Abstract
Summary
The NanoStringTM nCounter® is a platform for the targeted quantification of expression data in biofluids and tissues. While software by the manufacturer is available in addition to third parties packages, they do not provide a complete quality control (QC) pipeline. Here, we present NACHO (‘NAnostring quality Control dasHbOard’), a comprehensive QC R-package. The package consists of three subsequent steps: summarize, visualize and normalize. The summarize function collects all the relevant data and stores it in a tidy format, the visualize function initiates a dashboard with plots of the relevant QC outcomes. It contains QC metrics that are measured by default by the manufacturer, but also calculates other insightful measures, including the scaling factors that are needed in the normalization step. In this normalization step, different normalization methods can be chosen to optimally preprocess data. Together, NACHO is a comprehensive method that optimizes insight and preprocessing of nCounter® data.
Availability and implementation
NACHO is available as an R-package on CRAN and the development version on GitHub https://github.com/mcanouil/NACHO.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The NanoStringTM nCounter® platform enables the quantification of expression data in biofluids and tissues with prebuild panels (e.g. miRNA panel). Before any data can be analyzed and interpreted, data has to undergo quality control (QC) to identify poor measurements, samples and perform adequate normalization. The manufacturer’s software nSolverTM contains basic normalization and correction methods but lacks flexibility for data visualization. Other NanoStringTM quality-control solutions such as NanoStringDiff (Wang, 2016) or NanoStringNorm (Waggott, 2012) also lack insightful graphical representations. To address the lack of a complete, visual QC and normalization pipeline, we developed an R package that we called NACHO (‘NAnostring quality Control dasHbOard’). It allows the user to systematically perform QC on miRNA, CodeSet or PlexSet panels from NanoStringTM nCounter® platform.
2 Approach
NACHO consists of three subsequent quality steps, 1.summarize, 2.visualize and 3.normalize.
1. summarize A typical analysis of nCounter® starts with parsing of the raw data from RCC files and pre-processing to obtain quality metrics. The arguments that need to be given are the directory of the data, the samplesheet path, the column within the sample sheet that represents the file names and which housekeeping genes to use. For the latter, one can use all default housekeeping genes, provide a subset of the default housekeeping genes or identify custom housekeeping genes (Mestdagh, 2009).
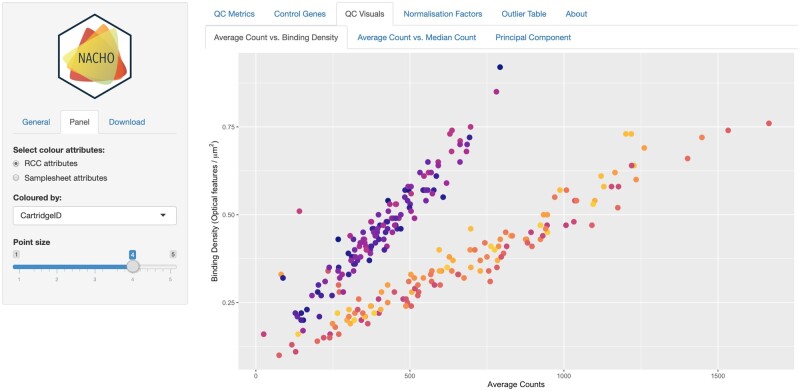
2. visualize Next, one can assess the quality of the data on an interactive web application based on Rstudio’s shiny and ggplot2 (Wickham, 2016) using the visualize function. The web application is initiated with the visualize function that only requires the object generated with the summarize function. The web application dashboard consists of six tabs (Fig. 1) QC Metrics, Control Genes, QC Visuals, Normalization Factors, Outlier table and About. Under the QC Metrics tab four metrics are shown, Binding Density, Imaging, Positive Control Linearity and Limit of Detection (also see Supplementary Methods). Binding density is the number of optical features per square micron. Imaging or Field of View is a metric that indicates how many sections of a lane are successfully processed. Positive control linearity is the Pearson’s correlation coefficient of the observed counts against the known positive control concentrations. Finally, limit of detection reflects the lower boundary of the detection spectrum.
Fig. 1.
Screenshot of the dashboard of the NACHO interface. Data used are miRNA levels of individuals with nasopharyngeal carcinoma (Bruce, 2015). Figure shows the binding density against the average counts colored by Cartridge ID
To further provide insight into the quality of the spike-in control probes, on the Control Genes tab plots are shown of the counts against the known concentrations of the positive control probes, the negative control probes, the predicted or default housekeeping genes (e.g. ACTB, B2M, GAPDH, RPL19, RPLP0).
The QC Visuals tab is subdivided in three sub tabs: Average Counts versus Binding Density, Average Counts versus Median Counts and Principal Components. The first two sub tabs are mainly focused on providing overall insight of the data. The usefulness hereof is illustrated using the public miRNA data of individuals with nasopharyngeal carcinoma (GEO, GSE70970), where two distinct groups of individuals are observed (Fig. 1). Of note, these two groups were analyzed separately in the original manuscript (Bruce, 2015). On the last tab of the QC Visuals, principal components of the data can be plotted against an outcome of interest.
3. normalize In the last step of the QC of nCounter® data, outliers are removed and the data is normalized. The effect of normalization on the data and the outliers can be found on the Normalization Factors—and the Outliers tab. Different methods are implemented in NACHO. For the housekeeping genes, the default genes or predicted housekeeping genes can be used (Mestdagh, 2009; Vandesompele, 2002). The normalization based on the positive control spike-ins can be done based on either the geometric mean (NanoString, 2018) or using a general linearized model (Wang, 2016). The raw counts, normalization factors, normalized counts and settings used are returned to the user as a list.
3 Conclusion and future prospects
The NACHO package is a complete pipeline to process nCounter® data by providing insight in the data quality, removal of poor samples and normalization of the data. In future versions of NACHO, new normalization methods will be added including a recently published normalization method (Molania, 2019).
Funding
This study was supported by grants from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement No 115881 (RHAPSODY). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation programme and EFPIA. This work is supported by the Swiss State Secretariat for Education, Research and Innovation (SERI) under contract number 16.0097-2. The opinions expressed and arguments employed herein do not necessarily reflect the official views of these funding bodies. This study was supported by grants for funding of scientific research conducted in France and within the European Union: Centre National de la Recherche Scientifique, Université de Lille, Institut Pasteur de Lille, Contrat de Plan Etat-Région, Agence Nationale de la Recherche, ‘Fédération de Recherche’ 3508 Labex EGID (European Genomics Institute for Diabetes; ANR-10-LABX-46), ANR EQUIPEX LIGAN-MP (ANR-10-EQPX-07-01), European Research Council GEPIDIAB-294785.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Mickaël Canouil, Université de Lille, CNRS, Institut Pasteur de Lille, UMR 8199 - EGID, F-59000 Lille, France.
Gerard A Bouland, Department of Cell and Chemical Biology, Leiden University Medical Center, 2333 ZC Leiden, The Netherlands.
Amélie Bonnefond, Université de Lille, CNRS, Institut Pasteur de Lille, UMR 8199 - EGID, F-59000 Lille, France; Department of Medicine, Section of Genomics of Common Disease, Imperial College London, London SW7 2AZ, UK.
Philippe Froguel, Université de Lille, CNRS, Institut Pasteur de Lille, UMR 8199 - EGID, F-59000 Lille, France; Department of Medicine, Section of Genomics of Common Disease, Imperial College London, London SW7 2AZ, UK.
Leen M ’t Hart, Department of Cell and Chemical Biology, Leiden University Medical Center, 2333 ZC Leiden, The Netherlands; Department of Epidemiology and Biostatistics, Amsterdam Public Health Institute, Amsterdam UMC, VU University Medical Center, Amsterdam 1081 HV, The Netherlands; Molecular Epidemiology Section, Department of Biomedical Data Sciences, Leiden University Medical Center, Leiden 2333 ZC, The Netherlands.
Roderick C Slieker, Department of Cell and Chemical Biology, Leiden University Medical Center, 2333 ZC Leiden, The Netherlands; Department of Epidemiology and Biostatistics, Amsterdam Public Health Institute, Amsterdam UMC, VU University Medical Center, Amsterdam 1081 HV, The Netherlands.
References
- Bruce J.P. et al. (2015) Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget, 6, 4537–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P. et al. (2009) A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol., 10, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molania R. et al. (2019) A new normalization for Nanostring nCounter gene expression data. Nucleic Acids Res., 47, 6073–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NanoString (2018) Analysis Software User Manual nSolver 4.0. NanoString Technologies, Inc., Seattle, USA.
- Vandesompele J. et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol., 3, research0034.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggott D. et al. (2012) NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics, 28, 1546–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. (2016) NanoStringDiff: a novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic Acids Res., 44, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.