Andre Buret and Thibault Allain discuss the mechanisms that regulate gut microbiota biofilms integrity and highlight the importance of studying phenotypic biofilm damage, beyond the characterization of relative bacterial abundance.
Abstract
Gut microbiota contain communities of viruses, bacteria, fungi, and Eukarya, and live as biofilms. In health, these biofilms adhere to the intestinal mucus surface without contacting the epithelium. Disruptions to the equilibrium between these biofilms and the host may create invasive pathobionts from these commensal communities and contribute to disease pathogenesis. Environmental factors appear to dominate over genetics in determining the shifts in microbiota populations and function, including when comparing microbiota between low-income and industrialized countries. The observations discussed herein carry enormous potential for the development of novel therapies targeting phenotype in microbiota dysbiosis.
Gut microbiota integrity is key to the maintenance of gastrointestinal and overall body homeostasis. The regulatory nature of this interaction has become the topic of intense research activities. In nature, bacteria can live as planktonic swimmers or sessile biofilms. Biofilms are multi-species microbial communities encased in their extracellular matrix containing exopolysaccharides, proteins, extracellular DNA, and environmental and host components. The biofilm mode of growth has been linked to a variety of medical and industrial concerns (Costerton et al., 1995; Cámara et al, 2022; Pannekens et al., 2019; Yuan et al., 2022; Yin et al, 2019), such as: colonization on medical devices and surgical implants, where biofilms can cause severe infections that require removal of the device; chronic infections, including in the lungs of patients with cystic fibrosis, in burn wounds, or in gum diseases; colonization of food products, where biofilm colonization and persistence are responsible for spoilage and food-borne diseases; colonization of air-conditioning and ventilation ducts (where they contribute to the spread of pathogenic bioaerosols), of ship hulls, oil wells and pipelines, where they are responsible for malfunction and corrosion.
Due to their extracellular matrix, biofilms are extremely resilient, and are shielded from ultraviolet radiation, extreme temperature, extreme pH, high salinity, high pressure, poor nutrients, and antimicrobials. The enormous economic and health costs associated with biofilms in nature offer a solid rationale for translational research across many sectors where biofilm growth should be inhibited. In contrast, growth as a biofilm promotes homeostasis at various mucosal surfaces, and disruptions of this mode of growth in such settings are detrimental to health. Microbial biofilms naturally colonize various surfaces of the body, including the gastrointestinal tract, the lungs, the vagina, and the skin (Flemming and Wuertz, 2019; Motta et al., 2021a). Recent discoveries illustrate how homeostatic maintenance of these complex microbial ecosystems is critical to health, and how their disruptions are a direct cause for pathophysiology (Flemming and Wuertz, 2019; Motta et al., 2021a; Zoetendal et al., 2002). In the gut, the microbiome is made up of mixed communities of viruses, bacteria, fungi, and Eukarya that co-habit with mucus layers. Microbial colonization, diversity, and density vary along the length of the gastrointestinal tract, with the lowest numbers of microbes only forming scattered biofilm fragments in the stomach and upper gut, whereas a rich and dense microbial biofilm lines the large intestinal mucosa (Flemming and Wuertz, 2019; Motta et al., 2021a). Not surprisingly therefore, there are well-established differences between the composition, genetics, and behavior of fecal versus mucosal microbiota (Motta et al., 2021a; Zoetendal et al., 2002). Under some circumstances, sessile microbiota constituents may leave these gut biofilms and behave as planktonic, swimming microorganisms (Buret et al., 2019). So, do gut microbiota normally live as swimming microbes or as biofilms?
Gut microbiota: Planktonic versus biofilm phenotype
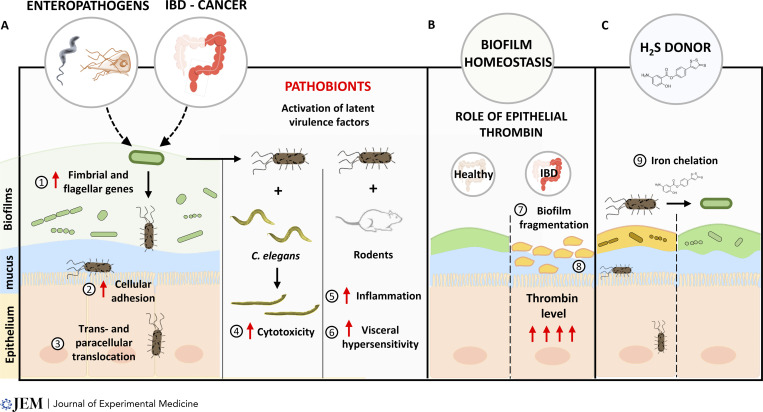
The data available today, including quantitative evidence, clearly support the hypothesis that the biofilm mode of growth is the dominant phenotype in the gut, as it is elsewhere in nature (Flemming and Wuertz, 2019; Motta et al., 2021a). However, it also has become apparent that mature biofilms may release planktonic bacteria that may then further colonize other sites. Disrupted gut microbiota biofilms may also generate the release of planktonic bacteria that can become pathogenic, adherent, and invasive pathobionts (Motta et al., 2021a; Buret et al., 2019). Pathobionts are temporarily benign microbes, or commensals, that under environmental or host pressure may cause disease. These observations stem, at least in part, from gut microbiota biofilm characterization in human tissues from patients with inflammatory bowel diseases or colorectal cancer (Motta et al., 2021a; Dejea et al., 2018; Kalischuk et al., 2009). Interestingly, it is only recently that the critical relevance of the gut microbiota biofilm phenotype integrity, and its regulatory processes, has become a focal point of research. The biofilm mode of growth is influenced by a variety of environmental and host factors, including proteases, fatty acids, oxygen, nutrients, or iron (Motta et al., 2021a). Recent observations also demonstrated that enteropathogens were hitherto unrecognized determinants of the microbiota biofilm phenotype. Indeed, using models of Campylobacter jejuni and Giardia sp. infections, findings have first demonstrated that enteric infections may activate latent virulence genes in otherwise commensal microbiota, and fragment the biofilm structure (Buret et al., 2019). C. jejuni or Giardia infection promotes bacterial translocation, an event that is associated with elevated expression of fimbrial and flagellar genes (Kalischuk et al., 2009; Gerbaba et al., 2015). Atomic force microscopy revealed that these “activated” bacteria are indeed able to stick to the epithelial surface with increased force of adherence, and that these events worsened colitis (Reti et al., 2015; O’Hara et al., 2012). Further research then showed that Giardia induces similar changes in human microbiota, which in turn causes inflammation in germ-free mice, and lethality in Caenorhabditis elegans (Beatty et al., 2017; Gerbaba et al., 2015). The observation that a Protozoan parasite may alter the behavior of microbiota bacteria points to novel trans-kingdom communications that warrant further research. The invasion of pathobionts during the acute phase of giardiasis was found to be directly responsible for gastrointestinal hypersensitivity, consistent with the common reports of post-infectious irritable bowel syndrome following enteric infections (Halliez et al., 2016). Recent findings uncovered a key role for epithelial thrombin in keeping microbiota biofilms at bay, at least in part by proteolytically cleaving components of the biofilm matrix, thus emphasizing the host’s role in regulating gut biofilm structures (Motta et al., 2019). The findings also demonstrated how the presence of commensal microbiota directly regulated mucosal thrombin, and that such epithelia-biofilm interactions also occurred in the skin, the lungs, and the bladder (Motta et al., 2019). Recent evidence also implicated elevated thrombin levels in necrotizing enterocolitis (NEC) and Crohn’s disease, where the protease was a direct cause of inflammation and tissue damage (Namachivayam et al., 2020; Motta et al., 2021b). While studies in NEC demonstrated that increased production of tissue factor by macrophages was a key trigger for elevated thrombin (Namachivayam et al., 2020), the regulatory pathways of epithelial thrombin, and their consequence on health and disease, remain obscure. Regardless, these observations reveal the undeniable significance of gut microbiota biofilms in health, and that disruptions of this phenotype cause disease pathogenesis at a variety of mucosal surfaces.
Therapeutic opportunities
Despite the great complexity that comes with characterizing gut microbiota biofilms in situ, attempts at developing novel therapies for diseases which result from microbiota dysbiosis should assess phenotypic biofilm damage, beyond the common characterization of relative bacterial abondance. Recent research in models of inflammatory bowel disease found that the formation of pathobionts required free environmental iron; and indeed, an experimental hydrogen sulfide–releasing drug with potent anti-colitis effects was further bolstered if the compound had iron-chelating properties that inhibited pathobiont formation (Motta et al., 2018). Imbalances in iron homeostasis are also implicated in biofilm-induced respiratory diseases, such as chronic obstructive pulmonary disease, cystic fibrosis, and lung cancer (Thomsen et al., 2022; Neves et al., 2019). In view of the mitigating benefits of hydrogen sulfide in mucosal iron imbalances, the findings have led to a heightened interest in the investigation of the toxicity versus therapeutic benefits of exogenous hydrogen sulfide (Buret et al., 2022). Hydrogen sulfide confers a broad range of health properties, in addition to the recently discovered stabilizing properties on the mucosal microbiome. Hydrogen sulfide is known to inhibit platelet aggregation, to promote vasodilation, and to open ATP-sensitive ion channels, all of which casting a promising therapeutic light in the context of cardiovascular disorders (Streeter et al., 2013; Wallace and Wang, 2015). The effects may also contribute to the benefits of hydrogen sulfide in inflammatory bowel diseases, where it was found to stabilize the mucus lining, to have anti-inflammatory properties, to generate cytoprotective effects, and to promote the resolution of tissue injury at least in part by activating sulfur-containing amino acid metabolism, inhibiting oxidative stress, and stimulating angiogenesis (Buret et al., 2019; Motta et al., 2018; Wallace et al., 2018). Indeed, hydrogen sulfide is a known free radical scavenger, neutralizing reactive oxygen species and enhancing the potency of endogenous antioxidant molecules (Buret et al., 2022; Streeter et al., 2013; Wallace and Wang, 2015). Stabilization of the gut mucus lining and protection of the gut microbiome via an iron-chelating effect therefore add to the long list of health benefits generated by exogenous administration hydrogen sulfide. Together, observations made to date emphasize the importance of environmental factors in defining features of the gut microbiota.
Gut microbiota biofilm disruption and pathogenesis: a role for pathobionts. (A) Enteric infections (e.g., Giardia sp., C. jejuni) and intestinal diseases (inflammatory bowel disease [IBD], colorectal cancer, etc.) are associated with a disruption of intestinal microbiota biofilms, which may ultimately lead to the generation and release of pathogenic planktonic bacteria. These pathobionts show (1) elevated expression of fimbrial and flagellar genes, (2) increased adherence to epithelial surfaces, and (3) increased trans- and paracellular translocation, causing gastrointestinal hypersensitivity and post-infectious disorders. Pathobionts dispersed from the biofilms adhere to and invade the epithelium, (4) are lethal in models of C. elegans nematodes, (5) induce inflammation, and (6) cause visceral hypersensitivity in rodent models of gastrointestinal infections. (B) Serine proteases such as epithelial thrombin constrain biofilms away from the epithelium by cleaving elements of the biofilm matrix (7). This spatial segregation no longer exists when thrombin expression is altered as observed in IBD, allowing adhesion of biofilm fragments onto mucosal surfaces (8). (C) The findings pave the way towards strategies targeting pathobiont formation for the treatment of gastrointestinal diseases associated with microbiota dysbiosis. (9) Hydrogen sulfide–releasing drugs functioning as iron chelators by reducing free mucosal iron levels may inhibit the formation of pathobionts and reduce their virulence.
Inheritance versus environmental pressures in human gut microbiota communities
Comparative research in wild apes and humans from Africa and the northern hemisphere has shed light on our understanding of environmental influences on gut microbiota composition. The findings established that the major gut microbiota families have evolved from a common ancestor for more than 15 million yr (Moeller et al., 2016). The significant Phyla shifts we observe today in people of the industrialized world versus those in low-income countries may have arisen due to various factors, including adaptation to diverse diets, incidence of enteric infections and diseases, and general habitat features. Overall, environmental factors seem to trump genetics in defining microbiota constituents. Regardless of the differences in microbiota composition, the propensity of microbiota to grow as a biofilm appears to be conserved. Indeed, bacterial communities in the honeybee’s ileum are characterized with genes associated with the formation of adherent biofilms; these genes encode for Type IV pili, adhesins, flagella, motility, intracellular trafficking, and biofilm-associated proteins (Engel et al., 2012). The presence of these genes carries the potential that the honeybee too may alter its gut biofilm phenotype in response to environmental triggers, and, in this way, perhaps cause disease via the production of invasive pathobionts from previously commensal gut microbiota. The conserved aspects of at least some of the key microbiome biofilm properties is at least in part what allows researchers to use a variety of animal models systems for this field of research, including rodents, non-human primates, or pigs, but also non-mammalian species such as C. elegans, Drosophila melanogaster, and zebrafish (Leulier et al., 2017).
While microbiota will directly modulate host factors, a variety of host factors are also able to affect the gut microbiome, including proteases, fatty acids, oxygen, nutrients, or iron (Motta et al., 2021a; de Vos et al., 2022). The gut microbiota activates the expression and production of inflammasome proteins that modulate health and disease in the gut (de Vos et al., 2022). Conversely, recent evidence also indicates that inflammasomes contribute to shaping the biogeography, diversity, and functions of the gut microbiome to regulate gut function, metabolism, as well as gut–brain homeostasis (Manko-Prykhoda et al., 2020; Privitera et al., 2022; Man, 2018). Since birth, the complex interactive co-development between the host and the microbiome will help establish functional homeostasis, while disruptions of this process will predispose to possibly life-long inflammatory and metabolic diseases. Physical exercise, age, medication, stress, hygiene, gender, diet, smoking, environment, enteric infections, geographical location, and birth and lactation methods are known regulators of microbiome structure and function (Pirr and Viemann, 2020; Lkhagva et al., 2021; Olm et al., 2022; Cani et al., 2021). As such, the development of these complex interactions are cornerstones of the maintenance of human health.
In view of the critical role played by the gut microbiota biofilm phenotype in health, more research is warranted on a number of fronts. Clearly, the iron- and thrombin-dependent regulatory mechanisms that maintain biofilm integrity must be further explored. Also, little is known about the effects for healthy gut microbiota constituents to transit through upper and lower mucus layers, as well as through strata with different oxygen tension. Similarly, more research is needed to characterize host immune responses and associated inflammatory consequences when triggered by homeostatic commensals versus pathobionts. Finally, metatranscriptomic, metagenomic, and metabolomic analyses, as well as research on sex-dependent microbiota regulations, will help guide our efforts towards targeted therapies. In view of the dominating effect of environmental factors over genetics in determining gut microbiota composition, and the relatively conserved biofilm phenotype in health, it is possible that similar therapeutics attempts aiming at preserving a homeostatic microbiota phenotype may be applied across various genetic backgrounds.
Conclusion
In homeostatic conditions, microbiota biofilms are the dominant phenotype of the human gut microbiota, and the relative abundance of their constituents is driven more by environmental than host genetic factors. In the gut, the mere presence of gut microbiota biofilms does not represent a detrimental force as it does in the broad range of medical and industrial conditions listed in the introduction. Rather, it is the disruption of this mucosal biofilm phenotype and its abnormal adherence to the epithelial surface that may cause pathophysiology. Diet, habitat, enteric infections, and disease for example may all alter the integrity of these biofilms, and/or promote the release of pathobionts. Causal studies of great clinical relevance are emerging and reveal several commonalities in the mechanisms initiated by enteropathogenic infections, inflammatory bowel disease, and colorectal cancer. Clearly, the early view of “good” versus “bad” bacteria is a vast oversimplification of the complexities that dictate host–microbiota interactions. The field of microbiome research needs to take into account the significant pathogenic potential of commensal microbes that, under environmental pressures, have escaped the microbiota biofilm and become invasive planktonic organisms. Despite the many challenges associated with characterizing the gut biofilm phenotype, these new observations may offer opportunities for a paradigm shift in our understanding of what a healthy biofilm is, and better educate our attempts at restoring a healthy microbiome for therapeutic purposes.
Disclosures: A.G. Buret is a co-founder of Antibe Therapeutics Inc. No other disclosures were reported.
Acknowledgments
We are grateful to Dr. Olivia Sosnowski for the helpful comments and edits to this Viewpoint.
Parts of this work were supported by grants from the Natural Sciences and Engineering Research Council of Canada (Discovery Grant 69-0446, and Collaborative Research and Training Experience grant 1001893)), from the Canadian Institutes of Health Research (20130401), from Crohn’s and Colitis Canada (2014-05), from the Canadian Association of Gastroenterology (1001124), and the University of Calgary Eyes High Postdoctoral Fellowship program (1002347).
References
- Beatty, J.K., et al. 2017. Int. J. Parasitol. 10.1016/j.ijpara.2016.11.010 [DOI] [Google Scholar]
- Buret, A.G., et al. 2019. J. Biomed. Sci. 10.1186/s12929-018-0495-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buret, A.G., et al. 2022. Antioxid. Redox Signal. 10.1089/ars.2021.0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara, M., et al. 2022. NPJ Biofilms Microbiomes. 10.1038/s41522-022-00306-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani, P.D., et al. 2021. Microorganisms. 10.3390/microorganisms9061302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton, J.W., et al. 1995. Annu. Rev. Microbiol. 10.1146/annurev.mi.49.100195.003431 [DOI] [PubMed] [Google Scholar]
- Dejea, C.M., et al. 2018. Science. 10.1126/science.aah3648 [DOI] [Google Scholar]
- de Vos, W.M., et al. 2022. Gut. 10.1136/gutjnl-2021-326789 [DOI] [Google Scholar]
- Engel, P., et al. 2012. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1202970109 [DOI] [Google Scholar]
- Flemming, H.C., and Wuertz S.. 2019. Nat. Rev. Microbiol. 10.1038/s41579-019-0158-9 [DOI] [PubMed] [Google Scholar]
- Gerbaba, T.K., et al. 2015. Am. J. Physiol. Gastrointest. Liver Physiol. 10.1152/ajpgi.00335.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliez, M.C.M., et al. 2016. Am. J. Physiol. Gastrointest. Liver Physiol. 10.1152/ajpgi.00144.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalischuk, L.D., et al. 2009. Gut Pathog. 10.1186/1757-4749-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier, F., et al. 2017. Cell Metabol. 10.1016/j.cmet.2017.02.001 [DOI] [Google Scholar]
- Lkhagva, E., et al. 2021. Microorganisms. 10.3390/microorganisms9122520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manko-Prykhoda, A., et al. 2020. Int. J. Parasitol. 10.1016/j.ijpara.2019.12.011 [DOI] [PubMed] [Google Scholar]
- Man, S.M. 2018. Nat. Rev. Gastroenterol. Hepatol. 10.1038/s41575-018-0054-1 [DOI] [Google Scholar]
- Moeller, A.H., et al. 2016. Science. 10.1126/science.aaf3951 [DOI] [Google Scholar]
- Motta, J.P., et al. 2018. Inflamm. Bowel Dis. 10.1093/ibd/izy116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta, J.P., et al. 2019. Nat. Commun. 10.1038/s41467-019-11140-w [DOI] [Google Scholar]
- Motta, J.P., et al. 2021a. Nat. Rev. Gastroenterol. Hepatol. 10.1038/s41575-020-00397-y [DOI] [Google Scholar]
- Motta, J.P., et al. 2021b. J. Crohn’s Colitis. 10.1093/ecco-jcc/jjaa229 [DOI] [Google Scholar]
- Namachivayam, K., et al. 2020. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1912357117 [DOI] [Google Scholar]
- Neves, J., et al. 2019. Pharmaceuticals. 10.3390/ph12010005 [DOI] [Google Scholar]
- O’Hara, J.R., et al. 2012. Infect. Immun. 10.1128/IAI.06066-11 [DOI] [Google Scholar]
- Olm, M.R., et al. 2022. Science. 10.1126/science.abj2972 [DOI] [Google Scholar]
- Pannekens, M., et al. 2019. N. Biotechnol. 10.1016/j.nbt.2018.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirr, S., and Viemann D.. 2020. Front. Immunol. 10.3389/fimmu.2020.584288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privitera, G., et al. 2022. Front. Cell. Infect. Microbiol. 10.3389/fcimb.2021.806680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reti, K.L., et al. 2015. Infect. Immun. 10.1128/IAI.00970-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter, E., et al. 2013. Med. Gas Res. 10.1186/2045-9912-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen, K., et al. 2022. Biomedicines. 10.3390/biomedicines10092064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace, J.L., and Wang R.. 2015. Nat. Rev. Drug Discov. 10.1038/nrd4433 [DOI] [PubMed] [Google Scholar]
- Wallace, J.L., et al. 2018. Am. J. Physiol. Gastrointest. Liver Physiol. 10.1152/ajpgi.00249.2017 [DOI] [Google Scholar]
- Yin, W., et al. 2019. J. Int. J. Mol. Sci. 10.3390/ijms20143423 [DOI] [Google Scholar]
- Yuan, L., et al. 2022. Front. Microbiol. 10.3389/fmicb.2022.1023428 [DOI] [Google Scholar]
- Zoetendal, E.G., et al. 2002. Appl. Environ. Microbiol. 10.1128/AEM.68.7.3401-3407.2002 [DOI] [Google Scholar]