Abstract
Motivation
The National Human Genome Research Institute Catalog of Published Genome-Wide Association Studies (GWAS) Catalog has collected, curated and made available data from over 7100 studies. The recently developed GWAS Catalog representational state transfer (REST) application programming interface (API) is the only method allowing programmatic access to this resource.
Results
Here, we describe gwasrapidd, an R package that provides the first client interface to the GWAS Catalog REST API, representing an important software counterpart to the server-side component. gwasrapidd enables users to quickly retrieve, filter and integrate data with comprehensive bioinformatics analysis tools, which is particularly critical for those looking into functional characterization of risk loci.
Availability and implementation
gwasrapidd is freely available under an MIT License, and can be accessed from https://github.com/ramiromagno/gwasrapidd.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The National Human Genome Research Institute (NHGRI) Catalog of published GWAS Catalog, created in 2014, is a publicly available, manually curated, database of all published genome-wide association studies (GWAS) (Welter et al., 2014). Its latest data release [date July 12, 2019] includes data from 4054 publications and 143 963 unique SNP-trait associations for human diseases. Currently, these data can be accessed by three methods: (i) via the web graphical user interface (GUI), (ii) by downloading database dumps, or, more recently, (iii) via the GWAS catalog representational state transfer (REST) application programming interface (API), which provides direct programmatic access and hence is the preferred method for bioinformatics analyses.
We developed the first R package (R Core Team, 2017) allowing programmatic access to the GWAS catalog REST API: gwasrapidd. This package provides a simple interface for querying catalog data, abstracting away the informatic details of the REST API. In addition, retrieved data are mapped to in-memory relational databases of tidy data tables, allowing prompt integration with tidy-verse packages for subsequent transformation, visualization and modelling of data (Wickham et al., 2014; Wickham and Grolemund, 2017).
2 Results
2.1 Retrieving data from the GWAS Catalog REST API
The GWAS Catalog REST API is an EBI service hosted at https://www.ebi.ac.uk/gwas/rest/api/. The REST API uses hypermedia with resource responses following the JSON hypertext application language (HAL) format (Kelly, 2016). Response data are, therefore, provided as hierarchical data in JSON format, can be paginated (i.e. split into multiple responses) and can also be embedded (i.e. have other resources contained within them), adding extra complexity to the returned JSON format [Additional File 1: Supplementary Table S1, and (NHGRI-EBI GWAS Catalog Team, 2019)].
To ease the conversion from the hierarchical to the relational tabular format—the preferred format for data analysis in R (Wickham and Grolemund, 2017), and to abstract away the informatic details associated with the HAL format, we developed a set of retrieval functions (Fig. 1A). Since the REST API data are organized around four core data entities —studies, associations, variants and traits (NHGRI-EBI GWAS Catalog Team, 2019)— we implemented four corresponding retrieval functions that encapsulate the technical aspects of resource querying and format conversion: get_studies(), get_associations(), get_variants() and get_traits() (Fig. 1A). These functions simplify the querying of GWAS entities, by providing a complete and consistent interface to the Catalog. For example, to query for studies, the user needs only to know the function get_studies(), whereas the REST API itself exposes a set of disparate resource URL endpoints for studies following the available search criteria [Additional file 1: Supplementary Table S1, and (NHGRI-EBI GWAS Catalog Team, 2019)]. Moreover, the user can choose from any number of available search criteria exposed by the REST API directly as arguments to the retrieval functions (Fig. 1B). All arguments are vectorized, meaning that multiple queries are promptly available from a single function call. Results obtained from multiple queries can be combined in an OR or AND fashion with the set_operation parameter. If set_operation is set to OR (default behavior), results are collated while removing duplicates, if any. If set_operation is set to AND, only entities that concomitantly match all criteria are returned. If finer control is needed on combining results, the following functions can be used: bind(), union(), intersect(), setdiff() and setequal(). These are S4 methods that work with the S4 classes created in gwasrapidd (Additional File 2: Figure S1). An example of a case study can be found in Additional File 3.
Fig. 1.
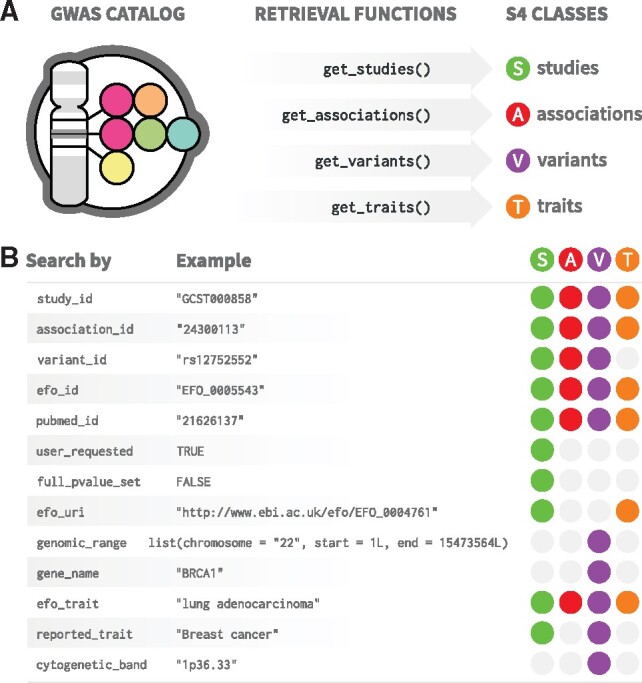
gwasrapidd retrieval functions. (A) Functions for retrieving data from the GWAS Catalog: get_studies(), get_associations(), get_variants() and get_traits(). (B) gwasrapidd search criteria (function parameters) to be used with retrieval functions. Colored circles indicate which entities can be retrieved by which criteria
2.2 Representation of GWAS Catalog entities
All S4 classes share the same design principles that makes them relational databases: (i) each slot corresponds to a table (data frame in R), (ii) the first slot corresponds to the main table that lists observations of the respective GWAS Catalog entity, e.g. studies and, (iii) all tables have a primary key, the identifier of the respective GWAS Catalog entity: study_id, association_id, variant_id or efo_id (Additional File 2: Supplementary Fig. S1). For easy consultation of the variables in the tables, we provide a cheat-sheet (Additional File 4: gwasrapidd cheat-sheet); for the detailed description the user can issue the following commands to open the help page about each class: class?studies, class?associations, class?variants or class?traits.
2.3 Improvements and limitations
Compared to the exposed REST API, we have augmented the search possibilities in gwasrapidd in two ways: (i) by allowing searches for variants by cytogenetic region (as is possible with the web GUI) and (ii) by allowing searching variants by EFO identifier (efo_id), indirectly via EFO traits get_traits(). The first was implemented by embedding a dataset of genomic ranges of the human cytogenetic bands in gwasrapidd, so that queries made by cytogenetic band can be translated into searches by genomic range (genomic_range). Additionally, gwasrapidd also provides a set of helper functions to easily browse linked web resources, such as PubMed [open_in_pubmed()], dbSNP [open_in_dbsnp()] and GTEx project [open_in_gtex()].
Currently, the limitations of the REST API when compared to the web GUI are: (i) it is not possible to perform free text searches, and (ii) it is not possible to search traits using child trait terms automatically, they need to be included explicitly. To find the trait child terms, we provide the function get_child_efo().
3 Conclusion
We have developed the first R client to the GWAS Catalog REST API, thus greatly facilitating programmatic access to the database. The main features of gwasrapidd are: (i) abstracting away the REST API informatic details by providing a simple and consistent interface, and (ii) a tidy data representation of the GWAS entities, i.e. of studies, associations, variants and traits in the form of in-memory relational databases. This improves data mining from within R, accelerating the integration of GWAS data into further genomic and biomedical/clinical studies.
Supplementary Material
Acknowledgements
The authors would like to thank the GWAS Catalog team, particularly Daniel Suveges, for all the help and support with the REST API throughout the entire development of gwasrapidd. They also thank also the remaining of the Cancer Functional Genomics lab members for feedback on the user experience of gwasrapidd.
Funding
This work was supported by national Portuguese funding through FCT—Fundação para a Ciência e a Tecnologia and CRESC ALGARVE 2020: UID/BIM/04773/2013 ‘CBMR’, POCI-01-0145-FEDER-022184 ‘GenomePT’ and ALG-01-0145-FEDER-31477 ‘DEvoCancer’. Publication costs were kindly supported by Município de Loulé via de Algarve Biomedical Center (ABC).
Conflict of Interest: none declared.
Contributor Information
Ramiro Magno, Centre for Biomedical Research (CBMR); Algarve Biomedical Center (ABC).
Ana-Teresa Maia, Centre for Biomedical Research (CBMR); Algarve Biomedical Center (ABC); Department of Biomedical Sciences and Medicine (DCBM), Universidade do Algarve, Faro 8005-139, Portugal.
References
- Kelly M. (2016) JSON Hypertext Application Language (4 May 2019, date last accessed).
- NHGRI-EBI GWAS Catalog Team. (2019) GWAS Catalog API Guide (4 May 2019, date last accessed).
- R Core Team. (2017) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Welter D. et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res., 42, D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. et al. (2014) Tidy data. J. Stat. Software, 59, 1–23. [Google Scholar]
- Wickham H., Grolemund G. (2017) R for Data Science: Import, Tidy, Transform, Visualize, and Model Data, 1st edn. O’Reilly Media. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


