Abstract
Ferroptosis is a unique type of cell death that is hallmarked with the imbalanced redox homeostasis as triggered by iron-dependent lipid peroxidation. Cysteines often play critical roles in proteins to help maintain a healthy cellular environment by dynamically switching between their reduced and oxidized forms, however, how the global redox landscape of cysteinome is perturbed upon ferroptosis remains unknown to date. By using a quantitative chemical proteomic strategy, we systematically profiled the dynamic changes of cysteinome in ferroptotic cells and identified a list of candidate sites whose redox states are precisely regulated under ferroptosis-inducing and rescuing conditions. In particular, C106 of the protein/nucleic acid deglycase DJ-1 acts as an intriguing sensor switch for the ferroptotic condition, whose oxidation results in the disruption of its interaction with the 20S proteasome and leads to a marked activation in the proteasome system. Our chemoproteomic profiling and associated functional studies reveal a novel functional link between ferroptosis and the proteasome-mediated protein degradation. It also suggests proteasome as a promising target for developing treatment strategies for ferroptosis-related diseases.
Subject terms: Proteomics, Biochemistry
Introduction
Ferroptosis is a newly discovered iron-dependent cell death, which is closely related to many diseases, such as cancer [1], ischemic necrosis [2] and neurodegenerative diseases [3]. As a major hallmark of ferroptosis, accumulation of lipid hydroperoxides that is presumably coupled with the iron-catalyzed Fenton reaction, results in a catastrophically increased production of reactive oxygen species (ROS) [4], leading to destructive damages to cellular components. To date, several mechanisms related to cellular redox regulation have been explored for ferroptosis, for example, the glutathione (GSH)-glutathione peroxidases (GPX) axis could reduce lipid peroxides and relieve cellular oxidative stress [5]. As a functional cellular lipophilic antioxidant, ubiquinol was found to trap the lipid peroxyl radicals during ferroptosis, and FSP1 and GCH1 contribute to ubiquinol production by reducing coenzyme Q10 [6–8] or synthesizing 6(R)-L-erythro-5,6,7,8-tetrahydrobiopterin(BH4) to promote the formation of ubiquinol [9, 10]. Moreover, multiple proteins that function in the desaturation of lipids or oxidation of the unsaturated fatty acids are also extensively involved in regulation of ferroptosis, including LPCAT3 [11], ACSL4 [12], POR [13] and CYB5R1 [14]. However, how the cell fate of ferroptosis is determined upon the imbalanced shock on the cellular redox system remains elusive [15].
As a “pivot” residue in maintaining redox homeostasis, cysteine possesses a highly active sulfhydryl sidechain group, which could participate in a redox reaction by being self-oxidized in order to protect cellular components from damaging ROS [16]. Increasing evidences have suggested important roles of cysteines involved in ferroptosis. For example, the synthesis of antioxidant metabolites, GSH and CoA, are critically dependent on the availability of cysteines. It has been shown that inhibiting the import of oxidized cysteines through system Xc- could induce ferroptosis in pancreatic ductal adenocarcinoma (PDAC) [17]. It was also found that targeting active-site cysteines of thioredoxins could effectively kill cancer cells that are resistant to chemotherapy [18]. In addition to redox regulation, cysteines can also act as catalytic residues in a diversified array of enzymes including proteases, acyltransferases and dehydrogenases etc. In particular, both E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases have cysteines in their active sites and help degrade oxidized proteins with loss of function via the ubiquitin proteasome system (UPS) when the oxidative stress exceeds the homeostatic level [19]. Thus, investigation of how the landscape of functional cysteinome is perturbed during ferroptosis will be informative to help decipher this complexed cell death pathway.
Quantitative chemoproteomic strategies have been developed to profile reactive and functional cysteines in complexed biological systems [20]. Notably, the isoTOP-ABPP method, which is an advanced derivative of Activity-Based Protein Profiling (ABPP) [21], employs a cysteine-reactive iodoacetamide probe with a bioorthogonal alkyne handle (IAyne) and a pair of isotopically encoded cleavable azide-biotin tags to enable quantification of intrinsic cysteine reactivity as well as sensitivity to modifications by other reactive metabolites in native proteomes [22–24]. Recently, we further enhanced the pipeline to incorporate the reductive dimethylation for triplex quantitation [25] and applied the so-called “rdTOP-ABPP” method to globally profile cysteines that are targeted by (1S,3R)-RSL3, a potent ferroptosis-inducing compound. We also applied rdTOP-ABPP with an aniline-based probe to globally analyze protein carbonylations in ferroptotic cells, which constitute a major class of modifications on cysteines by increasing amounts of electrophilic metabolites derived from peroxidation of membrane lipids [26]. The successful applications of such chemoproteomic strategies to ferroptosis-related systems inspired us to perform a more general functional cysteinome profiling in ferroptotic cells, aiming to discover new cysteine-based sensors or switches that play important regulatory roles in ferroptosis.
In the current study, we applied rdTOP-ABPP to globally quantify the functional states of cysteines in proteomes from both ferroptosis-inducing and ferroptosis-rescuing cells. We found multiple cysteines in enriched pathways of defense of cellular oxidative stress and protein degradation, whose redox status fluctuate along the ferroptotic process. We chose one of these sites in the protein/nucleic acid deglycase (DJ-1) protein, an important regulatory protein in response to oxidative stress, for further functional characterization. The results showed that C106 in DJ-1 is oxidized during ferroptosis, which in turn hinders its interaction with the 20S proteasome and unleash the activity of the protein-degrading machinery. Our studies reveal a novel functional link between ferroptosis and the UPS system and suggest that accelerated protein degradation might facilitate the progress of ferroptosis.
Results
Quantitative profiling of the perturbed cysteinome in ferroptosis
To profile cysteines that are perturbed in ferroptotic cells, we used (1S,3R)-RSL3, a potent GPX4 inhibitor [5], to induce ferroptosis in HT1080 cells (1.0 μM, 6 h). Considering RSL3 can covalently react with cysteines in proteomes, we added a “rescuing” condition where HT1080 cells were treated simultaneously with RSL3 and liproxstatin-1 [27] (600 nM), a ligand that can effectively quench lipid ROS and inhibit ferroptosis. We reason that if a cysteine is perturbed by the increasingly oxidized environment during ferroptosis, it will partially lose the labeling by the IAyne probe, which can be rescued by adding liproxstatin-1. However, RSL3’s alkylation would cause permanent competition on cysteine’s labeling by IAyne. By comparing the extent of IAyne labeling among these three conditions (“normal”, “ferroptotic” and “rescued”), we could specifically distinguish cysteine switches that sense the environmental changes during ferroptosis from those directly targeted by RSL3.
After confirming the overall cysteinome remains largely intact by in-gel fluorescence (Supplementary Fig. 1), we performed rdTOP-ABPP experiments to identify specific cysteine residues that were perturbed during ferroptosis (Fig. 1a). Proteomes from normal, ferroptotic and rescued cells were labeled by the IAyne probe and then conjugated with an acid-cleavable azide-biotin tags by (CuAAC) [28]. After enrichment with streptavidin and on-bead trypsin digestion, the probe-adducted peptides from the normal, ferroptotic and rescued cells were isotopically labeled by light, medium and heavy dimethylation regents, respectively. The combined samples were subjected to acid cleavage, and the released peptides were analyzed by LC-MS/MS and quantified by CIMAGE2.0 [29]. Peptides that were quantified in all three replicates were kept for further analysis. In total, we detected the reactivity of more than 4000 cysteines (Supplementary Table S1) and obtained quantitative information of IAyne’s labeling on 897 cysteines under the three treatment conditions with high confidence (Fig. 1b).
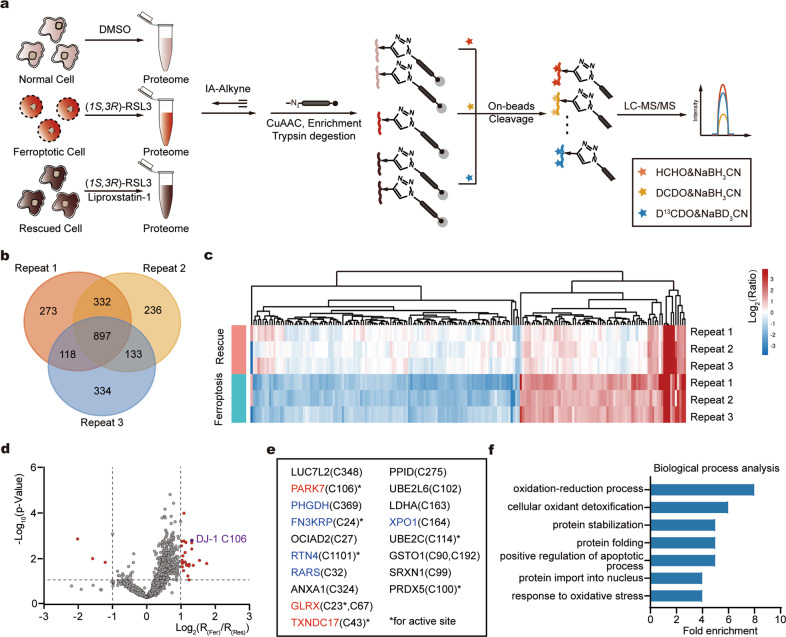
Fig. 1. Quantitative profiling of the perturbed cysteinome in ferroptosis.
a The scheme of quantitative profiling of reactive cysteines in ferroptotic proteomes by rdTOP-ABPP. Equal number of HT1080 cells were treated with DMSO, RSL3 and with or without Liproxstatin-1, respectively, for 6 h. The whole proteomes were labeled with cysteine-reactive IAyne probes, and then subjected to the rdTOP-ABPP procedures. b A venn diagram showing the number of reactive cysteines quantified from three biological replicates. c Hierarchical clustering analysis of perturbed cysteines based on their quantified rdTOP-ABPP ratios. Each row in the heat map represents the data from each individual profiling experiment. d Volcano plot of the rdTOP-ABPP ratios for each cysteine site quantified in the ferroptotic cells as compared to that in the rescued cells. In the upper left and right quadrants are reactive cysteines with significantly changed reactivity during ferroptosis (n = 3 per group, p-value < 0.05, 0.5 <R(Fer)/R(Fer) <2). Highlighted in red are cysteines with significantly suppressed reactivity during ferroptosis. e List of cysteines with suppressed reactivity during ferroptosis. Proteins in red are redox regulated proteins and proteins in blue are carbonylated proteins. * indicates the active-site cysteine in the host proteins. f Gene ontology analysis of the cysteines with suppressed reactivity during ferroptosis in terms of biological processes.
Two ratios were calculated for each cysteine identified from the profiling experiments. The light/medium (“normal/ferroptosis”) ratio R(Fer) reflects how much IAyne’s labeling was lost upon cell ferroptosis induced by RSL3 while the light/heavy (“normal/rescued”) ratio R(Res) indicates whether the loss of IAyne labeling could be reversed by co-treatment with liproxstatin-1. Clustering of these cysteines by their quantitative ratios demonstrated that they formed distinct categories, including those with increased chemical reactivity or with reversible/irreversible inhibition upon ferroptosis (Fig. 1c). We analyzed the statistical difference between R(Fer) and R(Res) by the student’s t-test (p-value) for each cysteine and drew a volcano plot against the ratio of R(Fer)/R(Res) (Fig. 1d). After applying the cutoffs of p-value <= 0.05 and R(Fer)/R(Res) >= 2, we obtained a list of 20 cysteines from 18 proteins whose reduced states are significantly compromised upon ferroptosis and are effectively reversed when cells are rescued by liproxstatin-1 (Fig. 1e).
According to the gene ontology (GO) analysis, these 18 proteins are enriched in biological processes including oxidation/reduction and cellular oxidant detoxification (Fig. 1f). For example, glutaredoxin (GLRX), glutathione S-transferase omega 1 (GSTO1) and sulfiredoxin1 (SRXN1) play important roles in GSH transsulfation and cellular redox regulation (Fig. 1e). Query of the Uniprot database shows that many of these perturbed cysteines are annotated as the active-site residues of their host proteins. Satisfyingly, some of the identified cysteines have been reported to be involved in the ferroptosis regulation, such as peroxiredoxin 5 (PRDX5) [30], thioredoxin (TXNDC17) [18] and lactate dehydrogenase A (LDHA) [31]. In addition, several of them have also been identified to undergo carbonylation during ferroptosis according to our previous study [26], such as D-3-phosphoglycerate dehydrogenase (PHGDH), Ketosamine-3-kinase (FN3KRP), Arginine-tRNA ligase (RARS), Reticulon-4 (RTN4) and Protein Exportin 1 A (XPO1), suggesting that they are subjected to reversible PTMs under ferroptosis.
C106 of DJ-1 plays a key role in regulation of ferroptosis
Among the identified functional cysteine residues, C106 in protein/nucleic acid deglycase (DJ-1) caught our attention, since the protein acts as an important regulator in response to oxidative stress in cells, and its abnormal function is closely related to the pathogenesis of neurodegenerative diseases and cancer [32, 33]. DJ-1 is a multifunctional protein [34], which could protect cells from death induced by toxicity such as oxidants and metals [34, 35]. The protein has previously been linked with ferroptosis through a potential mechanism of transsulfuration in human H1299 cell lines [36]. There are three cysteine residues in DJ-1, two of which, C46 and C106, were quantified by rdTOP-ABPP (Fig. 2a). Based on our profiling data, C106 of DJ-1 was an outstanding target with an averaged R(Fer) of 2.63 and R(Res) of 1.05 (Fig. 2a). In contrast, C46 was quantified with much less perturbation.
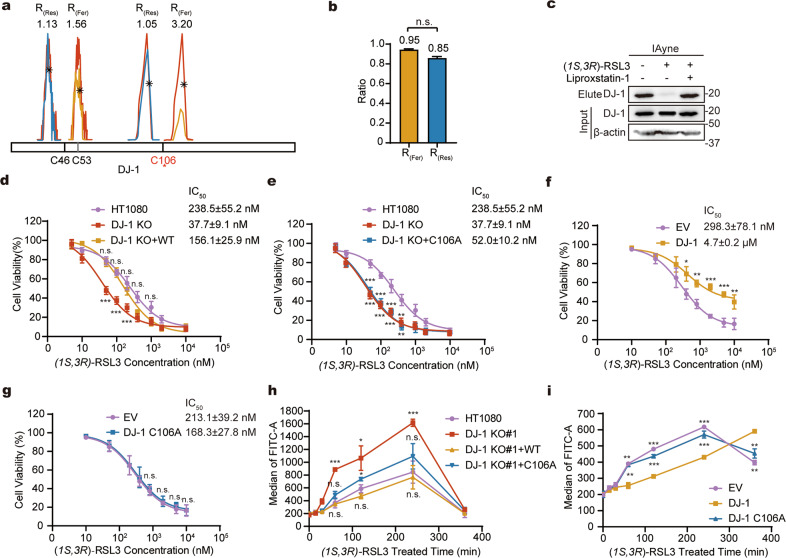
Fig. 2. Cys106 of DJ-1 is involved in regulation of ferroptosis.
a Extracted ion chromatograms (EICs) of the IAyne labeled peptides containing C46 or C106 from DJ-1. The peptide EICs from the normal, ferroptotic and rescued cells are colored in red, yellow and blue, respectively. The quantified ratios are shown above. b Quantified ratios of DJ-1 comparing the protein abundance between the normal and ferroptotic cells or between the normal and rescued cells. c Immunoblotting shows selective labelling of DJ-1 in normal, ferroptotic and rescued cells by IAyne. IAyne failed to pull down DJ-1 from the ferroptotic cells. d, e Knockout of DJ-1 sensitized cells to ferroptosis that can only be reversed by the overexpression of the WT but not mutant DJ-1. Dose-dependent cell death induced by RSL3 were measured by MTT. DJ-1 knockout subclones (DJ-1 KO) were generated from HT1080 cells by CRISPR/Cas9 using corresponding guide RNAs. f, g Stable expression of the WT but not mutant DJ-1 exerts protection of cells from ferroptosis. Dose-dependent cell death induced by RSL3 were measured by MTT. Cells were stably transfected with DJ-1 WT/ C106A expression plasmids or empty vehicle (EV). h Knockout of DJ-1 increased the level of lipid ROS in ferroptotic cells that can only be suppressed by the overexpression of the WT but not the C106A mutant DJ-1. i Stable expression of WT but not the C106A mutant DJ-1 suppresses the level of lipid ROS in the early stage of ferroptosis. In h, i time-course measurement of lipid ROS in RSL3-induced ferroptotic cells were performed with the fluorescent probe C11-BODIPY by flow cytometry using FITC 510 nM. Error bars in b, d–i indicate standard deviations from triplicates. Data was analyzed by student’s t-test (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; n.s., not significant, n = 3).
Interestingly, C106 is annotated as the active-site residue of DJ-1 [37], mutation of which results in the loss of the protein’s enzymatic activity and oxidation sensor function [38]. Based on the chemoproteomic profiling results, we hypothesized that DJ-1 might be functionally involved in regulating ferroptosis via C106. To confirm that the observed variation on R(Fer) is not caused by the reduced protein expression, we performed a whole-proteome analysis of the normal, ferroptotic and rescued cells by quantitative triplex dimethyl labeling and the results showed no significant difference in the abundance of DJ-1 among these three conditions (Fig. 2b and Supplementary Table S2). Consistent with both the rdTOP-ABPP and whole-proteome analysis, the IAyne probe could only label and enrich DJ-1 from the lysates of the normal and rescued but not ferroptotic cells (Fig. 2c), confirming that the chemical reactivity of C106 is compromised in ferroptosis.
We next evaluated how the expression of different forms of DJ-1 affects the sensitivity of HT1080 cells to ferroptosis. Knockout of DJ-1 by CRISPR-Cas9 could effectively sensitize HT1080 cells to RSL3-induced ferroptosis (Fig. 2d and Supplementary Fig. 2a–c). Interestingly, such extra sensitivity to ferroptosis could be reversed by exogenous expression of only the wild-type (WT) DJ-1 but not the C106A mutant (Fig. 2d, e and Supplementary Fig. 2a–d). Consistent with these results, stable expression of DJ-1 WT but not C106A showed a profound protection for HT1080 cells against ferroptosis (Fig. 2f, g and Supplementary Fig. 3). Similar results were observed when ferroptosis were induced by Erastin [39] or ML162 [40] (Supplementary Figs. 4 and 5). To further confirm the observed protective effect is specific to ferroptosis, we treated HT1080 cells with six reagents that are commonly used to induce different types of cell death including staurosporine, rapamycin, etoposide, hydrogen peroxide, methylglyoxal and vinblastine sulfate. Stable expression of DJ-1 could not perturb the cell death induced by any of these reagents except etoposide (Supplementary Fig. 6). Even for etoposide, the protective effect is not attributed to its C106 as stable expression of DJ-1 WT and C106A have similar effects. These results collectively suggested that C106 plays a critical role in DJ-1 to protect cells from ferroptosis, which seems to be quite specific to this unique type of cell death.
Previous studies have shown that knocking out DJ-1 accelerates the production of phospholipid peroxidation in H1299 cells treated with Erastin [36]. We, therefore, proceeded to assess the level of lipid peroxidation in RSL3-treated HT1080 cells with DJ-1 knockout or overexpression at different time points. Consistent with the aforementioned study, knockout of DJ-1 induced a profound increase in lipid ROS in cells, peaking at 4 h after RSL3 treatment, which could be better suppressed by overexpressing DJ-1 WT than C106A (Fig. 2h, Supplementary Fig. 7 and 8). Similarly, overexpression of DJ-1 WT in normal HT1080 cells effectively suppressed the elevated level of lipid ROS in the early stage of RSL3-induced ferroptosis while the effect was hardly observed in cells overexpressing the C106A mutant (Fig. 2i). Collectively, these data suggested that C106 plays an important role in DJ-1 to reduce the level of lipid ROS and inhibit ferroptosis induced by RSL3.
C106 of DJ-1 regulates ferroptosis by altering the interaction with proteasome
In addition to serving as the active-site residue of DJ-1, it has been reported that C106 interacts with the 20S proteasome [41, 42]. Given that protein stabilization and protein folding are also functionally enriched in our cysteinome profiling, we decided to test whether the interaction between DJ-1 and the 20S proteasome would be affected in ferroptosis. We pulled down the interacting proteins of endogenous DJ-1 via immunoprecipitation. Immunoblotting of the three representative proteasome subunits (PSMB1 and PSMA4 in the 20S proteasome, and PSMD1 in the 19S proteasome) showed that their interactions with DJ-1 were significantly disrupted under the ferroptosis-inducing condition and completely restored by addition of the Liproxstatin-1 (Fig. 3a–c). We next overexpressed DJ-1 WT with an N-terminal FLAG tag in HT1080 cells and immunoprecipitation of FLAG-DJ-1 recapitulated similar results as that of endogenous DJ-1 (Fig. 3d–f and Supplementary Fig. 9). Interestingly, DJ-1 C106A showed significantly reduced interaction with PSMB1 and PSMA4 (but not PSMD1) at the basal condition, which did not get further reduced upon ferroptosis (Fig. 3g–i and Supplementary Fig. 10). These results collectively suggested that C106 is one of the major sites contributing to the interaction between DJ-1 and the 20S proteasome and there might be another C106-independent mechanism to regulate the DJ-1’s interaction with the 19S proteasome.
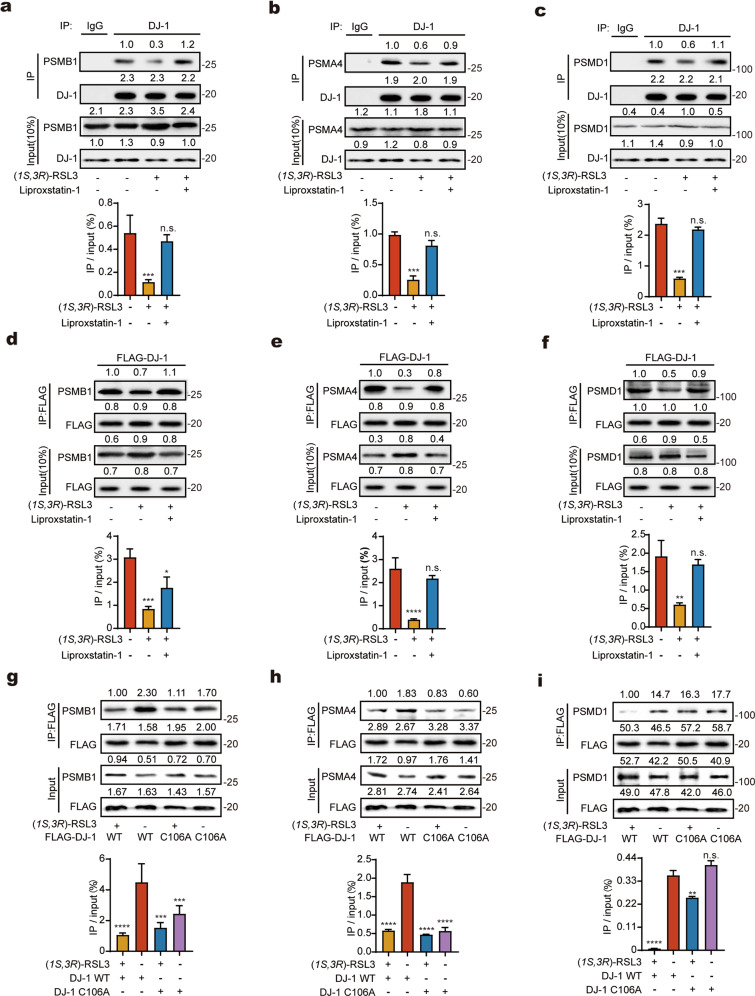
Fig. 3. C106 of DJ-1 regulates the interaction with proteasome during ferroptosis.
a–c Interaction between the endogenous DJ-1 with proteasome is abolished upon ferroptosis and is restored upon rescue. Whole-cell lysates were immunoprecipitated (IP) with the control IgG or anti-DJ-1 antibodies, and the precipitated proteins were detected by anti-PSMA4, anti-PSMB1 and anti-PSMD1 antibodies, respectively. d–f Interaction between the overexpressed DJ-1 and proteasome is disrupted upon ferroptosis and is restored upon rescue. g–i Mutation of C106A in DJ-1 abolishes its interaction with PSMB1 and PSMA4 but not with PSMD1. The whole-cell lysates in d–i were immunoprecipitated with anti-FLAG and precipitated proteins were detected by anti-PSMA4, anti-PSMB1 and anti-PSMD1. In a–i intensities of immunoblotting signals were quantified by ImageJ (NIH), and the quantitative ratios were calculated by dividing the intensity from the IP lane by that of the input lane. Relative band intensities are shown above each lane. Data was analyzed by student’s t-test. Error bars indicate standard deviations from triplicates. (**p < 0.01; ***p < 0.001; ****p < 0.0001; n.s. not significant, n = 3).
Releasing of 20S proteasome from DJ-1 accelerates the proteolytic process in ferroptosis
We next tested if the activity of the 20S proteasome was affected during ferroptosis. By using a fluorescently labeled substrate peptide Suc-LLVT-AMC, we measured that the chymotrypsin-like activity of proteasome was significantly increased after the RSL3-induced ferroptosis and was recovered by co-treatment with Liproxstatin-1 (Fig. 4a). Such an activity spike of proteasome induced by RSL3 could be suppressed by stable expression of the DJ-1 WT but not C106A (Fig. 4a), suggesting that the functional C106 acts as a regulatory switch for the proteasome activity during ferroptosis. Consistently, knockout of DJ-1 resulted in a similar activity spike of the proteasome, which could be reverted by exogenous expression of only DJ-1 WT but not C106A (Fig. 4b and Supplementary Fig. 11). Similar effects were observed no matter the cells were treated with ferroptosis inducers or not (Fig. 4b and Supplementary Figs. 11, 12). These results suggest that DJ-1 is an important regulator of proteasome activity during ferroptosis with its C106 serving as a critical functional switch.
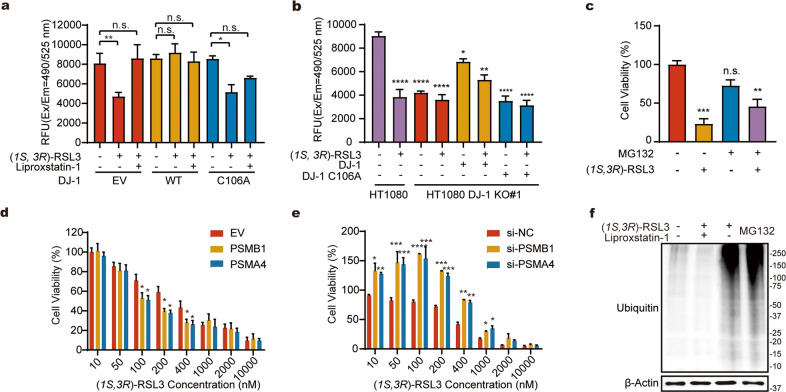
Fig. 4. Release of 20S proteasome from DJ-1 accelerates the proteolytic process in ferroptosis.
a The activity of proteasome is increased in ferroptosis that can be reversed by stable expression of WT but not the C106A DJ-1. b Knockout of DJ-1 increased the activity of proteasome that can be reversed by the overexpression of WT but not the C106A mutant DJ-1. In a, b A proteasome activity assay kit (AAT Bioquest) was used to evaluate proteasome activity in the RSL3-induced ferroptotic cells or liproxstatin-1-induced rescued cells. c Co-treatment of cells with RSL3 (300 nM) and MG132 (200 nM) partially rescue the RSL3-induced ferroptosis as measured by MTT. d Overexpression of PSMB1 and PSMA4 in HT1080 cells can sensitize cells to ferroptosis. e Knockdown of PSMB1 and PSMA4 exerts protection of HT1080 cells from ferroptosis. In d, e Dose dependent cell death induced by RSL3 was measured by MTT. In a–e Data was analyzed by student’s t-test. Error bars indicate standard deviations from triplicates (**p < 0.01, ***p < 0.001, ****p < 0.0001; n.s. not significant, n = 3). f Overall level of ubiquitination in proteomes is increased in cells treated with RSL3 treatment as detected by Anti-Ub antibody. MG132 is included as a positive control. EV Empty vehicle, siNC negative control siRNA, RFU Relative Fluorescence Units.
To further test the effect of 20S proteasome on ferroptosis, we co-treated cells with RSL3 and MG132, the latter of which is a well-established inhibitor of the 20S proteasome known to arrest cell cycles and lead to chronic cell death [43]. The results showed that co-treatment with MG132 (300 nM) was able to partially but significantly rescue the ferroptotic death induced by RSL3 alone (Fig. 4c) and the counteractive effect between RSL3 and MG132 was also evidenced by the hump in the titration curve of each compound when the other one was co-treated (Supplementary Fig. 13a, b). Considering the intrinsic cytotoxicity of MG132, we next proceeded to assess the cell viability in RSL3-treated HT1080 cells with overexpression or knockdown of PSMA4 and PSMB1. While we found that overexpression of either subunit could slightly sensitize cells to ferroptosis (Fig. 4d and Supplementary Fig. 14a), knockdown PSMA4 or PSMB1 could significantly protect cells from ferroptosis as dose-dependently induced by RLS3 (Fig. 4e, Supplementary Fig. 13c and Supplementary Fig. 14b, c).
We further investigate whether the active proteasome would lead to altered levels of ubiquitination in ferroptotic proteome. RSL3-induced ferroptosis resulted in higher ubiquitination levels which could be reversed by co-treatment with Liproxstatin-1 (Fig. 4f). Collectively, these data suggested that after release from DJ-1, the 20S proteasome is activated during the process of ferroptosis with the increase of global ubiquitination and inhibition of the 20S proteasome genetically or pharmacologically could slow down the ferroptosis.
Oxidation of C106 disrupts DJ-1’s interaction with the 20S proteasome
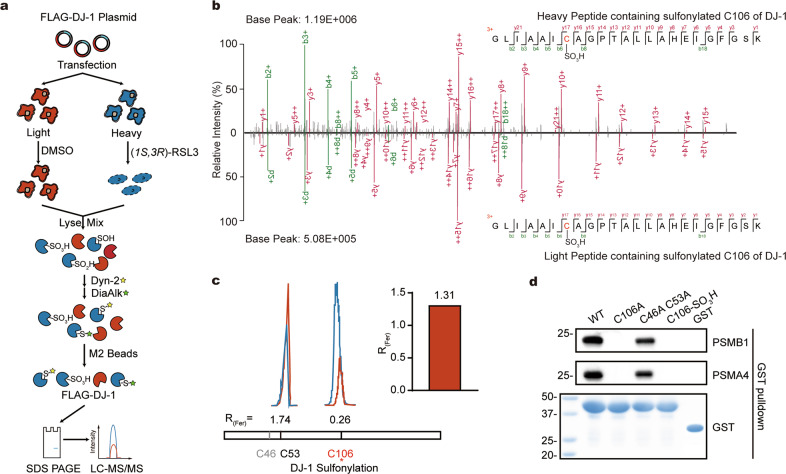
C106 of DJ-1 has been reported to undergo oxidations under several conditions related to oxidative stress [44]. We hypothesize that such modifications may also happen during ferroptosis and abrogate its interaction with proteasome. To confirm the oxidation on C106 of DJ-1, we overexpressed the DJ-1 protein with an N-terminal Flag-tag in HT1080 cells and after treatment of the cells with RSL3, DJ-1 was immunoprecipitated, in-gel digested by trypsin and analyzed by LC-MS/MS (Supplementary Fig. 15a). By setting several common types of oxidation and carbonylation (Supplementary Fig. 15b) as the variable modifications during the database search, we detected sulfonylation as the dominant modification (Supplementary Fig. 15c) with the corresponding spectral count far more than the rest of modifications (Supplementary Fig. 15d). To quantify the level of sulfonylation on C106 of DJ-1, we cultured HT1080 cells in SILAC media, transiently transfected DJ-1 overexpression plasmids and treated the light and heavy cells with DMSO and RLS3, respectively (Fig. 5a). To further rule out the possibility that the observed sulfonylation on C106 was due to the over-oxidation of existing sulfenylation or sulfinylation during the sample preparation, we used DYn-2 [45] and DiaAlk [46], two probes that are known to react with the sulfenylation and sulfinylation, respectively, to block these two modifications in advance (Fig. 5a). Consistently, the sulfonylation on C106 of DJ-1 could be confirmed by the MS/MS spectra in both light and heavy cells (Fig. 5b) and the SILAC heavy/light (“normal/ferroptosis”) ratio of the peptide containing the sulfonylated C106 was calculated to be 0.26, corresponding to an about 4-fold increase in the level of sulfonylation on this specific cysteine upon cell ferroptosis (Fig. 5c). In contrast, the sulfonylation level on C53 was only slightly decreased (Fig. 5c) after the RSL3 treatment, suggesting that the oxidation event occur specifically to C106.
Fig. 5. Oxidation of C106 disrupts DJ-1’s interaction with the 20S proteasome.
a The scheme of identifying the modifications on DJ-1 by SILAC-based quantitative proteomics. Equal number of SILAC light (red) and heavy(blue) HT1080 cells were treated with DMSO or RSL3, respectively, for 6 h. The whole proteomes were labeled with the sulfenylation probe, Dyn-2 and the sulfinylation probe, DiaAlk. DJ-1 were immunoprecipitated by the anti-DJ-1 antibody, in-gel digested by trypsin and quantified by LC-MS/MS. b Representative MS/MS spectra supporting the sulfonylation on C106 of DJ-1. The spectrum obtained from the heavy peptide in ferroptotic cells (top) is head-to-head compared with that obtained from the light peptide in normal cells (bottom). c Extracted ion chromatograms (EICs) of DJ-1’s peptides containing sulfonylated C53 or C106. The peptide EICs from the normal and ferroptotic cells are colored in red and blue, respectively. The quantified SILAC ratios are shown above. The red barplot shows the ratio of DJ-1 expression in HT1080 cells with DMSO vs RSL3 treatment. d Mutation or oxidation of C106 in DJ-1 abolishes its interaction of with proteasome. WT or mutant DJ-1 proteins were expressed with GST tags and immunoprecipitated PSMB1 and PSMA4 after GST pulldown were detected by corresponding antibodies.
We next tested whether the sulfonylation of C106 causes the dissociation of DJ-1 and the 20S proteasome. We recombinantly purified the wild-type DJ-1 and two mutants, C46A-C53A and C106A, with GST tags and investigated how they interacted with the 20 S proteasome via the GST-pulldown assay. Both the wild-type and the C46A-C53A double mutant could interact with the immunoprecipitated PSMA4 and PSMB1 while the C106A mutant could not pull down these two 20 S proteasome subunits at all (Fig. 5d), suggesting that C106 plays a critical role in mediating DJ-1’s interaction with the 20S proteasome. We then oxidized the C46A-C53A double mutant with hydrogen peroxide to produce a DJ-1 proteoform whose C106 was oxidized. As expected, the oxidation of C106 completely disrupted the interaction between DJ-1 and the 20 S proteasome subunits (Fig. 5d). Collectively, these results suggested a model that C106 of DJ-1 gets sulfonylated under the peroxidative environment of ferroptosis and the oxidation on C106 leads to the dissociation of DJ-1 and the 20S proteasome, which further upregulates the activity of proteasome. Our work established a link between ferroptosis and the UPS system with the functional cysteine 106 of DJ-1 as a critical regulator.
Discussion
The functional relevance of cysteine in ferroptosis has been implicated previously in several studies [17, 47, 48]. However, which cysteine mediators are important to regulate cellular ferroptosis remains unknown. In this study, we performed a chemoproteomic profiling of functional cysteines in proteomes during ferroptosis and identified C106 of DJ-1 as a redox sensor to suppress ferroptosis. The proteolytic function of the 20S proteasome was regulated by C106 of DJ-1 and the activity of proteasome in turn affects the process of ferroptosis (Supplementary Fig. 16). Our findings demonstrate a novel function of C106 of DJ-1 in mediating the protein degradation system in response to ferroptosis.
Ferroptosis has been recognized as an oxidative cell death with imbalance of cellular redox homeostasis in response to the accumulation of lipid peroxides and iron-dependent Fenton reaction [49, 50]. Since the thiol groups of cysteine residues are with tunable chemical reactivity and often function in redox sensing, a global survey of changes in reactive cysteinome upon ferroptosis is warranted. Here, we employed a triplex rdTOP-ABPP strategy to quantify the cysteinome changes between the normal state, the ferroptosis induction state and the ferroptosis rescuing state. Recent developments in chemical proteomics technologies with higher throughputs and multiplexity have been reported such as SLC-ABPP [51] and a more comprehensive profiling with the enhanced temporal resolution can be generated in future to provide more dynamic information on how cysteinome is perturbed along the course of ferroptosis. Such profiling technologies can also be readily applied to other types of cell death (e.g., apoptosis, pyroptosis, autophagy etc.) and relevant biological systems so that novel functional residues such as cysteine, lysine [52, 53] and tyrosine [54] can be discovered.
Our profiling identified that the reactivity of C106 of DJ-1 was significantly inhibited in ferroptotic cells. C106 is a well-known nucleophilic cysteine for DJ-1 which acts as oxidative stress sensor to protect cells from various oxidative environments (e.g., maintenance of mitochondrial ROS level [55], peroxide-induced cell death [56], metal toxicity [35]). Our data showed that loss of DJ-1 dramatically increased the level of lipid peroxidation as compared to that of normal cells. In addition, overexpression of DJ-1 WT seemed to “over-suppress” the level of lipid peroxidation to an even lower state than the normal cells. Interestingly, overexpression of C106A also showed partial compensation effect, which suggested that DJ-1 might also be able to reduce lipid peroxides through other pathways in a C106-independent manner.
More recently, DJ-1 was reported to suppress the ferroptosis by reserving the activity of S-adenosyl homocysteine hydrolase (SAHH) and it was demonstrated that DJ-1 could reduce Erastin-induced ferroptotic events in human H1299 cells [36]. Although the connection between DJ-1 and ferroptotic responses have been postulated, the specific regulatory residue remains to be found in the aforementioned study. Consistent with their results, we also found DJ-1 could suppress ferroptosis in RSL3-treated HT1080 cells. However, overexpression of DJ-1 WT or C106A could equally reduce the interaction between SAHH and ACHYL1 (Supplementary Fig. 17), suggesting these two mechanisms linking DJ-1 to ferroptosis might operate independently from each other. In addition to the reported interaction of DJ-1 with the 20S proteasome [41] and its intrinsic redox-sensitive protease activity [57], our data revealed a novel mechanism in which the oxidation of C106 of DJ-1 establishes a unique link between the 20S proteasome and ferroptosis. The discovery of such a mechanistic link of DJ-1 suggests the protein itself may serve as a promising target for developing a tailored therapy for the ferroptosis-related diseases.
MG132 is a pan-inhibitor of chymotrypsin activity for proteasome, which has been known to effectively affect the function of proteasome in cells. In 2021, Dixon and colleagues used the STACK profiling strategy to study the relationship between various small molecules and ferroptotic inducers from which they found that proteasome inhibitors could temporarily rescue the ferroptosis [58]. In our experiment, we not only verified this phenomenon, but also found that protein ubiquitination was significantly increased (Fig. 4f). These results suggest that in ferroptotic cells, upregulation of ubiquitination and proteasome activity are in the interests of accelerating the degradation of abnormal proteins. However, it should be noted that our titration curve also exhibited a hump at certain combinations of RSL3 and MG132 treatment. While inhibition of the proteasome activity by MG132 could rescue ferroptosis at higher RSL3 concentrations, it seemed to also sensitize cells to ferroptosis under the condition with lower RSL3 concentrations. The latter result is more similar to a recent study showing that suppressing the activity of proteasome by Cfz could accelerate the process of Fe-induced death of cancer cells [59]. These data together suggested a rather complexed relationship between the UPS pathway and ferroptosis, which warrants further investigation in future.
The cellular degradation system is responsible for removal of damaged and dysfunctional macromolecules to maintain the homeostasis. Recent studies have revealed that ubiquitination is involved in regulating protein stability and metabolic responses on key mediators of ferroptosis, including SLC7A11, GPX4 and Nrf2 [60]. Another evidence indicated that over-activation of selective autophagy, such as ferritinophagy and lipophagy, drive the development of ferroptosis by promoting the release of oxidative metabolites [61]. These studies suggested that both the degradation machinery and the redox systems are elicited in the ferroptotic response. In the current study, we established a link between the redox sensing system and the protein degradation machinery in ferroptosis by identifying C106 of DJ-1 as the functional switch. Upon its oxidation, DJ-1 released its interaction with the 20S proteasome and caused the activation of the protein degradation pathway. It will be of great interest to explore in future if any FDA-proved drugs that target the proteasome complex could be used to therapeutically intervene ferroptosis-related diseases.
Materials and methods
Cell lines and culture conditions
HT1080 cells were obtained from ATCC and cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco). Complete medium is supplemented with 10% (vol/vol) fetal bovine serum (FBS, PAN), and 1% (vol/vol) penicillin-streptomycin (P/S, Gibco). All cell lines were cultured in a humidified atmosphere at 37 °C with 5% CO2.
Antibodies and reagents
The antibodies used for immunoblotting were rabbit polyclonal anti-DJ-1 antibody (cat. no. 11681-1-AP, Proteintech), rabbit polyclonal anti-PSMB1 antibody (cat. no. 11943-2-AP, Proteintech) rabbit polyclonal anti-PSMA4 antibody (cat. no. 11681-1-AP, Proteintech), rabbit polyclonal anti-PSMD1 antibody (ab140682, Abcam), rabbit polyclonal anti-ACHYL1 antibody (cat. no. 10658-3-AP, Proteintech), mouse monoclonal anti-SAHH antibody (cat. no. 66019-1-Ig, Proteintech), HRP-conjugated FLAG tag antibody (cat. no. HRP-66008, Proteintech) and mouse monoclonal anti-beta-actin antibody (cat. no. 01002, Proteintech). Acid-cleavable azide-biotin tags (DADPS Biotin Azide, cat. no. 1330-5) and Rhodamine azide (AZ109) were purchased from Click Chemistry Tools. (1 S,3 R)-RSL3 (cat. no. S8155), liproxstatin-1(cat. no. S7699) MG132 (cat. no. S2619), rapamycin (cat. no. S1039), etoposide (cat. no. S1225), staurosporine (cat. no. S1421), liproxstatin-1(cat. no. S7699), puromycin (cat. no. S7417) and vinblastine sulfate (cat. no. S4505) were purchased from Selleck. Methylglyoxal solution (cat. no. M0252), anti-FLAG M2 affinity agarose gel (cat. no. A2220) were purchased from Sigma. C11-BODIPY (cat. no. D3581), Lipofectamine™ 2000 Transfection Reagent (cat. no. 11668030) and Lipofectamine™ RNAiMAX Transfection Reagent (cat. no. 13778030) were purchased from Invitrogen.
Plasmid construction
PARK7, PSMB1, PSMA4, PSMD1 genes were amplified from cDNAs by PCR, and subcloned into the pcDNA3 expression vector as entry clones. The FLAG tag was constructed at the N-terminal. For purification of DJ-1 protein, GST-tagged DJ-1was cloned into pQlinkGx vector. All site-specific mutants were generated using the PCR mutagenesis method and verified by sequencing. Primer sequences are designed as following:
DJ-1-F: ATGGCTTCCAAAAGAGCTCTGG
DJ-1-R: CGTCTTTAAGAACAAGTGGAGCCT
FLAG-DJ-1-kpn1-F: GGGGTACCGCCGCCATGGATTACAAGGATGACGACGATAAGGCTTCCAAAAGAGCTCTGG
HA-DJ-1-xba1-R: CTAGTCTAGACTAAGCGTAGTCTGGGACGTCGTATGGGTAGTCTTTAAGAACAAGTGGAG
DJ-1-C106A-F: TGATAGCCGCCATCGCTGCAGGTCCT
DJ-1-C106A-R: GCGATGGCGGCTATCAGGCCCTTCC
DJ-1-C46A-F: TACAGGCTAGCCGTGATTAGCCGTGATGTGGTCATTTGT
DJ-1-C46A-R: ATCACGGCTAGCCTGTACTGGGTCTGTACTGGGTCTTTTCCAGCC
DJ-1-C53A-F: TGGTCATTGCTCCTGATGCTCCTGATGCCAGCCTTGAAG
DJ-1-C53A-R: CATCAGGAGCAATGACCACAATGACCACATCACGGCTACACT
DJ-1-C46-C53-DOUBLE-F: AGCCGTGATGTGGTCATTGCTCCTGATGCCAGCCTTGAAG
DJ-1-C46-C53-DOUBLE-R: ATGACCACATCACGGCTAGCCTGTACTGGGTCTTTTCCAGCC
PSMB1-F: ATGTTGTCCTCTACAGCCATGT
PSMB1-R: TCAGTCCTTCCTTAAGGAAACAGT
PSMA4-F: ATGTCTCGAAGATATGACTCCAGG
PSMA4-R: CTCTGATTCTATTTATCCTTTTCTTTCTG
PSMD1-F: CTATTTATCCTTTTCTTTCTGTTCTTTTTC
PSMD1-R: TTAATCATCAATATACTCAAATGGTTCTG
FLAG-PSMB1-kpn1-F: GGGGTACCGCCGCCATGGATTACAAGGATGACGACGATAAGTTGTCCTCTACAGCCATGTATTC
HA-PSMB1-xbal-R: CTAGTCTAGATCATACCCATACGATGTTCCAGATTACGCTTAGGTCCTTCCTTAAGGAAACAGTTTC
FLAG-PSMA4-kpn1-F: GGGGTACCGCCGCCATGGATTACAAGGATGACGACGATAAGTCTCGAAGATATGACTCCAGG
HA-PSMA4-xba1-R: CTAGTCTAGATCATACCCATACGATGTTCCAGATTACGCTTAGTTTATCCTTTTCTTTCTGTTCTTTTTC
FLAG-PSMD1-kpn1-F: GGGGTACCGCCGCCATGGATTACAAGGATGACGACGATAAGATCACCTCGGCCGCTGGAAT
HA-PSMD1-xbal1-R: CTAGTCTAGATCATACCCATACGATGTTCCAGATTACGCTTAGATCATCAATATACTCAAATGGTTCTG
Construction of HT1080 DJ-1 KO cell lines
To generate HT1080 DJ-1 knockout (KO) cell lines, sgRNA sequences were ligated into LentiCRISPRv2 plasmid and then co-transfected with viral packaging plasmids (psPAX2 and pVSVg) into HEK293T cells. The cells were replaced with fresh medium at time 6 h. HT1080 cells were plated in 6 cm dishes. Two days post transfection, viral supernatant was filtered through 0.45 mm strainer. HT1080 cells were infected by viral supernatant and selected by 1 mg/mL puromycin for 2 weeks. The single positive population was sorted by FACS and clonally expanded. Individual cell clones were screened for DJ-1 deletion using standard PCR analysis. Loss of DJ-1 protein expression was confirmed by immunoblotting. LenitiCRISPRv2 plasmid, psPAX2 and pVSVg-based plasmid were generously gifted by Dr. Wensheng Wei at Peking University.
The sgRNA sequences targeting DJ-1 were designed by CRISPR designer at http://crispr.mit.edu/. The guide sequences targeting Exon 2 and Exon 3 of human PARK7 are shown.
DJ-1#1: 50-AGTACAGTGTAGCCGTGATG-30
DJ-1#2: 50-AGATGTCATGAGGCGAGCTG-30
For DJ-1-rescued cells, pcDNA3-DJ-1 or pcDNA3-DJ-1 C106A were transfected in DJ-1 KO cells. Cells were harvested after 48 h of transfection.
In-gel fluorescence analysis
HT1080 cells treated with DMSO, (1 S,3 R)-RSL3 (1 μM) and/or Liproxstatin-1(600 nM) for 6 h were harvested, suspended in PBS with 0.1% Triton X-100 (Sigma-Aldrich) and sonicated on ice. The soluble lysates were collected after ultracentrifugation at 20,000 g for 30 mins at 4 °C. Protein concentration of soluble lysates were detected by using the BCA protein assay Kit (Pierce) on a microplate reader (Bio-Rad) and adjusted to 2 mg/mL. The lysate (200 μL per aliquot) was labeled with the IAyne probe (100 μM) for 1 h at 25 °C. The probe-labeled lysates were reacted with 300 μM rhodamine-N3, 1 mM Tris(2-carboxyethyl) phosphine (TCEP, Sigma-Aldrich), 100 μM Tris- (benzyltriazolylmethyl) amine (TBTA) (Sigma-Aldrich), and 1 mM CuSO4 (Sigma-Aldrich) at room temperature with constant shaking on a thermo mixer. After reaction for 1 h, samples were mixed with 5 × SDS-PAGE loading buffer at 95 °C for 5 min and were separated by 10% SDS-PAGE. The image of rhodamine fluorescence was acquired on a ChemiDoc MP (Bio-rad) imaging system. The gels were then stained by Coomassie staining (CBB) as loading control.
Quantitative profiling of perturbed cysteinome in ferroptosis
For quantifying functional states of cysteines in proteomes from normal, ferroptosis-inducing and ferroptosis-rescuing cells, 107 HT1080 cells per well were seeded in 15 cm dishes to culture overnight and were treated with DMSO or 1 μM (1 S,3 R)-RSL3 or/and 600 nM Liproxstatin-1 for 6 h in serum-free media (DMEM). The cells were collected, washed by PBS for three times and centrifugated at 3000 rpm for 4 min. The cells were lysed in 700 μL of PBS with 0.1% Triton X-100 (Sigma-Aldrich) containing EDTA-free Pierce HaltTM protease inhibitor cocktail. After sonication on ice, the cell lysates were centrifugated for 20000 g, 30 min at 4 °C to remove the sediment. Protein concentration was detected by using the BCA protein assay Kit and adjusted to 2 mg/mL. 1 mL cell lysates were then reacted with 1 mM CuSO4, 100 μM TBTA ligand, 100 μM acid-cleavable azide-biotin tag and 1 mM TCEP for 1 h on thermo mixer at room temperature. The products of click-labeled lysates were centrifuged for 8000 g, 5 min at 4 °C and washed three times with 1 mL cold methanol.
The proteomes were resuspended in 1 mL PBS with 1.2% SDS. The samples were sonicated and heated at 90 °C for 10 min to dissolve the proteins, followed by centrifuging at 19000 g, 2 min at room temperature to remove the remaining cupric ion. The solution was diluted five times with 5 mL of PBS containing 100 μL of prewashed streptavidin Beads (Thermo Fisher Scientific) and incubated for 3 h at 29 °C on a rotator. After washing in 5 mL PBS and 5 ml distilled water for three times orderly, the resulting beads were resuspended in 500 μL PBS with 6 M urea and 10 mM DTT at 37 °C for 30 min and alkylated by addition of 20 mM iodoacetamide at 35 °C for 30 min in the dark. The beads were collected by centrifugation (2000 g, 3 min) and resuspended in 200 μL of 100 mM triethylammonium bicarbonate (TEAB) buffer (Sigma) with 2 M urea, 1 mM CaCl2 and 10 ng/μL trypsin (Promega). Trypsin digestion was performed at 37 °C in a thermomixer overnight.
The beads were collected by centrifugation and washed with 1 mL PBS, 1 mL H2O and 100 mM TEAB buffer for twice orderly, followed by suspending in 100 μL TEAB buffer. The peptides were reacted with 8 μL of 4% D13CDO (Sigma) for samples from ferroptosis-rescuing cells, DCDO (Sigma) for samples from ferroptosis-inducing cells, or HCHO (Sigma) for samples from normal cells, respectively. 8 μL of 0.6 M NaBD3CN was added to the samples of normal and ferroptosis-inducing cells and 8 μL of 0.6 M NaBD3CN was added to the samples of ferroptosis-rescuing cells. The reaction was incubated for 1 h at room temperature and the beads were washed with 1 mL H2O for twice. The labeled samples were combined and subjected for on-beads cleavage by using 200 μL of 2% formic acid solution (sigma) for twice. The eluant containing the peptides was collected for LC-MS/MS analysis.
Preparation of SILAC cells
The HT1080 cells were passaged 8 times in SILAC DMEM (Thermo Fisher Scientific) with 10% SILAC FBS (Thermo Fisher Scientific), 1% penicillin-streptomycin (Thermo Fisher Scientific), 100 μg/mL of regular L-arginine-HCl and L-lysine-HCl (Sigma-Aldrich) or [13C6,15N4]L-arginine-HCl and [13C6, 15N2]L-lysine-HCl (Cambridge Isotope Laboratory) were added to make the light or heavy media, respectively. 20 µg of lysates of light and heavy cells were mixed equally and lysated in 6 M urea/PBS. After reduction with 1 mM DTT at 37 °C for 30 mins and alkylation with 2 mM iodoacetamide at 35 °C for 30 mins in dark, the sample was diluted with PBS to 2 M urea/PBS and subjected to trypsin digestion at 37 °C in a thermomixer overnight (2 μg of trypsin with 1 mM of calcium chloride). The digested peptide sample was acidified with 0.1% formic acid and analyzed by LC-MS/MS. The distribution of SILAC ratios was checked to be centered around 1.0.
Identifying modifications on DJ-1 by SILAC-based quantitative proteomics
SILAC-HT1080 cells expressing FLAG-tagged DJ-1 and treated the light and heavy cells with DMSO and 1 μM RSL3 for 6 h, respectively. Cells were washed twice with ice-cold PBS and harvested. The cells were lysed in RIPA buffer (50 mM triethanolamine, pH 7.4, 150 mM NaCl, 1% (vol/vol) NP-40, 1% sodium deoxycholate, 0.1% (vol/vol) SDS) containing 1 × EDTA-free complete mini-protease inhibitors (Roche), catalase (200 U/mL, C4963, Sigma). After 1 h on ice (with frequent mixing), lysates were centrifugated at 20000 g at 4 °C for 30 min. Protein concentration was detected by using the BCA protein assay Kit. Light and heavy lysates (1 mg each) were mixed in a 1:1 ratio. 5 mM DYn-2 probe was added to the lysates. Cell lysates were then treated with 4-DPS (50 mM) at room temperature for 1 h to block free thiols and subsequently were buffer exchanged with one Micro Bio-Spin P-30 column (89890, Thermo) preequilibrated with 100 mM HEPES, pH 8.5, and 100 mM NaCl. DPS-free lysates were then reacted with DiaAlk (1 mM) in the presence of 0.5% (vol/vol) SDS in the dark at room temperature with rotation for 1 h. After reduction with 1 mM DTT at 35 °C for 30 mins the sample was passed through one detergent-removal spin column (87777, Pierce) preequilibrated with 20 mM Tris-HCl, pH 7.4. The remaining lysates were incubated with anti-Flag M2 affinity gel beads (Sigma) at 4 °C overnight. The resulting beads were washed with BC100 buffer (100 mM NaCl, 20 mM Tris (pH 7.3), 20% glycerol, 0.2% NP-40) for three times and mixed with 5 × SDS-PAGE loading buffer at 95 °C. The supernatant protein samples were separated by SDS- PAGE and subjected to in-gel trypsin digestion as previously described [62]. Protein digests were analyzed by LC-MS/MS.
LC-MS/MS and data analysis
Samples were analyzed by LC-MS/MS on Q Exactive-plus series Orbitrap mass spectrometers (Thermo Fisher Scientific) coupled with EasyNano-LC. Mobile phase A was 0.1% FA in H2O, and mobile phase B was 0.1% FA in ACN. The flow rate was 3 μL/min for loading and 0.3 μL/min for eluting. Labeled peptide samples were loaded onto a 100 μm fused silica column packed with 15 cm × 3 μm C18 resin. Under the positive-ion mode, full-scan mass spectra were acquired over the m/z range from 350 to 1800 using the Orbitrap mass analyzer with mass resolution of 70,000. MS/MS fragmentation is performed in a data-dependent mode, of which the TOP 20 most intense ions are selected for MS2 analysis a resolution of 17,500 using collision mode of HCD. Other important parameters: isolation window, 2.0 m/z units; default charge, 2+; normalized collision energy, 28%; maximum IT, 50 ms; and dynamic exclusion, 20.0 s.
LC-MS/MS data were analyzed by ProLuCID [63] with static modification of cysteine carbamidomethylation (+57.0215 Da). For rdTOP-ABPP data, additional modifications include N-terminus dimethyl labeling and lysine residue dimethyl labeling (+28.03130 Da (light), + 32.05641 Da (medium), and +36.07567 Da (heavy)) and variable oxidation of methionine (+15.9949 Da). For SILAC data, cysteine nitrosylation (+28.99016 Da), sulfenylation (+15.99491 Da), sulfinylation (+31.98983 Da), sulfonylation (+47.98474 Da), ONE ( + 154.09938 Da), HNE ( + 156.11503 Da), HHE ( + 128.08373 Da), OHE ( + 126.06808 Da), ACR ( + 56.02621 Da) were chosen as variable modifications. The ratios of SILAC-Labelling and rdTOP-ABPP were quantified by the CIMAGE 2.0 [29] software as described previously.
Statistical analysis
Unless specified, results are expressed as mean±s.d. for fold changes compared to the control samples in three independent biological replicates. Student’s t-test was used to evaluate statistical significance. We analyzed the data in GraphPad Prism (GraphPad Software), using the unpaired, two-tailed t-test module. Statistical significance was considered when a p value was below 0.05. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. n.s. not significant.
Clustering and heatmap were generated by using default settings available in the Morpheus suite (https://software.broadinstitute.org/morpheus/). To account in an unbiased way for differences in the overall reactivity of each cell samples, all difference values were Z scored separately for each sample query across all tested reacted cysteines. Z scores were hierarchically clustered in an unsupervised manner.
Co-immunoprecipitation assay
To examine the interaction between DJ-1 and proteasome exogenously. 107 HT1080 cells per well were seeded in 15 cm dishes to culture overnight and transiently transfected with wide-type or C106A mutant DJ-1 with N-terminal FLAG tag at time 0 h. The cells were replaced with fresh medium at time 8 h and added with DMSO or 1 μM (1 S,3 R)-RSL3 or 1 μM ML162 or 10 μM Erastin or/and 600 nM Liproxstatin-1 in serum-free media (DMEM) at time 18 h. At time 24 h, cells were collected, washed by PBS for three times and centrifugated at 3000 rpm for 4 min. The cells were lysed in 500 μL BC100 buffer containing EDTA-free Pierce HaltTM protease inhibitor cocktail. 5% or 10% of the lysates were collected as input samples. The remaining lysates were incubated with anti-Flag M2 affinity gel beads (Sigma) at 4 °C overnight. The resulting beads were washed with BC100 buffer for three times and mixed with 5 × SDS-PAGE loading buffer at 95 °C. The supernatant protein samples were separated by SDS- PAGE and immunoblotted with corresponding antibodies as indicated.
For immunoprecipitation of endogenous DJ-1, the cells were treated with DMSO or 1 μM (1 S,3 R)-RSL3 or/and 600 nM Liproxstatin-1 for 6 h in serum-free media (DMEM) and lysed with BC100 buffer. 10% of the cell lysates was collected as input samples and the remaining lysates were incubated with 1 μg anti-PARK7 or normal rabbit IgG (Santa Cruz Biotechnology) at 4 °C overnight. Protein A/G agarose beads (Santa Cruz Biotechnology) were added to the lysates and incubated for 6 h at 4 °C. Agarose beads were washed three times with BC100 buffer and mixed with 5 × SDS-PAGE loading buffer at 95°C. The supernatant protein samples were separated by SDS- PAGE and immunoblotted with corresponding antibodies as indicated.
For immunoprecipitation of endogenous ACHYL1, the DJ-1 KO cells were transfected with DJ-1 WT/C106A or empty vector plasmids as indicated and harvested after 48 h transfection. Cells were then lysed with BC100 buffer and incubated with 1 μg anti-ACHYL1 or normal rabbit IgG (Santa Cruz Biotechnology) at 4 °C overnight. Protein A/G agarose beads (Santa Cruz Biotechnology) were added to the lysates and incubated for 6 h at 4 °C. Agarose beads were washed three times with BC100 buffer and mixed with 5 × SDS-PAGE loading buffer at 95 °C. The supernatant protein samples were separated by SDS- PAGE and immunoblotted with corresponding antibodies as indicated.
Immunoprecipitation
For immunoprecipitation of PSMA4 and PSMB1, 107 HT1080 cells per well were seeded in 15 cm dishes to culture overnight and transiently transfected with PSMA4 and PSMB1 with N-terminal FLAG tag at time 0 h. The cells were replaced with fresh medium at time 8 h. After 36 h transfection, cells were collected, washed by PBS for three times and centrifugated at 3000 rpm for 4 min. The cells were lysed in 800 μL BC200 buffer (200 mM NaCl, 20 mM Tris (pH 7.3), 20% glycerol, 0.2% NP-40) containing EDTA-free Pierce HaltTM protease inhibitor cocktail. 5% or 10% of the lysates were collected as input samples. The remaining lysates were incubated with anti-Flag M2 affinity gel beads (Sigma) at 4 °C overnight. The resulting beads were washed with BC200 buffer for two times and BC100 buffer for three times. 20 μL 10 × FLAG peptide were used to elute the enriched proteins. The mixer was incubated with FLAG peptide for 12 h at 4 °C rotator. The supernatant protein samples were prepared for following GST-pulldown assay.
Cell viability assays
1.0 × 104 HT1080 cells were plated in 96-well plates with 100 μL of pre-warmed complete DMEM medium per well (six technical replicates). For experiments that require knockdown, 0.08 μg indicated siRNA were transfected in HT1080 cells for 48 h. For experiments that require overexpression, 0.1 μg indicated plasmids were transfected for 36 h. For experiments that require generation of stable cell lines, indicated DJ-1 plasmids were stably transfected in HT1080 cells. Cells were treated with each of the compounds in serum-free medium (DMEM) at 37 °C for 24 h. Cell viability was then detected by using MTS reagent (CellTiter 96® AQueous One Solution Cell Proliferation Assay). Cells were incubated in 100 μL serum-free medium (DMEM) containing 1/6 of the MTS reagent at 37 °C for 40 min and the absorbance at 490 nm was measured with a microplate reader (Bio-Rad). The cell viability under tested conditions were reported as a percentage value relative to the DMSO-treated condition. For data presentation, the mean and standard deviation for the three biological replicates of each data point in a representative experiment were plotted in Prism 8 (Graphpad) and presented. The IC50 curves were fitted with regression and IC50 values calculated by “Inhibitor vs Response” in Graphpad Prism software.
Flow cytometry analysis
2 × 105 HT1080 cells per well were seeded into 6-well dishes. For experiments that require overexpression, 2 μg indicated plasmids were transfected for 36 h. For experiments that require generation of stable cell lines, indicated DJ-1 plasmids were stably transfected in HT1080 cells. Indicated cells were treated with 1 μM (1 S,3 R)-RSL3 and/or 600 nM Liproxstatin-1 for 15 min, 30 min, 1 h, 2 h, 4 h and 6 h respectively in serum-free medium (DMEM) at 37 °C. BODIPY-C11 (Invitrogen) was used to detect reactive oxygen species in membrane lipids. The medium was removed and cells were washed once with PBS. After being labeled in 1 mL PBS containing 5 μM BODIPY 581/591 C11 and incubated at 37 °C for 15 min, the labeled cells were redistributed in 1 mL of fresh PBS and analyzed by using a flow cytometer (FACS Calibur or Accuri C6, BD Biosciences) equipped with 510 nm laser for excitation. A minimum of 10000 cells were analyzed in per tube. Each experiment was independently performed for three biological replicates. Data analysis was performed using the FlowJo6 software. An example gating strategy is demonstrated in Supplementary Fig. 4.
RNA interference
The siRNA constructs as listed below were synthesized by GenePharma (Shanghai, China). siRNA were transfected into HT1080 cells and the method of transfection were in accordance with the manufacturer’s instructions using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen). Transfection was repeated twice with an interval of 24 h to achieve maximal knock down efficiency.
PSMB1 No.1: CGGAGGUACUAUACUGGCAAUUGCUdTdT
PSMB1 No.2: GCAAUGCUGUCUACAAUCCUGUAUUdTdT
PSMA4 No.1: CCAAUACCUUGUGAGCAGUUGGUUAdTdT
PSMA4 No.2: CGCUGUGUGAUAUCAAACAAGCUUAdTdT
Proteasome chymotrypsin-like activity analysis
1.0 × 104 HT1080 cells were plated in 96-well plates with 100 μL of pre-warm complete DMEM medium per well (six technical replicates). For experiments that require overexpression, 0.1 μg indicated plasmids were transfected for 36 h. For experiments that require generation of stable cell lines, indicated DJ-1 plasmids were stably transfected in HT1080 cells. Cells were treated with each of the compounds in serum-free medium (DMEM) at 37 °C for 6 h. Proteasomal chymotrypsin-like activity was measured by an Amplit fluorimetric proteasome 20S activity assay kit (AAT-13456, AAT Bioquest, Sunnyvale, CA, USA). The absorbance was measured using a fluorescence spectrometer (with an emission wavelength of 525 nm and excitation wavelength of 490 nm).
Recombinant expression of DJ-1 in E. coli
Expression plasmids were first transformed into E.coli BL21. Cells were grown (100 ml LB medium) at 37 °C to an absorbance of 0.6–0.8 at 600 nm and induced by 0.8 mM IPTG at 18 °C for 16 h. Cells were lysed by high pressure homogenization at 800 bar for 20 mins in BC500 buffer (500 mM NaCl, 20 mM Tris (pH 7.3), 20% glycerol, 0.2% NP-40) containing 1 mM PSMF (AMRESCO). The mixture was centrifuged at 21500 rpm for 1 h. The abundance of DJ-1 in supernatant were analyzed by 10% SDS-PAGE.
GST pulldown
DJ-1 proteins with GST fusions were recombinantly expressed in BL21 cells. After being immobilized on Glutathione Sepharose 4B beads (cat. No. 17-0756-01, GE), GST fusion proteins were incubated with the immunoprecipitated PSMA4 and PSMB1 with N-teminal FLAG tag for 2 h. The eluted samples were subjected to SDS-PAGE and analyzed by western blotting. The GST fusion proteins were visualized by Coomassie blue staining.
Supplementary information
Acknowledgements
We thank the Computing Platform of the Center for Life Science for supporting the LC-MS/MS proteomics data analysis. We gratefully acknowledge Prof. Jing Yang at National Center for Protein Sciences for generously providing the DiaAlk probes and advice on the assay to identify sulfinylation. We thank Dr. Nan Chen for providing the DYn-2 and IAyne probes. We thank Prof. Wensheng Wei at Peking University for providing plasmids for making DJ-1 knockout. This work is supported by the National Natural Science Foundation of China (No. 21925701, No.91953109 and No. 92153301) to CW.
Author contributions
CW conceived the project and supervised the study. YW conducted the experiments. YW and CW analyzed the data and wrote the manuscript together.
Data availability
The proteomics data (ID: PXD029874) has been deposited at ProteomeXchange. Full and uncropped western blots can be found in Supplementary Materials. Request for the data and materials that support the findings of this study should be addressed to C.W at chuwang@pku.edu.cn.
Competing interests
The authors declare no competing interests.
Ethical approval
This study does not include human participants, human data, or human tissue.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01050-8.
References
- 1.Chu B, Kon N, Chen D, Li T, Liu T, Jiang L, et al. ALOX12 is required for p53-mediated tumour suppression through a distinct ferroptosis pathway. Nat Cell Biol. 2019;21:579–91. doi: 10.1038/s41556-019-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.She X, Lan B, Tian H, Tang B. Cross Talk Between Ferroptosis and Cerebral Ischemia. Front Neurosci. 2020;14:776. doi: 10.3389/fnins.2020.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou J, Jin Y, Lei Y, Liu T, Wan Z, Meng H, et al. Ferroptosis Is Regulated by Mitochondria in Neurodegenerative Diseases. Neurodegener Dis. 2020;20:20–34. doi: 10.1159/000510083. [DOI] [PubMed] [Google Scholar]
- 4.Yang WS, Stockwell BR. Ferroptosis: Death by Lipid Peroxidation. Trends Cell Biol. 2016;26:165–76. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doll S, Freitas FP, Shah R, Aldrovandi M, da Silva MC, Ingold I, et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–8. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 7.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–92. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao C, Liu X, Zhang Y, Lei G, Yan Y, Lee H, et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–90. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraft VAN, Bezjian CT, Pfeiffer S, Ringelstetter L, Muller C, Zandkarimi F, et al. GTP Cyclohydrolase 1/Tetrahydrobiopterin Counteract Ferroptosis through Lipid Remodeling. ACS Cent Sci. 2020;6:41–53. doi: 10.1021/acscentsci.9b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soula M, Weber RA, Zilka O, Alwaseem H, La K, Yen F, et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat Chem Biol. 2020;16:1351–60. doi: 10.1038/s41589-020-0613-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon SJ, Winter GE, Musavi LS, Lee ED, Snijder B, Rebsamen M, et al. Human Haploid Cell Genetics Reveals Roles for Lipid Metabolism Genes in Nonapoptotic Cell Death. ACS Chem Biol. 2015;10:1604–9. doi: 10.1021/acschembio.5b00245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan H, Li X, Zhang X, Kang R, Tang D. Identification of ACSL4 as a biomarker and contributor of ferroptosis. Biochem Biophys Res Commun. 2016;478:1338–43. doi: 10.1016/j.bbrc.2016.08.124. [DOI] [PubMed] [Google Scholar]
- 13.Zou Y, Li H, Graham ET, Deik AA, Eaton JK, Wang W, et al. Cytochrome P450 oxidoreductase contributes to phospholipid peroxidation in ferroptosis. Nat Chem Biol. 2020;16:302–9. doi: 10.1038/s41589-020-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan B, Ai Y, Sun Q, Ma Y, Cao Y, Wang J, et al. Membrane Damage during Ferroptosis Is Caused by Oxidation of Phospholipids Catalyzed by the Oxidoreductases POR and CYB5R1. Mol Cell. 2021;81:355–69. doi: 10.1016/j.molcel.2020.11.024. [DOI] [PubMed] [Google Scholar]
- 15.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell. 2017;171:273–85. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace NJ, Weerapana E. Diverse functional roles of reactive cysteines. ACS Chem Biol. 2013;8:283–96. doi: 10.1021/cb3005269. [DOI] [PubMed] [Google Scholar]
- 17.Badgley MA, Kremer DM, Maurer HC, DelGiorno KE, Lee HJ, Purohit V, et al. Cysteine depletion induces pancreatic tumor ferroptosis in mice. Science. 2020;368:85–9. doi: 10.1126/science.aaw9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llabani E, Hicklin RW, Lee HY, Motika SE, Crawford LA, Weerapana E, et al. Diverse compounds from pleuromutilin lead to a thioredoxin inhibitor and inducer of ferroptosis. Nat Chem. 2019;11:521–32. doi: 10.1038/s41557-019-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pajares M, Jimenez-Moreno N, Dias IHK, Debelec B, Vucetic M, Fladmark KE, et al. Redox control of protein degradation. Redox Biol. 2015;6:409–20. doi: 10.1016/j.redox.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bak DW, Bechtel TJ, Falco JA, Weerapana E. Cysteine reactivity across the subcellular universe. Curr Opin Chem Biol. 2019;48:96–105. doi: 10.1016/j.cbpa.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–5. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan T, Desai HS, Boatner LM, Yen SL, Cao J, Palafox MF, et al. SP3-FAIMS Chemoproteomics for High-Coverage Profiling of the Human Cysteinome. Chembiochem. 2021;22:1841–51. doi: 10.1002/cbic.202000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinogradova EV, Zhang X, Remillard D, Lazar DC, Suciu RM, Wang Y, et al. An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell. 2020;182:1009–26. doi: 10.1016/j.cell.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang F, Gao J, Che J, Jia G, Wang C. A Dimethyl-Labeling-Based Strategy for Site-Specifically Quantitative Chemical Proteomics. Anal Chem. 2018;90:9576–82. doi: 10.1021/acs.analchem.8b02426. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Liu Y, Lan T, Qin W, Zhu Y, Qin K, et al. Quantitative Profiling of Protein Carbonylations in Ferroptosis by an Aniline-Derived Probe. J Am Chem Soc. 2018;140:4712–20. doi: 10.1021/jacs.8b01462. [DOI] [PubMed] [Google Scholar]
- 27.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–91. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl. 2002;41:2596–9. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Gao J, Liu Y, Yang F, Chen X, Cravatt BF, Wang C. CIMAGE2.0: An Expanded Tool for Quantitative Analysis of Activity-Based Protein Profiling (ABPP) Data. J Proteome Res. 2021;20:4893–900. doi: 10.1021/acs.jproteome.1c00455. [DOI] [PubMed] [Google Scholar]
- 30.Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, et al. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep. 2019;9:16185. doi: 10.1038/s41598-019-52837-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zdralevic M, Vucetic M, Daher B, Marchiq I, Parks SK, Pouyssegur J. Disrupting the ‘Warburg effect’ re-routes cancer cells to OXPHOS offering a vulnerability point via ‘ferroptosis’-induced cell death. Adv Biol Regul. 2018;68:55–63. doi: 10.1016/j.jbior.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Cao J, Chen X, Ying M, He Q, Yang B. DJ-1 as a Therapeutic Target Against Cancer. Adv Exp Med Biol. 2017;1037:203–22. doi: 10.1007/978-981-10-6583-5_13. [DOI] [PubMed] [Google Scholar]
- 33.Dias V, Junn E, Mouradian MM. The role of oxidative stress in Parkinson’s disease. J Parkinsons Dis. 2013;3:461–91. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, et al. RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci. 2008;105:10244–9. doi: 10.1073/pnas.0708518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bjorkblom B, Adilbayeva A, Maple-Grodem J, Piston D, Okvist M, Xu XM, et al. Parkinson disease protein DJ-1 binds metals and protects against metal-induced cytotoxicity. J Biol Chem. 2013;288:22809–20. doi: 10.1074/jbc.M113.482091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao J, Chen X, Jiang L, Lu B, Yuan M, Zhu D, et al. DJ-1 suppresses ferroptosis through preserving the activity of S-adenosyl homocysteine hydrolase. Nat Commun. 2020;11:1251. doi: 10.1038/s41467-020-15109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richarme G, Mihoub M, Dairou J, Bui LC, Leger T, Lamouri A. Parkinsonism-associated protein DJ-1/Park7 is a major protein deglycase that repairs methylglyoxal- and glyoxal-glycated cysteine, arginine, and lysine residues. J Biol Chem. 2015;290:1885–97. doi: 10.1074/jbc.M114.597815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci. 2004;101:9103–8. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moosmayer D, Hilpmann A, Hoffmann J, Schnirch L, Zimmermann K, Badock V, et al. Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162. Acta Crystallogr D Struct Biol. 2021;77:237–48. doi: 10.1107/S2059798320016125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moscovitz O, Ben-Nissan G, Fainer I, Pollack D, Mizrachi L, Sharon M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat Commun. 2015;6:6609. doi: 10.1038/ncomms7609. [DOI] [PubMed] [Google Scholar]
- 42.Saito Y, Akazawa-Ogawa Y, Matsumura A, Saigoh K, Itoh S, Sutou K, et al. Oxidation and interaction of DJ-1 with 20S proteasome in the erythrocytes of early stage Parkinson’s disease patients. Sci Rep. 2016;6:30793. doi: 10.1038/srep30793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/S0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 44.Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;317:722–8. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 45.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2011;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akter S, Fu L, Jung Y, Conte ML, Lawson JR, Lowther WT, et al. Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat Chem Biol. 2018;14:995–1004. doi: 10.1038/s41589-018-0116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao M, Monian P, Quadri N, Ramasamy R, Jiang X. Glutaminolysis and Transferrin Regulate Ferroptosis. Mol Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirschhorn T, Stockwell BR. The development of the concept of ferroptosis. Free Radic Biol Med. 2019;133:130–43. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu Q, Zhang Y, Lou H, Ou Z, Liu J, Duan W, et al. GPX4 and vitamin E cooperatively protect hematopoietic stem and progenitor cells from lipid peroxidation and ferroptosis. Cell Death Dis. 2021;12:706. doi: 10.1038/s41419-021-04008-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun WY, Tyurin VA, Mikulska-Ruminska K, Shrivastava IH, Anthonymuthu TS, Zhai YJ, et al. Phospholipase iPLA2beta averts ferroptosis by eliminating a redox lipid death signal. Nat Chem Biol. 2021;17:465–76. [DOI] [PMC free article] [PubMed]
- 51.Kuljanin M, Mitchell DC, Schweppe DK, Gikandi AS, Nusinow DP, Bulloch NJ, et al. Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat Biotechnol. 2021;39:630–41. doi: 10.1038/s41587-020-00778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Abbasov ME, Kavanagh ME, Ichu TA, Lazear MR, Tao Y, Crowley VM, et al. A proteome-wide atlas of lysine-reactive chemistry. Nat Chem. 2021;13:1081–92. doi: 10.1038/s41557-021-00765-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hacker SM, Backus KM, Lazear MR, Forli S, Correia BE, Cravatt BF. Global profiling of lysine reactivity and ligandability in the human proteome. Nat Chem. 2017;9:1181–90. doi: 10.1038/nchem.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hahm HS, Toroitich EK, Borne AL, Brulet JW, Libby AH, Yuan K, et al. Global targeting of functional tyrosines using sulfur-triazole exchange chemistry. Nat Chem Biol. 2020;16:150–9. doi: 10.1038/s41589-019-0404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Junn E, Jang WH, Zhao X, Jeong BS, Mouradian MM. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J Neurosci Res. 2009;87:123–9. doi: 10.1002/jnr.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekito A, Koide-Yoshida S, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. DJ-1 interacts with HIPK1 and affects H2O2-induced cell death. Free Radic Res. 2006;40:155–65. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- 57.Matsuda N, Kimura M, Queliconi BB, Kojima W, Mishima M, Takagi K, et al. Parkinson’s disease-related DJ-1 functions in thiol quality control against aldehyde attack in vitro. Sci Rep. 2017;7:12816. doi: 10.1038/s41598-017-13146-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conlon M, Poltorack CD, Forcina GC, Armenta DA, Mallais M, Perez MA, et al. A compendium of kinetic modulatory profiles identifies ferroptosis regulators. Nat Chem Biol. 2021;17:665–74. doi: 10.1038/s41589-021-00751-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bordini J, Morisi F, Cerruti F, Cascio P, Camaschella C, Ghia P, et al. Iron Causes Lipid Oxidation and Inhibits Proteasome Function in Multiple Myeloma Cells: A Proof of Concept for Novel Combination Therapies. Cancers (Basel). 2020;12:970. [DOI] [PMC free article] [PubMed]
- 60.Wang X, Wang Y, Li Z, Qin J, Wang P. Regulation of Ferroptosis Pathway by Ubiquitination. Front Cell Dev Biol. 2021;9:699304. doi: 10.3389/fcell.2021.699304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Yu C, Kang R, Kroemer G, Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28:1135–48. doi: 10.1038/s41418-020-00728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu M, Liu Y, Yu K, Liu X. Decreasing the amount of trypsin in in-gel digestion leads to diminished chemical noise and improved protein identifications. J Proteom. 2014;109:16–25. doi: 10.1016/j.jprot.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 63.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, et al. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J Proteom. 2015;129:16–24. doi: 10.1016/j.jprot.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The proteomics data (ID: PXD029874) has been deposited at ProteomeXchange. Full and uncropped western blots can be found in Supplementary Materials. Request for the data and materials that support the findings of this study should be addressed to C.W at chuwang@pku.edu.cn.