Abstract
Despite updated recommendations for weight-based isoniazid dosing in children with drug-susceptible tuberculosis (TB) and higher dose isoniazid in regimens for adults with drug-resistant TB, individual pharmacokinetic variability can lead to sub-target isoniazid exposure. Host pharmacogenetics and isoniazid exposure remain understudied, especially in the East African population. We therefore employed a real-time polymerase chain reaction (qPCR) assay system to test genomic DNA extracted from saliva samples targeting the NAT2 gene responsible for isoniazid metabolism to describe the frequency of human single nucleotide polymorphisms in NAT2 within populations of children and adults in Tanzania, ascribe those polymorphisms to acetylator phenotype, and correlate to serum isoniazid exposures. In adults treated with higher dose isoniazid, genotypes with a predicted allelic phenotype of slow or intermediate acetylation were able to achieve a 0.41 μg/mL higher Cmax (p = 0.018) and a 2.9h*μg/mL higher AUC0–12 (p = 0.003) per mg/kg increase in isoniazid dosage versus adults with rapid acetylation phenotype. A similar relationship was not found in the younger age population as predicted by timing of NAT2 maturation. This saliva based qPCR assay was fieldable to guide personalized isoniazid dosing in adults but not young children that may not have full NAT2 maturation and activity.
Keywords: Isoniazid, Pharmacogenetics, Pharmacokinetics, Acetylation, N-acetyltransferase
1. Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis is among the top 10 causes of death globally and routinely ranks as the number one cause of global infectious disease mortality. In 2020, TB incidence was approximately 10 million, including a decline a notification of new cases as a result of COVID-19 related disruptions in routine services, yet mortality was estimated to have increased at ~1.5 million deaths [1]. Despite the global burden of mortality attributed to TB, curative antibiotic regimens have existed for decades. Increasingly pre-clinical and clinical studies have determined that a considerable portion of TB treatment failure and mortality is driven by suboptimal pharmacokinetics/pharmacodynamics (PK/PD) [2].
A cornerstone drug to the conventional anti-TB regimen is isonicotinic acid hydrazide (isoniazid, INH). INH is selective in spectrum to mycobacteria, primarily inhibiting mycolic acid synthesis and disrupting cell wall construction, and exerting bactericidal effect against populations of M. tuberculosis in rapid growth phases [3]. The bactericidal activity of INH is concentration-dependent whereby higher serum peaks (Cmax) or total area under the concentration time curve (AUC) associate with improved microbial kill. However, oral INH is rapidly absorbed and peaks early in the daily dosing interval [4], and much of the total exposure relates to its rapidity of metabolism by the arylamine N-acetyltransferase encoded on the human NAT2 gene [5]. NAT2 genotypes can vary considerably by population of study, and NAT2 maturation may not reach full activity until later childhood [6,7].
A new abundance of work has determined target serum thresholds for minimum exposure for anti-TB drugs including INH [2,8]. Yet despite WHO recommended weight-based dosing for INH in both drug-susceptible TB and higher dose INH for MDR-TB, we and others have observed considerable pharmacokinetic variability [9,10]. For instance, we previously reported that in a study of Tanzanian children undergoing TB treatment, children receiving increased dosing per revised World Health Organization (WHO) recommendations saw increased serum exposure of rifampin and pyrazinamide but not INH or ethambutol as compared to children receiving non-revised dosages [11]. Given that very little is known with regard to the pharmacogenetic epidemiology of NAT2 genotype distribution in East Africa more broadly and Tanzania specifically, we sought to describe the frequency of human single nucleotide polymorphisms (SNPs) in NAT2 within populations of children and adults in Tanzania, ascribe those polymorphisms to acetylator phenotype, and correlate to serum INH exposures.
Leveraging existing molecular diagnostics capacities at the Kilimanjaro Clinical Research Institute, we employed a real-time polymerase chain reaction (qPCR) based platform for characterizing high fidelity SNPs within NAT2 and leveraging DNA extraction from simple saliva collection. This pharmacogenetics work was nested within a larger study of the effects of pharmacokinetic variability and TB drug resistance on treatment outcomes which conveniently allowed comparison of populations with a wide range of mg/kg dosing treated for both drug- susceptible and MDR-TB (clinicaltrials.gov identifier NCT03559582).
2. Materials and methods
Patient population:
Patients and/or guardians signed written informed consent for a protocol approved by the institutional review boards for human subjects research at the National Institute for Medical Research in Tanzania (NIMR/HQ/R.8a/Vol.IX/2120) and the University of Virginia (UVA-HSR-IRB #18452). Adults starting treatment for MDR-TB were recruited from Kibong’oto Infectious Diseases Hospital in the Kilimanjaro region of Northern Tanzania, and children less than 15 years of age starting treatment for drug-susceptible TB were recruited from Haydom Lutheran Hospital in the Manyara region of Northern Tanzania. All adults were confirmed to have pulmonary disease from M. tuberculosis with rpoB mutation by GeneXpert MTB/RIF (Cepheid, Sunnyvale, CA USA) and putative rifampin resistance before starting on an individualized MDR-TB regimen. Children either had microbiologically confirmed M. tuberculosis by Xpert MTB/RIF or unconfirmed TB defined by the updated National Institutes of Health Consensus Clinical Case Definitions for TB research in children, and were started on weight based fixed dose combinations of rifampin, INH, pyrazinamide and ethambutol in an initiation phase of 2 months followed by rifampin and INH for another 4 months of continuation phase [12].
Nucleic Acid Extraction from Saliva:
Nucleic acid extractions and quantitative PCRs (described below) were carried out at the Kilimanjaro Clinical Research Institute in Moshi, Tanzania. DNA from saliva samples was collected using DNA Genotek Oragene kits (DNA Genotek Inc., Kanata, Ontario, Canada) and extracted using the PrepIT-L2P kit (DNA Genotek) at enrollment and within 3 days of treatment initiation, with a procedure modified from that of the manufacturer. Briefly, saliva samples were first incubated at 50 °C in a dry bath for 1 h 500 μL of heated sample was then mixed with 20 μL PrepIT-L2P reagent. Samples were then centrifuged and resulting supernatants were mixed with 100% ethanol before pelleting DNA by further centrifugation. DNA pellets were then washed with 70% ethanol before allowing the pellet to dry before resuspension with 50 μL of TE solution (10 mM Tris, 1 mM EDTA, buffered to pH8). An “extraction blank” in which nuclease-free water was utilized as the sample was included with each extraction set was used in order to monitor for contamination.
2.1. qPCR-based genotyping
Assays for ADME SNP targets were procured from Thermo Fisher (Thermo Fisher, Waltham, MA, USA) in a custom-designed plate with primers and probes pre-assembled in each well. NAT2 targets included: c.341 (C/T; rs1801280), c.590 (A/G; rs1799930), and c.803 (A/G; rs1208), high fidelity SNP targets deemed most predictive based on prior studies and in order to incorporate into a fieldable and later scalable assay [13].
Each 10 μL qPCR reaction included 5 μL 2x TaqPath ProAmp Master Mix (Thermo Fisher), 1 μL Nuclease-Free water, and 4 μL extracted nucleic acid. One column in each plate was reserved for no-template control (nuclease-free water as sample) and per testing session, one column was reserved for positive controls consisting of pooled control DNAs obtained from the Coriell Institute (Coriell Institute for Medical Research, Camden, New Jersey, USA). Control DNAs included NA20294, NA19189, NA19023, and NA19351. Cycling conditions were as follows: 30 s 60 °C, 5 min 95 °C, 40 cycles 15 at 95 °C and 1 min at 60 °C, with a final incubation of 30 s at 60 °C. Reactions were carried out in the Applied Biosystems ViiA 7 Real-Time PCR system (Thermo Fisher).
2.2. Conversion of NAT2 genotypes into NAT2 haplotypes and predicted phenotypes
Individual NAT2 genotypes at positions c.803, c.341 and c.590 were combined into haplotypes and then converted to a NAT2 predicted acetylator phenotypes of “rapid” or “slow” according to the indicated phenotypes available at the database of human NAT2 alleles hosted at the Democritus University of Thrace (https://nat.mbg.duth.gr/). Phenotypes derived from c.341 and c.590 haplotypes were predicted as driving phenotype over the c.803 haplotype in the individuals from this study. Thus, only pairs of c.341 and c.590 haplotype derived phenotypes thereafter were combined to give an overall predicted acetylator phenotype for each individual with <Rapid, Rapid ≥ Rapid, <Slow, Slow> = Slow, and <Rapid, Slow ≥ Intermediate.
2.3. Assaying for pharmacokinetic exposure
Blood collections occurred at weeks 2, 4, 8, and 24 after the start of medication. Collections were performed at 1, 2, 6 and 12 h after observed medication administration for adults and 1, 2, and 6 h after observed medication administration for children. Serum was stored at −80 °C prior to transport and testing. Serum drug concentrations were measured at the University of Florida Infectious Diseases Pharmacokinetics Laboratory. INH drug concentrations were measured by a validated LC MS MS assay [11]. Cmax and estimated AUC over 12 h (AUC0–12) were determined by noncompartmental analysis using Phoenix WinNonlin version 8.0 (Certara USA, Princeton, New Jersey), which allowed AUC0–12 estimates in those with 6-h values within the elimination phase. Minimum targets for Cmax (3 μg/mL) and AUC (52 h*μg/mL) were drawn from studies in drug-susceptible TB and have not been fully established for varying levels of INH resistance as occur in RR/MDR-TB [8].
Prior to this study, the expected drug content was validated for each of the fixed-drug concentration formulations. Briefly, individual tablets were crushed, weighed, and diluted in 1:1 methanol/water. The contents of each tablet were spiked into serum samples and assayed using high performance liquid chromatography as described above [11].
2.4. Comparison of NAT2 genotypes against INH pharmacokinetic exposures
Cmax and AUC0–12 were only considered for the time period in which the participant’s prescribed mg/kg dosage remained constant during the period in which there was also no significant weight change prompting a change in dosage. The Cmax and AUC0–12 values were averaged over that time and compared to their genotype distribution at each individual NAT2 locus. Further analyses were carried out by combining genotypes at c.341 and c.590 into their associated NAT2 alleles (i.e. NAT2*4, NAT2*5, or NAT2*6, [14]) and combining alleles into a predicted overall acetylation phenotype as detailed above. The pharmacokinetic data were stratified according to whether the participant possessed a predicted Rapid phenotype or a Slow/Intermediate phenotype. Data were further stratified by “age category” as a predictor of NAT2 enzyme maturity [15]. Participants were classified as age category 1 if they were younger than 5.3 years old (NAT2 enzyme immature) at baseline and age category 2 if they were 5.3 years old or older at baseline (NAT2 enzyme mature). Analyses were carried out in Microsoft Excel v16.16.27 and R 4.0.2.
3. Results
3.1. Genotype/Allele frequencies and predicted NAT2 acetylator phenotypes
The parent study aimed to enroll 50 children receiving standard anti-tuberculosis treatment for drug-susceptible TB and 125 adults with MDR-TB. Nucleic acids were successfully extracted from all saliva samples collected from 50 children and 124 adults. The characteristics of the source populations are described in Table 1. Briefly, the median age of adults was 42 years while that of children was 2.3 years. The majority of the adult population were males while the pediatric population was nearly evenly split between males and females. Both populations reported having approximately 90 days of preceding illness at the time of TB diagnosis. Underlying health conditions were largely experienced by the adults with 38.3% living with human immunodeficiency virus (HIV) and 32.5% reporting ever having smoked (Table 1).
Table 1.
Demographic information for the study participants.
| Overall | Kilimanjaro Tanzania, Adult MDR | Manyara Tanzania, Pediatric | |
|---|---|---|---|
|
| |||
| Participants | 174 | 124 | 50 |
| Median Age at baseline, years (range) | 34 (0.3–80) | 42 (18–80) | 2.3 (0.3–14.8) |
| Sex, %M | 60.7 | 65.3 | 48 |
| Median Height at baseline, cm (range) | 157 (59.5–184) | 164.5 (133.6–184) | 79.9 (59.5–151) |
| Median Mass at baseline, kg (range) | 44.1 (5–81) | 49 (27–81) | 8.6 (5–29.3) |
| Median days ill at baseline (range) | 90 (5–400) | 91 (7–370) | 90 (5–400) |
| HIV+, % | 27.2 | 38.3 | 0 |
| Diabetes, % | 1.7 | 2.4 | 0 |
| Ever smoked, % | 23.7 | 32.5 | 2.0 |
Successful NAT2 genotyping was achieved for all extracted DNA samples. Despite geographically proximal sites of enrollment, there were some differences between the pediatric and adult populations when examining NAT2 genotype frequencies. At c.341, the pediatric population had a higher proportion of individuals homozygous for the reference T variant as compared to the adult population (54.0% vs. 41.9%, respectively; Table 2a). The pediatric population also had a lower proportion of heterozygous individuals (genotype CT, 30.0%) versus the adults (46.8%). The two populations also differed from one another at the c.590 locus, with the pediatric population having fewer individuals homozygous for the reference G variant (38.0% vs. 52.4%) and more individuals heterozygous AG at this position (60.0% vs. 47.6%) as compared to the adult population. Despite the differences in genotype proportions at individual NAT2 loci, differences nearly disappeared upon conversion to predicted acetylator phenotypes with 14.5% and 16% having a predicted Rapid phenotype and 85.5% and 84% having a Slow or Intermediate phenotype in the adult and pediatric populations, respectively (Table 2b). These proportions were somewhat similar to those previously documented in Africa (Table S1).
Table 2A.
Genotype frequenciesat NAT2 loci tested.
| Kilimanjaro Tanzania, Adult MDR n = 124 | Manyara Tanzania, Pediatric n = 50 | |
|---|---|---|
|
| ||
| rs1801280, c.341 T- > C | ||
| TT | 41.9 | 54.0 |
| TC | 46.8 | 30.0 |
| CC | 11.3 | 16.0 |
| rs1799930, c.590 G- > A | ||
| GG | 52.4 | 38.0 |
| GA | 47.6 | 60.0 |
| AA | 0 | 2.0 |
| rs2282143, c.803 A- > G | ||
| AA | 29.8 | 34.0 |
| AG | 50.0 | 46.0 |
| GG | 20.2 | 20.0 |
Table 2B.
Frequencies of predicted NAT2 acetylation phenotypes.
| NAT2 Phenotype | Northern Tanzania, Adult, MDR, n = 124 | Northern Tanzania, Pediatric, n = 50 |
|---|---|---|
|
| ||
| Rapida | 18 (14.5) | (16.0) |
| Intermediate-Slowa | 106 (85.5)b | 42 (84.0)b |
NAT2 *4/*4 = Rapid; *4/*5 and *4/*6 = Intermediate; *5/*5, *5/*6 and *6/*6 = Slow.
Includes participants whose exact allelic makeup could not be determined due to heterozygosity at both c.341 and c.590.
3.2. Relationship of NAT2 genotype alone and INH pharmacokinetics by age category
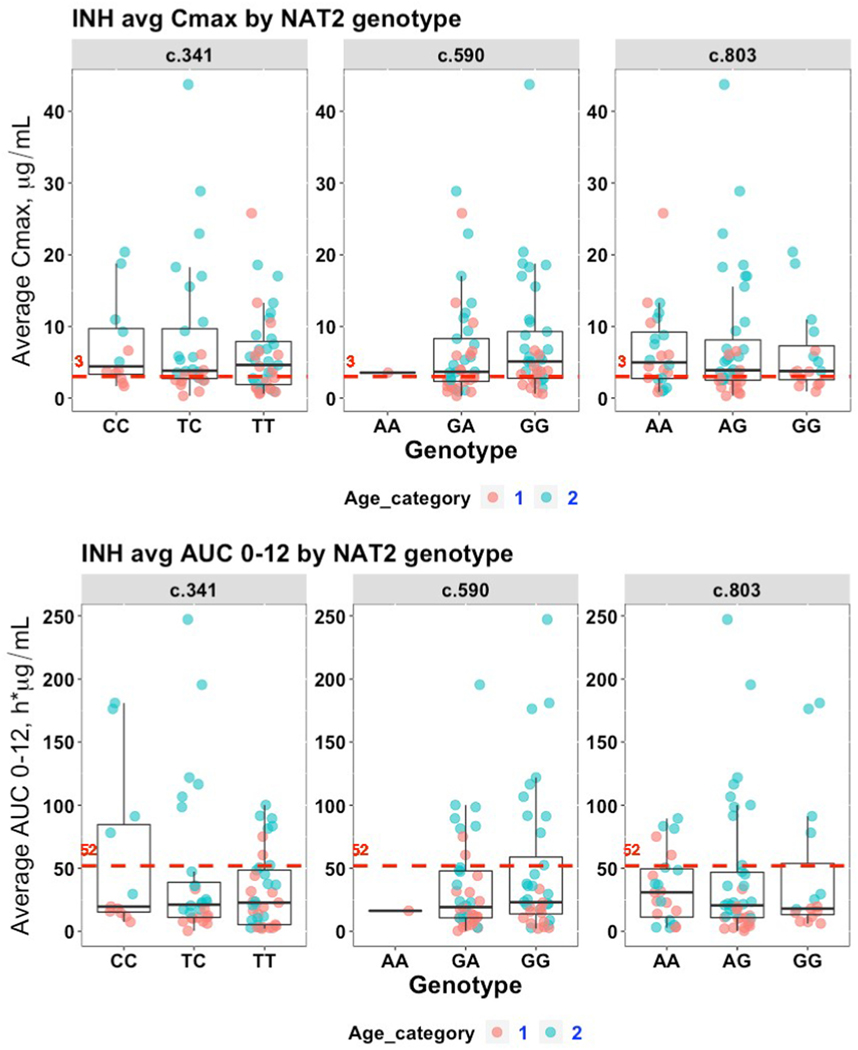
Among the 124 adults with MDR-TB and genotyping data, there were 30 participants treated with high dose INH and included in the comparative pharmacokinetic analysis. The median INH dosage for the pediatric population was 5.9 mg/kg (range 2.1–18.4) while the median dosage for the adult population was 18.3 mg/kg (range 8.1–36.8). As shown in Fig. 1A, the majority of participants (n = 55, 66.3%) achieved a minimum Cmax target of 3 μg/mL averaged over the treatment duration. Yet 36/55 (65.5%) of participants achieving this target were from Age Category 2. Only 19 of 80 (23.8%; Fig. 1B) of participants were able to achieve a 52 h*μg/mL target AUC value, with 17/19 (89.5%) coming from Age Category 2. However there were no statistically significant differences in Cmax or AUC0–12 values across individual loci (Tables S2A and S2B).
Fig. 1. INH exposure analyzed by genotypes at three NAT2 loci.
Available average Cmax (μg/mL) and average AUC0–12 (h*μg/mL) values were analyzed by genotype at each of three NAT2 SNP loci. Each marker represents an individual participant. Those having a non-mature NAT2 phenotype as measured by age less than 5.3 years of age (“Age Category 1”) represented by red markers and blue markers representing individuals 5.3 years of age or older (“Age Category 2”). A) Cmax; B) AUC0–12. The red, dashed line in each subplot represents the target expected exposure level for adult patients.
3.3. Relationship of predicted acetylator phenotype and INH pharmacokinetics
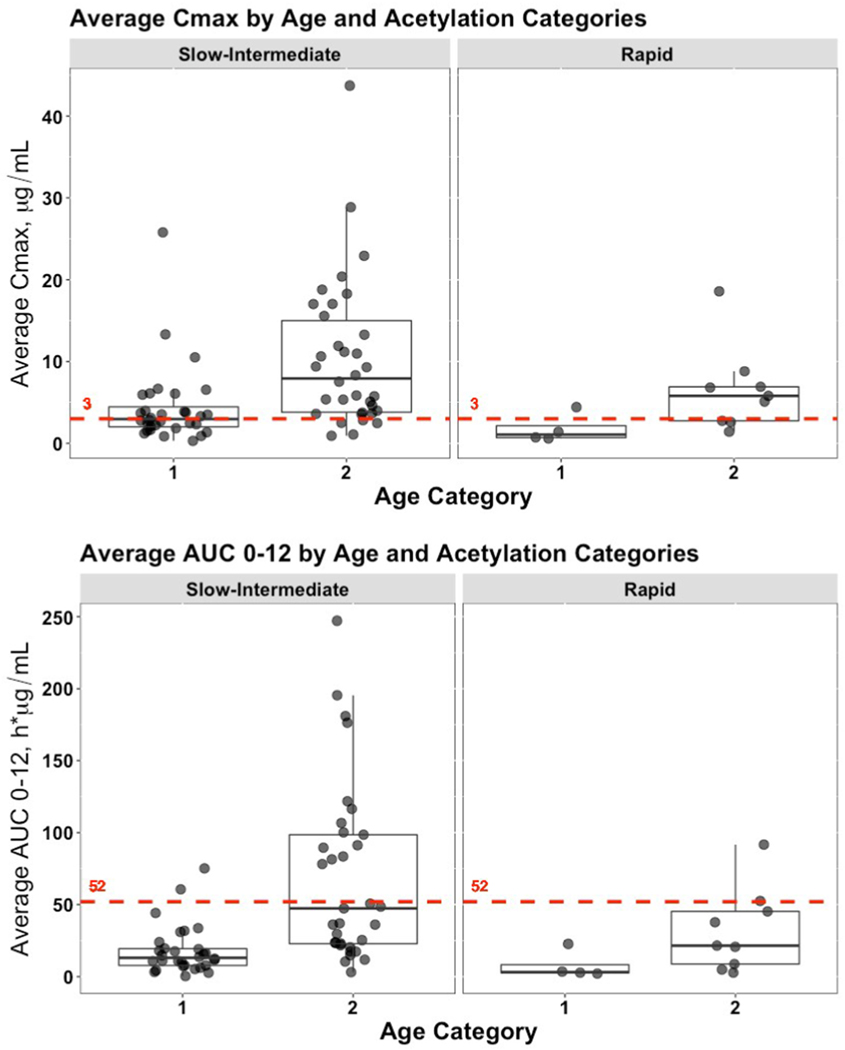
When analyzing the entire population without age categorization, the median Cmax for participants with a Rapid acetylation phenotype compared to Slow/Intermediate phenotypes (4.41 vs. 3.98 μg/mL, respectively), and median AUC0–12 (20.74 vs. 22.93 h*μg/mL, respectively) demonstrated no statistically significant differences. However, as shown in Fig. 2A and B, the median Cmax and AUC0–12 values for those individuals in Age Category 2 were higher with both acetylation types. These differences were statistically significant (Tables 3A and 3B).
Fig. 2. INH exposure analyzed by predicted NAT2 acetylator phenotype.
Genotype information at c.341 and c.590 were converted to NAT2 haplotypes/alleles and predicted NAT2 acetylator phenotypes and then used to analyze available average Cmax (μg/mL) and average AUC0–12 (h*μg/mL) values. Data was further stratified according to the same age categories used in Fig. 1. Each marker represents an individual participant. A) Cmax; B) AUC0–12. The red, dashed line in each subplot represents the target expected exposure level for adult patients.
Table 3A.
Frequencies and summary statistics for cmax as analyzed by predicted acetylator phenotype and age category.
| Predicted Acetylator Phenotype | Age Category | Participants achieving <3 μg/mL | Participants achieving ≥3 μg/mL | Median Cmax, μg/mL | IQR, μg/mL | p-value, Wilcoxon Rank Sum Test for Age Category |
|---|---|---|---|---|---|---|
|
| ||||||
| Slow-Intermediate | 1 | 16 | 16 | 2.97 | 2.44 | 8.89×10−5 |
| 2 | 5 | 29 | 7.91 | 11.19 | ||
| Rapid | 1 | 3 | 1 | 1.07 | 1.46 | 0.020 |
| 2 | 3 | 6 | 5.78 | 4.14 | ||
The p-value for the Wilcoxon Rank Sum Test comparing the overall Cmax difference between predicted acetylation categories is 0.35.
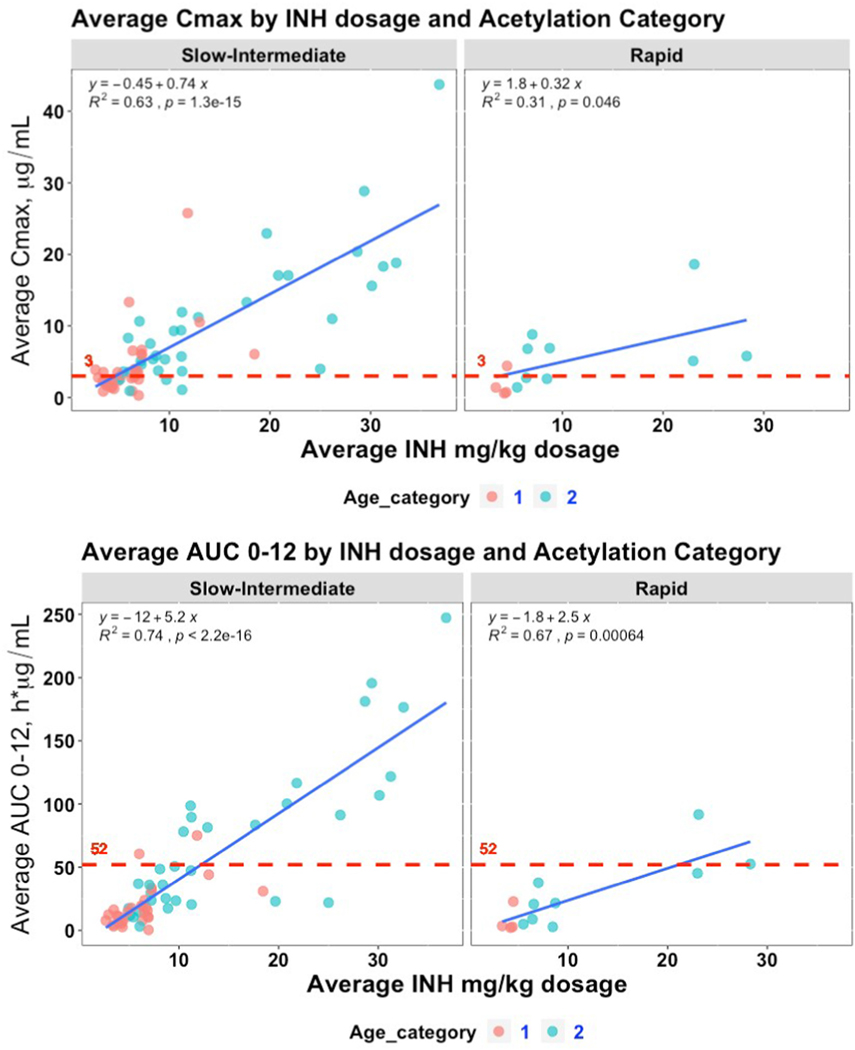
Understanding that one of the primary drivers of PK exposure is mg/kg dosing, we incorporated the mg/kg dosage along with visualizing the age category to determine if NAT2 alleles and predicted phenotypes influenced the slope of expected increase in PK exposure with increasing mg/kg dosage. When Cmax vs. mg/kg dosage and AUC0–12 vs. mg/kg dosage are stratified by predicted phenotype, as shown in Fig. 3A and B, the slope of the lines generated by Slow and Intermediate acetylators are significantly steeper than those for Rapid acetylators. As visualized in Fig. 3A and B, these differences are driven largely by individuals in the older age category. Slow and Intermediate acetylators achieve 0.41 μg/mL higher Cmax (p = 0.018) and a 2.9h*μg/mL higher AUC0–12 (p = 0.003) per mg/kg increase in INH dosage compared Rapid acetylators (Table 4).
Fig. 3. INH exposure analyzed by mg/kg dosage and predicted NAT2 acetylator phenotype.
Average Cmax (μg/mL) and average AUC0–12 (h*μg/mL) values were first stratified by predicted NAT2 acetylator phenotype and then plotted against the averaged mg/kg INH dosage given to each patient. Each marker represents an individual participant with individuals with a non-mature NAT2 phenotype as measured by age less than 5.3 years of age (“Age Category 1”) represented by red markers and blue markers representing individuals 5.3 years of age or older (“Age Category 2”). A) Cmax; B) AUC0–12. The red, dashed line in each subplot represents the target expected exposure level for adult patients.
Table 4.
Comparison of Pharmacokinetic Exposures Explained by mg/kg Dosage and Predicted NAT2 Acetylation Phenotype.
| Site | Slope | P value, ANCOVA Dosage*phenotype |
|---|---|---|
|
| ||
| Cmax, μg/mL per mg/kg change | ||
| Overall | 0.67 | |
| Slow-Intermediate | 0.73 | 0.018 |
| Rapid | 0.32 | |
| AUC0–12, h*μg/mL per mg/kg change | ||
| Overall | 5.0 | |
| Slow-Intermediate | 5.4 | 0.003 |
| Rapid | 2.5 | |
4. Discussion
The major findings of this study were that qPCR NAT2 genotypes from saliva samples found a predicted acetylator phenotype distribution similar to those in other locations previously reported from Africa (Table S1), and that predicted allelic phenotypes of Slow/Intermediate or Rapid influenced the slope increase in INH Cmax and AUC0–12 with increasing mg/kg dosage. Importantly however, age also had a significant impact on INH exposure, likely related to higher mg/kg doses in adults and NAT2 maturation status, whereby acetylator phenotype contributed most to INH exposure for adults receiving higher mg/kg dosages. These results are consistent with prior studies in adult populations including those in South Africa, India, Poland, and China with respect to the influence of genotype/phenotype and INH exposure parameters [16–19].
We suspect that the specific sub-analysis of the pediatric cohort failing to show that predicted acetylator phenotype to have a significant effect on INH exposures was due to the median childhood age of 2.2 years, which was below the 5.3 year old cutoff previously demonstrated to be associated with NAT2 maturity and adult-like activity [15]. These results from our pediatric population would appear to be consistent with two studies of South African children in which age, NAT2 maturation status, and dosage had a larger influence on INH kinetics than NAT2 genotype and predicted acetylator status alone [16,20]. Interestingly, a study of Ghanaian children showed a pharmacogenetic/pharmacokinetic relationship more akin to that of adults [21]. This might be explained by the higher median age in the Ghanian study and increased INH mg/kg dosage compared to that of the children in the present study [22].
Given that inter-individual differences in ADME physiology and other external environmental or host factors make it difficult to successfully implement a one-size-fits-all strategy to weight-based dosing, understanding an individual’s pharmacogenetic background can importantly account for one fixed factor when considering the dose and medication composition of long-term regimens. Furthermore, while sub-target PK exposure can adversely impact treatment efficacy, exposure-related adverse events are also major causes of side effects and treatment interruption. Notably, individuals classified as having the slow acetylator phenotype are at increased risk of INH-induced hepatotoxicity [23]. Yet with pre-dosing knowledge of NAT2 genotype, investigators from Japan were able to demonstrate that adjusting INH dosage according to NAT2 genotype resulted in minimizing treatment failures while avoiding INH-induced liver injury [24]. Given the increasing availability of qPCR capacity, as well as the ease and non-invasive nature of saliva collection, NAT2 genotype-based dosing may ultimately obviate the need for assays requiring more invasive serum collection or labor-intensive PK assays that utilize chromatography or mass spectrometry. This would cut down the need for cold transport and shipment and provide a simpler screening mechanism to identify individuals requiring more specialized testing.
Instructively, our initial examinations of individual NAT2 loci or even whole NAT2 haplotypes did not reveal any statistically significant relation to pharmacokinetic parameters. While the trend of increased Cmax and AUC0–12 INH levels amongst Slow and Intermediate acetylators versus Rapid acetylators was consistent with prior studies in South African [16] and Polish populations [18], only after integrating mg/kg dosage did we demonstrate statistically significant differences in the rates of change of both Cmax and AUC0–12 per mg/kg increase in INH. These lower rates of change in both Cmax and AUC0–12 parameters would be consistent with prior research demonstrating that individuals with a predicted Rapid acetylation phenotype clear INH at an increased rate as compared with predicted Intermediate or Slow acetylation phenotypes [25]. In addition, Kinzig-Schippers et al. previously demonstrated that as much as 88% of INH clearance variability could be explained by NAT2 genotypes, albeit this study was performed in adult Caucasians [26]. We chose to compare predicted acetylator phenotypes to Cmax and AUC rather than clearance, as Cmax and AUC are most associated with microbial kill and prevention of resistance, and target exposures have previously been described.
A key limitation of the study was the sample size drawn from a convenient cohort. Post-hoc power calculations based on observed distributions of Rapid and Slow/Intermediate acetylators for prediction of AUC0–12 revealed a power of 0.72 which is lower than the often targeted 0.80 beta-value. Furthermore, the relatively small number of genetic loci examined could have been explored more comprehensively with a sequencing approach.
In summary, NAT2 genotyping utilizing a qPCR platform from saliva samples found a genotype distribution in Tanzania where predicted allelic phenotype significantly influenced INH PK exposure for adults on higher mg/kg doses. A screening test for NAT2 genotype may therefore guide personalized dosing of INH now currently used in numerous regimens for drug-susceptible and MDR-TB.
Supplementary Material
Table 3B.
Frequencies and summary statistics for AUC0–12 as analyzed by predicted acetylator phenotype and age category.
| Predicted Acetylator Phenotype | Age Category | Participants achieving <52 h*μg/mL | Participants achieving ≥52 h*μg/mL | Median AUC0–12, μg/mL | IQR, μg/mL | p-value, Wilcoxon Rank Sum Test for Age Category |
|---|---|---|---|---|---|---|
|
| ||||||
| Slow-Intermediate | 1 | 28 | 2 | 13.10 | 11.71 | 1.30×10− 6 |
| 2 | 19 | 14 | 47.33 | 75.63 | ||
| Rapid | 1 | 4 | 0 | 3.15 | 5.70 | 0.076 |
| 2 | 7 | 2 | 21.56 | 36.46 | ||
The p-value for the Wilcoxon Rank Sum Test comparing the overall AUC0–12 difference between predicted acetylation categories is 0.14.
Funding statement
This work was supported by Award Number U01 AI115594 from the National Institutes of Health (NIH). This work was also partly supported by Award Number D43 TW006578 from the Fogarty International Center (FIC) of the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of FIC or the NIH.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.tube.2022.102246.
References
- [1].WHO. Global tuberculosis report 2021. 2021. [Google Scholar]
- [2].Pasipanodya JG, McIlleron H, Burger A, Wash PA, Smith P, Gumbo T. Serum drug concentrations predictive of pulmonary tuberculosis outcomes. J Infect Dis 2013; 208:1464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Timmins GS, Deretic V. Mechanisms of action of isoniazid. Mol Microbiol 2006;62: 1220–7. [DOI] [PubMed] [Google Scholar]
- [4].Alsultan A, Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs 2014;74:839–54. [DOI] [PubMed] [Google Scholar]
- [5].Hong BL, D’Cunha R, Li P, Al-Shaer MH, Alghamdi WA, An G, Peloquin C. A systematic review and meta-analysis of isoniazid pharmacokinetics in healthy volunteers and patients with tuberculosis. Clin Therapeut 2020. 10.1016/j.clinthera.2020.09.009. [DOI] [PubMed] [Google Scholar]
- [6].Sabbagh A, Darlu P, Crouau-Roy B, Poloni ES. Arylamine N-acetyltransferase 2 (NAT2) genetic diversity and traditional subsistence: a worldwide population survey. PLoS One 2011;6:e18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sabbagh A, Langaney A, Darlu P, Gerard N, Krishnamoorthy R, Poloni ES. Worldwide distribution of NAT2 diversity: implications for NAT2 evolutionary history. BMC Genet 2008;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Alffenaar JC, Gumbo T, Dooley KE, Peloquin CA, McIlleron H, Zagorski A, Cirillo DM, Heysell SK, Silva DR, Migliori GB. Integrating pharmacokinetics and pharmacodynamics in operational research to end tuberculosis. Clin Infect Dis 2020;70:1774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alkabab Y, Keller S, Dodge D, Houpt E, Staley D, Heysell S. Early interventions for diabetes related tuberculosis associate with hastened sputum microbiological clearance in Virginia, USA. BMC Infect Dis 2017;17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ramachandran G, Hemanth Kumar AK, Bhavani PK, Poorana Gangadevi N, Sekar L, Vijayasekaran D, Banu Rekha VV, Ramesh Kumar S, Ravichandran N, Mathevan G, Swaminathan S. Age, nutritional status and INH acetylator status affect pharmacokinetics of anti-tuberculosis drugs in children. Int J Tubercul Lung Dis 2013;17:800–6. [DOI] [PubMed] [Google Scholar]
- [11].Justine M, Yeconia A, Nicodemu I, Augustino D, Gratz J, Mduma E, Heysell SK, Kivuyo S, Mfinanga S, Peloquin CA, Zagurski T, Kibiki GS, Mmbaga B, Houpt ER, Thomas TA. Pharmacokinetics of first-line drugs among children with tuberculosis in rural Tanzania. J Pediatric Infect Dis Soc 2020;9:14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Graham SM, Cuevas LE, Jean-Philippe P, Browning R, Casenghi M, Detjen AK, Gnanashanmugam D, Hesseling AC, Kampmann B, Mandalakas A, Marais BJ, Schito M, Spiegel HM, Starke JR, Worrell C, Zar HJ. Clinical case Definitions for classification of intrathoracic tuberculosis in children: an update. Clin Infect Dis 61Suppl 2015;3:S179–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hein DW, Doll MA. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics 2012. Jan;13(1):31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hein DW, Doll MA, Fretland AJ, Leff MA, Webb SJ, Xiao GH, Devanaboyina US, Nangju NA, Feng Y. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol Biomarkers Prev 2000;9:29–42. [PubMed] [Google Scholar]
- [15].Rogers Z, Hiruy H, Pasipanodya JG, Mbowane C, Adamson J, Ngotho L, Karim F, Jeena P, Bishai W, Gumbo T. The non-linear child: ontogeny, isoniazid concentration, and NAT2 genotype modulate enzyme reaction kinetics and metabolism. EBioMedicine 2016;11:118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Naidoo A, Chirehwa M, Ramsuran V, McIlleron H, Naidoo K, Yende-Zuma N, Singh R, Ncgapu S, Adamson J, Govender K, Denti P, Padayatchi N. Effects of genetic variability on rifampicin and isoniazid pharmacokinetics in South African patients with recurrent tuberculosis. Pharmacogenomics 2019;20:225–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hemanth Kumar AK, Ramesh K, Kannan T, Sudha V, Haribabu H, Lavanya J, Swaminathan S, Ramachandran G. N-acetyltransferase gene polymorphisms & plasma isoniazid concentrations in patients with tuberculosis. Indian J Med Res 2017;145:118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zabost A, Brzezinska S, Kozinska M, Blachnio M, Jagodzinski J, Zwolska Z, Augustynowicz-Kopec E. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. BioMed Res Int 2013:853602. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jing W, Zong Z, Tang B, Wang J, Zhang T, Wen S, Xue Y, Chu N, Zhao W, Huang H. Population pharmacokinetic analysis of isoniazid among pulmonary tuberculosis patients from China. Antimicrob Agents Chemother 2020;64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhu R, Kiser JJ, Seifart HI, Werely CJ, Mitchell CD, D’Argenio DZ, Fletcher CV. The pharmacogenetics of NAT2 enzyme maturation in perinatally HIV exposed infants receiving isoniazid. J Clin Pharmacol 2012;52:511–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dompreh A, Tang X, Zhou J, Yang H, Topletz A, Adu Ahwireng E, Antwi S, Enimil A, Langaee T, Peloquin CA, Court MH, Kwara A. Effect of genetic variation of NAT2 on isoniazid and SLCO1B1 and CES2 on rifampin pharmacokinetics in Ghanaian children with tuberculosis. Antimicrob Agents Chemother 2018;62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Antwi S, Yang H, Enimil A, Sarfo AM, Gillani FS, Ansong D, Dompreh A, Orstin A, Opoku T, Bosomtwe D, Wiesner L, Norman J, Peloquin CA, Kwara A. Pharmacokinetics of the first-line antituberculosis drugs in Ghanaian children with tuberculosis with or without HIV coinfection. Antimicrob Agents Chemother 2017; 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lee SW, Chung LS, Huang HH, Chuang TY, Liou YH, Wu LS. NAT2 and CYP2E1 polymorphisms and susceptibility to first-line anti-tuberculosis drug-induced hepatitis. Int J Tubercul Lung Dis 2010;14:622–6. [PubMed] [Google Scholar]
- [24].Azuma J, Ohno M, Kubota R, Yokota S, Nagai T, Tsuyuguchi K, Okuda Y,Takashima T, Kamimura S, Fujio Y, Kawase I, Pharmacogenetics-based tuberculosis therapy research g. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy.Eur J Clin Pharmacol 2013;69:1091–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Parkin DP, Vandenplas S, Botha FJ, Vandenplas ML, Seifart HI, van Helden PD, vander Walt BJ, Donald PR, van Jaarsveld PP. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med 1997;155:1717–22. [DOI] [PubMed] [Google Scholar]
- [26].Kinzig-Schippers M, Tomalik-Scharte D, Jetter A, Scheidel B, Jakob V, Rodamer M, Cascorbi I, Doroshyenko O, Sorgel F, Fuhr U. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother 2005;49:1733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.