Abstract
Background
Extracellular vesicles (EVs) play an important role in cell–cell communication, and tumor-derived EVs circulating in patient blood can serve as biomarkers. Here, we investigated the potential role of plasma EVs in meningioma patients for tumor detection and determined whether EVs secreted by meningioma cells reflect epigenetic, genomic, and proteomic alterations of original tumors.
Methods
EV concentrations were quantified in patient plasma (n = 46). Short-term meningioma cultures were established (n = 26) and secreted EVs were isolated. Methylation and copy number profiling was performed using 850k arrays, and mutations were identified by targeted gene panel sequencing. Differential quantitative mass spectrometry was employed for proteomic analysis.
Results
Levels of circulating EVs were elevated in meningioma patients compared to healthy individuals, and the plasma EV concentration correlated with malignancy grade and extent of peritumoral edema. Postoperatively, EV counts dropped to normal levels, and the magnitude of the postoperative decrease was associated with extent of tumor resection. Methylation profiling of EV-DNA allowed correct tumor classification as meningioma in all investigated cases, and accurate methylation subclass assignment in almost all cases. Copy number variations present in tumors, as well as tumor-specific mutations were faithfully reflected in meningioma EV-DNA. Proteomic EV profiling did not permit original tumor identification but revealed tumor-associated proteins that could potentially be utilized to enrich meningioma EVs from biofluids.
Conclusions
Elevated EV levels in meningioma patient plasma could aid in tumor diagnosis and assessment of treatment response. Meningioma EV-DNA mirrors genetic and epigenetic tumor alterations and facilitates molecular tumor classification.
Keywords: cfDNA, exosome, liquid biopsy, meningioma, microvesicle
Key Points.
Extracellular vesicles (EVs) are increased meningioma patient plasma.
Meningioma methylation profiles, mutations, and copy number variations are reflected in EV-DNA.
Meningioma-associated EV proteins may facilitate enrichment of tumor-derived vesicles.
Importance of the Study.
Meningiomas represent the most common primary intracranial neoplasms. Following tumor resection, meningioma patients require long-term monitoring to detect possible tumor recurrences. The tumor biomarker potential of meningioma-derived extracellular vesicles (EVs) has not yet been explored. Here, we first demonstrate that levels of circulating EV are increased in the blood of meningioma patients, correlate with malignancy grade and edema, and decline postoperatively in association with extent of tumor resection. Furthermore, DNA extracted from EVs secreted by meningioma cells faithfully reflects tumor-specific methylation profiles, mutations, and copy number variations, thereby allowing molecular tumor classification. Proteomic profiling identifies meningioma-associated EV proteins that have the potential to serve enrichment protocols for the isolation of tumor-derived EVs from plasma to enhance the detection sensitivity for tumor-specific epigenetic and genetic alterations. Translation of our findings into the clinical practice could assist in preoperative tumor assessment and monitoring meningioma patients throughout their disease.
Meningiomas are the most common primary intracranial neoplasms, accounting for 38% of all central nervous system tumors.1 The incidence of meningiomas increases with age, and with the overall aging population these tumors are becoming increasingly more prevalent in neuro-oncology.2 Meningiomas are primarily treated with surgical resection, and with the increasingly interdisciplinary concepts of adaptive hybrid surgery, preoperative knowledge about the expected aggressiveness of a meningioma would be of practical value for surgical risk adaptation. This is particularly important as approximately 20% of tumors display aggressive behavior with early recurrence, requiring repeated surgery, radiotherapy, chemotherapy, or experimental molecular-targeted treatment, resulting in significant morbidity and mortality.3,4 In addition, for the further clinical course, having an easily accessible and reliable biomarker would be highly valuable, as monitoring during years-long follow-up currently relies on serial MRI examinations, which are expensive, time-consuming, and provide no information on molecular alterations, indicative of progression towards a more aggressive tumor. Therefore, the development of noninvasive liquid biopsy techniques that allow continuous monitoring of the disease state, including early detection of tumor recurrence with information on genetic and epigenetic changes, is desirable.
Cancer cells secrete extracellular vesicles (EVs), such as exosomes, microvesicles, and large oncosomes into the tumor microenvironment and bloodstream. EVs carry complex biologically active molecules, including proteins, RNA, DNA, and lipids, which are protected from degradation and fragmentation by the surrounding EV membrane, rendering EVs an attractive target for liquid biopsy in cancer patients.5,6 We and others demonstrated that levels of circulating EVs are elevated in the blood of glioblastoma patients and glioma-bearing mice.7–10 Increased plasma EV concentrations in glioblastoma patients drop after surgery but rise again upon recurrence, suggesting that EV dynamics can reflect the disease state.8 We recently described that genome-wide genetic and epigenetic alterations present in glioblastomas are detectable by analysing the DNA of EVs secreted by cultured glioblastoma cells.11
Little is known about the biomarker potential and properties of EVs in meningioma patients. The rare published studies focused on the micro RNA or protein composition of meningioma EVs.12,13 To our knowledge, no studies on meningioma EV-DNA have yet been performed. The present study shows that circulating EVs are elevated in the blood of meningioma patients and EV levels correlate with malignancy grade as well as peritumoral edema. We performed methylation array analysis and gene panel sequencing of meningioma cell-derived EV-DNA, and by using the Heidelberg brain tumor methylation classifier we demonstrate that all EV samples were correctly assigned to the brain tumor methylation class meningioma, and in almost all cases also the methylation subclass was assigned correctly. Genome-wide copy number variations (CNVs) and mutations present in original tumor tissue were accurately reflected in EV-DNA. Proteomic profiling identified meningioma-associated proteins that could potentially be utilized for enriching meningioma EVs from biofluids.
Materials and Methods
Human Specimens
Meningioma tissue and nontumorous temporal lobe tissue from patients undergoing epilepsy surgery were obtained as approved by the medical ethics committee of the Hamburg chamber of physicians (PV4904). Informed written consent was obtained from all patients. Blood samples (EDTA) were collected before the operation or between 1 and 6 days postoperatively.
Volumetric Measurement of Tumors
MRI images were analyzed by a single practitioner using the cranial planning software Brainlab and the Smartbrush tool. Tumor volume was measured by manually delineating the contrast-enhancing tumor area on sequential gadolinium-enhanced T1-weighted MRI images, followed by three-dimensional reconstruction. Similarly, FLAIR hyperintensity on T2-weighted sequences was outlined by hand at every level, creating a realistic three dimensional model of peritumoral edema. Tumor and edema volumes were calculated in cm3 by the software.
Cell Culture
Meningioma cell cultures were established, following a protocol for the generation of serum-free human glioblastoma stem-like cell cultures,14 as detailed in the Supplementary Methods. Collections of supernatants for EV preparation were initiated at the second medium change when red blood cells and debris had been removed, and were continued over about 2–4 weeks (depending on the growth rate), after which meningioma cells were harvested for analysis.
Isolation and Size Analysis of EVs
EVs were isolated from plasma and conditioned medium of primary meningioma cultures by differential centrifugation and were analyzed by nanoparticle tracking analysis (NTA) as described previously15 and as detailed in the Supplementary Methods.
IFCM
Imaging Flow Cytometry (IFCM) analysis of EVs was performed as described previously11 and as detailed in the Supplementary Methods.
Transmission Electron Microscopy (TEM)
The morphology of EVs was analyzed by TEM as described before11 and as detailed in the Supplementary Methods.
Methylation Array Analysis
Infinium MethylationEPIC arrays (850k) were used to obtain genome-wide DNA methylation profiles as described previously.11 DNA was isolated from EVs using the XCF exosomal DNA isolation kit (SBI), and DNA from cells and tissue was isolated using the Nucleospin tissue kit (Macherey Nagel). Forty to 100 ng DNA quantified by Qubit (Invitrogen) were used for analysis. Bisulfite treatment, whole-genome DNA amplification, hybridization and single-base extension, fluorescence staining, and scanning of the chips were performed following the manufacturer’s instructions (Illumina). Tumor methylation classification and per-sample CNV plotting were performed using the DKFZ brain tumor classifier version 11b4 (https://www.molecularneuropathology.org/mnp) as described.11,16 Data processing, normalization and CNV analysis was performed using the R packages ChAMP (v.2.24.0), conumee (v.1.28.0), ggplot2 (v.3.3.5), and custom scripts.11 T-distributed stochastic neighbor embedding (t-SNE) analysis was performed using the R packages Rtsne (v.0.15) and ggplot2 (v.3.3.5) as described.17 For t-SNE analysis, a publicly available dataset of 82 defined brain tumor classes containing 2801 individual tumor samples was used as reference.16
Mutation Analysis
Gene panel sequencing of the exonic regions of 169 genes known for frequent alterations in brain tumors (Supplementary Table 1) was performed on a NextSeq sequencer 500 (Illumina) as described before18 and as detailed in the Supplementary Methods.
Differential Quantitative Label-Free Mass Spectrometry-based Proteomics
Samples were analyzed by differential quantitative proteomics, using a LC-MS/MS system as described before11 and as detailed in the Supplementary Methods.
Statistical Analysis
One-way ANOVA with post hoc Bonferroni, or Kruskal–Wallis test with Dunn’s correction were conducted to compare multiple groups with normal or non-Gaussian sample distribution. Comparisons between two groups were conducted using paired and unpaired t-test, or Mann–Whitney, and Wilcoxon matched-pairs signed rank test, for normal or non-Gaussian distributed samples. Two-way repeated measures ANOVA with Bonferroni correction or Friedman test with Dunn's correction were used for timepoint comparisons between different groups. Statistical analyses were performed using Graph Pad Prism 9.
Results
Meningioma Patients Have Elevated Levels of Plasma EVs
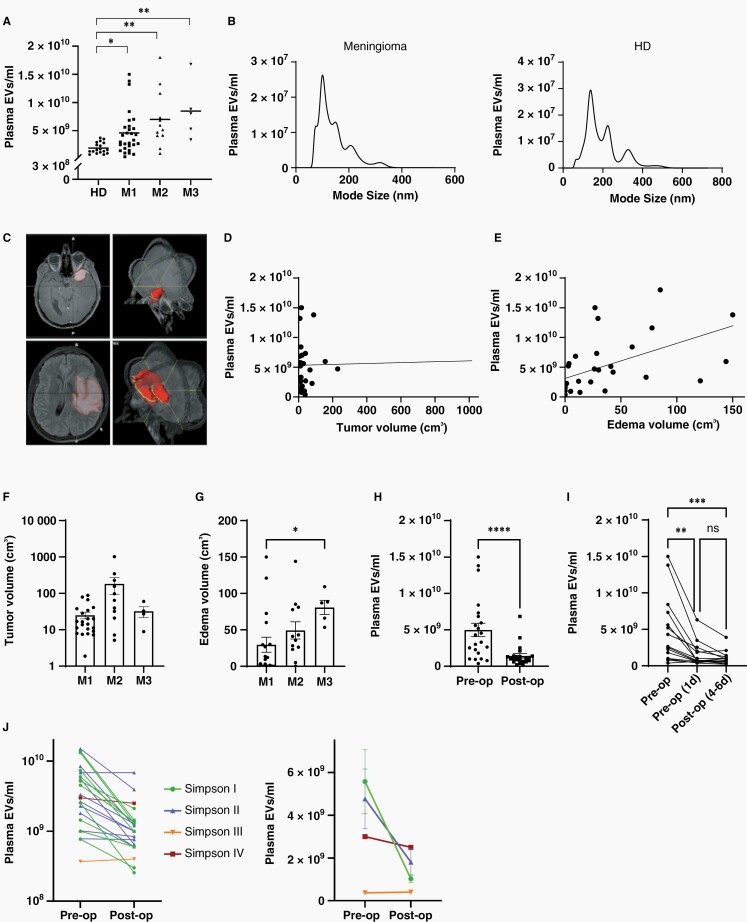
To quantify circulating EVs in meningioma patients, we isolated EVs preoperatively from the plasma of patients with meningiomas WHO grade 1 (M1, n = 29), grade 2 (M2, n = 12), and grade 3 (M3, n = 5) as well as from age-matched healthy donors (HD, n = 18). EV levels were significantly increased in meningioma patients compared to healthy controls (Figure 1A). Mean concentrations were elevated 2.4-fold in M1, 3.7-fold in M2, and 4.5-fold in M3 patients and correlated with histological malignancy grade (r = 0.4082, P = .0049, Spearman). The EV size spectrum did not differ significantly between meningioma patients and healthy donors and fell within the expected range of 50–150 nm, as confirmed by NTA (Figure 1B).
Fig. 1.
Concentration of plasma EVs in meningioma patients. (A) EV levels in the plasma of meningioma patients (WHO grade 1–3) are increased compared to healthy donors (HD), as determined by nanoparticle tracking analysis, (Kruskal–Wallis, Dunn's; horizontal lines represent means). (B) Size distribution of plasma EVs. (C) Volumetric measurement of tumor size (top) and peritumoral edema (bottom) with 3D reconstruction (right). (D) Relationship between tumor volume and plasma EV concentration (r2 = 0,0009). (E) Association between peritumoral edema volume and plasma EV concentration (r2 = 0.2806). (F) Tumor size of meningiomas of different WHO grades. (G) Association of edema with malignancy grade (Kruskal–Wallis, Dunn's). (H) Reduction of EV concentrations to normal levels 4–6 days postoperatively (n = 22, Wilcoxon signed-rank). (I) EV reduction occurred within the first day after the operation (n = 15, Friedman, Dunn's). (J) Comparison of EV concentration changes (4–6 days postoperatively vs. preoperatively) in individual patients with different extent of tumor resection, as defined by Simpson grading (left panel, n = 22). Mean EV concentration changes in patient groups with different extent of resection (right panel). Postoperative EV reduction was only significant in the Simpson grade I group (P =.0093, 2-way ANOVA, Bonferroni). Values in (F)–(H) are means ± SEM. P values are defined as * <.05, ** <.01, *** <.001, and **** <.0001.
Next, we investigated whether EV concentrations were associated with tumor size or extension of peritumoral edema. Tumor volume was quantified by measuring the contrast-enhancing mass on T1-weighted images, and the extent of peritumoral edema was determined as volume of FLAIR hyperintense lesions (Figure 1C). EV plasma concentrations were not associated with tumor size (Figure 1D) but correlated with the extent of peritumoral edema (r = 0.6316, P <.0001, Spearman), (Figure 1E), and correlations with edema were also significant for the MI and MII subgroups, with a similar trend for MIII (Supplementary Fig. 1). Furthermore, tumor size was not associated with WHO grade, whereas increased edema correlated with higher malignancy grade (r = 0.4852, P =.0031, Spearman, Figure 1F, G).
To assess whether the presence of the tumor was responsible for the elevated EV concentration in the blood of meningioma patients, we compared plasma samples obtained preoperatively with paired samples collected 4–6 days after the operation (n = 17 M1, 5 M2). Levels of plasma EVs were significantly reduced in the postoperative samples (Figure 1H), suggesting that the presence of the tumor is responsible for the elevated circulating EVs in meningioma patients. In 15 of these patients (11 M1, 4 M2), additional samples collected one day postoperatively were available, and their analysis showed that the major drop in circulating EVs occurred already at this early timepoint (Figure 1I), indicating that the majority of tumor-associated EVs are cleared rapidly from the circulation.
Finally, we analyzed whether the extent of resection, defined by Simpson grading with MRI confirmation,19 affected the postoperative decline in circulating EVs. The postoperative reduction of circulating EVs was strongest in the group of completely resected tumors (Simpson grade I, P = .0093, n = 10, Figure 1J). A trend towards reduced postoperative EVs in Simpson grade II resected tumors (dural attachment coagulated but not removed), did not reach significance (P = .1336, n = 10), and single cases of Simpson grade III and IV tumors showed little or no postoperative reduction. These findings suggest that the extent of tumor resection affects the degree of postoperative EV reduction in patient blood, and further supports the notion that the tumor is the main source of elevated circulating EVs in meningioma patients.
Isolation and Characterization of EVs Secreted by Meningioma Cells
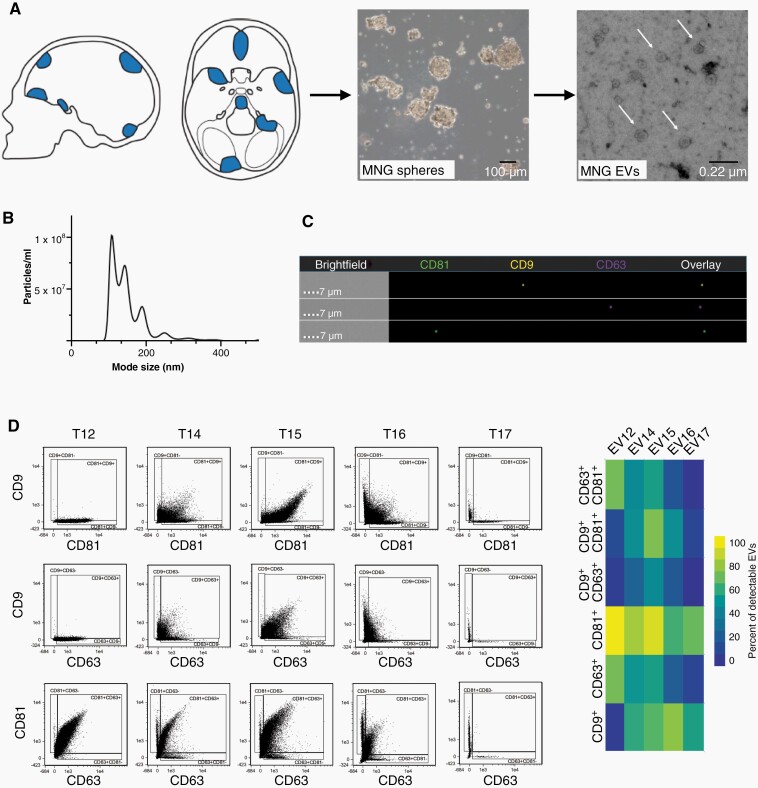
In preliminary experiments we observed that in most cases meningioma cells displayed improved in vitro growth and prolonged viability when cultured as spheres under neural stem cell conditions, compared to adherent monolayers cultured with serum (not shown). To obtain tumor cell-derived EVs, we therefore established primary cell cultures from freshly resected meningioma tissue using sphere conditions (Figure 2A). Collection of supernatants for EV preparation was initiated at the second medium change and repeatedly continued over 2–4 weeks, after which meningioma cells were harvested for analysis. Supernatants were pooled and EVs were isolated by differential ultracentrifugation. TEM showed that meningioma EVs exhibited the typical cup-shaped form that morphologically defines EVs,20 (Figure 2A). EVs fell within the expected size range of exosomes and microvesicles (Figure 2B) and expressed the tetraspanin markers CD9, CD63, and CD81 (Figure 2C, D).
Fig. 2.
Meningioma EV isolation and analysis. (A) Meningioma (MNG) tissue was taken into culture using neurosphere conditions and EVs secreted by the tumor cells were analyzed. Electron microscopy demonstrates the typical cup-shaped morphology of EVs (arrows). (B) Size spectrum of EVs isolated from a meningioma culture. (C) Detection of tetraspanin markers by imaging flow cytometry, representative image. (D) Quantification of the percentage of tetraspanin single-positive and double-positive EVs.
DNA Methylome Analysis of Meningioma EVs
To determine whether meningioma EV-DNA reflects the global DNA methylation pattern of parental cells and original tumors, we purified DNA from EVs secreted by cultured meningioma cells (n = 15), as well as from the corresponding cultured tumor cells (n = 14) and original tumors (n = 16). In one case, recurrent tumor samples from 3 operations were available, and matching EVs were obtained from 2 cultures (04-1_CL, 04-2_CL), however, one culture (01-1_CL) was eventually lost for analysis (Supplementary Table 2). Methylation profiling was performed using Infinium MethylationEPIC BeadChip Arrays.
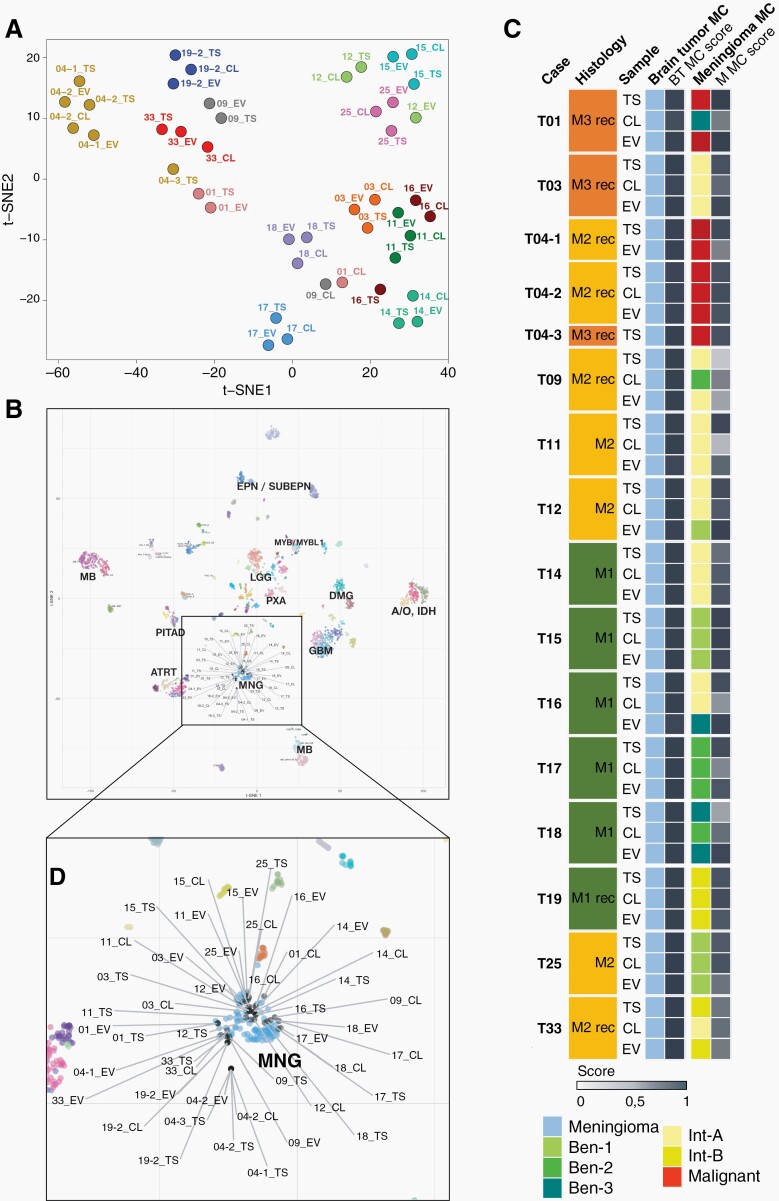
Using t-distributed stochastic neighbor embedding dimensionality reduction (t-SNE) of normalised methylation intensities in EVs, cells, and tissue, we found that EVs mostly mapped in close proximity to their corresponding parental cells and tumor tissue (Figure 3A). We then aligned our samples to the CNS tumor reference database, including over 2800 reference samples, representative of more than 80 tumor methylation classes.16 All our samples of EVs, cells, and tissue mapped to the meningioma cluster (Figure 3B).
Fig. 3.
DNA methylation profiling. (A) t-SNE analysis of genome-wide methylation profiles of EV-DNA, corresponding cells (CL), and tumor tissue (TS). (B) Comparison of samples with the Heidelberg CNS tumor reference cohort. All EV samples, corresponding cells, and tumor tissue (grey) map to the meningioma (MNG) cluster (light blue). (C) Summarized results of methylation profiling reports. In cases where tumor tissue, cells, or EVs displayed mixed epigenetic signatures, corresponding to more than one methylation class, the colour code represents the class with the higher match score.
We next submitted the methylation data to the Heidelberg brain tumor methylation classifier, which assigns unknown CNS tumors to different tumor methylation classes and subclasses based on prediction scores.16 All samples of EVs, cells, and tumors were correctly identified as meningioma with a high match score of >0.9 (Figure 3C). In addition, samples were subclassified into 6 meningioma methylation classes, including 3 benign subclasses (ben-1, ben-2, ben-3), two subclasses with intermediate outcome (int-A, int-B), and one malignant class with highly aggressive outcome (Figure 3C).21 EVs were correctly assigned to the methylation subclass of the corresponding original tumor in 13 of 15 samples that were available for paired analysis. In all except one (case 09) of the matching EV samples, the match score was at least 0.6. In case 09, the original tumor displayed a mixed epigenetic signature, partially corresponding to int-A (score 0.63) and to int-B (score 0.27), and for EV-DNA the int-A and int-B scores were 0.45 and 0.42, reflecting the biological duality of the tumor. In two other cases (12 and 16) the EV methylation class did not match the cells and original tumors. In case 12, the EV-DNA quality was low according to the CNV profile, which can serve as a quality control (Supplementary Fig. 2). In case 16, the cultured cells exhibited a relatively low match score of 0.52 for int-A compared to a high score of 0.92 for the tumor and EVs were classified as ben-3, suggesting that tumor heterogeneity and/or sampling variation or in vitro cell selection may account for the divergence.
In 4 of the 15 cases (01, 09, 18, and 33), the methylation subclass of the cultured cells did not match that of the corresponding tumor. In cases 01 and 09 the copy number variation (CNV) profiles of the cells were considerably flatter than those of the corresponding tumors and EVs (Supplementary Fig. 2), suggesting that tumor cells had eventually become senescent and overgrown by fibroblasts, which is common to occur in meningioma primary cultures22 and consistent with our microscopic impression of some cultures, although we did not systematically document the microscopic findings of the cultures over time. Of note, the collection of supernatants for EV preparation was initiated after the first medium change and continued over several weeks, after which the cells were finally harvested. Conditioned media from different collection time points were pooled for EV isolation, so that a substantial proportion of EV-DNA was derived from earlier cultures, explaining why in several cases EV methylation profiles were more consistent with original tumors than cell profiles. In cases 18 and 33 the original tumors exhibited mixed epigenetic signatures with features of both ben-3 (score 0.45) and ben-2 (score 0.33) in case 18, and of int-B (score 0.72) and int-A (score 0.18) in case 33, whereas in cell cultures the signature with the respective lower score had become dominant, suggesting adaptation and cell selection after prolonged culturing.
Taken together, our findings show that the methylation pattern of meningiomas is usually maintained in EV-DNA, allowing unequivocal classification of meningioma in all cases and meningioma subclassification in the vast majority of cases.
CNV Analysis of Meningioma EVs
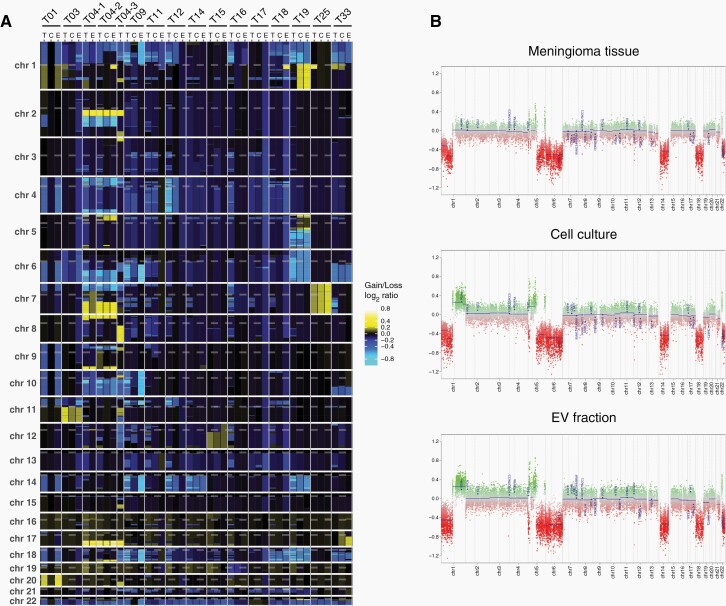
Copy number alterations were determined by analysing the methylation array data,23 (Supplementary Fig. 2). Heatmap representation of gains and losses highlights the CNV similarities between EV-DNA, cells, and matching tissues (Figure 4A). Hemizygous deletions of 22q were present in 14 of the 15 original tumors and were all also detected in corresponding EV-DNA. Losses involving 1p were found in 12 tumors and were also visible in EV samples except for case 12, in which the DNA quality was low. Other recurrent large deletions in tumor tissue affected chromosome arms 6q (8 tumors), 18q (7 tumors), 10q (6 tumors), 14q (5 tumors) 4p (5 tumors), and 11p (5 tumors), and all of these losses were also detectable in EV-DNA except for case 12. In addition, two tumors (T04-1 and T04-2) harbored hemizygous deletions of CDKN2A/B, which were also found in EV-DNA.
Fig. 4.
CNV analysis of meningioma EVs. (A) Heatmap representation of genome-wide copy number gains and losses inferred from the DNA methylation analysis. (B) Example of CNV profiles for tumor T19 with corresponding cells and EVs.
Chromosomal gains were infrequent and displayed no recurrent pattern, however, in all cases in which gains were present in tumors they were also present in corresponding EVs (cases 01, 03, 04-1, 04-2, 25). In case 19, gain of 1p newly occurred in cell culture and was also detected in EVs, suggesting that the original tumor was either genetically heterogeneous or the gain was newly acquired during culturing (Figure 4B). Likewise, loss on 7q in case 33 was exclusively present in the tumor, suggesting that a tumor subclone without this alteration was dominant in vitro.
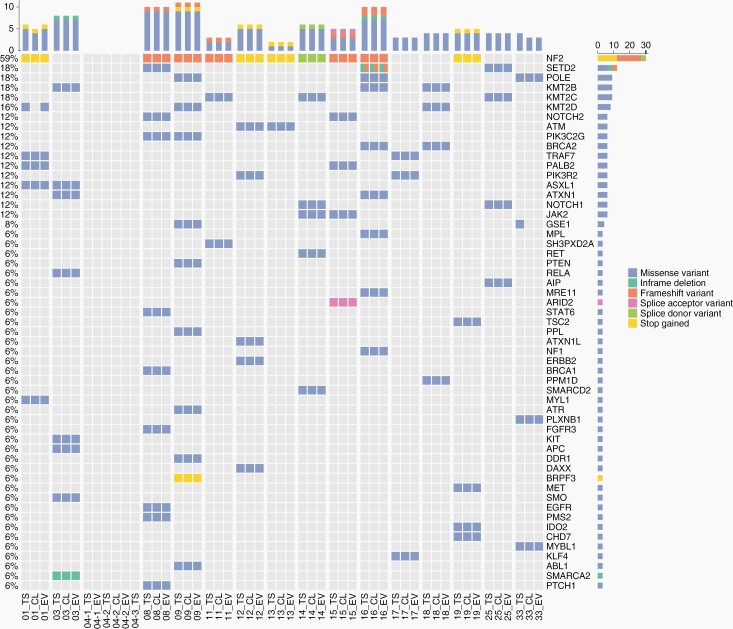
Mutation Analysis of Meningioma EVs
All samples that underwent methylation profiling, as well as two additional cases in which matching EVs and cells were available (case 08 and 13), were analyzed by gene panel sequencing to investigate 169 genes that are recurrently altered in meningiomas or other brain tumors. The somatic origin of DNA sequence alterations was determined through public databases, and details on detected alterations, including variant allele frequencies, are provided in Supplementary Table 3. The oncoprint diagram highlights the high congruence of single nucleotide variants (SNVs) and indels detected in tumors, EVs, and cells (Figure 5). NF2 mutations were found in 10 of 18 tumors, and were all detectable in corresponding EV-DNA and cells (Figure 5). Mutations in TRAF7 are second most common in meningiomas,21 and alterations in this gene were found in two tumors as well as corresponding EVs. SMO and PTEN are also recurrently mutated in meningiomas, and alterations in these genes each occurred in one tumor as well as in matching EVs (Figure 5). No mutations were found in other meningioma associated genes, such as KLF4, AKT1, POLR2A, PIK3CA, SUFU, or the TERT promoter. Of note, the SNV in KMT2D found in the tumor and EVs of case 01 was not detected in cells, which can be explained by eventual overgrowth by fibroblasts. In case 33, the alteration in GSE1 was only present in the tumor, providing further evidence for clonal heterogeneity which was observed in the CNV analysis.
Fig. 5.
Oncoprint of mutational frequencies and types of alterations detected by gene panel sequencing.
Proteomic Profiles of Meningioma EVs
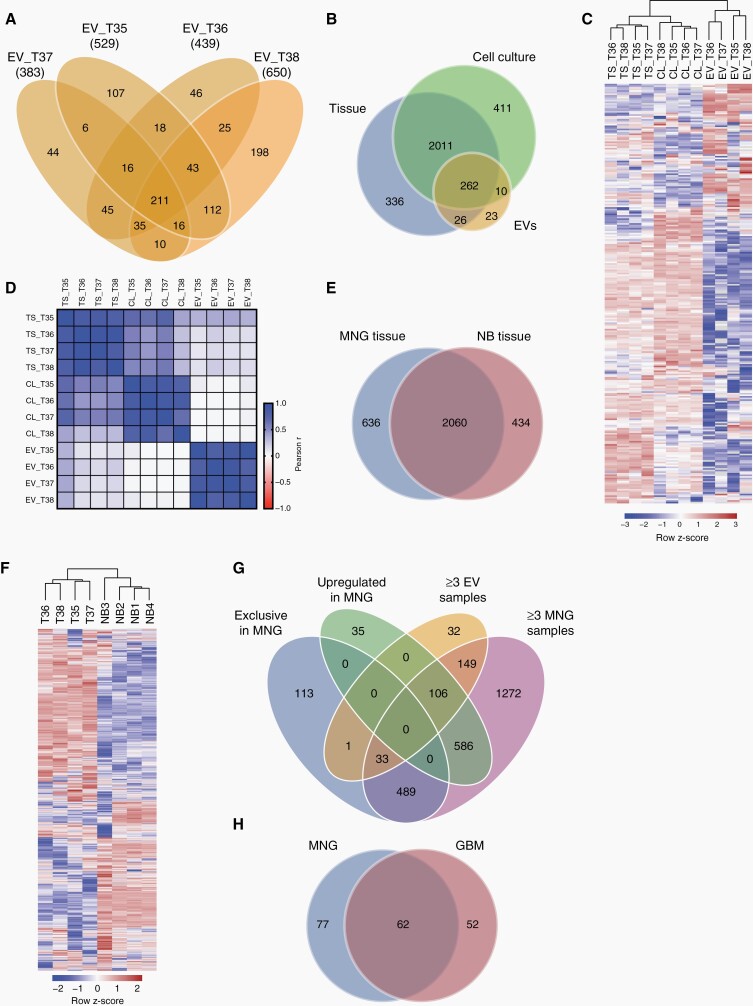
To investigate whether meningioma protein signatures are reflected in EVs, we analyzed the protein composition of tissues, cells, and EVs from 4 meningioma patients by differential quantitative proteomics. The total number of detected proteins in EV samples ranged from 383 (case 37) to 650 (case 38), (Figure 6A, Supplementary Table 4). Of 321 proteins that were present in at least 3 of 4 EV specimens, 262 were also found in at least 3 of 4 tumor and cell samples (Figure 6B, Supplementary Fig. 3A).
Fig. 6.
Proteomic profiles of meningioma EVs. (A) Numbers of proteins detected in EVs from cultured meningioma cells by differential quantitative proteomics. (B) Proteins detected in at least 3 of 4 samples each of EVs, cells, and original tumors. (C) Unsupervised clustering based on 262 proteins present in all three sample types. (D) Pearson correlation analysis of the proteomes of EVs, cells (C), and tumor tissue (T). (E) Overlap between proteins presents in ≥3 meningioma (MNG) tissue samples and in ≥3 samples of normal brain (NB). (F) Unsupervised clustering based on all proteins detected in ≥3 MNG samples and ≥3 NB samples. (G) Proteins either exclusively detected in MNG tissue vs NB, upregulated >2-fold in MNG vs NB, present in ≥3EV samples, and in ≥3 MNG tissue samples. (H) Overlap between proteins exclusive or upregulated in MNG tissue and present in ≥3EV samples as well as ≥3 MNG tissue samples, and proteins exclusive or upregulated in glioblastoma (GBM) tissue and present in at least 3 of 4 GBM EV samples as well as 3 of 4 GBM tissue samples (re-analyzed GBM data are from Maire et al.11).
Unsupervised hierarchical clustering of these proteins showed that tumor tissue, cells, and EVs each formed separate clusters and that tumors and cells shared more in common than they shared with EVs (Figure 6C). The similarity within the different groups of samples was confirmed by correlation analysis, however, no consistent correlations between EVs and corresponding cells or tumors were found (Figure 6D, Supplementary Fig. 3B and C). Instead, all EV samples were most closely related to tumor T35 and also the 4 cell cultures displayed highest similarity with this tumor.
In order to identify proteins that are specifically enriched in meningioma-derived EVs, we first compared the proteins contained in meningioma tissue samples to 4 samples of normal brain, which could be another source of the increased plasma EVs in meningioma patients, given the tumor-associated disruption of the blood–brain barrier. In total, 636 proteins were exclusively detected in meningiomas (Figure 6E, Supplementary Table 5). Of note, somatostatin receptor type 2 (SSTR2), a well-known target for meningioma imaging with radiolabeled SSTR2 ligands,3,24 was among these proteins. In addition to proteins that were exclusively found in meningioma samples, we further identified 727 proteins that were upregulated >2-fold in meningiomas vs normal brain (Figure 6F, Supplementary Table 6).
We then compared proteins that were (1) either exclusively found in meningioma tissue, or (2) >2-fold upregulated vs. normal brain, and (3) found in ≥3 EV samples, as well as (4) ≥3 meningioma tissue samples (Figure 6G). Of those proteins that were either exclusive or upregulated in meningioma tissue, 139 (106 + 33) were also detected in ≥3 EV and tumor samples (Figure 6G, Supplementary Table 7). Gene ontology analysis revealed that many of these proteins were related to exosomes/vesicles or extracellular matrix (ECM) interactions (Supplementary Fig. 4A, B). In a previous study we had analyzed EVs secreted by glioblastoma cells (n = 4) using the same proteomics approach, and had identified 114 proteins as exclusive or upregulated in glioblastoma tissue vs normal brain and present in ≥3 EVs and tumors.11 We therefore compared the 139 proteins identified in meningioma EVs with those identified in glioblastoma EVs. A third of all proteins overlapped between the two entities and 77 proteins were exclusive to meningioma EVs (Figure 6H, Supplementary Table 8). Among the latter was desmoplakin, a well-known meningioma marker.25 Other proteins that were specifically associated with meningioma EVs included periostin, matrix gla protein, tenascin x, and tetranectin, all of which are known to be normally involved in connective tissue and bone formation.
Collectively, these findings demonstrate that while the EV proteome does not allow to identify the precise tumor of origin, it can identify proteins which are potentially useful for enriching meningioma EVs.
Discussion
In summary, our study presents the following major findings: (1) Levels of circulating EVs are elevated in meningioma patients, and plasma EV counts correlate with the extent of tumor edema and malignancy grade; (2) the magnitude of the postoperative decline in EV numbers is associated with the extent of tumor resection; (3) methylation profiling of EV-DNA unequivocally allows accurate tumor classification as meningioma, and in almost all cases also correct methylation subclass assignment; (4) tumor-specific genome-wide copy number alterations are identifiable by meningioma EV-DNA methylome analysis; (5) targeted gene panel sequencing of EV-DNA facilitates the detection of meningioma-specific mutations, including potentially targetable driver mutations; (6) proteomic EV profiling does not permit original tumor identification but recognizes tumor-associated proteins potentially valuable for enriching meningioma EVs from biofluids.
Several studies have previously reported that the number of EVs is increased in the blood of glioblastoma patients compared with healthy donors.7–10 Osti et al. observed a 3.3-fold increase in circulating EVs in glioblastoma patients and suggested that this increase is specific for glioblastomas and does not pertain to other brain tumors.8 However, only 6 meningioma patients (of undescribed WHO grade) had been included in the investigation and results for meningiomas were pooled with other extra-axial tumors. Our study shows that about half of the patients with M1 displayed EV counts above those in healthy individuals and that EV concentrations increased with malignancy grade, up to a fold change of 5.4 in M3 which is even higher than reported for glioblastomas.8,9 EV levels were not associated with tumor size per se, but correlated with the extent of tumor-associated edema, suggesting that leakiness of the blood–brain barrier could primarily be responsible for the elevated peripheral EVs. Notably, other factors are also likely to affect levels of circulating EVs, since for example three M1 patients with highest EV counts displayed a rather broad range of tumor and edema volumes (8.0–150.0 cm3 and 7.9–88.2cm3, respectively). The rapid postoperative decline in circulating EVs and the association of the magnitude of reduction with the extent of resection strongly suggest that the presence of the tumor is responsible for the observed vesiclemia in meningioma patients. However, with currently available methods the precise origin of EVs in the bloodstream cannot be defined, and they may be derived from the tumor itself as well as from the tumor environment. Nonetheless, our data show that increased numbers of circulating EVs can serve as a biomarker indicative of tumor presence especially in higher grade meningiomas, which could potentially be useful in monitoring disease status and treatment response.
Genome-wide methylation profiling is a powerful tool for the classification of brain tumors. Methylation array analysis can predict patient outcome and recurrence more reliably than histological tumor grade.21,26 Integration of methylomic findings with WHO grading, CNV, mutation, and mRNA data can further refine prognostic accuracy.27,28 Our analysis of the methylomic, CNV, and mutational profiles of meningioma EVs shows that tumor-specific alterations are detectable in EV-DNA with high fidelity. Methylation profiles of all EV samples were highly similar to their corresponding original tumors, and all samples were correctly identified as meningioma by the Heidelberg classifier. By subclass analysis, all except two of the 15 EV samples were correctly assigned to the methylation subclass of the tumor. In the two nonmatching cases, the divergence could be attributed to either low EV-DNA quality (case 12) or in vitro cell selection (case 16). CNVs present in tumors were also detected in EV-DNA with high accuracy, including CDKN2A/B deletions. In two cases, the cultured cells (and EVs) differed slightly from the original tumor, by either acquiring gain of 1p (case 19) or failing to show loss on 7q (case 33). In case 33, sequencing revealed that also a missense alteration affecting the GSE1 gene was lost in vitro, indicating tumor heterogeneity and sampling variation. In all other cases, tumor mutations were faithfully reflected in EV-DNA, including in particular known meningioma driver mutations in NF2 and TRAF7. Taken together our results show that EVs reflect the epigenetic and genomic aberrations present in meningiomas with high accuracy and that methylomic tumor subclassification can be achieved via EV-DNA analysis.
Ideally, liquid biopsy could one day become a means to noninvasively monitor disease dynamics, including information on temporal heterogeneity, that is molecular changes over time that may prompt early therapeutic intervention. To that effect, Nassiri et al. reported that sequencing of methylated cell-free DNA (cfDNA) from patient plasma can in principle separate different groups of brain tumors, including meningiomas, IDH-mutant and IDHwt gliomas, and others.29 However, since the proportion of tumor-derived circulating DNA is low in the blood of patients with brain tumors,30,31 it is questionable that more subtle analyses of tumor subclasses and mutations will be feasible based on complete cfDNA analysis. While cfDNA is usually highly fragmented, EV-DNA is of higher molecular weight,32 which may be advantageous, provided that tumor-derived EVs can be enriched from patient plasma for molecular analyses. We therefore aimed to identify meningioma-specific proteins associated with EVs that could help to specifically capture and enrich tumor-derived EVs.
By differential quantitative proteomic analyses of meningioma EVs we could not identify the precise tumors or cells of origin. Instead, all EVs and also cells were most similar to the same original tumor (T35), suggesting that the proteome of this tumor most closely resembles the in vitro signature. However, we identified 139 proteins that were upregulated in meningiomas vs normal brain and also present in EVs, and might thus be useful for EV enrichment. Of note, despite being upregulated in meningioma tissue samples, the SSTR2 was not among these proteins and was completely nondetectable in EVs. The comparison with glioblastoma EVs left 77 proteins exclusive to meningioma EVs. Interestingly, many of these were also previously found to be upregulated in meningioma tissue vs normal brain, including desmoplakin, transgelin, filamin-B, vinculin, and others, validating our current findings.33 Most notably, desmoplakin is a well-known marker for the diagnosis of meningioma, and a major component of desmosomal intercellular junctions between meningioma cells,25 suggesting that this protein might be a candidate for enriching meningioma EVs for subsequent molecular analyses. Additional potential candidates identified by us include periostin, matrix gla protein, tenascin x, or tetranectin.
In conclusion, our study demonstrates that the number of circulating EVs is increased in meningioma patients and that epigenetic and genomic profiling of meningioma EV-DNA allows methylomic classification as well as correct identification of mutations and CNVs. Future studies are necessary to translate these findings to patients and investigate how circulating EVs can be leveraged for tumor classification and long-term disease monitoring, and to explore whether EV enrichment via capture of meningioma-associated proteins can support this task.
Supplementary Material
Acknowledgments
We thank the FACS core facility at the University Medical Center Hamburg-Eppendorf for support with the imaging flow cytometry.
Contributor Information
Franz L Ricklefs, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Cecile L Maire, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Kathrin Wollmann, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Lasse Dührsen, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Krystian D Fita, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Felix Sahm, Department of Neuropathology, University Hospital Heidelberg, Heidelberg, Germany.
Christel Herold-Mende, Division of Neurosurgical Research, Department of Neurosurgery, University Hospital Heidelberg, Heidelberg, Germany.
Andreas von Deimling, Department of Neuropathology, University Hospital Heidelberg, Heidelberg, Germany.
Katharina Kolbe, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Mareike Holz, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Leonie Bergmann, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Marceline M Fuh, Department of Biochemistry and Molecular Cell Biology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Hartmut Schlüter, Institute of Clinical Chemistry and Laboratory Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Malik Alawi, Bioinformatics Core, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Rudolph Reimer, Heinrich-Pette-Institut, Leibniz Institute for Experimental Virology, Hamburg, Germany.
Sven Peine, Institute of Clinical Chemistry and Laboratory Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Markus Glatzel, Institute of Neuropathology, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Manfred Westphal, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Katrin Lamszus, Department of Neurosurgery, University Medical Center Hamburg-Eppendorf, Hamburg, Germany.
Funding
This study was supported by a collaborative grant of the Deutsche Krebshilfe to K.L., F.L.R., M.W., F.S., C.H.M and A.v.D. (grant number 70112956) and by a grant from the Deutsche Forschungsgemeinschaft to F.L.R. and K.L. (RI2616/3-1).
Conflict of interest statement. None declared.
Authorship statement. K.L. and F.L.R. conceived the study, designed and analyzed experiments, and prepared the manuscript. C.L.M., K.W., K.K., M.H. and L.B. performed experiments and analyzed data. L.D. and S.P. acquired clinical data and provided human samples. F.S., C.H.M., S.F. and A.v.D. performed methylation array analyses and gene panel sequencing. M.M.F. and H.S. performed proteomic analyses. K.D.F. and M.A. performed bioinformatic analyses. R.R. performed electron microscopy. M.G. and M.W. provided scientific input.
References
- 1. Ostrom QT, Patil N, Cioffi G, et al. . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013-2017. Neuro Oncol. 2020; 22(12 Suppl 2):iv1–iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Suppiah S, Nassiri F, Bi WL, et al. . Molecular and translational advances in meningiomas. Neuro Oncol. 2019; 21(Suppl 1):i4–i17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldbrunner R, Stavrinou P, Jenkinson MD, et al. . EANO guideline on the diagnosis and management of meningiomas. Neuro Oncol. 2021; 23(11):1821–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Preusser M, Brastianos PK, Mawrin C. Advances in meningioma genetics: novel therapeutic opportunities. Nat Rev Neurol. 2018; 14(2):106–115. [DOI] [PubMed] [Google Scholar]
- 5. EL Andaloussi S, Mager I, Breakefield XO, et al. . Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013; 12(5):347–357. [DOI] [PubMed] [Google Scholar]
- 6. Westphal M, Lamszus K. Circulating biomarkers for gliomas. Nat Rev Neurol. 2015; 11(10):556–566. [DOI] [PubMed] [Google Scholar]
- 7. Cumba Garcia LM, Peterson TE, Cepeda MA, et al. . Isolation and analysis of plasma-derived exosomes in patients with glioma. Front Oncol. 2019; 9:651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Osti D, Del Bene M, Rappa G, et al. . Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019; 25(1):266–276. [DOI] [PubMed] [Google Scholar]
- 9. Ricklefs FL, Maire CL, Reimer R, et al. . Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019; 8(1):1588555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabbagh Q, Andre-Gregoire G, Alves-Nicolau C, et al. . The von Willebrand factor stamps plasmatic extracellular vesicles from glioblastoma patients. Sci Rep. 2021; 11(1):22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maire CL, Fuh MM, Kaulich K, et al. . Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 2021; 23(7):1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dobra G, Bukva M, Szabo Z, et al. . Small extracellular vesicles isolated from serum may serve as signal-enhancers for the monitoring of CNS tumors. Int J Mol Sci. 2020; 21(15):5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Negroni C, Hilton DA, Ercolano E, et al. . GATA-4, a potential novel therapeutic target for high-grade meningioma, regulates miR-497, a potential novel circulating biomarker for high-grade meningioma. EBioMedicine. 2020; 59:102941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gunther HS, Schmidt NO, Phillips HS, et al. . Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008; 27(20):2897–2909. [DOI] [PubMed] [Google Scholar]
- 15. Ricklefs FL, Alayo Q, Krenzlin H, et al. . Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018; 4(3):eaar2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Capper D, Jones DTW, Sill M, et al. . DNA methylation-based classification of central nervous system tumours. Nature. 2018; 555(7697):469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neumann JE, Spohn M, Obrecht D, et al. . Molecular characterization of histopathological ependymoma variants. Acta Neuropathol. 2020; 139(2):305–318. [DOI] [PubMed] [Google Scholar]
- 18. Sahm F, Schrimpf D, Jones DT, et al. . Next-generation sequencing in routine brain tumor diagnostics enables an integrated diagnosis and identifies actionable targets. Acta Neuropathol. 2016; 131(6):903–910. [DOI] [PubMed] [Google Scholar]
- 19. Combs SE, Edler L, Burkholder I, et al. . Treatment of patients with atypical meningiomas Simpson grade 4 and 5 with a carbon ion boost in combination with postoperative photon radiotherapy: the MARCIE trial. BMC Cancer. 2010; 10:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rikkert LG, Nieuwland R, Terstappen L, et al. . Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J Extracell Vesicles. 2019; 8(1):1555419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sahm F, Schrimpf D, Stichel D, et al. . DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017; 18(5):682–694. [DOI] [PubMed] [Google Scholar]
- 22. Baia GS, Slocum AL, Hyer JD, et al. . A genetic strategy to overcome the senescence of primary meningioma cell cultures. J Neurooncol. 2006; 78(2):113–121. [DOI] [PubMed] [Google Scholar]
- 23. Capper D, Stichel D, Sahm F, et al. . Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience. Acta Neuropathol. 2018; 136(2):181–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brastianos PK, Galanis E, Butowski N, et al. . Advances in multidisciplinary therapy for meningiomas. Neuro Oncol. 2019; 21(Suppl 1):i18–i31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akat K, Mennel HD, Kremer P, et al. . Molecular characterization of desmosomes in meningiomas and arachnoidal tissue. Acta Neuropathol. 2003; 106(4):337–347. [DOI] [PubMed] [Google Scholar]
- 26. Nassiri F, Mamatjan Y, Suppiah S, et al. . DNA methylation profiling to predict recurrence risk in meningioma: development and validation of a nomogram to optimize clinical management. Neuro Oncol. 2019; 21(7):901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maas SLN, Stichel D, Hielscher T, et al. Integrated molecular-morphologic meningioma classification: a multicenter retrospective analysis, retrospectively and prospectively validated. J Clin Oncol. 2021; 39(34):3839–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nassiri F, Liu J, Patil V, et al. . A clinically applicable integrative molecular classification of meningiomas. Nature. 2021; 597(7874):119–125. [DOI] [PubMed] [Google Scholar]
- 29. Nassiri F, Chakravarthy A, Feng S, et al. . Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat Med. 2020; 26(7):1044–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bettegowda C, Sausen M, Leary RJ, et al. . Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014; 6(224):224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wan JCM, Heider K, Gale D, et al. . ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci Transl Med. 2020; 12(548):eaaz8084. [DOI] [PubMed] [Google Scholar]
- 32. Thakur BK, Zhang H, Becker A, et al. . Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014; 24(6):766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sharma S, Ray S, Mukherjee S, et al. . Multipronged quantitative proteomic analyses indicate modulation of various signal transduction pathways in human meningiomas. Proteomics. 2015; 15(2-3):394–407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.