Abstract
BACKGROUND:
Prenatal cadmium (Cd) exposure has been implicated in both placental toxicity and adverse neurobehavioral outcomes. Placental microRNAs (miRNAs) may function to developmentally program adverse pregnancy and newborn health outcomes in response to gestational Cd exposure.
METHODS:
In a subset of the Rhode Island Child Health Study (RICHS, n=115) and the New Hampshire Birth Cohort Study (NHBCS, =281), we used small RNA sequencing and trace metal analysis to identify Cd-associated expression of placental miRNAs using negative binomial generalized linear models. We predicted mRNAs targeted by Cd-associated miRNAs and relate them to neurobehavioral outcomes at birth through the integration of transcriptomic data and summary scores from the NICU Network Neurobehavioral Scale (NNNS).
RESULTS:
Placental Cd concentrations are significantly associated with the expression level of five placental miRNAs in NHBCS, with similar effect sizes in RICHS. These miRNA target genes overrepresented in nervous system development, and their expression is correlated with NNNS metrics suggestive of atypical neurobehavioral outcomes at birth.
CONCLUSIONS:
Gestational Cd exposure is associated with the expression of placental miRNAs. Predicted targets of these miRNAs are involved in nervous system development and may also regulate placental physiology, allowing their dysregulation to modify developmental programming of early life health outcomes.
INTRODUCTION
Toxic metal exposure during pregnancy can influence fetal development and may have lifelong consequences to offspring health. Ranked 7th on the Agency for Toxic Substances and Disease Registry list of environmental toxicants, cadmium (Cd) remains a prominent public health concern. Cd is a naturally occurring, ubiquitous environmental toxicant with no known biological role in humans1. As a result of various industrial and agricultural processes, Cd exposure has dramatically increased over the last several decades1. Exposure primarily occurs through the consumption of contaminated foods and drinking water, with well-water posing an increased risk of exposure, in addition to the inhalation of cigarette smoke and other occupational-related exposures2. Chronic exposure to Cd is known to negatively impact overall cardiovascular health, renal function, and increases overall risk of mortality resulting from various cancers3–5.
Prenatal Cd exposure poses a particularly unique health risk as Cd can accumulate within the placenta itself and does not readily enter fetal circulation, potentially influencing both fetal and maternal health through placental impacts6. Resting at the interface of the maternal and fetal environment, the placenta participates in a variety of molecular processes, including gas and waste exchange, nutrient transport, immunomodulation, and endocrine signaling7. As a central vascular organ of fetal growth and development, proper functionality of the placenta remains critical to successful gestational outcomes.
Gestational Cd exposure is associated with adverse pregnancy and birth outcomes including decreased overall placental efficiency, and increased risk of preeclampsia and low birth weight6,8–10. A growing body of evidence suggests that the atypical neurobehavioral outcomes and intellectual deficits seen in children exposed to Cd in utero are linked to Cd levels in cord blood and maternal circulation during pregnancy10–14. Given the placenta’s ability to produce neurotransmitters and its involvement in supporting fetal brain development, these data suggest that gestational Cd may affect exposure as a potential mediator of neurodevelopment by acting through the placenta15–17. In support of this hypothesis, our group has demonstrated that a doubling of placental Cd concentration was associated with an over 2-fold increased risk of a newborn being classified in a NICU Network Neurobehavioral Network Scales (NNNS) profile that is considered atypical14. While the exact molecular mechanisms that underlie these relationships remain unclear, extensive research has suggested Cd-associated changes to epigenetic landscapes may play a pivotal role in the developmental programming of pregnancy and birth outcomes through disruption of placental physiology and function8,18–21.
While Cd-related changes in DNA methylation patterns have been thoroughly outlined in developmentally relevant tissues through human cohort studies, this same level of characterization has yet to be defined in the context of other epigenetic forms of regulation, namely placental microRNAs (miRNA)20–22. MiRNAs are small RNA molecules (~22 nucleotides) capable of post-transcriptional regulation of gene expression that utilize base-pairing to bind target mRNAs resulting in either translational repression or mRNA degradation. The dysregulation of placental miRNA expression has previously been implicated in adverse pregnancy conditions, such as preeclampsia and fetal growth restriction, highlighting their potential role as modifiers of newborn and maternal health outcomes in response to environmental perturbations in utero23–27. However, many of these studies largely focused on newborn outcomes as they associate with placental miRNA expression as opposed to the relationship between prenatal environmental factors and expression of placental miRNA23,24,26,28,29.
In order to broaden our understanding of how gestational Cd exposure influences the placental miRNA landscape, we utilized placental miRNA sequencing data, as well as placental trace metal quantification, from the Rhode Island Child Health Study (RICHS, n=115), and the New Hampshire Birth Cohort Study (NHBCS, n=281). We also integrated specific NNNS summary scores to examine if Cd-associated miRNA were also associated with newborn neurobehavioral performance.
RESULTS
This study analyzed miRNA sequencing and trace metal data from 115 placentae from RICHS and 281 placentae from the NHBCS. The demographics of the participants are outlined in Table 1. Generally, placentae collected in these studies were from full term pregnancies (≥37 weeks) from relatively healthy mothers who did not experience serious pregnancy complications. The most notable differences between the RICHS and NHBCS cohorts are with regards to the racial diversity of participants and infant birth size. The RICHS cohort is slightly more racially diverse than the NHBCS cohort (Table 1). RICHS, by design, was over-sampled for infants born large for gestational age (LGA) and small for gestational age (SGA). Additionally, NHBCS participants are from more rural regions, and utilize private water systems. Placental samples from the RICHS cohort exhibited higher mean Cd concentrations (4.386 ng/g) than the NHBCS sample mean (2.932 ng/g), with each cohort exhibiting approximately log-normally distributed concentrations (Fig. S1).
Table 1:
Demographic characteristics of RICHS and NHBCS participants included in the miRNA sequencing analysis
| MATERNAL CHARACTERISTICS | RICHS (n=115) | NHBCS (n=281) |
|---|---|---|
| Age (yrs) – mean (range) | 30 (18–40) | 31 (18–44) |
| Race (White) – % (n) | 72 (83) | 100 (281) |
| Pregnancy Smoking – % (n) | 10 (12) | 13 (37) |
| Pre-Pregnancy Body Mass Index (BMI) (kg/m2) – mean (range) | 26.5 (16.0 – 45.4) | 26.1 (16.9 – 49.9) |
| Pre-Pregnancy BMI Group | ||
| Normal and Underweight – % (n) | 47.8 (55) | 47.6 (134) |
| Overweight – % (n) | 29.5 (34) | 30.6 (86) |
| Obese – % (n) | 22.6 (26) | 18.5 (52) |
| INFANT CHARACTERISTICS | ||
| Placental Cd2+ (ng/g) mean (standard deviation) | 4.39 (2.52) | 2.93 (1.99) |
| Sex (Female) – % (n) | 46.9 (54) | 51.6 (145) |
| Gestational Age (wks) – mean (range) | 39.0 (37 – 41) | 39.5 (31.4 – 42.1) |
| Birthweight (g) – mean (range) | 3610 (2160–5465) | 3469 (1380–5216) |
| Birthweight Group | ||
| Small for Gestational Age (SGA) – % (n) | 15.6 (18) | 4.3 (12) |
| Average for Gestational Age (AGA) – % (n) | 45.2 (52) | 85.1 (242) |
| Large for Gestational Age (LGA) – % (n) | 39.1 (45) | 9.6 (27) |
Cd Differential Expression Analyses
In order to analyze associations between placental Cd concentrations and placental miRNA expression, we performed differential expression analyses using negative binomial generalized linear models on placental miRNA sequencing data. Transcript abundances were regressed against log2-transformed placental Cd concentrations while adjusting for gestational age, birth weight percentile, maternal pregnancy smoking status, RNA integrity estimates, and unknown confounders via surrogate variables in each cohort30,31. To further characterize the robustness of the differential expression analysis results, we utilized a meta-analytical approach where we combined parameter estimates and their respective standard-errors from each cohort using an inverse variance weighted fixed effects meta-analysis32. The NHBCS cohort-level analysis revealed five DEmiRs (Differentially Expressed miRNA) whose expression significantly associated with placental Cd concentrations at a False Discovery Rate (FDR) < 0.1, (miR-10b-3p, miR-10b-5p, miR-193b-5p, miR-506-3p and miR-509-3p) (Table 2 & Table S2). One of these five miRNAs, miR-509-3p, met a strict Bonferroni test correction threshold (p < 6.435e-05). Considering the potential influence of tobacco-related Cd exposure, we performed the NHBCS differential expression analysis in a population where all reported smokers were excluded (Fig. S2). The effect sizes resulting from the original model proved to be consistent with that of the exclusionary model where reported smokers were excluded (R2 = 0.86; Fig. S3A). Additionally, 100% of the coefficients from the self-reported exclusionary model are within the estimated 95% CI’s of the smoking-adjusted model (Fig. S3B & S3C). In the RICHS cohort-level analysis, we did not observe any miRNAs associated with placental Cd concentrations at an FDR<0.1 (Table S1). The meta-analysis emphasized the robustness of the miR-509-3p and miR-193b-5p association with Cd, and further revealed miR-1248 as consistently differentially expressed with respect to placental Cd concentrations, all at an FDR < 0.15 (Fig. 1A, Table 2 & Table S3).
Table 2:
RICHS, NHBCS and Meta-Analysis Results Summary
| miRNA | Study | Log2(Fold-Change) | Standard Error | p-value | −Log10(pvalue) | FDR |
|---|---|---|---|---|---|---|
| hsa-miR-10b-3p | RICHS | 0.1808 | 0.1449 | 0.2121 | 0.6734 | 0.9971 |
| hsa-miR-10b-3p | NHBCS | −0.3126 | 0.0843 | 0.0002 | 3.6831 | 0.0537* |
| hsa-miR-10b-5p | RICHS | 0.0993 | 0.1255 | 0.4289 | 0.3677 | 0.9971 |
| hsa-miR-10b-5p | NHBCS | −0.2866 | 0.0771 | 0.0002 | 3.6926 | 0.0537* |
| hsa-miR-1248 | Meta | −0.2395 | 0.0665 | 0.0003 | 3.5022 | 0.1128 |
| hsa-miR-1248 | RICHS | −0.2221 | 0.1409 | 0.1149 | 0.9396 | 0.9971 |
| hsa-miR-1248 | NHBCS | −0.2444 | 0.0754 | 0.0012 | 2.9268 | 0.1381 |
| hsa-miR-193b-5p | Meta | 0.1496 | 0.0431 | 0.0005 | 3.2888 | 0.1229* |
| hsa-miR-193b-5p | RICHS | 0.0844 | 0.0841 | 0.3158 | 0.5007 | 0.9971 |
| hsa-miR-193b-5p | NHBCS | 0.1728 | 0.0502 | 0.0006 | 3.2444 | 0.0885* |
| hsa-miR-506-3p | Meta | 0.2275 | 0.0711 | 0.0014 | 2.8645 | 0.1632 |
| hsa-miR-506-3p | RICHS | 0.0735 | 0.1160 | 0.5264 | 0.2787 | 0.9971 |
| hsa-miR-506-3p | NHBCS | 0.3201 | 0.0899 | 0.0004 | 3.4305 | 0.0721* |
| hsa-miR-509-3p | Meta | 0.1924 | 0.0522 | 0.0002 | 3.6480 | 0.1128* |
| hsa-miR-509-3p | RICHS | 0.0654 | 0.0838 | 0.4352 | 0.3613 | 0.9971 |
| hsa-miR-509-3p | NHBCS | 0.2728 | 0.0666 | 0.0000 | 4.3706 | 0.0331 |
Indicates FDR significance determined by analysis-specific threshold (RICHS & NHBCS: FDR ≤ 10%, Meta-analysis: FDR ≤ 15%
Figure 1: Placental Cd has associations with specific miRNA expression consistent across two independent cohorts.
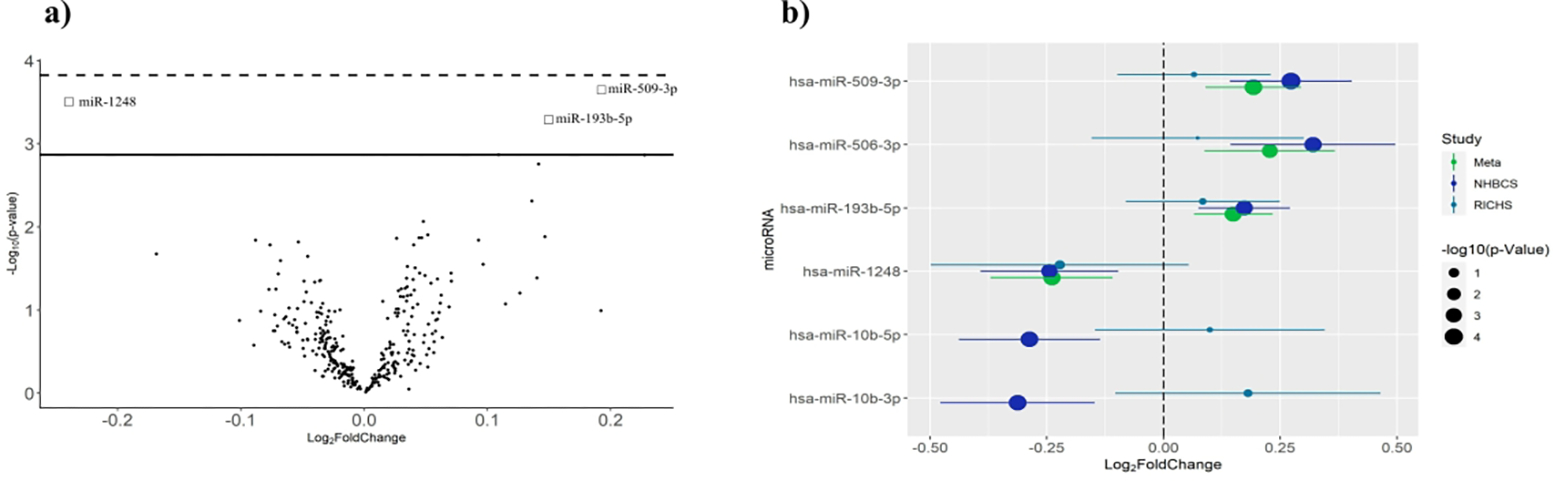
(a) Volcano plot representing the results of the NHBCS and RICHS meta-analysis. The y-axis shows the −log10(p-values) in the association of each miRNA with log2-transformed placental cadmium concentrations. The x-axis displays the effect estimates in units of log2 fold change in each miRNA’s transcript abundance per doubling of placental cadmium. 3 miRNAs are associated with placental cadmium accumulation at an FDR < 0.15; however, none are significant following Bonferroni correction (p-value < 1.50e-04). (b) Estimates of log2 fold change of miRNA transcript abundances of Cd-associated DEmiRs identified compared across the RICHS and NHBCS cohort analyses and meta-analysis. MiR-10b-3p and miR-10b-5p do not hold meta-analysis estimates considering the opposing direction of effect estimates identified in the cohort level analyses. Error bars in each plot represent the estimated 95% confidence interval, size of the point represents the −log10(p-value) of the estimate, and the color indicates the study from which the data were generated.
The effect size estimates of Cd-associated DEmiRs were largely consistent between the cohort-level and meta-analysis with the exception of miR-10b-3p and miR-10b-5p, which displayed opposing directions of effect between the RICHS and NHBCS analyses and were therefore excluded from the meta-analysis (Table 2, Fig. 1B).
Cd-DEmiR Target Prediction and Overrepresentation Analyses
Bioinformatic targets of each Cd-associated miRNA from the NHBCS cohort-level analysis were predicted using the miRNA Data Integration Portal (miRDIP)33. MiRDIP predicted 833 targets across all five differentially expressed miRNAs. However, perfect complementarity is not required between a miRNA and the target transcript, leaving in silico miRNA target prediction prone to the production of false positive interactions that may not occur biologically. To further enhance our pool of predicted targets, a strict filtering approach was taken. We removed predicted transcripts that are not shown to be expressed within RICHS placentae. After this filtering step, 806 predicted targets remained (Table S4). Targets were then utilized in a pathway overrepresentation analysis within ConsensusPathDB (CPDB), where only a single DEmiR’s list of predicted targets were analyzed at a time to highlight DEmiR-specific pathway dysregulation. These Cd-associated miRNA targets were largely revealed to participate in biological pathways relevant to cellular transcription, cellular metabolism and broader developmental signaling34,35. The predicted targets of miiR-509-3p and miR-193b-5p, which were robustly associated with Cd concentrations across the NHBCS and meta-analyses were enriched among nervous system development pathways (q-value < 0.001) (Fig. 2, Table S5 & S6).
Figure 2. Predicted targets of Cd-associated miRNAs miR-509-3p and miR-193b-5p are overrepresented in biological pathways relevant to nervous system development.
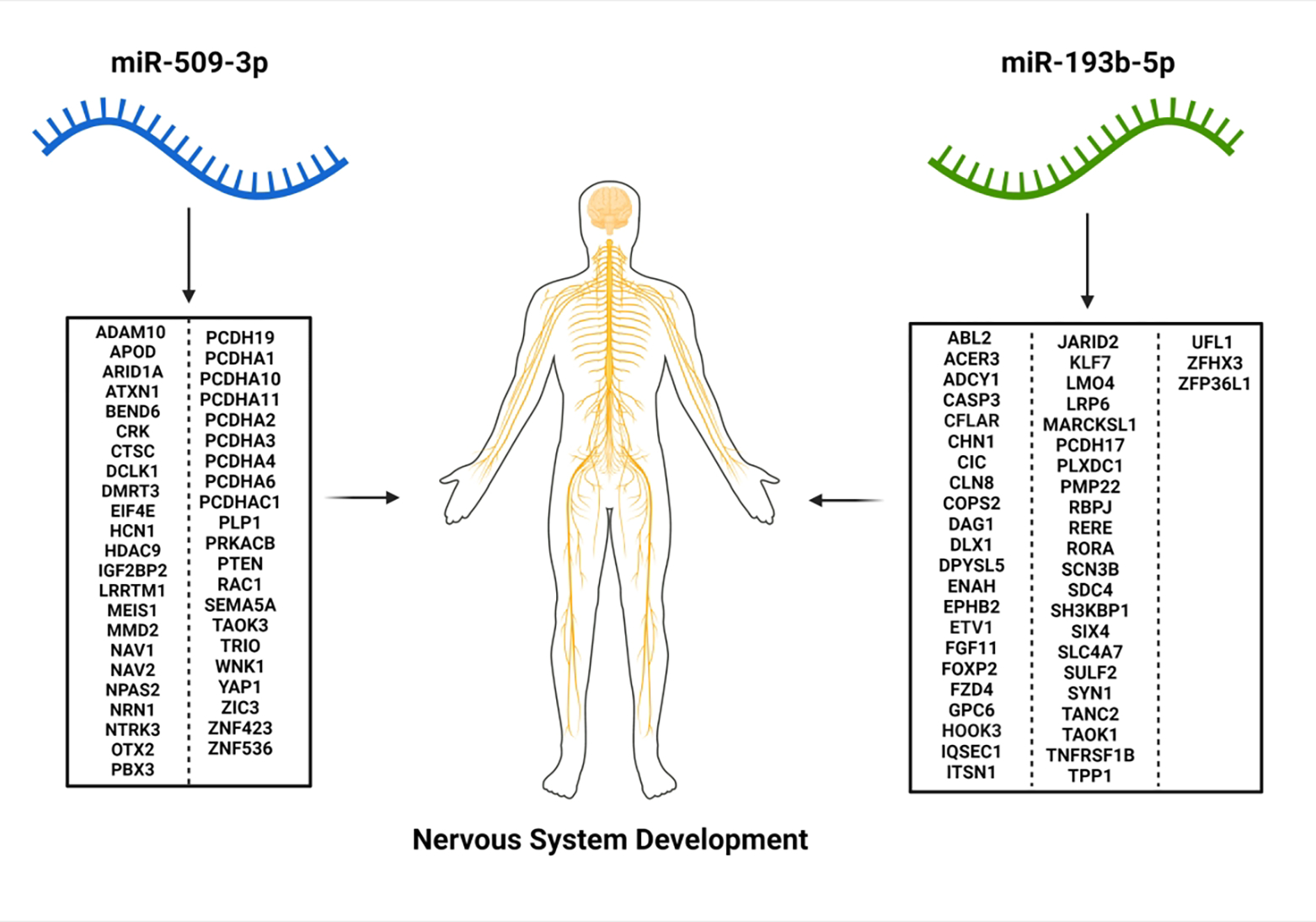
The predicted targets of miR-509-3p and miR-193b-5p commonly reported a potential influence toward nervous system development (q-value < 0.001). MiR-193b was predicted to target 47 genes involved in nervous system development while miR-509-3p was predicted to target 44 unique targets. While all genes listed contribute to the overrepresentation of nervous system development, no predicted gene is a predicted target of multiple miRNA. Of the 91 total mRNA targets involved in nervous system development, 72 were found to be expressed at >1cpm in the RICHS placental total RNA sequencing dataset. Created with www.BioRender.com.
NNNS and Cd-DEmiR Bivariate Association Analyses
Given the over-representation of target genes involved in nervous system development, we explored whether the Cd DEmiRs were associated with neurobehavioral phenotypes. Previous work from our group has constructed neurobehavioral profiles based on NICU Network Neurobehavioral Scale (NNNS) data from RICHS infants14. NNNS scores describe 13 characteristics of newborn neurobehavior. Tung et al14, utilized Latent Profile Analysis to group infants based on their component scores from this assessment. Among these predicted groups, the most atypical profile, which was also found to associate with a greater mean placental Cd concentration, was characterized by extreme scores in arousal level, quality of movement, stress abstinence, excitability and self-regulation capacity14. We investigated these scales for potential associations with normalized transcript abundances of miR-509-3p and miR-193b-5p in RICHS newborns (n=114). Simple linear regression analyses revealed nominally significant, negative associations between quality of movement scores and normalized transcript abundances of Cd-associated miR-509-3p (β = −0.47 ± 0.38, p ≤ 0.01) and miR-193b-5p (β = −0.278 ± 0.287, p ≤ 0.1) (Fig. 3A &B, Table S8). Additionally, excitability scores appear to be positively associated with only miR-193b-5p transcript abundance (β = 0.27 ± 0.3, p ≤ 0.1) (Fig. 3C, Table S8). Arousal, stress abstinence and self-regulation capacity showed no relationship to these Cd-associated miRNAs (Table S8).
Figure 3. Quality of Movement and Excitability at birth are correlated with the expression of miR-509-3p and miR-193b-5p.
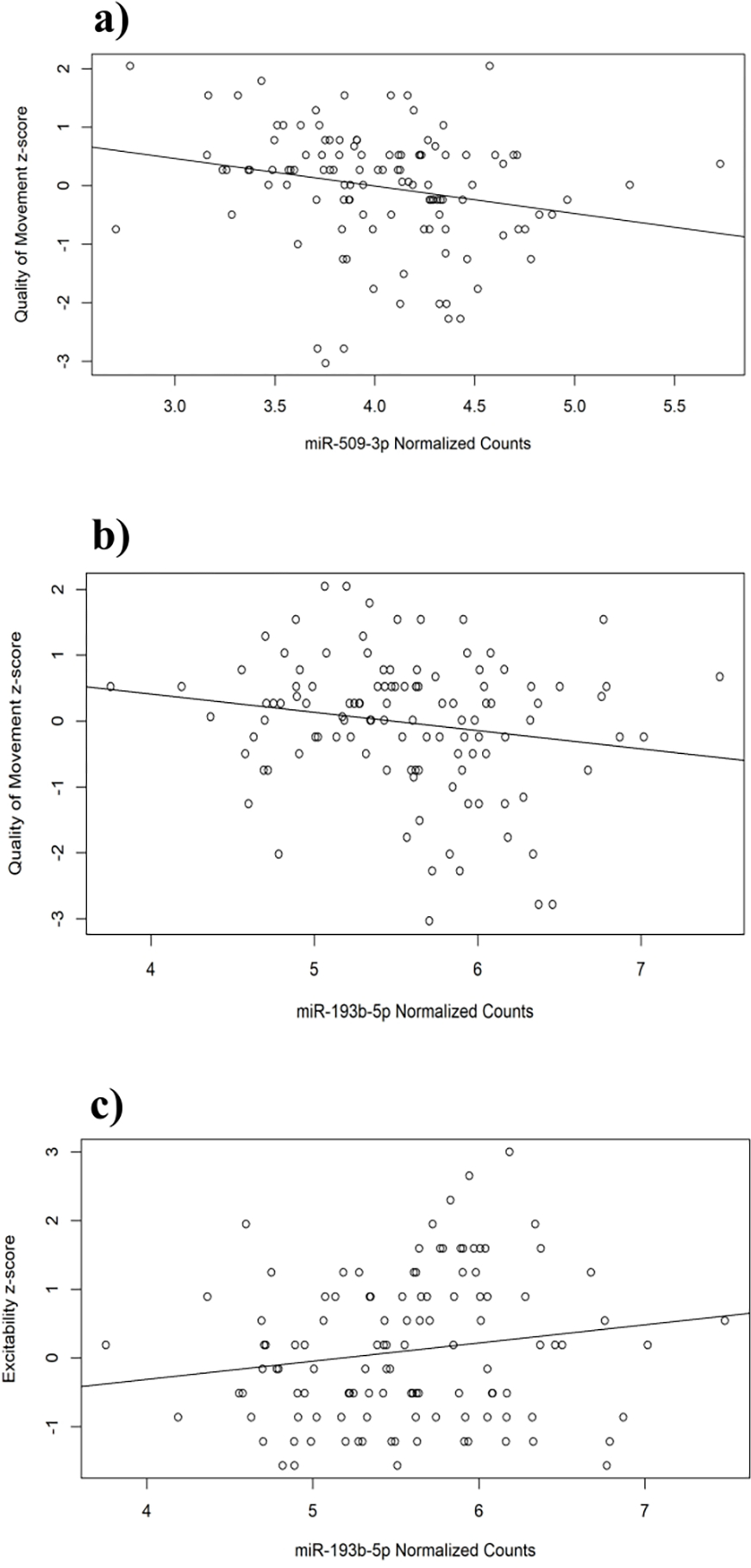
Scatterplots depicting the relationship between normalized counts of miR-509-3p and miR-193b-5p with respect to individual, z-transformed NNNS metrics (y-axes) (n=114) as determined by simple linear regression. (A) There is a reported β = −0.47 ± 0.380 attenuation in quality of movement scores per-cpm change in miR-509-3p transcript abundance (p ≤ 0.01). (B) There is a reported β = −0.278 ± 0.287 attenuation in quality of movement scores per-cpm change in miR-193b-5p transcript abundance (p = 0.06). (C) There is a reported β = 0.27 ± 0.3 attenuation in excitability measurements per-cpm change in miR-193b-5p transcript abundance (p =0.09). Error for each analysis is reported as the estimated 95% confidence interval, results are considered nominally significant at a p-value threshold of p ≤ 0.1.
DISCUSSION
The developmental and reproductive toxicity of Cd is well documented and extensively studied, however, the relationship between Cd accumulation in the placenta, its impacts on the expression of placental miRNA and, in turn, effects on early neurobehavioral outcomes remains poorly understood6–10,21.
Here we have conducted an association study of mother-infant dyads from two independent cohorts to examine relationships between placental Cd concentrations and miRNA expression from human placentae. The methods for quantifying miRNA expression and Cd concentrations were identical in both cohorts, and the levels of Cd are low compared to the range of concentrations previously reported in other placenta-based studies conducted in humans (1.2ng/g to 53.3ng/g)36. We successfully identified five Cd-associated miRNAs within the NHBCS cohort-level analysis with adjustment for covariates at an FDR <0.1 (Fig. 1, Table S2). Within the RICHS cohort-level analysis we did not identify any associations between placental Cd concentrations and miRNA expression at FDR<0.1 (Table S1). However, the association of two of the five Cd-associated miRNAs identified in NHBCS were consistent in a fixed-effects meta-analysis; (miR-509-3p and miR-193b-5p, FDR < 0.15). Importantly, the effect size estimates of Cd-associated miRNAs remain consistent between cohorts with the exception of miR-10b-3p and miR-10b-5p (Fig. 3A & 3B). The inability to detect associations in the RICHS cohort alone may be related to the more modest sample size and the lower variability in the Cd concentrations present within RICHS placentae, even though the mean concentration was greater than that in NHBCS.
Relevance of miR-509-3p and miR-193b-5p to placental physiology
A small number of miRNAs are known to play critical roles in placental function and physiology, but the function of most miRNA, particularly in the placenta, is unclear.
Cd-associated placental miRNA miR-509-3p is clinically relevant to the progression of medulloblastomas as well as in ovarian cancer37–39. The expression of miR-509-3p is positively correlated with favorable disease outcomes while downregulation of miR-509-3p is associated with cellular invasion and migratory properties and poor disease prognosis37–39. MAP3K8 (Mitogen Activated Kinase Kinase Kinase 8), regulates oncogenic pathways in cancers, including renal cancers, and contains a canonical 509-3p binding sit in its 3’UTR, which is thought to play a role in allowing miR-509-3p to exert its tumor suppressive properties38. Mitogen-activated protein kinases (MAPKs) in general participate in the development of the trophectoderm in early placentation, but also contribute to the differentiation of trophoblasts, which is an ongoing process that plays a major role in placental maintenance and remodeling throughout development. Thus, miR-509-3p expression could contribute to similar functions involving MAPK-mediated invasion and migratory processes, as well as angiogenic processes seen in placentation40,41.
Originally identified as a hypoxia-inducible miRNA, miR-193b-5p is also implicated in various cancers where its upregulation is associated with improved disease prognosis resulting from decreased proliferation, migration and invasion of cancerous cells in both lung and breast cancers42–45, similar to the roles played by miR-509-3p. The expression of miR-193b-5p has also been implicated in preeclampsia and in cases of intrauterine growth restriction (IUGR) which are thought to arise from a potential miR-193b-5p-mediated dysregulation of TGFβ−2 signaling, a gene shown to be directly regulated by miR-193b-5p and which oversees many facets of migration and invasion of trophoblasts28,40,46–48.
In the context of our study, both miR-509-3p and miR-193b-5p are upregulated in association with respect to placental Cd. Their function as tumor suppressors in various cancers suggests their participation in overseeing analogous processes critical to proper placental function and successful birth outcomes; proliferation, migration, invasion, angiogenesis and differentiation37,38,40,42–44. Being critical to normal placental development and function, dysregulation of these cellular processes often manifest as various placental insufficiencies, such as preeclampsia, or IUGR, and thus may contribute to adverse maternal and/or fetal health outcomes.
Relationship between placental function and neurobehavioral outcomes
While responsible for the transfer of nutrients and gases, the placenta is also involved in the production of various neurotransmitters, such as serotonin, dopamine, norepinephrine and epinephrine which may then circulate and directly influence fetal brain development16,49. Poorer neurobehavioral outcomes, in general, are known to arise more frequently in infants born SGA, which suggests proper placental function may underlie various neurobehavioral outcomes50. Additionally, alterations in placental miRNA expression are associated with both attention scores and quality of movement scores at birth, suggesting placental miRNA expression may contribute to the developmental origins of early childhood neurobehavioral outcomes51.
Bioinformatic target prediction and pathway analysis highlighted potential additional functional impacts of miR-509-3p and miR-193b-5p. Of their combined 370 unique predicted mRNA targets, these miRNAs were revealed to potentially target genes enriched for nervous system development pathways (Fig. 2, Table S5 & S6). Our group has previously identified an association between NNNS atypical neurobehavioral profiles and placental Cd concentrations in RICHS participants, but the molecular mechanisms underlying this specific association have yet to be defined14. We identified negative, though not statistically significant, relationships between the expression of Cd-associated DEmiRs (509-3p and 193b-5p) and NNNS quality of movement scores (Fig. 3, Table S8), and a positive, though not statistically significant, relationship between miR-193b-5p and NNNS excitability scores. The quality of movement metric is a measure of motor control, where higher scores suggest a greater quality of movement indicated by smoother movements with little or no jitteriness or tremors52. The excitability metric is a measure of high levels of motor, state and physiologic reactivity, with higher scores indicating infants who become and remain irritable during the examination, whereas lower scores are signature for infants that display lower irritability and fewer state changes during the exam53.
Lower quality of movement, and higher excitability scores are generally associated with higher risk pregnancies, including those associated with maternal drug use, and are predictive of long-term neurobehavioral deficits54. Deficiencies in the performance of both quality of movement and excitability were also characteristic within the atypical neurobehavioral profile previously characterized in RICHS14. These relationships suggest gestational Cd exposure may contribute to abnormal neurobehavioral outcomes at birth through dysregulation of placental miRNA expression. While RICHS does not have available long-term data to examine the persistence of these effects, other cohorts have linked NNNS atypical-neurobehavioral profiles to persistent adverse neurobehavioral performance through age 4.5, suggesting that Cd exposure, through placental miRNA, could have long-term developmental programming implications54,55. Considering these changes in miRNA expression are detectable at birth, this may allow for the development of early interventional methods to mitigate long-term developmental deficits.
There are some limitations of our study which need to be noted. This is an observational study specifically sampling full-term placentae from live births. The cross-sectional design of this study cannot address the issue of temporality, as we cannot conclude that our results are representative of miRNA and mRNA associations throughout all of development. Although we have attempted to address relevant confounders through direct modeling and the inclusion of surrogate variables, it is still possible that unmeasured sources of confounding had biased our findings. Additionally, even though we report significant associations between placental Cd levels, miRNA abundance and neurobehavioral outcomes, these findings may not represent causal relationships. Mechanism-based research making use of careful experimental design is needed to more formally establish the links between toxic metal exposure, placental miRNA expression, and neurobehavioral outcomes. Lastly, both RICHS and NHBCS consist of predominantly healthy, white mothers from the New England region of the United States, potentially limiting the ability of these results to be generalized to a more diverse population.
CONCLUSION
Placental miRNAs are critical molecules in post-transcriptional gene regulation. Their regulatory activities have been linked to overall physiology and function of the placenta throughout gestation and offspring developmental outcomes, marking them as essential to successful gestational outcomes. Here we have identified the abundance of miR-509-3p and miR-193b-5p to be associated with placental Cd, and with NNNS metrics indicative of atypical neurobehavior. Targets of these miRNAs are predicted to be involved in nervous system development and the transcript abundance of these Cd-associated miRNAs are also associated with individual NNNS summary scores, including quality of movement and excitability. These Cd-associated placental miRNAs may influence neurobehavioral outcomes at birth through their role in overseeing placental physiology throughout gestation, allowing for them to serve as early life indicators of long-term neurodevelopmental outcomes.
METHODS
Cohorts
New Hampshire Birth Cohort Study (NHBCS)
The New Hampshire Birth Cohort Study (NHBCS) is an ongoing birth cohort initiated in 2009 of mother-infant pairs. Pregnant women between the ages of 18 and 45 were recruited from participating prenatal care clinics in New Hampshire, USA. Women were enrolled in this study only if they reported if their primary source of drinking water was from an unregulated residential well since their previous menstrual period and had no plans to move before delivery. All participants provided written informed consent in accordance with the requirements of the Committee for the Protection of Human Subjects, the Institutional Review Board (IRB) of Dartmouth College. Placental gross measures such as placental diameter (cm) and placental weight (g) were collected after delivery. Interviewer administered questionnaires and medical record abstraction were utilized to collect sociodemographic, lifestyle and anthropometric data. Data provided by this study include placental microRNA transcript abundance and placental trace metal concentrations (n=281).
The Rhode Island Child Health Study (RICHS)
The Rhode Island Child Health Study (RICHS) is a cohort of mother-infant pairs from the Women & Infants Hospital in Providence, Rhode Island, enrolled between September 2010 and February 2013. All mothers were at least 18 years of age, had no life-threatening conditions, and delivered singletons free of congenital/chromosomal abnormalities at or after 37 weeks of gestation. Infants who were born small for gestational age (≤ 10th birth weight percentile) or large for gestational age (≥ 90th birth weight percentile) were oversampled. Infants who were adequate for gestational age (between 10th and 90th birth weight percentile) that matched on gestational age and maternal age were coincidentally enrolled. All participants provided written informed consent and all protocols were approved by the IRBs at the Women & Infants Hospital of Rhode Island and Emory University, respectively. Data provided by this study include placental microRNA transcript abundance and placental trace metal concentrations (n=115). Interviewer-administered questionnaires were utilized to collect sociodemographic and lifestyle data. Structured medical record review was used to collect anthropometric and medical history data.
Data Collection
Anthropometric Measures and Potential Covariates
Birth weight (g), head circumference (cm), and birth length (cm) were taken from medical records. Z-Scores were calculated for each, standardized by gestational age and infant sex, via Fenton growth curves56. Infants with standardized birth weight percentiles below the 10th percentile as small for gestational age (SGA), those above the 95th percentile as large for gestational age (LGA), and those between the 10th and 90th percentiles as adequate for gestational age (AGA).
Potential Covariates
Maternal smoking during pregnancy was defined as any self-reported smoking during any point of pregnancy versus those who reported no smoking during pregnancy. Fetal sex was obtained from medical records. Only samples with complete covariate, placental Cd, and placental miRNA sequencing data were utilized in this study.
Tissue Collection
Each cohort utilized the same sampling protocol. Placental parenchyma samples were collected within 2 hours of birth; sections were obtained two centimeters from the umbilical cord insertion side and were free of maternal decidua. One sample of collected tissue was immediately placed into RNA later solution (Life Technologies, Grace Island, NY) for at least 48 hours, then blotted dry and stored at −80°C. Another sample, for trace metal analysis, was immediately snap frozen and stored at −80°C.
Cadmium Quantification
Trace element concentrations were quantified in both RICHS and NHBCS placental samples at the Dartmouth Trace Elements Analysis core through inductively coupled plasma mass spectrometry (ICP-MS); details of this analysis have been previously described57. Given the right-tailed skewed nature of the Cd concentration distributions in each cohort, concentrations were log2 transformed for normalization to be used in differential expression analyses (Fig. S1).
NNNS Assessment
The NICU Network Neurobehavioral Scale (NNNS) is a standardized assessment which comprehensively examines the neurological function, behavioral functioning and signs of stress in newborns52. The NNNS yields summary scores on 13 individual behaviors and functions including habituation, attention, arousal, self-regulation, handling, quality of movement, excitability, lethargy, non-optimal reflexes, asymmetry reflexes, hypertonicity, hypotonicity, and stress/abstinence. Z-transformed measurements of individual NNNS metrics that are highly variable between atypical and neuro-typical constructed neurobehavioral profiles were utilized in downstream associative modeling (arousal level, quality of movement, stress abstinence, excitability, and self-regulation capacity)14.
Within RICHS, NNNS was administered after the first 24 hours of life, and prior to discharge by certified psychometrists. Of the 840 enrolled participants in RICHS, 625 were assessed with NNNS, however, 114 (99%) of these participants overlap with our placental miRNA sequencing and trace metal datasets.
NHBCS microRNA isolation and sequencing
Total RNA was extracted from fixed placenta using the Qiagen miRNeasy Mini Kit and a TissueLyser LT (Qiagen, Frederick, MD) following the manufacturer’s protocol. Briefly, 25–35mg of placental tissue was placed into a 2 ml round bottom tube with 700 μl of Qiazol Lysing Reagent and one 5 mm stainless steel bead. The tissue was homogenized on the TissueLyser LT for 2 minutes at 40 Hz. The resulting homogenate was processed with the Qiagen miRNeasy Mini Kit and eluted in 30 μl RNase-free water. The RNA was quantitated on a NanoDrop 2000 (Thermo Fisher, Waltham, MA) and quality checked on Agilent Bioanalyzer using the Agilent RNA 6000 Nano Kit (Agilent, Santa Clara, CA). Single end, 1 × 75 bp next-generation sequencing of placental microRNA was performed by Qiagen Genomic Services (Frederick, Maryland).
RICHS microRNA isolation and sequencing
Total RNA was extracted from placenta using the Qiagen miRNeasy Mini Kit and a TissueLyser LT (Qiagen, Germantown, MD) following manufacturer’s protocol. Briefly, 25–35 mg of frozen, powdered placental tissue was placed in a 2 ml round bottom tube with 700 μl of Qiazol Lysing Reagent and one 5 mm stainless steel bead. The tissue was homogenized in a pre-chilled tube holder on the TissueLyser LT for two, 5-minute cycles at 30 Hz. The resulting homogenate was processed with the Qiagen miRNeasy Mini Kit with on-column DNAse digestion and eluted in 50 μl RNase-free water. The RNA was quantitated on a NanoDrop (Thermo Fisher, Waltham, MA) and quality checked on Agilent Bioanalyzer using the Agilent RNA 6000 Nano kit (Agilent, Santa Clara, CA). Single end, 1 × 50 bp next generation sequencing of placental miRNA was performed by Omega Bioservices (Norcross, Georgia) as previously described58.
Small RNA-seq processing and quality control
Raw FASTQ reads obtained from a total of 115 RICHS and 281 NHBCS samples (n=396) were subject to adaptor trimming with cutadapt v1.1634. The 3’ adaptor sequence were trimmed (TGGAATTCTCGGGTGCCAAGG) and then four bases were trimmed from each end of the read following vendor’s recommendation (BIOO scientific, Austin TX). We then used trimmed reads and miRDeep2 to quantify microRNA59. miRDeep2 was used to first perform alignment using bowtie1 with human genome hg3860. The ‘Quantifier’ module in miRDeep2 was used to obtain raw counts of microRNAs with miRBase version 2261.
Transcript Filtering and Normalization
Raw miRNA counts were imported into DESeq2 for normalization and differential expression analysis. Only miRNA transcripts with more than one count per million in at least 10 percent of samples were included, leaving 778 miRNA transcripts to be analyzed of the initial 2656 sequenced transcripts. Dispersion estimates were then calculated, followed by generation of median ratio size factor estimates to normalize counts for analysis with in DESeq262. Normalized counts were then exported from DESeq2 for Surrogate Variable Analysis (SVA). The Variance Stabilization Transformation (VST) was applied to count matrices to yield approximately normalized and log2-transformed abundances, which were utilized in bivariate association analyses discussed below58.
RICHS total RNA Isolation, Sequencing, Processing and Quality Control
Total RNA was extracted from fixed, pulverized placenta using the RNeasy Mini Kit (Qiagen) and stored at −80°C until analysis. We quantified RNA using a Nanodrop Spectrophotometer (Thermo Scientific), assessed RNA integrity via Agilent Bioanalyzer (Agilent), removed ribosomal RNA with a Ribo-Zero Kit63, performed cDNA conversion using random hexamers (Thermo Scientific), followed by transcriptome-wide 50bp single-end RNA sequencing via the HiSeq 2500 platform (Illumina)64. Initial quality control was performed on raw reads using the FastQC software. Reads passing the quality control metrics were then mapped to the human reference genome (hg19) utilizing the Spliced Transcripts Alignment to a Reference (STAR) aligner65. Parametric estimates of dispersion were calculated, and the median ratio method was used to estimate size factors for normalization for modelling of transcripts with DESeq262. Count matrices normalized by the Variance Stabilizing Transformation (analogous to normalization and log2-transformation) were exported from DESeq2 for utilization in bioinformatic target prediction filtering and gene ontology analysis.
Statistical Analyses
Surrogate Variable Analysis
To adjust for unknown confounders, such as cell-type heterogeneity and unmeasured technical variation, surrogate variables were estimated for normalized miRNA transcript reads using the sva package30,31. In the svaseq function, the iteratively re-weighted least squares algorithm was used to estimate surrogate variables based on empirically derived control transcripts. The full model, for each RICHS and NHBCS estimations, (mod argument) contained the following covariates: log2[Cd2+], gestational age (wks), birth weight percentile, maternal pregnancy smoking status, and RNA integrity estimates. The null model, for each RICHS and NHBCS, contained all previously listed covariates while excluding log2[Cd2+]. For each cohort, one surrogate variable was included as a covariate in the final model of each respective analysis.
Differential expression analyses
MiRNA transcript counts were modeled using a negative binomial generalized linear model to identify differentially expressed transcripts in DESeq231. For each of the 778 individual miRNA transcripts which passed strict filtering and quality criteria, miRNA counts were regressed on log2-transformed placental Cd concentrations. The following covariates were included in each independent model: gestational age (wks), birth weight percentile, maternal pregnancy smoking status, RNA integrity estimates and the first estimated surrogate variable30,31. MiRNAs with a false discovery rate (FDR) less than 10% were considered to be a differentially expressed miRNA (DEmiR) with respect to log2[Cd2+]. Example code for the cohort-specific differential expression analyses can be found at https://doi.org/10.15139/S3/KHXJ2G.
Meta-Analysis
A fixed effects meta-analysis was performed, utilizing the inverse variance-weighted method implemented in METAL using the standard error based method32. Study-specific summary statistics from each differential expression analysis were utilized, allowing for adjustment for study-specific parameters prior to the final meta-analysis. From this meta-analysis, only miRNAs with consistent direction of effects across both the independent RICHS and NHBCS differential expression analyses, leaving 333 miRNAs to be investigated. Only those miRNAs displaying an FDR < 0.15 were considered to be meaningfully dysregulated with respect to log2-transformed placental Cd concentrations.
Target prediction and filtering
Potential targets of DEmiRs were identified using miRDIP33. Only targets within the top 1% of confidence scores were returned. To further enhance the biological relevance of target mRNAs we then utilized existing whole transcriptome RNA-seq data from RICHS to only allow mRNAs that were expressed >1cpm in at least 10% of RICHS whole transcriptome RNA-seq samples to be considered a potential biological target58.
Gene Ontology and Pathway analysis
Predicted and filtered DEmiR targets were then tested for pathway overrepresentation within ConsensusPathDB (CPDB)34,35, against all genes that passed general QC filtering in RICHS whole transcriptome RNA-seq analysis34,35,58. CPDB utilizes 12 separate biological pathway databases and calculates an enrichment p-value from the hypergeometric distribution of genes in the list of miRNA targets and the pathway gene set. False discovery rates were calculated from the enrichment p-values, and a q-value ≤0.001 was considered a significant enrichment of miRNA targets in the tested pathway/ontology group.
Bivariate Association Analyses
Simple linear regression was used to characterize the relationship between VST counts of Cd-associated placental miRNAs and z-transformed individual NNNS metrics from RICHS infants (Fig. S4) that are highly variable across estimated neurobehavioral profiles, previously described by Tung et al14. The metrics analyzed included: arousal level, quality of movement, stress abstinence, excitability, and self-regulation capacity. Among the NNNS assessed infants, 114 (99%) of these participants overlap with between the placental miRNA sequencing data and trace metal measurements. Associations were reported as nominally significant with a p-value ≤ 0.1.
Supplementary Material
IMPACTS:
This research aims to address the poor understanding of the molecular mechanisms governing adverse pregnancy and newborn health outcomes in response to Gestational cadmium (Cd) exposure.
Our results outline a robust relationship between Cd-associated placental microRNA expression and NICU Network Neurobehavioral Scales (NNNS) at birth indicative of atypical neurobehavior.
This study utilized healthy mother-infant cohorts to describe the role of Cd-associated dysregulation of placental microRNAs as a potential mechanism by which adverse neurobehavioral outcomes are developmentally programmed.
ACKNOWLEDGEMENTS:
We acknowledge the cooperation of the participants enrolled in the RICHS and NHBCS, and the contributions of the research staff who also worked on RICHS and NHBCS.
FINANCIAL SUPPORT:
This work was supported by the National Institutes of Health (NIH-NIGMS T32GM008490, NIH-NIEHS R24ES028507, NIH-NIEHS R01ES025145, NIH-NIEHS P30ES019776; NIH-NIGMS P20GM104416 and NIH-NIMHD R01MD011698).
Footnotes
DISCLOSURE STATEMENT:
The authors declare they have no competing interests or personal relationships that would potentially influence the work presented in this paper.
ETHICAL STANDARDS:
All participants provided written, informed consent and all protocols were approved by the IRBs at the Women & Infants Hospital of Rhode Island, Dartmouth College and Emory University, respectively.
DATA AVAILABILITY:
The data that support the findings of this study, and example code used in the current study, are available publicly from the UNC Dataverse repository at https://doi.org/10.15139/S3/KHXJ2G.
Summary level results from the analyses conducted in the current study are included within the published article, and its supplemental information files.
REFERENCES
- 1.WHO. Exposure to Cadmium : A Major Public Health Concern. (2010).
- 2.Jarup L & Akesson A Current Status of Cadmium as an Environmental Health Problem. Toxicol Appl Pharmacol 238, 201–208 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Adams SV, Passarelli MN & Newcomb PA Cadmium Exposure and Cancer Mortality in the Third National Health and Nutrition Examination Survey Cohort. Occup Environ Med 69, 153–156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters JL, Perlstein TS, Perry MJ, McNeely E & Weuve J Cadmium Exposure in Association with History of Stroke and Heart Failure. Environ Res 110, 199–206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suwazono Y et al. Benchmark Dose for Cadmium-Induced Renal Effects in Humans. Environ Health Perspect 114, 1072–1076 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Punshon T et al. Placental Metal Concentrations in Relation to Placental Growth, Efficiency and Birth Weight. Environ Int 126, 533–542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross JC et al. Genes, Development and Evolution of the Placenta. Placenta 24, 123–130 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Hussey MR et al. Placental Lncrna Expression Associated with Placental Cadmium Concentrations and Birth Weight. Environ Epigenet 6, dvaa003 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laine JE et al. Placental Cadmium Levels Are Associated with Increased Preeclampsia Risk. PLoS One 10, e0139341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hudson KM et al. Transcriptomic, Proteomic, and Metabolomic Analyses Identify Candidate Pathways Linking Maternal Cadmium Exposure to Altered Neurodevelopment and Behavior. Scientific Reports 11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J et al. Interaction of Prenatal Bisphenols, Maternal Nutrients, and Toxic Metal Exposures on Neurodevelopment of 2-Year-Olds in the Apron Cohort. Environ Int 155, 106601 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Tian LL et al. Effects of Gestational Cadmium Exposure on Pregnancy Outcome and Development in the Offspring at Age 4.5 Years. Biol Trace Elem Res 132, 51–59 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Wang Y et al. Effects of Prenatal Exposure to Cadmium on Neurodevelopment of Infants in Shandong, China. Environ Pollut 211, 67–73 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Tung PW et al. Association between Placental Toxic Metal Exposure and Nicu Network Neurobehavioral Scales (Nnns) Profiles in the Rhode Island Child Health Study (Richs). Environ Res 204, 111939 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lester BM & Marsit CJ Epigenetic Mechanisms in the Placenta Related to Infant Neurodevelopment. Epigenomics 10, 321–333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenfeld CS The Placenta-Brain-Axis. J Neurosci Res 99, 271–283 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shallie PD & Naicker T The Placenta as a Window to the Brain: A Review on the Role of Placental Markers in Prenatal Programming of Neurodevelopment. Int J Dev Neurosci 73, 41–49 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Vilahur N, Vahter M & Broberg K The Epigenetic Effects of Prenatal Cadmium Exposure. Curr Environ Health Rep 2, 195–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu S, Dai J, Qu T & He Z Emerging Roles of Micrornas and Long Noncoding Rnas in Cadmium Toxicity. Biol Trace Elem Res 195, 481–490 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Cowley M et al. Effects of Cadmium Exposure on DNA Methylation at Imprinting Control Regions and Genome-Wide in Mothers and Newborn Children. Environ Health Perspect 126, 037003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everson TM et al. Cadmium-Associated Differential Methylation Throughout the Placental Genome: Epigenome-Wide Association Study of Two U.S. Birth Cohorts. Environ Health Perspect 126, 017010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appleton AA, Jackson BP, Karagas M & Marsit CJ Prenatal Exposure to Neurotoxic Metals Is Associated with Increased Placental Glucocorticoid Receptor DNA Methylation. Epigenetics 12, 607–615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Na Q, Song WW & Song GY Altered Expression of Mir-518b and Mir-519a in the Placenta Is Associated with Low Fetal Birth Weight. Am J Perinatol 31, 729–734 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Thamotharan S et al. Differential Microrna Expression in Human Placentas of Term Intra-Uterine Growth Restriction That Regulates Target Genes Mediating Angiogenesis and Amino Acid Transport. PLOS ONE 12, e0176493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesseur C et al. Tissue-Specific Leptin Promoter DNA Methylation Is Associated with Maternal and Infant Perinatal Factors. Mol Cell Endocrinol 381, 160–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashijima A et al. Characterization of Placenta-Specific Micrornas in Fetal Growth Restriction Pregnancy. Prenat Diagn 33, 214–222 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Awamleh Z, Gloor GB & Han VKM. Placental Micrornas in Pregnancies with Early Onset Intrauterine Growth Restriction and Preeclampsia: Potential Impact on Gene Expression and Pathophysiology. BMC Medical Genomics 12, 91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Östling H, Kruse R, Helenius G & Lodefalk M Placental Expression of Micrornas in Infants Born Small for Gestational Age. Placenta 81, 46–53 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Meng M et al. Whole Genome Mirna Profiling Revealed Mir-199a as Potential Placental Pathogenesis of Selective Fetal Growth Restriction in Monochorionic Twin Pregnancies. Placenta 92, 44–53 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Leek JT Svaseq: Removing Batch Effects and Other Unwanted Noise from Sequencing Data. Nucleic Acids Res 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W & Anders S Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biology 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willer CJ, Li Y & Abecasis GR Metal: Fast and Efficient Meta-Analysis of Genomewide Association Scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokar T et al. Mirdip 4.1-Integrative Database of Human Microrna Target Predictions. Nucleic acids research 46, D360–D370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamburov A et al. Consensuspathdb: Toward a More Complete Picture of Cell Biology. Nucleic Acids Res 39, D712–717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamburov A, Wierling C, Lehrach H & Herwig R Consensuspathdb--a Database for Integrating Human Functional Interaction Networks. Nucleic Acids Res 37, D623–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esteban-Vasallo MD, Aragonés N, Pollan M, López-Abente G & Perez-Gomez B Mercury, Cadmium, and Lead Levels in Human Placenta: A Systematic Review. Environmental health perspectives 120, 1369–1377 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahir BK, Elias NM & Lakka SS Sparc Overexpression Alters Microrna Expression Profiles Involved in Tumor Progression. Genes Cancer 8, 453–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su Z et al. Microrna-509-3p Inhibits Cancer Cell Proliferation and Migration by Targeting the Mitogen-Activated Protein Kinase Kinase Kinase 8 Oncogene in Renal Cell Carcinoma. Mol Med Rep 12, 1535–1543 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Pan Y et al. Mir-509-3p Is Clinically Significant and Strongly Attenuates Cellular Migration and Multi-Cellular Spheroids in Ovarian Cancer. Oncotarget 7, 25930–25948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daoud G et al. Erk1/2 and P38 Regulate Trophoblasts Differentiation in Human Term Placenta. J Physiol 566, 409–423 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang K & Zheng J Signaling Regulation of Fetoplacental Angiogenesis. J Endocrinol 212, 243–255 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H, Li S, Liu J & Ni B Microrna-193b Modulates Proliferation, Migration, and Invasion of Non-Small Cell Lung Cancer Cells. Acta Biochimica et Biophysica Sinica 44, 424–430 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Hulin J-A et al. Mir-193b Regulates Breast Cancer Cell Migration and Vasculogenic Mimicry by Targeting Dimethylarginine Dimethylaminohydrolase 1. Scientific Reports 7, 13996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin CH et al. Regulation of Plk1 through Competition between Hnrnpk, Mir-149-3p and Mir-193b-5p. Cell Death & Differentiation 24, 1861–1871 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pineles BL et al. Distinct Subsets of Micrornas Are Expressed Differentially in the Human Placentas of Patients with Preeclampsia. Am J Obstet Gynecol 196, 261 e261–266 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Awamleh Z, Gloor GB & Han VKM. Placental Micrornas in Pregnancies with Early Onset Intrauterine Growth Restriction and Preeclampsia: Potential Impact on Gene Expression and Pathophysiology. BMC Med Genomics 12, 91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X et al. The Aberrantly Expressed Mir-193b-3p Contributes to Preeclampsia through Regulating Transforming Growth Factor-Β Signaling. Scientific Reports 6, 19910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Awamleh Z & Han VKM Potential Pathophysiological Role of Microrna 193b-5p in Human Placentae from Pregnancies Complicated by Preeclampsia and Intrauterine Growth Restriction. Molecular Biology Reports 47, 6531–6544 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Rosenfeld CS Placental Serotonin Signaling, Pregnancy Outcomes, and Regulation of Fetal Brain Development†. Biol Reprod 102, 532–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Figueras F et al. Neurobehavior in Term, Small-for-Gestational Age Infants with Normal Placental Function. Pediatrics 124, e934–941 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Maccani MA, Padbury JF, Lester BM, Knopik VS & Marsit CJ Placental Mirna Expression Profiles Are Associated with Measures of Infant Neurobehavioral Outcomes. Pediatr Res 74, 272–278 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lester BM, Tronick EZ & Brazelton TB The Neonatal Intensive Care Unit Network Neurobehavioral Scale Procedures. Pediatrics 113, 641–667 (2004). [PubMed] [Google Scholar]
- 53.Boukydis CF, Bigsby R & Lester BM Clinical Use of the Neonatal Intensive Care Unit Network Neurobehavioral Scale. Pediatrics 113, 679–689 (2004). [PubMed] [Google Scholar]
- 54.Liu J et al. Neonatal Neurobehavior Predicts Medical and Behavioral Outcome. Pediatrics 125, e90–98 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sucharew H, Khoury JC, Xu Y, Succop P & Yolton K Nicu Network Neurobehavioral Scale Profiles Predict Developmental Outcomes in a Low-Risk Sample. Paediatr Perinat Epidemiol 26, 344–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fenton TR & Kim JH A Systematic Review and Meta-Analysis to Revise the Fenton Growth Chart for Preterm Infants. BMC Pediatr 13, 59 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Punshon T et al. Placental Metal Concentrations in Relation to Maternal and Infant Toenails in a U.S. Cohort. Environ Sci Technol 50, 1587–1594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kennedy EM et al. Placental Microrna Expression Associates with Birthweight through Control of Adipokines: Results from Two Independent Cohorts. Epigenetics, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedlander MR, Mackowiak SD, Li N, Chen W & Rajewsky N Mirdeep2 Accurately Identifies Known and Hundreds of Novel Microrna Genes in Seven Animal Clades. Nucleic Acids Res 40, 37–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffiths-Jones S, Saini HK, van Dongen S & Enright AJ Mirbase: Tools for Microrna Genomics. Nucleic Acids Res 36, D154–158 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anders S & Huber W Differential Expression Analysis for Sequence Count Data. Genome Biology 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang R et al. An Rna-Seq Strategy to Detect the Complete Coding and Non-Coding Transcriptome Including Full-Length Imprinted Macro Ncrnas. PLoS One 6, e27288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bentley DR et al. Accurate Whole Human Genome Sequencing Using Reversible Terminator Chemistry. Nature 456, 53–59 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dobin A et al. Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study, and example code used in the current study, are available publicly from the UNC Dataverse repository at https://doi.org/10.15139/S3/KHXJ2G.
Summary level results from the analyses conducted in the current study are included within the published article, and its supplemental information files.


