SUMMARY
Pathogens evade host humoral responses by accumulating mutations in surface antigens. While variable, there are conserved regions that cannot mutate without compromising fitness. Antibodies targeting these conserved epitopes are often broadly protective but remain minor components of the repertoire. Rational immunogen design leverages a structural understanding of viral antigens to modulate humoral responses to favor these responses. Here, we report an epitope-enriched immunogen presenting a higher copy number of the influenza hemagglutinin (HA) receptor-binding site (RBS) epitope relative to other B cell epitopes. Immunization in a partially humanized murine model imprinted with an H1 influenza shows H1-specific serum and >99% H1-specific B cells being RBS-directed. Single B cell analyses show a genetically restricted response that structural analysis defines as RBS-directed antibodies engaging the RBS with germline-encoded contacts. These data show how epitope enrichment expands B cell responses toward conserved epitopes and advances immunogen design approaches for next-generation viral vaccines.
In brief
Caradonna et al. use structure-guided design to engineer an influenza immunogen enriched for a conserved viral site. In partially humanized mice, this immunogen expands serum and B cell responses toward the conserved epitope. Biochemical and structural characterization show expansion of multiple antibody lineages that make germline-encoded contacts.
Graphical Abstract
INTRODUCTION
A primary mechanism by which rapidly evolving viruses can evade host immune responses is accumulation of mutations in key surface proteins.1–3 Certain regions on these proteins, however, have structural or functional constraints, and mutations in those regions would compromise viral fitness.4–6 For influenza virus, the hemagglutinin (HA) receptor-binding site (RBS), stem, and “interface” are such conserved regions. Antibodies recognizing these regions can be cross-reactive and confer broad protection, but they generally make up a small fraction of the overall repertoire and appear to immunologically subdominant.7–11 Indeed, elicited humoral responses from vaccination or infection preferentially target variable immunodominant epitopes in which even single mutations can disrupt large fractions of an overall antibody response.12–15
Altering immunodominance patterns to favor responses targeting these conserved, broadly protective epitopes may result in more effective vaccines.16 Current immunogen design strategies to elicit stem-directed antibodies, for example, include removal or masking of head epitopes, germline targeting, and multivalent display of heterologous wild-type HAs or headstem chimeras.17–21 For the interface epitope, strategies such as hyperglycosylating HA have shown to be a promising approach to focus immune responses to this region.22–24 Preferentially eliciting antibodies to the conserved RBS, however, remains challenging.25–27
A significant hurdle for many immunogen design strategies targeting the RBS is how to selectively expand humoral responses to this site. We previously developed “resurfaced” HAs (rsHAs) to focus immune responses to the conserved RBS.27 These rsHAs presented an H1 RBS on antigenically distinct non-circulating avian HA scaffolds. We isolated RBS-directed antibodies with historical H1 breadth, but we did not see increased frequency of RBS-directed antibodies. A different approach used mosaicnano particles to express multiple antigenically distinct H1 HA heads.28,29 This approach immunofocused to the lateral patch epitope, and it showed how multivalent display of heterologous antigens could immunofocus to a shared epitope.28,29 A possible explanation for this observation is that B cells recognizing conserved epitopes are thought to have a competitive advantage due to enhanced antigen avidity.16,21,28 Therefore, enriching an immunogen for a single epitope should in theory give this putative advantage only to enriched epitope-specific B cells.
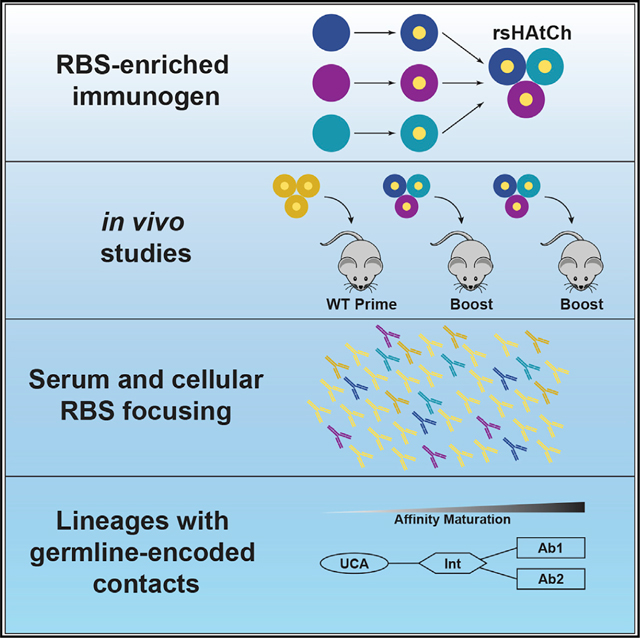
To explicitly test this hypothesis, we expanded our resurfacing approach to develop an RBS epitope-enriched immunogen composed of three heterologous HA head-only scaffolds, each resurfaced with the H1 RBS epitope. Disulfides engineered at the trimer interfaces resulted in a covalently stabilized heterotrimeric species “rsHAtCh,” or rsHA trimeric chimera. Due to the antigenic distance between the HA head-only scaffolds comprising the rsHAtCh, there is a higher frequency of the RBS epitope relative to other epitopes. We tested in vivo the recombinant rsHAtCh in IGHV1–2 HC2 mice, where antibodies use the IGHV1–2 sequence but have human-like diversity in the HCDR3 loop, which is the principal source of B cell receptor (BCR) diversity.30,31 We observe a robust expansion of H1-directed RBS responses in both the humoral and cellular compartments. Structural analysis showed a class of RBS-directed antibodies that engage the H1 RBS epitope through germline-encoded contacts. Epitope enrichment may be a more generalizable approach to preferentially expand otherwise diverse immune responses toward conserved epitopes.
RESULTS
Design and characterization of an RBS-enriched immunogen
In our previous work, we leveraged structural conservation across HA subtypes to graft the H1 Solomon Islands/03/2006 (H1 SI-06) RBS epitope onto non-circulating avian H4 and H14 HAs.27 A first step to create our RBS-enriched heterotrimer was to identify a third, antigenically distinct HA scaffold to present the H1 SI-06 RBS. We identified potential scaffolds by analyzing sequence identity between the H1 SI-06 RBS and other group 1 and 2 HAs (Figures 1A and 1B). Given the success of grafting onto group 2 HAs, we selected the H3 subtype as a third scaffold. H3 Hong Kong/4801/2014 (HK-14) is 35% identical to H1 SI-06 and 53% and 54% identical to the H4 and H14 scaffolds, respectively. Despite these differences, the H3 HK-14 accommodated the identical set of grafted residues for the H1 RBS (Figures S1A and S1B). The rsH3 HK-14 construct bound a panel of conformational specific H1 RBS-directed antibodies with uniformly high affinity (Figure S1C). These three head-only constructs (residues 37–319, H3 numbering), rsH14 WI-10, rsH3 HK-14, and rsH4 NB-10, each presenting the H1 SI-06 RBS, form the components of our RBS-enriched immunogen, rsHAtCh.
Figure 1. Design and characterization of rsHAtCh.
(A) Structure of the hemagglutinin RBS epitope and sequence alignment of the four primary RBS boundaries across various influenza A subtypes. “.” denotes amino acid identity with H1 SI-06, and “_” denotes a skipped residue.
(B) Matrix showing percentage of amino acid identified between H1 SI06 head and representative rsHA heads from different subtypes of influenza A.
(C) Size-exclusion chromatography traces showing relative efficiency of cystine bond formation for the six possible interfaces between rsH3, rsH4, and rsH14 head heterodimers. Dimeric (D) and monomeric (M) peaks labeled. “***” denotes high efficiency, “**” denotes ~50% efficiency, and “*” denotes low efficiency.
(D) Schematic and sequences of an NC2-derived heterotrimerization tag used to selectively express heterotrimeric rsHAtCh. Yellow bar represents an intratag disulfide bond.
(E) Assembly of a polycistronic “plug and play” DNA cassette composed of each component of rsHAtCh containing tPA (signal sequence), rsHA (insert), 3C (protease cleavage site), NC2 strand (heterotrimerization tag), and orthogonal affinity chromatography tag (AT1–3; His-tag, FLAG tag, or SBP tag, respectively), each separated by 2A peptides. “**” denotes a double stop codon.
(F) SDS-PAGE analysis of purified rsHAtCh under non-reducing (1) and reducing (2) conditions. Trimer (T) and component monomer (M) bands shown.
(G) BLI analysis of purified rsHAtCh and H1 SI-06 head, screened against a panel of H2227, H2526, CH67, 641 I-9, K03.12, C05, and HRV41.1 (negative control Ab), at 10 and 5 μM conditions.
(H) Design process for generating epitope-enriched immunogens.
Related to Figure S1.
The rsHAs have two conserved surface-exposed regions: the grafted H1 RBS and an interface region (Figure S1D). Because this latter region might “distract” from expanding RBS-directed responses, we occluded the region by introducing cysteines at positions 212 and 216. These modifications were previously shown to “lock” adjacent protomers in the prefusion conformation of full-length HA (Figure S1E).32 Subsequent covalent stabilization of the three rsHAs into a single heterotrimer brings each protomer into proximity to its neighbors. However, as these heterologous HAs never coevolved, some interfaces between the rsHA components may not be compatible. We therefore determined the efficiency of disulfide bond formation at each of the six possible paired interfaces by expressing individual rsHA heterodimers, where each rsHA had either Cys212 or Cys216 (Figure 1C). We determined the relative abundance of dimeric versus monomeric species using size-exclusion chromatography. rsH14-rsH3, rsH14-rsH4, and rsH3-rsH14 interfaces favored disulfide formation, while the rsH3-rsH4, rsH4-rsH14, and rsH4-rsH3 did not. Based on these results, only one of two possible orderings of the three rsHAs would result in a cross-linked heterotrimer: rsH14-rsH3-rsH4.
We next developed a heterotrimerization tag from the non-collagenous domain 2 (NC2) of human collagen IX to position the rsHAs in the order needed for optimal disulfide bond formation (Figure 1D).33,34 Appending an NC2-like coil to each rsHA was sufficient to form cystine-stabilized homotrimers in solution (Figure S1F). Further modifying each strand with a unique affinity tag (8xHis, FLAG, or streptavidin-binding peptide [SBP] motif), allowed for the selection of heterotrimeric tagged species. Following protease cleavage of the affinity and heterotrimerization tags, a final round of size-exclusion chromatography ensured the selective purification of a fully cystine-stabilized heterotrimer from monomer and dimers with incomplete cystine bond formation (Figure S1G). Finally, to promote stoichiometric expression of each component, we generated a polycistronic DNA cassette containing all three rsHAs with their NC2 and affinity tags, each separated by a P2A peptide for recombinant expression in mammalian cells (Figure 1E).35,36
SDS-PAGE analysis of the rsHAtCh construct after removal of the trimerization and affinity tags shows a single band of ~105 kDa protein under non-reducing conditions and three distinct bands under reducing conditions—the latter confirming the presence of disulfides (Figure 1F). The apparent molecular weight differences of the three rsHAs are likely due to differential glycosylation. To ensure rsHAtCh accurately presented the H1 RBS epitope, we tested the binding of six human H1 RBS-directed broadly neutralizing antibodies (bnAbs) using biolayer interferometry (BLI). This panel had comparable affinity binding to rsHAtCh and its components relative to wild-type H1 SI-06, indicating that the grafted RBS epitope had not been perturbed (Figure 1G). The generation of epitope-enriched immunogens such as rsHAtCh can be split into two steps: epitope grafting to establish a common epitope, and heterotrimerization to enrich that shared epitope (Figure 1H).
rsHAtCh immunization focuses serum and memory B cell responses to the H1 RBS
We next tested in vivo our RBS-enriched immunogen in IGHV1–2 HC2 mice; VH1–2 is the gene used for the RBS-directed CH65 lineage.30,37 We primed two cohorts with H1 SI-06 full-length soluble ectodomain (FLsE) HA and sequentially boosted them with H1 SI-06 FLsE (control) or rsHAtCh (Figure 2A). Serum samples from both cohorts were taken 12 days after the final boost at day 56. To identify the portion of serum Abs that engaged the H1 RBS epitope, we performed serum ELISAs, comparing titers against H1 SI-06 FLsE and H1 SI-06 ΔRBS FLsE; this latter construct has a glycan positioned within the RBS epitope and allows binning of RBS-directed responses (Figure S2A).38,39 Control-immunized mice showed no significant difference in serum titers between H1 SI-06 and H1 SI-06 ΔRBS, suggesting that RBS-directed Abs make up a minority of the H1-directed serum titer. However, rsHAtCh cohort sera showed a ~60-fold lower titer against H1 SI-06 ΔRBS than against H1 SI-06, as well as very low (~102) titers against H1 SI-06 ΔRBS (Figure 2B). These data suggest that most of the H1-directed titer in rsHAtCh-boosted sera is RBS directed.
Figure 2. Serum and IgG+ B cell analysis of rsHAtCh immunization.
(A) Schematic representation of immunization regimen in IGHV1–2 HC2 mice. Serum harvested 2 weeks after final immunization. Two replicates of each cohort immunized.
(B) Serum ELISA titers against H1 SI-06 FLsE and H1 SI-06 ΔRBS FLsE, where the RBS is sterically occluded by an N-linked glycan a position 190, for control cohort (N = 10) and rsHAtCh cohort (N = 8) mice. Statistical significance was determined using the Mann-Whitney U test (***p < 0.001).
(C) rsHAtCh cohort serum ELISA titers against five historical H1 FLsEs. Statistical significance was determined using the Mann-Whitney U test (***p < 0.001).
(D) Frequency of class-switched IgG+ B cells engaging RBS (ΔRBS non-binders) or non-RBS epitopes (ΔRBS binders); total numbers of cells sorted pooled across each cohort listed.
Related to Figure S2.
rsHAtCh cohort serum samples were also screened against a panel of historical H1 HA FLsEs that spanned ~20 years of antigenic evolution to determine breadth (Figure 2C). There were similar titers for H1 SI-06 and the related H1 NC-99 but ~20-to 50-fold lower titers against pre-1999 strains and the pandemic H1 CA-09—likely due to sequence differences within the RBS segments of these strains (Figure S2B). Control cohort serum had greater breadth against the same panel of historical H1 HAs (Figure S2C). Given the lack of RBS focusing in the control cohort serum, the enhanced breadth is likely due to responses targeting other conserved H1 epitopes across the selected panel of HAs (e.g., stem, lateral patch) rather than breadth against a single epitope (Figure S2D).
To characterize the expanded H1-directed cell population, class-switched H1-directed immunoglobulin G (IgG)+ B cells were pooled within each cohort and binned as RBS directed or non-RBS directed based on binding to H1 SI-06 ΔRBS probes (Figures S2E and S2F). H1+ IgG+ B cells from control mice were ~57% RBS directed compared with 99% from rsHAtCh cohort mice (Figure 2D). Furthermore, across all rsHAtCh cohort mice, only a single non-RBS-directed IgG+ B cell was identified (Figure S2G). These results indicate that relative to a wild-type H1 SI-06 FLsE immunization, rsHAtCh selectively enriches for RBS-directed responses within both the H1-directed serum and expanded IgG+ B cells. Comparison of the H1+ B cells within the class-switched IgG+ population showed a qualitatively lower frequency in the rsHAtCh cohort (Figure S2H). Class-switched “memory” B cells and the polyclonal Abs secreted from plasma cells form distinct pools with respect to affinity and target specificity, at least in mice.40 This suggests that while the two pools will have correlative features, we should also not expect complete overlap.
We also applied the H1 flow cytometry probes to measure the frequency of germline B cells specific for the RBS within the repertoire of IGHV1–2 HC2 mice compared with wild-type (WT) C57Bl/6 (Figure S2I). Within the spleen, which is the relevant secondary lymphoid organ for intraperitoneal (i.p.) injection, we find that the frequency of germline H1+ B cells targeting outside the RBS is similar between the two genotypes (0.037% and 0.034%), whereas the frequency of RBS-specific germline B cells is slightly elevated in C57Bl/6 (0.057% versus 0.018%; Figure S2J). Hence, IGHV1–2 HC2 mice provide a highly diverse germline repertoire that does not skew for the RBS, and thus the observed expansion of the H1-directed response toward the RBS is due to the immunogen and not inherent repertoire bias.
rsHAtCh immunization elicits B cells enriched for specific gene usages and features
We next obtained sequences of RBS-directed IgG+ B cells elicited by rsHAtCh and found a biased, genetically restricted response (Figures 3A and 3B). The entire RBS-directed population used JH3 alleles with ~85% of heavy-chain sequences containing DH7–27. Light chains were similarly enriched, with VK3–12 and JK2 found in over 70% of obtained sequences. Analysis of somatic hypermutation levels showed a majority of B cells with a mutational frequency greater than ~2.5%, suggesting a memory recall response with further affinity maturation (Figure 3C). Sequence analysis showed an enrichment for a Gly-X-Pro-Asp motif in the HCDR3 region that spans the DH-JH junction and Arg96 in the LCDR3 of VK gene-containing B cells (Figures 3D and 3E). Furthermore, all isolated B cells had HCDR3 and LCDR3 lengths of 14 and 9 amino acids, respectively. These data suggest that rsHAtCh immunization preferentially elicits B cells with specific sequence features from a germline repertoire containing human-like HCDR3 diversity.30,31
Figure 3. Sequence analysis of RBS-directed IgG+ B cells.
(A and B) Heavy- (A) and light-chain (B) gene usages from sorted RBS-directed cells elicited by rsHAtCh immunization; N = 71 pooled from 4 mice.
(C) VH gene mutation frequency among sequenced RBS-directed B cells.
(D and E) Sequence logos for HCDR3 (D) and LCDR3 (E) segments.
Related to Figure S3.
Lineage analysis of rsHAtCh-elicited B cells
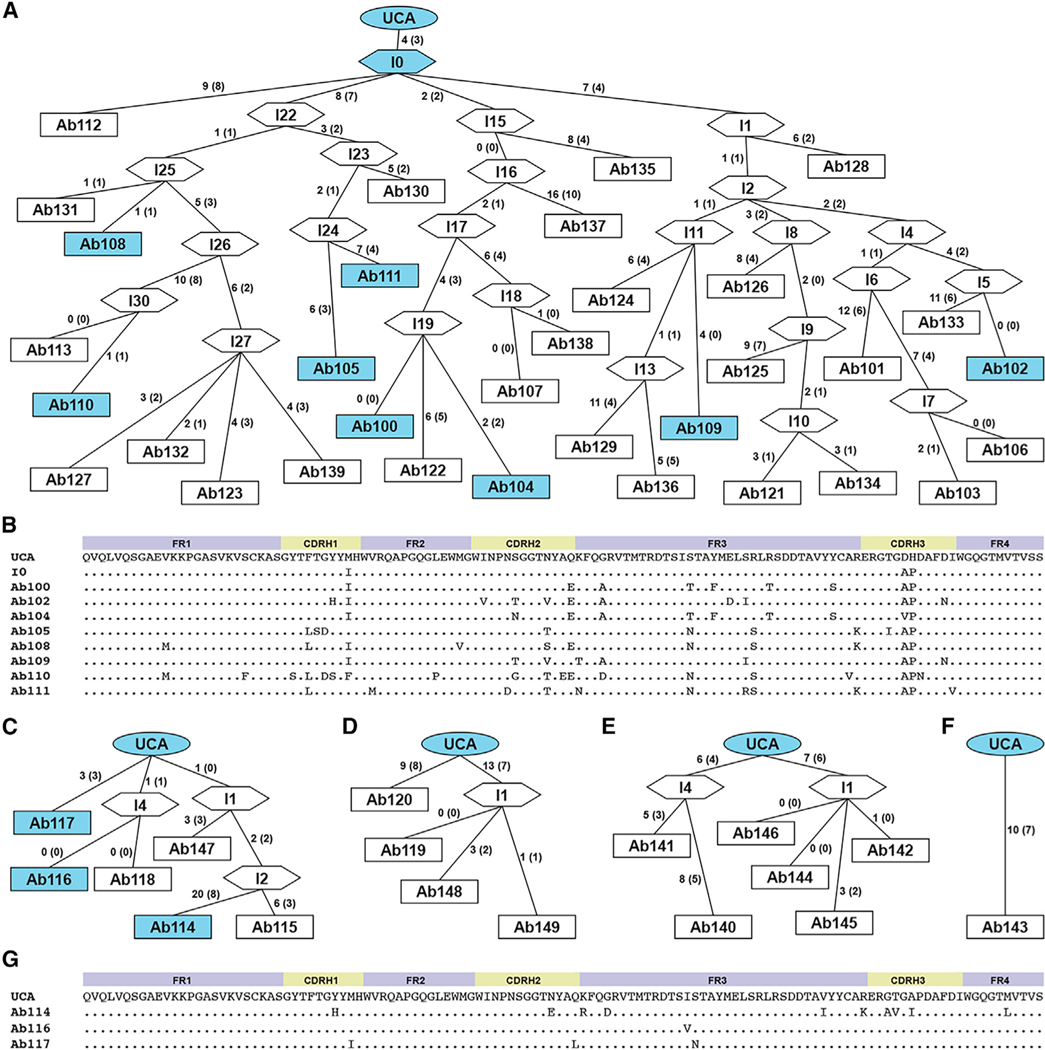
We identified several clonal lineages of rsHAtCh-elicited RBS-directed B cells (Figures S3A–S3D) and inferred their unmutated common ancestors (UCAs). All had human (hu)DH7–27 and huJH3 heavy chains paired with mouse (ms) VK3–12 and msJK2 light chains. In addition to identical gene usage, multiple UCAs from different mice had very similar sequences. Indeed, lineage 114 and 143 and 119 and 140 UCAs have identical heavy- and light-chain sequences (Figure S3E). Lineage 100 was the largest clonal population isolated from a single mouse, with 33 members, containing an average of 18 ± 4 amino acid mutations across its heavy and light chains relative to the UCA (Figures 4A, 4B, S4A, and S4B). Lineages 114, 119, and 140 had lower numbers of mutations with 7 ± 4, 10 ± 2, and 11 ± 4 amino acid changes relative to their UCAs, respectively (Figures 4C–4G and S4C–S4G). Lineage 143 contains a single orphan B cell, which has 10 amino acid changes relative to its UCA. Similar gene usages and identical UCAs from multiple mice further support the finding that boosting immunization with rsHAtCh reactivates genetically restricted populations of RBS-directed memory B cells in H1-primed (imprinted) mice.
Figure 4. Heavy-chain affinity maturation of elicited RBS-directed lineages.
Sequence analysis of RBS-directed lineages isolated from three mice immunized with rsHAtCh.
(A) Genealogy tree of lineage 100 antibody heavy chains. Nucleotide mutations between each branchpoint labeled, with corresponding number of amino acid mutations in parentheses. Lineage UCA, computationally inferred intermediates, and isolated IgG+ B cells shown as ovals, hexagons, and rectangles, respectively. Antibodies in blue were expressed recombinantly for binding analysis.
(B) Alignment of VH amino acid sequences of lineage 100 antibodies, including the UCA and I-0 branchpoint; “.” denotes conservation to the UCA.
(C–F) Genealogy trees of lineages (C) 114, (D) 119, (E) 140, and (F) 143.
(G) Alignment of VH amino acid sequences of lineage 114 antibodies and UCA.
Related to Figure S4.
RBS-directed Ab binding reflects bulk serum data
We next expressed 11 RBS-directed rsHAtCh-elicited Abs as Fabs and obtained binding affinities to a panel of HAs containing 8 historical H1 HAs and the 3 rsHAtCh components (Table 1). Ten of the eleven Abs bound H1 SI-06, H1 NC-99, and the three rsHAs with low- or submicromolar affinity. Additionally, Ab109 and Ab114, from lineages 100 and 114, respectively, had lower but measurable affinities for the historical H1 MA-90 and FL-93. While the affinity of an Ab for each of the rsHAtCh components can differ by up to two orders of magnitude, all still had affinities in the low- to submicromolar range. We also observed a wide range of affinities between lineage UCAs and our HA panel. Lineage 119, 140, and 143 UCAs had submicromolar affinities as Fabs for two of the three rsHAtCh components and H1 SI-06, from which the graft is based, while the lineage 100 UCA had undetectable affinity for any of the HAs tested. The observed variability in affinities for the affinity-matured Abs and lineage UCAs was primarily a consequence of differences in dissociation rate; the association rates were similar (Figures S5A and S5B). These data suggest that RBS-directed B cells with a range of affinities for H1 SI-06 or rsHAtCh are initially recruited. Furthermore, the overall pattern of high affinities for H1 NC-99 and H1 SI-06, and low affinity for other historical H1 isolates, reflects the bulk serum reactivity profiles, indicating that similar RBS-directed responses are present in both the serum and the expanded B cell compartments.
Table 1.
Breadth of rsHAtCh-elicited RBS-directed antibodies
| USSR77 | KW86 | MA90 | FL93 | BE95 | NC99 | SI06 | CA09 | rsH3 | rsH4 | rsH14 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab100 | >100 | >100 | >100 | >100 | >100 | 0.76 | 0.82 | >100 | 1.2 | 1.2 | 0.10 |
| Ab102 | >100 | >100 | >100 | >100 | >100 | <0.02 | <0.02 | >100 | 0.05 | 0.14 | <0.02 |
| Ab104 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 | >100 |
| Ab105 | >100 | >100 | >100 | >100 | >100 | 0.19 | 0.19 | >100 | 0.57 | 3.9 | 0.22 |
| Ab108 | >100 | >100 | >100 | >100 | >100 | 0.23 | 0.16 | >100 | 0.38 | 0.93 | 0.05 |
| Ab109 | >100 | >100 | 48 | >100 | >100 | 0.04 | 0.03 | >100 | 0.10 | 0.57 | 0.03 |
| Ab110 | >100 | >100 | >100 | >100 | >100 | 0.04 | 0.06 | >100 | 0.14 | 0.38 | 0.04 |
| Ab111 | >100 | >100 | >100 | >100 | >100 | 0.03 | 0.02 | >100 | 0.06 | 0.62 | 0.02 |
| Ab114 | >100 | >100 | >100 | 22 | >100 | 0.06 | 0.49 | >100 | 0.43 | 1.1 | <0.02 |
| Ab116 | >100 | >100 | >100 | >100 | >100 | 1.1 | 2.8 | >100 | 1.0 | 2.0 | 0.11 |
| Ab117 | >100 | >100 | >100 | >100 | >100 | 0.12 | 0.12 | >100 | 0.47 | 1.0 | 0.17 |
| Lin100 UCA | N.D. | N.D. | >100 | >100 | >100 | >100 | >100 | N.D. | >100 | >100 | >100 |
| Lin100 I-0 | N.D. | N.D. | >100 | >100 | >100 | 41 | >100 | N.D. | 30 | >100 | 11 |
| Lin114 UCA | N.D. | N.D. | >100 | >100 | >100 | 23 | 36 | N.D. | 23 | >100 | 5.8 |
| Lin119 UCA | N.D. | N.D. | >100 | >100 | >100 | 0.26 | 0.44 | N.D. | 0.69 | 1.5 | 0.09 |
| Lin140 UCA | N.D. | N.D. | >100 | >100 | >100 | 0.29 | 0.5 | N.D. | 0.67 | 1.6 | 0.1 |
| Lin143 UCA | N.D. | N.D. | >100 | >100 | >100 | 0.53 | 0.7 | N.D. | 0.67 | 6.8 | 0.37 |
rsHAtCh-elicited antibodies and computationally inferred lineage UCAs assayed for binding to a panel of historical H1s. KD values are for Fabs and monomeric HA heads expressed in μM. Lineages of clonally related mature antibodies labeled. N.D. is not determined, and >100 μM is beyond the limit of detection; Fabs with affinity are in bold for ease of viewing. At least four independent concentrations of analyte were used to obtain reported KD values. Related to Figure S5.
A dominant B cell lineage engages the H1 RBS through germline-encoded contacts
To understand how Abs from the largest clonal lineage engaged the RBS, we determined cryoelectron microscopy (cryoEM) structures of Ab109 and Ab111 bound to H1 SI-06 HA FLsE at ~3.4 and ~5 Å, respectively (Figures 5A, 5B, and S6A). Both Abs engage the RBS similarly. We restricted our subsequent analyses to the higher-resolution Ab109 structure. Ab109 engages H1 SI-06 with a paratope footprint that encompasses all four segments of the grafted RBS epitope. The LCDR1 spans residues across the 130, 150, and 220 loops, and both the LCDR3 and HCDR3 engage the 190 helix (Figure 5C). There are several key binding interactions: Arg96 in the LCDR3 and Glu99 in the HCDR3 engage HA residues Asp190 and Arg192, respectively. These interactions result in salt bridges that “straddle” the 190 helix, while LCDR1 makes a series of hydrogen-bonding contacts within the 150 loop (Figures 5D and 5E). The tip of the HCDR3 loop (GTGAP) has few contacts with HA; the conserved proline introduces a kink in the loop conformation, which, along with the small surrounding side chains, may help prevent steric clashes between HCDR3 and HA. Comparing the binding footprints of Ab109 with sialic acid as well as two human bnAbs, H2526 and 641I-9, show a distinct feature of RBS engagement. While sialic acid and human bnAbs contact the conserved core of the RBS pocket, Ab109 does not contact the invariant core residues (Figures 5F–5I).
Figure 5. rsHAtCh-elicited antibodies engage the RBS through germline-encoded contacts.
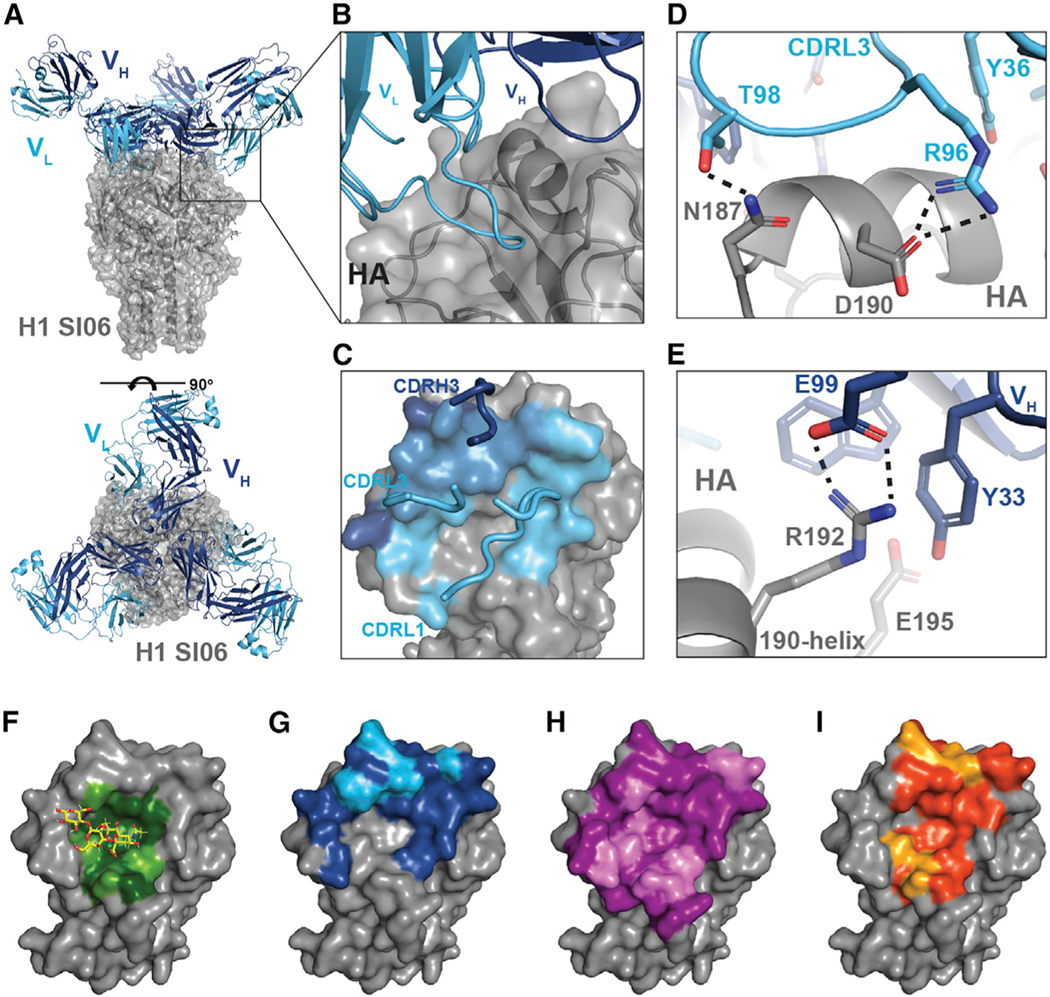
(A) cryoEM structure of Ab109 Fab in complex withH1 SI06 FLsE.
(B) Ab109-H1 SI-06 interface.
(C) Cartoon representation of Ab109 footprint on hemagglutinin (HA); heavy-chain contacts shown in dark blue, both heavy- and light-chain contacts shown in light blue, and light-chain contacts shown in cyan.
(D and E) Key contacts made between the CDRL3 (C) and CDRH3 (D) loops and the HA 190-helix. CDRH1 loop removed for clarity. Light-chain residues shown in cyan, heavy-chain residues shown in dark blue, and HA shown in gray. Salt bridges shown as dotted lines.
(F–I) Footprints of (F) sialic acid, (G) Ab109, (H) H2526, and (I) 641I-9; colored residues are located at the RBS-ligand interface, with pale green/cyan/violet/yellow residues forming hydrogen bonds or salt bridges with the corresponding ligand. All images generated using PyMol.
RBS-directed Abs neutralize strain-matched H1 virus and confer protection
A subset of the RBS-directed Fabs screened in BLI were then selected for microneutralization analysis against a panel of historical H1 viruses ranging from 1934 to 2009 (Figure 6A). Five of the seven Abs neutralized H1 SI-06, four at concentrations of <10 μg/mL. There was no detectable neutralization of new pandemic or pre-2001 viruses for the Abs tested, indicating that rsHAtCh immunization elicits Abs that neutralize only strain-matched viruses. The positions of the LCDR1 and LCDR3 of Ab109, relative to the G189R, G156E/K, and K133a mutations found in H1 MA-90, H1 BE-95, and H1 CA-09, suggest that the observed lack of binding and neutralization breadth is likely due to steric clashes in the RBS periphery (Figure 6B).
Figure 6. rsHAtCh-elicited RBS-directed antibodies neutralize strain-matched virus and partially protect.
(A) Microneutralization titers of seven antibodies tested against a panel of eight historical H1 virus isolates ranging from 1934 to 2009. Titers are reported as the maximal concentration (ug/mL) of each antibody at which complete neutralization was observed.
(B) Cartoon representation of Ab109 footprint on HA; heavy-chains contact shown in dark blue, both heavy- and light-chain contacts shown in light blue, and light-chain contacts shown in cyan. CDR L1, L3, and H3 loops are labeled. Residue changes across historical H1 isolates expected to abrogate binding shown in red, labeled with key mutations found in either H1 MA-90, H1 FL-93, H1 BE-95, or H1 CA-09.
(C) Protection studies following NC-99 infection in cDC-IrF5−/− knockout mice (N = 5 PBS control, N = 6 Ab111 cohorts). The y axes show percentage of weight change from initial weight (morbidity, left) and percentage of survival (mortality, right); *p < 0.01 compared with PBS control using the Mantle-Cox test.
We also carried out in vivo protection studies in mice against lethal H1 NC-99 virus challenge. As H1 NC-99 infection is not normally lethal in mice, we carried out these experiments in a recently characterized cDC-IRF5 knockout mouse line that is susceptible to H1 NC-99 lethal challenge.41 Passive transfer of Ab111 as murine IgG2a or IgG2a with the effector-function-ablating LALA-PG Fc mutations resulted in partial protection against an otherwise lethal infection (Figure 6C).42 IgG2a was selected over IgG2c, despite their significant homology, due to the extensive body of work demonstrating the efficacy of effector-function-ablating mutations in the IgG2a isotype.43 Notably, we found that Ab111 failed to protect as IgG1 (Figures 6B and 6C). This suggested isotype-dependent protection that was also independent of the LALA-PG mutation.
DISCUSSION
We show here that host immune responses against H1 HA, in the context of preexisting exposure to a WT H1, can be expanded to target a single site using an epitope-enriched immunogen; this expansion occurred in both the humoral and cellular compartments. The functional benefit of presenting multiple copies of the grafted RBS likely stems from an avidity advantage for RBS-directed B cells relative to those that recognize non-enriched epitopes. The low non-RBS-directed H1 serum titer and epitope focusing on the immunogen-expanded H1+/IgG+ B cell repertoire indicate altered patterns of dominance rather than a magnification of the WT response to reach the observed RBS-directed titer. Isolated RBS-directed Abs were members of similar, and in some cases identical, clonal lineages, which made germline-encoded heavy- and light-chain contacts with the grafted H1 RBS epitope. These Abs neutralized strain-matched virus and conferred partial protection against a lethal H1 NC-99 influenza challenge. The lack of protection from the IgG1 isotype and the equivalent protection seen with the IgG2a isoforms, with and without effector-function-ablating mutations, suggests a mechanism of protection that is both isotype dependent and effector-function independent within the context of the assay.42,43
While the HC2 locus provides human-like diversity in HCDR3, our selection of IGHV1–2 as the repertoire VH sequence may have inadvertently played a role in biasing the RBS-directed response.30,31 Although the entire naive repertoire contained the VH1–2 gene of CH65, a human sialic acid (SA)-mimicking RBS-directed Ab, and the full complement of human DH and JH genes, we did not identify any Abs with canonical sequence features of SA mimicry: both long HCDR3s with the characteristic hydrophobic-acidic dipeptide and huJH6 genes were absent in the immunogen-expanded repertoire.10,44–47 Despite observing an SA-mimicking Pro-Asp HCDR3 motif in most rsHAtCh-elicited Abs, structural studies showed primarily light-chain contacts within the grafted RBS; heavy-chain contacts were made on the opposite side of the 190 helix. Selection of H1 SI-06 as the RBS graft may explain the failure to elicit JH6-containing Abs, as JH6 encodes a poly-tyrosine motif that engages the Lys133a residue found in many H1 antigenic clusters but not in SI-06.10,11 As the two key salt-bridging contacts in the HCDR3 and LCDR3 were germline encoded, all B cells containing the VH1–2 and VK3–12 genes had the potential to make similar contacts. Thus, the combination of RBS-directed precursors having both increased avidity against the RBS-enriched immunogen and increased affinity due to germline-encoded contacts is the most likely explanation for the dominance of RBS-directed responses in this model system.16,30 Importantly, our results also showed that the precursor frequency of RBS-specific Abs was low in the IGHV1–2 HC2 mouse and similar to the value found in WT C57Bl/6. This provides proof of concept that structure-based epitope enrichment can be applied as an immunogen design strategy to focus otherwise low-frequency responses to the conserved RBS.
Contemporary immunogen design strategies attempt to engineer antigens that preferentially target conserved subdominant epitopes.17,24,27,28,48,49 A design strategy that has become increasingly prevalent is heterologous display, where multiple unique antigens are presented on a single immunogen.21,28,50,51 B cells engaging conserved epitopes between heterologous antigens have a putative avidity advantage relative to those specific for only a single antigen and thus may have a competitive edge in the germinal center (GC) reaction. However, when simultaneously presenting several WT proteins, there are many additional epitopes with varying degrees of conservation, which can lead to “off-target” epitope focusing. Indeed, previous work has shown that heterologous display of WT H1 HA heads on mosaic nanoparticles focuses not to the RBS but rather to the lateral patch, a similarly conserved epitope on the HA head.28,29 Both the RBS and lateral patch epitopes were conserved, yet responses preferentially targeted the lateral patch, suggesting that enrichment of multiple epitopes may impact the extent of immunofocusing to any specific one. While mosaic nanoparticles show significant promise as next-generation vaccine candidates, heterologous display may be more specifically applied to focus to a single region. Epitope enrichment, as demonstrated here, is simply a form of heterologous display optimized for a single epitope.
Preexisting immunity remains a key immunologic hurdle to overcome in the development of next-generation influenza vaccines, as previous exposures influence subsequent immune responses.1,52–56 Our use of rsHAtCh after an initial exposure to a WT HA is a proof of concept for epitope-enriched immunogens in the context of preexisting immunity. Immunologic imprinting from WT HAs, from vaccination or infection, imparts not just Ab reactivity but also an epitopic distribution, in which conserved epitopes like the RBS are often subdominant. Specific expansion of RBS-directed B cells suggests that epitope enrichment can be used to elevate subdominant responses within a previously imprinted repertoire. While the rsHAtCh elicited scaffold-specific H3-, H4-, and H14-directed responses, we observe expansion specifically of H1+ RBS-directed B cells rather than other H1-directed responses imprinted by the prime. The antigenically distinct HAs are analogous to orthogonal protein scaffolds used to present neutralizing epitopes, as shown previously for respiratory syncytial virus (RSV) F protein.57,58
Epitope focusing is a necessary step for an eventual improved vaccine, as influenza virus can readily escape monoclonal pressure from even potent bnAbs.4,59 Epitope enrichment represents a viable design strategy moving forward as rsHAtCh demonstrates a level of immunofocusing to any HA epitope including the stem that, to our knowledge, has not been previously achieved. However, as previous studies and our current data have shown, engaging the RBS epitope does not guarantee significant breadth.60 Thus, future RBS-eliciting immunogens should not only expand RBS-directed populations but should also confer breadth. To that end, in an accompanying study, we complement the initial characterization of rsHAtCh described here to additionally analyze primary and secondary GCs following rsHAtCh immunization in partially humanized JH6 mice. The enrichment of RBS-directed B cells in primary GCs, and the elicitation in secondary GCs of receptor-mimicking RBS-directed B cells with enhanced breadth, illustrates how multiple rounds of exposure to an epitope-enriched immunogen can both expand and increase the breadth of responses. Understanding how antigenic imprinting alters the effects of epitope enrichment is a goal of future studies.
We have established an adaptable platform for generating RBS-enriched heterotrimeric HAtCh constructs using antigenically distinct HAs as molecular scaffolds to present the RBS epitope. This first-generation construct illustrates how rationally designed epitope-enriched immunogens can focus humoral and cellular responses to subdominant epitopes while preventing reactivity to off-target, canonically dominant ones. Broadly protective or universal influenza vaccines will likely need to elicit Ab responses against multiple conserved epitopes. Epitope enrichment, as described here, is likely translatable to other HA epitopes, such as the conserved stem and head interface. More broadly, this approach can be extended to relevant conserved sites on antigens from other rapidly evolving pathogens.
Limitations of the study
The RBS-directed repertoire preferentially elicited by rsHAtCh immunization contained a population of B cells with germline-encoded affinity for the rsHA components of the rsHAtCh immunogen, which complicates determining the degree of immunofocusing induced without a germline-encoded bias. By extension, the degree to which broadly reactive, RBS-directed Abs are elicited by an RBS-enriched immunogen in this partially humanized mouse model cannot be fully assessed. Furthermore, characterizing how such an epitope-enriched immunogen might elicit RBS-focused immune responses in the context of complex influenza immune histories imprinted by repeated vaccinations or infections will be necessary.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aaron G. Schmidt (aaron.schmidt@mgh.harvard.edu).
Materials availability
All unique/stable reagents generated in this study will be made available on request, but we may require a payment and/or a completed materials transfer agreement if there is potential for commercial application. For non-commercial use, all unique/stable reagents generated in this study are available from the lead contact with a completed materials transfer agreement. There are restrictions to the availability of the transgenic mice due to an MTA with Bristol-Myers Squibb.
Data and code availability
CryoEM data have been deposited in the Electron Microscopy DataBase and PDB and are available as of the date of publication. B cell receptor sequences have been deposited in GenBank. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| HRP-conjugated rabbit anti-mouse IgG antibody | Abcam | CAT#ab97046 |
| Ly-6G-APCe780 | ThermoFisher | CAT#14-5931-82 |
| F4/80-APCe780 | ThermoFisher | CAT#47-4801-82 |
| CD3-BV785 | BioLegend | CAT#100231 |
| CD19-BV421 | BioLegend | CAT#115549 |
| IgM-BV605 | BioLegend | CAT#406523 |
| IgG-PerCP/Cy5.5 | BioLegend | CAT#405314 |
| msIgG1 isoforms of Ab100, Ab102, Ab109, Ab110, Ab111, Ab114, Ab116 | This paper | N/A |
| Ab111 IgG2a isoform | This paper | N/A |
| Ab111 IgG2a LALA-PG isoform | This paper | N/A |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| A/Michigan/45/2015 (H1N1) | Duke RBL | Lot 1483-180124 |
| A/California/07/2009 (H1N1) | Duke RBL | Lot 13659-20141211 |
| A/Solomon Islands/3/2006 (H1N1) | Duke RBL | Lot 331-05132014 |
| A/Hawaii/15/2001 (H1N1) | Duke RBL | Lot FR-394-20200928 |
| A/New Caledonia/20/1999 (H1N1) | IRR / Duke RBL | Lot FR-395-20200928 |
| A/Beijing/262/1995 (H1N1) | Duke RBL | Lot 12277-16524 |
| A/Texas/36/1991 | Duke RBL | Lot 1096-20200807 |
| A/Puerto Rico/8/1934 | Duke RBL | Lot CR-05282014 |
|
| ||
| Chemicals peptides and recombinant proteins | ||
|
| ||
| rsHAtCh (rsH4, rsH3, rsH14 heterotrimer) | This paper | N/A |
| rsH14 monomeric head domain | Produced in house; Bajic et al.27 | N/A |
| rsH3 monomeric head domain | This paper | N/A |
| rsH4 monomeric head domain | Produced in house; Bajic et al.27 | N/A |
| GABH heterodimeric coiled coil (acidic and basic strands) | Produced in house Keating et al.61 |
N/A |
| H2227 | Produced in house Schmidt et al.56 |
N/A |
| H2526 | Produced in house Schmidt et al.56 |
N/A |
| CH67 | Produced in house Whittle et al.37 |
N/A |
| 641 I-9 | Produced in house Schmidt et al.56 |
N/A |
| C05 | Produced in house Ekiert et al.47 |
N/A |
| HRV 41.1 | Produced in house Schmidt et al.56 |
N/A |
| Full length soluble ectodomains of H1 MA-90, H1 BE-95, H1 NC-99, H1 SI-06, and H1 CA-09 | Produced in house | N/A |
| H1 SI-06 FLsE ΔRBS | Produced in house Lingwood et al.39 |
N/A |
| Monomeric HA head domains of H1 USSR-77, KW-86, MA-90, FL-93, BE-95, NC-99, SI-06, CA-09 | Produced in house Schmidt et al.56 |
N/A |
| Ab100 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251202 / OP251203 |
| Ab102 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251204 / OP251205 |
| Ab104 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251206 / OP251207 |
| Ab105 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251208 / OP251209 |
| Ab108 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251210 / OP251211 |
| Ab109 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251212 / OP251213 |
| Ab110 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251214 / OP251215 |
| Ab111 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251216 / OP251217 |
| Ab114 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251218 / OP251219 |
| Ab116 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251220 / OP251221 |
| Ab117 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251222 / OP251223 |
| Lin100 UCA (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251224 / OP251225 |
| Lin100 (VH/VL sequence from IGHV1-2 HC2 mouse)I-0 | This paper | OP251226 / OP251227 |
| Lin114 UCA (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251228 / OP251229 |
| Lin119 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251230 / OP251231 |
| Lin140 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251232 / OP251233 |
| Lin143 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251234 / OP251235 |
| Pierce HRV 3C protease | ThermoScientific | CAT#88946 |
| Sigma Adjuvant System | SigmaAldrich | CAT#S6322 |
| Streptavidin-PE conjugate | Invitrogen | CAT#12-4317-87 |
| Aqua Live/Dead amine-reactive dye | ThermoFisher | CAT#L34957 |
| Calcein, AM, cell-permeant dye | ThermoFisher | CAT#C3100MP |
| Streptavidin-APC/Cy7 conjugate | BioLegend | CAT#405208 |
| Streptavidin-BV650 conjugate | BioLegend | CAT#405232 |
| StrepTactin-PE conjugate | IBA Lifesciences | Item#6-5000-001 |
| StrepTactin-APC conjugate | IBA Lifesciences | Item#6-5010-001 |
| RNaseOUT | ThermoFisher | CAT#10777019 |
| TALON Metal Affinity Resin | Takara | CAT#635652 |
| Pierce Protein G Agarose | ThermoFisher | CAT#20399 |
| 1-Step ABTS substrate | ThermoFisher | Prod#37615 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Ni-NTA Biosensors | ForteBio | Item#18-5101 |
| SuperScript IV VILO MasterMix | ThermoFisher | CAT#11756050 |
| MagMax mirVana Total RNA isolation kit | ThermoFisher | CAT#A27828 |
|
| ||
| Deposited data | ||
|
| ||
| Electron density map, Ab109 and H1 SI-06 FLsE | This paper | EMDB: EMD-26605 |
| Structure, Ab109 and H1 SI-06 FLsE | This paper | PDB: 7UMM |
| Electron density map, Ab111 and H1 SI-06 FLsE | This paper | EMDB: EMD-26604 |
| Isolated B cell antibody sequences | This paper | GenBank: OP251202 - OP251235 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID: CVCL_D615 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| cDC-IrF5−/− mice | Yousif et al.41 | N/A |
| IGHV1-2 HC2 mice | Sangesland et al.30 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Murine B cell receptor sequencing primers | Tiller et al.; Rohatgi et al.,62,63 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10 | TreeStar |
https://www.flowjo.com; RRID: SCR_008520 |
| Prism v9 | GraphPad |
https://www.graphpad.com; RRID: SCR_002798 |
| IMGT | International ImMunoGeneTics Information System |
http://www.imgt.org; RRID: SCR_012780 |
| Cloanalyst | Kepler et al.64 | https://www.bu.edu/computationalimmunology/research/software/ |
| RELION | Scheres et al.65 | https://relion.readthedocs.io/en/release-3.1/ |
| Chimera | Pettersen et al.66 | https://www.cgl.ucsf.edu/chimera/ |
| PHASER | McCoy et al.67 | http://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| PHENIX | Adams et al.68 | https://www.phenix-online.org/ |
| COOT | Emsley et al.69 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMol | Schrödinger | http://www.pymol.org |
| MolProbity | Chen et al.70 | http://molprobity.biochem.duke.edu |
| XDS | Kabsch et al.71 | https://xds.mr.mpg.de |
|
| ||
| Other | ||
|
| ||
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cytiva 28-9909-44 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
IGHV1–2 HC2 mice were deployed for immunization experiments and were a previously established transgenic model in which antibody VH usage is constrained to IGHV1–2*02, while allowing for recombination with diverse human D and J segments, enabling a human-like HCDR3 repertoire.30 For passive transfer of antibodies, cDC-IrF5−/− mice were used.41 Female and male animals aged 6–10 weeks were used in our study and were maintained within Ragone Institute’s HPPF barrier facility. Animal experiments were conducted with institutional IACUC approval (MGH protocol 2014N000252).
Cell lines
Mammalian cell lines FreeStyle 293F cells (Thermo Fisher Cat#R79007, female) and Expi293F cells (Thermo Fisher Cat#A14527, female) were cultured in accordance with the manufacturer’s instructions. Insect cell lines Hi5 (BTI-TN-5B1–4, female) (Trichoplusia ni) and Sf9 (Spodoptera frugiperda, female) were cultured in EX-CELL 405 SFM (Sigma, Cat# 14405C) and Sf-900 II SFM (Gibco, Cat# 10902096) respectively following the manufacturer’s instructions.
METHOD DETAILS
Immunogen expression and purification
rsH3, rsH4, and rsH14 HA head constructs were derived from A/long-tailed duck/Wisconsin/10OS3918/2010 (H14 WI-10), A/American black duck/New Brunswick/00464/2010(H4 NB-10), and A/HongKong/4801/2014 (H3 HK14) scaffolds resurfaced with the H1 SI-06 RBS described previously.27 All constructs were codon optimized by Integrated DNA Technologies (IDT) and purchased as gBlocks that were subsequently cloned into pVRC and sequence confirmed via Sanger sequencing from Genewiz. To generate the rsHAtCh construct, each rsHA component was subcloned into a pVRC protein expression vector containing individual components of the non-collagenous domain 2 (NC2)-derived heterotrimerization motif. Each component contained a rsHA, strand of the hetero-trimerization tag, and either 8xHis, FLAG, or streptavidin binding protein (SBP) tag. Fragments were amplified via PCR with primers to introduce overlapping P2A sequence regions, and all three fragments were combined with linearized pVRC vector via Gibson assembly into the final expression construct.
HEK293F cells (ThermoFisher) were used to express rsHAtCh. Transfections were performed with PEI per the manufacturer’s protocol. rsHAtCh protein was expressed via transient transfection in HEK293F cells for 5 days, and subsequently purified at 4°C. Culture supernatants were clarified via centrifugation and purified over Cobalt-TALON affinity resin (Takara), with subsequent size exclusion chromatography (SEC) purification over a Superdex 200 Increase (Cytiva) column equilibrated in a running buffer of 10mM Tris, 150mM NaCl, pH 7.5. rsHAtCh-containing fractions were then passed over G1 anti-FLAG-agarose resin (Genscript) equilibrated in 10mM Tris, 150mM NaCl, pH 7.5, incubated for 1 h while rotating, and eluted with 50mM Tris, 150mM NaCl, containing 160μg/mL 1xFLAG (DYKDDDDK) peptide (ApexBio) pH 7.5 following a 1-h incubation again with mild agitation. The sample was then purified a third time over Streptavidin-agarose resin (Pierce) preequilibrated in 10mM Tris, 150mM NaCl, pH 7.5, incubated for 1 h rotating, washed with 20mM HEPES, 150mM NaCl, 5% glycerol, pH 7.0, and eluted with 20mM HEPES, 150mM NaCl, 4mM biotin, 5% glycerol pH 7.0 after a 1-h incubation. The sample was concentrated via centrifugation and all affinity and heterotrimerization tags were removed via HRV 3C protease (Thermo) cleavage overnight. The sample was purified a final time via SEC equilibrated in PBS, and the trimeric peak was harvested and concentrated to 2.0mg/mL for immunizations.
HA expression and purification
rHA ‘head’ and full length soluble ectodomain (FLsE) constructs were cloned into pFastBac expression vectors for insect cell (Hi5) production; HA ΔRBS FLsE constructs were cloned into pVRC protein expression for expression in HEK 293F or Expi293F cells. FLsE constructs contain a 3C-cleavable foldon, 8xHis, and streptavidin binding protein (SBP) tag, HA head constructs contain a 3C-cleavable 8xHis and SBP tag. All constructs were sequenced through Genewiz Inc. All constructs were purified from clarified supernatants using Cobalt-TALON resin (Takara) and size exclusion chromatography over a Superdex 200 Increase (Cytiva) equilibrated in 10mM Tris pH 7.5, 150 mM NaCl. Affinity tags were removed via HRV 3C protease (ThermoScientific) cleavage, and HA was repurified using additional resin to remove un-cleaved protein, tags, and 3C protease. rHAs used in this study include H1 A/Solomon Islands/03/2006, H1 A/Massachusetts/1/1990, H1 A/Florida/2/1993, H1 A/Beijing/262/1995, H1 A/New Caledonia/20/1999, H1 A/Solomon Islands/03/2006, H1 A/California/07/2009, H4 A/American black duck/New Brunswick/00464/2010, H14 A/mallard/Wisconsin/10OS3941/2010, H2 A/Japan/305/1957, H3 A/Hong Kong/4801/2014, H5 A/gyrfalcon/Washington/41088–6/2014, and H6 A/chicken/CA/S0403106/2004.
Fab and IgG expression and purification
The variable heavy and light chain genes for all antibodies studied were codon optimized by Integrated DNA Technologies (IDT), purchased as gblocks, and cloned into pVRC constructs containing the desired constant domains as previously detailed.10,56 The Fab heavy chain vector has a C-terminal HRV 3C-cleavable 8xHis tag. The same transfection and purification protocol for rHAs was used for Fabs/IgGs.
Biolayer interferometry
Biolayer interferometry (BLI) experiments were performed on a BLItz instrument (Fortebio) loaded with Ni-NTA biosensors (Fortebio). All proteins were diluted in 10mM Tris, 150mM NaCl, pH 7.5. HA proteins were immobilized to the biosensors via their 8xHis tags, and 3C-cleaved Fabs were used as the analytes. To determine binding affinities, measurements at four of more concentrations were performed. Vendor-supplied software was used to generate a final KD estimate via a global fit model with a 1:1 binding isotherm. All binding studies reported were carried out in 10mM Tris, 150mM NaCl, pH 7.5 at room temperature.
Immunizations
IGHV1–2 HC2 mice were immunized intraperitoneally, first with H1 SI-06 FLsE and then 2x sequentially with either H1 SI-06 FLsE or rsHAtCh, each at Days 21 and 42. Mice were randomly assigned into either the control immunization or experimental immunization cohort. At each immunization point, the animals were inoculated with 15ug of H1 SI-06 FLsE or equimolar rsHAtCh in a 100ul inoculum containing 50% w/v Sigma adjuvant, as performed previously.30,31,72 The animals were bled before the immunization and then during the regimen, at days 14, 35, and 56.
Serum ELISAs
Serum ELISAs were performed using 96-well, clear, flat-bottom, high bind microplates (Corning). These plates were coated with 100 mL of protein, which were adjusted to a concentration of 5 mg/mL (in PBS). Plates were incubated overnight at 4°C. After incubation, plates had their coating solution removed and were blocked using 1% BSA in PBS with 1% Tween. This was done for 60 min nutating at room temperature. This blocking solution was removed, and sera was diluted 40-fold in PBS. A 4-fold serial dilution was then performed. CH67 IgG, similarly serially diluted (4-fold) from a 5 mg/mL starting concentration, was used as a positive control. 40 μL of primary antibody solution was used per well. Following this, samples were incubated for 90 min at room temperature. Plates were rinsed three times using PBS-Tween. 150 μL of HRP-conjugated rabbit anti-mouse IgG antibody, sourced commercially from Abcam (1:20,000 dilution in PBS), was used for the secondary incubation, performed for one hour, similarly nutating at room temperature. Plates were then rinsed three times using PBS-Tween. 150 μL of 1xABTS development solution (ThermoFisher) was used according to the manufacturer’s protocol, and stopped after 30 min using 100 μL of a 1% SDS solution, and plates were read using a SectraMaxiD3 plate reader (Molecular Devices) at 405 nm.
Protection studies
cDC-IrF5−/− mice41 were infused with 5mg/kg of antibody via intraperitoneal injection. One hour later, the mice were anesthetized by isoflurane inhalation and then intranasally infected with a 2x LD50 dose of H1 New Caledonia/20/1999 (H1 NC-99) influenza virus. H1 NC-99 propagates within C57Bl/6 mice, however, it has not been possible to generate >1xLD50 within the WT genotype.30,72,73 However, we find that a 2xLD50 infecting dose of H1 NC-99 can be achieved in cDC-IrF5−/− mice (TCID50 = 108.94) and consequently we deployed the cDC-IrF5−/− genotype to evaluate antibody-based protection against this influenza viral strain. Animal body weight was measured daily, and the mice were humanly euthanized when they reached 20% body weight loss.
Flow cytometry and B cell sorting
Mouse spleens were obtained from both naive and immunized mice (day 56), suspended in PBS and then gently sieved through a 70μM cell strainer. The red blood cells were then lysed via suspension in ACK lysis buffer (Lonza) and the remaining splenocytes were washed in PBS. To isolate the antigen specific IgG B cells expanded by vaccination, the splenocytes were first stained with Aqua live/dead amine-reactive dye along with calcein. After incubation, the cells were further stained with an fluorescent antibody panel [CD3-BV785; IgM-BV605; CD19 BV421; IgG- PerCP/cy5.5] that also contained 0.25ug of two positive selector H1 FLsE probes [H1SI(06)-PE; H1SI(06)-APC-Cy7] and 0.25ug of a negative selector H1 FLsE [(H1 SI(06) ΔRBS-APC, where the RBS was sterically occluded by an N-linked glycan a position 19039)]. Each of H1 FLsE probes contained the Y95F mutation to prevent binding to cell surface sialyl-oligosaccharide.38 In immunized animals, the immunogen-expanded IgG B cells specific for the RBS were defined as CD3−/CD19+/IgM−/IgG+/H1-PE+/H1-APC-Cy7+/H1ΔRBS-APC−. In naive animals off-target germline B cells were defined as CD3−/CD19+/IgM+/IgG−/H1-PE+/H1ΔRBS-APC+ and on-target germline B cells were defined as CD3−/CD19+/IgM+/IgG−/H1-PE+/H1-ΔRBS-APC−/H1-APC-Cy7+. In all cases, single cells were sorted (FACS Aria Fusion Sorter, BD Biosciences) into 96 well plates containing RLT lysis buffer with 1% BME and immediately frozen and stored at −80°C. A representative gating strategy is shown in Figure S2C.
BCR sequencing
Single cell BCRs were sequenced according to our established method.30,31,74 We first generated BCR libraries from whole transcriptome amplification (WTA) products using the Smart-Seq2 protocol.74 BCR sequences from heavy and light chains were enriched from single B cells using a pool of partially degenerate V region specific primers against all possible human IGHV or mouse IGLV and IGKV segments (FR1 region) and reverse primers against the heavy or light chain constant regions.30 Step out PCR was then applied to add cellular barcodes and Illumina sequencing adapters (based on Nextera XT Index Adapters) to each heavy and light chain.30 The samples were then pooled and sequenced using paired end V2–500 cycle Illumina Miseq System (250 × 250 read and 8 × 8 index reads). The reads were demultiplexed and the BCR heavy and light chains were reconstructed and paired using PandaSeq,75 and aligned against the human IMGT database76 with sequencing error correction using MigMAP, a wrapper for IgBlast (https://github.com/mikessh/migmap). Consensus VH and VL/VK chain for each single cell was determined by collapsing all reads with the same CDR3 sequence and calling the top heavy and light chain sequences by frequency.
Lineage and UCA analysis
Clonal lineages were assessed as previously described. Briefly, we used Cloanalyst77 to annotate paired heavy-light VDJ/VJ sequences, to infer the unmutated common ancestor (UCA) and intermediates to reconstruct the phylogenetic trees for clonal lineages.
CryoEM grid preparation
Complexes of H1N1 A/Solomon Island/3/2006 Q226 HA with Ab109 or Ab111 Fab were formed by combining each component at 1.3 mg/mL (1.3-fold excess of binding sites) in a buffer composed of 10 mM Tris pH 7.5 with 150 mM NaCl. Octyl β-D-glucoside (OG) was added at a final concentration of 0.1% (w/v) to correct the HA orientation bias. HA·Fab complexes were incubated for 30 min on ice before application to C-flat 1.2–1.3 400 Cu mesh grids (Protochips). Grids were glow discharged (PELCO easiGlow) for 30 s at 15 mA and prepared with a Gatan Cryoplunge 3 by applying 3.5 uL of sample and blotting for 4.0 s in the chamber maintained at a humidity between 86% and 90%.
CryoEM image recording
Images for HA·Ab109-Fab complexes were recorded on a Titan Krios microscope operated at 300 keV with a Gatan BioQuantum GIF/K3 direct electron detector. Automated image acquisition was performed with Serial EM.78 Specifications and statistics for images are reported in Table S1.
CryoEM image analysis and 3D reconstruction
Image analysis for was carried out in RELION. Beam-induced motion correction of micrograph movies was performed with UCSF MotionCor2 followed by contrast transfer function estimation with CTFFIND-4.1, both as implemented in RELION. Particles were picked from motion corrected micrographs by a general model using crYOLO. Extracted particles were down sampled by a factor of 3 and lowpass filtered with an estimated CTF resolution of 5 Å. An initial round of 2D classification revealed most particles were members of a 2D lattice. An initial 3D model was prepared with RELION and C3 symmetry was imposed. Models resulting from 3D classification suffered from an orientation bias and exhibited anisotropic resolution.
Particles from the HA·Ab109-Fab dataset were separated based on the orientation determined in 2D classification and a subset from each view was selected based on resolution to provide a roughly equal number of particles from each view. Several rounds of 3D classification provided a subset of particles that contributed to a Nyquist-limited 3D reconstruction at the down sampled pixel size. Refined particles were re-extracted at the full pixel size and subjected to further 3D classification and refinement leading to a 4.3Å reconstruction. CTF refinement was performed to correct beam tilt, trefoil, anisotropic magnification, and per particle defocus in RELION. Bayesian polishing was also performed in RELION leading to a 3.9Å reconstruction following 3D refinement. The final map was improved to 3.4Å resolution with B-factor sharpening (Figure S6). A similar procedure was followed for the HA·Ab111-Fab dataset, however, including the full resolution pixel size did not extend the resolution beyond 5Å.
CryoEM density fitting and model building
Heavy and light chains of PDBs 4IML and 5FB8 were aligned and extracted to make an initial model for the Fab. A protomer of H1 Solomon Islands was extracted from PBD 5UGY. These initial models were ridged body fit into the Ab109 CryoEM density map using Chimera. The gross positions of the initial models were further refined by ridged body real-space refinement with Phenix. The sequences of the initial antibody variable domains were corrected to match the actual sequence in COOT. The coordinates were further real space refined with Phenix. Model building was also performed with ISOLDE.79 Iterative cycles of model adjustment and refinement were performed to eliminate Ramachandran outliers and non-covalent interatomic distances of less than 2.2Å. Map-to-model statistics and FSC curves were prepared with Phenix (Table S1 and Figure S6). The resolution of the Ab111 Cryo-EM density was too low to permit model building.
Microneutralization assay
Neutralization endpoint titers (EPT) were determined using the microneutralization assay on Madin-Darby Canine Kidney (MDCK) cells (London Line, FR-58). Briefly, monoclonal murine IgG1 Abs were diluted to 100μg/mL and evaluated as duplicate two-fold dilutions series. Samples were co-incubated with 100 TCID50 influenza virus for one hour, and this mixture was then transferred to MDCK cells. 24 h after infection, monolayers were fixed in acetone and immunostained. Absorbance was measured with a Synergy H1 microplate reader (BioTek Instruments Inc., Winooski, Vermont). Influenza viruses were grown and passaged in MDCK (NBL-2) (ATCC CCL-34TM). Reagents obtained through BEI Resources, NIAID, NIH include: A/Michigan/45/2015 (H1N1) (IRR, FR-1483), A/California/07/2009 (H1N1) (BEI, NR-13663), A/Solomon Islands/3/2006 (H1N1) (BEI, NR-41798), A/Hawaii/15/2001 (H1N1) (IRR, FR-394), A/New Caledonia/20/1999 (H1N1) (BEI, NR-9692), A/Beijing/262/1995 (H1N1) (BEI, NR-3571), A/Texas/36/1991 (H1N1) (BEI, NR-3223), and A/Puerto Rico/8/1934 (H1N1) (BEI, NR348).
QUANTIFICATION AND STATISTICAL ANALYSIS
Curve fitting and statistical analyses were performed with GraphPad Prism (version 9). Non-parametric statistics were used throughout where feasible. Tests, numbers of animals, and statistical comparison groups are indicated in each of the Figure Legends. The Mann-Whitney U test was used to compare two populations without consideration for paired samples. A p value <0.05 was considered significant.
Supplementary Material
Highlights.
Rationally design an immunogen enriched for the HA receptor-binding site
Immunogen boosting expands serum and B cell responses to the RBS after H1 prime
Sequence analysis shows multiple RBS-directed lineages preferentially expanded
cryoEM characterization of immunogen-elicited mAbs shows germline-encoded contacts
ACKNOWLEDGMENTS
We thank Stephen Harrison, Kevin R. McCarthy, Goran Bajic, and Adam Johnson for helpful discussions. We acknowledge support from the NIH through R01AI146779 (A.G.S.), P01AI89618-A1 (A.G.S.), 2P30AI060354 (D.L.), R01AI137057 (D.L.), DP2DA042422 (D.L.), R01AI124378 (D.L.), R01AI153098 (D.L.), and T32 GM007753 (T.M.C.). This research has been funded in whole or in part with federal funds under a contract from the National Institute of Allergy and Infectious Diseases, NIH, contract 75N93019C00050 (A.G.S.).
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111628.
DECLARATION OF INTERESTS
T.M.C., M.K., and A.G.S. have filed a provisional patent for the described rsHAtCh immunogen.
REFERENCES
- 1.Bedford T, Suchard MA, Lemey P, Dudas G, Gregory V, Hay AJ, McCauley JW, Russell CA, Smith DJ, and Rambaut A. (2014). Integrating influenza antigenic dynamics with molecular evolution. Elife 3, e01914. 10.7554/eLife.01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova VN, and Russell CA (2018). The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 16, 47–60. 10.1038/nrmicro.2017.118. [DOI] [PubMed] [Google Scholar]
- 3.Webster RG, Laver WG, Air GM, and Schild GC (1982). Molecular mechanisms of variation in influenza viruses. Nature 296, 115–121. 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 4.Doud MB, Lee JM, and Bloom JD (2018). How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin. Nat. Commun. 9, 1386. 10.1038/s41467-018-03665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JM, Huddleston J, Doud MB, Hooper KA, Wu NC, Bedford T, and Bloom JD (2018). Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc. Natl. Acad. Sci. USA 115, E8276–E8285. 10.1073/pnas.1806133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipsitch M, and O’Hagan JJ (2007). Patterns of antigenic diversity and the mechanisms that maintain them. J. R. Soc. Interface 4, 787–802. 10.1098/rsif.2007.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andrews SF, Huang Y, Kaur K, Popova LI, Ho IY, Pauli NT, Henry Dunand CJ, Taylor WM, Lim S, Huang M, et al. (2015). Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 7, 316ra192. 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. (2011). A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856. 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 9.Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. (2016). Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166, 596–608. 10.1016/j.cell.2016.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao H-X, Moody MA, Haynes BF, and Harrison SC (2015). Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034. 10.1016/j.cell.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, and Harrison SC (2018). Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 48, 174–184.e9. 10.1016/j.immuni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GCM, Vervaet G, Skepner E, Lewis NS, Spronken MIJ, Russell CA, et al. (2013). Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342, 976–979. 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 13.Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. (2017). Defining B cell immunodominance to viruses. Nat. Immunol. 18, 456–463. 10.1038/ni.3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuraoka M, Schmidt AG, Nojima T, Feng F, Watanabe A, Kitamura D, Harrison SC, Kepler TB, and Kelsoe G. (2016). Complex antigens drive permissive clonal selection in germinal centers. Immunity 44, 542–552. 10.1016/j.immuni.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, et al. (2008). Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671. 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angeletti D, and Yewdell JW (2018). Understanding and manipulating viral immunity: antibody immunodominance enters center stage. Trends Immunol. 39, 549–561. 10.1016/j.it.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 17.Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, et al. (2019). Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio 10, e02810–18. 10.1128/mBio.02810-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nachbagauer R, Liu WC, Choi A, Wohlbold TJ, Atlas T, Rajendran M, Solórzano A, Berlanda-Scorza F, García-Sastre A, Palese P, et al. (2017). A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2, 26. 10.1038/s41541-017-0026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yassine HM, Boyington JC, McTamney PM, Wei C-J, Kanekiyo M, Kong W-P, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. (2015). Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med. 21, 1065–1070. 10.1038/nm.3927. [DOI] [PubMed] [Google Scholar]
- 20.Eggink D, Goff PH, and Palese P. (2014). Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 88, 699–704. 10.1128/JVI.02608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyoglu-Barnum S, Ellis D, Gillespie RA, Hutchinson GB, Park Y-J, Moin SM, Acton OJ, Ravichandran R, Murphy M, Pettie D, et al. (2021). Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 592, 623–628. 10.1038/s41586-021-03365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, et al. (2019). Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177, 1124–1135.e16. 10.1016/j.cell.2019.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, et al. (2019). A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177, 1136–1152.e18. 10.1016/j.cell.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, and Schmidt AG (2019). Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25, 827–835.e6. 10.1016/j.chom.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krammer F, and Palese P. (2015). Advances in the development of influenza virus vaccines. Nat. Rev. Drug Discov. 14, 167–182. 10.1038/nrd4529. [DOI] [PubMed] [Google Scholar]
- 26.Skehel JJ, and Wiley DC (2000). Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569. 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 27.Bajic G, Maron MJ, Caradonna TM, Tian M, Mermelstein A, Fera D, Kelsoe G, Kuraoka M, and Schmidt AG (2020). Structure-guided molecular grafting of a complex broadly neutralizing viral epitope. ACS Infect. Dis. 6, 1182–1191. 10.1021/acsinfecdis.0c00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanekiyo M, Joyce MG, Gillespie RA, Gallagher JR, Andrews SF, Yassine HM, Wheatley AK, Fisher BE, Ambrozak DR, Creanga A, et al. (2019). Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 20, 362–372. 10.1038/s41590-018-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, and Harrison SC (2018). Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc. Natl. Acad. Sci. USA 115, 168–173. 10.1073/pnas.1715471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sangesland M, Ronsard L, Kazer SW, Bals J, Boyoglu-Barnum S, Yousif AS, Barnes R, Feldman J, Quirindongo-Crespo M, McTamney PM, et al. (2019). Germline-encoded affinity for cognate antigen enables vaccine amplification of a human broadly neutralizing response against influenza virus. Immunity 51, 735–749.e8. 10.1016/j.immuni.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sangesland M, Yousif AS, Ronsard L, Kazer SW, Zhu AL, Gatter GJ, Hayward MR, Barnes RM, Quirindongo-Crespo M, Rohrer D, et al. (2020). A single human V(H)-gene allows for a broad-spectrum antibody response targeting bacterial lipopolysaccharides in the blood. Cell Rep. 32, 108065. 10.1016/j.celrep.2020.108065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godley L, Pfeifer J, Steinhauer D, Ely B, Shaw G, Kaufmann R, Suchanek E, Pabo C, Skehel JJ, Wiley DC, et al. (1992). Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68, 635–645. 10.1016/0092-8674(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 33.Boudko SP, Zientek KD, Vance J, Hacker JL, Engel J, and Bächinger HP (2010). The NC2 domain of collagen IX provides chain selection and heterotrimerization. J. Biol. Chem. 285, 23721–23731. 10.1074/jbc.M110.128405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boudko SP, and Bächinger HP (2016). Structural insight for chain selection and stagger control in collagen. Sci. Rep. 6, 37831. 10.1038/srep37831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Chen O, Wall JBJ, Zheng M, Zhou Y, Wang L, Vaseghi HR, Qian L, and Liu J. (2017). Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 7, 2193. 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JH, Lee S-R, Li L-H, Park H-J, Park J-H, Lee KY, Kim M-K, Shin BA, and Choi S-Y (2011). High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6, e18556. 10.1371/journal.pone.0018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whittle JRR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. (2011). Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA 108, 14216–14221. 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittle JRR, Wheatley AK, Wu L, Lingwood D, Kanekiyo M, Ma SS, Narpala SR, Yassine HM, Frank GM, Yewdell JW, et al. (2014). Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057. 10.1128/jvi.03422-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lingwood D, McTamney PM, Yassine HM, Whittle JRR, Guo X, Boyington JC, Wei CJ, and Nabel GJ (2012). Structural and genetic basis for development of broadly neutralizing influenza antibodies. Nature 489, 566–570, nature11371 [pii]. 10.1038/nature11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viant C, Weymar GHJ, Escolano A, Chen S, Hartweger H, Cipolla M, Gazumyan A, and Nussenzweig MC (2020). Antibody affinity shapes the choice between memory and germinal center B cell fates. Cell 183, 1298–1311.e11. 10.1016/j.cell.2020.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yousif AS, Ronsard L, Shah P, Omatsu T, Sangesland M, Bracamonte Moreno T, Lam EC, Vrbanac VD, Balazs AB, Reinecker H-C, and Lingwood D. (2021). The persistence of interleukin-6 is regulated by a blood buffer system derived from dendritic cells. Immunity 54, 235–246.e5. 10.1016/j.immuni.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes JM, Zhang Y, Lin YL, Chung S, Dennis MS, Zuchero YJY, et al. (2017). Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J. Biol. Chem. 292, 3900–3908. 10.1074/jbc.M116.767749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saunders KO (2019). Conceptual approaches to modulating antibody effector functions and circulation half-life. Front. Immunol. 10, 1296. 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, and Wilson IA (2014). Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat. Commun. 5, 3614. 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, and Crowe JE Jr. (2011). A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J. Virol. 85, 10905–10908. 10.1128/JVI.00700-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr., and Wilson IA (2013). A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat. Struct. Mol. Biol. 20, 363–370. 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, et al. (2012). Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532. 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caradonna TM, and Schmidt AG (2021). Protein engineering strategies for rational immunogen design. NPJ Vaccines 6, 154. 10.1038/s41541-021-00417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu WC, Nachbagauer R, Stadlbauer D, Soló rzano A, Berlanda-Scorza F, García-Sastre A, Palese P, Krammer F, and Albrecht RA (2019). Sequential immunization with live-attenuated chimeric hemagglutinin-based vaccines confers heterosubtypic immunity against influenza A viruses in a preclinical ferret model. Front. Immunol. 10, 756. 10.3389/fimmu.2019.00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen AA, Gnanapragasam PNP, Lee YE, Hoffman PR, Ou S, Kakutani LM, Keeffe JR, Wu H-J, Howarth M, West AP, et al. (2021). Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741. 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen AA, Yang Z, Gnanapragasam PNP, Ou S, Dam K-MA, Wang H, and Bjorkman PJ (2021). Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS One 16, e0247963. 10.1371/journal.pone.0247963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fonville JM, Wilks SH, James SL, Fox A, Ventresca M, Aban M, Xue L, Jones TC, Le NMH, Pham QT, et al. (2014). Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000. 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hensley SE (2014). Challenges of selecting seasonal influenza vaccine strains for humans with diverse pre-exposure histories. Curr. Opin. Virol. 8, 85–89. 10.1016/j.coviro.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, et al. (2013). Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 210, 1493–1500. 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, et al. (2011). H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6, e25797. 10.1371/journal.pone.0025797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt AG, Do KT, McCarthy KR, Kepler TB, Liao H-X, Moody MA, Haynes BF, and Harrison SC (2015). Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 13, 2842–2850. 10.1016/j.celrep.2015.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sesterhenn F, Galloux M, Vollers SS, Csepregi L, Yang C, Descamps D, Bonet J, Friedensohn S, Gainza P, Corthésy P, et al. (2019). Boosting subdominant neutralizing antibody responses with a computationally designed epitope-focused immunogen. PLoS Biol. 17, e3000164. 10.1371/journal.pbio.3000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sesterhenn F, Yang C, Bonet J, Cramer JT, Wen X, Wang Y, Chiang CI, Abriata LA, Kucharska I, Castoro G, et al. (2020). De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science 368, eaay5051. 10.1126/science.aay5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu NC, Thompson AJ, Lee JM, Su W, Arlian BM, Xie J, Lerner RA, Yen H-L, Bloom JD, and Wilson IA (2020). Different genetic barriers for resistance to HA stem antibodies in influenza H3 and H1 viruses. Science 368, 1335–1340. 10.1126/science.aaz5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zost SJ, Lee J, Gumina ME, Parkhouse K, Henry C, Wu NC, Lee C-CD, Wilson IA, Wilson PC, Bloom JD, and Hensley SE (2019). Identification of antibodies targeting the H3N2 hemagglutinin receptor binding site following vaccination of humans. Cell Rep. 29, 4460–4470.e8. 10.1016/j.celrep.2019.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Keating AE, Malashkevich VN, Tidor B, and Kim PS (2001). Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils. Proc. Natl. Acad. Sci. USA 98, 14825–14830. 10.1073/pnas.261563398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiller T, Busse CE, and Wardemann H. (2009). Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350, 183–193. 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Rohatgi S, Ganju P, and Sehgal D. (2008). Systematic design and testing of nested (RT-)PCR primers for specific amplification of mouse rearranged/expressed immunoglobulin variable region genes from small number of B cells. J. Immunol. Methods 339, 205–219. 10.1016/j.jim.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 64.Kepler TB, Munshaw S, Wiehe K, Zhang R, Yu JS, Woods CW, Denny TN, Tomaras GD, Alam SM, Moody MA, et al. (2014). Reconstructing a B-cell clonal lineage. II. Mutation, selection, and affinity maturation. Front. Immunol. 5, 170. 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scheres SHW (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 67.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Emsley P, and Cowtan K. (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 70.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kabsch W. (2010). Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amitai A, Sangesland M, Barnes RM, Rohrer D, Lonberg N, Lingwood D, and Chakraborty AK (2020). Defining and manipulating B cell immunodominance hierarchies to elicit broadly neutralizing antibody responses against influenza virus. Cell Syst. 11, 573–588.e9. 10.1016/j.cels.2020.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Glaser L, Conenello G, Paulson J, and Palese P. (2007). Effective replication of human influenza viruses in mice lacking a major alpha2, 6 sialyltransferase. Virus Res. 126, 9–18. 10.1016/j.virusres.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 74.Trombetta JJ, Gennert D, Lu D, Satija R, Shalek AK, and Regev A. (2014). Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–4.22.17. 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masella AP, Bartram AK, Truszkowski JM, Brown DG, and Neufeld JD (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 13, 31. 10.1186/1471-2105-13-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi B, Ma L, He X, Wang X, Wang P, Zhou L, and Yao X. (2014). Comparative analysis of human and mouse immunoglobulin variable heavy regions from IMGT/LIGM-DB with IMGT/HighV-QUEST. Theor. Biol. Med. Model. 11, 30. 10.1186/1742-4682-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kepler TB (2013). Reconstructing a B-cell clonal lineage. F1000Res. 2, 103. 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 79.Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530. 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CryoEM data have been deposited in the Electron Microscopy DataBase and PDB and are available as of the date of publication. B cell receptor sequences have been deposited in GenBank. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| HRP-conjugated rabbit anti-mouse IgG antibody | Abcam | CAT#ab97046 |
| Ly-6G-APCe780 | ThermoFisher | CAT#14-5931-82 |
| F4/80-APCe780 | ThermoFisher | CAT#47-4801-82 |
| CD3-BV785 | BioLegend | CAT#100231 |
| CD19-BV421 | BioLegend | CAT#115549 |
| IgM-BV605 | BioLegend | CAT#406523 |
| IgG-PerCP/Cy5.5 | BioLegend | CAT#405314 |
| msIgG1 isoforms of Ab100, Ab102, Ab109, Ab110, Ab111, Ab114, Ab116 | This paper | N/A |
| Ab111 IgG2a isoform | This paper | N/A |
| Ab111 IgG2a LALA-PG isoform | This paper | N/A |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| A/Michigan/45/2015 (H1N1) | Duke RBL | Lot 1483-180124 |
| A/California/07/2009 (H1N1) | Duke RBL | Lot 13659-20141211 |
| A/Solomon Islands/3/2006 (H1N1) | Duke RBL | Lot 331-05132014 |
| A/Hawaii/15/2001 (H1N1) | Duke RBL | Lot FR-394-20200928 |
| A/New Caledonia/20/1999 (H1N1) | IRR / Duke RBL | Lot FR-395-20200928 |
| A/Beijing/262/1995 (H1N1) | Duke RBL | Lot 12277-16524 |
| A/Texas/36/1991 | Duke RBL | Lot 1096-20200807 |
| A/Puerto Rico/8/1934 | Duke RBL | Lot CR-05282014 |
|
| ||
| Chemicals peptides and recombinant proteins | ||
|
| ||
| rsHAtCh (rsH4, rsH3, rsH14 heterotrimer) | This paper | N/A |
| rsH14 monomeric head domain | Produced in house; Bajic et al.27 | N/A |
| rsH3 monomeric head domain | This paper | N/A |
| rsH4 monomeric head domain | Produced in house; Bajic et al.27 | N/A |
| GABH heterodimeric coiled coil (acidic and basic strands) | Produced in house Keating et al.61 |
N/A |
| H2227 | Produced in house Schmidt et al.56 |
N/A |
| H2526 | Produced in house Schmidt et al.56 |
N/A |
| CH67 | Produced in house Whittle et al.37 |
N/A |
| 641 I-9 | Produced in house Schmidt et al.56 |
N/A |
| C05 | Produced in house Ekiert et al.47 |
N/A |
| HRV 41.1 | Produced in house Schmidt et al.56 |
N/A |
| Full length soluble ectodomains of H1 MA-90, H1 BE-95, H1 NC-99, H1 SI-06, and H1 CA-09 | Produced in house | N/A |
| H1 SI-06 FLsE ΔRBS | Produced in house Lingwood et al.39 |
N/A |
| Monomeric HA head domains of H1 USSR-77, KW-86, MA-90, FL-93, BE-95, NC-99, SI-06, CA-09 | Produced in house Schmidt et al.56 |
N/A |
| Ab100 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251202 / OP251203 |
| Ab102 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251204 / OP251205 |
| Ab104 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251206 / OP251207 |
| Ab105 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251208 / OP251209 |
| Ab108 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251210 / OP251211 |
| Ab109 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251212 / OP251213 |
| Ab110 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251214 / OP251215 |
| Ab111 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251216 / OP251217 |
| Ab114 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251218 / OP251219 |
| Ab116 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251220 / OP251221 |
| Ab117 (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251222 / OP251223 |
| Lin100 UCA (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251224 / OP251225 |
| Lin100 (VH/VL sequence from IGHV1-2 HC2 mouse)I-0 | This paper | OP251226 / OP251227 |
| Lin114 UCA (VH/VL sequence from IGHV1-2 HC2 mouse) | This paper | OP251228 / OP251229 |
| Lin119 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251230 / OP251231 |
| Lin140 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251232 / OP251233 |
| Lin143 (VH/VL sequence from IGHV1-2 HC2 mouse)UCA | This paper | OP251234 / OP251235 |
| Pierce HRV 3C protease | ThermoScientific | CAT#88946 |
| Sigma Adjuvant System | SigmaAldrich | CAT#S6322 |
| Streptavidin-PE conjugate | Invitrogen | CAT#12-4317-87 |
| Aqua Live/Dead amine-reactive dye | ThermoFisher | CAT#L34957 |
| Calcein, AM, cell-permeant dye | ThermoFisher | CAT#C3100MP |
| Streptavidin-APC/Cy7 conjugate | BioLegend | CAT#405208 |
| Streptavidin-BV650 conjugate | BioLegend | CAT#405232 |
| StrepTactin-PE conjugate | IBA Lifesciences | Item#6-5000-001 |
| StrepTactin-APC conjugate | IBA Lifesciences | Item#6-5010-001 |
| RNaseOUT | ThermoFisher | CAT#10777019 |
| TALON Metal Affinity Resin | Takara | CAT#635652 |
| Pierce Protein G Agarose | ThermoFisher | CAT#20399 |
| 1-Step ABTS substrate | ThermoFisher | Prod#37615 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Ni-NTA Biosensors | ForteBio | Item#18-5101 |
| SuperScript IV VILO MasterMix | ThermoFisher | CAT#11756050 |
| MagMax mirVana Total RNA isolation kit | ThermoFisher | CAT#A27828 |
|
| ||
| Deposited data | ||
|
| ||
| Electron density map, Ab109 and H1 SI-06 FLsE | This paper | EMDB: EMD-26605 |
| Structure, Ab109 and H1 SI-06 FLsE | This paper | PDB: 7UMM |
| Electron density map, Ab111 and H1 SI-06 FLsE | This paper | EMDB: EMD-26604 |
| Isolated B cell antibody sequences | This paper | GenBank: OP251202 - OP251235 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: FreeStyle 293F | Thermo Fisher | Cat#R79007; RRID: CVCL_D603 |
| Human: Expi293F | Thermo Fisher | Cat#A14527; RRID: CVCL_D615 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| cDC-IrF5−/− mice | Yousif et al.41 | N/A |
| IGHV1-2 HC2 mice | Sangesland et al.30 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Murine B cell receptor sequencing primers | Tiller et al.; Rohatgi et al.,62,63 | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10 | TreeStar |
https://www.flowjo.com; RRID: SCR_008520 |
| Prism v9 | GraphPad |
https://www.graphpad.com; RRID: SCR_002798 |
| IMGT | International ImMunoGeneTics Information System |
http://www.imgt.org; RRID: SCR_012780 |
| Cloanalyst | Kepler et al.64 | https://www.bu.edu/computationalimmunology/research/software/ |
| RELION | Scheres et al.65 | https://relion.readthedocs.io/en/release-3.1/ |
| Chimera | Pettersen et al.66 | https://www.cgl.ucsf.edu/chimera/ |
| PHASER | McCoy et al.67 | http://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| PHENIX | Adams et al.68 | https://www.phenix-online.org/ |
| COOT | Emsley et al.69 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMol | Schrödinger | http://www.pymol.org |
| MolProbity | Chen et al.70 | http://molprobity.biochem.duke.edu |
| XDS | Kabsch et al.71 | https://xds.mr.mpg.de |
|
| ||
| Other | ||
|
| ||
| Superdex 200 Increase 10/300 GL | GE Healthcare | Cytiva 28-9909-44 |