Abstract
Pathoadaptive mutations improve the fitness of pathogenic species by modification of traits that interfere with factors (virulence and ancestral) required for survival in host tissues. A demonstrated pathoadaptive mutation is the loss of lysine decarboxylase (LDC) expression in Shigella species that have evolved from LDC-expressing Escherichia coli. Previous studies demonstrated that the product of LDC activity, cadaverine, blocks the action of Shigella enterotoxins and that the gene encoding LDC, cadA, was abolished by large chromosomal deletions in each Shigella species. To better understand the nature and evolution of these pathoadaptive mutations, remnants of the cad region were sequenced from the four Shigella species. These analyses reveal novel gene arrangements in this region of the pathogens' chromosomes. Insertion sequences, a phage genome, and/or loci from different positions on the ancestral E. coli chromosome displaced the cadA locus to form distinct genetic linkages that are unique to each Shigella species. Hybridization studies, using an E. coli K-12 microarray, indicated that the genes displaced to form the novel linkages still remain in the Shigella genomes. None of these novel gene arrangements were observed in representatives of all E. coli phylogenies. Collectively, these observations indicate that inactivation of the cadA antivirulence gene occurred independently in each Shigella species. The convergent evolution of these pathoadaptive mutations demonstrates that, following evolution from commensal E. coli, strong pressures in host tissues selected Shigella clones with increased fitness and virulence through the loss of an ancestral trait (LDC). These observations strongly support the role of pathoadaptive mutation as an important pathway in the evolution of pathogenic organisms.
Classical models of evolution held that generation of new bacterial species was accomplished by accumulation of small changes in individual genes. Recently, however, two opposite mechanisms that shape the development and expression of novel traits in prokaryotes have gained recognition. Horizontal gene transfer is a widely acknowledged gain-of-function pathway used to confer new traits that enhance fitness in a current niche, or enable access to and survival in new environments (13, 25). A complementary loss-of-function system, termed antagonistic pleiotropy, fine tunes the fitness of organisms through mutation of ancestral genes encoding factors that interfere with the expression or function of traits, ancestral and newly acquired, necessary for success in an environment (6, 26). These complementary mechanisms are evident in the evolution of pathogenic bacteria.
Highly adapted, virulent bacteria evolve from commensal organisms through a series of genome modifications. Traits that permit commensal organisms access to new host environments, which often results in host disease, are incorporated into the genome via horizontal gene transfer of multiple linked genes contained on pathogenicity islands, bacteriophage, or plasmids. Selective pressures that confront the recently evolved pathogen in the new environment may be significantly different from those encountered in the ancestral niche. It has been suggested that the new pathogen is not optimally fit for the novel environment as it expresses both virulence factors (required for success in host tissues) and the full complement of ancestral traits (evolved for life in the ancestral niche) (40). Recent studies have demonstrated that, while a subset of ancestral traits may be neutral in the new environment, other traits, encoded by antivirulence genes, may negatively affect fitness by interfering with expression or function of ancestral or virulence factors required for survival within host tissues (2, 6, 16). The loss of ancestral traits encoded by antivirulence genes through negative selection has been termed pathoadaptive mutation (40). These alterations in the newly evolved pathogen genome may be generated through point mutations, which abolish protein function (non-sense mutations) or alter protein function (missense mutation), as well as deletion of antivirulence genes.
This expanded model of pathogen evolution can be traced in the generation of the shigellae. The four Shigella species, S. boydii, S. dysenteriae, S. flexneri, and S. sonnei, are closely related to the nonpathogenic enteric commensal organism Escherichia coli. Numerous studies have demonstrated that the two genera can be classified as a single species since their chromosomes are more than 90% homologous and largely colinear (3, 36). This observation is striking when one considers the very different habitats of the two closely related organisms. While nonpathogenic E. coli generally inhabit the lumen of the mammalian colon and do not cause disease, the shigellae are agents of bacillary dysentery, an acute inflammatory disease of the human colonic mucosa that results from invasion and intercellular spread of the pathogenic bacteria (29). It has been proposed that the salient factor required to transform the E. coli commensal to the Shigella phenotype is horizontal transfer of a large virulence plasmid that encodes an enterotoxin and factors required for invasion of, and intercellular spread between, host cells (23, 32).
Antagonistic pleiotropy predicts that, since E. coli and the shigellae occupy very different environments, when Shigella evolved from its E. coli ancestor, loss of a subset of ancestral traits was required for optimal fitness and success of the pathogen in host tissues. Consistent with this idea, a number of traits commonly expressed in E. coli are not expressed by any Shigella species. Among these traits are nicotinic acid prototrophy and the ability to metabolize a number of carbon sources including melibiose, allose, and lysine. For example, greater than ninety percent of all E. coli isolates metabolize lysine via decarboxylation. However, none of the shigellae catabolize lysine due to a lack of lysine decarboxylase (LDC) activity (38). Restoration of LDC activity in S. flexneri, by supplying the gene encoding lysine decarboxylase (cadA) in trans, and examination of the strain's virulence phenotypes revealed that the product of LDC activity, cadaverine, blocks the action of Shigella enterotoxins (16). The inhibition of this virulence factor's function defines cadA as an antivirulence gene. Therefore, we proposed that mutation of the cadA locus in Shigella species represents a pathoadaptive mutation since elimination of LDC activity enhances enterotoxin activity and, presumably, fitness of the pathogens in host tissues. Interestingly, we showed that absence of LDC activity in each Shigella species was due to deletion of a large region (up to 90 kb) of each species' chromosome surrounding cadA. Furthermore, the end points of the deletions in each Shigella species appeared to vary, and at least one locus, proP, that mapped to the middle of the deleted region in the corresponding K-12 chromosome was retained somewhere in the shigellae genomes.
Several studies support the hypothesis that the four Shigella species are derived from different E. coli ancestors and do not form a subgroup of E. coli (27, 31, 32, 34). Our observation that the boundaries of the cadA deletion are different in each species of Shigella suggested that excision of the antivirulence gene occurred independently in each species after the split from an ancestral E. coli. Thus, lack of LDC activity in each Shigella species may reflect convergent evolution since the fitness of each newly evolved Shigella clone would be optimized through deletion of the cadA antivirulence locus. We therefore hypothesized that if the cadA pathoadaptive mutation occurred independently in each pathogen, the end points and nature of the cadA deletion would be distinct in each Shigella species. This report describes the distinct nature of the cadA deletions in the four Shigella species as determined through sequence analysis of remnants of the cadA region in each Shigella chromosome. Our findings indicate convergent evolution of the Shigella cadA pathoadaptive mutations and suggest that, in order to optimize virulence and fitness in host tissues, each newly evolved Shigella clone abolished an ancestral trait (LDC) in response to selective pressures encountered in the virulence niche. These results provide further evidence of the important role of pathoadaptive mutations in the evolution of pathogenic bacteria.
MATERIALS AND METHODS
Bacterial strains and growth media.
Strains and species of Shigella and E. coli used in this study are described in Table 1. The four Shigella species employed, and the respective derivatives, are representatives of four separate, previously identified, Shigella lineages (32). Shigella species and E. coli strains were routinely cultured in Luria-Bertani broth at 30°C in a shaking water bath or on Luria-Bertani agar plates supplemented with the appropriate antibiotics in a 37°C incubator. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 100 μg/ml; naladixic acid, 40 μg/ml; spectinomycin, 100 μg/ml; tetracycline, 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant information | Source or reference |
|---|---|---|
| Shigella strains | ||
| 2457T | Wild type S. flexneri 2a (cluster III Shigella lineage) | 10 |
| 60R | Wild type S. dysenteriae 1 Tetr (clonal S. dysenteriae 1 lineage) | 22 |
| BS109 | 2457T galU::Tn10 Tetr | Laboratory stock |
| BS600 | BS109 (ΔrecB ptr recC recD)::Plac bet exo cat galU::Tn10 (P1L4 transductant from KM32) | This study |
| BS601 | Wild-type S. sonnei Nalr (clonal S. sonnei lineage) | Laboratory stock |
| BS649 | S. boydii 18 galU::Tn10 Tetr (cluster I Shigella lineage) | Laboratory stock |
| BS650 | BS649::pEBD221 Spcr Tetr | This study |
| BS660 | BS600 dsbD::ery Ermr | This study |
| BS661 | 60R::pEBD221 Spcr Tetr | This study |
| BS663 | BS601::pEBD221 Spcr Nalr | This study |
| E. coli strains | ||
| MG1655 | K-12 reference strain | 11 |
| χMZ115 | MG1655 zii-215::Tn10 CamErmRCP2 | 1 |
| χMZ2125 | MG1655 zjh-225::Tn10 SpcRCP2 | 1 |
| DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 (ΔlacIZYA-argF)U169 deoR (φ80 dlacΔ(lacZ) M15) | 12 |
| DH5α λpir | λpir lysogen of DH5α | Laboratory stock |
| KM32 | K-12 (ΔrecB ptr recC recD)::Plac bet exo cat | 20 |
| SM10λpir | thi-1 thr-1 leuB6 tonA21 lacY1 supE44 recA::RP4-2-Tc::Mu Kanr λpir | 39 |
| ATM558 | pEBD221 transformant of SM10λpir | This study |
| EC07 | ECOR strain, group A phylogeny | 18 |
| EC12 | ECOR strain, group A phylogeny | 18 |
| EC15 | ECOR strain, group A phylogeny | 18 |
| EC30 | ECOR strain, group B1 phylogeny | 18 |
| EC40 | ECOR strain, group D phylogeny | 18 |
| EC42 | ECOR strain, group E phylogeny | 18 |
| EC47 | ECOR strain, group D phylogeny | 18 |
| EC54 | ECOR strain, group B2 phylogeny | 18 |
| EC59 | ECOR strain, group B2 phylogeny | 18 |
| EC72 | ECOR strain, group B1 phylogeny | 18 |
| Plasmids | ||
| pUC19 | Cloning vector, ColE1 ori, Apr | NEB |
| pGP704 | Suicide vector, R6K ori, RP4 mob, Apr | 19 |
| pEBD173 | S. flexneri 2a yjdC-dsbD region cloned into XbaI/HindIII sites of pUC19, Apr | This study |
| pEBD176 | Derivative of pEBD173, ery marker and restriction sites of Tn10CamErmRCP2 cloned into unique BglII site in dsbD, Apr Ermr | This study |
| pEBD194 | 8.5-kb XmaI fragment of BS660 chromosome (S. flexneri 2a) cloned into XmaI site of pUC19, Ermr Apr | This study |
| pEBD218 | Derivative of pEBD173, spc gene and restriction sites of Tn10SpcRC cloned into unique EcoRV site located 32 bp from the yjdC stop codon, Apr Spcr | This study |
| pEBD221 | Derivative of pGP704, yjdC'-spc allele of pEBD218 cloned into FspI/SmaI sites of the suicide vector, Spcr | This study |
| pEBD249 | 14-kb self-ligated BglII fragment of BS661 (S. dysenteriae 1) chromosome, Spcr | This study |
| pEBD273 | 32-kb self-ligated BglII fragment of BS650 (S. boydii 18) chromosome, Spcr | This study |
| pEBD303 | 26-kb self-ligated XmaI fragment of BS663 (S. sonnei) chromosome, Spcr | This study |
Molecular methods.
Plasmids generated in the course of these studies are described in Table 1. Analyses of DNA, plasmid constructions, genomic extractions, and electroporations of E. coli were performed using manufacturers' suggested conditions or standard protocols described elsewhere (35). All restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.). Amplification of products less than 4 kb long employed AccuPOL DNA polymerase (PGC Scientifics, Frederick, Md.); amplification of products greater than 4 kb employed Long Taq Plus DNA polymerase (Stratagene, La Jolla, Calif.). Each thermostable DNA polymerase was used according to the manufacturer's protocols. Templates for DNA sequencing were prepared using the ABI Prism Dye Terminator cycle Sequencing Kit. Products were analyzed on an ABI Prism 377 DNA sequencer in the Uniformed Services University of the Health Sciences (USUHS) Biomedical Instrumentation Center. Oligonucleotide synthesis was performed using Applied Biosystems automated solid-phase synthesis with standard chemistry at the USUHS Biomedical Instrumentation Center.
Cloning of the S. flexneri 2a cadA deletion region.
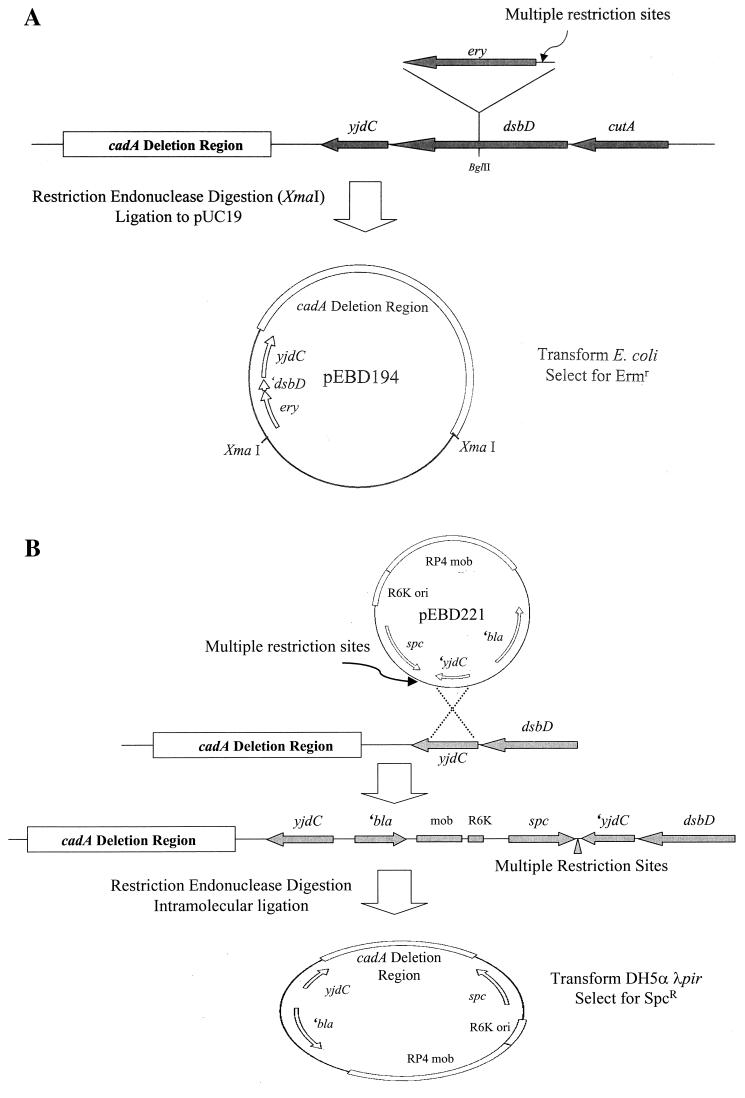
To clone regions counterclockwise (CCW) to yjdC in the S. flexneri 2a chromosome the dsbD locus was tagged with a cassette carrying an antibiotic resistance marker and multiple restriction sites. PCR amplification of the yjdC-dsbD region from the S. flexneri 2a chromosome was accomplished using primers corresponding to the 3′ end of yjdC and the 5′ end of dsbD. Employing restriction sites engineered on the 5′ ends of each primer the product was cloned into XbaI/HindIII-digested pUC19 to generate pEBD173. The ery gene from transposon Tn10CamErmRCP2, which contains a multiple restriction site cassette immediately upstream of the erythromycin resistance locus (14), was amplified from E. coli strain χMZ115 and cloned into the unique BglII site in dsbD on pEBD173. The resulting plasmid was designated pEBD176. Replacement of the S. flexneri 2a wild type dsbD allele with the dsbD::ery allele on pEBD176 was accomplished by linear DNA transformation. Briefly, the pEBD176 insert was PCR amplified using primers derived from vector sequences outside the multiple cloning site, and the products were digested with DpnI to degrade the plasmid template, concentrated via ethanol precipitation, and electroporated into BS600, a S. flexneri strain competent for linear DNA substrates. BS660, a double crossover recombinant that harbors the dsbD::ery allele, was identified by its resistance to erythromycin and scored for ampicillin sensitivity. The recombinant allele structure of BS660 was confirmed by subsequent PCR analysis. BS660 genomic DNA was then completely digested with XmaI, which cuts upstream of ery and CCW in the cadA deletion region (not in dsbD or yjdC). The products were ligated to pUC19, and electroporated into E. coli DH5α (Fig. 1A). Selection of ampicillin and erythromycin resistant transformants permitted recovery of pEBD194 which contained DNA CCW to yjdC in the S. flexneri 2a chromosome. The insert in pEBD194 was sequenced by primer walking.
FIG. 1.
Strategy used to clone regions CCW of yjdC. (A) To clone the regions CCW of yjdC from the S. flexneri 2a chromosome the ery gene from Tn10dCamErmRCP2 was introduced at the BglII site of dsbD oriented so that, following replacement of the wild-type dsbD with the marked allele (see Materials and Methods), restriction with endonucleases that cut within the multiple restriction site upstream of ery retains linkage of the resistance marker with regions CCW to yjdC. The ery-tagged fragment was cloned into pUC19, transformed into E. coli, and selected by plating for Ermr. (B) To clone the regions CCW to yjdC from the S. boydii 18, S. dysenteriae 1, and S. sonnei chromosomes, the circular suicide plasmid pEBD221 was introduced into each Shigella species by conjugation. pEBD221 integrates into the Shigella chromosome at yjdC by single-crossover recombination to generate the spectinomycin resistance allele depicted in the second chromosome. Restriction with endonucleases that cut within the multiple restriction site downstream of the spc resistance marker and at a CCW site in the cadA deletion region generates a self-replicating fragment containing the regions CCW to yjdC, the R6K ori, and the spc selectable marker. Self-ligated plasmids are recovered in a host carrying the pir gene (see Materials and Methods).
Cloning of the S. boydii 18, S. dysenteriae 1, and S. sonnei cadA deletion regions.
To clone regions CCW to yjdC in S. boydii 18, S. dysenteriae 1, and S. sonnei, a modification of the above strategy was employed. The spc gene from transposon Tn10SpcRCP2, which contains a multiple restriction site cassette immediately downstream of the spectinomycin resistance locus (14), was amplified from E. coli strain χMZ2125 and cloned into the unique EcoRV site of pEBD173 (see above), located 32 bp from the 3′ end of yjdC. The insert was oriented such that the restriction sites were situated between the yjdC′ gene fragment and the spc resistance marker to generate pEBD218. To construct a suicide plasmid that would integrate into the Shigella chromosome at yjdC and enable retrieval of regions CCW to yjdC, the yjdC′-spc allele from pEBD218 was cloned as a PvuII fragment into the FspI-SmaI sites of the R6K-based suicide vector pGP704 (19), generating pEBD221 (Fig. 1B). pEBD221 was introduced into E. coli SM10λpir and delivered to S. boydii 18, S. dysenteriae 1, and S. sonnei by conjugation. Selection for single crossover recombinants harboring pEBD221 integrated at the yjdC locus was made by plating for resistance to spectinomycin. Counterselection against the E. coli donor utilized antibiotics appropriate for each recipient Shigella strain. The recombinant allele structure of each strain (i.e., integration of pEBD221 at yjdC) was confirmed using PCR. Genomic DNA extracted from the recombinants BS650, BS661, and BS663 was completely digested with BglII or XmaI, which cut downstream of spc and CCW in the cadA deletion region (not in the integrated suicide vector or yjdC). The products were self-ligated, and electroporated into E. coli DH5αλpir, a K-12 strain that encodes the π protein and permits replication of plasmids carrying the R6K ori. This strategy permitted recovery of plasmids pEBD249, pEBD273, and pEBD303 that contain chromosomal DNA CCW of yjdC in S. dysenteriae 1, S. boydii 18, and S. sonnei, respectively (Table 1). Sequencing of all inserts was accomplished by primer walking.
Microarray hybridization and analysis.
Microarray nylon membranes containing all 4290 open reading frames (ORFs) from E. coli K-12 were purchased from Sigma-Genosys Biotechnologies (Woodland, Tex.). Hybridization, signal detection, and stripping of microarray filters were performed as directed by the manufacturer. Radiolabeled DNA probes derived from the Shigella genomes were generated using random hexamer primers (Roche Molecular Biochemicals, Indianapolis, Ind.) and Klenow DNA polymerase (New England Biolabs) to incorporate [33P]dCTP into newly synthesized products. Unincorporated nucleotides were removed using G-50 Sephadex spin columns (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). Microarray filters were prehybridized with 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 2% sodium dodecyl sulfate (SDS), 1× Denhardt's, and sheared salmon sperm DNA (100 mg/ml) for 2 h at 65°C. Hybridization with 3 ml of probe in fresh hybridization solution was performed overnight (18 h) at 65°C. Unbound and nonspecifically bound probe was removed by washing the filters three times in 0.5× SSPE–0.2% SDS for 10 min at 25°C followed by three washes in the same buffer for 20 min at 65°C. Washed filters were air dried and exposed overnight to a phosphorimager screen which was then scanned on a Storm 860 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) at a resolution of 50 μm. The filters were stripped to allow rehybridization by incubation in 50% formamide–0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS for 30 min at 65°C and rinsing in 0.1× SSC–0.1% SDS for 30 min at 65°C. Removal of bound probe was assessed by scanning the stripped filters overnight with the phosphorimager.
Analysis of the scanned microarray autoradiographs was performed as described by Ochman and Jones (24).
Nucleotide sequence accession numbers.
The sequences of the S. flexneri 2a, S. boydii 18, S. dysenteriae 1, and S. sonnei cadA deletion regions (pathoadaptive mutations) have been deposited in GenBank under accession numbers AF417476, AF417477, AF417478, and AF417480, respectively.
RESULTS
yjdC is a conserved anchor locus linked to the Shigella cadA deletion region.
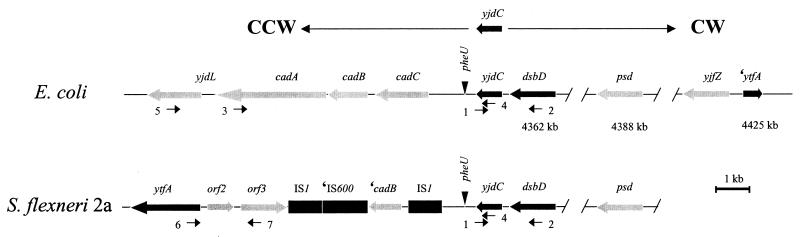
Our previous studies suggested that, relative to E. coli K-12, the Shigella chromosomes contain large (up to 90 kb) deletions surrounding the cadA antivirulence locus (16). As a first step to characterize this region of the shigellae chromosomes, we determined the end points of the deletion for each species chromosome. dsbD, a gene whose role in Shigella virulence was previously studied (42), is located 4,689 bp clockwise (CW) from cadA in E. coli K-12 (http://genolist.pasteur.fr/Colibri/genome.cgi). Since this earlier report demonstrated the presence of dsbD in the S. flexneri 2a genome, a PCR-based analysis was performed to determine whether the linear arrangement of genes neighboring this locus is similar to that found in E. coli K-12. The generation of identically sized PCR products from both S. flexneri 2a and E. coli K-12 chromosomes, using overlapping primer sets, demonstrated that linkage of dsbD to CW loci as far away as psd (located roughly 30 kb in the CW direction) is identical in S. flexneri 2a and E. coli K-12 and indicated colinearity of the two chromosomes in this region (Fig. 2 and Table 2). Moreover, since dsbD is located just CW of cadA in E. coli K-12, we reasoned that dsbD may be located close to the edge of the S. flexneri 2a cadA deletion. Therefore, PCR-based linkage analysis was performed to determine the distance between dsbD and the S. flexneri 2a cadA deletion border. Consistent with our previous findings, colinearity was lost CCW of yjdC, the locus immediately adjacent to dsbD (Table 2). No products were amplified from the S. flexneri 2a chromosome using a yjdC primer with primers derived from cadC, cadB, cadA, or yjdL (Table 2 and data not shown). PCR analysis further demonstrated that linkage of loci CW of the cadA deletion region (from yjdC to dsbD) is identical in all other species of Shigella examined, suggesting that yjdC is a conserved anchor point that maps to the edge of the cadA deletion region in the shigellae (Table 2).
FIG. 2.
Alignment of chromosomal regions adjacent to yjdC in E. coli and S. flexneri 2a based on sequence comparison of pEBD194 with E. coli K-12 and PCR-based linkage analyses. Orientation of the depicted region relative to yjdC of E. coli K-12 is indicated above the figure: CW, clockwise; CCW, counterclockwise. Gene loci are depicted as arrows, insertion sequences are depicted as black rectangles, the pheU tRNA locus is depicted as an inverted triangle; truncated ORFs and insertion sequences are indicated by a single quotation mark. The two chromosomal maps are aligned at the yjdC locus to facilitate comparison. Locations of primers used in the linkage analyses (Table 2) are depicted below the appropriate ORF. The physical map locations (in kilobase pairs) of dsbD, psd, and ′ytfA on the E. coli K-12 chromosome are indicated below each ORF.
TABLE 2.
PCR-based linkage analysis of loci adjacent to yjdC in E. coli and S. flexneri 2aa
| Strain | Site of amplification product
|
|||||
|---|---|---|---|---|---|---|
| dsbD-psd (primer walk)b | yjdC-dsbD (pr1-pr2) | cadA-yjdC (pr3-pr4) | yjdL-yjdC (pr5-pr4) | ytfA-yjdC (pr6-pr4) | ytfA-orf3 (pr6-pr7) | |
| S. flexneri 2a | 26.7 | 1.6 | NPAc | NPA | 7.5 | 1.8 |
| S. boydii 18 | 26.7 | 1.6 | NPA | NPA | NPA | NPA |
| S. dysenteriae 1 | 26.7 | 1.6 | NPA | NPA | NPA | NPA |
| S. sonnei | 26.7 | 1.6 | NPA | NPA | NPA | 1.8 |
| E. coli K-12 | 26.7 | 1.6 | 6.4 | 8.0 | NPA | NPA |
| EC07 | NDd | ND | 6.4 | 8.0 | ND | ND |
| EC12 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC15 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC30 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC40 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC42 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC47 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC54 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC59 | ND | ND | 6.4 | 8.0 | ND | ND |
| EC70 | ND | ND | 6.4 | 8.0 | ND | ND |
Loci examined for linkage are provided as gene pairs; loci located between the gene pairs are not indicated. Primers (pr1, pr2, etc.) used in PCR analyses, derived from the gene pairs examined, are indicated below each pair of genes and their positions are depicted in Fig. 2. Sizes of amplification products (in kilobase pairs) are indicated below each pair of genes analyzed.
Overlapping primer sets were used in long PCRs to demonstrate linkage of these loci.The product reported (26.7 kb) is the sum of amplification products.
NPA, no product amplified.
ND, not done.
Composition of the S. flexneri 2a cadA deletion region indicates rearrangement.
To gain a better understanding of the nature of the cadA deletion in S. flexneri 2a, an 8.5-kb region of the S. flexneri 2a chromosome, CCW to yjdC and corresponding to the cadA deletion, was cloned (as pEBD194) and sequenced (Fig. 1A; Materials and Methods). Analysis of the region revealed that all remnants of the cad operon, except for a fragment of the cadB gene, were absent and were replaced with insertion sequences linked to novel ORFs not found in the E. coli K-12 genome (Fig. 2). Homology between the S. flexneri 2a and E. coli K-12 chromosomes is reestablished two-thirds of the way through the ytfA gene, which is present as a gene fragment in the K-12 genomic sequence (http://genolist.pasteur.fr/Colibri/genome.cgi). BLASTp searches using the S. flexneri 2a ytfA ORF revealed significant global homology (25% identity and 47% similarity) to the transcription factor TetR (30). The relatedness of the predicted YtfA and TetR proteins is highest in the N-terminal domain that contains a helix-turn-helix motif required for DNA binding in the TetR/AcrR family of transcription factors (41). The observation that this conserved region is missing in the truncated E. coli K-12 ytfA ORF indicates that the K-12 chromosome is rearranged at this locus and suggests that E. coli K-12 (of E. coli group A phylogeny) is not a likely ancestor for S. flexneri 2a. Proteins encoded by orf2 and orf3 have most significant global homology to Mesorhizobium loti 3-oxoacyl-acyl carrier protein reductase (43% identity), which is involved in fatty acid elongation, and an M. loti tagatose 3-epimerase (31% identity), which is involved in carbohydrate catabolism, respectively.
Several studies have demonstrated that Shigella lineages or species have evolved from other E. coli phylogenies (groups B1, B2, D, or E). To determine whether the K-12 genomic sequence in the cadA region is representative of linkages in other E. coli strains that may be ancestors of Shigella species, gene order in the cad region was examined in representatives of the five E. coli phylogenies. Ten strains of E. coli, representing all E. coli phylogenies, were obtained from the ECOR collection (18), and the gene order in the cad region of these strains' chromosomes relative to the K-12 cad region was examined by PCR. Amplification of identically sized products from all E. coli strains examined, in both directions relative to cad, demonstrates the colinearity in this region of all 10 E. coli strains (Table 2). Moreover, colinearity of the cadA region in representatives of all five E. coli phylogenies suggests that disruptions of the cadA antivirulence gene are unique to the Shigella species and have not occurred in other E. coli strains.
To determine whether the novel gene arrangements in the S. flexneri 2a cadA deletion region were present in the cadA pathoadaptive mutations of other Shigella species, PCR analysis was conducted using primers derived from a number of loci present in the region (Fig. 2). These analyses demonstrate the novelty of the S. flexneri 2a cadA deletion region since similar linkage (specifically, ytfA to yjdC) was not observed in other Shigella species (Table 2). However, linkage of the complete ytfA allele to orf3 was observed in S. sonnei suggesting the likelihood that this linkage existed in E. coli phylogenies that are ancestral to S. flexneri 2a and S. sonnei. Collectively, these observations indicate that the S. flexneri 2a cadA deletion region is unique to this species and demonstrate rearrangement in this region of the chromosome.
Distinct composition of the cadA regions in each Shigella species.
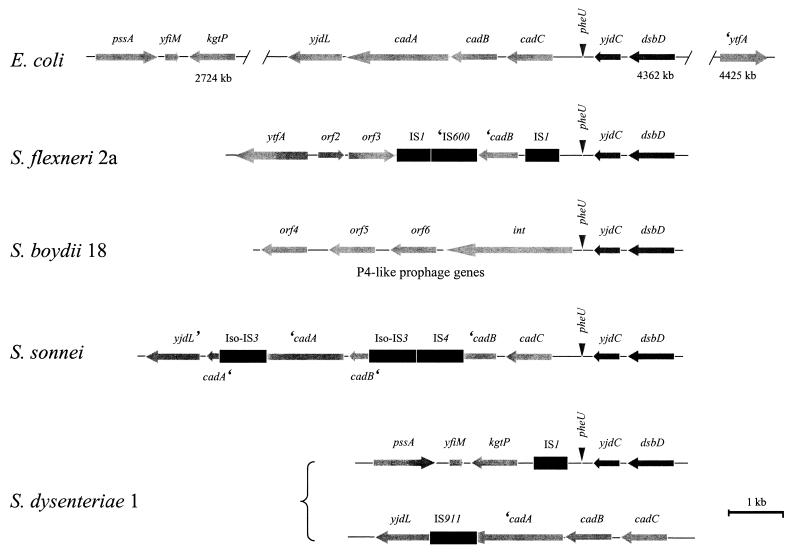
In order to compare the cadA deletion of each Shigella species, a strategy was devised that simplified cloning of regions CCW to the yjdC anchor (Fig. 1B; Materials and Methods). This strategy permitted recovery of regions CCW to yjdC in S. boydii 18, S. dysenteriae 1, and S. sonnei via linkage to a suicide plasmid (pEBD221) integrated at yjdC. The strains selected to represent each Shigella species also represent three different Shigella lineages as described by Pupo et al. (see Table 1) (32). As was the case for S. flexneri 2a, comparison of Shigella DNA sequences CCW of yjdC to the E. coli cad region revealed the nature of the pathoadaptive mutations (Fig. 3). In S. boydii 18, all remnants of the cad operon are replaced by genes homologous to those encoding a P4-like prophage that has integrated immediately CCW of the pheU (phenylalanine tRNA) region. While the presence of the prophage did not exclude the possibility that the cad operon existed CCW to the phage genome, hybridization analysis using an E. coli K-12 microarray demonstrated that the cad genes were not present in the S. boydii 18 genome (Fig. 4; see below). Preliminary downstream sequence data indicate that this region of the S. boydii 18 genome is allelic with, and identical to, the recently described SHI-3 pathogenicity island which is comprised of not only P4-like prophage loci but also encodes an iron-uptake system (33). In S. dysenteriae 1 the cad operon is replaced by an IS1 element. Homology between the S. dysenteriae 1 and E. coli K-12 chromosomes is re-established upstream of kgtP, a locus found at kb 2724 on the E. coli K-12 physical map. The novel linkage CCW of yjdC in the S. dysenteriae 1 chromosome indicates extensive rearrangement in this region. In S. sonnei, the entire cad operon linked to yjdL (CCW) and yjdC (CW) was recovered indicating that no rearrangement had occurred in this region of the chromosome relative to E. coli. The cadA pathoadaptive mutation in S. sonnei resulted from an iso-IS3 element inserted in cadA 259 bp from the stop codon. Two other IS elements, IS4 and iso-IS3, were present in this region and disrupt the cadB ORF.
FIG. 3.
Alignment of chromosomal regions CCW to yjdC in E. coli and the four Shigella species. All features are depicted as in Fig. 2. The chromosomal maps are aligned at the yjdC locus to facilitate comparison. The locations (in kilobase pairs) of kgtP, dsbD, and ′ytfA on the E. coli K-12 chromosome are indicated below each ORF. The S. dysenteriae 1 cad operon, which is displaced and not linked to yjdC, is depicted below the region CCW to yjdC.
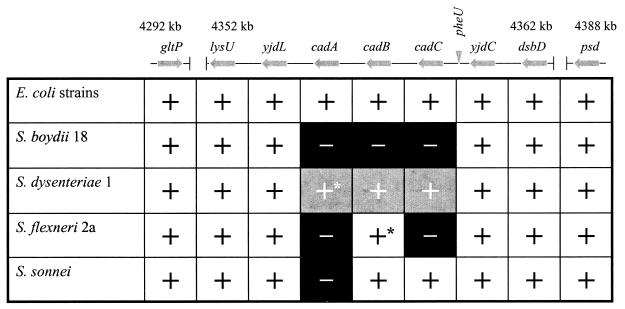
FIG. 4.
Microarray hybridization signals of loci adjacent to or displaced from the Shigella cadA regions. Gene loci (not drawn to scale) adjacent to yjdC, as ordered in this region of the E. coli K-12 chromosome, are depicted as arrows; the pheU tRNA locus is depicted as an inverted triangle. Loci between gltP and lysU and between dsbD and psd (indicated by the gapped line) demonstrated positive hybridization signals with DNA probes from all six genomes. The locations in kilobases on the E. coli K-12 chromosome are indicated above boundary loci. The plus symbol indicates the presence of a hybridization signal; the minus symbol on a black background indicates the absence of a hybridization signal. The plus symbol on a gray background indicates rearrangement of these loci; ∗, nonfunctional ORF (see text and Fig. 3). Identical hybridization patterns for this chromosomal region were obtained with genomic probes derived from two different strains of E. coli, MG1655 (K-12, group A E. coli phylogeny) and EC47 (group D E. coli phylogeny).
Rearrangement in the cadA region is not accompanied by deletion of neighboring loci.
Disruption of yjdL-yjdC linkage and the disparity in loci linked to the yjdC anchor in three of the Shigella species examined suggested that these chromosomes were extensively rearranged in this region relative to their respective ancestral E. coli. Our previous studies suggested that a number of loci CCW of yjdC were deleted from each Shigella genome, including S. sonnei (16). To determine whether the novel compositions observed CCW to yjdC in the shigellae were accompanied by deletion of neighboring, intervening, or displaced loci, a microarray containing all of the E. coli K-12 ORFs spotted in duplicate on nylon membranes was screened with labeled genomic DNA from each Shigella species. Hybridizations were also performed with genomic DNA from ECOR47, a group D E. coli (18). MG1655, the E. coli K-12 from which the microarray was generated, was used as a positive control. Figure 4 is a schematic representation of array signals obtained with each genome probe corresponding to loci present CCW of yjdC in E. coli K-12. The pattern of hybridization signals indicated the presence of all non-cad operon loci CCW to yjdC in each Shigella genome examined, suggesting that none of the rearrangements shown (Fig. 3) resulted in deletion of loci previously linked to cadA. Therefore, these displaced loci are present elsewhere in the S. boydii 18, S. dysenteriae 1, and S. flexneri 2a genomes, unlinked to yjdC. In contrast, hybridization signals for the cad operon varied between Shigella species. Consistent with sequence data CCW to yjdC in S. flexneri 2a, a hybridization signal was observed for cadB alone. Hybridization signals were observed for both cadB and cadC in S. sonnei. No signal was observed for the cadA ORF. These data are inconsistent with sequence analysis of the S. sonnei cad operon (Fig. 3). However, amplification of the interrupted cadA ORF from two different S. sonnei isolates (data not shown) substantiates our sequence data and indicates that the hybridization screenings are not as reliable (i.e., may yield false negatives) as sequence data. Sequence data of the region CCW to yjdC in S. boydii 18 did not suggest a fate for cadA in this species (Fig. 3). However, the absence of hybridization signals for any of the genes in the cad operon indicated that insertion of the phage genome at the pheU locus did not simply displace the cad genes. Rather, deletion of the cad operon may have preceded, coincided with, or occurred after integration of the prophage. Surprisingly, hybridization signals were observed for cadC, cadB, and cadA with the S. dysenteriae 1 probe, suggesting that the rearrangement resulting in the novel linkage CCW of yjdC was not accompanied by deletion of these genes. A positive S. dysenteriae 1 hybridization signal to yjdL, which is located immediately CCW of cadA in all E. coli phylogenies (Table 2), suggested that the apparently intact cad operon may retain CCW linkages present in the species ancestor. Using primers internal to yjdL and cadB, a PCR product of 3.0 kb was amplified from the S. dysenteriae 1 genome. Sequence analysis of the product indicated that the cadA ORF was interrupted by an IS911 insertion sequence 146 bp from the stop codon (Fig. 3). Together with sequence data obtained from the yjdC region of the S. dysenteriae 1 chromosome, these results indicate that the intact cad operon was moved to another region of the chromosome and contains a mutation in cadA. Collectively, our results demonstrate that the cadA deletion regions are unique in each Shigella species. Moreover, these findings strongly suggest that mutation of the cadA anti-virulence gene was accomplished by different mechanisms in each species.
DISCUSSION
It has been proposed that Shigella species evolved from commensal E. coli that acquired the Shigella virulence plasmid via horizontal transfer and, with it, the ability to occupy a new host niche (inside colonocytes) (23, 32). Pathoadaptive mutations arose within the genome of the new pathogen in response to selective pressure for optimal fitness in host tissues. This mutation pathway eliminates or modifies traits that inhibit fitness (i.e., antivirulence genes). Recent insights into the evolution of the shigellae suggest that these organisms may serve as a model for the study of pathoadaptive mutations and the evolution of pathogenic bacteria. Several investigators have suggested that the evolution of new pathogen lineages expressing the Shigella phenotype has occurred multiple times (27, 31, 34). These observations predict that the four Shigella species evolved from separate E. coli ancestors and do not form an individual E. coli subgroup. Most recently, Pupo et al. identified seven separate Shigella lineages (32). Three lineages (cluster I [which includes S. boydii 18], II, and III [which includes S. flexneri 2a]) are comprised of several pathogenic strains identified by metabolic characteristics and/or serotype. The remaining four lineages are comprised of single clones, including S. dysenteriae 1, and S. sonnei. An important implication of these observations is that expression of traits unique to and shared by all Shigella species reflects convergent evolution, whether the traits were acquired through gain-of-function (via horizontal gene transfer) or loss-of-function (pathoadaptive and random mutation) mutations.
We hypothesized that deletion of the cadA antivirulence locus occurred independently in each species or lineage of Shigella and was accomplished by different mechanisms. To test this hypothesis, the studies we report here examine the nature of the cadA pathoadaptive mutation in four Shigella species representative of four separate lineages through sequence analysis of what was once the cadA region of these organisms' chromosomes. Implicit in this analysis is the axiom that the regions studied are, in fact, remnants of the disrupted cadA regions. Data from the linkage studies support this assumption. First, linkage analysis of the cad region to yjdC in ten ECOR strains demonstrated that gene order in this region is conserved in all E. coli phylogenies. Since the shigellae evolved from commensal E. coli, it is reasonable to assume that yjdC was linked to the cad operon in ancestors of the shigellae. Second, the shigellae and E. coli chromosomes are colinear in the region CW to yjdC. Since colinearity between E. coli and each Shigella chromosome is disrupted immediately CCW to yjdC, this gene is positioned at the edge of the cadA pathoadaptive mutation in each pathogen's chromosome. Consequently, our studies focused on characterization of the regions CCW to yjdC that are linked to remnants of the cad operon in each Shigella chromosome.
The discrete compositions of loci present in this region of each Shigella chromosome support our hypothesis. These dissimilar linkages argue that deletion of the cadA antivirulence gene occurred independently, and by different mechanisms, in each species. The disparity in the remnants of the Shigella cadA regions also supports the idea that each Shigella species evolved independently and that lack of LDC activity in the shigellae results from convergent evolution. Collectively, these observations reinforce the concept of cadA as an antivirulence gene in the shigellae and that ablation of LDC activity in these pathogens occurred as a pathoadaptive mutation. Indeed, our findings suggest that each time an E. coli commensal acquired the Shigella enterotoxin gene senA (encoded on the virulence plasmid), it encountered strong selective pressure for deletion of the cadA antivirulence locus as LDC-deficient clones would permit optimal enterotoxin activity. The action of the toxin expressed in each new Shigella clone (21) likely enhanced dissemination of the pathogens into the environment where they may infect new hosts.
Our previous study reported that removal of the cadA locus from each Shigella chromosome was accomplished by deletion of distinct large (>90 kb) regions of the chromosomes (16). The novel linkages to yjdC in each Shigella chromosome raised the possibility that the previously reported large deletions resulted from rearrangements that generated the observed linkages. This possibility was particularly plausible for S. dysenteriae 1 and S. flexneri 2a, in which different sections of the E. coli chromosome are linked to yjdC. However, our data indicate that rearrangement at cadA was not accompanied by large deletions outside the cad operon. Hybridization screens of an E. coli K-12 microarray demonstrated that essentially all non-cad operon loci displaced to make way for the observed linkages are still present in the Shigella genomes. This observation indicates rearrangement of at least one large region of the S. dysenteriae 1 and S. flexneri 2a chromosomes. Similar observations have been reported by Shu et al. as pulsed field analysis revealed rearrangement of the S. dysenteriae 1 and S. flexneri 2a chromosomes that map close to the cadA deletion regions (37). While the extent and nature of rearrangements vary between S. dysenteriae 1 and S. flexneri 2a lineages, both rearrangements involve cadA and are associated with gene inactivation. No evidence of rearrangement was observed in any of the five E. coli phylogenies. In fact, gene order in this region of the commensal's chromosomes is highly conserved, suggesting that disruption of the shigella cadA regions, through insertion (IS) elements or rearrangement, did not first occur in an E. coli ancestor. Rather, we propose that disruption of cadA was selected in each Shigella clone following acquisition of the enterotoxin gene senA encoded on the virulence plasmid.
All four pathoadaptive mutations characterized in these studies involve insertion of heterologous elements in the form of IS elements or a prophage genome. The remnant cadA region in S. dysenteriae 1 contains two IS elements. In S. flexneri 2a and S. sonnei the remnant cadA region contains three IS elements. The ubiquitous presence of these mobile genetic elements suggests that each, particularly the IS elements, may have played an important role in generation of the cadA pathoadaptive mutation. Several studies have demonstrated the presence of many IS elements in the Shigella genomes and one report estimated that over 300 IS elements exist in the S. sonnei genome (5, 15, 28). While many of these elements are present in the Shigella virulence plasmid (4), most are located in the pathogens' chromosomes. Transposition of IS elements into new areas of the Shigella chromosome, such as cadA, may provide an effective means of niche adaptation, as interruption of an ORF with an IS element is more likely to abolish the trait associated with that ORF than random accumulation of mutations, which may occur at a lower rate than transposition and often form silent or conservative missense mutations. It is reasonable to speculate that the original cadA pathoadaptive mutation in S. flexneri 2a, S. sonnei and perhaps S. dysenteriae 1, was accomplished by an IS element. This DNA sequence may have provided a region of homology with other portions of the chromosome containing identical IS elements that could recombine to rearrange the chromosome and form the novel linkages we observed (7, 9, 17). This scenario requires two discrete steps, including an intermediate ancestral clone that harbors cadA interrupted by IS elements, to generate the novel linkages observed in these most virulent Shigella lineages. PCR analysis of the yjdC linkages in five other cluster III Shigella strains (that share a common ancestor with S. flexneri 2a) revealed identical gene arrangements consistent with the clonal nature of these strains (data not shown). The failure to identify an intermediate strain harboring a cadA::IS allele linked to yjdC suggests that either the cadA deletion that occurred in evolution of this lineage was accomplished by a one-step mechanism or rearrangement of the chromosome generated a strain that was more fit than the ancestor harboring only the virulence plasmid and an interrupted cadA.
The niche occupied by a pathogenic species is often very different than the environment of the ancestral organism. Not surprisingly, fitness in the ancestral niche does not guarantee fitness in the new environment. A subset of traits (encoded by antivirulence genes) antagonizes the function of virulence and ancestral factors required for growth and survival in host tissues (6, 40). Our previous studies defined cadA as an antivirulence gene for the shigellae. The results we report here indicate convergent evolution of cadA mutations in the shigellae and reinforce the important role of pathoadaptive mutations in the evolution of pathogenic bacteria. Thus, the mechanism of pathoadaptive mutation is one by which antivirulence genes are removed from the genomes of newly evolved pathogens through selection of clones for optimal fitness in the new host environment. These observations suggest criteria for the identification of antivirulence genes: the antivirulence gene must be present (and expressed) in closely related or ancestral species occupying the nonvirulent ancestral niche but absent from pathogenic clones living in host tissues. Most importantly, expression of the antivirulence gene by the pathogen in host tissues must attenuate virulence and inhibit fitness. These criteria, which essentially constitute the converse of molecular Koch's postulates (8), have been demonstrated for the cadA antivirulence locus in the shigellae. Application of converse Koch's postulates to additional loci in Shigella and other pathogenic organisms holds great promise for identification of antivirulence genes lost by pathoadaptive mutation. These new insights into the ecology of bacterial pathogens will identify factors, proteins, or products of biochemical pathways (such as the product of LDC activity, cadaverine) that block the activity of traits required for pathogen survival in host tissues. These natural agents, which are expressed by nonpathogenic ancestral organisms, may aid in the development of novel pathogen-specific therapies.
ACKNOWLEDGMENTS
This work was supported by grant AI24656 from the National Institute of Allergy and Infectious Diseases and grant RO7385 from the USUHS.
We thank Thomas Whittam for the gift of the ECOR strains; Michael N. Flora and the USUHS Biomedical Instrumentation Center for DNA sequencing and oligonucleotide synthesis services; and Rachel Binet, Colleen Kane, and Raymond Schuch for critical reading of the manuscript.
REFERENCES
- 1.Bloch C A, Rode C K, Obreque V H, Mahillon J. Purification of Escherichia coli chromosomal segments without cloning. Biochem Biophys Res Commun. 1996;223:104–111. doi: 10.1006/bbrc.1996.0853. [DOI] [PubMed] [Google Scholar]
- 2.Boucher J C, Yu H, Mudd M H, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner D J, Fanning G R, Johnson K E, Citarella R V, Falkow S. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol. 1969;98:637–650. doi: 10.1128/jb.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchrieser C, Glaser P, Rusniok C, Nedjari H, d'Hauteville H, Kunst F, Sansonetti P, Parsot C. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 5.Bustos-Martinez J A, Gomez-Eichelmann M C. Frequency of IS1-mediated molecular events in different members of the family Enterobacteriaceae. J Bacteriol. 1987;169:4946–4949. doi: 10.1128/jb.169.11.4946-4949.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper V S, Lenski R E. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- 7.Daveran-Mingot M L, Campo N, Ritzenthaler P, Le Bourgeois P. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J Bacteriol. 1998;180:4834–4842. doi: 10.1128/jb.180.18.4834-4842.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falkow S. Molecular Koch's postulates applied to microbial pathogenicity. Rev Infect Dis. 1988;10(Suppl. 2):S274–S276. doi: 10.1093/cid/10.supplement_2.s274. [DOI] [PubMed] [Google Scholar]
- 9.Fang Z, Doig C, Kenna D T, Smittipat N, Palittapongarnpim P, Watt B, Forbes K J. IS6110-mediated deletions of wild-type chromosomes of Mycobacterium tuberculosis. J Bacteriol. 1999;181:1014–1020. doi: 10.1128/jb.181.3.1014-1020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Formal S B, Dammin G J, LaBrec E H, Schneider H. Experimental Shigella infections: characteristics of a fatal infection produced in guinea pigs. J Bacteriol. 1958;75:604–610. doi: 10.1128/jb.75.5.604-610.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guyer M S, Reed R R, Steitz J A, Low K B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Lawrence J G, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahillon J, Rode C K, Leonard C, Bloch C A. New ultrarare restriction site-carrying transposons for bacterial genomics. Gene. 1997;187:273–279. doi: 10.1016/s0378-1119(96)00766-4. [DOI] [PubMed] [Google Scholar]
- 15.Matsutani S, Ohtsubo E. Distribution of the Shigella sonnei insertion elements in Enterobacteriaceae. Gene. 1993;127:111–115. doi: 10.1016/0378-1119(93)90624-c. [DOI] [PubMed] [Google Scholar]
- 16.Maurelli A T, Fernández R E, Bloch C A, Rode C K, Fasano A. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc Natl Acad Sci USA. 1998;95:3943–3948. doi: 10.1073/pnas.95.7.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonough M A, Butterton J R. Spontaneous tandem amplification and deletion of the shiga toxin operon in Shigella dysenteriae 1. Mol Microbiol. 1999;34:1058–1069. doi: 10.1046/j.1365-2958.1999.01669.x. [DOI] [PubMed] [Google Scholar]
- 18.Milkman R. Electrophoretic variation in E. coli from natural sources. Science. 1973;182:1024–1026. doi: 10.1126/science.182.4116.1024. [DOI] [PubMed] [Google Scholar]
- 19.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires ToxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy K C. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nataro J P, Seriwatana J, Fasano A, Maneval D R, Guers L D, Noriega F, Dubovsky F, Levine M M, Morris J G., Jr Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect Immun. 1995;63:4721–4728. doi: 10.1128/iai.63.12.4721-4728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Brien A D, Thompson M R, Gemski P, Doctor B P, Formal S B. Biological properties of Shigella flexneri 2A toxin and its serological relationship to Shigella dysenteriae 1 toxin. Infect Immun. 1977;15:796–798. doi: 10.1128/iai.15.3.796-798.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochman H, Groisman E A. The evolution of invasion by enteric bacteria. Can J Microbiol. 1995;41:555–561. doi: 10.1139/m95-074. [DOI] [PubMed] [Google Scholar]
- 24.Ochman H, Jones I B. Evolutionary dynamics of full genome content in Escherichia coli. EMBO J. 2000;19:6637–6643. doi: 10.1093/emboj/19.24.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H, Lawrence J G, Groisman E A. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 26.Ochman H, Moran N A. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1098. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- 27.Ochman H, Whittam T S, Caugant D A, Selander R K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 28.Ohtsubo H, Nyman K, Doroszkiewicz W, Ohtsubo E. Multiple copies of iso-insertion sequences of IS1 in Shigella dysenteriae chromosome. Nature. 1981;292:640–643. doi: 10.1038/292640a0. [DOI] [PubMed] [Google Scholar]
- 29.Parsot C, Sansonetti P J. Invasion and pathogenesis of Shigella infections. In: Miller V L, editor. Bacterial invasiveness. New York, N.Y: Springer-Verlag; 1996. pp. 25–42. [DOI] [PubMed] [Google Scholar]
- 30.Postle K, Nguyen T T, Bertrand K P. Nucleotide sequence of the repressor gene of the Tn10 tetracycline resistance determinant. Nucleic Acids Res. 1984;12:4849–4863. doi: 10.1093/nar/12.12.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pupo G M, Karaolis D K, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. J Bacteriol. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pupo G M, Lan R, Reeves P R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purdy G E, Payne S M. The SHI-3 iron transport island of Shigella boydii O-1392 carries the genes for aerobactin synthesis and transport. J Bacteriol. 2001;183:4176–4182. doi: 10.1128/JB.183.14.4176-4182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rolland K, Lambert-Zechovsky N, Picard B, Denamur E. Shigella and enteroinvasive Escherichia coli are derived from distinct ancestral strains of E. coli. Microbiology. 1998;144:2667–2672. doi: 10.1099/00221287-144-9-2667. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sanderson K E. Genetic relatedness in the family Enterobacteriaceae. Annu Rev Microbiol. 1976;30:327–349. doi: 10.1146/annurev.mi.30.100176.001551. [DOI] [PubMed] [Google Scholar]
- 37.Shu S, Setianingrum E, Zhao L, Li Z, Xu H, Kawamura Y, Ezaki T. I-CeuI fragment analysis of the Shigella species: evidence for large-scale chromosome rearrangement in S. dysenteriae and S. flexneri. FEMS Microbiol Lett. 2000;182:93–98. doi: 10.1111/j.1574-6968.2000.tb08880.x. [DOI] [PubMed] [Google Scholar]
- 38.Silva R M, Toledo M R, Trabulsi L R. Biochemical and cultural characteristics of invasive Escherichia coli. J Clin Microbiol. 1980;11:441–444. doi: 10.1128/jcm.11.5.441-444.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 40.Sokurenko E V, Hasty D L, Dykhuizen D E. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 1999;7:191–195. doi: 10.1016/s0966-842x(99)01493-6. [DOI] [PubMed] [Google Scholar]
- 41.Wissmann A, Baumeister R, Muller G, Hecht B, Helbl V, Pfleiderer K, Hillen W. Amino acids determining operator binding specificity in the helix-turn-helix motif of Tn10 Tet repressor. EMBO J. 1991;13:4145–4152. doi: 10.1002/j.1460-2075.1991.tb04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu J. Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 1998;66:3909–3917. doi: 10.1128/iai.66.8.3909-3917.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]