Summary
Stigma exsertion rate (SER) of the male sterile line is a key limiting factor for hybrid seed production in rice. Although a large number of quantitative trait loci associated with SER have been reported, few genes have been molecularly cloned and functionally characterized, severely hindering the genetic improvement of SER of the male sterile line and the breeding efficiency of hybrid rice. In this study, we identified three grain shape regulatory genes, GS3, GW8 and GS9, as potential candidate genes for targeted manipulation of grain shape and SER. We show that simultaneously knocking out these three genes could effectively increase SER by increasing the ratio of spikelet length/spikelet width and length of stigma and style, without negative impacts on other agronomic traits. Cellular examination and transcriptomic analyses revealed a role of these genes in coordinated regulation of transverse and longitudinal cell division in the pistils. Moreover, we demonstrate that targeted manipulation of these grain shape genes could significantly improve the outcrossing rate in both the ZH11 (a japonica variety) and Zhu6S (an indica male sterile line) backgrounds. Our results provide new insights into the mechanisms of rice SER regulation and develop an effective strategy to improve SER and out‐crossing rate in rice, thus facilitating hybrid rice production.
Keywords: hybrid rice, stigma exsertion rate, GS3, GW8, GS9
Introduction
Rice is a staple crop for more than half of the world's population (FAO Statistical Databases, 2021). Asian cultivated rice (Oryza sativa L.) is domesticated from the wild rice (Oryza rufipogon Griff.) and is differentiated into two major ecotypes, japonica and indica (Huang et al., 2012). Since the 1970s, the development of “three‐line” and “two‐line” hybrid systems has contributed significantly to increased rice production and securing global food supply (Peng et al., 1999; Yuan and Virmani, 1988; Zhu, 2016). During commercial production of hybrid seeds, plants of the female parent (usually a male sterile line) are planted in rows side‐by‐side with plants of the male parent (normally 5–6 rows of female parent:1 row of male parent), and pollination is aided by humans or mechanicals (such as unmanned‐helicopters). To be commercially profitable, hybrid seed yield should reach no less than 150 kg/mu, and higher yield is desirable for reducing the cost of hybrid seeds for selling to the farmers.
Several characteristics of the female and male parental lines affect the yield of hybrid seed production, including the interval of anthesis between the male and female parents, the stigma exsertion rate (SER) and stigma vigour of the female parental lines, and pollen number/vigour of the male parental lines (Marathi and Jena, 2015; Virmani et al., 1982). Among them, SER of the female parental line is believed to be a crucial one. The rice SER is defined as the frequency of exserted stigmas which stay outside after closing of the lemma and palea, and could be gauged by the single stigma exsertion rate (SSE), the dual stigma exsertion rate (DSE) and the total stigma exsertion rate (TSE) (Rahman et al., 2017b). As the exserted stigmas could stay viable for 4–6 days to be pollinated, the male sterile lines with high SER would extend the cross‐pollination opportunity by trapping more pollens and overcome non‐synchronous anthesis between the two parental lines, thus elevating the outcrossing ability of the male sterility lines and promoting hybrid rice seed production (Kato and Namai, 1987; Lou et al., 2014; Tian, 1993; Wen et al., 2009; Yan, 1999; Yuan, 2002). Thus, higher SER (normally above 40–50% TSE) is a preferred trait for the development of a commercially viable male sterile line for hybrid seed production. However, the average SER is <25% in indica, <15% in tropical japonica, and about 5% in temperate japonica (Ling and Xu, 1988; Uga et al., 2003; Ying and Zhang, 1989; Zhou et al., 2017). Thus, it is often a laborious and time‐consuming process to develop male sterile lines with high SER, which severely limits the breeding efficiency of hybrid rice.
The rice SER is a complex quantitative trait and is easily affected by environmental factors (such as temperature) (Yan et al., 2009). Previous studies have shown that SER is largely determined by stigma length (STL), style length (SYL), and the sum of stigma and style length (TSSL) (Dang et al., 2020), and thus extensive genetic analyses have been performed to identify SER‐associated loci through phenotyping STL, SYL and/or TSSL. Although more than 40 quantitative trait loci (QTL) for SER have been reported, none of them has been molecularly cloned and functionally characterized, largely due to the difficulty in precise phenotyping and the small additive effects of the individual locus (typically less than 10%) (Bakti and Tanaka, 2019; Li et al., 2001, 2014; Liu et al., 2015, 2019; Marathi and Jena, 2015; Rahman et al., 2017a,b; Tan et al., 2020; Uga et al., 2003; Zhang et al., 2018). Recently, genome‐wide association studies (GWAS) analyses were utilized to identify SER‐associated genes. Notably, three rice grain size regulatory genes, Grain Size 3 (GS3, encodes a G‐protein γ subunit, Mao et al., 2010), Grain Width 2 (GW2, encodes a RING‐type E3 ubiquitin ligase, Song et al., 2007) and Grain Width 5 (GW5, encodes a novel calmodulin‐binding protein, Liu et al., 2017), were identified (Dang et al., 2020; Zhou et al., 2017). These observations suggest that SER is tightly linked with grain shape. However, the detailed roles of these genes in regulating SER and whether these genes could be utilized to improve SER of the male parent line for hybrid seed production have not been meticulously evaluated.
In this study, we selected three grain shape regulatory genes, GS3, GW8 and GS9, as potential candidate genes for targeted manipulation of grain shape and SER. We show that these genes could act synchronously to modulate the spikelet length, spikelet width, stigma length, and style length. Their individual or double knockout plants exhibit improved SER, but the gs3/gw8/gs9 triple mutant exhibit the most dramatic improvement in SER (TSE reaching ~60%). Moreover, we show that simultaneously knocking out these three genes could effectively improve the outcrossing rate in both indica and japonica backgrounds. The potential utility of these genes in boosting hybrid seed production, especially in indica–japonica hybrid seed production, is discussed.
Results
Identification of SER regulatory genes by analysing rice grain shape genes
Previous studies have shown that in rice, SER is largely determined by stigma length (STL), style length (SYL), and the sum of stigma and style length (TSSL) (Dang et al., 2020) and tightly associated with grain shape (Uga et al., 2003, 2010; Zhou et al., 2017). Thus, we speculated that these traits might be regulated by a common set of genes. To substantiate this notion, we examined the developmental processes of the spikelet and pistil (containing style and stigma) of ZH11 throughout the S8b ~ S12 stages (Zhang et al., 2006, 2011). We found that the spikelet and pistil exhibited synchronous growth, with a sharp increase from S11a to S12 (Figure 1a,b). Pearson's correlation analysis showed that the sum of stigma and style length was significantly and positively correlated with the spikelet area, spikelet length and width (Figure 1b; Figure S1). We then performed a detail analysis of more than 50 rice grain size regulatory genes according to their functions and their genetic effects on grain shape‐related traits (Li et al, 2019) (Table S1). Three genes, GS3, GW8 and GS9, were selected as they are negative regulators of grain length and/or positive regulators of grain width, and their loss‐of‐function alleles could increase grain quality and confer no adverse effect on other agronomic traits in rice (Fan et al., 2006; Wang et al., 2012; Zhao et al., 2018). Reverse‐ transcription quantitative PCR (RT‐qPCR) assay showed that GS3, GW8 and GS9 displayed similar expression patterns in the developing pistils, with highest expression at the S11b stage (Figure 1c), coinciding with the maximum growth rate of the spikelet and pistil at this stage. Therefore, we speculated that knocking out these genes may confer increased grain length/width ratio, increased pistil length and consequently increased SER in rice.
Figure 1.
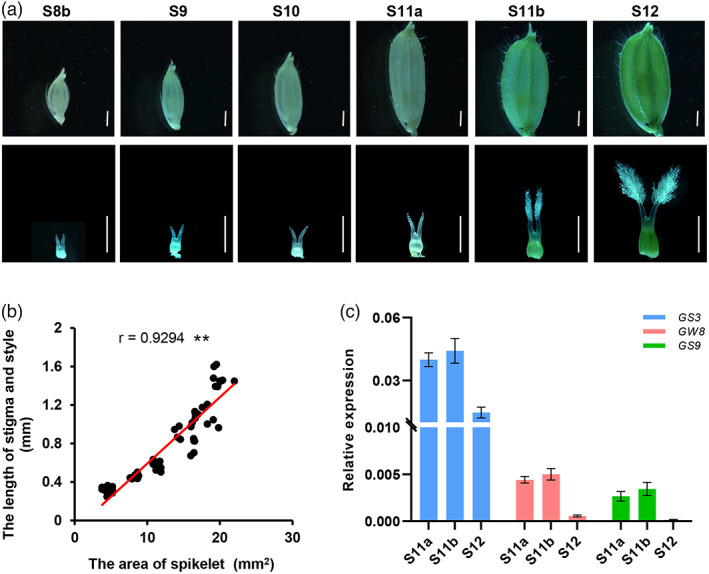
Identification of GS3, GW8 and GS9 as stigma exsertion regulatory genes in rice. (a) Dynamic change of spikelet and pistil for the ZH11 during spikelet development. S8b‐S12 stages represent different developmental stages based on the reference by Zhang et al. (2006) and Zhang et al. (2011). Bars = 1 mm. (b) Pearson's correlation between the spikelet area and the total length of stigma and style. r, Pearson's correlation coefficient. **P < 0.01. (c) RT‐qPCR analysis of GS3, GW8 and GS9 in pistils at S11a, S11b and S12 stages. Data are shown as means ± SEM (n = 3).
GS3 , GW8 and GS9 synchronously regulate glume shape, pistil growth and the SER in rice
We thus designed a CRISPR/Cas9 construct aimed to generate various single, double and triple knockout mutants of GS3, GW8 and GS9 in the japonica variety Zhonghua11 (ZH11) background (Figure S2). Multiple single, double and triple mutants harbouring base insertions or deletions were identified through sequencing analysis (Figure S2b–d). Next, we examined the phenotype of spikelet and pistil in various knockout mutants. Compared with the wild‐type (WT) plant ZH11, all the homozygous mutants exhibited increased spikelet length (except for the gw8 single mutant) but decreased glume width (except for the gs3 single mutant) (Figure 2a–h,q,r; Figure S3a–f,m,n). Notably, the spikelet length/width ratio was gradually elevated from the single mutant to the triple mutant, suggesting that GS3, GW8 and GS9 function additively in regulating grain shape in rice (Figure 2s; Figure S3o). In line with the increased spikelet length/width ratio, all the knockout mutants displayed increased stigma length and style length (except for the style length of the gw8/gs9 double mutant), thus leading to a gradual increase of the total stigma and style length from the single mutant to the triple mutant (Figure 2i–p,t–v; Figure S3g–l,p–r). Consistently, Pearson's correlation analysis showed that the stigma exsertion rate was positively correlated with spikelet length/width ratio and spikelet length, but negatively correlated with spikelet width (Figure S4). As expected, phenotype analysis showed that at anthesis, all the mutated lines exhibited increased SSE, DSE (except for the gs3 and gs9 single mutant) and TSE (Figure 2w–y; Figure S3s–u). In comparison to ZH11 with a very low TSE (<5%), the average TSE was increased by 54.92% in gs3/gw8/gs9#1 and 64.17% in gs3/gw8/gs9#2 (Figure 2w–y; Figure S3s–u). Noteworthy, the triple mutant produced more slender grains than ZH11with decreased 1000‐grain weight (Figure S5a–d), whereas the number of tillers and grains per plants in triple mutant were significantly increased compared to wild‐type ZH11 (Figure S5k,l). All the improved traits are beneficial for the hybrid seed production. No significant differences were observed for the panicle architecture (including panicle length, number of primary branches and number of secondary branches) and plant height between the triple mutants and wild‐type ZH11 (Figure 5e–j). Together, these findings clearly demonstrate that knocking out these grain shape genes could confer increased SER in rice.
Figure 2.
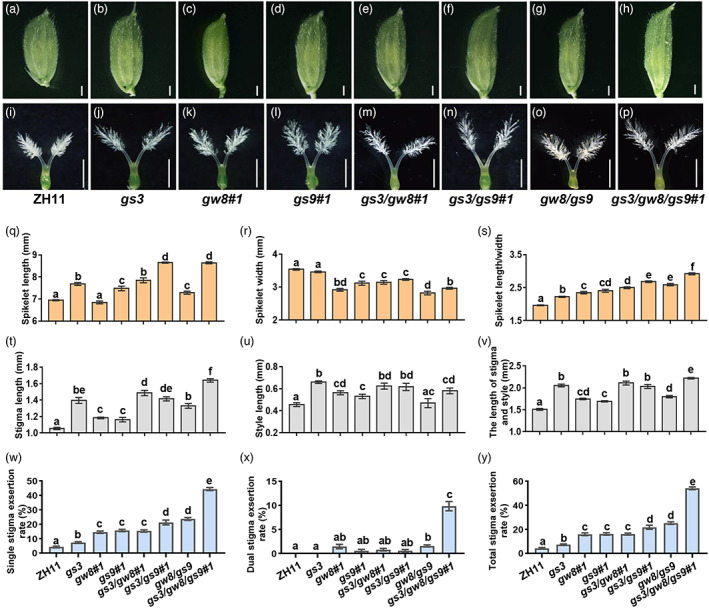
GS3, GW8 and GS9 synchronously regulate the spikelet, pistil growth and stigma exsertion rate in rice. (a–h) Comparison of spikelet shape among ZH11 and its various knockout combinations of gs3, gw8 and gs9. Bar = 1 mm. (i–p) Comparison of pistil shape among ZH11 and its various knockout combinations of gs3, gw8 and gs9. Bar = 1 mm. (q–v) Statistical analyses of spikelet length (q), spikelet width (r), spikelet length/spikelet width (s), stigma length (t), style length (u) the length of stigma and style (v), single stigma exsertion (w), dual stigma exsertion (x), total stigma exsertion (y) of ZH11 and its various knockout mutants. Letters above the bars indicate significant differences (P < 0.05) as determined by one‐way ANOVA with Tukey's post‐hoc analysis. Data are shown as means ± SEM (n = 10).
Figure 5.
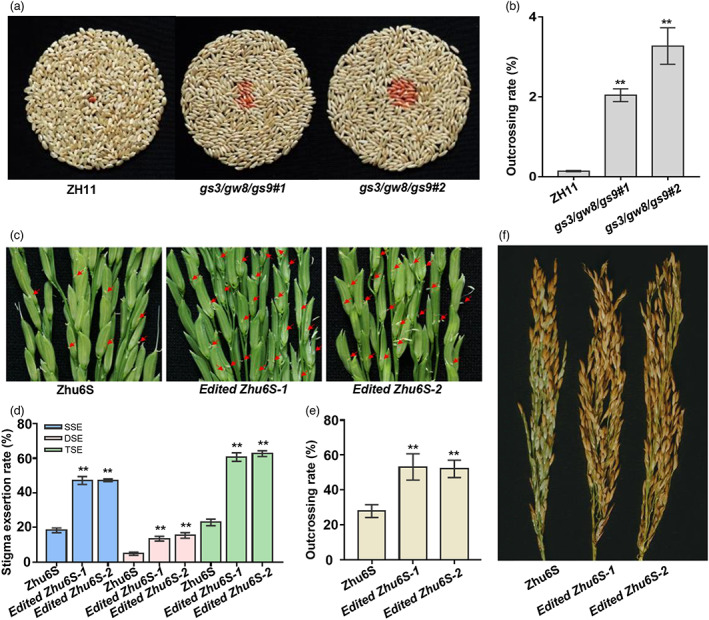
Knocking out GS3, GW8 and GS9 significantly increase outcrossing rates of ZH11 and Zhu6S. (a) The F1 seeds from the crossings of ZH11 or gs3/gw8/gs9 and the orange‐red‐grained astaxanthin rice. The orange‐red grains represent outcrossing seeds. (b) Measurement of the outcrossing rate of ZH11 and gs3/gw8/gs9. Bars indicate SEM (n = 4). (c) The stigma exsertion phenotype of Zhu6S and two edited lines (with GW8 and GS9 knockout). The red arrows indicate the exserted stigmas. (d) Statistical analysis of stigma exsertion rate shown in (c). Bars indicate SEM (n = 10). (e) Measurement of the outcrossing rate of Zhu6S and two edited lines after natural cross‐pollination with Tianfeng B. Bars indicate SEM (n = 3). (f) Panicle morphologies of Zhu6S and two edited lines after natural cross‐pollination with Tianfeng B. Asterisks indicate significant differences (**P < 0.01) from ZH11 or Zhu6S, as determined by two‐tailed Student's t‐test. Data are shown as means ± SEM.
GS3 , GW8 and GS9 regulate grain and pistil traits by altering cell division
To further examine the cellular changes that occurred in the spikelet and pistil of the gs3/gw8/gs9 triple mutant, we performed cytological observations in more detail. Scanning electron microscopy (SEM) observations showed that the length of the outer epidermal cells of spikelet glumes in gs3/gw8/gs9#1 was decreased by ~10% (Figure 3a–c), but an approximate 13% increase in longitudinal cell number compared with the wild‐type ZH11 (Figure 3d), indicating that the increased grain length in gs3/gw8/gs9#1 is likely resulted from increased cell division in the longitudinal direction. Additionally, transverse sections of palea and lemma showed that gs3/gw8/gs9#1 contained fewer inner parenchyma cells than the wild‐type ZH11, which may contribute to the decreased transverse spikelet perimeter (by 19%) of the gs3/gw8/gs9#1 hulls (Figure 3e–h).
Figure 3.
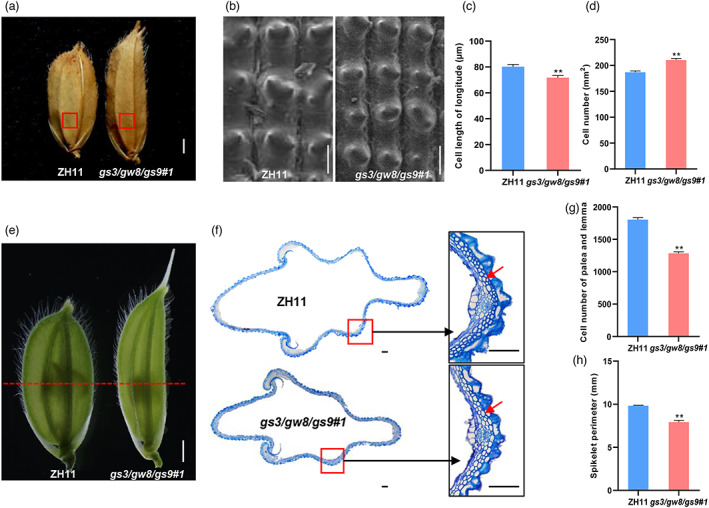
Histological comparison of the spikelet glumes between ZH11 and the gs3/gw8/gs9#1 triple mutant. (a, b) Scanning electron microscopy analyses of the glume of ZH11 and gs3/gw8/gs9#1. (b) is magnified views (enlarged by 200‐fold) of the red boxes areas in (a). (c) The cell length in longitude indicated with red boxes in (a). Bars indicate SEM (n = 5). (d) The cell number in the areas indicated with red boxes in (a). Bars indicate SEM (n = 5). (e) Spikelet morphology of ZH11 and gs3/gw8/gs9#1 before anthesis. The yellow dashed line indicates the position of the cross‐sections shown in (f). Bar = 1 cm. (f) Transverse sections of spikelet. The right‐hand images show magnified views of the red boxed region. The red arrow indicates lower epidermal cells. Bar = 100 μm. (g, h) Quantification analyses of cell number (g) and spikelet perimeter (h) of palea and lemma in ZH11 and gs3/gw8/gs9#1, Bars indicate SEM (n = 3). Asterisks indicate significant differences (**P < 0.01) between ZH11 and gs3/gw8/gs9#1 by two‐tailed Student's t‐test. Data are shown as means ± SEM.
Next, we compared the cellular changes within pistil between gs3/gw8/gs9#1 and wild‐type ZH11. Transverse section analysis of the joint region between stigma and style showed that the number of inner parenchyma cells in gs3/gw8/gs9#1 was significantly decreased compared with ZH11 (Figure 4a,b,e), thus leading to an approximate 22% decrease in transverse area of the joint region in gs3/gw8/gs9#1 (Figure 4f). Additionally, transmission electron microscopy (TEM) analysis showed no significant difference in the average cell length and longitudinal cell number of the style between gs3/gw8/gs9#1 and ZH11 (Figure 4c,d,g,h), implying that the increased style length in gs3/gw8/gs9#1 is also likely resulted from increased cell division in the longitudinal direction. These results suggest that GS3, GW8 and GS9 act together to inhibit cell division in the longitudinal direction and promote cell division in the transverse direction of style in rice.
Figure 4.
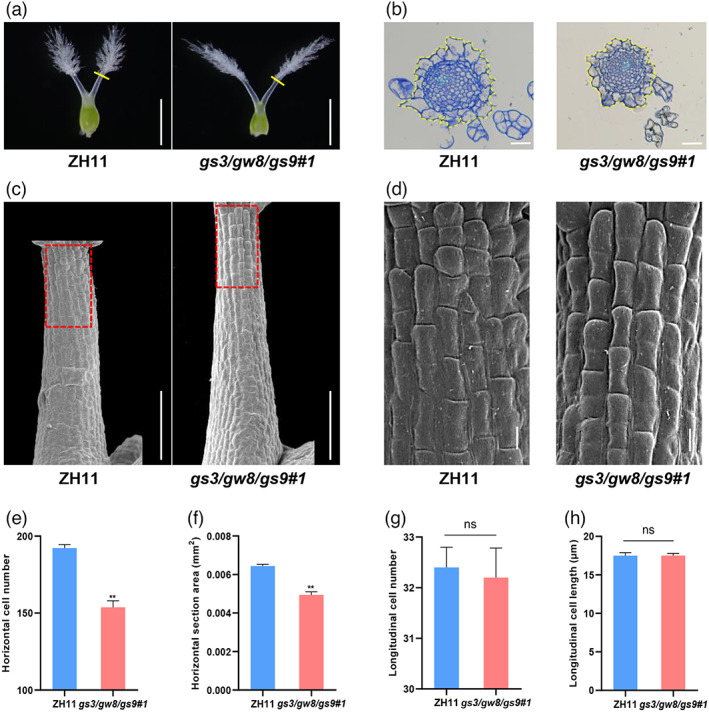
Histological comparison of the styles between ZH11 and gs3/gw8/gs9#1. (a) Pistil morphology of ZH11 and gs3/gw8/gs9#1. The yellow lines indicate the position of the cross‐sections shown in (b). Bar = 1 mm. (b) Transverse sections of ZH11 and gs3/gw8/gs9#1 styles. Bar = 20 μm. (c) Scanning electron microscopy analyses of ZH11 and gs3/gw8/gs9#1 styles. Bar = 100 μm. The dashed boxes highlight the region for comparison of the cell number and cell length in the style between ZH11 and gs3/gw8/gs9#1. (d) The magnified views of the boxed regions in (c). Bar = 10 μm. (e, f) The horizontal cell number (e) and area (f) indicated with dashed lines in (b). Bars indicate SEM (n = 4). (g, h) Quantification analyses of average parenchymal cell number (g) and cell length (h) shown in (d). Bars in (g) indicate SEM (n = 5) and Bars in (h) indicate SEM (n = 120). Asterisks indicate significant differences (**P < 0.01) between ZH11 and gs3/gw8/gs9#1 by two‐tailed Student's t‐test. Data are shown as means ± SEM.
Transcriptome analysis reveals enrichment of cell proliferation‐related genes in the triple mutant
To further probe the molecular mechanism of action of GS3, GW8 and GS9 in regulating pistil growth, we performed RNA sequencing (RNA‐seq) to compare the global gene expression profiling between the gs3/gw8/gs9 triple mutant and ZH11. As the stigma and style of gs3/gw8/gs9 triple mutant started to display a differential growth rate with that of ZH11 from the S11b to S12 stages (Figure S6), we used the pistils at the S11b stage for transcriptome analysis. Pearson's correlation analysis showed significant correlation between two biological replicates for each sample (Figure S7a). A total of 1498 differentially expressed genes (DEGs) were identified based on the criteria of a 2‐fold change and false discovery rate (FDR) < 0.05 (Dataset S1). Among them, 861 genes were up‐regulated and 637 genes were down‐regulated in the gs3/gw8/gs9 mutant when compared to ZH11 (Figure S7b). Gene Ontology (GO) term enrichment analysis and KEGG pathway enrichment analysis showed that these DEGs were mainly enriched in multiple processes (Figure S7c,d; Dataset S1, S3). Notably, a large proportion of the DEGs were clustered into cell proliferation–related terms such as ‘cell division’, ‘microtubule binding’, ‘microtubule motor activity’, and ‘cell wall remodeling’ (Figure S7c), consistent with the alteration of cell division detected in the gs3/gw8/gs9 mutant. We further confirmed the expression profiles of several cell division‐related genes by RT‐qPCR (Figure S7b). Our results suggest that GS3, GW8 and GS9 cooperatively regulate cell division during pistil development.
Knocking out of GS3 , GW8 and GS9 significantly increase outcrossing rates of ZH11 and Zhu6S
To test whether the increased SER in the gs3/gw8/gs9 mutants may promote out‐crossing, we further carried out a field experiment by crossing ZH11 and gs3/gw8/gs9 with an orange‐red‐grained astaxanthin rice (Zhu et al., 2018) as the male parent. The outcrossing rate was evaluated by calculating the percentage of orange‐red grain in the F1 seeds (the self‐pollinated seeds are white and the cross‐pollinated hybrid seeds are orange‐red). As expected, both lines of the gs3/gw8/gs9 triple mutants exhibited significantly higher outcrossing seed setting rates compared with ZH11, when pollinated by the orange‐red‐grained astaxanthin rice (Figure 5a,b). To further evaluate the potential application of loss‐of‐function alleles of gs3, gw8 and gs9 in hybrid seed production, we selected an elite indica male sterile line Zhu6S for genetic improvement. Zhu6S is known to have a low SER (~20%), and thus is not suitable for hybrid seed production. As Zhu6S contained a loss‐of‐function allele of gs3 (Figure S8a), we simultaneously knocked out GW8 and GS9 using the CRISPR/Cas9 technology (Figure S8b). As expected, the edited Zhu6S lines (with gw8 and gs9 knockout) exhibited significantly increased SSE, DSE and TSE when compared to the original Zhu6S (Figure 5c,d). The average TSE was increased from 22.87% in the original Zhu6S to 60.65% in the Edited Zhu6S‐1 and 62.62% in Edited Zhu6S‐2 (Figure 5d). We further performed a natural outcrossing test by planting the original Zhu6S and its edited lines together with a male fertile rice variety Tianfeng B. As both the original Zhu6S and the edited Zhu6S are male sterile lines, the seeds produced on these plants were hybrid seeds derived from natural cross‐pollination between the sterile lines and Tianfeng B. Notably, the seed setting rate of the edited Zhu6S reached ~50%, which is about two times higher than that of the original Zhu6S (~25%) (Figure 5e,f). These results indicate that manipulation of GS3, GW8 and GS9 could effectively improve the outcrossing rate of male sterile lines and thus hybrid rice seed production.
Discussion
The SER trait of the male sterile line is a key limiting factor for hybrid seed production in rice. However, few SER‐related genes have so far been cloned and characterized, severely hindering the genetic improvement of rice SER and the efficacy of hybrid rice breeding. In this study, we demonstrate that besides the previously reported GS3 (Takano‐Kai et al., 2009; Zhou et al., 2017), GW8 and GS9 are also common regulators of grain shape and SER and that targeted manipulation of these genes could be used to effectively improve SER and out‐crossing rate in rice, thus facilitating hybrid rice production. Our phenotypic analysis showed that there are about 1.7‐fold, 3.7‐fold and 3.8‐fold increases of SER in gs3, gw8, gs9 single mutant separately in comparison to ZH11, suggesting that GW8 and GS9 play more predominant roles than GS3 in regulating SER. SER of the double mutants (16.1–25.1%) is obviously higher than that of the single mutants (7.3–16.1%). Notably, SER of the gs3/gw8/gs9 triple mutant (54.1–64.2%) is significantly higher than that of the gw8/gs9 double mutant (16.1–25.1%) (Figure 2y; Figure S3u), suggesting that GS3, GW8 and GS9 may function in a synergistic fashion to regulate SER. Further studies are required to clarify their genetic interaction in SER regulation. In addition, it is worth noting that stigma viability is also an important factor for the success of out‐crossing seed setting (Qi and Wu, 2022). However, whether GS3, GW8 and GS9 affect stigma vitality also awaits detailed evaluation in future studies. Regardless, our findings hold great potential in targeted improvement of SER for the available male sterile lines as well as facilitating the breeding of new male sterile lines with the superior combining ability and hybrid vigour, thus promoting the utilization and commercialization of hybrid rice.
In addition, as most of the currently planted hybrid rice varieties were intra‐subspecific hybrids of the indica subspecies, and their yields have reached a plateau due to the narrow genetic diversity of the parental lines. The indica–japonica inter‐subspecific hybrid rice has stronger heterosis than intra‐subspecific hybrids of the indica subspecies, and has the potential to boost the yield by an additional 20–30% (Peng et al., 1999; Yuan and Virmani, 1988). However, utilization of the strong indica‐japonica inter‐subspecific hybrid vigour is currently restrained by several bottlenecks, including low fertility of the F1 plants due to reproduction barriers between the indica and japonica subspecies, tall plant height/lodging, prolonged life cycle, poor grain quality and low production of hybrid seeds. In particularly, the extremely low level of SER (about 5%) in japonica varieties is a major constraint for indica–japonica inter‐subspecific hybrid seed production. Our findings that both SER and outcrossing rate could be significantly improved in ZH11 (a japonica variety) and Zhu6S (an indica male sterile line) illustrate an effective approach to overcome this barrier, and thus contributing to the development and commercialization of indica–japonica inter‐subspecific hybrids in the future.
Besides being an important agronomic trait for hybrid seed production in rice, stigma exsertion is also a key trait of rice domestication. The wild rice ancestor O. rufipogon is known to have high SER (80%) and superior outcrossing habit (allogamous), whereas the Asian cultivated rice has greatly reduced SER (on average below 15% in indica rice and ~5% in japonica rice), rendering the cultivated rice become a strictly selfing (autogamous) species (Xu and Sun, 2021). However, the biological significance and regulatory mechanisms underlying such a transition remain largely mysterious. A recent study demonstrated that GS3 and GW5 likely contributed to this change. Based on population genetic analyses, it was inferred that wild rice should have the wild combination of GW5/GS3 and that in the process of rice domestication, gain of GS3 function and loss of GW5 function may have contributed greatly to the change of outcrossing habit of rice to selfing (Zhou et al., 2017). In this study, we showed that knocking out GW8 and GS9 could significantly increase both SER and outcrossing rate of cultivated rice. Thus, it will be of great interest to explore the genetic diversity of these genes (GS3, GW5, GW8 and GS9) in wild rice and examine how their haplotype combinations have been artificially selected during rice domestication and modern breeding process. Such analyses may lead to the identification of superior haplotype combinations that confer male sterile lines with desired grain shape/grain quality and high SER for hybrid seed production, with minimal detrimental effects on other agronomic traits, thus contributing to improved breeding efficiency and yield of hybrid rice.
Methods
Plant materials and growth conditions
The GS3, GW8 and GS9 gene were knocked out in the Oryza sativa cv. Japonica cultivar Zhonghua 11 (ZH11) and the indica thermo‐sensitive genic male sterile line Zhu6S through the CRISPR/cas9 technology. The orange‐red‐grained astaxanthin rice was kindly provided by Zhu et al. (2018). All rice plants were cultivated in the experimental field at the South China Agricultural University in Guangzhou (23°7′N, 113°15′E) from March to November in 2021 and 2022, and in Lingshui, Hainan (18°22′N, 109°45′E) from November to April in 2021 and 2022.
Investigation of spikelet shape, pistil‐related traits and agronomic traits
Spikelets of ZH11 and gs3/gw8/gs9 at different developmental stages were captured and photographed under a light microscope (Zeiss Stemi 508). For investigating the pistil‐related traits, the pistils were carefully separated from the spikelets and photographed under a light microscope (Zeiss Stemi 508). The length and width of the spikelet and the length of stigma and style were measured using the ImageJ software.
To analyse the agronomic traits, the wild type ZH11 and two gs3/gw8/gs9 lines were grown at the experimental field in Guangzhou from March to July 2021, and eight plants of ZH11 and each mutant line were randomly selected for measurement of grain length, grain width, 1000‐grain weight, panicle length, number of primary branches, number of second branches, plant height, tiller number and grain number.
To calculate the stigma exsertion rate (SER), SER could be gauged by three categories: the single stigma exsertion rate (SSE), the dual stigma exsertion rate and the total stigma exsertion rate (TSE). After anthesis, 10 main panicles from 10 individuals of ZH11, Zhu6S and each mutant line were used to calculate the number of spikelets with single exserted stigma (SES), dual exserted stigma (DES), and no exserted stigma (NES). SER is calculated using the following formulas:
Histological analysis
Fresh young spikelet hulls and pistils were firstly fixed in FAA and dehydrated through a graded series of ethanol, then were embedded in epoxy resin (Pon812 Epoxy Embedding Kit; Sigma‐Aldrich, Saint Louis, MO, USA) and polymerized. 3‐μm‐thick sections for pistils and 8‐μm‐thick sections for spikelet hulls were stained with filtered 1% toluidine blue and examined under a light microscope (Nikon, Y‐TV55). The cell number and cell area were measured using ImageJ. To analyse the surface cells of pistils, scanning electron microscopy (SEM) examination was performed as described by Juarez et al. (2004) with some modifications. Fresh pistils were fixed in a glutaraldehyde fixative solution (2.5% glutaraldehyde in 0.08 M phosphoric acid buffer) for 24 h at 4°C and then dehydrated through a graded ethanol series (30%, 50%, 70%, 95% and 100%). Dehydrated samples were then dried by a critical point dryer with liquid CO2. Finally, the samples were coated with gold palladium using a Desk II sputter coater (Denton Vacuum, Moorestown, NJ) for 45 s before observation under a Hitachi S‐3400N SEM (Hitachi, Kyoto, Japan). To analyse the surface cells of the glumes, mature spikelet glumes were dried and coated with gold palladium, then were observed with a Hitachi S‐3400N SEM with an accelerating voltage of 5 kV.
Total RNA extraction, RT‐qPCR analyses and RNA‐seq
Total RNA was extracted from the S11b stage pistil using the TRIzol reagent (Thermo Fisher). For RT‐qPCR analysis, approximate 1 μg total RNA of each sample was converted to cDNA using a Hifair® III 1st Strand cDNA Synthesis SuperMix Kit (Yeasen Biotechnology, Shanghai, China) according to the manufacturer's instructions. RT‐qPCR was performed using the Hieff UNICON® qPCR SYBR Green Master Mix (Yeasen Biotechnology) with a LightCycler® 96 System. The Actin1 gene (LOC_Os03g50885) was used as a reference gene. All primer sequences are listed in Table S2.
For RNA‐seq, total RNA of fresh pistils was extracted using Trizol according to the manufacture's protocol. Two biological replicates were used for each sample. A total of 2.0 μg RNA per sample was used to construct cDNA libraries using the mRNA‐seq V3 Library Prep Kit (Vazyme, Nanjing, China) according to the manufacturer's instruction. The Bioanalyzer 2100 (Agilent, Palo Alto, CA, USA) was used to quantify and assess the quality of the RNA samples and cDNA libraries. The paired‐end 2 × 150‐base sequencing was performed on Illumina HiSeq X sequencing platform.
The connectors and low‐quality reads were filtered using Cutadapt (V1.9.1) (Martin, 2011). The processed reads were mapped to the rice Nipponbare reference genome and genes (downloaded from ftp://ftp.ensemblgenomes.org/pub/) using Hisat2 (V2.0.1) (Kim et al., 2015). HTSeq (V0.6.1) (Putr et al., 2022) was used to count the numbers of reads mapped to each gene. EdgeR Bioconductor package was used to screen differential expression genes (DGEs) at the thresholds of false discovery rate (FDR) ≤ 0.05 and log2 Fold‐Change ≥1. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were analysed using an online tool DAVID (https://david.ncifcrf.gov/). Significantly enriched GO terms and KEGG pathway were selected with an empirical P value ≤0.05, and the enrichment results were plotted using the ggplot2 package. All DEGs are listed in Dataset S1. Enriched GO terms and KEGG pathway are listed in Dataset S1 and S3, respectively.
Evaluation of outcrossing rate
To compare the outcrossing rate between ZH11 and the gs3/gw8/gs9 mutants, ZH11 and two gs3/gw8/gs9 lines were selected as the female parent, while orange‐red‐grained astaxanthin rice as the male parent. The female and male lines were planted in a 2:4 row ratio, and two rows of female parents were planted in the middle encompassed by four rows of each male line. During the flowering stage, artificial supplementary pollination was performed twice per day. After approximately 30 days, the seeds of the ZH11 and two gs3/gw8/gs9 mutants were harvested. The outcrossing rate was evaluated by calculating the percentage of orange‐red grain in the F1 seeds (including white self‐pollinated seeds and orange‐red hybrid seeds). Four replicates were conducted for each combination.
For the natural outcrossing test, the unedited‐Zhu6S and two edited‐Zhu6S were planted as the female parents together with Tianfeng B (an indica variety with a similar heading day as Zhu6S) as the male parent. The female and male lines were planted in a 1:4 row ratio, and one row of female parents was planted in the middle encompassed by four rows of each male line. The crosses between the male and female naturally occurred without being aided by artificial pollination. After approximately 30 days, the seeds of the unedited‐ and edited‐Zhu6S were harvested. The natural outcrossing rate was evaluated by calculating the seed setting rate of the female lines. Six replicates were conducted for each combination.
Accession numbers
The gene sequence data from this article can be found in the Rice MSU Genome Annotation Release 7 under the following accession numbers: GW8 (LOC_Os08g41940), GS3 (LOC_Os04g56400.1), GS9 (LOC_Os09g27590), CYCIaZm (LOC_Os01g59120), CSLD4 (LOC_Os12g36890), CYCB2;2 (LOC_Os06g51110), CDKB2 (LOC_Os08g40170), CYCA2;1 (LOC_Os12g31810), PME1 (LOC_Os03g19610), PME2 (LOC_Os01g20980), PME31 (LOC_Os11g08750) and KIN7J (LOC_Os09g35890).
Conflict of interest
The authors declare they have no conflict of interest.
Author contributions
R.S. and H.W. conceived and designed the project. X.Z., and Y.G. performed the research. W.D., Y.L., D.Z., X.L., C.L. and C.W. participated in some experiments. X.Z., Y.G., and Y.H. analysed the data. R.S. and Y.H. wrote the article. H.W. revised the article.
Supporting information
Dataset S1 Gene list and FPKM of DEGs in RNA‐Seq.
Dataset S2 Gene Ontology (GO) term enrichment analyses of differentially expressed genes (DEGs).
Dataset S3 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms of differentially expressed genes (DEGs).
Figure S1 Correlation relationship between stigma and style length and spikelet traits.
Figure S2 GS3, GW8 and GS9 mutation sites in various knockout lines generated by the CRISPR/Cas9 technology.
Figure S3 GS3, GW8 and GS9 synchronously regulate glume, pistil growth and stigma exsertion in rice.
Figure S4 Correlation relationship between stigma exsertion rate and spikelet traits.
Figure S5 The effect of GS3, GW8 and GS9 on several agronomic traits in rice.
Figure S6 The dynamic change of spikelet and pistil for the ZH11 and gs3/gw8/gs9#1 during the stages of spikelet development.
Figure S7 Transcriptome analysis of the pistil of ZH11 and the gs3/gw8/gs9 mutant at stage 11.
Figure S8 GW8 and GS9 mutation in Zhu6S generated by the CRISPR/Cas9 technology.
Table S1 List of genes involved in rice grain shape controls.
Table S2 List of primers used in this study.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31921004, 32172056) and the Natural Science Foundation of Guangdong Province (2019B1515120061).
Contributor Information
Haiyang Wang, Email: whyang@scau.edu.cn.
Rongxin Shen, Email: shenrongxin@scau.edu.cn.
References
- Bakti, C. and Tanaka, J. (2019) Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession ‘W0120’. Breeding Science. 69, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, X. , Yang, Y. , Zhang, Y. , Chen, X. , Fan, Z. , Liu, Q. , Ji, J. et al. (2020) OsSYL2 AA , an allele identified by gene‐based association, increases style length in rice (Oryza sativa L.). Plant J. 104, 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Xing, Y. , Mao, H. , Lu, T. , Han, B. , Xu, C. , Li, X. et al. (2006) GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations Agriculture Databases (2021) FAO. https://www.fao.org/worldfoodsituation/csdb/en/
- Huang, X. , Kurata, N. , Wei, X. , Wang, Z. , Wang, A. , Zhao, Q. , Zhao, Y. et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez, M. , Twigg, R. and Timmermans, M. (2004) Specification of adaxial cell fate during maize leaf development. Development 131, 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kato, H. and Namai, H. (1987) Floral characteristics and environmental factors for increasing natural outcrossing rate for F1 hybrid seed production of rice (Oryza sativa L.). Japanese Journal of Breeding 37, 318–330. [Google Scholar]
- Kim, D. , Langmead, B. and Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Sun, C. , Mu, P. , Chen, L. and Wang, X. (2001) QTL analysis of anther length and ratio of stigma exsertion, two key traits of classification for cultivated rice (Oryza sativa L.) and common wild rice (Oryza rufipogon Griff.). Acta Genetica Sinica 28, 746–751. [PubMed] [Google Scholar]
- Li, P. , Zhang, Q. and He, Y. (2014) Genetic mapping and validation of quantitative trait loci for stigma exsertion rate in rice. Mol. Breeding 34, 2131–2138. [Google Scholar]
- Li, N. , Xu, R. and Li, Y. (2019) Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 435–463. [DOI] [PubMed] [Google Scholar]
- Ling, Z. and Xu, B. (1988) Study on stigma exsertion after anthesis in rice I. variation of stigma exsertion. Acta Agriculturae Universitatis Pekinensis 14, 388–392. [Google Scholar]
- Liu, Q. , Qin, J. , Li, T. , Liu, E. , Fan, D. , Edzesi, W. , Liu, J. et al. (2015) Fine mapping and candidate gene analysis of qSTL3, a stigma length‐conditioning locus in rice (Oryza sativa L.). PLoS One 10, e127938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Chen, J. , Zheng, X. , Wu, F. , Lin, Q. , Heng, Y. , Tian, P. et al. (2017) GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3, 17043. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Zhang, A. , Wang, F. , Kong, D. and Luo, L. (2019) Fine mapping a quantitative trait locus, qSER‐7, that controls stigma exsertion rate in rice (Oryza sativa L.). Rice 12, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, J. , Yue, G. , Yang, W. , Mei, H. and Lu, H. (2014) Mapping QTLs influencing stigma exertion in rice. Bulgarian J. Agr. Sci. 20, 1450–1456. [Google Scholar]
- Mao, H. , Sun, S. , Yao, J. , Wang, C. , Yu, S. , Xu, C. , Li, X. et al. (2010) Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. 107, 19579–19584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathi, B. and Jena, K. (2015) Floral traits to enhance outcrossing for higher hybrid seed production in rice: present status and future prospects. Euphytica 201, 1–14. [Google Scholar]
- Martin, M. (2011) Cutadapt removes adapter sequences from high‐throughput sequencing reads. Embnet J. 17, 10–12. [Google Scholar]
- Peng, S. , Cassman, K. , Virmani, S. , Sheehy, J. and Khush, G. (1999) Yield potential trends of tropical rice since the release of IR8 and the challenge of increasing rice yield potential. Crop. Sci. 39, 1552–1559. [Google Scholar]
- Putr, G. , Anders, S. , Py, P. , Pimanda, J. and Zanini, F. (2022) Analysing high‐throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38, 2938–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, B. and Wu, C. (2022) Potential roles of stigma exsertion on spikelet fertility in rice (Oryza sativa L.) under heat stress. Front. Plant Sci. 13, 983070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. , Zhang, Y. , Zhang, K. , Rahman, M. , Barman, H. , Riaz, A. , Chen, Y. et al. (2017a) Genetic dissection of the major quantitative trait locus (qSE11), and its validation as the major influence on the rate of stigma exsertion in rice (Oryza sativa L.). Front Plant Sci. 8, 1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman, M. , Zhang, Y. , Sun, L. , Zhang, K. , Rahman, M. , Wu, W. , Zhan, X. et al. (2017b) Genetic mapping of quantitative trait loci for the stigma exsertion rate in rice (Oryza sativa L.). J. Integr. Agric. 16, 1423–1431. [Google Scholar]
- Song, X. , Huang, W. , Shi, M. , Zhu, M. and Lin, H. (2007) A QTL for rice grain width and weight encodes a previously unknown RING‐type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. [DOI] [PubMed] [Google Scholar]
- Takano‐Kai, N. , Jiang, H. , Kubo, T. , Sweeney, M. , Matsumoto, T. , Kanamori, H. , Padhukasahasram, B. et al. (2009) Evolutionary history of gs3, a gene conferring grain length in rice. Genetics 182, 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, Q. , Zou, T. , Zheng, M. , Ni, Y. , Luan, X. , Li, X. , Yang, W. et al. (2020) Substitution mapping of the major quantitative trait loci controlling stigma exsertion rate from Oryza glumaepatula. Rice 13, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D. (1993) Studies on mechanism of outcrossing rate in hybrid rice seed production. Hybrid Rice 8, 12–14. [Google Scholar]
- Uga, Y. , Fukuta, Y. , Cai, H.W. , Iwata, H. , Ohsawa, R. , Morishima, H. and Fujimura, T. (2003) Mapping QTLs influencing rice floral morphology using recombinant inbred lines derived from a cross between Oryza sativa L. and Oryza rufipogon Griff. Theor. Appl. Genet. 107, 218–226. [DOI] [PubMed] [Google Scholar]
- Uga, Y. , Siangliw, M. , Nagamine, T. , Ohsawa, R. , Fujimura, T. and Fukuta, Y. (2010) Comparative mapping of QTLs determining glume, pistil and stamen sizes in cultivated rice (Oryza sativa L.). Plant Breed. 129, 657–669. [Google Scholar]
- Virmani, S. , Aquino, R. and Khush, G. (1982) Heterosis breeding in rice (Oryza sativa L.). Theor. Appl. Genet. 63, 373–380. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Wu, K. , Yuan, Q. , Liu, X. , Liu, Z. , Lin, X. , Zeng, R. et al. (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. [DOI] [PubMed] [Google Scholar]
- Wen, G. , Yong, L. , Agrama, H. , Luo, D. , Gao, F. , Lu, X. and Ren, G. (2009) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol. Breed. 24, 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , and Sun, C. (2021) What happened during domestication of wild to cultivated rice. Crop J. 9, 564–576. [Google Scholar]
- Yan, A. (1999) Manual for hybrid rice seed production and MS line multiplication. Beijing, China: China Agriculture Press. [Google Scholar]
- Yan, W.G. , Li, Y. , Agrama, H.A. , Luo, D. , Gao, F. , Lu, X. , and Ren, G. (2009) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol. Breed. 24, 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, C. and Zhang, S. (1989) Studies on the character of stigma exsertion among some of Oryza species. Chin. J. Rice Sci. 3, 62–66. [Google Scholar]
- Yuan, L. (2002) Hybrid Rice. Beijing, China: China Agriculture Press. [Google Scholar]
- Yuan, L. and Virmani, S.S. (1988) Status of Hybrid Rice Research and Development. Hybrid Rice. Proceedings of the International Symposium on Hybrid Rice. Manila, Philippines: International Rice Research Institute. [Google Scholar]
- Zhang, Z. , Lu, Y. , Liu, X. , Feng, J. and Zhang, G. (2006) Cytological mechanism of pollen abortion resulting from allelic interaction of F1 pollen sterility locus in rice (Oryza sativa L.). Genetica 127, 295–302. [DOI] [PubMed] [Google Scholar]
- Zhang, D. , Xue, L. and Zhu, L. (2011) Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38, 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Zhang, Y. , Wu, W. , Zhan, X. , Anis, G. , Rahman, M. , Hong, Y. et al. (2018) qSE7 is a major quantitative trait locus (QTL) influencing stigma exsertion rate in rice (Oryza sativa L.). Sci. Rep. 8, 14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D. , Li, Q. , Zhang, C. , Zhang, C. , Yang, Q. , Pan, L. , Ren, X. et al. (2018) GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9, 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. , Li, P. , Xie, W. , Hussain, S. , Li, Y. , Xia, D. , Zhao, H. et al. (2017) Genome‐wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol. Plant 10, 634–644. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. (2016) Fifty years of hybrid rice research in China. Chin. Sci. Bull. 61, 3740–3747. [Google Scholar]
- Zhu, Q. , Zeng, D. , Yu, S. , Cui, C. , Li, J. , Li, H. , Chen, J. et al. (2018) From golden rice to aSTARice: bioengineering astaxanthin biosynthesis in rice endosperm. Mol. Plant 11, 1440–1448. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset S1 Gene list and FPKM of DEGs in RNA‐Seq.
Dataset S2 Gene Ontology (GO) term enrichment analyses of differentially expressed genes (DEGs).
Dataset S3 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms of differentially expressed genes (DEGs).
Figure S1 Correlation relationship between stigma and style length and spikelet traits.
Figure S2 GS3, GW8 and GS9 mutation sites in various knockout lines generated by the CRISPR/Cas9 technology.
Figure S3 GS3, GW8 and GS9 synchronously regulate glume, pistil growth and stigma exsertion in rice.
Figure S4 Correlation relationship between stigma exsertion rate and spikelet traits.
Figure S5 The effect of GS3, GW8 and GS9 on several agronomic traits in rice.
Figure S6 The dynamic change of spikelet and pistil for the ZH11 and gs3/gw8/gs9#1 during the stages of spikelet development.
Figure S7 Transcriptome analysis of the pistil of ZH11 and the gs3/gw8/gs9 mutant at stage 11.
Figure S8 GW8 and GS9 mutation in Zhu6S generated by the CRISPR/Cas9 technology.
Table S1 List of genes involved in rice grain shape controls.
Table S2 List of primers used in this study.


