Abstract
Sulfated glycosaminoglycans (GAGs), or synthetic mimetics thereof, are not favorably viewed as orally bioavailable drugs owing to their high number of anionic sulfate groups. Devising an approach for oral delivery of such highly sulfated molecules would be very useful. This work presents the concept that conjugating cholesterol to synthetic sulfated GAG mimetics enables oral delivery. A focused library of sulfated GAG mimetics was synthesized and found to inhibit the growth of a colorectal cancer cell line under spheroid conditions with a wide range of potencies ( 0.8 to 46 μM). Specific analogues containing cholesterol, either alone or in combination with clinical utilized drugs, exhibited pronounced in vivo anticancer potential with intraperitoneal as well as oral administration, as assessed by ex vivo tertiary and quaternary spheroid growth, cancer stem cell (CSC) markers, and/or self-renewal factors. Overall, cholesterol derivatization of highly sulfated GAG mimetics affords an excellent approach for engineering oral activity.
Introduction
Glycosaminoglycans (GAGs)1−3 form an important class of natural products that are completely different from the traditional classes, such as the alkaloids, polyketides, steroids/terpenoids, and others. Although very few drugs are known to be GAG-based, a large number of drugs have been derived from the latter class of natural products. Yet, in terms of the sheer number of unique structures, GAGs are arguably the most diverse class of natural products. GAGs also bind to a huge number of proteins (>800) that have important roles in a number of diseases such as thrombosis, cancer, inflammation, Alzheimer’s, microbial infection, etc.4−9 More importantly, many proteins targeted by GAGs have been validated as critical mediators of diseases. Thus, GAGs present a rich and unexploited natural product resource for discovering medicinal agents.
Despite this advantage, GAGs are not traditionally thought of as medicinal agents. They are highly sulfated and polymeric. As available in nature, they possess unrivaled heterogeneity.1,2 Their chemical or chemoenzymatic synthesis is extremely challenging and very difficult to scale up.10 Also difficult is computationally designing GAG structures, especially sequences that display high level of selectivity for their targets.11 Finally, their polyanionic character makes GAGs orally unavailable.12 These barriers have led to the accepted perception that it is extremely difficult to convert GAGs, and their oligosaccharide mimetics, into a clinically viable agent, especially with oral activity.
Over the past decade, we have pursued the approach of developing non-saccharide GAG mimetics (NSGMs) to overcome some of the limitations described above. NSGMs are much smaller than natural GAGs. They are based on the aromatic scaffold, which contributes nonionic forces in binding to proteins.13,14 The nonsaccharide scaffold also makes NSGMs fully synthetic, thereby enabling better access to multiplicity of analogue structures that are homogeneous. Importantly, NSGMs are polysulfated, which is the primary reason for their ability to mimic GAGs in terms of binding to GAG-binding sites on proteins. This makes NSGMs very promising mimics of functions of GAGs, e.g., activation of antithrombin.15 This also affords NSGMs the ability to structurally mimic some GAG sequences, as demonstrated for a hexasaccharide sequence of heparan sulfate (HS).16 Interestingly, the ability to bind in GAG-binding sites of proteins also affords new functions to NSGMs, such as inhibition of thrombin17,18 and factor XIa.19,20
Some time ago we identified an NSGM, named G2.2 (Figure 1), as an inhibitor of cancer stem cells (CSCs) from a library of 53 synthetic members.21 CSCs represent a subpopulation of tumor cells that display intrinsic properties of self-renewal, differentiation, and tumorigenicity, which are fundamentally linked to cancer relapse. Recent work has shown that parenterally administered G2.2 inhibits the growth of colon xenografts without any toxicity to vital organs or bleeding consequences.22 Further, G2.2 inhibited the growth of xenografts developed from cells that survive current clinically used drugs against colon cancer, oxaliplatin, and 5-fluorouracil. Thus, G2.2 exhibits considerable promise as the first-in-class agent of the NSGM type against colorectal cancer. In this work, we report that the anti-CSC and in vivo antitumor activities of G2.2 can be significantly improved through lipid modification of its scaffold. We reasoned that conjugating cholesterol to G2.2 will introduce oral bioavailability because cholesterol is rapidly absorbed from the gut and may help carry its highly sulfated G2.2 cargo. More specifically, a structurally defined cholesterol derivative of G2.2 was designed and shown to possess very good in vivo anticancer potential when administered orally. This result leads to a paradigm that synthetic mimetics of GAGs can be induced to display oral bioavailability, a consequence that could have a major impact on the pursuit of GAGs and GAG mimetics as medicinal agents.
Figure 1.
Rationale behind the study of G2.2 analogues as potential anti-CSC agents. G2.2 was identified in a high-throughput screen of a library of 53 NSGMs, which contained 12 different types of scaffolds including linear monomer, linear dimer, and linear trimer NSGMs. Only linear dimers, especially G2.2, were active as inhibitors of colon CSCs, whereas related monomers and dimers were inactive. This work sought to first explore the importance of linker L (=(CH2)3) and 3′-substituent (=OSO3Na) in G2.2, which led to the synthesis of MQD1, MMD1, and MAD1 (L = 1,4-DMC; 1,4-dimethylenecyclohexyl group). A second objective was to conjugate G2.2 with a lipid (=cholesterol) to induce oral bioactivity, which led to the synthesis of G2C, G5C, and G8C.
Results
Rationale Behind the Design of G2.2 Analogues
G2.2 is structurally and functionally unique. First, G2.2 can be thought of as a dimer with four sulfate groups per monomer (Figure 1). The repeating building blocks of GAGs, e.g., heparin/heparan sulfate (Hp/HS), are also dimeric with an average of 3–4 anionic groups. G2.2 is linear in the manner of the helical, linear form of Hp/HS oligosacharides.23 In fact, detailed computational studies indicated that G2.2 mimics a hexasaccharide sequence of Hp/HS (see Figure S1).16 Second, structurally related agents belonging to the monomer and trimer class (Figure 1), e.g., G4.1 and G1.4 (see Figure S2), did not inhibit colon CSCs at all.21 Likewise, very closely related analogue G2.1 (see Figure S2) was found to exhibit weaker anti-CSC activity than G2.2, alluding to a fairly tight structure–activity relationship.21 Third, G2.2 inhibits the growth of cancer cells in the spheroidal state, but not in the monolayer state. More interestingly, G2.2-treated spheroids continue to exhibit inhibitory phenotype even in secondary (2°) and tertiary (3°) spheroidal growth in the absence of G2.2,22 which indicates its unique ability to target the self-renewal property of CSCs (and not just proliferative property of cancer cells).
Although highly promising, G2.2 is not orally bioavailable. This arises from its high number of sulfate groups. Yet, not all of the eight sulfate groups of G2.2 could be critical. We reasoned that analogues with fewer sulfate groups may make better lead candidates. It may also be possible to introduce oral bioavailability through lipid conjugation at less critical sites on the G2.2 scaffold. Thus, we designed two series of G2.2 analogues—one with variations in linker L and sulfate groups, i.e., MQD1, MMD1, and MAD1, and another with cholesterol conjugation at 3′-position, i.e., G2C, G5C, and G8C (Figure 1). Briefly, the former group contains 1,4-dimethylenecyclohexyl (1,4-DMC) linker (L) instead of the trimethylene linker of G2.2, while also containing substituent changes at the 3-, 2′-, and 3′-positions. This induces some rigidity in L while simultaneously altering distribution and/or density of sulfate groups on the scaffold. The latter group invokes cholesterol conjugation through an alkyl chain of varying length (2 to 8 carbons) at the 3′-position. Cholesterol modification has been suggested as a viable alternative for enhancing oral bioavailability24 and we reasoned that replacing a noncritical sulfate group with this moiety may serve the dual purpose of reducing charge density and enhancing lipophilicity. Thus, our focused small library of G2.2 analogues affords the possibility of addressing multiple concepts, especially the criticality or redundancy of sulfate groups and the value of lipid conjugation, if any.
Synthesis, Anti-CSC, and Cytotoxic Activity of the First Series of G2.2 Analogues
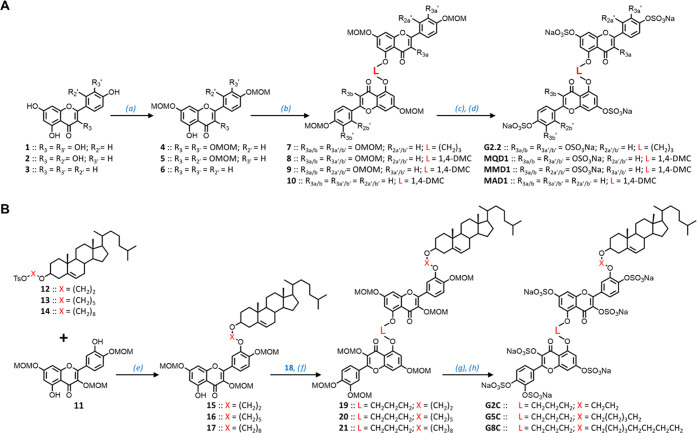
The synthesis of G2.2 analogues followed the protocol established for the parent molecule earlier,21 except for the use of the 1,4-dimethylene-cyclohexyl (1,4-DMC) linker between two flavonoid moieties (Figure 2A). Briefly, flavonoids quercetin (1), morin (2), or apigenin (3) were first O-protected to yield 4–6, each of which carries a free OH group at the 5-position. These were then dimerized to corresponding MOM-protected dimers 8–10 having the 1,4-DMC linker. Deprotection of all MOM groups proved a bit challenging with traditional agents such as HCl in methanol; however, para-toluenesulfonic acid (PTSA) aided by high temperature yielded corresponding polyphenols in good yields. These were persulfated under microwave conditions with trialkylamine-sulfur trioxide complex using the microwave method developed earlier in our lab.25,26 Microwaves are necessary for this reaction, especially for sulfating crowded phenolic groups because sulfation of one disfavors sulfation of an adjacent—OH owing to electronic repulsion. Despite this, the persulfated products G2.2, MQD1, MMD1, and MAD1 were the dominant products with minor amounts of a des-sulfated product, as quantified in Ultrahigh-performance liquid chromatography–electrospray ionization–tandem mass spectrometry (UPLC-ESI-MS) (Figures S3 and S4). The overall yields of these NSGMs from the parent flavonoids were 10–30% with apigenin analogues containing fewer sulfate groups affording higher yields. The overall yields are likely to improve with optimization, especially of the microwave-based sulfation step.
Figure 2.
Chemical synthesis of G2.2 and its analogues. (A) Synthesis of G2.2, MQD1, MMD1, and MAD1. (a) MOMCl (2–4 equiv), N,N-diisopropylethylamine (DIPEA), dichloromethane (DCM), 12 h, 50–60%; (b) TsO-L-OTs (where L = (CH2)3 or CH2C6H10 (1,4-DMC)), K2CO3 (3 equiv), N,N-dimethylformamide (DMF), rt, 12 h, 50–60%; (c) PTSA (4 equiv/OH), methanol, reflux, 12–24 h, 70–90%; (d) SO3/Me3N (6 equiv/OH), Et3N (10 equiv/OH), CH3CN, microwave, 90–100 °C, 4–8 h, 60–90%. (B) Synthesis of G2C, G5C and G8C. (e) K2CO3 (1.5 equiv), DMF, rt, 12 h, 40–70%; (f) 18 (1.2 equiv) and K2CO3 (1.5 equiv), DMF, rt, 12 h, 50–60%; (g) PTSA (4 equiv/OH), methanol, reflux, 12–24 h, 70–90%; (h) SO3/Me3N (6 equiv/OH), Et3N (10 equiv/OH), CH3CN, microwave, 90–100 °C, 7 h, 60–90%. Note: Synthesis of 12–14 and 18 is described in the Supplementary Materials.
To elucidate the anti-CSC activity of these analogues, we utilized spheroidal growth inhibition assay. This assay relies on the observation that CSCs are highly enriched in spheroid culture compared to monolayer culture.21,27 Spheroidal growth is characterized by increased expression of CSC marker proteins, e.g., LGR5, CD133, and CXCR4, as well as aberrant growth factors and morphogens involved in signaling, such as IGF-1R and Wnt/β-catenin,21,28,29 compared to monolayer growth. This implies that a molecule that simultaneously inhibits spheroidal growth, but not the monolayer growth, displays a preference for targeting CSCs, which drive cancerous growth. Thus, to elucidate the anticancer properties of our NSGMs, we performed inhibition of colon cancer cell line HT-29 under spheroidal and monolayer growth conditions.21 In line with the results described for G2.2 earlier, MQD1, MMD1, and MAD1 did not display significant cytotoxicity under monolayer conditions (see Figure S5). In contrast, spheroid growth inhibition profiles were much different (Figure 3A). The three molecules displayed different levels of CSC inhibition potential. Although MAD1 was about 1.6-fold weaker than G2.2, MMD1 and MQD1 were 1.7- and 7.3-fold better, respectively (Figure 3B). Few conclusions can be derived from these results noting that the second-generation analogues are structurally fairly similar to the parent G2.2. First, the second-generation analogues contain the 1,4-DMC linker, which appears to be an important reason for enhancing the anti-CSC activity. Second, changing the scaffold from quercetin (G2.2 with 3′,4′-substitution) to morin (MMD1 with 2′,4′-substitution) seems to improve anti-CSC potential but the improvement is lower than that for MQD1, which implies that the former flavonoid is a better scaffold. Finally, reducing the number of sulfate groups from eight (MQD1) to four (MAD1) dramatically lowers potency (11.4-fold).
Figure 3.
Inhibition profiles of colon cancer cell line HT-29 (p53 mutant, KRAS wild type, microsatellite stable) spheroid growth (A, C) and the corresponding potencies (B, D) of G2.2 and analogues. MQD1, MMD1, and MAD1 are shown in (A) and (B); G2C, G5C, and G8C are shown in (C) and (D). Data at each concentration were compiled (n ≥ 3) as percent of vehicle-treated cells. Solid lines show nonlinear regression using the standard dose–response equation (normalized % inhibition) to derive IC50 values. IC50’s were calculated to be 29.6 ± 3.4 μM (G2.2), 4.0 ± 0.8 μM (MQD1), 17.9 ± 5.0 μM (MMD1), 46 ± 10 μM (MAD1), 5.5 ± 1.4 μM (G2C), 11.5 ± 3.4 μM (G5C), and 0.8 ± 0.2 μM (G8C). (E) Profile of relative inhibition of human cancer organoids vs matched normal intestinal organoids derived from a patient undergoing surgery for colon cancer without any prior treatment and representing treatment-naïve cancer cultures. Error bars represent ±1 SE. # ptrend < 0.06; * p < 0.05. FUOX dose (1.25 μM OX + 25 μM FU).
Synthesis, Anti-CSC, and Cytotoxic Activity of Cholesterol-Modified Analogues of G2.2
The synthesis of cholesterol-conjugates of G2.2 utilized the reduced reactivity of 3′- and 5- phenols of quercetin in comparison to other positions.30,31 Although strong hydrogen bonding to 4-keto makes 5-phenol the least reactive, the 3′-OH is also hydrogen-bonded to 4′-OH, making it a weaker nucleophile than the remaining phenolic groups. This differential reactivity was exploited in synthesizing the 3,7,4′-triprotected quercetin 11 in reasonable yields. Simultaneously, cholesterol was tosylated and then reacted with α,ω-alkylene diols to primarily yield 3β-derivatives, which were further tosylated to synthesize 12–14 in very good overall yields (see Figures S6 and S7). Each of these was coupled with 11, wherein nucleophilic displacement of the tosyl group arose exclusively from the 3′-phenol to yield 15–17 (Figure 2B). The chemistry for transforming these into the final sulfated analogues G2C, G5C, and G8C utilized the methodology developed for G2.2(21) and also described above. Although this technology is now fairly well established owing to the construction of a large number of different scaffolds and derivatives, installation of cholesteryl moiety onto the G2.2 scaffold does introduce some challenges. UPLC-ESI-MS analysis identified the presence of a des-sulfated product in minor proportion in each case (0.5–5%) (Figures S8–S11), which formed irrespective of the multiple microwave conditions explored in the current study. The analysis also identified that the first step also yielded a tiny proportion of α-substituted cholesterol with the β-form being the predominant species. Finally, difficulties in synthesis and isolation tended to be higher for G8C in comparison to G2C. Despite these challenges, persulfated G2C, G5C, and G8C were synthesized with 95–99% purity in reasonably good overall yields.
Figure 3C shows the HT-29 spheroid growth inhibition profiles for lipid analogues of G2.2. All three molecules inhibited spheroidal growth very well. Regression analysis using normalized growth profiles led to IC50’s of 5.5, 11.5, and 0.8 μM for G2C, G5C, and G8C, respectively (Figure 3D). This implies an improvement of 5.3- to 37-fold in potency over the parent molecule as the length of the linker increases from two methylene units to eight. This represents a robust improvement in potency, especially for spheroid growth cultures, which are enriched in chemotherapy-resistant CSCs.29 To assess selectivity toward CSCs, cytotoxicity against HT-29 monolayer culture was also studied. The dose dependence profiles of these analogues using the MTT assay showed G2C and G8C were significantly cytotoxic at ∼100 μM, while G5C was essentially benign toward monolayer cancer cell culture even at ∼400 μM (Figure S12). This implied that the selectivity of targeting CSCs increases from ∼18.7- to ∼126-fold as linker length increases, the majority of which was attributable to the increase in anti-CSC potency.
Effect of the Second Series of G2.2 Analogues on Primary Human Cancer Organoids
It is increasingly recognized that the in vitro models involving cell lines are far removed from the primary in vivo tumor from which they were derived. Hence, we sought to further elucidate the relative efficacy of lipid analogues of G2.2 using primary human colon cancer organoids, which are better translational models of CSCs.32 We generated matched organoids from colon tumor as well as adjacent normal mucosa from a patient (see Figure S13) undergoing surgery for his colon cancer under an Institutional Review Board of the McGuire VAMC approved protocol (Number 02140). The subject had received no prior cancer treatment before surgery. The second passage normal and tumor organoids showed a striking differential response to FUOX (5-fluorouracil and oxaliplatin), the most common colon anticancer chemotherapy, compared to representative NSGMs. While FUOX significantly inhibited normal intestinal organoids, it had minimal impact on tumor organoids at the dose tested (Figure 3E). On the other hand, each NSGM inhibited tumor organoids, while largely sparing normal intestinal organoids. Among the NSGMs, the cholesterol-modified analogues were several-fold better than the parent G2.2, which further implied that cholesterol modification introduced higher potency against colonic CSCs, while retaining the ability to spare normal stem/progenitor cells. Overall, the two studies (cancer stem cells and human cancer organoids) led to two key principles including (a) cholesterol-based enhancement in potency and (b) the role of linker length in targeting CSCs.
Risk of Bleeding Arising from Cholesterol Modification of G2.2
Highly sulfated mimetics of GAGs carry the potential to induce bleeding, which could limit their use as anticancer agents. The potential to induce bleeding is routinely assessed using a variety of assays including plasma clotting time, rodent tail bleeding, and others.33,34 Because the activated partial thromboplastin time (APTT) is more sensitive than the prothrombin time (PT) to heparins, we used the former to evaluate the bleeding potential of G2.2 analogues. The dose-dependent variation in the APTT of pooled, normal, human plasma induced by G2.2, G2C, G5C, and G8C is shown in Figure 4. Each NSGM prolonged the clotting time as expected; however, the effect was most pronounced with G8C. A 2-fold increase in APTT (∼34 to ∼68 s) of normal human plasma required 900 μM of G2.2 (Figure 4A). For G2C, G5C, and G8C, these concentrations were calculated to be nearly 300, 200, and 37 μM (Figure 4B–D). Yet, these concentrations pale in comparison to APTT prolongation induced by enoxaparin, which doubled the time at only 3 μM (Figure 4E). Thus, in comparison to the bleeding risk of an established clinically used drug enoxaparin, the bleeding risk from the NSGMs is likely to be lower.
Figure 4.
Effect of G2.2 and its analogues on human plasma clotting time (A–D) and mouse tail bleeding parameters (F–I). Also shown are the corresponding data for the clinically used anticoagulant enoxaparin (E, J). Solid lines in (A–E) are trend lines from which the concentration necessary to the double human plasma clotting time was calculated. For mouse tail bleeding studies, the NSGMs, enoxaparin, or vehicle were administered IV at noted dosages (mg/kg), the tail clipped (see the Experimental Section), and the blood loss (in mg) was measured. Note the major difference in dosage level between enoxaparin (0.224 mg/kg) and NSGMs (1–100 mg/kg). (J) # represents p < 0.05.
To better evaluate the bleeding risk, we performed mouse tail bleeding studies, as described earlier in our works.20,35 We studied several in vivo concentrations reaching levels as high as 100 mg/kg to bleeding risk arising from acute dosing. Each NSGM was administered through the tail vein of wild-type ICR mice as a simple formulation in water followed by tail clipping. Figure 4F–I shows the blood loss before bleeding cessation. As with APTT studies, blood loss gradually increased from that observed with the vehicle; however, none of the agents induced major blood loss. Even G8C, displayed similar blood loss at the highest dosage studied as other NSGMs indicating a probable disconnect between APTT and tail bleeding. However, enoxaparin induced significantly higher blood loss at a much lower dose (224 μg/kg; Figure 4J), more than 400-fold lower than that of NSGMs. Thus, despite being highly sulfated, these NSGMs are not likely to display high bleeding consequences, which should serve them well as anticancer agents.
In Vivo Anticancer Stem Cell Potential of G5C
Although all three cholesterol-modified analogues of G2.2 were found to be reasonably safe in terms of bleeding, G8C did display a slight propensity for prolonging the clotting time (Figure 4E). Thus, we focused our attention on G2C and G5C with regard to their anti-CSC potential. Unfortunately, assessing the anti-CSC potential in vivo is not easy. CSCs comprise only a small population of tumor cells, even in the xenograft tumors generated through injection of CSC-enriched cancer cells,22 which implies that their influence on the overall tumor parameters, such as volume or weight, are less marked than those of bulk tumor cells. However, the influence of CSCs can be observed through assessment of characteristic properties such as quantitation of CSC markers and self-renewal proteins and/or ex vivo spheroidal growth. In fact, G2.2 had earlier been identified as a CSC-specific anticancer agent through a detailed assessment of these tools.21,22 Briefly, G2.2 inhibited the expression of CD133+, CXCR4+, DCAMLK1, and/or LGR5 markers, which are traditionally known as selective markers of colon CSC.22 Likewise, G2.2 induced significant reduction in the expression of BMI-1, a well-established marker of CSC self-renewal.22 Finally, G2.2’s influence on CSCs’ self-renewal could also be discerned through repeated growth under spheroid growth conditions ex vivo long after the cessation of its treatment. Thus, we employed these assays to evaluate the anti-CSC potential of cholesterol-modified analogues of G2.2.
In the first model, we utilized a dual CSC enrichment strategy to study G5C in vivo. In the first step of this model, HT-29 colon cancer cells were sorted for CD133+ and CXCR4+ surface markers using FACS to produce an enriched population of Dual Hi (CD133+/CXCR4+) CSCs that were then injected subcutaneously to form xenografts in NCr nude mice (Figure 5A). In the second step, the generation of palpable xenografts was followed by the administration of FUOX (25 mg/kg 5-fluorouracil and 2 mg/kg oxaliplatin) intraperitoneally for 3 weeks. Following this, mice were randomized into three cohorts, which were treated with vehicle, FUOX (IP), or G5C (100 mg/kg IP). We used FUOX as a positive reference because (a) it is the most common colon cancer chemotherapy drug combination used in the clinic today36 and (b) it is known to spare, or possibly even enrich, CSCs.22 Thus, the application of the dual CSC enrichment strategy may also provide insights into in vivo anti-CSC properties of G5C.
Figure 5.
In vivo CSC targeting by cholesterol-modified G2.2. (A) Schematic of the study. 1 × 105 CD133+/CXCR4+ (Dual Hi) HT-29 cells were injected subcutaneously to generate xenografts in NCr nude mice. Once the palpable tumors were observed, the animals were treated with vehicle (Veh) or FUOX (25 mg/kg 5-fluorouracil (FU) & 2 mg/kg oxaliplatin (OX); IP weekly for 3 weeks) followed by randomization to vehicle (Veh), FUOX (25 mg/kg FU & 2 mg/kg OX IP weekly), or G5C (100 mg/kg three times/week × 4.6 weeks (14 doses)). Animals were sacrificed at the end of treatment and tumors (n = 2 per group) were analyzed for CSCs. (B) Profile of change in tumor volume as the function of the second treatment (Veh, FUOX, or G5C). (C) Animal weight measured in grams at least three times a week. Although weight was measured since initiation of tumors, data is shown only following randomization to NSGM or FUOX. (D) Profile of ex vivo growth of 1, 2, and 3° spheroids from xenograft cells. (E) Quantitation of Dual Hi (CD133+/CXCR4+) cells in xenografts by flow cytometry. (F) Profile of LGR5, DCAMLK1 (CSC markers), and BMI-1 (self-renewal factor) proteins in lysate from xenografts using Western analysis. *p < 0.01 compared to vehicle controls; ¶ p < 0.01 compared to FUOX. Error bars represent 1 ± standard error of the mean (SEM).
Figure 5B–E presents in vitro analyses of HT-29 xenografts from animals. Both FUOX and G5C demonstrated similar effects in terms of withholding the progression of tumor progression. Yet, the two molecules were significantly different in terms of their influence on CSCs. Repeated cultures of xenograft cells under spheroid conditions showed that 3° spheroids lose their inhibition potential for animals exposed to FUOX, while G5C-derived 3° spheroids maintain inhibition (Figure 5D). This indicates that G5C preferentially targets CSCs present in xenografts. Interestingly, 3° spheroidal growth for FUOX was more than that observed for vehicles alluding to the possibility that FUOX treatment may increase, rather than decrease, CSC proportion in xenografts. In fact, FACS sorting using CD133+ and CXCR4+ surface markers shows that the proportion of CSCs increases from 11 to 26% for FUOX, whereas G5C reduces it to 8% (Figure 5E). Finally, quantifying other CSC markers also indicates the selective action of G5C against CSCs. A robust decrease in LGR5 and DCAMKL1 (two CSC markers) as well as BMI-1 (a self-renewal factor) was observed for G5C (Figure 5F). Thus, G5C treatment resulted in the inhibition of CSC phenotype in vivo.
In Vivo Anti-CSC Potential of G2C in Combination with Conventional Chemotherapy
As described above, FUOX is a conventional chemotherapy used for colon cancer treatment.36 We hypothesized that the antitumor potential of G2C (and G5C) may be enhanced when combined with FUOX because of simultaneous action on CSCs by the former and on bulk cancer cells by the latter. To assess this, we utilized a single in vivo enrichment strategy in which NCr nude mice carrying palpable HT-29 tumors were administered FUOX (25 mg/kg 5-fluorouracil & 2 mg/kg oxaliplatin) intraperitoneally for 3 weeks (Figure 6A). This strategy has been shown to enrich CSCs in xenografts and offers a fine method to increase the proportion of CSCs in tumors.22 Following this treatment, the animals were randomized (n = 4 in each arm) to receive the vehicle, FUOX, G2C (50 mg/kg), or G2C+FUOX combination, intraperitoneally for 3 weeks.
Figure 6.
In vivo CSC targeting by cholesterol analogues of G2.2 combined with standard colon cancer chemotherapy FUOX. (A) Schematic of the study. 1 × 106 HT-29 cells were injected subcutaneously to generate xenografts in NCr nude mice. Once the palpable tumors were observed, the animals were treated with FUOX (25 mg/kg FU & 2 mg/kg OX; IP weekly for 3 weeks) followed by randomization to vehicle, G2C (50 mg/kg three times/week; 8 doses), FUOX (25 mg/kg FU & 2 mg/kg OX; IP weekly for 3 weeks), or G2C+FUOX, as indicated. The animals were sacrificed at the end of treatment (n = 2 per group) and tumors were analyzed for CSC phenotype (spheroids and CSC markers). (B) Xenograft growth curves showing tumor volume inhibition. * p < 0.05. (C) Profile of ex vivo growth of 1, 2, and 3° spheroids from xenograft cells. * p < 0.01 compared to vehicle controls; ¶ p < 0.01 compared to the respective FUOX treatment. (D) Western blots showing G2C and G2C+FUOX treatment significantly reduced DCAMLK1 (CSC marker) and BMI-1 (self-renewal factor) protein levels in the xenograft lysate. (E) Photomicrograph of intestinal organoids suggesting no significant change in the self-renewal and proliferation of normal intestinal stem/progenitors as a result of G2C (alone or in combination with FUOX). Crypts were harvested and cultured in Matrigel in the presence of IntestiCult media. Error bars represent ±1 SEM.
Figure 6 presents the results of this study. At the end of the treatment, the mice treated with the combination therapy G2C+FUOX showed better reduction in tumor volume compared to the two treatments alone (Figure 6B). More importantly, G2C treatment either alone or in combination with FUOX showed significant inhibition of self-renewing CSCs, as determined by 3 and 4° spheroid formation from xenograft-derived cells in CSC media compared to the vehicle controls (Figure 6C). This is in line with their ability to target self-renewing CSCs, i.e., continued growth inhibition despite cessation of in vivo treatment. On the other hand, FUOX treatment alone resulted in a higher number of spheroids ex vivo suggesting enrichment of CSCs by traditional chemotherapy. The expression of self-renewal and CSC markers also supports these conclusions. Both G2C alone and G2C+FUOX combination showed inhibition in the expression of DCAMKL1 (CSC marker) and BMI-1 (self-renewal marker), which supports morphological inhibition of spheroid growth observed with the respective treatment (Figure 6D). With regard to toxicity, G2C alone produced no significant change in animal weight when administered alone or in combination with FUOX (Figure S14). Interestingly, in direct contrast to its effects on self-renewing CSCs, G2C (or G2C+FUOX) did not inhibit ex vivo growth of organoids derived from intestinal crypts (Figure 6E). Hence, it can be concluded that lipid-modified NSGMs, but not FUOX, inhibit CSCs while also sparing normal stem/progenitors.
Oral Antitumor Activity of G2C
Having ascertained that cholesterol analogues of G2.2 targeted CSCs present in tumor xenografts, we sought to investigate whether their antitumor could be realized through oral administration. For this, we utilized the dual enrichment strategy described above in which the first step involved generation of palpable tumors in NCr nude mice from Dual Hi HT-29 CSCs (CD133+/CXCR4+) enriched in vitro. The second step of CSC enrichment was accomplished in vivo by administering FUOX intraperitoneally (Figure 7A), as described earlier.22 Animals were then randomized to receive either the vehicle or G2C orally (experiment 1), or vehicle, G2.2 parenterally or G2C orally (experiment 2). Because the pharmacokinetics parameters, including metabolism, % bioavailability, and clearance, of G2C are yet to be determined, we used a higher dose while also increasing the frequency of dosing (Figure 7A).
Figure 7.
In vivo CSC targeting by orally administered cholesterol analogues of NSGMs. (A) Schematic of the study. 1 × 105 CD133+/CXCR4+ (Dual Hi) HT-29 cells were injected subcutaneously to generate xenografts. Once the palpable tumors were observed, the animals were treated with FUOX (50 mg/kg FU + 5 mg/kg OX; IP weekly for 3 weeks) followed by randomization to either vehicle (Veh) or G2C (100 mg/kg PO; 5 times/week for 3 weeks) [Experiment 1], or vehicle, G2.2 (100 mg/kg IP; 3 times/week for 3 weeks) or G2C (100 mg/kg PO; 5 times/week for 3 weeks) [Experiment 2]. (B, C) Ex vivo analyses of tumor xenograft for anti-CSC vs anti-NSC (normal stem cell) properties. (B) Profile of ex vivo growth of 1, 2, and 3° spheroids from xenograft cells. (C) Profile of ex vivo growth of intestinal crypt-derived organoids cultured in Matrigel in the presence of IntestiCult media. (D) Relative anti-CSC efficacy of parental G2.2 and oral G2C. (E) Growth curves showing tumor volume inhibition by oral G2C and parental G2.2. Quantitation of Dual Hi (CD133+/CXCR4+) and LGR5+ cells (CSCs) in xenografts by flow cytometry. # ptrend < 0.1, * p < 0.05, ** p < 0.01 compared to vehicle control. Error bars represent ±1 SEM.
Figure 7B shows the results of experiment 1. Oral G2C significantly inhibited Dual Hi cell-induced tumor growth as well as 3° spheroid formation from xenograft-derived cells in CSC media, compared to the vehicle control (see Figure S15 also). To the best of our knowledge, this result is the first demonstration of in vivo antitumor activity of a GAG mimetic following oral administration. More importantly, despite the possibility of significantly higher and direct exposure of intestinal progenitors to G2C through the oral route, it did not reduce intestinal crypt-derived organoid growths (Figure 7C). This suggests that G2C spares normal stem/progenitors, in spite of its anti-CSC effects.
To better define the G2C’s relative efficacy, in experiment 2 we studied intraperitoneal treatment of G2.2 in parallel with oral G2C. Despite daily administration of G2C, the animals did not lose weight (Figure S16) and showed no obvious sign of major bleeding either at the site of injection or in organs (not shown). Both G2.2 and G2C seemed to reduce tumor growth compared to vehicle control. However, oral G2C showed a higher growth inhibition (Figure 7D). One reason for this could be the higher relative dose intensity of G2C (∼350%) in experiments of Figure 7D than those of Figure 6B. This realized ∼70% (Figure 7D) versus 30% (Figure 6B) inhibition of xenografts by G2C compared to the vehicle controls. Intriguingly, the proportion of CD133+/CXCR4+ (Dual Hi) and LGR5 cells was significantly lower in the xenografts from oral G2C-treated animals (Figure 7E) confirming that the molecule did not lose anti-CSC property upon oral administration. Finally, oral G2C also did not produce any significant change in blood chemistry compared to vehicle control (Figure S17) suggesting that a fairly high dose of G2C could be tolerated well by the animals.
Discussion
Although GAGs are known to bind and modulate the function of hundreds of proteins, very few GAGs, or oligosaccharides thereof, have been introduced in the clinic as drugs. Fondaparinux is the only homogeneous GAG sequence used in the clinic,6 while pixatimod is the only homogeneous sulfated sequence in clinical trials for the treatment of advanced solid tumors.37,38 As presented in the Introduction section, the field of GAGs, or sulfated sequences like GAGs, as drugs is plagued by a number of difficulties. One major difficulty is their poor oral bioavailability arising from the high number of anionic sulfate groups present on GAGs or mimetics thereof. Other challenges include their polymeric (or oligomeric) character, compositional heterogeneity, and difficult chemical synthesis. This is the reason GAGs are generally viewed poorly as drugs.
Yet, GAGs, or oligosaccharides thereof, continue to be pursued as potential therapeutics, especially as anticancer agents. For example, ODSH, M402, roneparstat, dociparstat, PI-88, and pixatimod have been (or are being) pursued in a number of clinical trials to evaluate their potential against cancers of blood, lung, liver, colon, prostate, and pancreas (see NCT00097851, NCT01402908, NCT01621243, NCT01764880, NCT01843634, NCT02042781, NCT04571645, NCT05061017).39 A number of clinical trials are also being pursued on GAG-derived molecules as treatments against viral infection and thrombosis. Yet, other than pixatimod, these GAGs or oligosaccharides are complex mixtures that are heterogeneous and/or polydisperse. Such mixtures present a significant barrier for regulatory approval. Interestingly, pixatimod, a homogeneous sulfated tetrasaccharide, carries a cholestanol moiety at the reducing end.37,38 Yet, as with most oligosaccharides, the total synthesis of pixatimod has been reported to be challenging.40
Overcoming the barrier of oral bioavailability will enhance the applicability of GAGs and GAG mimetics as drugs. None of the above GAG-based molecules, including pixatimod,38 in clinical trials are being administered orally.38 Although the intravenous (IV) and subcutaneous (SC) routes are efficacious and viable for highly sulfated molecules, these do have limitations. For example, the IV route restricts usage to hospital settings, whereas repeated SC injections at home become challenging for many patients. In fact, many clinical trials with GAG-like molecules have been terminated early because of recruitment difficulties. Thus, opening up an oral route would greatly facilitate the development of GAG-like molecules as drugs.
This work presents the paradigm that conjugating cholesterol enables oral delivery of a synthetic GAG mimetic. An added benefit of this approach appears to be enhancement in the anti-CSC potency against a colon cancer cell line. The intracellular mechanism of action (MOA) of the parent NSGM, G2.2, has been identified as activation of p38 MAP kinase,22 which is likely to arise from the engagement of one of the cell surface receptors, e.g., receptor tyrosine kinases (RTKs). A number of RTKs are known to mediate the intracellular effects of heparin/heparan sulfates (Hp/HS),41−43 which suggests that our GAG mimetics may also function through modulation of one of the cell surface RTKs.
Cholesterol modification of 5-fluorouracil (FU) and other small drugs has been reported.44,45 Although none of these molecules appear to have been studied under oral administration, a consistent observation in these studies was the increase in in vitro anticancer cell line potency. One reason for this observation is that cholesterol improved the uptake of the cytotoxic drugs. Yet, the molecular size of cholesterol in comparison to the small hydrophobic drug, e.g., FU, is substantial (∼50%). This may be a cause for concern. In comparison, cholesterol modification of NSGMs, e.g., G2C or G5C, changes the size by a much smaller proportion because the sulfated scaffold has a much larger molecular mass. Finally, a cholesteryl moiety on an NSGM may also extend its half-life in vivo because polysulfated molecules are believed to be cleared from circulation faster.
Although this work presents a successful approach involving cholesterol modification of an NSGM, a broader aspect of this report is the development of the technology of cholesterol modification of highly sulfated molecules. In effect, this approach should also be feasible for native GAGs. In fact, earlier work on a series of native GAGs led to the observation that colorectal CSCs were inhibited preferentially by a heparan sulfate hexasaccharide (HS06).46 More specifically, screening oligosaccharides of different chain lengths (HS02 → HS36) led to the observation that inhibition of colorectal spheroids was highest with HS hexasaccharides, and not as much with tetra- or octa-saccharides. Further, HS06 was recently found to directly inhibit autoactivation of insulin-like growth factor–1 receptor (IGF-1R),47 a key cell surface receptor modulating the functions of CSC growth. In contrast, the polymer HS36 was devoid of such an inhibitory effect on IGF-1R.47 This chain length selectivity is unique and conveys the value in pursuing HS06 as a drug. Unfortunately, HS06 is very expensive to produce, is not homogeneous, and also not orally bioavailable. In contrast, synthetic NSGMs, e.g., G2.2,16,22 that structurally and functionally mimic HS06 are more promising anti-CSC agents. This implied that conjugating cholesterol to G2.2 would present a novel avenue to enhance its drug characteristics.
Despite the simplicity of this concept, its application will have to involve measured and thoughtful considerations. First, multiple sites on the GAG or NSGM scaffold are available for the introduction of cholesterol. For example, one of eight sulfate positions in G2.2 could have been modified. This presents a large number of theoretical possibilities. Second, the length of linker between the NSGM and lipid will have to be optimized. For example, the analogues G2C, G5C, and G8C studied here and having two, five, and eight methylene units, respectively, display different anti-CSC activity (Figure 3D). Yet, as described above, the biological activity alone may not determine the choice of linker, as evident in the case of G8C, which was found to be synthetically more challenging than G2C and G5C. Third, the type of linker is also expected to be important. Although the methylene unit-based linker ((CH2)n) presented excellent results, a number of other linkers are available, e.g., ethylene glycol, which may offer better potential. Fourth, in addition to cholesterol, a number of natural lipids could be conjugated, including fatty acids and bile acids,12 which may be sufficient to enhance hydrophobicity for oral delivery. Finally, the design of a potentially orally bioavailable GAG mimetic will also have to consider the possibility of bleeding consequences. Although G2C, G5C, and G8C did not exhibit much tail bleeding phenotype, G8C did exhibit some potential to prolong the plasma clotting time (Figure 4), which negated the improvement in its anti-CSC potency to some extent. In effect, the cholesterol conjugation technology presents multiple opportunities for the discovery of GAG mimetics with oral bioavailability.
In this work, we surveyed several structural modifications to identify a better anti-CSC lead candidate. The first series of analogues included variations in linker L and sulfate groups (MQD1, MMD1, and MAD1), while the second series presented cholesterol conjugation at 3′-position at varying lengths (G2C, G5C, and G8C). Of the former, MQD1 containing the 1,4-DMC linker presents a promising candidate with an IC50 gain of ∼9-fold (Figure 3B). This implies that conjugating cholesterol onto MQD1 scaffold may further enhance anti-CSC activity and introduce oral bioavailability. Unfortunately, reducing the level of sulfation drastically (four sulfate groups; 50%), as in MAD1, severely impacted anti-CSC potency. This does not automatically imply all eight sulfate groups are critical. Perhaps an analogue with an intermediate level of sulfation may present better parameters. Thus, additional scaffolds and sulfation patterns will have to be studied to fine-tune the pharmacophore and functional profile.
G2C and G5C, designed in this work, represent promising anticancer molecules. Both molecules were synthesized in less than eight steps and are essentially homogeneous. Both molecules target colorectal CSCs, in a manner very similar to G2.2, which implies that the two lipid analogues may also inhibit CSCs through a similar mechanism, i.e., activation of p38 MAPK.22 It is likely that these molecules exhibit their effects by targeting a receptor tyrosine kinase, e.g., IGF-1R, which is a known target of HS06,47 the native GAG parent of G2.2. Such preferential targeting ability is infrequently observed among highly sulfated GAGs and could possibly arise due to the aromatic scaffold of G2.2 and its lipid analogues. In contrast, anticancer agents, such as PG545 and PI88, do not possess aromatic groups and also do not appear to display a high level of target selectivity. Also, PG545 and PI88 have not been shown to exhibit much oral bioavailability as yet. Co-incidentally, the addition of cholesterol moiety to the G2.2 scaffold also appears to enhance half-life. In preliminary experiments, G5C displayed an almost 50% increase in half-life over G2.2 (data not shown). Thus, appropriately designed nonsaccharide mimetics of GAGs, e.g., G2C and G5C, represent a major advance in GAG-based anticancer drug discovery.
Overall, this work establishes the paradigm that cholesterol derivatization of GAG mimetics, especially synthetic agents such as NSGMs, is an excellent approach to invoke oral activity. This approach appears to present an added advantage of improved potency and/or biological activity.
Experimental Section
Materials for Synthesis
Anhydrous organic solvents were purchased from Sigma-Aldrich or Fisher and used as such. Flavonoids were purchased from Indofine (Hillsborough, NJ). Other chemicals were of reaction grade as used as received from Sigma-Aldrich, Fisher, or TCI America. Column chromatography silica gel (200–400 mesh, 60 Å) was from Sigma-Aldrich. Flash chromatography utilized disposable normal silica cartridges of 30–50 μ particle size, 230–400 mesh size, and 60 Å pore size. SP Sephadex–Na cation exchange chromatography was used for replacing quaternary ammonium counterions of sulfate groups. Each compound was characterized using 1H and 13C NMR spectroscopy, which was performed on a Bruker 400 MHz spectrometer in CDCl3, CD3OD, acetone-d6, or D2O. Each new sulfated GAG mimetic, especially G2.2, G2C, G5C, and others, was also characterized using the RP-IP UPLC-ESI-MS method (below) and found to be >95% pure.
Materials for Liquid Chromatography–Mass Spectrometry (LC–MS)
High-performance liquid chromatography (HPLC)-grade solvents were purchased from Fisher Scientific. n-Hexylamine and 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) for ion-pairing UPLC were from Alfa Aesar. A Waters Acquity TQD ESI-MS spectrometer in the positive-ion mode was used for nonsulfated compounds, whereas negative-ion mode was used for sulfated compounds. Samples were dissolved in either acetonitrile or water and infused at a rate of 20–100 μL/min. For nonsulfated compounds, the capillary voltage was varied between 3 and 4 kV and cone voltage ranged from 20 to 230 V. Ionization conditions were optimized for each compound to maximize the ionization of the parent ion. Generally, the extractor voltage was set to 3 V, the Rf lens voltage was 0.1 V, the source block temperature was set to 150 °C, and the desolvation temperature was about 250 °C. A reversed-phase Waters BEH C18 column of particle size 1.7 μm and 2.1 mm × 50 mm dimensions at 30 ± 2 °C was used for the separation of sulfated compounds. Solvent A consisted of 15 mM n-hexylamine in water containing 0.1% (v/v) formic acid and 100 mM HFIP, while solvent B consisted of 15 mM n-hexylamine and 100 mM HFIP in 85% v/v acetonitrile–water mixture containing 0.1% (v/v) formic acid. Resolution of sulfated molecule into distinct peaks, if any, was achieved with a flow rate of 500 μL/min and a linear gradient of solvent B was implemented. The sample was first monitored for absorbance in the range of 190–400 nm and then directly introduced into the mass spectrometer. ESI-MS detection was performed in positive-ion mode for which the capillary voltage was 4 kV, cone voltage was 20 V, desolvation temperature was 350 °C, and nitrogen gas flow was maintained at 650 L/h. Mass scans were collected in the range of 320–1900 amu within 0.25 s, and several of these added to enhance signal-to-noise ratio.
Synthesis of 4–6
To a solution of quercetin (1), morin (2), or apigenin (3) (1 equiv) in dichloromethane (DCM), N,N-diisopropylethylamine (DIPEA) (4–8 equiv) and methoxymethyl chloride (MOMCl) (4 equiv for quercetin and morin, and 2 equiv for apigenin) were added under nitrogen atmosphere. After vigorous stirring at 0 °C for 1 h, the reaction mixture was allowed to warm to room temperature for over 2 h and the stirring was maintained for 12 h. The resulting mixture was diluted with water (100 mL), extracted with ethyl acetate (200 mL), and then the organic layer was dried over Na2SO4, concentrated under reduced pressure, and purified by flash column chromatography to afford tetraprotected quercetin (4), tetraprotected morin (5) as yellow solids in yields of 50–55%, and diprotected apigenin (6) as a cream solid in yields of 50–60%.
2-(3,4-Bis(methoxymethoxy)phenyl)-5-hydroxy-3,7-bis-(methoxymethoxy)-4H-chromen-4-one (4)
1H NMR (400 MHz, CDCl3) δ 12.47 (s, 1H), 7.84 (d, J = 2.1 Hz, 1H), 7.64 (dd, J = 8.7, 2.1 Hz, 1H), 7.25–7.11 (m, 1H), 6.54 (d, J = 2.2 Hz, 1H), 6.39 (d, J = 2.1 Hz, 1H), 5.24 (d, J = 7.4 Hz, 4H), 5.17 (s, 2H), 5.11 (s, 2H), 3.47 (d, J = 4.6 Hz, 6H), 3.42 (s, 3H), 3.17 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 178.60, 162.94, 161.91, 156.67, 156.49, 149.65, 146.60, 135.63, 124.42, 123.92, 117.74, 115.66, 106.63, 99.70, 97.81, 95.64, 95.10, 94.22, 94.14, 57.72, 56.41, 56.35. MS (ESI) calculated for C23H26O11 [(M + H)]+, m/z 479.15, found for [(M + H)]+, m/z 479.08.
2-(2,4-Bis(methoxymethoxy)phenyl)-5-hydroxy-3,7-bis(methoxymethoxy)-4H-chromen-4-one (5)
1H NMR (400 MHz, CDCl3) δ 12.52 (s, 1H), 7.39 (d, J = 8.52 Hz, 1H), 6.90 (d, J = 2.20 Hz, 1H), 6.77 (d, J = 10.84 Hz, 1H), 6.49 (d, J = 2.16 Hz, 1H), 6.44 (d, J = 2.12 Hz, 1H), 5.18 (s, 2H), 5.17 (s, 2H), 5.15 (s, 2H), 4.98 (s, 2H), 3.46 (s, 3H), 3.44 (s, 6H), 2.95 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 178.63, 162.91, 162.11, 160.36, 157.55, 156.67, 136.75, 132.01, 114.40, 108.77, 107.08, 103.76, 99.53, 97.59, 94.74, 94.29, 77.30, 77.18, 76.98, 76.66, 56.62, 56.34, 56.24, 56.21. MS (ESI) calculated for C23H26O11 [(M + H)]+, m/z 479.15, found for [(M + H)]+, m/z 479.12.
5-Hydroxy-7-(methoxymethoxy)-2-(4-(methoxymethoxy)-phenyl)-4H-chromen-4-one (6)
1H NMR (400 MHz, CDCl3) δ 12.70 (s, 1H), 7.81–7.77 (m, 2H), 7.13–7.08 (m, 2H), 6.61 (d, J = 2.2 Hz, 1H), 6.54 (s, 1H), 6.43 (d, J = 2.2 Hz, 1H), 5.20 (d, J = 3.0 Hz, 4H), 3.46 (d, J = 1.2 Hz, 6H). 13C NMR (100 MHz, CDCl3) δ 182.48, 164.01, 163.00, 162.15, 160.27, 157.61, 127.99, 124.69, 116.61, 104.72, 100.10, 94.37, 94.30, 56.34, 56.21. MS (ESI) calculated for C19H18O7 [(M + H)]+, m/z 359.11, found for [(M + H)]+, m/z 359.17.
Ditosylation of Alcoholic Linkers L(OTs)2
To a solution of cyclohexane-1,4-diyldimethanol (1 equiv), diethylene glycol (1 equiv), or triethylene glycol (1 equiv) in DCM, 4-toluenesulfonylchloride (2.5 equiv) and potassium hydroxide (8 equiv) were added and stirred for 12 h. The resulting mixture was diluted with water (100 mL), extracted with ethyl acetate (200 mL), and the organic layer was dried over Na2SO4, concentrated under reduced pressure, and purified by flash column chromatography to afford the tosylated glycol as a white powder in yields of 85–90%.
Cyclohexane-1,4-diylbis(methylene)bis(4-methylbenzenesulfonate)
1H NMR (400 MHz, CDCl3) δ 7.77–7.68 (m, 4H), 7.28–7.25 (m, 4H), 3.79 (d, J = 7.16 Hz, 3H), 3.72 (d, J = 6.32 Hz, 1H), 2.38 (m, 6H), 1.77–1.76 (m, 1H), 1.66 (d, J = 7.16 Hz, 1H), 1.50 (s, 1H), 1.41–1.35 (m, 3H), 1.22–1.15 (m, 3H), 0.86–0.80 (m, 1H). 13C NMR (100 MHz, CDCl3) δ 144.77, 133.12, 130.11, 127.85, 77.32, 77.01, 76.69, 74.79, 72.71, 36.99, 34.43, 28.05, 24.55, 21.62, 14.19. MS (ESI) calculated for C22H28O6S2 [(M + Na)]+, m/z 475.12, found for [(M + Na)]+, m/z 475.10.
Oxybis(ethane-2,1-diyl)bis(4-methylbenzenesulfonate)
1H NMR (400 MHz, acetone-d6) δ 7.79 (d, J = 8.32 Hz, 4H), 7.47 (d, J = 8.44 Hz, 4H), 4.12–4.10 (m, 4H), 3.61–3.59 (m, 4H), 2.45 (s, 6H). 13C NMR (100 MHz, CDCl3) δ 144.23, 143.96, 143.66, 131.96, 131.81, 131.51, 129.18, 129.14, 128.88, 128.62, 128.58, 128.34, 127.16, 127.12, 126.89, 126.56, 125.58, 76.37, 76.25, 76.05, 75.73, 71.33, 68.18, 68.01, 68.69, 67.69, 67.54, 60.64, 20.61, 20.45. MS (ESI) calculated for C18H22O7S2 [(M + H)]+, m/z 415.09, found for [(M + H)]+, m/z 415.12.
(Ethane-1,2-diylbis(oxy))bis(ethane-2,1-diyl)bis(4-methylbenzenesulfonate)
1H NMR (400 MHz, CDCl3) δ 7.79 (d, J = 8.32 Hz, 4H), 7.34 (d, J = 8.04 Hz, 4H), 4.15–4.12 (m, 4H), 3.66–3.64 (m, 4H), 3.52 (s, 4H), 2.44 (s, 6H). 13C NMR (100 MHz, CDCl3) δ 143.83, 132.01, 128.83, 126.95, 76.31, 76.20, 75.99, 75.68, 69.69, 68.17, 67.74, 20.61. MS (ESI) calculated for C20H26O8S2 [(M + Na)]+, m/z 481.09, found for [(M + Na)]+, m/z 481.22.
Synthesis of 7–10
To a solution of 1 equiv tetraprotected quercetin (4), tetraprotected morin (5), or diprotected apigenin (6) in N,N-dimethylformamide (DMF) was added K2CO3 (3 equiv), and the mixture was stirred for 2 min. This was followed by the addition of ditosylated or dibromo linker (0.5 equiv) and vigorous stirring for 12 h at room temperature. After the reaction completion as indicated by TLC, the reaction mixture was diluted with a mixture of ethyl acetate/H2O (50 mL; 1:1 mixture). The organic layer was separated, and the aqueous phase was further extracted with ethyl acetate (2 × 25 mL). The organic layer was then washed with saturated NaCl solution (25 mL). The three organic layers were combined, dried over anhydrous Na2SO4, and concentrated under reduced pressure to afford the crude intermediates which were further purified using flash chromatography on silica gel. The pure intermediates, 7–10, were obtained as white to cream solids in yields of 50–60%.
5,5′-(Propane-1,3-diylbis(oxy))bis(2-(3,4-bis(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4H-chromen-4-one) (7)
1H NMR (CDCl3, 400 MHz): 7.88 (d, J = 2 Hz, 2 H), 7.66 (d, J = 6.6 Hz, 2 H), 7.24 (d, J = 8.6 Hz, 4 H), 6.63 (d, J = 2 Hz, 2 H), 6.53 (d, J = 2 Hz, 2 H), 5.29 (s, 4 H), 5.28 (s, 4 H), 5.20 (s, 4 H), 5.17 (s, 4 H), 4.45 (t, J = 5.4 Hz, 4 H), 3.53 (s, 6 H), 3.52 (s, 6 H), 3.46 (s, 6 H), 3.19 (s, 6 H), 2.5 (t, J = 5.4 Hz, 4 H). 13C NMR (DMSO-d6, 400 MHz): 173.63, 161.43, 160.48, 158.43, 153.18, 149.09, 146.51, 137.88, 125.03, 123.65, 117.72, 115.68, 110.09, 97.92, 97.81, 95.66, 95.18, 94.25, 94.31, 65.65, 57.54, 56.37, 56.33, 56.31, 21.5. MS (ESI) calculated for C49H56O22, [(M + H)]+, m/z 997.33, found for [(M + H)]+, m/z 997.263.
5,5′-((Cyclohexane-1,4-diylbis(methylene))bis(oxy))bis(2-(3,4-bis(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4H-chromen-4-one) (8)
1H NMR (400 MHz, CDCl3) δ 7.88 (d, J = 2.08 Hz, 2H), 7.70–7.68 (m, 2H), 7.23 (s, 2H), 6.67–6.66 (m, 2H), 6.45–6.42 (m, 2H), 5.29 (d, 8H), 5.25 (s, 4H), 5.20 (s, 4H), 4.02 (d, J = 6.84 Hz, 2H), 3.89 (d, J = 6.60 Hz, 2H), 3.54 (s, 6H), 3.53 (s, 6H), 3.51 (s, 6H), 3.20 (s, 6H), 2.13 (d, J = 7.11 HZ, 2H), 2.03–2.01 (m, 2H), 1.75 (m, 2H), 1.25–1.20 (m, 4H). 13C NMR (100 MHz, CDCl3) δ 168.2, 163.7, 159.3, 157.6, 155.4, 151.9,149.0, 146.8, 136.3, 122.1, 108.5, 107.4, 97.1, 96.2, 95.6, 91.8, 79.4, 55.6, 55.3, 38.7, 27.3. MS (ESI) calculated for C54H64O22 [(M + H)]+, m/z 1065.38, found for [(M + H)]+, m/z 1065.42.
5,5′-((Cyclohexane-1,4-diylbis(methylene))bis(oxy))bis(2-(2,4-bis(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4H-chromen-4-one) (9)
1H NMR (400 MHz, CDCl3) δ 7.43 (d, J = 8.52 Hz, 2H), 6.91 (d, J = 2.2 Hz, 2H), 6.79 (d, J = 10.72 Hz, 2H), 6.66–6.59 (m, 2H), 6.43 (d, J = 2.16 Hz, 2H), 5.20 (d, J = 2.76 Hz, 8H), 5.17 (s, 4H), 5.03 (s, 4H), 4.01 (d, J = 6.96 Hz, 1H), 3.89 (d, J = 6.60 Hz, 2H), 3.48–3.46 (m, 16H), 2.90–2.89 (m, 5H), 2.25–2.19 (m, 1H), 2.15 (d, J = 7.52 Hz, 2H), 2.04 (s, 2H), 1.79–1.70 (m, 3H), 1.25–1.20 (m, 6H). 13C NMR (100 MHz, CDCl3) δ 161.24, 160.95, 159.89, 159.17, 156.57, 154.03, 138.92, 132.08, 115.13, 108.70, 103.82, 97.55, 95.33, 94.71, 94.43, 77.30, 76.67, 56.35, 56.19, 56.13, 37.44, 29.12. MS (ESI) calculated for C54H64O22 [(M + H)]+, m/z 1065.38, found for [(M + H)]+, m/z 1065.30.
5,5′-((Cyclohexane-1,4-diylbis(methylene))bis(oxy))bis(7-(methoxymethoxy)-2-(4-(methoxymethoxy)phenyl)-4H-chromen-4-one) (10)
1H NMR (400 MHz, CDCl3) δ 7.88 (d, J = 8.96 Hz, 4H), 7.14 (d, J = 8.92 Hz, 6H), 6.80–6.79 (m, 2H), 6.50 (d, J = 2.20 Hz, 2H), 5.30–5.27 (m, 4H), 5.24 (s, 4H), 3.91 (d, J = 6.32 Hz, 4H), 3.53–3.52 (m, 4H), 3.49 (s, 8H), 2.95 (s, 1H), 2.88 (s, 1H), 2.13–2.10(m, 4H), 1.79–1.75(m, 4H). 13C NMR (100 MHz, CDCl3) δ 177.75, 162.95, 12.85, 160.84, 160.79, 160.53, 159.76, 128.39, 124.01, 116.61, 108.14, 105.57, 98.47, 95.34, 94.44, 94.22, 84.58, 74.95, 56.62, 56.32, 37.43, 29.70, 29.01, 25.41. MS (ESI) calculated for C46H48O14 [(M + H)]+, m/z 825.31, found for [(M + H)]+, m/z 825.35.
MOM Deprotection of 7–10
To a solution of MOM-protected molecule in dry methanol, PTSA (4 equiv/OH) was added. The reaction mixture was stirred at 75 °C under reflux conditions for 12–24 h. The deprotection was monitored using UPLC-MS until completion. After that, ethyl acetate (25 mL) was added to precipitate the polyphenol product from the reaction mixture. The precipitate was filtered, washed with excess ethyl acetate to remove the excess PTSA, and dried to obtain pure MOM-deprotected polyphenols as a yellow to orange solid, which was used in subsequent reactions without further purification.
5,5′-(Propane-1,3-diylbis(oxy))bis(2-(3,4-dihydroxyphenyl)-3,7-dihydroxy-4H-chromen-4-one)
1H NMR (DMSO-d6, 100 MHz): 10.6 (bs, 2H), 8.4-9.0 (bs, 3H), 7.55 (d, J = 2.1 Hz, 2 H), 7.41 (d, J = 2.1 Hz, 2 H), 6.80 (d, J = 8.4 Hz, 2 H), 6.38 (d, J = 1.9 Hz, 2 H), 6.30 (d, J = 2 Hz, 2 H), 4.29 (t, J = 5.8 Hz, 4 H), 2.26-2.22 (m, 2H). 13C NMR (DMSO-d6, 400 MHz): 171.04, 162.36, 159.74, 157.87, 146.90, 145.01, 141.88, 137.15, 122.28, 119.09, 115.56, 114.48, 105.21, 96.32, 94.54, 64.82, 28.54. MS (ESI) calculated for C33H24O14 [(M + Na)]+, m/z 667.54, found [(M + H)]+, m/z 645.21.
5,5′-((Cyclohexane-1,4-diylbis(methylene))bis(oxy))bis(2-(3,4-dihydroxyphenyl)-3,7-dihydroxy-4H-chromen-4-one)
1H NMR (400 MHz, DMSO-d6) δ 7.64 (d, J = 2.0 Hz, 2H), 7.49–7.47 (m, 2H), 6.87 (d, J = 8.44 Hz, 2H), 6.44 (s, 2H), 6.39 (s, 2H), 4.00 (d, J = 6.68 Hz, 4H), 2.08–1.99 (m, 2H), 1.71 (m, 8H). 13C NMR (100 MHz, DMSO-d6) δ 170.7, 161.0, 158.2, 147.2, 146.1, 145.4, 135.6, 121.7, 121.3, 117.1, 114.9, 105.2, 101.4, 96.2, 93.4, 74.2, 38.6, 27.2. MS (ESI) calculated for C38H32O14 [(M + Na)]+, m/z 735.17, found for [(M + Na)]+, m/z 735.23.
2-(2,4-Dihydroxyphenyl)-5-((4-(((2-(3,5-dihydroxyphenyl)-3,7-dihydroxy-4-oxo-4H-chromen-5-yl)oxy)methyl)cyclohexyl)methoxy)-3,7-dihydroxy-4H-chromen-4-one
1H NMR (400 MHz, DMSO-d6) δ 7.47 (d, J = 8.08 Hz, 3H), 7.19 (d, J = 8.40 Hz, 1H), 7.11 (d, J = 7.84 Hz, 3H), 6.37–6.30 (m, 3H), 2.29 (s, 5H), 2.03–1.99 (m, 3H), 1.82–1.68 (m, 2H), 1.29–1.17 (s, 4H). 13C NMR (100 MHz, DMSO-d6) δ 162.08, 158.54, 156.56, 145.69, 143.39, 137.57, 131.46, 128.01, 125.46, 106.67, 103.01, 40.13, 39.92, 38.87, 20.74. MS (ESI) calculated for C38H32O14 [(M + H)]+, m/z 713.18, found for [(M + H)]+, m/z 713.13.
7-Hydroxy-5-((4-(((7-hydroxy-2-(3-hydroxyphenyl)-4-oxo-4H-chromen-5-yl)oxy)methyl)cyclohexyl)methoxy)-2-(4-hydroxyphenyl)-4H-chromen-4-one
1H NMR (400 MHz, DMSO-d6) δ 7.84 (d, J = 1.96 Hz, 4H), 6.91 (d, J = 8.72 Hz, 4H), 6.51–6.35 (m, 6H), 2.02–1.99 (m, 4H), 1.81 (s, 2H), 1.61 (s, 1H), 1.47 (s, 1H), 1.19–1.16 (m, 6H). 13C NMR (100 MHz, DMSO-d6) 175.4, 163.5, 162.4, 158.6, 156.2, 155.4, 129.8, 122.4, 117.9, 115.1, 104.6, 104.2, 96.2, 93.2, 74.7, 38.2, 27.6. MS (ESI) calculated for C38H32O10 [(M + H)]+, m/z 649.21, found for [(M + H)]+, m/z 649.35.
Synthesis of G2.2, MQD1, MMD1, and MAD1
To a stirred solution of polyphenol in anhydrous CH3CN (∼3 mL) at room temperature, Et3N (10 equiv/–OH group) and SO3/Me3N complex (6 equiv/–OH) were added. The reaction vessel was sealed and microwaved (CEM Discover, Cary, NC) for 4–8 h at 90 °C. The reaction mixture was cooled and concentrated in vacuo at a temperature of <30 °C. The reaction mixture was then purified on a Combiflash RF system using CH2Cl2/CH3OH mobile system to obtain the persulfated molecules. The fractions containing the desired molecule were pooled together, concentrated in vacuo, and reloaded onto an SP Sephadex C-25 column for sodium exchange. Desired fractions containing sodium salts of the persulfated molecules were pooled, concentrated in vacuo, and lyophilized to obtain a fluffy white powder. All sulfation reactions were quantitative with >65% yield.
Sodium 4-(5-(3-((2-(3,4-Bis(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)propoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,2-phenylene Bis(sulfate) (G2.2)
1H NMR (DMSO-d6, 400 MHz): 8.07–7.99 (m, 4 H), 7.56 (d, J = 9 Hz, 2 H), 6.98 (d, J = 1.6 Hz, 2 H), 6.66 (s, 2 H), 4.2 (s, 4 H), 2.28 (s, 2 H). 13C NMR (DMSO-d6, 100 MHz): 159.07, 157.02, 146.44, 142.92, 135.31, 124.50, 123.56, 119.81, 109.56, 66.37, 42.41. MS (ESI) calculated for C33H16Na8O38S8 [(M – 8Na + 10HxA)]2+, m/z 1148.50, found [(M – 8Na + 8HxA) + 2HxA]2+, m/z 1149.09.
Sodium 4-(5-((4-(((2-(3,4-Bis(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)methyl)cyclohexyl)methoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,2-phenylene Bis(sulfate) (MQD1)
1H NMR (400 MHz, D2O) δ 8.10 (s, 2H), 8.04–8.02 (m, 2H), 7.60 (d, J = 8.96 Hz, 2H), 6.98–6.97 (m, 2H), 6.65 (s, 2H), 3.98–3.97 (m, 4H), 2.34–2.25 (m, 1H), 2.09 (m, 4H), 1.84 (s, 1H), 1.23 (s, 4H). 13C NMR (100 MHz, D2O) 176.43, 162.62, 162.04, 158.84, 158.82, 156.40, 156.28, 145.32, 145.29, 144. 98, 144.96, 135.31, 128.25, 128.24, 122.46, 122.43, 122.42, 121.68, 107.92, 107.76, 98.64, 97.82, 97.80, 96.32, 79.42, 37.81, 27.70, 27.69. MS (ESI) calculated for MS (ESI) calculated for C38H24Na8O38S8 [(M – 2Na)]2–, m/z 740.85, found for [(M – 2Na)/2]−, m/z 740.81.
Sodium 4-(5-((4-(((2-(2,4-Bis(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)methyl)cyclohexyl)methoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,3-phenylene Bis(sulfate) (MMD1)
1H NMR (400 MHz, D2O) δ 7.82 (d, J = 8.56 Hz, 2H), 7.54 (s, 2H), 7.44–7.42 (m, 2H), 7.18 (s, 2H), 7.03 (s, 2H), 4.22 (d, J = 7.12 Hz, 2H), 4.11 (d, J = 6.40 Hz, 2H), 2.27 (s, 2H), 1.79–1.68 (m, 4H), 1.23 (s, 4H). 13C NMR (100 MHz, D2O) 175.32, 160.16, 159.04, 158.62, 158.40, 157.82, 157.15, 149.82, 149.80, 144.74, 144.68, 133.93, 128.25, 128.24, 122.46, 122.43, 122.42, 121.68, 109.45, 109.30, 99.04, 98.84, 98.27, 97.84, 78.69, 39.29, 32.56, 29.76. MS (ESI) calculated for C38H24Na8O38S8 [(M – 2Na)]2–, m/z 740.8554, found for [(M – 2Na)]2–, m/z 740.8500.
Sodium 4-Oxo-5-((4-(((4-oxo-7-(sulfonatooxy)-2-(4-(sulfonatooxy)phenyl)-4H-chromen-5-yl)oxy)methyl)cyclohexyl)methoxy)-2-(4-(sulfonatooxy)phenyl)-4H-chromen-7-yl Sulfate (MAD1)
1H NMR (400 MHz, D2O) δ 7.83–7.79 (m,4H), 7.33–7.30 (m, 4H), 7.02 (s, 2H), 6.83–6.77 (m, 2H), 6.52 (s, 2H), 3.89–3.84 (m, 4H), 1.99–1.97 (m, 4H), 1.70–1.64 (m, 2H), 1.11 (s, 4H). 13C NMR (100 MHz, D2O) 177.26, 177.24, 166.32, 166.31, 160.70, 158.60,157.42, 156.71, 154.28, 154.26, 132.62, 122.79, 121.46, 118.54, 118.32, 108.28, 105.42, 104.94, 98.56, 97.36, 96.42, 95.62, 79.45, 78.38, 42.64, 32.22, 32.04. MS (ESI) calculated for C38H28Na4O22S4 [(M – 2Na)]2–, m/z 504.99, found for [(M – 2Na)]2–, m/z 505.01.
General Procedure for Attachment of Tosylated Lipids 12–14 to Triprotected Quercetin 11
To a solution of triprotected quercetin (1 equiv) in anhydrous DMF were added K2CO3 (1.5 equiv) and 12–14 (1 equiv). The solution was stirred under an atmosphere of nitrogen for 48 h. The solution was quenched with dilute HCl and extracted with ethyl acetate. The organic layer was washed with saturated brine and water and dried over anhydrous Na2SO4. The solvent was removed using vacuum, and the product was purified using flash chromatography to obtain pure compounds, 15–17, in 40–70% yield.
2-(3-(2-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)ethoxy)-4-(methoxymethoxy)phenyl)-5-hydroxy-3,7-bis(methoxymethoxy)-4H-chromen-4-one (15)
1H NMR (400 MHz, CDCl3) δ 12.53 (s, 1H), 7.69–7.62 (m, 2H), 7.23 (d, J = 8.52 Hz, 2H), 6.61 (d, J = 2.12 Hz, 1H), 6.46 (d, J = 2.12 Hz, 1H), 5.37–5.33 (m, 1H), 5.28 (s, 2H), 5.23 (s, 2H), 5.17 (s, 2H), 4.24 (t, J = 5.20 Hz, 2H), 3.90 (t, J = 5.22 Hz, 2H), 3.53 (s, 3H), 3.50 (s, 3H), 3.33–3.26 (m, 1H), 3.22 (s, 3H), 2.44–2.37 (m, 1H), 2.29–2.19 (m, 1H), 2.02–1.78 (m, 4H), 1.63–1.30 (m, 14H), 1.20–0.95 (m, 14H), 0.91 (d, J = 6.52 Hz, 4H), 0.87 (dd, J = 2.77 Hz, 6H), 0.68 (s, 3H). MS (ESI) calculated for C50H70O11, [(M + H)]+, m/z 847.49, found for [(M + H)]+, 847.37.
2-(3-((5-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)pentyl)oxy)-4-(methoxymethoxy)phenyl)-5-hydroxy-3,7-bis(methoxymethoxy)-4H-chromen-4-one (16)
1H NMR (400 MHz, CDCl3) δ 12.53 (s, 1H), 7.66–7.59 (m, 2H), 7.22 (d, J = 8.52 Hz, 2H), 6.61 (d, J = 2.16 Hz, 1H), 6.46 (d, J = 2.16 Hz, 1H), 5.35–5.32 (m, 1H), 5.28 (s, 2H), 5.23 (s, 2H), 5.17 (s, 2H), 3.54 (s, 3H), 3.49 (s, 3H), 3.22 (s, 3H), 3.17–3.08 (m, 1H), 2.39–2.30 (m, 1H), 2.23–2.14 (m, 1H), 2.03–1.78 (m, 8H), 1.70–1.30 (m, 18H), 1.25–0.95 (m, 12H), 0.91 (d, J = 6.52 Hz, 3H), 0.87 (dd, J = 2.77 Hz, 6H) 0.68 (s, 3H). MS (ESI) calculated for C53H76O11, [(M + H)]+, m/z 889.54, found for [(M + H)]+, 889.32.
2-(3-((8-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)octyl)oxy)-4-(methoxymethoxy)phenyl)-5-hydroxy-3,7-bis(methoxymethoxy)-4H-chromen-4-one (17)
1H NMR (400 MHz, CDCl3) δ 12.53 (s, 1H), 7.66–7.59 (m, 2H), 7.24 (d, J = 8.52 Hz, 2H), 6.61 (d, J = 2.16 Hz, 1H), 6.46 (d, J = 2.16 Hz, 1H), 5.36–5.33 (m, 1H), 5.28 (s, 2H), 5.23 (s, 2H), 5.17 (s, 2H), 3.53 (s, 3H), 3.49 (s, 3H), 3.48–3.42 (m, 2H), 3.22 (s, 3H), 3.16–3.08 (m, 1H), 2.39–2.30 (m, 1H), 2.23–2.14 (m, 1H), 2.03–1.78 (m, 7H), 1.63–1.26 (m, 23H), 1.28–0.94 (m, 12H), 0.91 (d, J = 6.52 Hz, 3H), 0.87 (dd, J = 2.77 Hz, 6H), 0.67 (s, 3H). MS (ESI) calculated for C56H82O11, [(M + Na)]+, m/z 953.57, found for [(M + Na)]+, 953.65.
Synthesis of 19–21
To a solution of 15–17 (1 equiv) in anhydrous DMF were added K2CO3 (1.5 equiv) and 18 (1 equiv), and the reaction was stirred under an atmosphere of nitrogen with heating at 60 °C for 48 h. The reaction was quenched with diluted HCl and extracted with ethyl acetate. The organic layer was washed with saturated brine and water, dried under Na2CO3, and the solvent was removed by vacuum. The product was purified using flash chromatography to obtain the pure compounds in yields of 50–60%.
2-(3,4-Bis(methoxymethoxy)phenyl)-5-(3-((2-(3-(2-(((10R,13R)-10,13-dimethyl-17-(6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)ethoxy)-4-(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-bis(methoxymethoxy)-4H-chromen-4-one (19)
1H NMR (400 MHz, CDCl3) δ 7.87 (d, J = 2.05 Hz, 1H), 7.69–7.64 (m, 2H), 7.57–7.54 (m, 1H), 7.25–7.17 (m, 2H), 6.66–6.62 (m, 2H), 6.56–6.52 (m, 2H), 5.35–5.32 (m, 1H), 5.30 (s, 2H), 5.28 (s, 2H), 5.26 (s, 2H), 5.21 (s, 2H), 5.18 (s, 2H), 5.17 (s, 2H), 4.46–4.42 (m, 4H), 3.54 (s, 3H), 3.54–3.51 (m, 6H), 3.48 (s, 6H), 3.20 (s, 3H), 3.17 (s, 3H), 2.56–2.47 (m, 2H), 2.39–2.30 (m, 1H), 2.23–2.14 (m, 1H), 2.03–1.78 (m, 7H), 1.70–1.29 (m, 22H), 1.21–1.03 (m, 7H), 0.99 (s, 6H), 0.91 (d, J = 6.52 Hz, 3H), 0.87 (dd, J = 2.77 Hz, 6H), 0.68 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 178.29, 163.85, 159.71, 159.48, 149.27, 149.05, 146.93, 140.82, 136.42, 122.19, 111.62, 108.45, 104.72, 98.71, 95.68, 83.27, 69.46, 65.02, 56.52, 50.82, 47.20, 38.19, 37.95, 35.82, 32.06, 30.73, 29.57, 28.12, 23.69, 22.74, 21.92, 19.32. MS (ESI) calculated for C76H100O22, [(M + H)]+, m/z 1366.62, found for [(M + H)]+, 1366.59.
2-(3,4-Bis(methoxymethoxy)phenyl)-5-(3-((2-(3-((5-(((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)pentyl)oxy)-4-(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-bis(methoxymethoxy)-4H-chromen-4-one (20)
1H NMR (400 MHz, CDCl3) δ 7.87 (d, J = 2.05 Hz, 1H), 7.69–7.64 (m, 2H), 7.57–7.54 (m, 1H), 7.25–7.17 (m, 2H), 6.66–6.62 (m, 2H), 6.56–6.52 (m, 2H), 5.35–5.32 (m, 1H), 5.30 (s, 2H), 5.28 (s, 2H), 5.26 (s, 2H), 5.21 (s, 2H), 5.18 (s, 2H), 5.17 (s, 2H), 4.46–4.42 (m, 4H), 3.54 (s, 3H), 3.54–3.51 (m, 6H), 3.48 (s, 6H), 3.20 (s, 3H), 3.17 (s, 3H), 2.56–2.47 (m, 2H), 2.39–2.30 (m, 1H), 2.23–2.14 (m, 1H), 2.03–1.78 (m, 7H), 1.70–1.29 (m, 22H), 1.21–1.03 (m, 7H), 0.99 (s, 6H), 0.91 (d, J = 6.52 Hz, 3H), 0.87 (dd, J = 2.77 Hz, 6H), 0.68 (s, 3H). MS (ESI) calculated for C79H100O22, [(M + H)]+, m/z 1407.72, found for [(M + H)]+, 1407.63.
2-(3,4-Bis(methoxymethoxy)phenyl)-5-(3-((2-(3-((8-(((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)octyl)oxy)-4-(methoxymethoxy)phenyl)-3,7-bis(methoxymethoxy)-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-bis(methoxymethoxy)-4H-chromen-4-one (21)
1H NMR (400 MHz, CDCl3) δ 7.88 (d, J = 2.05 Hz, 1H), 7.69–7.63 (m, 2H), 7.58–7.55 (m, 1H), 7.25–7.17 (m, 2H), 6.66–6.62 (m, 2H), 6.56–6.52 (m, 2H), 5.35–5.32 (m, 1H), 5.29 (s, 2H), 5.28 (s, 2H), 5.26 (s, 2H), 5.21 (s, 2H), 5.18 (s, 2H), 5.16 (s, 2H), 4.46–4.42 (m, 4H), 4.07 (t, J = 2.05 Hz, 2H), 3.54–3.51 (m, 9H), 3.47 (s, 6H), 3.19 (s, 3H), 3.16 (s, 3H), 2.56–2.47 (m, 2H), 2.39–2.30 (m, 1H), 2.23–2.14 (m, 1H), 2.05–1.75 (m, 6H), 1.70–1.28 (m, 24H), 1.25 (s, 6H), 1.19–1.01 (m, 6H), 0.99 (s, 4H), 0.91 (d, J = 6.52 Hz, 3H), 0.87 (dd, J = 2.77 Hz, 6H), 0.68 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 172.65, 160.45, 159.49, 157.48, 157.45, 125.58, 152.20, 148.13, 148.02, 147.50, 145.55, 140.19, 136.80, 136.83, 124.16, 124.05, 122.67, 120.97, 120.38, 116.79, 115.43, 114.75, 113.23, 109.11, 96.96, 96.88, 96.82, 94.70, 94.45, 94.23, 94.18, 93.27, 77.97, 68.32, 67.09, 64.68, 56.56, 56.54, 55.81, 55.36, 55.31, 55.19, 49.25, 41.34, 38.82, 38.52, 38.24, 36.32, 35.92, 35.20, 34.78, 30.96, 30.92, 29.22, 28.69, 28.44, 28.37, 28.20, 27.90, 27.51, 27.22, 27.00, 25.18, 24.98, 23.29, 22.83, 21.79, 21.54, 20.07, 18.37, 17.72, 10.85.
Synthesis of Phenolic Precursors of G2C, G5C, and G8C
The MOM groups of 19–21 were completely deprotected by PTSA (4 equiv/OH) in methanol. Briefly, MOM-protected molecule was dissolved in about 2 mL of dry dichloromethane and methanol was subsequently added. PTSA was added and the solution was refluxed for 12–24 h, and deprotection was monitored using MS until the reaction reached completion. Ethyl acetate (25 mL) was added to precipitate the polyphenol product from the reaction mixture. The precipitate was filtered, washed with excess ethyl acetate to remove the excess p-toluenesulfonic acid, and dried to obtain the corresponding polyphenols as yellow to orange solids, which were used in subsequent reactions without further purification.
2-(3,4-Dihydroxyphenyl)-5-(3-((2-(3-(2-(((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)ethoxy)-4-hydroxyphenyl)-3,7-dihydroxy-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-dihydroxy-4H-chromen-4-one
1H NMR (400 MHz, CDCl3) δ 10.64 (d, J = 11.16 Hz, 2H), 9.56–9.34 (m, 2H), 9.20 (bs, 1H), 8.71 (d, J = 19.15 Hz, 2H), 7.73 (d, J = 1.76 Hz, 1H), 7.57–7.54 (m, 2H), 7.47 (dd, J = 8.53 Hz, J = 2.31 Hz, 1H), 6.92 (d, J = 8.48 Hz, 1H), 6.85 (d, J = 8.48 Hz, 1H), 6.49 (d, J = 1.84 Hz, 1H), 6.44 (d, J = 1.88 Hz, 1H), 6.38–6.35 (m, 2H), 5.20 (bs, 1H), 4.34 (s, 4H), 4.10–4.00 (m, 2H), 3.46–3.37 (m, 2H), 3.08–2.97 (m, 1H), 2.37–2.20 (m, 3H), 1.95–1.63 (m, 8H), 1.60–1.21 (m, 14H), 1.15–0.92 (m, 8H), 0.91–0.72 (m, 15H), 0.59 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 182.34, 178.13, 165.24, 156.80, 152.41, 149.04, 148.07, 145.62, 141.82, 135.65, 122.48, 112.48, 109.72, 98.62, 93.06, 83.24, 73.05, 66.38, 65.92, 58.03, 56.52, 42.78, 39.87, 38.72, 38.19, 30.36, 29.52, 26.31, 25.29, 23.64, 21.12, 19.37, 12.05. MS (ESI) calculated for C62H72O15, [(M + H)]+, m/z 1057.49, found for [(M + H)]+, 1057.38.
2-(3,4-Dihydroxyphenyl)-5-(3-((2-(3-((5-(((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)pentyl)oxy)-4-hydroxyphenyl)-3,7-dihydroxy-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-dihydroxy-4H-chromen-4-one
1H NMR (400 MHz, CDCl3) δ 10.64 (d, J = 11.16 Hz, 2H), 9.56–9.34 (m, 2H), 9.20 (bs, 1H), 8.71 (d, J = 19.15 Hz, 2H), 7.73 (d, J = 1.76 Hz, 1H), 7.57–7.54 (m, 2H), 7.47 (dd, J = 8.53 Hz, J = 2.31 Hz, 1H), 6.92 (d, J = 8.48 Hz, 1H), 6.85 (d, J = 8.48 Hz, 1H), 6.49 (d, J = 1.84 Hz, 1H), 6.44 (d, J = 1.88 Hz, 1H), 6.38–6.35 (m, 2H), 5.20 (bs, 1H), 4.34 (s, 4H), 4.10–4.00 (m, 2H), 3.46–3.37 (m, 2H), 3.08–2.97 (m, 1H), 2.37–2.20 (m, 3H), 1.95–1.63 (m, 8H), 1.60–1.21 (m, 14H), 1.15–0.92 (m, 8H), 0.91–0.72 (m, 15H), 0.59 (s, 3H). MS (ESI) calculated for C65H78O15, [(M + H)]+, m/z 1099.54, found for [(M + H)]+, 1099.49.
2-(3,4-Dihydroxyphenyl)-5-(3-((2-(3-((8-(((8S,9S,10R,13R,14S,17R)-10,13-dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)octyl)oxy)-4-hydroxyphenyl)-3,7-dihydroxy-4-oxo-4H-chromen-5-yl)oxy)propoxy)-3,7-dihydroxy-4H-chromen-4-one
1H NMR (400 MHz, CDCl3) δ 10.66 (d, J = 9.73 Hz, 2H), 9.51–9.37 (m, 2H), 9.22 (bs, 1H), 8.75 (d, J = 13.08 Hz, 2H), 7.71 (d, J = 1.96 Hz, 1H), 7.66–7.59 (m, 2H), 7.47 (dd, J = 8.71 Hz, J = 2.40 Hz, 1H), 6.94 (d, J = 8.48 Hz, 1H), 6.85 (d, J = 8.52 Hz, 1H), 6.50 (d, J = 1.96 Hz, 1H), 6.44 (d, J = 1.96 Hz, 1H), 6.38–6.35 (m, 2H), 5.21 (bs, 1H), 4.35 (t, J = 5.02 Hz, 4H), 4.00–4.12 (t, J = 6.60 Hz, 2H), 3.08–2.96 (m, 1H), 2.37–2.20 (m, 3H), 2.06–1.97 (m, 1H), 1.92–1.64 (m, 7H), 1.54–1.20 (m, 20H), 1.14–0.74 (m, 23), 0.59 (s, 3H). MS (ESI) calculated for C68H84O15, [(M + H)]+, m/z 1141.58, found for [(M + H)]+, 1141.72.
Synthesis of G2C, G5C, and G8C
Sulfation of polyphenols was performed using microwave-assisted chemical protocol. Briefly, to a stirred solution of polyphenol in anhydrous CH3CN (∼3 mL) at room temperature, Et3N (10 equiv/–OH group) and SO3/Me3N complex (6 equiv/–OH) were added. The reaction vessel was sealed and microwaved (CEM Discover, Cary, NC) for 7 h at 90 °C. The reaction mixture was cooled and concentrated in vacuo at a temperature < 30 °C. The reaction mixture was then purified on a Combiflash RF system using CH2Cl2/CH3OH mobile system (6:4) to obtain persulfated molecules. The fractions containing the desired molecule were pooled together, concentrated in vacuo, and reloaded onto an SP Sephadex C-25 column for sodium exchange. Desired fractions containing sodium salts of the persulfated molecules were pooled, concentrated in vacuo, and lyophilized to obtain a fluffy white powder.
Sodium 4-(5-(3-((2-(3-(2-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)ethoxy)-4-(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)propoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,2-phenylene Bis(sulfate) (G2C)
1H NMR (400 MHz, DMSO-d6) δ 8.07 (d, J = 2.32 Hz, 1H), 8.03–7.98 (m, 1H), 7.95 (s, 1H), 7.62–7.54 (m, 3H), 7.07 (d, J = 1.84 Hz, 1H), 6.99 (d, J = 1.88 Hz, 1H), 6.70 (dd, J = 15.12 Hz, J = 1.92 Hz, 1H), 5.28 (bs, 1H), 4.32–4.21 (m, 4H), 4.12–4.07 (m, 2H), 3.73–3.65 (m, 2H), 3.19–3.10 (m, 1H), 2.37–2.20 (m, 3H), 2.14–2.00 (m, 1H), 1.95–1.66 (m, 4H), 1.53–0.86 (m, 24H), 0.82 (d, J = 6.33 Hz, 4H), 0.78 (dd, J = 6.60 Hz, J = 1.8 2Hz, 7H), 0.58 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 172.91, 159.07, 158.29, 157.02, 153.28, 152.96, 148.41, 146.45, 152.21, 142.93, 140.56, 135.32, 124.52, 123.60, 121.04, 119.86, 109.58, 78.66, 56.17, 55.56, 49.56, 41.85, 36.66, 35.64, 35.16, 31.43, 31.36, 28.00, 27.76, 27.36, 23.85, 23.16, 22.64, 22.37, 20.59, 19.06, 18.54, 11.87. MS (ESI) calculated for C62H65Na7O36S7, [(M – 2Na)]2–, m/z 862.04, found for [(M + H)]+, 862.28.
Sodium 4-(5-(3-((2-(3-((5-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)pentyl)oxy)-4-(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)propoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,2-phenylene Bis(sulfate) (G5C)
1H NMR (400 MHz, DMSO-d6) δ 8.07 (d, J = 2.24 Hz, 1H), 8.03–7.91 (m, 2H), 7.62–7.52 (m, 3H), 7.07 (d, J = 1.80 Hz, 1H), 6.99 (d, J = 1.76 Hz, 1H), 6.74–6.65 (m, 2H), 5.23 (bs, 1H), 4.32–4.21 (m, 4H), 3.98 (t, J = 6.24 Hz, 2H), 3.42–3.32 (m, 2H), 3.07–2.93 (m, 1H), 2.36–2.20 (m, 3H), 2.07–1.63 (m, 6H), 1.53–0.80 (m, 32H), 0.77 (dd, J = 6.60 Hz, J = 1.82 Hz, 7H), 0.58 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 172.98, 159.08, 158.33, 157.06, 153.31, 153.08, 148.75, 146.46, 145.19, 142.94, 140.65, 135.36, 135.32, 124.56, 123.65, 120.90, 120.44, 119.95, 119.52, 118.89, 114.70, 109.59, 100.63, 99.45, 99.22, 78.15, 68.50, 67.02, 66.09, 56.17, 55.57, 49.59, 41.84, 36.71, 36.30, 35.63, 35.16, 31.41, 31.35, 29.44, 28.70, 28.12, 27.75, 27.35, 23.83, 23.17, 22.63, 22.37, 22.32, 20.57, 19.06, 18.54, 11.65. MS (ESI) calculated for C65H71Na7O36S7, [(M – 2Na)]2–, m/z 883.06, found for [(M + H)]+, 883.13.
Sodium 4-(5-(3-((2-(3-((8-(((8S,9S,10R,13R,14S,17R)-10,13-Dimethyl-17-((R)-6-methylheptan-2-yl)-2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren-3-yl)oxy)octyl)oxy)-4-(sulfonatooxy)phenyl)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-5-yl)oxy)propoxy)-4-oxo-3,7-bis(sulfonatooxy)-4H-chromen-2-yl)-1,2-phenylene Bis(sulfate) (G8C)
1H NMR (400 MHz, DMSO-d6) δ 8.08 (d, J = 2.28 Hz, 1H), 8.03–7.90 (m, 2H), 7.62–7.54 (m, 3H), 7.07 (d, J = 1.88 Hz, 1H), 6.99 (d, J = 1.84 Hz, 1H), 6.74–6.65 (m, 2H), 5.23 (bs, 1H), 4.32–4.21 (m, 4H), 4.03–3.91 (m, 2H), 3.04–2.93 (m, 1H), 2.37–2.20 (m, 3H), 2.05–1.58 (m, 8H), 1.53–0.96 (m, 25H), 0.95–0.84 (m, 7H), 0.82 (d, J = 7.14 Hz, 4H), 0.77 (dd, J = 6.60 Hz, J = 1.82 Hz, 7H), 0.58 (s, 3H). 13C NMR (100 MHz, DMSO-d6) δ 173.00, 159.08, 158.33, 157.01, 153.31, 153.11, 148.79, 146.47, 145.20, 142.95, 140.64, 135.35, 135.31, 124.55, 123.67, 120.89, 120.00, 118.92, 109.60, 100.65, 99.45, 78.09, 68.57, 52.82, 49.60, 41.83, 36.71, 36.30, 35.63, 35.15, 31.41, 31.34, 29.71, 28.90, 28.12, 27.74, 27.34, 25.72, 25.49, 23.83, 23.17, 22.62, 22.36, 20.58, 19.05, 18.53, 11.65. MS (ESI) calculated for C68H77Na7O36S7, [(M – 2Na)]2–, m/z 904.08, found for [(M – 2Na)]2–, 904.35.
Materials for Cell Culture
HT-29 cancer cell line was a kind gift by Dr. Majumdar (Wayne State University). The cells were maintained in 10 cm tissue culture-treated plates (USA Scientific) in the monolayer form in Dulbecco’s modified Eagle’s medium—Nutrient Mixture F-12 (DMEM:F-12) (Gibco) Complete growth medium was prepared by addition of 10% fetal bovine serum (FBS; Gibco) and 1% streptomycin/penicillin (AA; Gibco). The cells were passaged using trypsin-containing ethylenediaminetraacetic acid (EDTA, Gibco) before they reached 70% confluency.
Spheroid Inhibition Assay
For spheroid formation, cells (no more than 6 passages) were plated in nontreated, low-adhesion 96-well plates (USA Scientific) at a cell density ranging from 100 to 300 cells/100 μL/well depending on optimal density needed for cell growth. Cells were plated in stem cell media (SCM) that consisted of DMEM:F12:AA (Gibco), supplemented with 1X B27 (Gibco; 20 ng/mL epidermal growth factor (EGF; Sigma), 10 ng/mL fibroblast growth factor (FGF; Sigma)). After a brief period of incubation, vehicle (control) or test compound at the desired concentration(s) was aliquoted into each well. Following desired growth period (typically 3–7 days), the numbers of spheroids in the range of 50–150 μm diameter were counted using a phase contrast microscope. These were used to calculate percent inhibition compared to control. Growth inhibition was calculated as percent of control (vehicle). The half-maximal inhibition concentration (IC50) was calculated using the standard normalized sigmoidal regression analysis to calculate IC50. A minimum of three replicates were performed for each condition.
Cell Proliferation Assay
Cell proliferation was evaluated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Approximately 2.5 × 103 cells/100 μL/well were plated in 96-well tissue culture-treated plates. After overnight incubation of cells at 37 °C with 5% CO2 vehicle (control), the test compound was added at the desired concentration(s) and allowed to incubate further for 60 to 72 h. Following incubation, 30 μL of 5 mg/mL MTT solution (Sigma) made in phosphate-buffered saline (PBS; Gibco) was added to each well and incubated for a minimum of 2–3 h until crystal formation was observed. The cell culture media and MTT solution were then discarded and 100 μL of dimethyl sulfoxide (DMSO, Sigma) was added dropwise to each well, the mixture gently aspirated to ensure dissolution of crystals and the absorbance at 590 nm recorded in a spectrophotometer. Growth inhibition was calculated as percent of control (vehicle). A minimum of three replicates were performed for each condition.
Materials for Bleeding Studies
Pooled normal human plasma for coagulation assays was purchased from Valley Biomedical (Winchester, VA). Activated partial thromboplastin time reagent containing elagic acid (APTT-LS), thromboplastin-d, and 25 mM CaCl2 were obtained from Fisher Diagnostics (Middletown, VA). CaCl2 was obtained from Haemoscope Corporation (Niles, IL).
Human Plasma Clotting Time Studies
A standard one-stage recalcification assay in a BBL Fibrosystem fibrometer (Becton–Dickinson, Sparks, MD) was used to measure clotting times in human plasma as reported earlier.20,35 For the activated partial thromboplastin time (APTT) assay, a 10 μL of test compound was mixed with 90 μL of citrated human plasma and 100 μL of prewarmed APTT reagent. After incubation for 4 min at 37 °C, clotting was initiated by adding 100 μL of prewarmed 25 mM CaCl2, and the time to clot was noted. The data were fit to a quadratic trend line, which was used to determine the concentration of inhibitor required to double the clotting time. Clotting time in the absence of test compound was determined in a similar manner using 10 μL of high-purity water and was found to be 34 ± 2 s.
Tail Bleeding Studies
Procedures involving mice were performed following approval by VCU IACUC. Wild-type ICR male mice (n ≥ 3; weight 29–50 g) were anesthetized with an intraperitoneal injection of pentobarbital (50 mg/kg). Following anesthesia, the animal was placed on a 37 °C heating pad and a test compound in 50 μL of PBS or vehicle (PBS alone) was injected into the lateral tail vein with a syringe. Two minutes later, the tail tip (1–2 mm) was transected and immediately immersed in an Eppendorf tube containing 1.7 mL of PBS prewarmed to 37 °C. The animal was monitored for a duration of 15 min during which the total bleeding times (defined as the sum of all bleeding events occurring within the 15 min) and the weight of blood loss were recorded. The animal was sacrificed prior to recovering from anesthesia.
Tumor Xenograft Studies
Experiments involving animals were approved by the animal component of the research protocol (ACORP) guideline of the Hunter Holmes Veterans Affairs McGuire Medical Center (Richmond, VA). In vivo CSC enrichment was performed prior to NSGM treatment with FUOX (5-fluorouracil & oxaliplatin), the most common colon cancer chemotherapy combination36,45 as described in our earlier publication.22 Additionally, in some models, dual CSC enrichment was performed by first sorting HT-29 cells for CD133+ and CXCR4+ surface markers using FACS, as described earlier.22 Xenografts were generated by injecting 106 bulk or 105 Dual Hi (CD133+/CXCR4+; dual enrichment) HT-29 cells subcutaneously in NCr nude mice. Once the palpable tumors formed (average >50 mm3), we performed CSC enrichment in vivo by administering FUOX intraperitoneally (IP) weekly for 3 weeks (oxaliplatin (2 mg/kg) and 5-fluorouracil (25 mg/kg) once a week). Following FUOX treatment for 3 weeks, the animals were randomized to receive vehicle, FUOX, NSGM (3 times a week IP or 5 times a week oral gavage), and/or a combination of NSGM + FUOX. IP injections were carried out over several weeks during which tumor volume was measured three times a week with Vernier calipers. At the end of study, the animals were sacrificed as per IACUC-approved methods of euthanasia. The tumor tissue was finely chopped and digested with 400 μg/ml collagenase type IV. Single-cell suspension was filtered with 70 μm cell strainer (BD Biosciences, San Jose, CA) and was subjected to various analyses. These would typically include the CSC phenotype of xenografts including (a) ability to form spheroids in vitro (primary (1°) through quaternary (4°)) and (b) proportion of CSCs via flow cytometry for Dual Hi cells (CD133+/CXCR4+) and/or LGR5+ cells or western blotting for LGR5 antibody.
Briefly, the spheroid formation ability was assessed, as described above. Cells from xenograft were plated in nontreated, low-adhesion plates in stem cell media (SCM) in the presence of EGF and FGF (see above). Following a defined growth period, the numbers of spheroids with 50–150 μm dimensions were counted using a phase contrast microscope. A minimum of three replicates were performed for each condition. For the analysis of CSC markers on cells within the xenograft, single cells were incubated with an appropriate antibody for 30 min at 4 °C and washed with PBS. We used the CXCR4 (anti-mouse CD184)-PE-conjugated clone 2B11 antibody (Cat # 12-9991-82, eBioscience, San Diego, CA; dilution 1:50) and the anti-CD133 mouse mAb (Cat # 130-090-826, Miltenyi Biotec, Auburn, CA; dilution 1:33) for quantifying CXCR4+ and CD133+ cells as well as anti-LGR5 mouse-PE antibody (Cat # TA400001, Origene, Rockville, MA; dilution 1:50). Cell sorting was performed using FACS Aria II High-Speed Cell Sorter (BD Biosciences, San Jose, CA), and data were analyzed using FCS Express 4 flow research edition software.
Studies on Intestinal Organoids
Mice intestinal pieces (2–4 cm) from vehicle, NSGM, or FUOX-treated animals were subjected to chelation and dissociation into crypts using previously described methods.22 Approximately 200 to 500 crypts were resuspended in 100 μL of Matrigel per well of a 96-well plate and overlaid with 100 μL of IntestiCult Organoid Growth Medium (Mouse; Stem Cell Technologies) after allowing for Matrigel polymerization and cultured in a CO2 incubator (37 °C, 5% CO2). The intestinal organoids were counted with a phase contrast microscope after 5 to 7 days of plating.
Human Cancer and Normal Intestinal Organoid Cultures
Human tissue collection was carried out under an Institutional Review Board-approved study (McGuire VAMC, Principal Investigator: Dr. Bhaumik B. Patel; Study Protocol #02140). Human colorectal cancer as well as normal appearing colonic mucosa >2 cm from the tumor was obtained via a punch biopsy soon after resection of the colon cancer. The tissue samples were finely chopped and digested with a mixture of 400 μg/mL Collagenase Type IV (STEMCELL Technologies) and 20 μg/mL hyaluronidase in PBS. Single-cell suspension was filtered with a 70 μm cell strainer. Approximately 500 cells were then resuspended in 100 μL of matrigel and overlayed with 100 μL of IntestiCult Organoid Growth Medium (Human Stem Cell Technologies). For secondary passage, the Matrigel was dissolved with cold PBS and organoids were dissociated with trypsin (0.05%), EDTA (0.1%), and mechanical dissociation as per 2–4° spheroid formation process described above. The treatment of organoids and assessment also followed a similar protocol as for the spheroid inhibition assay above.
Acknowledgments
This work was supported in part by VA Merit Award 5I01BX000837 and 1I01BX004584, and Commonwealth Health Research Board grant 811-04-19 to B.B.P.; NIH grants HL107152, HL141954, CA241951, and HL151333 to U.R.D.; and Massey Cancer Center Pilot Project Fund A35365 to B.B.P. and U.R.D.
Glossary
Abbreviations Used
- CSC
cancer stem cell
- FU
5-fluorouracil
- GAGs
glycosaminoglycans
- Hp
heparin
- HS
heparan sulfate
- IV
intravenous
- MOA
mechanism of action
- NSGMs
nonsaccharide GAG mimetics
- OX
oxaliplatin
- PO
per os (by mouth)
- RTKs
receptor tyrosine kinases
- SC
subcutaneous
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c01511.
Overlay of HS06 and G2.2; structures of G1.4, G4.1, and G2.1; UPLC-ESI-MS analysis of MQD1; UPLC-ESI-MS analysis of MMD1; cytotoxicity potential of G2.2, MQD1, MMD1, and MAD1; synthesis of tosylated intermediates CE-OTs, CP-OTs, and CO-OTs; synthesis of intermediate 18; RP-IP UPLC ESI-MS of G2.2; RP-IP UPLC ESI-MS of G2C; RP-IP UPLC ESI-MS of G5C; RP-IP UPLC ESI-MS of G8C; cytotoxicity potential of G2.2, G2C, G5C, and G8C; analysis of toxicity at the body weight level (G2C and FUOX); tumor growth curve for oral G2C; analysis of toxicity at the body weight level; and effect of G2.2 (IP) and G2C (PO) treatments on mouse blood chemistry (PDF)
Molecular formula strings (CSV)
Author Contributions
∇ S.M., O.R., C.O.H., and R.B. contributed equally to this work. These authors got permission from the corresponding authors to list their name first in their CVs.
The authors declare no competing financial interest.
Supplementary Material
References
- Gandhi N. S.; Mancera R. L. The structure of glycosaminoglycans and their interaction with proteins. Chem. Biol. Drug Des. 2008, 72, 455–482. 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- Rabenstein D. L. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 2002, 19, 312–331. 10.1039/b100916h. [DOI] [PubMed] [Google Scholar]
- Capila I.; Linhardt R. J. Heparin-protein interactions. Angew. Chem. Int. Ed. 2002, 41, 391–412. . [DOI] [PubMed] [Google Scholar]
- Sasisekharan R.; Shriver Z.; Venkataraman G.; Narayanasami U. Role of heparan-sulfate glycosaminoglycans in cancer. Nat. Rev. Cancer 2002, 2, 521–528. 10.1038/nrc842. [DOI] [PubMed] [Google Scholar]
- Afratis N.; Gialeli C.; Nikitovic D.; Tsegenidis T.; Karousou E.; Theocharis A. D.; Pavão M. S.; Tzanakakis G. N.; Karamanos N. K. Glycosaminoglycans: key players in cancer cell biology and treatment. FEBS J. 2012, 279, 1177–1197. 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- Desai U. R. New antithrombin–based anticoagulants. Med. Res. Rev. 2004, 24, 151–181. 10.1002/med.10058. [DOI] [PubMed] [Google Scholar]
- Farrugia B. L.; Lord M. S.; Melrose J.; Whitelock J. M. The role of heparan sulfate in inflammation, and the development of biomimetics as anti-inflammatory strategies. J. Histochem. Cytochem. 2018, 66, 321–336. 10.1369/0022155417740881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maïza A.; Chantepie S.; Vera C.; Fifre A.; Huynh M. B.; Stettler O.; Ouidja M. O.; Papy-Garcia D. The role of heparan sulfates in protein aggregation and their potential impact on neurodegeneration. FEBS Lett. 2018, 592, 3806–3818. 10.1002/1873-3468.13082. [DOI] [PubMed] [Google Scholar]
- Iravani S.; Varma R. S. Important roles of oligo- and polysaccharides against SARS-CoV-2: Recent advances. Appl. Sci. 2021, 11, 3512. 10.3390/app11083512. [DOI] [Google Scholar]
- Mende M.; Bednarek C.; Wawryszyn M.; Sauter P.; Biskup M. B.; Schepers U.; Bräse S. Chemical synthesis of glycosaminoglycans. Chem. Rev. 2016, 116, 8193–8255. 10.1021/acs.chemrev.6b00010. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan N. V.; Nagarajan B.; Desai U. R. So you think computational approaches for understanding glycosaminoglycan–protein interactions are too dry and too rigid? Think again!. Curr. Opin. Struct. Biol. 2018, 50, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G.; Tang B. Advanced delivery strategies facilitating oral absorption of heparins. Asian J. Pharm. Sci. 2020, 15, 449–460. 10.1016/j.ajps.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson G. T.; Desai U. R. Interaction of Designed sulfated flavanoids with antithrombin: Lessons on the design of organic activators. J. Med. Chem. 2002, 45, 4460–4470. 10.1021/jm020132y. [DOI] [PubMed] [Google Scholar]
- Desai U. R. The promise of sulfated synthetic small molecules as modulators of glycosaminoglycan function. Future Med. Chem. 2013, 5, 1363–1366. 10.4155/fmc.13.117. [DOI] [PubMed] [Google Scholar]
- Al-Horani R. A.; Liang A.; Desai U. R. Designing non-saccharide, allosteric activators of antithrombin for accelerated inhibition of factor Xa. J. Med. Chem. 2011, 54, 6125–6138. 10.1021/jm2008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan B.; Sankaranarayanan N. V.; Patel B. B.; Desai U. R. A molecular dynamics-based algorithm for evaluating the glycosaminoglycan mimicking potential of synthetic, homogenous, sulfated small molecules. PLoS One 2017, 12, e0171619 10.1371/journal.pone.0171619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu P. S.; Abdel Aziz M. H.; Sarkar A.; Mehta A. Y.; Zhou Q.; Desai U. R. Designing allosteric regulators of thrombin. Exosite 2 features multiple sub-sites that can be targeted by sulfated small molecules for inducing inhibition. J. Med. Chem. 2013, 56, 5059–5070. 10.1021/jm400369q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu P. S.; Liang A.; Mehta A. Y.; Abdel Aziz M. A.; Zhou Q.; Desai U. R. Rational design of potent, small, synthetic allosteric inhibitors of thrombin. J. Med. Chem. 2011, 54, 5522–5531. 10.1021/jm2005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Horani R. A.; Ponnusamy P.; Mehta A. Y.; Gailani D.; Desai U. R. Sulfated pentagalloylglucoside is a potent, allosteric, and selective inhibitor of factor XIa. J. Med. Chem. 2013, 56, 867–878. 10.1021/jm301338q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Horani R. A.; Abdelfadiel E. I.; Afosah D. K.; Morla S.; Sistla J. C.; Mohammed B.; Martin E. J.; Sakagami M.; Brophy D. F.; Desai U. R. A synthetic heparin mimetic that allosterically inhibits factor XIa and reduces thrombosis in vivo without enhanced risk of bleeding. J. Thromb. Haemostasis 2019, 17, 2110–2122. 10.1111/jth.14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. J.; Karuturi R.; Al-Horani R. A.; Baranwal S.; Patel J.; Desai U. R.; Patel B. B. Synthetic, non-saccharide glycosaminoglycan mimetics selectively target colon cancer stem cells. ACS Chem. Biol. 2014, 9, 1826–1833. 10.1021/cb500402f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothello R. S.; Patel N. J.; Sharon C.; Abdelfadiel E. I.; Morla S.; Brophy D. B.; Lippmann R. H.; Desai U. R.; Patel B. B. A Unique Nonsaccharide Mimetic of Heparin Hexasaccharide Inhibits Colon Cancer Stem Cells via p38 MAP Kinase Activation. Mol. Cancer Ther. 2019, 18, 51–61. 10.1158/1535-7163.MCT-18-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulloy B.; Forster M. J. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 2000, 10, 1147–1156. 10.1093/glycob/10.11.1147. [DOI] [PubMed] [Google Scholar]
- Irby D.; Du C.; Li F. Lipid-drug conjugate for enhancing drug delivery. Mol. Pharmaceutics 2017, 14, 1325–1338. 10.1021/acs.molpharmaceut.6b01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman A.; Riaz M.; Hindle M.; Desai U. R. Rapid, high-yielding microwave-assisted per-sulfation of organic scaffolds. Tetrahedron Lett. 2007, 48, 6754–6758. 10.1016/j.tetlet.2007.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Horani R. A.; Karuturi R.; Verespy S. III; Desai U. R.. Synthesis of glycosaminoglycan mimetics through sulfation of polyphenols. In Methods in Molecular Biology; Springer: New York, 2015; Vol. 1229, pp 49–67. 10.1007/978-1-4939-1714-3_7. [DOI] [PubMed] [Google Scholar]
- Ishiguro T.; Ohata H.; Sato A.; Yamawaki K.; Enomoto T.; Okamoto K. Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications. Cancer Sci. 2017, 108, 283–289. 10.1111/cas.13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar S. S.; Yu Y.; Nautiyal J.; Patel B. B.; Majumdar A. P. The Wnt/beta-catenin pathway regulates growth and maintenance of colonospheres. Mol. Cancer 2010, 9, 212. 10.1186/1476-4598-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon C.; Baranwal S.; Patel N. J.; Rodriguez-Agudo D.; Pandak W. M.; Majumdar A. P.; Krystal G.; Patel B. B. Inhibition of insulin-like growth factor receptor/AKT/mammalian target of rapamycin axis targets colorectal cancer stem cells by attenuating mevalonate-isoprenoid pathway in vitro and in vivo. Oncotarget 2015, 6, 15332–15347. 10.18632/oncotarget.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouktaib M.; Lebrun S.; Atmani A.; Rolando C. Hemisynthesis of all the O-monomethylated analogues of quercetin including the major metabolites, through selective protection of phenolic functions. Tetrahedron 2002, 58, 10001–10009. 10.1016/S0040-4020(02)01306-6. [DOI] [Google Scholar]
- Rao K. V.; Owoyale J. A. Partial methylation of quercetin: direct synthesis of tamarixetin, ombuin and ayanin. J. Heterocycl. Chem. 1976, 13, 1293–1295. 10.1002/jhet.5570130629. [DOI] [Google Scholar]
- Wuputra K.; Ku C. C.; Kato K.; Wu D. C.; Saito S.; Yokoyama K. K. Translational models of 3-D organoids and cancer stem cells in gastric cancer research. Stem Cell Res. Ther. 2021, 12, 492 10.1186/s13287-021-02521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonar R. A.; Lippi G.; Favaloro E. J.. Overview of hemostasis and thrombosis and contribution of laboratory testing to diagnosis and management of hemostasis and thrombosis Disorders. In Methods in Molecular Biology; Springer: New York, 2017; Vol. 1646, pp 3–27. 10.1007/978-1-4939-7196-1_1. [DOI] [PubMed] [Google Scholar]
- Johansen P. B.; Henriksen L.; Andresen P. R.; Lauritzen B.; Jensen K. L.; Juhl T. N.; Tranholm M. Automated registration of tail bleeding in rats. Thromb. Haemostasis 2008, 99, 956–962. 10.1160/TH07-12-0738. [DOI] [PubMed] [Google Scholar]
- Mehta A. Y.; Mohammed B. M.; Martin E. J.; Brophy D. F.; Gailani D.; Desai U. R. Allosterism-based simultaneous, dual anticoagulant and antiplatelet action: allosteric inhibitor targeting the glycoprotein Ibα-binding and heparin-binding site of thrombin. J. Thromb. Haemostasis 2016, 14, 828–838. 10.1111/jth.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson B.; Carlsson G.; Machover D.; Petrelli N.; Roth A.; Schmoll H. J.; Tveit K. M.; Gibson F. A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin. Colorectal Cancer. 2015, 14, 1–10. 10.1016/j.clcc.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Ferro V.; Liu L.; Johnstone K. D.; Wimmer N.; Karoli T.; Handley P.; Rowley J.; Dredge K.; Li C. P.; Hammond E.; Davis K.; Sarimaa L.; Harenberg J.; Bytheway I. Discovery of PG545: a highly potent and simultaneous inhibitor of angiogenesis, tumor growth, and metastasis. J. Med. Chem. 2012, 55, 3804–3813. 10.1021/jm201708h. [DOI] [PubMed] [Google Scholar]
- Dredge K.; Brennan T. V.; Hammond E.; Lickliter J. D.; Lin L.; Bampton D.; Handley P.; Lankesheer F.; Morrish G.; Yang Y.; Brown M. P.; Millward M. A Phase I study of the novel immunomodulatory agent PG545 (pixatimod) in subjects with advanced solid tumours. Br. J. Cancer 2018, 118, 1035–1041. 10.1038/s41416-018-0006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trials located from the Clinical Trials web resource www.clinicaltrials.gov.
- Chhabra M.; Wimmer N.; He Q. Q.; Ferro V. Development of improved synthetic routes to pixatimod (PG545), a sulfated oligosaccharide-steroid conjugate. Bioconjugate Chem. 2021, 32, 2420–2431. 10.1021/acs.bioconjchem.1c00453. [DOI] [PubMed] [Google Scholar]
- Lanzi C.; Cassinelli G. Receptor tyrosine kinases and heparan sulfate proteoglycans: Interplay providing anticancer targeting strategies and new therapeutic opportunities. Biochem. Pharmacol. 2020, 178, 114084 10.1016/j.bcp.2020.114084. [DOI] [PubMed] [Google Scholar]
- Ferguson H. R.; Smith M. P.; Francavilla C. Fibroblast growth factor receptors (FGFRs) and noncanonical partners in cancer signaling. Cells 2021, 10, 1201. 10.3390/cells10051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp L. E.; Mulloy B.; Gherardi E. Signalling by HGF/SF and Met: the role of heparan sulphate co-receptors. Biochem. Soc. Trans. 2006, 34, 414–417. 10.1042/BST0340414. [DOI] [PubMed] [Google Scholar]
- Radwan A. A.; Alanazi F. K. Design and synthesis of new cholesterol-conjugated 5-fluorouracil: A novel potential delivery system for cancer treatment. Molecules 2014, 19, 13177–13187. 10.3390/molecules190913177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radwan A. A.; Alanazi F. K. Targeting cancer using cholesterol conjugates. Saudi Pharm. J. 2014, 22, 3–16. 10.1016/j.jsps.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N. J.; Sharon C.; Baranwal C.; Boothello R. S.; Desai U. R.; Patel B. B. Heparan sulfate hexasaccharide selectively inhibits cancer stem cells by activating p38 MAP kinase. Oncotarget 2016, 7, 84608–84622. 10.18632/oncotarget.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothello R. S.; Sankaranarayanan N. V.; Sistla J. C.; Nagarajan B.; Sharon C.; Chittum J. E.; Niyaz R. Y.; Roy S.; Nandi A.; O’Hara C. P.; Navaz Gangji R.; Afosah D. K.; Ongolu R.; Patel B. B.; Desai U. R. Glycan modulation of insulin-like growth factor–1 receptor. Angew. Chem., Int. Ed. 2022, 61, e202211320 10.1002/anie.202211320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.