CLDN18 (M)‐expressing tumors. 27 , 29 CLDN18 is a promising therapeutic target in gastric and gastroesophageal adenocarcinoma due to its high expression on the surface of tumor cells. Our findings suggest that patients with CLDN18 (M)‐positive ECA may be candidates for CLDN18 (M)‐targeted therapy.
An interesting observation in this study was CLDN18 (N) immunoreactivity in ECAs. This detailed report provides the first description of a close association between CLDN18 (N) expression and ECA, independent of CLDN18 (M) expression in these tumors. In this study, CLDN18 (N) expression was found to be associated with Silva pattern A, which was associated with a good prognosis.
Studies have shown that claudins localize to the nucleus. When CLDNs are fused to a strong NLS sequence, the cytoplasmic fraction disappears, and the protein is completely localized in the nucleus. 30 Despite the absence of the NLS sequence, claudins may utilize the PDZ domain or other mechanisms for nuclear transportation. 31 Furthermore, claudin‐23 is highly expressed in the nucleus of pancreatic cancer cells as an intracellular signaling molecule. 32 Recently, Yu Takahashi and colleagues demonstrated that nuclear expression of CLDN18 was independently associated with gastric cancers and could be a novel diagnostic marker for gastric cancer. CLDN18 (N) is a new immunohistochemical early diagnostic marker for well‐differentiated gastric adenocarcinoma. In Irene Coati's study, strong nuclear positivity was observed in 22.5% of primary esophagogastric adenocarcinomas. 19 Similarly, in our study, among primary HPV‐associated ECA samples, 14 (28%) had nuclear positivity, and high nuclear CLDN18 expression was associated with the Silva pattern, with a higher incidence of positive expression in Silva pattern A cancers. These findings may indicate that CLDN18 translocates to the nucleus under certain cancerous conditions and might have a different function from CLDN18 (M).
In conclusion, we found CLDN18 (M) expression in ECAs, particularly in HPV‐independent ECAs, suggesting that CLDN18 may be a novel potential therapeutic target in ECA. Moreover, nuclear staining of CLDN18 is a new immunohistochemical marker for ECAs showing Silva pattern A, which have a good prognosis. Although shuttling of CLDN18 into the nucleus occurs and there is one putative NLS sequence in CLDN18, 13 little is known about the mechanism that activates this event. Although the existing data support that membranous and nuclear CLDNs may serve not only as TJ transmembrane proteins but also as intracellular signaling molecules that interact with transcriptional regulators, further studies are needed to identify the pathophysiological function of intracellular translocation of CLDN18. In this study, we found not only membranous and nuclear localization of CLDN18 expression but also different subclonal patterns in ECA tissues. However, whether CLDN18 subclonal expression patterns are related to CLDN18 genetic alterations has not been explored. Furthermore, studies are needed to investigate the diagnostic, prognostic, or therapeutic usefulness of membranous and nuclear staining of CLDN18 and its subclonal patterns and to develop clinical tests for ECAs.
AUTHOR CONTRIBUTIONS
Xiuzhen Du: Study design, data collection, data analysis, and manuscript writing. Yanjiao Hu: Pathological diagnosis and pathology review. Xiaoyu Ji: Data collection and results analysis. Qingmei Zheng: Research funding support and manuscript proofreading. Lei Sui: Pathological data collection. Kejuan Song: Study conception and clinical data collection. Teng Lv: Clinical data collection. Yulong Chen: Clinical data collection. Han Zhao: Immunohistochemical staining. Shuzhen Dai: Guidance for the study design. Peng Zhao: Pathological diagnosis and pathology review. Qin Yao: Study design and conception, manuscript editing and proofreading, and acquisition of research funding. All authors have read and approved the final manuscript.
FUNDING INFORMATION
The study received financial support from Science and Technology Bureau of Qingdao West Coast New Area (grant number 2019‐55).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
Our study was conducted in accordance with the principles of the Declaration of Helsinki and approved by the Institutional Review Committee of The Affiliated Hospital of Qingdao University. The clinicopathological information was collected from the patients' medical records and pathology reports. All data were anonymized prior to histology and immunohistochemistry.
Du X, Hu Y, Ji X, et al. Membranous and nuclear staining of CLDN18 in HPV‐independent and HPV‐associated endocervical adenocarcinomas. Cancer Med. 2023;12:1441‐1450. doi: 10.1002/cam4.5029
Contributor Information
Peng Zhao, Email: zhaopengciq@126.com.
Qin Yao, Email: dr_yaoqin@qdu.edu.cn.
DATA AVAILABILITY STATEMENT
Information about the data provided in the article is available.
REFERENCES
- 1. Smith HO, Tiffany MF, Qualls CR, Key CR. The rising incidence of adenocarcinoma relative to squamous cell carcinoma of the uterine cervix in the United States—a 24‐year population‐based study. Gynecol Oncol. 2000;78(2):97‐105. [DOI] [PubMed] [Google Scholar]
- 2. Adegoke O, Kulasingam S, Virnig B. Cervical cancer trends in the United States: a 35‐year population‐based analysis. J Womens Health (Larchmt). 2012;21(10):1031‐1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bray F, Carstensen B, Møller H, et al. Incidence trends of adenocarcinoma of the cervix in 13 European countries. Cancer Epidemiol Biomarkers Prev. 2005;14(9):2191‐2199. [DOI] [PubMed] [Google Scholar]
- 4. Stolnicu S, Barsan I, Hoang L, et al. International Endocervical Adenocarcinoma Criteria and Classification (IECC): a new pathogenetic classification for invasive adenocarcinomas of the endocervix. Am J Surg Pathol. 2018;42(2):214‐226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park KJ. Cervical adenocarcinoma: integration of HPV status, pattern of invasion, morphology and molecular markers into classification. Histopathology. 2020;76(1):112‐127. [DOI] [PubMed] [Google Scholar]
- 6. Chen L, Niu Y, Wan X, et al. Clinicopathological features and outcomes in gastric‐type of HPV‐independent endocervical adenocarcinomas. BMC Cancer. 2021;21(1):1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hodgson A, Olkhov‐Mitsel E, Howitt BE, Nucci MR, Parra‐Herran C. International Endocervical Adenocarcinoma Criteria and Classification (IECC): correlation with adverse clinicopathological features and patient outcome. J Clin Pathol. 2019;72(5):347‐353. [DOI] [PubMed] [Google Scholar]
- 8. Song F, Jia M, Yu S, Cao L, Sun PL, Gao H. PD‐L1 expression and immune stromal features in HPV‐independent cervical adenocarcinoma. Histopathology. 2021;79(5):861‐871. [DOI] [PubMed] [Google Scholar]
- 9. Halimi SA, Maeda D, Shinozaki‐Ushiku A, et al. Claudin‐18 overexpression in intestinal‐type mucinous borderline tumour of the ovary. Histopathology. 2013;63(4):534‐544. [DOI] [PubMed] [Google Scholar]
- 10. Li WT, Jeng YM, Yang CY. Claudin‐18 as a marker for identifying the stomach and pancreatobiliary tract as the primary sites of metastatic adenocarcinoma. Am J Surg Pathol. 2020;44(12):1643‐1648. [DOI] [PubMed] [Google Scholar]
- 11. Matsuda M, Sentani K, Noguchi T, et al. Immunohistochemical analysis of colorectal cancer with gastric phenotype: claudin‐18 is associated with poor prognosis. Pathol Int. 2010;60(10):673‐680. [DOI] [PubMed] [Google Scholar]
- 12. Halimi SA, Maeda D, Ushiku‐Shinozaki A, et al. Comprehensive immunohistochemical analysis of the gastrointestinal and Mullerian phenotypes of 139 ovarian mucinous cystadenomas. Hum Pathol. 2021;109:21‐30. [DOI] [PubMed] [Google Scholar]
- 13. Hagen SJ. Non‐canonical functions of claudin proteins: beyond the regulation of cell‐cell adhesions. Tissue Barriers. 2017;5(2):e1327839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi Y, Tsutsumi Y, Takeuchi C, et al. Nuclear staining of claudin‐18 is a new immunohistochemical marker for diagnosing intramucosal well‐differentiated gastric adenocarcinoma. Pathol Int. 2020;70(9):644‐652. [DOI] [PubMed] [Google Scholar]
- 15. Kiyokawa T, Hoang L, Pesci A, et al. Claudin‐18 as a promising surrogate marker for endocervical gastric‐type carcinoma. Am J Surg Pathol. 2021;46:628‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Asaka S, Nakajima T, Ida K, et al. Clinicopathological and prognostic significance of immunophenotypic characterization of endocervical adenocarcinoma using CLDN18, CDH17, and PAX8 in association with HPV status. Virchows Arch. 2021;480:269‐280. [DOI] [PubMed] [Google Scholar]
- 17. Asaka S, Nakajima T, Kugo K, et al. Immunophenotype analysis using CLDN18, CDH17, and PAX8 for the subcategorization of endocervical adenocarcinomas in situ: gastric‐type, intestinal‐type, gastrointestinal‐type, and Mullerian‐type. Virchows Arch. 2020;476(4):499‐510. [DOI] [PubMed] [Google Scholar]
- 18. Pirog EC. Cervical adenocarcinoma: diagnosis of human papillomavirus‐positive and human papillomavirus‐negative tumors. Arch Pathol Lab Med. 2017;141(12):1653‐1667. [DOI] [PubMed] [Google Scholar]
- 19. Coati I, Lotz G, Fanelli GN, et al. Claudin‐18 expression in oesophagogastric adenocarcinomas: a tissue microarray study of 523 molecularly profiled cases. Br J Cancer. 2019;121(3):257‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Espinoza JA, Riquelme I, Sagredo EA, et al. Mucin 5B, carbonic anhydrase 9 and claudin 18 are potential theranostic markers of gallbladder carcinoma. Histopathology. 2019;74(4):597‐607. [DOI] [PubMed] [Google Scholar]
- 21. Kang EY, Cheasley D, LePage C, et al. Refined cut‐off for TP53 immunohistochemistry improves prediction of TP53 mutation status in ovarian mucinous tumors: implications for outcome analyses. Mod Pathol. 2021;34(1):194‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Singh N, Piskorz AM, Bosse T, et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol. 2020;250(3):336‐345. [DOI] [PubMed] [Google Scholar]
- 23. Ejegod DM, Hansen M, Christiansen IK, et al. Clinical validation of the Cobas 4800 HPV assay using cervical samples in SurePath medium under the VALGENT4 framework. J Clin Virol. 2020;128:104336. [DOI] [PubMed] [Google Scholar]
- 24. Roma AA, Diaz de Vivar A, Park KJ, et al. Invasive endocervical adenocarcinoma: a new pattern‐based classification system with important clinical significance. Am J Surg Pathol. 2015;39(5):667‐672. [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi M, Fujinaga Y, Ota H. Reappraisal of the immunophenotype of pancreatic Intraductal Papillary Mucinous Neoplasms (IPMNs)‐gastric pyloric and small intestinal immunophenotype expression in gastric and intestinal type IPMNs. Acta Histochem Cytochem. 2014;47(2):45‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hashimoto I, Oshima T. Claudins and gastric cancer: an overview. Cancers (Basel). 2022;14(2):290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sahin U, Türeci Ö, Manikhas G, et al. FAST: a randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first‐line treatment of advanced CLDN18.2‐positive gastric and gastro‐oesophageal adenocarcinoma. Ann Oncol. 2021;32(5):609‐619. [DOI] [PubMed] [Google Scholar]
- 28. Sahin U, Schuler M, Richly H, et al. A phase I dose‐escalation study of IMAB362 (Zolbetuximab) in patients with advanced gastric and gastro‐oesophageal junction cancer. Eur J Cancer. 2018;100:17‐26. [DOI] [PubMed] [Google Scholar]
- 29. Tureci O, Sahin U, Schulze‐Bergkamen H, et al. A multicentre, phase IIa study of zolbetuximab as a single agent in patients with recurrent or refractory advanced adenocarcinoma of the stomach or lower oesophagus: the MONO study. Ann Oncol. 2019;30(9):1487‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. French AD, Fiori JL, Camilli TC, et al. PKC and PKA phosphorylation affect the subcellular localization of claudin‐1 in melanoma cells. Int J Med Sci. 2009;6(2):93‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikari A, Watanabe R, Sato T, et al. Nuclear distribution of claudin‐2 increases cell proliferation in human lung adenocarcinoma cells. Biochim Biophys Acta. 2014;1843(9):2079‐2088. [DOI] [PubMed] [Google Scholar]
- 32. Wang W, Tan X, Zhou L, Gao F, Dai X. Involvement of the expression and redistribution of claudin‐23 in pancreatic cancer cell dissociation. Mol Med Rep. 2010;3(5):845‐850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Information about the data provided in the article is available.


