Abstract
Glioblastoma multiforme (GBM) is acknowledged as the most aggressive primary brain tumor in adults. It is typically characterized by the high heterogeneity which corresponds to extensive genetic mutations and complex alternative splicing (AS) profiles. Known as a major repressive splicing factor in AS, polypyrimidine tract-binding protein 1 (PTBP1) is involved in the exon skipping events of multiple precursor mRNAs (pre-mRNAs) in GBM. However, precise mechanisms that modulate the expression and activity of PTBP1 remain to be elucidated. In present study, we provided evidences for the role of a long intergenic noncoding RNA (LINREP) implicated in the regulation of PTBP1-induced AS. LINREP interacted with PTBP1 and human antigen R (HuR, ELAVL1) protein complex and protected PTBP1 from the ubiquitin-proteasome degradation. Consequently, a broad spectrum of PTBP1-induced spliced variants was generated by exon skipping, especially for the skipping of reticulon 4 (RTN4) exon 3. Interestingly, LINREP also promoted the dissociation of nuclear UPF1 from PTBP1, which increased the binding of PTBP1 to RTN4 transcripts, thus enhancing the skipping of RTN4 exon 3 to some extent. Besides, HuR recruitment was essential for the stabilization of LINREP via a manner dependent on N6-methyladenosine (m6A) formation and identification. Taken together, our results demonstrated the functional significance of LINREP in human GBM for its dual regulation of PTBP1-induced AS and its m6A modification modality, implicating that HuR/LINREP/PTBP1 axis might serve as a potential therapeutic target for GBM.
Subject terms: Cancer genetics, CNS cancer
Introduction
Alternative splicing (AS) is an evolutionarily conserved post-transcriptional cellular process that drives the diversity of transcriptome and proteome in eukaryote [1, 2]. Apart from being a vital mechanism during tissue development and cell differentiation, AS is also involved in multitudinous pathological processes, especially in cancers [3, 4]. Emerging evidence indicates that each of the hallmarks of cancer is related to a switch in AS, which starts to be identified as a significant signature for tumor progression and therapeutic interventions [5]. Glioblastoma multiforme (GBM), known for its high heterogeneity and rapid clinical progression, is represented as the most common and aggressive intracranial primary malignancy in adults [6]. Despite advances in treatment over the past decades, the median survival time of GBM patients is still only 14–16 months under the current standard-of-care therapy [7, 8]. In GBM, aberrant spliced isoforms function as oncogenic drivers and contribute to a high degree of intratumoral heterogeneity as well as the malignant phenotypic shift [9]. Therefore, it is crucial to unveil the underlying mechanisms of AS events in GBM to find new-targeted therapeutic strategies.
Polypyrimidine tract-binding protein 1 (PTBP1), a member of the hnRNPs family, has been extensively characterized as a key regulator of pre-mRNA splicing by inducing exon skipping. Several studies have confirmed the multifarious biological functions of PTBP1 in GBM tumorigenesis and progression, including malignant transformation, invasion and adherence [10, 11]. However, new factors and precise mechanisms that modulate the expression or activity of PTBP1 are not well understood in GBM. Long noncoding RNAs (lncRNAs), the major epigenetic regulators, were aberrantly expressed across all cancer types and were involved in various oncogenic processes [12–14]. With respect to aberrant AS in GBM, the functional roles and biological significance of most lncRNAs remain elusive currently. Hence, exploring how lncRNAs interplay with PTBP1 to regulate AS is vital for a better understanding of the carcinogenic mechanisms in human GBM.
In this study, we identified a novel lncRNA LINREP with N6-methyladenosine (m6A) modification. LINREP impeded the degradation of PTBP1 from the ubiquitin-proteasome pathway. Therefore, the PTBP1-mediated AS events, especially for the exon 3 skipping in RTN4 transcripts were enhanced. Besides, LINREP promoted the dissociation of nuclear UPF1 from PTBP1, which led to the intensified RNA-binding affinity of PTBP1 to RTN4 transcripts. Accordingly, the dual roles of LINREP in the PTBP1-induced AS contributed to the proliferation, migration and invasion of GBM cells.
Materials and methods
Clinical specimens and cell lines
A total of 104 primary glioma tissue samples (Table S1) were collected from the Department of Neurosurgery at Shandong Provincial Qianfoshan Hospital (Jinan, China). The study was approved by the Research Ethics Committee of Shandong University and was performed in accordance with the ethical guidelines of World Medical Association Declaration of Helsinki. All human glioblastoma cell lines (U87, T98G and U251) and human embryonic kidney cell line 293 (HEK293) were obtained from Shanghai Institutes for Biological Sciences Cell Resource Center (Shanghai, China). Normal human astrocytes (NHA) were obtained from the Sciencell Research Laboratories (Carlsbad, CA, USA). Cell authentication was verified by short tandem repeat profiling. All the cells were maintained in the humidified incubator at 37 °C with a 5% CO2 humidified atmosphere and cultured in DMEM medium (BI, NY, USA) supplemented with 10% fetal bovine serum (FBS, BI, NY, USA), 100 units/mL penicillin and 100 μg/mL streptomycin (BI, NY, USA).
Transient transfection and lentivirus infection
siRNAs and the 2-O-methyl RNA/DNA antisense oligonucleotides (ASOs) modified by changing the five nucleotides at the 5′ and 3′ ends into 2′-O-methyl, were designed and synthesized by RiboBio (Guangzhou, China). Transient transfections of siRNAs, ASOs and plasmids were performed by virtue of reagent Lipofectamine 2000 (Thermo Fisher Scientific, IL, USA). The LINREP overexpressing lentivirus was constructed by cloning the full length of LINREP sequence into the Lenti-Overexpression vector (GenePharma, Shanghai, China). For LINREP ΔPTBP1 overexpression, the PTBP1-binding region (620–690 nt) of LINREP was excluded. For LINREP ΔHuR overexpression, the HuR-binding region (420–530 nt) of LINREP was excluded. According to the manufacturer’s instructions, U87 cells were infected with the lentivirus and selected in media containing 2 μg/mL puromycin (Solarbio, Beijing, China) for next two weeks. All constructs were verified by sequencing. The sequences of siRNAs, ASOs and plasmids used are listed in Table S2.
Co-Immunoprecipitation (Co-IP)
Cells were lysed in the pre-mixed Pierce® IP Lysis Buffer (Thermo Fisher Scientific, IL, USA) with the protease inhibitor (Solarbio, Beijing, China). Total cell lysates were incubated with primary antibodies (3 μg) or IgG (3 μg) overnight at 4 °C, and then incubated with Protein A/G magnetic beads (Thermo Fisher Scientific, IL, USA) for 2 h at room temperature. Beads were washed for several times to obtain the immunoprecipitated complexes, followed by western blot or mass spectrometry analysis. The antibodies used are described in Table S4.
Nuclear/cytoplasmic protein fractionation and western blot
Protein lysates from whole-cell pellets samples were generated using the pre-mixed RIPA Lysis Buffer (Beyotime, Shanghai, China) with the protease inhibitor. The separation and preparation of nuclear and cytoplasmic extracts were conducted by utilizing the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Protein concentration was quantified using the BCA Protein Quantification Kit (Vazyme, Nanjing, China). GAPDH and Lamin B1 acted as loading controls for cytoplasmic and nuclear protein extracts, respectively. The antibodies used for western blot are listed in Table S4.
LncRNA microarray profiling and RNA-seq analysis
To detect the candidate lncRNAs in human GBM tissues, total RNA was first extracted with TRIzol® Reagent (Invitrogen, CA, USA) from the tissues of five GBM patients (five GBM samples, four paracancerous samples and five normal brain samples in total). The label and hybridization to the Affymetrix GeneChip Human Exon Arrays-Gminix lncRNA-WT (Gminix, Shanghai, China) based on extracted RNA was then conducted. The lncRNAs were carefully constructed using the publicly available transcriptome databases (RefSeq, UCSC Known Genes, Ensemble, NONCODE, etc.). After the quantile normalization of raw signal, 249 lncRNAs with the significant differences among GBM, paracancerous and normal brain tissues (FDR <0.01) were chosen for further data analysis.
For RNA-seq analysis, mRNA sequencing was conducted by the application of Illumina HiSeq Platform (PE150). Briefly, total RNA was isolated and subjected to a cDNA library construction, in which the clean (high-quality) reads were aligned to Human Genome Reference (GRCh38). The way of gene expression normalization was determined by Fragments per Kilobase per Million Mapped Fragments (FPKM). Gene annotation (GO) and analysis of spliced events were dealt with Metascape (http://metascape.org/).
AS events, including mutually exclusive exon (MXE), alternative 3’ss exon (A3SS), alternative 5’ss exon (A5SS), retained intron (RI) and skipped exon (SE), were quantified through rMATS [15]. The AS events with FDR < 0.05 and inclusion level difference values (IncLevelDifference) >0.2 or <−0.2 were selected for further validation.
Measurement of m6A-modified LINREP level
To quantify the level of m6A-modified LINREP, methylated RNA immune-precipitation assays were conducted. Protein A/G magnetic beads were mixed with 5 µg anti-m6A or anti-lgG antibodies overnight at 4 °C before the addition of cell lysates (a total of approximately 150 µg RNA for each sample), RNase inhibitor and protease inhibitor. The RNA was then eluted from immunoprecipitated complex with elution buffer and purified for further analysis via qRT-PCR assays. The antibodies used are listed in Table S4.
Tumor xenograft model
In the orthotopic xenografts, a total of 5 × 105 firefly luciferase-expressing U87 cells were implanted into the frontal lobes of 4-week-old male BALB/c nude mice. BALB/c nude mice (male) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, China). Tumor growth was monitored at 6, 12 and 24 days via bioluminescence imaging (IVIS Spectrum in vivo imaging system, PerkinElmer, USA). After the mice were sacrificed, tumor tissues were embedded in paraffin, sectioned and stained with hematoxylin and eosin (H&E) or IHC stained with the antibodies against PTBP1 or RTN4A according to previously reported protocols [16]. The experimental animal study was approved by Animal Care and Use Committee of the Shandong Provincial Qianfoshan Hospital, Shandong University.
Statistical analysis
Data was presented as the mean ± SEM of at least three experiments. Statistical significance was assessed by Student’s t-test (two-tailed). R 4.0, GraphPad Prism 8 and Adobe Illustrator CC 2018 software was used for the statistical analyses and production of figures. Survival analyses were performed by the Kaplan–Meier method and compared using the log-rank test. P values less than 0.05 were considered statistically significant (*P < 0.05, **P < 0.01, and ***P < 0.001).
Results
Screening and identification of PTBP1-interacting lncRNAs
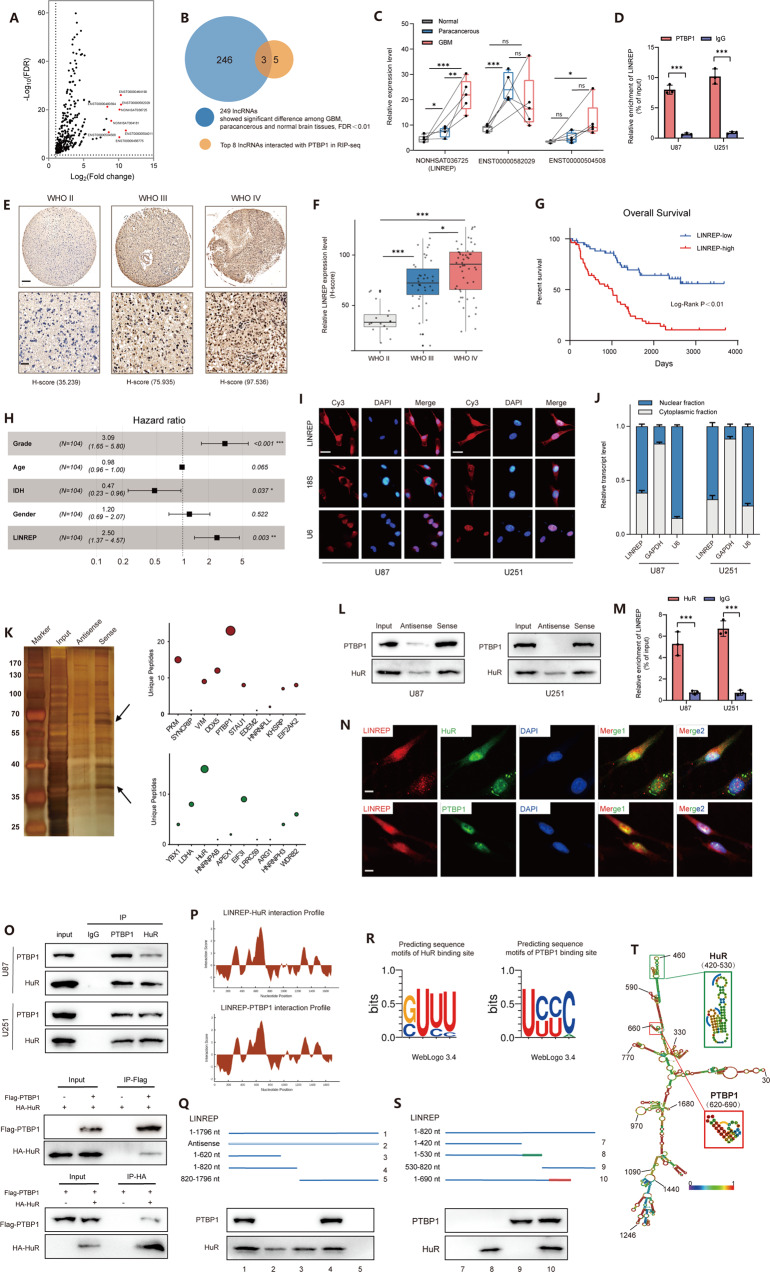
To explore the role of PTBP1-interacting lncRNAs, we initiated this study by RIP-seq experiments and screened out a number of candidates for PTBP1-binding lncRNAs (Fig. 1A and Additional Table 1). The top eight PTBP1-binding lncRNAs (fold change >2, FDR < 0.01) were then integrated with the differentially expressed lncRNAs (FDR < 0.01) detected by our group. Three PTBP1-interacting lncRNAs were identified (Fig. 1B and Fig. S1C). Of these lncRNAs, NONHSAT036725 (renamed as LINREP) was found to increase more than 4-fold in the GBM samples compared with normal tissues (Fig. 1C). RIP assays showed significant enrichment of LINREP by PTBP1 (Fig. 1D). LINREP has no coding potential in humans (Fig. S1D–F). RNA-ISH assays verified that LINREP was remarkably expressed in high-grade glioma tissues (n = 50; GBM, WHO grade IV) compared with low-grade glioma tissues (n = 54; LGG, WHO II-III) (Fig. 1E, F). The upregulation of LINREP predicted a prominently shorter overall survival (OS) and was an independent prognostic factor in glioma patients (Fig. 1G, H). FISH and subcellular fractionation assays further confirmed that LINREP was mainly localized to GBM cell nucleus (Fig. 1I, J).
Fig. 1. Identification and characterization of LINREP in GBM.
A RIP-seq experiments were performed to identify PTBP1-binding lncRNAs. Volcano plot showed the differentially expressed lncRNAs upon PTBP1 immunoprecipitation. Red dots marked top eight upregulated lncRNAs (fold change >2, FDR < 0.01). B Screening strategy was used to find key PTBP1-binding lncRNAs in GBM. C Box-plot analysis of PTBP1-binding lncRNAs in the lncRNA microarrays. D RIP assays were performed in U87 and U251 cells. n = 3 independent experiments, two-tailed Student’s t-test. E, F Representative images (E) and box-plot analysis (F) of RNA-ISH for LINREP expression on WHO grade II–IV glioma tissues. scale bar: 200 μm (top) or 50 μm (bottom). G Kaplan–Meier analysis of overall survival of glioma patients based on LINREP expression (n = 104). H Multivariate analysis for glioma patients using the COX regression model. I, J Identification of LINREP cytoplasmic and nuclear distribution by RNA-FISH (I) and qRT-PCR assays (J). Cy3 dye and DAPI stain, scale bar: 20 μm. *P < 0.05, **P < 0.01, ***P < 0.001. K RNA pulldown assay using LINREP sense and antisense RNAs in U251 cells. L Western blot validation of biotin-labelled antisense and sense LINREP pulldown. M RIP assays were performed in U87 and U251 cells. n = 3 independent experiments, two-tailed Student’s t-test. N Fluorescence assessment of LINREP and HuR or PTBP1 colocalization in U251 cells. Scale bar: 10 μm. O Western blot analysis of co-IP assays were performed on lysates prepared from GBM cells, and HEK293 cells transfected with HA-HuR and Flag-PTBP1. P Interaction profiles of HuR or PTBP1 with LINREP predicted by catRAPID. R POSTAR2 prediction of sequence motifs of HuR or PTBP1 binding sites. Q, R Serial deletions of LINREP were utilized in RNA pulldown assays to identify regions required for the LINREP and HuR or PTBP1 interactions. T The secondary structure of LINREP was predicted by RNAfold. The inset framed in green or red indicated the binding stem-loop structures of HuR or PTBP1 in LINREP, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
Interestingly, according to the unique peptides from the MS identification in RNA pulldown assays, we found that two RNA binding proteins (RBPs), HuR and PTBP1, were the highest-ranked target proteins (Fig. 1K). Consistently, LINREP was proved to interact directly with HuR and PTBP1 through the RNA pulldown and western blot assays in U87 and U251 cell lines (Fig. 1L). RIP assays showed significant enrichment of LINREP by HuR (Fig. 1M). Furthermore, LINREP was colocalized with HuR and PTBP1, where the colocalization was in both nucleus and cytoplasm for HuR, and only in the nucleus of U251 cells for PTBP1, respectively (Fig. 1N). HuR and PTBP1 were positively correlated in the three glioma cohorts (Fig. S1G). Endogenous and exogenous Co-IP assays verified the interaction of PTBP1 with HuR (Fig. 1O). Moreover, the interaction between PTBP1 and HuR was in an RNA-dependent manner (Fig. S1H).
To determine the regions of LINREP that bound to HuR or PTBP1, we firstly used the catRAPID online algorithm [17] to predict their interaction propensities (Fig. 1P). RNA pulldown assays showed that the 1–820 nt and 620–820 nt region of LINREP was required separately for its interaction with HuR and PTBP1 (Fig. 1Q). Next, according to the sequence motifs and structural preferences of RBP binding sites for HuR and PTBP1 provided by POSTAR2 [18, 19] (Fig. 1R), we constructed another series of LINREP truncated plasmids. Serial deletion analysis demonstrated that 420–530 nt region of LINREP was indispensable for the interaction with HuR, whereas PTBP1 bound to the 620–690 nt region of LINREP (Fig. 1S, T).
LINREP promoted growth and motility of GBM cells
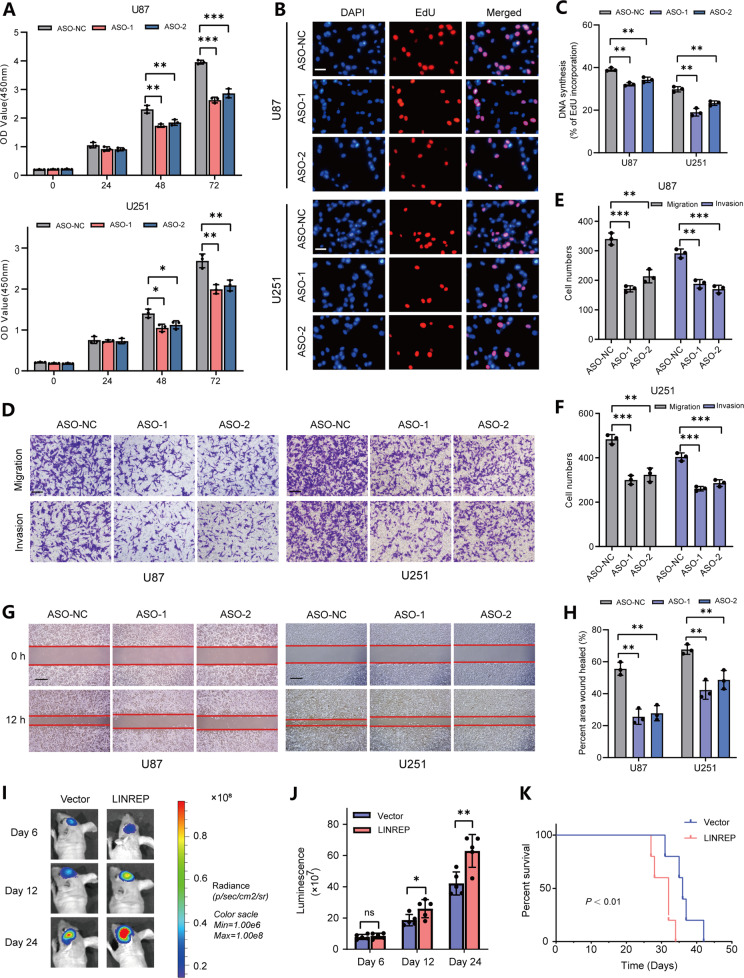
To clarify the biological functions of LINREP in GBM, we first validated the expression levels of LINREP in GBM cell lines and chose U87 and U251 cell lines for further study (Fig. S2A–C). As indicated by EdU and CCK-8 proliferation assays, LINREP knockdown suppressed cell growths (Fig. 2A–C), whereas LINREP upregulation had the opposite effect (Fig. S2D–G). The transwell and scratch wound healing assays additionally showed that knockdown of LINREP significantly diminished the migration and invasion abilities of GBM cells (Fig. 2D–H), whereas increased expression of LINREP notably enhanced the cell migration and invasion (Fig. S2H–L). In vivo results confirmed that LINREP upregulation significantly promoted the orthotopic tumor formation and shortened the survival time of tumor-bearing mice (Fig. 2I–K).
Fig. 2. LINREP knockdown impaired the proliferation, invasion and migration of GBM cells.
A Quantification of CCK-8 proliferation assays of U87 and U251 cells following LINREP knockdown. B, C Representative images (B) and quantification (C) of EdU assays in U87 and U251 cells after LINREP knockdown. Scale bar: 50 μm. D-F Representative images (D) and quantification (E, F) of transwell invasion and migration assays in U87 and U251 cells after LINREP knockdown. Scale bar: 100 μm. G, H Representative images (G) and quantification (H) of scratch wound healing assays in U87 and U251 cells following LINREP knockdown. Scale bar: 200 μm. Statistical significance was assessed using two-tailed Student’s test. Error bars represented the SD of three independent experiments. I, J Representative bioluminescence images (I) and histogram analysis (J) of Vector or LINREP-overexpressing U87 cells derived xenografts (n = 5). K Kaplan–Meier survival analysis of survival data from the xenografted mice. Log-rank test, P < 0.01. *P < 0.05, **P < 0.01, ***P < 0.001.
LINREP protected PTBP1 from the ubiquitin-proteasome degradation
Next, we investigated whether LINREP could affect the expression of HuR and PTBP1. However, LINREP depletion or upregulation obviously altered the levels of PTBP1 protein regardless of HuR/PTBP1 mRNA and HuR protein levels in U87 and U251 cells (Fig. 3A and Fig. S3A–C). As shown in Fig. S2M and S2N, upregulation of LINREP also increased the protein expression of PTBP1 in GBM tissues of tumor-bearing mice. Importantly, the level of LINREP expression was positively correlated with PTBP1 expression in the human glioma tissues (Fig. S3D). The RIP assays demonstrated that LINREP bound chiefly to the full-length PTBP1 (Fig. 3B and Fig. S3E).
Fig. 3. LINREP was involved in the proteasomal degradation of PTBP1.
A Western blot for HuR and PTBP1 in the LINREP-modified U87 and U251 cells. B Deletion mapping to identify the LINREP binding domain of PTBP1 by RIP-qPCR using Flag-tagged full length or the truncated mutants of PTBP1. n = 3 independent experiments. C, D Western blot of PTBP1 expression in LINREP knockdown (C) or overexpressed (D) U87 and U251 cells after treatment with MG132 (50 μmol/L). E, F Western blot to detect PTBP1 after 0, 4, 8 and 12 h of cycloheximide (100 μg/mL) treatment in LINREP knockdown (E) or overexpressed (F) U87 and U251 cells. G Western blot to detect the ubiquitination of PTBP1 in the LINREP-modified U87 and U251 cells.
The MG132 dosing assays showed that MG132 reversed the effect of LINREP on stability of PTBP1 in GBM cell lines (Fig. 3C, D and Fig. S3F, G). We further blocked the de novo protein synthesis using cycloheximide (CHX) (Fig. S3H). Comparatively, when LINREP-modified U87 and U251 cells were treated with CHX, western blot analyses indicated that knockdown of LINREP significantly expedited the degradation rate of PTBP1 (Fig. 3E and Fig. S3I). Conversely, upregulation of LINREP prolonged the half-life of PTBP1 (Fig. 3F and Fig. S3J). Co-IP assays confirmed that the ubiquitination of PTBP1 was attenuated in cells overexpressing LINREP while silencing of LINREP had the opposite effects (Fig. 3G). Taken together, these results demonstrated that LINREP protected PTBP1 from ubiquitin-proteasome degradation.
LINREP regulated the AS of transcripts through its association with PTBP1
Considering that PTBP1 acts as a classical splicing factor [20–22], we employed RNA-seq to investigate the changes of AS during the interaction between LINREP and PTBP1 (Fig. S4A-C). With respect to the AS analysis by RMATs [15], spliced changes on the basis of FDR < 0.05 and |IncLevelDifference| > 0.2 were selected. We found that LINREP or PTBP1 knockdown could affect all five types of AS events. Among these, SE events were the most affected and positively regulated (Fig. 4A, B). Importantly, subsequent analysis indicated that LINREP and PTBP1 regulated 175 common spliced genes from the SE type (Fig. 4C and Table S6). GO analyses revealed that these genes were mainly involved in the cancer cell motility and growth (Fig. 4D). Next, we selected the top differentially spliced genes and found five representative genes (RTN4, MAP4K4, PICALM, TPM1 and RAPGEF1) with similar exon usage patterns in GBM cells by deletion of LINREP or PTBP1 (Fig. 4E and Fig. S4D). Furthermore, the qRT-PCR assays using specific designed primers showed that the RTN4, MAP4K4, PICALM, TPM1 and RAPGEF1 exons were frequently retained following a restraint of either LINREP or PTBP1 (Fig. 4F and Fig. S4E). In particular, RTN4 remained the most significant spliced changes among five genes, and it has been recognized as a PTBP1-mediated splicing target before [11].
Fig. 4. LINREP modulated AS of RTN4 via interaction with PTBP1.
A The amount of significantly changed AS events in U251 cells with LINREP or PTBP1 knockdown. FDR < 0.05 and IncLevelDifference >0.2 or <−0.2. B The relative fraction of each AS event affected either positively or negatively by LINREP or PTBP1. C Venn diagram of splicing events regulated by LINREP and PTBP1. D Gene annotation and analysis of the common genes alternatively spliced by LINREP and PTBP1. E IGV plot illustrating the exon coverage regulated by LINREP and PTBP1. F qRT-PCR analysis of AS events in each target gene regulated by LINREP or PTBP1. Data was normalized to the control. n = 3 independent experiments, two-tailed Student’s t-test. G Schematic illustration of the RTN4 transcripts showing the exon 3-retained RTN4A and exon 3-skipped RTN4B isoforms. The exon 3 of RTN4 is 2400 bp long. H Box-plot analysis of RTN4A and RTN4B expression (normalized RSEM) in GBM and normal brain tissues by TSVdb. I Schematic diagrams of each exon usage of RTN4 sorted by PTBP1 mRNA expression viewed in GBM by TSVdb. J Western blot for RTN4A and RTN4B in U87 and U251 cells following LINREP knockdown. K, L qRT-PCR (K) and western blot analyses (L) of AS events of RTN4 in LINREP-knockdown U87 and U251 cells following reintroduction of PTBP1. n = 3 independent experiments, two-tailed Student’s t-test. M Correlation between RTN4 exon 3 skipping and LINREP expression in GBM tissues. n = 50, pearson correlation. N Representative images of RNA-ISH and RTN4A IHC staining in GBM tissues. Scale bar: 50 μm. O RIP assay performed using anti-PTBP1 or anti-IgG following LINREP knockdown. IgG served as the negative control. n = 3 independent experiments, two-tailed Student’s t-test. *P < 0.05, **P < 0.01, ***P < 0.001.
As shown in Fig. 4G, the splicing regulation of RTN4 exon 3 created two different RTN4 gene products—RTN4A (retained form) and RTN4B (skipped form). We then analyzed the AS of RTN4 exon 3 in GBM and normal brain tissues via TSVdb (http://www.tsvdb.com) [23]. The normalized expression levels of RTN4A were lower in GBM, while RTN4B was highly expressed in GBM compared with the normal (Fig. 4H). RNA-seq data from TCGA database showed that RTN4 acted as an oncogene, which enhanced the poor prognosis of GBM (Fig. S5A, B). In addition, RTN4A upregulation did prominently diminish the proliferation, migration and invasion of GBM cells (Fig. S5C–M).
The TSVdb demonstrated that high levels of PTBP1 expression were relevant to the increased skipping of RTN4 exon 3 as well as the other four genes in GBM (Fig. 4I and Fig. S6A–E). The relevance between LINREP expression and RTN4 exon 3 skipping was also confirmed by isoform-specific western blot analyses (Fig. 4J and Fig. S4F). Furthermore, the decreased skipping of RTN4 exon 3 induced by silencing of LINREP was rescued to some extent with PTBP1 reintroduction in U87 and U251 cells (Fig. 4K, L and Fig. S4G). Consistently, the skipping of RTN4 exon 3 was correlated with the LINREP expression in GBM tissues (Fig. 4M, N). RIP assays indicated that loss of LINREP impaired the binding of PTBP1 to RTN4 transcripts both in U87 and U251 cells (Fig. 4O and Fig. S4H–I), suggesting that LINREP might regulate skipping of RTN4 exon 3 in another way.
Several studies have reported that the PTBP1 binding sites on its target genes were generally located on the sequences upstream of the spliced exon (on the previous intron to be exact) [24, 25]. The RIP assays showed that PTBP1 indeed bound to the pre-RTN4 (Fig. S4J, K). The mutants of the binding sites on the intron 2 of pre-RTN4 predicted by several bioinformatic tools were subsequently constructed and the RNA pulldown assays demonstrated that PTBP1 indeed interacted with the intron 2 of pre-RTN4 (Fig. S4M, N).
LINREP promoted the dissociation of UPF1 from PTBP1 and enhanced RTN4 exon 3 skipping
Several studies have demonstrated that protein–protein interactions affected the RNA-binding ability of splicing-factors [26, 27]. Therefore, we speculated that LINREP might regulate the interaction of PTBP1 with RTN4 transcripts via altering the interplay between PTBP1 and its protein partners. Based on the screening criteria of the MS: confidence ≥ 95% and unique peptides ≥ 1, we discovered 148 and 123 putative proteins from the ASO-NC and ASO-LINREP group, respectively (Fig. 5A). The top eight proteins (UPF1, LRPPRC, FAM120A, TRAJ56, HNRNPU, FXR1, NCL and MOV10) with differential expressions between the ASO-NC and ASO-LINREP group were selected (Table S7). Of these, UPF1 was significantly increased in the LINREP-depleted group. It has conclusively been shown that PTBP1 directly bound to UPF1 and could protect mRNAs from nonsense-mediated decay (NMD) [28, 29]. Intriguingly, we found that both UPF1 and PTBP1 was mainly located in the nuclei of U251 cells from HPA (www.proteinatlas.org) (Fig. S7A, B). UPF1 and PTBP1 colocalized in the nucleus of U87 cells by immunostaining (Fig. 5B). Co-IP assays also confirmed that UPF1 directly interacted with PTBP1 in GBM and HEK293 cells (Fig. 5C, D).
Fig. 5. LINREP regulated the interaction between PTBP1 and UPF1.
A Co-IPs performed using anti-PTBP1 or anti-IgG and analyzed by Coomassie blue staining and mass spectrometry. B Fluorescence assessment of UPF1 and PTBP1 colocalization in U87 cells. Scale bar: 10 μm. C, D Western blot of co-IPs performed on lysates prepared from GBM cells (C), and HEK293 cells (D) transfected with His-UPF1 and Flag-PTBP1. E Western blot analysis of co-IPs performed on lysates prepared from HEK293 cells transfected with His-UPF1 alone or together with Flag-tagged full length or the truncated mutants of PTBP1. F Top, the schematic structures of UPF1 proteins and three His-tagged truncated mutants of UPF1 used in this study. Bottom, western blot analysis of co-IPs performed on lysates prepared from HEK293 cells transfected with Flag-PTBP1 alone or together with His-tagged full length or the truncated mutants of UPF1. G The top 10 biological enrichment analyses of gene ontology in UPF1-knockdown U251 cells. H, J qRT-PCR (H)and western blot (J) analysis of the exon 3 skipping of RTN4 in GBM cells following UPF1 knockdown or upregulation. n = 3 independent experiments, two-tailed Student’s t-test. I IGV plot illustrating the exon 3 coverage of RTN4 regulated by UPF1. K RIP assays performed using anti-PTBP1 or anti-IgG following UPF1 knockdown. IgG served as the negative control. n = 3 independent experiments, two-tailed Student’s t-test. L Western blot analysis of the exon 3 skipping of RTN4 in LINREP-knockdown U87 and U251 cells following depletion of UPF1. M Western blot analysis of co-IPs performed with anti-Flag and lysates derived from HEK293 cells transfected with adding amounts of LINREP, Flag-PTBP1 and His-UPF1. N Western blot analysis of co-IPs performed with anti-PTBP1 and cell lysates prepared from LINREP-modified U251 cells. IgG groups served as the negative controls. *P < 0.05, **P < 0.01, ***P < 0.001.
Serial deletion analysis demonstrated that UPF1 interacted mainly with full-length PTBP1 (Fig. 5E). UPF1 contains two conserved domains: the CH domain (residues 123–213) and the helicase core domain (residues 295–914) [30]. As shown in Fig. 5F, Flag-PTBP1 was only present in co-IPs when the helicase core domain of UPF1 was present, indicating that helicase core domain was crucial for the integration between PTBP1 and UPF1.
The top 10 biological enrichment analyses by RNA-seq demonstrated that UPF1 was involved in the cell motility and cell growth (Fig. 5G, Fig. S7C and Table S8). Silencing UPF1 led to the skipping of RTN4 exon 3, while UPF1 upregulation had the opposite effect without changes of PTBP1 mRNA or protein expression (Fig. 5H, J and Fig. S7E–H). Consistent with these results, IGV indicated that deletion of UPF1 promoted the exon skipping in U251 cell lines (Fig. 5I and Fig. S7I). RIP assays further demonstrated that the binding of PTBP1 to RTN4 transcripts was increased in U87 and U251 cells following UPF1 knockdown (Fig. 5K). As shown in Fig. 5L and Fig. S7J-K, the skipping of RTN4 exon 3 responding to LINREP depletion was rescued to some extent in GBM cells following UPF1 knockdown. We found that UPF1 did not bind to the RTN4 transcripts. Neither knockdown of LINREP or PTBP1 promoted the binding of UPF1 to RTN4 transcripts (Fig. S7L, M), suggesting that UPF1 did not compete with PTBP1 for the interaction with RTN4, but mediated alternative splicing via restraining the RNA binding ability of PTBP1 in part.
Changes in UPF1 mRNA and protein levels were inconspicuous regardless of the LINREP expression levels in GBM cells (Fig. S7N–P). However, the upregulation of LINREP led to less His-UPF1 protein in precipitation with Flag-PTBP1 in HEK293 cells (Fig. 5M and Fig. S7Q). Consistently, less UPF1 protein was precipitated with PTBP1 upon the elevated expression of LINREP, whereas loss of LINREP had the opposite effect in U87 and U251 cells (Fig. 5N and Fig. S7R-S). Interestingly, exogenous expression of LINREP enhanced the transportation of UPF1 from nucleus to cytoplasm in GBM cells, indicating that LINREP promoted the dissociation of UPF1 from PTBP1 and ultimately led to nucleocytoplasmic transportation of UPF1 (Fig. S7T, U).
LINREP-induced PTBP1 upregulation promoted the growth and invasion of GBM cells
To clarify whether PTBP1 was required for LINREP-induced tumorigenesis, we reintroduced PTBP1 into GBM cells following LINREP depletion. The results showed that reducing LINREP expression significantly diminished the proliferation and invasion of GBM cells, while ectopic PTBP1 abrogated the effects in vitro (Fig. 6A–G). Furthermore, in vivo assays revealed that the U87-LINREP groups exhibited larger-volume tumors and shorter survival times, which could be abolished by PTBP1 knockdown (Fig. 6H–J). IHC demonstrated that the protein expression levels of PTBP1 increased with upregulation of LINREP. In addition, PTBP1 knockdown rescued the attenuation of RTN4A induced by LINREP upregulation (Fig. 6K and Fig. S8A).
Fig. 6. LINREP interacted with PTBP1 to affect the proliferation, invasion and migration of GBM cells.
A Quantification of CCK-8 assays in LINREP-depleted GBM cells following reintroduction of PTBP1. B, C Representative images (B) and quantification (C) of EdU assays in LINREP-depleted U87 and U251 cells following reintroduction of PTBP1. Scale bar: 50 μm. D, E Representative images (D) and quantification (E) of transwell invasion and migration assays in LINREP-depleted U87 and U251 cells following reintroduction of PTBP1. Scale bar: 100 μm. F, G Representative images (F) and quantification (G) of scratch wound healing assays in LINREP-depleted GBM cells following reintroduction of PTBP1. Scale bar: 200 μm. Statistical significance was assessed using two-tailed Student’s test. Error bars represented the SD of 3 independent experiments. H, I Representative bioluminescence images (H) and histogram analysis (I) of either LINREP overexpressed or PTBP1 knockdown U87 cells derived xenografts (n = 5). J Kaplan–Meier survival analysis of survival data from the xenografted mice. Log-rank test, P < 0.01. K Representative images of H&E and IHC staining of PTBP1 and RTN4A expression in xenografts. Scale bars: 50 μm. L, M qRT-PCR (L) and western blot analyses (M) of AS events of RTN4 in LINREP-knockdown U87 and U251 cells following reintroduction of LINREP WT or LINREP ΔPTBP1. n = 3 independent experiments, two-tailed Student’s test. *P < 0.05, **P < 0.01, ***P < 0.001.
In support of the above notion, we first constructed the LINREP truncated plasmid by mutating the PTBP1-binding region (LINREP ΔPTBP1). The following RNA pulldown assays revealed that this region was indeed indispensable for LINREP to interact with PTBP1 (Fig. S8B). When LINREP ΔPTBP1 was induced in the GBM cells depleted of endogenous LINREP, the attenuated level of PTBP1 and the decreased skipping of RTN4 exon 3 could not be rescued in comparison to its wild-type counterpart (Fig. 6L–M and Fig. S8C). In addition, the capacities of proliferation, migration and invasion were almost abrogated in these cells (Fig. S8D–K). In vivo assays also revealed that LINREP ΔPTBP1 dramatically inhibited the xenograft tumor growth of tumor-bearing mice (Fig. S8L–N). Collectively, it was concluded that the effects of LINREP on PTBP1 were indeed associated with their interaction.
HuR enhanced LINREP stability via an m6A-dependent manner
HuR, which is highly expressed in gliomas and related to poor prognosis (Fig. 7A and Fig. S9A), has been demonstrated to preferentially bind and stabilize the m6A-modified RNAs [31–33]. Interestingly, knockdown of HuR significantly suppressed LINREP expression and reduced the half-time of LINREP as well (Fig. 7B and Fig. S9B–D). We constructed the LINREP truncated plasmid mutated in the binding region of HuR (LINREP ΔHuR). The RNA pulldown assays showed that LINREP ΔHuR lost its interaction with HuR, while the binding to PTBP1 was not affected (Fig. S9E). When LINREP ΔHuR was introduced to the LINREP-knockdown GBM cells, the decreased protein level of PTBP1, decreased skipping of RTN4 exon 3 as well as reduced cell proliferation and motility induced by silencing of LINREP were rescued to some extent. However, these effects were still impaired compared with LINREP-knockdown GBM cells overexpressed with LINREP WT (Fig. S9F–H and S9J–Q). As expected, the half-time of LINREP ΔHuR was significantly reduced (Fig. S9I). Therefore, we speculated that the regulatory role of LINREP in PTBP1 might have been compromised with the deficiency of the HuR-binding region.
Fig. 7. HuR enhanced LINREP stability via an m6A-dependent manner.
A Box-plot analysis of HuR expression (normalized TPM) in LGG and GBM by GEPIA2. B Western blot for HuR and qRT-PCR analysis of LINREP in U87 and U251 cells following HuR knockdown. C Enrichment of m6A-modified LINREP detected by MeRIP-qPCR assays in U87 and U251 cells. D Western blot for METTL3 and qRT-PCR analysis of LINREP in U87 and U251 cells following METTL3 knockdown. E Western blot for ALKBH5 and qRT-PCR analysis of LINREP in GBM cells following ALKBH5 knockdown. F Enrichment of m6A-modifed LINREP detected by MeRIP-qPCR assays in U87 and U251 cells following METTL3 knockdown. G Half-life of LINREP in METTL3-depleted U87 and U251 cells after treated with actinomycin D (Act D). H Fluorescence assessment of m6A and HuR colocalization in U87 cells. Scale bar: 10 μm. I qRT-PCR analysis of LINREP and HuR in either HuR knockdown or METTL3-enhanced U251 cells. J RIP assays performed using anti-HuR or anti-lgG in U251 cells following METTL3 knockdown. K Schematic representation of the position of m6A motifs within LINREP. L Half-life of LINREP in HEK293 cells transfected with plasmids overexpressing LINREP Δm6A/WT or its m6A site mutations (LINREP Δm6A/741, Δm6A/1185 and Δm6A/1256) after treated with Act D. M Enrichment of m6A-modified LINREP detected by MeRIP-qPCR assays in HEK293 cells transfected with plasmids overexpressing LINREP Δm6A/WT or its m6A sites mutants (LINREP Δm6A/741, Δm6A/1185 and Δm6A/741 + 1185). Statistical significance was assessed using two-tailed Student’s test. Error bars represented the SD of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Moreover, we verified the m6A modification on LINREP by Me-RIP assays in U87 and U251 cell lines (Fig. 7C). The process of m6A methylation is invertible and is dynamically modulated by the methyltransferases (writers), demethylases (erasers) and effector proteins (readers) [34]. Silencing of endogenous methyltransferase like 3 (METTL3) decreased LINREP expression whereas knockdown of AlkB homolog 5 (ALKBH5) increased LINREP expression in GBM cells (Fig. 7D, E and Fig. S10A–D). In addition, METTL3 deficiency contributed to a decreased m6A level on LINREP as well as the decay of LINREP, indicating that METTL3 was a major m6A methylase for LINREP in GBM (Fig. 7F, G). However, when we knocked down METTL3 in the GBM cell lines, the protein level of PTBP1 and skipping of exon 3 in the RTN4 transcripts was not significantly attenuated even though the deficiency of METTL3 decreased LINREP expression (Fig. S10E–G). We speculated that METTL3, the main methyltransferase in the GBM cells, had various downstream target transcripts, which might counteract the regulatory role of LINREP on PTBP1 and ultimately lead to the unvaried levels and activity of PTBP1 [35].
To clarify whether HuR stabilized LINREP in an m6A-dependent manner, we further detected the distribution of m6A and HuR in the U87 cells. The confocal imaging showed that m6A colocalized with HuR (Fig. 7H). The capacity of METTL3 to stabilize LINREP transcripts was attenuated in HuR-silenced U251 cells (Fig. 7I). Besides, RIP assays exhibited significantly reduced binding ability of HuR to LINREP in the METTL3-knockdown U251 cells (Fig. 7J).
There was a total of three m6A motifs (GGAC) on LINREP predicted by SRAMP (Fig. 7K and Fig. S10H, I). As shown in Fig. 7L, LINREP Δm6A/741 mutations and LINREP Δm6A/1185 mutations instead of LINREP Δm6A/1256 mutations (A-G transition mutation), could substantially lead to the reduced half-life of LINREP. Moreover, Me-RIP assays demonstrated that the high abundance of m6A modification in LINREP was impaired in the LINREP Δm6A/741 mutations or Δm6A/1185 mutations, and it was impaired more seriously in the LINREP Δm6A/741 + 1185 double mutations (Fig. 7M). Therefore, the m6A modification at 741 nt and 1185 nt of LINREP did play a congenerous role in mediating the stability of LINREP. We further upregulated LINREP WT or LINREP Δm6A/741 + 1185 in GBM cells. The results showed that skipping of RTN4 exon 3 as well as cell proliferation and motility in LINREP Δm6A/741 + 1185 overexpressed GBM cells was decreased compared with the LINREP WT (Fig. S10J-S), suggesting that the m6A modification of LINREP itself was necessary for its biological functions in GBM.
All together, these results demonstrated that HuR and m6A modification played a complementary role in stabilizing LINREP in GBM cells.
Discussion
Accumulating evidences have demonstrated that GBM cells possessed aberrant AS profiles that significantly increased the complexity of oncogenic networks and contributed to the tumorigenesis of GBM [36]. Currently, lncRNAs play an indispensable part in diverse pathological processes in GBM genesis, implying their potential roles in the regulation of GBM-associated AS. In the present study, we identified a brand-new lncRNA LINREP from several PTBP1-interacting lncRNAs that might be the key regulator of AS in GBM. We found that the LINREP upregulation was significant in GBM, and exhibited correlations with the increasing tumor grade and poor prognosis in human gliomas. LINREP overexpression endowed the GBM cells with aggressive growth capability both in vitro and in vivo.
As a splicing factor, extensive studies have been devoted to elucidate the biochemical and oncogenic roles of PTBP1 in RNA processing, but exact mechanisms for the regulation of PTBP1-induced AS remain elusive in GBM. Here, our data demonstrated that LINREP interacted physically with PTBP1 and regulated the AS by protecting PTBP1 from ubiquitin proteasome degradation. As a consequence, a broad spectrum of relevant genes involved in the cancer cell motility and growth were discomposingly expressed, such as RTN4, MAP4K4, PICALM, TPM1 and RAPGEF1 [37–40]. Of these, skipping of RTN4 exon 3 was the most significant, giving rise to the aberrant upregulation of RTN4B (skipped form) or the lack of RTN4A (retained form) expression. All these findings suggested that several AS events in GBM could be regulated by LINREP/PTBP1 interaction, revealing a novel crosstalk between lncRNAs and AS.
UPF1 has been acknowledged as a central effector of NMD, which could recognize mRNAs harboring a premature translation termination codon (PTC) or even normal mRNA transcripts in the cytoplasm and induce them to decay [41, 42]. Even though few studies have indicated that UPF1 could be localized into nucleus and exert functions in an RNA surveillance mechanism, such as the transcription elongation of nascent transcripts and DNA-damage repair [43–45], the underlying complex mechanisms of intra-nuclear UPF1 were poorly understood in the mass. Here, through subcellular localization analysis, high-throughput sequencing and functional studies, we found that UPF1 was mainly located in the nucleus of GBM cells and could interact with PTBP1 to regulate AS by altering its RNA binding ability towards the targeted downstream genes. Interestingly, LINREP took part in the formation of PTBP1/UPF1 complex, which revealed the dual roles of LINREP in regulating PTBP1-induced AS.
Previous research utilizing high-throughput sequencing has established single-nucleotide resolution mapping of the position of m6A modification in mRNAs and noncoding RNAs throughout transcriptome [46]. Up to now, the correlation between m6A mRNA methylation and tumorigenesis of GBM has been extensively explored [47]. However, the underlying mechanisms of m6A in lncRNAs were poorly understood in GBM. Here, our data demonstrated that the METTL3-induced upregulation of m6A modification increased LINREP expression, where recruitment of HuR, the other associated protein of LINREP, was indispensable. Interestingly, we found that there was no overlap between the m6A modified sites and the HuR-binding position of LINREP, suggesting HuR as an indirect m6A binding protein [48, 49]. In terms of its regulatory mechanism, Huang H recently reported that HuR was recruited by IGF2BPs to prevent the m6A-modified mRNAs from degradation [50]. Thus, we speculated that HuR might interact with different protein partners or other m6A readers to form a functional complex that collaboratively recognize m6A motifs of RNAs.
In conclusion, our investigation against GBM uncovered a functional link between lncRNAs and m6A modification in the PTBP1-induced AS (Fig. 8). These findings might provide an attractive target for new cancer biomarkers and therapeutic agents.
Fig. 8. A schematic model for the mechanisms of LINREP in the development of GBM.
m6A modification induced by METTL3 promoted recruitment of HuR and stability of LINREP expression. LINREP impeded the degradation of PTBP1 from the ubiquitin-proteasome pathway. Thus, the PTBP1-mediated AS events, especially for the exon 3 skipping in RTN4 transcripts were enhanced. Besides, LINREP promoted the dissociation of UPF1 from PTBP1, which led to the intensified RNA-binding affinity of PTBP1 to RTN4 transcripts. Accordingly, the dual roles of LINREP in the PTBP1-induced AS contributed to the proliferation, migration and invasion of GBM cells.
Supplementary information
Acknowledgements
We thank Mengqi Wang, Xiaowei Chen and Wenjie Zhu (Shandong University) for providing advice and technical assistance.
Author contributions
TX and QL designed the project. XJ, ZL and JG conducted the experiments. XJ, ZL and DH participated in data analysis and figure preparation. XJ, ZL and JG wrote the manuscript. XB, DH, WL and YW contributed to the clinical sample collection and pathological analysis. YW, YX and FZ helped with the animal study. MH, XL, ZW and XB extracted the information from databases. TX, QL and XB reviewed the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by Natural Science Foundation of China (81972340, 81871196 and 81471517), Key project of Shandong Provincial Natural Science Foundation (ZR202010300086), Science and Technology Project of Jinan city (201907048), Key Projects of Natural Science Foundation of Jiangxi Province (20192ACB20011), Shandong Province Key Research and Development Program (2019GSF107046) and Academic promotion program of Shandong First Medical University (2019LJ005).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Ethics statement and consent to participate
The research was approved by the Research Ethics Committee of Shandong University and performed in accordance with the ethical guidelines of World Medical Association Declaration of Helsinki. Written informed consent was obtained from all patients. All of the animal experiments were approved by the Animal Care and Use Committee of Shandong Provincial Qianfoshan Hospital, Shandong University.
Footnotes
Edited by A. Degterev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xiaoshuai Ji, Zihao Liu, Jiajia Gao.
Contributor Information
Qian Liu, Email: cardioqian@sdu.edu.cn.
Tao Xin, Email: drxintao@yeah.net.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01045-5.
References
- 1.Black D. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–43. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnal S, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol. 2020;17:457–74. doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 5.Goodall G, Wickramasinghe V. RNA in cancer. Nat Rev Cancer. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 6.Lapointe S, Perry A, Butowski N. Primary brain tumours in adults. Lancet. 2018;392:432–46. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 7.Sulman E, Ismaila N, Armstrong T, Tsien C, Batchelor T, Cloughesy T, et al. Radiation therapy for glioblastoma: American society of clinical oncology clinical practice guideline endorsement of the American society for radiation oncology guideline. J Clin Oncol. 2017;35:361–9. doi: 10.1200/JCO.2016.70.7562. [DOI] [PubMed] [Google Scholar]
- 8.Stupp R, Hegi M, Mason W, van den Bent M, Taphoorn M, Janzer R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 9.Pavlyukov M, Yu H, Bastola S, Minata M, Shender V, Lee Y, et al. Apoptotic cell-derived extracellular vesicles promote malignancy of glioblastoma via intercellular transfer of splicing factors. Cancer Cell. 2018;34:119–35. doi: 10.1016/j.ccell.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrarese R, Harsh G, Yadav A, Bug E, Maticzka D, Reichardt W, et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Investig. 2014;124:2861–76. doi: 10.1172/JCI68836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung H, Hai T, Zhu W, Baggerly K, Tsavachidis S, Krahe R, et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain: a J Neurol. 2009;132:2277–88. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Chen Y, Liu N, Shi Q, Han X, Gan W, et al. Low expression of TRAF3IP2-AS1 promotes progression of NONO-TFE3 translocation renal cell carcinoma by stimulating N-methyladenosine of PARP1 mRNA and downregulating PTEN. J Hematol Oncol. 2021;14:46. doi: 10.1186/s13045-021-01059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katsushima K, Lee B, Kunhiraman H, Zhong C, Murad R, Yin J, et al. The long noncoding RNA lnc-HLX-2-7 is oncogenic in Group 3 medulloblastomas. Neuro-Oncol. 2021;23:572–85. doi: 10.1093/neuonc/noaa235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B, et al. SChLAP1long noncoding rna forms a growth-promoting complex with HNRNPL in human glioblastoma through stabilization of ACTN4 and activation of NF-κB signaling. Clin Cancer Res. 2019;25:6868–81. doi: 10.1158/1078-0432.CCR-19-0747. [DOI] [PubMed] [Google Scholar]
- 15.Shen S, Park J, Lu Z, Lin L, Henry M, Wu Y, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA. 2014;111:E5593–601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji X, Ding F, Gao J, Huang X, Liu W, Wang Y, et al. Molecular and clinical characterization of a novel prognostic and immunologic biomarker FAM111A in diffuse lower-grade glioma. Front Oncol. 2020;10:573800. doi: 10.3389/fonc.2020.573800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Armaos A, Colantoni A, Proietti G, Rupert J, Tartaglia G. catRAPID omics v2.0: going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49:W72–W9. doi: 10.1093/nar/gkab393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, Xu G, Yang Y, Xu Z, Chen X, Shi B, et al. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 2019;47:D203–D11. doi: 10.1093/nar/gky830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Yang Y, Huang Y, Zhu Y, Lu Z. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 2017;45:D104–D14. doi: 10.1093/nar/gkw888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huan L, Guo T, Wu Y, Xu L, Huang S, Xu Y, et al. Hypoxia induced LUCAT1/PTBP1 axis modulates cancer cell viability and chemotherapy response. Mol cancer. 2020;19:11. doi: 10.1186/s12943-019-1122-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bielli P, Panzeri V, Lattanzio R, Mutascio S, Pieraccioli M, Volpe E, et al. The splicing factor PTBP1 promotes expression of oncogenic splice variants and predicts poor prognosis in patients with non-muscle-invasive bladder cancer. Clin Cancer Res. 2018;24:5422–32. doi: 10.1158/1078-0432.CCR-17-3850. [DOI] [PubMed] [Google Scholar]
- 22.Yap K, Mukhina S, Zhang G, Tan J, Ong HS, Makeyev EV. A short tandem repeat-enriched rna assembles a nuclear compartment to control alternative splicing and promote cell survival. Molecular cell. 2018;72:525–40.e13. doi: 10.1016/j.molcel.2018.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun W, Duan T, Ye P, Chen K, Zhang G, Lai M, et al. TSVdb: a web-tool for TCGA splicing variants analysis. BMC genomics. 2018;19:405. doi: 10.1186/s12864-018-4775-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen L, Lei S, Zhang B, Li S, Huang L, Czachor A, et al. Skipping of exon 10 in Axl pre-mRNA regulated by PTBP1 mediates invasion and metastasis process of liver cancer cells. Theranostics. 2020;10:5719–35. doi: 10.7150/thno.42010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calabretta S, Bielli P, Passacantilli I, Pilozzi E, Fendrich V, Capurso G, et al. Modulation of PKM alternative splicing by PTBP1 promotes gemcitabine resistance in pancreatic cancer cells. Oncogene. 2016;35:2031–9. doi: 10.1038/onc.2015.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao W, Adhikari S, Dahal U, Chen Y, Hao Y, Sun B, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol cell. 2016;61:507–19. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 27.Xu P, Zhang L, Xiao Y, Li W, Hu Z, Zhang R, et al. UHRF1 regulates alternative splicing by binding to splicing factors and U snRNAs. Hum Mol Genet. 2021;30:2110–22. doi: 10.1093/hmg/ddab178. [DOI] [PubMed] [Google Scholar]
- 28.Fritz S, Ranganathan S, Wang C, Hogg J. The RNA-binding protein PTBP1 promotes ATPase-dependent dissociation of the RNA helicase UPF1 to protect transcripts from nonsense-mediated mRNA decay. The. J Biol Chem. 2020;295:11613–25. doi: 10.1074/jbc.RA120.013824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge Z, Quek B, Beemon K, Hogg J. Polypyrimidine tract binding protein 1 protects mRNAs from recognition by the nonsense-mediated mRNA decay pathway. Elife. 2016;5:e11155. doi: 10.7554/eLife.11155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 31.Visvanathan A, Patil V, Arora A, Hegde A, Arivazhagan A, Santosh V, et al. Essential role of METTL3-mediated mA modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 32.Chang Y, Chai R, Pang B, Chang X, An S, Zhang K, et al. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-κB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021;511:36–46. doi: 10.1016/j.canlet.2021.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol cancer. 2019;18:142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608–24. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- 35.Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117. [DOI] [PMC free article] [PubMed]
- 36.Wang L, Shamardani K, Babikir H, Catalan F, Nejo T, Chang S, et al. The evolution of alternative splicing in glioblastoma under therapy. Genome Biol. 2021;22:48. doi: 10.1186/s13059-021-02259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín-Granado V, Ortiz-Rivero S, Carmona R, Gutiérrez-Herrero S, Barrera M, San-Segundo L, et al. C3G promotes a selective release of angiogenic factors from activated mouse platelets to regulate angiogenesis and tumor metastasis. Oncotarget. 2017;8:110994–1011. doi: 10.18632/oncotarget.22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bharadwaj S, Thanawala R, Bon G, Falcioni R, Prasad G. Resensitization of breast cancer cells to anoikis by tropomyosin-1: role of Rho kinase-dependent cytoskeleton and adhesion. Oncogene. 2005;24:8291–303. doi: 10.1038/sj.onc.1208993. [DOI] [PubMed] [Google Scholar]
- 39.Zhang F, Wang H, Yu J, Yao X, Yang S, Li W, et al. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol cancer. 2021;20:6. doi: 10.1186/s12943-020-01299-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han L, Lai H, Yang Y, Hu J, Li Z, Ma B, et al. A 5’-tRNA halve, tiRNA-Gly promotes cell proliferation and migration via binding to RBM17 and inducing alternative splicing in papillary thyroid cancer. J Exp Clin Cancer Res. 2021;40:222. doi: 10.1186/s13046-021-02024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leeds P, Peltz S, Jacobson A, Culbertson M. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–14. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 42.Kurosaki T, Popp MW, Maquat LE. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol. 2019;20:406–20. doi: 10.1038/s41580-019-0126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong D, Park T, Jeong S. Nuclear UPF1 is associated with chromatin for transcription-coupled RNA surveillance. Molecules cells. 2019;42:523–9. doi: 10.14348/molcells.2019.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh A, Choudhury S, De S, Zhang J, Kissane S, Dwivedi V, et al. The RNA helicase UPF1 associates with mRNAs co-transcriptionally and is required for the release of mRNAs from gene loci. Elife. 2019;8:e41444. doi: 10.7554/eLife.41444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ngo G, Grimstead J, Baird D. UPF1 promotes the formation of R loops to stimulate DNA double-strand break repair. Nat Commun. 2021;12:3849. doi: 10.1038/s41467-021-24201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linder B, Grozhik A, Olarerin-George A, Meydan C, Mason C, Jaffrey S. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767–72. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Geng X, Li Q, Xu J, Tan Y, Xiao M, et al. m6A modification in RNA: biogenesis, functions and roles in gliomas. J Exp Clin Cancer Res. 2020;39:192. doi: 10.1186/s13046-020-01706-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S, et al. Cross-talk among writers, readers, and erasers of mA regulates cancer growth and progression. Sci Adv. 2018;4:eaar8263. doi: 10.1126/sciadv.aar8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.