Summary
Antigen cross-presentation, wherein dendritic cells (DC) present exogenous antigen on major histocompatibility class I (MHC-I) molecules, is considered the primary mechanism by which DCs initiate tumor-specific CD8+ T cell responses. Here, we demonstrate that MHC-I cross-dressing, an antigen presentation pathway in which DCs acquire and display intact tumor-derived peptide:MHC-I molecules, is also important in orchestrating anti-tumor immunity. Cancer cell MHC-I expression was required for optimal CD8+ T cell activation in two subcutaneous tumor models. In vivo acquisition of tumor-derived peptide:MHC-I molecules by DCs was sufficient to induce antigen-specific CD8+ T cell priming. Transfer of tumor-derived human leukocyte antigen (HLA) molecules to myeloid cells was detected in vitro and in human tumor xenografts. In conclusion, MHC-I cross-dressing is crucial for anti-tumor CD8+ T cell priming by DCs. In addition to quantitatively enhancing tumor antigen presentation, MHC cross-dressing might also enable DCs to more faithfully and efficiently mirror the cancer cell peptidome.
eTOC blurb
Although antigen cross-presentation is important for DCs to orchestrate anti-tumor CD8+ T cell responses, the role of alternative antigen presentation pathways is unclear. MacNabb et al show that CD8+ T cell priming can be mediated by DCs that acquire and present tumor-derived MHC-I complexes – a phenomenon known as MHC cross-dressing.
Introduction
The anti-tumor immune response has been intensely studied over the past several decades (Vesely et al., 2011), leading to the development and widespread administration of cancer immunotherapies in the clinic (Baumeister et al., 2016; Waldman et al., 2020) However, cancer immunotherapy is not universally effective (Dougan et al., 2019), and progress is hindered by persisting gaps in our understanding of the fundamental mechanisms required for the orchestration of effective anti-cancer immune responses, particularly at the level of early T cell priming. While the mechanisms by which CD8+ T cells are primed against human cancers remain unclear, murine models have provided critical insights into these questions. Recently, the professional antigen presenting cells (APCs) involved in anti-tumor T cell priming have been identified (Roberts et al., 2016; Salmon et al., 2016); however, the specific pathways utilized by these APCs to acquire and present tumor antigens in vivo remain incompletely defined.
Migratory CD103+ and CD11b+ dendritic cells (DCs) exist in tissues throughout the body, constitutively acquiring proteins from surrounding cells before trafficking to draining lymph nodes, where they present derivative peptide antigens to T cells in the context of MHC molecules (Merad et al., 2013; Worbs et al., 2017). Migratory DCs within tumors similarly acquire and subsequently present cancer cell-derived antigens to T cells in tumor-draining lymph nodes (tdLNs) (Wculek et al., 2020). Recent evidence indicates that tumor antigens are exclusively transported to tdLNs by migratory CD103+ DCs (Ruhland et al., 2020; Salmon et al., 2016), and that these DCs are primarily responsible for anti-tumor CD8+ T cell priming in vivo (Roberts et al., 2016; Salmon et al., 2016). Migratory CD103+ DCs, along with lymph node resident CD8α+ DCs, comprise the BATF3- and IRF8-dependent conventional type 1 DC (cDC1) lineage (Anderson et al., 2021; Edelson et al., 2010; Hildner et al., 2008). Because antigen cross-presentation is a canonical function of cDC1 (Dudziak et al., 2007; Hildner et al., 2008), and because numerous studies have demonstrated a requirement for cDC1 in the activation of anti-tumor CD8+ T cell responses (Fuertes et al., 2011; Hildner et al., 2008; Wculek et al., 2020), prevailing thought is that tumor-specific CD8+ T cells are primed exclusively through antigen cross-presentation (Lee et al., 2020; Sánchez-Paulete et al., 2017).
However, an alternative antigen presentation mechanism has been described, in which DCs acquire and present intact peptide:MHC (pMHC) complexes captured directly from neighboring cells (André et al., 2004; Dolan et al., 2006; Herrera et al., 2004; Russo et al., 2000; Wolfers et al., 2001). This phenomenon, known as MHC cross-dressing, has been implicated in antigen presentation in various contexts ranging from viral infection (Wakim and Bevan, 2011) and vaccination (Li et al., 2012) to thymic selection (Koble and Kyewski, 2009; Kroger et al., 2017; Perry et al., 2018), graft rejection (Liu et al., 2016), and peripheral tolerance to maternal microchimerism (Bracamonte-Baran et al., 2017). By necessity, conclusions regarding the role of MHC cross-dressing in antigen presentation by DCs have largely arisen from in vitro studies and in vivo models of MHC-mismatched bone marrow chimeric mice (Dolan et al., 2006; Koble and Kyewski, 2009; Wakim and Bevan, 2011) or solid organ transplantation (Liu et al., 2016), due to difficulty in controlling for cross-presentation without entirely ablating antigen presentation on MHC-I. Indeed, while MHC cross-dressing by DCs is sufficient to induce T cell priming in various contexts, the necessity of this antigen presentation pathway in mediating in vivo T cell activation has never been conclusively demonstrated in syngeneic hosts.
In this study, we employed two syngeneic murine tumor models expressing distinct model antigens presented in the context of the MHC-I molecule, H-2Kb (Kb), in order to determine the extent to which MHC-I cross-dressing was involved in antigen-specific CD8+ T cell priming. While the magnitude of the effect varied, CD8+ T cell priming against Kb-restricted tumor antigens was impaired in mice harboring Kb−/− tumors, despite the cross-presentation pathway being fully intact. Furthermore, cancer cell-derived MHC-I molecules were readily observed within and on the surface of tumor-resident APCs, and MHC-I-deficient CD103+ cDC1 isolated from tdLNs of mice bearing Kb-sufficient tumors stimulated antigen-specific CD8+ T cells ex vivo. The importance of WDFY4-dependent antigen cross-presentation (Theisen et al., 2018) in mediating anti-tumor CD8+ T cell responses differed across experimental models, but in some cases was dispensable for in vivo CD8+ T cell priming. Finally, we observed that APCs became cross-dressed with human leukocyte antigen class I (HLA-I) molecules upon co-culture with HLA-mismatched tumor cells, and in tumors xenografted in immunodeficient mice. Acquisition of tumor cell-derived HLA-I molecules correlated with uptake of tumor antigens in vitro. Similarly, APCs isolated from murine tumors acquired both tumor-derived MHC-I and fluorescent antigen in vivo, suggesting that MHC-I cross-dressing and internalization of tumor material may be linked processes. Taken together, our results demonstrate that MHC-I cross-dressing is central to the ability of cDC1 to orchestrate anti-tumor CD8+ T cell responses.
Results
Loss of H-2Kb does not impact model antigen expression in cancer cells.
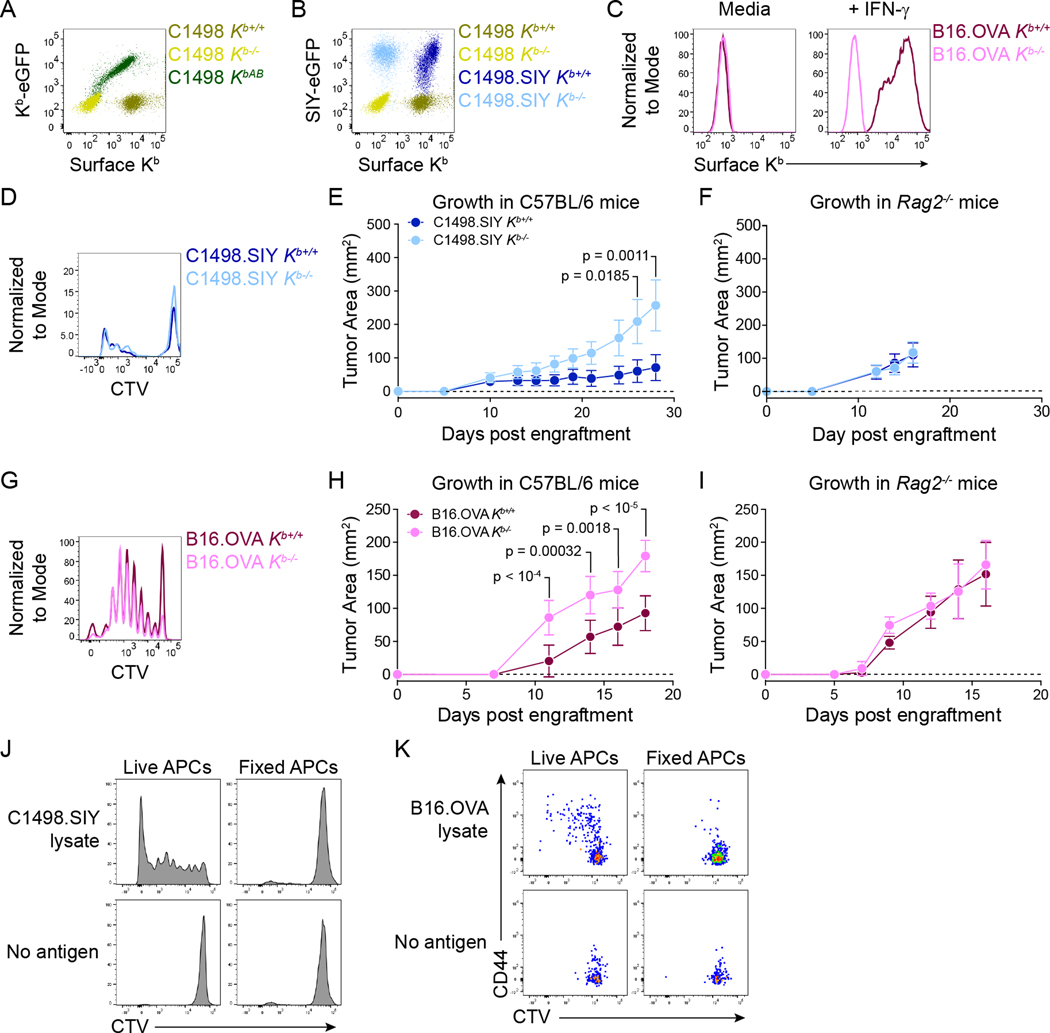
In order to define the impact of cancer cell MHC-I expression on anti-tumor CD8+ T cell priming, two tumor models were utilized, C1498 leukemia and B16.F10 melanoma (both H-2b). These models were selected due to differences in cell of origin and baseline MHC-I expression, and both have been extensively characterized, with well-established growth kinetics in syngeneic C57BL/6 mice (LaBelle et al., 2002; Overwijk and Restifo, 2000). Moreover, the timing and magnitude of endogenous antigen-specific CD8+ T cell responses raised against these tumors have been defined (Kline et al., 2008; Zhang et al., 2013). To facilitate tracking of tumor antigen-specific CD8+ T cell responses, C1498 cells expressing a Kb-restricted model peptide antigen, SIYRYYGL (SIY) (Kline et al., 2018; Zhang et al., 2013), and B16.F10 cells expressing the C-terminal domain (amino acids 161–385) of chicken ovalbumin (OVA), including its derivative Kb-restricted peptide antigen, SIINFEKL (OVA257–264) (de Witte et al., 2006), were employed. Kb-deficient (Kb−/−) parental C1498, C1498.SIY and B16.OVA cell lines were generated (Figure 1A–C), which allowed for a direct comparison of antigen-specific CD8+ T cell priming in mice bearing Kb+/+ versus Kb−/− tumors. Additionally, Kb was re-expressed in C1498 Kb−/− cells (C1498 KbAdd-back; KbAB) with a C-terminal eGFP tag (Hein et al., 2014), so that the localization of tumor-derived Kb molecules could be assessed within host APCs (Figure 1A).
Figure 1: Validation of Kb−/− cancer cell lines. A).
Surface anti-Kb antibody staining and Kb-eGFP expression in C1498 Kb+/+, Kb−/−, and KbAB cells. B) Surface anti-Kb antibody staining and SIY-eGFP expression in C1498 and C1498.SIY Kb+/+ and Kb−/− cells. C) Surface anti-Kb antibody staining on B16.OVA Kb+/+ and Kb−/− cells with and without IFN-γ treatment. D) Histogram showing CTV dilution by 2C T cells after a 72-hour co-culture with BMDCs and C1498.SIY Kb+/+ or Kb−/− cell lysates. E and F) Growth of C1498.SIY Kb+/+ and Kb−/− tumors in C57BL/6 (E) and Rag2−/− mice (F). G) Histogram showing CTV dilution by OT-I T cells following 72-hour co-culture with BMDCs and B16.OVA Kb+/+ or Kb−/− cell lysates. H and I) Growth of B16.OVA Kb+/+ and Kb−/− tumors in C57BL/6 (H) and Rag2−/− (I) mice. J) Histograms showing CTV dilution by 2C T cells following 72-hour co-culture with live or fixed CD11c+ splenocytes and C1498.SIY cell lysates. K) Flow cytometry plots showing CTV dilution and CD44 expression by OT-I T cells following 72-hour co-culture with live or fixed BMDCs and B16.OVA cell lysates. Statistical significance for tumor growth experiments was determined by two-way ANOVA with Sidak’s post-hoc test for multiple comparisons. Tumor growth data are depicted as mean ± s.d. and are pooled from two independent experiments (n = 10 mice per group in E and H; n = 5 mice per group in F) or from one experiment (I, n = 5 mice per group). Flow cytometry plots are representative of at least three independent experiments.
In C1498.SIY cells, the SIY peptide was expressed in-frame at the C-terminus of eGFP, which allowed for monitoring of SIY antigen expression (eGFP fluorescence), and for assessing uptake of C1498.SIY-derived proteins by tumor-resident APC populations. Importantly, SIY-eGFP expression in C1498.SIY Kb+/+ and C1498.SIY Kb−/− cells was similar, indicating that Kb deletion did not affect overall SIY antigen expression (Figure 1B). Additionally, bone marrow-derived dendritic cells (BMDCs) cultured with C1498.SIY Kb+/+ or C1498.SIY Kb−/− cell lysates were similarly capable of presenting SIY to antigen-specific, T cell receptor transgenic (TCR-tg) 2C CD8+ T cells in vitro (Figure 1D). As expected, subcutaneously (s.c.) implanted C1498.SIY Kb−/− tumors progressed more rapidly than C1498.SIY Kb+/+ tumors in C57BL/6 mice (Figure 1E), as the former could not be directly targeted for lysis by CD8+ T cells specific for Kb-restricted antigens. Conversely, C1498.SIY Kb+/+ and C1498.SIY Kb−/− tumors exhibited similar growth in Rag2−/− mice (Figure 1F), indicating that growth rate differences between Kb−/− and Kb+/+ C1498.SIY tumors in C57BL/6 mice were due to impaired effector responses by adaptive immune cells in the absence of cancer cell Kb expression.
Consistent with observations in C1498 cells, Kb deficiency did not affect OVA antigen expression in B16.F10 cells, as evidenced by equivalent in vitro activation of OVA257–264-specific TCR-tg OT-I CD8+ T cells by BMDCs cultured with B16.OVA Kb+/+ versus B16.OVA Kb−/− cell lysates (Figure 1G). B16.OVA Kb−/− tumors also grew more rapidly than B16.OVA Kb+/+ tumors in C57BL/6 mice (Figure 1H), while again, the two tumors grew similarly in Rag2−/− mice (Figure 1I). As expected, paraformaldehyde-fixed APCs presented neither C1498-derived SIY nor B16.F10-derived SIINFEKL in vitro (Figure 1J–K). Thus, we generated two tumor models with different baseline MHC-I expression and distinct Kb-restricted antigens in which to assess the role of cancer cell-derived MHC-I in anti-tumor CD8+ T cell priming.
CD8+ T cell priming against C1498.SIY tumors is dependent on cancer cell MHC-I expression
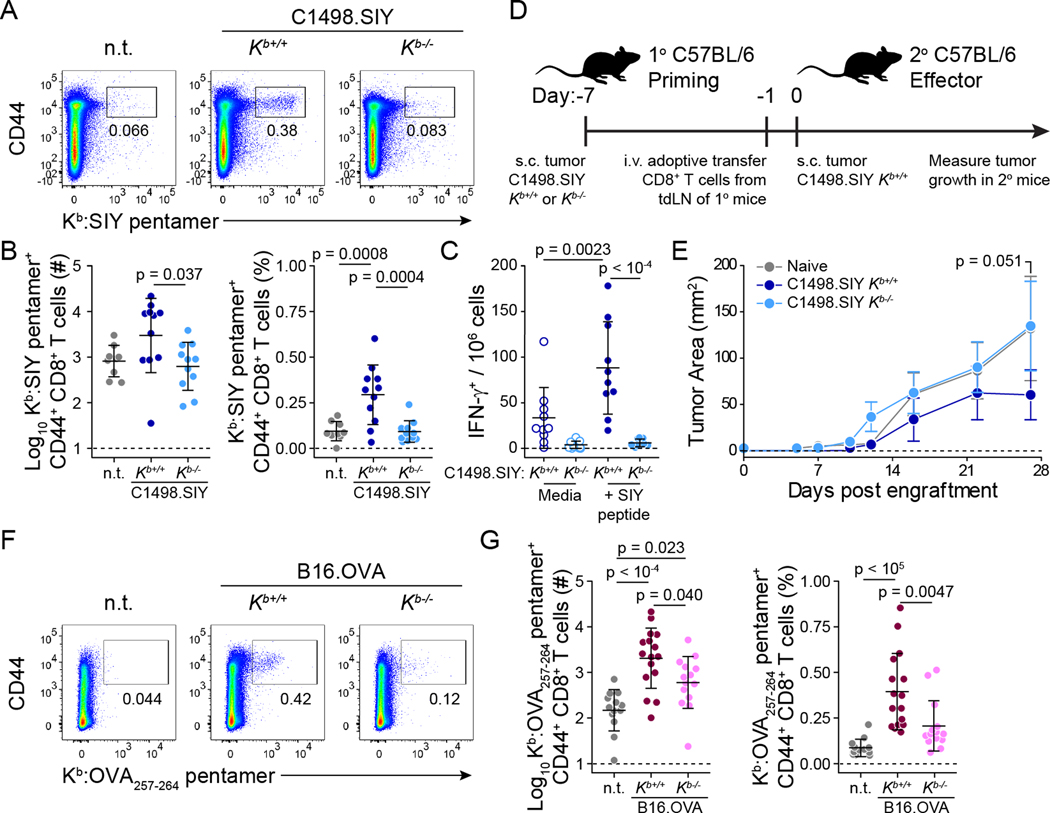
The degree to which MHC-I expression by cancer cells affected antigen-specific CD8+ T cell priming in tdLNs was determined by assessing the expansion of CellTrace violet (CTV)-labeled, adoptively transferred 2C CD8+ T cells six days following s.c. inoculation of C1498.SIY Kb+/+ or C1498.SIY Kb−/− cells in congenic C57BL/6 hosts (Figure 2A). In tdLNs of mice bearing C1498.SIY Kb+/+ tumors, 2C T cells proliferated extensively and accumulated both in terms of overall number and as a proportion of all tdLN CD8+ T cells (Figure 2B–E). In contrast, 2C T cell priming was almost completely abrogated in tdLNs of mice with C1498.SIY Kb−/− tumors, where 2C T cell frequencies and numbers were similar to those in analogous cutaneous lymph nodes (cLN) of tumor-free controls (Figure 2B–E). Additionally, 2C T cells that proliferated in response to C1498.SIY Kb+/+ tumors also produced the effector cytokines IFN-γ and TNF-α, as well as the cytolytic protein, granzyme B (Figure 2F–I). Thus, expression of Kb by C1498.SIY cells is required for functional 2C T cell priming.
Figure 2: Cancer cell Kb expression is required for optimal activation of Kb-restricted antigen-specific TCR-tg CD8+ T cells. A-E).
106 CTV-labeled CD45.1+ 2C T cells were transferred into C57BL/6 mice (CD45.2+) on day -1. Recipient mice were inoculated with 106 C1498.SIY Kb+/+ (n = 8) or Kb−/− (n = 9) cells or no tumor (n.t.; n = 5) on day 0. 2C T cell proliferation was assessed on day 6. Experimental design is depicted in (A). Representative flow cytometry plots depicting the identification of (B) and CTV dilution within (C) 2C T cells in tdLNs. Number (left) and frequency (right) of tdLN 2C T cells are quantified in (D). 2C T cell proliferation, quantified as percent divided (left), and the geometric mean fluorescence intensity (gMFI) of CTV within the population (right) in (E). F-I) CD45.1+ 2C T cells were transferred into C57BL/6 mice (CD45.2+) inoculated with 106 C1498.SIY Kb+/+ (n = 9) or Kb−/− (n = 9) cells or n.t. (n = 5) as in (A). 2C T cells were re-stimulated ex vivo with SIY peptide for 5 hours on day 6 and assessed for production of IFN-γ, TNF-α, and granzyme B. Representative flow cytometry plots are depicted in (F). Number and frequency of proliferated 2C T cells producing IFN-γ (G), TNF-a (H), and granzyme B (I). J-N) 106 CTV-labeled CD90.1+ OT-I T cells were adoptively transferred into C57BL/6 mice (CD90.2+) on day -1. Recipient mice were inoculated with 106 B16.OVA Kb+/+ (n = 11) or Kb−/− (n = 11) cells or n.t. (n = 5) on day 0. OT-I proliferation was assessed on day 6. Representative flow cytometry plots depicting the identification of (K) and CTV dilution by (L) OT-I T cells in tdLNs. Quantification of the number and frequency of OT-I T cells in tdLNs (M), and proliferation of OT-I T cells as percent divided and the gMFI intensity of CTV within the population (N). Statistical significance was determined by one-way ANOVA with post-hoc Tukey’s HSD for multiple comparisons. All summary plots are depicted is mean ± s.d. Data are pooled from two (F-I) or three (A-E; J-N) independent experiments.
A similar experiment was performed in which CTV-labeled OT-I CD8+ T cells were transferred into congenic C57BL/6 host mice challenged s.c. with B16.OVA Kb+/+ or Kb−/− cells the following day (Figure 2J). By day six, OT-I T cells proliferated and expanded in tdLNs of mice with B16.OVA tumors regardless of Kb expression, although a non-statistically significant reduction in priming was observed in mice bearing B16.OVA Kb−/− tumors (Figure 2K–N).
We next sought to determine the impact of cancer cell MHC-I expression on endogenous anti-tumor CD8+ T cell activation. C57BL/6 mice received C1498.SIY Kb+/+ or Kb−/− cells, and numbers of antigen-specific, tdLN CD8+ T cells were measured by Kb:SIY pentamer staining six days later. Consistent with observations in TCR-tg adoptive transfer experiments, endogenous SIY-specific CD8+ T cells expanded in response to C1498.SIY Kb+/+ tumors, while numbers of SIY-specific CD8+ T cells recovered from tdLNs of mice bearing C1498.SIY Kb−/− tumors were comparable to those from analogous cLNs of tumor-free controls (Figure 3A and B). Furthermore, numbers of functional, SIY antigen-specific T cells, as measured by IFN-γ ELISpot, were significantly reduced in tdLNs of mice with C1498.SIY Kb−/− tumors (Figure 3C).
Figure 3: Endogenous antigen-specific CD8+ T cell responses against Kb-restricted tumor antigens are reduced against Kb-deficient tumors. A and B).
Kb:SIY pentamer stain of endogenous CD8+ T cells from tdLNs of C57BL/6 mice bearing C1498.SIY Kb+/+ (n = 11) or Kb−/− (n = 11) tumors or n.t. (n = 8) at day 6. Representative flow cytometry plots are shown in (A), and data are quantified in (B). C) IFN-γ ELISpot on tdLN cells of C57BL/6 mice bearing C1498.SIY Kb+/+ or Kb−/− tumors (n = 10 each) at day 6. D and E) Primary C57BL/6 mice received s.c. challenge with C1498.SIY Kb+/+ or Kb−/− cells on day –7. On day –1, 5 x 106 CD8+ T cells from tdLNs of primary mice, or analogous cutaneous LNs (cLN) of tumor-free mice (n = 8 per group) were transferred into naïve secondary C57BL/6 mice. At day 0, secondary mice were challenged with 106 C1498.SIY Kb+/+ tumor cells, and tumor growth was measured. Experimental design is shown in (D) and the growth of C1498.SIY Kb+/+ tumors in secondary mice is depicted in (E). F and G) Identification (F) and quantification (G) of endogenous OVA257–264-specific CD8+ T cells in tdLNs of mice bearing B16.OVA Kb+/+ (n = 16) or Kb−/− (n = 14) tumors, or n.t. (n = 13) at day 6 by Kb:OVA257–264 pentamer stain. Statistical significance for tumor growth in (E) was determined by two-way ANOVA with Sidak’s post-hoc test for multiple comparisons. For all other comparisons, statistical significance was determined by one-way ANOVA with post-hoc Tukey’s HSD. Tumor growth in (E) is shown as mean ± s.e.m.; other plots are depicted is mean ± s.d. Data are pooled from two (E), three (B and C) or four (G) independent experiments. n.t. = no tumor.
To verify that tumor-derived Kb molecules were required specifically at the level of CD8+ T cell priming, and that diminished CD8+ T cell activation against Kb−/− tumors had functional consequences regarding control of tumor growth, we performed an experiment in which the priming and effector phases of the anti-tumor CD8+ T cell response were uncoupled. CD8+ T cells were isolated from tdLNs of mice six days post injection (d.p.i.) of C1498.SIY Kb+/+ or Kb−/− cells, such that initial priming occurred against a Kb-sufficient or -deficient tumor. CD8+ T cells were also isolated from analogous cLNs of tumor-free controls. Equal numbers of CD8+ T cells were then transferred into cohorts of naive C57BL/6 mice subsequently challenged with C1498.SIY Kb+/+ cells (Figure 3D). Adoptive transfer of CD8+ T cells initially primed against a Kb-sufficient tumor provided superior control of C1498.SIY Kb+/+ tumors in secondary recipients compared with CD8+ T cells initially primed against a Kb-deficient tumor, which provided no greater protection than did CD8+ T cells transferred from cLN of tumor-free mice (Figure 3E). Thus, Kb expression by C1498.SIY tumor cells is required for early priming of functional SIY-specific CD8+ T cell responses.
To determine the role of tumor-derived MHC-I in endogenous CD8+ T cell priming in a second model, C57BL/6 mice were inoculated with B16.OVA Kb+/+ or B16.OVA Kb−/− cells, and OVA257–264-specific, tdLN CD8+ T cell responses were analyzed at six d.p.i. Kb:OVA257–264 pentamer staining revealed expansion of endogenous antigen-specific CD8+ T cells in tdLNs of mice bearing B16.OVA Kb+/+ tumors, and that this expansion was significantly reduced in mice bearing B16.OVA Kb−/− tumors (Figure 3F and G). This result contrasts with those in Figure 2K–N, in which OT-I CD8+ T cell priming largely occurred independently of Kb expression by B16.OVA cells, likely because OT-I CD8+ T cells are not representative of the overall pool of endogenous OVA257–264-specific CD8+ T cells, which have varying affinities for Kb:OVA257–264 (Schober et al., 2020). Additionally, while endogenous OVA257–264-specific CD8+ T cell priming was reduced against Kb−/− B16.OVA tumors, it was not completely abrogated as was SIY-specific CD8+ T cell priming against C1498.SIY Kb−/− tumors. Together, these results highlight the differential impact of cancer cell Kb expression on anti-tumor CD8+ T cell responses mounted against Kb-restricted antigens in distinct tumor models. Nevertheless, Kb expression by cancer cells is required for optimal endogenous CD8+ T cell priming against in both tumor models.
Tumor-resident DCs acquire and present cancer cell-derived MHC-I
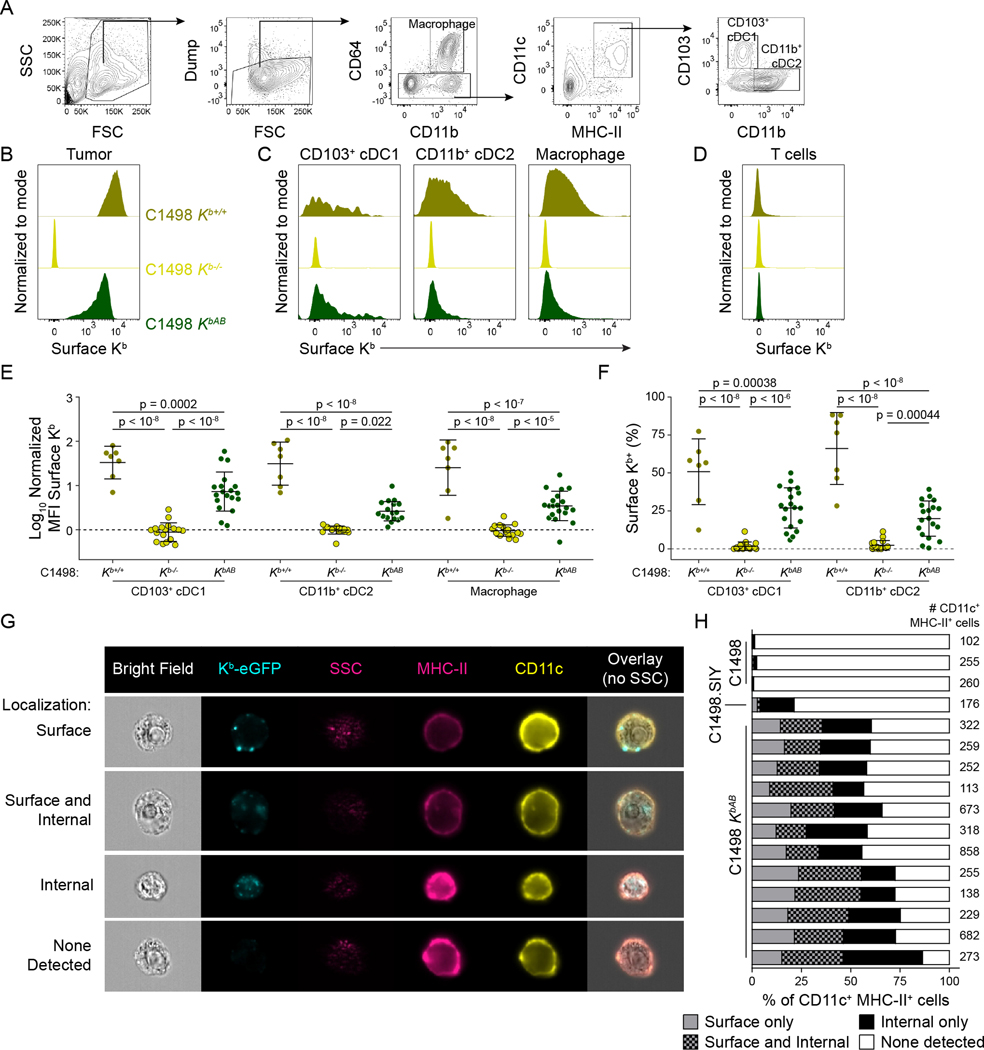
DCs have been shown to present exogenous MHC-I molecules in MHC-mismatched models of in vivo transplantation (Herrera et al., 2004; Liu et al., 2016) and maternal microchimerism (Bracamonte-Baran et al., 2017), as well as in vitro models (Dolan et al., 2006; Kroger et al., 2017; Russo et al., 2000; Wolfers et al., 2001). The above data suggest that MHC-I transfer between cancer cells and APCs occurs in the syngeneic tumor context as well. To determine the extent to which cancer cell-derived MHC-I molecules were acquired by tumor-resident APCs, C1498 Kb+/+, Kb−/−, or KbAB cells were inoculated into MHC-I-deficient (Kb−/−Db−/−) mice. Figure 4A shows the flow cytometry gating strategy utilized to identify tumor-resident macrophage and DC populations. Anti-Kb cell surface staining revealed that DC and macrophage populations isolated from C1498 Kb+/+ and KbAB tumors had broadly acquired cancer cell-derived Kb molecules to a similar degree (Figure 4B–F). Further, C1498 cell-derived Kb molecules were not detected on Kb−/−Db−/− tumor infiltrating T cells (Figure 4D), indicating that acquisition of tumor-derived MHC-I molecules was restricted to phagocytic cells. However, C1498-derived Kb molecules were not detectable on migratory or resident APC populations in tdLNs (Supplemental Figure S1A and B), likely due to limited sensitivity of conventional flow cytometric analysis.
Figure 4: Acquisition of C1498-derived Kb molecules by APC populations in the tumor. A-F).
Kb−/−Db−/− mice were challenged with C1498 Kb+/+ (n = 7), Kb−/− (n = 17) or KbAB (n = 19) cells. Tumors were analyzed at day 6–10 by surface Kb staining and flow cytometry. Gating strategy is depicted in (A). Representative histograms are shown for cancer cells (B), APCs (C), and T cells (D) isolated from the tumor. Data are quantified as MFI (E) or % Kb+ (F). G and H) ImageStream cytometry of CD11c+ MHC-II+ APCs from C1498.SIY (n = 1), C1498 Kb+/+ (n = 3) or KbAB (n = 12) tumors in C57BL/6 hosts at day 6–10. Representative images are shown in (F), and quantification of intracellular versus surface Kb localization is shown in (G), with each bar representing one mouse. Statistical significance for (D) and (E) was determined by two-way ANOVA with post-hoc Tukey’s HSD test. Graphs are depicted as mean ± s.d. Data are pooled from five (A-E) or three (F-G) independent experiments. Also see Supplemental Figures S1–S3.
To assess the localization of C1498-derived Kb molecules on or within APCs, and to verify that the presentation of C1498-derived Kb molecules by APCs was not an artefact resulting from the absence of endogenous MHC-I in Kb−/−Db−/− mice, over 4,000 CD11c+ MHC-II+ cells from C1498 KbAB tumors engrafted in 12 C57BL/6 hosts were visualized using ImageStream cytometry (Figure 4G). Here, most APCs from C1498 KbAB tumors had internalized Kb-eGFP molecules, and only a small subset displayed Kb-eGFP exclusively at the cell membrane (Figure 4H), suggesting that MHC-I cross-dressing may be associated with tumor antigen uptake. Importantly, eGFP fluorescence was absent in CD11c+ MHC-II+ cells isolated from control C1498 Kb+/+ tumors lacking eGFP expression, and eGFP fluorescence in CD11c+ MHC-II+ cells isolated from C1498.SIY Kb+/+ tumors was almost exclusively classified as internal in a system in which any acquired eGFP fluorescence must be internal (Figure 4H).
To test the hypothesis that MHC-I cross-dressing and tumor antigen uptake may be linked processes, C1498.SIY Kb+/+ or Kb−/− tumors were raised in Kb−/−Db−/− mice, and tumor-resident APCs were analyzed by imaging flow cytometry for acquisition of tumor antigen (SIY-eGFP) and cancer cell-derived Kb using an intracellular anti-Kb antibody stain. Here, APCs that acquired cancer cell-derived MHC-I molecules had also internalized the model tumor antigen SIY-eGFP (Supplemental Figure S2A and B), often with at least some colocalization in fluorescent signal between SIY-eGFP and internalized Kb molecules. These results demonstrate that APCs acquire C1498-derived MHC-I molecules in the tumor environment and suggest that this MHC-I cross-dressing is correlated with—and might occur through—tumor antigen uptake.
Attempts to replicate these results in the B16.OVA model were hindered by very low Kb expression on B16 cells in vitro (Figure 1C; Böhm et al., 1998; Kline et al., 2012; Seliger et al., 2001), and while Kb is upregulated on B16 cells in vivo in response to IFN-γ (Kline et al., 2012), this did not occur in Kb−/−Db−/− mice (Supplemental Figure S3A), presumably due to their reduced pool of endogenous CD8+ T cells. As a result, no Kb acquisition was observed on Kb−/−Db−/− APCs in B16.OVA Kb+/+ tumors (Supplemental Figure S3B and C). Thus, B16.OVA Kb+/+ and B16.OVA Kb−/− cells were pre-treated with IFN-γ in vitro for 48 hours prior to inoculation into Kb−/−Db−/− mice. IFN-γ treatment induced Kb upregulation on B16.OVA Kb+/+ cells, but not on B16.OVA Kb−/− cells (Figure 1C). Tumor analysis at 6–10 d.p.i. revealed that Kb-sufficient B16.OVA cells retained IFN-γ-induced Kb expression in vivo, and as in the C1498 system, B16.OVA tumor-resident APCs were broadly capable of acquiring and presenting tumor cell-derived Kb molecules (Supplemental Figure S3D–F). Thus, the ability of APCs to become cross-dressed with cancer-derived MHC-I is not restricted to a single tumor model.
Presentation of tumor-derived pMHC complexes by CD103+ DCs is sufficient for antigen-specific CD8+ T cell priming ex vivo
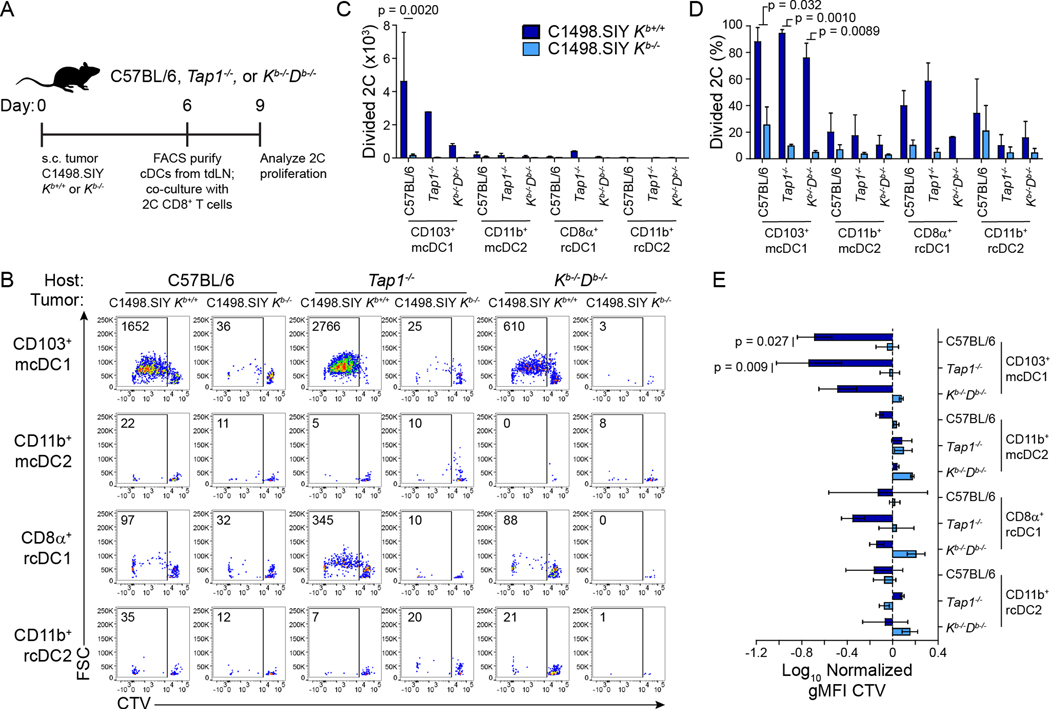
Considering the necessity for tumor derived Kb molecules in optimal tumor antigen-specific CD8+ T cell priming, the known requirement of cDC1 in this process, and the ability of APCs to acquire and present tumor-derived MHC-I, the sufficiency of MHC-I cross-dressing by DCs as a means of tumor antigen presentation was assessed. Accordingly, C1498.SIY Kb+/+ and C1498.SIY Kb−/− tumors were inoculated in wild type, Kb−/−Db−/−, and Tap1−/− mice. Due to defective peptide transport from the cytosol into the endoplasmic reticulum where MHC-I loading occurs, Tap1−/− mice are largely incapable of classical antigen cross-presentation (Androlewicz et al., 1993; Van Kaer et al., 1992). Six days later, tdLN-resident and migratory cDC1 and cDC2 populations were separately purified by fluorescence-activated cell sorting (FACS) and co-cultured directly ex vivo with CTV-labeled 2C T cells (Figure 5A). The ability to mediate 2C T cell activation was restricted almost exclusively to migratory CD103+ cDC1 from mice bearing C1498.SIY Kb+/+ tumors (Figure 5B–E), which was expected given the established role of CD103+ cDC1 in presenting tumor antigens to CD8+ T cells (Roberts et al., 2016; Salmon et al., 2016). Cross-presentation of the SIY antigen in this context was dispensable for 2C T cell activation, as evidenced by the fact that priming was mediated similarly by Tap1−/−, Kb−/−Db−/−, and wild type CD103+ cDC1 (Figure 5B–E). Further, migratory CD103+ cDC1 from tdLNs of wild type mice bearing C1498.SIY Kb−/− tumors were incapable of 2C T cell stimulation (Figure 5B–E), consistent with data presented in Figures 2 and 3, further emphasizing the requirement for cancer cell-derived MHC-I in anti-tumor CD8+ T cell priming. This result demonstrates that acquisition and presentation of C1498 cell-derived Kb:SIY molecules by CD103+ cDC1 is both necessary and sufficient for tumor-specific CD8+ T cell priming.
Figure 5: Presentation of C1498.SIY-derived pMHC complexes by CD103+ cDC1 is sufficient for 2C T cell priming ex vivo. A).
Experimental design. cDC populations isolated from tdLNs of C57BL/6, Tap1−/−, and Kb−/−Db−/− mice bearing 6-day C1498.SIY Kb+/+ or Kb−/− tumors were co-cultured with CTV-labeled 2C T cells for 72 hours. B) Representative flow cytometry plots showing 2C T cell proliferation. C-E) Bar graphs (mean + s.d) quantifying data from (B) as number (C) and frequency (D) of divided 2C T cells, and the normalized gMFI of CTV within 2C T cells (E). Data are pooled from two independent experiments. Statistical significance was determined by three-way ANOVA with post-hoc Tukey’s HSD test.
Generation and characterization of Wdfy4−/− mice
After establishing that migratory CD103+ cDC1 use MHC-I cross-dressing as a means of antigen presentation, we sought to determine the extent to which anti-tumor CD8+ T cell priming occurred independently of classical cross-presentation in mice with C1498.SIY tumors. Here, we utilized Wdfy4−/− mice, in which cross-presentation of cell-derived antigens is defective in cDC1 (Theisen et al., 2018). We generated Wdfy4−/− mice by deleting exon 4 using CRISPR/Cas9, resulting in a frameshift and premature stop codon after 146 amino acids (aa; compared to the 3,184 aa full-length protein; see STAR Methods for full details, scheme depicted in Supplemental Figure S4A). Deletion of exon 4 from Wdfy4 was confirmed by PCR amplification of genomic DNA and subsequent Sanger sequencing (Supplemental Figure S4B). Furthermore, cDNA was generated from RNA isolated from Wdfy4+/+ and Wdfy4−/− splenocytes. Regions of the Wdfy4 transcript were PCR amplified using four different primer pairs spanning multiple exons, including exon 4. The removal of exon 4 and resulting frameshift mutation were also confirmed by Sanger sequencing and depicted with the corresponding aa sequence from the mutation through the premature stop codon (Supplemental Figure S4C–E). Although no validated antibodies are available to directly measure murine WDFY4 protein, these results indicate that there is no functional WDFY4 protein in the Wdfy4−/− mice generated herein.
In order to validate our Wdfy4−/− mice in a tumor model where WDFY4 deficiency is associated with a known phenotype, Wdfy+/+ and Wdfy4−/− littermate mice were inoculated with 1969 sarcoma cells that generate tumors that are typically rejected spontaneously by wild type mice in a CD8+ T cell-dependent manner. As previously observed (Theisen et al., 2018), 1969 tumors were uniformly rejected by Wdfy4+/+ mice but grew progressively in Wdfy4−/− mice (Supplemental Figure S4F). In fact, 1969 tumor growth in Wdfy4−/− mice tended to be more rapid than in Batf3−/− mice which are largely devoid of cDC1 (Supplemental Figure S4F). This result confirms the requirement for WDFY4-dependent cross-presentation in mounting a productive CD8+ T cell response against 1969 sarcoma, while also highlighting that although MHC-I cross-dressing contributes to anti-tumor CD8+ T cell priming in some tumor models, classical cross-presentation dominates in others.
No defects were observed in T cell numbers in secondary lymphoid organs in Wdfy4−/− animals (data not shown). Importantly, WDFY4 deficiency did not affect numbers, proportions, or expression of costimulatory molecules by DC populations in cLNs (Supplemental Figure S5). Further, DC numbers and phenotype were broadly similar in various organs in Wdfy4+/+ and Wdfy4−/− mice, except for mesenteric LNs, where a reduction in CD103+ CD11bneg migratory cDC1 was observed in Wdfy4−/− mice (Supplemental Figure S6).
WDFY4-dependent cross-presentation is dispensable for CD8+ T cell priming against C1498.SIY tumors
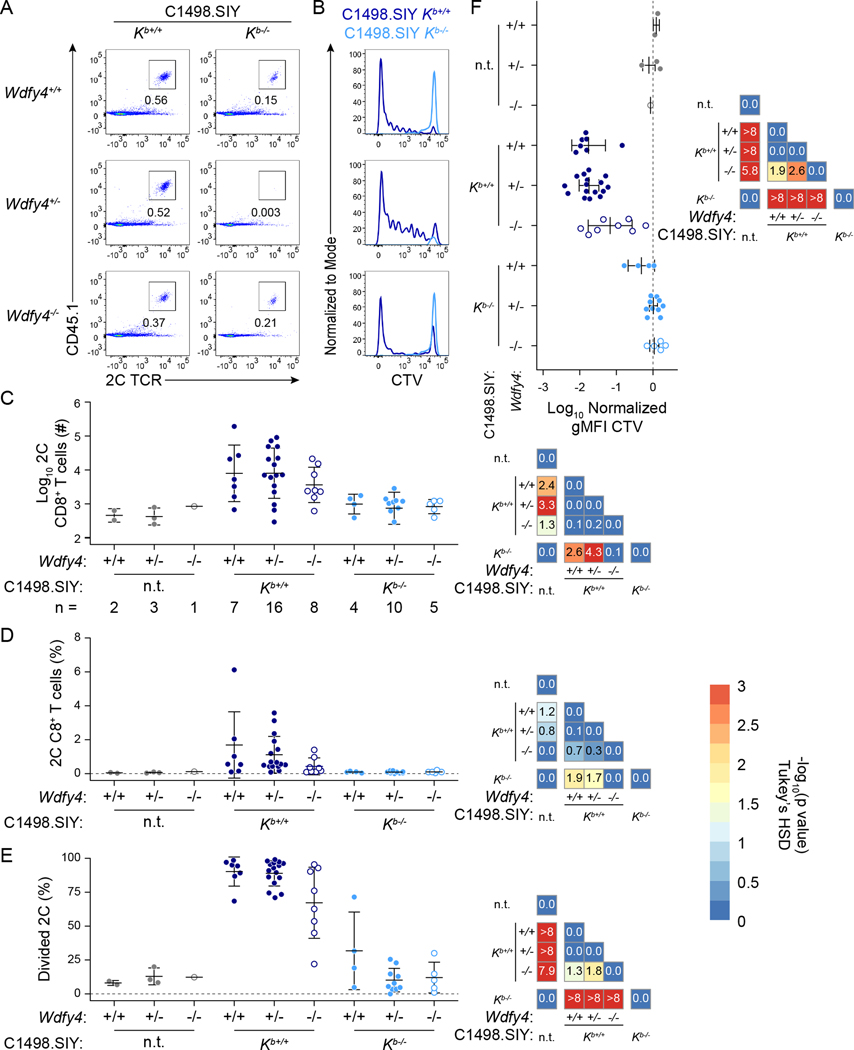
To test the sufficiency of MHC-I cross-dressing in anti-tumor antigen-specific CD8+ T cell priming in mice unable to cross-present cell-associated antigens, CTV-labeled CD45.1+ 2C CT cells were adoptively transferred into CD45.2+ Wdfy4+/+, Wdfy4+/−, and Wdfy4−/− mice, which were inoculated with C1498.SIY Kb+/+ or Kb−/− cells the following day. Six days later, the expansion of 2C T cells was assessed in tdLNs and analogous cLNs from non-tumor-bearing mice. As expected, minimal 2C T cell proliferation or expansion occurred in cLNs of tumor-free control mice or in mice bearing C1498.SIY Kb−/− tumors, regardless of Wdfy4 genotype (Figure 6A–F). For mice bearing C1498.SIY Kb+/+ tumors, 2C proliferation was observed in tdLNs of Wdfy4+/+ and Wdfy4+/− mice and was only slightly reduced in Wdfy4−/− mice (Figure 6A–F), with statistical significance reached when analyzing 2C T cell CTV dilution (Figure 6E and F), but not by 2C T cell number or frequency (Figure 6C and D). At the same time, 2C T cell proliferation was greater in Wdfy4−/− mice bearing C1498.SIY Kb+/+ tumors than in any mice bearing C1498.SIY Kb−/− tumors and in tumor-free control mice by the same measures (Figure 6C–F), demonstrating that WDFY4-dependent cross-presentation is dispensable for CD8+ T cell priming against C1498.SIY tumors. This result also suggests that classical cross-presentation contributes minimally to overall SIY antigen presentation by cDC1, or alternatively that WDFY4 plays a role in antigen presentation by cDC1 via MHC-I cross-dressing.
Figure 6: In vivo 2C T cell priming occurs in Wdfy4−/− mice bearing C1498.SIY Kb+/+ tumors.
2C T cell priming was assessed in tdLNs of Wdfy4+/+, Wdfy4+/−, and Wdfy4−/− mice bearing C1498.SIY Kb+/+ or Kb−/− tumors at day 6. A) Representative flow cytometry plots showing 2C T cell frequency among CD8+ T cells. B) Representative histograms of CTV dilution by 2C T cells. C-F) Summary plots (left) and heatmaps (right) depicting p-values for pairwise comparisons of the number (C), frequency (D), % divided (E), and geometric mean fluorescence intensity of CTV (F) of tdLN 2C T cells. Statistical significance was determined by one-way ANOVA with a Tukey’s post-hoc HSD test. The –log10 of the adjusted p-value is indicated in each tile of the heatmap. p < 0.05 (–log10(p) > 1.30) was considered statistically significant. Data are pooled from three independent experiments; group sizes are indicated in (C). n.t. = no tumor. Also see Supplemental Figures S4–S7.
To determine if WDFY4 might be directly involved in MHC-I cross-dressing, Wdfy4−/− mice were crossed with Kb−/−Db−/− mice. C1498 Kb+/+ or Kb−/− tumors were raised in littermate Wdfy4+/+ Kb−/−Db−/−, Wdfy4+/− Kb−/−Db−/−, and Wdfy4−/− Kb−/−Db−/− mice, and the ability of APCs to present C1498-derived Kb molecules was assessed as in Figure 4. Both Wdfy4+/+ Kb−/−Db−/− and Wdfy4+/− Kb−/−Db−/− mice (hereafter referred to as Wdfy4+) were included in the Wdfy4+ Kb−/−Db−/− group, as Wdfy4 heterozygotes were phenotypically identical to homozygous wild type mice (Supplemental Figures S5 and S6) and were equally capable of mounting anti-tumor CD8+ T cell responses (Figure 6). APCs isolated from Kb−/−Db−/− mice harboring C1498 Kb+/+ tumors similarly acquired and presented C1498-derived Kb molecules regardless of Wdfy4 genotype (Supplemental Figure S7), indicating that MHC-I cross-dressing is a WDFY4-independent process.
APCs acquire and present HLA-I molecules derived from human cancer cells
The above data indicate that MHC I cross-dressed DCs have a critical, non-redundant role in mediating anti-tumor CD8+ T cell responses in murine cancer models. We next sought to ascertain the extent to which HLA-I cross-dressing occurred through in vitro assays and xenograft models. First, HLA-A*02neg monocyte-derived macrophages (MDM) were co-cultured with CTV-labeled HLA-A*02pos CD19+ OCI-Ly8 diffuse large B cell lymphoma cells. MDM were utilized due to ease of acquisition and culture, as well as our observation that, like DCs, macrophages acquire and present cancer cell-derived MHC-I molecules in murine tumors (Figure 4). OCI-Ly8 lymphoma cells were chosen due to high expression of HLA-A*02, a common HLA-A allele for which a specific antibody exists.
Based on data presented in Figure 4, which suggest that MHC-I cross-dressing and internalization of tumor antigens are linked processes, a CD47-blocking antibody (α-CD47) was included in MDM and OCI-Ly8 cell cultures to induce phagocytosis by disrupting CD47-SIRPα interactions. MDMs treated with an isotype control antibody acquired neither lymphoma cell-derived CTV fluorescence nor captured HLA-A*02 molecules from OCI-Ly8 lymphoma cells; conversely, MDMs treated with α-CD47 acquired an abundance of both (Figure 7A, top), and displayed acquired HLA-A*02 on their cell surface. Consistent with observations in murine models (Figure 4), MDMs that acquired the most CTV fluorescence also presented the most OCI-Ly8 cell-derived HLA-A*02 (Figure 7A, top). Thus, human APCs can acquire and present human cancer cell-derived HLA molecules. Also notable was the fact that the MDM that acquired the highest OCI-Ly8-derived CTV and HLA-A*02 also displayed lymphoma-derived CD19 on their cell surface (Figure 7A, middle and bottom), suggesting that the ability of a surface protein to transfer from one cell to another is not limited to MHC and HLA molecules, and potentially is a broader phenomenon.
Figure 7: APCs cross-dress with human HLA molecules.
A) Monocyte-derived macrophages (MDMs) were differentiated from the blood of an HLA-A*02neg donor and co-cultured with CTV-labeled HLA-A*02pos OCI-Ly8 lymphoma cells for 4 hours with or without an α-CD47 blocking antibody. Representative flow cytometry plots of MDM for acquired OCI-Ly8-derived HLA-A*02 versus CTV (top), CD19 versus CTV (middle), and CD19 versus HLA-A*02 (bottom). B-F) OCI-Ly1 (blue) and OCI-Ly8 (green) tumors were xenografted s.c. into NSG mice (n = 9 each). Representative histograms showing cell surface staining for tumor-derived HLA-I molecules on DCs are shown in (B). Data are quantified as % HLA class I+ among cDC1 (C) and cDC2 (D), as well as normalized MFI of HLA class I for cDC1 (E) and cDC2 (F). Flow cytometry plots in (A) are from one experiment, representative of three independent experiments. Data in (B-F) are pooled from two independent experiments. Statistical significance was determined by one-way ANOVA with post-hoc Tukey’s HSD test.
To determine whether HLA molecules from human tumor cells are also transferred to APCs in vivo, lymphoma xenograft models were utilized. Here, human OCI-Ly1 and OCI-Ly8 lymphoma cells were inoculated s.c. into immune-deficient NOD.PrkdcscidIl2rgnull (NSG) mice. Tumor resident DCs isolated from lymphoma xenografts displayed lymphoma-derived HLA-I molecules on their cell surface (Figure 7B–F). Comparatively, HLA-I was not detected on splenic DCs in tumor-bearing NSG mice (Figure 7B–F). Together, these data indicate that MHC and HLA-I cross-dressing by tumor APCs is conserved in mice and humans.
Discussion
The finding that MHC-I cross-dressing by migratory CD103+ cDC1 is a critical antigen presentation pathway for mounting anti-tumor CD8+ T cell responses challenges our current understanding of the mechanisms by which DCs present cancer antigens. Our observations also raise the following question: Why does MHC-I cross-dressing appear to play such an important role in tumor antigen presentation when cross-presentation is a hallmark cDC1 function? Any definitive answer will require additional investigation; however, we can say that the contribution of MHC cross-dressing to antigen presentation varies across antigens and systems. Although our results highlight a key role of MHC-I cross-dressing in anti-tumor immunity, they certainly do not negate the importance of classical cross-presentation. For example, whereas MHC-I cross-dressing was absolutely required for antigen-specific CD8+ T cell priming against C1498.SIY tumors, both MHC-I cross-dressing and classical cross-presentation contributed to CD8+ T cell priming in the B16.OVA model. Moreover, the fact that immunogenic 1969 regressor tumors grew progressively in Wdfy4−/− mice, coupled with data indicating that MHC-I cross-dressing occurred independently of WDFY4, suggests that cross-presentation of 1969 cell-derived antigens is necessary for tumor rejection in this model. Although MHC-I cross-dressing occurred in the C1498 and B16 tumor environments, its contribution to CD8+ T cell priming in the tdLN differed, opening avenues for future study into questions regarding specific contexts in which MHC-I cross-dressing contributes to antigen presentation.
A recent study has also identified MHC cross-dressing as an important tumor antigen presentation pathway (Duong et al., 2021). Using MC57 regressor tumors, Duong et al. demonstrate that while CD103+ cDC1 present MC57-derived model antigens through cross-presentation and MHC-I cross-dressing, a subset of type I IFN-activated cDC2 (ISG+ cDC2) present MC57 antigens exclusively via MHC-I cross-dressing. Consistent with our observations, the absence of tumor cell MHC-I expression led to decreased CD8+ T cell priming, although to not the same degree as did the absence of cDC1, suggesting that the impact of MHC-I cross-dressing on overall tumor antigen presentation is context dependent. Moreover, that cDC2 are capable of presenting tumor antigens through MHC-I cross-dressing in the study by Doung et al. but not in ours may have been due to use of distinct tumor models, or to analysis of DCs isolated from different sites. Doung et al. assess the ability of tumor-resident DCs to present tumor antigen via MHC-I cross-dressing. In contrast, we tested the sufficiency of MHC-I cross-dressing in mediating CD8+ T cell activation ex vivo using DCs isolated from the tdLN, where initial anti-tumor CD8+ T cell priming occurs. It is unclear the extent to which the ISG+ cDC2 characterized by Doung et al traffic to the tdLN, and although migratory cDC2 in tdLNs of mice bearing C1498.SIY tumors were unable to present SIY antigen to CD8+ T cells ex vivo, we did observe that cDC2 were cross-dressed with cancer cell MHC-I in the tumor environment. It is therefore possible that tumor-resident cDC2 present antigens via MHC-I cross-dressing in the C1498 and B16 tumor models. Regardless, the two studies serve to highlight the impact of MHC-I cross-dressing on tumor antigen presentation.
Numerous factors likely determine the extent to which MHC-I cross-dressing occurs and contributes to T cell priming. Intuitively, the abundance of donor cell MHC-I must be important, as MHC-I cross-dressing did not occur in B16.OVA tumors in the absence of MHC-I expression on tumor cells — DCs cannot become cross-dressed with non-existent MHC-I molecules. Beyond the tumor context, reports of MHC-I cross-dressing have primarily focused on MHC-mismatched transplantation models (Herrera et al., 2004; Liu et al., 2016) and thymic tolerance (Koble and Kyewski, 2009; Kroger et al., 2017; Perry et al., 2018), environments with robust MHC expression and abundant cell death, the latter of which can promote MHC-I cross-dressing in vitro (Dolan et al., 2006). We cannot comment on the impact of cell death on MHC-I cross-dressing based on the experiments performed here, but tumors are certainly not devoid of cells undergoing various forms of cell death.
It is also possible that the nature of the antigen itself dictates whether its presentation occurs primarily through MHC-I cross-dressing or cross-presentation. Expression of different proteasomal subunits within immune and non-immune cells generates distinct peptidomes for presentation on MHC molecules (Murata et al., 2007). It is thus possible that certain peptide antigens displayed on MHC-I in a tumor cell are not efficiently generated by professional APCs. This explanation does not account for the fact that both SIY and SIINFEKL antigens can be displayed by DCs through cross-presentation (Spiotto et al., 2002; Theisen et al., 2018), despite the demonstrated importance of MHC-I cross-dressing for their presentation in the tumor context. For many antigens, a more likely explanation is that MHC-I cross-dressing simply increases the density and overall number of MHC-I molecules loaded with donor cell-derived peptide antigens. Thus, the contribution of MHC-I cross-dressing to antigen presentation could simply be the result of the quantity, rather than the quality, of antigens presented in this manner.
Given the varied nature of the models in which MHC-I antigen presentation via cross-dressing has been implicated, a particularly appealing hypothesis is that MHC-I cross-dressing is a default mechanism by which DCs present tissue antigens. Direct transfer of pMHC-I molecules from surrounding cells through phagocytosis, receptor-mediated endocytosis, trogocytosis, or exosome capture represents a rich source of intact pMHC-I molecules, all of which would contain a peptide that is necessarily generated from a donor cell-derived protein. We envisage that MHC-I cross-dressing occurs constitutively and silently under steady-state conditions, unobserved because all host cells express identical MHC-I molecules. Importantly, the occurrence of MHC-I cross-dressing would not preclude classical cross-presentation functioning as the dominant antigen presentation pathway in response to external stimuli, such as detection of necrotic cell death via the C-type lectin receptor DNGR-1 (Canton et al., 2021; Hanč et al., 2015; Sancho et al., 2009; Zhang et al., 2012) or pathogen-associated molecular patterns via toll-like receptors (TLRs; Nair-Gupta et al., 2014)—both of which would generate a bias toward classical cross-presentation due to the release of cellular antigens into the extracellular matrix upon lytic or necrotic cell death. Again, we can only speculate that the contributions of MHC-I cross-dressing and cross-presentation to overall presentation of a given antigen are a matter of degrees, and that MHC-I cross-dressing is dependent on the density of pMHC molecules present on the donor cell surface.
The mechanism by which MHC-I cross-dressing occurs is disputed. Super-physiological concentrations of purified exosomes are a sufficient source of pMHC-I complexes to facilitate MHC-I cross-dressing in vivo (Bracamonte-Baran et al., 2017; Herrera et al., 2004; Liu et al., 2016). Conversely, in vitro experiments have demonstrated that MHC-I cross-dressing requires direct cell-cell contact (Ruhland et al., 2020; Wakim and Bevan, 2011), implicating trogocytosis or contact-mediated transfer of synaptic vesicles. It is possible that the mechanism by which MHC-I cross-dressing occurs is context dependent. Our data strongly suggest that MHC-I cross-dressing is linked with tumor antigen uptake, which has been previously reported (Koble and Kyewski, 2009; Ruhland et al., 2020). We hypothesize that APCs internalize donor pMHC-I complexes, some of which are recycled to the plasma membrane via the endogenous MHC-I recycling pathway. This mechanism would account for the observed correlation between MHC-I cross-dressing and tumor antigen uptake, as MHC-I cross-dressing would be a useful byproduct of a DC’s habitual sampling of its surroundings. Indeed, should a DC internalize tumor cargo in which the original membrane topology is maintained, membrane fusion between this cargo and the endosome containing it would result in exposure of the cytosolic face of acquired MHC-I molecules to trafficking chaperones, as well as release of antigenic proteins to the cytosol, simultaneously promoting antigen presentation through both MHC-I cross-dressing and cross-presentation. Unless acquired MHC-I molecules were routinely ubiquitinated, there is no reason they would not be presented by an APC after export to the plasma membrane from an early recycling endosome (Reid and Watts, 1990; van Endert, 2016); APCs are capable of MHC-I recycling (Montealegre and van Endert, 2019; Radhakrishna and Donaldson, 1997) and are likely not able to discriminate between endogenous and exogenous MHC-I molecules. Our data and this proposed model neither discriminate between MHC-I internalization and trogocytosis as the putative mechanism of MHC-I cross-dressing, nor discount extracellular vesicles as a source of pMHC-I. While scavenging receptors such as CD36 contribute to this process (Perry et al., 2018), it is unlikely that any one receptor functions as the master regulator of MHC-I cross-dressing. APC subsets express a varied and often non-overlapping array of phagocytic receptors, yet all are capable of MHC-I cross-dressing in vivo. Interruption of CD47-SIRP-α signaling was required for in vitro HLA-I cross-dressing by MDMs, but CD47-SIRP-α interactions cannot regulate in vivo MHC-I cross-dressing by cDC1, which do not express SIRP-α. Redundancies in scavenging receptors within an individual APC further decrease the likelihood that any individual receptor would specifically promote MHC-I cross-dressing.
A related conundrum is why migratory CD103+ cDC1 exclusively prime anti-tumor CD8+ T cell responses via MHC-I cross-dressing in the tdLN when all APC populations are MHC-I cross-dressed in the tumor. Here, the answer may be inferred from the underlying cell biology and principal functions of DC subsets. For instance, while most APCs generate an acidic endosomal pH to promote elimination of pathogens and for efficient generation of peptides for loading onto MHC-II, cDC1 actively maintain a neutral to alkaline endosomal pH conducive to the partial protection of intact antigens for export to the cytosol for proteasomal degradation and eventual display via classical cross-presentation (Canton et al., 2021; Delamarre et al., 2005; Joffre et al., 2012; Savina et al., 2006; Savina et al., 2009). This fundamental cDC1 property may also promote MHC-I cross-dressing by increasing the likelihood that donor pMHC-I molecules are recycled to the cell surface intact, as pMHC-I complexes are unstable under acidic conditions (Chefalo and Harding, 2001; Reich et al., 1997). This theory fits well with the reported role of migratory CD103+ cDC1 in transporting intact, pH-sensitive antigenic proteins acquired in the tumor environment to the tdLN (Roberts et al., 2016; Ruhland et al., 2020; Salmon et al., 2016). Indeed, migratory CD103+ cDC1 can transfer both intact tumor-derived fluorescent antigens and MHC-I complexes to other APC subsets in the tdLN (Ruhland et al., 2020). That said, more work must be done to fully elucidate the cDC1-intrinsic mechanism of antigen presentation via MHC-I cross-dressing.
Finally, MHC-I cross-dressing needs to be examined in human cancers. Our data and published observations of others (Russo et al., 2000) demonstrate that APCs are capable of cross-dressing with human HLA molecules. However, the significance of this finding regarding downstream activation of anti-tumor T cell responses in cancer patients remains unknown. For all the same reasons that studying MHC-I cross-dressing in syngeneic mouse models has been difficult, investigating this process in humans is extremely challenging. A human tumor and its infiltrating immune cells always express identical HLA molecules, with the exception of HLA-mismatched bone marrow transplantation, which might represent one human cancer context in which HLA-I cross-dressing could be explored.
In conclusion, we found that cDC1 utilize acquired cancer cell-derived pMHC-I molecules to prime anti-tumor CD8+ T cell responses, even when classical cross-presentation is intact. These observations challenge the current dogma and raise additional questions about the mechanisms that underlie anti-tumor CD8+ T cell priming. Future studies are necessary to gain a complete understanding of the processes through which MHC-I cross-dressing occurs and its impact on antigen presentation and adaptive immune responses in cancer and in other contexts.
Limitations of Study
A limitation of our results is reliance on transplantable tumors and model antigens. It is possible that the impact of MHC-I cross-dressing on anti-tumor CD8+ T cell priming is over-estimated when analyzing responses against immunodominant antigens. MHC-I cross-dressing and its effect on antigen-specific CD8+ T cell activation will need to be examined in more physiological models in future studies; however, the nature of the questions addressed in the current study necessitated the use of these models. Also, as discussed above, the extent to which MHC-I cross-dressing and its impact on tumor-specific CD8+ T cell priming is operational in human cancers was not addressed in our experiments and remains unknown.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will befulfilled by the lead contact, Justin Kline (jkline@medicine.bsd.uchicago.edu)
Materials availability
B16.OVA Kb−/−, C1498 Kb−/−, and C1498 KbAB cell lines, as well as Wdfy4−/− mice were generated in this study. All will be made available from the lead contact upon request. Wdfy4−/− mice will also be made available through the Knockout Mouse Project (KOMP).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
In vivo animal studies
C57BL/6 (B6, H-2b), Ly5.1 (B6.SJL-PtprcaPepcb/BoyJ), OT-I (C57BL/6-Tg[TcraTcrb]1100Mjb/J), Tap1−/− (B6.129S2-Tap1tm1Arp/J), Thy1.1 (B6.PL-Thy1a/CyJ), and NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in our facility. Kb−/−Db−/− (B6.129P2-H-2K1tm1Bpe H-2D1tm1Bpe/DcrJ) mice were provided by Dr. A. Bendelac, University of Chicago. 2C (B6.Cg-Cd8a<a> Tg(Tcra2C,Tcrb2C)1Dlo) mice were provided by Dr. T. Gajewski, University of Chicago. Wdfy4−/− mice were generated in this study at the University of Chicago as described below. All mouse strains were bred and housed in a specific pathogen-free facility at the University of Chicago and used in accordance with protocols approved by the university’s Institutional Animal Care and Use Committee following guidelines established by the National Institutes of Health. Six- to fourteen-week-old sex-matched littermate controls were used for all experiments unless otherwise specified. Littermate mice of the same sex were randomly assigned to experimental groups.
Cell lines
The C1498 murine leukemia cell line and the OCI-Ly1 and OCI-Ly8 human lymphoma cell lines were purchased from American Type Culture Collection, and the C1498.SIY cell line was previously generated in our laboratory (Zhang et al., 2013). The B16.OVA cell line was provided by Dr. T. Schumacher (Netherlands Cancer Institute, Amsterdam, Netherlands). Kb−/− and KbAB cell lines were generated as described below. Surface expression of Kb protein was periodically assessed on all cell lines by flow cytometry. SIY antigen expression in cultured C1498.SIY and C1498.SIY Kb−/− cell lines was periodically monitored by flow cytometry to ensure equivalent eGFP fluorescence. 1969 sarcoma cells were provided by Dr. T. Gajewski (University of Chicago). C1498 and B16-F10 cell lines were grown in DMEM supplemented with 10% fetal bovine serum (FBS), 2-mercaptoethanol (2-ME), essential amino acids, and penicillin/streptomycin (complete DMEM). 1969 cells were grown in RPMI1640 supplemented with 10% fetal bovine serum (FBS), 2-mercaptoethanol (2-ME), essential amino acids, and penicillin/streptomycin (complete RPMI). Cells were cultured in a 37°C incubator with 5% CO2. All cell lines used were routinely monitored for mycoplasma contamination using Venor GeM Mycoplasma PCR-Based Detection Kit.
MDM co-cultures
Peripheral blood mononuclear cells (PBMCs) were extracted through Ficoll density gradient centrifugation using the peripheral blood of healthy HLA-A*02neg human donors. Monocytes were isolated by adhering PBMCs to culture plates for two hours at 37°C. Non-adherent cells were removed by washing. MDM were generated by culturing adherent monocytes in complete RPMI media containing 50 ng/mL M-CSF for 7 days. Subsequently, MDM were harvested by incubation in EDTA/Trypsin for 5 minutes followed by gentle scraping. 105 MDM were plated into each well of a 24-well plate, rested for 24 hours, and then co-cultured with 105 CTV-labeled HLA-A*02pos OCI-Ly8 lymphoma cells for 2 hours in the presence of 5 ug/mL of an isotype (clone BE0083, Bioxcell) or anti-CD47 antibody (clone B6H12, Bioxcell). MDM were then harvested from each well, identified by CD14 antibody staining, and assessed for expression of lymphoma cell-derived CTV, HLA-A*02, and CD19 by flow cytometry.
METHOD DETAILS
Generation of Kb−/− and KbAB cell lines
H-2Kb was deleted in C1498, C1498.SIY, and B16.OVA cells using CRISPR/Cas9 targeting of exon 1 (sequence 5’-AACAGCAGGAGCAGCGTGCACGG-3’) as previously described (Kline et al., 2018). A guide RNA was generated using the following primers (forward, 5’-CACCGAACAGCAGGAGCAGCGTGCA-3’; reverse, 5’-AAACTGCACGCTGCTCCTGCTGTTC-3’) and was subsequently subcloned into the Lenti_v2 vector, which also contains the cDNA encoding for Cas9. Cell lines were transduced with lentivirus and selected in culture with puromycin for 1 week. Kb−/− cell lines were subsequently established following 3 rounds of FACS purification of Kb-negative cells using surface staining with an antibody recognizing Kb. Kb was re-expressed with a C-terminal eGFP tag in C1498 Kb−/− cells to generate C1498 KbAB (Kb Add-back) cells using standard lentiviral transduction. The Kb-eGFP lentiviral construct (Hein et al., 2014) was provided by Dr. S. Springer (Jacobs University Bremen, Germany).
Generation of Wdfy4−/− mice
Wdfy4−/− mice were generated using CRISPR/Cas9. Guide (g) RNAs flanking exon 4 of the Wdfy4 gene were designed using the IDT design tool. Excising exon 4 results in a frame-shift in Wdfy4 after residue 116 (out of 3024) and a premature stop codon 30 amino acids later, as previously described (Theisen et al., 2018). Alt-R crRNA guides (sequences: ATGCATCACCAACGAGCTTT and AGCACCTGGGAACACCTTCG) and tracrRNA were purchased from IDT, and gRNA was assembled from the crRNA and tracrRNA according to the manufacturer’s protocol. Embryo-grade water containing 20 ng/μL gRNA and 60 ng/μL Cas9 protein was injected into the nuclei of C57BL/6J embryos at the University of Chicago Transgenics/ES Cell Technology Mouse Core Facility. Genotyping PCR primers (fwd: GCCTTGAGGTACATGGGCAA, rev: GGTTACACACAGCTCGTCCAT) were designed up- and down-stream of predicted cut sites. A male Wdfy4−/− founder mouse was backcrossed with a C57BL/6J female. F1 offspring were genotyped by PCR, and wild type and mutant bands were excised from the gel. DNA was purified using a QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s protocol. Finally, PCR-amplified DNA was sequenced at the University of Chicago Comprehensive Cancer Center DNA Sequencing & Genotyping Facility. Sanger sequencing confirmed a 144 bp deletion containing the entirety of exon 4. F1 mice were backcrossed with C57BL/6J for two further generations. Subsequently, Wdfy4+/− heterozygous mice were bred in order to generate Wdfy4+/+, Wdfy4+/−, and Wdfy4−/− littermate mice that were used for experimentation. In order to validate the deletion of exon 4, frameshift mutation, and premature stop codon at the RNA level, RNA was purified from splenocytes isolated from Wdfy4+/+ and Wdfy4−/− littermate mice via trizol/chloroform extraction. cDNA was generated from the RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s protocol. Partial Wdfy4 cDNA was amplified by PCR (cycling conditions: 98°C for 3 minutes, followed by 34 cycles of 98°C for 10 seconds, 64°C for 30 seconds, and 72°C for 30 seconds, and a 10 minute final extension at 72°C using four different primer combination; the two forward primers bound in exon 1 in the 5’-UTR and in exon 2 downstream of the ATG start codon, while the two reverse primers bound in exons 6 and 7 (primer sequences are listed in the Key Resources Table below), such that all resulting bands spanned multiple exons — including the deleted exon 4, predicted frameshift mutation, and premature stop codon — while also excluding the possibility of mistakenly amplifying potential genomic DNA contaminants. PCR products were purified from cut gel bands using a QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer’s protocol. Finally, the PCR-amplified DNA was sequenced at the University of Chicago Comprehensive Cancer Center DNA Sequencing & Genotyping Facility, and sequences were aligned to the full-length Wdfy4 transcript sequence using the NCBI’s nucleotide BLAST alignment tool.
Key resources Table
| Reagent or Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| CD116/32 - unconjugated (clone: 2.4g2) | University of Chicago Cytometry and Antibody Technology Facility | |
| CD3 - Biotin (clone: 17A2) | Biolegend | Cat# 100244 |
| CD3 - FITC (clone: 17A2) | Biolegend | Cat# 100204 |
| CD4 - Biotin (clone: GK1.5) | Biolegend | Cat# 100404 |
| CD4 - PE/Cy7 (clone: GK1.5) | Biolegend | Cat# 100422 |
| CD4 - APC (clone: GK1.5) | Biolegend | Cat# 100412 |
| CD8a - Biotin (clone: 53-6.7) | Biolegend | Cat# 100704 |
| CD8a - Pacific Blue (clone: 53-6.7) | Biolegend | Cat# 100725 |
| CD8a - BV605 (clone: 53-6.7) | Biolegend | Cat# 100744 |
| CD8a - FITC (clone: 53-6.7) | Biolegend | Cat# 100706 |
| CD8a - PerCP/Cy5.5 (clone: 53-6.7) | Biolegend | Cat# 100734 |
| CD8a - PE (clone: 53-6.7) | Biolegend | Cat# 100708 |
| CD8a - PE/Cy7 (clone: 53-6.7) | Biolegend | Cat# 100722 |
| CD8a - APC (clone: 53-6.7) | Biolegend | Cat# 100712 |
| CD8a - APC/Cy7 (clone: 53-6.7) | Biolegend | Cat# 100714 |
| CD8b - FITC (clone: YTS156.7.7) | Biolegend | Cat# 126606 |
| CD11b - Biotin (clone: M1/70) | Biolegend | Cat# 101204 |
| CD11b - BV510 (clone: M1/70) | Biolegend | Cat# 101263 |
| CD11b - PerCP/Cy5.5 (clone: M1/70) | Biolegend | Cat# 101228 |
| CD11c - Biotin (clone: N418) | Biolegend | Cat# 117304 |
| CD11c - PE/Cy7 (clone: N418) | Biolegend | Cat# 117318 |
| CD11c - APC (clone: N418) | Biolegend | Cat# 117310 |
| CD19 - Biotin (clone: 6D5) | Biolegend | Cat# 115504 |
| CD24 - FITC (clone: M1/69) | Biolegend | Cat# 101806 |
| CD44 - Pacific Blue (clone: IM7) | Biolegend | Cat# 103020 |
| CD44 - PE/Cy7 (clone: IM7) | Biolegend | Cat# 103030 |
| CD45.1 - Pacific Blue (clone: A20) | Biolegend | Cat# 110722 |
| CD45.1 - FITC (clone: A20) | Biolegend | Cat# 110706 |
| CD45.1 - PE (clone: A20) | Biolegend | Cat# 110708 |
| CD45.1 - APC (clone: A20) | Biolegend | Cat# 110714 |
| CD45.2 - Pacific Blue (clone: 104) | Biolegend | Cat# 109820 |
| CD45.2 - FITC (clone: 104) | Biolegend | Cat# 109806 |
| CD45.2 - PE/Cy7 (clone: 104) | Biolegend | Cat# 109830 |
| CD45.2 - APC (clone: 104) | Biolegend | Cat# 109814 |
| CD64 - FITC (clone: X54-5/7.1) | Biolegend | Cat# 139316 |
| CD64 - PE/Cy7 (clone: X54-5/7.1) | Biolegend | Cat# 139314 |
| CD80 - FITC (clone: 16-10A1) | Biolegend | Cat# 104706 |
| CD80 - PE (clone: 16-10A1) | Biolegend | Cat# 104708 |
| CD86 - BV605 (clone: GL-1) | Biolegend | Cat# 105037 |
| CD86 - FITC (clone: GL-1) | Biolegend | Cat# 105006 |
| CD86 - PE (clone: GL-1) | Biolegend | Cat# 105008 |
| CD90.1 - Pacific Blue (clone: OX-7) | Biolegend | Cat# 202522 |
| CD90.1 - FITC (clone: OX-7) | Biolegend | Cat# 202504 |
| CD90.2 - FITC (clone: 30-H12) | Biolegend | Cat# 105306 |
| CD90.2 - APC (clone: 30-H12) | Biolegend | Cat# 105312 |
| CD90.2 - PE/Cy7 (clone: 30-H12) | Biolegend | Cat# 105314 |
| CD103 - BV421 (clone: 2E7) | Biolegend | Cat# 121422 |
| CD103 - FITC (clone: 2E7) | Biolegend | Cat# 121420 |
| CD103 - PE (clone: 2E7) | Biolegend | Cat# 121406 |
| B220 - Biotin (clone: RA3-6B2) | Biolegend | Cat# 103204 |
| B220 - FITC (clone: RA3-6B2) | Biolegend | Cat# 103206 |
| F4/80 - BV421 (clone: BM8) | Biolegend | Cat# 123137 |
| F4/80 - PE/Cy7 (clone: BM8) | Biolegend | Cat# 123112 |
| Gr-1 - Biotin (clone: RB6-8C5) | Biolegend | Cat# 108404 |
| Granzyme B - PerCP/Cy5.5 (clone: QA16A02) | Biolegend | Cat# 372211 |
| H-2Kb - PE (clone: AF6-88.5) | eBioscience | Cat# 12-5958-82 |
| H-2Kb - APC (clone: AF6-88.5) | Biolegend | Cat# 116518 |
| H-2Kb:SIINFEKL - APC (clone: 25-D1.16) | Biolegend | Cat# 141606 |
| I-A/I-E - Biotin (clone: M5/114.15.2) | Biolegend | Cat# 107604 |
| I-A/I-E - Pacific Blue (clone: M5/114.15.2) | Biolegend | Cat# 107619 |
| I-A/I-E - FITC (clone: M5/114.15.2) | Biolegend | Cat# 107606 |
| I-A/I-E - PerCP/Cy5.5 (clone: M5/114.15.2) | Biolegend | Cat# 107626 |
| IFN-γ - APC (clone: XMG1.2) | BD Pharmingen | Cat# 554413 |
| NK1.1 - Biotin (clone: PK136) | Biolegend | Cat# 108704 |
| SIRP-a - PE (clone: P84) | Biolegend | Cat# 144012 |
| TCRb - Biotin (clone: H57-597) | Biolegend | Cat# 109204 |
| TCRb - FITC (clone: H57-597) | Biolegend | Cat# 109205 |
| TCRb - PerCP/Cy5.5 (clone: H57-597) | Biolegend | Cat# 109228 |
| TCR Va2 - PE (clone: B20.1) | Biolegend | Cat# 127808 |
| TCR Va2 - APC (clone: B20.1) | Biolegend | Cat# 127810 |
| TNF-α - PE (clone: MP6-XT22) | Invitrogen | Cat# 12-7321-82 |
| 2C TCR - Biotin (clone: 1B2) | University of Chicago Cytometry and Antibody Technology Facility | |
| hCD14 - PerCP/Cy5.5 (clone: 63D3) | Biolegend | Cat# 367110 |
| hCD19 - APC (clone: HIB19) | Biolegend | Cat# 302212 |
| hCD19 - FITC (clone: HIB19) | Biolegend | Cat# 302206 |
| hCD20 - PerCP/Cy5.5 (clone: 2H7) | Biolegend | Cat# 302326 |
| HLA-A/B/C - PE (clone: W6/32) | Biolegend | Cat# 311406 |
| HLA-A*02 - PE (clone: BB7.2) | Abcam | Cat# ab79523 |
| Anti-CD47 – unconjugated (clone: B6.H12) | Bio X Cell | Cat# BE0019-1 |
| Mouse IgG1 Isotype control – unconjugated (clone MOPC-21) | Bio X Cell | Cat# BE0083 |
| Chemicals, peptides, and recombinant proteins | ||
| Kb:SIY pentamer – PE | ProImmune | Cat# 1803 |
| Kb: SIINFEKL pentamer - PE | Proimmune | Cat# 93 |
| LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit | Invitrogen | Cat# L10119 |
| Pacific Orange succinimidyl ester | Invitrogen | Cat# P30254 |
| Golgi Plug | BD | Cat# 555029 |
| CellTrace Violet Cell Proliferation Kit | Invitrogen | Cat# C34557 |
| FOXP3/Transcription Factor Staining Buffer Set | Invitrogen | Cat# 00-5523-00 |
| Recombinant mouse Flt3L (carrier-free) | Biolegend | Cat# 550706 |
| Recombinant human M-CSF | Peprotech | Cat# 216-MC-025/CF |
| Dnase I | Roche | Cat# 10104159001 |
| Collgenase IV | Sigma | Cat# C5138 |
| 2X Taq RED Master Mix | Apex | Cat# 42-138B |
| High Capacity cDNA Reverse Transcription Kit | Applied Biosystems | Cat# 4368814 |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0530S |
| Critical commercial assays | ||
| Venor GeM Mycoplasma Detection Kit, PCR-based | Sigma | Cat# MP0025-1KT |
| QIAquick Gel Extraction Kit | QIAGEN | Cat# 28704 |
| Experimental models: cell lines | ||
| C1498 | ATCC | |
| B16.OVA | Schumacher Lab, Netherlands Cancer Institute | |
| OCI-Ly1 | ATCC | |
| OCI-Ly8 | ATCC | |
| Experimental models: organisms/strains | ||
| Mouse: C57BL/6 | Jackson Laboratories, Bar Harbor, Maine | Strain # 000664 |
| Mouse: Ly5.1: B6.SJL-Ptprca Pepcb/BoyJ | Jackson Laboratories, Bar Harbor, Maine | Strain # 000664 |
| Mouse: Thy1.1: B6.PL-Thy1a/CyJ | Gajewski Lab, University of Chicago | JAX strain # 000406 |
| Mouse: Tap1−/−: B6.129S2-Tap1tm1Arp/J | Jackson Laboratories, Bar Harbor, Maine | Strain # 002944 |
| Mouse: Kb−/−Db−/−: B6.129P2-H2-K1tm1Bpe H2-D1tm1Bpe/DcrJ | Bendelac Lab, University of Chicago | JAX strain # 019995 |
| Mouse: Wdfy4−/− | This paper | |
| Mouse: OT-I: C57BL/6-Tg(TcraTcrb)1100Mjb/J | Gajewski Lab, University of Chicago | JAX strain # 003831 |
| Mouse: 2C: B6-Tg(Tcra2C,Tcrb2C)1Dlo | Gajewski Lab, University of Chicago | |
| Mouse: NSG: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | Jackson Laboratories, Bar Harbor, Maine | Strain # 005557 |
| Oligonucleotides | ||
| Alt-R CRISPR-Cas9 crRNA Wdfy4 5’ Sense gRNA1 – CATGTAGCCTTGAGGTACAT | IDT | |
| Alt-R CRISPR-Cas9 crRNA Wdfy4 5’ Antisense gRNA1 – CTCCAGGGCTATTAACCTGG | IDT | |
| Alt-R CRISPR-Cas9 crRNA Wdfy4 3’ Sense gRNA2 – CAGGCCTCGAAGGTGTTCCC | IDT | |
| Alt-R CRISPR-Cas9 crRNA Wdfy4 3’ Antisense gRNA2 - GTCCCCTTTCCTCATAGACT | IDT | |
| Wdfy4 genotyping fwd primer – GCCTTGAGGTACATGGGCAA | IDT | |
| Wdfy4 genotyping rev primer – GGTTACACACAGCTCGTCCAT | IDT | |
| Wdfy4 transcript ex1 fwd primer – CTGGTGTAGCTTGTGAAGGGT | IDT | |
| Wdfy4 transcript ex2 fwd primer – TTCACTAGAAGGGCAGTCGC | IDT | |
| Wdfy4 transcript ex6 rev primer – CCTCCAGACCCTGAGATTCG | IDT | |
| Wdfy4 transcript ex7 rev primer – CCCCGTTCTCAAACTCCAGG | IDT | |
| Recombinant DNA | ||
| Kb-eGFP | Springer Lab (Hein et al., 2014) | |
| Software and algorithms | ||
| FlowJo v. 10 | BD | |
| IDEAS | Amnis | |
| ImmunoSpot | Cellular Technology Limited | |
| Prism v. 7 | GraphPad | |
| R v. 3.6.3 | R Foundation for Statistical Computing | |
| Rstudio v. 1.2.1335 | Rstudio, Inc | |
| Illustrator | Adobe |
Differentiation of BMDCs
Bone marrow was isolated from mice by flushing femurs, tibias, and pelvic bones with sterile PBS. Bone marrow cells were plated at 106 cells/mL in sterile complete RPMI media plus 100 ng/mL recombinant Flt3 ligand (Flt3L). Cells were cultured for 8 days, with media and Flt3L replacement at days 3 and 6. Differentiation of BMDCs was assessed on day 8 by flow cytometry following cell staining with fluorescent-conjugated antibodies directed against CD11c, MHC-II, CD24, and SIRP-α. BMDCs were considered to be differentiated if > 80% of the cells in culture were CD11chi MHC-IIhi, and the ratio of cDC1-like (CD24+ SIRP-αneg) to cDC2-like (CD24neg SIRP-α+) BMDCs was also noted.
CD8+ T cell isolation for adoptive transfer experiments and in vitro cultures
Splenic TCR-tg 2C and OT-I-tg CD8+ T cells were isolated from TCR-tg mice by pressing harvested spleens through a 70 μm filter into sterile PBS containing 2 mM EDTA and 0.5% bovine serum albumin (BSA). Cells were then labeled with anti-CD8α microbeads (Miltenyi) and positively selected by magnetic separation according to the manufacturer’s protocol. Next, isolated cells were washed three times with sterile PBS, followed by labeling with CTV (Invitrogen) according to the manufacturer’s protocol. For adoptive transfer experiments, CTV-labeled TCR-tg CD8+ T cells were resuspended in sterile PBS at 107 cells/mL, and 106 cells were injected intravenously (i.v.) through the lateral tail vein. For in vitro and ex vivo co-culture experiments, CTV-labeled TCR-tg CD8+ T cells were resuspended to a concentration of 106 cells/mL in complete RPMI, and 10,000 cells were added to each culture well.
In vitro assessment of SIY and OVA antigen expression
BMDCs were resuspended in complete RPMI at 106 cells/mL, and 30,000 cells/well were plated in a 384-well plate following 8 days of differentiation, along with 10,000 CTV-labeled TRC-tg 2C or OT-I-tg CD8+ T cells. Kb+/+ and Kb−/− C1498.SIY or B16.OVA cells were washed with sterile PBS and resuspended to a concentration of 3 × 107 cells/mL in microcentrifuge tubes. Cells were lysed by five cycles of rapid freezing and thawing, during which tubes were alternatively transferred between dry ice and a 37°C water bath. The cells were centrifuged at 10,000 x g for 5 min at 4° C, and 3 uL (equivalent lysate from 30,000 cells) of the supernatant was added to each well. TCR-tg CD8+ T cell proliferation was assessed by flow cytometry to assess for CTV dilution following a 72–96 hour co-culture. TCR-tg CD8+ T cells were identified by antibody staining for CD90, CD8β, and either the 2C TCR or Vα2 (the TCRα chain of OT-I).
Subcutaneous (s.c.) tumor inoculations
Cultured tumor cells were washed twice with PBS, counted, and resuspended in PBS at a concentration such that the injection volume was 100 μL (2 × 107 cells/mL for experiments directly investigating in vivo pMHC-I transfer, 4 × 107 cells/mL for ex vivo DC priming experiments, and 107 cells/mL for all other experiments). Mice were injected s.c. in the right flank (both flanks were injected for ex vivo DC priming experiments) with 100 μL of cells using a 27-gauge needle.
In vivo tumor growth experiments
Indicated tumor cells (106) were inoculated s.c. into the right flank of each host mouse as described above. Tumor area was measured using a caliper every 2–3 days starting at day 5 by a lab member who was blinded to the experimental setup. Mice were euthanized when they reached the humane endpoint established in the IACUC-approved protocol.
LN sample preparation
Mice were euthanized, and the indicated LNs were harvested. For experiments involving analysis of DC populations, LNs were incubated in RPMI 1640 containing 2% FBS, 1 mg/mL collagenase IV, and 20 μg/mL DNase I for 30 min at 37oC. After DNase/collagenase digestion (for DC experiments) or immediately after isolation (all other experiments) LNs were pressed through 70 μm filters to create single-cell suspensions. For experiments in which cell counts of a population were required, 5,000 counting beads were spiked into each sample to normalize counts across samples and experiments. Samples were stained and analyzed by flow cytometry as described below.
Tumor sample preparation
Mice were euthanized; tumors were harvested, minced with either razor blades or dissection scissors, and incubated in RPMI 1640 containing 2% FBS, 1 mg/mL collagenase IV, and 20 μg/mL DNase I for 30 min at 37°C. Each tumor was then pressed through a 70 μm filter in order to generate a single cell suspension before proceeding with antibody staining and analysis as described below.
Antibody staining and flow cytometry
Fc receptors were blocked with an anti-FcγRII/FcγRIII antibody (clone 2.4g2) in FACS buffer (PBS containing 2% FBS, 0.02 mM EDTA, and 0.03% sodium azide) for 15 min on ice. Antibody staining was performed for 20–30 min on ice in FACS buffer. After washing, secondary staining was performed for 20–30 min on ice if necessary. Samples were washed with FACS buffer and then with PBS. When possible, cells were then stained with a Near-IR live-dead dye according to the manufacturer’s protocol. Samples were fixed with 2% paraformaldehyde in PBS for 30 min at 4° C and subsequently washed. For the experiment in Supplemental Figure S2 in which MHC-I cross-dressing was assessed alongside tumor antigen uptake, cells were permeabilized using 0.1% Triton X-100 in PBS for 30 min on ice, followed by one PBS wash, one wash with FACS buffer, and antibody staining in FACS buffer at room temperature for one hour in the dark. Finally, cells were washed, resuspended in FACS buffer and analyzed on an LSR Fortessa (4–15 or X-20 5–18) flow cytometer (BD) or Amnis Image Stream X cytometer at the University of Chicago Cytometry and Antibody Technology Core facility. Flow cytometry data were analyzed using FlowJo v.10 (BD) and ImageStream data were analyzed using IDEAS (Amnis). See the Key Resources Table for a complete list of the antibodies used in this study.
Intracellular cytokine staining
tdLNs and analogous inguinal LNs from non-tumor-bearing mice were harvested and homogenized as described above. These samples were divided in three and the fractions were cultured in 200 μL complete RPMI containing anti-CD28 (1 μg/mL) and plate-bound anti-CD3 (1 μg/mL), 100 nM SIY peptide, or media alone in a 96-well u-bottom plate for five hours at 37°C. Golgi Plug (BD) was added to each well one hour into the culture according to the manufacturer’s protocol. Cells were then stained for cell surface markers as described above. Immediately after staining with a live/dead dye, cells were fixed and permeabilized using a FOXP3/Transcription Factor staining kit (Invitrogen) according to the manufacturer’s protocol, and stained overnight at 4°C with antibodies recognizing IFN-γ, TNF-α, and granzyme B.
In vivo TCR-tg CD8+ T cell proliferation experiments
106 CTV-labeled TCR-tg CD8+ T cells were adoptively transferred intravenously (i.v.) into congenic hosts at day -1 via the lateral tail vein. 106 tumor cells were injected s.c. into the right flank of each mouse at day 0. Mice were euthanized at day 6, and the right inguinal LN—which drains the tumor—of each mouse was harvested and processed as described above. The geometric mean fluorescence intensity (gMFI) of CTV in TCR-tg CD8+ T cells was normalized across experiments by dividing the gMFI of each sample by the mean of the gMFIs of the non-tumor bearing control mice for each experiment, such that the group mean gMFI of TCR-tg CD8+ T cells from non-tumor bearing mice = 1 for each experiment. Due to the large number of groups for experiments involving Wdfy4−/− mice, it was not possible to have sex-matched littermates in all groups. These experiments were designed such that every Wdfy4−/− mouse had sex-matched, co-housed, wild type and heterozygous littermates with the same tumor. Multiple age-matched litters were used in each experiment, and male and female mice were used in balanced numbers across groups.
Assessment of in vivo endogenous CD8+ T cell response by pMHC-I pentamer staining
106 tumor cells were injected s.c. into the right flank of each mouse at day 0. Mice were euthanized at day 6, and tdLNs were harvested and processed as described above. In order to minimize scavenging of pMHC-I pentamers, samples were labeled with anti-CD4, -CD19, -B220, and -Gr-1 biotin-conjugated antibodies in FACS buffer for 15 min on ice, followed by washing and incubation with biotin-binding Dynabeads (Invitrogen) for 10 min and subsequent magnetic depletion. Samples were then stained with 5 μL pMHC-I pentamer (SIY:Kb or OVA257–264:Kb, Proimmune) in 50 μL of FACS buffer for 15 min in the dark at room temperature. Next, 50 μL FACS buffer containing antibodies recognizing CD90.2, CD8β, and CD44 was added to each sample, and the cells were incubated for a further 15 min in the dark at room temperature. Cells were then washed with FACS buffer, followed by PBS, fixed, and analyzed as described above.
Assessment of the endogenous anti-tumor CD8+ T cell response by ELISpot
C57BL/6 mice were inoculated s.c. with 106 C1498.SIY Kb+/+ or Kb−/− cells on day 0. Mice were euthanized at day 6 and tdLNs were isolated and processed. 106 cells from each sample were restimulated overnight with media alone or with 100 nM SIY peptide, and IFN-γ producing cells were identified using an IFN-γ ELISpot kit (BD Biosciences) according to the manufacturer’s protocol. ELISpot plates were read using an ImmunoSpot Series 3 Analyzer, and data were analyzed with ImmunoSpot software.
Polyclonal CD8+ T cell transfer
106 C1498.SIY Kb+/+ or Kb−/− cells were injected s.c. into the right flank of primary C57BL/6 mice on day -7. Primary tumor-bearing and tumor-free control mice were euthanized on day -1, and tdLNs harvested from 5 mice were pooled into each sample. CD8+ T cells were isolated by magnetic separation using anti-CD8α microbeads (Miltenyi) according to the manufacturer’s protocol. CD8+-enriched cells were resuspended in sterile PBS, and 5 × 106 cells were adoptively transferred into secondary C57BL/6 mice i.v. through the lateral tail vein. On day 0, all secondary mice were inoculated s.c. in the right flank with 106C1498.SIY Kb+/+ cells. Tumors were measured as described above.
In vivo MHC-I transfer experiments
C57BL/6 and Kb−/−Db−/− mice were inoculated s.c. in the flank with 2 × 106 tumor cells on day 0. Mice were monitored for tumor growth and were euthanized when tumors became palpable in the majority of experimental animals (between days 6 and 10). Tumors were isolated (in some experiments tdLNs were also isolated) and processed for analysis by flow cytometry or ImageStream cytometry as described above. B16.OVA Kb+/+ and Kb−/− cells were treated with 100 ng/mL recombinant IFN-γ in culture for 48 hours prior to s.c. inoculation in mice.
Ex vivo DC priming assay
C57BL/6, Tap1−/−, and Kb−/−Db−/− mice were inoculated s.c. with 4 × 106 C1498.SIY Kb+/+ or Kb−/− cells in each flank on day 0. On day 6, tumor-draining inguinal, axillary, and brachial LNs were harvested and pooled by experimental group. Pooled LN samples were incubated with sterile RPMI 1640 containing 2% FBS, 1 mg/mL collagenase IV, and 20 μg/mL DNase I for 30 min at 37°C. Each sample was pressed through a 70 μm filter and washed with RPMI containing 10% FBS (R10). Samples were then washed with and resuspended in PBS supplemented with 0.5% bovine serum albumin and 2 mM EDTA. Samples were incubated with biotin-conjugated antibodies recognizing CD3, CD19, B220, Gr-1, and NK1.1 for 20 min on ice. After washing, cells were incubated with biotin-binding Dynabeads (Invitrogen) for 10 min, and the bead-bound cells were depleted by magnetic separation. Cells were then stained with fluorescent-conjugated antibodies, as well as streptavidin-conjugated APC-Cy7 on ice for 20 min in the dark. The samples were washed with PBS and stained with a Near-IR live-dead dye for 10 min at room temperature in the dark. Samples were washed and resuspended in R10 media and CD103+ migratory cDC1, CD11b+ migratory cDC2, CD8α+ resident cDC1, and CD11b+ resident cDC2 were FACS purified on a BD FACSAria II flow sorter in the University of Chicago Cytometry and Antibody Technology Core facility. Purified DC samples were resuspended in complete R10 media containing gentamicin (50 μg/mL) and amphotericin B (2.5 μg/mL) at a concentration of of 106 cells/mL, and 30,000 cells were plated per well in a 384-well plate, along with 10,000 CTV-labeled TRC-tg 2C CD8+ T cells. Cells were co-cultured for 72 hours and proliferation of 2C CD8+ T cells was determined by flow cytometry. All buffers and media used in these experiments were sterile, passed through 0.2 μm filters.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance for tumor growth experiments was determined by two-way ANOVA with Sidak’s post-hoc test using GraphPad Prism 7. For all other experiments, statistical significance was determined by ANOVA with a post-hoc Tukey HSD test using R version 3.6.3 to allow for multiple comparisons. The number of factors (categorical variables) determined the type of ANOVA used in each specific case, and the type of ANOVA used is indicated in the figure legends. For example, the ex vivo 2C T cell stimulation experiment depicted in Figure 5 used a three-way ANOVA to account for the three categorical variables in the experimental design: tumor, host genotype, and the APC population in the co-culture. Rounded adjusted p-values are indicated when 10−4 ≤ p-adj. < 0.1. When 10−8 < p-adj < 10−4, it is indicated as being less than the next largest power of 10 (e.g. p = 0.0000085 would be displayed as p < 10−5). Any p-adj < 10−8 are indicated as p < 10−8. Whenever a p-value is not indicated, p > 0.1. Adjusted p-values < 0.05 were considered statistically significant. All summary plots are depicted as mean ± s.d. unless indicated otherwise and sample size for each experiment is provided in the figure legends. Excluding tumor growth curves for which only the mean and s.d. are plotted, each dot represents the indicated data collected from one mouse.
DATA AND CODE AVAILABILITY
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Supplementary Material
Highlights.
Optimal anti-tumor CD8+ T cell priming requires MHC-I expression on cancer cells
Tumor-resident DCs and macrophages dress with cancer cell MHC-I in vivo
MHC-I cross-dressing is sufficient for antigen-specific CD8+ T cell priming ex vivo
The impact of MHC-I cross dressing on CD8+ T cell priming differs between tumor models
Acknowledgements
We would like to thank Drs. T. Gajewski, A. Bendelac, T. Schumacher, and S. Springer for sharing mice, cell lines, and constructs central to this research. Special thanks to L. Degenstein of the University of Chicago Transgenics/ES Cell Technology Mouse Core Facility for help with microinjections. Thanks also to Dr. P. Savage for careful review of the manuscript and insightful conversations during the project. The authors have no financial interests to disclose. This work was funded by R01 CA16670 to J.K. and used common resources managed by the University of Chicago Cytometry and Antibody Technology core facility, supported by P30 CA014599 to the University of Chicago Comprehensive Cancer Center. B.W.M., D.S.K., and D.E.K. were supported by the Immunology Training Grant at the University of Chicago (T32 AI007090). B.W.M. was supported by a Careers in Immunology Fellowship from the American Association of Immunologists.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Anderson DA, Dutertre C-A, Ginhoux F, Murphy KM, 2021. Genetic models of human and mouse dendritic cell development and function. Nat. Rev. Immunol 21, 101–115. 10.1038/s41577-020-00413-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- André F, Chaput N, Schartz NEC, Flament C, Aubert N, Bernard J, Lemonnier F, Raposo G, Escudier B, Hsu D-H, Tursz T, Amigorena S, Angevin E, Zitvogel L, 2004. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J. Immunol 172, 2126–2136. 10.4049/jimmunol.172.4.2126 [DOI] [PubMed] [Google Scholar]
- Androlewicz MJ, Anderson KS, Cresswell P, 1993. Evidence that transporters associated with antigen processing translocate a major histocompatibility complex class I-binding peptide into the endoplasmic reticulum in an ATP-dependent manner. Proc. Natl. Acad. Sci. U. S. A 90, 9130–9134. 10.1073/pnas.90.19.9130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH, 2016. Coinhibitory Pathways in Immunotherapy for Cancer. Annu. Rev. Immunol 34, 539–573. 10.1146/annurev-immunol-032414-112049 [DOI] [PubMed] [Google Scholar]
- Böhm W, Thoma S, Leithäuser F, Möller P, Schirmbeck R, Reimann J, 1998. T cell-mediated, IFN-gamma-facilitated rejection of murine B16 melanomas. J. Immunol 161, 897–908. [PubMed] [Google Scholar]
- Bracamonte-Baran W, Florentin J, Zhou Y, Jankowska-Gan E, Haynes WJ, Zhong W, Brennan TV, Dutta P, Claas FHJ, van Rood JJ, Burlingham WJ, 2017. Modification of host dendritic cells by microchimerism-derived extracellular vesicles generates split tolerance. Proc. Natl. Acad. Sci. U. S. A 114, 1099–1104. 10.1073/pnas.1618364114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton J, Blees H, Henry CM, Buck MD, Schulz O, Rogers NC, Childs E, Zelenay S, Rhys H, Domart M-C, Collinson L, Alloatti A, Ellison CJ, Amigorena S, Papayannopoulos V, Thomas DC, Randow F, Reis E Sousa C, 2021. The receptor DNGR-1 signals for phagosomal rupture to promote cross-presentation of dead-cell-associated antigens. Nat. Immunol 22, 140–153. 10.1038/s41590-020-00824-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefalo PJ, Harding CV, 2001. Processing of Exogenous Antigens for Presentation by Class I MHC Molecules Involves Post-Golgi Peptide Exchange Influenced by Peptide-MHC Complex Stability and Acidic pH. J. Immunol 167, 1274–1282. 10.4049/jimmunol.167.3.1274 [DOI] [PubMed] [Google Scholar]
- de Witte MA, Coccoris M, Wolkers MC, van den Boom MD, Mesman EM, Song J-Y, van der Valk M, Haanen JBAG, Schumacher TNM, 2006. Targeting self-antigens through allogeneic TCR gene transfer. Blood 108, 870–877. 10.1182/blood-2005-08-009357 [DOI] [PubMed] [Google Scholar]
- Delamarre L, Pack M, Chang H, Mellman I, Trombetta ES, 2005. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science (80-. ). 307, 1630–1634. 10.1126/science.1108003 [DOI] [PubMed] [Google Scholar]
- Dolan BP, Gibbs KDJ, Ostrand-Rosenberg S, 2006. Dendritic Cells Cross-Dressed with Peptide MHC Class I Complexes Prime CD8 + T Cells. J. Immunol 177, 6018–6024. 10.4049/jimmunol.177.9.6018 [DOI] [PubMed] [Google Scholar]
- Dougan M, Dranoff G, Dougan SK, 2019. Cancer Immunotherapy: Beyond Checkpoint Blockade. Annu. Rev. Cancer Biol 3, 55–75. 10.1146/annurev-cancerbio-030518-055552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee H-W, Park CG, Steinman RM, Nussenzweig MC, 2007. Differential Antigen Processing by Dendritic Cell Subsets in Vivo. Science (80-. ). 315, 107–111. 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Duong E, Fessenden TB, Lutz E, Dinter T, Yim L, Blatt S, Bhutkar A, Wittrup KD, Spranger S, 2021. Type I interferon activates MHC class I-dressed CD11b+ conventional dendritic cells to promote protective anti-tumor CD8+ T cell immunity. Immunity 1–16. 10.1016/j.immuni.2021.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson BT, KC W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung S-SJ, Murphy TL, Hildner K, Murphy KM, 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med 207, 823–836. 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes MB, Kacha AK, Kline J, Woo S-R, Kranz DM, Murphy KM, Gajewski TF, 2011. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J. Exp. Med 208, 2005–2016. 10.1084/jem.20101159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanč P, Fujii T, Iborra S, Yamada Y, Huotari J, Schulz O, Ahrens S, Kjær S, Way M, Sancho D, Namba K, Reis e Sousa C, 2015. Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens. Immunity 42, 839–849. 10.1016/j.immuni.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein Z, Uchtenhagen H, Abualrous ET, Saini SK, Janßen L, Van Hateren A, Wiek C, Hanenberg H, Momburg F, Achour A, Elliott T, Springer S, Boulanger D, 2014. Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J. Cell Sci 127, 2885–2897. 10.1242/jcs.145334 [DOI] [PubMed] [Google Scholar]
- Herrera OB, Golshayan D, Tibbott R, Salcido Ochoa F, James MJ, Marelli-Berg FM, Lechler RI, 2004. A novel pathway of alloantigen presentation by dendritic cells. J. Immunol 173, 4828–4837. 10.4049/jimmunol.173.8.4828 [DOI] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM, 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science (80-. ). 322, 1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre OP, Segura E, Savina A, Amigorena S, 2012. Cross-presentation by dendritic cells. Nat. Rev. Immunol 12, 557–569. 10.1038/nri3254 [DOI] [PubMed] [Google Scholar]
- Kline DE, MacNabb BW, Chen X, Chan W-C, Fosco D, Kline J, 2018. CD8α(+) Dendritic Cells Dictate Leukemia-Specific CD8(+) T Cell Fates. J. Immunol 201, 3759–3769. 10.4049/jimmunol.1801184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline J, Brown IE, Zha Y-Y, Blank C, Strickler J, Wouters H, Zhang L, Gajewski TF, 2008. Homeostatic proliferation plus regulatory T-cell depletion promotes potent rejection of B16 melanoma. Clin. Cancer Res 14, 3156–3167. 10.1158/1078-0432.CCR-07-4696 [DOI] [PubMed] [Google Scholar]
- Kline J, Zhang L, Battaglia L, Cohen KS, Gajewski TF, 2012. Cellular and molecular requirements for rejection of B16 melanoma in the setting of regulatory T cell depletion and homeostatic proliferation. J. Immunol 188, 2630–2642. 10.4049/jimmunol.1100845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koble C, Kyewski B, 2009. The thymic medulla: a unique microenvironment for intercellular self-antigen transfer. J. Exp. Med 206, 1505–1513. 10.1084/jem.20082449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger CJ, Spidale NA, Wang B, Tisch R, 2017. Thymic Dendritic Cell Subsets Display Distinct Efficiencies and Mechanisms of Intercellular MHC Transfer. J. Immunol 198, 249–256. 10.4049/jimmunol.1601516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBelle JL, Hanke CA, Blazar BR, Truitt RL, 2002. Negative effect of CTLA-4 on induction of T-cell immunity in vivo to B7–1+, but not B7–2+, murine myelogenous leukemia. Blood 99, 2146–2153. 10.1182/blood.v99.6.2146 [DOI] [PubMed] [Google Scholar]
- Lee MY, Jeon JW, Sievers C, Allen CT, 2020. Antigen processing and presentation in cancer immunotherapy. J. Immunother. Cancer 8. 10.1136/jitc-2020-001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Kim S, Herndon JM, Goedegebuure P, Belt BA, Satpathy AT, Fleming TP, Hansen TH, Murphy KM, Gillanders WE, 2012. Cross-dressed CD8α+/CD103+ dendritic cells prime CD8+ T cells following vaccination. Proc. Natl. Acad. Sci. U. S. A 109, 12716–12721. 10.1073/pnas.1203468109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rojas-Canales DM, Divito SJ, Shufesky WJ, Stolz DB, Erdos G, Sullivan MLG, Gibson GA, Watkins SC, Larregina AT, Morelli AE, 2016. Donor dendritic cell-derived exosomes promote allograft-targeting immune response. J. Clin. Invest 126, 2805–2820. 10.1172/JCI84577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M, Sathe P, Helft J, Miller J, Mortha A, 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol 31, 563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre S, van Endert PM, 2019. Endocytic recycling of MHC class I molecules in non-professional antigen presenting and dendritic cells. Front. Immunol 9, 1–11. 10.3389/fimmu.2018.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Sasaki K, Kishimoto T, Niwa S, Hayashi H, Takahama Y, Tanaka K, 2007. Regulation of CD8 T Cell Development by Thymus-Specific Proteasomes. Science (80-. ). 316, 1349–1353. [DOI] [PubMed] [Google Scholar]
- Nair-Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y, Banerjee M, Overholtzer M, Roche PA, Tampé R, Brown BD, Amsen D, Whiteheart SW, Blander JM, 2014. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell 158, 506–521. 10.1016/j.cell.2014.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overwijk WW, Restifo NP, 2000. B16 as a Mouse Model for Human Melanoma, in: Current Protocols in Immunology. pp. 20.1.1–20.1.29. 10.1002/0471142735.im2001s39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry JSA, Russler-Germain EV, Zhou YW, Purtha W, Cooper ML, Choi J, Schroeder MA, Salazar V, Egawa T, Lee B-C, Abumrad NA, Kim BS, Anderson MS, DiPersio JF, Hsieh C-S, 2018. Transfer of Cell-Surface Antigens by Scavenger Receptor CD36 Promotes Thymic Regulatory T Cell Receptor Repertoire Development and Allo-tolerance. Immunity 48, 923–936.e4. 10.1016/j.immuni.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna H, Donaldson JG, 1997. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol 139, 49–61. 10.1083/jcb.139.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich Z, Altman JD, Boniface JJ, Lyons DS, Kozono H, Ogg G, Morgan C, Davis MM, 1997. Stability of empty and peptide-loaded class II major histocompatibility complex molecules at neutral and endosomal pH: comparison to class I proteins. Proc. Natl. Acad. Sci. U. S. A 94, 2495–2500. 10.1073/pnas.94.6.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid PA, Watts C, 1990. Cycling of cell-surface MHC glycoproteins through primaquine-sensitive intracellular compartments. Nature 346, 655–657. 10.1038/346655a0 [DOI] [PubMed] [Google Scholar]
- Roberts EW, Broz ML, Binnewies M, Headley MB, Nelson AE, Wolf DM, Kaisho T, Bogunovic D, Bhardwaj N, Krummel MF, 2016. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 30, 324–336. 10.1016/j.ccell.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhland MK, Roberts EW, Cai E, Mujal AM, Marchuk K, Beppler C, Nam D, Serwas NK, Binnewies M, Krummel MF, 2020. Visualizing Synaptic Transfer of Tumor Antigens among Dendritic Cells. Cancer Cell 37, 786–799.e5. 10.1016/j.ccell.2020.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo V, Zhou D, Sartirana C, Rovere P, Villa A, Rossini S, Traversari C, Bordignon C, 2000. Acquisition of intact allogeneic human leukocyte antigen molecules by human dendritic cells. Blood 95, 3473–3477. [PubMed] [Google Scholar]
- Salmon H, Idoyaga J, Rahman A, Leboeuf M, Remark R, Jordan S, Casanova-Acebes M, Khudoynazarova M, Agudo J, Tung N, Chakarov S, Rivera C, Hogstad B, Bosenberg M, Hashimoto D, Gnjatic S, Bhardwaj N, Palucka AK, Brown BD, Brody J, Ginhoux F, Merad M, 2016. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 44, 924–938. 10.1016/j.immuni.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Paulete AR, Teijeira A, Cueto FJ, Garasa S, Pérez-Gracia JL, Sánchez-Arráez A, Sancho D, Melero I, 2017. Antigen cross-presentation and T-cell cross-priming in cancer immunology and immunotherapy. Ann. Oncol 28, xii44–xii55. 10.1093/annonc/mdx237 [DOI] [PubMed] [Google Scholar]
- Sancho D, Joffre OP, Keller AM, Rogers NC, Martínez D, Hernanz-Falcón P, Rosewell I, Reis e Sousa C, 2009. Identification of a dendritic cell receptor that couples sensing of necrosis to immunity. Nature 458, 899–903. 10.1038/nature07750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Duménil A-M, Seabra MC, Raposo G, Amigorena S, 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218. 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Savina A, Peres A, Cebrian I, Carmo N, Moita C, Hacohen N, Moita LF, Amigorena S, 2009. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8(+) dendritic cells. Immunity 30, 544–555. 10.1016/j.immuni.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Schober K, Voit F, Grassmann S, Müller TR, Eggert J, Jarosch S, Weißbrich B, Hoffmann P, Borkner L, Nio E, Fanchi L, Clouser CR, Radhakrishnan A, Mihatsch L, Lückemeier P, Leube J, Dössinger G, Klein L, Neuenhahn M, Oduro JD, Cicin-Sain L, Buchholz VR, Busch DH, 2020. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat. Immunol 21, 434–441. 10.1038/s41590-020-0628-2 [DOI] [PubMed] [Google Scholar]
- Seliger B, Wollscheid U, Momburg F, Blankenstein T, Huber C, 2001. Characterization of the major histocompatibility complex class I deficiencies in B16 melanoma cells. Cancer Res. 61, 1095–1099. [PubMed] [Google Scholar]
- Spiotto MT, Yu P, Rowley DA, Nishimura MI, Meredith SC, Gajewski TF, Fu YX, Schreiber H, 2002. Increasing tumor antigen expression overcomes “ignorance” to solid tumors via crosspresentation by bone marrow-derived stromal cells. Immunity 17, 737–747. 10.1016/s1074-7613(02)00480-6 [DOI] [PubMed] [Google Scholar]
- Theisen DJ, Davidson JT 4th, Briseño CG, Gargaro M, Lauron EJ, Wang Q, Desai P, Durai V, Bagadia P, Brickner JR, Beatty WL, Virgin HW, Gillanders WE, Mosammaparast N, Diamond MS, Sibley LD, Yokoyama W, Schreiber RD, Murphy TL, Murphy KM, 2018. WDFY4 is required for cross-presentation in response to viral and tumor antigens. Science (80-. ). 362, 694–699. 10.1126/science.aat5030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Endert P, 2016. Intracellular recycling and cross-presentation by MHC class I molecules. Immunol. Rev 272, 80–96. 10.1111/imr.12424 [DOI] [PubMed] [Google Scholar]
- Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S, 1992. TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4–8+ T cells. Cell 71, 1205–1214. 10.1016/s0092-8674(05)80068-6 [DOI] [PubMed] [Google Scholar]
- Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ, 2011. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol 29, 235–271. 10.1146/annurev-immunol-031210-101324 [DOI] [PubMed] [Google Scholar]
- Wakim LM, Bevan MJ, 2011. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature 471, 629–632. 10.1038/nature09863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman AD, Fritz JM, Lenardo MJ, 2020. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol 20, 651–668. 10.1038/s41577-020-0306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek SK, Cueto FJ, Mujal AM, Melero I, Krummel MF, Sancho D, 2020. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol 20, 7–24. 10.1038/s41577-019-0210-z [DOI] [PubMed] [Google Scholar]
- Wolfers J, Lozier A, Raposo G, Regnault A, Théry C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, Angevin E, Amigorena S, Zitvogel L, 2001. Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming. Nat. Med 7, 297–303. 10.1038/85438 [DOI] [PubMed] [Google Scholar]
- Worbs T, Hammerschmidt SI, Förster R, 2017. Dendritic cell migration in health and disease. Nat. Rev. Immunol 17, 30–48. 10.1038/nri.2016.116 [DOI] [PubMed] [Google Scholar]
- Zhang J-G, Czabotar PE, Policheni AN, Caminschi I, San Wan S, Kitsoulis S, Tullett KM, Robin AY, Brammananth R, van Delft MF, Lu J, O’Reilly LA, Josefsson EC, Kile BT, Chin WJ, Mintern JD, Olshina MA, Wong W, Baum J, Wright MD, Huang DCS, Mohandas N, Coppel RL, Colman PM, Nicola NA, Shortman K, Lahoud MH, 2012. The Dendritic Cell Receptor Clec9A Binds Damaged Cells via Exposed Actin Filaments. Immunity 36, 646–657. 10.1016/j.immuni.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Zhang L, Chen X, Liu X, Kline DE, Teague RM, Gajewski TF, Kline J, 2013. CD40 ligation reverses T cell tolerance in acute myeloid leukemia. J. Clin. Invest 123, 1999–2010. 10.1172/JCI63980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.