Abstract
Background
Oocyte components are maternally provided, solely determine oocyte quality, and coordinately determine embryo quality with zygotic gene expression. During oocyte maturation, maternal organelles are drastically reorganized and specialized to support oocyte characteristics. A large number of maternal components are actively degraded after fertilization and gradually replaced by zygotic gene products. The molecular basis and the significance of these processes on oocyte/embryo quality are not fully understood.
Methods
Firstly, recent findings in organelle characteristics of other cells or oocytes from model organisms are introduced for further understanding of oocyte organelle reorganization/specialization. Secondly, recent progress in studies on maternal components degradation and their molecular mechanisms are introduced. Finally, future applications of these advancements for predicting mammalian oocyte/embryo quality are discussed.
Main findings
The significance of cellular surface protein degradation via endocytosis for embryonic development, and involvement of biogenesis of lipid droplets in embryonic quality, were recently reported using mammalian model organisms.
Conclusion
Identifying key oocyte component characteristics and understanding their dynamics may lead to new applications in oocyte/embryo quality prediction and improvement. To implement these multidimensional concepts, development of new technical approaches that allow us to address the complexity and efficient studies using model organisms are required.
Keywords: autophagy, endocytosis, oocyte organelles, oocyte quality, ubiquitin‐proteasome system
During oocyte maturation, maternal organelles are drastically reorganized and specialized to support oocyte characteristics, whereas large amounts of maternal components are actively degraded after fertilization and gradually replaced by zygotic gene products. The significance of cellular surface protein degradation via endocytosis for embryonic development, and involvement of biogenesis of lipid droplets in embryonic quality, were recently reported using mammalian model organisms. Identifying key oocyte component characteristics and understanding their dynamics may lead to new applications in oocyte/embryo quality prediction and improvement.
1. INTRODUCTION
Currently, a major cause of infertility is the declining ability of oocytes to fertilize and develop because of oocyte aging, which correlates with an increase in the age of first childbearing. 1 However, the biological basis underlying changes in oocyte quality needs to be fully clarified. Chromosomes in unfertilized oocytes and pre‐implantation embryos occupy a large volume fraction of the cell. Failure of chromosome segregation is correlated with reduced developmental ability, depending on whether aneuploidy occurs during meiosis or mitosis. 2 However, especially in recent years, membrane structures such as organelles have been actively discussed as essential contributors to embryonic quality. This recent progress in the study of oocyte components other than chromosomes may be because active intervention is difficult in chromosomal abnormalities, as they lead to genetic mutations. In contrast, abnormalities of maternal components other than chromosomes are easier to diagnose and treat.
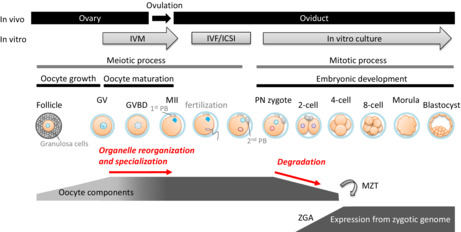
To understand the significance of oocyte components in appropriate development, differences between the oocyte and spermatozoon should be considered. First, similar to other oogamy‐type reproductive species, the mass of mammalian oocytes is huge relative to spermatozoon mass, generally accounting for more than 99.9% of the volume of the zygotes. In addition, spermatozoa have an extremely compact morphology with little cytoplasm. 3 , 4 , 5 , 6 Second, some sperm‐borne organelles introduced into the oocyte, such as mitochondria, are degraded by specific mechanisms. 7 , 8 Finally, prior to fertilization, the oocyte arrests in a quiescent state with extremely depressed transcriptional and translational activity, 9 , 10 , 11 while the spermatozoa replace histones, which provides structural support to the chromosome, with protamine to reduce chromosomal volume, consequently losing transcriptional or translational activity. 12 , 13 These facts indicate that de novo protein synthesis is severely depressed during terminal differentiation of mammalian gametes and immediately after fertilization, strongly suggesting that the content, interactions, enzymatic activities, and dynamics of oocyte components significantly determine embryonic quality. Importantly, maternal organelles that play essential roles in various cellular functions, increase quantitatively during follicular development and changes qualitatively during oocyte maturation, indicated by localization and activities (Figure 1). 14 , 15
FIGURE 1.
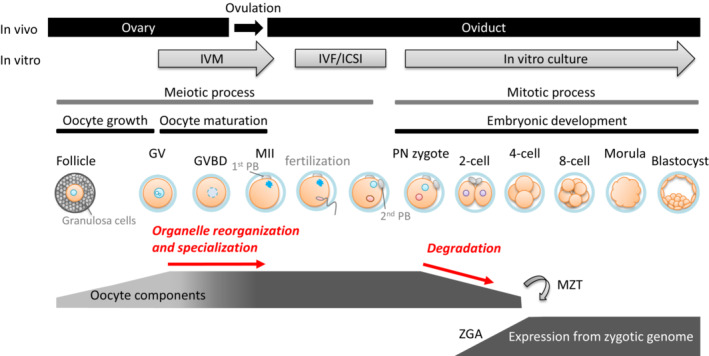
Changes of oocyte components during oocyte maturation and embryogenesis. In vivo and in vitro mammalian oocyte growth/maturation and embryonic development depicting corresponding stages of follicular development, oocyte maturation, and embryonic development. The changes of oocyte components prior to MZT are described, and the processes discussed in this review is shown in red letters. During the reorganization and specialization, oocyte components changes its localization and characteristics drastically (indicated by color gradation), and large amounts of oocyte components are actively degraded prior to ZGA. GV, germinal vesicle; GVBD, germinal vesicle breakdown; ICSI, intracytoplasmic sperm injection; IVF, in vitro fertilization; IVG, in vitro growth; IVM, in vitro maturation; MII, oocyte stops in metaphase of the second meiosis; MZT, maternal‐zygotic transition; PN, pronuclear; ZGA, zygotic gene activation
Although gene expression from the zygotic genome and subsequent de novo protein synthesis are important, these are induced mainly after the two‐cell stage. 10 , 11 On the other hand, various oocyte component degradative systems are activated before or at the two‐cell stage. For instance, large‐scale degradation of oocyte‐derived mRNA occurs among various organisms, 16 , 17 and autophagic activity increases immediately after fertilization. 18 Protein degradation by the ubiquitin–proteasome system (UPS) is also activated prior to the first mitotic cleavage. 19 , 20 , 21 These processes are essential for embryonic development. The initiation of zygotic gene expression accompanied by the disruption of oocyte components is referred to as the maternal–zygotic transition (MZT). The correlative timing of oocyte component degradation suggests its essential contribution towards providing resources prior to de novo protein/DNA synthesis via zygotic gene expression (Figure 1).
To better understand oocyte organelle reorganization and specialization, we first discuss recent updates in organelle biology research for oocyte/embryo and somatic cells. Especially, research on interactions among cellular organelles (and other structures) have progressed considerably in recent years. Comparing mammalian oocytes with other cells highlights the uniqueness of the oocyte organelles and common characteristics that can improve cellular quality. Specialization of cytoplasmic content in oocyte, including the formation of specific secretory granules and large‐scale intracellular organelle reorganization, occurs during the oogenesis and the oocyte maturation. Additionally, new perspectives on interactions between maternal components within the oocyte are introduced. Second, the regulatory mechanisms of intracellular degradative systems are discussed. In recent years, studies have indicated molecular mechanisms that degrade cytoplasmic components, such as autophagy and the UPS, and the significance of the degradation of cellular surface proteins, which has not yet received considerable attention during mammalian embryogenesis. 22 Furthermore, oocyte lipid droplets (LDs) that are prominent structures in optical microscopy, were recently confirmed to exhibit quantitative constancy and intense dynamics during early embryonic development. 23 Additionally, their relationship to embryonic quality was suggested.
The scope of this review is to summarize recent progress in understanding the mechanisms of oocyte organelle reorganization/specialization during oocyte maturation, and degradation of oocyte‐derived components during MZT. We also discuss the applications of these advancements for oocyte quality assessment. Gaining a molecular biological understanding of these progresses and identifying key factors is required to translate the benefits of these findings to applied research such as oocyte/embryo quality prediction and improvement. In this article, we focus on mammalian oocyte components other than chromosomes from the germinal vesicle (GV) stage to early pre‐implantation embryogenesis, where dramatic changes are observed. Most findings have been obtained from analysis using mice, other mammalian models, and human patients. However, highly conservative and cutting‐edge biological insights have also been gained from studies on non‐mammalian models.
2. COMPONENTS OF THE MAMMALIAN OOCYTE
Previous reports have reviewed changes in overall intracellular structures during oogenesis in mammalian oocytes, including human oocytes. 14 , 15 This section focuses on cytoplasmic components of that novel functions and dynamics in the oocyte or other cells were reported and attracted considerable attention in recent years. Here, we introduce the basic function and knowledge for each of the cytoplasmic components, followed by the recent findings.
2.1. Chromosomes and other cytoplasmic components
Although not the main topic of this review, we first give an overview of nuclear dynamics in oocytes, including chromosome segregation during oocyte differentiation and its relation to the progression of cytoplasmic division. In mammals, a fully grown primary oocyte has an obvious germinal vesicle (GV), arrests at the first meiotic division (MI), is ovulated, and releases the first polar body during or after transition from the ovary to the oviduct. The ovulated oocyte of mice and humans stops in metaphase of the second meiotic division (MII) to await the sperm in the oviduct. Therefore, unfertilized oocytes are commonly called MII oocytes. 24 , 25 , 26 The GV to MII oocyte maturation can be induced in vitro in a process called in vitro maturation (IVM). The major difference between sperm and oocyte differentiation in mammals is that spermatozoa complete meiosis to be haploid in the testis, whereas the oocyte does not complete meiosis in the ovary. On the other hand, the generation of the female haploid genome is completed by extruding a second polar body after fertilization in the oviduct. Upon fertilization, diploid and zygotic chromosomes are formed after fusion of the sperm and oocyte pronuclei. Thereafter, the mitosis phase begins (Figure 1).
Recent studies have shown that the abnormal chromosomal segregation that is observed prominently in aged mice occurs mainly at the time of first polar body release (i.e., during ovulation). 27 , 28 Interestingly, however, Eppig et al. 29 found that more than 50% of MI oocytes that failed to extrude the first polar body during IVM (despite the presence of twice the amount of chromosomes) develop to the two‐cell stage and form a blastocyst. Mayer et al. 30 showed that inducing a double‐strand break, which is a typical DNA damage, with the radiomimetic neocarzinostatin increased chromosome fragmentation but did not lead to delayed anaphase initiation. These findings suggest that neither excess nor irregular chromosomes are a critical suppressing factor in oocyte maturation or early embryogenesis. They are, to some extent, mutually independent phenomena. Notably, interventions for treating chromosomal aberrations themselves pose a risk that may lead to genetic manipulation. In this sense, oocyte chromosomes is distinct from other oocyte cytoplasmic components discussed in this review that are relatively easier to manipulate by intervention.
2.2. Endoplasmic reticulum
The endoplasmic reticulum (ER) that retains ribosomes on cytoplasmic surface is called the rough endoplasmic reticulum (rER). The rER is directly continuous with the nuclear envelope, serves as the site of protein translation from RNA to protein, or is the origin of protein‐membrane transport in the cell. In contrast, ER without the ribosomes is referred to as smooth ER (sER) and is widely distributed in the cytoplasm. The sER constitutes a meshwork of disk‐like or tubular structures that play an important role in the synthesis and storage of lipids, mainly cholesterol and phospholipids, and in new cellular membrane production.
In somatic cells, the ER has been recently reported to play a significant role in inter‐organelle communication as a center of networks consisting of various membrane structures. In particular, contact sites between the mitochondria and the ER (MERCS) are spread over almost the entire cytoplasm in some cells. At MERCS, the mitochondria and the ER actively exchange ions and nutrient substrates such as calcium and glucose and contribute to the involvement of the ER in regulating mitochondrial fusion and fission. In neurons, MERCS dysplasia is closely linked to the development of diseases such as hereditary spastic paraplegias. 31
The molecular mechanisms of autophagosomal membrane formation in the ER have recently been revealed. Noda and colleagues revealed that Autophagy‐related 2 (Atg2) and Atg9 proteins cooperate as lipid transporters and lipid scramblases, respectively, to promote autophagosomal membrane formation by delivering phospholipids from the ER membrane. 32 , 33 Because these recently noted roles of the ER are mainly independent from de novo protein synthesis, they may function even in MII stage oocytes with silenced gene translation machinery. Additionally, autophagy is an important proteolytic mechanism in mammalian embryos during early development, as described below.
The morphogenesis of the oocyte ER was described in detail by Kline et al. and FitzHarris et al. in live mouse eggs using confocal 3D observation with the specific fluorescent marker 1,1′‐dioctadecyl‐3,3,3′,3′‐tetramethylindocarbocyanine perchlorate (DiI). 34 , 35 The results of DiI staining, which can stain viable cells, in oocytes was consistent with the results of immunostaining with antibodies against ER‐specific proteins. In this context, the dynamics of the ER during oocyte maturation was reported as follows. First, in the GV oocyte, the ER is distributed throughout the cytoplasm, whereas ER accumulation around the GV and some isolated cytoplasmic accumulations are observed. Second, during the breakdown of the GV (GVBD), the ER is concentrated, forming a prominent ring structure surrounding the GV, and is then dispersed throughout the cytoplasm after GVBD completion. Finally, in MII oocytes, which do not have nuclear membranes, most of the ER is uniformly distributed in the cytoplasm, although signals around the chromosome spindles are relatively higher. However, a striking difference in MII oocytes is the formation of strong bright puncta (1–2 μm in diameter) just below the plasma membrane (PM), the so‐called PM cortex, except in the vicinity of the chromosome spindle (Figure 2). This group, called the cortical ER cluster, appears independent of cell cycle progression to MII and disappears during the release of the second polar body after fertilization. 34 FitzHarris et al. showed that treatment with nocodazole, which inhibits microtubule polymerization, and an inhibitory antibody (70.1 Ab) to dynein, a motor protein that binds to microtubules, blocked the formation of an ER perinuclear ring in the cytoplasm during GVBD. Conversely, the addition of latrunculin A, which inhibits actin cytoskeleton polymerization, prevents the formation of cortical ER clusters in MII oocytes. 36 These results indicate the significance of cytoskeletal proteins, actin, and tubulin, in the morphogenesis and distribution of the oocyte ER.
FIGURE 2.
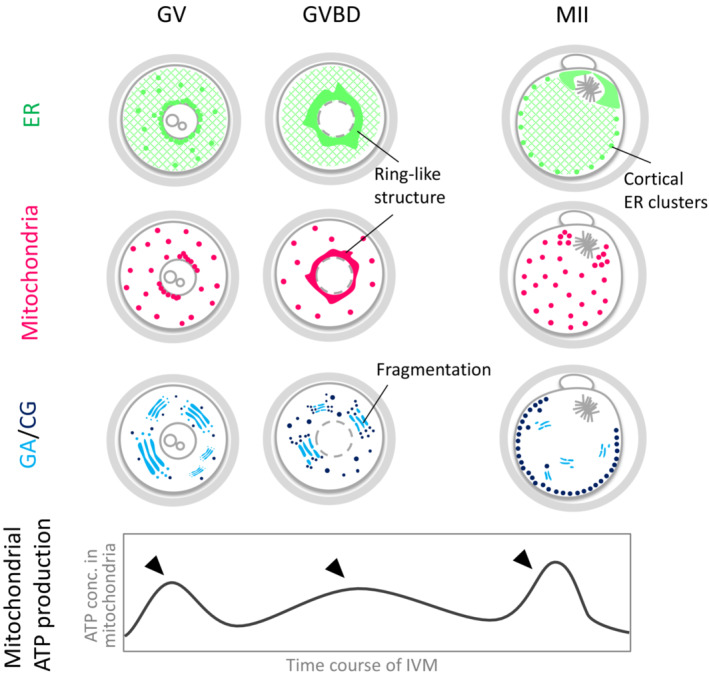
Changes in maternal organelles during oocyte maturation. The dispersed ER network is indicated as a meshed pattern. Arrowheads indicate three peaks (bursts) of mitochondrial ATP production (increase of mitochondrial ATP concentration) during in vitro maturation of mouse oocytes. CG, cortical granule; ER, endoplasmic reticulum; GA, Golgi apparatus
One of the most important roles of the oocyte ER may be as a source of Ca2+ in Ca2+ oscillations, which are the steep rise and fall of cytosolic Ca2+ concentrations over several hours in response to sperm stimulation after fertilization. Ca2+ oscillations induce a series of oocyte activation events, such as reactivation of cell cycle progression and activation of the polyspermy blocking system, which may also determine the success of embryogenesis. 37 The most essential Ca2+ release occurs through IP3 receptor‐type calcium channels on the cortical ER cluster, suggesting the significance of cortical ER cluster maturation. 35 , 38 , 39
In recent years, studies using cultured mammalian cells have raised a new concept regarding the mechanism of subcellular sER localization. Zheng et al. 40 showed that transmembrane proteins such as cytoskeleton‐linked membrane protein 63, ribosome binding protein 1, and kinectin 1 on sER membranes represent distinct affinities to mono‐ or poly‐glutamylated tubulin. These differences in tubulin modification, referred to as the tubulin code, and the localization of tubulin‐binding sER proteins regulate ER dispersion in the cell. The tubulin code includes glutamylation, acetylation, methylation, and other types of modifications. 41 However, its significance in mammalian oocytes has yet to be studied.
2.3. Mitochondria
The mitochondria are comprised of two separate phospholipid bilayers—an outer mitochondrial membrane (OMM) and an inner mitochondrial membrane (IMM). The IMM forms intricate folds called cristae in which respiratory chain enzymes are embedded to produce adenosine triphosphate (ATP). The space between the OMM and IMM is called the intermembrane space (IMS) and the lumen inside the IMM is called the mitochondrial matrix. Generally, the main function of the mitochondria is ATP production. Reactive oxygen species (ROS) are cell‐damaging byproducts of ATP production that arise from respiratory chain activity. However, healthy mitochondria have a mechanism for ROS elimination and degradation. In contrast, impaired mitochondrial function can lead to ROS accumulation and a cascade of ROS‐induced ROS release and other events that can disrupt the entire cell. 42 , 43
Mitochondrial ATP production depends on the Ca2+ concentration inside the mitochondria. A mitochondrial Ca2+ concentration higher than that of the cytosol promotes respiratory chain activity. Low mitochondrial Ca2+ concentration is correlated with diseases such as Parkinson's disease. Recently, abnormal Ca2+ concentrations in the mitochondria have become a therapeutic target for these diseases. 44 , 45
In recent years, functional and localization linkage between mitochondria and the ER, including MERCS formation, has attracted considerable attention. Furthermore, studies on the relationship between mitochondrial function and morphology have progressed. Mitofusin 1/2 (MFN1/2) proteins, which are exposed externally on the OMM, promote mitochondrial fusion. Optic atrophy 1 (OPA1), which is present on the IMM and is exposed to the IMS, regulates cristae formation by promoting fusion between inner membranes. Dynamin‐related protein 1 (DRP1) is present at the fission face on the OMM and promotes mitochondrial fission. MFN1/2, OPA1, and DRP1 are GTPases that degrade GTP to exert their activity. Deletion or mutation of these genes leads to mitochondrial dysfunction through altered morphology and causing a wide range of systemic diseases, as reviewed by Navaratnarajah et al. 46
The number of mitochondria in the mammalian oocyte estimated from mitochondrial DNA (mtDNA) copy numbers continues to increase during oogenesis. The mtDNA copy number is 1000 times higher in the MII stage than in the primordial germ cell (PGC). However, since the mtDNA copy number in the post‐implantation epiblast (as a whole) is not higher than that of the zygote stage, mtDNA replication during early embryonic development is predicted to be arrested. 47 , 48
The morphology of oocyte mitochondria changes dramatically during oogenesis. Mitochondria in the growing oocyte during differentiation from PGCs to follicular oocytes have a diameter of 1–1.5 μm with abundant cristae, whereas mitochondria in the GV and MII stages have a smaller diameter (0.4–0.6 μm) and fewer cristae. 49 , 50 The morphology of MII oocytes is maintained at least until the pronuclear stage. Since the surface of the cristae is where the respiratory chain produces ATP, mitochondrial activity in the MII oocytes was predicted to be reduced.
Yu et al. 51 examined mitochondrial respiratory activity from the GV to MII stage using the mouse IVM assay system. They also performed imaging analysis with different fluorescent indicators, that is, MitoTracker for mitochondrial dynamics and luciferase luminescence for ATP concentration in the mitochondria or cytosol (Figure 2). Mitochondrial ATP productivity did not decrease during IVM, but three peaks of increased ATP production were observed. The third peak corresponded to the timing of the first polar body extrusion, which is the last step of oocyte maturation. Furthermore, Wakai and Fissore 52 recently reported that GV oocytes undergo spontaneous Ca2+ oscillation, in which Ca2+ is periodically released into the cytoplasm through inositol 1,4,5‐trisphosphate receptor 1 (IP3R1) channels on the ER membrane before GVBD. Then, the released Ca2+ is absorbed and stored by the mitochondria, resulting in high ATP production capacity. This is an example of functional coordination between the ER and mitochondria. Additionally, these results indicate that microstructural changes do not necessarily correlate with ATP production capacity.
Mitochondrial oxidative phosphorylation, which is measured as the oxygen consumption rate (OCR), is generally the largest contributor to cellular ATP demand during preimplantation development. OCR has been reported to be positively correlated with human oocyte maturation, 53 morphological evaluation during early development, pregnancy, and delivery rates in bovine animals. 54
Studies on the dynamics of mitochondrial localization during oocyte maturation showed that the mitochondria form a ring‐like structure surrounding the GV and redistribute throughout the cytoplasm before the MII stage. 51 , 55 , 56 This is very similar to the ER localization dynamics described above, suggesting a functional link between the ER and mitochondria during oocyte maturation (Figure 2). This link continues to be examined with respect to the mitochondria. Udagawa et al. 57 showed that oocyte‐specific genetic knockout of the Drp1 gene in mice results in severely low fertility due to decreased oocyte fertilization and developmental ability. The mitochondria in these oocytes become unevenly distributed and, importantly, the ER also becomes unevenly distributed. Moreover, cortical granules and peroxisomes localize abnormally. In recent years, several phenotypes in Mfn1/2 gene knockout mice have been reported. In Mfn1‐gene‐deficient oocytes, the mitochondria and the ER aggregate, leading to oocyte apoptosis in the first or second follicular phase, causing the mice to become infertile. 58 , 59 However, Carvalho et al. 58 propose that MFN1 and MFN2 play distinct roles, because simultaneous Mfn1 and Mfn2 knockout alleviates the failure of follicle formation due to MFN1 deficiency, ameliorates abnormal mitochondrial localization, oocyte growth, and ovulation in double‐knockout females. The differences in MFN1 and MFN2 functions in oocytes were also demonstrated in overexpression experiments by Wakai et al. 60 in which RNAs encoding Mfn1, Mfn2, Opa1, and Drp1 were injected into mouse GV oocytes before IVM. MFN1 overexpression causes extremely high mitochondria and ER accumulation in the perinuclear region. This accumulation is retained even after polar body extrusion and causes abnormal regulation of the first polar body extrusion. In contrast, MFN2 overexpression causes transient mitochondria and ER accumulation, but the organelles are partly re‐distributed through the cytoplasm in the MII stage. No significant effect has been observed in the case of OPA1 and DRP1 overexpression. Although other reviews have discussed mitochondrial morphogenesis‐related factors and metabolic abnormalities, 61 the above findings suggest that regulating mitochondrial size and localization affects other intracellular organelles and plays important roles in oogenesis/oocyte maturation. Thus, mitochondrial size and location may be pivotal for estimating oocyte quality.
2.4. Lipid droplets
Lipid droplets (LDs) mainly function as a cellular reservoir for lipids. The main pathway for LD biogenesis is suggested to be initiated by lipid aggregation, either spontaneously or by lipid‐binding proteins, between the two single phospholipid layers of the ER membrane. As lipid aggregation proceeds, a lens structure is formed, which separates from the ER by budding, which forms an LD. Thus, LD membranes have a single phospholipid layer. LDs have unique proteins on this single membrane and the size of LDs can change after separation from the ER. LDs become larger by fusion or form a large aggregate by adhering to each other.
The main LD contents are triglycerides and cholesterol. Triglycerides are hydrolyzed to free fatty acids and glycerol. Fatty acids undergo β‐oxidation in the mitochondria and are processed via the citric acid cycle, whereas glycerol is processed via the glycolytic system. Both cycles are efficient ATP production systems for ATP. Thus, LDs are generally recognized as an energy resource. However, it was recently reported that certain lipid species such as dihydrosphingosine and dihydroceramide can cause dysfunction of other organelles, such as mitochondria and the ER, or can activate apoptotic cascades when present in excess, 62 , 63 , 64 suggesting that LDs play a role in sequestering these lipid species from the cytoplasm. Furthermore, during excessive ROS accumulation, for instance in the case of retinal diseases such as age‐related macular degeneration and brain diseases such as Alzheimer's disease and Parkinson's disease, a group of fatty acid transport proteins and apolipoproteins (Apo) likely play a role in preventing damage to the cytoplasm and other organelles. Increased ApoE protein levels have been shown to prevent ROS‐induced cellular damage by enhancing LD biogenesis and providing LDs as a direct target for oxidative damage. 65 , 66 These results indicate that LDs play a broad role in preventing toxicity in cells.
Moreover, recent studies have revealed the existence of subpopulations of LDs with different compositions within a single cell, suggesting the existence of specialized LDs required by specific cell types or local function. 67 Evidence shows that LDs form LD–organelle contact sites with the ER and diverse organelles such as mitochondria, lysosomes, and peroxisomes. Abnormalities in these contact sites are suggested to be associated with disease. For instance, a dysfunctional mutation in the SEIPIN gene, of which gene product involves in formation of specific ER–LD contact sites, causes progressive encephalopathy. Mutations in the SPASTIN gene cause hereditary spastic paraparesis, possibly by decreasing LD–peroxisome contact sites. 68 , 69
Lipid droplets in oocytes are the most prominent clear, yellow, or brown structures observed in bright‐field microscopy, depending on the organism or conditions of oocyte collection. Rather than being enlarged and appearing as a few vacuole‐like structures (like in adipocytes), oocyte LDs exhibit a granular morphology ranging from a few hundred nanometers to a few micrometers in diameter. LDs often appear to be dispersed in the cytoplasm, especially in unfertilized oocytes. Thus, LDs are used as an indicator for estimating cytoplasmic fluidity. 70 For more specific evaluation of oocyte LDs, specific LD dyes have been used, including non‐fluorescent dyes such as Lipophilic Nile Red or fluorescent dyes such as BODIPY. The content and size of oocyte LDs vary considerably among organisms. For example, the LD content is so high that it interferes with clear observation of other cytoplasmic contents in pig oocytes, but that is not so high in mice and humans. 71
Porcine oocyte LDs can be physically removed by combining relatively mild centrifugation and simple micromanipulation, as reported by Nagashima et al. 72 LD removal is called delipidation, which can preserve embryonic developmental ability after cryopreservation. This suggests that removing oocyte LDs does not fatally impair development. However, Aizawa et al. 23 recently developed an approach to almost completely remove LDs by treating mouse oocytes with solutions of different osmolarities and two‐step centrifugation. This group showed that unfertilized oocytes delipidated by this method had a significantly lower fertilization rate during in vitro fertilization (IVF) and that approximately half of the LDs were regenerated by the 2‐cell stage. Furthermore, treating embryos derived from delipidated oocytes with triacsin C to inhibit long‐chain acyl‐CoA synthetase (ACSL), which is involved in LD formation, strongly inhibited early development, especially after the morula stage. These results strongly suggest the importance of oocyte LDs and that LD homeostasis supports mammalian early development.
Moreover, most recently, Aizawa et al. 73 conducted an experiment in which mice were fed either a high‐fat diet (HFD) or a low‐fat diet for 3 days before ovulation (mice have a 4‐day estrous cycle). They analyzed the amount and lipid composition of LDs isolated by their delipidation method and found that a HFD significantly increased the amount of oocyte LDs. This result suggests an unexpectedly acute and tight relationship between maternal diet (nutrition) and oocyte quality.
2.5. Golgi apparatus and cortical granules
The Golgi apparatus (GA) functions as a delivery center that receives proteins and lipids produced in the ER and exports these molecules to endosomes, lysosomes, or the PM/extracellular space. The GA also serves as a factory for protein glycosylation and processing. In most vertebrate cells, the GA forms multiple cisternal layers comprised of cis (near the ER), medial (central), and trans (the most distal) cisternae. These compartments are mostly located near the centrosome. Cargo proteins from the ER are transported to the cis‐Golgi network, are passed through Golgi cisternae, and are then exported from the trans‐Golgi network. The GA in mammalian cells is a dynamic structure maintained by active vesicular trafficking and cisternal maturation. During somatic cell division, the GA disassembles into small membrane vesicles that reassemble in each daughter cell to form the functional GA with typical morphology. This dynamic process is repeated during mitosis and is referred to as GA reassembly. GA dynamics are confirmed by the regression of the GA upon inhibition of vesicular transport between the ER and the GA by Brefeldin A, which inhibits an activity of ARF1 guanine nucleotide exchange factor and indirectly blocks ER‐Golgi transport. 74 The membrane compartment between the ER and the cis‐side of the GA is referred to as the ER‐Golgi intermediate compartment (ERGIC). Unlike the GA, the ERGIC does not disappear even when membrane transport between the ER and Golgi is inhibited by Brefeldin A.
Electron microscopy (EM) suggest that the GA in the GV oocyte is dispersed throughout the cytoplasm. These GA structures are suggested to be functional with multiple cisternae surrounded by small vesicles. However, similar to GA reassembly in somatic mitosis where GA fragmentations occur upon nuclear envelope breakdown, the GA in mammalian oocytes drastically fragments during GVBD. 75 Notably, the typical centriole structure is also lost at these timings of GA fragmentation. A prominent centriole disappears in the oocyte, while centrosomes are replicated and bipolarized to form the spindle assembly in somatic cells. In bovines, the GA in MII oocytes is identified as a small, fragmented vesicle adjacent to the ER and appears to be vestigial. 76 After fertilization, the adjacency between GA‐derived vesicles and the ER is broken as the centriole structure becomes distinct and the GA localizes into the pericentriolar region.
Payne et al. indicated that inhibiting ER‐Golgi transport causes an increase in misshapen blastomeres and membrane blebbing, poorly defined cleavage furrows at the 8‐cell stage, and halted development before compaction. In rodents and humans, de novo protein synthesis from the zygotic genome is particularly essential after the 4‐cell stage. The above result in bovine oocytes suggests the importance of GA reassembly upon MZT to support the drastic increase in protein transport and protein modification.
Simultaneously with the regression of oocyte GA during the transition from the GV to MII stage, cortical granules (CGs) are formed from the GA. CG formation during oocyte maturation was confirmed by EM, which showed the formation of high electron density membranous granules adjacent to the GA. 77 Subsequently, most CGs localize in the cortex area just below the oocyte PM, while the cytoskeleton appears to prevent a direct contact between CGs and the PM (Figure 2). CGs are secretory granules that release their contents extracellularly in response to Ca2+ oscillation induced by fertilization, which was suggested to be important for preventing polyspermic fertilization. Mammalian CGs bind specifically to Lens culinaris agglutinin (LCA) lectin and can be detected by fluorescently labeled LCA such as LCA‐fluorescein isothiocyanate (LCA‐FITC). 78
The suggested mechanism underlying polyspermy prevention by CG secretion is that exocytosis (secretion) of the CG contents, referred as the cortical reaction (CR), alters the properties of the zona pellucida (ZP) and hardens the ZP to prevent excess sperm entry into the perivitelline space. Quantification of CGs using LCA markers suggested that CG secretion can also be induced by artificial Ca2+ influx into the oocyte cytoplasm and that CG secretion can occur multiple times in response to stimuli. 79 , 80 , 81 Barkurt et al. identified the enzyme responsible for ZP transformation as astacin‐like metalloendopeptidase (Astl, which is also called ovastacin) in CGs. Astl partially degrades ZP2 protein, the major ZP component that is responsible for binding to the sperm, preventing the entire ZP from binding to the sperm, and avoiding sperm passage. 82 In addition, although the Astl protein itself does not bind LCA, LCA lectin‐positive membrane granules completely disappear and only a few wheat germ agglutinin lectin‐positive granules remained in the oocytes of mice lacking the Astl gene. This result suggests that the Astl protein also plays essential roles in CG formation.
Recently, Satouh et al. succeeded in quantifying the progress of mouse oocyte CR induced by fertilization using a novel live imaging technique in which LCA‐FITC was supplemented extracellularly. This approach was used to assess the correlation between the number of Ca2+ spikes (oscillations) and the completion of CR. Both CR signal and ZP2 cleavage reach a plateau approximately 40 min after fertilization after the first two or three Ca2+ spikes were completed. The surface LCA‐FITC signal was indeed undetectable, and spermatozoa accumulate in perivitelline spaces of oocytes from Astl‐deficient mice. However, no polyspermy has been confirmed in in vivo fertilized oocytes. 83 , 84 These findings lead to an important suggestion that ZP hardening occurs within the first 40 min and that CR for the polyspermy block in mammals, at least in mice, is dispensable in in vivo fertilization but is important in IVF.
Regarding ZP hardening, interestingly, fetuin B, a liver‐derived plasma protein, prevents premature ZP hardening during oogenesis. Female mice lacking the fetuin B gene are completely infertile. 85 These findings indicate that a protein from a different organ regulates oocyte fertilization, and that premature release of the CG contents may affect oocyte fertilizing ability during oocyte maturation, even in vivo. Fetuin B (but not fetuin A) was further shown to bind directly to Astl to inhibit ZP2 digestion activity.
A different aspect of the CR is now attracting attention with respect to the release of oocyte cellular Zn2+. To understand this concept, the essentiality of intracellular Zn2+ for MII arrest should be clarified first. Suzuki et al. 86 reported that depleting oocyte Zn2+ by treating unfertilized mouse oocytes with the Zn2+ chelator, N,N,N′,N′‐tetrakis‐(2‐pyridylmethyl)‐ethylenediamine can induce successful development to full term without Ca2+ oscillations. Although the success rate of embryonic development to the blastocyst stage is only approximately 50% of that of sperm fertilization, this finding indicates that Ca2+ oscillations themselves are not essential for initiating early development and suggests that the release of Zn2+ from the cells is more important.
Woodruff's group recently reported a series of important analyses of Zn2+ storage and release in oocytes. 87 , 88 , 89 They analyzed oocyte Zn2+ with scanning transmission electron microscopy with energy‐dispersive spectroscopy to map biological zinc directly at the ultrastructural level, X‐ray fluorescence nanoprobe microscopy to quantify zinc within compartments, three‐dimensional X‐ray fluorescence tomography, and live imaging using their newly developed and highly sensitive Zn2+ fluorescence labeling reagent, ZincBY‐1, to map and quantify Zn2+ in oocytes. This analysis revealed that the granules just beneath the oocyte PM, which are not clearly defined as CGs, contain high Zn2+ content that is released at the time of spikes in Ca2+ oscillations. 87 , 88 , 89 This Zn2+ release, which can be visualized using an extracellularly supplemented Zn2+ fluorescent indicator, is called the Zn2+ burst. This process can predict the developmental potential of early embryos. 90 These results indicate that Zn2+ release, which is essential for oocyte activation, is induced or is strongly related to the CR.
3. IMPORTANCE OF OOCYTE PROTEOLYTIC SYSTEMS
3.1. Ubiquitin–proteasome system
The UPS is an intracellular proteolytic pathway in which cytosolic proteins are primarily targeted. The substrates are degraded upon binding of ubiquitin to lysine residues as a result of sequential processing by ubiquitin (Ub)‐activating (E1), Ub‐conjugating (E2), and Ub‐ligase (E3) enzymes. Primarily, E3 regulates substrate specificity and the timing of Ub modification. A recent bioinformatics survey of Ub enzymes suggests that 8 E1, 41 E2, and 634 E3 enzymes are encoded in the human genome. 91 Ubiquitinated substrates are delivered to the 26S proteasome, where they are unfolded and enzymatically degraded in an ATP‐dependent process.
Although ubiquitination is associated with the UPS and various intracellular phenomena, Shin et al. showed that polyubiquitinated proteins were highly accumulated from GV oocytes to the 2‐cell stage embryos, but disappear by the 4‐cell stage embryos in mice. More precisely, the ubiquitinated protein level is retained until the first cleavage, followed by a sharp decrease after the late 2‐cell stage (36 h post‐fertilization; hpf). In conjunction with this decrease, chymotrypsin‐like peptidase activity, which is likely derived from proteasomes, specifically and rapidly increases around 24 hpf, immediately after the first cleavage. Additionally, same group identified a zygote‐specific proteasome assembly chaperone protein (ZPAC) and observed that chymotrypsin‐like peptidase activity was almost completely abolished in zygotes in which ZPAC, or ubiquitin‐mediated‐proteolysis 1 (Ump1) expression was suppressed. Importantly, suppressing the Ump1 gene resulted in 90% of embryos halting before the first cleavage (Figure 3). 92 These results indicate both that ZPAC and UMP1 regulate the entire UPS system in the oocyte and early embryos and that the UPS degradation pathway is essential for mammalian embryonic development before ZGA.
FIGURE 3.
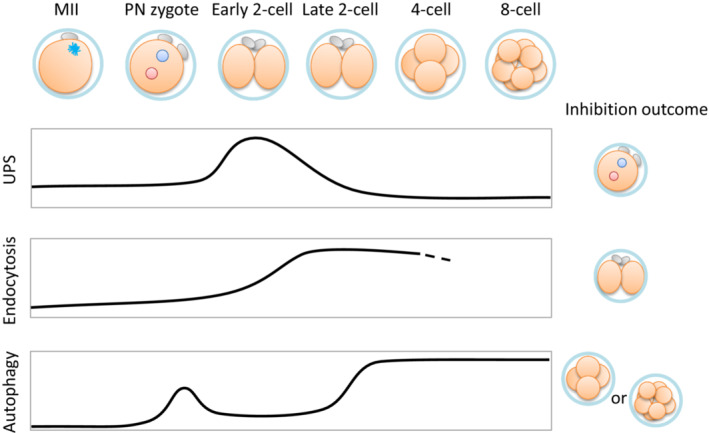
Oocyte proteolytic systems in early embryogenesis. Relative activities of proteolytic systems at each developmental stage are indicated. These stages are mainly studied in mice. Inhibition outcomes are results from gene deletion, gene suppression, or inhibitor assays. Endocytic activity after the 4‐cell stage was not assayed (partly shown with dotted line). Inhibition results using Pitstop2 are indicated. UPS, ubiquitin‐proteasome system
Mtango et al. used rhesus monkey oocytes as a nonhuman primate model to assess the correlation between UPS gene regulation and embryonic quality. They first categorized oocytes collected with different methods as exhibiting different oocyte qualities (potential developmental abilities). Oocytes collected with follicle‐stimulating hormone (FSH) only and matured in vitro were categorized as “low” quality, oocytes collected with FSH followed by stimulation with human chorionic gonadotropin (hCG) were “moderate”, and oocytes harvested without hormonal stimulation (naturally ovulated) were “high”. Second, they analyzed the oocytes or embryos with satisfactory morphologies at each stage, including GV, MII, and early developmental stages, using RT‐PCR to examine common or differential kinetics of mRNA encoding 53 UPS‐related genes (such as E1 ~ E3) at each stage. They reported that the “high” group showed a greater accumulation of specific mRNAs that are commonly observed in MII and that this group showed a greater alteration of kinetics during MII to the 2‐cell stage compared with the “low” or “moderate” categories. These results suggest the importance of silent but significant preparation for dynamic changes in UPS activity before ZGA for degradation of oocyte components. 18
Yang et al. 93 recently showed that RNF114, a UPS‐related E3 ubiquitin ligase, is predominantly expressed in MII‐stage oocytes. Silencing this gene results in a delay in TGF‐β activated kinase 1 (MAP3K7) binding protein 1 (TAB1) degradation and arrests embryonic development at the 2‐cell stage (prior to ZGA). Notably, RNF114 specifically targets TAB1, out of the 9000 candidate proteins they examined. These results strongly suggest that embryonic development potential may be determined by general UPS activity and by the degradation of specific key molecules by UPS.
3.2. Autophagy
Autophagy, like the UPS, is an intracellular degradation mechanism. However, autophagy differs from the UPS in that its substrates can degrade proteins, RNAs, and relatively larger structures, such as highly aggregated proteins and organelles (e.g., ER‐phagy, Mitophagy). 94 In macroautophagy, degradative substrates are surrounded by isolation membranes and are sequestered into autophagosomes that fuse with lysosomes to degrade the internal contents. This process can be selective, where the target is surrounded by adapter proteins, or non‐selective, in which the entire space can be taken up and degraded. 94 The mechanism by which relatively small organelles or cellular components are directly incorporated into lysosomes is also called microautophagy. As an example, direct integration of LCs into lysosomes in mammals was reported as LD microautophagy. 95
Starvation of intracellular nutrients is a major pathway for autophagy activation. For instance, amino acid deficiency, mainly detected by the mechanistic/mammalian target of rapamycin complex 1 (mTORC1), upregulates autophagic activity by suppressing negative regulation by mTORC1 itself. However, even without starvation, basal autophagy activity is important for various cellular functions and maintenance of intracellular circumstances.
Tsukamoto et al. generated an oocyte‐specific autophagy‐related 5 (Atg5) conditional knockout mouse line using the ZP3 promotor‐driven Cre transgenic line and indicated that abolishing autophagic activity in oocytes results in embryonic development arrest at the 4–8 cell stage. Autophagosome formation was visualized by green fluorescent protein‐tagged MAP1LC3 (GFP‐Microtubule‐associated protein light chain 3; GFP‐LC3) puncta formation, which demonstrated that autophagic activity is upregulated during PN formation and that the number of GFP‐LC3 puncta peaks mainly at the 4‐cell and later stages. 18 Furthermore, de novo protein synthesis, which is supposed to be derived from ZGA detectable at the 4‐ and 8‐cell stages, is reduced in Atg5‐depleted oocytes. These data strongly suggest that autophagic activity is necessary for embryogenesis, presumably to reserve amino acid resources prior to ZGA. Since mTORC1 inhibitors do not induce enhanced autophagy in early development, the molecular process of autophagy induction in early embryos remains unclear (Figure 3). 96
In addition, autophagy is used to degrade male‐derived cellular components. In the case of the model animal Caenorhabditis elegans, paternal mitochondria and membranous organelles (MOs), which are brought by spermatozoon, are selectively degraded by allophagy (allogeneic [non‐self] organelle autophagy) during embryogenesis. 97 Selective degradation of paternal mitochondria by allophagy prevents paternal mtDNA from transmitting to the offspring, leading to maternal mtDNA inheritance. In this process, allophagy‐1 (ALLO‐1) functions as an autophagy adaptor that is essential for autophagosome formation around paternal mitochondria and membranous organelles. ALLO‐1 directly binds to the worm LC3 homolog LGG‐1 and is required for autophagosomal formation around paternal organelles. ALLO‐1 also binds to a worm homolog of the mammalian inhibitor of NFκB kinase epsilon subunit (IKKE‐1), which is involved in ALLO‐1 phosphorylation and is important for paternal organelle clearance. 98 Although the physiological significance of allophagy remains unclear, it may prevent mtDNA derived from different genetic backgrounds from becoming a heteroplasmic state.
In mammals, the involvement of autophagy in the elimination of paternal mitochondria remains controversial. Rojansky et al. 99 observed degradation of fluorescently labeled sperm mitochondria in mouse zygotes/embryos and found that paternal mitochondria are degraded by the 8‐cell‐morula stage (84 hpf). Gene suppression assays using short hairpin RNA showed that sperm mitochondrial degradation in the 84 hpf embryo is significantly decreased by knocking down the p62 autophagy adapter, mitophagy‐related proteins such as PTEN‐induced putative kinase 1 (PINK1), or double knockdown of two ubiquitin ligases, Parkin and mitochondrial E3 ubiquitin‐protein ligase 1 (MUL1). However, Luo et al. 100 used two transgenic mouse strains, one bearing GFP‐LC3 for labeling autophagosomes (female) and the other bearing red fluorescent protein (RFP)‐labeled mitochondria (male) and reported that the GFP‐LC3 signal interacts with paternal mitochondria from the zygote to the 2‐cell stage, but dissociate at the 4‐cell stage. Furthermore, they examine whether the above result was obtained by GFP quenching, but not RFP, at low pH conditions in lysosomes and demonstrated that sperm mitochondria are not ingested into lysosomes labeled with LysoTracker Blue DND‐22. These results support the hypothesis that sperm mitochondria are degraded by more passive processes. Apparent discrepancies between these reports need further validation. The correlation between active degradation of sperm organelles and embryonic quality also remains to be elucidated.
3.3. Endocytosis
Endocytosis is a pivotal process that takes up extracellular proteins, fluid content, and PM components, and involved in various biological phenomena. 101 In this process, PM proteins are internalized from the cell surface by clathrin‐mediated or ‐independent endocytosis and are transported to early endosomes (EEs). Some PM proteins are recycled back to the PM via recycling endosomes or the trans‐Golgi network. Others are delivered to late endosomes (LEs) and are sorted into intralumenal vesicles of multivesicular bodies (MVBs) that finally fuse with lysosomes for degradation of their contents. Ubiquitinated membrane proteins are preferentially sorted into the intralumenal vesicles of MVBs via the endosomal sorting complexes required for transport (ESCRT) complex. Small GTPase Rab family proteins are required for endocytic transport and for endosomal maturation. Transport from the PM to lysosomes is mainly regulated by RAB5 and RAB7, whereas recycling pathways are regulated by RAB4 and RAB11. Immediate endocytosis also exists. For instance, compensatory endocytosis occurs in response to rapid and frequent exocytosis (secretion) of synaptic vesicles in neurons. 102
While the importance of the UPS and autophagy has been analyzed, the significance of endocytosis in mammalian embryonic development had rarely been studied. 103 Recently, several studies using C. elegans revealed that a subset of maternal PM proteins, such as caveolin homolog 1 (CAV‐1) and chitin synthase 1 (CHS‐1), are internalized from the PM and are transported to endosomes and lysosomes for degradation. 104 , 105 , 106 In zygotes, the accumulation of high levels of K63‐linked poly‐ubiquitin chains is transiently observed on endosomes approximately 25 min after fertilization, suggesting the involvement of ubiquitination machinery in the selective degradation of maternal PM proteins. Consistent with this observation, E2 ubiquitin conjugating enzymes such as UBC‐13 and UEV‐1 are involved in this process. In addition, this process is tightly linked to anaphase‐promoting complex/cyclosome (APC/C)‐dependent meiotic cell progression after fertilization, suggesting the involvement of maternal PM protein degradation via endocytosis in oocyte reactions to fertilization stimuli. 107
In this situation, we recently reported that multiple maternal PM proteins, including the glycine transporter GlyT1a, are selectively transported from the PM to the EE at the late 2‐cell stage and are degraded in lysosomes by the 4–8‐cell stage during mouse embryogenesis. 22 Interestingly, kinetic analysis of GlyT1a trafficking from EEs to LEs indicated that the transition of GlyT1a from EEs to LEs proceeds very slowly and takes more than 6 h. These results suggest that unique mechanisms underlie the regulation of endosomal maturation or trafficking during early development. Accumulation of a large number of ubiquitinated proteins in endosomes takes place in 2‐cell stage embryos, suggesting the involvement of ubiquitination in this process. Glyt1a‐mCherry partially overlapped with ubiquitin‐positive endosomes in late 2‐cell embryos. Mutations of lysine‐residues on Glyt1a, which are potential ubiquitination sites, caused significant retardation of GlyT1a degradation, suggesting the importance of ubiquitination in sorting maternal PM proteins into the lysosomal degradation pathway. Furthermore, pharmacological assays for GlyT1a endocytosis showed that protein kinase C (PKC) activation with phorbol 12‐myristate 13‐acetate or inactivation with staurosporine resulted in ectopic initiation or inhibition of endocytosis, respectively. More importantly, Pitstop2, an inhibitor of clathrin‐mediated endocytosis, completely blocked both GlyT1a endocytosis and embryogenesis at the 2‐cell stage. These results demonstrate that PKC‐dependent and clathrin‐mediated endocytosis of maternal membrane proteins is essential for embryonic transition from the 2‐ to 4‐cell stage (Figure 3).
With respect to the immediate response, Gomez‐Elias et al. recently demonstrated that glycoproteins, which are contained in CGs and are delivered to the PM by oocyte CRs are endocytosed from the PM immediately after fertilization. 108 When the glycoproteins targeted to the PM were labeled with extracellularly supplemented fluorescent‐labeled LCA, they were rapidly taken into the cytoplasm. Unlike Glyt1a endocytosis, this process was inhibited with the calcineurin inhibitor cyclosporine A or staurosporine, but not with Pitstop2. These results suggest that mammalian oocytes are equipped with a clathrin‐independent compensatory endocytic system for CR exocytosis.
These recent findings suggest that endocytic activity is already active immediately after fertilization and that the selective removal of oocyte contents from the PM can determine embryonic quality. Although endocytosed PM proteins are transported to lysosomes mainly after the 4‐cell stage, 22 it should be noted that many PM proteins exert their functions by residing on the cell surface. Thus, their functional significance is assumed to be lost following delivery into intracellular vesicles.
Importantly, autophagy and endocytosis share an important similarity in that proteins are degraded within lysosomes. Lysosomal associated membrane protein 2 (LAMP2) is a major protein constituent of lysosomal membranes. Lamp2‐knockout mice show increased postnatal mortality, indicating a significant contribution to lysosome formation itself. 109 Tsukamoto et al. showed that acidified lysosomes can be prominently detected by Lysotracker in embryos from the 2‐cell stage. Injecting a mixture of siRNAs against Lamp2 and Lamp1, a functionally related gene, results in the arrest of embryos at the 2‐cell stage. Tsukamoto et al. also analyzed the effects of multiple inhibitors such as E‐64d and bafilomycin A1 on lysosomal functions or maturation and showed that most of the inhibitors arrested embryonic development. These results suggest that lysosomes functionally mature and play pivotal roles during the development of the preimplantation embryo. 110 We examined the dynamics of EGFP‐human RAB5a and EGFP‐human RAB7a from unfertilized oocytes to the blastocyst stage to analyze the biogenesis of endosomes and lysosomes during mouse embryogenesis, since endosomes, especially LE, share commonalities with lysosomes. We found that RAB5a‐ and RAB7a‐positive puncta are completely co‐localized in the cortical punctate structure in unfertilized oocytes and at the 1‐cell stage. However, at the 2‐cell stage, RAB5a and RAB7a signals become redistributed to distinct punctate structures in the cytoplasm. At the 4‐cell stage, RAB7a‐positive LE are particularly increased in size and number. 22 These results suggest that endosomal/lysosomal maturation prior to or at the 2‐4‐cell stages may play a pivotal role in determining embryonic quality through degradation of oocyte components.
4. APPLICATIONS OF OOCYTE COMPONENTS STATUS TO OOCYTE QUALITY RESEARCH
4.1. Diagnosis of oocyte components and prediction of embryo quality
To establish predictive and diagnostic techniques for oocytes or embryonic quality using findings on oocyte component dynamics, a causal relationship first needs to be proved between the phenomena (or molecules) and embryo quality through basic research. In addition, non‐invasive or extremely low‐invasive observation and diagnostic techniques need to be developed that do not significantly affect full‐term development. To meet these requirements, studies need to use an analysis method that does not require introducing/supplementing a marker to the cell. Optimized detection methods with low invasiveness would also be preferable.
Extracellular flux analysis of the OCR, which has been recently used in cultured cells, small animal individuals, and tissue fragments, is a system that assays changes in the composition of the external fluid in which cells are cultured. Therefore, this approach shows very low invasiveness to the culture subject. In fact, Muller et al. 111 reported its application to various mammalian embryos. Thus, this extracellular flux analysis is expected to be a useful method of measuring mitochondrial activity for predicting embryonic quality. However, Tan et al. 112 reported that quantitative confocal imaging of the auto‐fluorescent cofactors reduced nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) and Flavin adenine dinucleotide (FAD) have advantages, especially when the target cell is accompanied by other cells, such as an oocyte in a cumulus oophorus complex, since it enables the analysis of individual cells. These findings imply the importance of using different options depending on the application and limitations.
LD can be tracked almost non‐invasively and multi‐dimensionally by visualization with label‐free coherent anti‐stokes Raman scattering (CARS). Bradley et al. 113 reported that CARS is less invasive and more useful than fluorescent‐labeled reagents in tracking mouse embryonic development and that it offers advantages such as quantifying the lipid content, type, and spatial distribution with sub‐micron resolution. As mentioned above, LD distribution and total amount vary widely among organisms. Tsukamoto et al. reported that differences were observed even among mouse strains. 114 To apply these findings in future embryo quality studies, it will be necessary to focus on whether the quantitative and qualitative trends in oocyte LDs are directly related to the quality of human embryos.
4.2. Molecular imaging with fluorescent probes
The introduction of fluorescent probes for target molecules, such as proteins or ions, and fluorescent imaging are accompanied by reagent toxicity, stress associated with introduction method such as microinjection, and phototoxicity during observation. However, fluorescent molecules are useful for quantifying the dynamics of molecules that may be related to embryonic quality. To address this dilemma, recent assays combined the use of fluorescent molecular probes with the development of experimental systems with lower invasiveness. As a result, certain prediction techniques using fluorescent markers have succeeded in identifying critical perspectives for predicting embryonic quality. For instance, Yamagata et al. visualized nuclei and microtubules with fluorescently tagged histone H2B and tubulin, respectively, to observe the timing and number of oocyte divisions during mitosis and the presence or absence of micronuclei. They found that the first mitotic division without micronucleus formation is particularly important in mouse full‐term development. 115 , 116 , 117 , 118 Similarly, visualization methods have been developed that quantify cytosolic Ca2+ by using Ca2+‐bound fluorescent protein genetically encoded Ca2+‐indicators for optical imaging (GECO) series with extremely low‐invasive confocal equipment. These methods have been used to determine the number of Ca2+ oscillatory spikes required to initiate oocyte development and reject polyspermic fertilization. 83 , 84 , 119 The developments of low‐invasive imaging approaches have additionally benefited basic biological studies on dynamics in oocytes before and after fertilization, which is important for understanding the fundamental requirement for these phenomena.
Regarding the degradation of oocyte component, Tsukamoto et al. focused on autophagic activity around the 4‐cell stage, which is important in early mammalian development, and concluded that full‐term development outcomes can be predicted by injecting mRNA for LC3‐GFP and quantifying autophagosome formation in embryos at the 4‐cell stage. 18 , 120 Tsukamoto et al. additionally reported a prediction study based on LD dynamics and showed that LD number constancy can be quantified by injecting mRNA for perilipin 2 protein fused with GFP. Additionally, they compared the LD content between mouse strains with different developmental competence and concluded that an imbalance between lipolysis activity, LD autophagy (lipophagy), and lipogenesis is manifested as LD depletion or aggregation, which both lead to poor embryonic quality. 23 , 73 , 114 , 121
According to a very recent report, a new type of super‐resolution microscopy is compatible with extremely low‐invasive observation of mouse embryonic development. This system is capable of detecting segregating chromosomes and counting the number of chromosomes. 122 Thus, fluorescent probes, which are important tools for analysis, allow precise tracing and/or quantification of specific molecules and multidimensional molecular biological analysis, such as three dimensional co‐localization analysis, time‐course analysis, and simultaneous analysis of multiple targets. Thus, development of fluorescent imaging will continue to be required, especially for basic embryonic quality studies using model animals.
5. SUMMARY AND FUTURE PERSPECTIVES
Oocytes have to prepare oocyte components with enough quantity and quality to satisfy the requirements for healthy embryonic development before fertilization and some oocyte components need to be degraded appropriately after fertilization. Understanding the criteria of oocyte and embryo that support embryonic development is beneficial for evaluating oocyte/embryo quality. As discussed in this review, further studies using oocytes to examine newly found concepts or those that remain cryptic, such as the huge mitochondria‐ER network, LD subpopulations, and cross‐talk among these concepts, are required. Additional cross‐talk may be found between unexpected factors. For example, Yue et al. 123 recently reported a system in which the accumulation of UPS components around the mitochondria disengages the quiescence of the mitochondrial respiratory as a common mechanism from fungi to mammalian culture cells. In this context, identifying the key factors responsible for oocyte quality deterioration from aging or genetic disorders may enable us to intervene to improve oocyte quality by adding or removing such molecules from the oocytes of patients.
However, as introduced in this review, several essential changes in oocyte components were analyzed as continuously dynamic processes that require sequential observations of oocyte maturation or early embryonic development. As studies provide further information on the entire process of oocyte maturation and embryonic development, finding critical steps in this process may simplify diagnostic techniques, such as examining the oocyte/embryo only at critical stages in a snapshot fashion. On the other hand, multidimensional concepts will be unavoidable to understand the biology of oocyte/embryo quality comprehensively, although practical options for quantification are limited by toxicity and technical limitations. Thus, further studies should aim towards decreasing the toxicity arising from the detection method and the dimensions of the targeted factor.
Attempts to substitute fluorescent‐marker‐based studies with other methods to make them less invasive may be useful for multidimensional analysis. For example, a transient increase in the velocity of cytoplasmic flow given by contractile movement of actomyosin in response to increased cytoplasmic Ca2+ during Ca2+ oscillations can be quantified (although not perfectly) by bright‐field microscopy, which was shown to be applicable for embryonic quality prediction. 124 However, in this approach, attention should be paid to uncertainty based on optical microscopic observations. For instance, ER aggregation (referred to as SERa or sER clustering) that can be observed by bright‐field microscopy after ICSI or IVF was first reported as an indicator of poor embryonic quality by Serhal et al. 125 Careful analysis of SERa assayed simultaneously with a specific fluorescent marker such as ER‐tracker showed that SERa+ human embryos did not result in live birth. 126 However, even in recent years, studies were reported showing that the presence of SERa in human embryo does not affect live‐birth rates. 127 Such seemingly contradictory opinions may be caused by unclear criteria for judging such SERa and other aggregates by optical microscopy. To address these problems, careful verification of new optical microscope methods with highly specific and reliable markers that will establish solid and stable criteria is required, such as verifying visualization of LDs by CARS.
The quantification and qualification of oocyte/embryo cellular surface proteins, which was shown to be significant for embryonic quality by endocytosis study, 22 may enable us to develop applications with lower invasiveness by avoiding the risk of introducing indicators into cells, which may also avoid technical difficulty. Furthermore, the endocytosis of GlyT1a, the only identified glycine transporter on the oocyte PM, indicates the conversion of embryonic amino acid intake capability. Pre‐implantation mammalian embryos are sensitive to increased osmolarity of the extracellular media, even within the physiological range, and the intake of amino acids, including glycine, betaine, and other amino acids from the extracellular media have been strongly suggested to be a primary player that confers resistance to increased osmolality. 128 , 129 Understanding of the dynamics of such transporter proteins will shed light on the development of proper and on‐demand nutrition supplementation methods for in vitro culture. In addition, since appropriate synthesis of oocyte components during follicular development and embryonic components by the zygotic genome is essential for ensuring embryo quality, the improvement of these processes could be helpful to obtain healthy embryo. Findings on such critical changes in surface proteins may be applicable to diagnosis of embryos after ZGA.
To address the overall complexity, machine learning applications such as deep learning technology were proposed by some groups. Tokuoka's study, for instance, indicates easier methods for determining the relationship between each of the observed parameters (e.g., timing and synchronicity of mitosis, brightness and distribution of fluorescent markers) and their correlation to embryonic developmental success. These approaches may also enable researchers to identify the importance of each factor in determining embryonic quality. 130
Finally, in the future, technologies that are less invasive and not mutually exclusive may be applied in combination for achieving synergistic effects. The development of these analytical technologies will become important for basic biological research targeting specific molecules mainly using model organisms. Beyond these studies, such technologies may also have clinical applications for human oocyte/embryo quantification.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
HUMAN AND ANIMAL RIGHTS
This article does not contain any study on human or animal participants that was performed by any of the authors.
ACKNOWLEDGMENTS
This study was supported by the Japan Society for the Promotion of Science KAKENHI (Grant Number JP22H01922 to YS, 19H05711 and 20H00466 to KS) and Mochida Memorial Foundation for Medical and Pharmaceutical Research to YS.
Satouh Y, Sato K. Reorganization, specialization, and degradation of oocyte maternal components for early development. Reprod Med Biol. 2023;22:e12505. 10.1002/rmb2.12505
Contributor Information
Yuhkoh Satouh, Email: yuhkohs@gunma-u.ac.jp.
Ken Sato, Email: sato-ken@gunma-u.ac.jp.
REFERENCES
- 1. Secomandi L, Borghesan M, Velarde M, Demaria M. The role of cellular senescence in female reproductive aging and the potential for senotherapeutic interventions. Hum Reprod Update. 2022;28(2):172–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vazquez‐Diez C, FitzHarris G. Causes and consequences of chromosome segregation error in preimplantation embryos. Reproduction. 2018;155(1):R63–76. [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev. 2009;89(1):193–277. [DOI] [PubMed] [Google Scholar]
- 4. Tscherner AK, Macaulay AD, Ortman CS, Baltz JM. Initiation of cell volume regulation and unique cell volume regulatory mechanisms in mammalian oocytes and embryos. J Cell Physiol. 2021;236(10):7117–33. [DOI] [PubMed] [Google Scholar]
- 5. Yaniz JL, Soler C, Santolaria P. Computer assisted sperm morphometry in mammals: a review. Anim Reprod Sci. 2015;156:1–12. [DOI] [PubMed] [Google Scholar]
- 6. Gu NH, Zhao WL, Wang GS, Sun F. Comparative analysis of mammalian sperm ultrastructure reveals relationships between sperm morphology, mitochondrial functions and motility. Reprod Biol Endocrinol. 2019;17(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sato M, Sato K. Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012;8(3):424–5. [DOI] [PubMed] [Google Scholar]
- 8. Sato K, Sato M. Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J Biochem. 2017;162(4):247–53. [DOI] [PubMed] [Google Scholar]
- 9. Susor A, Jansova D, Anger M, Kubelka M. Translation in the mammalian oocyte in space and time. Cell Tissue Res. 2016;363(1):69–84. [DOI] [PubMed] [Google Scholar]
- 10. Jukam D, Shariati SAM, Skotheim JM. Zygotic genome activation in vertebrates. Dev Cell. 2017;42(4):316–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen H, Einstein LC, Little SC, Good MC. Spatiotemporal patterning of zygotic genome activation in a model vertebrate embryo. Dev Cell. 2019;49(6):852–66 e857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vara C, Paytuvi‐Gallart A, Cuartero Y, Le Dily F, Garcia F, Salvà‐Castro J, et al. Three‐dimensional genomic structure and cohesin occupancy correlate with transcriptional activity during spermatogenesis. Cell Rep. 2019;28(2):352–67 e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okada Y. Sperm chromatin condensation: epigenetic mechanisms to compact the genome and spatiotemporal regulation from inside and outside the nucleus. Genes Genet Syst. 2022;97:41–53. [DOI] [PubMed] [Google Scholar]
- 14. Trebichalska Z, Kyjovska D, Kloudova S, Otevrel P, Hampl A, Holubcova Z. Cytoplasmic maturation in human oocytes: an ultrastructural study. Biol Reprod. 2021;104(1):106–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mao L, Lou H, Lou Y, Wang N, Jin F. Behaviour of cytoplasmic organelles and cytoskeleton during oocyte maturation. Reprod Biomed Online. 2014;28(3):284–99. [DOI] [PubMed] [Google Scholar]
- 16. Asami M, Lam BYH, Ma MK, Rainbow K, Braun S, VerMilyea MD, et al. Human embryonic genome activation initiates at the one‐cell stage. Cell Stem Cell. 2022;29(2):209–16 e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sha QQ, Zheng W, Wu YW, Li S, Guo L, Zhang S, et al. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte‐to‐embryo transition in humans. Nat Commun. 2020;11(1):4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321(5885):117–20. [DOI] [PubMed] [Google Scholar]
- 19. DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte‐to‐embryo transition. Trends Cell Biol. 2004;14(8):420–6. [DOI] [PubMed] [Google Scholar]
- 20. Mtango NR, Latham KE. Ubiquitin proteasome pathway gene expression varies in rhesus monkey oocytes and embryos of different developmental potential. Physiol Genomics. 2007;31(1):1–14. [DOI] [PubMed] [Google Scholar]
- 21. Verlhac MH, Terret ME, Pintard L. Control of the oocyte‐to‐embryo transition by the ubiquitin‐proteolytic system in mouse and C. elegans . Curr Opin Cell Biol. 2010;22(6):758–63. [DOI] [PubMed] [Google Scholar]
- 22. Morita A, Satouh Y, Kosako H, Kobayashi H, Iwase A, Sato K. Clathrin‐mediated endocytosis is essential for the selective degradation of maternal membrane proteins and preimplantation development. Development. 2021;148(14):dev199461. [DOI] [PubMed] [Google Scholar]
- 23. Aizawa R, Ibayashi M, Tatsumi T, Yamamoto A, Kokubo T, Miyasaka N, et al. Synthesis and maintenance of lipid droplets are essential for mouse preimplantation embryonic development. Development. 2019;146(22):dev181925. [DOI] [PubMed] [Google Scholar]
- 24. Zielinska AP, Bellou E, Sharma N, Frombach AS, Seres KB, Gruhn JR, et al. Meiotic kinetochores fragment into multiple lobes upon cohesin loss in aging eggs. Curr Biol. 2019;29(22):3749–65 e3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bakloushinskaya I. Chromosome changes in soma and germ line: heritability and evolutionary outcome. Genes (Basel). 2022;13(4):602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conti M, Franciosi F. Acquisition of oocyte competence to develop as an embryo: integrated nuclear and cytoplasmic events. Hum Reprod Update. 2018;24(3):245–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas C, Cavazza T, Schuh M. Aneuploidy in human eggs: contributions of the meiotic spindle. Biochem Soc Trans. 2021;49(1):107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakakibara Y, Hashimoto S, Nakaoka Y, Kouznetsova A, Hoog C, Kitajima TS. Bivalent separation into univalents precedes age‐related meiosis I errors in oocytes. Nat Commun. 2015;6:7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Eppig JJ, Schultz RM, O'Brien M, Chesnel F. Relationship between the developmental programs controlling nuclear and cytoplasmic maturation of mouse oocytes. Dev Biol. 1994;164(1):1–9. [DOI] [PubMed] [Google Scholar]
- 30. Mayer A, Baran V, Sakakibara Y, Brzakova A, Ferencova I, Motlik J, et al. DNA damage response during mouse oocyte maturation. Cell Cycle. 2016;15(4):546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Darios F, Mochel F, Stevanin G. Lipids in the physiopathology of hereditary spastic paraplegias. Front Neurosci. 2020;14:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol. 2019;26(4):281–8. [DOI] [PubMed] [Google Scholar]
- 33. Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020;27(12):1185–93. [DOI] [PubMed] [Google Scholar]
- 34. FitzHarris G, Marangos P, Carroll J. Cell cycle‐dependent regulation of structure of endoplasmic reticulum and inositol 1,4,5‐trisphosphate‐induced Ca2+ release in mouse oocytes and embryos. Mol Biol Cell. 2003;14(1):288–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kline D, Mehlmann L, Fox C, Terasaki M. The cortical endoplasmic reticulum (ER) of the mouse egg: localization of ER clusters in relation to the generation of repetitive calcium waves. Dev Biol. 1999;215(2):431–42. [DOI] [PubMed] [Google Scholar]
- 36. FitzHarris G, Marangos P, Carroll J. Changes in endoplasmic reticulum structure during mouse oocyte maturation are controlled by the cytoskeleton and cytoplasmic dynein. Dev Biol. 2007;305(1):133–44. [DOI] [PubMed] [Google Scholar]
- 37. Stricker SA. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev Biol. 1999;211(2):157–76. [DOI] [PubMed] [Google Scholar]
- 38. Fissore RA, Longo FJ, Anderson E, Parys JB, Ducibella T. Differential distribution of inositol trisphosphate receptor isoforms in mouse oocytes. Biol Reprod. 1999;60(1):49–57. [DOI] [PubMed] [Google Scholar]
- 39. Shiraishi K, Okada A, Shirakawa H, Nakanishi S, Mikoshiba K, Miyazaki S. Developmental changes in the distribution of the endoplasmic reticulum and inositol 1,4,5‐trisphosphate receptors and the spatial pattern of Ca2+ release during maturation of hamster oocytes. Dev Biol. 1995;170(2):594–606. [DOI] [PubMed] [Google Scholar]
- 40. Zheng P, Obara CJ, Szczesna E, Nixon‐Abell J, Mahalingan KK, Roll‐Mecak A, et al. ER proteins decipher the tubulin code to regulate organelle distribution. Nature. 2022;601(7891):132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roll‐Mecak A. The tubulin code in microtubule dynamics and information encoding. Dev Cell. 2020;54(1):7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zandalinas SI, Mittler R. ROS‐induced ROS release in plant and animal cells. Free Radic Biol Med. 2018;122:21–7. [DOI] [PubMed] [Google Scholar]
- 43. Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS‐induced ROS release. Physiol Rev. 2014;94(3):909–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dey K, Bazala MA, Kuznicki J. Targeting mitochondrial calcium pathways as a potential treatment against Parkinson's disease. Cell Calcium. 2020;89:102216. [DOI] [PubMed] [Google Scholar]
- 45. Ludtmann MHR, Abramov AY. Mitochondrial calcium imbalance in Parkinson's disease. Neurosci Lett. 2018;663:86–90. [DOI] [PubMed] [Google Scholar]
- 46. Navaratnarajah T, Anand R, Reichert AS, Distelmaier F. The relevance of mitochondrial morphology for human disease. Int J Biochem Cell Biol. 2021;134:105951. [DOI] [PubMed] [Google Scholar]
- 47. Jansen RP. Germline passage of mitochondria: quantitative considerations and possible embryological sequelae. Hum Reprod. 2000;15(Suppl 2):112–28. [DOI] [PubMed] [Google Scholar]
- 48. Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40(12):1484–8. [DOI] [PubMed] [Google Scholar]
- 49. Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15(Suppl 2):148–59. [DOI] [PubMed] [Google Scholar]
- 50. Motta PM, Nottola SA, Makabe S, Heyn R. Mitochondrial morphology in human fetal and adult female germ cells. Hum Reprod. 2000;15(Suppl 2):129–47. [DOI] [PubMed] [Google Scholar]
- 51. Yu Y, Dumollard R, Rossbach A, Lai FA, Swann K. Redistribution of mitochondria leads to bursts of ATP production during spontaneous mouse oocyte maturation. J Cell Physiol. 2010;224(3):672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wakai T, Fissore RA. Constitutive IP3R1‐mediated Ca2+ release reduces Ca2+ store content and stimulates mitochondrial metabolism in mouse GV oocytes. J Cell Sci. 2019;132(3):jcs225441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tejera A, Herrero J, de Los Santos MJ, Garrido N, Ramsing N, Meseguer M. Oxygen consumption is a quality marker for human oocyte competence conditioned by ovarian stimulation regimens. Fertil Steril. 2011;96(3):618–23 e612. [DOI] [PubMed] [Google Scholar]
- 54. Lopes AS, Greve T, Callesen H. Quantification of embryo quality by respirometry. Theriogenology. 2007;67(1):21–31. [DOI] [PubMed] [Google Scholar]
- 55. Dumollard R, Duchen M, Sardet C. Calcium signals and mitochondria at fertilisation. Semin Cell Dev Biol. 2006;17(2):314–23. [DOI] [PubMed] [Google Scholar]
- 56. Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234(2):339–51. [DOI] [PubMed] [Google Scholar]
- 57. Udagawa O, Ishihara T, Maeda M, Matsunaga Y, Tsukamoto S, Kawano N, et al. Mitochondrial fission factor Drp1 maintains oocyte quality via dynamic rearrangement of multiple organelles. Curr Biol. 2014;24(20):2451–8. [DOI] [PubMed] [Google Scholar]
- 58. Carvalho KF, Machado TS, Garcia BM, Zangirolamo AF, Macabelli CH, Sugiyama FHC, et al. Mitofusin 1 is required for oocyte growth and communication with follicular somatic cells. FASEB J. 2020;34(6):7644–60. [DOI] [PubMed] [Google Scholar]
- 59. Zhang M, Bener MB, Jiang Z, Wang T, Esencan E, Scott R III, et al. Mitofusin 1 is required for female fertility and to maintain ovarian follicular reserve. Cell Death Dis. 2019;10(8):560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wakai T, Harada Y, Miyado K, Kono T. Mitochondrial dynamics controlled by mitofusins define organelle positioning and movement during mouse oocyte maturation. Mol Hum Reprod. 2014;20(11):1090–100. [DOI] [PubMed] [Google Scholar]
- 61. Chiaratti MR. Uncovering the important role of mitochondrial dynamics in oogenesis: impact on fertility and metabolic disorder transmission. Biophys Rev. 2021;13(6):967–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–7. [DOI] [PubMed] [Google Scholar]
- 63. Tam AB, Roberts LS, Chandra V, Rivera IG, Nomura DK, Forbes DJ, et al. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev Cell. 2018;46(3):327–43 e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hoglinger D, Burgoyne T, Sanchez‐Heras E, et al. NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat Commun. 2019;10(1):4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu L, MacKenzie KR, Putluri N, Maletic‐Savatic M, Bellen HJ. The Glia‐neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 2017;26(5):719–37 e716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Den Brink DM, Cubizolle A, Chatelain G, Davoust N, Girard V, Johansen S, et al. Physiological and pathological roles of FATP‐mediated lipid droplets in Drosophila and mice retina. PLoS Genet. 2018;14(9):e1007627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Eisenberg‐Bord M, Mari M, Weill U, Rosenfeld‐Gur E, Moldavski O, Castro IG, et al. Identification of seipin‐linked factors that act as determinants of a lipid droplet subpopulation. J Cell Biol. 2018;217(1):269–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Herker E, Vieyres G, Beller M, Krahmer N, Bohnert M. Lipid droplet contact sites in health and disease. Trends Cell Biol. 2021;31(5):345–58. [DOI] [PubMed] [Google Scholar]
- 69. Rakotonirina‐Ricquebourg R, Costa V, Teixeira V. Hello from the other side: membrane contact of lipid droplets with other organelles and subsequent functional implications. Prog Lipid Res. 2022;85:101141. [DOI] [PubMed] [Google Scholar]
- 70. Swann K, Windsor S, Campbell K, Elgmati K, Nomikos M, Zernicka‐Goetz M, et al. Phospholipase C‐ζ‐induced Ca2+ oscillations cause coincident cytoplasmic movements in human oocytes that failed to fertilize after intracytoplasmic sperm injection. Fertil Steril. 2012;97(3):742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dunning KR, Russell DL, Robker RL. Lipids and oocyte developmental competence: the role of fatty acids and beta‐oxidation. Reproduction. 2014;148(1):R15–27. [DOI] [PubMed] [Google Scholar]
- 72. Nagashima H, Kashiwazaki N, Ashman RJ, Grupen CG, Nottle MB. Cryopreservation of porcine embryos. Nature. 1995;374(6521):416. [DOI] [PubMed] [Google Scholar]
- 73. Aizawa R, Ibayashi M, Hatakeyama T, Tatsumi T, Tsukamoto S. Impact of short‐term high‐fat feeding on lipid droplet content in mouse oocytes. J Reprod Dev. 2021;67(1):73–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Nakano A. The Golgi apparatus and its next‐door neighbors. Front Cell Dev Biol. 2022;10:884360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Moreno RD, Schatten G, Ramalho‐Santos J. Golgi apparatus dynamics during mouse oocyte in vitro maturation: effect of the membrane trafficking inhibitor brefeldin A. Biol Reprod. 2002;66(5):1259–66. [DOI] [PubMed] [Google Scholar]
- 76. Payne C, Schatten G. Golgi dynamics during meiosis are distinct from mitosis and are coupled to endoplasmic reticulum dynamics until fertilization. Dev Biol. 2003;264(1):50–63. [DOI] [PubMed] [Google Scholar]
- 77. Sathananthan AH, Ng SC, Chia CM, Law HY, Edirisinghe WR, Ratnam SS. The origin and distribution of cortical granules in human oocytes with reference to Golgi, nucleolar, and microfilament activity. Ann N Y Acad Sci. 1985;442:251–64. [DOI] [PubMed] [Google Scholar]
- 78. Hoodbhoy T, Talbot P. Characterization, fate, and function of hamster cortical granule components. Mol Reprod Dev. 2001;58(2):223–35. [DOI] [PubMed] [Google Scholar]
- 79. Ozil JP, Markoulaki S, Toth S, Matson S, Banrezes B, Knott JG, et al. Egg activation events are regulated by the duration of a sustained [Ca2+]cyt signal in the mouse. Dev Biol. 2005;282(1):39–54. [DOI] [PubMed] [Google Scholar]
- 80. Ducibella T, Schultz RM, Ozil JP. Role of calcium signals in early development. Semin Cell Dev Biol. 2006;17(2):324–32. [DOI] [PubMed] [Google Scholar]
- 81. Ducibella T, Fissore R. The roles of Ca2+, downstream protein kinases, and oscillatory signaling in regulating fertilization and the activation of development. Dev Biol. 2008;315(2):257–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Burkart AD, Xiong B, Baibakov B, Jimenez‐Movilla M, Dean J. Ovastacin, a cortical granule protease, cleaves ZP2 in the zona pellucida to prevent polyspermy. J Cell Biol. 2012;197(1):37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Satouh Y, Nozawa K, Yamagata K, Fujimoto T, Ikawa M. Viable offspring after imaging of Ca2+ oscillations and visualization of the cortical reaction in mouse eggs. Biol Reprod. 2017;96(3):563–75. [DOI] [PubMed] [Google Scholar]
- 84. Nozawa K, Satouh Y, Fujimoto T, Oji A, Ikawa M. Sperm‐borne phospholipase C zeta‐1 ensures monospermic fertilization in mice. Sci Rep. 2018;8(1):1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Dietzel E, Wessling J, Floehr J, Schäfer C, Ensslen S, Denecke B, et al. Fetuin‐B, a liver‐derived plasma protein is essential for fertilization. Dev Cell. 2013;25(1):106–12. [DOI] [PubMed] [Google Scholar]
- 86. Suzuki T, Yoshida N, Suzuki E, Okuda E, Perry AC. Full‐term mouse development by abolishing Zn2+‐dependent metaphase II arrest without Ca2+ release. Development. 2010;137(16):2659–69. [DOI] [PubMed] [Google Scholar]
- 87. Kim AM, Bernhardt ML, Kong BY, Ahn RW, Vogt S, Woodruff TK, et al. Zinc sparks are triggered by fertilization and facilitate cell cycle resumption in mammalian eggs. ACS Chem Biol. 2011;6(7):716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhang N, Duncan FE, Que EL, O'Halloran TV, Woodruff TK. The fertilization‐induced zinc spark is a novel biomarker of mouse embryo quality and early development. Sci Rep. 2016;6:22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Que EL, Duncan FE, Bayer AR, Philips SJ, Roth EW, Bleher R, et al. Zinc sparks induce physiochemical changes in the egg zona pellucida that prevent polyspermy. Integr Biol (Camb). 2017;9(2):135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Duncan FE, Que EL, Zhang N, Feinberg EC, O'Halloran TV, Woodruff TK. The zinc spark is an inorganic signature of human egg activation. Sci Rep. 2016;6:24737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liu L, Damerell DR, Koukouflis L, Tong Y, Marsden BD, Schapira M. UbiHub: a data hub for the explorers of ubiquitination pathways. Bioinformatics. 2019;35(16):2882–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shin SW, Shimizu N, Tokoro M, Nishikawa S, Hatanaka Y, Anzai M, et al. Mouse zygote‐specific proteasome assembly chaperone important for maternal‐to‐zygotic transition. Biol Open. 2013;2(2):170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yang Y, Zhou C, Wang Y, Liu W, Liu C, Wang L, et al. The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal‐to‐zygotic transition. EMBO Rep. 2017;18(2):205–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J. 2021;40(19):e108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Schulze RJ, Krueger EW, Weller SG, Johnson KM, Casey CA, Schott MB, et al. Direct lysosome‐based autophagy of lipid droplets in hepatocytes. Proc Natl Acad Sci USA. 2020;117(51):32443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yamamoto A, Mizushima N, Tsukamoto S. Fertilization‐induced autophagy in mouse embryos is independent of mTORC1. Biol Reprod. 2014;91(1):7. [DOI] [PubMed] [Google Scholar]
- 97. Sato M, Sato K. Degradation of paternal mitochondria by fertilization‐triggered autophagy in C. elegans embryos. Science. 2011;334(6059):1141–4. [DOI] [PubMed] [Google Scholar]
- 98. Sato M, Sato K, Tomura K, Kosako H. The autophagy receptor ALLO‐1 and the IKKE‐1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans . Nat Cell Biol. 2018;20(1):81–91. [DOI] [PubMed] [Google Scholar]
- 99. Rojansky R, Cha MY, Chan DC. Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife. 2016;5:e17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, Ouyang YC, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci USA. 2013;110(32):13038–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mukhopadhyay D, Riezman H. Proteasome‐independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–5. [DOI] [PubMed] [Google Scholar]
- 102. Gan Q, Watanabe S. Synaptic vesicle endocytosis in different model systems. Front Cell Neurosci. 2018;12:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Toralova T, Kinterova V, Chmelikova E, Kanka J. The neglected part of early embryonic development: maternal protein degradation. Cell Mol Life Sci. 2020;77(16):3177–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sato K, Sato M, Audhya A, Oegema K, Schweinsberg P, Grant BD. Dynamic regulation of caveolin‐1 trafficking in the germ line and embryo of Caenorhabditis elegans . Mol Biol Cell. 2006;17(7):3085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Sato M, Sato K. Dynamic regulation of autophagy and endocytosis for cell remodeling during early development. Traffic. 2013;14(5):479–86. [DOI] [PubMed] [Google Scholar]
- 106. Sato M, Konuma R, Sato K, Tomura K. Fertilization‐induced K63‐linked ubiquitylation mediates clearance of maternal membrane proteins. Development. 2014;141(6):1324–31. [DOI] [PubMed] [Google Scholar]
- 107. Sato K. Multiple roles of endocytosis and autophagy in intracellular remodeling during oocyte‐to‐embryo transition. Proc Jpn Acad Ser B Phys Biol Sci. 2022;98(5):207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gomez‐Elias MD, Fissore RA, Cuasnicu PS, Cohen DJ. Compensatory endocytosis occurs after cortical granule exocytosis in mouse eggs. J Cell Physiol. 2020;235(5):4351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lüllmann‐Rauch R, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP‐2‐deficient mice. Nature. 2000;406(6798):902–6. [DOI] [PubMed] [Google Scholar]
- 110. Tsukamoto S, Hara T, Yamamoto A, Ohta Y, Wada A, Ishida Y, et al. Functional analysis of lysosomes during mouse preimplantation embryo development. J Reprod Dev. 2013;59(1):33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Muller B, Lewis N, Adeniyi T, Leese HJ, Brison DR, Sturmey RG. Application of extracellular flux analysis for determining mitochondrial function in mammalian oocytes and early embryos. Sci Rep. 2019;9(1):16778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Tan TCY, Brown HM, Thompson JG, Mustafa S, Dunning KR. Optical imaging detects metabolic signatures associated with oocyte quality. Biol Reprod. 2022;107:1014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bradley J, Pope I, Masia F, Sanusi R, Langbein W, Swann K, et al. Quantitative imaging of lipids in live mouse oocytes and early embryos using CARS microscopy. Development. 2016;143(12):2238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ibayashi M, Aizawa R, Mitsui J, Tsukamoto S. Homeostatic regulation of lipid droplet content in mammalian oocytes and embryos. Reproduction. 2021;162(6):R99–109. [DOI] [PubMed] [Google Scholar]
- 115. Yamagata K, Suetsugu R, Wakayama T. Long‐term, six‐dimensional live‐cell imaging for the mouse preimplantation embryo that does not affect full‐term development. J Reprod Dev. 2009;55(3):343–50. [DOI] [PubMed] [Google Scholar]
- 116. Yamagata K, Suetsugu R, Wakayama T. Assessment of chromosomal integrity using a novel live‐cell imaging technique in mouse embryos produced by intracytoplasmic sperm injection. Hum Reprod. 2009;24(10):2490–9. [DOI] [PubMed] [Google Scholar]
- 117. Mizutani E, Yamagata K, Ono T, Akagi S, Geshi M, Wakayama T. Abnormal chromosome segregation at early cleavage is a major cause of the full‐term developmental failure of mouse clones. Dev Biol. 2012;364(1):56–65. [DOI] [PubMed] [Google Scholar]
- 118. Yamagata K, Ueda J. Long‐term live‐cell imaging of mammalian preimplantation development and derivation process of pluripotent stem cells from the embryos. Dev Growth Differ. 2013;55(4):378–89. [DOI] [PubMed] [Google Scholar]
- 119. Satouh Y, Nozawa K, Ikawa M. Sperm postacrosomal WW domain‐binding protein is not required for mouse egg activation. Biol Reprod. 2015;93(4):94. [DOI] [PubMed] [Google Scholar]
- 120. Tsukamoto S, Hara T, Yamamoto A, Kito S, Minami N, Kubota T, et al. Fluorescence‐based visualization of autophagic activity predicts mouse embryo viability. Sci Rep. 2014;4:4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tatsumi T, Takayama K, Ishii S, Yamamoto A, Hara T, Minami N, et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development. 2018;145(4):dev161893. [DOI] [PubMed] [Google Scholar]
- 122. Hatano Y, Mashiko D, Tokoro M, Yao T, Yamagata K. Chromosome counting in the mouse zygote using low‐invasive super‐resolution live‐cell imaging. Genes Cells. 2022;27(3):214–28. [DOI] [PubMed] [Google Scholar]
- 123. Yue S, Wang L, DeMartino GN, Zhao F, Liu Y, Sieber MH. Highly conserved shifts in ubiquitin‐proteasome system (UPS) activity drive mitochondrial remodeling during quiescence. Nat Commun. 2022;13(1):4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ajduk A, Ilozue T, Windsor S, Yu Y, Seres KB, Bomphrey RJ, et al. Rhythmic actomyosin‐driven contractions induced by sperm entry predict mammalian embryo viability. Nat Commun. 2011;2:417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Serhal PF, Ranieri DM, Kinis A, Marchant S, Davies M, Khadum IM. Oocyte morphology predicts outcome of intracytoplasmic sperm injection. Hum Reprod. 1997;12(6):1267–70. [DOI] [PubMed] [Google Scholar]
- 126. Otsuki J, Okada A, Morimoto K, Nagai Y, Kubo H. The relationship between pregnancy outcome and smooth endoplasmic reticulum clusters in MII human oocytes. Hum Reprod. 2004;19(7):1591–7. [DOI] [PubMed] [Google Scholar]
- 127. Gurunath S, Biliangady R, Sundhararaj UM, Gangadharswamy A, Gundlapalli S, Reddy GMM. Live birth rates in in vitro fertilization cycles with oocytes containing smooth endoplasmic reticulum aggregates and normal oocytes. J Hum Reprod Sci. 2019;12(2):156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Drabkova P, Andrlova L, Hampl R, Kandar R. Amino acid metabolism in human embryos. Physiol Res. 2016;65(5):823–32. [DOI] [PubMed] [Google Scholar]
- 129. Baltz JM, Zhou C. Cell volume regulation in mammalian oocytes and preimplantation embryos. Mol Reprod Dev. 2012;79(12):821–31. [DOI] [PubMed] [Google Scholar]
- 130. Tokuoka Y, Yamada TG, Mashiko D, Ikeda Z, Hiroi NF, Kobayashi TJ, et al. 3D convolutional neural networks‐based segmentation to acquire quantitative criteria of the nucleus during mouse embryogenesis. NPJ Syst Biol Appl. 2020;6(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]


