Abstract
The initial stages of dental plaque formation involve the adherence of early colonizing organisms such as Streptococcus gordonii and Actinomyces naeslundii to the saliva-coated tooth surface and to each other. The S. gordonii surface proteins SspA and SspB are known to play a role in adherence to salivary proteins and mediate coaggregation with other bacteria. Coaggregation is the adhesin receptor-mediated interaction between genetically distinct cell types and appears to be ubiquitous among oral isolates. To define the function of SspA and SspB separately on the surface of their natural host, we constructed and analyzed the coaggregation properties of an isogenic sspB mutant of S. gordonii DL1, an sspAB double mutant, and a previously described sspA mutant. A. naeslundii strains have been previously classified into six coaggregation groups based on the nature of their coaggregations with S. gordonii DL1 and other oral streptococci. Coaggregation assays with the sspA and sspB mutants showed that SspA and SspB are the streptococcal proteins primarily responsible for defining these coaggregation groups and, thus, are highly significant in the establishment of early dental plaque. SspA exhibited two coaggregation-specific functions. It participated in lactose-inhibitable and -noninhibitable interactions, while SspB mediated only lactose-noninhibitable coaggregations. Accordingly, the sspAB double mutant lacked these functions and allowed us to detect a third coaggregation interaction with one of these organisms. These proteins may play an important role in development of S. gordonii-A. naeslundii communities in early dental plaque. Understanding these adhesin proteins will aid investigations of complex microbial communities that characterize periodontal diseases.
A property of nearly all human oral bacteria is the ability to coaggregate with members of other oral bacterial genera or species (13, 17). These interactions are mediated by protein adhesins on one cell type and cognate receptors on genetically distinct partner cell types. Coaggregations are thought to be important in the development of the oral biofilm, as the ability of cells to bind the tooth-associated biofilm affords an opportunity to join the developing microbial community. For example, we recently demonstrated that the ability of two species of oral bacteria to interact facilitates luxuriant growth in a saliva-coated flow cell, an environment in which neither organism grows independently (21).
The initial stages of dental plaque formation involve the adherence of early colonizing organisms such as Streptococcus gordonii and Actinomyces naeslundii to the saliva-coated tooth surface and to each other. SspA and SspB, members of the antigen I/II family, are among the proteins involved in interactions between these organisms (11). These proteins are known to have multiple functions, including binding to human salivary agglutinin, collagen, and certain A. naeslundii strains (5, 6, 10, 12), suggesting that they are important for the development of the plaque community. SspB is also known to mediate interactions with Porphyromonas gingivalis (2). We have recently identified sspAB as genes that are induced in response to saliva (7), enforcing the importance of the products of these genes in the oral microbial community.
In previous studies of coaggregation between S. gordonii and A. naeslundii, we have defined six coaggregation groups of A. naeslundii strains (groups A through F) based on the heat and protease sensitivity of the surface molecules responsible for interactions and on the ability of spontaneous mutants to coaggregate with members of various groups of partner strains (14). Here we demonstrate that the ability of A. naeslundii strains to interact with S. gordonii proteins SspA and SspB almost entirely defines the coaggregation groups to which these strains are assigned. We describe the construction of an sspB mutant and an sspAB double mutant of S. gordonii strain DL1. By examining the coaggregation properties of these mutants and a previously described sspA mutant (12) with strains of A. naeslundii, we are the first to report independent coaggregation functions of SspA and SspB with respect to coaggregation with actinomyces strains and to detect a previously unrecognized lactose-inhibitable interaction between S. gordonii and members of actinomyces coaggregation group D (14).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. gordonii DL1 was grown in brain heart infusion (BHI) medium (Becton Dickinson Co., Sparks, Md.) anaerobically using the GasPak System (Becton Dickinson Co.) at 37°C. Escherichia coli cultures were grown in Luria-Bertani broth at 37°C with shaking. For the following organisms, antibiotics (Sigma Chemical Co., St. Louis, Mo.) were used at the indicated concentrations (in micrograms per milliliter): for E. coli, ampicillin, 100, and kanamycin, 100; for S. gordonii, kanamycin, 500, and erythromycin, 10. All streptococcus and actinomyces strains used as coaggregation partners of S. gordonii DL1 have been described (1, 4, 16) and were grown in medium containing (per liter): tryptone (5.0 g), yeast extract (5.0 g), Tween 80 (0.5 ml), and glucose (2.0 g) and buffered to pH 7.5 with K2HPO4 (5.0 g) (19). Fusobacterium nucleatum ATCC 10953 was grown in BHI medium supplemented with 0.25% ammonium glutamate. Veillonella atypica and Veillonella dispar were grown in modified Schaedler's medium (3) in which lactic acid (0.1 M) was substituted for glucose.
Mutagenesis of sspB.
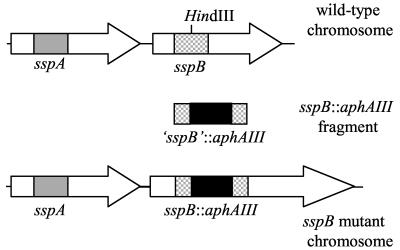
An isogenic sspB mutant of S. gordonii DL1 was constructed by gene replacement using a fragment of sspB that had been insertionally inactivated with the kanamycin resistance gene, aphAIII (Fig. 1). The sspB fragment was PCR amplified from genomic DNA purified from S. gordonii DL1 using primers based on the sequence of sspB from S. gordonii M5 (6). Because sspA and sspB share a high level of identity over much of the length of their sequences (5), the region of sspB chosen for targeting is the region known to be divergent among members of the antigen I/II family (11). The sequences of the primers F64 (5′-AGCGGCTTATCAGACAGAACTAGC-3′) and B74 (5′-AACAGGTTTCGTCGGAACCC-3′) correspond to bases 1697 to 1720 and bases 2776 to 2795, respectively, of the S. gordonii M5 sspB sequence (accession number U40026). The coding region of sspB starts at base 513 of this sequence. A 1,099-bp fragment was PCR amplified from S. gordonii DL1 and cloned into pUC18, yielding the plasmid pUC18-1099. The cloned S. gordonii DL1 sspB fragment was sequenced and found to be 99% identical to the corresponding region of S. gordonii M5. The kanamycin resistance gene, aphAIII, was PCR amplified from the plasmid pDL276 (8) using primers F3 (5′-GGGAAGCTTTGTGGTTTCAAAATCGGCTCC-3′) and B4 (5′-GGGAAGCTTGTATGGACAGTTGCGGATGTACTTC-3′), which incorporate HindIII sites (in boldface type) at the ends of the PCR product, and cloned into a unique HindIII site at position 619 of the sspB fragment. The sspB::aphAIII fragment was excised from the resulting plasmid (pUC18-1099::Kmr) and gel purified to eliminate the ampicillin resistance gene-containing vector. The linear DNA fragment was then transformed into S. gordonii DL1 and the sspA mutant OB220 (12) to generate the sspB (PK3246) and sspAB (PK3247) mutants, respectively. Transformants were plated on BHI medium containing kanamycin to select for isolates that had undergone double recombination. The mutations were verified by Southern hybridizations using the sspB fragment and kanamycin resistance gene as probes.
FIG. 1.
Strategy for mutagenesis of sspB. The gray and checkered boxes within sspA and sspB, respectively, represent the regions of divergent sequence of these genes. The remainder of the genes share 90% identity (5). The divergent region from sspB was PCR amplified and cloned into pUC18. The kanamycin resistance gene (aphAIII) was PCR amplified and cloned into the HindIII site in the sspB fragment. The sspB::aphAIII fragment was excised from the vector, gel purified, and transformed into strain DL1 to construct the sspB mutant PK3246 and into strain OB220 (12) to generate the sspAB double mutant PK3247.
Immunoblot analysis.
Surface proteins from S. gordonii strains were prepared by treatment with mutanolysin as described (5) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a 4 to 12% Bis-Tris gradient gel and MOPS (morpholinepropanesulfonic acid) buffer system (Invitrogen, Carlsbad, Calif.). Proteins were transferred to a nitrocellulose membrane, which was then blocked overnight with a solution of 10 mM Tris, 150 mM NaCl, 0.05% Tween 20, and 1.0% bovine serum albumin. The blot was then treated with a 1:2,000 dilution of rabbit anti-SspB immunoglobulin G (kindly supplied by D. Demuth, University of Pennsylvania) for 1 h, washed, and incubated for 30 min with a 1:3,000 dilution of alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Bio-Rad, Hercules, Calif.). Detection was accomplished using Western Blue substrate (Promega, Madison, Wis.).
Coaggregation assays.
The visual assay used to score coaggregation has been described previously (15). Briefly, cells were washed three times and suspended in coaggregation buffer (1 mM Tris [pH 8.0], 150 mM NaCl, 0.1 mM CaCl2, 0.1 mM MgCl2, 0.02% Na3N) to a cell density of 260 Klett units (about 109 cells/ml) using a Klett-Summerson colorimeter (Klett Manufacturing Co. Inc., New York, N.Y.). An equal volume (0.15 ml) of each cell type was combined in a glass tube (10 by 75 mm) and vortexed for 10 s. The coaggregation score was assigned as follows. A score of 4 indicates rapid and complete settling of large coaggregates, leaving a clear supernatant. A score of 3 is given when large coaggregates settle rapidly but the supernatant remains slightly cloudy; a score of 2 is given when coaggregates are formed immediately but remain suspended in a turbid background; a score of 1 indicates detectable but finely dispersed coaggregates; a negative coaggregation score (−) is given to an evenly turbid suspension, that is, one that does not change appearance after the two suspensions are vortexed. For testing inhibition by lactose, 50 μl of a 1 M lactose solution was added to the cell suspensions.
RESULTS AND DISCUSSION
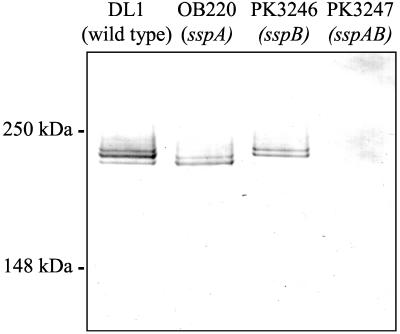
To identify independent coaggregation functions of SspA and SspB, we used the sspA mutant OB220 of S. gordonii DL1 (12) and we constructed an sspB mutant of S. gordonii DL1 and an sspAB double mutant carrying an antibiotic resistance cassette inserted in each gene. Immunoblot analysis using anti-SspB antiserum confirmed the absence of the SspA and SspB antigens in surface preparations of appropriate S. gordonii mutants (Fig. 2). The anti-SspB antiserum reacted with a doublet band for each protein (D. Demuth, personal communication). The upper bands correspond to 215 and 205 kDa for SspA and SspB, respectively (5, 6). In the wild-type strain, the doublet bands overlap, giving the appearance of a triplet band. The high level of identity between SspA and SspB is reflected by the reaction of anti-SspB antiserum with both proteins. Strain OB220 (sspA::ermAM) (12) possessed only the faster-migrating SspB. Strain PK3246 (sspB::aphAIII) expressed only SspA, and PK3247 (sspA::ermAM sspB::aphAIII) had neither protein. Thus, the single insertion mutants expressed either SspA or SspB and the double mutant expressed neither protein. Consequently, these mutants allowed us to separate the functions of SspA and SspB. Using an sspA mutant and a strain carrying mutations in both sspA and sspB, others have shown that the proteins encoded by these genes are involved in coaggregation with certain A. naeslundii strains (5, 12). The sspA mutant, OB220, shows less than 50% of wild-type ability to coaggregate with A. naeslundii T14V and PK606. In addition, a double mutant, which has an insertion of ermAM in sspA and deletions of portions of both sspA and sspB, has a further reduction of coaggregation with these actinomyces.
FIG. 2.
Immunoblot of S. gordonii surface proteins prepared from wild-type (DL1) and mutant S. gordonii strains reacted with antibody to SspB. Strain genotypes are indicated at the top. Lanes contained 10 μg of protein. Positions of molecular mass standards are shown at the left.
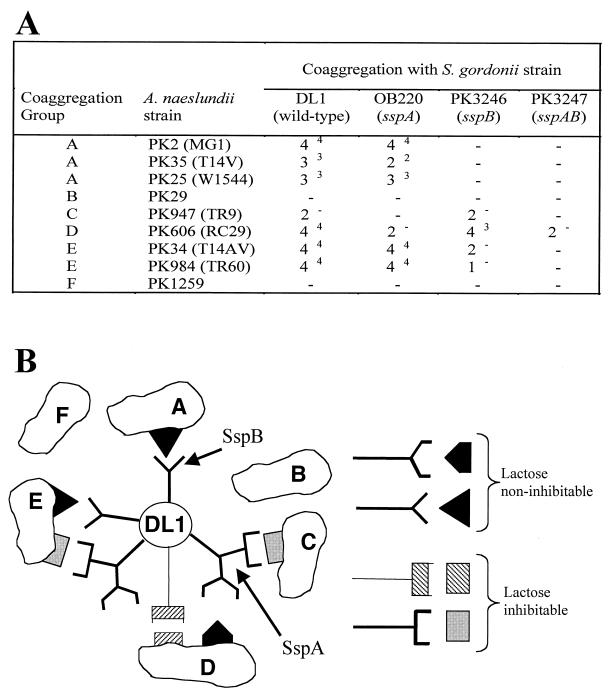
We analyzed the previously described sspA mutant (OB220) as well as the sspB (PK3246) and sspAB (PK3247) mutants described here with respect to their abilities to coaggregate with representative members of the six coaggregation groups of A. naeslundii strains (14). These six groups encompass more than 90% of the more than 300 strains of oral A. naeslundii tested (13, 14). The parent strain, S. gordonii DL1, coaggregates with four of the six coaggregation groups (groups A, C, D and E) (Fig. 3A). Only the coaggregation with strain PK947, the actinomyces group C representative, is lactose inhibitable. The sspA mutant was unaffected in coaggregation with A. naeslundii strains in coaggregation groups A and E. However, it was unable to coaggregate with group C and exhibited reduced and lactose-inhibitable coaggregation with group D. The sspB mutation resulted in a complete loss in the ability to coaggregate with the strains in group A, caused no change in coaggregation with groups C and D, and exhibited lactose-inhibitable coaggregation with strains of group E. Loss of both SspA and SspB in the sspAB double mutant resulted in the inability to coaggregate with any group except group D, with which it retained a weakened lactose-inhibitable coaggregation.
FIG. 3.
(A) Coaggregation of S. gordonii wild-type (DL1) and mutant strains with A. naeslundii strains and Actinomyces sp. strain WVA963 strain PK1259. Coaggregation group designations are shown at left. Coaggregation scores range from no coaggregation (−) to strongest coaggregation (4) (see text for details), in which large coaggregates settle out of suspension immediately, leaving a clear supernatant. Superscripts indicate the coaggregation score after addition of lactose. (B) Diagrammatic model of interactions of SspA and SspB with the six actinomyces coaggregation groups. At right, complementary sets of symbols used to represent interactions are depicted. The symbols with stems (left side) represent protein adhesins. Their cognates without stems (right side) represent receptors.
The above data are depicted in a diagrammatic model (Fig. 3B). The interactions between S. gordonii DL1 and the three coaggregation group A strains of A. naeslundii require SspB. This is consistent with the work of others showing that, when expressed on the surface of Enterococcus faecalis, SspB can mediate interactions with A. naeslundii T14V, one of the three coaggregation group A actinomyces (10). The coaggregation between S. gordonii DL1 and the group C A. naeslundii strain is lactose inhibitable. The sspA mutation abolished coaggregation with group C strain A. naeslundii PK947, suggesting that SspA is the lactose-inhibitable adhesin involved in this reaction. SspA also appeared to be involved in coaggregation with group D A. naeslundii PK606. Compared to strain DL1, coaggregation between the sspA mutant and PK606 was significantly weaker. In addition, the remaining activity was lactose inhibitable (cross-hatched rectangular symbols in Fig. 3B), indicating that S. gordonii possesses an additional adhesin that is recognized by this actinomyces. A lactose-inhibitable interaction between these strains has been reported previously (14). The absence of any contribution to this coaggregation by SspB was confirmed by the sspAB double mutant, which retained the lactose-inhibitable coaggregation observed between the sspA mutant and PK606. These results are simplest to interpret by proposing that SspA is a multifunctional adhesin (2, 11) depicted as a branched symbol in Fig. 3B. The protein may possess one domain that mediates strong lactose-noninhibitable coaggregation (coaggregation score of 4 with group D actinomyces). Another domain of SspA appears to mediate weaker lactose-inhibitable coaggregations (coaggregation score of 2 with group C and E strains).
Coaggregation results with the group E A. naeslundii strains indicated that both SspA and SspB were involved in coaggregation with these strains. SspB was required for a strong lactose-noninhibitable reaction as was observed in the coaggregation with group A actinomyces. Only the putative domain of SspA mediating weak lactose-inhibitable coaggregation appears to be involved in interactions with group E actinomyces. This lactose-inhibitable interaction between strain DL1 and group E was masked by the strength of the SspB-mediated lactose-noninhibitable interaction with the wild-type strain. Consequently, it was not recognized in previous studies (14). This lactose-inhibitable interaction was undetectable until an sspB mutant was constructed, allowing separation of SspA and SspB functions.
S. gordonii DL1 is also known to express an adhesin that allows a lactose-inhibitable intrageneric coaggregation with other streptococcus species, including Streptococcus oralis strains 34 and C104. Consistent with the results seen with the lactose-inhibitable interactions with the A. naeslundii group C strains, the sspA and sspAB mutants were unable to coaggregate with S. oralis strains 34 and C104. The sspB mutant retains the coaggregation phenotype of the wild type in these intrageneric coaggregations. Thus, sspA appears to mediate lactose-inhibitable coaggregation between S. gordonii and both streptococci and actinomyces.
In addition to participating in streptococcal adhesin-mediated coaggregation with A. naeslundii, S. gordonii can participate in coaggregations with other organisms in which the partner organism provides an adhesin that interacts with a cognate carbohydrate receptor present on S. gordonii. To determine if the coaggregation defect caused by the absence of SspA and/or SspB might affect the ability of S. gordonii to participate in coaggregations requiring its receptors, we tested coaggregation with F. nucleatum ATCC 10953, V. atypica PK1910, and V. dispar PK1950. The coaggregations between these organisms and all three S. gordonii mutants were the same as with the wild type (data not shown), suggesting that the sspA and/or sspB mutants do not have a generalized cell surface defect such as a change in cell surface charge or hydrophobicity.
Others have shown that SspA and SspB are involved in binding to immobilized salivary agglutinin and collagen (5, 6, 10, 12) as well as coadhesion with immobilized cells using actinomyces (5, 12) strains, P. gingivalis (18), and Candida albicans (9) as partner organisms. Here we examine interactions between suspended S. gordonii bearing these proteins and actinomyces strains also in suspension. This work extends previous studies by utilizing isogenic mutants of both sspA and sspB and by including representatives of each of the six previously described coaggregation groups of A. naeslundii. The results presented here have allowed us to separate the functions of SspA and SspB and to ascribe to these proteins the previously identified coaggregation properties of whole cells of S. gordonii (14). Since streptococci and actinomyces constitute more than 40% of the bacteria in dental plaque from healthy sites (20), the absence of SspA or SspB on a streptococcal cell surface could severely impair its ability to be retained under the stress of salivary flow in the human oral environment. These proteins are not only essential in interactions of S. gordonii and other early colonizing bacteria but they are also important in binding to salivary agglutinin and collagen (5, 6, 10, 12), suggesting a central role in the development of the oral biofilm community.
ACKNOWLEDGMENTS
We thank R. Andersen for her technical expertise, D. Demuth for providing the antiserum used in immunoblots and R. Lamont, D. Demuth, and H. Jenkinson for numerous helpful discussions. We thank J. Cisar, D. Blehert, and J. Foster for helpful comments in preparation of the manuscript.
REFERENCES
- 1.Andersen R N, Ganeshkumar N, Kolenbrander P E. Helicobacter pylori adheres selectively to Fusobacterium spp. Oral Microbiol Immunol. 1998;13:51–54. doi: 10.1111/j.1399-302x.1998.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 2.Brooks W, Demuth D R, Gil S, Lamont R J. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Celesk R A, London J. Attachment of oral Cytophaga species to hydroxyapatite-containing surfaces. Infect Immun. 1980;29:768–777. doi: 10.1128/iai.29.2.768-777.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cisar J O, Kolenbrander P E, McIntire F C. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect Immun. 1979;24:742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demuth D R, Duan Y, Brooks W, Holmes A R, McNab R, Jenkinson H F. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–413. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- 6.Demuth D R, Golub E E, Malamud D. Streptococcal-host interactions. Structural and functional analysis of a Streptococcus sanguis receptor for a human salivary glycoprotein. J Biol Chem. 1990;265:7120–7126. [PubMed] [Google Scholar]
- 7.Dû L D, Kolenbrander P E. Identification of saliva-regulated genes of Streptococcus gordonii DL1 by differential display using random arbitrarily primed PCR. Infect Immun. 2000;68:4834–4837. doi: 10.1128/iai.68.8.4834-4837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny G M, Lee L N, LeBlanc D J. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol. 1991;57:1194–1201. doi: 10.1128/aem.57.4.1194-1201.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes A R, Cannon R D, Jenkinson H F. Interactions of Candida albicans with bacteria and salivary molecules in oral biofilms. J Ind Microbiol. 1995;15:208–213. doi: 10.1007/BF01569827. [DOI] [PubMed] [Google Scholar]
- 10.Holmes A R, Gilbert C, Wells J M, Jenkinson H F. Binding properties of Streptococcus gordonii SspA and SspB (antigen I/II family) polypeptides expressed on the cell surface of Lactococcus lactis MG1363. Infect Immun. 1998;66:4633–4639. doi: 10.1128/iai.66.10.4633-4639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 12.Jenkinson H F, Terry S D, McNab R, Tannock G W. Inactivation of the gene encoding surface protein SspA in Streptococcus gordonii DL1 affects cell interactions with human salivary agglutinin and oral actinomyces. Infect Immun. 1993;61:3199–3208. doi: 10.1128/iai.61.8.3199-3208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolenbrander P E. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 14.Kolenbrander P E. Surface recognition among oral bacteria: multigeneric coaggregations and their mediators. Crit Rev Microbiol. 1989;17:137–159. doi: 10.3109/10408418909105746. [DOI] [PubMed] [Google Scholar]
- 15.Kolenbrander P E, Andersen R N. Characterization of Streptococcus gordonii (S. sanguis) PK488 adhesin-mediated coaggregation with Actinomyces naeslundii PK606. Infect Immun. 1990;58:3064–3072. doi: 10.1128/iai.58.9.3064-3072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolenbrander P E, Andersen R N. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect Immun. 1989;57:3204–3209. doi: 10.1128/iai.57.10.3204-3209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolenbrander P E, Andersen R N, Kazmerzak K M, Palmer J R J. Coaggregation and coadhesion in oral biofilms. In: Allison D G, Gilbert P, Lappin-Scott H M, Wilson M, editors. Community structure and co-operation in biofilms. Cambridge, United Kingdom: Cambridge University Press; 2000. pp. 65–85. [Google Scholar]
- 18.Lamont R J, Gil S, Demuth D R, Malamud D, Rosan B. Molecules of Streptococcus gordonii that bind to Porphyromonas gingivalis. Microbiology. 1994;140:867–872. doi: 10.1099/00221287-140-4-867. [DOI] [PubMed] [Google Scholar]
- 19.Maryanski J H, Wittenberger C L. Mannitol transport in Streptococcus mutans. J Bacteriol. 1975;124:1475–1481. doi: 10.1128/jb.124.3.1475-1481.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore W E, Moore L V. The bacteria of periodontal diseases. Periodontology. 2000;5:66–77. doi: 10.1111/j.1600-0757.1994.tb00019.x. [DOI] [PubMed] [Google Scholar]
- 21.Palmer R J, Kazmerzak K M, Hansen M C, Kolenbrander P E. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]