Abstract
Mycobacterium tuberculosis and Mycobacterium avium are facultative intracellular pathogens that are able to survive and replicate in mononuclear phagocytes. Human complement component C3 has previously been shown to mediate attachment and phagocytosis of these bacteria by mononuclear phagocytes. In this study, a C3 ligand affinity blot protocol was used to identify a 30-kDa C3-binding protein in M. tuberculosis and Mycobacterium smegmatis and a 31-kDa C3-binding protein in M. avium. The C3-binding proteins in M. tuberculosis and M. avium localized to the cell membrane fraction and partitioned to the detergent fraction during Triton X-114 phase partitioning. The C3-binding protein from M. tuberculosis was partially purified using a cation exchange column and was shown to bind concanavalin A. The N terminus and an internal fragment of the partially purified C3-binding protein were subjected to amino acid sequence analysis. The resulting amino acid sequences matched the M. tuberculosis heparin-binding hemagglutinin (HbhA) protein. Recombinant full-length HbhA and the C terminus of HbhA fused to maltose-binding protein, but not recombinant HbhA lacking the C-terminal region, bound human C3. Recombinant full-length HbhA coated on polystyrene beads, was found to enhance the adherence and/or phagocytosis of the coated beads to J774.A1 cells in both the presence and absence of human serum. The presence of complement-sufficient serum increased the adherence of the HbhA-coated beads to the J774.A1 cells in a C3-dependent manner. If HbhA within the bacterial cell membrane functions similarly to isolated HbhA, this protein may enhance the adherence and phagocytosis of M. tuberculosis and M. avium to mononuclear phagocytes through the binding of C3 and interaction with C3 receptors on mononuclear phagocytes.
Mycobacterium tuberculosis, the causative agent of tuberculosis, is a facultative intracellular pathogen that can be phagocytosed by human mononuclear phagocytes and survive and replicate inside these cells. This bacterium can bind to several types of receptors on the surface of mononuclear phagocytes (recently reviewed by Ernst [14]), including complement receptors (10, 20, 41, 42, 44, 47). Complement receptor one (CR1) (CD35) is a single-chain glycoprotein that binds complement fragments C3b and C4b (23). CR3 (CD11b/CD18) and CR4 (CD11c/CD18) are heterodimers belonging to the leukocyte β2-integrin family. These two receptors bind complement fragment C3bi and also contain a polysaccharide binding site (2, 24). M. tuberculosis can bind to the complement receptors via both complement-dependent and -independent pathways (10, 20, 41, 42, 44, 47) and is subsequently phagocytosed by the phagocytic cell. The presence of human serum containing active complement components was found to enhance the binding of M. tuberculosis to CR1, CR3, and CR4 on the surface of human monocytes and monocyte-derived macrophages (MDMS) (20, 41, 42). Complement component C3 was identified as the major component in human serum involved in enhancing the adherence and uptake of M. tuberculosis by mononuclear phagocytes (42).
Mycobacterium avium, a causative agent of opportunistic infections in immunocompromised individuals such as AIDS patients, is also a facultative intracellular pathogen that is able to survive and replicate in mononuclear phagocytes. M. avium is able to bind to several types of receptors on monocytes and macrophages in both the presence and absence of serum, including CR1 and CR3 (6, 7, 37). The presence of normal human serum (NHS) significantly enhances the adherence and phagocytosis of M. avium by MDMs and monocytes (7, 45), and C3 was found to be an important opsonin for the adherence and uptake of M. avium by MDMs (7).
C3-binding molecules on the surface of several intracellular pathogens have been identified. These molecules include major outer membrane protein (MOMP) from Legionella pneumophila (3), MOMP from Chlamydia trachomatis (17), lipophosphoglycan from Leishmania major promastigotes (35), gp63 from Leishmania mexicana promastigotes (39), gp72 from Trypanosoma cruzi epimastigotes (22), and phenolic glycolipid-1 from Mycobacterium leprae (43). In this study, we identified a C3-binding protein in M. tuberculosis, M. avium, and Mycobacterium smegmatis using a C3 ligand affinity blot protocol and further characterized these proteins in M. tuberculosis and M. avium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. tuberculosis H37Rv (ATCC 27294) was purchased from the American Type Culture Collection (ATCC). M. avium ATCC 49601 was provided by C. Jagganath at the University of Texas-Houston Medical School, and M. smegmatis mc2155 was provided by W. R. Jacobs at the Albert Einstein College of Medicine. Escherichia coli SURE2, E. coli TOPP3, and E. coli BL21(DE3) pLysS were obtained from Stratagene. M. tuberculosis was cultured with shaking at 37°C in Middlebrook 7H9 broth (Difco) containing 0.2% glycerol, 0.05% Tween 80, and 10% ADC enrichment (0.2% glucose, 0.5% bovine serum albumin [BSA] fraction V, 0.085% sodium chloride) for 14 days. In experiments where culture supernatants were examined, M. tuberculosis was first cultured as described above for 5 days, and the cells were centrifuged and washed three times with 7H9 broth. The washed cells were used to inoculate into 7H9 broth containing 0.2% glycerol and cultured with occasional agitation for 25 days at 37°C; the culture supernatant was then collected by centrifugation. Culture supernatant used for electrophoretic analysis was filtered through a 0.22-μm-pore-size filter and then concentrated 10-fold using a Centricon-10 concentrator (Amicon) per the manufacturer's instructions. M. avium was cultured at 37°C on Middlebrook 7H11 agar plates containing 10% OADC (Remel) for 19 days. M. smegmatis was cultured at 37°C for 2 days on Middlebrook 7H10 (Difco) agar plates containing 10% ADC enrichment. E. coli strains were cultured overnight at 37°C either on Luria-Bertani agar plates or in Luria-Bertani broth, with shaking; carbenicillin (50 μg/ml) and/or 0.5% glucose was added as needed for selection and enhancement of recombinant protein expression, respectively.
J774.A1 cell line and medium.
The murine macrophage-like cell line J774.A1 (ATCC TIB-67) was purchased from the ATCC and was cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with sodium bicarbonate (2.2. g/liter), HEPES (50 mg/liter), l-arginine (50 mg/liter), penicillin (50 mg/liter), gentamicin (50 mg/liter), and 10% heat-inactivated fetal bovine serum.
Electrophoretic techniques.
All protein samples prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were solubilized at 25°C in a solution containing 0.125 M Tris-HCl (pH 6.8), 10% glycerol, 5% 2-mercaptoethanol, 2.3% SDS, and 0.01% bromphenol blue. When mycobacterial cell lysates were electrophoresed, whole cells were lysed by ultrasonic disruption using a microtip probe (three times for 15 s each) and then solubilized in solubilization buffer. All protein samples were boiled for 5 min at 100°C prior to electrophoresis. Proteins were electrophoresed by the method of Laemmli (25) in SDS–12% polyacrylamide gels and then either stained with 0.25% Coomassie blue R-250 or transferred to polyvinylidene difluoride (PVDF) membranes (PVDF-PLUS; Micron Separations Inc.) (46). Preparations of the M. tuberculosis C3-binding protein were subjected to nonequilibrium pH gradient electrophoresis (NEPHGE) by the method of O'Farrell et al. (32) using ampholines with a pH of 3.5 to 10 (Pharmacia) and then to SDS-PAGE in 8 to 20% gradient polyacrylamide gels.
C3 ligand affinity blot.
The C3 ligand affinity blot protocol used to detect C3 bound to Mycobacterium proteins or to purified recombinant proteins was modified slightly from that used by Bellinger-Kawahara and Horwitz (3) to detect C3 bound to L. pneumophila MOMP. A PVDF membrane electroblot of SDS-PAGE separated proteins was incubated overnight at 4°C in 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBST) to block nonspecific binding. After the membrane was washed with PBST, it was incubated at 37°C for 40 min in 2.5% NHS obtained from a purified protein derivative-negative laboratory volunteer. In some experiments, the serum was heated at 56°C for 30 min to inactivate C3, treated with 25 mM methlyamine, or treated with 10 mM EDTA. The membrane was then washed four times for 15 min each in PBST. The membrane was incubated for 1 h in a mouse monoclonal antibody against human C3c (Quidel Corporation) diluted 1:20,000. This antibody recognizes C3, C3b, C3bi, and C3c but not C3a, C3d, or C3dg. The membrane was washed and then incubated for 1 h in a horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (Pierce) diluted 1:30,000. After washing, the membrane was reacted with chemiluminescent detection reagents (ECL; Amersham) according to the manufacturer's instructions and exposed to autoradiographic film.
Preparation of cell membrane fraction.
Fractions containing either M. tuberculosis or M. avium cytoplasmic membranes were prepared as described in Nikaido et al. (30) with modifications. M. tuberculosis or M. avium cells were resuspended in 20 mM sodium phosphate–10 mM EDTA (pH 7.0) containing a protease inhibitor cocktail (Sigma catalog no. P8849), incubated on ice for 30 min, and lysed by ultrasonic disruption using a microtip probe in a laminar-flow hood 10 times for 30 s each for M. tuberculosis or 6 times for 30 s each for M. avium with a 30-s incubation on ice between each 30-s burst. The cell lysate was centrifuged at 4°C in a microcentrifuge at 10,000 × g for 10 min to remove intact cells. The supernatant was transferred to a new tube and centrifuged again in the same manner. The supernatant, representing a clear cell lysate, was removed to a new tube and centrifuged at 4°C at 49,500 × g for 2 h. The supernatant (soluble fraction) was carefully removed to a new tube, and solubilization buffer was added to solubilize the proteins. The pellet (particulate fraction) was resuspended in solubilization buffer.
Triton X-114 phase partitioning.
The protocol used for Triton X-114 phase partitioning was modified from that of Radolf et al. (36). M. tuberculosis or M. avium cells were washed in 200 mM Tris-HCl (pH 8.0)–1 mM EDTA and resuspended in 200 mM Tris-HCl (pH 8.0)–1 mM EDTA–10% sucrose containing a protease inhibitor cocktail. The cells were lysed by ultrasonic disruption using a microtip probe in a laminar-flow hood two times for 2.5 min each on ice with a 1-min incubation on ice between bursts. The cell lysate was centrifuged at 4°C in a microcentrifuge at 10,000 × g for 10 min. The supernatant was transferred to a new tube and centrifuged again in the same manner to create a clear cell lysate. A 900-μl aliquot of the supernatant (clear lysate) was transferred to a new tube, and 100 μl of 10% Triton X-114 was added to the lysate to give a final concentration of 1% Triton X-114. The mixture was first incubated on ice for 30 min, then incubated in a 37°C waterbath for 10 min, and finally centrifuged at 37°C at 3,000 × g for 10 min. The two phases were separated and washed. The upper aqueous phase was washed by addition of 10% Triton X-114 to a final concentration of 1%, and the lower detergent phase was washed with 25 mM Tris-HCl, pH 8.0. The samples were again incubated on ice and at 37°C and centrifuged as described above. The washed aqueous phase and detergent phase preparations were removed to new tubes, and solubilization buffer was added to both phases to solubilize the proteins.
Ion exchange affinity chromatography.
M. tuberculosis cells were washed in 200 mM Tris-HCl (pH 8.0)–1 mM EDTA. The cells were resuspended in 200 mM Tris-HCl (pH 8.0)–1 mM EDTA–10% sucrose and lysed by ultrasonic disruption as described above for Triton X-114 phase partitioning. The cell lysate was made using ultrasonication in the absence of protease inhibitors. An M. tuberculosis H37Rv cell lysate was partitioned into an aqueous fraction and a detergent fraction using Triton X-114 as described above. The detergent fraction and the aqueous fraction were tested for the presence of the C3-binding protein using the C3 ligand affinity blot protocol. The protein was present in the aqueous fraction under these conditions.
Q-Sepharose (Pharmacia) and S-Sepharose (Pharmacia) gravity flow columns (250 μl bed volume) were generously provided by R. T. Owens at the Institute of Biosciences and Technology, Texas A & M University. The columns were equilibrated using 25 mM Tris-HCl, pH 8.0. The aqueous fraction was divided equally, with one half being loaded on the Q-Sepharose column and the other half being loaded on the S-Sepharose column. The flow-through was collected from each column, and the columns were washed with 25 mM Tris-HCl, pH 8.0, to elute proteins that bound nonspecifically. Bound proteins were eluted with a step gradient of NaCl in increasing concentrations (50, 100, 150, 250, 500, and 1,000 mM) in 2 mM Tris-HCl, pH 8.0. The distribution of the C3-binding protein was determined by testing the flowthrough, wash, and elution fractions from both columns using the C3 ligand affinity blot protocol.
Concanavalin A ligand affinity blot.
A protocol from Herrmann et al. (18) was used to detect binding of concanavalin A to mycobacterial proteins. PVDF electroblots containing SDS-PAGE separated, partially purified M. tuberculosis C3-binding protein were incubated in PBST with 5% BSA for 1 h to block nonspecific binding. The membrane was then incubated in peroxidase-conjugated concanavalin A from the jack bean (0.5 μg/ml; Sigma) in 5% BSA in PBST for 1 h. The membrane was washed three times in PBST and one time in PBS and then reacted with Amersham's ECL detection reagents according to the manufacturer's instructions and exposed to autoradiographic film.
Protease digestion and amino acid sequencing.
The C3-binding protein was excised from a Coomassie blue-stained two-dimensional gel and submitted to Richard G. Cook at the Baylor College of Medicine Protein Chemistry Core Facility for protease digestion and amino acid sequencing. The protein gel slice was digested with Lys-C purified from Achromobacter lyticus (Wako Chemicals), and the peptide fragments were isolated using reverse-phase high-performance liquid chromatography. The intact protein and one peptide fragment were subjected to Edman degradation and sequenced to ten amino acids and twenty amino acids, respectively.
Monoclonal antibody to HbhA.
Mouse monoclonal antibody 3921E4 against heparin-binding hemagglutinin (HbhA) was generously provided by M. J. Brennan at the Food and Drug Administration and has been described previously (38). This antibody was used in Western blot analysis at a dilution of 1:10,000, and horseradish peroxidase conjugated goat anti-mouse IgG antibody (Pierce) was used at a dilution of 1:20,000.
Expression and purification of recombinant M. tuberculosis HbhA.
M. tuberculosis H37Rv genomic DNA was isolated from liquid culture as described by Armitige et al. (1). PCR amplification was performed using the Thermalase PCR kit (Amresco) in a minicycler from MJ Research. A 644-bp fragment containing the entire coding region of hbhA, except for the ATG start codon, was amplified from M. tuberculosis genomic DNA using the forward primer 5BhbhA (5′-CTGGGATCCGCTGAAAACTCGAACATTGAT-3′) and the reverse primer 3hbhA (5′-GACAAGCTTACTCGGAGTCGATGGTGATTC-3′) (The underlined regions contain the BamHI and HindIII regions, respectively, and were added for cloning purposes [see below].) in the following PCR program: 96°C for 2 min, six cycles of denaturation at 94°C for 40 s, annealing at 60°C for 40 s, and extension at 72°C for 1 min, followed by 26 cycles at a higher annealing temperature of 65°C, and then by a final extension at 72°C for 10 min.
The 644-bp hbhA PCR product was used as template in another PCR to amplify a 492-bp fragment containing the first 474 bp of the coding region of hbhA, except for the ATG start codon. The forward primer used in the PCR was the 5BhbhA primer above, and the reverse primer used was primer 100 (5′-CCAAAGCTTCAGCTCGATGCCGACCAG-3′; the underlined region contain an added HindIII site). The PCR program was as follows: 96°C for 2 min, 27 cycles of denaturation at 94°C for 40 s, annealing at 60°C for 40 s, and extension at 72°C for 1 min, followed by a final extension at 72°C for 5 min.
For cloning purposes, 9-bp sequences (underlined above) were added to the 5′-ends to create a BamHI site in the 5BhbhA primer and a HindIII site in the 3hbhA primer and the 100 primer. The 644-bp and the 492-bp PCR products were treated with BamHI and HindIII (New England Biolabs) and ligated separately into the pQE30 expression vector (Qiagen) that had been treated with BamHI and HindIII. The resulting plasmids (pQEhbhA and pQE492hbhA) were separately transformed first into E. coli SURE2 as described in (40), and then supercoiled plasmid purified from SURE2 cells was used to transform E. coli TOPP3. The insert sequences of pQEhbhA and pQE492hbhA were verified prior to use for protein expression. Expression of recombinant HbhA (rHbhA) and truncated HbhA (trHbhA) was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM followed by incubation for 2 h at 37°C with vigorous shaking. rHbhA and trHbhA contain a polyhistidine tag at the N terminus encoded from the pQE30 vector and were purified using Ni-nitrilotriacetic acid agarose (Qiagen) columns according to the manufacturer's instructions. Protein purification was performed at 4°C and in the presence of a protease inhibitor cocktail (Sigma catalog no. P8849) to minimize the degradation of recombinant proteins.
The C terminus of HbhA was expressed from plasmid pMAL+3R1+2R2, which was kindly provided by F. D. Menozzi at the Pasteur Institute. This plasmid encodes the final 39 amino acids of HbhA, and its construction has been described (33). The plasmid was electroporated into E. coli BL21(DE3) pLysS. Expression of the C terminus of HbhA (cHbhA) was induced by the addition of IPTG at a final concentration of 1 mM followed by incubation for 2 h at 37°C with vigorous shaking. cHbhA is fused to the maltose-binding protein (MBP) at the N terminus, which is encoded from the pMAL-c vector (New England Biolabs). cHbhA was purified using an amylose resin (New England Biolabs) column according to the manufacturer's instructions at 4°C in the presence of a protease inhibitor cocktail (Sigma) to minimize the degradation of recombinant protein. Purified MBP was purchased from New England Biolabs.
Preparation of HbhA-coated beads.
Polystyrene latex beads were coated with protein, with minor modifications, following a protocol by Polysciences Inc. for coupling proteins to carboxylated microparticles. Polystyrene beads (carboxylate-modified, fluorescent beads, Sigma catalog no. L4530; 1.43 × 109 beads per reaction), 2 μm in diameter, were washed two times in carbonate buffer and three times in sodium phosphate buffer by centrifugation at 7,200 × g for 7 min. The beads were then incubated in a 1% carbodiimide solution on a rotator for 3.5 h at room temperature. The beads were centrifuged as above and washed three times in borate buffer. The beads were then incubated on a rotator overnight at 4°C in either borate buffer alone (uncoated beads), borate buffer containing 150 μg of BSA, or borate buffer containing 150 μg of rHbhA and a protease inhibitor cocktail. The next morning, the beads were centrifuged as above, and the supernatant was removed to a separate tube for protein concentration analysis. The beads were then incubated on a rotator for 2 hours at room temperature in a 5% BSA solution to block nonspecific binding sites, centrifuged as above, and resuspended in sodium phosphate buffer containing 0.5% BSA. The amount of protein coated on the beads was determined by subtracting the amount of protein left in the supernatant from the total amount of protein added to the beads. Using the bicinchoninic acid protein assay kit (Pierce) and BSA as a standard, approximately 120 μg of rHbhA was bound to the beads.
Adherence assay of HbhA-coated beads.
J774.A1 macrophage-like cells were plated on glass coverslips in 12-well tissue culture plates at 106 cells/ml. The cells were then cultured at 37°C for 24 h in DMEM containing supplements as described previously and 10% heat-inactivated fetal bovine serum. The next day the cells were washed once in warm PBS and then were incubated with 107 beads (either uncoated, BSA-coated, or HbhA-coated) in DMEM containing either no serum, 2.5% heat-inactivated human serum (HIS), or 2.5% NHS. The beads were allowed to incubate with the cells for 1 h at 37°C with gentle rotation at 100 rpm. The cells were washed three times with warm PBS to remove nonadherent beads, fixed with warm 10% buffered formalin for 10 min at 37°C, washed once with PBS, stained with 0.1% Evans Blue for 10 min, and washed twice with PBS. The mean number (± standard error) of beads per 100 cells for each serum treatment was determined by counting 100 cells in each of triplicate wells by light microscopy. Student's t test was used for statistical analysis.
Adherence assay of HbhA-coated beads in the presence of antibody to C3.
J774.A1 cells were plated in 24-well tissue culture plates at 106 cells/ml. The cells were then cultured as above for 24 h. The next day 107 beads (either uncoated or HbhA coated) were incubated at 37°C for 30 min with gentle shaking in DMEM containing either no serum, 2.5% HIS, or 2.5% NHS and either no antibody, purified normal goat F(ab′)2 (Accurate Chemical and Scientific Corp.) at a 1:300 dilution, or goat F(ab′)2 to human complement C3 (ICN Pharmaceuticals, Inc.) at a 1:300 dilution. The J774.A1 cells were washed once in warm PBS and incubated with the bead mixtures for 1 h as described above. The cells were washed, fixed, stained, and counted as described above.
Antibody production.
Antiserum to rHbhA was prepared in a New Zealand male rabbit using TiterMax (CytRx) (4, 29) as an adjuvant according to the manufacturer's instructions. TiterMax was provided by J. K. Actor at the University of Texas-Houston Medical School. The rabbit was immunized intramuscularly with 15 μg of purified rHbhA in each hind quadricep and subcutaneously with 15 μg of purified rHbhA under the scruff of the neck for a total immunization of 45 μg of rHbhA. The rabbit received a booster 5 weeks later with TiterMax as the adjuvant as above and was bled 5 days postboost. The antiserum was used in Western blot analysis at a 1:10,000 dilution.
RESULTS
Identification of mycobacterial proteins that bind C3.
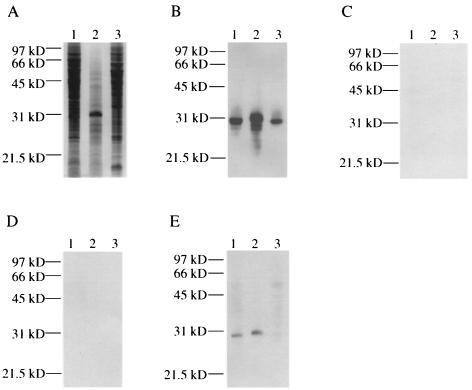
To determine if M. tuberculosis proteins bound human C3, we used a protocol modified from that used by Bellinger-Kawahara and Horwitz (3) to identify an L. pneumophila C3-binding protein. An M. tuberculosis H37Rv cell lysate was separated using SDS-PAGE and then transferred to a PVDF membrane. The membrane was incubated in 2.5% NHS to allow interaction of complement components with proteins on the membrane. After washing the membrane, the blot was incubated with a monoclonal antibody that recognizes C3, C3b, C3bi, and C3c. C3 or its derivatives bound to a 30-kDa protein band from M. tuberculosis H37Rv (Fig. 1B). Using the same protocol, we also found that a 31-kDa protein from M. avium and a 30-kDa protein from the nonpathogenic species M. smegmatis bound C3 (Fig. 1B). Several reactive bands below the 31-kDa reactive band are present in the M. avium lysate (Fig. 1B, lane 2). These bands may represent degradation products of the 31-kDa protein. Upon shorter exposure, the only reactive protein is the 31-kDa protein (data not shown). The anti-C3 antibody does not react to mycobacterial products since no reactive bands were observed if the blot was incubated in anti-C3 antibody alone in the absence of NHS (data not shown). No reactive bands were observed when NHS was heated at 56°C for 30 min to inactivate C3 prior to incubation with the Mycobacterium cell lysates or when 10 mM EDTA was added to the NHS (Fig. 1C and D, respectively). Treatment of serum with 10 mM EDTA prevents activation of complement through both the alternative and classical pathways (31). Methylamine reacts to the internal thioester of C3, thereby making the thioester of C3 unavailable for covalent binding to target molecules such as proteins and carbohydrates (31). Addition of 25 mM methylamine to the NHS significantly reduced the binding of C3 to the M. tuberculosis and M. avium C3-binding proteins and abolished the binding of C3 to the M. smegmatis C3-binding protein (compare Fig. 1E and B). Taken together, these results indicate that complement activation is required for the binding of C3 to the Mycobacterium C3-binding proteins and also suggest that the thioester of C3 plays a role in the binding of C3 to the Mycobacterium C3-binding proteins.
FIG. 1.
Detection of C3-binding proteins in cell lysates of Mycobacterium spp. by C3 ligand affinity blot analysis. Bacterial cell lysates were either stained with Coomassie blue R-250 (A) or transferred to PVDF membranes and reacted with either NHS (B), HIS (C), NHS containing 10 mM EDTA (D), or NHS containing 25 mM methylamine (E). Lane 1, M. tuberculosis; lane 2, M. avium; lane 3, M. smegmatis. Molecular mass markers are indicated on the left.
Cellular location of the C3-binding protein.
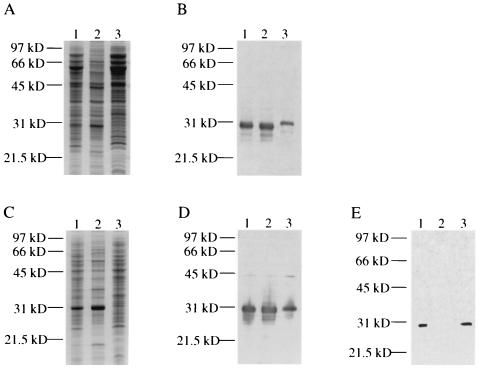
As a first step in determining the location of the C3-binding proteins of M. tuberculosis and M. avium, particulate and soluble fractions of both bacteria were prepared. In both organisms, the C3-binding proteins were present in both the particulate and soluble fractions, but a greater proportion of the C3-binding proteins was present in the particulate fractions (Fig. 2B and D). The soluble form of the M. tuberculosis C3-binding protein consistently migrated slightly higher in SDS-PAGE gels compared to the form found in the cell lysate or in the particulate fraction (Fig. 2B). A faint reactive band at 45 kDa was observed in the M. avium cellular fractions shown in Fig. 2D but has not been characterized further. We were unable to detect C3-binding activity in 10-fold-concentrated cell culture supernatants of M. tuberculosis (data not shown). Triton X-114 phase partitioning was performed using a M. tuberculosis cell lysate to determine if the C3-binding protein has hydrophobic properties. The protein localized to the detergent phase and was absent in the aqueous phase, indicating that the protein may be an integral membrane protein or a lipoprotein (Fig. 2E). Taken together, these results suggest that most of the cellular mycobacterial C3-binding proteins are membrane associated.
FIG. 2.
Cellular localization and hydrophobicity of C3-binding proteins from M. tuberculosis and M. avium, as determined by C3 ligand affinity blot analysis. M. tuberculosis cellular fractions were either stained with Coomassie blue R-250 (A) or transferred to a PVDF membrane and reacted with NHS (B). Lane 1, M. tuberculosis cell lysate; lane 2, M. tuberculosis particulate fraction; lane 3, M. tuberculosis soluble fraction. M. avium cellular fractions were either stained with Coomassie blue R-250 (C) or transferred to a PVDF membrane and reacted with NHS (D). Lane 1, M. avium cell lysate; lane 2, M. avium particulate fraction; lane 3, M. avium soluble fraction. (E) C3 ligand affinity blot of M. tuberculosis cell lysate following Triton X-114 phase partitioning. Lane 1, cell lysate; lane 2, aqueous phase; lane 3, detergent phase. Molecular mass markers are indicated on the left.
Characterization of the C3-binding protein.
Ion-exchange chromatography was utilized as a first step in the purification of the M. tuberculosis C3-binding protein. Cells were sonicated for a total of 5 min on ice in the absence of protease inhibitors to create a cell lysate, and then Triton X-114 phase partitioning was used to separate the cell lysate into a detergent phase and an aqueous phase. Under these conditions, the C3-binding protein localized to the aqueous phase. Sonication in the presence of a protease inhibitor cocktail resulted in partitioning of the protein to the detergent phase; the Mr of the protein was not affected by the presence or absence of protease inhibitors (data not shown). The C3-binding protein obtained in the presence of protease inhibitors bound irreversibly to column materials. For this reason, the aqueous phase preparation obtained in the absence of protease inhibitors was used in subsequent purification steps. In initial experiments, one half of the aqueous phase was loaded onto a Q-Sepharose column, and the other half was loaded onto an S-Sepharose column. The C3-binding protein did not bind specifically to the Q-Sepharose column (anion-exchange column), in that the protein was only present in the flowthrough and wash fractions of this column (data not shown). The C3-binding protein bound specifically to the S-Sepharose column (cation-exchange column) and eluted in 500 mM NaCl (data not shown).
Several glycoproteins, including a 28-kDa glycoprotein, have previously been described in M. tuberculosis and M. bovis (12, 13, 15, 16, 18, 19, 27). Using a protocol described by Herrmann et al. (18), we examined the ability of the partially purified C3-binding protein of M. tuberculosis to bind peroxidase-conjugated concanavalin A, a plant lectin that binds mannose and glucose in glycoproteins and polysaccharides. The 30-kDa C3-binding protein bound concanavalin A (data not shown), indicating that the protein may be a glycoprotein.
Two-dimensional gel electrophoresis using NEPHGE was performed to separate the proteins in the 500 mM NaCl S-Sepharose fraction. Three identical polyacrylamide gels were run in the second dimension using SDS-PAGE. One gel was silver stained (Fig. 3A), and the second gel was transferred to a PVDF membrane. The location of the C3-binding protein was identified using the C3 ligand affinity blot protocol (Fig. 3B), and the corresponding spot was excised from the third, Coomassie blue-stained gel. The protein in the gel slice was treated with the proteinase Lys-C, and the resulting peptides were isolated using reverse-phase high-performance liquid chromatography.
FIG. 3.
Demonstration of M. tuberculosis C3-binding protein partial purification by two-dimensional gel electrophoresis. An M. tuberculosis 500 mM NaCl S-Sepharose fraction was subjected to two-dimensional gel electrophoresis using NEPHGE in the first dimension and SDS-PAGE in the second dimension. (A) Silver-stained gel with the white arrow indicating the location of the C3-binding protein. (B) C3 ligand affinity blot of two-dimensional gel pattern identical to that shown in panel A. The basic end of the NEPHGE separation is on the left in both panels. Prestained molecular mass markers are indicated between panels.
The amino acid sequences of the N terminus of the C3-binding protein partially purified from a Triton X-114 aqueous phase and a selected Lys-C peptide from the S-Sepharose purified material were determined to be AENSNIDDIK and AAEGYLEAATSRYNELVERG, respectively. These two sequences matched perfectly to the previously described (27, 28) M. tuberculosis heparin-binding hemagglutinin protein (HbhA) beginning at the 2nd and 83rd amino acids, respectively.
The amino acid sequence of HbhA was examined using structural prediction programs. PSORT, which predicts protein localization sites, did not find an N-terminal signal sequence and predicted HbhA to be associated with the cytoplasmic membrane via a possible transmembrane domain at the N-terminus of the protein (data not shown). TMpred, which predicts protein hydrophobicity, also identified a 22-amino acid hydrophobic stretch at the N-terminus of HbhA (residues 14 to 35) (Fig. 7) and predicted the remainder of the protein to be hydrophilic. COILS predicted HbhA to have a possible coiled-coil region outside of the hydrophobic region at the N terminus (data not shown), and PLOTSTRUCTURE (Genetics Computer Group Wisconsin Package, version 10.0), which examines secondary structure, predicted HbhA to have a high alpha-helical content. The carboxy terminus of M. tuberculosis HbhA contains two types of lysine-alanine repeats. The repeat KKAAPA occurs three times in tandem and is followed by the repeat KKA(A/P)A, which also occurs three times in tandem (see Fig. 7).
FIG. 7.
Comparison of the amino acid sequences of M. tuberculosis HbhA and the M. avium and M. leprae HbhA homologs. The amino acid sequence of the M. avium HbhA homolog is 89% similar and 84% identical to M. tuberculosis HbhA (accession number F70742), and the M. avium homolog contains a 6-amino-acid insertion in the C terminus that is not found in M. tuberculosis HbhA. The amino acid sequence of the M. leprae HbhA homolog is 87% similar and 81% identical to M. tuberculosis HbhA, and the M. leprae homolog contains an 11-amino-acid deletion in the C terminus compared to M. tuberculosis HbhA. A consensus sequence is shown below each line of the alignment, with identical amino acids indicated by asterisks and gaps introduced for optimal alignment indicated by hyphens. The 22-amino-acid hydrophobic region present in the N terminus of each species is shown in bold, and the lysine-alanine repeats in the C terminus are boxed.
To further substantiate that the C3-binding protein was HbhA, a monoclonal antibody (3921E4) previously shown to react specifically to HbhA of M. tuberculosis (27, 28) was used in Western blots of an M. tuberculosis cell lysate and the C3-binding protein partially purified by S-Sepharose column chromatography. The antibody reacted with a 30-kDa protein in both lanes (data not shown), confirming that the C3-binding protein from M. tuberculosis is HbhA.
Expression and purification of recombinant HbhA.
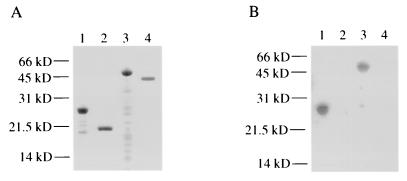
Full-length M. tuberculosis HbhA and truncated HbhA (encoding the first 159 amino acids of HbhA without the lysine-alanine repeats) were expressed in E. coli TOPP3 using the pQE30 expression vector and were purified by affinity chromatography (Fig. 4A). Purified full-length recombinant HbhA (rHbhA) is 1 to 2 kDa smaller than native M. tuberculosis HbhA (Fig. 4), which may be due to a lack of posttranslational modification in E. coli, and is susceptible to degradation, even in the presence of several protease inhibitors (Fig. 4A). Purified trHbhA migrates at approximately 21.5 kDa (Fig. 4A) but is estimated to be 17.6 kDa in size. The C terminus of HbhA (cHbhA)(encoding the final 39 amino acids of HbhA, including all of the lysine-alanine repeats) was expressed as a fusion protein to MBP in E. coli BL21(DE3) pLysS using the pMAL-c expression vector and was purified by affinity chromatography (Fig. 4A). All three of the recombinant HbhA proteins (rHbhA, trHbhA, and cHbhA) reacted to rabbit antiserum produced against full-length rHbhA (data not shown). Purified rHbhA was able to bind human C3, but purified trHbhA was not able to bind human C3 (Fig. 4B). cHbhA was also able to bind human C3, whereas purified MBP was not able to bind human C3 (Fig. 4B). These data indicate that the C terminus of HbhA participates in binding of C3 to HbhA.
FIG. 4.
Expression of HbhA and demonstration that full-length HbhA and the repeat region of HbhA, but not truncated HbhA, bind human C3, as determined by C3 ligand affinity blot analysis. Shown are a Coomassie-stained gel (A) and a C3 ligand affinity blot (B) of purified recombinant HbhA proteins. Lane 1, purified rHbhA; lane 2, purified trHbhA; lane 3, purified cHbhA; lane 4, purified MBP. Molecular mass markers are indicated on the left.
Adherence of HbhA-coated beads to J774.A1 cells.
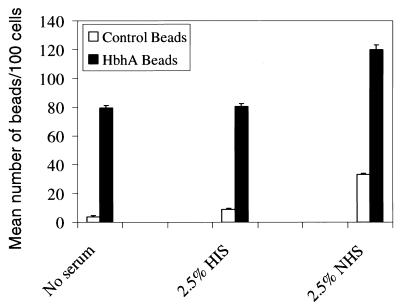
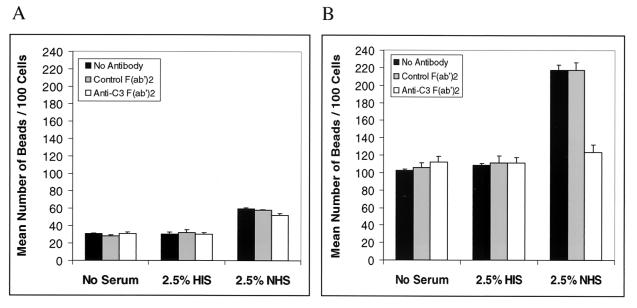
Polystyrene beads were coated with rHbhA to determine if HbhA could mediate the adherence of beads to macrophage-like cells in the presence of human serum. J774.A1 macrophage-like cells were incubated with either HbhA-coated beads or uncoated beads (at a 10:1 ratio of beads per cell) in either no serum, 2.5% HIS, or 2.5% NHS. The J774A.1 cells bound significantly more of the HbhA-coated beads compared to the uncoated beads under all three conditions (Fig. 5). The same experiment was performed comparing the adherence of BSA-coated beads to that of HbhA-coated beads. The J774.A1 cells bound 1.9-, 2.1- and 2.5-fold more HbhA-coated beads than BSA-coated beads in the absence of serum, in the presence of HIS, and in the presence of NHS, respectively (data not shown). The differences between the uncoated beads or BSA-coated beads and the HbhA-coated beads in binding to the J774.A1 cells was statistically significant under all conditions tested with P values of ≤0.003. The differences in binding between the HbhA-coated beads in the presence of HIS or NHS was also statistically significant with P values of ≤0.006. Although the presence of complement-sufficient serum (NHS) was not required for the HbhA-coated beads to bind to the J774.A1 cells, the presence of NHS did enhance the binding of HbhA-coated beads to the J774.A1 cells by 1.5-fold (Fig. 5), indicating a role for complement in the binding of HbhA to macrophage-like cells.
FIG. 5.
HbhA mediates adherence of latex beads to J774.A1 macrophage-like cells. Latex beads were coated with either rHbhA (black bars) or buffer only (white bars) and were incubated with J774.A1 cells with either no serum, HIS, or NHS. The mean number of beads per 100 cells ± SE (error bars) was determined by evaluating 100 cells in each of triplicate wells by light microscopy. This experiment was performed twice, and data shown are from one representative experiment.
To determine if human complement component C3 was responsible for the enhanced adherence of the HbhA-coated beads to J774.A1 cells in the presence of NHS, we performed the above experiment in the presence of normal goat F(ab′)2 as a control or goat anti-human C3 F(ab′)2. The F(ab′)2 form of these immunoglobulins was used to avoid possible confounding effects due to the complement activation or Fc receptor binding activities of the Fc portion of the antibodies. The control F(ab′)2 had no significant effect on the binding of uncoated or HbhA-coated beads to the J774.A1 cells (Fig. 6A and B). However, the anti-human C3 F(ab′)2 decreased the binding of HbhA-coated beads to the cells 43% in the presence of NHS compared to the control antibody (Fig. 6B). The anti-human C3 F(ab′)2 had no effect on the binding of HbhA-coated beads to the J774.A1 cells in the absence of serum or in the presence of HIS (Fig. 6B). The differences between the uncoated beads and the HbhA-coated beads in binding to the J774.A1 cells was statistically significant under all conditions tested with P values of ≤0.003. The differences in binding between the HbhA-coated beads in the presence of NHS and either the control F(ab′)2 or the anti-human C3 F(ab′)2 was also statistically significant (P = 0.004). These data indicate that C3 does play a role in the binding of HbhA-coated beads to macrophage-like cells in the presence of complement-sufficient human serum.
FIG. 6.
The presence of human C3 increases the adherence of HbhA-coated latex beads to J774.A1 macrophage-like cells. Latex beads were coated with either buffer only (A) or rHbhA (B) and were incubated with J774.A1 cells with either no serum, HIS, or NHS and either no antibody, goat F(ab′)2, or goat anti-human C3 F(ab′)2. The mean number of beads per 100 cells ± SE (error bars) was determined by evaluating 100 cells in each of triplicate wells by light microscopy. This experiment was performed twice, and data shown are from one representative experiment.
DISCUSSION
In this study, we have determined that the M. tuberculosis HbhA protein and a related protein in M. avium bind a derivative of human complement component C3 and that recombinant HbhA can mediate adherence of polystyrene beads to macrophage-like cells in both the presence and absence of human serum. HbhA was characterized previously in Mycobacterium bovis and M. tuberculosis and found to mediate attachment of these bacteria to epithelial cells (28). The nucleotide sequences of hbhA in M. bovis and M. tuberculosis are identical (27), and the gene is predicted to encode a 199-amino-acid protein. HbhA has a predicted molecular weight of 21.5 kDa and a predicted isoelectric point of 8.98, and the high isoelectric point would explain its behavior in cation- and anion-exchange columns. The predicted molecular mass is several kilodaltons smaller than the relative molecular mass of 28 to 30 kDa observed by us and by other groups (11, 27, 28). This difference in relative molecular mass is likely due to posttranslational modification and to the presence of a lysine-rich carboxy terminus. HbhA was previously shown by gas chromatography-mass spectrophotometry analysis to be a glycoprotein containing approximately 2.8% carbohydrate, consisting primarily of glucose with some xylose, mannose, arabinose, and galactose (27). Our result that the M. tuberculosis C3-binding protein binds concanavalin A is consistent with this finding.
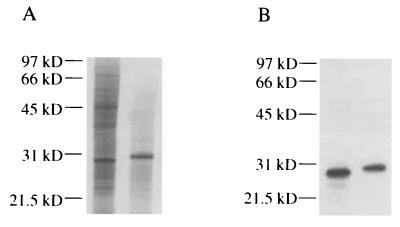
The genome of M. avium strain 104 is currently being sequenced by The Institute for Genomic Research, and the genome of M. leprae has been sequenced by the Sanger Center (8). A search of the available M. avium database (at www.tigr.org) revealed a HbhA homolog with 84% identity and 89% similarity to M. tuberculosis HbhA at the amino acid level. The M. avium HbhA homolog contains a 6-amino-acid insertion (QKAIAK) in the carboxy terminus (Fig. 7) that is not present in M. tuberculosis HbhA. The first set of lysine-alanine repeats (KKAAPA) in the carboxy terminus is not as well-conserved in M. avium HbhA as it is in M. tuberculosis HbhA: the first repeat of the set contains four changes, the second repeat of the set is interrupted by the 6-amino-acid insertion mentioned above, and the last repeat of the set is missing an alanine (Fig. 7). The second set of lysine-alanine repeats [KKA(A/P)A] in the carboxy terminus of M. avium HbhA is highly conserved (Fig. 7). M. avium HbhA has a predicted molecular mass of 22.2 kDa and a predicted isoelectric point of 9.54. We were able to demonstrate reactivity of a 31-kDa protein from an M. avium cell lysate to rabbit antiserum against recombinant M. tuberculosis HbhA (Fig. 8), which like M. tuberculosis HbhA, is larger than the predicted molecular mass for HbhA. Delogu and Brennan (11) purified a heparin-binding protein from M. avium that migrated at approximately 29 to 30-kDa compared to the 28 kDa of M. tuberculosis HbhA. A search of the Mycobacterium leprae database (at www.sanger.ac.uk) revealed an HbhA homolog with 81% identity and 87% similarity to M. tuberculosis HbhA at the amino acid level. The M. leprae HbhA homolog contains an 11-amino-acid deletion in the carboxy terminus (APAKKAAPAKK) compared to M. tuberculosis HbhA (Fig. 7). This deletion has largely disrupted the first set of lysine-alanine repeats (Fig. 7). The second set of lysine-alanine repeats [KKA(A/P)A] is highly conserved compared to M. tuberculosis or M. avium HbhA, with only a change of one lysine to an arginine in the first repeat (Fig. 7). To our knowledge HbhA has not been studied in M. leprae. Pethe et al. (34) attempted to purify an HbhA-like protein from M. smegmatis but were unable to identify such a protein in this species of Mycobacterium. Instead, this group identified a protein they named laminin-binding protein, which cross-reacts to a monoclonal antibody to HbhA but not to antiserum against a recombinant form of HbhA lacking the repeat region. We were able to demonstrate reactivity of a 33- to 34-kDa protein from an M. smegmatis cell lysate to rabbit antiserum against M. tuberculosis full-length HbhA (data not shown). Because of our data and those of Pethe et al. (34), we do not believe the M. smegmatis 30-kDa C3-binding protein to be an HbhA homolog.
FIG. 8.
Western blot analysis of a M. avium cell lysate demonstrating reactivity to antiserum to HbhA. (A) Coomassie-stained gel of M. tuberculosis cell lysate (left lane) and M. avium cell lysate (right lane). (B) Western blot of gel pattern identical to that shown in panel A reacted with antiserum to HbhA. Molecular mass markers in kilodaltons [kD] are indicated on the left.
Menozzi et al. (28) showed that M. tuberculosis HbhA is surface exposed using immunoelectron microscopy. One of the monoclonal antibodies (3921E4) used in those studies was mapped to the lysine-alanine repeat region of HbhA (33), indicating that the repeat region is surface exposed. They also provided evidence that HbhA is present in cell wall preparations of M. bovis and M. tuberculosis, and our cell fractionation and Triton X-114 studies further substantiate the association of HbhA with the cell membrane or the cell wall. Menozzi et al. (28) reported that HbhA was present in culture supernatants of M. bovis and M. tuberculosis, but we were unable to detect C3-binding activity or HbhA in M. tuberculosis culture supernatants (data not shown). HbhA does not contain a recognizable signal sequence for export out of the cytoplasm. The protein does, however, contain a hydrophobic region at the N terminus consisting of 22 amino acids (Fig. 7). This hydrophobic segment may insert into the cytoplasmic membrane and serve as an anchor for the protein. The remainder of the protein presumably extends externally toward the cell wall from the outer leaflet of the membrane, based on its surface exposure and hydrophilic nature. The computer programs PSORT and TMpred both predict HbhA to be a membrane protein anchored by this hydrophobic region. HbhA may become present in culture supernatants due to degradation of cells or due to HbhA possibly being present in membrane blebs or glycolipid droplets, but HbhA would not be expected to be actively secreted by M. tuberculosis. Computer secondary structure models predict HbhA to have a high alpha-helical content and a possible coiled-coil region just outside of the hydrophobic stretch at the N terminus. These data indicate that HbhA may have a linear rather than globular structure and thereby be accessible to antibody on the cell surface (28).
In our hands, HbhA fractionated with the detergent phase of Triton X-114 separations in the presence of protease inhibitors but in the aqueous phase in the absence of these inhibitors. We were unable to demonstrate any detectable change in SDS-PAGE migration properties in HbhA purified without the use of protease inhibitors. N-terminal amino acid sequencing of the aqueous phase version of this protein revealed that the N terminus was the same as that determined by Menozzi et al. (28). It is possible that proteolytic cleavage of the C-terminal region or modifications due to the protease inhibitors result in changes of Triton X-114 partitioning.
HbhA was able to mediate adherence and internalization of coated polystyrene beads to macrophage-like J774.A1 cells in both a C3-dependent and a C3-independent manner. The presence of NHS significantly increased the binding of HbhA-coated beads to J774.A1 cells compared to the binding in the absence of serum or in the presence of HIS. An F(ab′)2 antibody to human C3 was able to reduce the binding of HbhA-coated beads to J774.A1 cells in the presence of NHS to the level of binding seen in the absence of serum or in the presence of HIS. HbhA has a positively charged carboxy terminus, which may allow the protein to bind to negatively charged molecules on the surface of the macrophage-like cells in the absence of serum. HbhA has been shown to bind to several sulfated glycoconjugates, such as heparin, dextran sulfate, fucoidan, and chondroitin sulfate, and to decorin (11, 27, 28, 33). HbhA was found not to bind to fibronectin, BSA, ovalbumin, dextran, mannose, or galactose (11, 28). The increase in the adherence of the uncoated control beads to the J774.A1 cells was proportionately greater than that of the HbhA-coated beads in the presence of NHS (Fig. 5). This increase in adherence of the control beads in the presence of NHS was found to be C3-independent, unlike the HbhA-coated beads, because the anti-human C3 F(ab′)2 did not decrease the binding of the control beads to the J774.A1 cells in the presence of NHS (Fig. 6A).
It is important to note that the C3 ligand affinity blot protocol used in this study to identify HbhA as a C3-binding protein would not identify all Mycobacterium C3-binding molecules. For instance, glycolipids, which are abundant on the surface of mycobacteria, would not electrophorese in SDS-PAGE gels, and proteins that require a particular conformation for binding to C3 would not be detected due to denaturation by SDS. Using a protocol similar to an enzyme-linked immunosorbent assay, we have also previously demonstrated the ability of purified trehalose 6,6′-dimycolate and trehalose monomycolate from M. tuberculosis to bind C3 from NHS but not from HIS (S. L. Mueller, S. J. Norris, and A. R. Wanger, Abstr. 98th Gen. Meet. Am. Soc. Microbiol., abstr. U-98, p. 511–512, 1998). It is possible that M. tuberculosis and M. avium have more than one molecule on their surface that is able to bind C3 and mediate attachment of the bacteria to mononuclear cells and that HbhA is only one such molecule. We have recently identified a second C3-binding protein in M. tuberculosis and are in the process of characterizing this protein further (data not shown).
M. tuberculosis can bind to complement receptors on monocytes and macrophages by both C3-dependent and (to a lesser extent) C3-independent mechanisms (10, 20, 41, 42, 44, 47). Macrophages themselves produce and secrete complement proteins, so evidence that M. tuberculosis can bind to CR3 and CR4 in the absence of complement components came when CR3 and CR4 were individually transfected into CHO cells (9, 47). These transfected cells were shown to bind M. tuberculosis at a low level in the absence of serum. The molecule(s) on the surface of M. tuberculosis that binds directly to CR3 and CR4 is unknown but may contain a carbohydrate moiety, since these receptors have both C3bi and carbohydrate recognition sites (2, 24). Binding of M. tuberculosis to monocytes and macrophages is greatly enhanced in the presence of serum (20, 41, 42). This enhanced binding and uptake of M. tuberculosis is decreased in the presence of HIS (20, 42). Schlesinger et al. (42) found that destruction of serum complement activity through heat inactivation results in a 75% reduction in M. tuberculosis adherence to monocytes compared to adherence in the presence of fresh serum. Similarly, Hirsch et al. (20) found that heat inactivation of serum results in a 59 and 79% reduction in internalization by alveolar macrophages and blood monocytes, respectively, compared to internalization in the presence of fresh serum. Complement component C3 was identified as the major component in human serum involved in enhancing the adherence and uptake of M. tuberculosis by mononuclear phagocytes, in that the adherence of pre-opsonized M. tuberculosis to monocytes was reduced 71% in the presence of Fab fragments of anti-human C3 IgG. Therefore, C3 plays a major role in the binding and uptake of M. tuberculosis by mononuclear phagocytes (42).
Hu et al. (21) and Melo et al. (26) recently used CR3-deficient mice to examine the role of CR3 in M. tuberculosis infection. These groups found that macrophages from CR3-deficient mice phagocytized significantly less M. tuberculosis cells in the presence (21, 26) and absence (26) of serum than macrophages from wild-type mice. However, they found no difference in infection or disease outcome in CR3-deficient mice infected with M. tuberculosis compared to wild-type mice. Similar numbers of bacteria were found in the livers, spleens, and lungs of infected mice (21, 26).
M. avium can bind to CR1 and CR3 (6, 7, 37), and adherence and phagocytosis of M. avium by MDMs and monocytes is significantly enhanced in the presence of complement-sufficient human serum (7, 45). This enhanced adherence and phagocytosis was reduced 47% in the presence of HIS as compared to fresh serum (45). Preincubation of serum with antibody against C3 resulted in up to a 93% decrease in the binding of M. avium to MDMs (7), demonstrating the importance of C3 in the phagocytosis of M. avium by MDMs. Bermudez et al. (5) reported that CD18-deficient mice (lacking expression of CR3 and CR4) infected with M. avium have equal numbers of bacteria in the livers and spleens compared to wild-type mice.
HbhA may play a multifunctional role in promoting entry of Mycobacterium into host cells. HbhA has been shown by others to bind heparin and other sulfated sugars (11, 27, 28, 33), which may in turn mediate adherence to host cell and extracellular matrix components. HbhA mediates attachment of M. tuberculosis to Chinese hamster ovary (CHO) cells (28), and purified rHbhA binds to human A549 pneumocytes (33). We hypothesize that HbhA binds C3 in vivo, which then facilitates binding of the bacterium to complement receptors and hence phagocytosis by mononuclear phagocytes. The lysine-alanine repeats of HbhA have been shown to be responsible for heparin-binding (33), and our data indicates that this region is at least partially responsible for binding to C3 as well (Fig. 4B). It is possible that the sugars on native HbhA also participate in the binding of the native protein to C3, but since recombinant full-length HbhA and the repeat region of HbhA bind C3, the sugars are not necessary for binding to C3. It would be interesting to compare the C3-binding activity of a deglycosylated form of native HbhA to the fully glycosylated form. Isolation of mutants of M. tuberculosis deficient in HbhA expression should help to delineate the role of this protein in the internalization of mycobacteria by host cells and thus in pathogenesis.
ACKNOWLEDGMENTS
We thank R. T. Owens for advice and materials for the purification of native HbhA, C. Jagganath for providing the M. avium strain, W. R. Jacobs for providing the M. smegmatis strain, F. D. Menozzi for providing plasmid pMAL+3R1+2R2, J. K. Actor for providing the TiterMax adjuvant, M. J. Brennan for providing monoclonal antibody 3921E4, J. K. Howell for assistance in production of rabbit antiserum to HbhA, and M. R. Olsen for assistance in culturing the J774.A1 cells. We also thank J. M. Hardham and L. Y. Armitige for helpful discussions and suggestions.
This work was supported in part by the Gilson Longenbaugh Foundation of Houston, Texas.
REFERENCES
- 1.Armitige L Y, Jagannath C, Wanger A R, Norris S J. Disruption of the genes encoding antigen 85A and antigen 85B of Mycobacterium tuberculosis H37Rv: effect on growth in culture and in macrophages. Infect Immun. 2000;68:767–778. doi: 10.1128/iai.68.2.767-778.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaout M A. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75:1037–1050. [PubMed] [Google Scholar]
- 3.Bellinger-Kawahara C, Horwitz M A. Complement component C3 fixes selectively to the major outer membrane protein (MOMP) of Legionella pneumophila and mediates phagocytosis of liposome-MOMP complexes by human monocytes. J Exp Med. 1990;172:1201–1210. doi: 10.1084/jem.172.4.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett B, Check I J, Olsen M R, Hunter R L. A comparison of commercially available adjuvants for use in research. J Immunol Methods. 1992;153:31–40. doi: 10.1016/0022-1759(92)90302-a. [DOI] [PubMed] [Google Scholar]
- 5.Bermudez L E, Goodman J, Petrofsky M. Role of complement receptors in uptake of Mycobacterium avium by macrophages in vivo: evidence from studies using CD18-deficient mice. Infect Immun. 1999;67:4912–4916. doi: 10.1128/iai.67.9.4912-4916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bermudez L E, Parker A, Goodman J R. Growth within macrophages increases the efficiency of Mycobacterium avium in invading other macrophages by a complement receptor- independent pathway. Infect Immun. 1997;65:1916–1925. doi: 10.1128/iai.65.5.1916-1925.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bermudez L E, Young L S, Enkel H. Interaction of Mycobacterium avium complex with human macrophages: roles of membrane receptors and serum proteins. Infect Immun. 1991;59:1697–1702. doi: 10.1128/iai.59.5.1697-1702.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole S T, Eiglmeier K, Parkhill J, James K D, Thomson N R, Wheeler P R, Honore N, Garnier T, Churcher C, Harris D, Mungall K, Basham D, Brown D, Chillingworth T, Connor R, Davies R M, Devlin K, Duthoy S, Feltwell T, Fraser A, Hamlin N, Holroyd S, Hornsby T, Jagels K, Lacroix C, Maclean J, Moule S, Murphy L, Oliver K, Quail M A, Rajandream M A, Rutherford K M, Rutter S, Seeger K, Simon S, Simmonds M, Skelton J, Squares R, Squares S, Stevens K, Taylor K, Whitehead S, Woodward J R, Barrell B G. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 9.Cywes C, Godenir N L, Hoppe H C, Scholle R R, Steyn L M, Kirsch R E, Ehlers M R. Nonopsonic binding of Mycobacterium tuberculosis to human complement receptor type 3 expressed in Chinese hamster ovary cells. Infect Immun. 1996;64:5373–5383. doi: 10.1128/iai.64.12.5373-5383.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cywes C, Hoppe H C, Daffe M, Ehlers M R. Nonopsonic binding of Mycobacterium tuberculosis to complement receptor type 3 is mediated by capsular polysaccharides and is strain dependent. Infect Immun. 1997;65:4258–4266. doi: 10.1128/iai.65.10.4258-4266.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delogu G, Brennan M J. Functional domains present in the mycobacterial hemagglutinin, HBHA. J Bacteriol. 1999;181:7464–7469. doi: 10.1128/jb.181.24.7464-7469.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobos K M, Khoo K H, Swiderek K M, Brennan P J, Belisle J T. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. J Bacteriol. 1996;178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobos K M, Swiderek K, Khoo K H, Brennan P J, Belisle J T. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect Immun. 1995;63:2846–2853. doi: 10.1128/iai.63.8.2846-2853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J D. Macrophage receptors for Mycobacterium tuberculosis. Infect Immun. 1998;66:1277–1281. doi: 10.1128/iai.66.4.1277-1281.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fifis T, Costopoulos C, Radford A J, Bacic A, Wood P R. Purification and characterization of major antigens from a Mycobacterium bovis culture filtrate. Infect Immun. 1991;59:800–807. doi: 10.1128/iai.59.3.800-807.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbe T, Harris D, Vordermeier M, Lathigra R, Ivanyi J, Young D. Expression of the Mycobacterium tuberculosis 19-kilodalton antigen in Mycobacterium smegmatis: immunological analysis and evidence of glycosylation. Infect Immun. 1993;61:260–267. doi: 10.1128/iai.61.1.260-267.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall R T, Strugnell T, Wu X, Devine D V, Stiver H G. Characterization of kinetics and target proteins for binding of human complement component C3 to the surface-exposed outer membrane of Chlamydia trachomatis serovar L2. Infect Immun. 1993;61:1829–1834. doi: 10.1128/iai.61.5.1829-1834.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herrmann J L, P O G, Gallagher A, Thole J E, Young D B. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 1996;15:3547–3554. [PMC free article] [PubMed] [Google Scholar]
- 19.Hewinson R G, Michell S L, Russell W P, McAdam R A, Jacobs W J. Molecular characterization of MPT83: a seroreactive antigen of Mycobacterium tuberculosis with homology to MPT70. Scand J Immunol. 1996;43:490–499. doi: 10.1046/j.1365-3083.1996.d01-78.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirsch C S, Ellner J J, Russell D G, Rich E A. Complement receptor-mediated uptake and tumor necrosis factor-alpha-mediated growth inhibition of Mycobacterium tuberculosis by human alveolar macrophages. J Immunol. 1994;152:743–753. [PubMed] [Google Scholar]
- 21.Hu C, Mayadas-Norton T, Tanaka K, Chan J, Salgame P. Mycobacterium tuberculosis infection in complement receptor 3-deficient mice. J Immunol. 2000;165:2596–2602. doi: 10.4049/jimmunol.165.5.2596. [DOI] [PubMed] [Google Scholar]
- 22.Joiner K, Hieny S, Kirchhoff L V, Sher A. gp72, the 72 kilodalton glycoprotein, is the membrane acceptor site for C3 on Trypanosoma cruzi epimastigotes. J Exp Med. 1985;161:1196–1212. doi: 10.1084/jem.161.5.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita T, Seya T. Complement regulatory proteins on nucleated cells. In: Erdei A, editor. Medical intelligence unit: new aspects of complement structure and function. R. G. Austin, Tex: Landes Company; 1994. p. 36. [Google Scholar]
- 24.Kishimoto T K, Larson R S, Corbi A L, Dustin M L, Staunton D E, Springer T A. The leukocyte integrins. Adv Immunol. 1989;46:149–182. doi: 10.1016/s0065-2776(08)60653-7. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Melo M D, Catchpole I R, Haggar G, Stokes R W. Utilization of CD11b knockout mice to characterize the role of complement receptor 3 (CR3, CD11b/CD18) in the growth of Mycobacterium tuberculosis in macrophages. Cell Immunol. 2000;205:13–23. doi: 10.1006/cimm.2000.1710. [DOI] [PubMed] [Google Scholar]
- 27.Menozzi F D, Bischoff R, Fort E, Brennan M J, Locht C. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc Natl Acad Sci USA. 1998;95:12625–12630. doi: 10.1073/pnas.95.21.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menozzi F D, Rouse J H, Alavi M, Laude-Sharp M, Muller J, Bischoff R, Brennan M J, Locht C. Identification of a heparin-binding hemagglutinin present in mycobacteria. J Exp Med. 1996;184:993–1001. doi: 10.1084/jem.184.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman M J, Actor J K, Balusubramanian M, Jagannath C. Use of nonionic block copolymers in vaccines and therapeutics. Crit Rev Ther Drug Carrier Syst. 1998;15:89–142. doi: 10.1615/critrevtherdrugcarriersyst.v15.i2.10. [DOI] [PubMed] [Google Scholar]
- 30.Nikaido H, Kim S H, Rosenberg E Y. Physical organization of lipids in the cell wall of Mycobacterium chelonae. Mol Microbiol. 1993;8:1025–1030. doi: 10.1111/j.1365-2958.1993.tb01647.x. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson B, Ekdahl K N. Components of the alternative pathway. In: Rother K, Till G O, Hansch G M, editors. The complement system. Heidelberg, Germany: Springer-Verlag; 1998. pp. 23–49. [Google Scholar]
- 32.O'Farrell P Z, Goodman H M, O'Farrell P H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1142. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 33.Pethe K, Aumercier M, Fort E, Gatot C, Locht C, Menozzi F D. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J Biol Chem. 2000;275:14273–14280. doi: 10.1074/jbc.275.19.14273. [DOI] [PubMed] [Google Scholar]
- 34.Pethe K, Puech V, Daffe M, Josenhans C, Drobecq H, Locht C, Menozzi F D. Mycobacterium smegmatis laminin-binding glycoprotein shares epitopes with Mycobacterium tuberculosis heparin-binding haemagglutinin. Mol Microbiol. 2001;39:89–99. doi: 10.1046/j.1365-2958.2001.02206.x. [DOI] [PubMed] [Google Scholar]
- 35.Puentes S M, Sacks D L, da Silva R P, Joiner K A. Complement binding by two developmental stages of Leishmania major promastigotes varying in expression of a surface lipophosphoglycan. J Exp Med. 1988;167:887–902. doi: 10.1084/jem.167.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radolf J D, Chamberlain N R, Clausell A, Norgard M V. Identification and localization of integral membrane proteins of virulent Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent triton X-114. Infect Immun. 1988;56:490–498. doi: 10.1128/iai.56.2.490-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roecklein J A, Swartz R P, Yeager H., Jr Nonopsonic uptake of Mycobacterium avium complex by human monocytes and alveolar macrophages. J Lab Clin Med. 1992;119:772–781. [PubMed] [Google Scholar]
- 38.Rouse D A, Morris S L, Karpas A B, Mackall J C, Probst P G, Chaparas S D. Immunological characterization of recombinant antigens isolated from a Mycobacterium avium lambda gt11 expression library by using monoclonal antibody probes. Infect Immun. 1991;59:2595–2600. doi: 10.1128/iai.59.8.2595-2600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell D G. The macrophage-attachment glycoprotein gp63 is the predominant C3-acceptor site on Leishmania mexicana promastigotes. Eur J Biochem. 1987;164:213–221. doi: 10.1111/j.1432-1033.1987.tb11013.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schlesinger L S. Macrophage phagocytosis of virulent but not attenuated strains of Mycobacterium tuberculosis is mediated by mannose receptors in addition to complement receptors. J Immunol. 1993;150:2920–2930. [PubMed] [Google Scholar]
- 42.Schlesinger L S, Bellinger-Kawahara C G, Payne N R, Horwitz M A. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J Immunol. 1990;144:2771–2780. [PubMed] [Google Scholar]
- 43.Schlesinger L S, Horwitz M A. Phenolic glycolipid-1 of Mycobacterium leprae binds complement component C3 in serum and mediates phagocytosis by human monocytes. J Exp Med. 1991;174:1031–1038. doi: 10.1084/jem.174.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stokes R W, Thorson L M, Speert D P. Nonopsonic and opsonic association of Mycobacterium tuberculosis with resident alveolar macrophages is inefficient. J Immunol. 1998;160:5514–5521. [PubMed] [Google Scholar]
- 45.Swartz R P, Naai D, Vogel C W, Yeager H., Jr Differences in uptake of mycobacteria by human monocytes: a role for complement. Infect Immun. 1988;56:2223–2227. doi: 10.1128/iai.56.9.2223-2227.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaffran Y, Zhang L, Ellner J J. Role of CR4 in Mycobacterium tuberculosis-human macrophages binding and signal transduction in the absence of serum. Infect Immun. 1998;66:4541–4544. doi: 10.1128/iai.66.9.4541-4544.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]