Abstract
In the past decade, growing interest in microribonucleic acids (miRNAs) has catapulted these small, non-coding nucleic acids to the forefront of biomarker research. Advances in scientific knowledge have made it clear that miRNAs play a vital role in regulating cellular physiology throughout the human body. Perturbations in miRNA signaling have also been described in a variety of pediatric conditions – from cancer, to renal failure, to traumatic brain injury. Likewise, the number of studies across pediatric disciplines that pair patient miRNA-omics with longitudinal clinical data are growing. Analysis of these voluminous, multivariate data sets require understanding of pediatric phenotypic data, data science, and genomics. Use of machine learning techniques to aid in biomarker detection have helped decipher background noise from biologically meaningful changes in the data. Further, emerging research suggests that miRNAs may have potential as therapeutic targets for pediatric precision care. Here, we review current miRNA biomarkers of pediatric diseases and studies that have combined machine learning techniques, miRNA-omics, and patient health data to identify novel biomarkers and potential therapeutics for pediatric diseases.
Keywords: Biomarkers, Epigenetics, Pediatrics, Machine Learning, microRNA, Therapeutics
Category of study: Review
Introduction
MiRNA Physiology
Two decades ago, it was not clear that miRNAs played an integral role in human physiology, nor that they were dysregulated in pathologic states.1 Today, miRNA expression has been characterized in nearly every tissue and biofluid in the human body.2,3 Perturbations in miRNA expression have been described in the vast majority of organ systems.4 The relationship between miRNA physiology and human health and disease stems from the critical role of miRNAs in regulating translation of messenger RNA (mRNA) into proteins.5,6 Primary miRNAs are stem-loop structures initially transcribed from intergenic or intronic regions of the genome (Figure 1).7 They are cleaved into precursor miRNA in the nucleus (by the microprocessor complex), exported to the cytoplasm as a miRNA:miRNA duplex (by Exportin-5), and then cleaved again into mature miRNA by Dicer.8 Mature miRNAs are single-stranded, short RNA fragments (17–23 base-pair) that are incorporated into the RNA-induced silencing complex, where they can bind complementary segments of mRNA. Argonaute proteins within the RNA-induced silencing complex facilitate endonuclease cleavage of mRNA, or inhibition of translation, depending upon the degree of miRNA:mRNA base pairing.9 Pairing of mature miRNAs with mRNA targets depends upon the miRNA seed sequence, a 2–8 base pair region on the 5’ end of the miRNA. It is important to note that: 1) several miRNAs can have overlapping seed sequences; and 2) miRNAs do not require 100% complementarity with mRNA targets.5 This means that several miRNAs can target the same mRNA, and a single miRNA can target numerous mRNAs. Collectively, these traits make miRNAs potent regulators of cellular physiology, and ideal candidates for biomarker development and drug discovery.10,11
Figure 1. miRNA processing and activity.
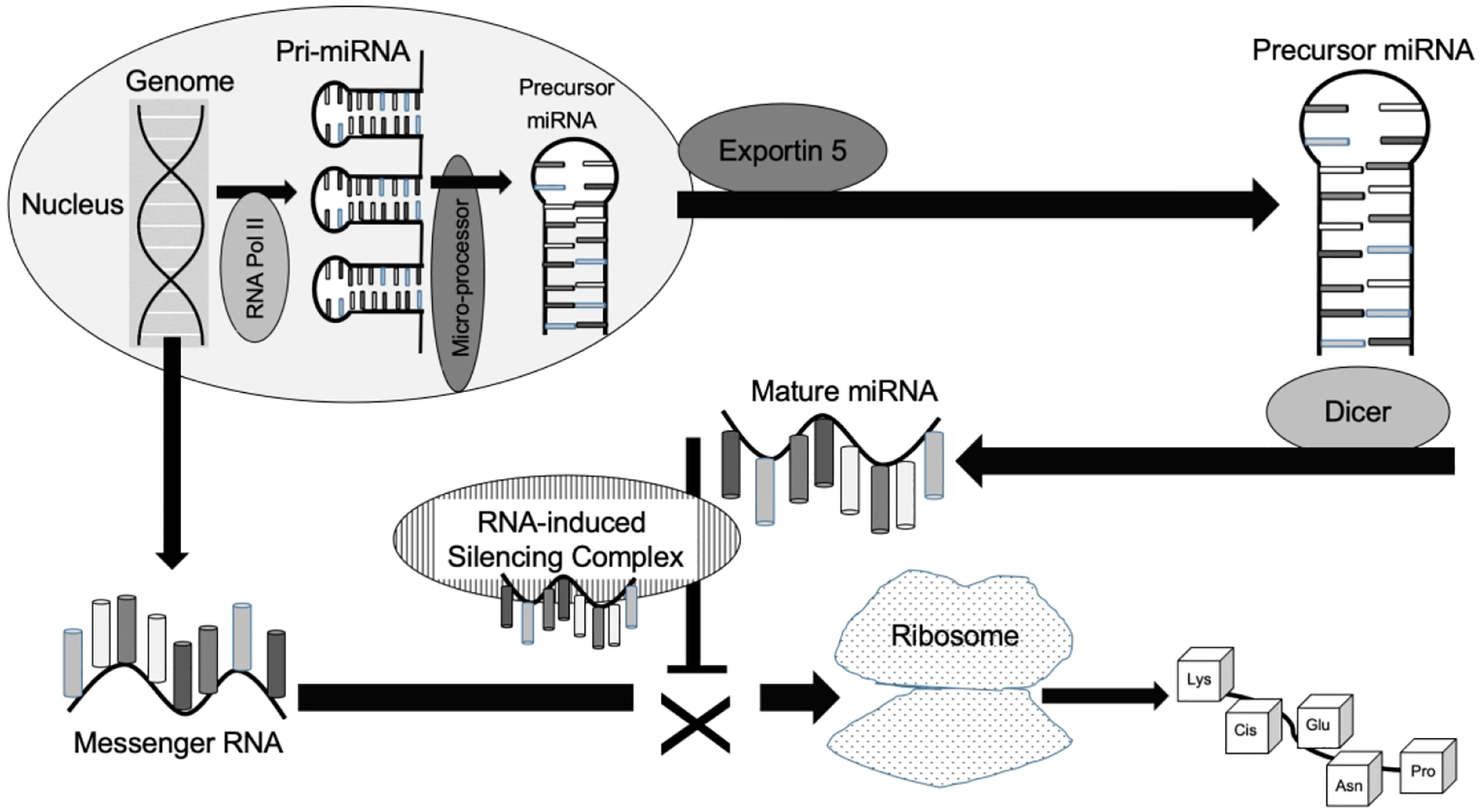
Both messenger RNA and pri-micro-ribonucleic acids (miRNAs) are transcribed from the genome inside the nucleus. After the microprocessor complex converts pri-miRNA to a precursor miRNA complex, it is exported outside the nucleus by Exportin 5. The enzyme, Dicer, processes precursor miRNA into its single-stranded mature form, it can be loaded into the RNA-induced silencing complex. This complex facilitates binding with complementary messenger RNA molecules, preventing translation into proteins by ribosomes.
MiRNAs as Diagnostic and Prognostic Biomarkers
Newborn screening identifies known mutations of metabolic enzyme deficiencies, specific hemoglobinopathies, various endocrine disorders, and hearing loss. In most regions of the United States, newborns are screened for over 60 conditions within hours of birth.12 In contrast, the list of biologic screening tools for non-Mendelian disorders presenting in childhood is relatively small13,14, compared with the enormous array of pathology that may arise. The development of novel molecular markers could enable early detection, targeted intervention, and personalized treatment of many pediatric conditions. Such an approach would not necessarily be limited to the newborn period, and could represent a significant advance from current testing paradigms. For example, erythrocyte sedimentation rate (ESR) is a measure of rouleaux formation that has frequently been used to screen for inflammation in diseases of childhood for the past century.15 ESR is elevated in infectious diseases, auto-immune conditions, neoplasms, kidney failure, and even pregnancy.16 Despite its non-specific nature, ESR continues to be widely used as a biomarker of pediatric pathology. Strategic replacement of ESR testing with targeted biomarkers for each of these disparate conditions could dramatically improve medical care.
Ideally, biomarkers should be inexpensive, collected through non-invasive techniques, rapidly measurable, and easily interpreted by general clinicians.17,18 Ideal biomarkers should also have high sensitivity, providing utility as a screening tool, and allowing early detection of emerging pathology. miRNA physiology satisfies many of these criteria.5,19 miRNAs are present in biofluids throughout the human body.2 Stability of miRNA levels within these fluids results from their small size, which helps protect them from degradation by ribonucleases.20 This characteristic also provides resistance to formalin fixation, yielding utility in pathologic specimens. Because miRNAs are so small, they can easily be transported across biofluid spaces, such as the blood brain barrier.21,22 This means that miRNAs originating from organs that otherwise require invasive sampling (e.g., the central nervous system) can be measured through non-invasive biofluid collection.23 Intercellular and cross-tissue transport is further enhanced by encapsulation of miRNAs within exosomes and other micro-vesicles.22 Protein signatures on vesicles may be used to confirm the “tissue-of-origin” for miRNA cargo, yielding tissue-specific biomarkers.23 Measurement of miRNAs can be achieved with standard lab assays, such as quantitative polymerase chain reaction (qPCR).24 qPCR can detect miRNAs at pico-molar levels, and is already utilized for many clinical assays. Because miRNA transcription occurs rapidly in response to environmental stimuli (minutes-to-hours),25,26 application of miRNA biomarkers is not limited to diagnoses. Tracking miRNA dynamics may ultimately serve as a valuable tool for prognostic and therapeutic monitoring as well.10
Current miRNA Biomarkers of Pediatric Diseases
There are numerous pediatric diseases that currently rely on subjective, invasive, or costly assessments, making them amenable to miRNA-based diagnostics. Here, we highlight seven conditions with miRNA biomarker candidates that have matured beyond initial pilot explorations to external validation studies. These seven conditions were chosen to underscore the potential of miRNA diagnostics to impact organ systems throughout the human body (Figure 2A).
Figure 2. Candidate miRNAs for the diagnosis and therapy of pediatric diseases.
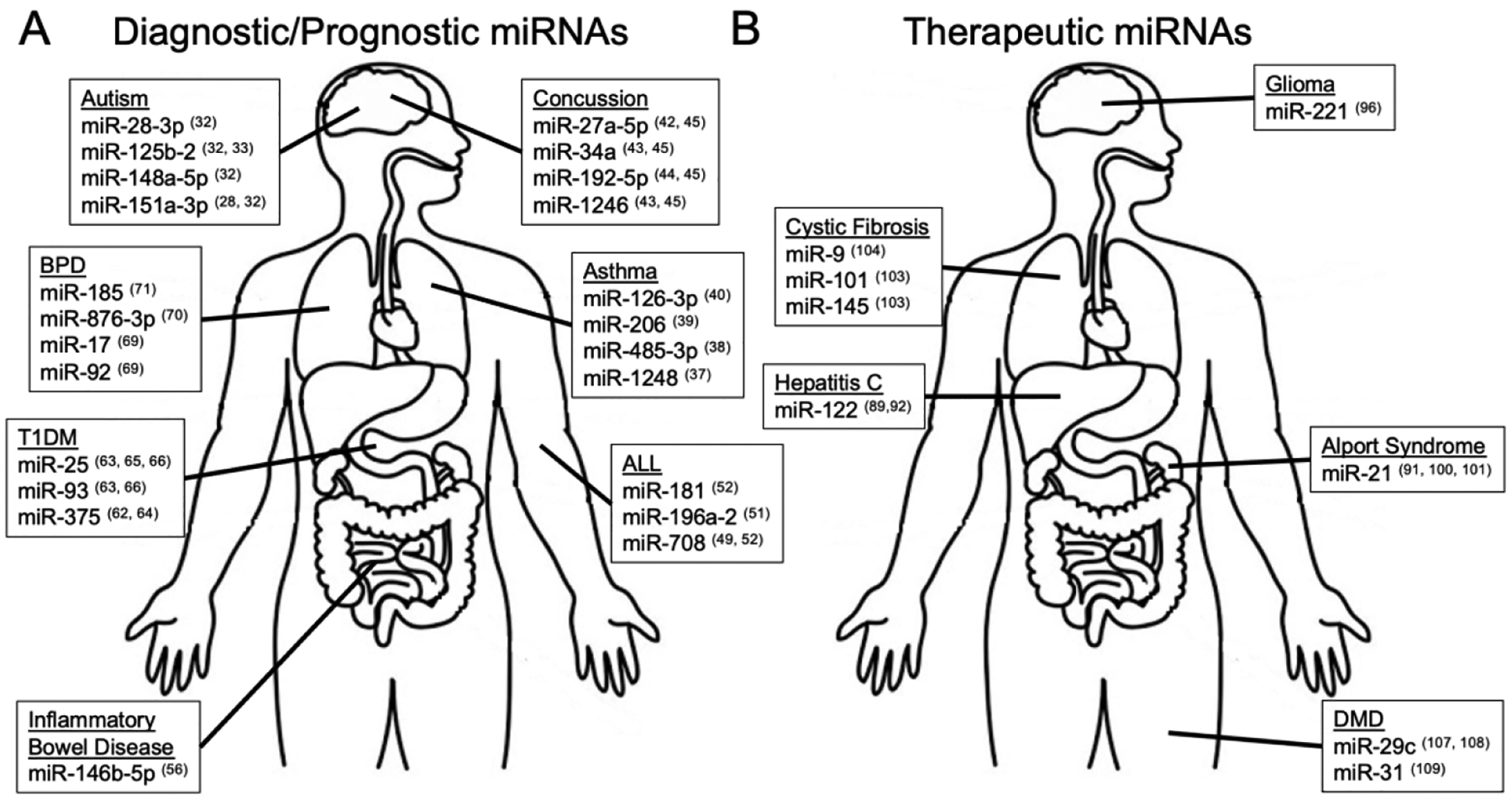
Top miRNA candidates for diagnosis/prognosis (A), and therapy (B) of pediatric diseases are displayed. Diseases of interest were selected based on their abundance of miRNA literature, and to reflect the ability of miRNAs to detect or treat pathology in organ systems throughout the body. In disease states where numerous miRNA candidates have been identified, ≤ 4 miRNAs are displayed based upon their detection across multiple studies, or their detection in studies with large sample sizes. The number after each miRNA denotes the reference paper in which it was reported. Abbreviations: Type 1 diabetes mellitus (T1DM); Bronchopulmonary dysplasia (BPD); Acute lymphoblastic leukemia (ALL); Duchene’s muscular dystrophy (DMD).
Autism Spectrum Disorder
The diagnostic workup for autism spectrum disorder (ASD) is currently relies on screening tools whose poor specificity contributes to long wait times for subjective assessments by developmental specialists.27 ASD is felt to arise from interactions between a child’s underlying genetics and environmental exposures.28 The ability of miRNAs to direct degradation of aberrant mRNAs, or to down-regulate mRNAs involved in key neurologic pathways after an environmental insult, makes miRNAs ideal biomarker candidates for ASD. Indeed, over 20 studies have now characterized dysregulated miRNA expression in the brain, blood, and saliva of children with ASD.29–31 The largest of these studies involved 443 children, ages 2–6 years, and demonstrated that a panel of four saliva miRNAs (miR-28-3p, miR-148a-5p, miR-151a-3p, miR-125b-2) could differentiate children with ASD from peers with typical development or developmental delay.32 This served as the basis for the first Certified Laboratory Improvement Amendment (CLIA) test utilizing miRNA levels (in parallel with other non-coding RNAs) to aid ASD assessment in children.33
Asthma
The diagnosis of asthma is based on clinical examination and expiratory airflow limitation on pulmonary function testing.34 However, the inability of young children to complete pulmonary function tests, is a major drawback to the current approach. In vitro studies utilizing respiratory epithelium, T-cells, and eosinophils have clearly demonstrated the importance of miRNAs in regulating the inflammatory milieu of asthma.35,36 Several studies have shown that serum levels (miR-1248)37 and blood levels (miR-221, miR-485)38 of certain miRNAs may be used to differentiate individuals with asthma from healthy controls. Serum miRNA profiles have also shown promise for prognosis (miR-146b, miR-206, and miR-720 associated with exacerbation occurrence),39 and therapeutic monitoring (miR-133, miR-155, miR-126-3p associated with regulation of Th2 endotype expression).40
Traumatic Brain Injury
School age children and adolescents suffer a disproportionate burden of concussions, and there is growing public awareness regarding the post-injury sequelae associated with traumatic brain injuries (TBI).41 Nonetheless, diagnosis of concussion relies predominantly on subjective symptom reports. Growing evidence suggests that miRNAs may aid concussion diagnoses, predict duration and character of symptoms, and inform decisions about safe return to activities.42–44 A study involving 538 adolescents and young adults demonstrated that ratios of seven salivary non-coding RNAs (including miRNAs) could differentiate participants with concussion from peers with concussion-like symptoms with similar accuracy to balance testing, cognitive testing, or subjective symptom reports.45 The seven non-coding RNAs included miR-34a-5p, miR-192-5p, miR-27a-5p, and miR-4510 (as well as several small nucleolar and piwi-interacting RNAs). Three of these miRNAs (miR-34a-5p, miR-192-5p, miR-27a-5p) have been identified in multiple studies of individuals with TBI. Although current guidelines for pediatric concussion assessment do not recommend the use of biomarkers in routine clinical practice, miRNAs may soon comprise a novel option in the concussion toolbox.
Pediatric Acute Lymphoblastic Leukemia
In the past century, significant improvement in mortality rates for pediatric acute lymphoblastic leukemia (ALL) have been driven by the ability to identify patient sub-types and stratify treatment approaches.46 Increasingly, sub-typing is being accomplished through high throughput RNA sequencing,47 which has opened a new window into previously unidentified mutations associated with ALL.48 MiRNA patterns may help differentiate ALL from acute myelogenous leukemia, aid identification of T-cell and B-cell lineage, and confer information about drug resistance, providing a prognostic tool for predicting risk of relapse.49 Further, the miR-181 family was shown to differentiate CNS-positive and CNS-negative ALL in pediatric CSF samples.50 Studies of miRNA expression in ALL are relatively large in number and scope, however, much of the research in this space is focused on harnessing miRNAs as therapeutic targets.51–53
Pediatric Inflammatory Bowel Disease
Inflammatory bowel disease (IBD), such as Crohn’s disease, relies on non-specific inflammatory markers for initial screening in children with gastrointestinal symptoms, and requires invasive procedures for a diagnosis that lacks a universal gold-standard.54 Nearly thirty studies have characterized miRNA expression in the blood, stool, and gastrointestinal mucosa of patients with IBD.55 The largest of these studies examined serum miRNA levels in 527 adult participants, and found levels of miR-146b-5p to be more accurate for differentiating IBD than C-reactive protein.56 There are fewer studies focused on miRNA levels in pediatric IBD,57 however, mounting evidence suggests miRNAs may provide valuable information about pathology location, severity, and treatment response.58–60
Type 1 Diabetes Mellitus
There has been a surprising increase in the prevalence of Type 1 Diabetes Mellitus (TD1M) among pediatric patients, with over 1.1 million children world-wide suffering from this disorder in 2017.61 Currently the diagnosis of T1DM relies on the emergence of symptoms, which can be sudden and fatal.10 A study of children with new onset T1DM identified circulating miRNAs that are perturbed with elevations in serum glucose (miR-375).62 Some miRNAs appear to reflect auto-antibody status (miR-93)63,64, oral glucose tolerance (miR-146b, miR-151a)63, and glycosylated hemoglobin (miR-25)65 in children at risk for T1DM. Urinary levels of several miRNAs have also demonstrated promise for detection of diabetic nephropathy, with specific miRNAs reflecting estimated glomerular filtration rate.65,66 Thus, miRNA levels may be used for early detection of T1DM, therapeutic monitoring, or even prediction of microvascular sequelae.
Bronchopulmonary Dysplasia
Bronchopulmonary dysplasia (BPD) is a common condition among extremely premature infants that carries high mortality and can lead to chronic lung dysfunction. The pathophysiology of BPD is not completely understood, which hinders accurate diagnosis, prognosis, and treatment. Levels of miRNAs in blood, plasma, tracheal aspirates, and saliva may be perturbed in neonates with BPD.67–69 Early studies suggest miRNA levels are associated with severity of BPD and can be used to predict long-term outcomes.70–72 It remains to be seen how these potential markers respond to therapy and whether they normalize over the course of recovery.
Machine Learning Methods for Pediatric Disease miRNA Biomarker Discovery
Despite the growing evidence that miRNAs play an essential role in pediatric diseases, relying completely on labor-intensive, and time-consuming wet lab experiments to discover miRNA biomarkers is a daunting task. With the advent of high-throughput miRNA expression profiling technology, classic machine learning approaches have streamlined the identification and functional characterization of miRNA biomarkers.73 These popular machine learning methods have focused on improving model classification performance measured by area under the receiver operating characteristic curve (AUROC) using features extracted from miRNA profiles. Variations of these linear techniques, such as support vector machine (SVM) and random forest, are the most widely used machine learning methods in miRNA research.74 These methods have been used in development of pediatric disease miRNA biomarkers across disciplines, such as ASD and developmental delay, hand-foot-mouth disease (HFMD), acute lymphoblastic leukemia (ALL), and celiac disease (CD). As discussed later, novel and advanced deep learning methods are beginning to appear in the field as well.
Traditional Machine Learning Techniques
AUROC, Radial Kernel SVM and LASSO
Although miRNAs exist in many human biofluids, there is strong parental preference for medical tests that measure miRNAs with non-invasive collection techniques. This is especially true in vulnerable populations, such as children with ASD. A study using non-invasive saliva collection in children with ASD harnessed radial kernel SVM to distinguish ASD (total of 456 children) by measuring five families of salivary RNAs (i.e., miRNA, piwi-interacting RNA, long non-coding RNA, ribosomal RNA, and microbial RNA).33 Across each of these 5 RNA types, 32 salivary RNA features (12 microbial taxa, 7 mature miRNAs, 4 precursor miRNAs, 8 piRNAs and 1 snoRNA) were used to differentiate 188 children with ASD from 184 non-ASD peers with typical development or non-ASD developmental delay. The 32 salivary RNA features were identified by pairing the top ranked RNAs from each category with transformed demographic data and fit to a stochastic gradient-boosted model. To verify that the classifier algorithm performance was not biased by patient characteristics, differences in age, sex, race, BMI and Vineland adaptive behavior social scales were assessed between correctly and incorrectly classified children. The least absolute shrinkage and selection operator (LASSO) regularized logistic regression methods75 was also applied to infer the functional association between known ASD-associated copy number variations, single nucleotide variants (SNPs), and 11 miRNA biomarker features (miR-106-5p, miR-10a, miR-125a-5p, miR-146a, miR-146b, miR-146b-5p, miR-361-5p, miR-378a-3p, miR-3916, miR-410, miR-92a-3p).
The diagnosis of HFMD is made clinically based on the appearance and location of oral exanthem. However, a biologic test may be warranted if there is diagnostic uncertainty. Min et al. used both traditional AUROC and radial kernel SVM methods to identify miRNA biomarkers in hospitalized pediatric patients with HFMD obtained via oral swab.76 Two ethnically diverse cohorts were collected: a cohort of HFMD and control participant samples from Singapore (n=58) and Taiwan (n=48). Using AUROC in the Singapore Cohort, miR-221-3p had the highest accuracy in diagnosing HFMD with a positive predictive value of 83% and a negative predictive value of 75%. In order to determine if diagnostic accuracy could be further improved by including the expression of multiple miRNAs, a training set of 75% of the Singapore Cohort was used and then applied to the entire cohort using radial kernel SVM. The model identified 6 miRNAs with a peak accuracy of 85% (miR-221-3p, miR-145-5p, miR-142-3p, miR-125a-5p, miR-324-3p, and miR-18b-5p).
Random Forest
Diagnostics of ALL are sometimes nebulous, as accumulating mutations contributing to carcinogenesis are often shared between childhood B-cell ALL and T-cell ALL.77 To help discriminate childhood T-cell and B-cell ALL, Almeida et al. proposed a random forest method using miRNA expression profiles from bone marrow and peripheral blood blast samples of 8 children exhibiting T-ALL and 8 children exhibiting B-ALL. Samples in both groups ranged in severity levels.78 They reported 16 miRNAs differentially expressed between T- and B-ALL. There were 10 miRNAs downregulated and 6 miRNAs upregulated in T-ALL. miR-29c-5p was the best predictor for leukemia subtypes, with an accuracy of 95% and a ROC area of 0.953. RT-qPCR validated that miR-29c-5p expression predicts ALL cell lineage, but is not associated with a specific subtype within a lineage, or a maturation stage.
Naïve Bayes
Autophagy has been thought to play a crucial role in various autoimmune diseases. In celiac disease (CD), it is thought that exogenous gliadin peptides may be targets of the autophagy clearance process.79 To further identify molecular players in this mechanism, Comincini et al developed a Naïve Bayes method to identify regulatory miRNAs associated with CD.80 Using RT-qPCR, miRNA levels were measured from pediatric CD patient intestinal biopsy samples. To predict the relative contribution of each miRNA in the model’s stratification accuracy, a nomogram analysis was performed. miR-17 and miR-30 were positive predictive features. Further, anti-miR-17 was shown to increase autophagy after in vitro experiments validated an increased presence of autophagic vesicles.
Deep Neural Network-Based Methods for miRNA Biomarker Discovery
Traditional machine methods are often limited to low true-positive rates due to their inability to capture complex nonlinear relationships from thousands of miRNA species. Deep neural network-based methods are designed to utilize multiple levels of transformation steps to extract higher-order features from the raw input data. These deep learning methods have been applied to pediatric diseases detection with significantly improved performance over traditional machine learning methods. For example, Casalino et al. proposed a multi-layer perceptron (MLP) for pediatric multiple sclerosis using miRNA expression profiles.81 This model achieved an overall classification accuracy of 94% using 40 features selected by Chi-squared testing. It also outperformed a model selected by human experts that used 42 features and achieved only 83% accuracy. More recently, Casalino et al. extended their approach with a more general feature selection process.82 They used a linear SVM model to select top-ranked miRNA features and then utilized an MLP with two hidden layers to perform the classification task. The MLP model achieved the highest classification accuracy, precision, and recall scores in the test set compared to random forest classifiers.83 Deep neural network models may even function as a feature extractors by augmenting the raw input data using their impressive capacity of capturing hidden, intricate, nonlinear relationships. Zheng et al. exploited a deep auto-encoder neural network (AE) for extracting feature representation and a random forest classifier for predicting potential miRNA–disease associations using miRNA profiles from multiple sources (including MISIM, MeSH, and HMDD datasets).84
Notably, these traditional machine learning methods are often linear or tree-based interpretable models such that the associations between pediatric diseases and miRNAs are clearly uncovered. On the other hand, deep learning methods generally outperform traditional machine learning methods, but they are notoriously hard to interpret. Fortunately, recent advances in interpretable machine learning have brought about an array of effective methods for explaining deep learning based prediction, such as knowledge distillation85, DeepLIFT86, Integrated Gradients87, and Adversarial Gradient Integration.88
MiRNAs in Pediatric Precision Care
Since 2012, there have been over 100 clinical trials studying miRNA therapeutics in pediatric pathology. Some of the more advanced studies involving miRNA therapeutics include clinical trials for hepatitis C89, solid tumors90, and Alport Syndrome91. Below, we discuss these conditions, and two additional disorders that may benefit from miRNA therapeutics (Figure 2B).
Although hepatitis C virus (HCV) infection is predominantly a disease of adulthood, HCV can be acquired in the perinatal period through vertical transmission, and there are few options for treating childhood HCV. One of the first miRNA therapies to reach clinical studies was Miravirsen (anti-miR-122).89,92 This miRNA antagonist prevents binding between miR-122 and HCV, which is critical for viral stabilization and accumulation. A phase II study involving adults with HCV demonstrated a sustained, dose-dependent decrease in HCV levels, concomitant with changes in the expression of ~100 hepatic gene products.89 Subsequently, a mutation in HCV mRNA was detected among patients whose HSV load rebounded following therapy, rendering this HCV strain insensitive to miR-122 antagonists. HCV mutations are also a common source of resistance to therapies involving protease inhibitors or polymerase inhibitors.93 Therefore, addition of miR-122 antagonists to these regimens might one day provide synergistic benefits.
Treatment success for solid tumors, such as glioblastoma multiforme, remains elusive, partly due to drug resistance.94 Emerging evidence suggests that co-treatment of solid tumors with miRNA therapeutics has the potential to overcome drug resistance.95 In gliomas, elevated levels of miR-221 are associated with invasiveness and temozolomide resistance. Co-delivery of temozolomide and a miR-221 antagonist induces apoptosis in temozolomide-resistant cells.96 In initial studies of hepatocellular carcinoma, miR-34a displayed tumor-suppressor potential through inhibition of key oncogenes.97 However, a subsequent study in hepatocellular carcinoma cells with β-catenin mutations suggested that miR-34a might actually have oncogenic effects, and a clinical trial of a miR-34a mimic was terminated following adverse events.98,99 Conversely, a phase I clinical trial for mesothelioma, which employed a miR-16 mimic, has shown promising early safety results (NCT02657460).90
Alport Syndrome is a rare condition that stems from genetic mutations in Type IV collagen genes and leads to progressive renal dysfunction (as well as disorders of the inner ears and eyes). In 27 patients with Alport Syndrome, levels of miR-21 were elevated in tubular epithelial cells and glomeruli, and demonstrated direct relationships with measures of disease severity (as measured by proteinuria, kidney function, and histopathology scores).100,101 Inhibition of miR-21 in Col4α3−/− mice using a miR-21 antagonist improved survival and reduced pathologic measures of glomerulosclerosis, interstitial fibrosis, and tubular injury. These results led to a planned Phase II, randomized, double-blind, placebo-controlled study (clinical trial number: NCT02855268).91
miRNA-based therapies also hold potential in cystic fibrosis (CF).102 There are several miRNAs that regulate expression of the CFTR gene, and many exert effects on inflammatory markers or gene networks implicated in CF pathology. Some of the most promising miRNA-based strategies in CF involve miRNA antagonists or target site blockers (molecules that bind to the 3’ untranslated region of a mRNA and prevent miRNA interaction). Transfection of target site blockers for miR-101 and miR-145 into bronchial epithelial cells from patients with CF has been shown to increase CFTR expression and activity.103 Similarly, target site blockers for miR-9, reduce interactions with ANO1 (an alternative chloride ion channel involved in airway surface maintenance), increasing cell migration and mucociliary clearance in animal models.104 Although clinical trials have not been initiated, it is possible that this therapy could eventually be delivered via intranasal administration, promoting tissue-specific effects, and augmenting existing CFTR modulator therapies.
Duchene’s muscular dystrophy (DMD) is a progressive neuromuscular condition that results from a mutation in the dystrophin gene.105 Recently, developments in gene therapy have shown promise in treating individuals with DMD.106 However, current gene therapy does not completely cure DMD, since it does not incorporate the full protein into the muscle. There is a need for novel co-treatments that can facilitate enhance the therapeutic response. MiRNAs play an important role in myoblast-mediated muscle regeneration. Levels of miR-29c are decreased in the myoblasts of children with DMD.107 Co-treatment of a mouse model of DMD using gene therapy and miR-29 suppressed fibrosis and restored muscle function.108 Conversely, levels of miR-31 are elevated in both a dystrophic mouse model and muscle biopsies from DMD patients.109 Inhibiting the binding of miR-31 to the dystrophin mRNA (in combination with exon skipping) increased production of the dystrophin protein in myoblasts from DMD patients. Together these studies highlight how miRNA therapeutics might ameliorate neuromuscular pathology in DMD and supplement the exciting advances taking place in gene therapy.
Promise and Potential of miRNA Therapeutics
The therapeutic potential of miRNAs stems from their ability to target multiple mRNAs within the same pathway.11,110 Thus, while cells may find ways to circumnavigate single gene therapies through alternate/redundant signaling mechanisms, delivery of a single miRNA (or miRNA cluster) may successfully inhibit an entire physiologic process.
This characteristic holds significant promise in neoplastic conditions, such as leukemia. For example, the miR-17/miR-92 cluster can be used to target cell proliferation through inactivation of c-Myc and E2F transcripts111; while the miR-23a/miR-27b/miR-24-2 cluster may inhibit hematopoiesis through degradation of Fox01, Ebf1, and Pax5 transcripts.112 Conversely, novel therapies may even target miRNA levels.113 Synthetic “antagomiRs” can be used to incapacitate specific miRNAs, addressing medical conditions with miRNA over-expression, or increasing translation of key mRNAs.114 For such therapeutics to become reality, it will be crucial to deliver miRNAs and miRNA antagonists to pathologic tissues with high-fidelity (avoiding off-target side effects in healthy cells).115 Fortunately, rapid advances in our understanding of exosomes are catalyzing the development of synthetic micro-vesicles that utilize cell- or tissue-specific docking proteins to facilitate targeted delivery.116 Though such therapeutics would likely be delivered directly into circulation by intravenous administration, intranasal or oral administration of miRNA therapeutics may also be possible for certain medical conditions.95
Challenges of miRNA Therapeutics
The same pleiotropic characteristics that make miRNAs an appealing class of novel therapeutics, also yield makes them prone to off-target effects and adverse reactions. For example, miR-21 cancer therapy might ultimately provide an advantage over therapy aimed at a specific gene, by simultaneously targeting multiple oncogenic transcripts.117 However, miR-21 is expressed throughout the human body, and suppression of key mRNAs in healthy tissues could cause unwanted apoptosis. One emerging strategy to reduce off-target effects involves the synthesis of target-specific extracellular vesicles (EVs) that can deliver miRNAs to specific cells. Synthesis of EVs with a CD47 marker may permit delivery to macrophages.118 However, many EVs doc at cells in a nonspecific manner, and synthetic vesicles may be eliminated from circulation by the innate immune system, resulting in unpredictable drug delivery.119 An alternative approach to synthetic EV generation, involves harvesting endogenous EVs from cell culture. Endogenous EVs may evade the host immune response by displaying membrane features that mimic natural EV composition.120 Currently, the processes necessary for cell culture, EV isolation, and EV storage inhibit large-scale drug production. These barriers will need to be overcome before endogenous EVs can be used to deliver miRNAs in the clinical setting.
Gaps in miRNA Biomarker Research
Gaps in miRNA biomarker research include widespread variability of post-analytical factors that can impact results.121 This is particularly true in next generation sequencing, where variations in the sensitivity of RNA alignment tools may lead to inaccurate detection of sequence errors, mismatches, and non-templated nucleotide additions that ultimately influence differential expression results.122 Normalization and scaling of RNA sequencing data post-alignment can also introduce intra-study variation in reported miRNA levels.123 Post-analytic issues are not unique to high throughput sequencing. In qPCR, miRNAs of interest must be quantified against a non-variant reference gene.124 Selected reference genes vary widely across studies,125,126 and few studies ensure that stability of chosen reference genes extends to external cohorts, across pediatric ages, or throughout the circadian clock.127 There is a paucity of information about how the miRNome (i.e., the totality of human miRNAs) fluctuates with age.128 Few studies have specifically determined how child traits (sex, ethnicity) and environment (prandial status, exercise status, stress) impact miRNA levels.129,130 However, it is clear that a subset of miRNAs display diurnal variation.127,131 This highlights the importance of recording and controlling for sample collection time when developing miRNA tests.
Conclusions
The future of pediatric disease diagnostics relies on our ability to discover robust biomarkers. MiRNAs serve as a prime option, given their established role in human health and disease, their presence in a variety of human biofluids, and their ability to regulate entire pathways. The body of literature that is utilizing traditional and neural-network based methods for miRNA biomarker discovery continues to grow, and may hopefully yield potential therapeutic targets for more pediatric diseases. The interest of pharmaceutical companies in synthetic oligonucleotides also continues to grow; over 1 billion dollars were pledged by investors for Ionis Pharmaceutical in 2018. In March 2022, Creyon Bio secured 40 million dollars to use machine learning approaches to identify oligonucleotide-based precision medicine therapies. As pre-clinical and clinical bodies of work use a multi-omic approach, there will be a need for researchers and physicians alike to understand how to interpret these large data sets, and what these data mean in the context of disease.
Impact Statement:
In the following review article, we summarized how recent developments in microRNA research may be coupled with machine learning techniques to advance pediatric precision care.
Funding
No financial assistance was received in support of this work. SDH’s time was supported by the National Institutes of Health (grant number R61HD105610 to SDH; R01NS115942 to SDH; 4R61HD105610-02 to SDH) and the Gerber Foundation (grant number 5295 to SDH).
Footnotes
Competing Interests
SDH serves as a scientific advisory board member for Quadrant Biosciences, a biotech company involved in the development of RNA-based diagnostics. Quadrant Biosciences had no role in drafting or editing the proposed review article. DZ, CL, and RES have no conflicts of interest to disclose.
References
- 1.Almeida MI, Reis RM & Calin GA MicroRNA history: Discovery, recent applications, and next frontiers. Mutat. Res. Mol. Mech. Mutagen 717, 1–8 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Weber JA et al. The microRNA spectrum in 12 body fluids. Clin. Chem 56, 1733–1741 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehlmann T, Ludwig N, Backes C, Meese E & Keller A Distribution of microRNA biomarker candidates in solid tissues and body fluids. RNA Biol. 13, 1084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esteller M Non-coding RNAs in human disease. Nat. Rev. Genet. 2011 1212 12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Mohr AM & Mott JL Overview of MicroRNA Biology. Semin. Liver Dis 35, 3 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newman MA & Hammond SM Emerging paradigms of regulated microRNA processing. Genes Dev. 24, 1086–1092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creugny A, Fender A & Pfeffer S Regulation of primary microRNA processing. FEBS Lett. 592, 1980–1996 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Fiorenza A & Barco A Role of Dicer and the miRNA system in neuronal plasticity and brain function. Neurobiol. Learn. Mem 135, 3–12 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi H & Tomari Y RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta - Gene Regul. Mech 1859, 71–81 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Paul S et al. Roles of microRNAs in chronic pediatric diseases and their use as potential biomarkers: A review. Arch. Biochem. Biophys 699, 108763 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Simonson B & Das S MicroRNA Therapeutics: the Next Magic Bullet? Mini Rev. Med. Chem 15, 467 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newborn Screening - Understanding Genetics - NCBI Bookshelf. Available at: https://www.ncbi.nlm.nih.gov/books/NBK132148/. (Accessed: 8th March 2022)
- 13.Wood SK & Sperling R Pediatric Screening: Development, Anemia, and Lead. Prim. Care Clin. Off. Pract 46, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Kanji A, Khoza-Shangase K & Moroe N Newborn hearing screening protocols and their outcomes: A systematic review. Int. J. Pediatr. Otorhinolaryngol 115, 104–109 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Tishkowski K & Gupta V Erythrocyte Sedimentation Rate. Lab. Hematol. Pract 638–646 (2021). doi: 10.1002/9781444398595.ch49 [DOI] [Google Scholar]
- 16.Clinical Utility of the Erythrocyte Sedimentation Rate - American Family Physician. Available at: https://www.aafp.org/afp/1999/1001/p1443.html. (Accessed: 8th March 2022) [PubMed]
- 17.Bock C Epigenetic biomarker development. Epigenomics 1, 99–110 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Phillips KA, Van Bebber S & Issa AM Diagnostics and biomarker development: priming the pipeline. Nat. Rev. Drug Discov 5, 463–469 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Kuzhandai Velu V, Ramesh R & Srinivasan AR Circulating MicroRNAs as Biomarkers in Health and Disease. J. Clin. Diagn. Res 6, 1791 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung M et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem 56, 998–1006 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto J, Stewart T, Banks WA & Zhang J The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Curr. Pharm. Des 23, 6206–6214 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Zhang J et al. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics. Proteomics Bioinformatics 13, 17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cha DJ et al. miR-212 and miR-132 Are Downregulated in Neurally Derived Plasma Exosomes of Alzheimer’s Patients. Front. Neurosci 13, 1208 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andrews WJ, Brown ED, Dellett M, Hogg RE & Simpson DA Rapid quantification of microRNAs in plasma using a fast real-time PCR system. Biotechniques 58, 244–252 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Scott GK, Mattie MD, Berger CE, Benz SC & Benz CC Rapid alteration of microRNA levels by histone deacetylase inhibition. Cancer Res. 66, 1277–1281 (2006). [DOI] [PubMed] [Google Scholar]
- 26.LaRocca D et al. Comparison of serum and saliva miRNAs for identification and characterization of mTBI in adult mixed martial arts fighters. PLoS One 14, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwaigenbaum L & Maguire J Autism screening: Where do we go from here? Pediatrics 144, (2019). [DOI] [PubMed] [Google Scholar]
- 28.Hicks SD & Middleton FA A comparative review of microRNA expression patterns in autism spectrum disorder. Front. Psychiatry 7, 176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozkul Y et al. A heritable profile of six miRNAs in autistic patients and mouse models. Sci. Rep 10, 9011–9011 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicks SD, Ignacio C, Gentile K & Middleton FA Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 16, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mor M, Nardone S, Sams DS & Elliott E Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hicks SD et al. Saliva MicroRNA Differentiates Children With Autism From Peers With Typical and Atypical Development. J. Am. Acad. Child Adolesc. Psychiatry 59, 296–308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicks SD et al. Validation of a Salivary RNA Test for Childhood Autism Spectrum Disorder. Front. Genet 9, 534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogt B, Falkenberg C, Weiler N & Frerichs I Pulmonary function testing in children and infants. Physiol. Meas 35, R59 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Simpson LJ et al. A miRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat. Immunol 15, 1162 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattes J, Collison A, Plank M, Phipps S & Foster PS Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. U. S. A 106, 18704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panganiban RPL et al. Differential microRNA epression in asthma and the role of miR-1248 in regulation of IL-5. Am. J. Clin. Exp. Immunol 1, 154 (2012). [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Qin HB, Xu B, Zhou H & Zhao DY Profiling of miRNAs in pediatric asthma: Upregulation of miRNA-221 and miRNA-485–3p. Mol. Med. Rep 6, 1178–1182 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Kho AT et al. Circulating microRNAs and prediction of asthma exacerbation in childhood asthma. Respir. Res 19, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendes FC et al. Development and validation of exhaled breath condensate microRNAs to identify and endotype asthma in children. PLoS One 14, e0224983 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lumba-Brown A et al. Centers for Disease Control and Prevention Guideline on the Diagnosis and Management of Mild Traumatic Brain Injury Among Children. JAMA Pediatr. 172, e182853–e182853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atif H & Hicks SD A Review of MicroRNA Biomarkers in Traumatic Brain Injury. J. Exp. Neurosci 13, 117906951983228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Pietro V et al. Unique diagnostic signatures of concussion in the saliva of male athletes: the Study of Concussion in Rugby Union through MicroRNAs (SCRUM). Br. J. Sports Med 55, 1395–1404 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson JJ et al. Association of Salivary MicroRNA Changes With Prolonged Concussion Symptoms. JAMA Pediatr. 172, 65–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hicks SD et al. Diagnosing mild traumatic brain injury using saliva RNA compared to cognitive and balance testing. Clin. Transl. Med 10, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee DK, Chang VY, Kee T, Ho CM & Ho D Optimizing Combination Therapy for Acute Lymphoblastic Leukemia Using a Phenotypic Personalized Medicine Digital Health Platform: Retrospective Optimization Individualizes Patient Regimens to Maximize Efficacy and Safety. SLAS Technol. 22, 276–288 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Wallaert A et al. Comprehensive miRNA expression profiling in human T-cell acute lymphoblastic leukemia by small RNA-sequencing. Sci. Reports 2017 71 7, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lilljebjörn H & Fioretos T New oncogenic subtypes in pediatric B-cell precursor acute lymphoblastic leukemia. Blood 130, 1395–1401 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Luan C, Yang Z & Chen B The functional role of microRNA in acute lymphoblastic leukemia: relevance for diagnosis, differential diagnosis, prognosis, and therapy. Onco. Targets. Ther 8, 2903 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Egyed B et al. MicroRNA-181a as novel liquid biopsy marker of central nervous system involvement in pediatric acute lymphoblastic leukemia. J. Transl. Med 18, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong N et al. Hsa-miR-196a2 polymorphism increases the risk of acute lymphoblastic leukemia in Chinese children. Mutat. Res. Mol. Mech. Mutagen 759, 16–21 (2014). [DOI] [PubMed] [Google Scholar]
- 52.de Oliveira JC, Brassesco MS, Scrideli CA, Tone LG & Narendran A MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL). Pediatr. Blood Cancer 59, 599–604 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Ultimo S et al. Roles and clinical implications of microRNAs in acute lymphoblastic leukemia. J. Cell. Physiol 233, 5642–5654 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Sellin JH & Shah RR The Promise and Pitfalls of Serologic Testing in Inflammatory Bowel Disease. Gastroenterol. Clin. North Am 41, 463–482 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Ghafouri-Fard S, Eghtedarian R & Taheri M The crucial role of non-coding RNAs in the pathophysiology of inflammatory bowel disease. Biomed. Pharmacother 129, 110507 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Chen P et al. Circulating microRNA146b-5p is superior to C-reactive protein as a novel biomarker for monitoring inflammatory bowel disease. Aliment. Pharmacol. Ther 49, 733–743 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Koukos G et al. MicroRNA-124 Regulates STAT3 Expression and is Downregulated in Colon Tissues of Pediatric Patients with Ulcerative Colitis. Gastroenterology 145, 842 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iborra M et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol 173, 250 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mohammadi A, Kelly OB, Smith MI, Kabakchiev B & Silverberg MS Differential miRNA Expression in Ileal and Colonic Tissues Reveals an Altered Immunoregulatory Molecular Profile in Individuals With Crohn’s Disease versus Healthy Subjects. J. Crohns. Colitis 13, 1459 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valmiki S, Ahuja V & Paul J MicroRNA exhibit altered expression in the inflamed colonic mucosa of ulcerative colitis patients. World J. Gastroenterol 23, 5324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patterson CC et al. Worldwide estimates of incidence, prevalence and mortality of type 1 diabetes in children and adolescents: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res. Clin. Pract 157, (2019). [DOI] [PubMed] [Google Scholar]
- 62.Marchand L et al. MiRNA-375 a Sensor of Glucotoxicity Is Altered in the Serum of Children with Newly Diagnosed Type 1 Diabetes. J. Diabetes Res 2016, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Åkerman L, Casas R, Ludvigsson J, Tavira B & Skoglund C Serum miRNA levels are related to glucose homeostasis and islet autoantibodies in children with high risk for type 1 diabetes. PLoS One 13, e0191067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertoccini L et al. Circulating miRNA-375 levels are increased in autoantibodies-positive first-degree relatives of type 1 diabetes patients. Acta Diabetol. 2019 566 56, 707–710 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Nielsen LB et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp. Diabetes Res 2012, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abdelghaffar S et al. MicroRNAs and Risk Factors for Diabetic Nephropathy in Egyptian Children and Adolescents with Type 1 Diabetes. Diabetes, Metab. Syndr. Obes. Targets Ther 13, 2485–2494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong XQ et al. Umbilical Cord Blood-Derived Exosomes From Very Preterm Infants With Bronchopulmonary Dysplasia Impaired Endothelial Angiogenesis: Roles of Exosomal MicroRNAs. Front. Cell Dev. Biol 9, 529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nardiello C & Morty RE MicroRNA in late lung development and bronchopulmonary dysplasia: the need to demonstrate causality. Mol. Cell. Pediatr. 2016 31 3, 1–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rogers LK et al. Attenuation of MIR-17~92 cluster in bronchopulmonary dysplasia. Ann. Am. Thorac. Soc 12, 1506–1513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lal CV et al. Exosomal microRNA predicts and protects against severe bronchopulmonary dysplasia in extremely premature infants. JCI Insight 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oji-Mmuo CN et al. Tracheal aspirate transcriptomic and miRNA signatures of extreme premature birth with bronchopulmonary dysplasia. J. Perinatol. 2020 413 41, 551–561 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Siddaiah R et al. MicroRNA Signatures Associated with Bronchopulmonary Dysplasia Severity in Tracheal Aspirates of Preterm Infants. Biomedicines 9, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, Xie D, Zhao Q & You ZH MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform 20, 515–539 (2019). [DOI] [PubMed] [Google Scholar]
- 74.Chen L, Heikkinen L, Wang C, Yang Huiyan Sun Y & Wong Correspondence author Garry Wong G Trends in the development of miRNA bioinformatics tools. doi: 10.1093/bib/bby054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Varma M et al. Outgroup Machine Learning Approach Identifies Single Nucleotide Variants in Noncoding DNA Associated with Autism Spectrum Disorder. Pac. Symp. Biocomput 24, 260 (2019). [PMC free article] [PubMed] [Google Scholar]
- 76.Min N et al. Circulating Salivary miRNA hsa-miR-221 as Clinically Validated Diagnostic Marker for Hand, Foot, and Mouth Disease in Pediatric Patients. EBioMedicine 31, 299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chiaretti S, Zini G & Bassan R Diagnosis and Subclassification of Acute Lymphoblastic Leukemia. Mediterr. J. Hematol. Infect. Dis 6, 2014073 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Almeida RS et al. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol. Oncol 37, 103–112 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Bozzola M et al. The Emerging Role of the Autophagy Process in Children with Celiac Disease: Current Status and Research Perspectives. Celiac Dis. - From Bench to Clin (2018). doi: 10.5772/INTECHOPEN.80692 [DOI] [Google Scholar]
- 80.Comincini S et al. Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children. Int. J. Mol. Sci 18, 391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casalino G et al. A Predictive Model for MicroRNA Expressions in Pediatric Multiple Sclerosis Detection. in Lecture Notes in Computer Science 177–188 (Springer Verlag, 2019). doi: 10.1007/978-3-030-26773-5_16 [DOI] [Google Scholar]
- 82.Casalino G, Castellano G, Consiglio A, Nuzziello N & Vessio G MicroRNA expression classification for pediatric multiple sclerosis identification. J. Ambient Intell. Humaniz. Comput 1, 1–10 (2021). [Google Scholar]
- 83.Geurts P, Ernst D & Wehenkel L Extremely randomized trees. Mach. Learn. 2006 631 63, 3–42 (2006). [Google Scholar]
- 84.Zheng K et al. MLMDA: A machine learning approach to predict and validate MicroRNA-disease associations by integrating of heterogenous information sources. J. Transl. Med 17, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li X, Zhu D & Levy P Predicting Clinical Outcomes with Patient Stratification via Deep Mixture Neural Networks. AMIA Summits Transl. Sci. Proc 2020, 367 (2020). [PMC free article] [PubMed] [Google Scholar]
- 86.Shrikumar A, Greenside P & Kundaje A Learning Important Features Through Propagating Activation Differences. PMLR 70, (2017). [Google Scholar]
- 87.Sundararajan M, Taly A & Yan Q Axiomatic Attribution for Deep Networks. PMLR 70, (2017). [Google Scholar]
- 88.Pan D, Li X & Zhu D Explaining Deep Neural Network Models with Adversarial Gradient Integration. Thirtieth Int. Jt. Conf. Artif. Intell (2021). [Google Scholar]
- 89.Van Der Ree MH et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 111, 53–59 (2014). [DOI] [PubMed] [Google Scholar]
- 90.van Zandwijk N et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 18, 1386–1396 (2017). [DOI] [PubMed] [Google Scholar]
- 91.Omachi K & Miner JH Alport Syndrome Therapeutics: Ready for Prime-Time Players. Trends Pharmacol. Sci 40, 803–806 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Ottosen S et al. In Vitro Antiviral Activity and Preclinical and Clinical Resistance Profile of Miravirsen, a Novel Anti-Hepatitis C Virus Therapeutic Targeting the Human Factor miR-122. Antimicrob. Agents Chemother 59, 599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kuntzen T et al. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naïve patients. Hepatology 48, 1769–1778 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pottoo FH, Javed MN, Rahman JU, Abu-Izneid T & Khan FA Targeted delivery of miRNA based therapeuticals in the clinical management of Glioblastoma Multiforme. Semin. Cancer Biol 69, 391–398 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Sukumar UK et al. Intranasal Delivery of Targeted Polyfunctional Gold-Iron Oxide Nanoparticles Loaded with Therapeutic microRNAs for Combined Theranostic Multimodality Imaging and Presensitization of Glioblastoma to Temozolomide. Biomaterials 218, 119342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertucci A et al. Combined Delivery of Temozolomide and Anti-miR221 PNA Using Mesoporous Silica Nanoparticles Induces Apoptosis in Resistant Glioma Cells. Small 11, 5687–5695 (2015). [DOI] [PubMed] [Google Scholar]
- 97.Cao C et al. The Long Intergenic Noncoding RNA UFC1, a Target of MicroRNA 34a, Interacts With the mRNA Stabilizing Protein HuR to Increase Levels of β-Catenin in HCC Cells. Gastroenterology 148, 415–426.e18 (2015). [DOI] [PubMed] [Google Scholar]
- 98.Gougelet A et al. Antitumour activity of an inhibitor of miR-34a in liver cancer with β-catenin-mutations. Gut 65, 1024–1034 (2016). [DOI] [PubMed] [Google Scholar]
- 99.Hong DS et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020 12211 122, 1630–1637 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guo J et al. Dysregulated Expression of microRNA-21 and Disease-Related Genes in Human Patients and in a Mouse Model of Alport Syndrome. https://home.liebertpub.com/hum 30, 865–881 (2019). [DOI] [PubMed] [Google Scholar]
- 101.Gomez IG et al. Anti–microRNA-21 oligonucleotides prevent Alport nephropathy progression by stimulating metabolic pathways. J. Clin. Invest 125, 141 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hunt AMD, Glasgow AMA, Humphreys H & Greene CM Alpha-1 Antitrypsin—A Target for MicroRNA-Based Therapeutic Development for Cystic Fibrosis. Int. J. Mol. Sci. 2020, Vol. 21, Page 836 21, 836 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Viart V et al. Transcription factors and miRNAs that regulate fetal to adult CFTR expression change are new targets for cystic fibrosis. Eur. Respir. J 45, 116–128 (2015). [DOI] [PubMed] [Google Scholar]
- 104.Sonneville F et al. MicroRNA-9 downregulates the ANO1 chloride channel and contributes to cystic fibrosis lung pathology. Nat. Commun. 2017 81 8, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aránega AE et al. MiRNAs and Muscle Regeneration: Therapeutic Targets in Duchenne Muscular Dystrophy. Int. J. Mol. Sci 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Odom GL, Gregorevic P & Chamberlain JS Viral-mediated gene therapy for the muscular dystrophies: Successes, limitations and recent advances. Biochim. Biophys. Acta 1772, 243 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang L et al. Loss of miR-29 in Myoblasts Contributes to Dystrophic Muscle Pathogenesis. Mol. Ther 20, 1222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heller KN, Mendell JT, Mendell JR & Rodino-Klapac LR MicroRNA-29 overexpression by adeno-associated virus suppresses fibrosis and restores muscle function in combination with micro-dystrophin. JCI Insight 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cacchiarelli D et al. miR-31 modulates dystrophin expression: new implications for Duchenne muscular dystrophy therapy. EMBO Rep. 12, 136–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bader AG, Brown D & Winkler M The promise of microRNA replacement therapy. Cancer Res. 70, 7027–7030 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gruszka R, Zakrzewski K, Liberski PP & Zakrzewska M mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17–92 Network in the Most Common Pediatric Brain Tumors. Int. J. Mol. Sci 22, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kurkewich JL et al. The Mirn23a and Mirn23b MicroRNA Clusters are Necessary for Proper Hematopoietic Progenitor Cell Production and Differentiation. Exp. Hematol 59, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Battistella M & Marsden PA Advances, Nuances, and Potential Pitfalls When Exploiting the Therapeutic Potential of RNA Interference. Clin. Pharmacol. Ther 97, 79–87 (2015). [DOI] [PubMed] [Google Scholar]
- 114.Bernardo BC, Ooi JYY, Lin RCY & Mcmullen JR miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. 10.4155/fmc.15.107 7, 1771–1792 (2015). [DOI] [PubMed] [Google Scholar]
- 115.Castanotto D & Rossi JJ The promises and pitfalls of RNA-interference-based therapeutics. Nature 457, 426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Murphy DE et al. Natural or synthetic RNA delivery: A stoichiometric comparison of extracellular vesicles and synthetic nanoparticles. Nano Lett. 21, 1888–1895 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hayes J, Peruzzi PP & Lawler S MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med 20, 460–469 (2014). [DOI] [PubMed] [Google Scholar]
- 118.Feng M et al. Macrophages eat cancer cells using their own calreticulin as a guide: Roles of TLR and Btk. Proc. Natl. Acad. Sci. U. S. A 112, 2145–2150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hanna J, Hossain GS & Kocerha J The potential for microRNA therapeutics and clinical research. Front. Genet 10, 478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elsharkasy OM et al. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev 159, 332–343 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Binderup HG et al. Quantification of microRNA levels in plasma – Impact of preanalytical and analytical conditions. PLoS One 13, e0201069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ziemann M, Kaspi A & El-Osta A Evaluation of microRNA alignment techniques. RNA 22, 1120–1138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Faraldi M et al. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Reports 2019 91 9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao H et al. Identification of valid reference genes for mRNA and microRNA normalisation in prostate cancer cell lines. Sci. Rep 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Serafin A et al. Identification of a set of endogenous reference genes for miRNA expression studies in Parkinson’s disease blood samples. BMC Res. Notes 7, 1–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang Y, Tang W, Peng L, Tang J & Yuan Z Identification and validation of microRNAs as endogenous controls for quantitative polymerase chain reaction in plasma for stable coronary artery disease. Cardiol. J 23, 694–703 (2016). [DOI] [PubMed] [Google Scholar]
- 127.Hicks SD et al. Diurnal oscillations in human salivary microRNA and microbial transcription: Implications for human health and disease. PLoS One 13, e0198288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fehlmann T et al. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hicks SD, Jacob P, Middleton FA, Perez O & Gagnon Z Distance running alters peripheral microRNAs implicated in metabolism, fluid balance, and myosin regulation in a sex-specific manner. Physiol. Genomics 50, 658–667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scipioni AM, Ornstein RM & Hicks SD Differential Expression Of Salivary Microrna In Anorexia Nervosa And Anxiety Disorders. J. Adolesc. Heal 64, S13 (2019). [Google Scholar]
- 131.Kinoshita C, Okamoto Y, Aoyama K & Nakaki T MicroRNA: A Key Player for the Interplay of Circadian Rhythm Abnormalities, Sleep Disorders and Neurodegenerative Diseases. Clocks & Sleep 2, 282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]


